Abstract
Objectives
Caspase-1 (casp1), a key protease involved during systemic inflammatory response syndrome (SIRS), controls the brain expression of a set of eight genes: Nos2 and Ptgs2 (nitric oxide synthase 2 and prostaglandin-endoperoxide synthase 2, two inducible enzymes), Cxcl1 and Cxcl10 (C-X-C motif chemokine ligand 1 and ligand 10), Tgtp and Gbp2 (T cell specific GTPase 1 and guanylate binding protein 2, two GTPases), Adamts1 (a disintegrin-like and metallopeptidase with thrombospondin type 1 motif, 1, a metalloprotease), and Il1rn (interleukin (IL)-1 receptor antagonist). Our objective was to ascertain whether casp1 also controlled the peripheral expression of these genes and, if so, to compare their central vs. peripheral patterns of gene expression in immune and endocrine tissues during SIRS.
Methods
Wild-type (wt) and casp1 knockout (casp1−/−) mice were injected with either saline or a high dose of endotoxin/lypopolysaccharide (LPS; 800μg/mice i.p). Saline-injected mice were immediately euthanized after injection, whereas LPS-injected mice were sacrificed 6 and 12h after LPS administration. Hippocampal, splenic and adrenal gene expressions were determined by real-time PCR.
Results
Overall, casp1−/− mice showed a lower inflammatory response than wt mice. The expression level of powerful proinflammatory factors such as Nos2 and Ptgs2 was reduced in casp1−/− mice. Moreover, a hierarchical clustering analysis aimed at studying patterns of gene co-expression revealed large alterations in the hippocampal pattern of casp1−/− mice. Surprisingly, the expression of Adamts1was increased in the hippocampus and adrenals of casp1−/− mice.
Conclusions
The resilience of casp1−/− mice to SIRS lethality is associated with a lower inflammatory response, loss of hippocampal gene co-expression patterns, and increased hippocampal Adamts1 gene expression. The latter might be beneficial for casp1−/− mice, since ADAMTS1 is likely to play a role in neuronal plasticity. The mechanisms described here may help the development of either novel biomarkers or therapeutic targets against SIRS/sepsis.
Keywords: Inflammation, LPS, transcription, Nos2, Cox2, Chemokines, Cxcl1, Cxcl10, Adamts1, co-expression
INTRODUCTION
Systemic inflammatory response syndrome (SIRS) defines a serious medical condition of extreme and severe generalized inflammation in the absence of infection, whereas infection is the underlying process in sepsis [1]. Both SIRS and sepsis are leading causes of death in intensive care units [2].
The molecular mechanisms underlying SIRS remain elusive, and consequently, effective therapeutic strategies have not yet been developed. Animal models have been developed to aid the elucidation of the pivotal role of the innate immune system on SIRS [3-10], such as the administration of endotoxin/lipopolysaccharide (LPS) from Gram-negative bacteria to genetically-engineered mouse strains in order to mimic the generalized inflammation present in SIRS [11, 12]. One of these strains is the caspase-1 knockout (casp1−/−) mouse that is resilient to LPS-induced lethality [11]. Casp1 is a cysteine protease that may be a crucial element in SIRS lethality. It cleaves pro-interleukin (IL)-1ß and pro-IL-18 into mature biologically active cytokines. By submitting casp1−/− mice to LPS-induced SIRS, we previously observed that casp1 orchestrates the differential expression of eight genes in the central nervous system (CNS): two inducible enzymes [nitric oxide synthase 2 (Nos2) and prostaglandin-endoperoxide synthase 2/cyclooxygenase 2 (Ptgs2)], two chemokines [C-X-C motif ligand 1 (Cxcl1) and 10 (Cxcl10)], two GTPases [T-cell specific GTPase (Tgtp) and guanylate nucleotide binding protein 2 (Gbp2)], one metalloprotease [A disintegrin-like and metallopeptidase with thrombospondin type 1 (Adamts1)] and the IL-1 receptor antagonist gene (Il1rn), an endogenous antagonist of IL-1 [12].
