Abstract
Artemisinin combination therapy (ACT) is the first line to treat uncomplicated Plasmodium falciparum malaria worldwide. Artemisinin treatment failures are on the rise in southeast Asia. Delayed parasite clearance after ACT is associated with mutations of the P. falciparum kelch 13 gene. Patients (N = 148) in five districts of northwest Ethiopia were enrolled in a 28-day ACT trial. We identified a unique kelch 13 mutation (R622I) in 3/125 (2.4%) samples. The three isolates with R622I were from Negade-Bahir and Aykel districts close to the Ethiopia–Sudan border. One of three patients with the mutant strain was parasitemic at day 3; however, all patients cleared parasites by day 28. Correlation between kelch 13 mutations and parasite clearance was not possible due to the low frequency of mutations in this study.
A report by World Health Organization (WHO) estimated that 3.3 billion people are at risk of contracting malaria. In 2013 alone, there were an estimated 198 million cases of malaria worldwide with 584,000 deaths. Ninety percent of the deaths occurred in sub-Saharan Africa where children are the main victims.1 According to the WHO, between 2000 and 2013, an expansion of malaria interventions helped to reduce malaria incidence by 30% globally and by 34% in Africa.1 Many countries have initiated malaria elimination programs.1 Artemisinin-based combination therapies (ACTs) have been highly effective first-line drugs for the treatment of uncomplicated malaria worldwide.2 However, the recent emergence of Plasmodium falciparum strains that are resistant to artemisinin in southeast Asian countries poses a huge challenge to the effectiveness of ACT.1,3 The possibility of dissemination to or independent emergence of artemisinin-resistant strains in Africa, where the majority of malaria-associated deaths occur, will have potentially catastrophic outcomes.
A recent study using genome sequencing of P. falciparum has demonstrated an association between in vivo delayed parasite clearance, in vitro artemisinin “resistance” in a ring-stage assay, and mutations (Y493H, R539T, I543T, and C580Y) in the propeller domain of the parasite kelch 13 gene located on chromosome 13 (PF3D7_1343700 or PF13_0238).4 Moreover, an artemisinin-resistant P. falciparum strain that was selected by dose-escalating in vitro culture harbored the mutation, M476I, in the kelch 13 gene. Another study on samples from different southeast Asian countries confirmed the association of kelch 13 mutations and increased parasite clearance half-life. It also demonstrated the independent emergence of these mutations in different geographical areas.5 Targeted genetic engineering of kelch 13 using zinc-finger nuclease technology significantly increases the ring-stage survival rate of the parasite in vitro.6 Thus, detection of mutations on kelch 13 can be used to track artemisinin resistance in places where clinical treatment failure has not been observed. Kelch 13 mutation screening studies of P. falciparum isolates collected across sub-Saharan African countries have not identified the genotypes associated with treatment failure. However, several other non-synonymous mutations were detected in these countries.7,8
Ethiopia, with a population of more than 90 million, is a malaria-endemic country situated in sub-Saharan Africa. Approximately 68% of the population of Ethiopia live in malarious areas and are at risk of contracting the disease.9 There has been a significant reduction in malaria cases in Ethiopia since the introduction of artemether–lumefantrine for the treatment of uncomplicated falciparum malaria in 2004.9,10 In this study, we aimed to assess the presence of kelch 13 propeller mutations in P. falciparum isolates from northwest Ethiopia as part of a 28-day ACT clinical trial. We identified a mutation in the P. falciparum kelch 13 gene that has not been observed in Asia and Africa before.
The study protocol was reviewed and approved by Research and Ethical Review Committee of School of Biomedical and Laboratory Sciences, College of Medicine and Health Sciences, University of Gondar (reference no.: SBMLS/344/05). The study was conducted on P. falciparum DNA samples obtained from malaria patients from five different health centers in Amhara Regional State, northwest Ethiopia, between October 2013 and April 2014. The five sites were Addis Zemen, Aykel, Maksegnet, Negade Bahir, and Sanja. A total of 148 confirmed P. falciparum malaria patients were enrolled in a 28-day ACT efficacy trial. Patients were treated with artemether–lumefantrine (IPCA Laboratories, Mumbai, India). A directly observed therapy model was used for the first and each morning dose thereafter. The evening doses were given to the patient/guardian for self-administration in the presence of health extension workers.
