Abstract
Overexpression experiments are sometimes considered as qualitative experiments designed to identify novel proteins and study their function. However, in order to draw conclusions regarding protein overexpression through association analyses using large-scale biological data sets, we need to recognize the quantitative nature of overexpression experiments. Here I discuss the quantitative features of two different types of overexpression experiment: absolute and relative. I also introduce the four primary mechanisms involved in growth defects caused by protein overexpression: resource overload, stoichiometric imbalance, promiscuous interactions, and pathway modulation associated with the degree of overexpression.
INTRODUCTION
Cellular functions are performed through cooperative actions of thousands of proteins. Intracellular levels of these proteins vary substantially (Kulak et al., 2014; Liebermeister et al., 2014; Figure 1), and the level of each protein has to be highly optimized to maximize cellular functionality (Zaslaver et al., 2004; Dekel and Alon, 2005; Wagner, 2005; Li et al., 2014). The abnormal expression of a protein can have a detrimental effect on cellular functions. Overexpression of some proteins is associated with human conditions, such as Down syndrome and cancer (Tang and Amon, 2013). Researchers have used artificial overexpression of proteins to identify novel proteins involved in biological processes of interest and examine the functions of their target proteins (Rine, 1991; Prelich, 2012). Detailed molecular mechanisms found in individual overexpression experiments are summarized by Prelich (2012). Here I focus on the results of large-scale overexpression experiments using the budding yeast Saccharomyces cerevisiae. I emphasize the often-overlooked quantitative nature of overexpression experiments, as shown by considering normal protein levels. I also discuss some primary mechanistic consequences of protein overexpression as demonstrated in large-scale experiments.
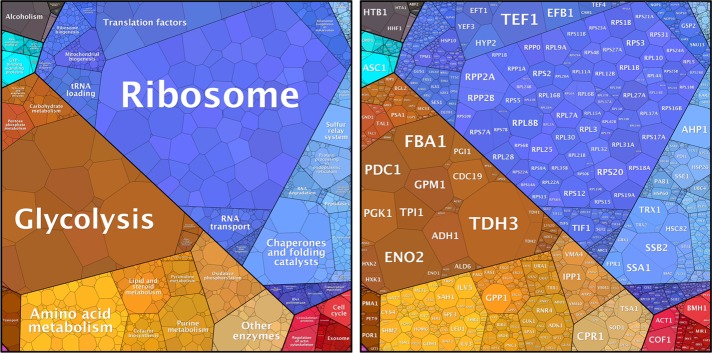
FIGURE 1:
Expression levels of yeast proteins and their allocations in the yeast proteome. Proteome data (Kulak et al., 2014; Supplemental Table S1) visualized using a proteomap (Liebermeister et al., 2014; www.proteomaps.net/). The area occupied by each process (left) or protein (right) is proportional to its relative protein amount in a cycling yeast population.
OVEREXPRESSION EXPERIMENTS ARE INHERENTLY QUANTITATIVE
Many large-scale overexpression experiments have been performed using transcriptional induction of strong promoters in yeast to identify the proteins causing growth defects when overexpressed (hereafter called dosage-sensitive proteins; Liu et al., 1992; Espinet et al., 1995; Akada et al., 1997; Stevenson et al., 2001; Boyer et al., 2004; Gelperin et al., 2005; Sopko et al., 2006; Niu et al., 2008; Yoshikawa et al., 2011; Douglas et al., 2012). In most cases, overexpression experiments are considered as qualitative screening methods to identify novel proteins involved in cell morphology and cellular processes, such as the cell cycle (Stevenson et al., 2001; Sopko et al., 2006; Niu et al., 2008). Recently the characteristics of proteins that are strongly associated with dosage-sensitive proteins have been surveyed in an attempt to uncover primary mechanistic consequences of protein overexpression (Gelperin et al., 2005; Sopko et al., 2006; Vavouri et al., 2009; Ma et al., 2010; Yoshikawa et al., 2011; Makanae et al., 2013; Tomala and Korona, 2013).
In gene deletion experiments, the expression of a target protein is stopped by removing the protein-coding regions from the genome. On the other hand, in overexpression experiments, the degree of overexpression of a target protein is quite diverse and varies with the methods and target proteins used. Therefore, to use data on dosage-sensitive proteins obtained by large-scale overexpression experiments for association studies, we need to recognize that the overexpression experiments are inherently quantitative. In the next sections, I discuss the quantitative nature of overexpression experiments, distinguishing them into two different types: absolute and relative.
ABSOLUTE OVEREXPRESSION USING PROMOTER SWAPPING
The GAL promoter is commonly used for strong induction of protein expression in the budding yeast. The promoter-swapped gene cassettes are sometimes cloned into multicopy 2-μm plasmids to further increase expression levels (Liu et al., 1992; Espinet et al., 1995; Akada et al., 1997; Stevenson et al., 2001; Boyer et al., 2004; Gelperin et al., 2005; Sopko et al., 2006; Niu et al., 2008; Yoshikawa et al., 2011; Douglas et al., 2012). In these experiments, target proteins are induced at similar high levels. This type of experiment is considered an absolute overexpression experiment. For further discussion, I use the dosage-sensitive protein data set obtained by Sopko et al. (2006) as a representative data set for absolute overexpression experiments. That study is the most extensive, widely cited analysis.