Because SIRS is characterized by an imbalance in the neuro-immuno-endocrine derangements leading to death [13-16], we focused our studies on neural (hippocampus), immune (spleen) and endocrine (adrenals) tissues of wild-type (wt; control) and casp1−/− mice. The hippocampus was chosen because it is a brain area that plays a decisive role in the regulation of the hypothalamic-pituitary-adrenal (HPA) axis during different type of stress [17], including immunological challenges [18, 19]. The adrenals are the effector organs of the HPA-axis and were studied because of their crucial role in the secretion of corticoids during SIRS, whereas the spleen was chosen for being a prominent immune organ.
Given the fact that many of the proinflammatory factors that are triggered during LPS-induced SIRS require de novo synthesis, we measured their mRNA expression before (time 0h), and after LPS injection (6 and 12h later). We hypothesized that their involvement in SIRS-induced lethality would be reflected by differential mRNA expression levels between wt (control) and casp1−/− mice (resilient to SIRS-induced lethality).
We also assessed gene-gene interactions in wt and casp1−/− mice, and compared them to the patterns of gene expression in the central nervous system (hippocampal). Herein, we present evidence that a lower inflammatory response, pattern disruption of hippocampal gene co-expression of the aforementioned set of eight genes, and activation of hippocampal Adamts1 may be key mechanisms underpinning resilience to SIRS lethality in casp1−/− mice.
MATERIALS AND METHODS
Animals
We used virus-and-antibody-free, eight to ten-week-old male wt and casp1−/− mice that were bred in the same genetic background (C57BL/6). Casp1−/− mice were created, characterized, and generously provided by BASF Bioresearch (Cambridge, MA, USA) [11]. Those mice are fertile, develop normally, and have no histopathological abnormalities. It is noteworthy that casp1−/− mice are resistant to the lethal effects of LPS [11], as well as to the anorectic effects exerted by the central action of LPS-induced cytokines [20]. Mice were kept in a light- (12h on/12h off) and temperature-controlled environment, with food and water ad libitum. All procedures were performed under established guidelines of humane care and use of animals; they were approved by the University of Miami Institutional Animal Care and Use, and the Australian National University Animal Experimentation Ethics Committee. Experiments were designed to avoid confounding variables such as infection and stress, which could alter mRNA levels.
Experimental protocol
Wt and casp1−/− mice (n=23 per genotype) were injected with either 0.9% NaCl (saline) or a lethal dose of LPS [800μg/mice intraperitoneally (ip) from E. coli serotype O111:B4 (Merk Millipore, VIC, Australia)] [11]. Control animals were immediately euthanized after saline injection ip [0h, wt (n=7) and casp1−/− (n=8) mice], whereas LPS-injected mice were euthanized at 6h [wt (n=13) and casp1−/− (n=12)], and 12h [wt and casp1−/− (n=3 per genotype)] after LPS injection. We chose to use the specific LPS serotype, dose and administration route because this regimen was previously shown to cause 100% lethality in wt mice during the first 36h post-injection [11]. Injections were given between 7:00 and 9:00 AM; mice were sacrificed by decapitation one at a time in a room distant from the animal holding area in a manner to avoid stress triggered by blood scent. Organs were extracted, snap-frozen, and stored at -80°C until processed.
RNA isolation and reverse transcription
We chose one brain area (hippocampus) and two peripheral tissues (spleen and adrenals), respectively, to determine variations in central and peripheral mRNA expression levels. Total RNA was extracted from individual tissues using RNeasy Lipid Mini Kit including DNase digestion (Qiagen, Germantown, Maryland, USA) and quantified using a Nanodrop ND-1000 spectrophotometer (Thermo-Fisher Scientific, Waltham, Massachusetts, USA); 1 μg of RNA was reverse transcribed to cDNA with the Omniscript RT Kit (Qiagen) using random hexamer primers (Invitrogen, Carlsbad, CA, USA).
PCR Primers
The genes of interest were selected from our previous microarray study [12]. To avoid amplification of genomic DNA traces, forward and reverse primers were designed in different exons with Primer Express Software (Applied Biosystems, Foster City, CA, USA). We used negative and positive controls (respectively, reverse transcription with genomic DNA and cDNA from mouse brain treated with LPS). Table 1 lists our primer pair sequences, which amplified cDNA only.
