Abstract
Mucosal leishmaniasis (ML) is a disfiguring manifestation of Leishmania (Viannia) infection. We evaluated parasite load (PL) over time as a potential biomarker of treatment outcome in ML. PL was assessed with kinetoplast DNA quantitative real-time polymerase chain reaction (kDNA-qPCR) at enrollment, days 14 and 21–28 of therapy and 3, 6, 12–18, and 18–24 months after treatment of ML and correlated to demographic, clinical, and parasitologic factors. Forty-four patients were enrolled: 30 men and 14 women. Enrollment PL differed significantly by causative species (P < 0.001), and was higher in patients with severe ML (nasal and laryngeal involvement) compared with those with only isolated nasal involvement (median = 1,285 versus 51.5 parasites/μg tissue DNA; P = 0.005). Two patterns of PL emerged: pattern 1 (N = 23) was characterized by a sequential decline in PL during and after therapy until kDNA was undetectable. Pattern 2 (N = 18) was characterized by clearance of detectable kDNA during treatment, followed by an increased PL thereafter. All patients who failed treatment (N = 4) demonstrated pattern 1. Leishmania (Viannia) braziliensis was overrepresented among those with pattern 2 (P = 0.019). PL can be quantified by cytology brush qPCR during and after treatment in ML. We demonstrate that treatment failure was associated with undetectable PL, and L. (V.) braziliensis infection was overrepresented in those with rebounding PL.
Introduction
In Latin America, mucosal leishmaniasis (ML) represents a disfiguring manifestation of long-term infection with members of the Leishmania (Viannia) complex, most commonly Leishmania (Viannia) braziliensis.1,2 Noninvasive cytology brush polymerase chain reaction (PCR) is a novel approach to the diagnosis of ML, and one that has been validated clinically.3 Moreover, this well-tolerated, easy-to-perform sampling method coupled with a platform such as quantitative real-time PCR (qPCR) could enable quantitation of Leishmania parasite load (PL) during treatment and follow-up of patients. Little human data exist on the clinical utility of repeated quantifications of the PL as a biomarker of treatment outcome. Studies on visceral leishmaniasis (VL) and cutaneous leishmaniasis (CL) patients demonstrate the applicability of quantifying PL for prognostic evaluation and monitoring the efficacy of antileishmanial therapy.4–8 Recently, we validated a qPCR assay targeting the multicopy minicircle kinetoplast DNA (kDNA) for quantifying L. (Viannia) parasites in human biopsy specimens, which was highly sensitive and accurate.9
We sought to determine if detectable PL by cytology brush kDNA-qPCR during therapy is correlated to clinical failure at 28 days or the requirement for multiple courses of therapy. In addition, we sought to determine if patients with detectable Leishmania PL during therapy or follow-up differed demographically or parasitologically from those with undetectable PL.
Methods
Study site and population.
The study was conducted at the Leishmaniasis Clinic of the Instituto de Medicina Tropical “Alexander von Humboldt,” Hospital Nacional Cayetano Heredia (HNCH), in Lima, Peru, between January 2011 and July 2013. Consecutive patients presenting to the clinic for evaluation of mucosal lesions were approached to participate and screened for eligibility criteria. History, physical examination, direct anterior rhinoscopy, and oropharyngoscopy were performed on all patients by clinic physicians. We included patients who were referred for suspected ML; had stigmata of mucosal disease, including nasal, palatal, or laryngeal infiltration, erosion, and/or erythema; and were able to provide informed consent. We excluded patients undergoing active treatment for CL or ML and those with any contraindication to mucosal biopsy. ML was classified as mild, moderate, or severe based on the extent of involvement, where mild ML consisted of isolated nasal involvement, moderate ML consisted of nasal and oral involvement, and severe ML consisted of nasal, laryngeal, and/or oral involvement. ML was further classified as primary versus classic (i.e., ML that occurred without a history and with a history of documented CL, respectively). Clinical cure was defined as complete resolution of mucosal lesions by 12 months posttreatment. Treatment was considered to have failed if there was no improvement in lesion appearance by end of 28-day treatment or if the lesion recurred or expanded at any point during the 12-month follow-up period posttreatment.
Ethics statement.
This study was approved by the Institutional Review Boards of HNCH and the University of Toronto. All patients provided written informed consent for the study procedures before enrollment.
Specimen collection and procedures.
Sterile and duplicate CerviSoft® (Puritan Medical Products, Guilford, ME) cytology brushes were collected as described.3 Brush tips were cut off with sterile scissors directly into 1.5-mL microcentrifuge tubes containing 700 μL of 100% ethanol and stored at −20°C. Mucosal lesions were then anesthetized with 20 mg/mL lidocaine spray and an incisional biopsy was obtained. Tissue was then divided into two, stored in 1.5-mL microcentrifuge tubes containing 700 μL of 100% ethanol at −20°C or placed in 10% formalin for histopathology with hematoxylin and eosin, Ziehl–Neelsen, and Giemsa staining. Sterile gauze was applied with pressure to the mucosa until hemostasis was achieved. Lesions were sampled by cytology brushes and punch biopsy at enrollment. Thereafter during treatment, lesions were sampled only by cytology brushes on days 14 and 21 or 28 and at 3, 6, 12–18, or 18–24 months after treatment.
Leishmanin skin test.
Leishmanin skin tests were performed using 0.1 mL in-house, sterile, heat-killed promastigote lysate in 0.005% thimerosal as read as previously described.4,10,11
DNA isolation and quantification.
Minced biopsy specimens and cytology brushes were lysed overnight with proteinase K, and DNA was isolated using the High Pure PCR Template Preparation Kit (Roche, Manheim, Germany), according to manufacturer's instructions. Isolated DNA was quantified by fluorometry using the Quant-iT Broad-Range DNA Assay Kit and the Qubit fluorometer (Invitrogen, Singapore).
Diagnostic PCR and species identification.
