Abstract
It has been long understood that mutation distribution across genomic space and in time is not completely random. Indeed, recent surprising discoveries identified multiple simultaneous mutations occurring in tiny regions within chromosomes, while the rest of the genome remains relatively mutation-free. Mechanistic elucidation of these phenomena called mutation showers, mutation clusters, or kataegis is ongoing, in parallel with findings of abundant clustered mutagenesis in cancer genomes. So far, the combination of factors most important for clustered mutagenesis is the induction of DNA lesions with unusually long and persistent single-strand DNA intermediates. In addition to being a fascinating phenomenon, clustered mutagenesis also became an indispensable tool for identifying a previously unrecognized major source of mutation in cancer – APOBEC cytidine deaminases. Future research on clustered mutagenesis carries a promise of shedding light onto important mechanistic details of genome maintenance, with potentially profound implications for human health.
Keywords: Mutagenesis, cancer, evolution, genome instability, mutation showers, kataegis
INTRODUCTION
Mutations generate the genetic diversity that enables biological evolution by natural selection and allelic drift (9; 38; 60). The discovery of radiation-induced mutagenesis, which preceded an understanding of the nature of genetic material, inspired the elegantly simple concept of random mutagenesis, analogous to stochastic radioactive decay, where each event is unpredictable in space and time (131). However, elucidating the physical nature of genes led to the realization that mutation is a result of complex DNA transactions (45), suggesting that there could be regions of the genome with higher propensity for sequence change. Not surprisingly, strong mutation hotspots were found in the very first large collection of mutations within a single gene (12). Later, the drastically unequal chances to find mutations in different regions of a gene or genome were demonstrated in multiple studies (110). An important biological consequence of non-uniform distribution of mutations is that a disproportionately large number of changes can accumulate in a limited section of a genome, forming a mutation cluster (Table 1). Crucially, such mutation clusters are abundant in human cancers (104; 105), afflictions which can be considered cases of highly accelerated evolution within somatic tissues. Recent studies have revealed various molecular mechanisms of clustered mutagenesis. These mechanisms, their biological implications, and the utility of clustered mutagenesis phenomena for dissecting mechanisms of genome instability are the subject of this review.
Table 1.
Clusters of simultaneous mutations.
| System | Cause of hypermutation | Hypermutable DNA substrate | Cluster size, (# of mutations) | Selected references |
|---|---|---|---|---|
| Complex mutations | Error by TLS-polymerase | Lesion or local secondary structure | 2–30 nt (2–16) | (49; 68; 85; 86; 123) |
| Mutation showers in Big Blue Mouse | Unknown | Unknown | 1–20 kb (2–6) | (15; 55; 132) |
| E. coli– proliferation in the presence of DNA damaging agent | DNA damage (EMS) | Lesions in dsDNA in front of replication fork | up to 1500 kb (up to 30) | (88) |
| Yeast-subtelomeric | Endogenous (APOBEC) or exogenous (UV, MMS, sufites, H2O2) DNA damage | Long ssDNA at uncapped telomeres | 2–17 kb (2–10) | (17; 24; 25; 36; 140) |
| Yeast – site-specific DSBs | Endogenous (APOBEC) or exogenous (MMS) DNA damage | Long ssDNA created by 5′→3′ resection around DSB or by break-induced replication | 1–170 kb (2–15) | (113; 128; 139; 140) |
| Yeast – proliferation in the presence of DNA damaging agent | Endogenous (APOBEC) or exogenous (MMS) DNA damage | Long ssDNA around DSB (resection and/or BIR), at dysfunctional replication forks or at R-loops | 1–200 kb (2–30) | (63; 108; 128; 129) |
| Retroelements | Endogenous (APOBEC) DNA damage | ssDNA generated by reverse transcription | Entire retroelements, 0.3–10 kb (up to 20–30) | (50; 100; 116) |
| SHM in Ig genes | Endogenous (AID) DNA damage | R-loops or transcription bubbles | 1–4 kb (>2a) | (70; 78; 90; 91; 122) |
| Cancer | Endogenous (AID/APOBEC) DNA damage, other (unidentified) | Long ssDNA | 1–50 kb (2–160) | (3; 14; 83; 91; 98; 99; 104; 107; 108) |
| Human germ line | Unknown | Unknown | 10–100 kb (2–5) | (20; 43; 79) |
While multiple mutations accumulate in SHM regions, they may result from AID stimulated mutagenesis in the unknown number of cell generations
LOCALIZED ACCUMULATION OF MUTATIONS OVER MULTIPLE GENERATIONS
We begin by briefly describing phenomena which should be taken into account in order to appreciate clustered mutagenesis in its proper context. Mutation clusters may occur in certain region(s) of the genome evolving over large numbers of germline or somatic generations; i.e., mutations are clustered in space, but not necessarily in time. Regional increase in accumulation of mutations may occur in genes under selection pressure towards higher diversity, such as structural genes for venoms (126) or peptide pheromones (134). Increased mutation rates also can be associated with the relief of stabilizing selection against deleterious mutations in duplicated genes (87). Local accumulation of unselected mutations also may be associated with region-specific increase in mutation rate. Comparison of human and chimpanzee genomes with an inferred common ancestor sequence highlighted several “human accelerated regions” (HAR) in which more mutations accumulated during ten million years of primate evolution than over the preceding hundred million-year mammalian genome evolution (13; 16; 93). One mechanism suggested for such mutation clustering posits higher mutation rates and/or biased gene conversion, resulting in faster accumulation and fixation of mutations in regions associated with hotspots of meiotic crossing-over (41; 42; 67; 82; 97). Increased mutation rate in the vicinity of DNA double-strand breaks (DSBs), observed in multiple studies (35; 51; 54; 74; 124; 140) also reviewed in (74)), may result in higher density of mutations around meiotic crossing-over hotspots as well as in other regions with increased strand breakage. Local increases of mutation density in primate evolution were detected in late replicating regions (121) as well as in human cancers (65). This phenomenon, confirmed in many biological systems, may result from complex interactions of replication and gene expression with accessibility to damage and repair, nuclear positioning, transcription and chromatin modification (101; 118).
In theory, multiple successive alterations accumulated over time in a single gene or regulatory region could result in significant fitness increase. However, it may be difficult or even impossible for an individual to acquire this advantageous combination of mutations within the time and space constraints of biological evolution. As initially noted by Wright (136) followed by others (61; 133), the vast majority of individual mutations, within a cluster conferring high fitness, would be neutral or even deleterious (sign epistasis). Thus, two states with high fitness often would be separated by a “valley” of lower fitness, which can be traversed by a combination of steps, although such stepwise traversals would be evolutionarily disfavored. Such “fitness valleys” impose a strong constraint on protein evolution by natural selection (21; 119). A frequent outcome in protein engineering and directed evolution studies is selection for alleles combining multiple mutations that together confer higher level of function, even as each individual mutation confers no effect or lower fitness (48; 76). Based on cross-species comparisons, sign epistasis and fitness valleys remain part of ongoing protein evolution trends (96). Given mutation rates in genomes of wild-type cells as low as <10−4 per gene in somatic (39) and <10−2 per gene in germline (43) generations, the probability of finding even one case of fitness-increasing cluster of multiple mutations in any given gene would be very low, if mutation rates are distributed uniformly over time and across genome space.
THE INCIDENCE AND UTILITY OF MUTATION CLUSTERS: EXAMPLES FROM NATURE
While mutation clustering by random mutagenesis is exceedingly rare, nature has evolved mechanisms that harness clustered mutagenesis to provide selective advantage and/or enhance immune function. As these processes are reviewed extensively elsewhere, we only describe them briefly here.
Retroelement Hypermutation
Life cycles of retroviruses and retrotransposons include reverse transcription of the element’s RNA, producing RNA-DNA hybrid duplexes. The RNA is degraded promptly, leaving a transient ssDNA minus strand intermediate, which in turn is the template for second (plus) DNA strand synthesis (reviewed in (30; 127)). Both retrovirus propagation and retrotransposon mobility are restricted by ssDNA-specific APOBEC cytidine deaminases ((52; 116) also reviewed in (7; 29; 100)). Multiple cytosines are converted into uracils, terminating propagation of the element by mutations inactivating essential functions and/or impeding synthesis of the second DNA strand. If the ssDNA intermediate with multiple deaminations is converted successfully into dsDNA and integrated into a chromosome, it carries multiple mutations (Table 1). As expected, strand-coordinated APOBEC mutation signatures are strongly biased to the minus strand. Curiously only C→T transitions, which do not require uracil excision by uracil glycosylase (UDG/UNG), were observed by several groups (see (100) and below). The relative importance of uracil excision is debatable, as some papers showed independence of hypermutation from UDG/UNG (56; 115), while in others, elimination of glycosylase activity enhanced hypermutation (102; 138). Given the low density of mutations genome-wide, mutations within a hypermutated retroelement represent a cluster. APOBEC clustered mutagenesis in mammalian retroelements is clearly detectable also on an evolutionary scale (6; 22). Ironically, human immunodeficiency virus (HIV) and possibly other retroviruses utilize hypermutation caused by APOBEC host defense system in the perpetual molecular arms race to defeat host defenses (2; 50).
Somatic Hypermutation of Immunoglobulin Genes
Another example of beneficial clustered mutagenesis is somatic hypermutation (SHM) – a critical component of adaptive immunity (Table 1; reviewed in (70; 78; 90; 130). Mammalian SHM occurs primarily in mature B-cells within germinal centers. Multiple mutations within the immunoglobulin variable (~1 kb) region (i.e., mutation clusters) occur within a single or a small number of consecutive cell divisions. The resulting antibodies can have increased affinity to an antigen by several orders of magnitude. Hypermutation is triggered via targeted cytosine deamination by the ssDNA-specific activation induced deaminase (AID) enzyme, which has homology to APOBEC deaminases (32). The uracils are excised enzymatically to generate abasic sites. Lesion bypass by specialized translesion DNA synthesis (TLS) polymerases and error-prone repair often result in fixation of multiple mutations (114). Unlike APOBEC mutagenesis of retroelements, there is at most a moderate strand bias in SHM, which has several potential explanations, including partial accessibility to AID of the DNA strand paired with RNA within transcriptional R-loop(s) (78; 122).
Sidebar: Complex Mutations.
