Abstract
The hotdog-fold enzyme 4-hydroxybenzoyl-coenzyme A (4-HB-CoA) thioesterase from Arthrobacter sp strain AU catalyzes the hydrolysis of 4-HB-CoA to form 4-hydroxybenzoate (4-HB) and coenzyme A (CoA) in the final step of the 4-chlorobenzoate dehalogenation pathway. Guided by the published X-ray structures of the liganded enzyme (Thoden J. B., Zhuang Z., Dunaway-Mariano D., Holden H. M. J. Biol. Chem. 2003 278, 43709-43716) a series of site-directed mutants were prepared for testing the roles of active site residues in substrate binding and catalysis. The mutant thioesterases were subjected to X-ray structure determination to confirm retention of the Native fold, and in some cases, to reveal changes in the active site configuration. In parallel, the wild-type and mutant thioesterases were subjected to transient and steady-state kinetic analysis, and to 18O-solvent labeling experiments. Evidence is provided that suggests that Glu73 functions in nucleophilic catalysis, that Gly65 and Gln58 contribute to transition-state stabilization via hydrogen bond formation with the thioester moiety and that Thr77 orients the water nucleophile for attack at the 4-hydroxybenzoyl carbon of the enzyme-anhydride intermediate. The replacement of Glu73 with Asp was shown to switch the function of the carboxylate residue from nucleophilic catalysis to base catalysis and thus, the reaction from a two-step process involving a covalent enzyme intermediate to a single-step hydrolysis reaction. The E73D/T77A double mutant regained most of the catalytic efficiency lost in the E73D single mutant. The results from 31P-NMR experiments indicate that the substrate nucleotide unit is bound to the enzyme surface. Kinetic analysis of site-directed mutants was carried out to determine the contributions made by Arg102, Arg150, Ser120 and Thr121 in binding the nucleotide unit. Lastly, we show by kinetic and X-ray analyses of Asp31, His64 and Glu78 site directed mutants that these three active site residues are important for productive binding of the substrate 4-hydroxybenzoyl ring.
Keywords: 4-hydroxybenzoyl-CoA thioesterase, nucleophilic catalysis, thioester hydrolysis, hotdog-fold family, divergent evolution, enzyme mechanism
Organic acids comprise ~20 % of the E. coli metabolome (1). The carboxylate group increases the solubility of the metabolite in the aqueous environment of the cell and it provides a source of binding energy needed for metabolite-protein association. Organic acids are activated for acyl transfer, Claisen condensation, Michael addition and β-elimination reactions by conversion to the corresponding thioester (2-4), a reaction that is catalyzed by ATP-dependent synthetases or ligases (5). The most common thiol unit used for thioester formation is the pantetheine phosphate moiety of coenzyme A (CoA)1 or holo-acyl carrier proteins (holo-ACP).
Thioesterases catalyze the hydrolysis of the thioester metabolite as needed to release the chemically transformed organic acid. Thioesterases also function in a housekeeping mode to release the CoA or holo-ACP from pathway intermediates that accumulate in stalled pathways (for examples see references 6-10). Thioesterases have primarily evolved within two large enzyme superfamilies families: the α/β-hydrolase-fold (11) family and the hotdog-fold family (12) (for a recent reviews see references 13 and 14). The well-studied α/β-hydrolase-fold family employs an Asp/Glu-His-Ser catalytic triad to catalyze the hydrolysis of amides and oxygen esters in addition to the comparatively less challenging thioester substrates (15). The hotdog-fold family contributes more than half of the cellular thioesterases, but hotdog-fold amidases or O-esterases have yet to be discovered.
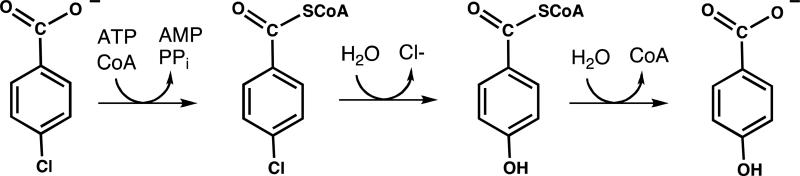
The hotdog-fold thioesterase is a small (~19 kDa) protein comprised of a 5 or 6- stranded β-sheet that wraps a long α-helix (16) (hence, the name hotdog-fold (17)). Two subunits associate to form the minimal catalytic unit. The active site is located at the subunit interface and is a composite of residues from each subunit. The thioesterase catalytic scaffold appears to have been invented at least twice during the evolution of the hotdog-fold family as evidenced by the 4-hydroxybenzoyl-CoA (4-HB-CoA) thioesterases from Pseudomonas sp strain CBS3 and Arthrobacter sp strain AU (UniProt accession # Q04416), which are the archetypes of two distinct clades of the family (16, 18, 19). Both thioesterases function in the final step of the 4-chlorobenzoate dehalogenation pathway (see Scheme 1), which is operative in specialized strains of soil-dwelling bacteria (20). Remarkably, these two 4-HB-CoA thioesterases are not homologous in sequence and they do not use the same catalytic machinery, yet their tertiary structures are highly similar.
Scheme 1.
The three chemical steps of the 4-chlorobenzoate dehalogenation pathway catalyzed by 4-chlorobenzoate: CoA ligase, 4-chlorobenzoyl-CoA dehalogenase and 4-hydrobenzoyl-CoA thioesterase, respectively.
The Arthrobacter sp strain AU 4-hydroxybenzoyl-CoA thioesterase clade, which is comprised of a wide variety of aryl- and acyl-CoA and aryl- and acyl-ACP thioesterases, is by-far the largest and most diversified of the two clades. The goal of the work reported herein was to determine the mechanisms by which the Arthrobacter sp strain AU 4-HB-CoA thioesterase binds 4-HB-CoA and catalyzes its hydrolysis. This particular thioesterase was selected as the mechanistic prototype because it proved to be an ideal system for structure determination, transient kinetic studies, 18O-solvent labeling experiments and site-directed mutagenesis.
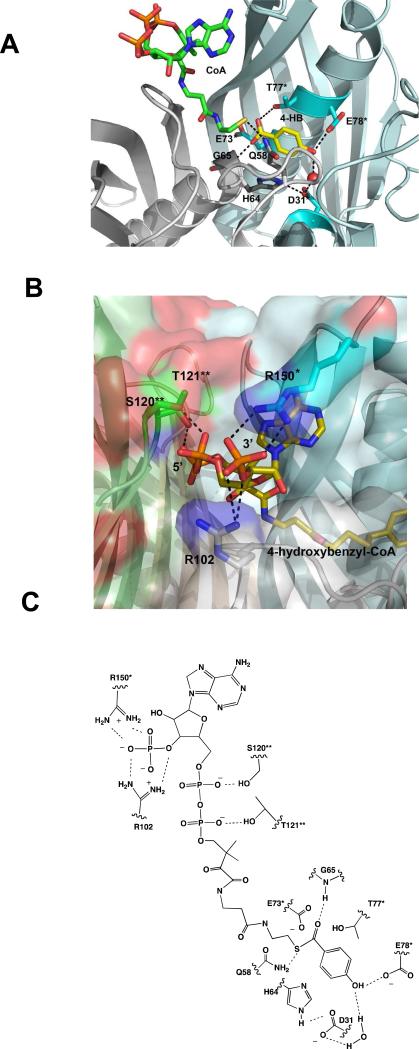
The starting point for our studies was the examination of the reported X-ray structures of the thioesterase bound with its products 4-hydroxybenzoate (4-HB) and CoA, with the inert substrate analog 4-hydroxybenzyl-CoA (CH2-S replaces O=C-S) or with the inert substrate analog 4-hydroxyphenacyl-CoA (4-HP-CoA; O=C-CH2-S replaces O=C-S) (18) (PDB accession codes 1Q4S, 1Q4U, 1Q4T respectively). The thioesterase catalytic site is comprised of invariant residues contributed from the central α-helix N-terminus (His64, Gly65) and ensuing loop (Gln58) from one subunit (subunit A) and the mid-section of the central α-helix from the opposing subunit (subunit B) (Glu73*, Thr77*, Glu78*) (Figure 1A). The back wall of the 4-hydroxybenzoyl binding pocket is formed by the N-terminal region of subunit A (Asp31). The 4-HB-CoA pantetheine arm snakes through a narrow hydrophobic tunnel that links the catalytic site to the protein surface. The nucleotide unit is located on the protein surface where subunits A (Arg102) and B (Arg150*) join and it is also in positioned to interact with subunit C (Ser120** and Thr121**) (Figure 1B). In this paper, we report on an in depth experimental investigation of the roles played by the key active site residues (depicted in Figure 1C) in substrate recognition and catalysis. We provide evidence that Arg102, Arg150, Ser120 and Thr121 contribute to the substrate binding energy through favorable electrostatic interaction with the phosphoryl groups, that Asp31, His64 and Glu78 assist in orienting the 4-hydroxybenzoyl ring, that Glu73 functions in nucleophilic catalysis, that Gly65 and Gln58 contribute to transition-state stabilization via hydrogen bond formation with the thioester moiety and that Thr77 orients the water nucleophile for attack at the 4-hydroxybenzoyl carbon of the anhydride intermediate. We also report that the replacement of Glu73 with Asp switches the function of the carboxylate residue from nucleophilic catalysis to base catalysis and thus, the reaction from a two-step process involving a covalent enzyme intermediate to a single-step hydrolysis reaction.2
Figure 1.
(A) The catalytic site of the Arthrobacter sp. strain SU 4-HB-CoA thioesterase bound with 4-HB and CoA (PDB accession #1Q4S). Only the AB dimer is shown. The nitrogen atoms are colored blue, the oxygen atoms red and the sulfur atom yellow. The subunit A carbon atoms are colored gray whereas the subunit B carbon atoms are colored cyan. (B) The nucleotide binding site of the Arthrobacter sp. strain SU 4-HB-CoA thioesterase bound with 4-hydroxybenzyl-CoA (PDB accession #1Q4U). The ABCD tetramer is shown. The coloring scheme is the same as that stated in (A) but also includes the green colored carbon atoms of subunit C and wheat colored carbon atoms of subunit D. (C) The ChemDraw depiction of the proposed binding interactions between the thioesterase and the 4-HB-CoA substrate. The residues from subunits B and C are marked with * and **, respectively and the hydrogen bonds are represented with dashed lines.
Materials and Methods
Materials
Wild-type recombinant Arthrobacter sp. strain SU 4-HB-CoA thioesterase was prepared as previously reported (23). The thioesterase site-directed mutants were prepared by using a PCR based strategy as is detailed in the Supporting Information. The substrates 4-HB-CoA (24) and the 4-hydroxy-dithiobenzoyl-CoA (25) were prepared according to published procedures, as were the inhibitors 4-hydroxybenzyl-CoA and 4-HP-CoA (18) and the radiolabeled substrate [14C]4-HB-CoA (benzoyl ring 14C=O) (23).
Steady-state kcat, Km and Ki determinations
The initial velocity of the 4-HB-CoA thioesterase-catalyzed hydrolysis of 4-HB-CoA was measured by monitoring the decrease in solution absorption at 300 nm (Δε = 11.8 mM−1 cm−1.). Reactions were carried out in 50 mM K+HEPES (pH 7.5, 25 °C) containing wild-type or mutant 4-HB-CoA thioesterase and varying concentrations of 4-HB-CoA (Km - 10Km). The inhibition data were measured using 200 and 400 μM 4-HB, 18 and 36 μM CoA or 2Ki concentration of 4-hydoxybenzyl-CoA and 4-hydoxyphenacyl-CoA (for which linear inhibition was previously demonstrated (18)). For all measurements, the initial velocity data were analyzed using equations (1), (2) or (3) and the computer program KinetAsyst (IntelliKinetics, PA):
| eq. (1) |
| eq. (2) |
| (eq. (3) |
where V = initial velocity, Vmax = maximum velocity, [S] = substrate concentration, Km= Michaelis constant, [I] = inhibitor concentration and Kis = the slope inhibition constant and Kii = the intercept inhibition constant. The kcat was calculated from Vmax/[E] where [E] is the enzyme concentration (determined using the Bradford method (26)).
Stopped-flow Uv-vis transient kinetic determinations of multiple and single turnover reactions
A DX.17MV sequential stopped-flow spectrometer (Applied Photophysics, Leatherhead, U. K.) with a light path of 10 mm and a mixing time of 3 ms was used to mix buffered wild-type or mutant thioesterase with 4-HB-CoA and to monitor the hydrolysis reaction at 300 nm (Δε = 11.8 mM−1 cm−1). For the wild-type thioesterase-catalyzed multiple turnover reactions, the reaction solutions initially contained 115 μM 4-HB-CoA and 16, 21 or 31 μM enzyme (in 0.15 M KCl/50 mM K+HEPES (pH 7.5, 25 °C). For the mutant thioesterase-catalyzed multiple turnover reactions, the reaction solutions initially contained 80 or 100 μM 4-HB-CoA and 20 or 30 μM enzyme (in 0.15 M KCl/50 mM K+HEPES (pH 7.5, 25 °C).
For single turnover reactions, the thioesterase and 4-HB-CoA were present at 40 μM and 20 μM, respectively. The data were fitted to a single (eq. (4)) or double exponential equation (eq. (5))
| eq. (4) |
| eq. (5) |
Yt and Y0 are the absorbance at time t and zero, A, A1 and A2 are amplitudes, and k, k1 and k2 are rate constants.
