Abstract
Several recent studies have demonstrated that virulence in Entamoeba histolytica is triggered in the presence of both pathogenic and nonpathogenic bacteria species using in vitro and in vivo experimental animal models. In this study, we examined samples aspirated from abscess material obtained from patients who were clinically diagnosed with amebic liver abscess (ALA) or pyogenic liver abscess (PLA). To determine the diversity of bacterial species in the abscesses, we performed partial 16S rRNA gene sequencing. In addition, the E. histolytica and Entamoeba dispar species were genotyped using tRNA-linked short tandem repeats as specific molecular markers. The association between clinical data and bacterial and parasite genotypes were examined through a correspondence analysis. The results showed the presence of numerous bacterial groups. These taxonomic groups constitute common members of the gut microbiota, although all of the detected bacterial species have a close phylogenetic relationship with bacterial pathogens. Furthermore, some patients clinically diagnosed with PLA and ALA were coinfected with E. dispar or E. histolytica, which suggests that the virulence of these parasites increased in the presence of bacteria. However, no specific bacterial groups were associated with this effect. Together, our results suggest a nonspecific mechanism of virulence modulation by bacteria in Entamoeba.
Introduction
In 1937, Westphal performed a highly controversial study to test the importance of associations between intestinal protozoa and bacteria in maintaining the virulence of Entamoeba histolytica.1 In the 1980s, Bracha and others reexamined this hypothesis and reported the increased expression of various E. histolytica virulence factors in human isolates when the ameba was co-cultured with different bacterial species.2 In 1983, the same authors published an extensive review of the possible mechanisms underlying the bacterial modulation of expression of E. histolytica genes encoding pathogenic factors.3 The complex mechanisms underlying the interaction between amebas, bacteria, and host cells are incompletely understood. However, it is know that certain bacterial groups contribute to the structure of the genome in some eukaryotic organisms, including Entamoeba, through horizontal gene transfer (HGT).4–6 Some of the first evidence of HGT in Entamoeba species was discovered by Rosenthal and others in 1997,7 and these conclusions were supported after sequencing of the E. histolytica HM1:IMSS genome.8,9 In addition, it has been demonstrated that amebas can either ingest bacteria or keep them attached to the amebic membrane in a mutualistic interaction that allows the transfer of genetic information to the Entamoeba parasite.2,3,10,11 Other studies have shown that in the absence of bacteria, E. histolytica does not express virulence factor–associated genes. Long-term axenic cultures of E. histolytica HM1:IMSS show loss of virulence in both in vitro (cytopathic and cytotoxic effects in cell cultures) and in vivo models of infection (amebic liver abscess [ALA] in hamsters).12,13 In 2008, Galván-Moroyoqui and others11 demonstrated that damage to epithelial cells caused by E. histolytica is dramatically increased in the presence of Escherichia coli and Shigella dysenteriae because of the increased expression of GalNAc lectin and cysteine proteases. Moreover, bacteria can transform aerobic environments into anaerobic environments, thereby increasing the production of free oxygen radicals because of the presence of peroxidases and catalases. This altered environment has been proposed to trigger the virulent behavior of E. histolytica.14,15
Several taxonomic groups of bacteria have been associated with the genus Entamoeba in human amebiasis. Among the most important of these bacteria are the Gammaproteobacteria, including Enterobacteriaceae E. coli, Shigella spp., Klebsiella spp., and Salmonella spp.16,17 Almost all of these bacteria express some type of mannose-linked lectins, which are essential for cell–cell binding.3,18 Other bacteria, such as Salmonella spp., express N-acetyl glucosamine as a surface antigen, which aids in attachment to E. histolytica through the GalNAc lectin complex.2,3 Gram-positive bacteria such as Streptococcus, Staphylococcus, and Pseudomonas, as well as other groups of bacteria, are highly efficient in triggering the virulence behavior of E. histolytica.16,19 Mirelman16 observed that only a few bacterial species have the capacity to attach to the amebic membrane or be phagocytosed by E. histolytica. Based on the epidemiology of diarrheal diseases, amebiasis-endemic areas are also commonly endemic for other enteropathogens causing diarrhea. In poor and crowded communities, mixed intestinal infections constitute a considerable number of diarrheal cases.20,21 In this scenario, almost all pathogenic enterobacteria can be cultured in the laboratory,22–25 in contrast to the intestinal microbiota, which generally includes nonculturable bacteria.26
Evidence of coinfections with both species of Entamoeba (E. histolytica and Entamoeba dispar) has been reported in patients with ALA.27 Of note, the molecular characterization of E. dispar in a patient with enteritis was recently reported in Italy.28 Furthermore, some patients with ALA present with mixed hepatic abscesses that are coinfected with pyogenic bacteria.
The goal of this study was to identify bacterial species in the aspirate material from patients with ALA as well as the protozoan species present in pyogenic abscesses. To this end, we collected aspirated abscess material from patients with ALA or pyogenic liver abscess (PLA) to genotype the bacterial species and Entamoeba present in the abscess. We also estimated the bacterial population diversity and determined the phylogenetic relationships between organisms. Furthermore, a correspondence analysis of clinical data with the etiology (pyogenic, amebic, or mixed) of the hepatic abscess and Entamoeba genotypes and bacterial groups was performed to test for significant associations.
Methods
Sample collection and DNA extraction.
The protocol for this study was submitted and approved by both the Ethics Committee of the Faculty of Medicine of National University of Mexico and the Committee of Hospital General de Mexico in Mexico City. Eleven Mexican patients, six with clinical diagnoses of ALA and five with clinical diagnoses of PLA, were included after they signed a letter of informed consent. Liver abscess patients underwent sonography-guided draining to reduce the risk of rupture, and the obtained material was kept at −80°C until DNA extraction. The DNA was extracted using the DNeasy Kit Blood and Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions.
Bacterial cloning and sequencing.
