Abstract
A new strategy for synthesizing spherical nucleic acid (SNA) nanostructures from biodegradable DNA block copolymers is reported. Multiple DNA strands are grafted to one end of a polyester chain (poly-caprolactone) to generate an amphiphilic DNA brush block copolymer (DBBC) structure capable of assembling into spherical micelles in aqueous solution. These novel DBBC-based micelle-SNAs exhibit a higher surface density of nucleic acids compared to micelle structures assembled from an analogous linear DNA block copolymer (DBC), which endows them with the ability to more efficiently enter cells without the need for transfection agents. Importantly, the new SNAs show effective gene regulation without observable cellular toxicity in mammalian cell culture.
Keywords: Spherical nucleic acid, DNA-brush block copolymer, Micelle, Cellular uptake, Gene regulation
1. Introduction
Spherical nucleic acids (SNAs) are an emerging class of nanostructure, typically consisting of a nanoparticle core densely functionalized with a nucleic acid shell.[1,2] These structures exhibit a wide variety of novel properties that are substantively different from their linear nucleic acid counterparts, making them especially attractive for intracellular applications.[3-5] Specifically, they are recognized by Class A scavenger receptors and naturally internalized by many cell types via caveolin-mediated endocytosis.[6] They have been used as novel probes for measuring intracellular genetic content and as potent gene regulation agents.[7,8] They are especially attractive as gene regulation materials since they do not need ancillary transfection agents such as viruses, peptides, lipids or cationic polymers to cross the cell membrane.[9] Thus far, SNAs have been made with a variety of core materials, including many inorganic compositions[1,2,10,11] and several polymer compositions,[12-24] and there are now a few examples of hollow SNA structures that are held together by cross-linked DNA or silica shells.[25,26] Although the nanoparticle core typically plays little role in determining the general chemical and biological properties of the corresponding SNAs, it can be a major concern when such structures are being considered as therapeutic candidates.[27,28] Indeed, a significant synthetic challenge for the chemistry community is to design totally biocompatible and biodegradable SNAs that exhibit the hallmark properties of conventional SNAs with inorganic cores. One approach has been to use liposomal architectures as cores,[29] but an alternative approach that may provide even greater tailorability is the use of polymer micelle architectures. Although a variety of nucleic acid polymer micelle architectures have been reported,[12-24] none have been developed and optimized with regard to general SNA features and intracellular biological activity. Indeed, a few studies involving polymer micelle structures for intracellular gene regulation have been undertaken, but they either required a cationic transfection agent (e.g. polyethyleneimine) for transfection or were constructed using non-degradable polymers.[18,30] To date, there are no biodegradable DNA block copolymer (DBC) based micelle structures that have been shown to exhibit the natural cellular uptake properties of SNAs, and therefore the use of such structures with respect to intracellular and therapeutic applications has been limited. Importantly, SNA-like properties are directly related to the orientation and high density of nucleic acids on the nanoparticle surface.[31]
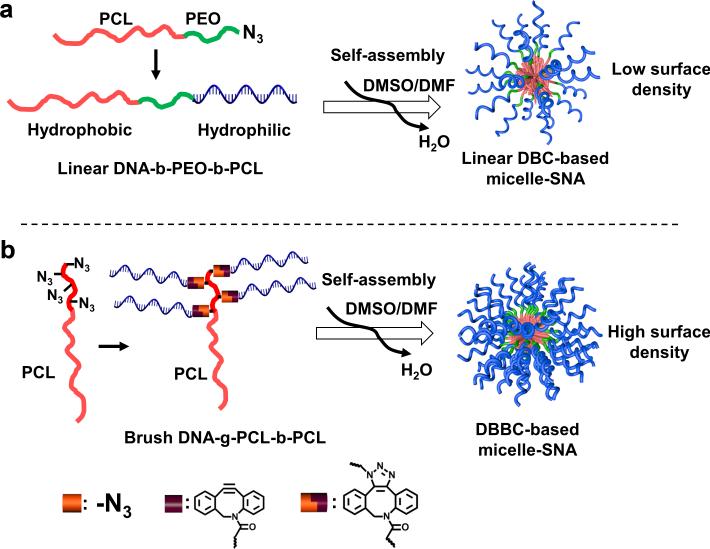
To overcome the problem of low cell internalization associated with DNA block copolymer micelle structures, we identified the need to synthesize micelle SNAs with a dense layer of nucleic acids on the polymer core surface. Herein, we describe a strategy for preparing DBC micelle-SNAs with a biodegradable core and a dense layer of highly oriented oligonucleotides projecting from the surface of the aforementioned core. Traditionally, approaches to synthesizing DBC micelle architectures have involved the linear coupling of nucleic acids to hydrophobic polymer blocks (Scheme 1a).[12] We have found this approach does not reliably yield structures with a nucleic acid density high enough and suitable for optimal SNA cellular transfection capabilities (vide infra). Therefore, our synthetic approach relies on the generation of a DNA-brush block copolymer (DBBC) based micelle structure (Scheme 1b), which was pioneered by Gianneschi et al. in the context of non-degradable constructs for materials assembly purposes.[32] In our synthetic approach, the key components of the DBBC micelle structures are prepared by grafting multiple DNA strands onto the terminal segment of a diblock copolymer consisting of polycaprolactone (PCL; PCL is a component in FDA-approved therapeutics[33]) and azide-modified PCL via copper-free click chemistry to form a DBBC macromolecule. Then, when transferred from an organic solvent mixture consisting of 1:1 DMSO/DMF into an aqueous solution, the as-synthesized amphiphilic DNA-brush block copolymers assemble into ~ 40 nm diameter spherical micelles (Scheme 1b). For comparative purposes, we also prepared a similarly sized micelle structure from a linear DBC component (Scheme 1a).
Scheme 1.