Qualitative Leishmania PCR targeting a conserved region of minicircle kDNA was performed using primers and conditions described previously.3,12 Parasites were identified to the species level using the algorithm reported elsewhere,13 which includes a combination of PCR assays and restriction fragment length polymorphism (RFLP) analysis, targeting mannose phosphate isomerase (mpi), cysteine proteinase b (cpb), and heat shock protein-70 (hsp70) genes. Mpi PCR distinguishes Leishmania (Viannia) peruviana, while cpb PCR-RFLP distinguishes L. (V.) braziliensis, and hsp70 PCR-RFLP differentiates Leishmania (Viannia) guyanensis from Leishmania (Viannia) lainsoni, which are the major endemic species of Peru, and most of which can cause ML. Hybrid or mixed L. (V.) braziliensis/L. (V.) peruviana infections are those that display RFLP banding characteristics of both species. Without genetic cloning of isolates, differentiation of genetic hybrids from mixed co-species infection is impossible.13 Thus, these types of isolates are reported as “hybrid or mixed infections” throughout. Causative species was considered “indeterminate” if insufficient amplification of genomic DNA at the mpi, cpb, and hsp70 genes to perform RFLP analysis occurred, or if the pattern of banding on RFLP analysis did not enable differentiation of causative species.
Isolates not identifiable by this algorithm underwent further molecular analyses as described below. Additional PCR assays were performed with proof-reading polymerase AccuPrime Pfx Supermix (Life Technologies) for all sequence-sensitive assays (RFLP and sequencing). Internal transcribed spacer 1 (ITS1) region was amplified with primers LITSR and L5.8S as described.14 PCR conditions were as follows: 95°C for 5 minutes followed by 38 cycles of denaturation at 95°C for 15 seconds, primer annealing at 52°C for 30 seconds, extension at 68°C for 1 minute, and a final extension step of 68°C for 5 minute (Veriti Fast Cycler; ABI). After confirmation of amplification indicative of an ∼350-bp band on 1% agarose gel, PCR product was digested in 1× FastDigest Green Buffer and 1 μL of HaeIII FastDigest enzyme (Thermo Scientific, Foster City, CA) for 1 hour at 37°C. Digested products were resolved in 1× Tris-borate EDTA (TBE) 2% agarose gel. PCR-RFLP analysis of the ITS1 region provides an initial identification of New World cutaneous species Leishmania amazonensis, Leishmania mexicana, and L. (Viannia) complex species.14 Further sequencing analyses of the ITS2 region are required to differentiate species within the L. (Viannia) subgenus and to provide confirmation of the species identified by initial ITS1 assay.
Thus, ITS2 region was amplified with primers LGITSF2 and R2 as described.15 PCR conditions were as follows: 95°C for 5 minutes followed by 45 cycles of denaturing at 95°C for 15 seconds, primer annealing at 60°C for 30 seconds, extension at 68°C for 1 minute, and a final extension step of 68°C for 5 minutes (Veriti Fast Cycler, ABI, Singapore). PCR products were confirmed on 1% agarose gel, then purified with QIAquick PCR Purification Kit (Qiagen, Germantown, MD). Sequence products were purified with BigDye XTerminator Purification Kit (Life Technologies, Carlsbad, CA), vortexed for 30 minutes and then centrifuged at 2,400 rpm for 2 minutes before loading into ABI 3130xl Genetic Analyzer. Sequences obtained from both primers were assembled, and BLAST searched for species homology and identification.
Leishmania PL quantification.
We applied a SYBR Green-based qPCR assay targeting kDNA minicircles to quantify L. (Viannia) parasites in clinical samples before, during, and after treatment of ML, as previously described.9 All clinical samples were run in duplicate; if replicates differed by a standard deviation (SD) of > 0.35 in Cq (quantification cycle) values (> 0.5 cycles), they were retested. Each kDNA–qPCR run included a standard curve ranging from 5 × 104 to 5 × 10−3 parasites/reaction (run in duplicate), a positive control with known amount of Leishmania parasites (run in duplicate), and a negative control (run in triplicate). Leishmania load was calculated as follows: [(parasite DNA equivalents per reaction)/amount of tissue DNA per reaction] × 103, and expressed as the number of Leishmania parasites per microgram of tissue DNA (tDNA).
Statistical analysis.
Descriptive statistics (mean, SD, median, range) were calculated for continuous variables, and differences were compared using two-tailed t testing or, in the case of non-normal distribution, Mann–Whitney rank-sum test, or for groups of variables, one-way analysis of variance (ANOVA) on ranks. Categorical variables were quantitated by proportions, and differences were compared using Yate's corrected χ2 analysis. Statistical analyses were performed using SigmaStat 2.03 software (SPSS Inc., Chicago, IL). Level of significance was set at P < 0.05.
Results
Forty-four patients were enrolled: 30 men and 14 women, with median age of 41.5 years (range = 12–89 years). Patients presented with a median lesion duration of 36 months (range = 1 month to 38 years). Mucosal involvement manifested after prior CL in 37 subjects (84.1%), whereas five patients (11.4%) denied prior CL with no suspicious scar identified on physical examination, and the remaining two participants (4.5%) had ML due to contiguity. Thus, 42/44 patients (95.5%) had metastatic ML, while only 4.5% had contiguous spread. Mild ML (only nasal involvement) was identified in 23 participants (52.3%), whereas moderate ML (nasal and oral) was identified in seven subjects (15.9%). Severe involvement (nasal and larynx ± oral involvement) was identified in 14 subjects (31.8%). Median duration of exposure in the risk area was 174 months (range = 2 days to 89 years) with the Departments of Junin (N = 8), Cusco (N = 7), Huanuco (N = 7), Madre de Dios (N = 5), Ayacucho (N = 3), and Ucayali (N = 3) being the most well-represented risk areas.
Thirty-seven patients (84.1%) reported a past history of CL, with 25/37 (74%) having received prior treatment for the primary skin disease. Two additional patients had probable past CL based on the presence of typical scars. In those with known past CL (N = 37), median latency between CL and the diagnosis of ML was 15 years. Of 25 patients with known prior CL treatment, 13 (52%) were treated with an antimony-containing regimen. Twelve patients were previously treated for CL with a systemic antimonial (48%), one with an intralesional antimonial (4%), five with chemical or thermal burns including acid application or cryotherapy (20%), and seven received some unknown treatment (28%). Of 25 patients with known past CL treatment, cure was achieved in 24 (96%), with only one patient progressing immediately to mucocutaneous leishmaniasis (MCL; concurrent CL and ML).