A fragment of approximately 500 base pairs (bp) from the 16S rRNA gene was amplified using the primers 27F-AGAGTTTGATCMTGGCTCAG (forward) and 1492R-ACCTTGTTACGACTT (reverse).29 The polymerase chain reaction (PCR) mixture (50 μL) was prepared with 1.5 μL of each primer (3 mM), 5 μL 10× buffer, 2 μL of 15 mM MgCl2, 4 μL dNTPs (10 mM), 34.8 μL water, and 1 U Taq DNA polymerase (Platinum Taq; Invitrogen Corporation, São Paulo, Brazil). The PCR cycling parameters were as follows: denaturation at 94°C for 3 minutes, followed by 30 cycles of 94°C for 1 minute, annealing at 61°C for 1 minute, and extension at 72°C for 1 minute and 30 seconds, with a final post-amplification incubation at 72°C for 10 minutes. The PCR products were analyzed on a 1% agarose gel containing ethidium bromide (0.5 μg/mL) and purified using Millipore Ultrafree-DA columns (Millipore Corporation, Bedford, Massachusetts). The purified products were cloned by ligation into the pCR 2.1-TOPO vector (Life Technologies Corporation, Carlsbad, CA) and used to transform competent E. coli (TOP10). Positive clones were identified by blue/white selection, and PCR was performed on DNA extracts prepared from bacterial (clone) colonies to confirm the insert (clone) size. Liquid cultures were grown in Luria medium containing 50 mg/mL kanamycin, and the DNA plasmids were purified for sequencing with the commercial Montage Miniprep kit (Millipore Corporation). Plasmids from each ligation were sequenced for both DNA strands using the 27F and 1492R universal primers. Sequencing was carried out by Sanger dye-terminator sequencing through high-throughput sequencing (High Throughput Genomics Center, Seattle, WA).
Short tandem repeat (STR) fragments were amplified from the genomic DNA by PCR using two E. histolytica/dispar-specific tRNA-linked STR primers (D-A and N-K2) under the previously described conditions,30 with the exception that PCR was performed in triplicate 20 μL reactions with Platinum Taq® polymerase (Invitrogen, Carlsbad CA). Thirty-five PCR cycles were used for annealing at a temperature of 50°C for D-A and 58°C for N-K2 primers. Triplicate PCR products were combined and concentrated to 30 μL. The amplified products were separated using a 2% agarose gel containing ethidium bromide (0.5 μg/mL) and purified using Ultrafree®-MC centrifugal filter devices (Millipore). After purification, the PCR products were sequenced through HTSeq as described above.
Data analysis.
The bacterial sequences were deposited in GenBank (accession nos. KP658212-KP658352). For the E. histolytica/dispar sequences please refer to the report of Zermeño and others from 2013.31 All sequences were assembled and analyzed using BioEdit Sequence Alignment Editor version 7.2.0 (Ibis Bioscience, Carlsbad, CA).32 The phylogenetic reconstructions were carried out by Bayesian probability analysis using MrBayes 3.2.33 The Markov chain Monte Carlo method was run for 1 million generations with sampling every 100 generations. The substitution model GTR + I + G was used. Finally, two “cold” chains and two “hot” chains were run with a temperature equal to 0.1. Of the sampled trees, 25% (1,250 trees) were used to build a consensus tree.
Multiple correspondence analysis.
Multiple correspondence analysis (MCA) was used to detect patterns of association between 1) the clinical diagnosis of pyogenic or amebic hepatic abscess and bacterial diversity (groups and genera); 2) the clinical diagnosis and the most relevant clinical characteristics (serum anti-amebic IgG antibodies and sonography); and 3) the E. histolytica or E. dispar genotypes (presence or absence of DA and STGA amebic molecular markers).
MCA is a multivariate analytic technique that simplifies complex sets of categorical variables, thus providing an exhaustive analysis that allows a detailed description of the data.34 The results of MCA are visualized in a bidimensional graphical output, which shows clouds of points representing categories of each variable. The pattern of association between variables is interpreted in terms of the relative position of their points along the dimensions. Points that are closer together are more strongly associated.
Results
Clinical features of the patients.
As previously mentioned, patients with ALA were included in the study after they signed a letter of consent. The physician in charge performed the clinical examination and the clinical diagnosis. Some variables analyzed in this study were obtained from the clinical files, including the sonography images suggestive of hepatic abscess and the results of enzyme-linked immunosorbent assays used to detect high levels of anti-amebic antibodies (cutoff value > 0.520 optical density [OD], as validated in a Mexican population).35 Diagnosis of PLA was based on the presence of metabolic disease and chronic degenerative or inflammatory disease and the absence of high levels of anti-amebic antibodies (cutoff value < 0.520 OD; Table 1). Serology can be used to effectively differentiate between PLA and ALA.
Table 1.
Key clinical symptoms of PLA and ALA
| Results | PLA | % | ALA | % | |
|---|---|---|---|---|---|
| (N = 5) | (N = 6) | ||||
| ELISA (OD) | < 0.52 | 5 | 100 | 0 | 0 |
| > 0.52 | 0 | 0 | 6 | 100 | |
| Leukocytes (mm3) | < 10,000 | 1 | 20 | 0 | 0 |
| > 10,000 | 4 | 80 | 6 | 100 | |
| One abscess | – | 2 | 40 | 3 | 50 |
| ≥ 2 abscess | – | 2 | 40 | 3 | 50 |
ALA = amebic liver abscess; ELISA = enzyme-linked immunosorbent assay; OD = optical density; PLA = pyogenic liver abscess.
OD cutoff value: 0.052.
Bacterial species and Entamoeba genotyping.
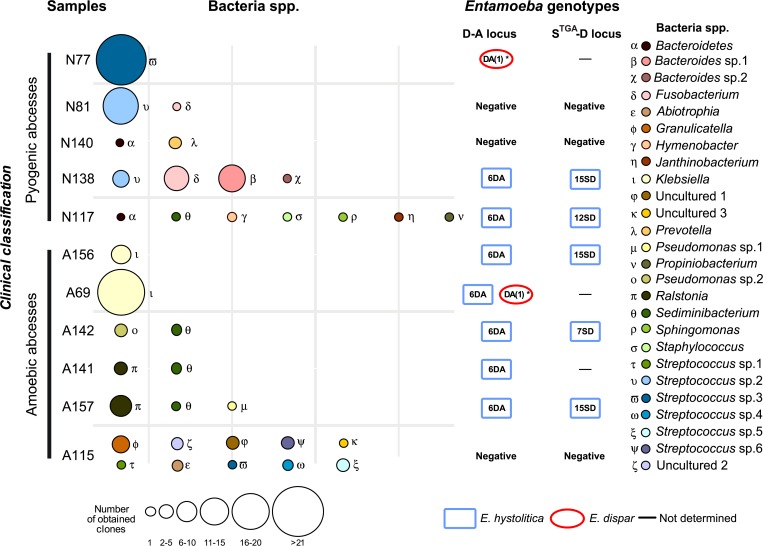
Twenty-seven bacterial groups were found in the 11 studied liver abscesses (Figure 1 ). The bacteria were identified to the genus level in most cases by comparison of their sequences to sequences in GenBank (Table 2). Three groups corresponded to uncultured bacteria, showing no taxonomic affiliation (uncultured sp. 1–3). A search of homologous sequences in the GenBank database showed that 16 of the 27 (60%) bacterial groups corresponded to the genera Streptococcus, Prevotella, Abiotrophia, and others. The identified bacteria may represent new genotypes because their partial 16S rDNA sequences did not show full sequence identity with any sequence in GenBank. Sequences corresponding to Hymenobacter showed a maximum identity of 96%, potentially indicating a new species (Table 2).
