Quantitative experiments and mathematical modeling show that the activity of β-catenin in the adherens junction pathway is regulated by interlocked network motifs consisting of a positive feedback loop, which restores the integrity of adherens junctions, and a negative feedback loop, which limits β-catenin–induced gene expression.
Abstract
The integrity of epithelial tissue architecture is maintained through adherens junctions that are created through extracellular homotypic protein–protein interactions between cadherin molecules. Cadherins also provide an intracellular scaffold for the formation of a multiprotein complex that contains signaling proteins, including β-catenin. Environmental factors and controlled tissue reorganization disrupt adherens junctions by cleaving the extracellular binding domain and initiating a series of transcriptional events that aim to restore tissue homeostasis. However, it remains unclear how alterations in cell adhesion coordinate transcriptional events, including those mediated by β-catenin in this pathway. Here were used quantitative single-cell and population-level in vitro assays to quantify the endogenous pathway dynamics after the proteolytic disruption of the adherens junctions. Using prior knowledge of isolated elements of the overall network, we interpreted these data using in silico model-based inference to identify the topology of the regulatory network. Collectively the data suggest that the regulatory network contains interlocked network motifs consisting of a positive feedback loop, which is used to restore the integrity of adherens junctions, and a negative feedback loop, which is used to limit β-catenin–induced gene expression.
INTRODUCTION
The integrity of epithelial tissue is established and maintained through extracellular homotypic protein–protein interactions called adherens junctions (Baum and Georgiou, 2011). One of the best-characterized members of the cadherin family of transmembrane proteins that provide the homotypic interactions that are central to adherens junctions is E-cadherin (Niessen et al., 2011). E-cadherins also provide a protein scaffold that links extracellular binding sites with the intracellular actin cytoskeleton through a multiprotein complex composed of members of the catenin family of proteins (McCrea et al., 1991; Reynolds et al., 1994). To facilitate these interactions, the cytoplasmic domain of cadherins contains two cadherin homology (CH) domains, CH2 and CH3. The CH2 domain is located proximal to the transmembrane region and interacts with either p120-catenin or δ-catenin (Yang et al., 2010). The CH3 domain is located distal to the transmembrane region and interacts with β-catenin. As illustrated in Figure 1, β-catenin is a pleiotropic transcriptional coactivator that also participates in the canonical Wnt signaling pathway (Clevers and Nusse, 2012) and, in the context of adherens junctions, couples E-cadherin with α-catenin (Aberle et al., 1994). In turn, α-catenin links this multiprotein complex to the actin cytoskeleton (Drees et al., 2005; Yamada et al., 2005). Whereas the role of β-catenin in the canonical Wnt pathway has been the subject of multiple studies that integrate experimental studies with mechanistic modeling and simulation (e.g., Lee et al., 2003; Lloyd-Lewis et al., 2013), quantitative studies into the role of β-catenin in adherens junctions have been primarily theoretical (e.g., van Leeuwen et al., 2007; Ramis-Conde et al., 2008; Basan et al., 2010; Chen et al., 2014).
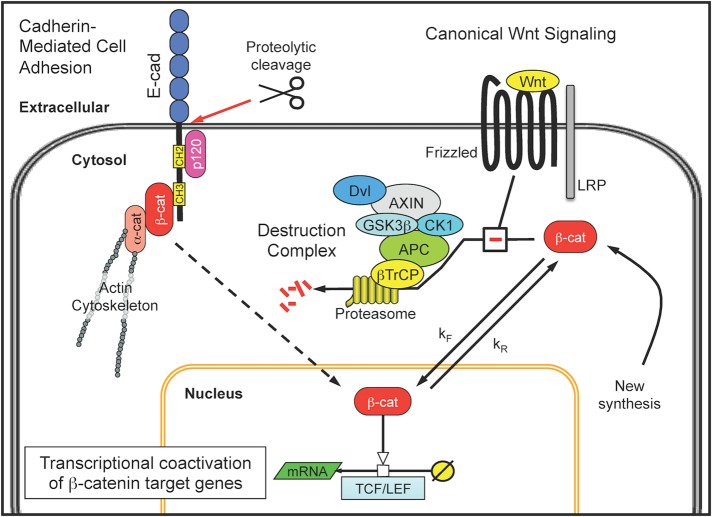
FIGURE 1:
β-Catenin participates in cadherin-mediated cell adhesion and is a transcriptional coactivator in the canonical Wnt signaling pathway. In epithelial cells, the best-characterized adherens junctions are formed around the single-pass transmembrane protein E-cadherin (E-cad). The cytoplasmic tail of E-cadherin provides a scaffold for a multiprotein complex that includes p120-catenin (p120) and β-catenin (β-cat) and links extracellular adhesion sites to the actin cytoskeleton through α-catenin (α-cat). The intracellular portion of the complex can be liberated through proteolytic cleavage of adherens junctions, although the functional role of this cytosolic complex is unclear. Free cytosolic β-catenin is also regulated by a destruction complex associated with the canonical Wnt signaling pathway. The destruction complex comprises the scaffold proteins axin and APC, the serine-threonine kinases CK1 and GSK3β, a Dishevelled segment polarity protein (Dvl), and the β-transducin repeat-containing protein (β-TrCP), which collectively bind, phosphorylate, and ubiquitinate β-catenin. The posttranslationally modified β-catenin is then shuttled to the proteasome for degradation. In the presence of a Wnt ligand, the destruction complex is disassembled by interacting with the Wnt receptor complex of Frizzled and an LRP protein. Disassembly of the destruction complex enables newly synthesized β-catenin to accumulate within the cytosol, translocate to the nucleus, and promote transcriptional coactivation of β-catenin target genes in conjunction with lymphocyte enhancer binding factors (LEF) and T-cell factor (TCF) transcription factors.
In addition to providing mechanical linkage between cells, adherens junctions also play roles in various signal transduction pathways (Fagotto and Gumbiner, 1996; Jamora and Fuchs, 2002). For instance, proteolytic cleavage of the extracellular portion of E-cadherin creates a fragment that contains the cytoplasmic portion, called the C-terminal fragment (CTF; Marambaud et al., 2002). These cytoplasmic fragments can be biologically active, as they can enter the nucleus, bind to catenins (Sadot et al., 1998), and modulate Kaiso-dependent transcriptional activity (Ferber et al., 2008). Creation of these fragments can occur via autocrine signaling mechanisms that use proteases, such as extracellular metalloproteases, γ-secretase, and caspase (Lochter et al., 1997; Steinhusen et al., 2001; Marambaud et al., 2002; Maretzky et al., 2005), which can act in concert—for example, as γ-secretase removes the transmembrane domain from the CTF after an initial cleavage of the extracellular domain by an extracellular metalloprotease (Fortini, 2002). Although this mechanism has been best characterized for the Notch signaling pathway (Schroeter et al., 1998; Struhl and Adachi, 1998; Gao and Pimplikar, 2001), similar mechanisms have been observed with cadherins. For instance, bone morphogenetic protein 4 (BMP4) stimulates the nuclear translocation of a CTF derived from N-cadherin that activates cyclin D1 transcription. This cytoplasmic fragment is formed by the BMP-induced release of a disintegrin and metalloprotease (ADAM) 10 and is subsequently cleaved by γ-secretase. Collectively this process promotes neural crest delamination during development (Shoval et al., 2007). Although many of these mechanistic insights were derived using genetic constructs transfected into cell lines, how alterations in cell adhesion dynamically coordinate transcriptional events that are associated with cadherin-mediated cell adhesion or with canonical Wnt signaling to control tissue organization, especially in an endogenous context, is an important question that remains unresolved (Niessen et al., 2011).
Here our aim was to clarify the endogenous regulatory network associated with β-catenin–induced gene expression in response to proteolytic disruption of E-cadherin–based adherens junctions. Identifying the regulatory network that controls the cellular response typically involves observing the dynamics of the system in response to perturbations (Purvis and Lahav, 2013). Toward this aim, we used trypsin to cleave the extracellular portion of E-cadherin and a small-molecule inhibitor of β-catenin to disrupt the regulatory network in different ways. The cellular copy number and spatial location of β-catenin and E-cadherin were quantified, and functional responses to nuclear localization of β-catenin were measured as a function of time. Creating a holistic view of how information flows within cellular signaling networks from a statistical analysis of experimental data, a process called statistical inference, relies on a variety of computational tools that vary in how they use prior knowledge (Klinke, 2014a). Although many details associated with canonical Wnt signaling and cadherin-mediated cell adhesion have been revealed through detailed biochemical studies, the dynamics and topology associated with how these two pathways are integrated remain uncertain. Mechanistic models can be used to represent this prior knowledge and test competing hypotheses regarding topology. Conventionally, the inference problem is still ill posed, as the uncertainty associated with the model parameters is convoluted with regard to the pathway topology. To address this problem, we use a computationally intensive approach to test whether the topology of the regulatory network is sufficient to explain the observed data for any plausible set of parameter values. Using this in silico model-based inference approach, we found that the transcriptional events associated with disruption of the adherens junctions are regulated by interlocked positive and negative feedback loops, a common network motif used to stabilize the basal level of a protein that is subject to constitutive turnover and that can promote its own expression.
RESULTS
B16F0 cells form E-cadherin–based adherens junctions and produce WISP1 after their proteolytic cleavage
To clarify the regulatory network associated with adherens junctions and canonical Wnt signaling, we selected a model system that relied on E-cadherin for cell-to-cell adhesion and exhibited a functional readout attributed to β-catenin–induced gene expression. In vitro, E-cadherin expression promotes the spontaneous formation of compact multicellular tumor spheroids from breast, colon, and renal carcinoma cell lines (St Croix et al., 1998; Shimazui et al., 2004; Ivascu and Kubbies, 2007). We used the mouse melanoma B16F0 cell line as a model system to study E-cadherin–based adherens junctions because they formed tight spheroids spontaneously (Figure 2A). Formation of spheroids was inhibited by the addition of an antibody against the extracellular domain of E-cadherin to the culture medium (Supplemental Figure S1). The B16F0 spheroids exhibited a tight morphology, as manual pipetting did not disrupt them, and contained metabolically active cells at the periphery and dead cells in the center (Figure 2B). Similar phenotypes were observed in Melan-A cells, an immortalized normal mouse melanocyte cell line, and BT474 and Hs578T cells, two human breast cancer cell lines (Supplemental Table S3 and Supplemental Figure S1).