Rapid quench transient kinetic determinations
A Rapid-quench instrument from KinTek Instruments was used to mix buffered wild-type or mutant thioesterase with [14C]4-HB-CoA and then with 0.2 M HCl to terminate the reaction. For the multiple turnover reactions, mixtures initially contained 10 μM enzyme and 50 μM [14C]4-HB-CoA in 0.15 M KCl/50 mM K+HEPES (pH 7.5, 25 °C), whereas for the single turnover reactions, enzyme and [14C]4-HB-CoA concentrations were 50 μM and 5 μM, respectively. The enzyme was precipitated from the quenched reaction solution by vigorous mixing with 200 μL CCl4 using a Vortex mixer, and then collected by (micro)centrifugation of the mixture (14,000 rpm for 1 min). The (upper) water phase was injected into an HPLC for separation of the [14C]4-HB-CoA and [14C]4-HB as described previously (27). The eluent was collected using a fraction collector and analyzed by liquid scintillation counting. Control reactions (lacking enzyme) were carried out in parallel to establish the background radioactivity associated with the [14C]4-HB fraction. The background radioactivity (~2%) was taken into account when calculating the concentrations of substrate and product remaining in each quenched reaction mixture.
The single turnover time course data were fitted to a single (eq. (4)) or double exponential equation (eq. (5)). Yt and Y0 represent the concentrations of [14C]4-HB-CoA or [14C]4-HB at time t and zero, respectively.
Kd determination
A FluoroMax-2 fluorometer (290 nm excitation; 334 nm emission) was used to monitor intrinsic protein fluorescence quenching for 50 mM K+HEPES/0.2 M KCl/1 mM DTT (pH 7.5, 25 °C) solutions containing wild-type or mutant 4-HB-CoA thioesterase and varying concentrations of 4-HB-CoA or 4-HP-CoA ligand. For a typical titration experiment, 1 μL aliquots of ligand were added to a 1 mL solution of 1 μM thioesterase, and the fluorescence intensity at 334 nm was measured following each addition. The fluorescence data, collected at ligand concentrations ranging from 0 to 12 μM, were fitted to equation (6) using the Kaleida Graph computer program for nonlinear regression analysis.
| eq. (6) |
where [S] is the total ligand concentration, [E] is the total enzyme concentration, Kd is the apparent dissociation constant of the enzyme-ligand complex, ΔF is the observed change in fluorescence intensity and ΔFmax is the maximum change in fluorescence intensity.
Determination of kcat pH rate profile in H2O and D2O
The kcat pH rate profiles for Arthrobacter sp. strain 4-HB-CoA thioesterase-catalyzed hydrolysis of 4-HB-CoA in H2O and D2O were measured by using a dual buffer system consisting of 50 mM acetate and 50 mM MES (pH 4.0 – 5.6); 50 mM MES and 50 mM HEPES (pH 5.6-8.0); 50 mM HEPES and 50 mM TAPS (pH 8.0-9.1); 50 mM TAPS and 50 mM CAPSO (pH 9.1-9.6). The enzyme activity at overlapping pH values was tested to ensure equal enzyme activity in the different buffer systems. The stability of the thioesterase at the acidic and basic pH values was verified by carrying out pre-incubation test experiments.
Steady-state multiple turnover time courses
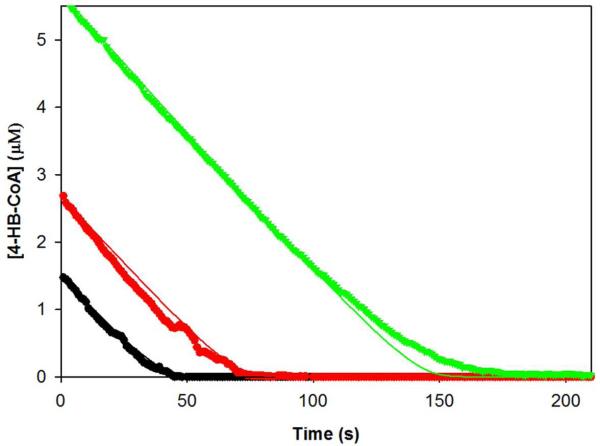
The time courses for 0.003 μM thioesterase-catalyzed 4-HB-CoA hydrolysis of 4-HB-CoA (1.5, 2.7 and 5.7 μM) in 50 mM K+HEPES (pH 7.5, 25 °C) were monitored at 300 nm (Δε = 11.8 mM−1s−1) to completion. The data were fitted by curve simulation using the KinTek Corporation Global Kinetic Explorer (www.kintek-corp.com/KGExplorer/index.php) (28).
Solvent18 O- labeling experiments
Stock solutions of wild-type or mutant thioesterase in 97% H218O (Spectra) containing 0.15 M KCl/10 mM K+HEPES (pH 7.5) were prepared by lyophilizing 2 mM or 0.02 mM thioesterase in 0.15 M KCl/10 mM K+HEPES (pH 7.5) and then dissolving the resulting powder with the desired volume of H218O. The thioesterase activity was measured to ensure that full activity was retained. The enzyme stock solution was combined with 4-HB-CoA in 97% H218O to generate a 200-400 μL reaction solution that for a typical experiment contained 2.0 mM thioesterase and 0.2 mM 4-HB-CoA (test reaction) or 0.02 mM thioesterase and 0.2 mM 4-HB-CoA thioesterase (control reaction), in 0.15 M KCl/10 mM K+HEPES (pH 7.5) at 25 °C. Following a 15 min incubation period the reaction solution was filtered using a 10 kDa micro separation filter (Pall Filtron). The filtrate was acidified by adding 5 μL of 6 N HCl and extracted four times with 1 mL portions of ethyl acetate. The ethyl acetate extracts were combined, dried over anhydrous sodium sulfate and the solvent was evaporated in-vacuo. The solid was dissolved in 100 μL of absolute methanol and subjected to GC-MS analysis using a Saturn GC-MS spectrometer equipped with a Restek XTI-5 column.
31P-NMR spectral determinations
The 31P NMR spectra were recorded on a Bruker AC-F 500 NMR spectrometer at 202.5 MHz. Samples were contained in 5 mm NMR tubes, and spectra were recorded at 5 °C under conditions of proton decoupling. The sample solution contained 10% D2O for use in signal locking. The instrument was calibrated using an 85% phosphoric acid standard.
X-ray structure determinations
Arthrobacter sp. strain 4-HB-CoA thioesterase crystals were grown as reported previously (18). Briefly, the mutant proteins were concentrated to approximately 17-20 mg/mL at pH 7 (10 mM HEPES) in the presence of 150 mM KCl and 1 mM DTT. Either 4-HP-CoA or 4-HB-CoA were added to a concentration of 1 mM. Crystals were grown at room temperature via hanging drop vapor diffusion using 15-20% PEG-3400 and 200 mM LiCl buffered at pH 7 with 100 mM MOPS. Crystals were transferred to cryoprotectant solutions containing 30% PEG-3400, 200 mM KCl, 250 mM LiCl, 15% ethylene glycol and 100 mM MOPS at pH 7. 4-HP-CoA or 4-HB-CoA were added to a concentration of 1 mM.
X-ray data were collected at 100K with a BRUKER HI-Star area detector system equipped with Goebel optics. The data were processed and scaled with SAINT (Bruker AXS Inc). The structures were solved using Fourier difference techniques and refined with REFMAC (29). Iterative model building was performed with COOT (30). When crystallized in the presence of 4-HB-CoA, the mutant proteins E73D, E73Q, and Q58A were able to hydrolyze the ligand, generating the various product complexes that were observed. Specific unit cell parameters, data collection and refinement statistics are presented in Table SI1.
Results and Discussion
Transient Kinetic Experiments Provide Evidence For a Two-step Chemical Pathway
Pre-steady-state multiple turnover reaction
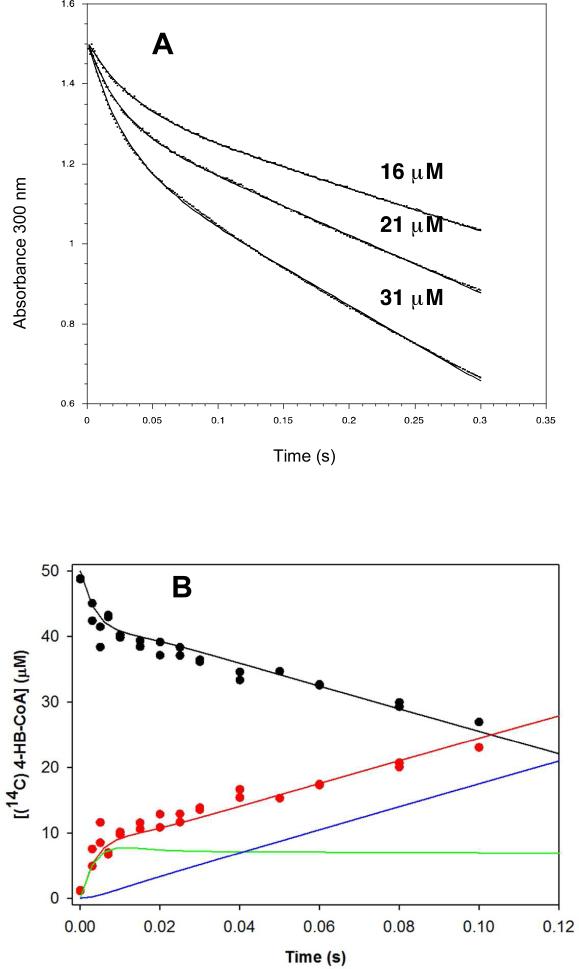
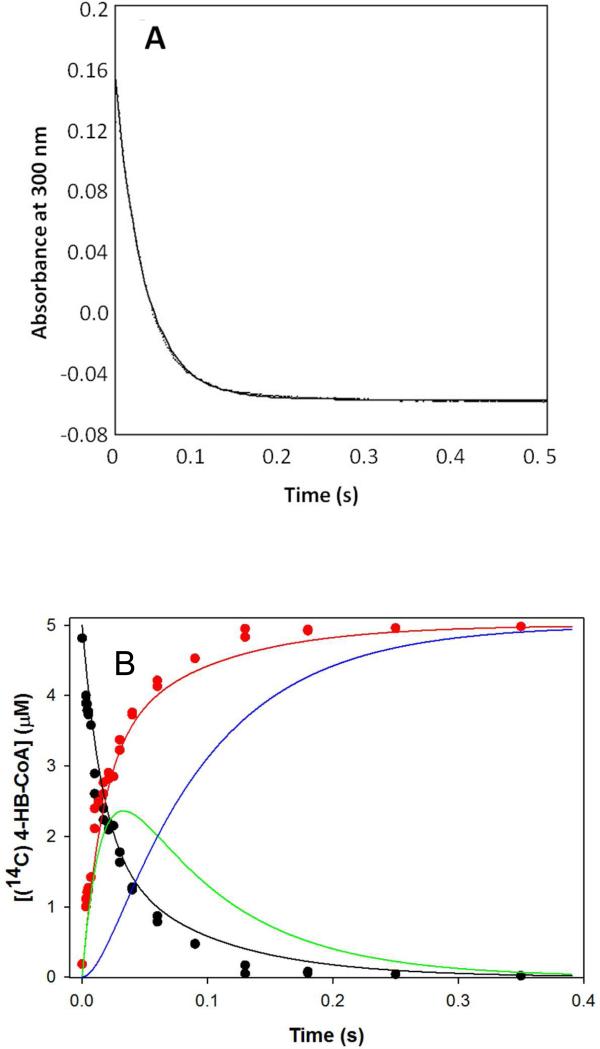
The time course for pre-steady-state wild-type thioesterase-catalyzed 4-HB-CoA hydrolysis was examined under multiple-turnover conditions so that the rate of the first turnover and that of subsequent turnovers might be compared. The reaction time courses were first determined by monitoring the decrease in solution absorbance at 300 nm (Δε = 11.8 mM−1 cm−1 for the thioester cleavage) with a stopped flow UV-vis spectrophotometer (Figure 2A). The stopped-flow absorbance traces for the reaction of 115 μM 4-HB-CoA with 16, 21 and 31 μM wild-type thioesterase are biphasic and are suggestive of a “burst” of substrate consumption approximating a single turnover of substrate on the enzyme (i.e., ~16, 21, 31 μM calculated from the amplitude derived from the fit to a double exponential equation) followed by a slow, linear phase that derives from the subsequent turnover events (rate constants derived from data fitting to a double exponential equation are reported in Table 1).
Figure 2.
(A) stopped-flow UV-vis absorbance traces of (16, 21 and 31 μM) wild-type thioesterase-catalyzed hydrolysis of (112 μM) 4-HB-CoA carried out under multiple turnover conditions and monitored at 300 nm (Δε = 11.8 mM−1 cm−1). The traces were fitted using a double exponential equation. (B) The time course for the multiple turnover reaction of 50 μM [14C]4 HB-CoA and 10 μM thioesterase monitored using rapid quench/HPLC techniques. The curves were simulated using the kinetic model shown in Scheme 2 and the microscopic rate constants k1 = 172 μM−1 s−1, k−1 = 30 s−1, k2 = 264 s−1, k−2 = 9 s−1, k3 = 25 s−1, k−3 = 0 s−1, k4 = 129 s−1 , k−4 = 5 μM−1 s−1, k5 = 368 μM−1 s−1 and k−5 = 5 μM−1 s−1. The simulated curve-coloring scheme is as follows: E-I(CoA) + E(4-HB)(CoA) + E(4-HB) + 4-HB (red); E(4-HB-CoA) + 4-HB-CoA (black); E-I(CoA) (green); E(4-HB)(CoA) + E(4-HB) + 4-HB (blue).
Table 1.