Figure 1.
Bacterial diversity and presence of amebic molecular markers in liver abscesses clinically classified as amebic or pyogenic.
Table 2.
Bacterial composition of the studied abscesses
| Sample ID (clones per sample) | Best blast hit* (accession number: percentage of similitude) | Previously reported source† |
|---|---|---|
| N-138 (1) | Uncultured (KF842135: 96) | Fecal sample |
| Hymenobacter (AY647897: 95) | ||
| A-115 (5) | Uncultured (KF842135: 100) | Fecal sample |
| Ganulicatella (FR822389: 99) | ||
| A-157 (1), A-142 (2), N-117 (1) | Sediminibacterium (FJ915149: 100) | Soil, water |
| A-117 (1) | Janthinobacterium (KF113922: 99) | Human pathogen |
| N-81 (1), N138 (9) | Uncultured (HM299064: 99) | Oropharyngeal pathogen |
| Fusobacterium (KF44237: 99) | ||
| A-115 (1) | Gibberella (AM946177: 100) | Associated to fungi |
| N-117 (1) | Uncultured (AJ609013: 99) | Soil |
| Sphingomonas (FJ362389: 98) | ||
| N-117 (1) | Propionibacterium (JQ194169: 100) | Soil, water |
| N-138 (11) | Bacteroides sp. 1. (KF842135: 100) | Human pathogen |
| N-138 (2) | Bacteroides sp. 2. (NR074839: 100) | Human pathogen |
| N-117 (1), N-140 (1) | Bacteroidetes (GU409054: 100) | Human pathogen |
| N-117 (1) | Pseudomonas sp. 1. (KF760556: 99) | Environmental |
| A-142 (5) A-157 (1) | Pseudomonas sp. 2. (KF263567:99) | Environmental |
| A-141 (3), A-157 (7) | Ralstonia (KJ149172: 100) | Human pathogen |
| N-140 (3) | Prevotella (JF214727: 99) | Human pathogen |
| A-69 (18), A-156 (6) | Klebsiella (KC692181: 99) | Human pathogen |
| N-117 (1) | Staphylococcus (KF575165: 100) | Human pathogen |
| A-115 (1) | Streptococcus sp.1 (GU374045: 99) | Human pathogen |
| A-115 (2) | Streptococcus sp.2 (GU797873: 99) | Human pathogen |
| A-115 (2) | Streptococcus sp.3 (GU907525: 99) | Human pathogen |
| A-115 (2) | Streptococcus sp.4 (HM075591: 99) | Human pathogen |
| N-81 (9), N-138 (4) | Streptococcus sp.5 (NR102797: 100) | Human pathogen |
| N-77 (22) | Streptococcus sp.6 (AF145244: 99) | Human pathogen |
| A-115 (5) | Uncultured sp.1 (HM268877: 99) | Skin pathogen |
| A-115 (2) | Uncultured sp.2 (JX473212: 100) | Fecal |
| A-115 (3) | Uncultured sp.3 (AY479308: 99) | Fecal |
Refers to the genus established by the best blast hit. When uncultured bacteria were the best blast hit, the second best blast hit was used (bold).
Source(s) where the genus has been previously described.
Figure 1 shows the genera of bacteria found in more than one abscess; these included Sediminibacterium, Fusobacterium, Bacteroidetes, Pseudomonas, Ralstonia, Klebsiella, and Streptococcus sp. 5. Sediminibacterium was present in four abscesses, three clinically diagnosed as ALA and one diagnosed as a PLA. The most abundant, but not necessarily the most frequent, genera were Klebsiella (24 clones: 18 in patient A69 and six in patient A156) and Streptococcus sp. 6 (22 clones from the PLA of patient N77). Additionally, 13 clones of Streptococcus sp. 5 were obtained: four from the PLA of patient N138 and nine from patient N81 (Figure 1). The most frequent, but not the most abundant, genus was Sediminibacterium, which was present in two ALA patients (A157 and A142) and one PLA patient (N117; Table 2). The abscesses with the greatest bacterial species diversity were from patients A115 (10 species) and N117 (7 species).
The samples from patients N138 and N81, both of whom had PLA, showed similarity in their bacterial composition, including Streptococcus sp. 2 and Fusobacterium. However, molecular techniques revealed the presence of E. histolytica DNA, demonstrating that patient N138 should be considered a mixed abscess case, whereas N81 had a pure PLA. These results indicate that the majority of the detected bacteria appeared in only one abscess, suggesting that bacterial diversity is unique to each abscess and that there is no specific bacterial group participating in invasion in liver abscess patients.
Table 2 shows the 20 bacterial groups detected in this study that have been previously reported in intestinal samples as mutualistic organisms or human pathogens. Nevertheless, Sediminibacterium, Sphingomonas, Propionibacterium, and Pseudomonas (sp. 1 and sp. 2; environmental) had not been previously reported in human clinical cases. Indeed, these genera have been previously isolated from soils, water bodies, and other environments but not from human specimens. In the uncultured bacteria, uncultured sp. 2 and 3 were previously isolated from fecal samples and uncultured sp. 1 was reported as a skin pathogen (Table 2).