Peripheral blood was collected by finger prick and thin and thick blood films were prepared on a microscope slide. Giemsa staining and microscopy was performed as described previously.11 Only those with mono-infection with P. falciparum were included in this study. Finger prick blood samples were collected on Whatman 903 filter paper (GE Healthcare, Mississauga, Canada) at the time of diagnosis, air-dried, individually inserted in a zip-lock bag, and transported to University of Calgary, Canada, for molecular analysis. Genomic DNA extraction from the filter paper blood spots was performed as described previously in a study.11 Plasmodium falciparum kelch 13 was amplified from the genomic DNA samples using nested polymerase chain reaction (PCR) with slight modification.4 The annealing temperature was set at 65.5°C for 45 seconds for the second step of the nested PCR to increase specificity.4,12 Sequencing of the kelch 13 propeller domain gene was performed using Applied Biosystems (Burlington, ON, Canada) 3730XL 96 capillary DNA analyzer. Bidirectional kelch 13 sequences from each patient sample and the reference kelch 13 gene sequence (PF3D7_1343700) (Genbank ID: AL844509.2) were aligned using Clustal Omega software (EMBL-EBI, Hinxton, UK). The sequences in the multiple sequences alignment were edited manually by Jalview software (Dundee, UK) to remove the gaps.13 Nested PCR was performed to confirm the species of Plasmodium as described previously in a study.11 Modeling of the predicted structure of the mutant kelch 13 propeller protein from Ethiopian isolates and its putative effect on the function of the protein was performed using Phyre2 (London, UK) (PDB ID: 2WOZ).14
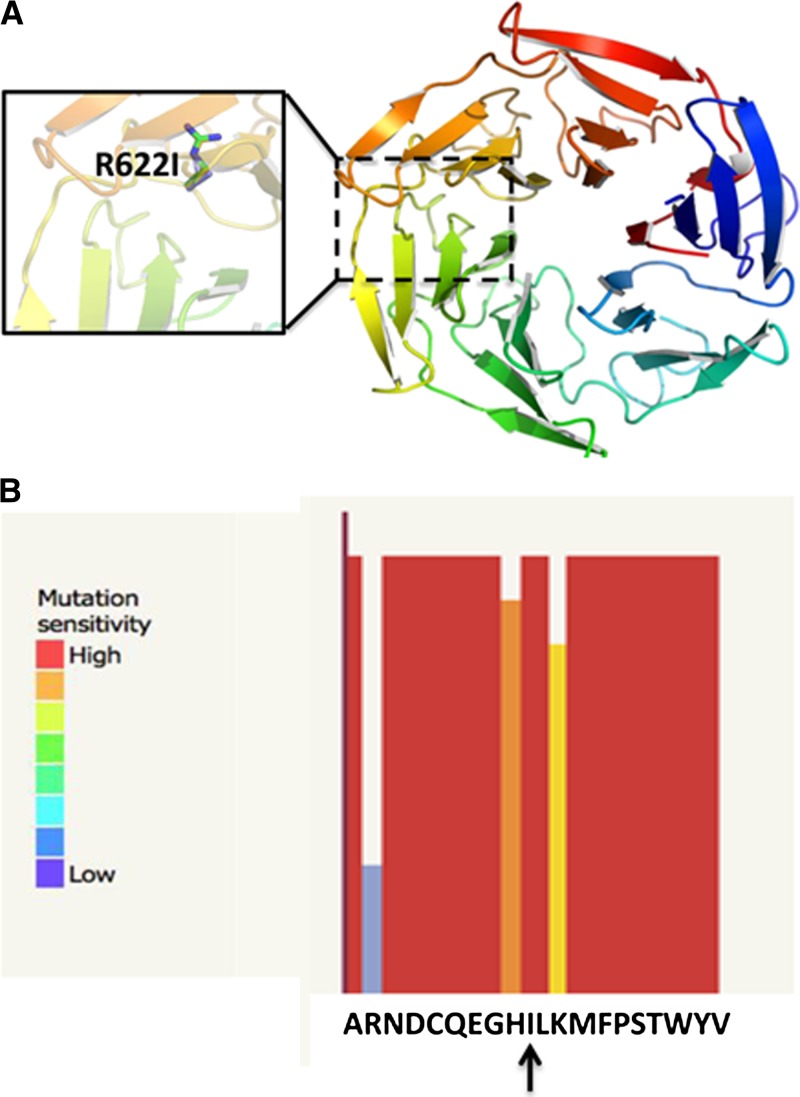
A total of 148 confirmed P. falciparum-infected malaria patients (age = 1–69 years) were enrolled in a 28-day ACT treatment trial. The kelch 13 propeller domain gene was amplified and sequenced from 125 of the 148 blood samples collected on filter paper. As shown in Table 1, the kelch 13 gene was successfully identified from 58%, 87.8%, 90.3%, 95.5%, and 100% of samples from Negade Bahir, Maksegnet, Addis Zemen, Sanja, and Aykel, respectively. Samples from three of 125 patients showed a single non-synonymous mutation at amino acid 622 (arginine [R] with isoleucine [I]). No synonymous mutation was detected in all the samples analyzed. Two of the mutations were seen on samples collected from Negade Bahir and one from Aykel, approximately 150 km from the Sudan border. The rate of R622I mutation in samples collected from Negade Bahir and Aykel was 6.7% and 11.1%, respectively (Table 1). One of the three patients that had a mutant strain was parasitemic based on Giemsa-stained microscopy on day 3 after the commencement of ACT. The R622I mutation is located on blade 5 of the kelch 13 propeller domain. Figure 1 shows the predicted model of kelch 13 propeller domain with the R622I mutation. The model predicts that the substitution from basic-to-aliphatic residue at this position is likely to impact the protein function. Confirmation of the species of the parasite in the samples where mutation was detected was performed using nested PCR to amplify the small subunit ribosomal RNA gene.15 Patients with mutant kelch 13 were infected with P. falciparum only.
Table 1.
Plasmodium falciparum kelch 13 gene nested PCR and mutation results from study sites in northwest Ethiopia
| Name of the site | Total no. of sample collected | K13 nested PCR positive N (%) | Non-synonymous mutation N (%) |
|---|---|---|---|
| Addis Zemen | 31 | 28 (90.3) | 0 (0) |
| Aykel | 15 | 15 (100) | 1 (6.7) |