Because expression levels of proteins can vary by more than 10,000-fold (Kulak et al., 2014; Figures 1 and 2), the degree of the overexpression in absolute overexpression experiments should differ by some orders of magnitude. The average expression level of target proteins expressed by genes under the control of the GAL1 promoter on a 2-μm plasmid has been estimated as ∼1% of the proteins expressed in the yeast (Tomala and Korona, 2013). According to recent proteome data (Kulak et al., 2014; Figure 1), ∼50,000,000 protein molecules are expressed in a budding yeast cell. Only eight glycolytic enzymes are expressed more than 1% of the total proteins (500,000 molecules/cell). Almost all proteins are thus “overexpressed” in the foregoing experiment. However, the degree of overexpression relative to the native level should vary strongly among the analyzed proteins (Figure 2). If the native level of a target protein is 100 molecules/cell, the degree of overexpression will be estimated as 50,000-fold. If the native level of a target protein is 100,000 molecules/cell, the overexpression degree will be 50-fold. Thus the absolute overexpression experiments analyze the consequences of comparably strong production of target proteins independently of their native expression levels. As shown in Figure 3A, proteins with natively low expression are preferentially isolated as the dosage-sensitive proteins in absolute overexpression experiments. If the native expression levels are strongly optimized (see later discussion), the huge degree of overexpression of low-expression proteins can become a bias leading to preferential isolation of those proteins.
FIGURE 2:
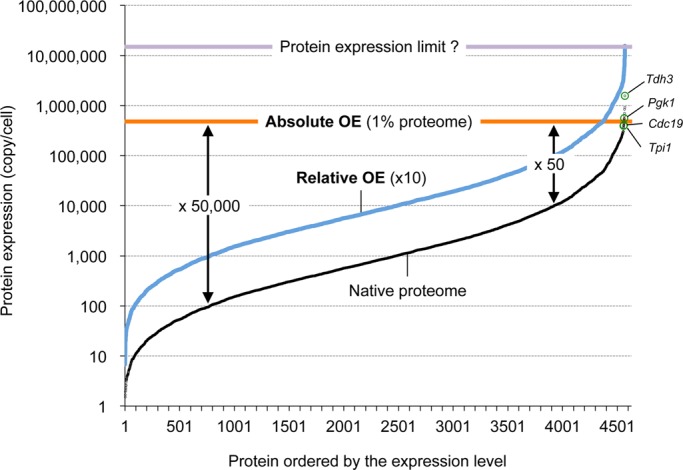
Conceptual representation of the quantitative nature of protein overexpression experiments. Native expression levels of yeast proteins (Kulak et al., 2014; Supplemental Table S1) are shown in black. Estimated levels of the overexpressed proteins in an absolute overexpression experiment (orange line) and a relative overexpression experiment (blue line). Estimated degrees of overexpression (fold increases) are shown. Highly expressed glycolytic enzymes analyzed in Table 1 are shown in green circles. The estimated maximal protein expression level (Table 1) is shown in purple.
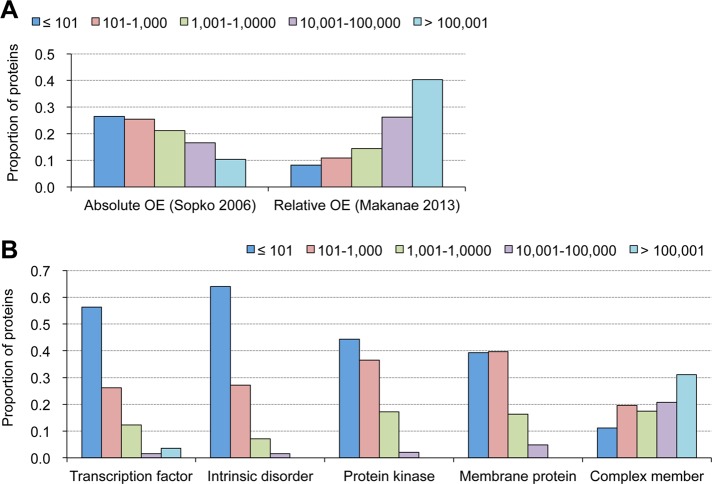
FIGURE 3:
Trends in native expression levels of dosage-sensitive proteins and of proteins within various categories. (A) Proteins isolated in absolute and relative overexpression experiments show opposite trends in their native expression levels. Absolute overexpression tends to isolate lowly expressed proteins, whereas relative overexpression tends to isolate highly expressed proteins. Proteins isolated as dosage-sensitive proteins in overexpression experiments are separated by their expression levels, and their proportions are shown. Absolute overexpression, 603 proteins (Sopko et al., 2006); relative overexpression, 682 proteins (Makanae et al., 2013). (B) Proteins within different categories show different trends in their native expression levels, which could be biases to isolate dosage-sensitive proteins in overexpression experiments. Proteins within indicated categories are separated by their expression levels, and their proportions are shown. Transcription factors, 125 proteins (Teixeira et al., 2014; www.yeastract.com/); intrinsic disorder, 439 proteins with GlobPlot score >200 (Linding et al., 2003; http://globplot.embl.de/); protein kinases, 114 proteins (Breitkreutz et al., 2010; www.yeastkinome.com/); membrane proteins; 255 proteins with TMHMM score >100 (www.cbs.dtu.dk/services/TMHMM/); and protein complex members, 1485 proteins (Pu et al., 2009). The original data are given in Supplemental Table S1.
