A mass spectrometry–based tandem mass tag 9-plex strategy was used to determine alterations in relative protein abundance due to three carbon sources—glucose, galactose, and raffinose. More than 4700 proteins were quantified across all nine samples; 1003 demonstrated statistically significant differences in abundance in at least one condition.
Abstract
The global proteomic alterations in the budding yeast Saccharomyces cerevisiae due to differences in carbon sources can be comprehensively examined using mass spectrometry–based multiplexing strategies. In this study, we investigate changes in the S. cerevisiae proteome resulting from cultures grown in minimal media using galactose, glucose, or raffinose as the carbon source. We used a tandem mass tag 9-plex strategy to determine alterations in relative protein abundance due to a particular carbon source, in triplicate, thereby permitting subsequent statistical analyses. We quantified more than 4700 proteins across all nine samples; 1003 proteins demonstrated statistically significant differences in abundance in at least one condition. The majority of altered proteins were classified as functioning in metabolic processes and as having cellular origins of plasma membrane and mitochondria. In contrast, proteins remaining relatively unchanged in abundance included those having nucleic acid–related processes, such as transcription and RNA processing. In addition, the comprehensiveness of the data set enabled the analysis of subsets of functionally related proteins, such as phosphatases, kinases, and transcription factors. As a resource, these data can be mined further in efforts to understand better the roles of carbon source fermentation in yeast metabolic pathways and the alterations observed therein, potentially for industrial applications, such as biofuel feedstock production.
INTRODUCTION
The budding yeast Saccharomyces cerevisiae is a model system for studying biological functions and pathways by both genomic and proteomic strategies. In industrial applications, S. cerevisiae has been used traditionally for brewing and baking, and more recently for biofuels (Raghavulu et al., 2011; Tang et al., 2013b). S. cerevisiae can grow on a variety of carbon sources, both fermentable (e.g., glucose, fructose, sucrose, galactose, maltose, raffinose) and nonfermentable (e.g., ethanol, glycerol, acetate, oleic acid) (Fendt and Sauer, 2010). We expect that culturing yeast on a particular carbon source would result in pronounced proteomic changes associated with metabolic perturbation (Gao et al., 2003). Here we investigate quantitatively the global proteomic alterations in wild-type S. cerevisiae following growth on minimal media supplemented with one of three carbon sources—galactose, glucose, or raffinose—using a versatile mass spectrometry–based multiplexing strategy.
For S. cerevisiae, minimal growth media are typically composed of three components: 1) yeast nitrogen base, 2) ammonium sulfate, and 3) a carbon source as a basal medium to which amino acids may be supplemented. Glucose (dextrose) is a monosaccharide preferred as an energy source by many organisms, including S. cerevisiae, and is a component of standard yeast extract–peptone–dextrose yeast media. Cells can sense glucose levels in the environment and can adapt central metabolic pathways to glucose availability (Towle, 2005). Central pathways of carbohydrate metabolism, including those in yeast, have evolved to efficiently process glucose for general metabolism. For example, glucose availability can induce the expression of glucose transporters such as certain HXT genes (Kim et al., 2013). Although glucose is the preferred carbon source, yeast can ferment other carbon sources, including galactose and raffinose. Similar to glucose, galactose is a monosaccharide sugar that is converted into a glycolytic metabolite, glucose 6-phosphate, via the Leloir pathway (Frey, 1996). As such, yeast growth on galactose induces the expression of proteins under GAL4-dependant promoters, for example, GAL1, GAL7, and GAL10 (Timsol, 2007). In contrast, raffinose is a trisaccharide composed of galactose, glucose, and fructose. Raffinose is gradually hydrolyzed extracellularly by invertase (SUC2) to monosaccharides that can typically enter the cell via SNF3, among other hexose transporters (Granot and Snyder, 1993). We expect extensive protein alterations, particularly of transport and metabolism-related proteins, when the carbon source is modified.
Multiplexing strategies applied to mass spectrometry–based quantitative proteomics have expanded the efficiency, depth, and throughput of comprehensive protein analyses. Employing isobaric labeling (Thompson et al., 2003; Ross et al., 2004) permits the quantification of protein samples from virtually any source, with the major limitation being the number of available labels. The maturation of mass spectrometry–based proteomic analysis currently permits the quantification of thousands of individual proteins, a level approaching the entire yeast proteome. The elucidation of a complete proteome allows not only for the study of individual proteins but also for investigation of various classes and pathways in which these proteins function. Previous work upon which this study builds has shown promise in elucidating the comprehensive proteome of S. cerevisiae via mass spectrometry–based techniques (King et al., 2006; Nagaraj et al., 2012; Picotti et al., 2013; Webb et al., 2013; Hebert et al., 2014; Paulo and Gygi, 2014).
Here we quantitatively explore the proteomic alterations in wild-type S. cerevisiae cultured in three different carbon sources: galactose, glucose, and raffinose. We use a tandem mass tag (TMT) 9-plex strategy to determine the relative protein abundance alterations due to the particular carbon source. This “3 × 3” strategy is advantageous, as nine samples (three samples per each carbon source) can be analyzed simultaneously in a single experiment, and statistical analyses can be performed with biological triplicates. We present the largest mass spectrometry–based analysis of the yeast S. cerevisiae to date using a TMT9-plex quantitative strategy on an Orbitrap Fusion mass spectrometer. These data can be mined further to develop a better understanding of yeast metabolic pathways under different carbon sources and potentially leverage these observed alterations for industrial applications.
RESULTS
More than 4700 proteins were quantified across nine S. cerevisiae cultures in a single experiment
Using the strategy we described above (outlined in Figure 1), we successfully characterized the proteome of S. cerevisiae when grown with galactose, glucose, and raffinose as the carbon sources. Growth curves for the cultures are presented in Supplemental Figure 1. As expected, below 10 OD600/ml, all cultures grew exponentially, with the galactose and raffinose cultures showing similar, slightly slower average doubling times (t1/2 = 123 and 126 min, respectively) than the glucose cultures (t1/2 = 108 min, see inset). The increase in time between cell divisions for the cells growing in galactose or raffinose could be attributed to the time and metabolic costs required to synthesize the additional enzymes needed to metabolize the noncanonical sugars. Our analysis was of the proteomes of cultures harvested at OD600≈0.6/ml.
FIGURE 1:
TMT9-plex analysis of S. cerevisiae grown on three carbon sources. The procedure was as follows: 1) Three starter cultures of minimal media with raffinose as the carbon source were each inoculated with a single colony. Cultures were grown overnight in raffinose media. Cultures were centrifuged, washed in deionized water, and diluted to an OD600 of 0.1/ml in growth media containing galactose, glucose, or raffinose. At OD600 of 0.6/ml, cultures were harvested, cells were lysed, and proteins were extracted via mechanical lysis and chloroform-methanol precipitation. 2) Proteins were digested with LysC and trypsin and labeled with TMT reagents. 3) The pooled samples were separated using BPRP chromatography. 4) Desalted peptides were subjected to HPLC and TMT-MS3–based mass spectrometry.
We confidently identified 175,417 peptides, of which 60,251 were nonredundant (unique) with a peptide false discovery rate (FDR) of 0.094% (Table 1). A total of 4873 proteins were identified with a protein FDR of 0.995%. Of these proteins, 4765 were confidently quantified across all nine cultures, representing the largest number of proteins quantified in S. cerevisiae using isobaric tagging. Supplemental Tables 1 and 2 list the proteins and peptides, respectively, as well as normalized TMT reporter ion intensities used for quantitative analysis.