| Maksegnet | 49 | 43 (87.8) | 0 (0) |
| Negade Bahir | 31 | 18 (58) | 2 (11.1) |
| Sanja | 22 | 21 (95.5) | 0 (0) |
PCR = polymerase chain reaction.
Figure 1.
Predicted model of the Plasmodium falciparum kelch 13 propeller protein with the R622I mutation. (A) Location of the mutation sites relative to the overall model of the protein. The beta-propeller domain of the btb-kelch protein Krp1 (PDB ID: 2WOZ) was used as the modeling template. (B) The colored bars indicate the probability that a mutation to the corresponding residue will have some effect on function of the protein (mutation sensitivity) or on the phenotype of the organism. The tall and red bars indicate that the likelihood is high. Short and blue bars indicate that the likelihood is low. The R622I substitution (vertical arrow) is predicted to impact protein function.
Mutations on the propeller domain of P. falciparum kelch 13 gene are strongly associated with artemisinin resistance as defined by a ring-stage assay in vitro and delayed clearance in patients from southeast Asia. The Y493H, R539T, I543T, and C580Y mutations as well as others are associated with delayed parasite clearance in vivo.4 The effect of these mutations has also been confirmed using targeted genetic engineering of kelch 13 gene using zinc-finger nuclease technology.6 Demonstration of these mutations on kelch 13 propeller gene could be used as an important surveillance tool to track the possible emergence of artemisinin treatment failure in places other than southeast Asia.16 So far, clinically significant kelch 13 mutations extend geographically only to Myanmar17 but not Bangladesh on the westernmost front.12
In this study, we identified a unique mutation in the kelch 13 propeller domain of P. falciparum isolates from northwest Ethiopia, close to the Sudan border. This mutation is different from those previously identified in southeast Asia or Africa.4,8,18,19 In contrast to our study, Kamau and others7 reported the absence of any single nucleotide polymorphism (SNP) on isolates from central Ethiopia. Predicted protein structure of the R622I mutant kelch 13 propeller gene indicates that the mutation is likely to disrupt the function of the protein (Figure 1B). This mutation is located on blade 5 of the kelch 13 propeller domain protein. Ashley and others16 have shown that mutations downstream of the 440 amino acid are significantly associated with increase in the parasite clearance half-life. Only one out of the three patients who had the R622I mutant strains showed day-3 positivity by Giemsa-stained microscopy. Interestingly, all 11/148 day-3 positive patients were located in Negade Bahir or Aykel where R622I was observed. Day-3 positivity is significantly associated with the overall kelch 13 mutations seen in a recent study conducted in Myanmar.20 On the basis of our study, the relative frequency of mutations does not make it possible to draw similar conclusions in this part of sub-Saharan Africa. Alternative explanations for day-3 positivity in this area include differences in technical expertise at parasite quantification, higher starting parasitemia, and different host immunity in this area, perhaps as a result of human immunodeficiency virus coinfection. Drug formulation and adherence are not likely contributing factors as these were controlled in the study. A limitation of this study was the inability to culture the mutant parasite to demonstrate increased inhibitory concentrations to artemisinin in vitro. Therefore, it remains to be seen whether kelch 13 SNPs can lead to delays in parasite clearance in Africa.