RELATIVE OVEREXPRESSION USING GENE COPY NUMBER INCREASE
Increasing the gene copy number for the target protein should induce overexpression of the target protein. Multicopy plasmids derived from the 2-μm plasmid are usually used for this purpose in budding yeast (Rine, 1991; Jones et al., 2008). If the expression of the protein is controlled by its native regulatory elements (i.e., promoter and terminator), the expression relative to its native level should increase as the copy number increases. If a multicopy plasmid with a copy number of 10 is used in the experiment, the expression of the proteins coded on the plasmid is expected to increase 10-fold (Figure 2, blue line). This type of experiment is considered a relative overexpression experiment. The genetic tug-of-war (gTOW) experiment (Moriya et al., 2006, 2011, 2012; Kaizu et al., 2010; Makanae et al., 2013) is an example. In the gTOW experiment, the relative overexpression levels (fold increases) causing cellular defects are estimated by measuring the copy number limits of the genes encoding the target proteins. For further discussion, I use the dosage-sensitive protein data set obtained by Makanae et al. (2013) as a representative data set for the relative overexpression experiments; their analysis is the most extensive available.
In relative overexpression experiments, levels of target proteins after overexpression vary, depending on the native expression levels. If a target protein is already highly expressed natively, the after overexpression, a much larger amount of the protein should be present in the cell. As shown in Figure 3A, proteins with high native expression are preferentially isolated as the dosage-sensitive proteins in the relative overexpression experiment. High-level protein expression can become a bias in the preferential isolation of such proteins in relative overexpression experiments. This is because the need for massive turnover of proteins causes cellular defects due to process overloads (discussed later).
A series of physiological analyses have been performed using disomic yeast strains with 1 of the 16 chromosomes duplicated in a haploid cell (Torres et al., 2007, 2008, 2010; Sheltzer et al., 2011, 2012; Oromendia et al., 2012; Thorburn et al., 2013; Dephoure et al., 2014; Blank et al., 2015; Bonney et al., 2015). In each disomic cell, almost all of the genes on the duplicated chromosome are overexpressed twofold (Torres et al., 2007). This can be also considered relative overexpression. Analysis of this simultaneous overexpression of hundreds of genes has led to many important insights into the consequences of protein overexpression (Torres et al., 2008; Sheltzer and Amon, 2011; Tang and Amon, 2013; Oromendia and Amon, 2014).
As discussed, absolute and relative overexpression experiments carry different biases due to their quantitative nature. These experiments contribute to the understanding of the consequences of protein overexpression. However, the biases have to be taken into consideration when investigating the mechanistic consequences of protein overexpression using association analyses of large-scale biological data sets. In the following section, I discuss the independent mechanisms of cellular defects upon overexpression of proteins in the context of the quantitative nature of overexpression experiments. Overexpression experiments have also been performed in other organisms, such as Schizosaccharomyces pombe, Drosophila melanogaster, Arabidopsis thaliana, and Homo sapiens (Prelich, 2012), all using promoter swapping, meaning that they are all absolute overexpression experiments.
CONSEQUENCES OF PROTEIN OVEREXPRESSION
The ways by which protein overexpression can cause cellular defects depend on the protein properties and functions (Prelich, 2012). Several mechanisms of cellular defects upon overexpression of proteins have been suggested from the analyses of dosage-sensitive proteins obtained in genome-wide absolute (Vavouri et al., 2009; Ma et al., 2010; Tomala et al., 2014) and relative overexpression experiments (Makanae et al., 2013) and from the analyses of physiologies of disomic yeast strains (Sheltzer and Amon, 2011; Tang and Amon, 2013). Here I focus on four primary mechanisms: resource overload, stoichiometric imbalance, promiscuous interactions, and pathway modulation (Figure 4). The growth defects are largely explained by overload and toxicity (Tomala and Korona, 2013). Overload is an abnormal cellular environment created by enhancement of normal activity or turnover of a protein caused by its overexpression. Toxicity is an effect causing cellular defects due to novel and unrelated properties generated by the overexpression of a protein. Overload is expressed as either resource overload or pathway modulation. Toxicity is triggered by promiscuous interactions. Stoichiometric imbalance can trigger both overload and toxicity.
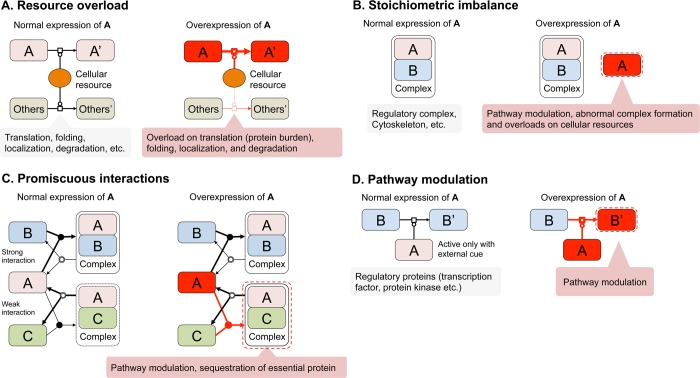
FIGURE 4:
Primary mechanisms of cellular defects after protein overexpression. (A) Resource overload. When a protein requires large amounts of cellular resources for translation, folding, localization, or degradation, the overexpression of the protein overloads those cellular resources. The protein burden effect is believed to be one of the overload of translation resources (i.e., ribosomes). (B) Stoichiometric imbalance. When a protein is a subunit of a protein complex, the overexpression of the protein disrupts the stoichiometry. The excess of subunits causes pathway modulation, abnormal complex formation, or overload of the cellular protein quality control resources. (C) Promiscuous interaction. Enhancement of protein–protein interaction upon the overexpression of ID region–containing proteins and aggregative proteins causes pathway modulation or sequestration of essential proteins. Under normal conditions, complex A–C is either scarce or nonexistent. (D) Pathway modulation. Overexpression of a regulatory protein causes pathway modulation. Pathway modulation can be triggered by stoichiometric imbalance and promiscuous interactions. Most regulators might have very large buffering ranges to avoid an untimely pathway modulation (see text). Diagrams are based on process description language level 1 of the systems biology graphical notation (www.sbgn.org/). An arrow with an open square refers to a process. A line that ends with a circle signifies catalysis for the process to which it attaches. A merging arrow with a filled circle signifies association, and a branching arrow with a double circle indicates dissociation. The thickness of each arrow reflects the rate of each process. Processes shown with black lines happen in normal growth conditions. Processes shown with red lines are triggered upon overexpression of A.