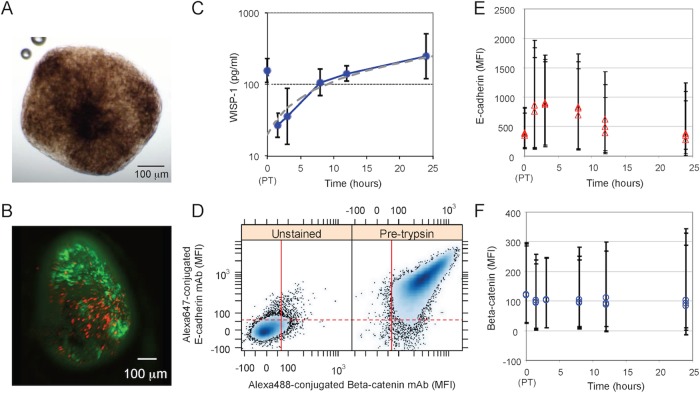
FIGURE 2:
B16F0 cells as a model for E-cadherin–based adherens junctions and their functional response to proteolytic disruption of adherens junctions. (A) Bright-field image of spheroids formed by B16F0 at day 13. (B) Fluorescence microscope image of B16F0 spheroids; metabolically active cells were stained with fluorescein diacetate (green), and dead cells were stained with propidium iodide (red). (C) In 2D culture, WISP1 was assayed by ELISA in B16F0 conditioned medium at the indicated times after trypsinization (mean ± SD, n = 3 biological replicates; representative of at least two independent experiments). A WISP1 concentration profile that corresponds to a constant cellular production rate was determined by linear regression (gray dotted line). (D) Total cellular levels of E-cadherin and β-catenin assayed by flow cytometry (left: unstained B16F0 cells as a negative control; right: B16F0 cells stained with both Alexa Fluor 647–conjugated E-cadherin mAb and Alexa Fluor 488–conjugated β-catenin mAb). Red lines correspond to data-driven fluorescence threshold; 95% of unstained flow cytometric events exhibited a mean fluorescence intensity (MFI) below this value. (E, F) Distribution in MFI associated with E-cadherin (E) and β-catenin (F) represented by symbols (median) and error bars that enclose 95% of the distribution. The results for three biological replicates shown at each data point are representative of at least two independent experiments. In C, E, and F, the 0-h time point corresponds to a sample taken just before trypsinization.
In two recent studies, WISP1 was found to be up-regulated in essentially all tumor tissue samples obtained from patients with invasive breast carcinomas (Klinke, 2014b) and at the invasive front of melanoma (Kulkarni et al., 2012). Given that WISP1 is induced after the nuclear localization of β-catenin (Xu et al., 2000), the production of WISP1 might be a functional response to the disruption of adherens junctions. To confirm this link, we characterized the dynamics of WISP1 production after disruption of E-cadherin–based adherens junctions in vitro using trypsin, a protease that has high activity and specificity (Olsen et al., 2004). Starting with B16F0 cells cultured in two dimensions (2D) at 80% confluence, cells were trypsinized, washed, and replated in new media. After this trypsinization protocol, the concentration of WISP1 in B16F0 cell media was measured by enzyme-linked immunosorbent assay (ELISA), and this measurement was repeated at multiple time points (Figure 2C). A sample of the conditioned medium was taken before and after the trypsinization protocol to establish the dynamic range of the assay. By 12 h, the concentration of WISP1 had recovered to the value observed before the trypsinization protocol, and it continued to increase thereafter for up to 24 h. WISP1 levels in B16F0-conditioned medium in a similar experiment were previously shown to remain relatively constant between 24 and 72 h (see Supplemental Figure S8 in Kulkarni et al., 2012). In an initial analysis of our data, the observed WISP1 dynamics was roughly consistent with WISP1 being produced by B16F0 cells at a constant rate (gray dotted line in Figure 2C). On closer inspection, however, the deviation between the constant-production model and the data suggested an underlying kinetics that deviates from expression at a constant rate, as the 1.5- and 3-h time points were below the values and the 8- and 12-h time points were above the values predicted if WISP1 is expressed at a constant rate. WISP1 was also observed in media conditioned by Melan-A and Hs578T cells but not BT474 cells (Supplemental Table S3).
WISP1 is expressed in response to the nuclear localization of β-catenin, and β-catenin is a component of the adherens junction protein complex. Thus we examined the levels of total cellular E-cadherin and β-catenin at equivalent time points after the proteolytic disruption of the adherens junctions. Flow cytometric analysis showed that B16F0 cells are positive for both β-catenin and E-cadherin (Figure 2D) and that the total cellular protein levels appear to remain relatively constant after trypsinization (Figure 2, E and F). However, although β-catenin remained at nearly constant levels, the levels of E-cadherin appeared to increase slightly after trypsinization and returned to baseline levels within 24 h. The E-cadherin antibody used recognizes a cytoplasmic epitope; thus the slight increase in total E-cadherin may reflect a change in kinetic processes associated with protein turnover, including the de novo synthesis of new E-cadherin and the proteolytic degradation of the cytoplasmic fragment of E-cadherin.
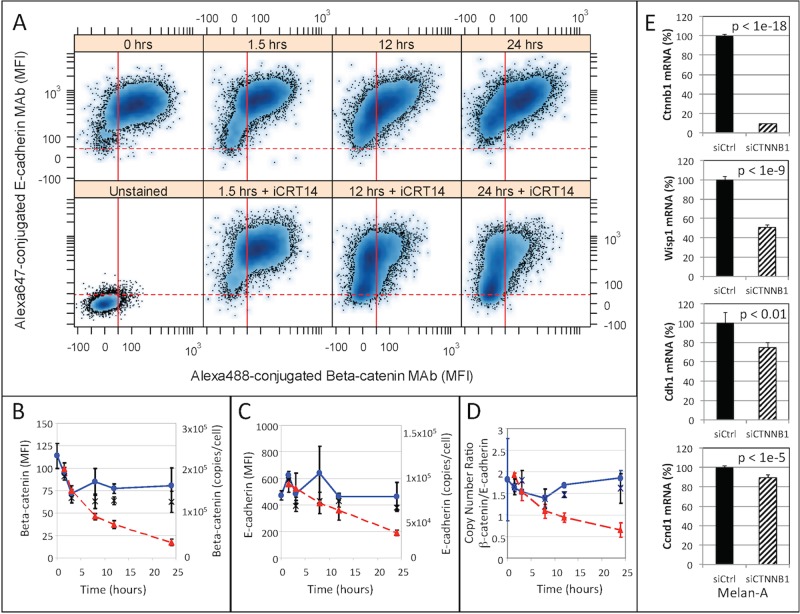
We next used confocal fluorescence microscopy to characterize the spatial distribution of β-catenin and E-cadherin within the cell as a function of time (Figure 3). Cells were visualized just before and 1.5, 3, 8, 12, and 24 h after trypsinization. After fixation and permeabilization, β-catenin and E-cadherin were visualized using antibodies. Spatial localization was assessed by image segmentation: 4′,6-diamidino-2-phenylindole (DAPI)–stained DNA and rhodamine-conjugated phalloidin–stained actin were used to establish the nuclear and whole-cell regions, respectively. The cytoplasmic regions were calculated by subtracting the nuclear regions from the whole-cell regions. Nuclear localization of both β-catenin and E-cadherin was observed at the 1.5-h time point and peaked at the 3-h time point. After 3 h, β-catenin and E-cadherin were distributed throughout the cytoplasm to the same extent as observed at the 0-h time point. The relative distribution between the cytoplasm and the nucleus, measured by the nuclear-to-cytoplasmic log-normalized ratio, was approximately the same for both E-cadherin and β-catenin at all time points (Figure 3, B and C). Trypsinization also altered the morphology of the cells by which cellular protrusions retracted to the center of mass. As discussed later, the results obtained using a small-molecule inhibitor of β-catenin argue against the possibility that the shift in nuclear-to-cytoplasmic localization is an indirect consequence of the change in cell morphology. Similar spatial distributions of E-cadherin and β-catenin were also found in cells stained with fluorophore-conjugated primary antibodies, which suggests that the pattern observed was not due to the cross-reactivity of the secondary antibodies (Supplemental Figure S2). Collectively the results suggest that adherens junction disruption initiates the transient translocation of both β-catenin and E-cadherin species labeled by the antibody (i.e., full-length E-cadherin and truncated variants containing the cytoplasmic fragment of E-cadherin) into the nucleus. This transient localization appears to be temporally coincident with total E-cadherin and the extracellular release of WISP1. Of interest, total cellular β-catenin remained constant throughout this process.
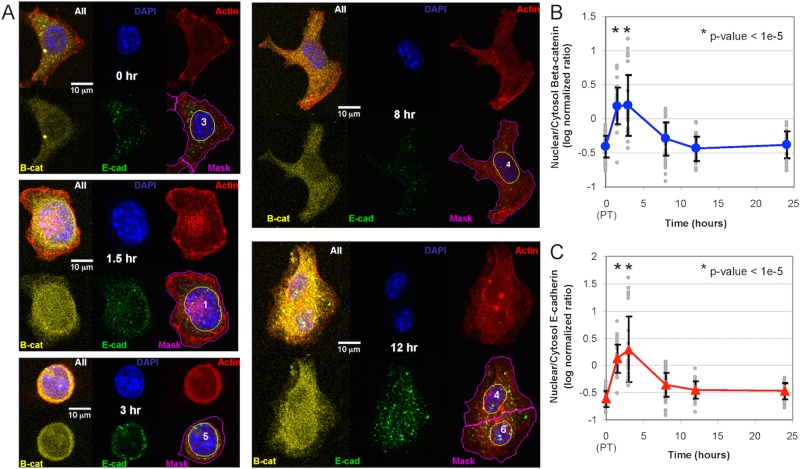
FIGURE 3:
Confocal fluorescence microscopy of β-catenin and the cytoplasmic fragment of E-cadherin after trypsinization. (A) Representative confocal fluorescence microscopy images of the localization of β-catenin and E-cadherin at different time points after trypsinization. Individual images show a composite image, DAPI staining (blue), actin cytoskeleton (red), β-catenin (yellow), and E-cadherin (green), and a composite image with a nuclear mask (yellow line) and a cell mask (magenta line). (B, C) Log10 ratio of nuclear-to-cytoplasmic localization of β-catenin (B) and cytoplasmic fragment of E-cadherin (C) at different times after trypsinization. Approximately 40 cells were analyzed at each time point (gray circles), and the symbols and error bars represent the means ± SD of the observed population. *p < 1e-5 relative to the pretrypsinized samples (0 h). The results are representative of at least two biological replicates.