Five patients (11.4%) received a prior course of amphotericin (N = 4; 9.1%) or miltefosine (N = 1; 2.3%) for ML, whereas 39 participants (88.6%) had not received prior treatment for ML.
Of 44 patients enrolled, final diagnosis was pure ML in 39 (89%), MCL in four (9%), and ML with basal cell carcinoma in one (2%). Histopathologic examination of biopsy tissue at enrollment demonstrated isolated granulomatous inflammation in 27 (61%), amastigotes within a parasitic granuloma in nine (20%), lymphocytic infiltrate in four (9%), macrophage infiltrate in one (2%), and mixed acute and chronic inflammation in one (2%). Histopathological results were unavailable in two (4.5%) patients. Leishmanin skin test was positive in 41 of 44 (93%) patients.
Causative species was identified in 27 of 32 (84%) patients (Table 1). In 12 patients (27%), insufficient amplifiable DNA for the genomic targets prevented species identification. Nineteen patients were infected with L. (V.) braziliensis, three with L. (V.) guyanensis, three with hybrid or mixed infections (L. (V.) braziliensis/L. (V.) peruviana), one with L. (V.) peruviana, and one with Leishmania (Viannia) panamensis. All non-braziliensis infections caused the mild phenotype, whereas 63% of L. (V.) braziliensis caused a moderate or severe phenotype (Table 2). All mixed or hybrid L. (V.) braziliensis/peruviana infections caused the severe phenotype. Similarly, all primary ML (i.e., which occurred in the absence of known prior CL) was caused by either L. (V.) braziliensis or the hybrid/mixed infections (Table 2).
Table 1.
Causative Leishmania species and pretreatment PL in tissue
| Species | Number | PL | |||
|---|---|---|---|---|---|
| Mean | SD | Median* | Range | ||
| Leishmania (Viannia) braziliensis | 19 | 10,484.67 | 34,271.38 | 260 | 3.19–143,000 |
| Leishmania (Viannia) guyanensis | 3 | 535,603.3 | 484,635.7 | 657,000 | 1,810–948,000 |
| Hybrid/mixed infection | 2§ | 1,662,123.7 | 865,673.7 | 1,662,123.7 | 1,050,000–2,274,247.5 |
| Leishmania(Viannia) peruviana | 1 | 128,465.35 | – | 128,465.35 | – |
| Leishmania (Viannia) panamensis | 1 | 45.1 | – | 45.1 | – |
| Indeterminate† | 5 | 95,542.27 | 210,449.83 | 1,170 | 1.33–472,000 |
| Not assessed‡ | 12 | 26.6 | 28.7 | 18.5 | 1.08–94.8 |
PL = parasite load; RFLP = restriction fragment length polymorphism; SD = standard deviation.
P < 0.001 by one-way analysis of variance on ranks (with Dunn's post hoc test) for all species.
Insufficient amplification of genomic DNA at mpi, cpb, and hsp70 genes to perform RFLP analysis, and definitively identify the causative species, or a pattern of banding on RFLP analysis that did not enable differentiation of causative species.
Not assessed because of insufficient amplifiable DNA in the primary sample.
Three patients were infected with hybrid/mixed species; however, sufficient DNA was available for quantitation in only two.
Table 2.
Species identification of causative Leishmania by phenotype and severity of ML
| ML phenotype and severity | Number with known species ID | Species Identity | ||||
|---|---|---|---|---|---|---|
| LB | LB/LP | LG | LP | LPAN | ||
| Phenotype | ||||||
| Primary ML | 3 | 2 | 1 | 0 | 0 | 0 |
| Classic ML | 24 | 17 | 2 | 3 | 1 | 1 |
| Severity | ||||||
| Mild | 12 | 7 | 0 | 3 | 1 | 1 |
| Moderate | 6 | 6 | 0 | 0 | 0 | 0 |
| Severe | 9 | 6 | 3 | 0 | 0 | 0 |
CL = cutaneous leishmaniasis; LB = Leishmania (Viannia) braziliensis; LB/LP = hybrid/mixed infection pattern for Leishmania (Viannia) braziliensis/peruviana; LG = Leishmania (Viannia) guyanensis; LPAN = Leishmania (Viannia) panamensis; ML = mucosal leishmaniasis.
Primary ML, which occurred in the absence of history of prior CL; classic ML, which occurred after a documented history of CL; mild, isolated nasal involvement; moderate, nasal and oral involvement; severe, nasal and laryngeal involvement with or without oral involvement.
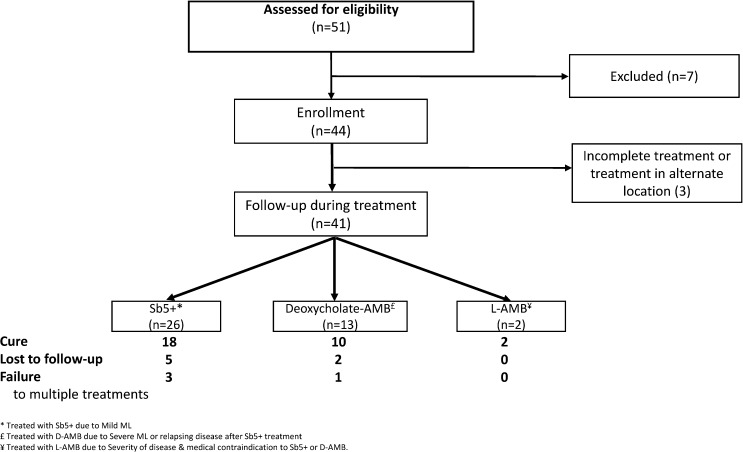
Of 44 patients enrolled, treatment regimen data were available for 41 (93%), and patient flow through the study over time is outlined in Figure 1 . Three patients (7%) were lost to follow-up immediately after enrollment. Of the 41 patients who received treatment at our center, 26 (63%) received a single 28-day course of sodium stibogluconate, while 13 (32%) received amphotericin B and two (5%) received liposomal amphotericin B (Figure 1). Cure was achieved in 30 patients (73%), while four patients (10%) failed one or multiple courses of therapy. Clinical outcome is unknown in seven patients (17%) who were lost to follow-up after completing therapy. There was no correlation between phenotype (primary versus classic ML), or severity of ML and clinical outcome (cure versus failure), or prior treatment of CL.
