Abstract
In order to localize the neural circuits involved in generating behaviors, it is necessary to assign activity onto anatomical maps of the nervous system. Using brain registration across hundreds of larval zebrafish, we have built an expandable open source atlas containing molecular labels and anatomical region definitions, the Z-Brain. Using this platform and immunohistochemical detection of phosphorylated-Extracellular signal-regulated kinase (ERK/MAPK) as a readout of neural activity, we have developed a system to create and contextualize whole brain maps of stimulus- and behavior-dependent neural activity. This MAP-Mapping (Mitogen Activated Protein kinase – Mapping) assay is technically simple, fast, inexpensive, and data analysis is completely automated. Since MAP-Mapping is performed on fish that are freely swimming, it is applicable to nearly any stimulus or behavior. We demonstrate the utility of our high-throughput approach using hunting/feeding, pharmacological, visual and noxious stimuli. The resultant maps outline hundreds of areas associated with behaviors.
Introduction
Zebrafish larvae possess a tiny brain, less than half a cubic millimeter, containing ~100,000 neurons. Despite such a compact nervous system, and being less than a week old, these animals are capable of producing a diversity of fascinating behaviors. These include swimming in three dimensions, escape maneuvers, visually-guided hunting, learning and sleep1. However, our knowledge of how the zebrafish brain is organized functionally, and how it produces behavior, is limited. To understand how the brain generates behavior we need to identify the neurons and networks relevant to particular tasks. This can begin through measurements of neural activity correlated with behavior. To explore the full range of natural behaviors and to avoid artifacts of manipulation, such measurements should ideally be performed in freely behaving animals. Imaging approaches can allow for nearly brain-wide imaging in larval zebrafish2,3, but are limited to head-fixed animals and behaviors that can be performed under a microscope. The recently developed CaMPARI integrative Ca2+ sensor can map activity in freely swimming fish4, but requires perturbation through exposure to bright blue/UV light, which causes aversive responses in adult fish5. Recording from unperturbed larval zebrafish is possible using aequorin bioluminescent imaging6, which can provide good temporal resolution, but spatial information is limited to the aequorin expression pattern.
Biochemical events that occur naturally as a consequence of neural activity can also be used to find the neurons that were active in a freely behaving animal at cellular resolution. In mammals, the expression of immediate early genes (IEGs), such as c-Fos and Arc, have localized neurons critical for diverse behaviors such as memory, sleep, fear, mating and drug addiction7. However, such techniques have relatively poor temporal resolution and suffer problems of low sensitivity. Indeed, the very low amount of baseline c-fos staining observed in zebrafish brains8,9, and the relatively slow time course of cFos activation of 15–30 min and 1–2 hrs for mRNA and protein responses respectively, in both mammalian and teleost neurons8,10–14, limits the applicability of c-fos to the study of natural behaviors in zebrafish larvae. Here we use a more permissive endogenous sensor: phosphorylated extracellular signal-regulated kinase (ERK, also known as Mitogen activated protein kinase)15–17 In response to depolarization, calcium influx through L-type voltage gated calcium channels activates the Ras-Erk pathway18 leading to the phosphorylation of transcription factors such as CREB and Elk, and IEG expression19. Therefore, activation/phosphorylation of Erk1/2 (pERK), can be used to localize active neurons15,16 including zebrafish12,20, and offers improved temporal resolution over IEGs as signals are created within 5 minutes of activation15,16,21.
Once created, activity maps are of limited utility unless they intersect with detailed neuroanatomical information22. Anatomical resources currently available for larval zebrafish are restricted to either maps of 2–4 day old embryos/larvae (ViBE-Z23) or to 2-dimensional images (zebrafishbrain.org and24), from which it can be difficult to infer 3-dimensional relationships. Therefore, understanding neuroanatomical features in an activity map is difficult and unstandardized. Here we leverage high-throughput confocal imaging and registration to create both a reference atlas and brain-wide activity maps.
Results
Z-Brain, a zebrafish reference brain atlas
We chose to create our atlas at the 6 days post fertilization (dpf) stage, lying in the middle of the often-studied 5–7dpf age range. Our goal was to include as many anatomical labels as possible, and a detailed segmentation. We registered confocal stacks of the brain to a template brain based on the expression of total-ERK/MAPK (tERK) (Fig. 1a). For registration, we used the Computational Morphometry Toolkit (CMTK)25,26. CMTK uses non-rigid registration/morphing algorithms to align imaging data, and can achieve an accuracy of 3–4 um26,27. To quantify our registration accuracy we used spinal backfills to label identifiable reticulospinal neurons in different brains (Fig. 1b). Measuring the position of axon emergence from four identified neurons (Mauthner and CaD neurons) yielded a 3D positioning error of ~ 1 cell body diameter across registered brains (4.6 um, mean absolute deviation, n=23 fish). As the variability in distance between these two neurons in individual fish before warping was 3.2 um, much of the estimated morphing error might reflect legitimate biological variability.
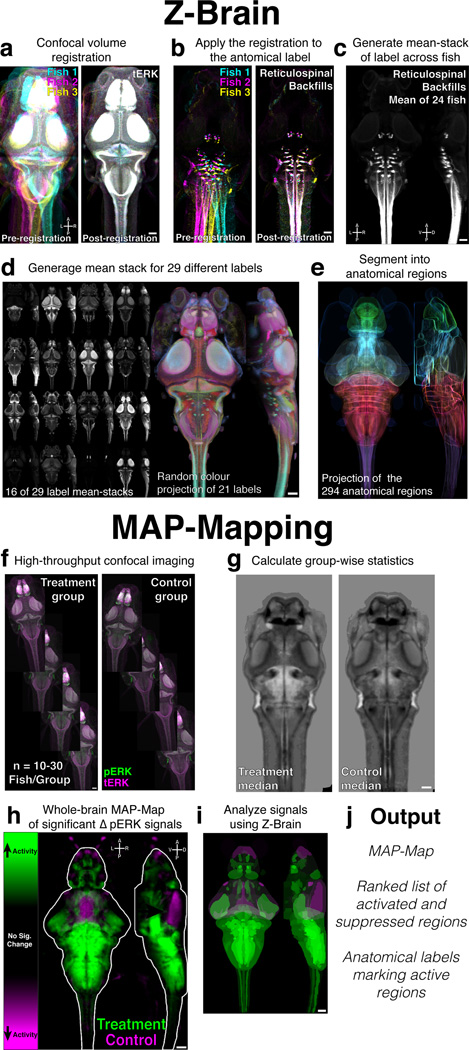
Figure 1. Analysis pipeline: creating the zebrafish reference brain atlas (Z-Brain) and whole-brain activity maps (MAP-Maps).
A) tERK Confocal stacks registered to a reference brain. Shown are three fish (cyan, magenta, yellow) as maximum intensity projections. B) Registrations are applied to an anatomical label (reticulospinal backfills). C) The mean across all registered fish is calculated. Shown are Z and X maximum intensity projections D) Mean-stacks from a total of 29 transgenic, antigenic, or dye labels were generated (Supplementary Table 1). Shown are maximum intensity Z and X projections of 16 different labels (left), and a color Z and X random color projection image for 21 labels (right) E) Z and X mean projections of the outlines of the segmented Z-Brain regions, drawn in colors biased towards green = Telencephalon, cyan = Diencephalon, yellow = Mesencephalon, red = Rhombencephalon, magenta = Spinal Cord, F) To create a MAP-Map, pERK/tERK confocal stacks are acquired for ~10–30 fish per condition. G) Stacks are registered to Z-Brain, and the pERK level statistics are calculated at each voxel. H) Voxels found exhibiting significantly higher (green) and lower (magenta) pERK levels in the stimulus group are localized to create a MAP-Map (Online Methods). Shown are mean Z and X projections for heat exposure (Fig. 4c). I, J) The MAP-Map is then analyzed using the Z-Brain (see Online Methods). Shown is the Z and X maximum intensity projections depicting the mean signal in the Z-Brain regions. Scale bars represent 50um. R = Right, L = Left, A = Anterior, P = Posterior, D = Dorsal, V = Ventral.
28 additional labels were registered into the reference brain, for a total of 899 brains (Supplementary Table 1). For each label we created a mean-across-fish stack, yielding pseudo-density maps highlighting the domains where labeled components typically reside (Fig. 1c,d, Supplementary Video 1). The accuracy of registration is again evident qualitatively through the overlap of labeled neurons, evident lamination in the tectal neuropil, and a convergence of synaptic markers at the Mauthner axon cap (Supplementary Fig. 1). These labels, along with Mueller and Wulliman’s classical reference atlas24, were used to guide the manual segmentation of our reference brain into 294 anatomical regions (Fig. 1e, Supplementary Video 2).