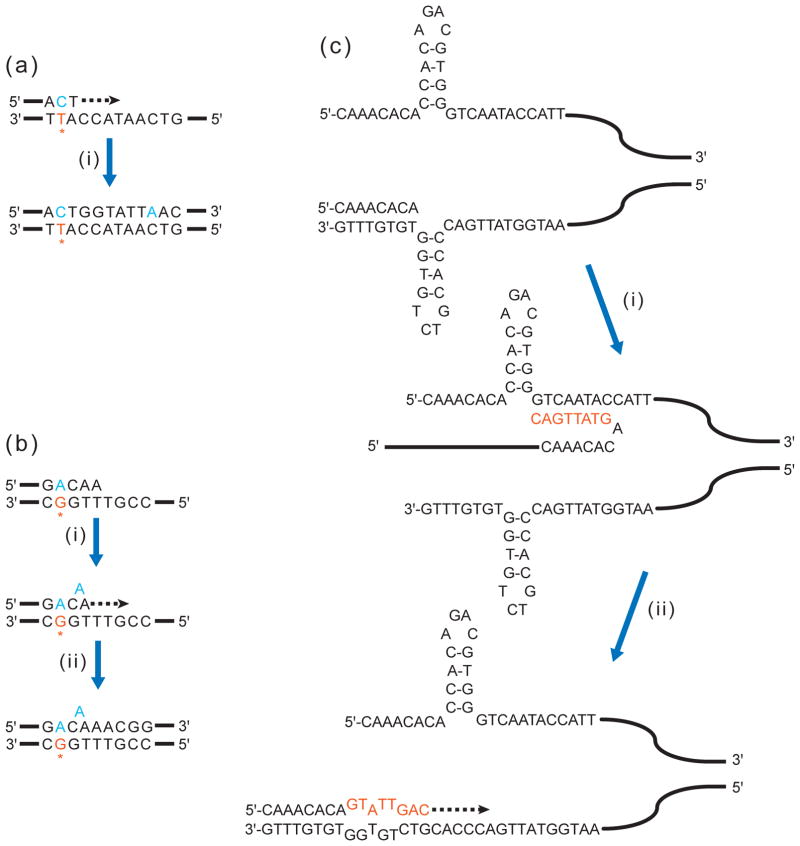
Single-gene mutation spectra often contain a small fraction of closely-spaced base substitutions and/or frameshifts, which are difficult to explain by mere coincidence of independent mutation events (40). Such “complex” mutations (Table 1) occur because error-prone TLS DNA polymerases (77; 117) copy undamaged template in an error-prone way after inserting a mismatch opposite a damaged template base (see Figure 1a, b). Thus, complex mutations might be considered the simplest form of clustered mutagenesis. Several hypothetical mechanisms involve dissociation of a nascent strand from a template and re-association up- or downstream with the same template (11) or more complicated events of template switching to imperfect direct or inverted repeats in the vicinity (ref. (103) and Figure 1c). Multiple mutations result from a combination of nucleotide incorporation errors, TLS and copying of a very short repeated sequence into the nascent strand. As noted by Shcherbakova and colleagues, misalignment and formation of complex mutations can be facilitated by secondary structure formed by inverted repeats in the template strand (85). Given these considerations, the individual base changes within a complex mutation should not be treated as independent mutational events.
Figure 1. Complex mutations can be generated by a single error or obstruction in DNA copying.
(a) Misincorporation by error-prone TLS polymerase. Initiating event: A TLS polymerase inserts a mismatched C (in blue) opposite a damaged T (in orange, with asterisk). Step (i): The TLS polymerase synthesizes several more bases and misincorporates another base (A opposite undamaged C), generating a complex mutation event.
(b) Slippage by error-prone TLS polymerase. Initiating event: A TLS polymerase inserts a mismatched A opposite a damaged G. Step (i): The polymerase synthesizes further, encountering a T homonucleotide run. Slippage within the run produces a single-nucleotide bulge. (ii) Synthesis continues beyond homonucleotide run, leaving the bulge uncorrected. Adapted from Figure 2 in (49).
(c) A replicase is blocked from further synthesis at a hairpin formed by short inverted repeat. Step (i): A TLS polymerase bypasses the blocking hairpin by switching template and synthesizing a short stretch (in orange). Step (ii): The newly extended nascent strand then realigns to original template, despite mismatches to the resolved hairpin (bulging nucleotides). A replicase continues synthesis beyond the mismatches, yielding a complex mutation event. Adapted from Figure 6 in (85).
THE PHENOMENOLOGY OF MUTATION CLUSTERS IN CANCERS
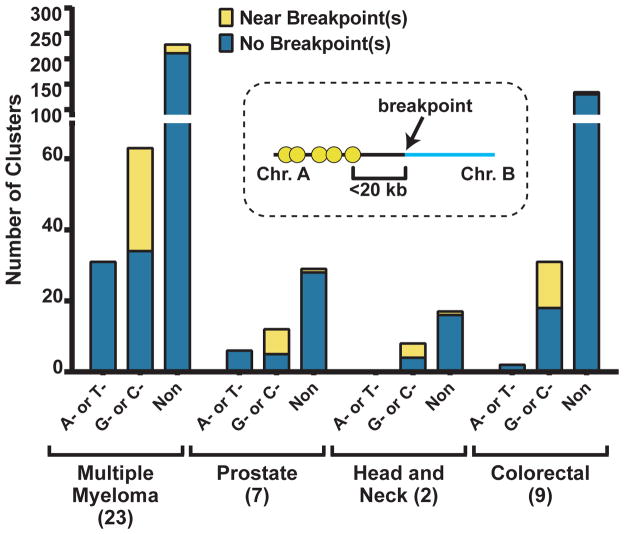
Sequencing of thousands of human cancer genomes and exomes produced datasets with millions of mutations, which provide statistical power for detecting characteristic patterns of mutagenic mechanisms in cancer. Several prominent mutation signatures consisting of mutated nucleotide and immediate (1–2 nucleotides) surrounding context (reviewed in (5; 10; 53; 94; 106)) have been identified. Another striking pattern of mutagenesis revealed by genome-wide sequencing of multiple samples of several cancer types was the presence of many mutation clusters (up to several hundred per sample, see Table 1; also reviewed in (104; 105)), identified by two independent and complementary analytical methods. Using an approach based on statistical evaluation of cluster formation probability and on similarities with general clustering patterns identified in yeast studies (see below), our group (108) detected clustered mutagenesis in all three types of cancers (multiple myeloma, prostate and head-and-neck) whose genome mutation catalogues were available for analysis at the time (Figure 2). Even if the most stringent criterion of strict strand-coordination was imposed, multiple clusters containing mutations originating from the same nucleotide type in the same DNA strand were found (Figure 3a). The most frequent were C- or G-coordinated clusters. These clusters also showed a high frequency of colocalization with breakpoints of chromosome rearrangements (Figure 2). Mutations in C- or G-coordinated clusters frequently occurred at tCw motifs (mutated base capitalized; w corresponds to A or T), characteristic of a subgroup of ssDNA-specific APOBEC cytidine deaminases (APOBEC1, APOBEC3A/B/C/D/F/H) (Figure 3a). Apparently these enzymes, which normally attack ssDNA of retroelements (see above), can cause mutation clusters in chromosomal DNA, if it persists sufficiently long in single-strand form. However, there were key differences in chromosomal clusters as compared with mutations in retroviruses and retrotransposons. Firstly, mutations at the motif for APOBEC3G (cC), which is the major restriction factor for HIV, were depleted in cancers, indicating this enzyme does not access chromosomal DNA. Secondly, in contrast to predominant C→T transitions in retroviral restriction, there were two equally prevalent types of mutations, C→T and C→G (Figure 3c).
Figure 2.
Mutation clusters are abundant across multiple cancer types. Clusters are categorized as: perfectly A- or T-coordinated, C- or G-coordinated, or non-coordinated. Note that nearly half of C- or G-coordinated clusters are found <20 kb from a breakpoint junction between two rearranged chromosomes. Adapted from (107; 108).
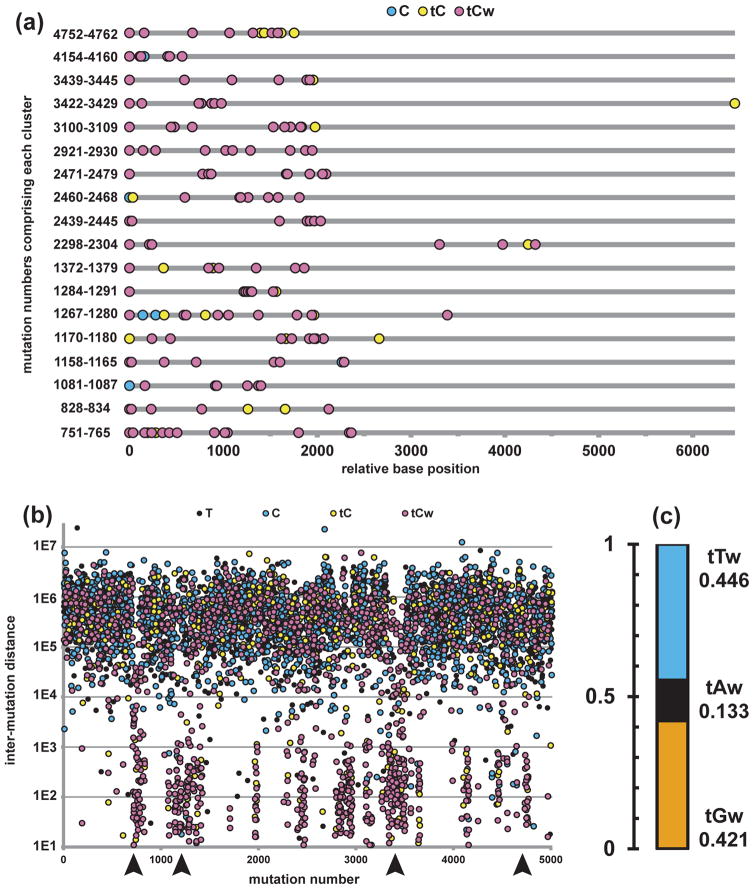
Figure 3. Signature of APOBEC mutagenesis in clusters of a breast cancer genome.
