Abstract
Historically, many mass spectrometry–based proteomic studies have aimed at compiling an inventory of protein compounds present in a biological sample, with the long-term objective of creating a proteome map of a species. However, to answer fundamental questions about the behavior of biological systems at the protein level, accurate and unbiased quantitative data are required in addition to a list of all protein components. Fueled by advances in mass spectrometry, the proteomics field has thus recently shifted focus toward the reproducible quantification of proteins across a large number of biological samples. This provides the foundation to move away from pure enumeration of identified proteins toward quantitative matrices of many proteins measured across multiple samples. It is argued here that data matrices consisting of highly reproducible, quantitative, and unbiased proteomic measurements across a high number of conditions, referred to here as quantitative proteotype maps, will become the fundamental currency in the field and provide the starting point for downstream biological analysis. Such proteotype data matrices, for example, are generated by the measurement of large patient cohorts, time series, or multiple experimental perturbations. They are expected to have a large effect on systems biology and personalized medicine approaches that investigate the dynamic behavior of biological systems across multiple perturbations, time points, and individuals.
INTRODUCTION
For quantitative systems biology, accurate and precise measurements of analyte concentrations across multiple conditions constitute a crucial requirement. This allows researchers to study human disease across large cohorts, compare multiple perturbations, or describe the dynamics of a transformation in a biological system. The data output of a typical systems biology experiment is generally a two-dimensional data matrix containing quantitative measurement values of specific analytes (first dimension) across multiple samples (second dimension; Figure 1a). For proteomic measurements, the analytes are typically peptides, modified peptides, or proteins inferred from peptide measurements. The comprehensiveness and accuracy of the data matrix mostly determine the success of the downstream data analysis, where both dimensions are of equal importance: the number of measured compounds, as well as the number of analyzed samples.
FIGURE 1:
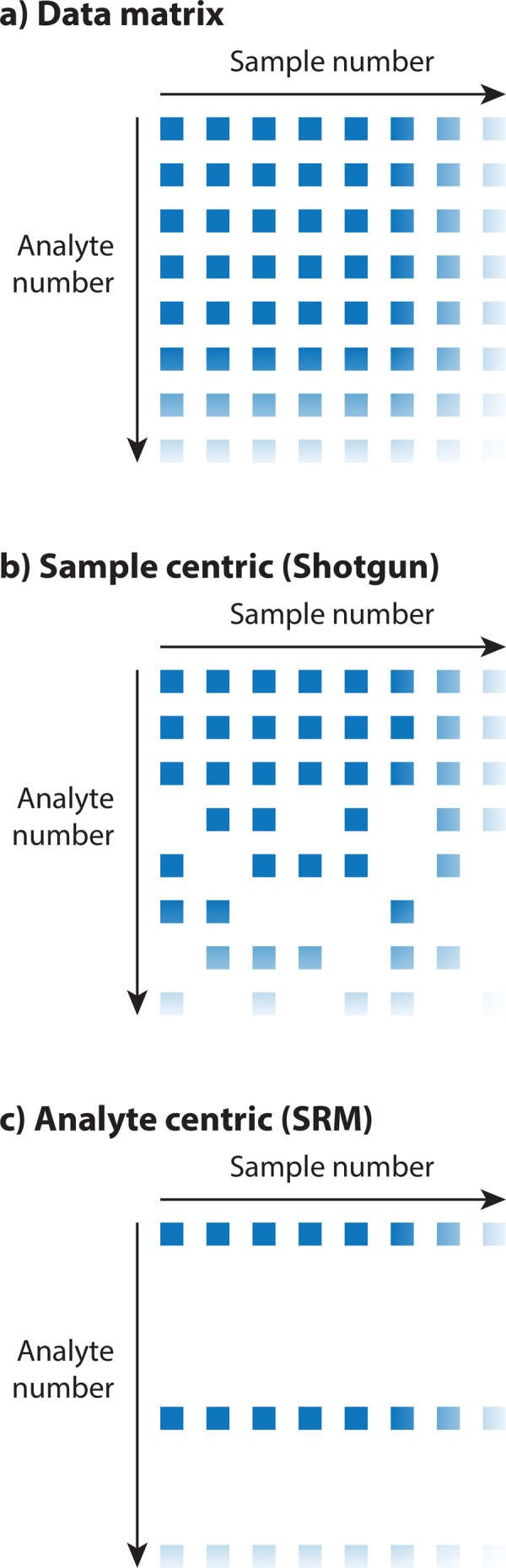
The proteotype data matrix as often found in proteomics experiments. (a) The data matrix contains quantitative values for different analytes (peptides or proteins) measured across multiple samples. One major goal in proteomics is to achieve high throughput (high number of quantified analytes) consistently quantified across many samples (experimental conditions, perturbations, or patient samples). (b) Sample-centric workflows (such as discovery proteomics or shotgun proteomics) place heavy emphasis on a high number of identifications in a single sample, which is achieved by data-dependent acquisition. However, the resulting data matrices often contain missing values due to undersampling issues, and in large studies, not all analytes can be quantified in every single sample. (c) In analyte-centric workflows (such as SRM and other low-throughput targeted proteomics techniques), the major focus is on achieving highly consistent quantification across many samples. The resulting data matrices are often devoid of missing values but only cover a few, carefully selected analytes.