pERK is a reporter of neural activity in zebrafish neurons
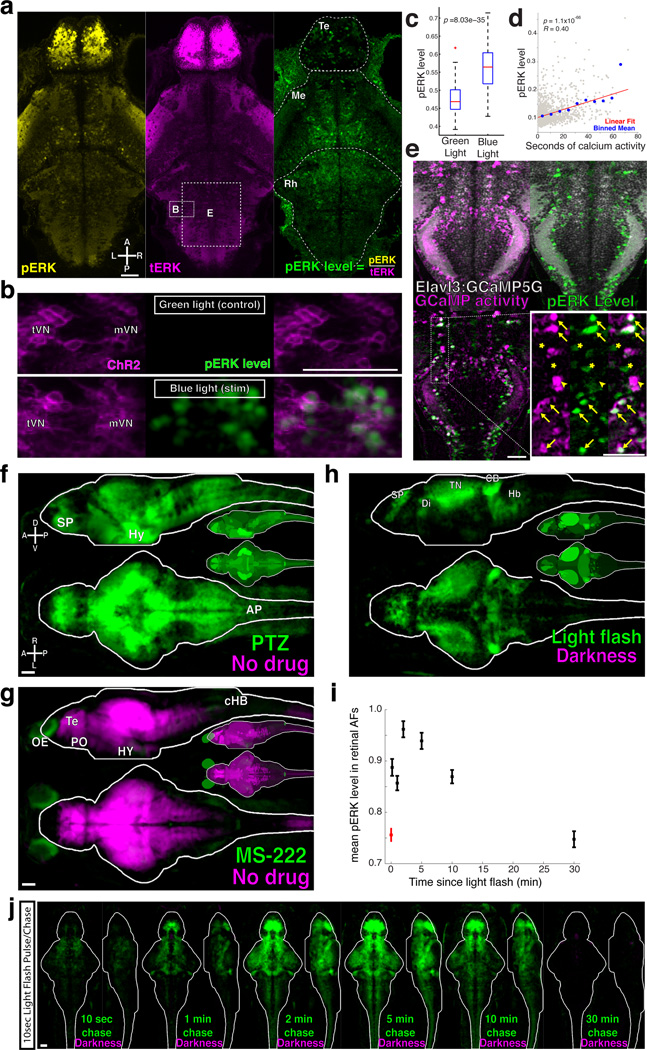
To assay for neural activity, we turned to ERK, a kinase which has previously been used to identify active neurons in the zebrafish brain12,20. Staining for phosphorylated-ERK (pERK) in 6 dpf larvae revealed enrichment in the nervous system. Punctate staining of individual cells was seen throughout the brain, with strongest staining in the telencephalon (Fig. 2a). This is in contrast to staining for total-ERK (tERK), which exhibited much more homogenous staining. Co-staining with both antibodies allowed us to calculate the pERK level normalized to tERK (pERK/tERK). To confirm that pERK levels are activity dependent, we optogenetically activated channel-rhodopsin (ChR2) expressing neurons and quantified pERK levels (Fig. 2b,c). Blue light stimulation significantly increased pERK levels within ChR2-expressing neurons, including neurons in vestibular nuclei, and in the habenula, hypothalamus, ventral hindbrain, and the spinal cord (p < 2×10−5 in all cases, ranksum test, data not shown).
Figure 2. pERK is a neural activity sensor in zebrafish neurons.
A) A confocal slice of a fish stained for phosphorylated-ERK (pERK, yellow) and total-ERK (tERK, magenta), from which we calculate the normalized ‘pERK level’ (pERK/tERK). Te = Telencephalon, Me = Mesencephalon, Rh = Rhombencephalon. Boxes depict the approximate x/y positions of neurons shown in panels B and E, but in a different z-plane. B) ChR2-YFP was driven in multiple neuron types in Tg(−6.7FRhcrtR:gal4VP16);Tg(14xUAS-E1b:hChR2(H134R)-EYFP); atoh7th241/th241; Tg(atoh7:GAP-RFP) larvae. Shown are neurons of the tangential and median vestibular nucleus (tVN and mVN), stimulated with either blue or green light. C) ChR2 activation significantly increased pERK levels (p = 8.03×10–35, ranksum test, n =1056 neurons from 20 larvae). In the boxplots, red line = median, blue box = 25th and 75th quartiles, whiskers extend to the most extreme non-outliers, red crosses mark points considered outliers. D) The total number of seconds an ROI was active during Ca2+ imaging plotted against the pERK level after fixation, revealing a significant correlation (p=1.1 × 10−66, R = 0.40, Pearson’s correlation, n=1771 ROIs). Plotted are all of the ROIs (grey), the mean in 7-second x-bins (blue), and the linear best-fit line (red). E) Cells exhibiting Ca2+ activity localized by correlation (Online Methods and27), compared their pERK level (green) reveals many instances of good signal correspondence (arrows), as well as potential false positives (asterisks) and false negatives (arrowheads). F) MAP-Map from exposure to 8.25mM pentylenetetrazol (PTZ) for 15 min. n=12/12 (PTZ/ no drug controls). Subpallium (SP), hypothalamus (Hy) and area postrema (AP). Smaller brain insets depict the mean signal within each Z-Brain region in this and other MAP-Map panels G) MAP-Map from exposure to 15mM MS-222 for 1hr. Telencephalon (Te) and preoptic area (PO), olfactory epithelium (OE), caudal hindbrain (cHB) and hypothalamus (Hy) n=12/12 (MS-222/no drug controls). H) MAP-Map from a 10 second light pulse delivered 30 seconds before fixation. n=15/16 (light pulse/darkness controls). Subpallium (SP), tectal neuropil (TN), cerebellum (CB) and hindbrain (HB). I) Mean pERK level (+/− SEM) in retinal arborization fields (AFs, 1–10) for dark adapted larvae (red), and at different chase-times after delivery of a 10 second light pulse (red), and J) resultant MAP-Maps. n = 8/13/12/13/13/13/13 (10sec/1min/2min/5min/10min/30min/Darkness). Scale bars represent 50um.
To determine how well pERK reports activity, we performed calcium imaging on fish with pan-neuronal GCaMP5G expression2, which were stimulated with forward moving gratings for 15–30min. Such stimulation is known to activate specific sets of midbrain and hindbrain neurons28,29. Fish were then quickly fixed and stained. The imaging plane was re-identified based on the Tg(Elavl3:GCaMP5G) transgene by morphing the live imaging data into the post-fixation confocal stack (Supplementary Fig. 2). Comparing the Ca2+ activity of a cell with its pERK level revealed a significant correlation (Fig. 2d), which was consistent across experiments (p<1.5×10−12 in 4 fish). In a different analysis of the same data, active neurons were highlighted based on the correlation among their pixels27. This analysis revealed clear overlap between high pERK levels and active neurons (Fig. 2e). Not all active neurons identified with 2-photon calcium imaging showed high pERK levels, hinting at either cell-type differences in pERK activation, or a non-linear relationship between neural activity and pERK staining not revealed by our experiments/analysis. There were also neurons with high pERK levels that were not active during Ca2+ imaging. These “false positives” may reflect neurons that were active outside of the imaging window (e.g. signals created during agarose dis-embedding), neurons whose activity is not detectable by or do not express GCaMP5G, or perhaps ERK signaling independent of neural activity30. We then analyzed the discriminability of pERK for active and inactive cells using the Receiver Operator Characteristic (ROC) (Supplementary Fig. 3a,b). This confirmed that pERK performs substantially better than a random model, and that the discriminability of the pERK indicator increases with increasing levels of activity. Finally, we observed strong pERK signals in glutamatergic, GABAergic, and glycinergic neurons (Supplementary Fig. 3c,d, Fig. 3f), indicating that none of the major cell classes is refractory to pERK activation. From these experiments we conclude that pERK levels increase within many active larval zebrafish neurons, and therefore can be used as a reporter of activity in larval zebrafish neurons.
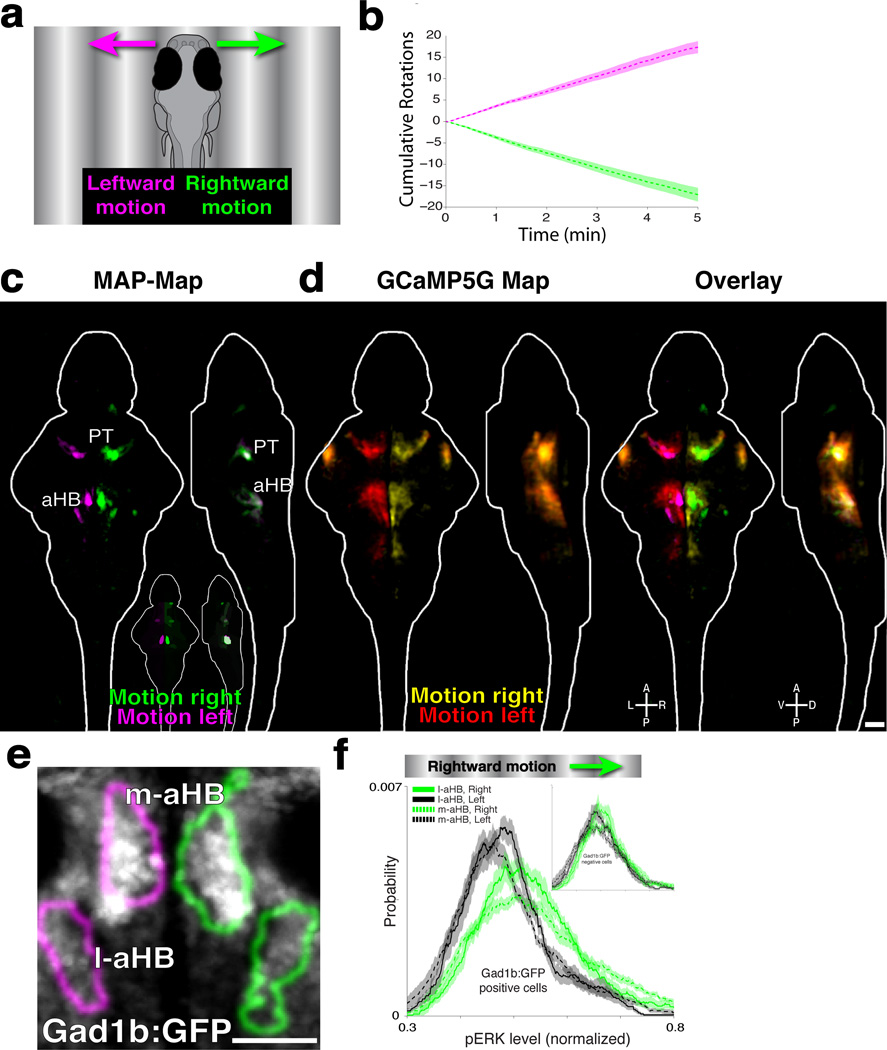
Figure 3. Neural activity underling the optomotor response.