TABLE 1:
Summary of mass spectrometry data.
| Number | |
|---|---|
| Identified proteins | 4873 |
| Unique peptides | 60,251 |
| Total peptides | 175,417 |
| Quantified proteinsa | 4765 |
| Proteins with significantly altered abundanceb | 1003 |
aProteins quantified across all nine TMT channels.
bA Bonferroni-corrected ANOVA p value < 0.01 was required for statistical significance.
We subjected the 4765 quantified yeast proteins to two-dimensional clustering (by normalizing to the average value for the glucose cultures) and then using the Euclidean distance metric (Figure 2A). The samples clustered as expected, as triplicates of each carbon source clustered together. Similarly, principal components analysis also reflected the tight clustering of these samples (Figure 2B), as the first two principal components explained greater than 90% of the variance. Principal component 1 explained nearly 60% of the variance and separated samples grown in glucose from galactose and raffinose. Similarly, principal component 2 explained 34% of the variance and separated cultures grown with galactose from those grown with raffinose. To examine the overall distribution of protein abundance alterations, we constructed histograms of the log2 ratios for each pair of carbon sources (Figure 2C). Agreeing with the clustering of the three carbon sources in the dendrogram in Figure 2A, the galactose versus raffinose histogram (Figure 2C, top) showed a narrower fold-change distribution compared with the glucose versus raffinose (Figure 2C, middle) and galactose versus glucose (Figure 2C, bottom) histograms. Collectively these results revealed that of the three comparisons, the global protein abundance differences between raffinose and galactose were more similar relative to that between either of these two sugars and glucose. In regard to the reproducibility of the biological triplicates, we determined that the Pearson correlation coefficients (r2) for each pair of replicates are all >0.99 (Supplemental Figure 2).
FIGURE 2:
Global protein expression analysis. (A) Heat map and dendrogram show the relative expression levels across the nine TMT channels and the clustering, respectively, of the 4765 quantified proteins. To the right of the heat map are three example proteins that show increased protein abundance in each of the three carbon sources investigated. The data were normalized to the average TMT relative abundance values from the glucose cultures. (B) Principal component analysis correlates well with the clustering of the replicates. (C) Histograms show the distribution of the fold changes between galactose and raffinose, glucose and raffinose, and galactose and glucose. Gal, galactose; Glu, glucose; Raf, raffinose; TMT RA, tandem mass tags relative abundance; *, p value < 0.01.
As examples, we highlighted three proteins: 1) HXT3, which showed higher abundance in cultures grown in glucose; 2) GAL10, which had higher abundance in those grown in galactose; and 3) SUC2, which was of higher abundance for cultures grown in raffinose. HXT3 is a glucose transporter, and its expression was induced in the presence of glucose, which correlated well with our data (Ko et al., 1993). GAL10, also known as UDP-glucose-4-epimerase and galactose mutarotase, is a bifunctional enzyme that functions in galactose catabolism and was up-regulated in the presence of galactose (De Robichon-Szulmajster, 1958). SUC2 (invertase) catalyzes the hydrolysis of di- and trisaccharides (such as raffinose) to produce monosaccharides, which eventually enter glycolysis (Trumbly, 1992). In agreement with our data, SUC2 expression has been shown to be up-regulated under low-glucose conditions (Guaragnella et al., 2013).
k-Means clustering of statistically significant proteins revealed metabolic pathways as being highly altered
We tested for statistical significance using a one-way analysis of variance (ANOVA) and corrected for multiple testing of proteins in each set of samples using the Bonferroni method (Dunn, 1961). With a threshold for the p value of < 0.01, we determined 1003 proteins as demonstrating a statistically significant difference in abundance due to the carbon source used. Using only proteins with altered abundance (n = 1003), we performed k-means clustering (Figure 3). Three major k-means clusters emerged: 1) proteins that were of higher abundance (n = 378) in the galactose-containing media (Figure 3A), 2) those that were of higher abundance (n = 289) in the glucose-containing media (Figure 3B), and 3) those that were of higher abundance (n = 337) in the raffinose-containing media (Figure 3C).
FIGURE 3:
k-Means clustering and associated KEGG pathways. k-Means clustering of protein subsets with statistically significant alterations in abundance from cultures grown in media containing (A) galactose, (B) glucose, and (C) raffinose. Listed on the right are the KEGG pathways that were represented by the proteins in the associated k-means cluster. Each trace represents the abundance profile of a single protein. Gal, galactose; Glu, glucose; Raf, raffinose.
Subsequently, we classified these clusters of proteins using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analysis (Wixon and Kell, 2000) through the STRING interface (Franceschini et al., 2013) and tallied the number of significantly altered proteins in given categories. Of particular interest was the metabolic pathways category, which had the highest number of proteins in each of the three clusters (Figure 3, right). In fact, the majority of pathways listed were specific metabolic processes that were directly or indirectly related to carbohydrate metabolism. We expected that metabolic pathways would be significantly altered, as carbon sources are tightly linked to metabolism.
Gene ontology classification of unaltered proteins due to carbon source (coefficient of variation < 5%) was primarily of nuclear function and localization
In addition to investigating the proteins with the highest differences in abundance, we also determined those that were relatively unaltered regardless of the carbon source. For this, we chose proteins with coefficients of variation < 5%, which resulted in a total of 114 proteins (Supplemental Table 3). For comparison, we selected the 114 proteins with the highest coefficients of variation among the three different carbon sources (Supplemental Table 4). We then used the Database for Annotation, Visualization and Integrated Discovery (DAVID; Huang da et al., 2009) to perform gene ontology analysis to determine which biological processes (Table 2, top) and cellular components (Table 2, bottom) were represented by these subsets of proteins.
TABLE 2:
Gene ontology classification.
| Number of proteins | ||
|---|---|---|
| Unchangeda | Changedb | |
| Biological function | ||
| Regulation of transcription | 23 | 0 |
| RNA processing | 22 | 0 |
| Transcription | 18 | 0 |
| Cellular macromolecular complex subunit organization | 16 | 0 |
| ncRNA processing | 13 | 0 |
| Chromatin organization | 11 | 0 |
| mRNA metabolic process | 11 | 0 |
| Generation of precursor metabolites and energy | 0 | 18 |
| Oxidation reduction | 0 | 17 |
| Monosaccharide metabolic process | 0 | 15 |
| Hexose metabolic process | 0 | 14 |
| Transmembrane transport | 0 | 14 |
| Energy derivation by oxidation of organic compounds | 0 | 13 |
| Cofactor metabolic process | 0 | 12 |
| Response to temperature stimulus | 0 | 10 |
| Cellular carbohydrate catabolic process | 0 | 10 |
| Carbohydrate catabolic process | 0 | 10 |
| Coenzyme metabolic process | 0 | 10 |
| Cellular component | ||
| Intracellular organelle lumen | 27 | 0 |
| Membrane-enclosed lumen | 27 | 0 |
| Organelle lumen | 27 | 0 |
| Nuclear lumen | 20 | 0 |
| Ribonucleoprotein complex | 18 | 0 |
| Nucleoplasm | 11 | 0 |
| Nucleoplasm part | 11 | 0 |
| Intrinsic to membrane | 0 | 37 |
| Integral to membrane | 0 | 33 |
| Mitochondrion | 0 | 29 |
| Plasma membrane | 0 | 22 |
| Insoluble fraction | 0 | 12 |
| Membrane fraction | 0 | 12 |
aProteins that were relatively unaltered in abundance (coefficient of variation < 5%, n = 114).
bProteins with the highest fold changes in abundance (n = 114).