Table 1.
List of real-time PCR primers used to determine the gene expression levels of all the eight selected genes.
| Gene | Forward | Reverse | NCBI Ref. Seq. |
|---|---|---|---|
| β-actin | 5-CCTAGGCACCAGGGTGTGAT-3 | 5-CATGTCGTCCCAGTTGGTAACA-3 | NM_007393 |
| Ptgs2 | 5-TGGGCCATGGAGTGGACTT-3 | 5-GGATGTGAGGAGGGTAGATCATCT-3 | NM_011198.3 |
| Nos2 | 5-AAGGCCACATCGGATTTCAC-3 | 5-TGTTCCTCTATTTTTGCCTCTTTAAAG-3 | NM_010927.3 |
| Cxcl1 | 5-ACCGAAGTCATAGCCACACTCA-3 | 5-CACCTTTTAGCATCTTTTGGACAA-3 | NM_008176.3 |
| Cxcl10 | 5-CTGGGTCTGAGTGGGACTCAA-3 | 5-TCATTGCCACGATGAAAAAGAAT-3 | NM_021274.1 |
| Tgtp | 5-CACCACTGCTGAGCTTC TACCATA-3 | 5-TCGATGTCTCTCAGTACCTTTTCAA-3 | NM_011579.3 |
| Gbp2 | 5-TTGAGAAGGGTGACAACCAGAA-3 | 5-TGGTTCCTATGCTGTTGTAGATGAA-3 | NM_010260.1 |
| Il-1ra | 5-CCAGCTGGAGGAAGTTAACATCA-3 | 5-ACGGTCAGCCTCTAGTGTTGTG-3 | NM_031167.5 |
| Adamts1 | 5-GACAATAACGGAAAAACGTTCAGA-3 | 5-GTGAGCTTGCACCTGTCCTTT-3 | NM_009621.4 |
Quantitative Real-Time PCR
We performed SYBR green-based real-time PCR to determine variations in mRNA expression profiles. Standard curves of pooled, serially diluted cDNAs were constructed for each primer pair. Experiments were performed using the 7900HT Fast Real-Time PCR System (Applied Biosystems). cDNA samples were run in triplicates; SYBR green cycle threshold (Ct) values were averaged for each sample, and the RNA input for each target gene was calculated using standard curves. Fold-changes were expressed as a ratio to β-actin expression for each sample. Each reaction included cDNA from 20 ng of RNA, 4.5 pmol of each primer, and 7.5 μL of SYBR green PCR Mastermix (Applied Biosystems) in a final volume of 15 μL. PCR parameters were: 50°C for 2 min, 95°C for 10 min, 40 cycles of 95°C for 15 sec, and 60°C for 1 min.
Statistical Analysis
Variance Analyses
Two-way ANOVA tests were performed to assess the effects of genotype (wt or casp1−/−) and time (0, 6, or 12 hours), individually and in combination, on gene expression levels in adrenal, hippocampus and spleen tissues. Tukey's honest significant difference (HSD) test was used within each gene/tissue combination to perform pair-wise comparisons maintaining the family-wise probability of coverage at 95%. We present 95% confidence intervals (CI) for the differences and family-wise-corrected P-values to assess statistical significance. All analyses were performed in R package version 3.0.2 (R Foundation for Statistical Computing, 2013).
Correlation analysis
We calculated pair-wise Pearson correlation coefficients to examine the relationships between gene expression in wt and casp1−/− mice. As a complementary strategy, nonparametric local regressions (LOESS) were fitted to the data to assess the relationship between pair-wise gene expression levels. Statistical comparison of the linear correlation coefficient was performed based on a normal distribution, and resulting P-values were corrected using the False Discovery Rate (FDR) [21].