(a) 18 perfectly C- or G-coordinated clusters with >6 mutations from breast cancer sample PD4103a (83) are lined up, with the first mutation assigned as relative base position 1. Mutations are numbered consecutively, from the first mutation in the short arm of chromosome 1 to the last mutation in the long arm of chromosome X. Numbers listed along vertical axis denote first and last mutations that comprise each cluster.
(b) All base substitutions from PD4103a are shown in a “rainfall” plot, where distance between adjacent mutations is plotted on a log scale vs. mutation number (mutation numbers defined as in (a)). Some regions of “kataegis” are indicated by arrowheads. There are many more clusters within the kataegic regions than shown in (a), including shorter and/or not perfectly C- or G-coordinated examples.
(c) C- or G-Coordinated clusters consist predominantly of tCw -> tGw (42.1%) or tCw -> tTw (44.6%) mutations. tCw -> tAw represent only 13.3%.
In a parallel study, Stratton and colelagues (83) noticed a striking non-uniformity of mutation distribution in several breast cancer genomes, when visualized by plotting inter-mutation distances (Figure 3b). Areas with relatively high mutation densities look like rainfall above genomic sections. These events were termed kataegis (thunder shower in Greek), which also underscores possible connection with the previously described phenomenon of “mutation showers” ((132) see Big Blue Mouse sidebar). Similar to our group, Stratton and colleagues noticed high prevalence of mutations in tCw APOBEC motifs, strand coordination of mutations at C- or in G-nucleotides of the same DNA strand and colocalization of kataegis with breakpoints of chromosome rearrangements.
Sidebar: Mutation Showers in Big Blue Mice.
The concept of localized hypermutation in a randomly chosen genomic region emerged from sequencing mutations accumulated in a construct with Lac-repressor carried by lambda bacteriophage, integrated into a mouse chromosome (named “Big Blue Mouse” after LacI mutant screen based on blue colony color (62)). Sommer and colleagues found that a fraction of LacI mutant alleles carried 2–5 mutations within small distances, indicating clustering (15; 55). Sequencing of the region surrounding LacI revealed even more mutations in multiple mutant alleles within the 20 kb region containing LacI. Sommer and colleagues proposed that mutations occurred during a short period of time, possibly in a single cell generation, likening this process to showers falling on a patch of the genome landscape (132). An excess of multiple mutations was also found in human cancer genes (27; 28; 57) and in cell lines from human kidney epithelium (31). The finding of mutation showers stimulated hypotheses about simultaneous (“chronocoordinate” in the terminology suggested by Sommer and colleagues) transient increase in mutability within a small section of the genome; however, neither the hypothesis of simultaneous incidence nor the mechanisms of transient localized hypermutation were supported by data at that time.
Altogether, both studies suggested that in many cancers a combination of two factors, formation of transient stretches of long ssDNA and access of APOBEC enzymes to ssDNA, results in mutation clusters. If so, colocalization of APOBEC-signature clusters with rearrangement breakpoints could be due to higher chance of ssDNA formation around DNA breaks and/or higher chance of strand breakage in ssDNA.
MOLECULAR MECHANISMS OF MUTATION CLUSTER FORMATION
There are pressing questions concerning the molecular origins of mutation clusters, especially in cancers. Several mechanisms underlying mutagenesis generating clusters of multiple mutations have been elucidated (Table 1). In principle, hypermutation can result from localized increase in synthesis error, increase in DNA lesions, decrease in lesion repair or from various combinations of these factors. Studies in model systems demonstrated the existence of clustered simultaneous mutations and revealed mechanistic sources, which for all cases so far appear to involve erroneous copying of damaged template usually by error-prone TLS polymerases (92) and with a single exception are associated with lesions in ssDNA. Such localized changes can be region-specific or occurr at random with respect to genomic position.
Replication Preceding Lesion Repair in Double-Strand (ds) DNA
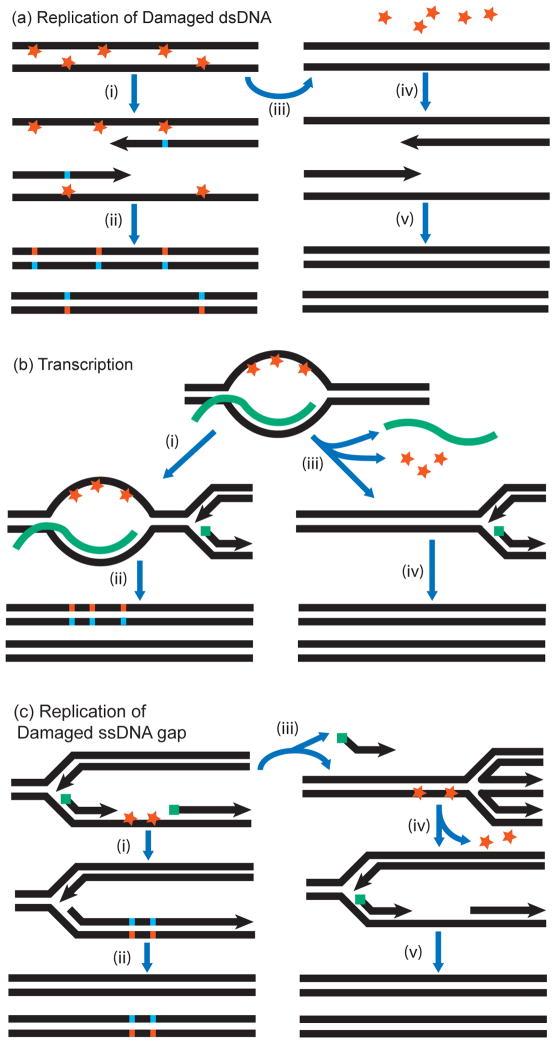
A mechanism of generating clusters of simultaneous mutations could rely on the lesions caused by acute mutagenesis, randomly distributed across the genome combined with reduced or absent repair in a subset of the genome. Potent repair mechanisms can correct lesions in dsDNA to the intact original sequence using the complementary strand as template for repair synthesis (Figure 4a). Repair mechanisms are so efficient that many thousands of lesions, spaced as densely as one lesion per several kb, can be repaired in a single cell cycle. However, if a section of the genome is replicated before repair can complete, TLS would introduce mutations into multiple positions of the nascent strand(s). Importantly, even though lesions were introduced into both DNA strands, clustered mutations in a daughter cell would result from lesions in only one strand. Thus, if lesions are nucleotide- or motif-specific, mutations in the cluster would be strand-coordinated, i.e., mutations of the same kind would be found in the same strand. These phenomena were documented in several whole-genome sequenced (WGS) clones of E. coli grown from cells treated acutely with ethyl methanesulfonate (EMS), a mutagen which is highly specific for inducing G→A changes (88). Cluster sizes were in the hundreds of kb, and mutations were either nearly all G→A or nearly all C→T (as expected if G→A changes occurred in the opposite strand). Clusters were more frequent in the regions that would replicate earlier after cells were restored to growth conditions. This work sets a precedent for clusters originating from locally impeded repair of globally induced lesions in dsDNA, but so far it is the only example of this kind.
Figure 4. Mutation clusters resulting from lesions that had opportunity for templated repair.
(a) Lack of lesion repair in a region of dsDNA. In presence of mutagen, base damage occurs in both strands of dsDNA. Step (i): Replication of unrepaired template strands results in TLS-mediated incorporation of mismatches opposite the lesions. Step (ii): Repair after replication fixes mutation clusters. If a mutagen is base-specific, there will be reciprocal strand-coordination in the two daughter duplexes. Step (iii): Repair occurs before replication. Step (iv): Undamaged templates are replicated. Step (v): Daughter duplexes contain no mutations.
(b) R-loops. A nascent mRNA (in green) remains annealed to its DNA template, generating an R-loop. The displaced ssDNA is damaged. Step (i): A persistent R-loop would block excision repair and replication would proceed on a damaged template top strand. Step (ii): TLS and excision repair fix mutation cluster in a daughter duplex. Step (iii): If the mRNA is removed, the DNA duplex re-anneals, enabling excision repair of damage. Step (iv) Replication of the repaired template is mutation-free.
(c) Lesions in ssDNA of the lagging strand. Perturbation of lagging strand synthesis exposes portion of template ssDNA. Green squares denote RNA primers. Exposed ssDNA sustains base damage. Step (i): Error-prone TLS. Step (ii) Subsequent excision repair fixes mutation cluster in a daughter duplex. Step (iii): Fork regression repositions lesions into dsDNA section, where excision repair can occur. Step (iv): Damage is repaired. Step (v): Replication occurs on undamaged template, resulting in mutation-free daughter duplexes.
Mutation Clusters Originating from Damaged Single-Strand (ss) DNA in Yeast Models
Cells can repair various kinds of DNA damage accurately even if simultaneous lesions have occurred in every 1–10 kb, which for the human genome would result in 105 – 106 lesions genome-wide (44; 69). Most repair pathways use the undamaged complementary strand as a template for accurate DNA synthesis. This could be an intact strand within an ssDNA gap formed around excised damage or within a DNA duplex undergoing strand invasion during template-switching damage avoidance or during recombination repair. While vast numbers of lesions in dsDNA can be repaired accurately, there is much less possibility of faithful repair in patches of ssDNA. Multiple unrepaired lesions could be copied by error-prone TLS (92) resulting in mutation clusters. Mutation clusters could also result from dense ssDNA-specific lesions (see below). Importantly, lesions in ssDNA would result in clustered mutations either because repair has not occurred before the next round of replication (Figure 4), or if accurate lesion repair was not possible because the second strand was unavailable (Figure 5). Recent work in yeast models highlighted several possible molecular mechanisms of ssDNA-associated clusters.