DAVID analysis revealed that the major biological processes for the proteins with high changes in abundance were categorized as metabolic and transmembrane transport. Such a result was expected, as the carbon source provided a direct input for metabolic processes and the expression of particular transmembrane transporters were influenced by the carbon source (Boles and Hollenberg, 1997). In contrast, the main biological processes represented by proteins that remained relatively constant in abundance regardless of carbon source were those involved in transcription and RNA processing. Moreover, the cellular component gene ontology classification results correlated well with those for biological process. In particular, proteins in the nuclear lumen, ribonucleoproteins, and nucleoplasm-localized proteins did not demonstrate a substantial change in abundance. Likewise, many highly altered proteins were classified as (plasma) membrane- or mitochondria-associated, which reflect changes in carbon source transport into the cell and metabolic consequences, respectively. Even with extensive alterations in metabolic processes and associated proteins induced by growth on different carbon sources, nuclear protein abundance levels were perturbed minimally. These gene ontology classifications were also consistent with those observed for the k-means clustering analysis described above.
Several dozen protein kinases, phosphatases, and transcription factors were quantified in our data set
The comprehensiveness of this data set allowed us to compile large subsets of protein families and therefore investigate further the differences in the abundance of proteins with similar or redundant functions. For example, we compared our data set with the protein kinases (Supplemental Figure 3A), phosphatases (Supplemental Figure 3B), and transcription factors (Supplemental Figure 3C) found in available databases (Hunter and Plowman, 1997; Teixeira et al., 2014). In all three categories, we quantified in our data set more than 80% of the categorized proteins, including a subset of at least 104 protein kinases, 36 phosphatases, and 128 transcription factors (Supplemental Table 5). Of these, 23 kinases, eight phosphatases, and 19 transcription factors show statistically significant alterations in protein abundance resulting from growth on different carbon sources. Kinases and phosphatases are integral members of cell signaling pathways, and perturbations thereof can dramatically alter cell function. Likewise, transcription factors can influence and thereby regulate protein expression. In the following paragraphs, we highlight three proteins within each of these subsets.
Among protein kinases with the highest fold changes due to the particular carbon source are KNS1, PRR2, and CKA2 (Supplemental Figure 3A, left). KNS1 (YLL019C) and PRR2 (YDL214C) both are of higher abundance in yeast cultures grown on raffinose. KNS1 is a putative protein kinase of unknown cellular role (Padmanabha et al., 1991), and as such, KNS1 could be part of a mechanism that facilitates growth on raffinose. The elevated expression of KNS1 and those of similar putative proteins may reveal hitherto unknown functions of such proteins. PRR2, a serine/threonine protein kinase, inhibits pheromone-induced signaling downstream of MAPK (Burchett et al., 2001) and has a significant role in many signaling events that may be linked indirectly to various metabolic pathways. CKA2 (YOR061W) is of higher abundance in yeast grown on glucose. CKA2, the alpha-catalytic subunit of casein kinase 2a, is a serine/threonine protein kinase with essential roles in cell growth and proliferation (Hermosilla et al., 2005). Changes in kinase abundance can alter phosphorylation equilibrium, and such alterations can have a major impact on cellular signaling and a number of downstream processes that promote growth on a particular carbon source.
Like kinases, some phosphatases also show statistically significant alterations in abundance. Among phosphatases with the highest fold changes due to the particular carbon source are PHO3, SER2, and PPZ2 (Supplemental Figure 3B, left). PHO3 (YBR092C) and SER2 (YGR208W) are of higher abundance in the glucose-containing cultures. PHO3 is an acid phosphatase that hydrolyzes thiamine phosphates in the periplasmic space (Blinnikova et al., 2002; Nosaka et al., 2005). The role of thiamine has been linked historically to fermentation, and evidence supports its function as carbon-source dependent (Suomalainen and Axelson, 1956). SER2 is a phosphoserine phosphatase of the phosphoglycerate pathway involved in serine and glycine biosynthesis (Qiu et al., 2009). This protein is of potential importance, given that glycine biosynthesis and glucose metabolism are linked tightly (Melcher and Entian, 1992). In contrast, PPZ2 (YDR436W) demonstrates higher expression in galactose and raffinose compared with glucose. PPZ2 is a serine/threonine protein phosphatase involved in the regulation of potassium transport that affects osmotic stability and cell cycle progression (Posas et al., 1995) and can enhance resistance to multiple alcohols (Lam et al., 2014). Such alterations in phosphatase abundance can influence cellular signaling and, as such, render a compensatory effect to sustain growth on different carbon sources.
Although we identified nuclear-related proteins to be among those with unchanged abundances, 19 transcription factors demonstrated statistically significant differences in abundance. Such a result may be expected, as we observed changes in abundance of hundreds of proteins. Supplemental Table 5 lists significantly altered transcription factors, the targets of which can be queried using the YEASTRACT database (Teixeira et al., 2014). Among the transcription factors with the highest fold changes due to the particular carbon source are XBP1, YGR067C, and MSN4 (Supplemental Figure 3C, left). XBP1 (YIL101C) is of higher abundance in raffinose compared with the other two carbon sources. XBP1 is a transcriptional repressor that binds to promoter sequences of the cyclin genes and therefore has a role in cell cycle regulation (Back et al., 2005). Conversely, we determined YGR067C to be significantly lower in abundance when grown on galactose. YGR067C is a putative zinc finger motif–containing protein of unknown function (Pir et al., 2006). Similar to KNS1, YGR067C may have a currently undefined function in yeast metabolism and, as such, merits further study. In contrast, MSN4 (YKL062W) is of higher abundance when grown on galactose compared with the two other carbon sources. MSN4 is activated under stress conditions, in which it translocates from the cytoplasm to the nucleus and contributes to altered transcription (Martinez-Pastor et al., 1996), which can potentially affect the expression of various proteins. Like kinases and phosphatases, transcription factors have important roles in maintaining cellular homeostasis. Alteration in transcription factor abundance can result in the activation or repression of certain genes with many potential downstream effects. Further dissection of the perturbed cellular pathways may reveal mechanisms that modulate or are regulated by the alterations in transcription factors, as well as kinases and phosphatases, resulting from cultures grown with different carbon sources.
DISCUSSION
In this study, we use a multiplexed quantitative mass spectrometry–based strategy to comprehensively investigate the proteomic alterations in S. cerevisiae resulting from growth on different carbon sources (i.e., galactose, glucose, or raffinose). We labeled our peptide samples using TMT9-plex reagents and used a 3 × 3 strategy to examine in triplicate the relative protein abundance alterations. In total, we identified 4873 proteins, of which 1003 demonstrated a statistically significant difference in abundance in at least one condition. Gene ontology analysis reveals that the majority of altered proteins function in metabolic processes and that these proteins are of plasma membrane and mitochondrial origin. We also examined a subset of proteins that remained relatively unchanged among the carbon sources used. This subset includes proteins related to nuclear processes, such as transcription and RNA processing. In total, we have quantified the largest number of yeast proteins and peptides in a single analysis via a TMT-based mass spectrometry strategy to date. However, our list of more than 4700 quantified proteins is not complete. Currently 5800 validated open reading frames (ORFs) are thought to be present in S. cerevisiae, although not all encode proteins (Cherry et al., 2012). As an estimate, ∼10–15% more proteome coverage is needed for a complete proteome of budding yeast. Our data set, however, does achieve ∼90% overlap with the second-largest proteomic analysis of S. cerevisiae (Kulak et al., 2014). It is worth noting that our search database included the 785 dubious ORFs, of which only one, YPR169W-A, was quantified. This protein identification was associated with only one peptide, and we determined the change in protein abundance as nonsignificant (p value = 1). Moreover, the percentage of dubious ORFs identified is 0.1%, which is below our 1% protein-level FDR.