Hierarchical clustering
We used hierarchical clustering to analyze gene co-expression in our samples. Hierarchical clustering uses an agglomerative algorithm that joins the most similar component features, and then joins the next most similar using the first aggregation as a single combined unit. In hierarchical clustering, the clusters were generated via complete linkage. To assess the uncertainty of the analysis, approximately unbiased (AU) and bootstrap probability (BP) P-values were calculated by pvclust [22], R package version 3.0.2. AU P-values were computed by multiscale bootstrap re-sampling, and are a better approximation to unbiased P-values than BP-based P-values (computed by normal bootstrap re-sampling).
RESULTS
LPS-induced differential temporal gene expression in peripheral and central tissues of wt and casp1−/− mice
Our study was conducted up to 12h after LPS injection. The health status of mice of both genotypes gradually worsened with the passage of time. Towards the last collection point, 12h post-LPS injection, wt mice were quite immobile but without signs of being moribund (e.g. hard-labored breathing), whereas casp1−/− mice were less active but still responsive to external stimuli such as a gentle tapping and/or shaking the cage. We corroborated the resilience of casp1−/− mice to the lethal effects of LPS in a separate experiment. We confirmed that the same LPS serotype and dose used in the present studies caused 100% lethality in wt mice (n=4) within 12–24h post-injection, whereas all casp1−/− mice (n=5) were alive at 24h, which confirmed previous results showing that 100% of casp1−/− survived for at least 30h after receiving the same i.p. dose and serotype of LPS injection [11] (Table 2). Figure 1 illustrates gene expression level changes in the hippocampus, spleen and adrenals.
Table 2.
Survival rate of wt and casp1−/− mice after receiving a LD100 of LPS. 100% of wt mice (n=4) died at 24h whereas 100% of casp1−/− (n=5) were still alive.
| n | 12h-Post LPS injection | 24h-Post LPS Injection | |
|---|---|---|---|
| Wt | 4 | 100% | 0% |
| Casp1−/− | S | 100% | 100% |
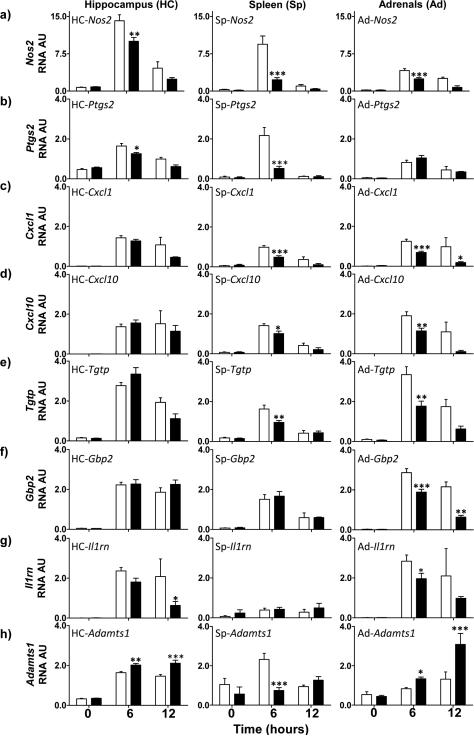
Figure 1. mRNA expression pattern of the eight selected genes in the hippocampus, spleen and adrenals of wt and casp1−/−mice.
In this figure, the time-dependent pattern of expression of each gene (rows a-h) is shown for hippocampal, (left panels), splenic (center panels) and adrenal tissues (right panels). The mean expression levels at 0, 6 and 12h post-LPS injection are represented by white and black columns, respectively for wt and casp1−/− mice; the line above represents one standard error of the mean (SEM). Statistically significant differences between wt and casp1−/− at the same time-point are shown: * = P<0.05, ** = P<0.01 and *** = P<0.001.