Schematic showing the synthesis of DNA grafted block copolymer-based micelle SNAs. (a) The synthesis of the linear DNA-b-PEO-b-PCL block copolymer and the corresponding formation of micelle-SNAs (LDBC-SNAs). (b) The synthesis of the brush DNA-g-PCL-b-PCL block copolymer and the formation of micelle SNAs (DBBC-SNAs) with a higher surface density of nucleic acids.
2. Results and Discussions
In a typical micelle-SNA synthesis (Figure S1), α-chloro-ε-caprolactone (α-Cl-εCL) prepared by the Baeyer-Villager oxidation of α-chlorocyclohexanone[34] was employed as a monomer for polymerization. The diblock copolymer poly(α-Cl-εCL-b-εCL) was synthesized from the monomer, α-Cl-εCL, and commercially available polycaprolactone (MW = ~ 14 kDa) as the macro-initiator (see Supporting Information). The pendant chlorides of the as-synthesized poly(α-Cl-εCL-b-εCL) were converted into azides by treatment with sodium azide in DMF overnight at room temperature.[35] 1H NMR spectroscopy was used to characterize the product, and the spectrum suggested the as-synthesized diblock copolymer contains on average 15 N3 groups, as determined by integration (peak A and G in Figure S2). In addition, FT-IR spectroscopy of the azide-containing PCL diblock copolymer showed the characteristic azide band at 2106 cm−1 (N=N=N stretch, Figure S3).[36] Gel permeation chromatography (GPC) analysis is also consistent with block copolymer formation as evidenced by an increase in molecular weight from ~ 14 to ~ 16 kDa (Figure S4). When substituted with azide groups, the poly(α-N3-εCL-b-εCL) exhibits a slightly shorter retention time than that of poly(α-Cl-εCL-b-εCL) in the GPC analysis. Finally, poly(α-N3-εCL-b-εCL) and an excess of cyclooctyne-terminated DNA strands were added to an organic solvent mixture (1:1 DMSO:DMF) to initiate the copper-free click reaction of DNA strands onto the azide-modified block, resulting in the formation of the DNA-g-PCL-b-PCL macromolecule. For comparative purposes, a linear DBC was also synthesized (Figure S1). Instead of using 14k PCL as the macro-initiator, an azide-terminated poly(ethylene oxide) with 4 ethylene glycol units was employed to initiate the caprolactone polymerization to synthesize a PEO-b-PCL block copolymer with a comparable molecular weight (~ 18 kDa based on 1H NMR integration, Figure S2). Again, click chemistry was used to conjugate DNA to the as-synthesized PEO-b-PCL block copolymer and generate the linear DNA-b-PEO-b-PCL block copolymer (Figure S2). After the DNA conjugation reaction, both the DNA brush block copolymer micelle-SNAs (DBBC-SNAs) and the linear DNA block copolymer micelle-SNAs (LDBC-SNAs) were prepared and purified according to literature protocols used for analogous linear non-biodegradeable structures.[13] In this process, excess DNA was removed by an ultrafiltration device with a MWCO of 50 kDa membrane. The amount of DNA conjugated to the polymer was determined by measuring the absorbance of the micelle-SNA solutions at 260 nm. Based on the total amount of polymer used for conjugation, it was determined that an average of 10 DNA strands were successfully conjugated onto each polymer chain. Moreover, polyacrylamide gel electrophoresis, under denaturing conditions, was used to analyze the resulting DNA-g-PCL-b-PCL macromolecules. As shown in Figure S5, the bands representing DBBC conjugates with multiple DNA strands can be clearly identified on the gel when excess DNA was used for the conjugation.
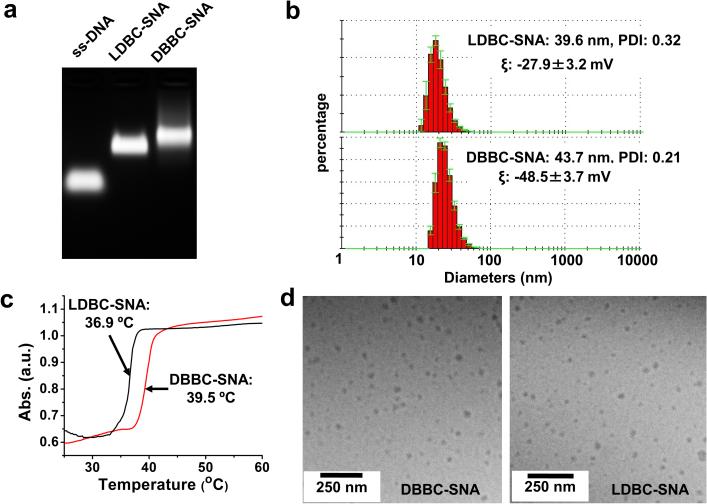
With purified samples, multiple techniques were employed to characterize these two polycaprolactone-based SNAs (Figure 1). Gel electrophoresis (1% agarose gel, Figure 1a) showed a single major band for each sample on the gel image, indicating the micelle-SNAs have a narrow size distribution. Compared to unmodified DNA of the same sequence, the electrophoretic mobilities of the micelle-SNAs are greatly decreased, consistent with the significant difference in size and overall charge. Notably, the electrophoretic mobility of SNA made from the brush architecture is slightly lower than that of one made from the linear structure, indicating that the former SNA is slightly larger. The size distributions of the micelle-SNAs were also probed by dynamic light scattering (DLS), which indicated an average hydrodynamic diameter of ~ 44 nm for the SNAs derived from the brush architecture and ~ 40 nm for ones derived from the linear architecture (Figure 1b), consistent with the gel analysis. Meanwhile, zeta potential analysis gave values of −48.5 ± 3.7 mV and −27.9 ± 3.2 mV, respectively (Figure S6), consistent with the micelle derived from the brush architecture having a higher density of DNA. Furthermore, the micelle-SNAs were cast on mica and imaged by atomic force microscopy in dry form (Figures S7); in both cases, spherical structures were readily apparent. To visualize the morphology of the micelle-SNAs in an environment closer to the one in which they are prepared, cryogenic transmission electron microscopy (Cryo-TEM) was used (Figure 1d). In this method, a very thin layer of micelle-SNA-containing solution was quickly frozen and directly imaged. The Cryo-TEM imaging avoids dehydration-induced artifacts, allowing the fully intact nanoparticles to be visualized. Indeed, round 30-50 nm diameter particles of both DBBC-SNAs and LDBC-SNAs were observed, which are consistent with the DLS size data.