The apparent rate constants for the wild-type and mutant thioesterase catalysis of multiple and single turnover hydrolysis of 4-HB-CoA as measured using stopped-flow UV-vis absorbance spectroscopy (sf) and rapid quench (rq) techniques. All entries that are not designated were measured using stopped-flow UV-vis absorbance spectroscopy. Biphasic time courses were fitted to a double exponential equation and single phase time courses were fitted to a single exponential equation.
| Thioesterase | Multiple Turnover | Single Turnover | ||
|---|---|---|---|---|
| Single phase | Double phase | |||
| k (s−1) | Fast phase k (s−1) | Slow phase k2 (s−1) | k (s−1) | |
| Wild-type | 40 ± 2 (sf)a 100 ± 30 (rq) |
6.9 ± 0.4 (sf)a 8.7 ± 0.3 (rq) |
28.7 ± 0.1 (sf) 36 ± 2 (rq) |
|
| E73D | 0.0614 ± 0.0002 (sf) 0.0774 ± 0.001 (rq) |
-------- -------- |
-------- -------- |
0.774 ± 0.001 -------- |
| T77A | -------- -------- |
19.7 ± 0.2 (sf) 47 ± 8 (rq) |
0.49 ± 0.01(sf) 1.38 ± 0.06 (rq) |
17.0 ± 0.1 (sf) 24 ± 2 (rq) |
| T77S | -------- | 18.9 ± 0.2 | 1.208 ± 0.006 | 19 s−1 |
| T77V | 0.152 ± 0.002 | -------- | -------- | -------- |
| E73D/T77A | 0.705 ± 0.001 | -------- | -------- | -------- |
| E73D/T77S | 0.0619 ± 0.0001 | -------- | -------- | -------- |
| Q58A | 0.0842 ± 0.0001 | -------- | -------- | -------- |
| Q58E | 0.288 ± 0.001 | -------- | -------- | -------- |
| E78A | 0.175 ± 0.001 | -------- | -------- | -------- |
| H64A | 0.0139 ± 0.0001 | -------- | -------- | -------- |
| H64Q | 0.0691 ± 0.0001 | -------- | -------- | -------- |
| D31A | 0.0120 ± 0.0001 | -------- | -------- | -------- |
| D31N | 0.420 ± 0.002 | -------- | -------- | -------- |
These error limits are based on the standard deviation of three individual measurements. All of the other error limits shown are based on the fit to a single representative data set.
Next, the time course for the multiple turnover of [14C]4-HB-CoA (50 μM) catalyzed by wild-type thioesterase (10 μM) was measured using rapid (acid) quench techniques coupled with HPLC separation of the 14C-labeled reactant and 4-HB product released from the denatured enzyme (Figure 2B). The time course shown in Figure 2B is biphasic, with the fast phase approximating a single turnover of substrate on the enzyme (i.e., ~10 μM) (rate constants derived from data fitting to a double exponential equation are reported in Table 1)
The burst-phase kinetics could be rationalized in terms of fast hydrolysis of the thioester in the enzyme-substrate complex followed by slow product release.3 Alternatively, the burst-phase kinetics might reflect fast conversion of the enzyme -substrate complex to a chemical intermediate in which the thioester chromophore is absent (stopped-flow) and which hydrolyzes to 4-HB in the acid quench solution (rapid quench), followed by slow regeneration of the catalytically active form of the enzyme.
Based on the X-ray structure of the thioesterase bound with its products 4-HB and CoA (Figure 1A), a likely candidate for a chemical intermediate is the anhydride formed by Glu73 displacement of CoA from 4-HB-CoA. Specifically, the structure shows that the Glu73 positioned for nucleophilic attack at the C=O, and that there is no room for a water nucleophile (19). The enzyme-anhydride intermediate lacks the ring-conjugated thioester group. Its formation would therefore account for initial drop in absorbance at 300 nm as was observed in the stopped-flow experiment (Figure 2A). In addition, an anhydride is chemically labile, and in the acid quench solution it is likely to undergo hydrolysis to form 4-HB, thus accounting for the burst of [14C]4-HB formation observed in the rapid quench experiment (Figure 2B). Consequently, our next experiments focused on testing the formation of this putative thioesterase-anhydride intermediate.
Solvent deuterium kinetic isotope effect
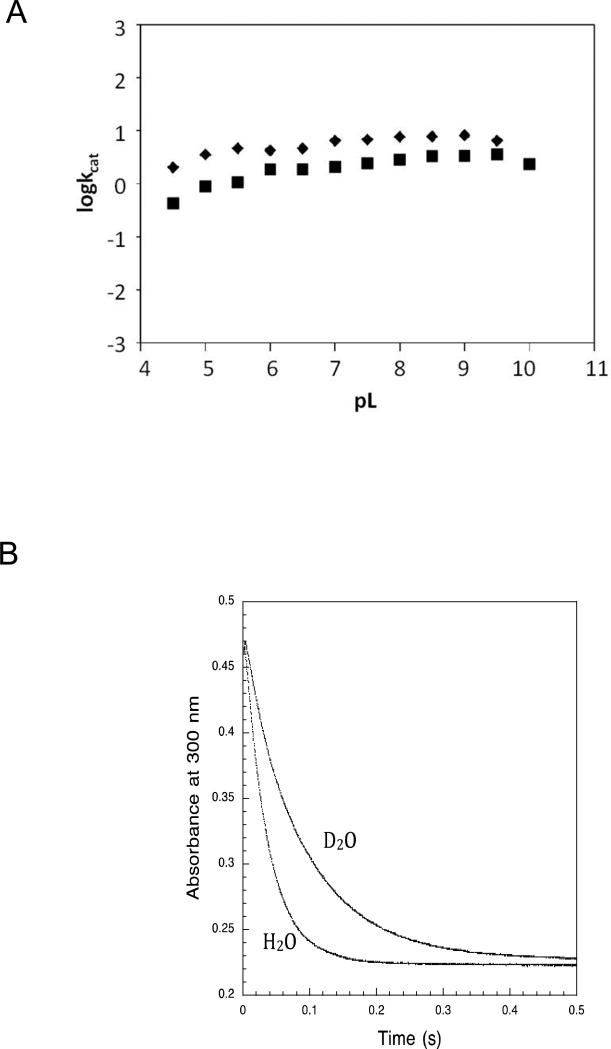
In order to distinguish between the posited two-step chemical pathway and a single-step chemical pathway with rate-limiting product release, we first tested whether the chemical reaction or product dissociation is rate-limiting.3 This was accomplished by measuring the solvent deuterium isotope effect (SKIE) on the steady-state kcat. General acid-mediated proton transfer to the CoA thiolate leaving group and/or general base-mediated proton abstraction from the water nucleophile will give rise to a SKIE if, and only if, the proton transfer occurs in a rate-limiting step. The kcat pH profiles for wild-type thioesterase-catalyzed 4-HB-CoA hydrolysis in H2O and D2O solvents are shown in Figure 3A. The pH profiles are essentially flat, and therefore determination of the pH independent C values, that one would obtain by fitting the data to a proton ionization equation was not possible. Consequently, the SKIE value was not calculated from the ratio of C values derived from fitting the respective pH profiles. Nonetheless, we took advantage of lack of kcat dependence on the reaction solution pH and made a direct comparison of the respective kcat values measured at pL 7.5. The ratio of the kcat measured in the H2O-solvent and the kcat measured in the D2O solvent at pL 7.5 is 6.8 s−1/2.4 s−1 = 2.8. This normal isotope effect suggests that the transfer of a solvent derived proton occurs in a rate-limiting step.
Figure 3.
(A) A plot of logkcat vs reaction solution pL measured for wild-type thioesterase-catalyzed hydrolysis of 4-HB-CoA in buffered H2O (◆) or buffered D2O (■). (B) A stopped-flow UV-vis absorbance trace (300 nm) of (40 μM) wild-type thioesterase-catalyzed hydrolysis of (20 μM) 4-HB-CoA carried out at 25 °C in buffered H2O or buffered D2O (pL = 7.5). The data were fitted with a single exponential equation.
Next, we measured the SKIE on the rate of a single turnover reaction so as to limit the steps queried to the chemical step(s) of the catalyzed reaction. The SKIE on the rate constant for a single turnover reaction was determined at the same pL = 7.5 by using stopped-flow UV-vis spectral techniques to measure the time course for a single turnover of 20 μM 4-HB-CoA catalyzed by 40 μM thioesterase (Figure 3B). The data were fitted with a single exponential equation to define the apparent rate constants 28.7 ± 0.1s−1 (H2O) and 12.5 ± 0.1 s−1 (D2O) from which the SKIE equal to 2.3 was calculated. This value compares favorably with the SKIE of kcat of 2.8, and therefore we conclude that product release is not rate-limiting and that the burst phase observed in Figures 2A and 2B to the enzyme-anhydride intermediate. Next, we set forth to trap this intermediate.
Solvent 18O-Labeling Experiments Provide Evidence for an Anhydride Intermediate
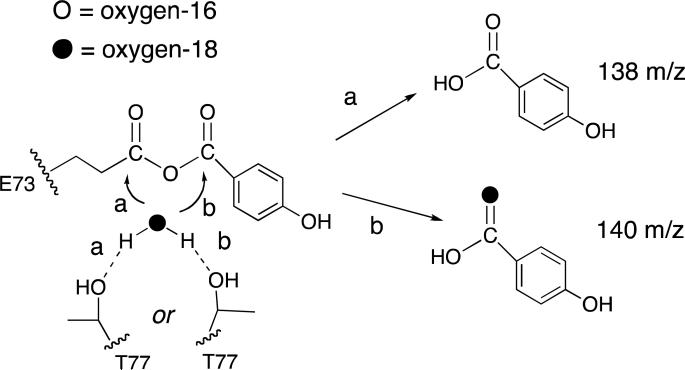
A priori, the putative anhydride intermediate can undergo hydrolytic cleavage at either one of its carbonyl carbon atoms (see Scheme 2). If a single turnover reaction is carried out in H218O, 18O will not be incorporated in the 4-HB product provided that the attack occurs at the Glu73 carbonyl carbon. Instead, the 16O derived from the Glu73 will be incorporated. On the other hand, if the attack occurs at the benzoyl carbon the solvent-derived 18O will be incorporated in the 4-HB product. This labeling pattern would also be obtained if the Glu73 functions in base catalysis rather than in nucleophilic catalysis. The incorporation of the Glu73-derived 16O can only occur in the event of nucleophilic catalysis, which provided the incentive to test this pathway by carrying out thioesterase-catalyzed single turnover reactions in H 182O.
Scheme 2.
The two possible regiochemistries for hydrolytic cleavage of the proposed anhydride intermediate formed in the thioesterase-catalyzed hydrolysis of 4-HB-CoA carried-out in oxygen-18 enriched water under single turnover conditions. In “pathway a” the Thr77 directs the water nucleophile to attack the Glu73 carbonyl carbon and therefore the 4-hydroxybenzoate product is not labeled with oxygen-18. In “pathway b” the Thr77 directs the water nucleophile to attack the 4-hydroxybenzoyl carbonyl carbon and therefore the 4-hydroxybenzoate product is labeled with oxygen-18.
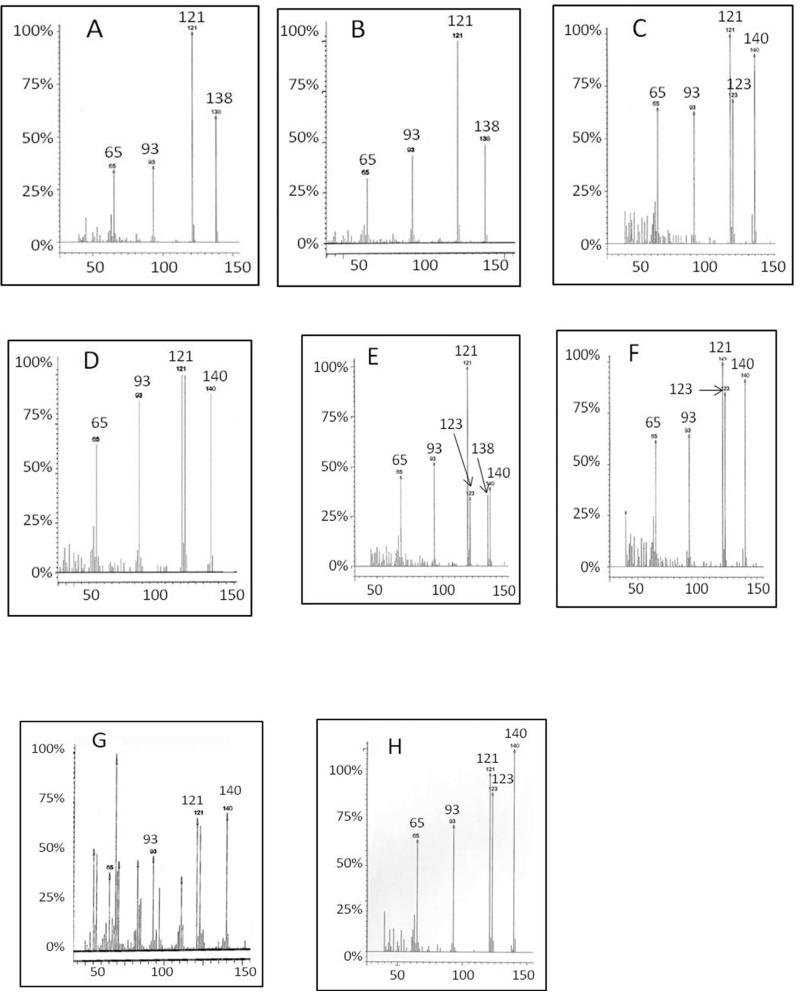
The oxygen transfer experiments employed reaction solutions initially containing 2.0 mM wild-type thioesterase and 0.2 mM 4-HB-CoA in 97% H218O. The acid form of the 4-HB product was analyzed by GC-MS. The mass spectrum of the 4-HB standard (Figure 4A) contains a parent ion (M*) peak at 138 m/z, parent ion fragment (M*-OH) peak at 121 m/z (note that the carboxylic acid OH is lost), parent ion fragment (M*-COOH) peak at 93 m/z and lastly, a parent ion fragment peak at 65 m/z. The parent ion (M*) and parent ion fragment (M*-OH) were used as reporters of 18O incorporation.