In relation to Entamoeba in eight of the 11 studied samples, the presence of this parasite was confirmed with at least one molecular marker. Entamoeba dispar was detected in two patients, N77 and A69, representing PLA and ALA cases, respectively. Furthermore, E. histolytica was identified in almost all amebic samples (Figure 1). As expected, in PLAs (patients N81 and N140), there were no evidence of E. histolytica or E. dispar according to both markers examined; however, the sample from one amebic abscess (patient A115) also did not show positive results for these two markers (Figure 1). The Entamoeba genotypes most frequently found in patient samples were E. histolytica HM1-IMSS (6DA) and E. dispar SAW-760 (DA (1)*). Three other E. histolytica genotypes (15SD, 12SD, and 7SD) were also found in the abscesses of patients N138, A156, A157, N117, and A142 using the STGA marker. All of these genotypes have been previously reported in Bangladesh and elsewhere.36 Both Entamoeba species (E. histolytica and E. dispar) were detected in one abscess (patient A69) using the DA molecular marker. The two liver abscesses showed the greatest diversity of bacteria (A115 and N117), and E. histolytica DNA was also detected in the abscess from patient A115 (Figure 1).
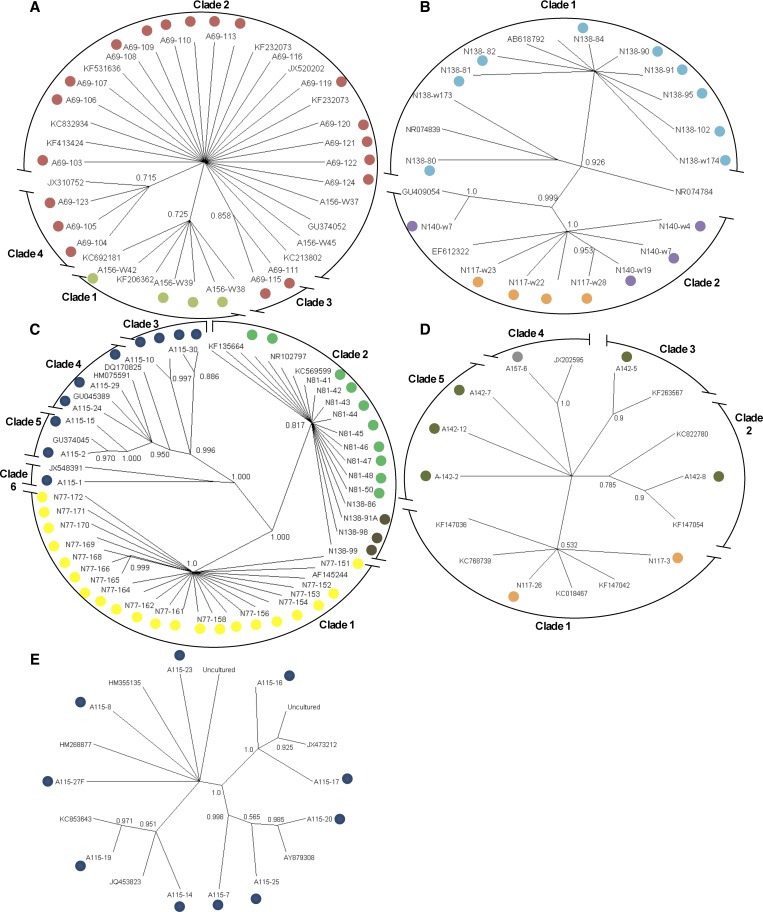
Phylogenetic analyses of the most abundant groups of bacteria.
Phylogenetic analyses were performed on the bacterial groups found in more than one abscess to determine the relationships with other species and genotypes previously reported in GenBank. The phylogenetic reconstruction showed that Klebsiella was represented by four genotypes; two genotypes were present in the abscesses of patient A156 (clade 1 and 2) and A69 ([clades 2, 3, and 4]; Figure 2A ). These four genotypes were closely associated with Klebsiella pneumonia (accession no. KC692181), which has been associated with human infections in some liver abscesses.37,38 The phylogenetic reconstruction separated Bacteroides and Bacteroidetes into two clades (Figure 2B). The two variants from Bacteroides were detected in the same patient with a pyogenic abscess (N138; clade 1), whereas Bacteroidetes was represented in two groups in the pyogenic abscesses of patients N140 and N117 (clade 2). These four variants of Bacteroides and Bacteroidetes showed low sequence identity (< 97% identity), which suggests that these variants may be considered separate species (Table 2).
Figure 2.
Phylogenetic reconstruction with the Bayesian method using the 16S rDNA sequences of five bacterial groups found in liver abscesses. The posterior probability from the MrBayes and GTR+I+G analysis is shown at each node. Groups: (A) Klebsiella, (B) Bacteroides and Bacteroidetes, (C) Streptococcus, (D) Pseudomonas, and (E) uncultured bacteria.
The genus Streptococcus was classed in six different clades; Streptococcus sp. 2 (clade 2) was found in two patients clinically diagnosed with PLAs (N81 and N138), both with the same genotype (Figure 2C). Streptococcus sp. 3 was found in patient N77 (clade 1), represented by only one genotype. The other genotypes of this genus were detected in only one patient, A115 (clades 3–6; Figure 2C).
Five genotypes from the Pseudomonas group were found in two patients diagnosed with ALA (A157 and A142) and in one patient diagnosed with PLA (N117; Figure 2D). Although the Pseudomonas phylogeny was not resolved, some unique genotypes were found in patients A142 and A157. The uncultured group, which includes multiple genotypes in each clade, was detected in patient A115 (Figure 2E).
In summary, some patients developed abscesses coinfected with more than one bacterial group; in addition, the same genotype could be found in more than one patient (Figures 1 and 2).
Bacteria–ameba association and clinical diagnosis.
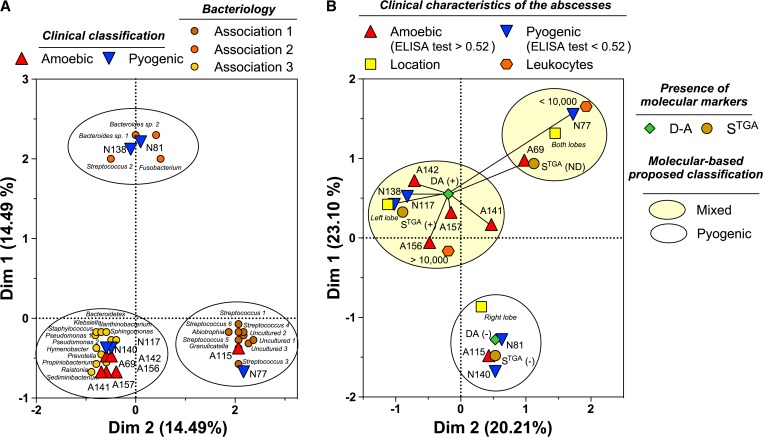
Regarding the association between isolated bacterial groups and the type of abscess, multivariate analysis showed the formation of three groups in the spatial distribution defined by MCA; however, as expected from the diversity and abundance of the bacterial species isolated from each liver abscess (Figure 1), the pattern of distribution described a limited association (14.49% for each dimension) among bacterial groups and the clinical diagnosis (Figure 3A ). No predictions regarding the type of abscess could be made based on any specific bacterial group. Only two of the PLAs (N81, N138) were clearly separated from the rest; the remaining pyogenic abscesses were included in groups with both types of abscess.