A) Larvae were presented with gratings moving to the right (green) or left (magenta). B) This induces the optomotor response (OMR) and turning in the direction of motion (n = 18 fish per group, mean +/− SEM). C) MAP-Map highlighting the differential activity of fish presented in B, revealing increased activity for rightward (green) and leftward (magenta) motion. Pretectum (PT) and anterior hindbrain (aHB). (n = 17/18, right/left). D) Two-photon GCaMP5G Ca2+ imaging data from 17 fish stimulated with moving gratings were registered into the Z-Brain, and compared to the MAP-Map. Shown are Z and X projections. E) Virtual colocalization analysis to Z-Brain labels, comparing the OMR-induced activity in the medial-aHB (m-aHB) and lateral-aHB (l-aHB) to the Tg(Gad1b:GFP) label. The MAP-Map activity patterns are shown as outlines of the activated areas. F) Tg(Gad1B:GFP) fish were presented with gratings moving to the right, to compare the pERK level within the Gad1B-positive cells in the m-aHB and l-aHB. Shown are pERK level probability histograms, revealing significantly increased pERK levels on the right side of the brain (p=4.3×10−25, and p=8.2×10−15 for the m-aHB and l-aHB respectively, ranksum test, n=8 fish). The inset shows the results for non-GFP labeled cells, which do not show such a strong (although still significant) shift in distribution (p=6.9×10−4, and p=3.6×10−3 for the m-aHB and l-aHB, respectively). Scale bars represent 50um.
Automated whole-brain neural activity mapping
High baseline pERK staining makes finding stimulus- or behavior-dependent changes in staining challenging. Additionally, the potential existence of false-positive signals (those arising in inactive neurons) necessitates careful comparisons to control samples to ensure that only truly activity-dependent signals are highlighted. For this reason we developed analyses to specifically localize sites of stimulus/behavior induced changes in pERK levels (Fig. 1f–h). Accurate registration of pERK- and tERK-stained brains to the Z-Brain using the tERK stain causes voxels belonging to equivalent physical locations in different fish to overlap. Thus, we can localize anatomical areas exhibiting differential activity by determining which voxels show significantly higher (color coded green) or lower (color coded magenta) pERK levels across treatment and control groups. These neural activity MAP-Maps (Mitogen Activated Protein kinase-Maps), are then analyzed using the Z-Brain by determining which anatomical regions exhibit activity and which Z-Brain labels best overlap with the activity signals. (Fig. 1i–j).
As a first test we MAP-Mapped fish treated with the GABAA receptor antagonist pentylenetetrazole (PTZ), which causes strong epileptic activity6,8,9. As expected, this analysis revealed widespread activation (Fig. 2f, Supplementary Video 3, and Data 1). Next we tested if we could find suppressive signals by exposing fish to the sodium-channel blocking anesthetic MS-222. Widespread suppression was seen after MS-222 treatment (Fig. 2g, Supplementary Video 4 and Data 1). This analysis also revealed activation in some parts of the MS-222 treated brain, indicating that MS-222 may act as an olfactory stimulant. These pharmacological experiments indicate that our approach can reveal areas of differential neural activity.
Neural activity underlying two visual behaviors
We then analyzed the response to visual stimuli. Dark-adapted fish were given a 10 second light pulse (Fig. 2h, Supplementary Video 5 and Data 1). As expected, we saw strong signals in the retinal projection fields. Areas of activity were also seen throughout the brain, including diencephalic areas containing Otpb labeled neurons, which may reflect signals arising from Opn4a expressing deep brain photoreceptors31. This light pulse experiment also allowed us to perform a pulse/chase experiment to measure the time course of the pERK signal. Consistent with previous results in rodent neurons16,21, we found that the signals peaked within 2–5 minutes of the stimulus, and returned to baseline within 30 min (Fig. 2I–J).
The next behavior we analyzed was the optomotor response (OMR), where fish follow the direction of motion when stimulated with moving gratings. Freely swimming fish were stimulated with leftward- or rightward-moving gratings, in closed loop, which induces consistent turning in the direction of motion (Fig 3a,b, and28,29). After 5 min of stimulation and behavioral recording, fish were fixed and MAP-Mapped (Fig. 3c, Supplementary Video 6 and Data 1). By comparing fish stimulated with gratings moving to the right vs. left we found symmetrical lateralized activity, with most prominent signals in the pretectum (PT) and in two clusters in the anterior hindbrain (aHB), which we refer to as the medial- and lateral-aHB. To determine how these regions compare with areas found by calcium imaging, we performed 2-photon calcium imaging on Tg(Elavl3:GCaMP5G) head-fixed larvae stimulated with leftward- or rightward-moving gratings. Registration of the Ca2+ activity onto the Z-Brain revealed the same PT and aHB regions as were found by MAP-Mapping (Fig. 3d), thus validating our MAP-Mapping results.
The activity in aHB overlapped with GABAergic cells labeled by Tg(Gad1b:GFP)32, in Gad1b Cluster 1 and Gad1b Stripe 1 of the Z-Brain (Fig. 3e). To confirm this result we performed cellular resolution analysis in Tg(Gad1b:GFP) larvae stimulated with rightward-moving gratings. Quantifying the pERK level in Gad1b:GFP cells revealed significant activation of these cells (Fig. 3f). Gad1b:GFP-negative neurons in these areas were not as strongly activated. These results describe motion responsive activity across the brain, and identify a novel population of hindbrain neurons that may mediate the biased turning behavior induced by whole-field motion.
Neural responses to aversive stimuli
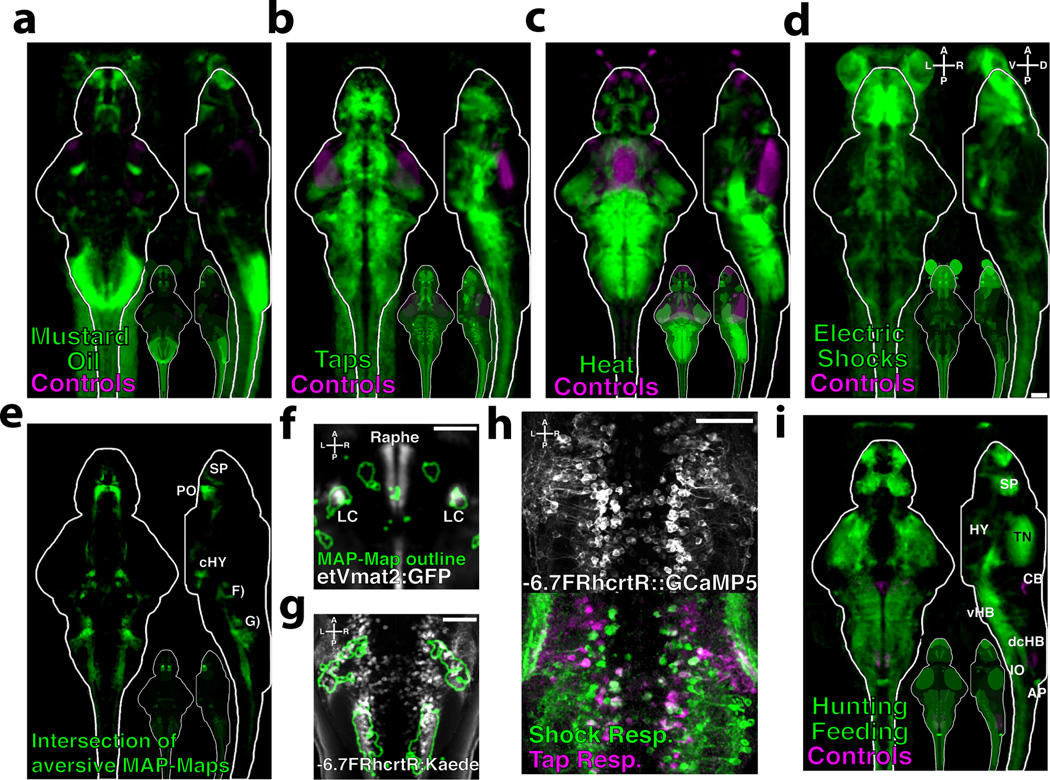
Zebrafish larvae exhibit aversive responses to electric shocks33, acoustic/vibrational stimuli such as dish-taps34, the chemical irritant mustard oil, and noxious heat35. We MAP-Mapped the response to a 15-minute exposure to each of these stimuli (Fig. 4a–d, Supplementary Videos 7–10 and Data 1). Since all four stimuli are aversive, we asked if there were regions in the brain that were co-activated or co-suppressed by all four stimuli (Fig. 4e, Supplementary Video 11 and Data 1). Areas of co-activation where found in all major brain regions, and were present more frequently than would be expected by chance (p<1×10−15, Chi Square Stat = 6.4×105), suggesting that there is a core-network commonly activated by these stimuli. Specifically, Z-Brain analyses revealed that the locus coeruleus was co-activated (Fig. 4f), a region that regulates arousal and is activated by aversive stimuli in mammals36. Co-activation was also seen in the spinal cord and hindbrain, which overlapped with neurons labeled in the Tg(−6.7FRhcrtR:Gal4) line37 (Fig. 4g). These are a grouping of uncharacterized neurons that are likely inhibitory since they lie in a domain containing largely GABAergic or glycinergic neurons, based on the Z-Brain database (data not shown). To confirm this result we performed Ca2+ imaging while delivering tap and shock stimuli (Fig 4h), which confirmed the strong activation of these cells. These experiments reveal activity induced by four aversive stimuli, and genetically identify a population of neurons that may mediate general aversion behavior.