Figure 4.
GC-MS spectra of 4-HB. The X and Y axes designate m/z units and relative intensity (%), respectively. (A) The 4-HB standard (parent ion m/z = 138). (B) The 4-HB produced by the (2.0 mM) wild-type thioesterase-catalyzed hydrolysis of 4-HB-CoA (0.2 mM) in natural isotopic abundance water. (C) The 4-HB produced by (2.0 mM) wild-type thioesterase-catalyzed hydrolysis of 4-HB-CoA (0.2 mM) in 97% H218O. (D) The 4-HB produced by (2.0 mM) E73D thioesterase-catalyzed hydrolysis of 4-HB-CoA (0.2 mM) in 97% H218O. (E) The 4-HB produced by (2.0 mM) T77A thioesterase-catalyzed 4-HB-CoA (0.2 mM) hydrolysis in 97% H218O. (F) The 4-HB produced by (0.02 mM) T77A thioesterase-catalyzed hydrolysis of 4-HB-CoA (0.2 mM) in 97% H218O. (G) The 4-HB produced by the (2.0 mM) T77S thioesterase-catalyzed hydrolysis of 4-HB-CoA (0.2 mM) in 97% H218O. (H) The 4-HB produced by (2.0 mM) E73D/T77A thioesterase-catalyzed hydrolysis of 4-HB-CoA (0.2 mM) in 97% H218O.
When the single turnover reaction was carried out in natural isotopic abundance water, the product mass spectrum obtained was identical to that of the 4-HB standard (Figure 4B). On the other hand, the mass spectrum of the 4-HB formed in 97% H218O contained parent ion peaks at 138 and 140 m/z (corresponding to the all- 16O parent ion and the parent ion labeled with one 18O at the carboxylic acid group) at the relative intensities of 1:9 (Figure 4C). The relative intensities of the parent ion fragment peaks (M*-OH) observed at 121 m/z and 123 m/z is 5:4. A duplicate experiment produced the same result.
The 10% 16O-incorrporation can be interpreted as a 9:1 preference for water attack at the 4-hydroxybenzoyl carbonyl carbon or alternatively, as the result of unanticipated contamination by H216O. In order to test the amount of contamination by H216O in the reaction solution the experiment was repeated using the E73D thioesterase in place of the wild-type thioesterase. The Asp side chain is shorter than the Glu side chain and therefore it is not expected to react with the substrate carbonyl carbon as needed to form the anhydride intermediate. Indeed, as will be reported at a later point in this paper, the X-ray structure of the E73D thioesterase(CoA) complex shows an ordered water molecule that forms a hydrogen bond to the Asp73 carboxylate group. The mass spectrum of 4-HB (Figure 4D) formed by E73D thioesterase-catalyzed hydrolysis of substrate in 97% H218O shows negligible 16O-incorrporation. This result suggested that the majority of the 10% 16O-incorrporation into the 4-HB product formed by the wild-type thioesterase derives from the Glu73 carboxylate group and not from the solvent.
In order to further test Glu73 participation in nucleophilic catalysis we carried out the T77A thioesterase-catalyzed single turnover reaction of 4-HB-CoA in 97% H218O. From the X-ray structures of the liganded wild-type thioesterase (see for example, Figure 1A) we gathered that the Thr77 side chain is positioned to bind and orient a water molecule for attack at the benzoyl carbonyl carbon of the putative anhydride intermediate (see Scheme 2). By replacing the Thr77 with Ala the potential influence of the Thr77 on the regiochemistry of the hydrolysis step is removed. The X-ray structure of the T77A mutant shows that the configuration of the active site residues is unchanged (vide infra). The mass spectrum of the 4-HB product contains the 138 and 140 m/z peaks at equal intensities (Figure 4E). Thus, 50% of the 4-HB contained no 18O, while 50% contained one 18O atom at carboxylic acid group. Because the two oxygen atoms of the 4-HB carboxylic acid group are lost with equal probability, the ratio of the peaks at 121 m/z and 123 m/z is calculated to be 3:1, which is consistent with the experimental peak ratio. In contrast, the GS-MS spectrum obtained for the multiple turnover reaction (T77A thioesterase 0.02 mM and 4-HB-CoA 0.2 mM) showed that ~92% of 4-HB was 18O-labeled (Figure 4F) which is consistent with the exchange of the Glu73 16O with solvent-derived 18O upon each catalytic cycle.
Lastly, the T77S thioesterase-catalyzed single turnover reaction of 4-HB-CoA in 97% H218O was carried out. The 16O-incorporation observed in the isolated 4-HB was negligible (see Figure 4G). This suggests that the Ser77 is more effective than is the Thr77 at positioning the water for attack at the 4-hydroxybenzoyl carbonyl carbon of the anhydride intermediate.
In summary, the amount of 16O incorporation into the 4-HB carboxylate group upon a single catalytic turnover varies depending on the residue at position 73 or 77: 10% for the wild-type thioesterase, 0% for the E73D mutant, 50% for the T77A mutant and 0% for the T77S mutant.
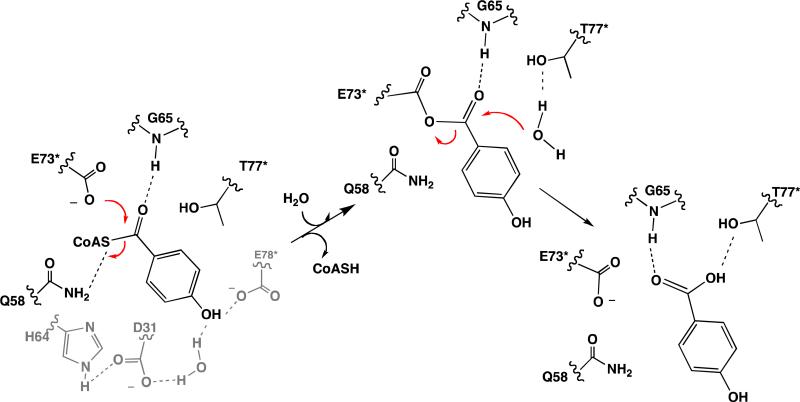
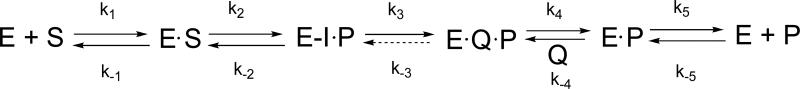
Kinetic Mechanism
Based on the (i) X-ray structural data, (ii) the burst phase kinetics coupled with the SKIE results and (iii) the observed T77A mutant-catalyzed 16O-incorporation in the 4-HB, we conclude that a majority, if not all, of the conversion of substrate to product proceeds via the mixed anhydride intermediate pictured in Figure 5. Our next step was to determine if independent kinetic data sets could be fitted to the simple kinetic model shown in Scheme 3. This model assumes ordered product release, as suggested by the X-ray structural data (Figure 1), with CoA departing first thereby allowing the 4-HB to exit through the tunnel that leads to the protein surface. Indeed, 4-HB showed competitive product inhibition (Kis = 240 ± 80 μM) whereas the CoA and desulfoCoA showed noncompetitive inhibition (Kis = 16 ± 2 μM, Kii = 29 ± 2 and Kis = 19 ± 6 μM, Kii =13 ± 3 μM, respectively) (Table 2 and Figure SI1).
Figure 5.
Depiction of the catalytic mechanism of the wild-type thioesterase.
Scheme 3.
The kinetic model for Arthrobacter sp. strain SU 4-HB-CoA thioesterase catalysis where E is enzyme, S is 4-HB-CoA, E-I is the enzyme-anhydride intermediate, P is CoA and Q is 4-HB.
Table 2.
CoA and 4-HB steady-state kinetic inhibition constants measured for wild-type and mutant thioesterases at pH 7.5 and 25 °C. See Materials and Methods for details.
| Thioesterase | CoA | 4-HB | |
|---|---|---|---|
| Kis (μM) | Kii (μM) | Kis (μM) | |
| Wild-type | 16 ± 2 | 29 ± 2 | 240 ± 80 |
| E73D | 32 ± 2 | -------- | 1700 ± 400 |
| S120A | 110 ± 30 | 110 ± 10 | -------- |
| T121A | 360 ± 50 | 580 ± 60 | -------- |
| R150A | 460 ± 80 | 700 ± 700 | -------- |
| R102A | 500 ± 100 | 1200 ± 300 | -------- |
The simulations were generated using the KinTek Corporation Global Kinetic Explorer software for the time courses reported in Figures 2B (pre-steady state multiple turnover reaction), 6B (single turnover reaction), and 7 (steady-state multiple turnover reaction). The values of the microscopic rate constants were varied individually, and in combination, until the simulated curves, shown in the respective figures, matched the experimental time course. The CoA and 4-HB inhibition constants were used to constrain the values of k4/k−4 and k5/k−5.
The Km is defined by:
The Km value ~0.13 M derived from the microscopic rate constants is smaller than that (Km = 1.5 M) estimated from steady-state initial velocity data. We consider the Km value derived from the initial velocity data to be less accurate because the measurement of initial velocities at such low substrate concentrations exceeds the sensitivity limit of the spectrophotometric assay.
The value of the rate constant for the formation of the anhydride intermediate (k2 confined to 44-264 s−1) exceeds that of the rate constant that governs the ensuing hydrolysis step (k3 confined to 17-25 s −1). Whereas the hydrolysis step appears to be irreversible (k−3 = 0), the anhydride intermediate can partition backward (k−2 confined to 9-14 s−1) as well as forward (k2 = 44-264 s−1). The simulated time course for the formation and consumption of the enzyme-anhydride intermediate during a single turnover ([E] >> [S]; Figure 6B) shows that at 20-30 ms the level of the enzyme-anhydride intermediate peaks at ~45% of the starting enzyme-substrate complex and then slowly declines in parallel with 4-HB formation. The simulated time course for the multiple turnover ([S] >> [E]; Figure 2B) indicates that at steady-state ~50% of the enzyme exists as the enzyme-anhydride intermediate.
Figure 6.
(A) stopped-flow UV-vis absorbance trace of (40 μM) wild-type thioesterase-catalyzed hydrolysis of 4-HB-CoA (20 μM) carried out under single turnover conditions and monitored at 300 nm (Δε = 11.8 mM−1 cm−1). (B) Time course for the single turnover reaction of 5 μM [14C]4-HB-CoA and 50 μM thioesterase monitored using rapid quench/HPLC techniques. The data were fitted to a double exponential equation. The curves were simulated using the kinetic model shown in Scheme 2 and the microscopic rate constants k1 = 194 μM−1 s−1, k−1 = 30 s−1, k2 = 44 s−1, k−2 = 14 s−1, k3 = 17 s−1, k−3 = 0 s−1, k4 = 129 s−, k−4 = 5 μM−1 s−1, k5 = 368 μM−1 s−1 and k−5 = 5 μM−1 s−1. The simulated curve-coloring scheme is as follows: E-I(CoA) + E(4-HB)(CoA) + E(4-HB) + 4-HB (red); E(4-HB-CoA) + 4-HB-CoA (black); E-I(CoA) (green); E(4-HB)(CoA) + E(4-HB) + 4-HB (blue).
Function Assignment to Catalytic Site Residues
The key active site residues include Gly65, Glu73, Thr77, Gln58, His64, Glu78, and Asp31. The roles that we posit for these residues, based on the results presented below, are depicted in Figure 5. Each of these residues was separately replaced by site-directed mutagenesis and each mutant thioesterase was subjected to biochemical analysis. X-ray structure determinations of the liganded E73A, E73D, E73Q, T77A, T77S, Q58A, H64A and E73A/Q58E mutants were carried out to evaluate the impact of each mutation on the configuration of the active site residues. The data collection and refinement statistics are reported in Table SI1 of Supporting Information and a list of the structures determined (along with their PDB accession codes) is provided in Table 3. Each structure was solved at 1.80 Å resolution with the exception of that of the E73Q thioesterase which was solved at 1.95 Å resolution. Each of the mutant thioesterase tetrameric structures superimpose on the wild-type thioesterase tertrameric structure with RMSD in the range of 0.2 to 0.35 Å. We thus conclude that the mutations did not alter the tertiary structure.
Table 3.
Summary of the X-ray structures of the mutant thioesterase ligand complexes reported in this paper.
| Thioesterase | Ligand(s) | PDB accession code |
|---|---|---|
| E73A | 4-HP-CoA | 3R32 |
| E73D | CoA | 3R34 |
| E73D | 4-HP-CoA | 3R35 |
| E73Q | 4-HB | 3R36 |
| E73Q | 4-HP-CoA | 3R37 |
| Q58A | 4-HB, CoA | 3R3A |
| Q58A | 4-HP-CoA | 3R3B |
| H64A | 4-HP-CoA | 3R3C |
| T77S | 4-HP-CoA | 3R3D |
| T77A | 4-HP-CoA | 3R3F |
| E73D/Q58A | 4-HP-CoA | 3TEA |
Glu73
The posited nucleophile Glu73 was separately replaced with Asp, Gln and Ala. Comparable Kd values (Table 4) for the complexes of the inert substrate analog 4-HP-CoA and wild-type thioesterase or E73A thioesterase indicates that the Glu73 carboxylate group contributes little to ligand binding. On the other hand, the turnover rate of the E73A thioesterase is estimated to be ~7 × 104-fold smaller than that of the wild-type thioesterase (Table 4). The superposition of the structures of wild-type and E73A thioesterase complexes of 4-HP-CoA shows that the positions of the 4-HP-CoA ligands and the active site residues are almost identical (Figure SI2). Therefore, we attribute the dramatic decrease in the kcat value to the inability of the Ala73 to contribute to catalysis. Notably, we did not find an ordered water molecule located in the space vacated by the Glu73 CH2COO- . The mechanism by which the slow hydrolysis occurs in the E73A thioesterase active site is presently not known.