In wt and casp1−/− mice, most of the studied genes displayed the highest expression levels (23 out of 24 measurements) at 6h post injection (Chi square, P<0.0001), except for adrenal Adamts1 which peaked at 12h. In casp1−/− mice, the highest mRNA levels (19 out of 24 measurements) were also seen at 6h post-injection (Chi square, P<0.01), except for splenic Il1rn, Ptgs2, Nos2, and Adamts1, and adrenal Adamts1, which peaked at 12h (Figure 1). We found that LPS induced significant increases (above baseline levels observed at time 0h) in the expression of most of the eight studied genes in the three tissues of both genotypes, except for the splenicexpression of Il1rn, Nos2, Ptgs2, and Adamts1 genes in casp1−/− mice, and splenic Il1rn gene in wt mice (Figure 1 and Table 3); therefore, four out of the eight genes were not activated in the spleen of casp1-deficient mice. Thus, it appears that the temporal expression of the studied genes mostly followed an early and sharp increase pattern similar to that described for pro-inflammatory elements during the initial stage of the inflammatory response. A summary of the two-way ANOVA significance probabilities for the differential expression of each gene within each organ is given in Table 3.
Table 3.
Two-way ANOVA summary. The significance of the major three components (genotype, time and interaction) is shown for the eight genes in each one of the tissues studied. P-values are familywise corrected.
| Organ | Gene | Component | WT | caspl-/- | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Time | Interaction | 6 vs. 0h | 12 vs. 0h | 12 vs. 6h | 6 vs. 0h | 12 vs. 0h | 12 vs. 6h | ||
| HC | Nos2 | *** | *** | ns | *** | ns | *** | *** | ns | ** |
| Ptgs2 | ** | *** | * | *** | ns | * | *** | ns | * | |
| Cxcl1 | ** | *** | ns | *** | *** | ns | *** | ns | ** | |
| Cxcl10 | ns | *** | ns | *** | *** | ns | *** | * | ns | |
| Tgtp | ns | *** | ns | *** | ** | ns | *** | ns | *** | |
| Gbp2 | ns | *** | ns | *** | *** | ns | *** | *** | ns | |
| Il1rn | *** | *** | * | *** | *** | ns | *** | ns | * | |
| Adamts1 | *** | *** | *** | *** | *** | ns | *** | *** | ns | |
| Spleen | Nos2 | *** | *** | ** | *** | ns | ** | ns | ns | ns |
| Ptgs2 | *** | *** | ** | *** | ns | ** | ns | ns | ns | |
| Cxcl1 | *** | *** | ** | *** | ns | ** | ** | ns | ns | |
| Cxcl10 | ** | *** | ns | *** | ns | *** | *** | ns | ** | |
| Tgtp | ** | *** | * | *** | ns | *** | ** | ns | ns | |
| Gbp2 | ns | *** | ns | *** | ns | ns | *** | ns | ns | |
| Il1rn | ns | ns | ns | ns | ns | ns | ns | ns | ns | |
| Adamts1 | *** | * | * | * | ns | ns | ns | ns | ns | |
| Adrenals | Nos2 | *** | *** | * | *** | ** | ns | *** | ns | * |
| Ptgs2 | ns | *** | * | ** | ** | ns | *** | ns | ns | |
| Cxcl1 | *** | *** | ** | *** | *** | ns | *** | ns | ns | |
| Cxcll0 | *** | *** | * | *** | * | ns | *** | ns | * | |
| Tgtp | *** | *** | * | *** | ns | ns | * | ns | ||
| Gbp2 | *** | *** | ** | *** | *** | ns | *** | ns | ** | |
| Il1rn | *** | *** | ns | *** | * | ns | *** | ns | ns | |
| Adamts1 | *** | *** | *** | ns | * | ns | *** | *** | *** | |
The comparison among time-points within genotypes is also shown for wt and casp1-/- mice. Statistically significant differences between time-points are shown:
P<0.05
P<0.01
P<0.001, ns = not significant. Bold “ns” show genes that did not show increased expression when compared with time 0h. For example, splenic expression of Nos2, Ptgs2 and Adamts1 remained significantly unchanged in casp1-/- mice, whereas the splenic expression of Il1rn remained significantly unchanged in both genotypes.