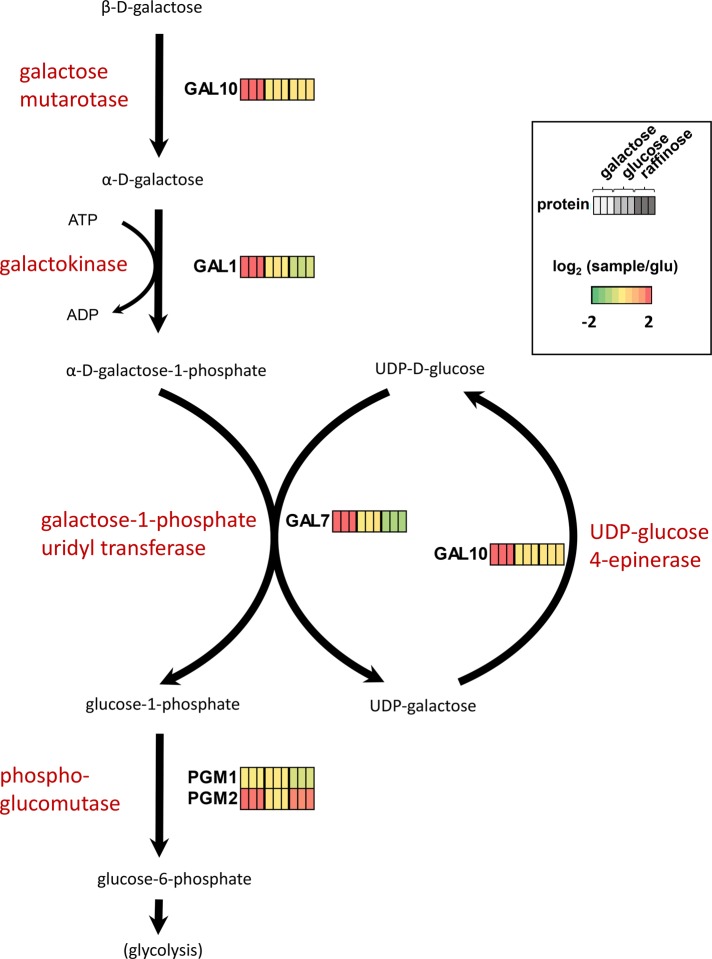
Our data set facilitated the expression mapping of protein alterations in complete metabolic pathways. With more than 4700 quantified proteins in our data set, the entire subset of proteins from many common biological pathways were quantified. As examples, we outline four metabolic pathways for which we quantified the complete set of proteins. All the enzymes involved in galactose catabolism (Figure 4), glucose fermentation (Supplemental Figure 4A), the tricarboxylic acid (TCA) cycle (Supplemental Figure 4B), and the galactose catabolism pathway (Supplemental Figure 4C) were quantified in our data set. As these were metabolic pathways, the corresponding enzymes were expected to be altered as their function was directly or indirectly linked to the carbon source on which the cultures were grown.
FIGURE 4:
All proteins comprising the galactose catabolism pathway were quantified in our data set. The colored bars indicate the relative abundance of the protein across the nine channels, with each triplicate of bars representing protein abundance in cultures grown in media containing galactose, glucose, or raffinose.
More specifically, we highlight the five proteins involved in galactose catabolism, also known as the Leloir pathway (Figure 4). In this pathway, GAL10 catalyzes β-d-galactose into α-d-galactose. Next, GAL1 catalyzes the stereospecific phosphorylation of α-d-galactose, producing α-d-galactose 1-phosphate (Howard and Heinrich, 1965). In a reaction catalyzed by galactose 1-phosphate uridyltransferase (GAL7), α-d-galactose 1-phosphate reacts with UDP-d-glucose to produce d-glucose 1-phosphate and UDP-galactose (Segawa and Fukasawa, 1979). In addition, UDP-glucose is regenerated from UDP-galactose via UDP-galactose 4-epimerase (GAL10; Fukasawa et al., 1980). The final stage in the pathway is the isomerization of glucose 1-phosphate to glucose 6-phosphate by phosphoglucomutase (PGM1, PGM2). GAL10, GAL1, and GAL7 are exclusive to this pathway, and the abundance of these proteins was substantially greater (∼30–50 times higher TMT signal) in yeast grown with galactose as the carbon source. These three enzymes are regulated coordinately at the transcriptional level in response to galactose (De Robichon-Szulmajster, 1958; Lohr et al., 1995). Likewise, PGM2 was of higher abundance when galactose was the carbon source, but only approximately twofold, as this protein is not exclusive to the Leloir pathway and also has a role in glycogen metabolism (Timsol, 2007). Further knowledge of the pathway and the manipulation thereof may have various applications. For example, although S. cerevisiae can ferment galactose into ethanol, its yield is significantly lower than from glucose. As galactose is one of the most abundant sugars in marine plant biomass, efficiently using it for growth and ethanol production is advantageous in the biofuels industry (Lee et al., 2011).
Likewise, our data set also quantified the enzymes involved in the glucose fermentation pathway (Supplemental Figure 4A). For glucose fermentation, glucose is phosphorylated by glucokinases (HXK1, HXK2, GLK1) to glucose 6-phosphate and then isomerized to fructose 6-phosphate by phosphoglucose isomerase (GPI1). Phosphofructokinase (PFK1, PFK2) then phosphorylates fructose 6-phosphate to fructose 1,6-bisphosphate. Other enzymes in the pathway include aldolase (FBA1), triosephosphate isomerase (TPI1), glyceraldehyde 3-phosphate dehydrogenase (TDH1, TDH2, TDH3), phosphoglycerate kinase (PGK1), phosphoglycerate mutase (GPM1), enolase (ENO1, ENO 2), and pyruvate kinase (CDC19, PYK2). The majority of these enzymes, however, were not exclusively associated with a particular carbon source, but changes in abundance were noted. For example, in the glucose fermentation cycle, ALD5, PDC1, and PDC5 were of higher abundance (∼3–15-fold) when glucose was used as the carbon source. In addition, ALD4, PDC6, ALD2, GLK1, and HXK1 demonstrated a statistically significant increase in abundance (approximately twofold) when raffinose was used. Further studies will be needed to understand fully the pathways underpinning these differences and to develop further such mechanisms for beneficial applications.
Similarly, all of the enzymes in the TCA cycle (Supplemental Figure 4B) were quantified in our data set (Blank and Sauer, 2004). The TCA cycle is an important source of biosynthetic building blocks used in gluconeogenesis, amino acid biosynthesis, and fatty acid biosynthesis. Occurring in the mitochondria, the TCA cycle oxidizes acetyl-CoA and extracts energy primarily as the reduced electron carriers NADH and FADH2 for use in the electron transport chain (Lowenstein, 1969). The enzymes quantified in our data set included: citrate synthase (CIT1, CIT2, CIT3), aconitrate hydratase (ACO1, ACO2), NAD-dependent isocitrate dehydrogenase (IDH1, IDH2), 2-ketogluterate dehydrogenase complex (KGD1, KGD2, LPD1), succinate dehydrogenase (LSC1, LSC2), fumarate hydrolase (FUM1), and malate dehydrogenase (MDH1, MDH2, MDH3). Our data showed that certain enzymes demonstrated statistically significant differences in abundance resulting from the carbon source utilized. In particular, CIT2, ACO2, and MDH2 were of higher abundance in cultures containing glucose, while KGD1, KGD2, LPD1, ACO1, SDH1, SDH2, SDH3, and LSC2 were of higher abundance in cultures grown in the raffinose-containing media. Moreover, fatty acid concentrations have been shown to increase via the modulation of enzymes involved in citrate turnover in the TCA cycle (Tang et al., 2013a). As such, these fatty acids may be potential substrates for the production of biofuel hydrocarbon molecules, which may benefit from alternative carbon sources in addition to genetic engineering techniques. Further studies will lead to a better understanding of the mechanisms governing carbon source–induced protein abundance alterations in these pathways.