Measurements primarily focusing on the first dimension (many analytes, one or few samples or conditions) may provide a useful overview of the sample and can generate an inventory of analytes present in the sample. These enumeration-oriented approaches, however, often lack the statistical power, number of conditions, or temporal resolution to observe subtle and nontrivial biological effects. For example, multiple consistent and reproducible measurements during a time-dependent system transformation are critical to understanding the time evolution of biological systems. To describe such a system’s response not only qualitatively but also quantitatively, dense sampling during the transition phase is important. Furthermore, to estimate confounding sources of error and variation in quantitative measurements and model them appropriately, repeat measurements of high reproducibility are required. In clinical studies, for example, large patient cohorts are critical to uncovering biological signal against a background of individual variation, which means that measurements need to be performed on dozens to hundreds of patient samples with high reproducibility.
Conversely, measurements focusing on the second dimension alone (few analytes, many samples) may suffer from bias and potentially miss important parts of the system’s behavior if they are not included in the data collection scheme. In proteomics, the proteins selected for measurement are often chosen based on the availability of measurement assays (frequently, assays based on affinity reagents) and the previous literature, leading to many experimental studies focusing on a few “popular” targets while leaving out a number of potentially crucial system components (Edwards et al., 2011; Reker and Malmström, 2012). Therefore these types of experiments are only suitable for later stages of a study, when the proteins that best describe a system and its behavior are well characterized. In practice, however, the optimal set of such target proteins can often be defined only by exactly the types of large-scale studies that generate a complete data matrix across conditions. This leads to a catch-22 situation in which, in order to perform large-scale proteomics studies, the targets need to be known in advance, but they can only be identified by such large-scale studies. Historically, for lack of methods to generate large-scale data matrices by direct proteomic measurement, target protein sets for systems studies were frequently extracted from the literature or inferred from surrogate measurements, for example, at the transcript level, with various levels of success.
For a truly comprehensive systems approach, both dimensions of the data matrix need to be given equal consideration. This would allow researchers to perform a single experiment to obtain information about which proteins are involved and the manner in which they participate in specific biological processes and their quantitative behavior. Specifically, proteomics could be used to study protein–protein interaction networks in their native and perturbed states and reveal how complex diseases such as cancer or diabetes rewire these networks (Lage et al., 2010; Collins et al., 2013). Furthermore, improved proteomic profiling could facilitate the search for new protein biomarkers in tissue and blood, since more samples and a larger number of proteins could be quantitatively compared across many patients (Liu et al., 2014, 2015). Applying proteomics techniques to signaling networks would require dense temporal sampling and accurate quantification of posttranslational modifications to capture fast-acting changes in, for example, phosphorylation states (Bodenmiller et al., 2010). This could improve our capacity to model the dynamics of these cellular signaling networks and lead to potential points for intervention to modulate these networks in disease states (Sabidó et al., 2012). Furthermore, accurate data matrices would allow a multitude of tools from statistics and machine learning to draw inferences about causal interactions among different proteomic compounds (Swan et al., 2013; Libbrecht and Noble, 2015). Applying such data-driven methods to biological problems might uncover important regulatory mechanisms and implicate novel proteins in well-studied biological processes, which could help researchers to better determine the behavior of the system. Finally, such matrices could foster integration with high-throughput data from other fields (such as genomics and other sequencing-based fields) in which comprehensive data matrices are already a standard experimental output. However, obtaining high-quality data matrices from proteomics data has historically been highly challenging.