Nuclear localization of β-catenin is required for WISP1 production
To test more directly whether the transient nuclear localization of β-catenin is associated with the extracellular release of WISP1, we used a small-molecule inhibitor to block β-catenin signal transduction. B16F0 cells were treated with iCRT14 before trypsinization. iCRT14 is a thiazolidinedione reported to block β-catenin binding to the T-cell factor (TCF) transcription factor to limit β-catenin–mediated transcription (Gonsalves et al., 2011). B16F0 cells were treated with iCRT14 before trypsinization, and confocal fluorescence microscopy was used to quantify the spatial localization of β-catenin and E-cadherin after trypsinization (Figure 4). Compared to untreated cells (Figure 4B and dotted black lines in Figure 4, C and D), the intracellular transport of β-catenin and E-cadherin in iCRT14-treated cells was inhibited, as both proteins remained at the cell periphery. A slight shift in distribution toward the nucleus was observed at 1.5 h compared with all of the other time points in iCRT14-treated cells (Figure 4, C and D). However, compared with untreated cells, nuclear localization was significantly reduced at 1.5, 3, and 8 h (compare solid to dotted lines in Figure 4, C and D). Because the disruption of the adherens junctions compressed the cytoplasmic area (as seen by comparing the 3- and 8-h time points in Figure 3A), the inability of β-catenin and E-cadherin species to enter the nucleus in cells treated with iCRT14 argues against the possibility that the observed nuclear localization is an indirect consequence of compression of the cytoplasmic area rather than an active transport process.
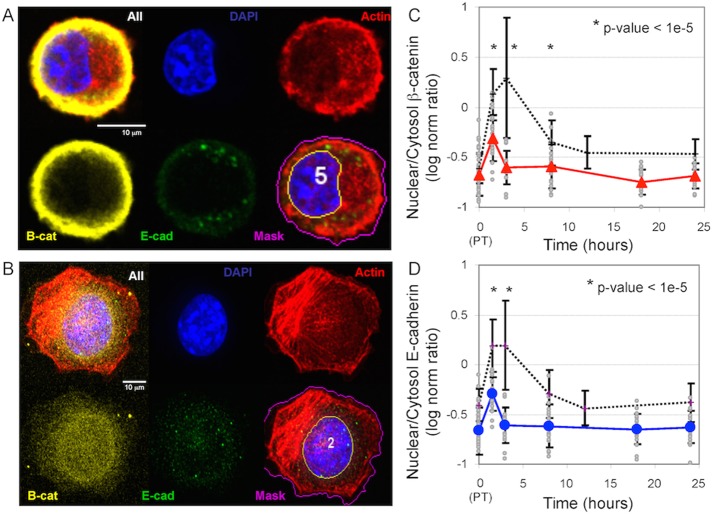
FIGURE 4:
Localization of the β-catenin complex after iCRT14 inhibitor treatment. (A, B) Representative confocal fluorescence microscopy images of the localization of β-catenin and E-cadherin 3 h after trypsinization in B16F0 cells pretreated with iCRT14 (A) or left untreated (B). Image montage shows a composite image, DAPI staining (blue), actin cytoskeleton (red), β-catenin (yellow), and E-cadherin (green), and a composite image with a nuclear mask (yellow line) and a cell mask (magenta line). (C, D) Log10 ratio of nuclear-to-cytoplasmic localization of β-catenin (C) and the cytoplasmic fragment of E-cadherin (D) at different times after trypsinization. Approximately 20 iCRT14-treated B16F0 cells were analyzed at each time point (gray circles), and the symbols and error bars represent the means ± SD of the observed population. Summary of results for untreated cells are shown for comparison (black dotted lines). *p < 1e-5 relative to the untreated cells observed at the same time point. The results are representative of at least two biological replicates.
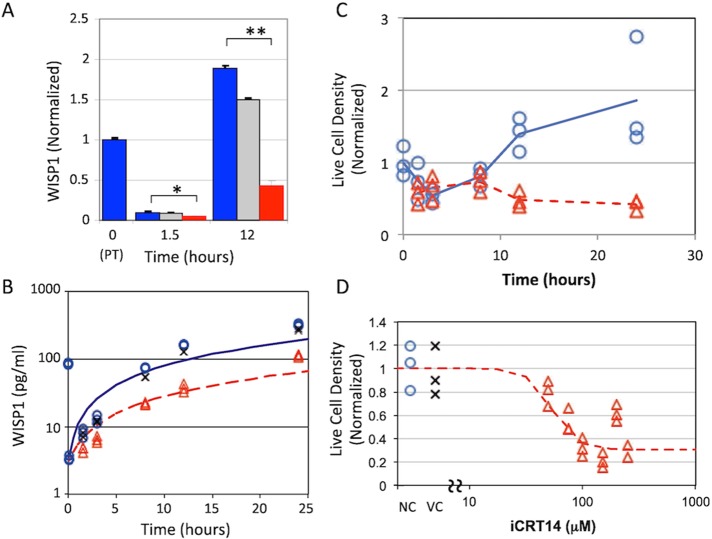
Of importance, the extracellular release of WISP1 was also inhibited in cells exposed to iCRT14 compared with untreated and vehicle-treated control cells (1.5 h, p < 0.05; 12 h, p < 0.0001; Figure 5A). The dynamic behavior of WISP1 among untreated, vehicle-treated, and iCRT14-treated cells was qualitatively similar, although its levels were consistently lower in the media of iCRT14-treated cells (Figure 5B). We also found that iCRT14 inhibited cell proliferation after trypsinization in a dose-dependent manner (Figures 5, C and D) but not cell viability (Supplemental Figure S3). For all treatment conditions, the experimental data deviated from a model in which WISP1 is produced constitutively and at a constant rate. Specifically, the WISP1 level was lower than expected at the 1.5- and 3-h time points and greater than expected at the 24-h time point (Figure 5B), suggesting that adherens junctions disruption results in the nuclear localization of β-catenin, which then induces the expression of WISP1.
FIGURE 5:
WISP1 production and cell density after iCRT14 inhibitor treatment. (A) WISP1 in conditioned medium was observed at 1.5 and 12 h posttrypsinization for the different treatment conditions (untreated, blue; DMSO, gray; iCRT14, red) and normalized to the initial value observed before trypsinization (t = 0). WISP1 was lower in medium conditioned by B16F0 cells pretreated with iCRT14 than in untreated cells. *p < 0.05, **p < 0.0001. Data represent mean ± SD with n = 3 biological replicates and are representative of at least two independent experiments. (B) B16F0 cells pretreated with iCRT14 (red triangles), pretreated with DMSO (vehicle control, x), or untreated (blue circles) were trypsinized (t = 0) and cultured for the indicated times. WISP1 in the cell-conditioned medium was quantified by ELISA (three biological replicates were acquired at each time point and are representative of at least two independent experiments). Untreated and iCRT14-treated results are compared against WISP1 concentration profiles that correspond to constant cellular production rates, as determined by linear regression (untreated, blue solid line; iCRT14, red dotted line). The density of live B16F0 cells was assessed by flow cytometry. (C) First, live-cell density was assayed as a function of time after trypsinization when cultured in sDMEM (blue circles) and sDMEM with 25μM iCRT14 (red triangles). (D) Second, live-cell density was assayed at 24 h when cultured in sDMEM (blue circles, negative control [NC]), sDMEM plus DMSO (x, vehicle control [VC]), and sDMEM plus increasing concentrations of iCRT14 (red triangles).
β-Catenin and E-cadherin levels also depend on β-catenin nuclear localization
Because iCRT14 inhibited the nuclear localization of β-catenin and cytoplasmic fragments of E-cadherin, we next investigated how β-catenin and E-cadherin responded to iCRT14 treatment. Flow cytometry analysis (Figure 6) showed that B16F0 cells were initially positive for both β-catenin and E-cadherin. Of interest, the median fluorescence intensity associated with both β-catenin and E-cadherin declined in iCRT14-treated cells but not in untreated or vehicle-treated control cells. The decline in E-cadherin–associated fluorescence occurred more slowly than that associated with β-catenin, which occurred almost exponentially. Using Quantum Simply Cellular calibration beads, we quantified the copy numbers of total β-catenin and E-cadherin in cells. In untreated cells, we observed between 150,000 and 250,000 β-catenin molecules and between 75,000 and 125,000 E-cadherin molecules per cell. On a per-cell basis, the copy number ratio of β-catenin to E-cadherin was ∼1.75 in untreated cells, whereas the ratio decreased from 1.75 to 0.5 in cells treated with iCRT14 in 24 h. The stoichiometry of β-catenin to E-cadherin within an adherens junction protein complex has been reported to be 1:1 (Huber et al., 2001); thus this difference in ratio in untreated cells could be due to the presence of another cadherin that we have not measured, the possibility that the efficiency of E-cadherin monoclonal antibody (mAb) labeling was not 100%, or the possibility that the excess β-catenin is involved in canonical Wnt signaling and protected by the adenomatous polyposis coli complex. Similarly, we also found that expression of Wisp1, E-cadherin, and cyclin-D1 mRNA was decreased in Melan-A cells after knockdown of β-catenin using small interfering RNA (siRNA; Figure 6E). Collectively the declines in β-catenin and E-cadherin in cells treated with iCRT14 or siRNA suggest that the de novo synthesis of β-catenin and E-cadherin in these cell models depends on the nuclear localization of β-catenin and the cytoplasmic fragment of E-cadherin. Moreover, the decline in the copy number ratio in cells treated with iCRT14 suggests that β-catenin transcription may be more responsive to the nuclear localization of these proteins than the transcription of E-cadherin.
FIGURE 6:
Effect of inhibition of β-catenin–mediated transcription on the levels of β-catenin and E-cadherin after trypsinization. (A) E-cadherin and β-catenin abundance after trypsinization of untreated B16F0 cells (top) and in B16F0 cells pretreated with iCRT14 (bottom) was assayed by flow cytometry at different time points. Red lines indicate data-driven gates for which 95% of the unstained B16F0 cells (lower left) exhibited a lower mean fluorescence intensity associated with β-catenin (solid red line) or with E-cadherin (dashed red line). Summary of the change in β-catenin (B) and E-cadherin (C) as a function of time after trypsinization. The means of the population median MFI are shown on the left y-axis and the median copies per cell on the right y-axis. The copies per cell were determined using Quantum Simply Cellular calibration microbeads. (D) Copy number ratio of β-catenin to E-cadherin in individual cells as estimated by flow cytometry. In B–D, results for untreated B16F0 cells are indicated by blue circles, results for B16F0 cells pretreated with DMSO are indicated by black x’s, and results for B16F0 cells pretreated with iCRT14 are indicated by red triangles. Data obtained from three biological replicates obtained at each time point (mean of the median for each replicate ± SD) are representative of at least two independent experiments. (E) Expression of Ctnnb1, Wisp1, Cdh1, and Ccnd1 was assayed in biological triplicate by qRT-PCR in Melan-A cells 24 h after transfection with either a nontargeting siRNA (siCtrl) or siRNA against human β-catenin (siCtnnb1). Expression of Gapdh was used as a loading control.