Figure 1.
Flow diagram of study.
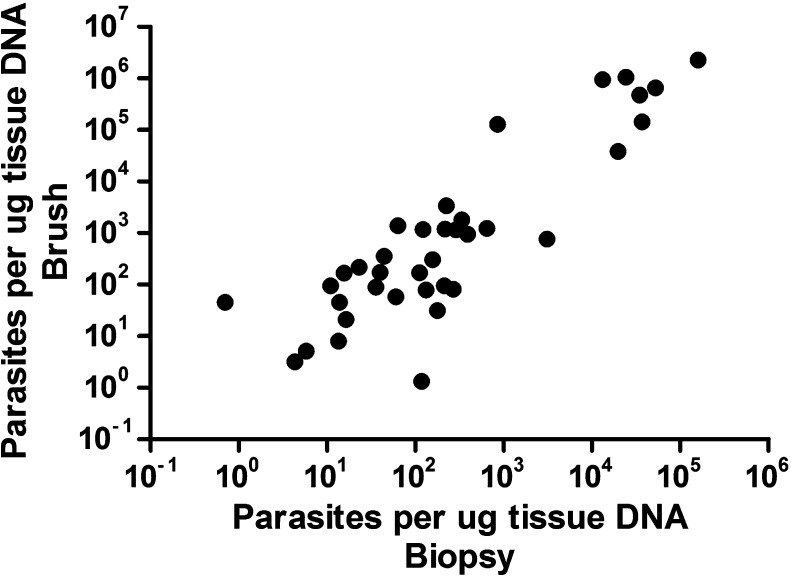
Mean PL at enrollment was 139,665.8 ± 65,641.9 parasites/μg tDNA (median = 172, range = 1.08–2,274,247.5). Enrollment PL differed significantly by causative species (P < 0.001 by one-way ANOVA on ranks), as illustrated in Table 1. Initial PL was higher in those for whom species identification was possible and definitive compared with those with indeterminate species identification and in those for whom species identification was not undertaken because of insufficient amplifiable DNA (median = 765 versus 20.85 parasites/μg tDNA; P = 0.002). Enrollment PL was also higher in those patients with the severe phenotype compared with those with only isolated nasal involvement (median = 1,285 versus 51.5 parasites/μg tDNA; P = 0.005) (Table 3). Enrollment PL did not differ by sex (P = 0.448), age (P = 0.248), region of CL/ML acquisition (P = 0.413), past treatment for CL (P = 0.895), or chronicity of ML, where a chronic lesion was defined as duration > 12 months (P = 0.741). Enrollment PL appeared to be lower in those who failed current therapy compared with those who cured (median = 5.2 versus 260 parasites/μg tDNA), though this difference was not significant (P = 0.103). Enrollment PL in biopsies and brushes was highly correlated (Spearman's ρ = 0.82 [95% confidence interval = 0.67–0.90], P < 0.0001) (Figure 2 ).
Table 3.
Pretreatment PL in tissue by ML phenotype and severity
| ML phenotype and severity | Number | PL | |||
|---|---|---|---|---|---|
| Mean | SD | Median* | Range | ||
| Phenotype | |||||
| Primary ML | 5 | 262,547.7 | 524,968.2 | 94.7 | 1.33–1,050,000 |
| Classic ML | 39 | 126,381.2 | 414,086.5 | 218.0 | 1.08–2,274,247.5 |
| Severity | |||||
| Mild | 23 | 78,976.2 | 240,004.2 | 51.508 | 1.08–948,000 |
| Moderate | 7 | 5,759.5 | 14,487.8 | 95.4 | 2.4–38,600 |
| Severe | 14 | 329,041.9 | 688,788.7 | 1,285.0 | 80.7–2,274,247.5 |
CL = cutaneous leishmaniasis; ML = mucosal leishmaniasis; PL = parasite load; SD = standard deviation.
Primary ML, which occurred in the absence of history of prior CL; classic ML, which occurred after a documented history of CL; mild, isolated nasal involvement; moderate, nasal and oral involvement; severe, nasal and laryngeal involvement with or without oral involvement.
P = 0.005 by one-way analysis of variance on ranks (with Dunn's post hoc test) for severity of ML.
Figure 2.
Correlation between kinetoplast DNA quantitative real-time polymerase chain reaction (kDNA-qPCR) measurements in biopsies and brushes. The number of Leishmania parasites at baseline could be quantified in 37 patients in both types of samples. We found a good correlation between both sampling methods: Spearman's ρ = 0.82 (95% confidence interval = 0.67–0.90) with P < 0.0001.
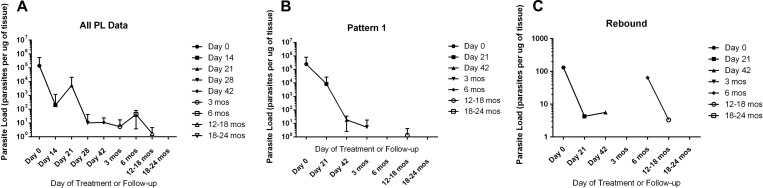
Figure 3A demonstrates the natural history of mean PL over time. Two patterns of PL kinetics during therapy and follow-up emerged: pattern 1 (“clearance”) (N = 23) was characterized by a mean PL of 240,605.7 ± 112,857.1 (median = 218, range = 1.33–2,274,247.5) at enrollment, with sequential decline in PL during and after therapy until kDNA was undetectable (Figure 3B). Pattern 2 (“rebound”) (N = 18) was characterized by mean PL of 464.7 ± 133.4 (median = 131.4, range = 1.08–1,810) at enrollment, with clearance of detectable kDNA during treatment, followed by an increased PL thereafter during treatment or follow-up, that became very low again at 12–18 or 18–24 months after treatment (Figure 3C). Initial PL did not differ significantly between those with patterns 1 and 2 (P = 0.287). All patients who failed treatment demonstrated sequential clearance (pattern 1). Of those with clinical cure (N = 30), 16 (53%) demonstrated sequential clearance (pattern 1), while 14 (47%) demonstrated pattern 2 or rebound of kDNA. All patients with primary ML demonstrated sequential kDNA clearance (pattern 1), compared with only 49% of those with classic ML; however, interpretation must be cautious due to the small number with primary ML (N = 5).