Figure 3.
Multiple correspondence analyses of the association between (A) the clinical diagnosis of liver abscesses and bacterial diversity and (B) the key clinical symptoms of the patient and Entamoeba genotype.
Figure 3B also shows the MCA results for relationships among clinical variables (Table 1) and ameba genotypes (Figure 3B). Interestingly, the clinical features of patients with ALAs showed no correspondence with any Entamoeba genotype. Serology is the most used indicator of acute amebiasis, and the serologic assays for all ameba variants are extraordinarily simple to perform and interpret, which might explain the variety of clinical pictures in both types of patients. Moreover, several signs and laboratory indicators (e.g., location of the abscess in the right or left hepatic lobule and high leukocytes counts) are common to ALA and PLA39–43; the definitive differential and etiological diagnosis of a mixed-type abscess or ALA is based on the results of PCR analysis targeting the D-A and STGA haplotypes of E. histolytica or E. dispar in the abscess aspirate. The MCA results (Figure 3) identified three discrete groups of abscesses arising from E. histolytica and/or E. dispar infection.
Discussion
The high prevalence of Entamoeba infection and other enteropathogens in underdeveloped countries has been recently reported.21,22,25,44,45 In most cases, efficient diagnostic technologies are not available, making diagnosis almost impossible in some communities.46 In our experience, molecular diagnosis is not yet available throughout the majority of health-care services even in technologically advanced countries. In most cases, these assays are performed in very specific research centers, and the technology is generally not commercially available. For these reasons, the clinical expertise of physicians is the key to diagnosis and successful treatment.
Efforts have been made in recent decades to design diagnostics based on molecular techniques to identify genetic variants of prokaryotic and eukaryotic microorganisms that are of importance to human health.30,38 Such tools would allow the reliable identification of the most prevalent etiologic agents in endemic areas of gastrointestinal infectious diseases. Mexico is one such endemic area, and the most vulnerable population is children younger than 15 years of age. Beyond the effects of amebiasis on the intestinal tract, this disease can also invade extraintestinal organs such as the liver.
Until now, it was thought that liver abscesses could be clinically differentiated into ALAs (caused by amebas exclusively), PLAs (caused by mainly anaerobic bacterial infections of intestinal origin), and mixed abscesses (originally caused by amebas and then infected by pyogenic bacteria). However, our results strongly suggest that most of the ALAs caused by E. histolytica are mixed abscesses (bacteria and protozoa). Indeed, the spectrum of outcomes of these infections could be related to the bacterial/protozoan density ratio. In a previous study,27 we hypothesized that the size of intestinal amebic ulcers and the local inflammatory response permits the exit of E. histolytica trophozoites and intestinal bacteria, some of which enter the phagocytic vacuoles of trophozoites.11,47,48 Our current results agree with our previous hypothesis and may explain the diversity of disease outcomes. Importantly, however, the causes of PLA and ALA are entirely different. PLA is primarily a consequence of chronic inflammatory disease or chronic degenerative disease.49–52 However, residence in or travel to amebiasis-endemic areas and exposure to multiple sources of infection could explain the presence of E. dispar in some PLA patients. Only patients N81 and N140 could be considered to have pure PLAs because they did not show molecular evidence of Entamoeba infection. Patient N115, clinically diagnosed with an ALA (Table 1), showed the greatest number of bacterial genera (10 genera); nevertheless, the PCR results (DA and STGA markers) were negative, which is a frequent outcome when working with DNA extracted from complex clinical samples. Unfortunately, no other molecular markers were tested to rule out the presence of PCR inhibitors in the sample. The other patients with liver abscesses were molecularly characterized as having mixed liver abscesses (Figure 1), suggesting that clinical diagnosis alone is not always reliable. Thus, when available, molecular techniques are an invaluable tool for specific etiological diagnosis; these methods aid in therapeutic decision making and improve the clinical evolution of patients.
One important observation from our study was that in most ALA abscesses, there was no specific group of bacteria associated with Entamoeba infection. In contrast, other studies have found that Klebsiella is the most common bacteria detected in PLA abscesses in America and Asia,53,54 although the majority of these studies were based on the analysis of culturable enteric bacteria.
In 1998, Brook and Frazier55 carried out a study of 116 pyogenic abscesses and showed that 76% were polymicrobial abscesses with more than one anaerobic, microaerophilic, or facultative anaerobic bacterial species. A study by Cosme and others found that most pyogenic cases show polymicrobial infection; however, E. coli and Streptococcus milleri were the most commonly found enterobacteria.56
Mixed infections include not only abscesses in which bacteria coexist with Entamoeba species but also coinfections by E. histolytica and E. dispar or more than one genotype of a particular species.27 Two abscesses from patients N77 and N69 from the previous study were included; the first showed E. dispar and one species of Streptococcus, while N69 showed E. histolytica and E. dispar as well as several genotypes of K. pneumoniae.
It is known that of the many individuals who carry asymptomatic intestinal infections with Entamoeba (90%), only a small number develop invasive intestinal or extraintestinal amebic disease. As previously mentioned, some individuals who would otherwise be asymptomatic cyst passers may develop invasive amebic disease on infection with a particular bacterial genus, species, or subspecies through the triggering of virulence-associated gene expression in Entamoeba trophozoites.11
Recent studies on the role of the intestinal microbiota in pathogenic gastrointestinal infections suggest that these organisms participate in the upregulation or downregulation of the virulence of parasites and bacterial pathogens.57,58 Upregulation of virulence-related gene expression in Entamoeba might lead to acute amebic colitis or dysentery, facilitating extraintestinal invasion. As mentioned previously, tissue invasion allows the exit of Entamoeba trophozoites and enterobacteria into the mesenteric and portal circulatory system for seeding into the hepatic parenchyma. Subsequent crucial steps in the host–parasite interaction in the liver mostly depend on the capacity of the parasite to adapt to an entirely new environment and the interaction between the trophozoites and the efficacy of the innate and adaptive immune response.