CURRENT APPROACHES IN PROTEOMICS
One of the primary objectives in the field of proteomics in recent decades has been the identification of peptide and protein species in complex biological samples (Sabidó et al., 2012). In contrast to nucleic acid sequencing–based approaches, particularly by next-generation sequencing (NGS), in proteomics, the analyte cannot be amplified, the dynamic range of protein abundances is substantially larger than that of transcripts (Schwanhäusser et al., 2011), and the number of analytes (peptides) from a complex sample by far exceeds the available sequencing cycles of even the most advanced instruments. Therefore most proteomics approaches rely on extensive biochemical fractionation methods that produce a (mostly) pure form of the analyte and then subsequently use highly sensitive analysis techniques to determine the nature of and quantify the analyte. Initially, fractionation was achieved on whole proteins using two-dimensional biochemical separation (2D-PAGE) by isoelectric focusing and apparent molecular mass separation, and subsequent identification of separated species was performed by Edman sequencing or mass spectrometry (MS). This approach was supplanted by a number of strategies based on online chromatographic peptide separation and subsequent gas-phase separation or isolation of selected peptide ions (precursor ions) in the gas phase.
Shotgun proteomics
Most high-throughput proteomics studies use so-called “bottom-up” liquid chromatography (LC) coupled to tandem mass spectrometry (LC-MS/MS), in which proteins are enzymatically cleaved to produce a mixture of homogeneous peptides and then separated by online LC coupled to MS/MS. In an effort to subject as many peptide precursors (molecular ions of a specific peptide entity) as possible to sequencing, the mass spectrometer selects the most intense peptide precursor for fragmentation at each time point, a process known as data-dependent acquisition or “shotgun proteomics.” This strategy is highly efficient in obtaining the fragment ion information necessary to identify the amino acid sequence of the respective peptide, since it samples precursor ions at positions with high MS1 intensity and thus has increased likelihood of obtaining a high-quality fragment ion spectrum (Aebersold and Mann, 2003; Domon and Aebersold, 2006). When applied to whole-cell lysates, shotgun proteomics provides fast enumeration of the most abundant protein species present in the sample, which enables exploratory data analysis and identification of previously unknown peptides. However, whereas shotgun proteomics allows discovery-driven research and offers high throughput, its sensitivity is strongly sample dependent, and it suffers from inconsistent identification reproducibility across samples. This is mainly due to the fact that for complex samples, the number of peptides by far exceeds the number of sequencing cycles provided by the mass spectrometer, leading to an undersampling of the proteome (Figure 1b; Michalski et al., 2011; Bruderer et al., 2015).
These challenges are substantially influenced by different sample preparation and quantification strategies. The undersampling issue can be alleviated by sample fractionation before LC-MS/MS analysis, albeit at the cost of sample throughput and increased complications in quantitative cross-run comparisons, because several repeat analyses are required per sample to achieve maximal coverage (Domon and Aebersold, 2010). Furthermore, each quantification strategy comes with its own challenges and provides different quantitative accuracy and throughput. Isotopic labeling approaches such as Isotope-coded affinity tag (ICAT), stable isotope labeling with amino acids in cell culture (SILAC), or dimethyl N-terminal labeling deliver high quantitative accuracy but increase sample complexity and further exacerbate the undersampling problem. On the other hand, isobaric labeling approaches like iTRAQ and TMT can increase multiplexing and decrease cross-sample variability on the MS1 level but at the cost of coupling quantification to fragmentation and thus accepting missing values for cases for which no fragmentation was triggered. Even though isotopic and isobaric labeling methods support multiplexing, the capacity is limited to a few (two to 10) channels per MS run, which still poses a substantial challenge in large-scale analyses, in which hundreds of samples may be analyzed. Finally, label-free approaches do not increase sample complexity but still suffer from undersampling, as well as from reduced quantitative accuracy due to the lack of an internal standard.