Below is a brief summary of gene expression changes for each gene:
Nos2
Nos2 gene expression was significantly lower at 6h in all three casp1−/− mice tissues when compared to wt ones; it was 30% lower in the hippocampus (P<0.01), 75% lower in the spleen (P<0.001), and 40% lower in the adrenals (P<0.001) (Figure 1a). The LPS-induced expression of Nos2 followed a similar pattern in both genotypes, but of decreased magnitude in casp1−/− mice. Both genotypes showed activation of gene expression at 6h, and a decrease towards baseline at 12h (except for Nos2 expression in wt adrenal, which remained elevated at 12h; P<0.01 vs. time 0h).
Ptgs2
The peak Ptgs2 mRNA level at 6h was significantly lower by 25% and 75% in casp1−/− mice in the hippocampus (P<0.05) and the spleen (P<0.001), respectively, whereas no significant changes were observed within the adrenals of both genotypes (Figure 1b). Ptgs2 transcriptional patterns were similar to those of Nos2, i.e. Ptgs2 gene expression was increased at 6h and returned to baseline values at 12h.
Cxcl1, Cxcl10 and Tgtp
In the two peripheral organs, casp1−/− mice showed significantly lower and variable magnitude of increases in these three genes than wt mice; casp1−/− mice displayed 30% to 80% lower mRNA levels (P <0.05 to <0.001) (Figures 1c-e). These genes showed similar transcriptional patterns in the three tissues/organs studied from both genotypes. At the CNS level, their hippocampal expression showed no significant differences between the two genotypes.
Gbp2
Adrenal casp1−/− mice mRNA expression of Gbp2 was significantly lower than wt mice by 35% at 6h (P<0.001) (Figure 1f); thereafter, Gbp2 expression decreased in both genotypes at 12h to similar levels. Hippocampal and splenic Gbp2 expression levels showed no genotype differences.
Il1rn
In the hippocampus of both genotypes, LPS elicited similarly large Il1rn expression increases at 6h. At 12h, Il1rn mRNA remained elevated in wt mice and declined by 70% in casp1−/− mice (P≤0.05) (Figure 1g). Adrenal Il1rn activation for both genotypes was similar to that of the hippocampus, reaching significantly larger increases of 40% at 6h (P<0.05) and of 115% at 12h (P<0.001) in wt mice. Splenic Il1rn mRNA expression was unchanged in both genotypes.
Adamts1
Surprisingly, hippocampal Adamts1 mRNA expression was of higher magnitude in casp1−/− mice, increased 23% at 6h (P≤0.01) and 45% at 12h (P<0.001) (Figure 1h), and followed a similar temporal pattern in wt and casp1−/− mice. Adrenal Adamts1 expression was also larger in casp1−/−mice: they were 50% higher at 6h (P<0.05), and 2.3-fold higher at 12h (P<0.001). Splenic Adamts1 mRNA expression increased significantly at 6h and returned to baseline levels at 12h in wt mice, whereas no changes above baseline were observed in casp1−/− mice. Genotype comparison showed that splenic Adamts1 mRNA levels were 70% lower in casp1−/− mice at 6h (P<0.001).
Correlation analysis
Hippocampus
All correlations between hippocampal mRNA expression levels of the genes studied were significant within each genotype; however, correlation coefficients comparing genotypes (wt vs. casp1−/− mice) showed significant differences only between Tgtp/Gbp2 (PFDR<0.05) and Tgtp/Adamts1 (PFDR<0.01) (Figure 1S).
Spleen
In wt mice, correlation analyses of splenic mRNA levels were significant, except for all Il1rn correlations (Figure 2S). In casp1−/− mice, most splenic mRNA level correlations were significant, and most Il1rn and Adamts1 correlations were not significant except for the correlation between Il1rn/Adamts1 which was significant (Figure 2S). Splenic correlation coefficients comparing genotype showed that only three coefficient pairs, all related to Adamts1, were significantly different: Ptgs2/Adamts1 (PFDR<0.001), Nos2/Adamts1 (PFDR=0.006) and Gbp2/Adamts1 (PFDR=0.018) (Figure 2S).
Adrenals
Correlation analysis of adrenal mRNA levels showed that all correlations were significant in wt mice, except for Nos2/Adamts1, whereas six pairs of correlations related to Adamts1 were not significant in casp1−/− mice: Ptgs2/Adamts1, Nos2/Adamts1, Cxcl1/Adamts1, Cxcl10/Adamts1, Tgtp/Adamts1 and Gbp2/Adamts1. Adrenal correlation coefficients comparing genotypes did not show any significant differences (Figure 3S).