Our data set also quantified the enzymes responsible for thiamine diphosphate (ThDP) biosynthesis (Supplemental Figure 4C). Across many organisms, ThDP serves as an essential cofactor for a variety of core metabolic reactions, often as a molecular handle for the biochemical manipulation of two-carbon compounds. In yeast, THI6 catalyzes the coupling of 4-amino-2methyl-5-hydrozymethylpyrimidine diphosphate to 2-(2-carboxy-4-methyl-thiazol-5-yl) ethyl phosphate to produce thiamine phosphate, which is ultimately phosphorylated by THI80 to produce ThDP. Two main branches feed the methylpyrimidine and thiazole substrates into THI6. In the pyrimidine branch, 4-amino-2-methyl-5-methylpyrimidine derived from either the pyrimidine de novo synthesis or the salvage pathway is sequentially doubly phosphorylated by THI20 or THI21 (Kawasaki et al., 2005). In yeast, THI4 catalyzes the entire thiazole synthesis branch, which requires five reactions across three enzymes in prokaryotes (Chatterjee et al., 2011; Hazra et al., 2011). THI4, however, is a suicide enzyme, capable of producing only one molecule of 2-(2-caboxy-4-methyl-thiazol-5-yl) ethyl phosphate at the cost of donating an active-site sulfide group (Chatterjee et al., 2011). Sustained up-regulation of the ThDP biosynthesis pathway would require continuous translation and proteolysis of THI4. All four of the ThDP biosynthesis pathway enzymes were strongly up-regulated specifically during growth on glucose. Our data suggest thiamine supplementation as potential approach to improve the efficiency of glucose-based growth for industrial applications, allowing more of the carbon source to be funneled into the desired end product, rather than spent on biomass production.
Characterizing all ORFs in the yeast S. cerevisiae has become an unexpectedly elusive goal. Optimistic estimates had predicted “known” functions for all ORFs by 2008 (Hughes et al., 2004). However, 706 ORFs remain uncharacterized (Cherry et al., 2012). The primary setback may be that “genome-wide” screens measure uncharacterized ORFs less often than those characterized, yielding a perpetually suppressed rate of data collection specific to uncharacterized ORFs (Pena-Castillo and Hughes, 2007). This caveat led Peña-Castillo and Hughes to propose that alternative growth conditions may be required to characterize the recalcitrant last ∼14% of yeast ORFs (Pena-Castillo and Hughes, 2007). Similar to previous studies, our data quantified the abundances of uncharacterized ORFs at a lower frequency (43.1%) than those characterized (87.1%), comprising only 6.8% of quantified proteins (Figure 5A). Supporting the conjecture of Peña-Castillo and Hughes, 68% of the uncharacterized ORFs displayed increased expression on galactose and raffinose when compared with glucose (Figure 5B). The majority of those proteins with altered expression in raffinose (61%) comprised a set enriched for integral membrane proteins (GO:0016021, p ∼ 0.03, hypergeometric test, multiple hypothesis-corrected Benjamini-Hochberg) (Ashburner et al., 2000; Hong et al., 2008; Cherry et al., 2012). We highlight three uncharacterized proteins that show significant alterations in expression: YMR122W-A, YDL218W, and YJR061W (Figure 5C). YMR122W-A is strongly and selectively expressed upon growth in raffinose. This protein is predicted to have a single, highly conserved transmembrane domain flanked by a cytoplasmic domain and a noncytoplasmic domain. YMR122W-A may interact with PMT1 and CBR1 and be involved in regulating cell wall protein glycosylation in the endoplasmic reticulum (ER; Huh et al., 2003), perhaps altering protein glycosylation in response to different environmental sugars. Similarly, YDL218W is strongly and selectively expressed upon growth in raffinose and is likely a transmembrane protein with a cytoplasmically exposed domain. High-throughput screens have shown induced expression under aerobic or stressful conditions, and interaction with the vacuole-targeting protein VPS8 suggests a possible role in vacuole regulation or autophagy (Wu et al., 2004). YJR061W demonstrates increased expression in galactose and raffinose. This protein consists of a short transmembrane domain and a large (∼100 amino acid) noncytoplasmic domain. Its paralogue, MNN4, has mannosyltransferase activity and increases expression as the cell walls thicken for late-log and stationary phase. YJR061W may glycosylate proteins in the ER, potentially regulating cell wall integrity (Lamb and Mitchell, 2003; Byrne and Wolfe, 2005). All 63 uncharacterized proteins that were quantified herein are listed in Supplemental Table 6. Future experiments of proteome-wide quantification across even more diverse growth conditions may prove more informative toward the elusive goal of a completely characterized yeast genome. Furthermore, improvements in instrument sensitivity, faster scan times, greater robustness, and higher selectivity will further expand the extent of identified and quantified yeast proteins.
FIGURE 5:
Comparing proteins from characterized and uncharacterized ORFs. (A) Bar chart comparing quantified and differentially expressed characterized and uncharacterized ORFs. (B) Proportion of differentially expressed uncharacterized ORFs (n = 63), which are up-regulated when grown on a particular sugar. (C) Examples of uncharacterized ORFs demonstrating statistically significant (p value < 0.01) differences in abundance with growth on different sugars within our data set. Error bars represent ± SD for triplicates. TMT RA, tandem mass tags relative abundance.
The use of multiplexed proteomic techniques can facilitate the high-throughput elucidation of comprehensive proteomes. The TMT9-plex strategy outlined herein required only 72 h of analysis, without which data collection for each individual sample could require weeks. Using the data herein, studies may be developed to investigate the influence of carbon sources on yeast proteomes using yeast deletion and overexpression strains and with other treatments or forms of cellular stress. Likewise, additional multiplexing can be achieved by incorporating the previously published 3 × 3 + 1 strategy to link multiple experiments (Paulo and Gygi, 2014; Paulo et al., 2014) or performing hyperplexing for higher-order multiplexing (Dephoure and Gygi, 2012). Furthermore, our data set contains more than 60,000 unique S. cerevisiae peptides, which may be useful for future targeted mass spectrometry–based experiments (Doerr, 2013). In summary, we have used a TMT9-plex strategy to quantitatively compare the proteomic profiles of yeast grown on three carbon sources (galactose, glucose, and raffinose) and have assembled one of the largest catalogues of yeast proteins to date in a single quantitative mass spectrometry–based experiment.
MATERIALS AND METHODS
Materials
TMT isobaric reagents were from Thermo Fisher Scientific (Waltham, MA). Water and organic solvents were from J.T. Baker (Center Valley, PA). Unless otherwise noted, all other chemicals were from Sigma-Aldrich (St. Louis, MO).
Media and growth
The yeast strain was BY4742, derived from S288c. The yeast minimal medium was composed of yeast nitrogenous base with amino acids, ammonium sulfate, and 2% final volume of the appropriate sugar (galactose, glucose, or raffinose). Three starter cultures were grown in raffinose-containing minimal media overnight from individual colonies. Each of the three starter cultures were spun down, washed with deionized water twice, and divided into three different cultures—containing either galactose, glucose, or raffinose—at OD 600 of 0.1/ml. Cultures were grown to reach an optical density (OD) of 0.6/ml and then harvested.