In the context of the systems biology data matrix, the data produced by shotgun proteomics thus pose significant challenges, since measurements are performed with high throughput and coverage but generally low comprehensiveness. Often the resulting data matrices are only complete for the most intense peptides of high abundance proteins but contain missing values for proteins of lower abundance (Figure 1b; Sabidó et al., 2012). In addition, the more samples are analyzed and the more biologically diverse the samples are, the lower is the number of complete rows; due to the intensity dependence of the sampling and undersampling issues for complex samples, the missing values will generally not be missing completely at random (Bruderer et al., 2015). Specifically, proteins that are variable across the experimental conditions will likely contain more missing values (with those conditions not quantified where abundance is low), whereas highly abundant, invariant proteins are faithfully sampled by the approach. It is therefore the efficiency of shotgun proteomics that produces maximal information on a single sample that is detrimental to the production of highly informative data matrices on multiple samples, since sampling more often occurs at noninformative positions, whereas information-rich processes with high variance are sparsely sampled.
Targeted proteomics
To address these problems, proteomic scientists have developed techniques that allow deterministic sampling across multiple conditions (Sabidó et al., 2012). The most prominent ones are “targeted proteomics” approaches, specifically selective reaction monitoring (SRM) and, more recently, parallel reaction monitoring, both of which can target multiple proteins (which need to be selected before the measurement) consistently across multiple conditions (Lange et al., 2008; Domon and Aebersold, 2010). In SRM mode, the mass spectrometer is programmed to deterministically record the signal at fixed coordinates across the chromatographic retention time. These coordinates (the assay) are specific to a peptide analyte and will reliably detect the analyte signal if present, similarly to a classical biochemical assay such as an antibody-based method. The acquisition of signal for multiple fragment ions (transitions) ensures high specificity (Sherman et al., 2009; Röst et al., 2012) and sensitivity. This deterministic acquisition strategy increases reproducibility and quantification consistency compared to shotgun approaches, where sampling is semistochastic and data acquisition for each single peptide depends on a multitude of factors. However, SRM is limited by throughput and can only monitor dozens to hundreds of peptides per run, since the deterministic sampling strategy implies acquiring signal even at time points at which no analyte elutes in order to collect complete chromatographic traces (Picotti et al., 2013).
Thus the data matrices obtained from SRM are much more complete than those produced by shotgun proteomics but generally contain one to two orders of magnitude fewer proteins (Figure 1c). Because the proteins to be measured have to be preselected, the measurements tend to be biased by prior hypotheses and may not cover all biologically relevant cellular processes and pathways. Therefore SRM has mostly been used in studies in which large sample numbers are required and only few proteins are under investigation (such as clinical biomarker studies; Cima et al., 2011; Hüttenhain et al., 2012; Drabovich et al., 2013; Li, 2013; Surinova et al., 2015a, b), for protein quantitative trait analysis, in which sets of protein are quantified across genetic reference strain collections (Picotti et al., 2013; Wu et al., 2014), or for systems biology studies, in which the response of a biological system to perturbations is measured (Sabidó et al., 2013).
PROTEOMICS FOR SYSTEMS BIOLOGY
For systems biology investigations, neither SRM nor shotgun approaches are fully satisfactory to generate the desired complete data matrix. Whereas shotgun proteomics places heavy emphasis on the analyte dimensions and successfully identifies many protein species, it is often challenging to trace analytes across the sample dimension (Figure 1b). Conversely, SRM is well able to quantify analytes across many MS runs but suffers from low throughput in the analyte dimension (Figure 1c). To allow proteomics to become a true systems science, efforts should be directed toward improving proteomics measurement with regard to both dimensions of the data matrix, which means that future improvements in measurement technology and analysis strategy should be evaluated by the quality of the data matrices they are able to produce. Although the field was highly successful in compiling extensive protein inventories in the past, future efforts should turn toward the generation of fully quantitative, high-quality data matrices.