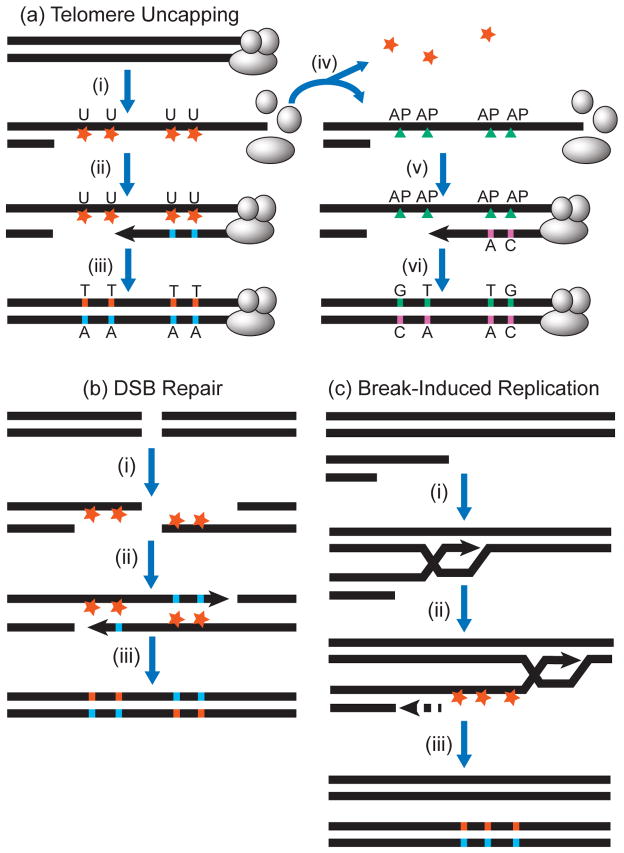
Figure 5. Mutation clusters resulting from lesions in ssDNA without an opportunity for templated repair.
(a) Clusters associated with 5′→3′ resection at uncapped telomeres. Step (i): The protective cap of a telomere dissociates. The uncapped telomere is recognized as a DSB and is resected to generate long 3′ ssDNA overhang. Base damage (orange stars) occurs within the exposed ssDNA, e.g., APOBEC generating uracils. Step (ii): If uracils are not excised, a replicase simply inserts adenines. Step (iii): Base excision repair excises uracils and uses the adenines to template repair, resulting in cluster of C to T transitions. Alternatively, DNA with U:A pairs can be copied without base excision repair, still producing a product with C to T transitions. Step (iv): If uracils created by C-deamination in ssDNA are excised by uracil DNA glycosylase, abasic (AP) sites are generated. Step (v): AP sites are bypassed in an error-prone manner by TLS polymerases, usually inserting either A or C across AP-sites. Step (vi): Excision repair fixes mutation cluster containing a mix of C to T transitions and C to G transversions.
(b) Clusters associated with 5′→3′ resection around a DSB. Step (i): A two-sided DSB is resected to generate 3′ ssDNA overhangs. Base damage at e.g. cytosines (base specificity shown by orange color) occurs within overhangs. Step (ii): Repair synthesis, using TLS polymerase(s) inserts bases opposite the lesions in an error-prone way. Step (iii): Excision repair fixes mutation cluster spanning the DSB region. Note the switch in base specificity of strand-coordination within these clusters.
(c) Clusters associated with ssDNA generated by break-induced replication (BIR). Step (i): The 3′ ssDNA overhang from a resected one-sided DSB invades a homologous sequence in a donor chromosome. Step (ii): Leading strand synthesis (solid arrow) proceeds, while lagging strand synthesis (dashed arrow) occurs later, exposing long stretches of ssDNA, which sustain base damage. Step (iii): Completion of lagging strand synthesis and excision repair fix mutation cluster in repaired chromosome. Note that BIR-associated clusters do not show a switch in base specificity of strand-coordination (compare (c) with (b))
R-loops
Transient stretches of ssDNA called R-loops are formed when nascent mRNA remains annealed to the transcribed strand, displacing a loop of non-coding strand DNA (1). Aguilera and colleagues constructed yeast models where R-loop formation was facilitated by boosting transcription from inducible promoters and further enhanced by genetic inactivation of the THO complex linking transcription and mRNA export (47). Concomitantly, they expressed activation-induced cytidine deaminase (AID), a strong ssDNA mutagen which normally converts cytosines to uracils during somatic hypermutation of immunoglobulin genes (70). In wild-type yeast, combining high transcription and an ssDNA-specific mutagen resulted in a 25-fold increase in mutation frequency. Genetic defects facilitating R-loop formation enhanced mutagenesis by 630-fold. Crucially, increased mutagenesis was observed despite the presence of base excision repair (BER), which should remove uracils if re-annealing of DNA strands occurs before DNA replication (Figure 4b). Interestingly in wild-type yeast, mutations induced by AID were not strand biased, suggesting that the DNA strand paired with RNA in a dynamic fashion. However in a THO-complex mutant, where R-loops should be more stable, AID-specific mutations were mostly in the displaced non-transcribed strand. Although multiple mutations were not reported, this work demonstrated that ssDNA lesions in R-loops could be a source of localized hypermutation. Indeed, hypermutation in Ig-genes correlates with transcription of the immunoglobulin variable region, with evidence suggesting a role for R-loops to provide ssDNA substrates for SHM (reviewed in (78)).
Resection at site-specific DSBs and uncapped telomeres
In yeast, kilobase stretches of ssDNA can be generated by 5′→3′ resection, i.e. strand-biased nucleolytic degradation starting at the ends of double-strand breaks (DSBs) or at unprotected “uncapped” telomeres (Figure 5a and (37; 81)). This ssDNA was suggested as a potential source of increased spontaneous mutation rates around DSBs due to a combination of more lesions and inaccurate DNA synthesis on ssDNA templates (51; 54; 74; 124). Despite high levels of spontaneous mutagenesis near DSBs, this was insufficient to cause clusters of simultaneous mutations (summarized in Fig. 1 of ref. (17)). However, when we and others applied acute DNA damage (ultraviolet light (UV), methyl-methanesulfonate (MMS), oxidative damage (H2O2), sulfites, or APOBEC3G cytidine deaminase) to yeast cells where site-specific DSBs or telomere uncapping were induced by environmental shift, clusters of multiple mutations were found in regions where ssDNA was expected to form (Table 1 and (17; 24; 25; 36; 139; 140)). Another important feature of these experimental systems was that restoration of intact dsDNA could be triggered “at will” after the damage had been inflicted to ssDNA. The systems were designed to restrict the region-specific ssDNA formation to occur within a single cell cycle, which excluded or severely reduced the chance of lesions in ssDNA being repaired from a template of the second DNA strand or from another homologous DNA region. Therefore, all mutations in a cluster likely only arose from TLS in the same long ssDNA region generated in the yeast chromosome. Indeed, hypermutation and cluster generation depended on Polζ, an essential component of yeast TLS.
For APOBEC-mediated mutagenesis, where uracils created from cytosines are first converted to abasic (AP) sites by uracil excision, the two most frequent kinds of mutations were C→T and C→G, and TLS-dependent (Figure 5a). This is consistent with previous studies on TLS at abasic sites (24) and the composition of APOBEC-signature clusters in cancers (4; 83; 107; 108). Also as expected, C→G mutations depended on catalytic activity of Rev1 TLS polymerase. The yeast model clusters carried the signature matching each specific mutagen used for acute treatment, when such a mutational specificity and signature was known a priori (17; 25; 139; 140). Moreover, mutation types and motifs were strand-coordinated and agreed completely with expectations for lesions in a strand left intact after resection (Figure 5a,b). For example the prevailing category of mutations caused by MMS were substitutions of cytosines in the strand oriented 3′→5′ towards the site-specific DSB. Strand-coordinated mutation clusters induced by ssDNA-specific cytidine deaminase APOBEC3G were also observed by Neuberger and colleagues (128). Whole genome-sequencing of several yeast isolates with clusters induced by UV in subtelomeric ssDNA revealed that a single cluster with a size <1% of the yeast genome can contain several times more mutations that in the remaining >99% of the genome that was double-stranded at the time of treatment (17). Significantly, most mutations agreed with the expected pattern of UV-mutagenesis mutating pyrimidines of the unresected strand. This lack of genome-wide hypermutation was due to high efficiency of nucleotide-excision repair, capable of repairing thousands of bulky UV lesions in a single cell cycle. As a result, hypermutation was observed only in regions where ssDNA was formed.
In summary, experiments with acute damage in resected DNA showed that many kilobases of locally formed ssDNA carrying multiple lesions can be restored efficiently to dsDNA with multiple mutations.
Break-induced replication at site-specific DSBs
Recently, another source of long persistent ssDNA stretches was identified in yeast: break-induced replication (BIR). BIR starts with strand invasion by a resected strand from a one-sided DSB into homologous sequence within a sister chromatid or homologous chromosome. BIR then extends by a migrating “bubble” of DNA synthesis to generate the continuous leading strand. Since this nascent leading strand serves as template for lagging strand synthesis, BIR is conservative, unlike S-phase replication which is semi-conservative (reviewed in (75)). Malkova and colleagues found that the two directions of conservative BIR are temporally decoupled, leaving long stretches of the leading strand in ssDNA form ((112) and Figure 5c). In a follow-up study, they showed that BIR-generated ssDNA is also vulnerable to induction of vast (up to 170 kb) clusters of simultaneous multiple (up to 15) mutations by acute methylation damage with MMS (113). Similar to DSB-associated clusters, BIR-associated clusters matched the expected MMS mutational specificity and the predicted strand-coordination. It is noteworthy that the mutation bias associated with BIR is expected to be distinct from that in the vicinity of a DSB. While a cluster associated with DSB repair (see Figure 5b) would switch strand-coordination across the DSB, no strand-coordination switch occurs in BIR-associated clusters (compare with Figure 5c). Intriguingly, clusters also were observed in about half of unselected BIR events. BIR has been recently demonstrated in model studies with mammalian cells (33; 135). It remains to determine if BIR is also a source of ssDNA for APOBEC-induced mutation clusters in cancers.
Chronic Damage to DNA in Proliferating Yeast Cells
Based on experiments in yeast, it became clear how the interplay between ssDNA formation, DNA damage specificity, lesion repair in dsDNA, and copying of damaged DNA can result in mutation cluster formation: (i) Some DNA transaction generates long, persistent ssDNA, which can accumulate multiple lesions after acute DNA damage; (ii) Cells have robust mechanisms to restore long ssDNA with multiple lesions into dsDNA with multiple mutations; (iii) Damage-induced mutations cluster in the region that underwent prolonged ssDNA exposure, either because lesions were ssDNA-specific or because most lesions in dsDNA were repaired accurately. Certainly, this combination of conditions could coincide in natural settings, e.g., in cells proliferating in the presence of chronic DNA damage, resulting in mutation clusters.