RESOURCE OVERLOAD
Turnover of a protein includes its production (translation), folding, localization, degradation, and posttranslational modifications. These processes require their own cellular resources. Strong expression of a protein might overstretch these resources by monopolizing them, leading to cellular defects (Figure 4A). Strong expression of unnecessary proteins causes growth defects (Snoep et al., 1995; Stoebel et al., 2008; McIsaac et al., 2011). These defects are created by a protein burden effect. The effect becomes apparent when the cellular resources for protein turnover, especially the ribosomes producing other proteins, are overstretched, monopolized by a strong expression of unnecessary proteins (Stoebel et al., 2008; Shachrai et al., 2010; Shah et al., 2013). Any otherwise harmless protein thus could cause growth defects due to the protein burden effect when it is ultimately highly expressed. In other words, proteins that have no harmful effects on cellular functions could be expressed at levels causing a protein burden. These levels are considered the maximum protein expression limit in a yeast cell and are almost the same for any protein (see later discussion).
What are the proteins expressed at these limits, and what is the maximum protein expression limit? We have presented evidence that the limits of overexpression for some highly expressed glycolytic enzymes (Tdh3, Pgk1, Cdc19, and Tpi1; Figure 2, green circles) are determined by the protein burden (Makanae et al., 2013). In gTOW overexpression experiments, these proteins should be expressed up to the levels causing cellular defects due to the protein burden. As shown in Table 1, we can estimate the expression levels by multiplying the native expression levels by the copy number limits of the glycolytic enzymes (the average level is shown as the purple line in Figure 2). The total proteome consists of ∼50,000,000 molecules/cell (Kulak et al., 2014). We can estimate the number of amino acids in the total proteome by multiplying the number of protein molecules by their amino acid lengths and adding the results. This estimate gives ∼16,000,000,000 amino acids/cell (Supplemental Table S1). Thus we estimate the expression levels of the aforementioned glycolytic enzymes as 14.2–56.5% of the total proteome in terms of the number of protein molecules and as 14.0–41.6% of the total proteome in terms of amino acid molecules (Table 1). Although these are very rough estimates, the residual protein production capacity of a yeast cell and the maximum expression limit for any protein can be inferred from these results.
TABLE 1:
Estimation of absolute overexpression levels of proteins in relative overexpression experiments.
| Gene name | ORF name | Protein molecules per cell (% of total proteome) (#1) | Gene copy number limit (#2) | Protein length (amino acids) (#3) | Product of #1 × #2 molecules (% of total proteome) | Product of #1 × #2 × #3 amino acids (% of total proteome) |
|---|---|---|---|---|---|---|
| TDH3 | YGR192C | 1,575,311 (3.3) | 4.3 | 332 | 6,801,949 (14.2) | 2,258,246,952 (14.0) |
| PGK1 | YCR012W | 561,265 (1.2) | 22.0 | 416 | 1,2358,595 (25.8) | 5,141,175,549 (31.9) |
| CDC19 | YAL038W | 404,162 (0.8) | 31.8 | 500 | 12,864,113 (26.9) | 6,432,056,484 (39.9) |
| TPI1 | YDR050C | 395,237 (0.8) | 68.5 | 248 | 27,054,398 (56.5) | 6,709,490,697 (41.6) |
| RPL9A | YGL147C | 265,169 (0.6) | 6.2 | 191 | 1,642,828 (3.4) | 313,780,172 (1.9) |
| RPS12 | YOR369C | 258,298 (0.5) | 6.2 | 143 | 1,611,136 (3.4) | 230,392,526 (1.4) |
| Total proteome | 47,900,214 | 16,130,396,439 |
The original data are given in Supplemental Table S1.
Because the overexpression of glycolytic enzymes might perturb the glycolytic flux, these estimates do not necessarily reflect the maximum expression limits for other proteins. However, if a protein causes a cellular defect at an expression level substantially below this limit, we can assume that mechanisms other than the protein burden effect are involved. Ribosomal proteins are generally highly expressed (Figure 1). Among them, RPL9A and RPS12 have been isolated as dosage-sensitive proteins because they have low copy number limits (Table 1). The estimated protein levels in the overexpression experiments are 1.4–1.9% of the total proteome in terms of amino acids (Table 1), substantially lower than the limit levels calculated using the glycolytic enzymes. We can assume that there is an unknown mechanism other than the protein burden causing the cellular defects upon the overexpression of ribosomal proteins.