This challenge has been recognized by the field, and multiple efforts toward this aim have been presented recently or are under way. In particular, recent advances in acquiring and analyzing data-independent acquisition mass-spectrometric data, such as SWATH-MS data, constitute a promising advance toward this goal (Gillet et al., 2012; Röst et al., 2014). In SWATH-MS, the mass spectrometer performs deterministic acquisition of fragment ion spectra but does not aim to target specific peptides explicitly by their intensity (as shotgun does) or by prior hypothesis (as SRM does). Instead, SWATH-MS records the complete fragment ion signal in a single experiment, essentially creating a complete digital representation of all fragment ion signals in a biological sample. This digitized sample can then be used to extract quantitative information for individual peptides after data acquisition. SWATH-MS features the same characteristics as SRM regarding specificity, reproducibility, and sensitivity but allows for high throughput and coverage of the analyzed proteome (Table 1; Gillet et al., 2012). Similar to SRM, in the sample dimension, SWATH-MS is able to reproducibly measure protein analytes across hundreds of samples. However, unlike SRM, SWATH-MS is capable of high throughput in the analyte dimension and achieves substantial proteomic coverage; in microbial samples, coverage reaches almost saturation even with single MS injections (Röst et al., 2014; Schubert et al., 2015b). However, one of the main limitations of SWATH-MS is the complexity of the resulting data, which consists of highly multiplexed fragment ion spectra that require novel algorithmic approaches for deconvolution. To assign signal to individual peptides and quantify analytes, multiple open-source tools using complementary algorithms are available, but further research is required to improve the underlying analysis approaches and fully exploit the potential of SWATH-MS.
TABLE 1:
Comparison of MS-based proteomics methods.
| Shotgun | SRM | Data-independent acquisition (SWATH-MS) | |
|---|---|---|---|
| Throughput | High | Low to medium | Medium to high |
| Reproducibility | Low | High | High |
| Identification specificity | High | Medium | Medium |
| Sensitivity | Low to medium | High to very high | Medium to high |
| Quantitative accuracy | Medium to high | High to very high | High |
| Acquisition method | Fragment spectra | Fragment chromatograms | Fragment spectra and chromatograms |
| Application | Protein enumeration and discovery | Reproducible quantification | Reproducible quantification in high throughput |
| Analysis software | Well established | Visual (manual) | Multiple tools available |
This table compares three major techniques used in mass spectrometry–based proteomics according to different performance criteria: shotgun proteomics, targeted proteomics or SRM, and data-independent acquisition or SWATH-MS. All three techniques have unique benefits and disadvantages; therefore different techniques need to be applied for different tasks.
Thus, SWATH-MS is a technology that addresses both dimensions of the data matrix at the same time and allows true systems analysis on protein measurements. It provides a valuable addition to the set of tools available to proteomics researchers and strikes a balance between throughput and reproducibility, making it an interesting option next to shotgun and targeted proteomics. Recent studies have shown the applicability of SWATH-MS to a multitude of problems in systems biology and medicine. These studies include investigations of the dynamics of microbial virulence with high proteomic coverage (Röst et al., 2014; Schubert et al., 2015b), the interrogation of the dynamics of the human interactome (Collins et al., 2013; Lambert et al., 2013), and the quantification of >2000 proteins in human and mouse tissue across multiple patient samples and experimental conditions (Bruderer et al., 2015; Guo et al., 2015). In addition, SWATH-MS measurements allowed the investigation of protein abundance of 342 human plasma proteins across >200 individuals, uncovering considerable variation of blood plasma protein levels across genetically identical twins and quantifying the relative contributions of heredity and environmental factors to the overall observed variability (Liu et al., 2015). Analysis of SWATH-MS samples was further facilitated by the recent development of multiple software tools to analyze the generated data sets (MacLean et al., 2010; Bernhardt et al., 2012; Röst et al., 2014, 2015a, b; Teleman et al., 2015; Tsou et al., 2015), the development of a step-by-step protocol to generate high-quality assay libraries (Schubert et al., 2015a), and the publication of SWATH-compatible assay libraries containing the measurement coordinates for >10,000 human proteins (Rosenberger et al., 2014). SWATH-MS is thus a promising technology that could help to provide the proteomics field with complete and accurate data matrices and may play a key role in investigating systems biology questions on the protein level.