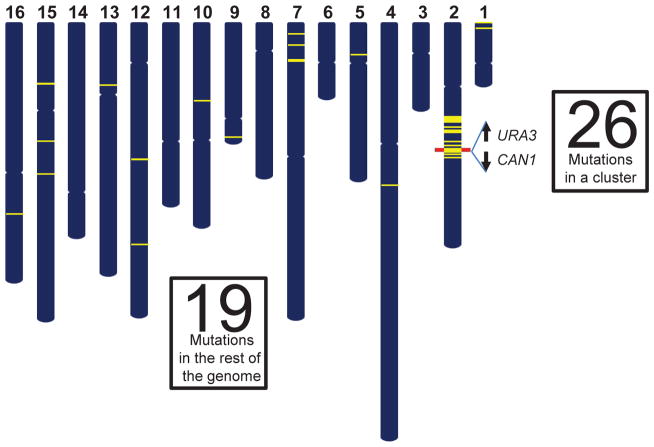
In order to increase the chance of detecting cells with clusters, we placed the URA3 and CAN1 reporter genes close together, allowing selection by 5-fluororotic acid and canavanine, respectively. Multiple mutations inactivating both genes would lead to double drug resistance (108). Indeed, many double-resistant isolates from yeast populations grown in the presence of MMS carried not just single mutations in each of the two genes, but clusters of multiple mutations spanning the double reporter (Figure 6). Mutations in clusters were different in composition from single mutations scattered over the genome and were strand-coordinated, in agreement with their expected origin from MMS alkylation of ssDNA strands with opposite orientation. Strand-coordination strongly implied that mutations in a cluster occurred simultaneously. Amazingly, in many cases the number of simultaneous mutations in a cluster exceeded the number of scattered mutations in the rest of the genome, which likely accumulated over 20–25 generations of growth in the presence of MMS.
Figure 6.
A mutation cluster caused by chronic damage to DNA of proliferating yeast cells (108). Whole genome sequencing of yeast exposed chronically to MMS revealed a large strand-coordinated cluster of 26 mutations, which extended for ~200 kb including the CAN1-URA3 double reporter gene region (red bar), illustrating the hypermutable nature of exposed ssDNA. Strikingly, the rest of genome harbored only 19 mutations.
From several possible mechanisms with a potential to generate ssDNA resulting in vast strand-coordinated clusters (Figures 4 and 5) R-loops appear unlikely, because individual clusters often included non-transcribed regions, and genes transcribed from different strands. Strand-coordinated clusters could have originated from long ssDNA formed by asymmetric, one-sided resection at DSBs or by unusual DNA synthesis during BIR (Figure 5b,c). Interestingly, about 10% of clusters showed a switch of strand bias expected for DSBs with long-range, two-sided resection (Figure 5b). In order to check if ssDNA formed at dysfunctional and/or uncoupled replication forks (Figure 4c) also could be involved in cluster formation, we repositioned the multiple mutation reporter to the other side of the nearest replication origin (108). We reasoned that formation of ssDNA should be biased towards either leading or lagging strand, depending on relative orientation to the origin. This asymmetry should result in strand bias of mutation spectra within strand-coordinated clusters. We did observe this strand bias, but only in yeast strains deleted for TOF1/CSM3 (homologs of human TIMELESS/TIPIN), which control replication fork integrity (59; 66), suggesting that in normal cells ssDNA within replication forks plays a lesser role than DSBs in cluster formation.
Based on results with MMS-induced mutation clusters we concluded, “Once a mutagen is present…the limiting factor in cluster formation appears to be the formation of ssDNA, where the length of ssDNA region and the time it persists are the key parameters determining the cluster’s mutation density and length” (108). For MMS, it was plausible that both ssDNA lesions and derived DSBs were induced by this agent – ssDNA damage by base methylation ((139) and therein) and DSBs by Ntg1/Ntg2 endonucleases introducing ssDNA nicks during BER of closely-opposed alkylated bases in dsDNA (72). Alternatively, breaks could have arisen from replication fork collision with alkylated bases, or with AP-site intermediates of BER. Telling, mutation clusters were found when yeast were grown in the presence of other forms of DNA damage, including ssDNA-specific cytidine deaminases, PmCDA1 from lamprey (63; 64) or human AID/APOBEC (128; 129). Cytosine deamination in ssDNA creates uracils, which are substrates for the yeast uracil DNA glycosylase Ung1 (example on Figure 5a and (25)). AP sites could in turn stimulate ssDNA formation via breakage and/or replication fork uncoupling. Indeed, wild-type yeast had greater numbers of mutation clusters than ung1Δ mutants (128). However in multiple studies, clusters also were observed in the yeast lacking UNG1, suggesting that ssDNA may occur if there is no increased AP-site formation. This could be via spontaneous breaks and/or uncoupled forks. Close examination of mutation distribution across the genome suggested that R-loops might be a secondary source of clusters, especially in tRNA genes (129). Regardless of specific sources of ssDNA and pathways of damage processing, experiments with yeast proliferating in the presence of chronic DNA damage indicated the feasibility of multiple mechanisms associated with lesions in ssDNA, summarized in Figures 4 and 5, as sources of mutation clusters.
Sidebar: Mutation Clusters in the Human Germline.
Meiosis is associated with higher mutation rates than mitotic divisions (73), which could be due to a general phenomenon of increased mutagenesis in the vicinity of DNA breaks (74). Increased density of polymorphisms in the vicinity of meiotic break hotspots have been documented in several studies (8; 82; 97; 120). This correlation was evident even with the most precise location of crossing-over hotspots and with the most conservative choice of only low frequency SNPs correlation analysis (97). However, small numbers of clusters found by three studies reporting de novo mutations in human germline identified by whole-genome sequencing of parent-child trios (20; 43; 79), were not associated with hotspots of meiotic crossovers. In the largest and most comprehensive study (43), clustered mutations showed some preference toward C→G base substitutions. The underlying mechanisms remain unidentified.
SIGNIFICANCE AND FUTURE PROSPECTS
Mutation clusters are a uniquely valuable analytical tool for dissecting mutagenic processes operating in human cancers (reviewed in (104; 106)). The APOBEC mutation signature (tCw→tTw or →tGw) derived from cluster analysis was used for statistical evaluation of APOBEC mutagenesis prevalence in many cancer types (19; 107). This analysis highlighted cervical, bladder, breast, head and neck, and lung cancers as the most enriched with APOBEC mutagenesis signature, which agreed well with hypothesis-independent mutation signature deciphering in multiple cancer types (3; 4). Significantly, even in cancers with low genome- or exome-wide prevalence of APOBEC mutagenesis, there were C- or G-coordinated clusters displaying APOBEC mutagenesis pattern and colocalization with rearrangement breakpoints (summarized in Table 1 of (104); also for kidney chromophobe cancer see Supplemental Figure 5 in (34)). Thus, ssDNA could be the rate-limiting factor while APOBEC activity is present in the background and makes mutations whenever ssDNA becomes available. Consistent with this hypothesis, the APOBEC mutagenesis signature and mutation clusters were detected in plasmids with engineered U/G or T/G mismatches, when such plasmids were transfected into cultured human cells (26), where ssDNA gaps were generated by mismatch-triggered excision repair. The mechanisms producing long, persistent stretches of ssDNA, and distribution thereof, in cancer genomes are important current questions.
Out of seven APOBEC enzymes with potential roles in chromosomal mutagenesis, APOBEC3A and APOBEC3B are considered prime suspects. The potential mutagenic role of APOBEC3B is inferred based on its high expression level breast cancers (18) as well as several other cancer types (19; 107). However, correlation of APOBEC3A expression with mutagenesis was also noted (107). To further complicate the issue, cancers from individuals heterozygous for germline deletion of APOBEC3B, resulting in a fusion transcript encoding for APOBEC3A polypeptide but carrying the 3′-UTR of APOBEC3B, can still have high prevalence of APOBEC-signature mutagenesis (84). In recent studies, the fusion transcript showed higher stability, which may contribute to increased mutagenesis (23) and probability of breast cancer incidence (71; 137).
There is little knowledge about sources and mechanisms of mutation clusters in cancers other than APOBEC. Some hematological cancers also displayed small numbers of clusters with AID mutation signature (wrC, w – A or T; r – purine; mutated nucleotide capitalized) (3; 14; 91; 98; 108). AID-signature clusters often were associated with immunoglobulin gene regions as well as with secondary AID genomic targets (70; 89). Secondary targets of AID mutagenesis and mutation clusters in this type of cancer have been correlated recently with 3D-linked topological domains formed by gene promoters and enhancers within the interphase nucleus (98). When AID secondary target regions known at the time of the study (108) were excluded from cluster analysis in multiple myelomas, we detected no significant AID mutation signature presence. Moreover, in contrast to high enrichment with APOBEC mutation signature, the AID-signature was depleted in C- or G-coordinated clusters, which supports the view that ssDNA per se is not sufficient for AID-hypermutation in vivo. Interestingly, the same study found significant numbers of A- or T-coordinated clusters, with preference to Tw mutation motifs, which is characteristic of DNA synthesis errors of TLS Polη in Ig-SHM region (111). Paradoxically, A- or T-coordinated clusters were not associated with AID-targeted regions of the myeloma genomes.
Non-coordinated clusters in cancers with high prevalence of APOBEC mutagenesis also show statistically significant enrichment with this mutation signature, probably because of mixed presence of mutations from other sources (107; 108). The latter also could be taking advantage of hypermutation in transient ssDNA. Further accumulation of genome-wide mutation catalogues for cancers that are not hypermutated by APOBEC or by AID may enable identification of new mutagens operating on ssDNA opportunstically during the history of cancer. While long ssDNA intermediates are not expected normally in nuclei, they might well be common during mitochondrial DNA replication (58). Interestingly, a strand-biased spectrum of MMS-induced mutations, consistent with action on ssDNA< has been reported recently (125), suggesting that mitochondrial DNA may be vulnerable to clustered mutagenesis.
So far most of the cases of clustered mutagenesis in model studies, as well as many observed in cancers, could be associated plausibly with damage in ssDNA. But other examples remain mechanistically poorly understood. It remains unclear if there are mechanisms other than damage in long persistent ssDNA that can result in cancer mutation clusters. Such mechanisms hypothetically could involve regions of the genome where lesion repair in dsDNA is severely inhibited. For example, there are indications that excision repair pathways are, at most, very weakly active in telomeres(46; 80; 95; 109). If other genomic regions are similarly impaired in key DNA repair activities, colocalization with cancer mutation clusters should be investigated. We and others look forward to tackling the tantalizing challenges of this field in the years to come.