Intracellular proteins are folded with the help of molecular chaperones, localized to specific cellular compartments by the translocation and transport machinery, and degraded by the proteasome. Overexpression of proteins with strong demands for those resources might cause resource overloads, leading to cellular defects (Figure 4A). Growth defects in disomic strains have been explained by the overload of the protein quality control machinery involved in protein folding and degradation (Torres et al., 2010; Oromendia et al., 2012). The protein degradation overload has been also tested by overexpression experiments using the model protein green fluorescent protein (GFP). The addition of a degradation signal to GFP increased its potential to cause cellular defects upon its overexpression (Makanae et al., 2013). As noted earlier, gTOW relative overexpression experiments preferentially isolate natively highly expressed proteins as dosage-sensitive proteins (Figure 3A). Because levels after overexpression of those proteins should be very high (Figure 2), they might include many proteins causing overloads of different cellular resources.
STOICHIOMETRIC IMBALANCE
The expression level of each protein (as shown in Figure 1) should be ultimately determined by various types of equilibrium: the balanced numbers of subunits within a protein complex and of protein complexes within a cellular module and balance among cellular modules. Appropriate resource allocation in cellular modules should maximize cellular performance (Liebermeister et al., 2014). For example, the production rate of each subunit of a complex is highly related to the molecular ratio (stoichiometry) of the subunit within the complex (Li et al., 2014). This indicates that cellular systems are optimized to keep protein expression in balance. Because overexpression of a protein complex member perturbs the equilibrium, stoichiometric imbalance is believed to be a primary mechanism of cellular defects upon protein overexpression (balance hypothesis; Papp et al., 2003; Veitia et al., 2008; Figure 4B). Examples of cellular defects caused by overexpression-triggered disruption of the stoichiometric balance in complexes have been reported. Overexpression of a catalytic subunit relative to a regulatory subunit causes constitutive activation/inactivation of the catalytic subunit (de Nadal et al., 1998; Kaizu et al., 2010; Moriya et al., 2011). Overexpression of a subunit in comparison with the expression of other subunits causes an abnormal toxic complex formation (Abruzzi et al., 2002). The disomic yeast can suffer from overloads of the protein quality control resources (most probably caused by unnecessary subunit production) due to stoichiometric imbalances (Torres et al., 2010; Oromendia et al., 2012).
Although stoichiometric imbalance explains the results of individual overexpression experiments, contradictory results have been obtained from large-scale experiments. About 30% of yeast proteins are protein-complex members (Pu et al., 2009). If stoichiometric imbalances of protein complexes are the primary mechanisms of cellular defects after protein overexpression, protein-complex members should be preferentially isolated as the dosage-sensitive proteins. Although initial results supported this hypothesis (Papp et al., 2003), the dosage-sensitive proteins isolated in the absolute overexpression experiment were not preferentially the members of protein complexes (Sopko et al., 2006; Vavouri et al., 2009). However, the dosage-sensitive genes isolated in the relative overexpression experiment preferentially include the complex members (Makanae et al., 2013). In that report, the balance hypothesis was further validated in rescue experiments with simultaneously overexpressed stoichiometric partner subunits. These contradictory results could be explained by differences in the quantitative nature of the experiments. To change the balance, the proteins should be overexpressed in comparison with the native levels. In absolute overexpression experiments, the large variation in the degrees of balance disruptions caused by the overexpressed proteins could hide the cellular defects triggered by stoichiometric imbalance. Protein-complex members tend to be natively highly expressed (Figure 3B). This might also create a bias against the isolation of complex members because the absolute overexpression experiment preferentially isolated proteins with natively low expression, as discussed earlier (Figure 3A).
Note that stoichiometric imbalance in a protein complex does not necessarily cause immediate cellular defects. As described earlier and shown in Figure 4B, cellular defects caused by stoichiometric imbalances in protein complexes depend on the functions and characteristics of the overexpressed proteins. For example, to overload cellular resources, proteins should be highly expressed. It is thus possible that stoichiometric imbalance in a protein with a natively low expression will not cause resource overload.
PROMISCUOUS INTERACTIONS
Proteins containing regions without stable structures are called intrinsically disordered proteins (IDPs; Habchi et al., 2014). The ID regions enable flexible protein–protein interactions (Liu and Huang, 2014; Supplemental Table S1). Although the flexible interactions are advantageous to the function of these proteins, there might be some unwanted interactions with nonphysiological partner proteins. The association analysis of absolute overexpression experiments has shown that these promiscuous interactions might be one of the primary mechanisms of cellular defects (Vavouri et al., 2009; Ma et al., 2010; Figure 4C). Despite the tendency of IDPs to have low native expression levels (Figure 3B; Gsponer et al., 2008), they also have been preferentially isolated as dosage-sensitive proteins in gTOW overexpression experiments (Makanae et al., 2013). The hypothetical involvement of promiscuous interactions in cellular defects is thus supported by both absolute and relative overexpression experiments.
Promiscuous interaction–triggered cellular defects are difficult to validate in vivo because of the diversity of the functions and characteristics of IDPs. We do not have good model proteins or model domains for testing such interactions. Regulatory proteins such as transcription factors and signaling molecules often contain ID regions (Supplemental Table S1). Systematic physiological analyses, such as transcriptome analyses of cells overexpressing these IDPs, might distinguish the consequences of promiscuous interactions (e.g., unexpected pathway activation and stress response) from pathway overloads (see later discussion). The degree of false-positive (unexpected) interactions in two-hybrid analyses could be an index for promiscuous interactions because strong promoters are normally used in these experiments (Uetz et al., 2000; Ito et al., 2001).