Table 4.
Steady-state kinetic constants determined for wild-type and mutant thioesterase catalyzed hydrolysis of 4-HB-CoA plus the 4-HP-CoA dissociation and/or competitive inhibition constants. Measurements were made at pH 7.5 and 25 °C. See Materials and Methods for details.
| Enzyme | kcat (s−1) | Km (μM) | kcat/ Km (M−1s−1) | Kd or Kia (μM) |
|---|---|---|---|---|
| Wild-Type | 6.7 ± 0.1 | 1.24 ± 0.06 | 5.4 × 106 | (7 ± 3) × 10−3 a(3.0 ± 0.4) × 10−3 |
| E73A | 1 × 10−4 | ------ | ------ | (6 ± 3) × 10−3 |
| E73Q | (1.7 ± 0.2) × 10−3 | (1.42 ± 0.06) × 101 | 1.2 × 102 | (2.7 ± 0.8) × 10−2 |
| E73D | (1.23 ± 0.03) × 10−1 | (1.8 ± 0.1) × 101 | 6.8 × 103 | (2.2 ± 0.5) × 10−3 |
| T77A | 2.09 ± 0.03 | 1.22 ± 0.05 | 1.7 × 106 | (3.7 ± 0.5) × 10−2 |
| T77S | 3.16 ± 0.04 | (7.7 ± 0.3) × 10−1 | 4.1 × 106 | (1.3 ± 0.3) × 10−2 |
| T77D | (1.03 ± 0.02) × 10−2 | (3.8 ± 0.3) × 10−1 | 2.7 × 104 | (2.7 ± 0.5) × 10−2 |
| T77V | (1.73 ± 0.05) × 10−1 | 5.5 ± 0.4 | 3.1 × 104 | (1.3 ± 0.4) × 10−2 |
| E73D/T77A | 1.54 ± 0.02 | 4.6 ± 0.1 | 3.4 × 105 | (1.1 ± 0.3) × 10−2 |
| E73D/T77S | (1.29 ± 0.09) × 10−1 | (1.3 ± 0.2) × 10 | 1.0 × 104 | (1.2 ± 0.4) × 10−2 |
| Q58E | (3.64 ± 0.06) × 10−1 | 4.0 ± 0.2 | 9.1 × 104 | (1.1 ± 0.2) × 10−2 a(4.7 ± 0.8) × 10−3 |
| Q58A | (1.27 ± 0.02) × 10−1 | 1.6 ± 0.1 | 7.9 × 104 | (3.7 ± 0.5) × 10−2 |
| Q58D | (1.3 ± 0.2) × 10−1 | (1.8 ± 0.4) × 102 | 7.2 × 102 | (2.1 ± 0.1) × 10−2 |
| Q58D/E73D | NA | ------- | ------- | (1.7 ± 0.1) × 10−1 |
| Q58E/E73Q | NA | ------- | ------- | (1.3 ± 0.2) × 10−2 |
| Q58E/E73A | NA | ------- | ------- | (9 ± 2) × 10−3 |
| H64A | (1.5 ± 0.2) × 10−1 | (5.1 ± 0.8) × 102 | 2.9 × 102 | (2.2 ± 0.6) × 10−1 |
| H64Q | (2.82 ± 0.04) × 10−1 | (2.37 ± 0.07) × 102 | 1.2 × 103 | (3.3 ± 0.2) × 10−1 |
| E78A | 1.08 ± 0.05 | (9.6 ± 0.8) × 101 | 1.1 × 104 | 2.5 ± 0.2 a(7 ± 1) × 10−1 |
| D31A | (4.17 ± 0.05) × 10−2 | 4.0 ± 0.1 | 1.0 × 104 | (7 ± 2) × 10−2 a(8 ± 3) × 10−2 |
| D31N | 1.15 ± 0.03 | (1.28 ± 0.08) × 101 | 9.0 × 104 | 1.2 ± 0.3 a(1.12 ± 0.06) × 10−1 |
| R150A | 9.5 ± 0.4 | (1.5 ± 0.1) × 101 | 6.3 × 105 | (5 ± 3) × 10−3 a(4 ± 0.2) × 10−2 |
| R102A | 9.9 ± 0.3 | (1.4 ± 0.1) × 101 | 7.1 × 105 | (1.4 ± 0.5) × 10−2 a(2.1 ± 0.1) × 10−2 |
| S120A | (1.01 ± 0.02) × 10 | 1.6 ± 0.1 | 6.3 × 106 | (5 ± 3) × 10−3 a(4.6 ± 0.2) × 10−3 |
| T121A | 9.7 ± 0.3 | 6.1 ± 0.4 | 1.6 × 106 | (1.6 ± 0.3) × 10−2 a(1.3 ± 0.1) × 10−2 |
The steady-state inhibition constant Kis.
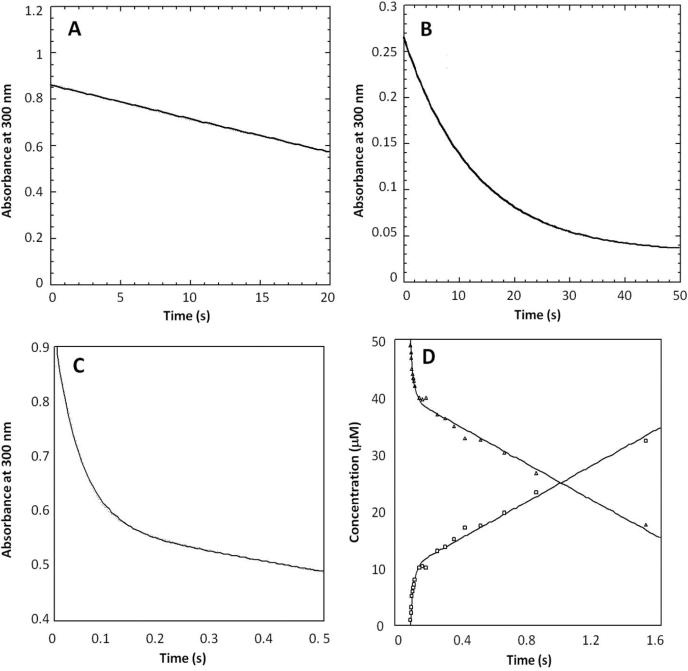
The structure and catalytic properties of the E73D thioesterase were examined to determine the impact of reducing the length of the Glu73 side chain. The kcat value of this mutant was 50-fold lower than that of the wild-type thioesterase (Table 4) whereas the E73D mutant binds 4-HP-CoA 3-fold tighter than does the wild-type thioesterase. Stopped-flow UV-vis techniques were used to determine the time course for the presteady-state multiple turnover (20 μM enzyme and 80 μM substrate) and the time course for the single turnover (40 μM enzyme and 20 μM substrate) of 4-HB-CoA catalyzed by the E73D mutant (Figures 8A and 8B). The time course data were fitted to a linear equation or a single exponential equation to define the apparent rate constants 0.0614 ± 0.0002 s−1 and 0.0774 ± 0.001 s−1, respectively (Table 1). The absence of the burst phase kinetics observed for the wild-type enzyme (Figure 2) plus the fact that the 10% 16O-incorrporation from the wild-type thioesterase into the 4-HB product (Figure 4C) was not observed for the E73D thioesterase-catalyzed single turnover reaction of 4-HB-CoA in H218O solvent (Figure 4D; vide supra), suggested a change in mechanism.
Figure 8.
(A) stopped-flow UV-vis trace of (20 μM) E73D thioesterase-catalyzed hydrolysis of (80 μM) 4-HB-CoA carried out under multiple turnover conditions and monitored at 300 nm (Δε = 11.8 mM−1 cm−1). The trace was fitted with a linear equation. (B) stopped-flow UV-vis trace of (40 μM) E73D thioesterase-catalyzed hydrolysis of (20 μM) 4-HB-CoA carried out under single turnover conditions and monitored at 300 nm. The trace was fitted with a single exponential equation. (C) stopped-flow UV-vis trace of (30 μM) T77A thioesterase-catalyzed hydrolysis of (100 μM) 4-HB-CoA carried-out under multiple turnover conditions and monitored at 300 nm. The trace was fitted with a double exponential equation. (D) Time course for the multiple turnover reaction of 50 μM [14C]4-HB-CoA and 10 μM T77A thioesterase monitored using rapid quench/HPLC techniques. The data were fitted to a double exponential equation.
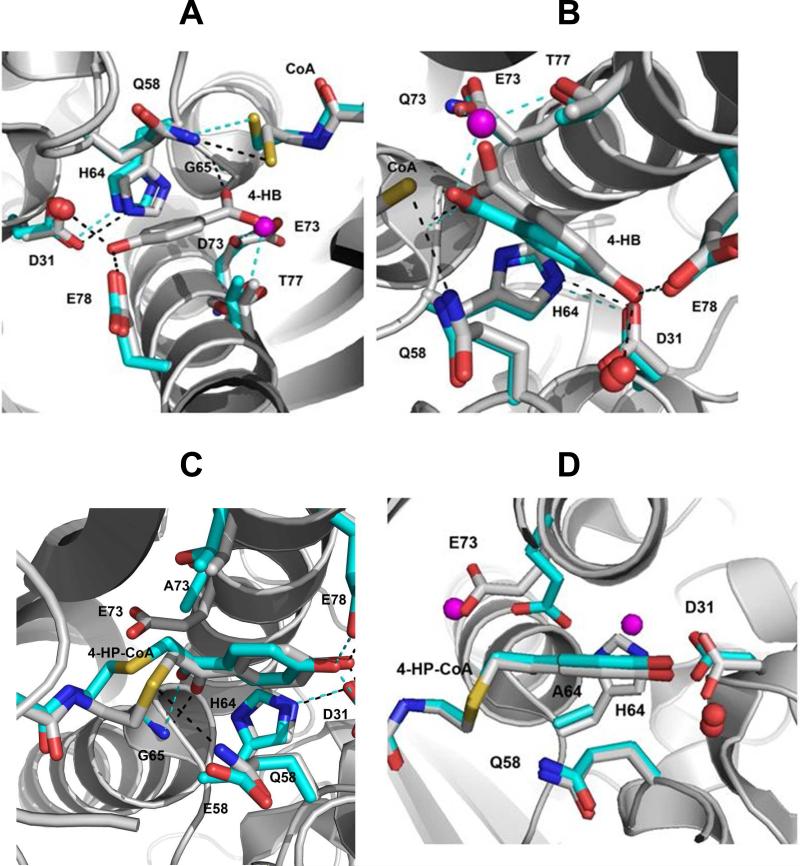
The X-ray structures of the E73D thioesterase(CoA) (Figure 9A) and E73D thioesterase(4-HP-CoA) (Figure SI3) complexes were determined to reveal that both structures retain the conformations of the active site side chains of the wild-type thioesterase. A notable change is the presence of an ordered water molecule located in the space vacated by the Glu73 carboxylate group. The superposition of the structure of the wild-type thioesterase(4-HB)(CoA) complex and the structure of the E73D thioesterase(CoA) complex (Figure 9A) places the water molecule on top of the 4-HB carboxylate oxygen opposite the oxygen atom that engages in hydrogen bond formation (2.9 Å) with the Gly65 backbone amide NH. The water molecule is located within hydrogen bond distance of the side chains of Thr77 (2.7 Å) and Asp65 (2.9 Å). Based on this structural information we posit that the E73D thioesterase retains a significant fraction of the wild-type thioesterase catalytic activity because the Asp73 carboxylate group can bind and activate the water for attack at the substrate thioester carbonyl carbon. In short, the replacement of Glu73 with Asp appears to have switched the catalytic role of the carboxylate group from that of nucleophile to Bronsted-Lowry base. Consistent with this conclusion is the observation that whereas 4-HB and CoA show competitive and noncompetitive product inhibition of the wild-type thioesterase, respectively both products show competitive inhibition (4-HB Kis =1.7 ± 0.4 mM; CoA Kis = 32 ± 2 μM; Table 2) towards the E73D mutant.
Figure 9.
(A) Superposition of the X-ray structure of the wild-type thioesterase(4-HB)(CoA) complex with that of the E73D thioesterase(CoA) complex. (B) Superposition of the X-ray structure of the wild-type thioesterase(4-HB)(CoA) complex with that of the E73Q thioesterase(4-HB) complex. (C). Superposition of the X-ray structure of the wild-type thioesterase(4-HP-CoA) complex with that of the E73A/Q58E thioesterase(4-HP-CoA) complex. (D). Superposition of the X-ray structure of the wild-type thioesterase(4-HP-CoA) complex with that of the H64A thioesterase(4-HP-CoA) complex. Only the backbone of the wild-type thioesterase is shown. The nitrogen atoms are colored blue, the oxygen atoms red and the sulfur atom yellow. The carbon atoms are colored gray for the wild-type thioesterase and cyan for the mutant thioesterase.