Figure 1.
Characterization of as-synthesized polycaprolactone-based micelle-SNAs. (a) 1% agarose gel electrophoresis; nucleic acids were stained with ethidium bromide; (b) A typical DLS measurement of micelle-SNAs derived from (top) linear and (bottom) brush block copolymer structures; (c) Melting transition behaviour for micelle-SNAs hybridized to complementary 15 nm AuNP-SNAs; (black trace) micelle-SNAs made from linear structures and (red trace) micelles made from the brush architecture; (d) Cryogenic TEM images of micelle-SNAs derived from brush block copolymer. Both DBBC-SNA and LDBC-SNA nanoparticles remain intact under cryogenic conditions and are relatively uniform in size with diameters ~ 30-50 nm.
Since the density of DNA on the particle surface is a key factor that leads to its SNA-like properties, it is important to determine nucleic acid surface coverage. In contrast with conventional AuNP-SNAs, where particle concentration can be easily determined by measuring the plasmon resonance peak associated with AuNPs, it is difficult to directly determine the micelle particle concentration using spectrophotometric methods. Alternatively, we used nanoparticle tracking analysis (NTA)[37,38] to determine the particle concentrations for micelle-SNAs (see details in Supplementary Information, Figure S8 and Table S2). Importantly, the DNA loading for the brush block copolymer based micelle-SNAs (302 strands/particle; 22.2 pmol/cm2) was significantly higher than the linear block copolymer based micelle-SNA (190 strands/particle; 15.6 pmol/cm2), which is comparable to AuNP-based SNAs of similar sizes (300 strands/particle for 30 nm AuNP cores; corresponding to a DNA density of 17.6 pmol/cm2).[39] Another important feature of SNA structures is their cooperative hybridization properties, arising from the association of multiple DNA strands between each particle. This results in a sharp melting transition during the thermal denaturation of hybridized complementary particles.[2,13] Generally, higher DNA loading results in higher melting temperatures and sharper transitions under comparable conditions. The thermal denaturation of micelle-SNAs was carried out by hybridizing both brush and linear block copolymer based micelle-SNAs with AuNP-SNAs containing complementary sticky ends, respectively, and then monitoring the absorbance change at 260 nm with gradual heating (Figure 1c and Figure S9). Around the melting temperature, extinction dramatically increased due to cooperative DNA dehybridization. As shown in Figure 1c, the melting curves for both types of micelle-SNAs when hybridized with AuNP-SNAs were sharp and elevated, with the Tm of the DBBC-based micelle-SNA (39.5 °C) being almost 3 degrees higher than the linear DBC-based micelle-SNA (36.9 °C). The higher melting temperature for the DBBC-based micelle-SNA is consistent with a structure with a higher surface density of nucleic acids.[40]
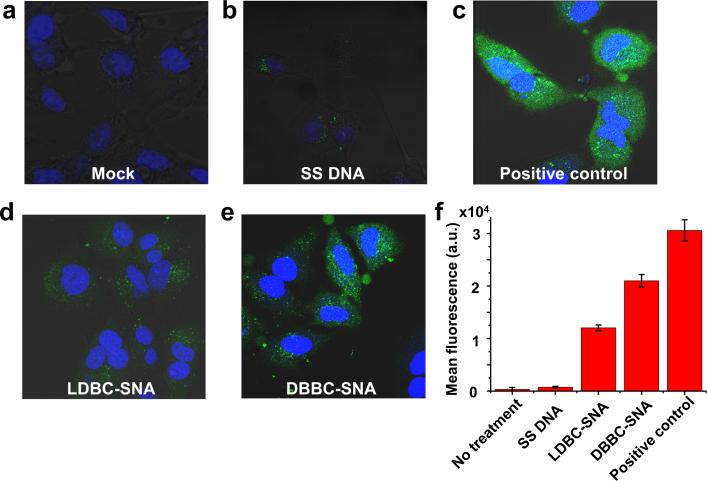
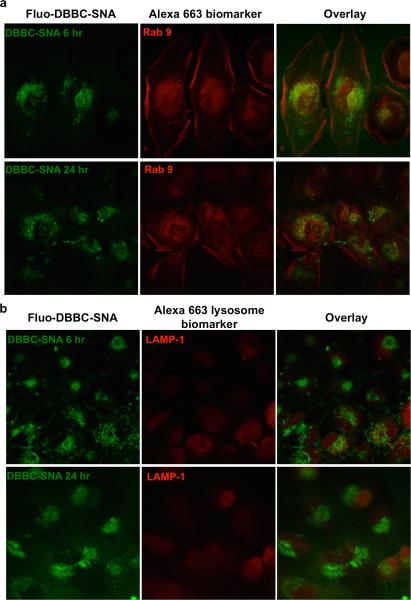
With well-characterized micelle-SNAs in hand, we evaluated their biological function with regard to in vitro cell uptake studies (Figure 2). Both micelle-SNAs exhibit the ability to enter cells without the assistance of cationic transfection reagents, but with different efficiencies. For example, confocal microscopy was employed to reveal that fluorescein-labelled micelle-SNAs (1 μM total DNA) enter HeLa cells after 16 h of incubation. Notably, the green fluorescence from micelle-SNAs consisting of brush block copolymers was significantly more intense than that of the linear block copolymer based micelle-SNAs (Figure 2d,e), indicating more efficient uptake of the former as opposed to the latter. This result was also confirmed by quantifying the fluorescence intensity of the cell population using flow cytometry. Although incubated with equal amounts of total DNA, the mean fluorescence intensity of cells treated with brush block copolymer based micelle-SNAs was almost twice that of brush block copolymer based micelle-SNAs (Figure 2f), presumably due to their higher surface density of nucleic acids. Compared to the positive control which utilized Lipofectamine® 2000 as the transfection agent, DBBC-SNAs show slightly lower but comparable transfection efficiency without any co-carrier. We further studied the intracellular location of DBBC-SNAs as a function of incubation time under conditions where HeLa cells were continuously incubated with fluorescein-labelled micelle-SNAs (1 μM total DNA). SNA-AuNP conjugates traffic through the endocytic pathway into late endosomes and reside there without accumulating in lysosomes.[28] It is hypothesized that the DBBC-SNAs follow the same route upon cellular entry due to their similar architecture. In these experiments, the DBBC-SNAs carry a green fluorophore (fluorescein), and the cells are stained with a complementary Alexa dye-labeled marker of interest (red). As shown in Figure 3, strong colocalization of SNAs with Rab9 (a protein which preferentially localizes in late endosomes) was observed after 6 h of incubation, and colocalization persisted 24 h post-incubation. Importantly, we did not observe appreciable colocalization between the fluorescent signals of DBBC-SNAs and markers for lysosomes (LAMP-1) over a 24 h period of time. From these data, we conclude that SNAs primarily remain inside late endosomes and do not migrate beyond this point to the lysosome. These observations confirm that within a typical cell doubling time (23-24 h for HeLa), the micelle-SNAs likely employ the same intracellular trafficking pathway as AuNP-based SNAs.