Strong expression of aggregative proteins, such as the disease-related heterologous proteins misfolded yellow fluorescent protein mutants, causes cellular defects in yeast cells (Geiler-Samerotte et al., 2011; Kaiser et al., 2013; Park et al., 2013). Some protein aggregates are toxic because they capture essential proteins and limiting chaperones into the aggregates (Treusch and Lindquist, 2012; Kaiser et al., 2013; Park et al., 2013). Protein aggregates could cause toxicity, lead to novel and unwanted protein–protein interactions, and overload protein quality control resources. Although overexpressed endogenous proteins sometimes form aggregates in yeast cells, they do not necessarily lead to cellular defects; they might even have a positive effect on environmental adaptation (Alberti et al., 2009). Their toxicities might depend on the proteins with which they interact. Association studies suggest that aggregative proteins tend to be dosage sensitive (Gsponer and Babu, 2012). However, it has not been experimentally demonstrated that protein aggregates are the primary cause of cellular defects after protein overexpression.
PATHWAY MODULATION
The expression level of each protein seems to be optimized to maximize cellular functionality. It is possible that the more the expression level is changed, the more cellular functions are negatively affected. If this were true, proteins with low native expression would be isolated as dosage-sensitive proteins in absolute overexpression experiments. In these experiments, such proteins tend to be overexpressed substantially above their native levels (Figures 2 and 3). Transcription factors, protein kinases, membrane proteins, and IDPs tend to be lowly expressed (Figure 3B). These proteins, except for IDPs, are preferentially isolated as dosage-sensitive proteins in absolute overexpression experiments (Sopko et al., 2006), but not in gTOW relative overexpression experiments (Makanae et al., 2013). One simple explanation is that they are isolated as dosage-sensitive proteins in absolute overexpression experiments because their levels of overexpression are far beyond their physiological concentrations.
Activities of regulatory molecules such as transcription factors and signaling molecules are usually modulated in response to changes in external and internal cellular conditions. Their biochemical characteristics, such as binding constants and enzymatic activities, should differ by some orders of magnitude to avoid unwanted activation/inactivation of target pathways in which they are involved. Due to a mass-action effect, overexpression of inactive forms of these proteins above the threshold might cause untimely activation/inactivation of their target pathways. If this causes cellular defects, these proteins could be dosage sensitive (pathway modulation; Figure 4D; Sheltzer and Amon, 2011). Such pathway modulations have been repeatedly observed in individual overexpression experiments (Prelich, 2012). However, regulatory proteins are not preferentially isolated as dosage-sensitive proteins in gTOW relative overexpression experiments (Makanae et al., 2013). This suggests that the pathway overloads happen only when regulatory proteins are expressed in large excess above their native levels; the responses of biological systems to changes in intracellular parameters are inherently robust (Alon et al., 1999; Little et al., 1999; von Dassow et al., 2000; Kitano, 2004). Further analyses will be required to confirm the pathway modulation mechanisms. Such modulations could be relatively easily detected using transcriptome analyses of the downstream genes specific for each pathway.
CONCLUSION
I have discussed the quantitative nature of overexpression experiments and some primary consequences of protein overexpression as revealed in the large-scale studies. Figure 2 is a conceptual presentation of absolute and relative overexpression experiments. It allows an intuitive understanding of the quantitative nature of overexpression and illustrates the importance of measurements of overexpressed protein levels. Some pioneering studies have revealed the diversity of protein expression levels even in absolute overexpression experiments (Gelperin et al., 2005; Tomala and Korona, 2013; Tomala et al., 2014). It is important to measure precisely the limits of protein expression to obtain a clear picture of the consequences of protein overexpression leading to cellular defects.
Another issue is the confirmation of the hypotheses described. The consequences of protein overexpression are complex because the functions and characteristics of proteins are diverse but interconnected. For example, IDPs include many transcription factors and protein kinases. Overexpression of these regulatory factors could cause both promiscuous interactions and pathway modulations. Overexpression of a protein-complex member could cause pathway activation and overloads. Further systematic analysis of cellular physiology using omics technologies and suppressor mutants (such as in the analyses of disomic strains; Torres et al., 2010) are needed. Systematic genetic interaction analyses (Sopko et al., 2006; Douglas et al., 2012) and analyses using model proteins (Geiler-Samerotte et al., 2011; Makanae et al., 2013; Park et al., 2013; Tomala et al., 2014) should help to dissect the consequences of protein overexpression. Ideally, we should find a biomarker or a reporter associated with each mechanism to assess it properly.
Acknowledgments
I am sincerely grateful to Takashi Makino, Tohoku University, Sendai, Japan, for various useful suggestions and comments on the manuscript. I am also grateful to Kouji Ishikawa, Okayama University, Okayama, Japan, for his help with data analysis.
Abbreviations used:
- GFP
green fluorescent protein
- ID
intrinsically disordered
- IDP
intrinsically disordered protein
- OE
overexpression
- ORF
open reading frame.