Figure 3.
(A) Parasite load (PL; parasites per microgram of tissue) by cytology brush kinetoplast DNA quantitative real-time polymerase chain reaction (kDNA PCR) over time. (B) PL of patients with sequential decrease in kDNA burden during treatment and follow-up. (C) PL of patients with rebound of detectable kDNA during treatment and/or follow-up.
Baseline PL, sex, age, exposure duration, past treatment for CL, symptom duration, lesion number, and ML location were not correlated to pattern of PL. Those with visible amastigotes histopathologically (N = 9) appeared more likely to demonstrate pattern 1 (clearance) (N = 7/9; 78%) than those without visible amastigotes (N = 15/33; 45%), though this difference was not significant (P = 0.15). Of those for whom species identification was possible, L. (V.) braziliensis was overrepresented in pattern 2 (rebound). Although L. (V.) braziliensis was the causative species for 70% of enrollees for whom species identification was possible, it caused 92% (N = 12/13) of ML demonstrating pattern 2 (rebound) of PL (P = 0.019).
Discussion
We have demonstrated, as proof of principle, that kDNA is detectable in ML lesions throughout treatment. PL in mucosal lesions can be quantified by commercial cytology brush sampling and kDNA-qPCR before, during, and after treatment, and may have future potential as a biomarker of outcome. We noted that PL differed by causative species, with L. (V.) guyanensis and hybrid or mixed infections demonstrating the highest PL. We also noted that PL was highest in patients with severe ML, perhaps suggesting poorer immunologic control of the disease. The observed trend toward lower enrollment PL in those who failed treatment warrants further investigation in a larger clinical study.
We observed two distinct patterns of PL during and after treatment. Given that drug treatment has been found not to eliminate parasites,16–19 the two patterns of PL kinetics observed here indicate that during and after treatment, parasites remain at the site involved, though at low level. It is noticeable that observed PL patterns were discordant with the clinical treatment outcome in most ML patients. On the one hand, detectable kDNA was observed in 47% of clinically cured ML during follow-up, which should be derived from viable amastigotes or from recently deceased parasites because of their ephemeral persistence inside macrophages.20 On the other hand, it is not clear why all four patients with documented treatment failure demonstrated a clearance pattern (i.e., undetectable) of PL during therapy. We hypothesize different but nonexclusive scenarios: 1) the amount of persistent parasites in mucosal lesions is below the detection limit of highly sensitive PCR assays21,22; 2) persistent parasites may be in a quiescent stage with reduced metabolic rate, thus exhibiting less susceptibility to drugs, as has been hypothesized previously21,22; 3) persistent parasites may have infected other tissue niches distant from the mucosal lesion that are inaccessible to drugs (e.g., skin), from where infection can spread again and eventually cause treatment failure21; and 4) there are host-related factors (e.g., immunosuppression) contributing to treatment failure. Previous reports that have assessed the value of kDNA PCR as a test of cure after therapy in VL demonstrated correlation of PCR with clinical treatment outcome, in that parasite DNA remained detectable in blood from cured individuals who experienced relapse, whereas parasitological cure was correlated with clinical cure of VL.23–25 In these studies, PCR result and clinical outcome discordance was exceptional, thereby indicating the limitations of PCR in predicting cure or relapse in all cases. Altogether, the observations made in VL studies and ours suggest that persistence of parasites in tissue (blood in VL, mucosa in ML) seems not to be an unequivocal indicator of future treatment failure. Indeed, treatment failure in leishmaniasis is a complex phenomenon, influenced by host, parasite, and pharmacokinetic/pharmacodynamic factors.21
The high percentage of patients with ML and history of CL in this study (84%) could suggest that persistent parasites after clinical cure of CL may be related to the progression to ML. Consistent with this, parasite persistence after clinical cure has been documented in CL.16–18,26 A study of recurrent disease in L. (V.) braziliensis-infected patients from Colombia provides strong evidence that persistent parasites could be the cause of reactivated CL.26 The mechanisms related to parasite persistence and dissemination, and the role that parasite-derived factors play in this process, warrant more investigation.
Leishmania (Viannia) braziliensis was overrepresented among ML patients with the rebound pattern of PL; the significance of this finding is unknown. Future longitudinal studies should evaluate host-related factors such as inflammatory and immune responses, which play a major role in the development and prognosis of ML.27–29 Studies comparing cellular and cytokine responses in peripheral blood mononuclear cells stimulated with L. (V.) braziliensis antigens during active disease and after clinical cure have identified possible beneficial immunological parameters associated with clinical cure of ML.28–30 In addition, immunologic studies of ML lesions from patients that evolved to cure or treatment failure have provided markers that may be useful for predicting treatment outcome of ML.31,32 Certainly, combining the assessment of human immune response markers, PL kinetics in tissue before, during, and after treatment, and parasitic factors that may contribute to clinical treatment failure (e.g., infecting Leishmania species, parasite adaptations such as virulence or quiescence [reviewed in 21]) will contribute to better understanding the prognosis of ML and to identifying host and parasite biomarkers of outcome.