Figure 2.
Cellular uptake of micelle-SNAs. (a-e) Fluorescence micrograph of HeLa cells incubated with different forms of nucleic acids at a total DNA concentration of 1 μM for 16 h. DNA strands were labelled at the 5’-end with fluorescein and the dye molecules are located at the outside terminus of the micelle-SNA structure. a) negative control, cells without DNA incubation; b) single stranded fluorescein-labelled DNA (Fluo-DNA); c) positive control, single stranded Fluo-DNA transfected with Lipofectamine® 2000; d) micelle-SNAs derived from linear block copolymer structures; e) micelle-SNAs derived from brush block copolymer structures. In the fluorescence images, micelle-SNAs assembled from the brush block copolymer show significantly higher uptake compared to those derived from the linear analog or component single stranded DNA; (f) Fluorescence-activated cell sorting (FACS) analysis of the cells when incubated with different forms of nucleic acids. FACS data also confirm the brush block copolymer based micelle-SNA has higher cell uptake efficiency than that of the linear block copolymer based micelle-SNA. Single stranded DNA and DNA transfected by conventional Lipofectamine® 2000 were used as controls.
Figure 3.
Confocal microscopy of fluorescein-labelled DBBC-SNA and immunofluorescence staining of organelle markers (red, labelled by Alexa Fluor 663). Biomarkers are Rab9 (for late endosomes) and LAMP-1 (for lysosomes). Note that most DBBC-SNAs colocalize with late endosomes during the incubation in HeLa cells. There is no significant colocalization of DBBC-SNAs with lysosomes, which is consistent with the behaviour of AuNP-SNAs.
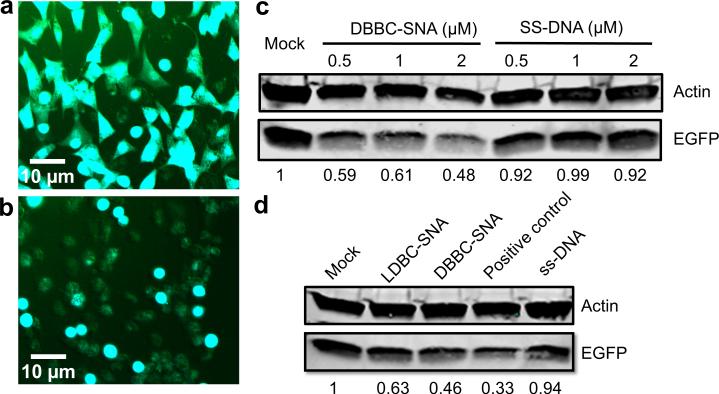
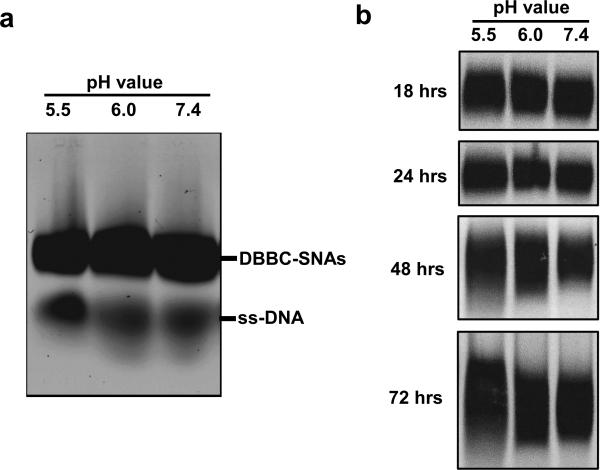
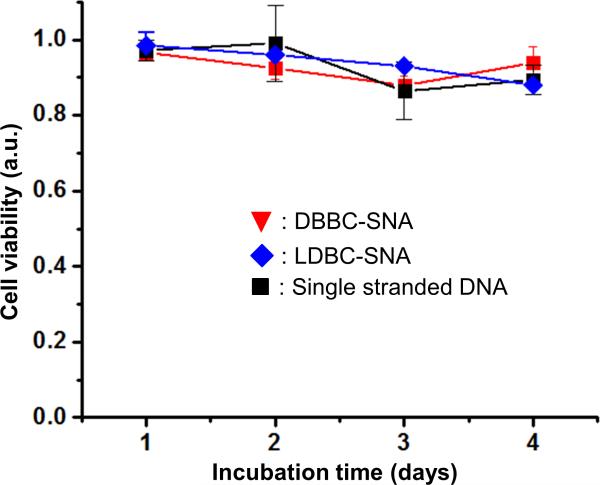
We further tested the intracellular gene regulation capacity of the DBBC-based micelle-SNAs in specialty C166 mouse endothelial cells over-expressing enhanced Green Fluorescent Protein (EGFP). The DNA strands on the micelle-SNAs were designed as an antisense sequence against the EGFP mRNA. After transfection and further incubation for 48 h, EGFP knockdown is readily apparent by fluorescence microscopy (Figure 4a,b). Importantly, the quantification of EGFP protein expression via western blot shows a concomitant ~ 52% knockdown (Figure 4c), consistent with the conclusion that the micelle-SNA can effectively bind the cytosolic mRNA target and alter the expression of its associated protein. Compared to LDBC-SNAs at the same condition, DBBC-SNAs are more effective at regulating gene expression via the antisense mechanism (Figure 4d). When the DBBC-based micelle-SNAs are modified with a scrambled DNA sequence, EGFP expression does not reduce at all concentrations tested (0.5-2 μM of total DNA), indicating the gene knockdown effect is sequence-specific (Figure S10). To further validate that our new constructs may have significant potential for biomedical applications, we tested the biodegradation and cellular toxicity of DBBC-SNAs. During the transfection and intracellular trafficking process, SNAs will encounter quite different chemical environments, such as varied pH values and the presence of different enzymes in serum, endosomes, and lysosomes. This may cause the PCL core of the micelle particles to degrade at a different rate depending on their location. To evaluate their biodegradability, we incubated the DBBC-SNAs in buffers of varied pH values to mimic different intracellular environments. As shown by agarose gel electrophoresis, component single-stranded DNA appeared after 24 h incubation, indicating the degradation of micelle-SNAs (Figure 5a). Meanwhile, gel bands representing the DBBC-SNAs became smeared and diffused along increased incubation time (Figure 5b). Notably, micelle-SNAs degraded faster in lower pH buffer conditions, pointing to the possibilities of their faster degradation when trafficking to intracellular vesicles with low pH, such as the endosomes.[28] The cellular toxicity of DBBC-based micelle-SNAs was analyzed using the standard MTT assay (Figure 6).[41] Even with an incubation time of up to 4 days, we found that cell viability remained essentially unchanged. This confirms that the as-synthesized micelle-SNAs with biocompatible and biodegradable polycaprolactone cores do not have measurable cellular toxicity under these conditions.