CONCLUSION
When evaluating proteomics techniques from the viewpoint of the quantitative proteotype data matrix, we can obtain a much clearer picture of data utility for systems biology studies. It becomes apparent that neither patchy matrices littered with missing values nor highly consistent measurements of a few proteins are sufficient for systems approaches to biology. Although shotgun and SRM are valuable for a multitude of purposes, new paradigms need to be developed in order to be able to apply unbiased, data-driven systems approaches in proteomics. The field should embrace this realization and increase efforts to establish novel experimental and computational methods able to produce data matrices with extensive proteome coverage and high comprehensiveness suitable for quantitative biology approaches.
Current technology and analysis software has matured enough by now to tackle the next major challenge in proteomics, namely the proteotype data matrix. Next-generation proteomics technologies, such as SWATH-MS, present promising solutions to address this challenge. They combine the strength of SRM (high reproducibility and quantitative accuracy) with the high throughput of shotgun proteomics, thus focusing on both analyte and sample dimension of the data matrix at the same time. Using SWATH-MS, proteomics technology can produce quantitatively accurate and qualitatively complete data matrices, allowing researchers to track protein quantities across many samples. These advances in the field will allow proteomics researchers to ask novel questions about ensembles of proteins and their behavior across many experimental conditions, time points, and individuals. Thus proteomics is expected to contribute significantly to the emerging fields of precision and personalized medicine, high-throughput screening, and analysis, as well as to systems biology and systems medicine.
Abbreviations used:
- 2D-PAGE
two-dimensional PAGE
- ICAT
Isotope-coded affinity tag
- iTRAQ
isobaric tags for relative and absolute quantitation
- LC
liquid chromatography
- MS
mass spectrometry
- NGS
next-generation sequencing
- SILAC
stable isotope labeling with amino acids in cell culture
- TMT
tandem mass tag.
Footnotes
REFERENCES
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- Bernhardt OM, Selevsek N, Gillet LC, Rinner O, Picotti P, Aebersold R, Reiter L. Spectronaut: a fast and efficient algorithm for MRM-like processing of data independent acquisition (SWATH-MS) data. 60th American Society for Mass Spectrometry Conference 2012. 2012 [Google Scholar]
- Bodenmiller B, Wanka S, Kraft C, Urban J, Campbell D, Pedrioli PG, Gerrits B, Picotti P, Lam H, Vitek O, et al. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal. 2010;3:rs4. doi: 10.1126/scisignal.2001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruderer R, Bernhardt OM, Gandhi , Miladinovic´ SM, Cheng LY, Messner S, Ehrenberger T, Zanotelli V, Butscheid Y, Escher C, et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol Cell Proteomics. 2015;14:1400–1410. doi: 10.1074/mcp.M114.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cima I, Schiess R, Wild P, Kaelin M, Schüffler P, Lange V, Picotti P, Ossola R, Templeton A, Schubert O, et al. Cancer genetics-guided discovery of serum biomarker signatures for diagnosis and prognosis of prostate cancer. Proc Natl Acad Sci USA. 2011;108:3342–3347. doi: 10.1073/pnas.1013699108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins BC, Gillet LC, Rosenberger G, Röst HL, Vichalkovski A, Gstaiger M, Aebersold R. Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat Methods. 2013;10:1246–1253. doi: 10.1038/nmeth.2703. [DOI] [PubMed] [Google Scholar]
- Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- Domon B, Aebersold R. Options and considerations when selecting a quantitative proteomics strategy. Nat Biotechnol. 2010;28:710–721. doi: 10.1038/nbt.1661. [DOI] [PubMed] [Google Scholar]