Our results suggest that a high percentage of known amebic liver abscesses are coinfected with nonculturable bacteria of the intestinal microbiota as well as potentially pathogenic bacteria. Moreover, we did not identify a specific group or groups of bacteria associated with any species or genotype of E. histolytica or E. dispar in ALA.
Although the number of patients in our study was small, the sample size was sufficient to demonstrate that extraintestinal invasion by E. histolytica is accompanied by the translocation of different ratios of intestinal bacteria. The Entamoeba–bacteria interaction seems to be entirely nonspecific, and in ALA, both species of Entamoeba might be present in the same liver abscess, along with a range of bacterial groups.
In the etiological diagnosis of hepatic abscesses, assays should be performed when a sample of aspirated material from the hepatic abscess is available for DNA extraction and the diagnostician has the capacity to perform specific PCRs for E. histolytica and E. dispar and enterobacteria. The results will give insight into the etiological agents of PLA and ALA and facilitate better therapeutic interventions for each patient.
On the basis of our correspondence analysis, serological results did not seem to be directly related to the type of liver abscess or clinical manifestations; however, in clinical practice, particularly when aspirated material is not available, the detection of high levels of serum anti-amebic antibodies remains a good and important clinical diagnostic indicator of ALA.
ACKNOWLEDGMENTS
We acknowledge the National University of Mexico (UNAM), Direction of General Affairs of Academic Personal (DGAPA) for the scholarship provided by the Postdoctoral Fellowships Program. We also thank the technical assistance of Martha Elena Zaragoza and Angeles Padilla.
Footnotes
Financial support: This work was partially supported by the Grant IN218214 and IN226511 from the Support Program to Research and Technological Development Projects (PAPIIT) from the National University of Mexico and Grant number 210-COI-140990 from the National Council of Science and Technology in Mexico.
Authors' addresses: Miriam E. Reyna-Fabián, Valeria Zermeño, Cecilia Ximénez, Janin Flores, Jesús Argueta, Patricia Moran, Alicia Valadez, and René Cerritos, Departamento de Medicina Experimental, Facultad de Medicina, Universidad Nacional Autónoma de México (UNAM), México City, México, E-mails: erandif@yahoo.com, valeria.zermeno.leon@gmail.com, cximenez@unam.mx, arlequin_flora@hotmail.comBiol, jadclear@yahoo.com, patricia_morans@yahoo.com.mx, avaladez17@yahoo.com.mx, and renecerritos@gmail.com. Miguel F. Romero and Daniel Diaz, Departamento de Biología Celular y Fisiología, Instituto de Investigaciones Biomédicas, Universidad Nacional Autónoma de México (UNAM), México City, México, E-mails: phyrexian.kavu@gmail.com and ddiaz@ciencias.unam.mx.
References
- 1.Westphal A. Betrachtungen und experimentelle untersucchungen zur virulenz der Entamoeba histolytica beim menschen. Arch Schiffs Tropenhyg Leipzig. 1937;41:262–279. [Google Scholar]
- 2.Bracha R, Kobiler D, Mirelman D. Attachment and ingestion of bacteria by trophozoites of Entamoeba histolytica. Infect Immun. 1982;36:396–406. doi: 10.1128/iai.36.1.396-406.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bracha R, Mirelman D. Adherence and ingestion of Escherichia coli serotype 055 by trophozoites of Entamoeba histolytica. Infect Immun. 1983;40:882–887. doi: 10.1128/iai.40.3.882-887.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nixon JE, Wang A, Field J, Morrison HG, McArthur AG, Sogin ML, Loftus BJ, Samuelson J. Evidence for lateral transfer of genes encoding ferredoxins, nitroreductases, NADH oxidase, and alcohol dehydrogenase 3 from anaerobic prokaryotes to Giardia lamblia and Entamoeba histolytica. Eukaryot Cell. 2002;1:181–190. doi: 10.1128/EC.1.2.181-190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nixon JE, Field J, McArthur AG, Sogin ML, Yarlett N, Loftus BJ, Samuelson J. Iron-dependent hydrogenases of Entamoeba histolytica and Giardia lamblia: activity of the recombinant entamoebic enzyme and evidence for lateral gene transfer. Biol Bull. 2003;204:1–9. doi: 10.2307/1543490. [DOI] [PubMed] [Google Scholar]
- 6.Field J, Rosenthal B, Samuelson J. Early lateral transfer of genes encoding malic enzyme, acetyl-CoA synthetase and alcohol dehydrogenases from anaerobic prokaryotes to Entamoeba histolytica. Mol Microbiol. 2000;38:446–455. doi: 10.1046/j.1365-2958.2000.02143.x. [DOI] [PubMed] [Google Scholar]
- 7.Rosenthal B, Zhiming M, Caplivski D, Ghosh S, de la Vega H, Graf T, Samuelson J. Evidence for the bacterial origin of genes encoding fermentation enzymes of the amitochondriate protozoa parasite Entamoeba histolytica. J Bacteriol. 1997;179:3736–3745. doi: 10.1128/jb.179.11.3736-3745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, Roncaglia P, Berriman M, Hirt RP, Mann BJ, Nozaki T, Suh B, Pop M, Duchene M, Ackers J, Tannich E, Leippe M, Hofer M, Bruchhaus I, Willhoeft U, Bhattacharya A, Chillingworth T, Churcher C, Hance Z, Harris B, Harris D. Jagels The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 9.Alsmark UC, Sicheritz-Ponten T, Foster GP, Hirt PR, Embley TM. Horizontal gene transfer in eukaryotic parasites: a case study of Entamoeba histolytica and Trichomonas vaginalis. Methods Mol Biol. 2009;532:489–500. doi: 10.1007/978-1-60327-853-9_28. [DOI] [PubMed] [Google Scholar]