Compared to the wild-type thioesterase, the E73Q thioesterase displayed a 4-fold increase in the 4-HP-CoA Kd value (Table 4). The E73Q thioesterase kcat value is 4000-fold smaller than that of the wild-type thioesterase, yet 14-fold larger than that of the E73A mutant. The superposition of the structures of the E73Q thioesterase(4-HP-CoA) and wild-type thioesterase(4-HP-CoA) complexes (Figure SI4) revealed a conserved active site configuration with the exception that the Gln73 side chain is rotated slightly relative to the Glu73 side chain. More informative, however is the comparison of the structures of the E73Q thioesterase(4-HB) and wild-type thioesterase(4-HB)(CoA) complexes (see Figure 9B). Notably, there is an ordered water molecule located between the Gln73 amide substituent and the 4-HB carboxylate group. It appears that in order to accommodate the water molecule the 4-HB has been tilted from the position seen in the active site of the wild-type thioesterase thus moving it ~1 Å further away from residue 73. The ordered water molecule is within hydrogen bond distance (2.4 Å) of the 4-HB carboxylate oxygen (opposite the oxygen atom that engages in hydrogen bond formation with the Gly65 backbone amide NH at 2.5 Å), the Gln73 amide NH2 (2.9 Å) and the Thr77 hydroxyl group (2.6 Å). The comparatively large reduction in the kcat value suggests that the accommodation of the water molecule imposes a tax on catalysis which adds to that associated with the loss of nucleophilic catalysis (used by the wild-type thioesterase) or base catalysis (used by the E73D thioesterase).4
Thr77
Thr77 is posited to orient the water nucleophile for attack at the 4-hydroxybenzoyl carbonyl carbon of the thioesterase anhydride intermediate (Figure 5). The Thr77 was separately replaced with Ser, Ala, Val and Asp. The Ser residue conserves the hydroxyl substituent required to bind the water nucleophile yet does not have the steric bulk of the Thr side chain. The Ala and Val substitution removes the hydroxyl substituent interaction and either reduces (Ala) or increases (Val) the size of the side chain. The Asp substitution replaces the hydroxyl group with a carboxylate substituent that could potentially function as a base to activate the water nucleophile but is more likely to perturb the local electrostatic environment. The mutations did not cause a large decrease in 4-HP-CoA binding affinity (2-5 fold) (Table 4). Likewise, the steady-state kcat values measured for T77A and T77S 4-HB-CoA thioesterase-catalyzed 4-HB-CoA hydrolysis are decreased only ~2-fold decrease (Table 4). In contrast, the kcat value of the T77V thioesterase is 40-fold smaller than that of the wild-type thioesterase and the kcat value of the T77D 4-HB-CoA thioesterase is 650-fold smaller. This indicates, as does the structure of the liganded enzyme (Figure 1A), that Thr77 does not function in 4-HB-CoA binding. The significant reduction in catalytic efficiency (200-fold) observed for the T77D mutant suggests that the charged carboxylate group of the Asp perturbs the electrostatic environment at the reaction site.
The time course for T77A thioesterase-catalyzed 4-HB-CoA hydrolysis under multiple turnover conditions is biphasic (Figures 8C and 8D). The time courses measured for the wild-type and mutant thioesterases were fitted with a double exponential equation for comparison of rates. In the case of the T77A mutant stopped-flow data, the rate of the fast phase is 19.7 ± 0.2 s−1 and the rate of the slow phase 0.49 ± 0.2 s−1 whereas in the case of the rapid quench data, the rate of the fast phase is 47 ± 8 s−1 and the rate of the slow phase is 1.38 ± 0.06 s−1 (Table 1). For the wild-type thioesterase stopped-flow data the rate of the fast phase is 28.3 ± 0.4 s−1 and the rate of the slow phase is 4.30 ± 0.2 s−1, and for the rapid quench data the rate of the fast phase is 100 ± 30 s−1 and the rate of the slow phase is 8.7 ± 0.3 s−1 (Table 1). Thus, in the case of the T77A mutant the fast phase (formation of the enzyme-anhydride) is slowed down 2-fold compared to that of the wild-type thioesterase, whereas the slow phase (hydrolysis of the enzyme-anhydride intermediate) is reduced in rate 6 to 14–fold. Similarly, the time course (measured by stopped-flow UV-vis) for (30 μM) T77S thioesterase-catalyzed (100 μM) 4-HB-CoA hydrolysis under multiple turnover conditions is biphasic (Figure SI5A). The rate of the fast phase is 18.9 ± 0.2 s−1 and that of the slow phase is 1.208 ± 0.006 s−1 (Table 1). In contrast, the time courses for T77V and T77D thioesterase-catalyzed 4-HB-CoA hydrolysis under multiple turnover conditions are monophasic (Figure SI5B and SI5C) and define turnover rates of 0.152 ± 0.002 s−1 and 0.0081 ± 0.0001 s−1 (Table 1), respectively.
The single turnover time courses for T77A thioesterase catalysis measured by stopped-flow UV-vis (20 μM 4-HB-CoA and 40 μM T77A thioesterase) or rapid quench (5 μM 4-HB-CoA and 50 μM T77A thioesterase) (Figure SI5D and SI5E) were fitted with a single exponential equation to define the apparent rate constant for a single turnover reaction of 17 s−1 and 24 s−1, respectively (Table 1). The stopped-flow UV-vis absorbance trace of (40 μM) T77S thioesterase-catalyzed (20 μM) 4-HB-CoA hydrolysis under single turnover conditions (Figure SI5F) was fitted with a single exponential equation to define the apparent rate constant for a single turnover reaction of 19 s−1 (Table 1) The superposition of the structures of the wild-type thioesterase(4-HP-CoA) and T77S thioesterase(4-HP-CoA) complexes (Figure SI6) shows that with the exception of a slight shift in the position of the residue 77 hydroxyl group, all other residues plus ligands are unchanged. The notable difference in the mutant structure is the absence of the steric confinement of the residue 77 side chain CH2OH, with is imposed by the T77 side chain methyl group because of its close contact with Ala91.
The structure of E73D thioesterase(CoA) complex revealed hydrogen bond formation between Thr77 and the putative water nucleophile (Figure 9A). By analogy, one could envision a Thr77-bound water molecule positioned for attack at the 4-hydroxybenzoyl carbonyl carbon of the wild-type thioesterase anhydride intermediate. The posited participation of Thr77 in the orientation of the water nucleophile is supported by the observed (i) 6 to 20-fold reduction in the rate hydrolysis step, (ii) the change in the ratio of attack at the benzoyl carbonyl carbon vs the Glu73 carbonyl carbon in the T77A (from 9:1 to 1:1) and T77S (from 9:1 to 1:0) (vide supra) and (iii) the reduction in the rate of the enzyme-anhydride intermediate in contrast to no reduction in the rate of the formation of the enzyme-anhydride intermediate.
The kinetic properties of the E73D/T77A and E73D/T77S double mutants were also examined. The X-ray structure of the E73D thioesterase(CoA) complex shows that both Thr77 and Asp73 form hydrogen bonds with the ordered water molecule posited to attack the 4-HB-CoA carbonyl carbon (Figure 9A). The E73D/T77A thioesterase on the other hand, would only be able to bind the water nucleophile with the Asp73. Strikingly, the kcat value of the E73D/T77A thioesterase is 10-fold larger that that of E73D thioesterase (only 5-fold smaller than that of the wild-type thioesterase) and there is no significant change in the 4-HP-CoA Kd (Table 4). In contrast, the steady-state kinetic constants of the E73D/T77S thioesterase are essentially the same as those of the E73D mutant (Table 4). The time courses for the E73D/T77A and E73D/T77S thioesterase-catalyzed hydrolysis of 4-HB-CoA measured under multi-turnover reactions using stopped-flow UV-vis techniques (Figure SI5F and SI5G) each showed a single phase and together defined apparent rate constants of 0.705 ± 0.001 s−1 and 0.0619 ± 0.0001 s−1, respectively (Table 1).
Lastly, the E73D/T77A thioesterase-catalyzed single turnover reaction of 4-HB-CoA in 97% H218O was carried out. The 16O-incorporation observed in the isolated 4-HB was negligible (see Figure 4H). If we compare these results with those obtained for the wild-type, T77A and T77S thioesterases an interesting trend emerges. Namely, residue 77 influences the isotopic labeling pattern observed in the product when residue 73 is Glu (vide supra) but not when it is Asp. Furthermore, the kinetic properties of the T77A and T77S thioesterases are essentially the same, whereas the E73D/T77A thioesterase is considerably more active than is the E73D/T77S thioesterase. Based on these findings we posit that in the case of the E73D thioesterase, hydrogen bond formation between the Thr77 hydroxyl group (or in the E73D/T77S thioesterase hydrogen bond formation between the Ser77 hydroxyl group) and the water nucleophile partially impairs the ability of the Asp73 to optimize the orientation of the water nucleophile for attack at the 4-HB-CoA carbonyl carbon. In contrast, in the case of the wild-type thioesterase the Thr77 contributes to catalysis by facilitating the attack of the water nucleophile on the anhydride intermediate.
Gln58
The structure of the wild-type thioesterase(4-HB)(CoA) complex shows that the Gln58 side chain NH2 is within hydrogen bond distance (3.5 Å) of the CoA thiol substituent (Figure 1A). A comparison of the steady-state inhibition constants measured for CoA (Kis = 16 ± 2 μM, Kii = 29 ± 2 μM) (vide infra) (Table 2) and desulfoCoA (Kis = 19 ± 6 μM, Kii =13 ± 3 μM) suggests that this interaction does not contribute significantly to the CoA binding affinity. Nevertheless, Gln58 might form a stronger hydrogen bond with the CoA thiolate anion that is forming in the transition state of the first partial reaction.
To further examine the contribution that Gln58 makes to substrate binding and catalysis the kinetic properties of the Q58A, Q58D, Q58E, Q58D/E73D, Q58E/E73Q and Q58E/E73Athioesterases were determined (Table 4). The kcat values of the Q58A, Q58D and Q58E mutants were reduced 53, 52 and 18-fold, respectively compared to that of the wild-type thioesterase. The dissociation constants (Kd) of the Q58A, Q58D and Q58E thioesterase complexes of 4-HP-CoA were increased 5, 3 and 2-fold, respectively. The 4-HP-CoA inhibition constant (Ki = 4.7 × 10−3 μM) measured with Q58E thioesterase is essentially the same as that of the wild-type thioesterase (Table 4). These results indicate that the Q58 contributes primarily to transition state binding (stabilization) rather than to the substrate binding.
The time course for (20 μM) Q58A thioesterase-catalyzed multiple turnover of (80 μM) 4-HB-CoA determined (Figure SI7A) using stopped-flow UV-vis absorbance techniques, was fitted with a single exponential to define k = 0.0842 ± 0.0001 (Table 1) which agrees with the steady-state kcat = 0.127 ± 0.002 (Table 4). The absence of a burst phase in the time course suggests that the anhydride-forming step is rate-limiting. A comparison of this rate to the rate of the burst phase (40 s−1) measured for the wild-type thioesterase (480-fold difference) indicates that Gln48 makes a sizable contribution to catalysis of the formation of the anhydride intermediate. The X-ray structures of the Q58A thioesterase complexes of 4-HB plus CoA (Figure SI8A) and of 4-HP-CoA (Figure SI8B) showed that the active site is unchanged except for the residue 58. Notably, the space vacated by the removal of the CH2C(=O)NH2 group was not filled by an ordered water molecule. Together, these findings are interpreted as evidence that Glu58 functions in catalysis by stabilizing the CoA thiolate-leaving group via hydrogen bonding (as depicted in Figure 5).
Amino acid replacements at both Gln58 and Glu73 were found to have a synergistic inhibitory effect on catalysis. Whereas the dissociation constants measured for the 4-HP-CoA complexes are submicromolar (albeit up to 20-fold larger than that of the wild-type thioesterase; Table 4) the Q58D/E73D, Q58E/E73Q and Q58E/E73A thioesterases displayed no activity above the detection limit of kcat = 1 × 10−5 s−1. The superposition of the X-ray structures of the Q58E/E73A thioesterase(4-HP-CoA) and wild-type thioesterase(4-HP-CoA) complexes (Figure 9C) shows that although the conformations of the residues that form the catalytic site are unchanged, the hydroxyphenacyl unit of the Q58E/E73A thioesterase bound 4-HP-CoA is substantially shifted in the direction of Ala73. In comparison, the E73A thioesterase(4-HP-CoA) structure depicts a much smaller shift in the position of the hydroxyphenacyl unit, suggesting that the Gln58 might play an essential role in the particular case of the E73D thioesterase catalysis by preventing the substrate reaction center from sliding out of position as allowed by the vacant space.
Gly65
The structure of the wild-type thioesterase bound with product (Figure 1A) shows that the 4-HB carboxylate group is in-line with the positive pole of the central helix and that it is engaged in hydrogen bond formation (2.8 Å) with the backbone amide NH of Gly65 located at the helix N-terminus. The findings from an earlier Raman study of the E73A thioesterase(4-HB-CoA) and wild-type thioesterase(4-HP-CoA) complexes indicated that the C=O of the respective ligand O=C-S and O=C-CH2S moieties are held in a rigid non-fluctuating environment which provides a modest hydrogen bonding strength of ~20 kJ/mol (19). The X-ray structure of the thioesterase bound with 4-HP-CoA reveals a short hydrogen bond (2.6 Å) between the oxygen atom of the O=C-CH2-S group and the Gly65 NH. In the X-ray structure of thioesterase(4-hydroxybenzyl-CoA) (CH2-S replaces O=C-S) complex, there is no hydrogen bond interaction with the Gly65 NH and consequently the CH2-S group, which is free to rotate, is observed in two different orientations. The 4-HB-CoA k−1/k1 ~ 0.16 μM compares with the competitive inhibition constant Kis = 0.6 ± 0.1 μM (18) determined for 4-hydroxybenzyl-CoA, consistent with a modest hydrogen bond interaction at the ground state.