We have documented that 52% of enrollees with past CL had been treated with an antimony-containing regimen, and still went on to develop ML over a decade after clinical cure. Of those with prior treatment of CL, 96% were considered cured. Our findings reiterate that good evidence to support a protective role of initial parenteral, oral, or local treatment of CL in the pathogenesis of ML is lacking.33,34
Species identification is important in countries such as Peru where several members of the L. (Viannia) subgenus are co-endemic and portend different clinical prognoses and response to therapy.35 Leishmania (Viannia) braziliensis is the most well-represented causative species in ML.1,35,36 We have demonstrated that related species including L. (V.) guyanensis, L. (V.) panamensis, and L. (V.) peruviana are implicated in noncontiguous metastatic mucosal disease as well, though in this study, the ML that they caused was confined to the nasal mucosa. Interestingly, all primary ML in this study was caused by either L. (V.) braziliensis or hybrid/mixed species. Case report and series level data implicating non-braziliensis Viannia species in ML are mounting,1–3,36,37 and in this study, we documented that at least 30% of ML cases presenting to one specialized leishmaniasis clinic in Peru are caused by non-braziliensis species of the L. (Viannia) complex.
The finding of a Peruvian patient with ML due to L. (V.) panamensis is novel, as this species is not known to circulate in Peru, although travel to a L. (V.) panamensis-endemic area in a neighboring country cannot be excluded in this case. PCR amplification of hsp70 followed by RFLP analysis is a standard approach to differentiating L. (V.) guyanensis from other major co-endemic species in Peru, notably L. (V.) braziliensis and L. (V.) peruviana.13 However, this classic assay is unable to distinguish L. (V.) guyanensis from L. (V.) panamensis, which requires either gene sequencing as described above or use of another restriction enzyme (BccI) after hsp70 PCR.38 Thus, it is possible that L. (V.) panamensis is another co-endemic L. (Viannia) species in Peru, the burden of which needs to be ascertained in future molecular epidemiologic studies allowing specific typing and comprehensive L. (Viannia) strain characterization.
The limitations of this analysis must be acknowledged. First, comparison of PL by species is biased by the low numbers of non-braziliensis species causing ML. Thus, differences in PL by species may be lost with comparison of equal numbers of causative species in each group. Second, heterogeneity of the patient population regarding treatment assignment and treatment response may have biased our interpretation of PL patterns. However, this study was exploratory in nature, and neither initial PL nor causative species identity was available on enrollment. Further studies that explore the contribution of PL to clinical failure in a larger cohort of patients are warranted. Third, our small sample size and numbers of PL assessments at each time point may influence perceived differences, and with greater numbers of enrollees, those observed differences may be lost. Finally, the clinical outcome in seven enrollees is unknown because of loss to follow-up. Despite these limitations, we have discovered two distinct kinetic patterns of PL during and after treatment in this cohort of patients with ML in Peru. Elucidating the clinical implications of our findings in a larger prospective and homogeneous cohort is justified.
In summary, ML is diagnostically and therapeutically challenging, and evidence-based biomarkers of clinical outcome are lacking. Sophisticated molecular platforms have enabled attribution of ML to species not previously known to cause mucosal disease, though the mechanisms by which the host interacts with different Leishmania species to cause mucosal involvement, often over a decade after initial CL, remain unknown. That PL differed by extent of clinical disease and causative species suggests that both intrinsic host and parasite factors contribute to the overall parasite burden in ML. Our observation of two distinct patterns of parasite clearance during treatment of ML offers a tantalizing foundation on which to build future studies of host–parasite interactions during treatment, and how these interactions may govern outcome.
ACKNOWLEDGMENTS
We would like to thank Sra. Ana Luz Quispe and Sra. Carmen Medina of the Instituto de Medicina Tropical “Alexander von Humboldt” (UPCH) for logistical support.
Disclaimer: This work was presented in abstract format at WorldLeish5, Porto de Gallinas, Brazil, May 2013, and at the 62nd Annual Meeting of the American Society of Tropical Medicine and Hygiene, Washington, DC, November 2013.
Footnotes
Financial support: This study was funded by the American Society of Tropical Medicine and Hygiene through the Gorgas Memorial Institute Research Award (Andrea K. Boggild). Personnel and facility support for the Arevalo molecular laboratory (Marlene Jara, Milena Alba, Vanessa Adaui, and Jorge Arevalo) was provided by the Institutional Collaboration Framework Agreement 3 from the Belgian Directorate-General for Development (project 95502). Marlene Jara was a Fogarty Scholar supported by NIH/Fogarty International Center Global Infectious Diseases Training Grant D43TW007120 during the study period.
Authors' addresses: Marlene Jara, Vanessa Adaui, Milena Alba, and Jorge Arevalo, Departmento de Bioquimica, Biologia Molecular y Farmacologia, Facultad de Ciencias, Universidad Peruana Cayetano Heredia, Lima, Peru, E-mails: jara_marlene@yahoo.es, vanessa.adaui@upch.pe, milenacollado@yahoo.es, and biomoljazz@gmail.com. Braulio Mark Valencia and Alejandro Llanos-Cuentas, Institute of Tropical Medicine Alexander von Humboldt, Universidad Peruana Cayetano Heredia, Lima , Peru, E-mails: braulio.valencia@upch.pe and alejandro.llanos.c@upch.pe. Rachel Lau, Public Health Ontario Laboratories, Toronto, Canada, E-mail: rachel.lau@oahpp.ca. Andrea K. Boggild, Tropical Disease Unit, Toronto General Hospital, Toronto, Canada, E-mail: andrea.boggild@utoronto.ca.