- 10.Padilla-Vaca F, Ankri S, Bracha R, Koole L, Mirelman D. Down regulation of Entamoeba histolytica virulence by monoxenic cultivation with Escherichia coli O55 is related to a decrease in expression of the light (35-kilodaltons) subunit of the Gal/GalNac lectin. Infect Immun. 1999;67:2096–2102. doi: 10.1128/iai.67.5.2096-2102.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galván-Moroyoqui JM, Domínguez-Robles MC, Franco E, Meza I. The interplay between Entamoeba and enteropathogenic bacteria modulates epithelial cell damage. PLoS Negl Trop Dis. 2008;2:e266. doi: 10.1371/journal.pntd.0000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wittner M, Rosembaum RM. Role of bacteria in modifying virulence of Entamoeba histolytica. Am J Trop Med Hyg. 1970;19:755–761. doi: 10.4269/ajtmh.1970.19.755. [DOI] [PubMed] [Google Scholar]
- 13.Pimenta PF, Diamond LS, Mirelman D. Entamoeba histolytica Schaudinn, 1903 and Entamoeba dispar Brumpt, 1925: differences in their cell surfaces and in the bacteria-containing vacuoles. J Eukaryot Microbiol. 2002;49:209–219. doi: 10.1111/j.1550-7408.2002.tb00525.x. [DOI] [PubMed] [Google Scholar]
- 14.Bracha R, Mirelmam D. Entamoeba histolytica trophozoites: effect of bacteria, microaerobic conditions and metronidazole. J Exp Med. 1984;160:353–369. doi: 10.1084/jem.160.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray HW, Aley SB, Scott WA. Susceptibility of Entamoeba histolytica to oxygen intermediates. Mol Biochem Parasitol. 1981;3:381–391. doi: 10.1016/0166-6851(81)90038-4. [DOI] [PubMed] [Google Scholar]
- 16.Mirelman D. Ameba-bacterium relationship in amebiasis. Micriobiol Rev. 1987;51:272–284. doi: 10.1128/mr.51.2.272-284.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basu S. Klebsiella pneumoniae: an emerging pathogen of pyogenic liver abscess. Oman Med J. 2009;24:131–133. doi: 10.5001/omj.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bar-Shavit Z, Goldman R, Ofek I, Sharon N, Mirelman D. Mannose-binding activity of Entamoeba histolytica: a determinant of attachment and ingestion of the bacteria by macrophages. Infect Immun. 1980;29:417–424. doi: 10.1128/iai.29.2.417-424.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu-Jang S, Yen-Chun L, Yu-Chia L, Yu-Hang Y. Treatment and prognosis of pyogenic liver abscess. Int J Emerg Med. 2010;3:381–384. doi: 10.1007/s12245-010-0232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramos F, Mora P, González E, García G, Ramiro M, Gómez A, DE León Mdel C, Melendro EI, Valadez A, Ximénez C. High prevalence rate of Entamoeba histolytica asymptomatic infection in a rural Mexican community. Am J Trop Med Hyg. 2005;73:87–91. [PubMed] [Google Scholar]
- 21.Ximénez C, Morán P, Rojas L, Valadez A, Gómez A. Reassessment of the epidemiology of amebiasis: state of the art. Infect Genet Evol. 2009;9:1023–1032. doi: 10.1016/j.meegid.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Lara R, Galindo E, Olarte J, Hernández G. Mixed infections by E. histolytica, Shigella and other enteropathogenic bacteria, found in children with diarrhea. Arch Invest Med (Mex) 1974;5:515–518. [PubMed] [Google Scholar]
- 23.Chatterjee BD, Thawani G, Sanyal SN. Etiology of acute childhood diarrhea in Calcutta. Trop Gastroenterol. 1989;10:158–166. [PubMed] [Google Scholar]
- 24.Orlandi PP, Silva T, Magalhães GF, Alves F, de Almeida Cunha RP, Durlacher R, Pereira da Silva LH. Enteropathogens associated with diarrheal disease in infants of poor urban areas of Porto Velho, Rondônia: a preliminary study. Mem Inst Oswaldo Cruz. 2001;96:621–625. doi: 10.1590/s0074-02762001000500005. [DOI] [PubMed] [Google Scholar]
- 25.Haque R, Mondal D, Kirkpatrick BD, Akther S, Farr BM, Sack RB, Petri WA. Epidemiologic and clinical characteristics of acute diarrhea with emphasis on Entamoeba histolytica infections in preschool children in an urban slum of Dhaka, Bangladesh. Am J Trop Med Hyg. 2003;69:398–405. [PubMed] [Google Scholar]
- 26.Lagier JC, Million M, Hugon P, Armougom F, Raoult D. Human gut microbiota: repertoire and variations. Front Cell Infect Microbiol. 2012;2:136. doi: 10.3389/fcimb.2012.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ximénez C, Cerritos R, Rojas L, Dolabella S, Morán P, Shibayama M, González E, Valadez A, Hernández E, Valenzuela O, Limón A, Partida O, Silva EF. Human amebiasis: breaking the paradigm? Int J Environ Res Public Health. 2010;7:1105–1120. doi: 10.3390/ijerph7031105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graffeo R, Archibusacci CM, Soldini S, Romano L, Masucci L. Entamoeba dispar: a rare case of enteritis in a patient living in a nonendemic area. Case Rep Gastrointest Med. 2014;2014:498058. doi: 10.1155/2014/498058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. New York, NY: Wiley; 1991. pp. 115–175. [Google Scholar]
- 30.Ali IKM, Zaki M, Clark CG. Use of PCR amplification of tRNA gene-linked short tandem repeats for genotyping Entamoeba histolytica. J Clin Microbiol. 2005;43:5842–5847. doi: 10.1128/JCM.43.12.5842-5847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zermeño V, Ximénez C, Morán P, Valadez A, Valenzuela O, Rascón E, Diaz D, Cerritos R. Worldwide genealogy of Entamoeba histolytica. An overview to understand haplotype distribution and infection outcome. Infect Genet Evol. 2013;17:243–252. doi: 10.1016/j.meegid.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 32.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 33.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 34.Costa PS, Santos CN, Cunha P, Cotter J, Sousa N. The use of multiple correspondence analysis to explore associations between categories of qualitative variables in healthy ageing. J Aging Res. 2013;2013:302163. doi: 10.1155/2013/302163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morán P, Gómez A, Valadez A, Ramos F, González E, García G, Limón A, Valenzuela O, Ramiro M, Hidalgo H, Melendro EI, Ximénez C. International Proceedings of the 5th European Congress on Tropical Medicine and International Health. Amsterdam, The Netherlands: 2007. Amebic and Pyogenic Liver Abscess: Importance of Differential Diagnosis in Endemic Areas of Amebiasis; pp. 57–64. [Google Scholar]