Overall, there was a loss of correlation for the Adamts1 gene in peripheral tissues in casp1-deficient mice.
Hierarchical clustering
This analysis was done to ascertain patterns of gene co-expression using clustering strategies by comparing: i) The same tissue between wt and casp1−/− mice, and ii) The three tissues within the same genotype (Figure 2).
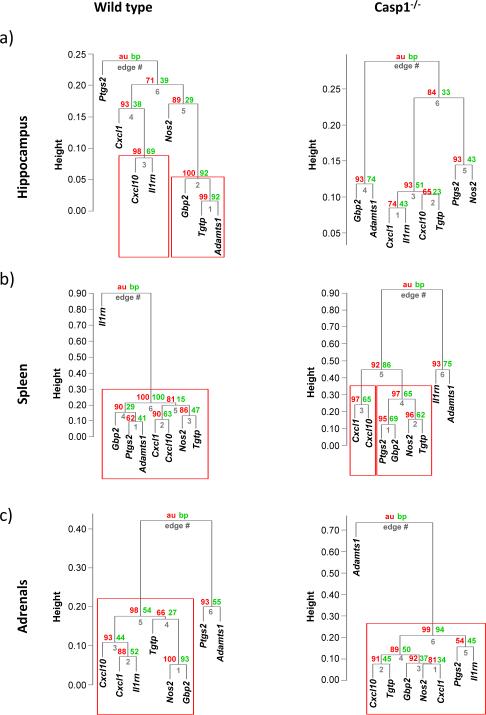
Figure 2.
Hierarchical cluster analysis of gene expression levels with AU- and BP-based P-values (in red and green, respectively). The vertical distance between the gene expression levels indicate how dissimilar they are. Results for (a) hippocampus, (b) spleen and (c) adrenal tissues are presented. The genes shown inside the red boxes clustered together according to an approximately unbiased (AU) probability set at ≥95% level, after bootstrap re-sampling.
Clustering comparison between wt and casp1−/− mice
In the hippocampus (Figure 2a), two clusters were supported at the 95% CI level in wt animals. The first cluster was comprised of the Cxcl10 and Il1rn genes, whereas the second was defined by the Gbp2, Tgtp and Adamts1 genes (Figure 2a). Interestingly, no clusters were supported at the 95% level in casp1−/− mice (Figure 2a). There were qualitative similarities between splenic data of both genotypes (Figure 2b). In the spleen of wt mice, all genes except the Il1rn gene formed one cluster (Figure 2b), whereas in casp1−/− mice, two clusters were supported at the 95% level (the Cxcl1 and Cxcl10 cluster and the Ptgs2, Gbp2, Nos2 and Tgtp cluster) (Figure 2b, right panel). High similarities were found between adrenal data from wt and casp1−/− mice: they had one cluster consisting of six and seven genes, respectively, at a 95% significance level. The Ptgs2 (in wt mice) and the Adamts1 (in wt and casp1−/−mice) were not included in the clusters (Figure 2c, right panel).
Clustering comparison among tissues
More similarities were found between the two peripheral organs (spleen and adrenals) than between those tissues and the hippocampus. In wt mice, five genes were co-expressed both within the spleen and adrenals (Gbp2, Tgtp, Cxcl1, Cxcl10, andNos2). In casp1−/− mice, six out of the seven genes that clustered together within the adrenal gland were divided into two clusters (one with two genes, Cxcl1 and Cxcl10, and another with four genes Ptgs2, Gbp2, Nos2 and Tgtp) in the spleen. Within the hippocampus of wt mice, there were two clusters. One of them showed a co-expression of two genes (Cxcl10 and Il1rn) that were also shown to be co-expressed in the adrenals of wt mice (Figure 2a and 2c). The other cluster within the brain of wt mice displayed three genes that were also shown to be co-expressed within the spleen (Figure 2a and 2b). Overall, at the 95% level, Adamts1 did not co-express with any gene in casp1−/− mice tissues, while in wt it co-expressed in 2 of the 3 tissues.