Figure 4. Activity induced by aversive stimuli and hunting/feeding.
Fish were exposed to aversive stimuli for 15 minutes, and MAP-Mapped A) 10uM Mustard oil vs. DMSO controls (n = 19/18 fish) B) Dish-taps vs. no taps controls (n = 28/29 fish) C) 37°C Heat vs. room temperature controls (n = 23/21 fish) D) Electric shocks vs. controls (n = 21/21 fish) E) The intersection of the MAP-Maps in A–D. Subpallium (SP), preoptic area (PO), caudal hypothalamic neural cluster (cHY). F) Z-Brain virtual colocalization reveals co-activation of the locus coeruleus (LC) labeled by Et(Vmat2:GFP), and G) cells labeled by the Tg(−6.7FRhcrtR:gal4VP16;UAS:Kaede) line in the caudal hindbrain H) 2-photon Ca2+ imaging of Tg(−6.7FRhcrtR:gal4VP16);Tg(UAS:GCaMP5G) transgenic larvae stimulated with dish-taps and electric shocks. Functional images depict fluorescence correlation with the stimuli. i) MAP-Map revealing the activity induced by a 1hr of paramecia exposure, mapped relative to a non-fed control group (n = 28/23). Tectal neuropil (TN), hypothalamus (Hy), area postrema (AP), subpallium (Sp), ventral hindbrain (vHB), inferior olive (IO), cerebellum (CB), dorsal-caudal hindbrain (dcHB). Scale bars represent 50um
Neural activity during hunting and feeding
One of the most sophisticated behaviors exhibited by larval zebrafish is their ability to hunt prey. Although zebrafish respond to and engage with small moving prey-like spots when restrained38, the complete behavior including capture events and feeding can only be studied in freely-swimming larvae. To reveal hunting/feeding-dependent activity, zebrafish were allowed to feed on paramecia for 1hr, and MAP-Mapped relative to a non-fed control group (Fig. 5a, Supplementary Video 12 and Data 1).
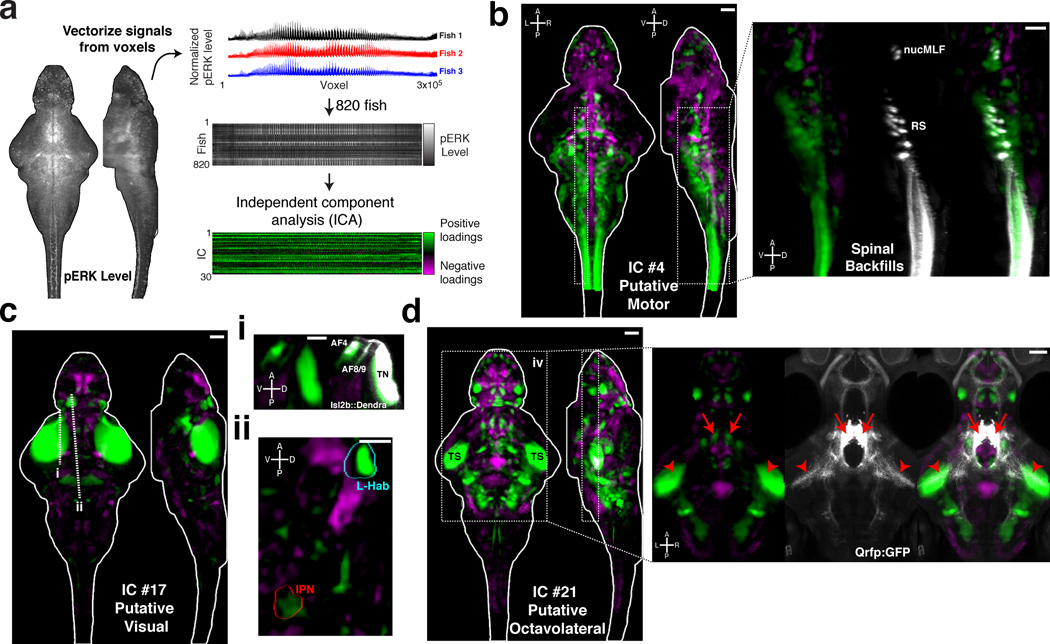
Figure 5. Spatial independent component analysis across fish as a method to localize functional brain networks.
A) The pERK level stack is reshaped into a vector, and the vectors from 820 fish are then combined into an array for independent component analysis (ICA) (see Online Methods) B–D) Voxels for each recovered independent component (IC) are painted with their intensity proportional to the z-score of the loadings of the ICA signal linearly mapped between z = 1–4, and are shown as maximum Z and X projections. B) IC #4 highlights a putative motor network, which associates regions overlapping with reticulospinal neurons (RS), the nucleus of the medial longitudinal fascicle (nucMLF), and the spinal cord. Overlap with the spinal backfill Z-Brain label is shown as an X-projection over the boxed area (right). C) IC #17 highlights a putative visual response network. i) This IC overlaps with areas of the retinal arborization fields (AF) 4, 8 and 9, and the tectal neuropil (TN) labeled by Tg(Isl2b:Gal4);Tg(uas:Dendra) in the Z-Brain. ii) Prominent signals are also observed in the left habenula (L-Hab) and interpeduncular nucleus (IPN). Dashed lines represent the position of the resliced views in ii and iii. D) IC #21 highlights a putative octavolateral network, since it contains prominent signals in the torus semicircularis (TS). Foci of signal in the rostral hypothalamus overlap with the cell bodies of Tg(Qrfp:GFP) labeled neurons (right panel, arrows), which send projections to the TS (arrowheads), implicating these cells in the network.
Despite the highly complex nature of this stimulus and behavior, a well-structured activity map was observed. Z-Brain analyses (Supplementary Fig 4) revealed activation of the tectal neuropil (TN) and the nucMLF MeL/R neurons and surrounding neuropil, which have previously been implicated in prey capture39. We observed particularly strong activation of the area postrema, which, to our knowledge, has not been implicated in feeding behavior in zebrafish, but is a well-known site of feeding regulation in mammals40. We also observed an interesting inhibitory pattern – overlapping with two regions in the dorsal-caudal hindbrain where glycinergic cells reside. The role of this putative dis-inhibition remains to be tested, but serves as an example of the many novel areas now implicated in this complex behavior.
Functional association analyses from pERK stained brains
In total, our experiments provided us with pERK data for 820 fish brains in a variety of pharmacological and behavioral conditions. We wondered if activity patterns across this array might reveal functional associations among brain regions. Inspired by functional connectivity analyses of fMRI and Ca2+ imaging datasets41–43, we explored this idea using (spatial) independent component analysis (ICA). ICA is designed to extract independent signals from linearly mixed observations44. In our context, we sought independent brain-wide activity patterns that combine to generate pERK signals (Fig. 5a). Our analysis yielded intricate anatomically structured maps associating both proximal and distal brain regions (Supplementary Fig. 5). Some maps reflected known anatomical or functional circuitry such as independent component (IC) that recapitulated a zebrafish motor network, including the reticulospinal system (RS), nucMLF, and spinal cord (Fig. 5b). Another IC contained the retinal arborization fields and may represent a visual network (Fig. 5c). This IC also contained prominent signals in the left habenula (L-Hab), and the interpeduncular nucleus (IPN), which are anatomically connected and responsive to light45,46. Structure observed in other ICs lead us to hypothesize novel functional associations in the zebrafish brain. For example, we observed associations between specific forebrain and hindbrain nuclei with the midbrain torus semicircularis (Fig. 5d), which receives octavolateral (lateral line and acoustic) input in teleosts24,47. Close inspection of one nucleus in the rostral hypothalamus revealed its overlap with Tg(Qrfp:GFP)48 expressing neurons. We found that these neurons send prominent projections to the torus semicircularis, anatomically validating this novel functional association between brain areas.
How precisely these ICA maps relates to anatomical and functional circuitry remains unclear, but our results foreshadow how large-scale unsupervised analyses of pERK stained fish might help to unravel the organizational principles of the zebrafish brain. Future work should also investigate how pharmacology and sensory stimuli modulate the independent components characterizing wild type and mutant animals.
Discussion
pERK as a marker of active neurons
pERK is an endogenous sensor that has been used to mark active neurons for over a decade15,16, and can provide improved temporal resolution over IEGs. (Fig. 2i,j, and16,21). Historically, pERK has been less widely used as an activity reporter than IEGs since it generally shows higher baseline staining in unstimulated brains. We have used this perceived weakness as a major advantage: baseline activity provides an opportunity to find areas that become suppressed in comparison to control brains.
Although we have demonstrated that pERK can serve as a very useful activity sensor, its relationship to activity is known to be complex and dependent on the frequency and repetition of neural firing, and perhaps on cell type21,49,50. It is clear that the pERK level is not a perfect representation of neural activity. For example we did not observe high pERK levels in all active neurons (Fig. 2d,e), or OMR/turning induced activity in the reticulospinal system, which has been observed by other methods28,29. This could indicate a negative bias in detecting activity signals from large-diameter neurons. While pERK signals may not capture the entire activity pattern in the brain, it is a powerful method to create cellular activity maps from freely behaving animals and identify candidate regions for further analyses.
Using image registration to create activity maps
By registering brains to a reference brain, MAP-Mapping can automatically detect differences in activity by localizing voxels that change in pERK levels. The power of this anatomically unbiased approach is highlighted by its ability to localize critical sites of action for a wide range of stimulus and behavior conditions, some of which we have validated by Ca2+ imaging.