- Drabovich AP, Dimitromanolakis A, Saraon P, Soosaipillai A, Batruch I, Mullen B, Jarvi K, Diamandis EP. Differential diagnosis of azoospermia with proteomic biomarkers ECM1 and TEX101 quantified in seminal plasma. Sci Transl Med. 2013;5:212ra160. doi: 10.1126/scitranslmed.3006260. [DOI] [PubMed] [Google Scholar]
- Edwards AM, Isserlin R, Bader GD, Frye SV, Willson TM, Frank HY. Too many roads not taken. Nature. 2011;470:163–165. doi: 10.1038/470163a. [DOI] [PubMed] [Google Scholar]
- Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.O111.016717. O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Kouvonen P, Koh CC, Gillet LC, Wolski WE, Röst HL, Rosenberger G, Collins BC, Blum LC, Gillessen S, et al. Rapid mass spectrometric conversion of tissue biopsy samples into permanent quantitative digital proteome maps. Nat Med. 2015;21:407–413. doi: 10.1038/nm.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhain R, Soste M, Selevsek N, Röst H, Sethi A, Carapito C, Farrah T, Deutsch EW, Kusebauch U, Moritz RL, et al. Reproducible quantification of cancer-associated proteins in body fluids using targeted proteomics. Sci Transl Med. 2012;4:142ra94. doi: 10.1126/scitranslmed.3003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage K, Møllgård K, Greenway S, Wakimoto H, Gorham JM, Workman CT, Bendsen E, Hansen NT, Rigina O, Roque FS, et al. Dissecting spatio-temporal protein networks driving human heart development and related disorders. Mol Syst Biol. 2010;6:381. doi: 10.1038/msb.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JP, Ivosev G, Couzens AL, Larsen B, Taipale M, Lin ZY, Zhong Q, Lindquist S, Vidal M, Aebersold R, et al. Mapping differential interactomes by affinity purification coupled with data-independent mass spectrometry acquisition. Nat Methods. 2013;10:1239–1245. doi: 10.1038/nmeth.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XJ, Hayward C, Fong PY, Dominguez M, Hunsucker SW, Lee LW, McLean M, Law S, Butler H, Schirm M, et al. A blood-based proteomic classifier for the molecular characterization of pulmonary nodules. Sci Transl Med. 2013;5:207ra142. doi: 10.1126/scitranslmed.3007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libbrecht MW, Noble WS. Machine learning applications in genetics and genomics. Nat Rev Genet. 2015;16:321–332. doi: 10.1038/nrg3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Buil A, Collins BC, Gillet LC, Blum LC, Cheng LY, Vitek O, Mouritsen J, Lachance G, Spector TD, et al. Quantitative variability of 342 plasma proteins in a human twin population. Mol Syst Biol. 2015;11:786. doi: 10.15252/msb.20145728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chen J, Sethi A, Li QK, Chen L, Collins B, Gillet LCJ, Wollscheid B, Zhang H, Aebersold R. Glycoproteomic analysis of prostate cancer tissues by SWATH mass spectrometry discovers n-acylethanolamine acid amidase and protein tyrosine kinase 7 as signatures for tumor aggressiveness. Mol Cell Proteomics. 2014;13:1753–1768. doi: 10.1074/mcp.M114.038273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski A, Cox J, Mann M. More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J Proteome Res. 2011;10:1785–1793. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- Picotti P, Clément-Ziza M, Lam H, Campbell DS, Schmidt A, Deutsch EW, Röst H, Sun Z, Rinner O, Reiter L, et al. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature. 2013;494:266–270. doi: 10.1038/nature11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reker D, Malmström L. Bioinformatic challenges in targeted proteomics. J Proteome Res. 2012;11:4393–4402. doi: 10.1021/pr300276f. [DOI] [PubMed] [Google Scholar]