ACKNOWLEDGMENTS
We thank Dea Shahinas for assistance with computational analysis. We also thank the patients who participated in the study and the laboratory technicians at the field sites.
Footnotes
Financial support: Funding for this study was provided in part by Calgary Laboratory Services and Department of Medicine, University of Calgary.
Authors' addresses: Abebe Genetu Bayih and Abu Naser Mohon, Department of Pathology, University of Calgary, Alberta, Canada, E-mails: abebegenetu@gmail.com and manmohon@ucalgary.ca. Gebeyaw Getnet, Abebe Alemu, and Sisay Getie, Department of Medical Parasitology, University of Gondar, Gondar, Ethiopia, E-mails: ggebeyaw@yahoo.com, yanbule@gmail.com, and sisaygetie@yahoo.com. Dylan R. Pillai, Department of Pathology and Laboratory Medicine, University of Calgary, Alberta, Canada, E-mail: drpillai@ucalgary.ca.
References
- 1.World Health Organization . World Malaria Report. Geneva, Switzerland: World Health Organization; 2014. [Google Scholar]
- 2.World Health Organization . Guidelines for the Treatment of Malaria. 3rd edition. Geneva, Switzerland: World Health Organization; 2015. [Google Scholar]
- 3.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Trent Herdman, An SS, Yeung S, Singhasivanon P, Day NPJ, Lindegardh N, Socheat D, White NJ. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois A-C, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Ménard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale JC, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Ménard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takala-Harrison S, Jacob CG, Arze C, Cummings MP, Silva JC, Dondorp AM, Fukuda MM, Hien TT, Mayxay M, Noedl H, Nosten F, Kyaw MP, Nhien NT, Imwong M, Bethell D, Se Y, Lon C, Tyner SD, Saunders DL, Ariey F, Mercereau-Puijalon O, Menard D, Newton PN, Khanthavong M, Hongvanthong B, Starzengruber P, Fuehrer HP, Swoboda P, Khan WA, Phyo AP, Nyunt MM, Nyunt MH, Brown TS, Adams M, Pepin CS, Bailey J, Tan JC, Ferdig MT, Clark TG, Miotto O, MacInnis B, Kwiatkowski DP, White NJ, Ringwald P, Plowe CV. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in southeast Asia. J Infect Dis. 2015;211:670–679. doi: 10.1093/infdis/jiu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Straimer J, Gnädig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, Dacheux M, Khim N, Zhang L, Lam S, Gregory PD, Urnov FD, Mercereau-Puijalon O, Benoit-Vical F, Fairhurst RM, Ménard D, Fidock DA. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamau E, Campino S, Amenga-Etego L, Drury E, Ishengoma D, Johnson K, Mumba D, Kekre M, Yavo W, Mead D, Bouyou-Akotet M, Apinjoh T, Golassa L, Randrianarivelojosia M, Andagalu B, Maiga-Ascofare O, Amambua-Ngwa A, Tindana P, Ghansah A, MacInnis B, Kwiatkowski D, Djimde AA. K13-Propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J Infect Dis. 2014;211:1352–1355. doi: 10.1093/infdis/jiu608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor SM, Parobek CM, DeConti DK, Kayentao K, Coulibaly SO, Greenwood BM, Tagbor H, Williams J, Bojang K, Njie F, Desai M, Kariuki S, Gutman J, Mathanga DP, Mårtensson A, Ngasala B, Conrad MD, Rosenthal PJ, Tshefu AK, Moormann AM, Vulule JM, Doumbo OK, Ter Kuile FO, Meshnick SR, Bailey JA, Juliano JJ. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis. 2015;211:680–688. doi: 10.1093/infdis/jiu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ministry of Health, The Federal Democratic Republic of Ethiopia . National Strategic Plan for Malaria Prevention Control and Elimination in Ethiopia 2011–2015. 2010. [Google Scholar]