Footnotes
REFERENCES
- Abruzzi KC, Smith A, Chen W, Solomon F. Protection from free beta-tubulin by the beta-tubulin binding protein Rbl2p. Mol Cell Biol. 2002;22:138–147. doi: 10.1128/MCB.22.1.138-147.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akada R, Yamamoto J, Yamashita I. Screening and identification of yeast sequences that cause growth inhibition when overexpressed. Mol Gen Genet. 1997;254:267–274. doi: 10.1007/s004380050415. [DOI] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U, Surette MG, Barkai N, Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- Blank HM, Sheltzer JM, Meehl CM, Amon A. Mitotic entry in the presence of DNA damage is a widespread property of aneuploidy in yeast. Mol Biol Cell. 2015;26:1440–1451. doi: 10.1091/mbc.E14-10-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonney ME, Moriya H, Amon A. Aneuploid proliferation defects in yeast are not driven by copy number changes of a few dosage-sensitive genes. Genes Dev. 2015;29:898–903. doi: 10.1101/gad.261743.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer J, Badis G, Fairhead C, Talla E, Hantraye F, Fabre E, Fischer G, Hennequin C, Koszul R, Lafontaine I, et al. Large-scale exploration of growth inhibition caused by overexpression of genomic fragments in Saccharomyces cerevisiae. Genome Biol. 2004;5:R72. doi: 10.1186/gb-2004-5-9-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekel E, Alon U. Optimality and evolutionary tuning of the expression level of a protein. Nature. 2005;436:588–592. doi: 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- de Nadal E, Clotet J, Posas F, Serrano R, Gomez N, Ariño J. The yeast halotolerance determinant Hal3p is an inhibitory subunit of the Ppz1p Ser/Thr protein phosphatase. Proc Natl Acad Sci USA. 1998;95:7357–7362. doi: 10.1073/pnas.95.13.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Hwang S, O’Sullivan C, Dodgson SE, Gygi SP, Amon A, Torres EM. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. Elife. 2014;3:e03023. doi: 10.7554/eLife.03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AC, Smith AM, Sharifpoor S, Yan Z, Durbic T, Heisler LE, Lee AY, Ryan O, Göttert H, Surendra A, et al. Functional analysis with a barcoder yeast gene overexpression system. G3 (Bethesda) 2012;2:1279–1289. doi: 10.1534/g3.112.003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinet C, de la Torre MA, Aldea M, Herrero E. An efficient method to isolate yeast genes causing overexpression-mediated growth arrest. Yeast. 1995;11:25–32. doi: 10.1002/yea.320110104. [DOI] [PubMed] [Google Scholar]
- Geiler-Samerotte KA, Dion MF, Budnik BA, Wang SM, Hartl DL, Drummond DA. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc Natl Acad Sci USA. 2011;108:680–685. doi: 10.1073/pnas.1017570108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsponer J, Babu MM. Cellular strategies for regulating functional and nonfunctional protein aggregation. Cell Rep. 2012;2:1425–1437. doi: 10.1016/j.celrep.2012.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science. 2008;322:1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habchi J, Tompa P, Longhi S, Uversky VN. Introducing protein intrinsic disorder. Chem Rev. 2014;114:6561–6588. doi: 10.1021/cr400514h. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones GM, Stalker J, Humphray S, West A, Cox T, Rogers J, Dunham I, Prelich G. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nat Methods. 2008;5:239–241. doi: 10.1038/nmeth.1181. [DOI] [PubMed] [Google Scholar]
- Kaiser CJ, Grötzinger SW, Eckl JM, Papsdorf K, Jordan S, Richter K. A network of genes connects polyglutamine toxicity to ploidy control in yeast. Nat Commun. 2013;4:1571. doi: 10.1038/ncomms2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizu K, Moriya H, Kitano H. Fragilities caused by dosage imbalance in regulation of the budding yeast cell cycle. PLoS Genet. 2010:6, e1000919. doi: 10.1371/journal.pgen.1000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. Biological robustness. Nat Rev Genet. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebermeister W, Noor E, Flamholz A, Davidi D, Bernhardt J, Milo R. Visual account of protein investment in cellular functions. Proc Natl Acad Sci USA. 2014;111:8488–8493. doi: 10.1073/pnas.1314810111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JW, Shepley DP, Wert DW. Robustness of a gene regulatory circuit. EMBO J. 1999;18:4299–4307. doi: 10.1093/emboj/18.15.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Huang Y. Advantages of proteins being disordered. Protein Sci. 2014;23:539–550. doi: 10.1002/pro.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Krizek J, Bretscher A. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics. 1992;132:665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Pang CN, Li SS, Wilkins MR. Proteins deleterious on overexpression are associated with high intrinsic disorder, specific interaction domains, and low abundance. J Proteome Res. 2010;9:1218–1225. doi: 10.1021/pr900693e. [DOI] [PubMed] [Google Scholar]
- Makanae K, Kintaka R, Makino T, Kitano H, Moriya H. Identification of dosage-sensitive genes in Saccharomyces cerevisiae using the genetic tug-of-war method. Genome Res. 2013;23:300–311. doi: 10.1101/gr.146662.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIsaac RS, Silverman SJ, McClean MN, Gibney PA, Macinskas J, Hickman MJ, Petti AA, Botstein D. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Mol Biol Cell. 2011;22:4447–4459. doi: 10.1091/mbc.E11-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya H, Chino A, Kapuy O, Csikasz-Nagy A, Novak B. Overexpression limits of fission yeast cell-cycle regulators in vivo and in silico. Mol Syst Biol. 2011:7, 556. doi: 10.1038/msb.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya H, Makanae K, Watanabe K, Chino A, Shimizu-Yoshida Y. Robustness analysis of cellular systems using the genetic tug-of-war method. Mol Biosystems. 2012;8:2513–2522. doi: 10.1039/c2mb25100k. [DOI] [PubMed] [Google Scholar]