SUMMARY POINTS.
Mutations are not completely random in genome space and in time and often form clusters of individual changes spaced very tightly as compared to mutation distribution in the rest of the genome.
Mutation clusters are formed on regular basis in specific areas of immunoglobulin genes in B-lymphocyte genomes by the action of activation-induced deaminase (AID). This process of somatic hypermutation (SHM) assures generation of antibodies with high affinity to an antigen.
Multiple mutations can be formed in retroelements by the action of APOBEC cytidine deaminases, enzymes which are the part of innate immunity system. ssDNA specific APOBEC enzymes attack ssDNA intermediate of a retroelement replication cycle, which results in hypermutation of integrated retroelement.
Mutation clusters are often found in tumor genomes of several cancer types. Many of these clusters carry clear mutation signature of APOBEC cytidine deaminases. Mutations are probably induced in accidentally formed unusually long and persistent stretches of ssDNA in cells where APOBEC enzymes gained access to chromosomal DNA.
The strongest cases of mutation clustering are so far associated with lesions in transient stretches of single strand DNA. There is only one report about clusters originated from lesions to dsDNA. There are no reports about multiple clustered errors in copying DNA template without lesions.
Lesions in ssDNA can be caused by ssDNA specific environmental agents or by endogenous sources, such as APOBEC/AID cytidine deaminases. Lesions could be also caused by agents acting on both ds and ssDNA. In this case, clustering is due to the lack of repair of the lesions in ssDNA, while lesions in dsDNA are repaired nearly completely. Error-prone TLS fixes ssDNA lesions into mutations in the course of generating a second strand on the damaged ssDNA template.
Clusters in damaged ssDNA can occur, if the lesions did not get a chance to be repaired using the existing complementary DNA strand as a template. Examples include clusters originated from uncoupled replication forks and R-loops
Clusters can occur, if multiple lesions are caused in long stretches of ssDNA formed by 5′→3′ resection at free end of double-strand breaks or uncapped telomeres as well as by break-induced replication.
FUTURE ISSUES.
What are sources of ssDNA in cancer clusters with APOBEC mutagenesis signature?
Which APOBEC enzyme(s) cause clusters in cancers?
Are there other prominent sources of lesions resulting in cancer mutation clusters?
Are there mutation clusters in mitochondrial DNA?
Are there mutation clusters in telomeric repeats and in subtelomeric regions?
Are there mutation clusters in meiosis?
Are there prominent mutation cluster-forming mechanisms originating from lesions in dsDNA?
Are there mutation clusters in somatic cells besides cancer?
Acknowledgments
We are grateful to Drs. Steven A. Roberts and William C. Copeland for comments on the manuscript. We would also like to apologize to those whose relevant contributions could not be cited because of space limitations. Authors acknowledge support by Intramural Research Program of the National Institutes of Health (NIH), National Institute of Environmental Health Sciences (project Z1AES103266, PI - D.A.G) as well as the Pathway to Independence Award 1K99-ES024424-01 to K.C. from the National Institute of Environmental Health Sciences (NIH/NIEHS)
ACRONYMS AND DEFINITIONS
- Mutation cluster
group of mutations with unusually close spacing, as compared to mutations in the bulk of the genome
- Strand-coordinated mutation cluster
mutation cluster where a type of base change occurs exclusively on one DNA strand (e.g., top strand has only C’s mutated)
- Kataegis
alternative description for clustered mutagenesis, often applied to clusters with other unusual features, such as strand-coordination or mutagenesis pattern
- Mutation shower
multiple mutations in a limited genomic space, proposed to represent a mutation cluster
- R-loop
DNA strand displaced by a transcript paired with transcribed strand behind transcription complex
- Retroviruses
viruses with reverse transcription life cycle step in which DNA strand is synthesized from RNA template
- Retrotransposons
mobile elements whose transposition mechanisms include reverse transcription step
- Retroelements
common name referring to both retroviruses and retrotransposons
- Somatic Hypermutation (SHM)
extremely high mutation rate confined to hypermutable regions of immunoglobulin genes in B-cells
- Translesion DNA synthesis (TLS)
error-prone synthesis by specialized DNA polymerase(s) to bypass lesions in damaged template, often resulting in mutations
- Replication error
insertion of mismatched nucleotide(s) into nascent strand by DNA polymerase copying undamaged template
- Resection
5′→3′ enzymatic degradation of DNA strand initiated at free DNA ends (double-strand breaks or uncapped telomeres)
- Break-induced replication (BIR)
replication initiated by strand-invasion from a free DNA end
- Base-excision repair (BER)
repair of base damage in dsDNA initiated by excision of a damaged base, while backbone is left intact
- AP-site (abasic site) in DNA
site in DNA strand from which a base was excised in the first step of base excision repair (BER)
- Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC)
family of cytidine deaminases converting cytidines into uridines in RNA or DNA.
- AID
cytidine deaminase from APOBEC family operating in somatic hypermutation (SHM)
- Mutation motif
preferred DNA sequence associated with certain exogenous or endogenous mutagenic factors
- Mutation signature
mutation motif with preference for certain kinds of base substitutions
- Mutagenesis pattern
increased presence and/or non-random distribution (e.g., clustering) of a mutation signature in the genome
Contributor Information
Kin Chan, Email: kin.chan@nih.gov.
Dmitry A. Gordenin, Email: gordenin@niehs.nih.gov.
LITERATURE CITED
- 1.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–24. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Albin JS, Hache G, Hultquist JF, Brown WL, Harris RS. Long-term restriction by APOBEC3F selects human immunodeficiency virus type 1 variants with restored Vif function. Journal of virology. 2010;84:10209–19. doi: 10.1128/JVI.00632-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell reports. 2013;3:246–59. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev. 2014;24:52–60. doi: 10.1016/j.gde.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anwar F, Davenport MP, Ebrahimi D. Footprint of APOBEC3 on the genome of human retroelements. Journal of virology. 2013;87:8195–204. doi: 10.1128/JVI.00298-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arias JF, Koyama T, Kinomoto M, Tokunaga K. Retroelements versus APOBEC3 family members: No great escape from the magnificent seven. Frontiers in Microbiology. 2012:3. doi: 10.3389/fmicb.2012.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Auton A, Fledel-Alon A, Pfeifer S, Venn O, Segurel L, et al. A fine-scale chimpanzee genetic map from population sequencing. Science. 2012;336:193–8. doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ayala FJ, Fitch WM. Genetics and the origin of species: An introduction. Proceedings of the National Academy of Sciences. 1997;94:7691–7. doi: 10.1073/pnas.94.15.7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bacolla A, Cooper DN, Vasquez KM. Mechanisms of base substitution mutagenesis in cancer genomes. Genes. 2014;5:108–46. doi: 10.3390/genes5010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bebenek K, Kunkel TA. Frameshift errors initiated by nucleotide misincorporation. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:4946–50. doi: 10.1073/pnas.87.13.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benzer S, Freese E. Induction of Specific Mutations with 5-Bromouracil. Proceedings of the National Academy of Sciences of the United States of America. 1958;44:112–9. doi: 10.1073/pnas.44.2.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berglund J, Pollard KS, Webster MT. Hotspots of biased nucleotide substitutions in human genes. PLoS biology. 2009;7:e26. doi: 10.1371/journal.pbio.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nature communications. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buettner VL, Hill KA, Scaringe WA, Sommer SS. Evidence that proximal multiple mutations in Big Blue transgenic mice are dependent events. Mutation research. 2000;452:219–29. doi: 10.1016/s0027-5107(00)00090-7. [DOI] [PubMed] [Google Scholar]
- 16.Burbano HA, Green RE, Maricic T, Lalueza-Fox C, de la Rasilla M, et al. Analysis of human accelerated DNA regions using archaic hominin genomes. PloS one. 2012;7:e32877. doi: 10.1371/journal.pone.0032877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burch LH, Yang Y, Sterling JF, Roberts SA, Chao FG, et al. Damage-induced localized hypermutability. Cell Cycle. 2011;10:1073–85. doi: 10.4161/cc.10.7.15319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–70. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;45:977–83. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell CD, Chong JX, Malig M, Ko A, Dumont BL, et al. Estimating the human mutation rate using autozygosity in a founder population. Nat Genet. 2012;44:1277–81. doi: 10.1038/ng.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camps M, Herman A, Loh E, Loeb LA. Genetic constraints on protein evolution. Crit Rev Biochem Mol Biol. 2007;42:313–26. doi: 10.1080/10409230701597642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carmi S, Church GM, Levanon EY. Large-scale DNA editing of retrotransposons accelerates mammalian genome evolution. Nature communications. 2011;2:519. doi: 10.1038/ncomms1525. [DOI] [PubMed] [Google Scholar]
- 23.Caval V, Suspene R, Shapira M, Vartanian JP, Wain-Hobson S. A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3′UTR enhances chromosomal DNA damage. Nature communications. 2014;5:5129. doi: 10.1038/ncomms6129. [DOI] [PubMed] [Google Scholar]
- 24.Chan K, Resnick MA, Gordenin DA. The choice of nucleotide inserted opposite abasic sites formed within chromosomal DNA reveals the polymerase activities participating in translesion DNA synthesis. DNA Repair (Amst) 2013 doi: 10.1016/j.dnarep.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan K, Sterling JF, Roberts SA, Bhagwat AS, Resnick MA, Gordenin DA. Base damage within single-strand DNA underlies in vivo hypermutability induced by a ubiquitous environmental agent. PLoS Genet. 2012;8:e1003149. doi: 10.1371/journal.pgen.1003149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Miller BF, Furano AV. Repair of naturally occurring mismatches can induce mutations in flanking DNA. Elife. 2014;3:e02001. doi: 10.7554/eLife.02001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Z, Feng J, Buzin CH, Sommer SS. Epidemiology of doublet/multiplet mutations in lung cancers: evidence that a subset arises by chronocoordinate events. PloS one. 2008;3:e3714. doi: 10.1371/journal.pone.0003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Z, Feng J, Saldivar JS, Gu D, Bockholt A, Sommer SS. EGFR somatic doublets in lung cancer are frequent and generally arise from a pair of driver mutations uncommonly seen as singlet mutations: one-third of doublets occur at five pairs of amino acids. Oncogene. 2008;27:4336–43. doi: 10.1038/onc.2008.71. [DOI] [PubMed] [Google Scholar]