Most striking is the tight binding of the 4-HP-CoA for which a competitive inhibition constant Kis = 0.003 ± 0.1 μM (19) and the Kd= 0.007 ± 0.003 μM (Table 4). The tight binding of the 4-HP-CoA ligand suggests that the additional methylene group somehow locks the carbonyl into a position that facilitates strong hydrogen bond formation between the C=O and the Gly65 NH.
Previous studies have shown that 4-HB dithioCoA, in which S=C-S replaces O=C-S, is less susceptible to base catalyzed hydrolysis than is 4-HB-CoA by one order of magnitude (34). In contrast, the steady-state kcat = 9.6 × 10−4 s−1 measured for thioesterase-catalyzed hydrolysis of 4-HB dithioCoA is almost four orders of magnitude smaller than that measured for 4-HB-CoA. This suggests that the steric and/or electronic requirements of the transition states of thioesterase catalysis are more stringent than the transition state of the base-catalyzed hydrolysis. Moreover, the hydrogen bond interaction with the Gly65 appears to be used for transition state stabilization and less so for substrate binding.
Asp31, His64 and Glu78
Based on the X-ray structural data (Figure 1A) these three residues appear to create a hydrogen bond network, which could facilitate the productive binding of the 4-hydroxybenzoyl ring. The Glu78 forms a hydrogen bond with the ring hydroxyl group and the hydroxyl group in turn forms a hydrogen bond with an Asp31 bound water molecule. The Asp31 engages the His64 in hydrogen bond formation and the His64 ring stacks against the edge of the 4-hydroxybenzoyl ring. The E78A, D31A, D31N, H64A and H64Q thioesterase mutants exhibit a 10, 160, 6, 45 and 24-fold decrease in kcat and a 357, 10, 171, 31 and 47-fold decrease in 4-HP-CoA binding affinity, respectively (Table 4). The stopped-flow UV-vis absorbance traces (Figures SI7B, SI7C, SI7D, SI7E and SI7F) measured for the (20 μM) D31A, D31N, H64A, H64Q and E78A thioesterase catalyzed hydrolysis of (80 μM) 4-HB-CoA under multiple turnover conditions were fitted to a linear equation to define the apparent rate constants of 0.012, 0.42, 0.014, 0.069 and 0.18 s−1, respectively (Table 1). The loss of burst phase kinetics suggests that formation of the anhydride intermediate rate-limiting for these mutants.
The lower kcat values observed for the mutants are likely due to changes in the local electrostatic environment and residue packing. The X-ray structure of the H64A thioesterase bound with 4-HP-CoA was solved to reveal that a water molecule and the Glu73 side chain have entered the space vacated by the His64 imidazole ring (Figure 9D), although the hydrogen bond network involving Asp31, H2O, the C(4)OH and Glu78 was left unchanged. Thus, His64 serves to restrict the conformational space of the Glu73 side chain (which is otherwise unconstrained) in addition to interacting with Asp31.
The CoA Binding Site
The CoA unit of the 4-HB-CoA extends from the 4-HB binding pocket through a tunnel that joins with a crater-like depression at the protein surface (Figure 1B). The pantothenate arm is bound in the tunnel were it engages in hydrogen bond interactions with backbone amide NHs and with ordered water molecules. The nucleotide moiety sits in the surface depression where the CoA 3’-phosphate (3’-P) forms two ion pairs with the positively charged Arg102 and Arg150 side-chains, and the 5’-pyrophosphate (5’-PP) forms hydrogen bonds with the Ser120 and Thr121 side chains (Figure 1B). The superposition of the structures of the wild-type and mutant thioesterases that contain bound 4-HP-CoA, 4-hydroxybenzyl-CoA or CoA shows that the conformation of the CoA unit is conserved. Moreover, the B factors (temperature factors) indicate that the atoms of the bound 4-HP-CoA and 4-hydroxybenzyl-CoA ligands experience little motion (see the liganded structure colored according to B factor in Figure SI9). These observations suggested that the thioesterase binds the nucleotide unit of its substrate. The experiments described below where carried out to test whether the CoA unit binding interactions indicated by the crystal structure actually occur in solution.
31P-NMR evidence for a protein surface nucleotide binding site
If the electrostatic interactions observed to occur between the 3’-P and 5’-PP and the electropositive side chains are preserved in the enzyme-ligand complex in solution, the 3’-P and 5’-PP is expected to experience an electronic environment different from that of pure solvent. Thus, a difference in the chemical shifts of the 3’-P and 5’-PP 31P-NMR resonances deriving from the solvated enzyme-ligand complex versus those deriving from the free ligand would signify association of the CoA nucleotide with the enzyme surface.
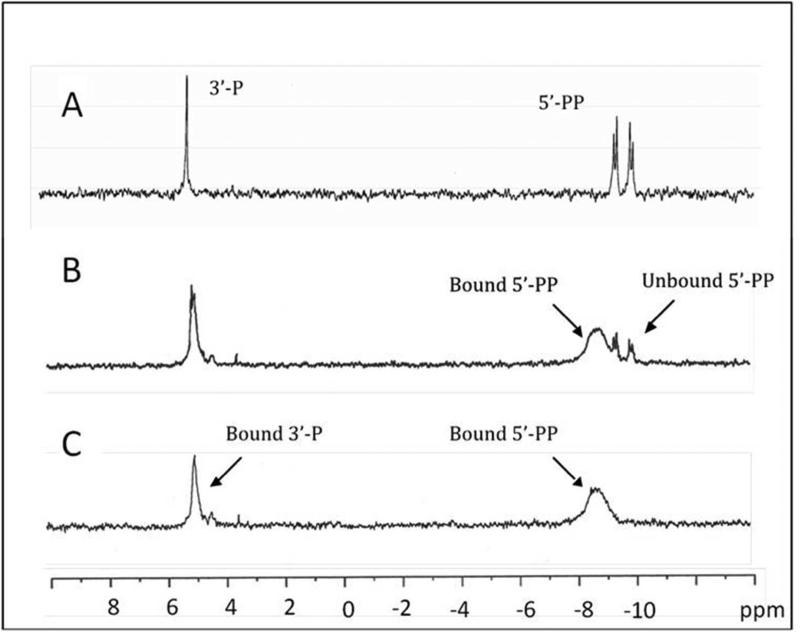
The proton-decoupled 31P-NMR spectrum of 4-HP-CoA in K+HEPES buffer (pH 7.5) is characterized by a singlet at +5.05 ppm and two doublets (J = 20 Hz) centered at –9.36 and – 9.90 ppm, respectively (panel A in Figure 10). The singlet was assigned to the CoA 3’-P, while the two doublets were assigned to the 5’-PP. The 31P-NMR resonance of the CoA 5’-PP observed in the spectrum of the thioesterase(4-HP-CoA) complex is characterized by a broad peak centered at –8.7 ppm, which is shifted ~1 ppm downfield from the two doublets of unbound 4-HP-CoA (panels B and C Figure 10). The 0.1 ppm upfield shift in the 3’-P resonance (from +5.05 ppm to +4.95 ppm) is comparatively small, possibly because it appears (from the X-ray structure, Figure 1B) to be more highly solvated and somewhat less restrained (Figure SI9). The peaks are broadened because of the slow tumbling rate of the enzyme coupled with the restricted motion in the 3’-P and 5’-PP groups. This finding suggests that the 3’-P and 5’-PP groups are bound to the protein surface.
Figure 10.
31P-NMR spectrum of (A) 1 mM 4-HP-CoA, (B) 1.5 mM 4-HP-CoA and 1 mM wild-type thioesterase and (C) 1 mM 4-HP-CoA and 1 mM wild-type thioesterase. See Materials and Methods for details.
Kinetic analysis of site-directed mutants
The contributions that the ion pairs and hydrogen bonds formed between the substrate 3’-P and 5’-PP groups and the Arg102, Arg150, Ser120 and Thr121 side-chains make to substrate binding and catalysis were accessed by separately replacing each residue with Ala and measuring the catalytic efficiency (kcat/Km) (Table 4), the 4-HP-CoA binding properties (Kis and Kd) (Table 4) and the CoA binding properties (Kis and Kii) (see Table 2) with the corresponding Ala mutants. Modest decreases in kcat/Km and modest increases in the 4-HP-CoA Kis (or Kd) were observed for the T121A, R150A, R102A thioesterases but not for the S120A thioesterase. The inhibition constants measured for CoA, on the other hand, showed substantial increases (23, 29, 31 and 7-fold, respectively). These findings are evidence for the binding interaction between the nucleotide unit and the thioesterase surface residues. The tight binding of the CoA (Kis = 16 μM; Kii =29 μM) suggests that the pantothenate arm also contributes to the binding energy possibly through desolvation effects and Van der Waals interactions.
Conclusions
Sequence homologues of the Arthrobacter 4-HB-CoA thioesterase which have known physiological substrates include the bacterial phenylacetyl-CoA thioesterase (34), benzoyl-CoA thioesterase (35), enterobactin biosynthetic pathway “housekeeper” thioesterase EntH (8, 36, 37), long chain acyl-CoA thioesterase II and III (37, 38) and fluoroacetyl-CoA thioesterase (39-41). Sequence alignments of these homologues show conservation of Thr77 yet substitutions of Glu73 with an Asp residue and Gln58 with either an Asn or Glu residue. This suggests that the catalytic mechanism employed might vary among clade members. To our knowledge, the catalytic mechanisms of Arthrobacter 4-HB-CoA thioesterase homologues have not been studied in depth. Rather, the reported posited mechanisms are based solely on the steady-state kinetic constants of site-directed mutants and/or on the liganded X-ray structures (32, 34, 36, 39, 41, 43). Nevertheless, each of the proposed mechanisms is different from that posited herein for the Arthrobacter 4-HB-CoA thioesterase, and are unique from one another.
The present work has shown that the Arthrobacter 4-HB-CoA thioesterase-catalyzed reaction can be switched by Asp replacement of Glu73, from the two-step pathway depicted in Figure 5 to a single step pathway in which the Asp activates a water molecule for direct attack on the substrate thioester moiety. Remarkably, the efficiency of this alternative reaction pathway was raised to near wild-type activity in the E73D/T77A mutant by the secondary replacement of Thr77 with Ala. This finding suggests that the hotdog-fold thioesterase catalytic scaffold has high plasticity, which might explain why the catalytic residues are not stringently conserved. The fact that thioester hydrolysis does not require a highly proficient enzyme catalyst (22) might in turn account for the plasticity of the hotdog-fold thioesterase catalytic scaffold.
The use of nucleophilic catalysis in enzyme catalyzed thioester hydrolysis would appear to be unnecessary and prompts the question of how this catalytic strategy evolved. The same question might be posed for the use of nucleophilic catalysis by the catalytic Asp of the crotonase family 3-hydroxyisobutyryl-CoA thioesterase (44). We suspect that the proximity of the carboxylate group and the substrate thioester C=O is the determining factor and that this in turn is simply a matter how the substrate happens to be oriented in the active site. The plasticity of the hot-dog thioesterase catalytic site suggests that the substrate specificity might be easily controlled by protein engineering and therefore that the thioesterases from this family might be ideal for synthetic biology applications, particularly in the area fatty alcohol biofuel production. Furthermore, the high degree of divergence in the hotdog-fold thioesterase active site structure and possibly, catalytic mechanism makes these enzymes excellent candidates for target specific inhibitor design.
Supplementary Material
Figure 7.
The time courses for the steady-state multiple-turnover reactions of 1.5 (green), 2.7 (red) or 5.7 (black) μM 4-HB-CoA catalyzed by 0.003 μM thioesterase in 50 mM K+HEPES (pH 7.5, 25 °C). The 300 nm absorbance traces (dotted lines) are shown in the context of the calculated concentration of 4-HB-CoA (ε = 11.8 mM−1 cm−1) present in the reaction mixture (y-axis) as a function of reaction time (x-axis). The simulated curves (solid lines) were generated using the kinetic model shown in Scheme 2 and rate constants: k1 = 192 μM−1 s−1, k−1 = 30 s−1, k2 = 44 s−1, k−2 = 14 s−1, k3 = 36 s−1, k−3 = 0 s−1, k4 = 129 s−1, k−4 = 5 μM−1 s−1, k5 = 368 μM s−1 and k−5 = −5 μM−1 s−1.
Footnotes
The coordinates for the X-ray crystal structures of the 4-hydroxybenzoyl-CoA thioesterase mutants are deposited in the protein database under the ID codes 3R32, 3R34, 3R35, 3R36, 3R37, 3R3A, 3R3B, 3R3C, 3R3D, 3TEA and 3R3F.
Abbreviations used are: 4-HB, 4-hydroxybenzoate; CoA, coenzyme A; 4-HB-CoA, 4-hydroxybenzoyl-coenzyme A; 4-HP-CoA, 4-hydroxyphenacyl-CoA; 4-CBA, 4-chlorobenzoate; PCR, polymerase chain reaction; K+HEPES, potassium salt of N-(2-hydroxyethyl)piperzine-N-’ 2-ethanesulfonic acid; DTNB, 5,5’-dithio-bis(2-nitrobenzoic acid); SDS-PAGE, sodium dodecyl-sulfate-polyacrylamide gel electrophoresis; IPTG, isopropyl-β-D-galactopyranoside; DTT, dithiothreitol; GC-MS, gas chromatography-mass spectrometry.