Data suggest that adherens junctions regulate β-catenin using two interlocked network motifs
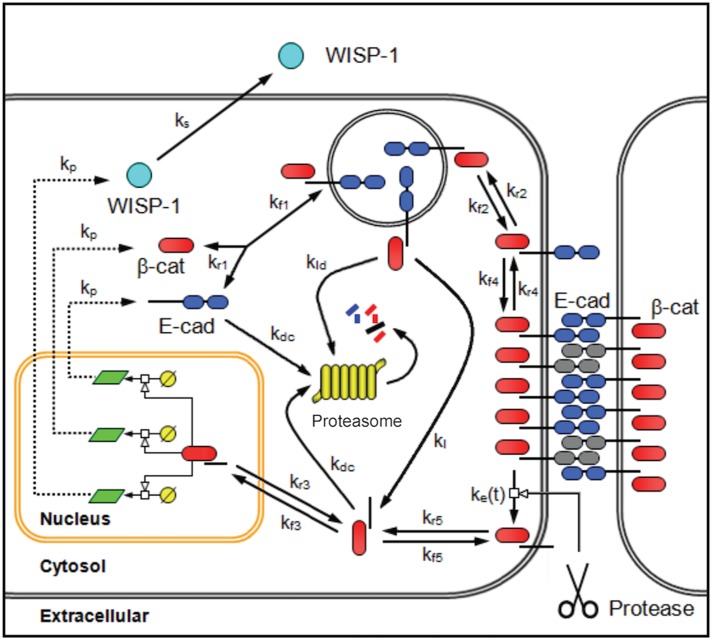
To integrate these experimental observations into a more coherent picture of this cellular system, we formulated a mechanistic model that contained the core relationships related to β-catenin and E-cadherin in adherens junctions observed by other groups (Figure 7). The topology of the model aims to capture the basic regulatory network rather than provide a comprehensive model of all of the protein interactions associated with adherens junctions, as summarized in Zaidel-Bar (2013). Briefly, our mechanistic model represents an average cell at a constant cell density and incorporates four compartments to represent the transport of species among the membrane region, cytoplasm, nucleus, and extracellular space, and it contained two dynamic network motifs. The first network motif is a positive feedback loop; it comprises the multiprotein complexes that contain β-catenin in the cytoplasm and are produced either through an intracellular recycling pathway or from the extracellular cleavage of adherens junctions. These can localize to the nucleus to induce the expression of β-catenin and E-cadherin. WISP1 is included as another gene product of this positive feedback loop that is secreted. The second network motif is a negative feedback loop involving proteasomal degradation of the multiprotein complexes in the cytoplasm that contain β-catenin. Multiprotein complexes that are endocytosed can be degraded (Yap et al., 2007). Alternatively, lysosomes also contain pH-activated proteases, including the cysteine protease cathepsin B. E-cadherin is a substrate of cathepsin B, suggesting that lysosomal degradation may also involve liberating the cytoplasmic portion of the multiprotein complex containing E-cadherin and β-catenin. As the fate of this multiprotein complex upon endocytosis is unclear, we included two alternative hypotheses in the model: one path in which endocytosed complex is degraded entirely, and an alternative path in which endocytosed complex provides a source of cytoplasmic complex. The specific equations used in the model are described in the Supplemental Methods.
FIGURE 7:
Schematic diagram of the network topology associated with the adherens pathway activity of β-catenin. Adherens junctions are formed by homotypic interactions between the extracellular cadherin domains of a single-pass transmembrane protein E-cadherin (E-cad, blue double oval). The cytoplasmic tail of E-cadherin provides a scaffold for a multiprotein complex that includes β-catenin (β-cat, red oval). On the proteolytic cleavage of adherens junctions, a cytoplasmic fragment of E-cadherin and associated catenins (tBE) are transported to the cytoplasm. In the cytoplasm, the cytoplasmic fragment of E-cadherin and associated catenins can either enter the nucleus or undergo proteasomal degradation. In the nucleus, this multiprotein complex promotes the transcription and translation of WISP1, β-catenin, and E-cadherin, among other factors. mRNA is represented by a light green parallelogram. Once synthesized, WISP1 (light blue circle) is secreted. Newly synthesized β-catenin and E-cadherin reform the multiprotein complex and are transported to the cell membrane to reestablish adherens junctions. If no E-cadherin binding sites are present, the multiprotein complex is internalized and degraded. The model also includes an alternative hypothesis regarding the fate of endocytosed multiprotein complex containing E-cadherin and associated catenin. Based on the presence of pH-activated proteases for which E-cadherin is a substrate, lysosomal degradation of the complex can provide an additional source of the cytoplasmic fragment of E-cadherin and associated catenins.
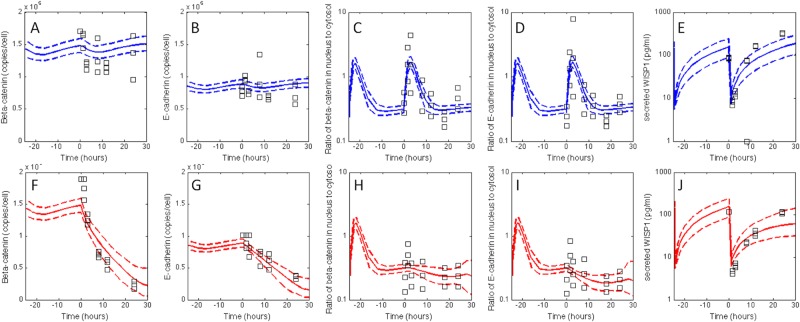
We used in silico model-based inference to determine whether the postulated topology of the mechanistic model was consistent with the observed behavior of WISP1, E-cadherin, and β-catenin in trypsinated B16F0 cells and in the presence of the Wnt-pathway inhibitor iCRT14. In silico model-based inference involves a random walk in parameter space to select parameter combinations that are consistent, within a certain probability, with the observed data (Klinke, 2009; Klinke et al., 2012). In theory, selecting appropriate parameter values is an overdetermined problem because 176 data points were used to constrain 29 model parameters, which are listed in Supplemental Tables S1 and S2. However, in practice, selecting appropriate parameter values is a more difficult problem computationally and conceptually (Klinke, 2014a). An adaptive Markov chain Monte Carlo approach based on a Metropolis–Hastings algorithm, which is suited to these types of issues, was used to provide this random walk in parameter space; the computational details of the approach are described in the Supplemental Methods. Converged segments from four independent Markov chains were used to establish the posterior distributions in the model predictions and establish an identifiable subset of model parameters (Supplemental Figures S4 and S5). The posterior distributions in the model predictions show good agreement with the observed experimental data (Figure 8). In most cases, the dynamic trends were consistent between the model predictions and experimental observations despite the fact that the experimental variability was greater than the posterior distribution in model predictions. The small variability in the model predictions is consistent with the in silico model-based inference approach; the experimental variability associated with each time point is averaged out by using a mechanistic model to integrate across the different time points. However, the model underestimates the concentration of WISP1 in cell-conditioned media at later times. This underprediction may be a consequence of assuming that the cell density remains constant during the experiment in the case of B16F0 cells cultured in media alone or assuming that the effect of iCRT14 also remains constant during the experiment. The posterior distribution in the model parameters also exhibited a complicated structure, with a subset of the model parameters exhibiting narrow-bounded distributions, including the maximum rate associated with proteasomal degradation (V mP), the rate constant for intracellular transport of a multiprotein complex containing β-catenin and the cytoplasmic fragment of E-cadherin from the membrane to the cytoplasm (kr5), and the number of extracellular binding sites for E-cadherin (Esites; Supplemental Figure S6). The relative values of kl (0.2 h−1) versus kld (0.009 h−1) suggest that pH-dependent proteolytic cleavage of the lysosomal multiprotein complex containing E-cadherin and β-catenin is a significant source of CTF–β-catenin complex in the cytoplasm (Supplemental Figure S7). In addition, a number of parameters were positively correlated and could not be independently identified, including the maximum rate of WISP1 (V mW), β-catenin (V mB), and E-cadherin (V mE) mRNA synthesis.
FIGURE 8:
The posterior distribution in the model predictions of the adherens junction pathway compared against the observed data. The experimental data (squares) for total β-catenin (A, F), total E-cadherin (B, G), nuclear-to-cytoplasmic ratio of β-catenin (C, H), and E-cadherin (D, I) and the level of secreted WISP1 (E, J) were compared with the posterior distributions in the model predictions. The cellular responses to trypsinization (A–E) and to trypsinization combined with iCRT14 treatment (F–J) are shown separately. The uncertainty in the model predictions was determined using an empirical Bayesian approach based on an adaptive Metropolis–Hastings algorithm, as described in Supplemental Methods S1. Using the converged segments of four Markov chains, the most likely predictions are represented by the solid line, and the dashed lines enclose the 95% confidence interval. Because the initial values for many of the biomolecular species are unknown and the values of the parameters also influence these initial values, the system was simulated using multiple trypsinization cycles, with a single cycle having a 25-h duration. Figure shows the second and third trypsinization cycles, where the third trypsinization cycle was used to compare against the observed data.