References
- 1.David CV, Craft N. Cutaneous and mucocutaneous leishmaniasis. Derm Ther. 2009;22:491–502. doi: 10.1111/j.1529-8019.2009.01272.x. [DOI] [PubMed] [Google Scholar]
- 2.Lucas CM, Franke ED, Cachay MI, Tejada A, Cruz ME, Kreutzer RD, Barker DC, McCann SH, Watts DM. Geographic distribution and clinical description of leishmaniasis cases in Peru. Am J Trop Med Hyg. 1998;59:312–317. doi: 10.4269/ajtmh.1998.59.312. [DOI] [PubMed] [Google Scholar]
- 3.Valencia BM, Veland N, Adaui V, Arevalo J, Low DE, Llanos-Cuentas A, Boggild AK. Non-invasive cytology brush PCR diagnostic testing in mucosal leishmaniasis: superior performance to conventional biopsy with histopathology. PLoS One. 2012;6:e26395. doi: 10.1371/journal.pone.0026395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mary C, Faraut F, Drogoul MP, Xeridat B, Schleinitz N, Cuisenier B, Dumon H. Reference values for Leishmania infantum parasitemia in different clinical presentations: quantitative polymerase chain reaction for therapeutic monitoring and patient follow-up. Am J Trop Med Hyg. 2006;75:858–863. [PubMed] [Google Scholar]
- 5.Verma S, Kumar R, Katara GK, Singh LC, Negi NS, Ramesh V, Salotra P. Quantification of parasite load in clinical samples of leishmaniasis patients: IL-10 level correlates with parasite load in visceral leishmaniasis. PLoS One. 2010;5:e10107. doi: 10.1371/journal.pone.0010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorlo TP, van Thiel PP, Schoone GJ, Stienstra Y, van Vugt M, Beijnen JH, de Vries PJ. Dynamics of parasite clearance in cutaneous leishmaniasis patients treated with miltefosine. PLoS Negl Trop Dis. 2011;5:e1436. doi: 10.1371/journal.pntd.0001436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudarshan M, Weirather JL, Wilson ME, Sundar S. Study of parasite kinetics with antileishmanial drugs using real-time quantitative PCR in Indian visceral leishmaniasis. J Antimicrob Chemother. 2011;66:1751–1755. doi: 10.1093/jac/dkr185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Meide WF, Peekel I, van Thiel PP, Schallig HD, de Vries HJ, Zeegelaar JE, Faber WR. Treatment assessment by monitoring parasite load in skin biopsies from patients with cutaneous leishmaniasis, using quantitative nucleic acid sequence-based amplification. Clin Exp Dermatol. 2008;33:394–399. doi: 10.1111/j.1365-2230.2007.02680.x. [DOI] [PubMed] [Google Scholar]
- 9.Jara M, Adaui V, Valencia BM, Martinez D, Alba M, Castrillon C, Cruz M, Cruz I, Van der Auwera G, Llanos-Cuentas A, Dujardin JC, Arevalo J. A real-time PCR assay for detection and quantification of Leishmania (Viannia) in skin and mucosal lesions: an exploratory study of parasite load and clinical parameters. J Clin Microbiol. 2013;51:1826–1833. doi: 10.1128/JCM.00208-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boggild AK, Valencia BM, Espinosa DE, Veland N, Ramos AP, Arevalo J, Llanos-Cuentas A, Low DE. Detection and species identification of Leishmania DNA from filter paper lesion impressions in patients with American cutaneous leishmaniasis. Clin Infect Dis. 2010;50:e1–e6. doi: 10.1086/648730. [DOI] [PubMed] [Google Scholar]
- 11.Sokal JE. Measurement of delayed skin-test responses. N Engl J Med. 1975;293:501–502. doi: 10.1056/NEJM197509042931013. [DOI] [PubMed] [Google Scholar]
- 12.Lopez M, Orrego C, Cangalaya M, Inga R, Arevalo J. Diagnosis of Leishmania via the polymerase chain reaction: a simplified procedure for field work. Am J Trop Med Hyg. 1993;49:348–356. doi: 10.4269/ajtmh.1993.49.348. [DOI] [PubMed] [Google Scholar]
- 13.Veland N, Boggild A, Valencia C, Valencia BM, Llanos-Cuentas A, Van der Auwera G, Dujardin JC, Arevalo J. Leishmania (Viannia) species identification on clinical samples from cutaneous leishmaniasis in Peru: assessment of a molecular step-wise approach. J Clin Microbiol. 2012;50:495–498. doi: 10.1128/JCM.05061-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schonian G, Nasereddin A, Dinse N, Schweynoch C, Schallig HDFH, Presber W, Jaffe CL. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis. 2003;47:349–358. doi: 10.1016/s0732-8893(03)00093-2. [DOI] [PubMed] [Google Scholar]
- 15.De Almeida ME, Steurer FJ, Koru O, Herwaldt BL, Pieniazek NJ, da Silva AJ. Identification of Leishmania spp. by molecular amplification and DNA sequencing analysis of a fragment of rRNA internal transcribed spacer 2. J Clin Microbiol. 2011;49:3143. doi: 10.1128/JCM.01177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendonça MG, de Brito ME, Rodrigues EH, Bandeira V, Jardim ML, Abath FG. Persistence of Leishmania parasites in scars after clinical cure of American cutaneous leishmaniasis: is there a sterile cure? J Infect Dis. 2004;189:1018–1023. doi: 10.1086/382135. [DOI] [PubMed] [Google Scholar]
- 17.Coutinho SG, Pirmez C, Da-Cruz AM. Parasitological and immunological follow-up of American tegumentary leishmaniasis patients. Trans R Soc Trop Med Hyg. 2002;96(Suppl 1):S173–S178. doi: 10.1016/s0035-9203(02)90072-6. [DOI] [PubMed] [Google Scholar]
- 18.Schubach A, Marzochi MC, Cuzzi-Maya T, Oliveira AV, Araujo ML, Oliveira AL, Pacheco RS, Momen H, Conceicao-Silva F, Coutinho SG, Marzochi KB. Cutaneous scars in American tegumentary leishmaniasis patients: a site of Leishmania (Viannia) braziliensis persistence and viability eleven years after antimonial therapy and clinical cure. Am J Trop Med Hyg. 1998;58:824–827. doi: 10.4269/ajtmh.1998.58.824. [DOI] [PubMed] [Google Scholar]
- 19.Schubach A, Haddad F, Oliveira-Neto MP, Degrave W, Pirmez C, Grimaldi G, Jr, Fernandes O. Detection of Leishmania DNA by polymerase chain reaction in scars of treated human patients. J Infect Dis. 1998;178:911–914. doi: 10.1086/515355. [DOI] [PubMed] [Google Scholar]