Cell lysis and protein digestion
Yeast cultures were harvested by centrifugation, washed twice with ice-cold deionized water, and resuspended at 4°C in a buffer containing 50 mM HEPES (pH 8.5), 8 M urea, 75 mM NaCl, and protease (complete mini, EDTA-free) and phosphatase (PhosphoStop) inhibitors (Roche, Basel, Switzerland). Cells were lysed using the MiniBeadbeater (Biospec, Bartlesville, OK) in microcentrifuge tubes at maximum speed for three cycles of 60 s each, with 1-min pauses between cycles to avoid overheating of the lysates. After centrifugation, lysates were transferred to new tubes. We determined the protein concentration in the lysate using the bicinchoninic acid protein assay (Thermo Fisher Scientific, Waltham, MA).
Proteins were subjected to disulfide reduction with 5 mM Tris (2-carboxyethyl)phosphine (room temperature, 25 min) and alkylation with 10 mM iodoacetamide (room temperature, 30 min in the dark). Excess iodoacetamide was quenched with 15 mM dithiothreitol (room temperature, 15 min in the dark). Methanol-chloroform precipitation was performed before protease digestion. In brief, four parts neat methanol was added to each sample and vortexed, one part chloroform was added to the sample and vortexed, and three parts water was added to the sample and vortexed. The sample was centrifuged at 14,000 rpm for 5 min at room temperature and after removal of the aqueous and organic phases subsequently washed twice with 100% methanol, before being air-dried.
Samples were resuspended in 50 mM HEPES (pH 8) and digested at room temperature for 16 h with LysC protease at a 100:1 protein-to-protease ratio. Trypsin was then added at a 100:1 protein-to-protease ratio, and the reaction was incubated 6 h at 37°C.
TMT labeling
TMT reagents (0.8 mg) were dissolved in anhydrous acetonitrile (40 μl), of which 10 μl was added to the peptides along with 30 μl of acetonitrile to achieve a final acetonitrile concentration of ∼30% (vol/vol). Following incubation at room temperature for 1 h, the reaction was quenched with hydroxylamine to a final concentration of 0.3% (vol/vol). The TMT-labeled samples were pooled at a 1:1:1:1:1:1:1:1:1 ratio. The sample was vacuum centrifuged to near dryness and subjected to C18 solid-phase extraction (Sep-Pak; Waters).
Off-line basic pH reversed-phase (BPRP) fractionation
We fractionated the pooled TMT-labeled peptide sample using BPRP high-pressure liquid chromatography (HPLC). We used an Agilent 1100 pump equipped with a degasser and a photodiode array detector (set at 220- and 280-nm wavelength) from Thermo Fisher Scientific (Waltham, MA). Peptides were subjected to a 50-min linear gradient from 5 to 35% acetonitrile in 10 mM ammonium bicarbonate (pH 8) at a flow rate of 0.8 ml/min over an Agilent 300Extend C18 column (5-μm particles, 4.6-mm inner diameter, and 250 mm in length). The peptide mixture was fractionated into a total of 96 fractions that were consolidated into 24. Samples were subsequently acidified with 1% formic acid and vacuum centrifuged to near dryness. Each eluted fraction was desalted via StageTip, dried via vacuum centrifugation, and reconstituted in 5% acetonitrile, 5% formic acid for liquid chromatography and tandem mass spectrometry (LC-MS/MS) processing.
LC-MS/MS
Our mass spectrometry data were collected using an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific, San Jose, CA) coupled to a Proxeon EASY-nLC II LC pump (Thermo Fisher Scientific). Peptides were fractionated on a 75-μm inner diameter microcapillary column packed with ∼0.5 cm of Magic C4 resin (5 μm, 100 Å, Michrom Bioresources) followed by ∼35 cm of GP-18 resin (1.8 μm, 200 Å; Sepax, Newark, DE). For each analysis, we loaded ∼1 μg onto the column.
Peptides were separated using a 3-h gradient of 6–26% acetonitrile in 0.125% formic acid at a flow rate of ∼350 nl/min. Each analysis used the multi-notch MS3-based TMT method (McAlister et al., 2014) on an Orbitrap Fusion mass spectrometer. The scan sequence began with an MS1 spectrum (Orbitrap analysis; resolution 120,000; mass range 400−1400 m/z; automatic gain control [AGC] target 2 × 105; maximum injection time 100 ms). Precursors for MS2/MS3 analysis were selected using the TopSpeed parameter of 2 s. MS2 analysis consisted of collision-induced dissociation (quadrupole ion-trap analysis; AGC 4 × 103; normalized collision energy (NCE) 35; maximum injection time 150 ms). Following acquisition of each MS2 spectrum, we collected an MS3 spectrum using our recently described method in which 10 MS2 fragment ions were captured in the MS3 precursor population using isolation waveforms with multiple frequency notches (McAlister et al., 2014). MS3 precursors were fragmented by high-energy collision-induced dissociation and analyzed using the Orbitrap (NCE 55; AGC 5 × 104; maximum injection time 150 ms, resolution was 60,000 at 400 Th).
Data analysis
Mass spectra were processed using a SEQUEST-based in-house software pipeline (Huttlin et al., 2010). Spectra were converted to mzXML using a modified version of ReAdW.exe. Database searching included all entries from the yeast SGD (Saccharomyces Genome Database, March 11, 2014). This database was concatenated with one composed of all protein sequences in the reversed order. Searches were performed using a 50-ppm precursor ion tolerance for total protein-level analysis. The product ion tolerance was set to 0.9 Da. These wide mass tolerance windows were chosen to maximize sensitivity in conjunction with Sequest searches and linear discriminant analysis (Beausoleil et al., 2006; Huttlin et al., 2010). TMT tags on lysine residues and peptide N termini (+229.163 Da) and carbamidomethylation of cysteine residues (+57.021 Da) were set as static modifications, while oxidation of methionine residues (+15.995 Da) was set as a variable modification.
Peptide-spectrum matches (PSMs) were adjusted to a 1% FDR (Elias and Gygi, 2007, 2010). PSM filtering was performed using a linear discriminant analysis, as described previously (Huttlin et al., 2010), while considering the following parameters: XCorr, ΔCn, missed cleavages, peptide length, charge state, and precursor mass accuracy. For TMT-based reporter ion quantitation, we extracted the signal-to-noise (S/N) ratio for each TMT channel and found the closest matching centroid to the expected mass of the TMT reporter ion. PSMs were identified, quantified, collapsed to a 1% FDR, and then collapsed further to a final protein-level FDR of 1%. Moreover, protein assembly was guided by principles of parsimony to produce the smallest set of proteins necessary to account for all observed peptides.
Proteins were quantified by summing reporter ion counts across all matching PSMs using in-house software, as described previously (McAlister et al., 2012, 2014). Briefly, a 0.003 Th window around the theoretical m/z of each reporter ion (126: 126.127 Th; 127N: 127.124 Th; 127C: 127.131 Th; 128N: 128.128 Th; 128C: 128.134 Th; 129N: 129.131 Th; 129C: 129.138 Th; 130N: 130.135 Th; 130C: 130.141 Th) was scanned for ions, and the maximum intensity nearest the theoretical m/z was used. PSMs with poor quality, MS3 spectra with more than six TMT reporter ion channels missing, MS3 spectra with TMT reporter summed S/N ratio < 100, quantitation isolation specificity of <0.7, or no MS3 spectra were excluded (McAlister et al., 2012). Protein quantification values were exported for further analysis in Excel or JMP. Each reporter ion channel was summed across all quantified proteins and normalized assuming equal protein loading of all nine samples. One-way ANOVA was then used to identify proteins that were differentially expressed across strains. We used the Bonferroni method as the multiple testing correction for our p values to minimize the probably of type I error (Dunn, 1961). The p values calculated for each comparison (i.e., protein) were multiplied by the number of comparisons (n = 4765 proteins), and this corrected p value was considered significant if p < 0.01.