Registration-based analyses may work better for some cell types than others, depending on the stereotypy of neuron positioning in different animals. For example, the position of the Mauthner and CaD neurons is precise to within 4um, while etVmat2:GFP-labelled neurons in the medial tectum are much more variably positioned. We find less than 20 of these cells randomly distributed within a section of > 5300um2, suggesting that over 90% of their possible territory is occupied by other cell types. A neuron’s positional variability determines over what spatial scale its activity signals are diluted, influencing both signal-to-noise ratio and the resolution of the activity signal. In cases where the position of a class of neurons is highly stereotyped, the resolution can be near-cellular (see Fig 4F). However, as all of the underlying confocal data used to generate MAP-Maps is at cellular resolution, the activated neurons within the MAP-Map signal can be readily identified in individual fish for cellular resolution analyses (Fig. 2b,c, 3f).
Interfacing activity and anatomy in a reference atlas
Measuring activity is only the first step towards describing neural circuitry. Next one needs to put the activity in the context of the neuroanatomy, and ideally identify relevant neurons. Using the Z-Brain atlas as our reference brain provides a precise language to describe regions of activity, allows for direct comparisons across experiments, can identify labels for candidate neurons mediating behavior, and can reveal brain-wide functional connectivity patterns. Such analyses will become even more powerful as more labels and activity maps are accumulated within the Z-Brain.
Our approach in creating the Z-Brain is conceptually similar to that taken for the ViBE-Z atlas23, but uses a simpler imaging setup, and the cross-platform CMTK registration software that can be implemented on a standard computer. Registration quality is accurate to ~1 cell body diameter and has been validated across species25–27, different microscopes, and even imaging modalities; we have successfully registered 2-photon imaging data into the Z-Brain (Fig. 3d), allowing us to directly compare MAP-Mapping, Ca2+ imaging, and anatomical data.
The Z-Brain currently contains 29 labels and 294 segmented regions. Because this many-channel atlas would be cumbersome to visualize using standard visualization software, we built a simple web interface to browse the image data (http://engertlab.fas.harvard.edu/Z-Brain/), and a Matlab-based visualization program that performs multicolor overlays of many channels, rapidly switches between channels, and performs click-to-define interrogation of the many anatomical regions (Supplementary Software). The Z-Brain is an expandable/editable platform which is open source and freely available. New regional definitions can be easily created, facilitating collaborative input and improvement. As any label of interest can be incorporated into the Z-Brain, the zebrafish community can rapidly expand the atlas by contributing additional anatomical or functional stacks.
In summary, our study presents the Z-Brain atlas, and MAP-Mapping as a technique to map neural activity from freely swimming fish. MAP-Mapping has several features that promise wide use in characterizing brain regions and neurons that mediate behavior: it is rapid, high-throughput, low cost and is performed on freely swimming unperturbed wild-type larvae and so can be applied to nearly any stimulus or behavior.
Online Methods
Subjects
Larvae were raised in E3 media, supplemented with 1mM HEPES buffer pH 7.0. Larvae and adult fish we maintained on a 14:10 hr light:dark cycle at 28°C. All protocols and procedures involving zebrafish were approved by the Harvard University/Faculty of Arts & Sciences Standing Committee on the Use of Animals in Research and Teaching (IACUC). For MAP-Mapping experiments, the fish to be compared were usually all siblings from the same clutch, grown at a density ~0.5 fish/mL, and distributed randomly into treatment groups. In the few cases where a single clutch did not yield sufficient numbers, 2 clutches were pooled and then distributed randomly to avoid any clutch specific effects. Transgenic fish used in this study (Supplementary Table 1) have been previously described2,32,37,48,51–64, with the exception of Tg(Elavl3:H2B-RFP). The transgenic construct was made by cloning H2B-RFP downstream of an AttR1-R2 cassette flanked by Tol2 arms, and then placing it under the control of elavl3 (HuC) cis-regulatory sequences via L/R recombination (Gateway cloning) with an AttL flanked elavl3 entry clone2.
Immunohistochemistry
Since pERK is a fast indicator, fish need to be fixed as rapidly and consistently as possible across treatment groups. To do this, we quickly collect the fish by funneling the plate through a fine-mesh sieve, and the sieve is then immediately dropped into 4% paraformaldehyde (PFA) in PBS + 0.25% Triton (PBT). Fish are then immunostained using standard procedures. Briefly, fish are washed in PBT, incubated in 150mM Tris-HCl ph9 for 15min at 70°C65, washed in PBT, permeabilized in 0.05% Trypsin-EDTA for 45min on ice, washed in PBT, blocked in PBT + 1% bovine serum albumin (BSA) + 2% normal goat serum (NGS) + 1% dimethyl sulfoxide (DMSO), and then incubated in primary and secondary antibodies overnight at 4°C in PBT + 1%BSA + 1%DMSO. The pERK antibody (Cell Signaling, #4370), and tERK antibody (Cell Signaling, #4696) were used 1:500. Other primary antibodies and dilutions can be found in Supplementary Table 1. Secondary antibodies conjugated with alexa-fluorophores (Life Technologies) were diluted 1:500.
Confocal Imaging
Imaging of stained tissue was performed using an upright confocal microscope (either Olympus FV1000, Zeiss LSM710 or LSM780), using a 20× 1.0NA water dipping objective. Fish are mounted dorsal-up in 2% (w/v) low melting agarose (Invitrogen) and imaged at ~0.8/0.8/2um voxel size (x/y/z). To cover the entire brain two imaging tiles are acquired, and stitched together using the ‘Pairwise Stitching’ plugin in FIJI66. All fish to be analyzed in a MAP-Mapping experiment were mounted together on a single imaging dish, and imaged in one run to ensure they were imaged with near-identical imaging conditions. As all imaging was performed from the dorsal aspect, image intensity and quality is somewhat degraded in the more ventral regions of the brain.
Two-photon calcium imaging
For Ca2+ imaging measurements and quantitative comparison with pERK levels (Fig 2D,E), 6dpf Tg(Elavl3:GCaMP5G) larvae were anesthetized in MS-222 and mounted in 2% low melting point agarose, and an anatomy stack acquired under anesthesia at 1×1×2um voxel size. The anesthetic was removed and fish were imaged in a single plane at 2Hz using a custom 2-photon microscope29,67, while presenting forward-moving gratings to activate neurons in the hindbrain. ROIs in the cell-body regions were generated using the watershed algorithm in Matlab. Activity within the ROIs was detected as events when the fluorescence Z-trace was above a threshold of 2, after applying a 1×5 moving average filter in time. The total number of frames above threshold was considered to be the total time that neuron was active during imaging. After imaging, the fish were rapidly removed from the microscope, dissected out of the agarose and fixed in 4%PFA. After staining for pERK and tERK, confocal imaging stacks were obtained of the pERK level and the residual GCaMP5G expression. This GCaMP expression was used for registering the fixed data into the anatomy stack acquired live (Supplementary Fig. 2). The correlation between total firing time and pERK level within each ROI was calculated using MATLAB’s ‘corr’ function. To visually highlight active neurons, the correlation between each pixel’s time series and the summed signal an approximately cell-sized (15×15 px) square surrounding that pixel was calculated (similar to:27), and overlayed with the pERK level in the registered imaging plane. The Receiver Operator Characteriscit (ROC) analysis and the area under the curve calculations were performed using Matlab’s ‘roc’ and ‘perfcurve’ functions.
For neural activity measurements related to routine turns evoked by leftward or rightward moving sinusoidal gratings (Fig 3D), 6dpf Tg(Elavl3:GCaMP5G) larvae were paralyzed in α-bungarotoxin (1mg/ ml for ~60 s, Invitrogen, USA) in E3 medium and embedded in 1.2% low melting point agarose (Invitrogen, USA). Two-photon image stacks were acquired with excitation pulsed Ti-sapphire laser tuned to 920 nm (Spectra Physics, USA), while a flickering stimulus was projected from below with a DLP projector, which allowed simultaneous visual stimulation and detection of green fluorescence. In each experiment, images were acquired at 2.75 Hz, minimum 4 repetitions per stimulus set and z-plane. The resulting image time and depth series were analyzed in Matlab to extract the positional origin of functional responses to left and rightward motion. Directional activity maps were generated by calculating the average fluorescence change during stimulus presentation in one direction divided by the average fluorescence change in the opposite direction, at each voxel. Data from 17 fish imaged in the vicinity of the pretectum and anterior hindbrain (between 2 and 39 imaging planes per fish, at 3um spacing) were registered to the Z-Brain. The activity maps from all fish were combined by calculating the average activity signal per pixel, and smoothed using the ‘Median_3D’ filter in FIJI (x,y,z = 3,3,2 pixels).
For tap and shock stimulation experiments (Fig 4H), 6dpf, Tg(−6.7FRhcrtR:gal4VP16);Tg(uas:GCaMP5G)37 larvae were paralyzed in α-bungarotoxin and mounted in 2% LMP agarose. Three repetitions of alternating tap and shock stimuli were given in 30sec intervals, repeated in z-planes to cover the majority of the hindbrain at 3um z-steps. Taps were delivered by striking the imaging platform with a solenoid (Guardian Electric 28P-I-12D). Electric shocks were 100ms pulses of 15V, delivered over a 6cm dish, with anode and cathode aligned to the head and tail of the fish to maximize responses33. Activity maps were created by correlating the signal in each voxel with a vector representing the stimulus delivery convolved with an exponential decay kernel approximating the GCaMP5G decay rate (half-life = 0.667s)68, and calculating the maximum intensity Z projection over 17 imaging planes (51um).