Figure 4.
Gene regulation by DBBC-based micelle-SNAs. (a) Fluorescence micrograph of C166 mouse endothelial cells that highly express the EGFP protein before DBBC-based micelle-SNA treatment. (b) Fluorescence micrograph of C166 mouse endothelial cells after DBBC-SNA treatment. The green fluorescence was significantly suppressed due to the EGFP gene knockdown. The micelle-SNAs were equipped with anti-EGFP sequence and the cells were cultured for another 2 days after SNA transfection. (c) Western blotting of EGFP expression in C166 cells after treatment with anti-EGFP DBBC-based micelle-SNAs and single stranded DNA samples under various total DNA concentrations. (d) Western blotting of EGFP expression in C166 cells after treatment with anti-EGFP DBBC-SNAs and control samples under total DNA concentrations of 2 μM. The positive control sample was antisense DNA transfected by conventional Lipofectamine® 2000. Actin was used as an internal reference.
Figure 5.
The pH-dependent degradation of DBBC-based micelle-SNAs. Samples were incubated in 50 mM MES buffer (pH 5.5), 50 mM HEPES buffer (pH 6.0), or 1X PBS (pH 7.4). Agarose gel electrophoresis was used to monitor the degradation process. a) Degradation of DBBC-based micelle-SNAs after 24 hour incubation under different pH buffer conditions. Note that a significant amount of single stranded DNA can be observed in the gel image; b) the mobility and shape of the DBBC-based SNA bands over incubation time under different buffer conditions. In the first 24 hours, the SNA bands look relatively sharp. However, as the incubation time increases, the bands become smeared and diffuse, indicating the gradual degradation of the entire micellar structures. Note that the micelles held at lower pH (5.5) suffer considerably more degradation after 72 hours.
Figure 6.
Cellular toxicity of DBBC-based micelle-SNAs analyzed by a standard MTT assay. Cells treated with DBBC-SNAs (red), LDBC-SNAs (blue), and single stranded DNA (black) at the same total DNA concentration (2 μM). Viable cells with active metabolism convert MTT into a purple colored formazan product with an absorbance maximum near 570 nm, and cell viability was quantified by normalization of the absorbance at 570 nm to non-treated cells. Error bar are standard deviation of absorbance at 570 nm from 3 independent wells of cells in a 96-well plate.
3. Conclusion
In conclusion, we have developed a novel strategy to construct a DNA-brush block copolymer based micelle-SNA with increased DNA surface density, which endows the new construct with properties similar to the well-established AuNP-SNAs. Compared to the micelle-SNAs constructed from linear block copolymers, the DBBC-based micelle-SNAs exhibit a higher surface density of nucleic acids, a more negatively charged surface, a higher melting temperature, a cooperative melting profile, and more effective transfection agent-free cellular uptake. Importantly, the micelle-SNAs derived from the DNA-brush block copolymer show effective target gene knockdown in vitro.
It is worthwhile noting that since the polymer core can gradually degrade under physiological conditions due to acid-catalyzed or esterase-catalyzed cleavage of ester bonds,[42] these constructs open new avenues towards the controllable, continuous release of nucleic acids as regulatory agents for intracellular biological processes. Generally, in addition to PCL, a wide variety of polyesters, such as PLA[43] and PLGA[44] can be employed to initiate polymerization and then form DBBC-based SNAs consisting of different types of polyester cores. As such, the rate of particle degradation and nucleic acid release may be tuned by judicious monomer selection. We anticipate that this synthetic approach can be extended to a wide variety of polyesters to generate a new class of nanostructured materials with highly tailorable properties and efficacies for various nucleic-acid based therapeutic approaches.