Data access
Supplemental Tables 1 and 2 list the proteins and peptides, respectively, as well as normalized TMT reporter ion intensities used for quantitative analysis. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (Vizcaino et al., 2014) via the PRIDE partner repository with the data set identifier PXD002875.
Supplementary Material
Acknowledgments
We thank the members of the Gygi lab at Harvard Medical School. This work was funded in part by National Institutes of Health grants K01 DK098285 (J.A.P.) and GM97645 (S.P.G.).
Glossary
Abbreviations used:
- AGC
automatic gain control
- ANOVA
analysis of variance
- BPRP
basic pH reversed-phase
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- ER
endoplasmic reticulum
- FDR
false discovery rate
- HPLC
high-pressure liquid chromatography
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- ORF
open reading frame
- PSM
peptide-spectrum matches
- S/N
signal-to-noise
- TCA
tricarboxylic acid
- ThDP
thiamine diphosphate
- TMT
tandem mass tag.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-07-0499) on September 23, 2015.
REFERENCES
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back SH, Schroder M, Lee K, Zhang K, Kaufman RJ. ER stress signaling by regulated splicing: IRE1/HAC1/XBP1. Methods. 2005;35:395–416. doi: 10.1016/j.ymeth.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- Blank LM, Sauer U. TCA cycle activity in Saccharomyces cerevisiae is a function of the environmentally determined specific growth and glucose uptake rates. Microbiology. 2004;150:1085–1093. doi: 10.1099/mic.0.26845-0. [DOI] [PubMed] [Google Scholar]
- Blinnikova EI, Mirjuschenko FL, Shabalin YA, Egorov SN. Vesicular transport of extracellular acid phosphatases in yeast Saccharomyces cerevisiae. Biochemistry (Mosc) 2002;67:485–490. doi: 10.1023/a:1015246325818. [DOI] [PubMed] [Google Scholar]
- Boles E, Hollenberg CP. The molecular genetics of hexose transport in yeasts. FEMS Microbiol Rev. 1997;21:85–111. doi: 10.1111/j.1574-6976.1997.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Burchett SA, Scott A, Errede B, Dohlman HG. Identification of novel pheromone-response regulators through systematic overexpression of 120 protein kinases in yeast. J Biol Chem. 2001;276:26472–26478. doi: 10.1074/jbc.M103436200. [DOI] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Abeydeera ND, Bale S, Pai PJ, Dorrestein PC, Russell DH, Ealick SE, Begley TP. Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase. Nature. 2011;478:542–546. doi: 10.1038/nature10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Gygi SP. Hyperplexing: a method for higher-order multiplexed quantitative proteomics provides a map of the dynamic response to rapamycin in yeast. Sci Signal. 2012;5:rs2. doi: 10.1126/scisignal.2002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robichon-Szulmajster H. Induction of enzymes of the galactose pathway in mutants of Saccharomyces cerevisiae. Science. 1958;127:28–29. doi: 10.1126/science.127.3288.28. [DOI] [PubMed] [Google Scholar]
- Doerr A. Mass spectrometry-based targeted proteomics. Nat Methods. 2013;10:23. doi: 10.1038/nmeth.2286. [DOI] [PubMed] [Google Scholar]
- Dunn OJ. Multiple comparisons among means. J Am Stat Assoc. 1961;56:52–64. [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol Biol. 2010;604:55–71. doi: 10.1007/978-1-60761-444-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendt SM, Sauer U. Transcriptional regulation of respiration in yeast metabolizing differently repressive carbon substrates. BMC Systems Biol. 2010;4:12. doi: 10.1186/1752-0509-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey PA. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 1996;10:461–470. [PubMed] [Google Scholar]
- Fukasawa T, Obonai K, Segawa T, Nogi Y. The enzymes of the galactose cluster in Saccharomyces cerevisiae. II. Purification and characterization of uridine diphosphoglucose 4-epimerase. J Biol Chem. 1980;255:2705–2707. [PubMed] [Google Scholar]
- Gao J, Opiteck GJ, Friedrichs MS, Dongre AR, Hefta SA. Changes in the protein expression of yeast as a function of carbon source. J Proteome Res. 2003;2:643–649. doi: 10.1021/pr034038x. [DOI] [PubMed] [Google Scholar]
- Granot D, Snyder M. Carbon source induces growth of stationary phase yeast cells, independent of carbon source metabolism. Yeast. 1993;9:465–479. doi: 10.1002/yea.320090503. [DOI] [PubMed] [Google Scholar]
- Guaragnella N, Zdralevic M, Lattanzio P, Marzulli D, Pracheil T, Liu Z, Passarella S, Marra E, Giannattasio S. Yeast growth in raffinose results in resistance to acetic-acid induced programmed cell death mostly due to the activation of the mitochondrial retrograde pathway. Biochim Biophys Acta. 2013;1833:2765–2774. doi: 10.1016/j.bbamcr.2013.07.017. [DOI] [PubMed] [Google Scholar]
- Hazra AB, Han Y, Chatterjee A, Zhang Y, Lai RY, Ealick SE, Begley TP. A missing enzyme in thiamin thiazole biosynthesis: identification of TenI as a thiazole tautomerase. J Am Chem Soc. 2011;133:9311–9319. doi: 10.1021/ja1110514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AS, Richards AL, Bailey DJ, Ulbrich A, Coughlin EE, Westphall MS, Coon JJ. The one hour yeast proteome. Mol Cell Proteomics. 2014;13:339–347. doi: 10.1074/mcp.M113.034769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermosilla GH, Tapia JC, Allende JE. Minimal CK2 activity required for yeast growth. Mol Cell Biochem. 2005;274:39–46. doi: 10.1007/s11010-005-3112-2. [DOI] [PubMed] [Google Scholar]
- Hong EL, Balakrishnan R, Dong Q, Christie KR, Park J, Binkley G, Costanzo MC, Dwight SS, Engel SR, Fisk DG, et al. Gene Ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 2008;36:D577–581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard SM, Heinrich MR. The anomeric specificity of yeast galactokinase. Arch Biochem Biophys. 1965;110:395–400. doi: 10.1016/0003-9861(65)90140-2. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Robinson MD, Mitsakakis N, Johnston M. The promise of functional genomics: completing the encyclopedia of a cell. Curr Opin Microbiol. 2004;7:546–554. doi: 10.1016/j.mib.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Hunter T, Plowman GD. The protein kinases of budding yeast: six score and more. Trends Biochem Sci. 1997;22:18–22. doi: 10.1016/s0968-0004(96)10068-2. [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki Y, Onozuka M, Mizote T, Nosaka K. Biosynthesis of hydroxymethylpyrimidine pyrophosphate in Saccharomyces cerevisiae. Curr Genet. 2005;47:156–162. doi: 10.1007/s00294-004-0557-x. [DOI] [PubMed] [Google Scholar]