ChR2 activation
Stimulated fish were 6dpf, Tg(−6.7FRhcrtR:gal4VP16);Tg(14xUAS-E1b:hChR2(H134R)-EYFP); atoh7th241/th241; Tg(atoh7:GAP-RFP)37,69,70. Blind atoh7 mutant fish were used to prevent retinal-driven neural responses to light, and were screened for a lack of reginal ganglion cells using the Tg(atoh7:GAP-RFP) label. Fish were first paralyzed with 1uM bungarotoxin (Invitrogen), and anesthetized with MS-222 for 30min to minimize background staining. Anesthetic solution was washed away and fish were stimulated for 5 min with either blue or green light from an arc lamp under a fluorescent dissecting scope followed by immediate fixation in 4% PFA in PBT.
Behavioral Stimuli
Light Flash: larvae were dark adapted by covering dishes with tinfoil-covered boxes for 3–4 hours. Light stimuli were delivered for 10 seconds using a fluorescent light source suspended above the dish, yielding a light level of ~2500 lux, and then fish were returned to darkness.
Optomotor stimuli: Fish were stimulated and recorded using the apparatus described previously29. The fish were stimulated for 5 minutes with sinusoidal light/dark gratings moving either to the right or left. The stimulus is presented in closed loop, such that the stimulus always moves in the same direction relative to the long axis of the fish. Fish were then quickly caught using a pipette, and fixed in 4% PFA for MAP-Mapping.
Aversive stimuli: In 85mm petri dishes, fish were exposed to mustard oil (10mM) or DMSO control (0.1%). For taps, the petri dish was placed on an acrylic platform, in a dark box. After 30min of dark-adaptation, the platform was struck with a solenoid (Guardian Electric 28P-I-12D) driven at 5V using a Teensy 2.0 microcontroller (https://www.pjrc.com/teensy/), which produces short and long-latency escape responses (data not shown). Taps were delivered every 10 seconds for 15 min. Controls were placed in the same dark box, but beside the tapping platform, and isolated from the tap stimuli. For heat stimulation fish were put in 45 mL of E3, in 50mL falcon tubes, and submerged into a 37°C water bath for 15min. The control falcon tube was placed beside the water bath. Finally, for shock experiments fish were placed in a 6cm petri dish, over which a 100msec, 15 volt (2.5 V/cm) shock was delivered every 10 seconds for 15 minutes. Current was delivered using alligator clips and the apparatus described previously71. Controls were placed beside the shocked dish, affixed with alligator clips, but not connected to the circuit.
Feeding/Hunting: An 85mm petri dish, containing the 6dpf fish that had not previously been fed, was inoculated with filtered paramecia. Fish were left to hunt/feed for 1hr (feeding was confirmed visually), and then fixed in PFA. Controls were inoculated with water over the same time period.
Image registration
Non-rigid image registration was performed using the Computational Morphometry Toolkit (CMTK, http://www.nitrc.org/projects/cmtk/), and user interface written by Jefferis and colleagues72 with the command string (-awr 010203 -T 8 -X 52 -C 8 -G 80 -R 3 -A '--accuracy 0.4' -W '--accuracy 1.6'). This setup performed registrations at <15min/fish on a 2 × 3.2 GHz Quad-Core Mac Pro (Apple). Our template brain is a 6dpf nacre mutant (mifta−/−) larvae73, stained with anti-tERK. Staining the fish to be registered with anti-tERK allows for direct registrations to the template. However, in cases where another anti-Mouse IgG1 antibody was used as a cell-type label, anti-tERK could not be easily co-stained. In these cases fish were indirectly registered into the template brain by staining with anti-Syt2/Znp1 and using the anti-Znp1 mean-stack as a template. To register the live 2-photon calcium imaging data during right/left OMR stimulation (Fig. 3D) we used a single registered Tg(Elavl3:GCaMP5G) fish as the template, and command string (-awr 010203 -T 2 -X 52 -C 8 -G 80 -R 4 -A '--accuracy 0.4' -W '--accuracy 0.8'). To register in the live-imaging 2-photon data of Tg(Elavl3:GCaMP5G) fish and the post-fixation data from the same fish to re-identify cells (Supplementary Fig. 2), the GCaMP5G channel imaged by confocal microscopy after fixation was registered into the anatomy stack taken live by 2-photon microscope was registered using command string (-awr 010203 -T 4 -X 200 -C 4 -G 160 -R 5 -A '--accuracy 0.4' -W '--accuracy 0.4')
Z-Brain creation
The mean-stack labels in the Z-Brain were created in Matlab by calculating the mean value across all registered fish stained with the given label, and normalizing to the maximum intensity value. Voxels that were not imaged in any given fish (for example, due to differential mounting or imaging coverage) were excluded from the mean calculation. These stacks were then integrated into a single HDF5 file: ‘AnatomyLabelDatabase.hdf5’.
The Z-Brain regions were drawn manually using either the ‘Segmentation Editor’ plugin in FIJI, or using our ‘ZBrainViewer.m’ Matlab function, which utilizes the ‘roipoly’ tool to draw 2-dimensional ROIs. In either case, 2D ROIs are drawn at every ~3 Z planes to define the outline of a region. These ROIs are then interpolated in the z-dimension, and then smoothed by convolution with an ~8um diameter sphere to create the 3D ROI volume defining an anatomical region. Regional mask definitions are contained in the file: ‘MaskDatabase.mat’. Finally, we also ranked all labels for their signal within each anatomical region, both for signal enrichment over local surrounding (Supplementary Fig. 12A) and mean signal within the region (Supplementary Fig. 12B), providing a quantitative anatomical description of each region.
When segmenting the Z-Brain we drew the regions at high resolution (maximally at cellular resolution). As a consequence, the Z-Brain contains extensive overlap between regions. This is in contrast to most neuroanatomical atlases, which contain exclusive boundaries (for example, Drosophila: http://www.virtualflybrain.org/, Mouse: http://atlas.brain-map.org/, Zebrafish:23,24). Even when looking within a single brain there is clear overlap between groupings of neurons labeled by different markers (Supplementary Fig. 1a), and there is substantial biological noise in the position of some neuron types across brains. For these reasons, we felt that allowing for overlap was a more accurate representation than the forcing of hard borders. Ideally, a probabilistic approach to regional definition74, should be developed for future versions of the Z-Brain, and for other high-resolution reference atlases. These segmentation efforts should also be computationally driven, rather than relying on hand-drawn regions. This should be possible using the staining statistics across the many Z-Brain labels, and could even incorporate activity information through MAP-Maps, Ca2+ imaging, and functional connectivity.
MAP-Map calculation
Analysis was performed using FIJI/ImageJ75 and Matlab (Mathworks, USA). To create MAP-Maps, post-warping pERK/tERK stacks are downsampled to x/y/z = 300/679/80 resolution, and smoothed with a 2D Gaussian filter (sigma = 2) using FIJI. This is done using the macro ‘PrepareStacksForMAPMapping.ijm’. In Matlab, the Mann-Whitney U statistic Z score is calculated for each voxel, comparing between the two groups. The significance threshold is set using an FDR-based method, where 0.005% of control pixels would be called as significant. This is done by creating a simulated control distribution of Z-scores using randomization of the voxel data into pseudo-groups over 500 iterations, and calculating the FDR threshold value. If the absolute value of the treatment vs. control Z-score is larger than this value it is considered significantly different across the two groups, and the pixel is assigned an intensity equal to the difference in the median values of the groups (0:0.5 deta-median maps to 0:65535 intensity values). The voxel is color coded according to sign (green = higher and magenta = lower, in the treatment group). This is analysis can be performed by running the ‘MakeTheMAPMap.m’ Matlab function. For the projection images shown in the figure panels the image lookup table was scaled linearly so as the saturation point is 60% of the maximum pixel intensity across the projection image. Un-manipulated MAP-Maps were used for all quantitative analyses and can be downloaded from the website.
The intersection of aversive MAP-Maps (Fig 4E) was calculated by identifying the voxels that were significantly activated or suppressed across all four individual MAP-Maps (Fig 4 A–D). The number of active voxels found was compared to chance levels of overlap using the Chi-Square statistic (Matlab).
Analyzing MAP-Maps with the Z-Brain atlas
To quantify the amount of MAP-Map signal in the Z-Brain regions the mean signal in each anatomical region is calculated, and the regions are ranked from highest to lowest average signal. This is done separately for the activation (positive) and suppression (negative) signals. In the regions that exhibit activity, we ask which Z-Brain label shows the most overlap with this signal by calculating the mean signal in the active voxels divided by the mean signal in the 50-voxels surrounding the region. Therefore, numbers greater than 1 indicate relative signal enrichment in the active voxels. The top five candidate cell type labels are then listed, along with their enrichment signals. This analysis can be performed by running the ‘ZBrainAnalysisOfMAPMaps.m’ Matlab function.
Independent component analysis
For independent component analysis (ICA) across fish, the registered tERK and pERK stack for each fish were downsampled to x/y/z = 300/679/80 resolution, and smoothed with a 2D Gaussian filter (sigma = 2), in the same way as is done for MAP-Mapping. The central brain (not including eyes, ganglia, or olfactory epithelia) from each fish was then downsampled into 4.7um3 sized voxels, yielding a pERK level vector for each fish. Fish in which any of the voxels was not imaged (due to incomplete coverage) were excluded from the analysis. Fish were normalized for overall brightness by dividing by the 10th percentile intensity value, and voxels normalized by subtracting the mean value across fish. The fish-by-voxel array was then analyzed for spatially independent components using FastICA (http://research.ics.aalto.fi/ica/fastica/, Version 2.5), treating each fish as a signal and each voxel as sample, using the symmetric approach, ‘pow3’ nonlinearity, retaining the first 30 principle components and calculating 30 independent components. Spatial (rather than temporal) ICA was chosen for three reasons: 1) the relatively slow nature of the pERK activity indicator, which will limit the temporal separation of signals, 2) the complex nature of the stimuli and experiences of the individual fish, which may not yield strictly temporally independent signals, and 3) the number of voxels dwarfs the number of observations (fish) in the dataset76. Independent component (IC) maps are displayed as the z-score values of the IC signals. Analyzed fish included those given no specific stimulus: control fish from all MAP-Maps presented here (Fig 2–4), as well as additional fish treated with vehicle controls in other experiments not presented here, fish sampled throughout different points of the day and night, and fish given one of various stimuli, including ‘treatment’ fish from all MAP-Maps (Fig 2–5), as well as additional fish stimulated with electric shocks, light flashes, moving gratings, heat, mustard oil, melatonin, clonidine, nicotine, cocaine, ethanol and d-amphetamine.