- 29.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annual review of immunology. 2008;26:317–53. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 30.Coffin JM, Hughes SH, Varmus H. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. p. xv.p. 843. [PubMed] [Google Scholar]
- 31.Colgin LM, Hackmann AF, Emond MJ, Monnat RJ., Jr The unexpected landscape of in vivo somatic mutation in a human epithelial cell lineage. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:1437–42. doi: 10.1073/pnas.032655699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Molecular biology and evolution. 2005;22:367–77. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 33.Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, et al. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis CF, Ricketts CJ, Wang M, Yang L, Cherniack AD, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26:319–30. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, et al. Break-induced replication is highly inaccurate. PLoS biology. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Degtyareva NP, Heyburn L, Sterling J, Resnick MA, Gordenin DA, Doetsch PW. Oxidative stress-induced mutagenesis in single-strand DNA occurs primarily at cytosines and is DNA polymerase zeta-dependent only for adenines and guanines. Nucleic acids research. 2013;41:8995–9005. doi: 10.1093/nar/gkt671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dewar JM, Lydall D. Similarities and differences between “uncapped” telomeres and DNA double-strand breaks. Chromosoma. 2012;121:117–30. doi: 10.1007/s00412-011-0357-2. [DOI] [PubMed] [Google Scholar]
- 38.Dobzhansky TG. Genetics and the Origin of Species. Columbia University Press; 1937. [Google Scholar]
- 39.Drake JW. The distribution of rates of spontaneous mutation over viruses, prokaryotes, and eukaryotes. Ann N Y Acad Sci. 1999;870:100–7. doi: 10.1111/j.1749-6632.1999.tb08870.x. [DOI] [PubMed] [Google Scholar]
- 40.Drake JW. Too many mutants with multiple mutations. Crit Rev Biochem Mol Biol. 2007;42:247–58. doi: 10.1080/10409230701495631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dreszer TR, Wall GD, Haussler D, Pollard KS. Biased clustered substitutions in the human genome: the footprints of male-driven biased gene conversion. Genome Res. 2007;17:1420–30. doi: 10.1101/gr.6395807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duret L, Arndt PF. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008;4:e1000071. doi: 10.1371/journal.pgen.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Francioli LC, Sunyaev S, et al. Genome-wide patterns and properties of de novo mutations in humans. Personal communication. 2015. [DOI] [PMC free article] [PubMed]
- 44.Friedberg E, Walker G, Siede W, Wood R, Schultz R, Ellenberger T. DNA Repair and Mutagenesis. 2. Washington, DC: ASM Press; 2005. [Google Scholar]
- 45.Friedberg EC. DNA damage and repair. Nature. 2003;421:436–40. doi: 10.1038/nature01408. [DOI] [PubMed] [Google Scholar]
- 46.Fumagalli M, Rossiello F, Clerici M, Barozzi S, Cittaro D, et al. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nature cell biology. 2012;14:355–65. doi: 10.1038/ncb2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gomez-Gonzalez B, Aguilera A. Activation-induced cytidine deaminase action is strongly stimulated by mutations of the THO complex. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8409–14. doi: 10.1073/pnas.0702836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guthrie VB, Allen J, Camps M, Karchin R. Network models of TEM beta-lactamase mutations coevolving under antibiotic selection show modular structure and anticipate evolutionary trajectories. PLoS computational biology. 2011;7:e1002184. doi: 10.1371/journal.pcbi.1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harfe BD, Jinks-Robertson S. DNA polymerase zeta introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol Cell. 2000;6:1491–9. doi: 10.1016/s1097-2765(00)00145-3. [DOI] [PubMed] [Google Scholar]
- 50.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–9. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 51.Harris RS, Longerich S, Rosenberg SM. Recombination in adaptive mutation. Science. 1994;264:258–60. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- 52.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Molecular Cell. 2002;10:1247–53. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 53.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nature reviews. Genetics. 2014;15:585–98. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–5. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hill KA, Wang J, Farwell KD, Scaringe WA, Sommer SS. Spontaneous multiple mutations show both proximal spacing consistent with chronocoordinate events and alterations with p53-deficiency. Mutation research. 2004;554:223–40. doi: 10.1016/j.mrfmmm.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 56.Kaiser SM, Emerman M. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. Journal of virology. 2006;80:875–82. doi: 10.1128/JVI.80.2.875-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kashuba VI, Pavlova TV, Grigorieva EV, Kutsenko A, Yenamandra SP, et al. High mutability of the tumor suppressor genes RASSF1 and RBSP3 (CTDSPL) in cancer. PloS one. 2009;4:e5231. doi: 10.1371/journal.pone.0005231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kasiviswanathan R, Collins TR, Copeland WC. The interface of transcription and DNA replication in the mitochondria. Biochimica et biophysica acta. 2012;1819:970–8. doi: 10.1016/j.bbagrm.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaufmann WK. Initiating the uninitiated: replication of damaged DNA and carcinogenesis. Cell Cycle. 2007;6:1460–7. [PubMed] [Google Scholar]
- 60.Kimura M. The neutral theory of molecular evolution. Cambridge University Press; 1984. [Google Scholar]
- 61.Kogenaru M, de Vos MG, Tans SJ. Revealing evolutionary pathways by fitness landscape reconstruction. Crit Rev Biochem Mol Biol. 2009;44:169–74. doi: 10.1080/10409230903039658. [DOI] [PubMed] [Google Scholar]
- 62.Kohler SW, Provost GS, Fieck A, Kretz PL, Bullock WO, et al. Spectra of spontaneous and mutagen-induced mutations in the lacI gene in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:7958–62. doi: 10.1073/pnas.88.18.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lada AG, Dhar A, Boissy RJ, Hirano M, Rubel AA, et al. AID/APOBEC cytosine deaminase induces genome-wide kataegis. Biol Direct. 2012;7:47. doi: 10.1186/1745-6150-7-47. discussion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lada AG, Stepchenkova EI, Waisertreiger ISR, Noskov VN, Dhar A, et al. Genome-Wide Mutation Avalanches Induced in Diploid Yeast Cells by a Base Analog or an APOBEC Deaminase. PLoS Genet. 2013;9:e1003736. doi: 10.1371/journal.pgen.1003736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leman AR, Noguchi E. Local and global functions of Timeless and Tipin in replication fork protection. Cell Cycle. 2012;11:3945–55. doi: 10.4161/cc.21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lercher MJ, Hurst LD. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 2002;18:337–40. doi: 10.1016/s0168-9525(02)02669-0. [DOI] [PubMed] [Google Scholar]
- 68.Levine JG, Schaaper RM, DeMarini DM. Complex frameshift mutations mediated by plasmid pKM101: mutational mechanisms deduced from 4-aminobiphenyl-induced mutation spectra in Salmonella. Genetics. 1994;136:731–46. doi: 10.1093/genetics/136.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol. 2000;65:127–33. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- 70.Liu M, Schatz DG. Balancing AID and DNA repair during somatic hypermutation. Trends in immunology. 2009;30:173–81. doi: 10.1016/j.it.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 71.Long J, Delahanty RJ, Li G, Gao YT, Lu W, et al. A common deletion in the APOBEC3 genes and breast cancer risk. J Natl Cancer Inst. 2013;105:573–9. doi: 10.1093/jnci/djt018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma W, Resnick MA, Gordenin DA. Apn1 and Apn2 endonucleases prevent accumulation of repair-associated DNA breaks in budding yeast as revealed by direct chromosomal analysis. Nucleic Acids Res. 2008;36:1836–46. doi: 10.1093/nar/gkm1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Magni GE, Von Borstel RC. Different Rates of Spontaneous Mutation during Mitosis and Meiosis in Yeast. Genetics. 1962;47:1097–108. doi: 10.1093/genetics/47.8.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Malkova A, Haber JE. Mutations arising during repair of chromosome breaks. Annu Rev Genet. 2012;46:455–73. doi: 10.1146/annurev-genet-110711-155547. [DOI] [PubMed] [Google Scholar]
- 75.Malkova A, Ira G. Break-induced replication: functions and molecular mechanism. Curr Opin Genet Dev. 2013;23:271–9. doi: 10.1016/j.gde.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mannervik B, Runarsdottir A. The quest for molecular quasi-species in ligand-activity space and its application to directed enzyme evolution. FEBS letters. 2010;584:2565–71. doi: 10.1016/j.febslet.2010.04.024. [DOI] [PubMed] [Google Scholar]
- 77.Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel TA. Low fidelity DNA synthesis by human DNA polymerase-eta. Nature. 2000;404:1011–3. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- 78.Maul RW, Gearhart PJ. AID and somatic hypermutation. Adv Immunol. 2010;105:159–91. doi: 10.1016/S0065-2776(10)05006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Michaelson JJ, Shi Y, Gujral M, Zheng H, Malhotra D, et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151:1431–42. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller AS, Balakrishnan L, Buncher NA, Opresko PL, Bambara RA. Telomere proteins POT1, TRF1 and TRF2 augment long-patch base excision repair in vitro. Cell Cycle. 2012;11:998–1007. doi: 10.4161/cc.11.5.19483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mimitou EP, Symington LS. DNA end resection--unraveling the tail. DNA Repair (Amst) 2011;10:344–8. doi: 10.1016/j.dnarep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nachman MW. Single nucleotide polymorphisms and recombination rate in humans. Trends Genet. 2001;17:481–5. doi: 10.1016/s0168-9525(01)02409-x. [DOI] [PubMed] [Google Scholar]
- 83.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–93. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nik-Zainal S, Wedge DC, Alexandrov LB, Petljak M, Butler AP, et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat Genet. 2014;46:487–91. doi: 10.1038/ng.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Northam MR, Moore EA, Mertz TM, Binz SK, Stith CM, et al. DNA polymerases ζ and Rev1 mediate error-prone bypass of non-B DNA structures. Nucleic Acids Research. 2014;42:290–306. doi: 10.1093/nar/gkt830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Northam MR, Robinson HA, Kochenova OV, Shcherbakova PV. Participation of DNA polymerase zeta in replication of undamaged DNA in Saccharomyces cerevisiae. Genetics. 2010;184:27–42. doi: 10.1534/genetics.109.107482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ohta T. Simulating evolution by gene duplication. Genetics. 1987;115:207–13. doi: 10.1093/genetics/115.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Parkhomchuk D, Amstislavskiy V, Soldatov A, Ogryzko V. Use of high throughput sequencing to observe genome dynamics at a single cell level. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20830–5. doi: 10.1073/pnas.0906681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pavri R, Nussenzweig MC. AID targeting in antibody diversity. Adv Immunol. 2011;110:1–26. doi: 10.1016/B978-0-12-387663-8.00005-3. [DOI] [PubMed] [Google Scholar]