Throughout this paper we treat the substitution reaction at the benzoyl carbonyl carbon as a concerted reaction. This assumption is based on published work, which showed that nucleophilic substitution reactions of esters do not proceed via a tetrahedral intermediate in cases where the leaving group is low in energy as indicated by the pKa of the conjugate acid (21). Earlier we demonstrated that for the base-catalyzed hydrolysis of para-substituted benzoyl-CoA thioesters that the plot of log k2 vs the σ value of the para-substituents was linear with a slope (ρ) of 1.5, thus indicating only a small accumulation of charge in the transition state (22) consistent with a concerted reaction.
No distinction is made between rate-limiting product dissociation and a rate-limiting thioesterase conformational change that must occur following product formation to regenerate the catalytically active form of the enzyme.
Base catalysis applied to an enzymatic hydrolysis reaction typically contributes less than 2-orders of magnitude to the kcat value (31). In the extreme case of the hotdog-fold fatty acyl-CoA thioesterase hTHEM2, base catalysis by the active site Asp65 contributes only 4-fold to the kcat value (as inferred from the comparison of the kcat values of the wild-type and D65N mutant enzyme) (32). If Glu73 functions in base catalysis then we would expect that the E73Q mutant would retain at least 1% of the activity of the wild-type thioesterase rather than only 0.025%.
Supporting Information Available: This material includes the protocol for preparation of the thioesterase mutants, a table listing the X-ray data collection and refinement statistics, graphics showing of X-ray structures liganded thioesterase mutants, stopped-flow UV-vis traces for mutant thioesterase-catalyzed 4-HB-CoA hydrolysis and steady-state inhibition plots. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Nobeli I, Ponstingl H, Krissinel EB, Thorton JM. A structure-base anatomy of the E. coli metabolome. J. Molecular Biology. 2003;334:697–719. doi: 10.1016/j.jmb.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Amyes TL, Richard JP. Generation and stability of a simple thiol enolate in aqueous solution. Amer. Chem. Soc. 1992;114:10297–10302. [Google Scholar]
- 3.D'Ordine RL, Bahnson BJ, Tonge PJ, Anderson VE. Enoyl-coenzyme A hydratase-catalyzed exchange of the alpha-protons of coenzyme A thiol esters: a model for an enolized intermediate in the enzyme catalyzed elimination? Biochemistry. 1994;33:14733–4142. doi: 10.1021/bi00253a011. [DOI] [PubMed] [Google Scholar]
- 4.Heath RJ, Rock CO. The Claisen condensation in biology. Nat. Prod. Rep. 2002;19:581–596. doi: 10.1039/b110221b. [DOI] [PubMed] [Google Scholar]
- 5.Schmelz S, Naismith JH. Adenylate-forming enzymes. Curr Opin Struct Biol. 2009;19:666–671. doi: 10.1016/j.sbi.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo ZF, Sun Y, Zheng S, Guo Z. Preferential hydrolysis of aberrant intermediates by the type II thioesterase in Escherichia coli nonribosomal enterobactin synthesis: substrate specificities and mutagenic studies on the active-site residues. Biochemistry. 2009;48:1712–1722. doi: 10.1021/bi802165x. [DOI] [PubMed] [Google Scholar]
- 7.Chen D, Wu R, Bryan TL, Dunaway-Mariano D. In vitro kinetic analysis of substrate specificity in enterobactin biosynthetic lower pathway enzymes provides insight into the biochemical function of the hot dog-fold thioesterase EntH. Biochemistry. 2009;48:511–513. doi: 10.1021/bi802207t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou Y, Meng Q, You D, Li J, Chen S, Ding D, Zhou X, Zhou H, Bai L, Deng Z. Selective removal of aberrant extender units by a type II thioesterase for efficient FR 008/candicidin biosynthesis in Streptomyces sp. strain FR 008. Appl. Environ. Microbiol. 2008;74:7235–7242. doi: 10.1128/AEM.01012-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claxton HB, Akey DL, Silver MK, Admiraal SJ, Smith JL. Structure and functional analysis of RifR, the type II thioesterase from the rifamycin biosynthetic pathway. J. Biol. Chem. 2009;284:5021–5029. doi: 10.1074/jbc.M808604200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koglin A, Löhr F, Bernhard F, Rogov VV, Frueh DP, Strieter ER, Mofid MR, Güntert P, Wagner G, Walsh CT, Marahiel MA, Dötsch V. Structural basis for the selectivity of the external thioesterase of the surfactin synthetase. Nature. 2008;454:907–911. doi: 10.1038/nature07161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, Sussman JL, Verschueren KHG, Goldman A. The alpha/beta hydrolase fold Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 12.Dillon SC, Bateman A. The Hotdog fold: wrapping up a superfamily of thioesterases and dehydratases. BMC Bioinformatics. 2004;5:109. doi: 10.1186/1471-2105-5-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cantu DC, Chen Y, Reilly P. Thioesterases: a new perspective based on their primary and tertiary structures. J. Protein Sci. 2010;2010;19:1281–1295. doi: 10.1002/pro.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brocker C, Carpenter C, Nebert DW, Vasiliou V. Evolutionary divergence and functions of the human acyl-CoA thioesterase gene (ACOT ) family. Hum Genomics. 2010;4:411–420. doi: 10.1186/1479-7364-4-6-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmquist M. Alpha/Beta-hydrolase fold enzymes: structures, functions and mechanisms. Curr Protein Pept Sci. 2000;1:209–235. doi: 10.2174/1389203003381405. [DOI] [PubMed] [Google Scholar]
- 16.Benning MM, Wesenberg G, Liu R, Taylor KL, Dunaway-Mariano D, Holden HM. The three-dimensional structure of 4-hydroxybenzoyl-CoA thioesterase from Pseudomonas sp. Strain CBS-3 J. Biol. Chem. 1998;273:33572–33579. doi: 10.1074/jbc.273.50.33572. [DOI] [PubMed] [Google Scholar]
- 17.Leesong M, Henderson BS, Gillig JR, Schwab JM, Smith JL. Structure of a dehydratase-isomerase from the bacterial pathway for biosynthesis of unsaturated fatty acids: two catalytic activities in one active site. Structure. 1996;4:253–264. doi: 10.1016/s0969-2126(96)00030-5. [DOI] [PubMed] [Google Scholar]
- 18.Thoden JB, Zhuang Z, Dunaway-Mariano D, Holden HM. The structure of 4-hydroxybenzoyl-CoA thioesterase from Arthrobacter sp. strain SU. J. Biol. Chem. 2003;278:43709–43716. doi: 10.1074/jbc.M308198200. [DOI] [PubMed] [Google Scholar]
- 19.Thoden JB, Holden HM, Zhuang Z, Dunaway-Mariano D. X-ray crystallographic analyses of inhibitor and substrate complexes of wild-type and mutant 4-hydroxybenzoyl-CoA thioesterase. J. Biol. Chem. 2002;2002;277:27468–2776. doi: 10.1074/jbc.M203904200. [DOI] [PubMed] [Google Scholar]
- 20.Dunaway-Mariano D, Babbitt PC. On the origins and functions of the enzymes of the 4-chlorobenzoate to 4-hydroxybenzoate converting pathway. Biodegradation. 1994;1994;5:259–276. doi: 10.1007/BF00696464. [DOI] [PubMed] [Google Scholar]
- 21.Hess RA, Hengge AC, Cleland WW. Isotope Effects on Enzyme-Catalyzed Acyl Transfer from p-Nitrophenyl Acetate: Concerted Mechanisms and Increased Hyperconjugation in the Transition State. J. Amer. Chem. Soc. 1998;120:2703–2709. [Google Scholar]
- 22.Song F, Zhuang Z, Dunaway-Mariano D. Structure-activity analysis of base and enzyme-catalyzed 4-hydroxybenzoyl coenzyme A hydrolysis Bioorg. Chem. 2007;35:1–10. doi: 10.1016/j.bioorg.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Zhuang Z, Gartemann KH, Eichenlaub R, Dunaway-Mariano D. Characterization of the 4-hydroxybenzoyl-coenzyme A thioesterase from Arthrobacter sp. strain SU. Appl Environ Microbiol. 2003;2003;69:2707–2711. doi: 10.1128/AEM.69.5.2707-2711.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo L, Taylor KL, Xiang H, Wei Y, Zhang W, Dunaway-Mariano D. Role of active site binding interactions in 4-chlorobenzoyl-coenzyme A dehalogenase catalysis. Biochemistry. 2001;40:15684–15692. doi: 10.1021/bi011536f. [DOI] [PubMed] [Google Scholar]
- 25.Dong J, Luo L, Liang P, Dunaway-Mariano D, Carey P. Raman difference spectroscopic studies of dithiobenzoyl substrate and product analogs binding to the enzyme dehologenase: pi-electron polarization is prevented by C=O to C=S substitution. J. Raman Spectroscopy. 2000;31:365–371. [Google Scholar]
- 26.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 27.Zhang W, Wei Y, Luo L, Taylor KL, Yang G, Dunaway-Mariano D, Benning MM, Holden HM. Histidine 90 function in 4-chlorobenzoyl-coenzyme a dehalogenase catalysis. Biochemistry. 2001;40:13474–82. doi: 10.1021/bi0114426. [DOI] [PubMed] [Google Scholar]
- 28.Johnson KA. Fitting enzyme kinetic data with KinTek Global Kinetic Explorer. Methods Enzymol. 2009;467:601–626. doi: 10.1016/S0076-6879(09)67023-3. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Fersht A. Structure and Mechanism in Protein Science. W.H. Freeman and Co.; 1998. pp. 62–65. [Google Scholar]
- 32.Cao J, Xu H, Zhao H, Gong W, Dunaway-Mariano D. The mechanisms of human hotdog-fold thioesterase 2 (hTHEM2) substrate recognition and catalysis illuminated by a structure and function based analysis. Biochemistry. 2009;48:1293–1304. doi: 10.1021/bi801879z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong J, Zhuang Z, Song F, Dunaway-Mariano D, Carey PR. A Thioester Substrate Binds to the Enzyme Arthrobacter Thioesterase in Two Ionization States; Evidence from Raman Difference Spectroscopy. J. Raman Spectroscopy. 2012;43:65–71. doi: 10.1002/jrs.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song F, Zhuang Z, Finci L, Dunaway-Mariano D, Kniewel R, Buglino JA, Solorzano V, Wu J, Lima CD. Structure, function, and mechanism of the phenylacetate pathway hot dog fold thioesterase PaaI. J. Biol. Chem. 2006;281:11028–11038. doi: 10.1074/jbc.M513896200. [DOI] [PubMed] [Google Scholar]
- 35.Ismail W. Benzoyl-coenzyme A thioesterase of Azoarcus evansii: properties and function. Arch Microbiol. 2008;190:451–460. doi: 10.1007/s00203-008-0393-3. [DOI] [PubMed] [Google Scholar]
- 36.Guo ZF, Sun Y, Zheng S, Guo Z. Preferential hydrolysis of aberrant intermediates by the type II thioesterase in Escherichia coli nonribosomal enterobactin synthesis: substrate specificities and mutagenic studies on the active-site residues. Biochemistry. 2009;2009;48:1712–1722. doi: 10.1021/bi802165x. [DOI] [PubMed] [Google Scholar]
- 37.Leduc D, Battesti A, Bouveret E. In vitro kinetic analysis of substrate specificity in enterobactin biosynthetic lower pathway enzymes provides insight into the biochemical function of the hot dog-fold thioesterase EntH. J Bacteriol. 2007;189:7112–7126. [Google Scholar]
- 38.Nie L, Ren Y, Janakiraman A, Smith S, Schulz H. A novel paradigm of fatty acid beta-oxidation exemplified by the thioesterase-dependent partial degradation of conjugated linoleic acid that fully supports growth of Escherichia coli. Biochemistry. 2008;47:9618–9626. doi: 10.1021/bi801074e. [DOI] [PubMed] [Google Scholar]
- 39.Wang F, Langley R, Gulten G, Wang L, Sacchettini JC. Identification of a type III thioesterase reveals the function of an operon crucial for Mtb virulence. Chem. Biol. 2007;2007;14:543–551. doi: 10.1016/j.chembiol.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Huang F, Haydock SF, Spitelle D, Mironenko T, Li TL, O'Hagan D, Leadlay PF, Spencer JB. The gene cluster for fluorometabolite biosynthesis in Streptomyces cattleya: a thioesterase confers resistance to fluoroacetyl-coenzyme A. Chem. Biol. 2006;2006;13:475–484. doi: 10.1016/j.chembiol.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 41.Dias MV, Huang F, Chirgadze DY, Tosin M, Spiteller D, Dry EF, Leadlay PF, Spencer JB, Blundell TL. The gene cluster for fluorometabolite biosynthesis in Streptomyces cattleya: a thioesterase confers resistance to fluoroacetyl-coenzyme A. J. Biol. Chem. 2010;2010;285:22495–22504. [Google Scholar]
- 42.Weeks AM, Coyle SM, Jinek M, Doudna JA, Chang MC. Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity. Biochemistry. 2010;49:9269–79. doi: 10.1021/bi101102u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kunishima N, Asada Y, Sugahara M, Ishijima J, Nodake Y, Sugahara M, Miyano M, Kuramitsu S, Yokoyama S, Sugahara MJ. Mol. Biol. 2005;352:212–228. doi: 10.1016/j.jmb.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 44.Wong BJ, Gerlt JA. Divergent function in the crotonase superfamily: an anhydride intermediate in the reaction catalyzed by 3-hydroxyisobutyryl-CoA hydrolase. J. Amer. Chem Soc. 2003;125:12076–12077. doi: 10.1021/ja037652i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.