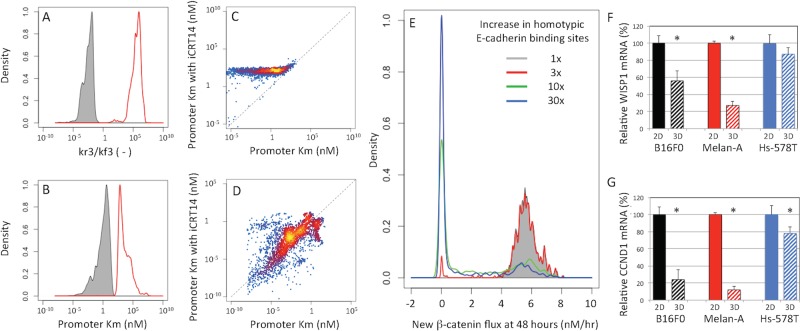
We also used in silico model-based inference to gain insight into the mechanism of action of iCRT14, a small-molecule inhibitor of the Wnt pathway. In analyzing the results of iCRT14 treatment, many of the model parameters were assumed to be the same as in the untreated conditions, and only a few kinetic parameters changed in response to iCRT14 treatment, which are listed in Supplemental Table S2. These parameters included the rate parameters associated with the nuclear localization of the multiprotein complex containing β-catenin (kf3 and kr3) and the Kms associated with the induction of mRNA synthesis for Wisp1, Ctnnb1, and Cdh1 (KmW, KmB, and KmE). The mechanism of action corresponds to how the posterior distributions in these rate parameters were shifted in the presence of iCRT14, as shown in the curves in red in Figure 9. According to the model, iCRT14 inhibited the nuclear localization of the multiprotein complex containing β-catenin as represented by a 108 shift in the ratio of rate constants for nuclear-to-cytoplasmic transport (Figure 9A). In addition, the parameter distributions suggest that iCRT14 independently increases the effective Km associated with the induction gene expression of β-catenin (Figure 9B) and WISP1 (Figure 9C). The inhibition of β-catenin gene expression was more pronounced than the inhibition of WISP1; the Kms were shifted by a factor of 105 for β-catenin compared with only a factor of 101 for WISP1. Of interest, the posterior distributions in the effective Km associated with the induction of E-cadherin gene expression suggest that iCRT14 does not independently inhibit E-cadherin gene expression once the multiprotein complex has entered the nucleus (Figure 9D).
FIGURE 9:
Model-based inference of the effect of the β-catenin small-molecule inhibitor iCRT14 and the contribution of feedback loops. Given the agreement between the model predictions and observed data shown in Figure 8, the model-based inference results also include distributions in model parameters that can be used to gain insight into the mechanism of action of the drug and the contribution of the positive and negative feedback loops. Posterior distributions in the ratio of rate constants associated with the nuclear localization of β-catenin (A) and in the Km associated with β-catenin gene expression (B) in untreated B16F0 cells (gray shaded) and in B16F0 cells treated with iCRT14 (red line). Scatter plots in the inferred Km in untreated cells (x-axis) vs. cells treated with iCRT14 (y-axis) for the induction of WISP1 (C) and E-cadherin (D) gene expression. The points are colored based on a blue/red/yellow density scale calculated using kernel density estimation. (E) Posterior distributions in the synthesis rate of new β-catenin at 48 h after trypsinization, expressed in terms of nM/h. The β-catenin synthesis rate was estimated for increasing levels of extracellular E-cadherin binding sites (Esites): 1× (gray shaded), 3× (red line), 10× (green line), and 30× (blue line). WISP1 (F) and CCND1 (G) mRNA was assayed by qRT-PCR in B16F0, Melan-A, and Hs-578T cells cultured in 2D tissue culture (solid bars) and 3D tissue spheroids (hashed bars). Gene expression was quantified using the delta/delta Ct methods using glyceraldehyde-3-phosphate dehydrogenase mRNA as a loading control and normalized to cells cultured in 2D tissue culture (*p < 0.05 relative to cells cultured in 2D, where n = 3).
Our results moreover suggest that these two network motifs are balanced to enable a stable basal level of β-catenin and E-cadherin gene expression. At 48 h after the enzymatic disruption of the adherens junctions, the molecular production, that is, molecular flux, through the positive feedback loop is nonzero and contributes, on average, 3 copies/s of newly synthesized β-catenin (gray curve in Figure 9E). Overall the dynamics of the system is governed by a balance between these competing feedback mechanisms (Supplemental Figure S8). Initially, the negative feedback loop dominates, as the net β-catenin flux is negative. De novo protein synthesis associated with the positive feedback loop introduces a delay and persists, such that the net β-catenin flux is positive 10 h after trypsinization and remains positive as the system approaches equilibrium at later times. Under these conditions, the adherens pathway activity of β-catenin can be considered constitutively active.
The model also suggests that the constitutive activity of β-catenin depends on the number of extracellular binding sites for E-cadherin. Using an ensemble of model parameters obtained from the converged segments of the Markov chains, we simulated the cell response upon exposure to increasing numbers of extracellular binding sites for E-cadherin. The simulation results suggest that, depending on the model parameters, the predicted responses segregate into one of two distinct states (Figure 9E). In one state, the adherens pathway activity of β-catenin is constitutively active, and in a second state, the pathway is inactive. As the number of extracellular binding sites increases, the inactivation of the adherens pathway activity of β-catenin becomes more prevalent, and the predicted cell response becomes less sensitive to variation in the model parameters. Because the model parameters represent the propensity for reaction among interacting molecular species, these values can vary among cells of a population due to stochastic fluctuations in molecular species or local factors that alter the molecular conformations that protein can access. These simulation results imply that as the number of extracellular binding sites increases, a cell is less likely to exhibit constitutive β-catenin adherens pathway activity irrespective of these stochastic fluctuations. Functionally, these results suggest that WISP1 secretion and cell proliferation would decrease as the number of homotypic E-cadherin binding sites increase. To test this prediction, we cultured B16F0, Melan-A, and Hs-578T cells in two different configurations—as a monolayer in conventional tissue culture plates (2D) and as tissue spheroids in hanging drops (three dimensions [3D])—and assayed Wisp1 (Figure 9F) and cyclin D1 (Ccnd1, Figure 9G) gene expression by quantitative reverse transcription PCR (qRT-PCR). In all three cell lines, we found that both Wisp1 and Cyclin D1 gene expression was decreased in cells cultured as 3D spheroids compared with a 2D monolayer configuration. In comparison to the B16F0 cell line, the phenotype was most pronounced in the Melan-A, whereas the response of the Hs-578T cell line was more subtle.
DISCUSSION
The presence of interlocked feedback loops that drive robust cellular responses to extracellular signals is a common theme in biological systems (Brandman et al., 2005; Amit et al., 2007; Tsai et al., 2008). Conceptually, the competition between positive and negative feedback loops can give rise to a variety of dynamic behaviors that elicit different cellular responses, including oscillations, switching behavior, and single transient bursts of signaling activity (Purvis and Lahav, 2013). Moreover, systems constructed from the same components can elicit different dynamics that cause different cellular responses, with β-catenin as an example of a pleiotropic component. A mechanistic model was used to represent prior knowledge of protein–protein interactions and intracellular transport of proteins reported to occur in this pathway and competing hypotheses regarding the network topology. Because the particular response is regulated by the values of the parameters associated with the regulatory network motifs, we used in silico model-based inference to sample statistically all possible parameter values and generate confidence intervals on the model predictions and parameter values based on the available data.
Effective cell–cell adhesion through cadherins involves the transport of newly synthesized E-cadherin to the cell membrane and the assembly of a multiprotein complex around its cytoplasmic portion to stabilize the cell-to-cell interaction. Using the cytoplasmic tail of E-cadherin as a scaffold, this multiprotein complex includes β-catenin, α-catenin, and p120-catenin. The formation of adherens junctions is a dynamic process, and protein turnover and trafficking play essential roles (Yap et al., 2007). Our analysis suggests that the rates of β-catenin and E-cadherin synthesis are designed to be stoichiometrically equivalent to reconstitute this multiprotein complex in response to its degradation. This suggestion is supported by the high positive correlation between the maximum rates for β-catenin (V mB) and E-cadherin (V mE) transcription and similar Km values, with β-catenin having a slightly higher value than E-cadherin. However, the biological mechanisms used to coregulate β-catenin and E-cadherin gene expression are different. In the case of β-catenin, p120-catenin binds to and inhibits the transcriptional repressor ZBTB33 (also called ZNF348/KAISO), which has a binding site 857 base pairs upstream from the β-catenin gene (Park et al., 2005; Del Valle-Perez et al., 2011). Release of the transcriptional repressor enables β-catenin to promote the expression of targeted genes, including β-catenin and cyclin D1, an important regulator of cell cycle progression (Brown et al., 1998; Shtutman et al., 1999). We observed that the de novo synthesis of β-catenin and cell proliferation in response to adherens junctions disruption are both inhibited by iCRT14. Collectively the induction of β-catenin gene expression requires the joint activities of p120-catenin and β-catenin within the nucleus. In contrast, nuclear β-catenin directly regulates the expression of E-cadherin and WISP1 by interacting with a number of other transcription factors, including LEF1. LEF1 has a binding site upstream of E-cadherin (TRANSFAC accession number T00930) and, together with another transcription factor, TCF1, has a transcription factor binding site 814 base pairs upstream (Motif M00978 at motifmap.ics.uci.edu) of the WISP1 gene.
In addition, these results suggest that the cytoplasmic fragment of E-cadherin functions as a scaffold to transport β-catenin and p120-catenin into the nucleus as a multiprotein complex. The proteolytic liberation of the cytoplasmic fragment of E-cadherin rather than the disruption of homotypic bonds between extracellular cadherin repeats appears to be required for the transcriptional activity of this complex (Ivanov et al., 2004; Kam and Quaranta, 2009). Once liberated, the nuclear localization of this complex is dependent on the presence of β-catenin, which has an importin α–dependent nuclear localization sequence (NLS; Kosugi et al., 2009). Moreover, the cytoplasmic fragment of E-cadherin does not have an importin α–dependent NLS (Kosugi et al., 2009). If dissociation of multiprotein complex occurred at the membrane and we assume that the cytosolic fragment of E-cadherin has an importin α–independent NLS, then iCRT14 would have to inhibit independently the nuclear localization of β-catenin and the cytoplasmic fragment of E-cadherin. The evidence provided to support the specificity of iCRT14 in inhibiting β-catenin activity argues against this possibility (Gonsalves et al., 2011). Although it may seem that the observation that β-catenin influences the nuclear localization of the complex contradicts the findings of other groups, the results are consistent with the transcriptional regulatory network that we identified. For instance, in one study (Ferber et al., 2008), the authors use an engineered cDNA construct to help clarify the biological role of the CTF of E-cadherin. A subtle but significant steady-state increase in nuclear localization of a transfected E-cadherin/CTF construct was observed in cells cotransfected with p120-catenin but not in cells cotransfected with β-catenin. This finding led the authors to conclude that p120-catenin promotes the nuclear localization of the E-cadherin CTF. Here we found that the nuclear localization of the endogenous E-cadherin CTF was inhibited by iCRT14, a small-molecule inhibitor of β-catenin. Although one explanation is that iCRT14 is nonspecific, a more realistic possibility is that p120-catenin inhibits the proteasomal degradation of E-cadherin (Xiao et al., 2003). Thus the model implies that an increase in p120-catenin would inhibit the negative feedback pathway and thereby increase the steady-state levels of the multiprotein complex containing the E-cadherin CTF, as similarly observed for β-catenin in the canonical Wnt pathway when the destruction complex is disassembled. Given that the multiprotein complex containing the E-cadherin CTF preferentially localizes to the nucleus and the number of membrane-bound complexes depends on the total number of extracellular homotypic binding sites, an increase in the steady-state levels of the multiprotein complex containing the E-cadherin CTF would increase nuclear localization when the pool of homotypic binding sites is saturated. In 2D tissue culture, the number of homotypic binding sites is limited due to the spatial arrangement of cells. Moreover, the knockdown of p120-catenin would inhibit the positive feedback pathway and enhance the negative feedback pathway, resulting in a loss of E-cadherin expression, as observed previously (Davis et al., 2003; Xiao et al., 2003).