- Kim JH, Roy A, Jouandot D, II, Cho KH. The glucose signaling network in yeast. Biochim Biophys Acta. 2013;1830:5204–5210. doi: 10.1016/j.bbagen.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King NL, Deutsch EW, Ranish JA, Nesvizhskii AI, Eddes JS, Mallick P, Eng J, Desiere F, Flory M, Martin DB, et al. Analysis of the Saccharomyces cerevisiae proteome with PeptideAtlas. Genome Biol. 2006;7:R106. doi: 10.1186/gb-2006-7-11-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CH, Liang H, Gaber RF. Roles of multiple glucose transporters in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:638–648. doi: 10.1128/mcb.13.1.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- Lam FH, Ghaderi A, Fink GR, Stephanopoulos G. Biofuels. Engineering alcohol tolerance in yeast. Science. 2014;346:71–75. doi: 10.1126/science.1257859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb TM, Mitchell AP. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:677–686. doi: 10.1128/MCB.23.2.677-686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Hong ME, Jung SC, Ha SJ, Yu BJ, Koo HM, Park SM, Seo JH, Kweon DH, Park JC, Jin YS. Improved galactose fermentation of Saccharomyces cerevisiae through inverse metabolic engineering. Biotechnol Bioeng. 2011;108:621–631. doi: 10.1002/bit.22988. [DOI] [PubMed] [Google Scholar]
- Lohr D, Venkov P, Zlatanova J. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 1995;9:777–787. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- Lowenstein J (ed.) Methods in Enzymology. vol. 13. Boston: Academic; 1969. Citric acid cycle. [Google Scholar]
- Martinez-Pastor MT, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, Rogers JC, Kuhn K, Pike I, Grothe RA, Blethrow JD, Gygi SP. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal Chem. 2012;84:7469–7478. doi: 10.1021/ac301572t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister GC, Nusinow DP, Jedrychowski MP, Wuhr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem. 2014;86:7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K, Entian KD. Genetic analysis of serine biosynthesis and glucose repression in yeast. Curr Genet. 1992;21:295–300. doi: 10.1007/BF00351686. [DOI] [PubMed] [Google Scholar]
- Nagaraj N, Kulak NA, Cox J, Neuhauser N, Mayr K, Hoerning O, Vorm O, Mann M. System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Mol Cell Proteomics. 2012;11:M111 013722. doi: 10.1074/mcp.M111.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosaka K, Onozuka M, Konno H, Kawasaki Y, Nishimura H, Sano M, Akaji K. Genetic regulation mediated by thiamin pyrophosphate-binding motif in Saccharomyces cerevisiae. Mol Microbiol. 2005;58:467–479. doi: 10.1111/j.1365-2958.2005.04835.x. [DOI] [PubMed] [Google Scholar]
- Padmanabha R, Gehrung S, Snyder M. The KNS1 gene of Saccharomyces cerevisiae encodes a nonessential protein kinase homologue that is distantly related to members of the CDC28/cdc2 gene family. Mol Gen Genet. 1991;229:1–9. doi: 10.1007/BF00264206. [DOI] [PubMed] [Google Scholar]
- Paulo JA, Gygi SP. A comprehensive proteomic and phosphoproteomic analysis of yeast deletion mutants of 14–3-3 orthologs and associated effects of rapamycin. Proteomics. 2014;15:474–486. doi: 10.1002/pmic.201400155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo JA, McAllister FE, Everley RA, Beausoleil SA, Banks AS, Gygi SP. Effects of MEK inhibitors GSK1120212 and PD0325901 in vivo using 10-plex quantitative proteomics and phosphoproteomics. Proteomics. 2014;15:462–473. doi: 10.1002/pmic.201400154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Castillo L, Hughes TR. Why are there still over 1000 uncharacterized yeast genes. Genetics. 2007;176:7–14. doi: 10.1534/genetics.107.074468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P, Clement-Ziza M, Lam H, Campbell DS, Schmidt A, Deutsch EW, Rost H, Sun Z, Rinner O, Reiter L, et al. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature. 2013;494:266–270. doi: 10.1038/nature11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pir P, Ulgen KO, Hayes A, Ilsen Onsan Z, Kirdar B, Oliver SG. Annotation of unknown yeast ORFs by correlation analysis of microarray data and extensive literature searches. Yeast. 2006;23:553–571. doi: 10.1002/yea.1375. [DOI] [PubMed] [Google Scholar]
- Posas F, Camps M, Arino J. The PPZ protein phosphatases are important determinants of salt tolerance in yeast cells. J Biol Chem. 1995;270:13036–13041. doi: 10.1074/jbc.270.22.13036. [DOI] [PubMed] [Google Scholar]
- Qiu H, Hu C, Hinnebusch AG. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol Cell. 2009;33:752–762. doi: 10.1016/j.molcel.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavulu SV, Goud RK, Sarma PN, Mohan SV. Saccharomyces cerevisiae as anodic biocatalyst for power generation in biofuel cell: influence of redox condition and substrate load. Bioresource Technol. 2011;102:2751–2757. doi: 10.1016/j.biortech.2010.11.048. [DOI] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Segawa T, Fukasawa T. The enzymes of the galactose cluster in Saccharomyces cerevisiae. Purification and characterization of galactose-1-phosphate uridylyltransferase. J Biol Chem. 1979;254:10707–10709. [PubMed] [Google Scholar]
- Suomalainen H, Axelson E. Effect of thiamine on the rate of fermentation of zymohexoses and of maltose by baker’s yeast. Biochim Biophys Acta. 1956;20:315–318. doi: 10.1016/0006-3002(56)90291-8. [DOI] [PubMed] [Google Scholar]
- Tang X, Feng H, Chen WN. Metabolic engineering for enhanced fatty acids synthesis in Saccharomyces cerevisiae. Metab Eng. 2013a;16:95–102. doi: 10.1016/j.ymben.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Tang X, Feng H, Zhang J, Chen WN. Comparative proteomics analysis of engineered Saccharomyces cerevisiae with enhanced biofuel precursor production. PLoS One. 2013b;8:e84661. doi: 10.1371/journal.pone.0084661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MC, Monteiro PT, Guerreiro JF, Goncalves JP, Mira NP, dos Santos SC, Cabrito TR, Palma M, Costa C, Francisco AP, et al. The YEASTRACT database: an upgraded information system for the analysis of gene and genomic transcription regulation in Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42:D161–D166. doi: 10.1093/nar/gkt1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- Timsol DJ. Galactose metabolism in Saccharomyces cerevisiae. Dyn Biochem Process Biotechnol Mol Biol. 2007;9:63–73. [Google Scholar]
- Towle HC. Glucose as a regulator of eukaryotic gene transcription. Trends Endocrinol Metab. 2005;16:489–494. doi: 10.1016/j.tem.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Trumbly RJ. Glucose repression in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1992;6:15–21. doi: 10.1111/j.1365-2958.1992.tb00832.x. [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, Dianes JA, Sun Z, Farrah T, Bandeira N, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb KJ, Xu T, Park SK, Yates JR., 3rd Modified MuDPIT separation identified 4488 proteins in a system-wide analysis of quiescence in yeast. J Proteome Res. 2013;12:2177–2184. doi: 10.1021/pr400027m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wixon J, Kell D. The Kyoto Encyclopedia of Genes and Genomes—KEGG. Yeast. 2000;17:48–55. doi: 10.1002/(SICI)1097-0061(200004)17:1<48::AID-YEA2>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Zhang N, Hayes A, Panoutsopoulou K, Oliver SG. Global analysis of nutrient control of gene expression in Saccharomyces cerevisiae during growth and starvation. Proc Natl Acad Sci USA. 2004;101:3148–3153. doi: 10.1073/pnas.0308321100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.