4. Experimental Section
Materials and Methods
All DNA synthesis was carried out on a BioAutomation MM48 DNA synthesizer according to the standard manufacturer trityl-on protocol. All reagents for oligonucleotide synthesis were purchased from Glen Research (Sterling, VA) and used following manufacturer's instructions. Ac-dC and dmf-dG phosphoramidites were used to enable room-temperature deprotection of the nucleobases. α-Chlorocyclohexanone, sodium azide, N,N-dimethylformamide (DMF), tin(II) octanoate, caprolactone (CL), and all other solvents were purchased from Sigma-Aldrich (St. Louis, MO) and used without further purification. M-chloroperoxybenzoic acid (mCPBA) was purchased from Fluka (Buchs, Switzerland).
Synthesis of Diblock Copolymer Poly(α-N3-εCL-b-εCL)
The monomer α-chloro-ε-caprolactone was prepared by the Baeyer-Villager oxidation of α-chlorocyclohexanone using excess mCPBA. After purification, 0.5 g monomer was mixed with 3.0 g commercially available polycaprolactone (MW = ~ 14 kDa, as macro-initiator) in dry toluene (10 mL). The mixture was dried by repeated (3×) azeotropic distillation with toluene. Then, the solution was heated to 130 °C in a preheated oil bath and stirred under N2. The reaction mixture was stirred for 5 min at 130 °C and then one drop of tin(II) octanoate was added via syringe. The temperature was maintained at 130 °C for another 3 hours. The resulting viscous mixture was rapidly cooled, upon which it solidified. The crude polymer was purified by repeated dissolution in dichloromethane and heptane precipitation (3×), resulting in a white solid after drying. Then the pendant chlorides of the as-synthesized poly(α-Cl-εCL-b-εCL) were converted into azides by treatment with sodium azide in DMF overnight at room temperature. After removal of DMF in vacuo, 15 mL of toluene was added, and the remaining NaN3 was removed by centrifugation (4000 rpm at 25 °C for 20 min). The diblock copolymer was recovered by precipitation in heptane. After washing with heptane multiple times, the solid precipitate was collected by vacuum filtration and dried under reduced pressure (see more details in Supporting Information).
Synthesis of Azide-terminated PEO-PCL Diblock Copolymer
Azide-terminated oligo(ethylene oxide) with 4 PEG units (C8H16O4N3, 109 mg, 0.5 mmol) was weighed into a dry flask and ε-caprolactone (8.5 g with M/I ratio of ~ 150:1) was subsequently added. The synthesis and purification of single azide-terminated PEO-PCL diblock copolymer follow the same procedures for poly(α-Cl-εCL-b-εCL) as mentioned above with similar isolated yield of the desired product.
DNA Conjugation
For each azide-containing polymer, the same procedure was followed. First, the polymer was dissolved in a 1:1 DMSO/DMF solution, and then a large excess of DBCO-DNA dissolved in DMSO was added to the polymer solution and incubated at 40 °C overnight. After conjugation, the resulting linear and brush DBC samples were used immediately for micelle formation.
Formation and Purification of Micelle-SNAs
Briefly, DBCs dissolved in an organic solvent (DMSO/DMF, 1:1) were loaded in a dialysis bag with a 50 kDa MW cutoff membrane and the solution was dialyzed against deionized water for 12 hours. After removal of large aggregates by filtration through a 0.2 μm syringe filter, the micelle-SNA conjugates were purified using an Amicon Ultra-15 ultrafiltration device (Millipore, MWCO 50 kDa) to remove excess free DNA.
Characterizations
1H NMR spectra were recorded on a Bruker Avance 400 MHz NMR spectrometer and referenced internally to residual proton signals in the denatured solvents. FTIR spectra were collected on a Perkin-Elmer Spectrum 100 FTIR spectrometer using disposable KBr plates. Gel permeation chromatography (GPC) was carried out in THF at 32 °C at 1 mL/min with a Viscotek TDAmax liquid chromatograph (Malvern Instruments, UK) equipped with a programmable autosampler and refractive index detector. AFM images were obtained with a Bruker Dimension Icon atomic force microscope in a tapping mode under ambient conditions. DLS hydrodynamic size and zeta potential measurements were collected on a Malvern Zetasizer Nano-ZS (Malvern Instruments, UK) with a laser wavelength of 633 nm. Cryogenic TEM images were collected on a JEM 1230 microscope (JEOL) at an accelerating voltage of 80 kV. NTA measurements were performed with a NanoSight LM10 (NanoSight, Amesbury, UK) equipped with a sample chamber with a 638 nm laser and a Viton fluoroelastomer O-ring. Confocal fluorescence images of the cells treated with micelle-SNAs were collected with a Zeiss LSM 510 inverted confocal scanning microscope.
Cell Culture, Flow Cytometry, and Western Blotting
Cells were cultured at 37 °C and 5% CO2 in DMEM supplemented with 10% FBS and 1% streptomycin/penicillin. For cellular uptake study, HeLa cells were seeded in a 6-well plate 24 h prior to treatment and incubated with micelle-SNAs derived from linear or brush block copolymer structures, respectively. Right before flow cytometry analysis, cells were trypsinized, washed, suspended in 0.5 ml 1× phosphate-buffered saline (PBS) and fixed by addition of 0.5 ml of 3.7% formaldehyde in PBS. The fluorescence intensity from fluorescein (excitation wavelength at 488 nm, emission wavelength at 520 nm) of 10,000 cells was collected using a BD LSR II flow cytometer. For western blotting, C166 cells over-expressing EGFP were transfected with micelle-SNAs (with 0.5, 1, and 2 μM of total DNA in OptiMEM) overnight and further incubate for another 2 days after medium change. Protein lysate with equal amount of total protein were fractionated by 4-20% Precast gradient gel (Bio-Rad). The intact gel was then transferred to a nitrocellulose membrane (Thermo Scientific) and blocked in odyssey blocking buffer (LI-COR Biosciences). Proteins were detected with primary antibodies against actin (1:500) (Santa Cruz Biotechnology), EGFP (1:1000) (Clontech Laboratories Inc) followed by IRDye 680 secondary antibodies (1: 10,000) (LI-COR Biosciences) diluted in PBST containing 5% non-fat milk. The desired bands were visualized using the Odyssey® CLx Infrared Imaging System (LI-COR Biosciences).