Mechanistic modeling is an important cognitive tool to help understand dynamic systems by integrating prior knowledge of system components and experimental observations of the system. In formulating a mechanistic model, the topology reflects two competing objectives: incorporating biochemical details to capture the complexity of the studied system versus simplifying the model to distill regulatory principles. Although biochemical details can be mined from the scientific literature, simplifying a model relies on mathematical analyses that assume that the topology of the model and the corresponding model parameters reflect the “truth” and that a steady-state analysis is relevant for the modeled system. Prior studies of the canonical Wnt pathway reflect these competing objectives. For instance, Lee et al. (2003) developed one of the first computational models of the canonical Wnt pathway, based on studies using Xenopus extracts. Although their model captures the biology as experimentally observed, a number of subsequent mathematical analyses found that this complicated model could be simplified (Krüger and Heinrich, 2004), even to a single ordinary differential equation for β-catenin (Mirams et al., 2010). These studies naturally raise the question of what level of complexity is supported in this study. Although the focus here is to determine whether the topology of the network describes the observed system, the data presented challenge a number of the assumptions associated with simplifying the model. First, consider whether the parameters reflect the “truth.” Conventionally, mathematical analyses are applied to a model and a single set of best-fit parameters, that is, their “true values” (Klinke, 2014a). The model-based inference results suggest that there are a number of different parameter combinations that provide predictions consistent with the observed data. Focusing our attention on Figure 9E, we see that the distribution in β-catenin production at 48 h is generated by simulating the particular conditions using different parameter combinations sampled from the converged segments of the Markov chains. As the number of extracellular binding sites increases, the distribution in flux of new β-catenin shifts from ∼6 nM/h to 0. However, there are still parameter sets predicting that, even if one increases the homotypic E-cadherin binding sites by 30 times, the flux of new β-catenin remains at 6 nM/h. This suggests that simplified models derived from time-scale analysis or stability analysis may be different depending on which particular sets of parameters one uses. Of greater importance, a single set of “true” parameter values is not supported by the experimental data. Second, consider whether a steady-state analysis is relevant for the modeled system. Experimentally, culturing adherent cells in vitro requires passaging the cells every couple of days such that the cells do not occupy the entire tissue culture surface, as cells subsequently start to detach and die. Passaging cells involves detaching them from the tissue culture surface using trypsin, diluting the cells in fresh media, and distributing the cells into multiple tissue culture flasks. In modeling the system, multiple trypsinization cycles were simulated, with a single cycle having a 25-h duration. Figure 8 shows the second and third trypsinization cycles, where the third trypsinization cycle was used to compare against the observed data. If just one cycle is used in the analysis, initial conditions can be selected that can achieve a fairly good fit to the data but provide a transient response. A transient response is inconsistent with our experience, as the cellular response is similar for cells at an early passage versus later passages. Given that the adherens junction pathway in adherent cells is reactivated every 24–48 h due to cell passaging, we felt that a steady-state analysis, although mathematically interesting, may be misleading. Collectively these observations motivated the presented mechanistic model.
In summary, we identified a transcriptional regulatory network composed of interlocked positive and negative feedback loops that maintain the expression of adherens junction proteins. This identified network provides a mechanism by which contextual cues present in the tissue microenvironment maintain cell fate, as represented by promotion of cell proliferation or changes in cell differentiation due to activation of the Wnt developmental pathway. The identified regulatory network may provide a valuable conceptual framework to interpret how genetic alterations and alterations in protein–protein interactions or protein abundance shift the preferred phenotype according to prevailing environmental conditions. This concept is especially important because the functional maintenance of adherens junctions is to suppress tumor formation and invasion (Berx and Van Roy, 2001), and loss of adherens junctions has been implicated in a variety of cancers (Guilford et al., 1998; van Roy and Berx, 2008; Corso et al., 2013). For instance, inactivation of mutations in the extracellular calcium-binding sites of E-cadherin, which would effectively decrease the number of homotypic binding sites, results in constitutive activity. Such mutations are observed in ∼50% of diffuse gastric carcinomas, occur early in oncogenesis, and are associated with increased cell proliferation (Becker et al., 1994). Understanding how genetic alterations functionally alter the dynamic behavior of the network is a necessary prerequisite for the rational design of therapies to restore tissue homeostasis.
MATERIALS AND METHODS
Antibodies and reagents
Alexa Fluor 647–conjugated mouse immunoglobulin G2a (IgG2a) anti-mouse E-cadherin (clone: 36/E-cadherin), Alexa Fluor 488–conjugated mouse IgG1 anti–β-catenin (clone: 14/β-catenin), rat-tail collagen I, BD Phosflow lyse/fix buffer 5X, and BD Phosflow Perm Buffer III were purchased from BD Biosciences (San Diego, CA). Alexa Fluor 488–conjugated donkey anti-goat IgG1 (A-11055), LIVE/DEAD Fixable violet dead cell stain, Prolong Gold with DAPI, Cytoseal 60, rhodamine–phalloidin, fluorescein diacetate, and propidium iodide were purchased from Life Technologies (Grand Island, NY). A WISP-1 DuoSet ELISA (DY1627) and polyclonal goat anti–β-catenin IgG antibody (AF1329) were purchased from R&D Systems (Minneapolis, MN). A rat monoclonal IgG1 anti–E-cadherin (clone: DECMA-1) was purchased from Thermo Scientific (Waltham, MA). The β-catenin pathway inhibitor iCRT14 was purchased from Sigma-Aldrich (St. Louis, MO). AccuCount Fluorescent Particles (8.0–2.9 μm) were purchased from Spherotech (Lake Forest, IL). Quantum Simply Cellular uniform microspheres conjugated to anti-mouse IgG were purchased from Bangs Laboratories (Fishers, IN). Trypsin-Versene mixture was purchased from BioWhittaker (Walkersville, MD). Dulbecco’s Phosphate-buffered saline (DPBS) 1X was purchased from Mediatech (Manassas, VA). Sodium azide was purchased from United States Biochemical Corp (Cleveland, OH). DMEM (Mediatech, Manassas, VA) was supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS; Hyclone, Logan, UT), 2 mM l-glutamine, and 100 IU/ml penicillin–streptomycin (1% vol/vol; Biowhittaker, Walkersville, MD) and is referred to as sDMEM. RPMI-1640 (Mediatech) was supplemented with 10% FBS, 2 mM l-glutamine, and 1% penicillin/streptomycin and is referred to as sRPMI.
Tissue culture and stimulation
The B16F0 and B16F10 cell lines were purchased from the American Type Culture Collection (ATCC, Bethesda, MD) and grown in 75-cm2 tissue culture flasks with sDMEM. An immortalized mouse melanocyte cell line, Melan-A, was kindly provided by V. Hearing (National Cancer Institute, Bethesda, MD). The Hs-578T, BT-474, MCF7, and T47D were kindly provided by J. M. Ruppert (West Virginia University, Morgantown, WV). Melan-A cells were maintained in DMEM supplemented with 10% non–heat-inactivated FBS, 20 mM hydrogen chloride, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Life Technologies), 2 mM l-glutamine, 1% penicillin/streptomycin, 200 nM 12-o-tetradecanoyl phorbol 13-acetate (Sigma-Aldrich), and 200 μM phenylthiourea (Sigma-Aldrich). Hs578T cells were maintained in sDMEM. BT-474, MCF7, and T47D cell lines were maintained in sRPMI. Cells were cultured at 37°C and 5% CO2. Before the start of the hanging-drop and time-course experiments, the cells were ∼80% confluent. The hanging-drop method was used to generate spheroids, with 25 μl of cells suspended at a density of 1 × 104 cells/ml in medium alone or with medium supplemented with an anti–E-cadherin mAb or an isotype control mAb and placed on the inside of a six-well plate lid. The lid was placed over wells containing phosphate-buffered saline (PBS) to maintain humidity. The time course was initiated by trypsinizing the cells at 37°C for 6 min or for sufficient time for the cells to detach from the tissue culture flask. At a cell density of 6 × 105 cells/ml, cells were replated in either 96-well plates for flow cytometry assays or six-well plates for confocal microscopy. Cells were then exposed to one of three treatment conditions (sDMEM, sDMEM plus 25 μM iCRT14, and sDMEM plus 0.4% dimethyl sulfoxide [DMSO; vehicle control]) and subsequently cultured at 37°C in 5% CO2 for the indicated times. At each time point, the plate was centrifuged at 1250 rpm for 5 min at 4°C. The supernatant from each well was collected and stored at −20°C for the subsequent quantification of WISP-1 by ELISA, which was performed according to the manufacturer’s instructions.
Flow cytometry
Stimulated B16F0 cells were fixed, permeabilized, and stained with fluorophore-conjugated antibodies specific for E-cadherin and β-catenin, as described previously (Finley et al., 2011). AccuCount calibration beads were added to each of the tubes containing stained B16F0 cells, which were subsequently analyzed using a FACSAria flow cytometer and FACSDiva, version 6.1.1, software (BD Biosciences). Unstained samples were used as negative flow cytometry controls. Single-stain controls were used to establish fluorescence compensation parameters. Cellular events were identified by forward and side scatter characteristics. Dead cells were excluded from subsequent analysis based on LIVE/DEAD Fixable violet staining. Spherotech AccuCount beads were used to calibrate the cellular density among replicates and time points. At least two independent experiments were performed, and three biological replicates were obtained at each time point. On average, 2 × 104 events were analyzed in each technical replicate at each time point. Flow cytometry data were exported as FCS3.0 files and analyzed using R/Bioconductor, as described previously (Klinke and Brundage, 2009). The statistical difference between two treatment conditions was assessed using a two-tailed Student’s t test that assumes unequal variance; p < 0.05 was considered statistically significant.