Code access and supplementary software Instructions
Analysis code is can be downloaded from the project website (http://engertlab.fas.harvard.edu/Z-Brain/). For reporting bugs and issues, please use the project github repository (https://github.com/owenrandlett/Z-Brain).
For running the Z-Brain viewer, ensure that the ‘MaskDatabase.mat’, ‘AnatomyLabelDatabase.hdf5’, and ‘ZBrainViewer.m’ files are in a folder contained in your Matlab path, and then run the function ‘ZBrainViewer.m’. Instructions on interacting with the data will be displayed on the screen.
Detailed instructions on the ZBrainViewer visualization options, as well as instructions on how to use the main MAP-Map creating and analysis functions (‘MakeTheMAPMap.m’, and ‘ZBrainAnalysisOfMAPMaps.m’) are commented as headers of the functions.
Supplementary Material
Acknowledgements
We are grateful to M. Wullimann (Ludwig Maximilian University) for his critical and detailed input regarding the identification of regions for the Z-Brain segmentation, to Y. Yoshihara (RIKEN Brain Science Institute) and T. Okuyama (University of Tokyo) for pointing us to the pERK antibody, to G. Jefferis (MRC Laboratory of Molecular Biology) for help with brain registrations, to M. Nikitchenko (Harvard) for help with computational and web resources, to A. Douglass (University of Utah) and J. Wortzman (Harvard) for creation of the Tg(UAS:GCaMP5G) line, to D. Prober (Caltech) for sharing the Tg(Hcrt:mRFP) and Tg(Qrfp:GFP) lines prior to publication, to M. Hasemeyer (Harvard) for many helpful discussions, and to the many members of the zebrafish community who shared their transgenic fish lines. Funding was provided by an HFSP Long-Term fellowship LT000772/2012-L (O.R.), the Agency for Science, Technology and Research, Singapore (C.L.W.), a Marie Curie Fellowship (E.A.N), the Swartz Foundation (J.E.F), NIH grants R01 HL109525 and U01 MH105960 (A.F.S.), R24 NS086601 and U01 NS090449 (F.E.).
Footnotes
Author Contributions
O.R., F.E. and A.F.S. conceived of the project. O.R. performed most experiments and data analysis. C.L.W., E.A.N., D.S. and A.L. also performed experiments. E.A.N., J.E.F. and R.P. also analyzed data. D.S. and C.R. created novel transgenic fish strains. O.N. built the website. O.R., F.E. and A.F.S. wrote the paper, with input from all authors. F.E. and A.F.S. supervised and contributed equally to the project.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Fero K, Yokogawa T, Burgess HA. Neuromethods. Vol. 52. Humana Press; 2010. pp. 249–291. [Google Scholar]
- 2.Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Meth. 2013;10:413–420. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- 3.Panier T, et al. Fast functional imaging of multiple brain regions in intact zebrafish larvae using selective plane illumination microscopy. Front Neural Circuits. 2013;7:65. doi: 10.3389/fncir.2013.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fosque BF, et al. Labeling of active neural circuits in vivo with designed calcium integrators. Science. 2015;347:755–760. doi: 10.1126/science.1260922. [DOI] [PubMed] [Google Scholar]
- 5.Nava SS, An S, Hamil T. Visual detection of UV cues by adult zebrafish (Danio rerio) Journal of Vision. 2011;11:2–2. doi: 10.1167/11.6.2. [DOI] [PubMed] [Google Scholar]
- 6.Naumann EA, Kampff AR, Prober DA, Schier AF, Engert F. Monitoring neural activity with bioluminescence during natural behavior. Nat Neurosci. 2010;13:513–520. doi: 10.1038/nn.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guzowski JF, et al. Mapping behaviorally relevant neural circuits with immediate-early gene expression. Curr. Opin. Neurobiol. 2005;15:599–606. doi: 10.1016/j.conb.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 8.Baraban SC, Taylor MR, Castro PA, Baier H. Pentylenetetrazole induced changes in zebrafish behavior, neural activity and c-fos expression. Neuroscience. 2005;131:759–768. doi: 10.1016/j.neuroscience.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 9.Ellis LD, Seibert J, Soanes KH. Distinct models of induced hyperactivity in zebrafish larvae. Brain Research. 2012;1449:46–59. doi: 10.1016/j.brainres.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 10.Okuyama T, et al. Induction of c-fos transcription in the medaka brain (Oryzias latipes) in response to mating stimuli. Biochemical and Biophysical Research Communications. 2011;404:453–457. doi: 10.1016/j.bbrc.2010.11.143. [DOI] [PubMed] [Google Scholar]
- 11.Xiu J, et al. Visualizing an emotional valence map in the limbic forebrain by TAI-FISH. Nat Neurosci. 2014 doi: 10.1038/nn.3813. [DOI] [PubMed] [Google Scholar]
- 12.Hussain A, et al. High-affinity olfactory receptor for the death-associated odor cadaverine. Proceedings of the National Academy of Sciences. 2013;110:19579–19584. doi: 10.1073/pnas.1318596110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovacs K. Measurement of Immediate-Early Gene Activation-c-fos and Beyond. Journal of neuroendocrinology. 2008 doi: 10.1111/j.1365-2826.2008.01734.x. [DOI] [PubMed] [Google Scholar]
- 14.Morgan JI, Cohen DR, Hempstead JL, Curran T. Mapping patterns of c-fos expression in the central nervous system after seizure. Science. 1987;237:192–197. doi: 10.1126/science.3037702. [DOI] [PubMed] [Google Scholar]
- 15.Cancedda L, et al. Patterned vision causes CRE-mediated gene expression in the visual cortex through PKA and ERK. J. Neurosci. 2003;23:7012–7020. doi: 10.1523/JNEUROSCI.23-18-07012.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji RR, Baba H, Brenner GJ, Woolf CJ. Nociceptive-specific activation of ERK in spinal neurons contributes to pain hypersensitivity. Nat Neurosci. 1999;2:1114–1119. doi: 10.1038/16040. [DOI] [PubMed] [Google Scholar]
- 17.Xia Z, Dudek H, Miranti CK, Greenberg ME. Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. The Journal of neuroscience. 1996;16:5425–5436. doi: 10.1523/JNEUROSCI.16-17-05425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosen LB, Ginty DD, Weber MJ, Greenberg ME. Membrane depolarization and calcium influx stimulate MEK and MAP kinase via activation of Ras. Neuron. 1994;12:1207–1221. doi: 10.1016/0896-6273(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 19.Thomas GM, Huganir RL. MAPK cascade signalling and synaptic plasticity. Nat Rev Neurosci. 2004;5:173–183. doi: 10.1038/nrn1346. [DOI] [PubMed] [Google Scholar]
- 20.Itoh M, Yamamoto T, Nakajima Y, Hatta K. Multistepped optogenetics connects neurons and behavior. Current Biology. 2014;24:R1155–R1156. doi: 10.1016/j.cub.2014.10.065. [DOI] [PubMed] [Google Scholar]
- 21.Dai Y, et al. Phosphorylation of extracellular signal-regulated kinase in primary afferent neurons by noxious stimuli and its involvement in peripheral sensitization. J. Neurosci. 2002;22:7737–7745. doi: 10.1523/JNEUROSCI.22-17-07737.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arrenberg AB, Driever W. Integrating anatomy and function for zebrafish circuit analysis. Front Neural Circuits. 2013;7:1–9. doi: 10.3389/fncir.2013.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronneberger O, et al. ViBE-Z: a framework for 3D virtual colocalization analysis in zebrafish larval brains. Nat Meth. 2012;9:735–742. doi: 10.1038/nmeth.2076. [DOI] [PubMed] [Google Scholar]
- 24.Mueller T, Wullimann MF. Atlas of Early Zebrafish Brain Development. 2005 doi: 10.1016/s0165-3806(02)00583-7. [DOI] [PubMed] [Google Scholar]
- 25.Rohlfing T, Maurer CR. Nonrigid image registration in shared-memory multiprocessor environments with application to brains, breasts, and bees. IEEE Trans. Inform. Technol. Biomed. 2003;7:16–25. doi: 10.1109/titb.2003.808506. [DOI] [PubMed] [Google Scholar]
- 26.Jefferis GSXE, et al. Comprehensive Maps of Drosophila Higher Olfactory Centers: Spatially Segregated Fruit and Pheromone Representation. Cell. 2007;128:1187–1203. doi: 10.1016/j.cell.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Portugues R, Feierstein CE, Engert F, Orger MB. Whole-Brain Activity Maps Reveal Stereotyped, Distributed Networks for Visuomotor Behavior. Neuron. 2014;81:1328–1343. doi: 10.1016/j.neuron.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orger MB, Kampff AR, Severi KE, Bollmann JH, Engert F. Control of visually guided behavior by distinct populations of spinal projection neurons. Nat Neurosci. 2008;11:327–333. doi: 10.1038/nn2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang K-H, Ahrens MB, Dunn TW, Engert F. Spinal projection neurons control turning behaviors in zebrafish. Curr. Biol. 2013;23:1566–1573. doi: 10.1016/j.cub.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearson G, et al. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 31.Fernandes AM, et al. Deep Brain Photoreceptors Control Light-Seeking Behavior in Zebrafish Larvae. Current Biology. 2012:1–6. doi: 10.1016/j.cub.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Satou C, et al. Transgenic tools to characterize neuronal properties of discrete populations of zebrafish neurons. Development. 2013;140:3927–3931. doi: 10.1242/dev.099531. [DOI] [PubMed] [Google Scholar]