Confocal Microscopy and Immunofluorescence
Cells were seeded in a 35 mm FluoroDish and incubated with fluorescein-labelled DBBC-SNAs (with total DNA 1 μmol) in complete DMEM for different time points. Cells were rinsed with PBS, fixed in 3.7% PFA in PBS for 15 min, and imaged under a Zeiss LSM 510 inverted confocal scanning microscope. The excitation wavelength of fluorescein-labelled DBBC-SNAs was 488 nm, and the corresponding emission filter was 500-550 nm. To track the colocalization of SNAs with intracellular proteins, after incubation with fluorescein-labelled DBBC-SNAs (with total [DNA] = 1 μmol) for different durations of time, cells were rinsed with PBS, fixed in 3.7% PFA in PBS, and permeated with 0.1% Triton X-100 for 3min. After blocking with 5% BSA in PBS for 1 h, cells were stained with a primary antibody against the protein marker of interest at 5 μg/mL (1% BSA in PBS) overnight at 4 °C. After rinses with 0.05% Tween-20 in PBS, cells were stained with an Alexa Fluor 633-labeled secondary antibody (Invitrogen Alexa Fluor 633 Goat Anti-Rabbit or mouse IgG (H+L) at 1 μg/mL (1% BSA in PBS) for 1 hour at RT. The excitation wavelength of the secondary antibody was 633 nm, and the corresponding emission filter was 660 - 710 nm. The primary antibodies include rabbit against Rab9 (Abcam ab179815), mouse against LAMP1 (Santa Cruz sc-20011).
Degradation of DBBC-SNAs
To visualize the amount of nucleic acids released from DBBC-SNAs under different pH conditions an agarose gel experiment was carried out. Briefly, fluorescein-labeled DBBC-SNAs containing 10 μM total DNA were diluted 1:1 with the following buffer solutions (100 mM each): HEPES buffer, pH 5.5; MES buffer, pH 6.0, PBS buffer, pH 7.4. As a control, a sample was prepared with the AuNP functionalized with the same oligonucleotides (anti-EGFP antisense DNA) diluted 1:1 with the buffers described above, respectively. The solutions were allowed to stand at RT for 18, 24, 48, and 72 hours, then were analyzed by agarose gel electrophoresis (1% agarose). The agarose gels were imaged on a Fujifilm FLA-5100 gel imager with a 473 nm laser to visualize the fluorescein-labeled DNA.
Supplementary Material
Acknowledgements
C. Zhang, L. Hao, C. M. Calabrese contributed equally to this work.
C.A.M. acknowledges support from the Center for Cancer Nanotechnology Excellence (CCNE) initiative of National Institutes of Health (NIH) Awards U54 CA119341 and U54 CA151880, Dixon Translational Research Grants Initiative, and the Defense Advanced Research Planning Agency Award N66001-11-1-4189. L.H. was a Howard Hughes Medical Institute International Student Research Fellow. C.H.J.C. acknowledges a postdoctoral research fellowship from the Croucher Foundation. The electron microscopy work was performed at the Biological Imaging Facility (BIF) and the Electron Probe Instrumentation Center (EPIC) at Northwestern University. We also acknowledge Dr. Natalia Chernyak for providing the azide-terminated oligo(ethylene oxide) initiator.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dr. Chuan Zhang, Department of Chemistry, International Institute for Nanotechnology, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208-3113, USA
Dr. Liangliang Hao, Department of Chemistry, International Institute for Nanotechnology, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208-3113, USA
Colin M. Calabrese, Department of Chemistry, International Institute for Nanotechnology, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208-3113, USA
Yu Zhou, Department of Materials Science and Engineering, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208-3113, USA.
Dr. Chung Hang J. Choi, Department of Chemistry, International Institute for Nanotechnology, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208-3113, USA
Dr. Hang Xing, Department of Chemistry, International Institute for Nanotechnology, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208-3113, USA
Prof. Chad A. Mirkin, Department of Chemistry, International Institute for Nanotechnology, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208-3113, USA; Department of Materials Science and Engineering, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208-3113, USA.