- 20.Prina E, Roux E, Mattei D, Milon G. Leishmania DNA is rapidly degraded following parasite death: an analysis by microscopy and real-time PCR. Microbes Infect. 2007;9:1307–1315. doi: 10.1016/j.micinf.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Vanaerschot M, Dumetz F, Roy S, Ponte-Sucre A, Arevalo J, Dujardin JC. Treatment failure in leishmaniasis: drug-resistance or another (epi-) phenotype? Expert Rev Anti Infect Ther. 2014;12:937–946. doi: 10.1586/14787210.2014.916614. [DOI] [PubMed] [Google Scholar]
- 22.Kloehn J, Saunders EC, O'Callaghan S, Dagley MJ, McConville MJ. Characterization of metabolically quiescent Leishmania parasites in murine lesions using heavy water labeling. PLoS Pathog. 2015;11:e1004683. doi: 10.1371/journal.ppat.1004683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurya R, Singh RK, Kumar B, Salotra P, Rai M, Sundar S. Evaluation of PCR for diagnosis of Indian kala-azar and assessment of cure. J Clin Microbiol. 2005;43:3038–3041. doi: 10.1128/JCM.43.7.3038-3041.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Disch J, Oliveira MC, Orsini M, Rabello A. Rapid clearance of circulating Leishmania kinetoplast DNA after treatment of visceral leishmaniasis. Acta Trop. 2004;92:279–283. doi: 10.1016/j.actatropica.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Nuzum E, White F, 3rd, Thakur C, Dietze R, Wages J, Grogl M, Berman J. Diagnosis of symptomatic visceral leishmaniasis by use of the polymerase chain reaction on patient blood. J Infect Dis. 1995;171:751–754. doi: 10.1093/infdis/171.3.751. [DOI] [PubMed] [Google Scholar]
- 26.Saravia NG, Weigle K, Segura I, Giannini SH, Pacheco R, Labrada LA, Goncalves A. Lancet. 1990;336:398–402. doi: 10.1016/0140-6736(90)91945-7. [DOI] [PubMed] [Google Scholar]
- 27.Carvalho EM, Johnson WD, Barreto E, Marsden PD, Costa JL, Reed S, Rocha H. Cell mediated immunity in American cutaneous and mucosal leishmaniasis. J Immunol. 1985;135:4144–4148. [PubMed] [Google Scholar]
- 28.Da-Cruz AM, Bittar R, Mattos M, Oliveira-Neto MP, Nogueira R, Pinho-Ribeiro V, Azeredo-Coutinho RB, Coutinho SG. T-cell-mediated immune responses in patients with cutaneous or mucosal leishmaniasis: long-term evaluation after therapy. Clin Diagn Lab Immunol. 2002;9:251–256. doi: 10.1128/CDLI.9.2.251-256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maretti-Mira AC, Bittner J, Oliveira-Neto MP, Liu M, Kang D, Li H, Pirmez C, Craft N. Transcriptome patterns from primary cutaneous Leishmania braziliensis infections associate with eventual development of mucosal disease in humans. PLoS Negl Trop Dis. 2012;6:e1816. doi: 10.1371/journal.pntd.0001816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dos Santos Nogueira R, Gomes-Silva A, de Cássia Bittar R, Silva Mendonça D, Amato VS, da Silva Mattos M, Oliveira-Neto MP, Coutinho SG, Da-Cruz AM. Antigen-triggered IFN-gamma and IL-10 pattern in cured mucosal leishmaniasis patients is shaped during the active phase of disease. Clin Exp Immunol. 2014;177:679–686. doi: 10.1111/cei.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuon FF, Gomes-Silva A, Da-Cruz AM, Duarte MI, Neto VA, Amato VS. Local immunological factors associated with recurrence of mucosal leishmaniasis. Clin Immunol. 2008;128:442–446. doi: 10.1016/j.clim.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 32.Maurer-Cecchini A, Decuypere S, Chappuis F, Alexandrenne C, De Doncker S, Boelaert M, Dujardin JC, Loutan L, Dayer JM, Tulliano G, Arevalo J, Llanos-Cuentas A, Chizzolini C. Immunological determinants of clinical outcome in Peruvian patients with tegumentary leishmaniasis treated with pentavalent antimonials. Infect Immun. 2009;77:2022–2029. doi: 10.1128/IAI.01513-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blum J, Lockwood DNJ, Visser L, Harms G, Bailey MS, Caumes E, Clerinx J, van Thiel PP, Morizot G, Hatz C, Buffet P. Local or systemic treatment for New World cutaneous leishmaniasis? Re-evaluating the evidence for the risk of mucosal leishmaniasis. Int Health. 2012;4:153–163. doi: 10.1016/j.inhe.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Hodiamont CJ, Kager PA, Bart A, de Vries HJ, van Thiel PP, Leenstra T, de Vries PJ, van Vugt M, Grobusch MP, van Gool T. Species-directed therapy for leishmaniasis in returning travellers: a comprehensive guide. PLoS Negl Trop Dis. 2014;8:e2832. doi: 10.1371/journal.pntd.0002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arevalo J, Ramirez L, Adaui V, Zimic M, Tulliano G, Miranda-Verástegui C, Lazo M, Loayza-Muro R, De Doncker S, Maurer A, Chappuis F, Dujardin JC, Llanos-Cuentas A. Influence of Leishmania (Viannia) species on the response to antimonial treatment in patients with American tegumentary leishmaniasis. J Infect Dis. 2007;195:1846–1851. doi: 10.1086/518041. [DOI] [PubMed] [Google Scholar]
- 36.Santrich C, Segura I, Arias AL, Saravia NG. Mucosal disease caused by Leishmania braziliensis guyanensis. Am J Trop Med Hyg. 1990;42:51–55. doi: 10.4269/ajtmh.1990.42.51. [DOI] [PubMed] [Google Scholar]
- 37.Osorio LE, Castillo CM, Ochoa MT. Mucosal leishmaniasis due to Leishmania (Viannia) panamensis in Colombia: clinical characteristics. Am J Trop Med Hyg. 1998;59:49–52. doi: 10.4269/ajtmh.1998.59.49. [DOI] [PubMed] [Google Scholar]
- 38.Montalvo AM, Fraga J, Monzote L, Montano I, De Doncker S, Dujardin JC, Van der Auwera G. Heat-shock protein 70 PCR-RFLP: a universal simple tool for Leishmania species discrimination in the New and Old World. Parasitology. 2010;137:1159–1168. doi: 10.1017/S0031182010000089. [DOI] [PubMed] [Google Scholar]