- 36.Ali IKM, Solaymani-Mohammadi S, Akhter J, Roy S, Gorrini C, Calderaro A, Parker SK, Haque R, Petri WA, Clark CG. Tissue invasion by Entamoeba histolytica: evidence of genetic selection and/or DNA reorganization events in organ tropism. PLoS Negl Trop Dis. 2008;2:e219. doi: 10.1371/journal.pntd.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sachdev DD, Yin MT, Horowitz JD, Mukkamala SK, Lee SE, Ratnerc AJ. Klebsiella pneumoniae K1 liver abscess and septic endophthalmitis in a U.S. resident. J Clin Microbiol. 2013;51:1049–1051. doi: 10.1128/JCM.02853-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Wang J, Jiang W. An increasing prominent disease of Klebsiella pneumoniae liver abscess: etiology, diagnosis, and treatment. Gastroenterol Res Pract. 2013;2013:258514. doi: 10.1155/2013/258514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benazzouz M, Afifi R, Ibrahimi A, Essaid FA, Sebti MF. Liver abscess: diagnosis and treatment. Study of a series of 22 cases. Ann Gastroenterol Hepatol (Paris) 1996;31:333–336. [PubMed] [Google Scholar]
- 40.Tazawa J, Sakai Y, Maekawa S, Ishida Y, Maeda M, Marumo F, Sato C. Solitary and multiple pyogenic liver abscesses: characteristics of the patients and efficacy of percutaneous drainage. Am J Gastroenterol. 1997;92:271–274. [PubMed] [Google Scholar]
- 41.Jamil B, Hamid SS. Abscesses in the liver: amebic or pyogenic? J Pak Med Assoc. 2002;52:495–496. [PubMed] [Google Scholar]
- 42.Ahsan T, Jehangir MU, Mahmood T, Ahmed N, Saleem M, Shahid M, Shaheer A, Anwer A. Amebic versus pyogenic liver abscess. J Pak Med Assoc. 2002;52:497–501. [PubMed] [Google Scholar]
- 43.Lodhi S, Sarwari AR, Muzammil M, Salam A, Smego RA. Features distinguishing amebic from pyogenic liver abscess: a review of 577 adult cases. Trop Med Int Health. 2004;9:718–723. doi: 10.1111/j.1365-3156.2004.01246.x. [DOI] [PubMed] [Google Scholar]
- 44.Valenzuela O, Ramos F, Moran P, Gonzalez E, Valadez A, Gómez A, Melendro EI, Ramiro M, Muñoz O, Ximénez C. Persistence of secretory antiamoebic antibodies in patients with past invasive intestinal or hepatic amebiasis. Parasitol Res. 2001;87:849–852. doi: 10.1007/s004360100418. [DOI] [PubMed] [Google Scholar]
- 45.Alam MA, Maqboo A, Nazir MM, Lateef M, Khan MS, Ahmed AN, Mughal Z, Lindsay DS. Prevalence of Entamoeba histolytica-like cysts compared to E. histolytica antigens detected by ELISA in the stools of 600 patients from three socioeconomic communities in the metropolitan city of Lahore, Pakistan. J Parasitol. 2014;101:236–239. doi: 10.1645/14-560.1. [DOI] [PubMed] [Google Scholar]
- 46.Tengku SA, Norhayati M. Public health and clinical importance of amebiasis in Malaysia: a review. Trop Biomed. 2011;28:194–222. [PubMed] [Google Scholar]
- 47.Nathaniel CVC, Sarah NB, William AP., Jr. Common pathways for receptor-mediated ingestion of Escherichia coli and LDL cholesterol by Entamoeba histolytica regulated in part by transmembrane kinase 39. Int J Parasitol. 2012;42:393–400. doi: 10.1016/j.ijpara.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Talamaìs-Lara D, Chaìvez-Munguiìa B, Gonzaìlez-Robles A, Talamaìs-Rohana P, Salazar-Villatoro L, Duraìn-Diìaz A, Martiìnez-Palomo A. Erythrophagocytosis in Entamoeba histolytica and Entamoeba dispar: a comparative study. BioMed Res Int. 2014;2014:626259. doi: 10.1155/2014/626259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ilan Y. Leaky gut and the liver: a role for bacterial translocation in nonalcoholic steatohepatitis. World J Gastroenterol. 2012;18:2609–2618. doi: 10.3748/wjg.v18.i21.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim JK, Chung DR, Wie SH, Lin YT, Wang FD, Wu PF, Fung CP. Klebsiella pneumoniae liver abscess in diabetic patients: association of glycemic control with the clinical characteristics. BMC Infect Dis. 2013;13:56. doi: 10.1186/1471-2334-13-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199:697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fung CP, Lin YT, Lin JC, Chen TL, Yeh KM, Chang FY, Chuang HC, Wu HS, Tseng CP, Siu LK. Klebsiella pneumoniae in gastrointestinal tract and pyogenic liver abscess. Emerg Infect Dis. 2012;18:1322–1325. doi: 10.3201/eid1808.111053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rahimian J, Wilson T, Oram V, Holzman RS. Pyogenic liver abscess: recent trends in etiology and mortality. Clin Infect Dis. 2004;39:1654–1659. doi: 10.1086/425616. [DOI] [PubMed] [Google Scholar]
- 54.Lok KH, Li KF, Szero ML. Pyogenic liver abscess: clinical profile, microbiological characteristics, and management in a Hong Kong hospital. J Microbiol Immunol Infect. 2008;41:483–490. [PubMed] [Google Scholar]
- 55.Brook I, Frazier EH. Microbiology of liver and spleen abscesses. J Med Microbiol. 1998;47:1075–1080. doi: 10.1099/00222615-47-12-1075. [DOI] [PubMed] [Google Scholar]
- 56.Cosme A, Ojeda E, Zamarreño I, Bujanda L, Garmendia G, Echeverría MJ, Benavente J. Pyogenic versus amebic liver abscesses. A comparative clinical study in a series of 58 patients. Rev Esp Enferm Dig. 2010;102:90–99. doi: 10.4321/s1130-01082010000200004. [DOI] [PubMed] [Google Scholar]
- 57.Sheh A, Fox JG. The role of the gastrointestinal microbiome in Helicobacter pylori pathogenesis. Gut Microbes. 2013;4:505–531. doi: 10.4161/gmic.26205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Halliez MC, Buret AG. Extra-intestinal and long term consequences of Giardia duodenalis infections. World J Gastroenterol. 2013;19:8974–8985. doi: 10.3748/wjg.v19.i47.8974. [DOI] [PMC free article] [PubMed] [Google Scholar]