Gene expression assay and siRNA transfection
Control siRNA was purchased from Thermo Scientific (siGENOME Non-Targeting siRNA #2, D-001210-02), and siRNA against human β-catenin was from Life Technologies (VHS50822). siRNA was transfected into Melan-A cells using Lipofectamine RNAiMax (Life Technologies) to reach a final concentration of 50 nM. Cells were harvested 24 h after transfection for RNA isolation and real-time RT-PCR. RNA samples were purified using a PicoPure RNA Isolation kit (Life Technologies) and reversely transcribed with High Capacity RNA-to-cDNA kit (Applied Biosystems). Quantitative PCR was carried out in triplicate on a StepOnePlus Real-time PCR System (Applied Biosystems) with Brilliant II SYBR Green QPCR Master Mix (Agilent Technologies). The delta/delta Ct method was used for data analysis. The primer sequences used are listed in Supplemental Table S4. The statistical significance between two samples was assessed using a two-sided Student’s t test assuming equal variance, with p < 0.05 considered significant.
Microscopy
For protein localization experiments, B16F0 cells were plated on rat-tail collagen I–coated coverslips (BD Biosciences), which were prepared according to the manufacturer’s instructions. Cells were untreated or treated with 25 μM iCRT14 for the indicated times, rinsed, and fixed with fresh 4% paraformaldehyde. After fixation, cells were permeabilized with 0.5% Triton-X 100 in PBS for 2 min, rinsed in PBS, and then blocked in 2% BSA and 20% goat serum in PBS for 30 min. Cells were then incubated with rhodamine-conjugated phalloidin (diluted 1:40 in PBS [RH-PH/PBS]), goat anti–β-catenin polyclonal antibody (1:4), and Alexa Fluor 647–conjugated mouse IgG2a anti–human E-cadherin monoclonal antibody (1:50) for 1 h at room temperature in the dark. After washing, cells were stained for 1 h in the dark at room temperature with Alexa Fluor 488 donkey anti-goat IgG that was diluted 1:500 in 0.5× RH-PH/PBS. Cells were rinsed and mounted in Prolong Gold with DAPI and sealed using Cytoseal 60 (Thermo Scientific). Confocal fluorescence microscopy was also performed with primary fluorophore-conjugated antibodies to confirm that the secondary antibody did not exhibit cross-reactivity.
For quantifying the cellular localization of β-catenin and the cytoplasmic portion of E-cadherin, 12-bit, 512 × 512–pixel confocal images were acquired with a Zeiss LSM510 confocal microscope using AIM software (Carl Zeiss MicroImaging, Thornwood, NY). Fluorescence images were scanned sequentially through the approximate center of the nucleus at a resolution of 3.83 pixels/μm with a 63×/1.4 numerical aperture Oil Plan-Apochromat objective. The image acquisition parameters for each channel (pinhole size, laser intensity, and gain) were adjusted to keep the fluorescence intensity within the dynamic range of the instrument. Acquired from a minimum of three independent fields, an average of 40 cells for untreated samples and 20 cells for iCRT14-treated samples were analyzed at each time point. At least two biological replicates were performed for each experiment. Each channel was exported as a 12-bit TIF image and analyzed using the EBImage package (Pau et al., 2010) in R (2.14.1; Gentleman et al., 2004). Nuclei were identified in the images by threshold-based segmentation using the DAPI fluorescence channel. Cellular regions of the image were identified using a cell mask that used an adaptive threshold applied to the geometric sum of the DAPI and rhodamine-conjugated phalloidin fluorescence channels. Using the nuclei as seeds, each cell was then identified using a Voronoi segmentation algorithm applied to the cell mask. The nuclear-to-cytoplasmic ratio of β-catenin and the cytoplasmic portion of E-cadherin were calculated as the ratio of total fluorescence intensity for each cell contained within the nucleus relative to the total fluorescence intensity for each cell excluding the nuclear region. Significant differences at each time point in the log10 nuclear/cytoplasmic ratio of β-catenin and E-cadherin between treatment conditions was assessed using a two-tailed Student’s t test assuming unequal variance; p < 0.05 was considered statistically significant.
To image live and dead regions of B16F0 spheroids, spheroids were washed three times with DPBS, stained using a solution containing 12 mM fluorescein diacetate and 3 mM propidium iodide for 15 min at room temperature in the dark, and then washed three times with DPBS. The spheroid was mounted with glycerol and immediately imaged using a Zeiss LSM510 confocal microscope using AIM software. Fluorescence images at 12-bit resolution were scanned sequentially through the approximate center of the spheroid at a resolution of 1.22 pixels/μm with a 10x/0.30 M27 EC Plan-Neofluar objective. Bright-field images of the spheroids were acquired using a color charge-coupled device camera attached to an Olympus MVX10 MacroView microscope with a 1×/0.25 Plan Apo objective (Olympus America, Center Valley, PA). Using exported 8-bit RGB TIF images and the EBImage package in R, cell density was estimated by the projected area identified using a threshold-based segmentation of the blue channel. The experiments were repeated at least twice, and, within an experiment, at least four biological replicates were imaged for each DECMA-1 concentration.
Models and inference
A deterministic multicompartmental mathematical model was formulated to describe the observed system and integrate existing knowledge and competing hypotheses about the adherens junction pathway activity of β-catenin. Postulated relationships among the observed biochemical species were expressed in terms of a mass-action formalism as ordinary differential equations. To represent the spatial organization of proteins within the cell, four different compartments were created to discriminate between species observed in the nuclear, cytoplasmic, membrane, or extracellular regions. The ordinary differential equations of the model represent the postulated causal relationships among the biochemical species represented in the model (Pearl, 2000). Geometrically, these causal relationships, that is, the model topology, represent a family of curves that trace all of the possible dynamic trajectories of the system in biochemical state space. In silico model-based inference was used to determine whether the observed data are consistent with the postulated model, given the uncertainty in the model parameters (Klinke, 2009, 2014a; Klinke et al., 2012). Details related to the formulation of the mathematical model and in silico model-based inference are described in the Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Emily Chambers McGinley, Neha Lal, and Amanda Strauch for assistance with experimental aspects of the study. This work was supported by grants from the National Science Foundation (CAREER 1053490) and the National Cancer Institute (R15CA123123). The flow cytometry core facility is supported by National Institutes of Health Grants GM103488, RR032138, RR020866, OD016165, and GM103434. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute, the National Institutes of Health, or the National Science Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations used:
- CTF
C-terminal fragment
- DMSO
dimethyl sulfoxide
- DPBS
Dulbecco’s phosphate-buffered saline
- sDMEM
supplemented DMEM
- WISP1
Wnt-inducible signaling pathway protein 1.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-02-0083) on July 29, 2015.
REFERENCES
- Aberle H, Butz S, Stappert J, Weissig H, Kemler R, Hoschuetzky H. Assembly of the cadherin-catenin complex in vitro with recombinant proteins. J Cell Sci. 1994;107:3655–3663. doi: 10.1242/jcs.107.12.3655. [DOI] [PubMed] [Google Scholar]
- Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, et al. A module of negative feedback regulators defines growth factor signaling. Nat Genet. 2007;39:503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- Basan M, Idema T, Lenz M, Joanny JF, Risler T. A reaction-diffusion model of the cadherin-catenin system: a possible mechanism for contact inhibition and implications for tumorigenesis. Biophys J. 2010;98:2770–2779. doi: 10.1016/j.bpj.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B, Georgiou M. Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. J Cell Biol. 2011;192:907–917. doi: 10.1083/jcb.201009141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KF, Atkinson MJ, Reich U, Becker I, Nekarda H, Siewert JR, Hofler H. E-cadherin gene mutations provide clues to diffuse type gastric carcinomas. Cancer Res. 1994;54:3845–3852. [PubMed] [Google Scholar]
- Berx G, Van Roy F. The E-cadherin/catenin complex: an important gatekeeper in breast cancer tumorigenesis and malignant progression. Breast Cancer Res. 2001;3:289–293. doi: 10.1186/bcr309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Ferrell JE, Li R, Meyer T. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science. 2005;310:496–498. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JR, Nigh E, Lee RJ, Ye H, Thompson MA, Saudou F, Pestell RG, Greenberg ME. Fos family members induce cell cycle entry by activating cyclin D1. Mol Cell Biol. 1998;18:5609–5619. doi: 10.1128/mcb.18.9.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Xie ZR, Wu Y. Computational modeling of the interplay between cadherin-mediated cell adhesion and Wnt signaling pathway. PLoS One. 2014;9:e100702. doi: 10.1371/journal.pone.0100702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Corso G, Carvalho J, Marrelli D, Vindigni C, Carvalho B, Seruca R, Roviello F, Oliveira C. Somatic mutations and deletions of the E-cadherin gene predict poor survival of patients with gastric cancer. J Clin Oncol. 2013;31:868–875. doi: 10.1200/JCO.2012.44.4612. [DOI] [PubMed] [Google Scholar]
- Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. J Cell Biol. 2003;163:525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle-Perez B, Casagolda D, Lugilde E, Valls G, Codina M, Dave N, de Herreros AG, Dunach M. Wnt controls the transcriptional activity of Kaiso through CK1epsilon-dependent phosphorylation of p120-catenin. J Cell Sci. 2011;124:2298–2309. doi: 10.1242/jcs.082693. [DOI] [PubMed] [Google Scholar]
- Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell. 2005;123:903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F, Gumbiner BM. Cell contact-dependent signaling. Dev Biol. 1996;180:445–454. doi: 10.1006/dbio.1996.0318. [DOI] [PubMed] [Google Scholar]