- Moriya H, Shimizu-Yoshida Y, Kitano H. In vivo robustness analysis of cell division cycle genes in Saccharomyces cerevisiae. PLoS Genet. 2006;2:1034–1045. doi: 10.1371/journal.pgen.0020111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Li Z, Zhan W, Iyer VR, Marcotte EM. Mechanisms of cell cycle control revealed by a systematic and quantitative overexpression screen in S. cerevisiae. PLoS Genet. 2008;4:e1000120. doi: 10.1371/journal.pgen.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oromendia AB, Amon A. Aneuploidy: implications for protein homeostasis and disease. Dis Model Mech. 2014;7:15–20. doi: 10.1242/dmm.013391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oromendia AB, Dodgson SE, Amon A. Aneuploidy causes proteotoxic stress in yeast. Genes Dev. 2012;26:2696–2708. doi: 10.1101/gad.207407.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pál C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Park SH, Kukushkin Y, Gupta R, Chen T, Konagai A, Hipp MS, Hayer-Hartl M, Hartl FU. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 2013;154:134–145. doi: 10.1016/j.cell.2013.06.003. [DOI] [PubMed] [Google Scholar]
- Prelich G. Gene overexpression: uses, mechanisms, and interpretation. Genetics. 2012;190:841–854. doi: 10.1534/genetics.111.136911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu S, Wong J, Turner B, Cho E, Wodak SJ. Up-to-date catalogues of yeast protein complexes. Nucleic Acids Res. 2009;37:825–831. doi: 10.1093/nar/gkn1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J. Gene overexpression in studies of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:239–251. doi: 10.1016/0076-6879(91)94019-9. [DOI] [PubMed] [Google Scholar]
- Shachrai I, Zaslaver A, Alon U, Dekel E. Cost of unneeded proteins in E. coli is reduced after several generations in exponential growth. Mol Cell. 2010;38:758–767. doi: 10.1016/j.molcel.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. Rate-limiting steps in yeast protein translation. Cell. 2013;153:1589–1601. doi: 10.1016/j.cell.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Amon A. The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends Genet. 2011;27:446–453. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A. Aneuploidy drives genomic instability in yeast. Science. 2011;333:1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Torres EM, Dunham MJ, Amon A. Transcriptional consequences of aneuploidy. Proc Natl Acad Sci USA. 2012;109:12644–12649. doi: 10.1073/pnas.1209227109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoep JL, Yomano LP, Westerhoff HV, Ingram LO. Protein burden in Zymomonas rnobilis: negative flux and growth control due to overproduction of glycolytic enzymes. Microbiology. 1995;141:2329–2337. [Google Scholar]
- Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Stevenson LF, Kennedy BK, Harlow E. A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc Natl Acad Sci USA. 2001;98:3946–3951. doi: 10.1073/pnas.051013498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoebel DM, Dean AM, Dykhuizen DE. The cost of expression of Escherichia coli lac operon proteins is in the process, not in the products. Genetics. 2008;178:1653–1660. doi: 10.1534/genetics.107.085399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Amon A. Gene copy-number alterations: a cost-benefit analysis. Cell. 2013;152:394–405. doi: 10.1016/j.cell.2012.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MC, Monteiro PT, Guerreiro JF, Gonçalves JP, Mira NP, dos Santos SC, Cabrito TR, Palma M, Costa C, Francisco AP, et al. The YEASTRACT database: an upgraded information system for the analysis of gene and genomic transcription regulation in Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42:D161–D166. doi: 10.1093/nar/gkt1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn RR, Gonzalez C, Brar GA, Christen S, Carlile TM, Ingolia NT, Sauer U, Weissman JS, Amon A. Aneuploid yeast strains exhibit defects in cell growth and passage through START. Mol Biol Cell. 2013;24:1274–1289. doi: 10.1091/mbc.E12-07-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomala K, Korona R. Evaluating the fitness cost of protein expression in Saccharomyces cerevisiae. Genome Biol Evol. 2013;5:2051–2060. doi: 10.1093/gbe/evt154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomala K, Pogoda E, Jakubowska A, Korona R. Fitness costs of minimal sequence alterations causing protein instability and toxicity. Mol Biol Evol. 2014;31:703–707. doi: 10.1093/molbev/mst264. [DOI] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch S, Lindquist S. An intrinsically disordered yeast prion arrests the cell cycle by sequestering a spindle pole body component. J Cell Biol. 2012;197:369–379. doi: 10.1083/jcb.201108146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138:198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- Veitia RA, Bottani S, Birchler JA. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet. 2008;24:390–397. doi: 10.1016/j.tig.2008.05.005. [DOI] [PubMed] [Google Scholar]
- von Dassow G, Meir E, Munro EM, Odell GM. The segment polarity network is a robust developmental module. Nature. 2000;406:188–192. doi: 10.1038/35018085. [DOI] [PubMed] [Google Scholar]
- Wagner A. Energy constraints on the evolution of gene expression. Mol Biol Evol. 2005;22:1365–1374. doi: 10.1093/molbev/msi126. [DOI] [PubMed] [Google Scholar]
- Yoshikawa K, Tanaka T, Ida Y, Furusawa C, Hirasawa T, Shimizu H. Comprehensive phenotypic analysis of single-gene deletion and overexpression strains of Saccharomyces cerevisiae. Yeast. 2011;28:349–361. doi: 10.1002/yea.1843. [DOI] [PubMed] [Google Scholar]
- Zaslaver A, Mayo AE, Rosenberg R, Bashkin P, Sberro H, Tsalyuk M, Surette MG, Alon U. Just-in-time transcription program in metabolic pathways. Nat Genet. 2004;36:486–491. doi: 10.1038/ng1348. [DOI] [PubMed] [Google Scholar]