- 90.Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, et al. The biochemistry of somatic hypermutation. Annual review of immunology. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 91.Pettersen HS, Galashevskaya A, Doseth B, Sousa MM, Sarno A, et al. AID expression in B-cell lymphomas causes accumulation of genomic uracil and a distinct AID mutational signature. DNA Repair (Amst) 2015;25:60–71. doi: 10.1016/j.dnarep.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 92.Plosky BS, Woodgate R. Switching from high-fidelity replicases to low-fidelity lesion-bypass polymerases. Curr Opin Genet Dev. 2004;14:113–9. doi: 10.1016/j.gde.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 93.Pollard KS, Salama SR, King B, Kern AD, Dreszer T, et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2:e168. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Poon S, McPherson J, Tan P, Teh B, Rozen S. Mutation signatures of carcinogen exposure: genome-wide detection and new opportunities for cancer prevention. Genome Medicine. 2014;6:24. doi: 10.1186/gm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pope-Varsalona H, Liu F-J, Guzik L, Opresko PL. Polymerase η suppresses telomere defects induced by DNA damaging agents. Nucleic Acids Research. 2014;42:13096–109. doi: 10.1093/nar/gku1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Povolotskaya IS, Kondrashov FA. Sequence space and the ongoing expansion of the protein universe. Nature. 2010;465:922–6. doi: 10.1038/nature09105. [DOI] [PubMed] [Google Scholar]
- 97.Pratto F, Brick K, Khil P, Smagulova F, Petukhova GV, Camerini-Otero RD. DNA recombination. Recombination initiation maps of individual human genomes. Science. 2014;346:1256442. doi: 10.1126/science.1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qian J, Wang Q, Dose M, Pruett N, Kieffer-Kwon KR, et al. B Cell Super-Enhancers and Regulatory Clusters Recruit AID Tumorigenic Activity. Cell. 2014;159:1524–37. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rebhandl S, Huemer M, Gassner FJ, Zaborsky N, Hebenstreit D, et al. APOBEC3 signature mutations in chronic lymphocytic leukemia. Leukemia. 2014;28:1929–32. doi: 10.1038/leu.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Refsland EW, Harris RS. The APOBEC3 family of retroelement restriction factors. Curr Top Microbiol Immunol. 2013;371:1–27. doi: 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rhind N, Gilbert DM. DNA replication timing. Cold Spring Harbor perspectives in biology. 2013;5:a010132. doi: 10.1101/cshperspect.a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Richardson SR, Narvaiza I, Planegger RA, Weitzman MD, Moran JV. APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. 2014 doi: 10.7554/eLife.02008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ripley LS. Frameshift mutation: determinants of specificity. Annu Rev Genet. 1990;24:189–213. doi: 10.1146/annurev.ge.24.120190.001201. [DOI] [PubMed] [Google Scholar]
- 104.Roberts SA, Gordenin DA. Clustered and genome-wide transient mutagenesis in human cancers: Hypermutation without permanent mutators or loss of fitness. BioEssays : news and reviews in molecular, cellular and developmental biology. 2014 doi: 10.1002/bies.201300140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Roberts SA, Gordenin DA. eLS. John Wiley & Sons, Ltd; 2014. Clustered Mutations in Human Cancer. Number of. [Google Scholar]
- 106.Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer. 2014;14:786–800. doi: 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–6. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roberts SA, Sterling J, Thompson C, Harris S, Mav D, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46:424–35. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rochette PJ, Brash DE. Human telomeres are hypersensitive to UV-induced DNA Damage and refractory to repair. PLoS Genet. 2010;6:e1000926. doi: 10.1371/journal.pgen.1000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rogozin IB, Pavlov YI. Theoretical analysis of mutation hotspots and their DNA sequence context specificity. Mutat Res-Rev Mutat. 2003;544:65–85. doi: 10.1016/s1383-5742(03)00032-2. [DOI] [PubMed] [Google Scholar]
- 111.Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nature immunology. 2001;2:530–6. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 112.Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, et al. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502:389–92. doi: 10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sakofsky CJ, Roberts SA, Malc E, Mieczkowski PA, Resnick MA, et al. Break-induced replication is a source of mutation clusters underlying kataegis. Cell reports. 2014;7:1640–8. doi: 10.1016/j.celrep.2014.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sale JE. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harbor perspectives in biology. 2013;5:a012708. doi: 10.1101/cshperspect.a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schumacher AJ, Hache G, Macduff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. Journal of virology. 2008;82:2652–60. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schumacher AJ, Nissley DV, Harris RS. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9854–9. doi: 10.1073/pnas.0501694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shcherbakova PV, Fijalkowska IJ. Translesion synthesis DNA polymerases and control of genome stability. Frontiers in bioscience : a journal and virtual library. 2006;11:2496–517. doi: 10.2741/1985. [DOI] [PubMed] [Google Scholar]
- 118.Sima J, Gilbert DM. Complex correlations: replication timing and mutational landscapes during cancer and genome evolution. Curr Opin Genet Dev. 2014;25:93–100. doi: 10.1016/j.gde.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Smith JM. Natural selection and the concept of a protein space. Nature. 1970;225:563–4. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- 120.Spencer CC, Deloukas P, Hunt S, Mullikin J, Myers S, et al. The influence of recombination on human genetic diversity. PLoS Genet. 2006;2:e148. doi: 10.1371/journal.pgen.0020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stamatoyannopoulos JA, Adzhubei I, Thurman RE, Kryukov GV, Mirkin SM, Sunyaev SR. Human mutation rate associated with DNA replication timing. Nat Genet. 2009;41:393–5. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Steele EJ. Mechanism of somatic hypermutation: critical analysis of strand biased mutation signatures at A:T and G:C base pairs. Molecular immunology. 2009;46:305–20. doi: 10.1016/j.molimm.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 123.Stone JE, Lujan SA, Kunkel TA. DNA polymerase zeta generates clustered mutations during bypass of endogenous DNA lesions in Saccharomyces cerevisiae. Environmental and Molecular Mutagenesis. 2012;53:777–86. doi: 10.1002/em.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Strathern JN, Shafer BK, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–72. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stumpf JD, Copeland WC. MMS exposure promotes increased MtDNA mutagenesis in the presence of replication-defective disease-associated DNA polymerase gamma variants. PLoS Genet. 2014;10:e1004748. doi: 10.1371/journal.pgen.1004748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sunagar K, Fry BG, Jackson TN, Casewell NR, Undheim EA, et al. Molecular evolution of vertebrate neurotrophins: co-option of the highly conserved nerve growth factor gene into the advanced snake venom arsenalf. PloS one. 2013;8:e81827. doi: 10.1371/journal.pone.0081827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tang H, Kuhen KL, Wong-Staal F. Lentivirus replication and regulation. Annu Rev Genet. 1999;33:133–70. doi: 10.1146/annurev.genet.33.1.133. [DOI] [PubMed] [Google Scholar]
- 128.Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, et al. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Taylor BJ, Wu YL, Rada C. Active RNAP pre-initiation sites are highly mutated by cytidine deaminases in yeast, with AID targeting small RNA genes. Elife. 2014:3. doi: 10.7554/eLife.03553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Teng G, Papavasiliou FN. Immunoglobulin somatic hypermutation. Annu Rev Genet. 2007;41:107–20. doi: 10.1146/annurev.genet.41.110306.130340. [DOI] [PubMed] [Google Scholar]
- 131.Timoféeff-Ressovsky N, Zimmer K, Delbrück M. Über die Natur der Genmutation und der Genkostruktur (The nature of genetic mutations and structure of the gene) Nachrichten aus der Biologie der Gesellschaft der Wissenschaften Göttingen. 1935;1:189–245. [Google Scholar]
- 132.Wang J, Gonzalez KD, Scaringe WA, Tsai K, Liu N, et al. Evidence for mutation showers. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8403–8. doi: 10.1073/pnas.0610902104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Weinreich DM, Watson RA, Chao L. Perspective: Sign epistasis and genetic constraint on evolutionary trajectories. Evolution. 2005;59:1165–74. [PubMed] [Google Scholar]
- 134.Wilburn DB, Bowen KE, Doty KA, Arumugam S, Lane AN, et al. Structural insights into the evolution of a sexy protein: novel topology and restricted backbone flexibility in a hypervariable pheromone from the red-legged salamander, Plethodon shermani. PloS one. 2014;9:e96975. doi: 10.1371/journal.pone.0096975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Willis NA, Chandramouly G, Huang B, Kwok A, Follonier C, et al. BRCA1 controls homologous recombination at Tus/Ter-stalled mammalian replication forks. Nature. 2014;510:556–9. doi: 10.1038/nature13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wright S. The roles of mutation, inbreeding, crossbreading and selection in evolution. Proc. Proceedings of The Sixth International Congress on Genetics; 1932; pp. 356–66. [Google Scholar]
- 137.Xuan D, Li G, Cai Q, Deming-Halverson S, Shrubsole MJ, et al. APOBEC3 deletion polymorphism is associated with breast cancer risk among women of European ancestry. Carcinogenesis. 2013;34:2240–3. doi: 10.1093/carcin/bgt185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yang B, Chen K, Zhang C, Huang S, Zhang H. Virion-associated uracil DNA glycosylase-2 and apurinic/apyrimidinic endonuclease are involved in the degradation of APOBEC3G-edited nascent HIV-1 DNA. The Journal of biological chemistry. 2007;282:11667–75. doi: 10.1074/jbc.M606864200. [DOI] [PubMed] [Google Scholar]
- 139.Yang Y, Gordenin DA, Resnick MA. A single-strand specific lesion drives MMS-induced hyper-mutability at a double-strand break in yeast. DNA Repair (Amst) 2010;9:914–21. doi: 10.1016/j.dnarep.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 2008;4:e1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]