- Ferber EC, Kajita M, Wadlow A, Tobiansky L, Niessen C, Ariga H, Daniel J, Fujita Y. A role for the cleaved cytoplasmic domain of E-cadherin in the nucleus. J Biol Chem. 2008;283:12691–12700. doi: 10.1074/jbc.M708887200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley SD, Gupta D, Cheng N, Klinke DJ. Inferring relevant control mechanisms for interleukin-12 signaling within naive CD4+ T cells. Immunol Cell Biol. 2011;89:100–110. doi: 10.1038/icb.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini ME. Gamma-secretase-mediated proteolysis in cell-surface-receptor signalling. Nat Rev Mol Cell Biol. 2002;3:673–684. doi: 10.1038/nrm910. [DOI] [PubMed] [Google Scholar]
- Gao Y, Pimplikar SW. The gamma -secretase-cleaved C-terminal fragment of amyloid precursor protein mediates signaling to the nucleus. Proc Natl Acad Sci USA. 2001;98:14979–14984. doi: 10.1073/pnas.261463298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalves FC, Klein K, Carson BB, Katz S, Ekas LA, Evans S, Nagourney R, Cardozo T, Brown AM, DasGupta R. An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc Natl Acad Sci USA. 2011;108:5954– 5963. doi: 10.1073/pnas.1017496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilford P, Hopkins J, Harraway J, McLeod M, McLeod N, Harawira P, Taite H, Scoular R, Miller A, Reeve AE. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392:402–405. doi: 10.1038/32918. [DOI] [PubMed] [Google Scholar]
- Huber AH, Stewart DB, Laurents DV, Nelson WJ, Weis WI. The cadherin cytoplasmic domain is unstructured in the absence of beta-catenin. A possible mechanism for regulating cadherin turnover. J Biol Chem. 2001;276:12301–12309. doi: 10.1074/jbc.M010377200. [DOI] [PubMed] [Google Scholar]
- Ivanov AI, Nusrat A, Parkos CA. Endocytosis of epithelial apical junctional proteins by a clathrin-mediated pathway into a unique storage compartment. Mol Biol Cell. 2004;15:176–188. doi: 10.1091/mbc.E03-05-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivascu A, Kubbies M. Diversity of cell-mediated adhesions in breast cancer spheroids. Int J Oncol. 2007;31:1403–1413. [PubMed] [Google Scholar]
- Jamora C, Fuchs E. Intercellular adhesion, signalling and the cytoskeleton. Nat Cell Biol. 2002;4:E101–E108. doi: 10.1038/ncb0402-e101. [DOI] [PubMed] [Google Scholar]
- Kam Y, Quaranta V. Cadherin-bound beta-catenin feeds into the Wnt pathway upon adherens junctions dissociation: evidence for an intersection between beta-catenin pools. PLoS One. 2009;4:e4580. doi: 10.1371/journal.pone.0004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinke DJ. An empirical Bayesian approach for model-based inference of cellular signaling networks. BMC Bioinformatics. 2009;10:371. doi: 10.1186/1471-2105-10-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinke DJ. In silico model-based inference: a contemporary approach for hypothesis testing in network biology. Biotechnol Prog. 2014a;30:1247–1261. doi: 10.1002/btpr.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinke DJ. Induction of Wnt-inducible signaling protein-1 correlates with invasive breast cancer oncogenesis and reduced type 1 cell-mediated cytotoxic immunity: a retrospective study. PLoS Comput Biol. 2014b;10:e1003409. doi: 10.1371/journal.pcbi.1003409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinke DJ, Brundage KM. Scalable analysis of flow cytometry data using R/Bioconductor. Cytometry A. 2009;75:699–706. doi: 10.1002/cyto.a.20746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinke DJ, Cheng N, Chambers E. Quantifying crosstalk among interferon-γ, interleukin- 12, and tumor necrosis factor signaling pathways within a TH1 cell model. Sci Signal. 2012;5 doi: 10.1126/scisignal.2002657. ra32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci USA. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger R, Heinrich R. Model reduction and analysis of robustness for the Wnt/beta-catenin signal transduction pathway. Genome Inform. 2004;15:138–148. [PubMed] [Google Scholar]
- Kulkarni YM, Chambers E, McGray AJ, Ware JS, Bramson JL, Klinke DJ. A quantitative systems approach to identify paracrine mechanisms that locally suppress immune response to Interleukin-12 in the B16 melanoma model. Integr Biol (Camb) 2012;4:925–936. doi: 10.1039/c2ib20053h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Salic A, Kruger R, Heinrich R, Kirschner MW. The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol. 2003;1:E10. doi: 10.1371/journal.pbio.0000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Lewis B, Fletcher AG, Dale TC, Byrne HM. Toward a quantitative understanding of the Wnt/β-catenin pathway through simulation and experiment. Wiley Interdiscip Rev Syst Biol Med. 2013;5:391–407. doi: 10.1002/wsbm.1221. [DOI] [PubMed] [Google Scholar]
- Lochter A, Galosy S, Muschler J, Freedman N, Werb Z, Bissell MJ. Matrix metallopro-teinase stromelysin-1 triggers a cascade of molecular alterations that leads to stable epithelial-to-mesenchymal conversion and a premalignant phenotype in mammary epithelial cells. J Cell Biol. 1997;139:1861–1872. doi: 10.1083/jcb.139.7.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marambaud P, Shioi J, Serban G, Georgakopoulos A, Sarner S, Nagy V, Baki L, Wen P, Efthimiopoulos S, Shao Z, et al. A presenilin-1/gamma-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. EMBO J. 2002;21:1948–1956. doi: 10.1093/emboj/21.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretzky T, Reiss K, Ludwig A, Buchholz J, Scholz F, Proksch E, de Strooper B, Hartmann D, Saftig P. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and beta-catenin translocation. Proc Natl Acad Sci USA. 2005;102:9182–9187. doi: 10.1073/pnas.0500918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea PD, Turck CW, Gumbiner B. A homolog of the armadillo protein in Drosophila (plakoglobin) associated with E-cadherin. Science. 1991;254:1359–1361. doi: 10.1126/science.1962194. [DOI] [PubMed] [Google Scholar]
- Mirams GR, Byrne HM, King JR. A multiple timescale analysis of a mathematical model of the Wnt/beta-catenin signalling pathway. J Math Biol. 2010;60:131–160. doi: 10.1007/s00285-009-0262-y. [DOI] [PubMed] [Google Scholar]
- Niessen CM, Leckband D, Yap AS. Tissue organization by cadherin adhesion molecules: dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol Rev. 2011;91:691–731. doi: 10.1152/physrev.00004.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Ong SE, Mann M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol Cell Proteomics. 2004;3:608–614. doi: 10.1074/mcp.T400003-MCP200. [DOI] [PubMed] [Google Scholar]
- Park JI, Kim SW, Lyons JP, Ji H, Nguyen TT, Cho K, Barton MC, Deroo T, Vleminckx K, Moon RT, McCrea PD. Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Dev Cell. 2005;8:843–854. doi: 10.1016/j.devcel.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Pau G, Fuchs F, Sklyar O, Boutros M, Huber W. EBImage—an R package for image processing with applications to cellular phenotypes. Bioinformatics. 2010;26:979–981. doi: 10.1093/bioinformatics/btq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl J. Causality: Models, Reasoning, and Inference. Cambridge, UK: Cambridge University Press; 2000. [Google Scholar]
- Purvis JE, Lahav G. Encoding and decoding cellular information through signaling dynamics. Cell. 2013;152:945–956. doi: 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramis-Conde I, Drasdo D, Anderson AR, Chaplain MA. Modeling the influence of the E-cadherin-beta-catenin pathway in cancer cell invasion: a multiscale approach. Biophys J. 2008;95:155–165. doi: 10.1529/biophysj.107.114678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AB, Daniel J, McCrea PD, Wheelock MJ, Wu J, Zhang Z. Identification of a new catenin: the tyrosine kinase substrate p120cas associates with E-cadherin complexes. Mol Cell Biol. 1994;14:8333–8342. doi: 10.1128/mcb.14.12.8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadot E, Simcha I, Shtutman M, Ben-Ze’ev A, Geiger B. Inhibition of beta-catenin-mediated transactivation by cadherin derivatives. Proc Natl Acad Sci USA. 1998;95:15339–15344. doi: 10.1073/pnas.95.26.15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- Shimazui T, Schalken JA, Kawai K, Kawamoto R, van Bockhoven A, Oosterwijk E, Akaza H. Role of complex cadherins in cell-cell adhesion evaluated by spheroid formation in renal cell carcinoma cell lines. Oncol Rep. 2004;11:357–360. [PubMed] [Google Scholar]
- Shoval I, Ludwig A, Kalcheim C. Antagonistic roles of full-length N-cadherin and its soluble BMP cleavage product in neural crest delamination. Development. 2007;134:491–501. doi: 10.1242/dev.02742. [DOI] [PubMed] [Google Scholar]
- Shtutman M, Zhurinsky J, Simcha I, Albanese C, D’Amico M, Pestell R, Ben-Ze’ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Croix B, Sheehan C, Rak JW, Florenes VA, Slingerland JM, Kerbel RS. E-Cadherin-dependent growth suppression is mediated by the cyclin-dependent kinase inhibitor p27(KIP1) J Cell Biol. 1998;142:557–571. doi: 10.1083/jcb.142.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhusen U, Weiske J, Badock V, Tauber R, Bommert K, Huber O. Cleavage and shedding of E-cadherin after induction of apoptosis. J Biol Chem. 2001;276:4972–4980. doi: 10.1074/jbc.M006102200. [DOI] [PubMed] [Google Scholar]
- Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- Tsai TY, Choi YS, Ma W, Pomerening JR, Tang C, Ferrell JE. Robust, tunable biological oscillations from interlinked positive and negative feedback loops. Science. 2008;321:126–129. doi: 10.1126/science.1156951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen IM, Byrne HM, Jensen OE, King JR. Elucidating the interactions between the adhesive and transcriptional functions of beta-catenin in normal and cancerous cells. J Theor Biol. 2007;247:77–102. doi: 10.1016/j.jtbi.2007.01.019. [DOI] [PubMed] [Google Scholar]
- van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci. 2008;65:3756–3788. doi: 10.1007/s00018-008-8281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Allison DF, Buckley KM, Kottke MD, Vincent PA, Faundez V, Kowalczyk AP. Cellular levels of p120 catenin function as a set point for cadherin expression levels in microvascular endothelial cells. J Cell Biol. 2003;163:535–545. doi: 10.1083/jcb.200306001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Corcoran RB, Welsh JW, Pennica D, Levine AJ. WISP-1 is a Wnt-1- and beta- catenin-responsive oncogene. Genes Dev. 2000;14:585–595. [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang I, Chang O, Lu Q, Kim K. Delta-catenin affects the localization and stability of p120-catenin by competitively interacting with E-cadherin. Mol Cells. 2010;29:233–237. doi: 10.1007/s10059-010-0030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap AS, Crampton MS, Hardin J. Making and breaking contacts: the cellular biology of cadherin regulation. Curr Opin Cell Biol. 2007;19:508–514. doi: 10.1016/j.ceb.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R. Cadherin adhesome at a glance. J Cell Sci. 2013;126:373–378. doi: 10.1242/jcs.111559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.