- Rosenberger G, Koh CC, Guo T, Röst HL, Kouvonen P, Collins BC, Heusel M, Liu Y, Caron E, Vichalkovski A, et al. A repository of assays to quantify 10,000 human proteins by SWATH-MS. Sci Data. 2014;1:140031. doi: 10.1038/sdata.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röst HL, Rosenberger G, Navarro P, Gillet L, Miladinovic´ SM, Schubert OT, Wolski W, Collins BC, Malmström J, Malmström L, et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat Biotechnol. 2014;32:219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- Röst H, Malmström L, Aebersold R. A computational tool to detect and avoid redundancy in selected reaction monitoring. Mol Cell Proteomics. 2012;11:540–549. doi: 10.1074/mcp.M111.013045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röst HL, Rosenberger G, Aebersold R, Malmström L. Efficient visualization of high-throughput targeted proteomics experiments: Tapir. Bioinformatics. 2015a;31:2415–2417. doi: 10.1093/bioinformatics/btv152. [DOI] [PubMed] [Google Scholar]
- Röst HL, Schmitt U, Aebersold R, Malmström L. Fast and efficient XML data access for next-generation mass spectrometry. PLoS One. 2015b;10:e0125108. doi: 10.1371/journal.pone.0125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabidó E, Selevsek N, Aebersold R. Mass spectrometry-based proteomics for systems biology. Curr Opin Biotechnol. 2012;23:591–597. doi: 10.1016/j.copbio.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Sabidó E, Wu Y, Bautista L, Porstmann T, Chang C-Y, Vitek O, Stoffel M, Aebersold R. Targeted proteomics reveals strain-specific changes in the mouse insulin and central metabolic pathways after a sustained high-fat diet. Mol Syst Biol. 2013;9:681. doi: 10.1038/msb.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert OT, Gillet LC, Collins BC, Navarro P, Rosenberger G, Wolski WE, Lam H, Amodei D, Mallick P, MacLean B, et al. Building high-quality assay libraries for targeted analysis of SWATH MS data. Nat Protoc. 2015a;10:426–441. doi: 10.1038/nprot.2015.015. [DOI] [PubMed] [Google Scholar]
- Schubert OT, Ludwig C, Kogadeeva M, Zimmermann M, Rosenberger G, Gengenbacher M, Gillet LC, Collins BC, Röst HL, Kaufmann SH, et al. Absolute proteome composition and dynamics during dormancy and resuscitation of mycobacterium tuberculosis. Cell Host Microbe. 2015b;18:96–108. doi: 10.1016/j.chom.2015.06.001. [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Sherman J, McKay MJ, Ashman K, Molloy MP. Unique ion signature mass spectrometry, a deterministic method to assign peptide identity. Mol Cell Proteomics. 2009;8:2051–2062. doi: 10.1074/mcp.M800512-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surinova S, Choi M, Tao S, Schüffler PJ, Chang CY, Clough T, Vysloužil K, Khoylou M, Srovnal J, Liu Y, et al. Prediction of colorectal cancer diagnosis based on circulating plasma proteins. EMBO Mol Med. 2015a;7:1166–1178. doi: 10.15252/emmm.201404873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surinova S, Radová L, Choi M, Josef S, Brenner H, Vitek O, Hajdúch M, Aebersold R. Non-invasive prognostic protein biomarker signatures associated with colorectal cancer. EMBO Mol Med. 2015b;7:1153–1165. doi: 10.15252/emmm.201404874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan AL, Mobasheri A, Allaway D, Liddell S, Bacardit J. Application of machine learning to proteomics data: classification and biomarker identification in postgenomics biology. OMICS. 2013;17:595–610. doi: 10.1089/omi.2013.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman J, Röst H, Rosenberger G, Schmitt U, Malmström L, Malmström J, Levander F. DIANA—algorithmic improvements for analysis of data-independent acquisition MS data. Bioinformatics. 2015;31:555–562. doi: 10.1093/bioinformatics/btu686. [DOI] [PubMed] [Google Scholar]
- Tsou C-C, Avtonomov D, Larsen B, Tucholska M, Choi H, Gingras A-C, Nesvizhskii AI. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat Methods. 2015;12:258–264. doi: 10.1038/nmeth.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Williams EG, Dubuis S, Mottis A, Jovaisaite V, Houten SM, Argmann CA, Faridi P, Wolski W, Kutalik Z, et al. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell. 2014;158:1415–1430. doi: 10.1016/j.cell.2014.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]