- 10.Ministry of Health, The Federal Democratic Republic of Ethiopia . Malaria Diagnosis and Treatment, Guideline for Health Workers in Ethiopia. 2010. [Google Scholar]
- 11.Sema M, Alemu A, Bayih A, Getie S, Getnet G, Guelig D, Burton R, LaBarre P, Pillai DR. Evaluation of non-instrumented nucleic acid amplification by loop-mediated isothermal amplification (NINA-LAMP) for the diagnosis of malaria in northwest Ethiopia. Malar J. 2015;14:44. doi: 10.1186/s12936-015-0559-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohon AN, Alam MS, Bayih AG, Folefoc A, Shahinas D, Haque R, Pillai DR. Mutations in Plasmodium falciparum K13 propeller gene from Bangladesh (2009–2013) Malar J. 2014;13:431. doi: 10.1186/1475-2875-13-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 14.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 15.Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, Thaithong S, Brown KN. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- 16.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NTT, Thanh NV, Phu NH, Htut Y, Han K-T, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, Rahman MR, Hasan MM, Islam A, Miotto O, Amato R, MacInnis B, Stalker J, Kwiatkowski DP, Bozdech Z, Jeeyapant A, Cheah PY, Sakulthaew T, Chalk J, Intharabut B, Silamut K, Lee SJ, Vihokhern B, Kunasol C, Imwong M, Tarning J, Taylor WJ, Yeung S, Woodrow CJ, Flegg JA, Das D, Smith J, Venkatesan M, Plowe CV, Stepniewska K, Guerin PJ, Dondorp AM, Day NP, White NJ, for the Tracking Resistance to Artemisinin Collaboration Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tun KM, Imwong M, Lwin KM, Win AA, Hlaing TM, Hlaing T, Lin K, Kyaw MP, Plewes K, Faiz MA, Dhorda M, Cheah PY, Pukrittayakamee S, Ashley EA, Anderson TJ, Nair S, McDew-White M, Flegg JA, Grist EP, Guerin P, Maude RJ, Smithuis F, Dondorp AM, Day NP, Nosten F, White NJ, Woodrow CJ. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet Infect Dis. 2015;15:415–421. doi: 10.1016/S1473-3099(15)70032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torrentino-Madamet M, Fall B, Benoit N, Camara C, Amalvict R, Fall M, Dionne P, Ba Fall K, Nakoulima A, Diatta B, Diemé Y, Ménard D, Wade B, Pradines B. Limited polymorphisms in K13 gene in Plasmodium falciparum isolates from Dakar, Senegal in 2012–2013. Malar J. 2014;13:472. doi: 10.1186/1475-2875-13-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Escobar C, Pateira S, Lobo E, Lobo L, Teodosio R, Dias F, Fernandes N, Arez AP, Varandas L, Nogueira F. Polymorphisms in Plasmodium falciparum K13-propeller in Angola and Mozambique after the introduction of the ACTs. PLoS One. 2015;10:e0119215. doi: 10.1371/journal.pone.0119215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nyunt MH, Hlaing T, Oo HW, Tin-Oo L-LK, Phway HP, Wang B, Zaw NN, Han SS, Tun T, San KK, Kyaw MP, Han ET. Molecular assessment of artemisinin resistance markers, polymorphisms in the K13 propeller, and a multidrug-resistance gene in the eastern and western border areas of Myanmar. Clin Infect Dis. 2014;60:1208–1215. doi: 10.1093/cid/ciu1160. [DOI] [PubMed] [Google Scholar]