References
- 1.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 2.Cutler JI, Auyeung E, Mirkin CA. J. Am. Chem. Soc. 2012;134:1376. doi: 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- 3.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Science. 2006;312:1027. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 4.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. J. Am. Chem. Soc. 2009;131:2072. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jensen SA, Day ES, Ko CH, Hurley LA, Luciano JP, Kouri FM, Merkel TJ, Luthi AJ, Patel PC, Cutler JI, Daniel WL, Scott AW, Rotz MW, Meade TJ, Giljohann DA, Mirkin CA, Stegh AH. Sci. Transl. Med. 2013;5:209ra152. doi: 10.1126/scitranslmed.3006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi CHJ, Hao L, Narayan SP, Auyeung E, Mirkin CA. Proc. Natl. Acad. Sci. U.S.A. 2013;110:7625. doi: 10.1073/pnas.1305804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prigodich AE, Seferos DS, Massich MD, Giljohann DA, Lane BC, Mirkin CA. ACS Nano. 2009;3:2147. doi: 10.1021/nn9003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giljohann DA, Seferos DS, Daniel WL, Massich MD, Patel PC, Mirkin CA. Angew. Chem. Int. Ed. 2010;49:3280. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Massich MD, Giljohann DA, Schmucker AL, Patel PC, Mirkin CA. ACS Nano. 2010;4:5641. doi: 10.1021/nn102228s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cutler JI, Zheng D, Xu XY, Giljohann DA, Mirkin CA. Nano Lett. 2010;10:1477. doi: 10.1021/nl100477m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang C, Macfarlane RJ, Young KL, Choi CHJ, Hao L, Auyeung E, Liu G, Zhou X, Mirkin CA. Nature Mater. 2013;12:741. doi: 10.1038/nmat3647. [DOI] [PubMed] [Google Scholar]
- 12.Schnitzler T, Herrmann A. Acc. Chem. Res. 2012;45:1419. doi: 10.1021/ar200211a. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Zhang Y, Fullhart P, Mirkin CA. Nano Lett. 2004;4:1055. [Google Scholar]
- 14.Kwak M, Herrmann A. Angew. Chem. Int. Ed. 2010;49:8574. doi: 10.1002/anie.200906820. [DOI] [PubMed] [Google Scholar]
- 15.Kedracki D, Safir I, Gour N, Ngo KX, Vebert-Nardin C. Adv. Polym. Sci. 2013;253:115. [Google Scholar]
- 16.Alemdaroglu FE, Alemdaroglu NC, Langguth P, Herrmann A. Adv. Mater. 2008;20:899. [Google Scholar]
- 17.Rush AM, Thompson MP, Tatro ET, Gianneschi NC. ACS Nano. 2013;7:1379. doi: 10.1021/nn305030g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rush AM, Nelles DA, Blum AP, Barnhill SA, Tatro ET, Yeo GW, Gianneschi NC. J. Am. Chem. Soc. 2014;136:7615. doi: 10.1021/ja503598z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong JH, Park TG. Bioconjugate Chem. 2001;12:917. doi: 10.1021/bc010052t. [DOI] [PubMed] [Google Scholar]
- 20.Yang CYJ, Pinto M, Schanze K, Tan WH. Angew. Chem. Int. Ed. 2005;44:2572. doi: 10.1002/anie.200462431. [DOI] [PubMed] [Google Scholar]
- 21.Peng L, Wu CS, You MX, Han D, Chen Y, Fu T, Ye M, Tan WH. Chem. Sci. 2013;4:1928. doi: 10.1039/C2SC21198J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fong RB, Ding ZL, Long CJ, Hoffman AS, Stayton PS. Bioconjugate Chem. 1999;10:720. doi: 10.1021/bc980151f. [DOI] [PubMed] [Google Scholar]
- 23.Oishi M, Nagasaki Y, Itaka K, Nishiyama N, Kataoka K. J. Am. Chem. Soc. 2005;127:1624. doi: 10.1021/ja044941d. [DOI] [PubMed] [Google Scholar]
- 24.Isoda K, Kanayama N, Miyamoto D, Takarada T, Maeda M. React. Funct. Polym. 2011;71:367. [Google Scholar]
- 25.Cutler JI, Zhang K, Zheng D, Auyeung E, Prigodich AE, Mirkin CA. J. Am. Chem. Soc. 2011;133:9254. doi: 10.1021/ja203375n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young KL, Scott AW, Hao L, Mirkin SE, Liu GL, Mirkin CA. Nano. Lett. 2012;12:3867. doi: 10.1021/nl3020846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonavane G, Tomoda K, Makino K. Colloids Surf., B. 2008;66:274. doi: 10.1016/j.colsurfb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Wu XA, Choi CHJ, Zhang C, Hao L, Mirkin CA. J. Am. Chem. Soc. 2014;136:7726. doi: 10.1021/ja503010a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banga RJ, Chernyak N, Narayan SP, Nguyen ST, Mirkin CA. J. Am. Chem. Soc. 2014;136:9866. doi: 10.1021/ja504845f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SH, Mok H, Lee Y, Park TG. J Control Release. 2011;152:152. doi: 10.1016/j.jconrel.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 31.Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Nano Lett. 2007;7:3818. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chien MP, Rush AM, Thompson MP, Gianneschi NC. Angew. Chem. Int. Ed. 2010;49:5076. doi: 10.1002/anie.201000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodruff MA, Hutmacher DW. Prog. Polym. Sci. 2010;35:1217. [Google Scholar]
- 34.Lenoir S, Riva R, Lou X, Detrembleur C, Jerome R, Lecomte P. Macromolecules. 2004;37:4055. [Google Scholar]
- 35.Riva R, Schmeits S, Jerome C, Jerome R, Lecomte P. Macromolecules. 2007;40:796. [Google Scholar]
- 36.Lin-Vien D, Colthup NB, Fateley WG, Grasselli JG. Handbook of Infrared and Raman Characteristic Group Frequencies. Academic Press; London: 1991. p. 221. [Google Scholar]
- 37.Filipe V, Hawe A, Jiskoot W. Pharm. Res. 2010;27:796. doi: 10.1007/s11095-010-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Domingos RF, Baalousha MA, Ju-Nam Y, Reid MM, Tufenkji N, Lead JR, Leppard GG, Wilkinson KJ. Environ. Sci. Technol. 2009;43:7277. doi: 10.1021/es900249m. [DOI] [PubMed] [Google Scholar]
- 39.Hurst SJ, Lytton-Jean AKR, Mirkin CA. Anal. Chem. 2006;78:8313. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin R, Wu G, Li Z, Mirkin CA, Schatz GC. J. Am. Chem. Soc. 2003;125:1643. doi: 10.1021/ja021096v. [DOI] [PubMed] [Google Scholar]
- 41.Roehm NW, Rodgers GH, Hatfield SM, Glasebrook AL. J. Immunol. Methods. 1991;142:257. doi: 10.1016/0022-1759(91)90114-u. [DOI] [PubMed] [Google Scholar]
- 42.Lassalle V, Ferreira ML. Macromol. Biosci. 2007;7:767. doi: 10.1002/mabi.200700022. [DOI] [PubMed] [Google Scholar]
- 43.Soppimath KS, Aminabhavi TM, Kulkarni AR, Rudzinski WE. J. Control. Release. 2001;70:1. doi: 10.1016/s0168-3659(00)00339-4. [DOI] [PubMed] [Google Scholar]
- 44.Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Preat V. J. Control. Release. 2012;161:505. doi: 10.1016/j.jconrel.2012.01.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.