- 33.Tabor KM, et al. Direct activation of the Mauthner cell by electric field pulses drives ultra-rapid escape responses. Journal of Neurophysiology. 2014 doi: 10.1152/jn.00228.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimmel CB, Patterson J, Kimmel RO. The development and behavioral characteristics of the startle response in the zebra fish. Dev Psychobiol. 1974;7:47–60. doi: 10.1002/dev.420070109. [DOI] [PubMed] [Google Scholar]
- 35.Prober DA, et al. Zebrafish TRPA1 Channels Are Required for Chemosensation But Not for Thermosensation or Mechanosensory Hair Cell Function. J. Neurosci. 2008;28:10102–10110. doi: 10.1523/JNEUROSCI.2740-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amaral DG, Sinnamon HM. The locus coeruleus: neurobiology of a central noradrenergic nucleus. Progress in neurobiology. 1977;9:147–196. doi: 10.1016/0301-0082(77)90016-8. [DOI] [PubMed] [Google Scholar]
- 37.Lacoste AMB, et al. A Convergent and Essential Interneuron Pathway for Mauthner-Cell-Mediated Escapes. Current Biology. 2015:1–10. doi: 10.1016/j.cub.2015.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bianco IH, Kampff AR, Engert F. Prey capture behavior evoked by simple visual stimuli in larval zebrafish. Front Syst Neurosci. 2011;5:101. doi: 10.3389/fnsys.2011.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gahtan E. Visual Prey Capture in Larval Zebrafish Is Controlled by Identified Reticulospinal Neurons Downstream of the Tectum. J. Neurosci. 2005;25:9294–9303. doi: 10.1523/JNEUROSCI.2678-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edwards GL, Ritter RC. Ablation of the area postrema causes exaggerated consumption of preferred foods in the rat. Brain Research. 1981;216:265–276. doi: 10.1016/0006-8993(81)90129-3. [DOI] [PubMed] [Google Scholar]
- 41.van de Ven VG, Formisano E, Prvulovic D, Roeder CH, Linden DEJ. Functional connectivity as revealed by spatial independent component analysis of fMRI measurements during rest. Hum. Brain Mapp. 2004;22:165–178. doi: 10.1002/hbm.20022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vladimirov N, et al. mapping brain activity at scale with cluster computing. Nat Meth. 2014:1–17. doi: 10.1038/nmeth.3041. [DOI] [PubMed] [Google Scholar]
- 43.McKeown MJ, et al. Analysis of fMRI data by blind separation into independent spatial components. Hum. Brain Mapp. 1998;6:160–188. doi: 10.1002/(SICI)1097-0193(1998)6:3<160::AID-HBM5>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hyvärinen A, Oja E. Independent component analysis: algorithms and applications. Neural Netw. 2000;13:411–430. doi: 10.1016/s0893-6080(00)00026-5. [DOI] [PubMed] [Google Scholar]
- 45.Dreosti E, Llopis NV, Carl M, Yaksi E, Wilson SW. Left-Right Asymmetry Is Required for the Habenulae to Respondto Both Visual and Olfactory Stimuli. Current Biology. 2014:1–6. doi: 10.1016/j.cub.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bianco IH, Wilson SW. The habenular nuclei: a conserved asymmetric relay station in the vertebrate brain. Philosophical Transactions of the Royal Society. B. Biological Sciences. 2009;364:1005–1020. doi: 10.1098/rstb.2008.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wullimann MF, Rupp B, Reichert H. Neuroanatomy of the Zebrafish Brain. Birkhauser; 1996. [DOI] [PubMed] [Google Scholar]
- 48.Liu J, et al. Evolutionarily conserved regulation of hypocretin neuron specification by Lhx9. Development. 2015 doi: 10.1242/dev.117424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu GY, Deisseroth K, Tsien RW. Spaced stimuli stabilize MAPK pathway activation and its effects on dendritic morphology. Nat Neurosci. 2001;4:151–158. doi: 10.1038/83976. [DOI] [PubMed] [Google Scholar]
- 50.Ha S, Redmond L. ERK mediates activity dependent neuronal complexity via sustained activity and CREB-mediated signaling. Dev Neurobiol. 2008;68:1565–1579. doi: 10.1002/dneu.20682. [DOI] [PubMed] [Google Scholar]
Methods-only references
- 51.Scott EK, et al. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Meth. 2007 doi: 10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- 52.Wen L, et al. Visualization of monoaminergic neurons and neurotoxicity of MPTP in live transgenic zebrafish. Developmental Biology. 2008;314:84–92. doi: 10.1016/j.ydbio.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 53.McLean DL, Fan J, Higashijima S-I, Hale ME, Fetcho JR. A topographic map of recruitment in spinal cord. Nature. 2007;446:71–75. doi: 10.1038/nature05588. [DOI] [PubMed] [Google Scholar]
- 54.Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J. Neurosci. 2000;20:206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fredj NB, et al. Synaptic Activity and Activity-Dependent Competition Regulates Axon Arbor Maturation, Growth Arrest, and Territory in the Retinotectal Projection. J. Neurosci. 2010;30:10939–10951. doi: 10.1523/JNEUROSCI.1556-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shin J, Park H-C, Topczewska JM, Mawdsley DJ, Appel B. Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods Cell Sci. 2003;25:7–14. doi: 10.1023/B:MICS.0000006847.09037.3a. [DOI] [PubMed] [Google Scholar]
- 57.Lambert AM, Bonkowsky JL, Masino MA. The Conserved Dopaminergic Diencephalospinal Tract Mediates Vertebrate Locomotor Development in Zebrafish Larvae. J. Neurosci. 2012;32:13488–13500. doi: 10.1523/JNEUROSCI.1638-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fujimoto E, Stevenson TJ, Chien C-B, Bonkowsky JL. Developmental Biology. Developmental Biology. 2011;352:393–404. doi: 10.1016/j.ydbio.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coffey CM, et al. Novel Oxytocin Gene Expression in the Hindbrain Is Induced by Alcohol Exposure: Transgenic Zebrafish Enable Visualization of Sensitive Neurons. PLoS ONE. 2013;8:e53991. doi: 10.1371/journal.pone.0053991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lillesaar C, Stigloher C, Tannhäuser B, Wullimann MF, Bally-Cuif L. Axonal projections originating from raphe serotonergic neurons in the developing and adult zebrafish, Danio rerio, using transgenics to visualize raphe-specific pet1expression. J. Comp. Neurol. 2009;512:158–182. doi: 10.1002/cne.21887. [DOI] [PubMed] [Google Scholar]
- 61.Parsons MJ, et al. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mechanisms of Development. 2009;126:898–912. doi: 10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scott EK, Baier H. The cellular architecture of the larval zebrafish tectum, as revealed by gal4 enhancer trap lines. Front Neural Circuits. 2009;3:13. doi: 10.3389/neuro.04.013.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kimmel CB, Powell SL, Metcalfe WK. Brain neurons which project to the spinal cord in young larvae of the zebrafish. J. Comp. Neurol. 1982;205:112–127. doi: 10.1002/cne.902050203. [DOI] [PubMed] [Google Scholar]
- 64.Bae Y-K, et al. Developmental Biology. Developmental Biology. 2009;330:406–426. doi: 10.1016/j.ydbio.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 65.Inoue D, Wittbrodt J. One for All—A Highly Efficient and Versatile Method for Fluorescent Immunostaining in Fish Embryos. PLoS ONE. 2011;6:e19713. doi: 10.1371/journal.pone.0019713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahrens MB, et al. Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature. 2012;485:471–477. doi: 10.1038/nature11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen T-W, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zolessi FR, Poggi L, Wilkinson CJ, Chien C-B, Harris WA. Polarization and orientation of retinal ganglion cells in vivo. Neural Dev. 2006;1:2. doi: 10.1186/1749-8104-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kay JN, Finger-Baier KC, Roeser T, Staub W, Baier H. Retinal ganglion cell genesis requires lakritz, a Zebrafish atonal Homolog. Neuron. 2001;30:725–736. doi: 10.1016/s0896-6273(01)00312-9. [DOI] [PubMed] [Google Scholar]
- 71.Valente A, Huang KH, Portugues R, Engert F. Ontogeny of classical and operant learning behaviors in zebrafish. Learning & Memory. 2012;19:170–177. doi: 10.1101/lm.025668.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ostrovsky A, Cachero S, Jefferis G. Clonal Analysis of Olfaction in Drosophila: Image Registration. Cold Spring Harbor Protocols. 2013;2013 doi: 10.1101/pdb.prot071738. pdb.prot071738–pdb.prot071738. [DOI] [PubMed] [Google Scholar]
- 73.Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126:3757–3767. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- 74.Amunts K, Schleicher A, Zilles K. Cytoarchitecture of the cerebral cortex—More than localization. NeuroImage. 2007;37:1061–1065. doi: 10.1016/j.neuroimage.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 75.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Meth. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Calhoun VD, Adali T, Pearlson GD, Pekar JJ. Spatial and temporal independent component analysis of functional MRI data containing a pair of task-related waveforms. Hum. Brain Mapp. 2001;13:43–53. doi: 10.1002/hbm.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.