DISCUSSION
We used LPS-induced SIRS in casp1-deficient mice to detail patterns of gene expression that may contribute to their resilience to LPS-induced lethality, which could lead to the development of novel therapeutic targets for SIRS. In this study, we described central and peripheral changes in the expression of a set of eight genes in the context of casp1-deficiency and a lethal dose of LPS, and showed that endotoxin elicited an inflammatory cascade of much lower magnitude in casp1−/− mice. Overall, these changes occurred in a time-dependent manner, with peak mRNA expression of most genes occurring 6 hours post LPS injection, and declining to baseline levels by 12 hours. In previous studies, we have shown evidence suggesting that activated transcripts during inflammation show 2 types of temporal patterns: transient and sustained [23]. With exception of Il1rn, it appears that within the spleen most of the genes followed a transient pattern of increase by augmenting at 6h and returning to baseline at 12h, whereas most of the genes seemed to follow a more sustained pattern of activation in the hippocampus and adrenals, which lasted for at least 12h.
The most contrasting changes in gene expression between both genotypes occurred in the spleen. After LPS injection, the splenic expression of three genes, namely Nos2, Ptgs2, and Adamts1, was largely increased in wt mice, whereas it was not significantly altered in casp1−/− mice. There are a couple of exceptions to the generalized idea that the inflammatory cascade elicited by LPS was of a lower magnitude in casp1−/−. For instance, we found that the expression of Adamts1 remained significantly increased in the hippocampus and adrenals of casp1-deficient mice.
Our previous work suggested that the expression of these eight genes was controlled by casp1 [12]. In order to study gene-gene interactions, we evaluated their relationship by performing correlation and hierarchical clustering analyses. There was an overall loss of correlation for the Adamts1 gene in peripheral tissues of casp1−/− mice. Differences observed in the hierarchical clustering analyses seem to support that casp1 deficiency altered the co-expression of these genes in a more significant manner within the brain than in the peripheral organs. Indeed, these analyses revealed similar clusters within the adrenals and the spleen of wt and casp1−/− mice, whereas no similarities were evident in the hippocampus of both genotypes. In addition, co-expression analyses showed more similarities between peripheral organs (spleen and adrenals) than between these peripheral organs and the hippocampus; there was also a tendency of co-expression loss for Adamts1 in casp1−/− mice.
Products of three out of the eight genes reported here, specifically the Nos2, Ptgs2 and Il1rn genes, have been pharmacologically targeted in preclinical and clinical trials of SIRS/sepsis. The inhibition of nitric oxide (NO) synthesis, a free radical largely synthesized by NOS2 during SIRS/sepsis, improved hemodynamic parameters in immunologically-challenged rodents and septic patients [24-30]. However, multicenter randomized placebo-controlled studies showed that the mortality rate of septic shock patients treated with NOS inhibitors was not improved [29, 30]. Similarly, preclinical studies largely support the concept that pharmacological inhibition of cyclooxygenase and/or genetic ablation of PTGS2 resulted in beneficial effects on mortality [31]. Unfortunately, the beneficial effects of cyclooxygenase inhibition observed in preclinical studies have not been convincingly and consistently translated into the clinical setting so far [32-34]. Only a sub-study from one clinical trial showed a significant reduction in the 30-day mortality rate of sepsis [35]. Studies involving IL-1ra also yielded promising outcomes in preclinical trials [36]. Likewise casp1−/−, mice overexpressing IL-1ra have also shown resilience to LPS-induced lethality [36]. In both casp1−/− and IL-1ra overexpressing mice, the resilience to LPS-induced lethality was observed for at least 6 and 7 days, respectively, indicating that in these two strains of mice the improvement in survival was long-lasting [11, 36]. However, these results could not be effectively translated into the clinical setting as rhIL1ra failed to reduce sepsis mortality [37, 38].
The contribution of the products of the other five genes that were also differentially expressed between wt and casp1−/− mice has not been clearly ascertained in preclinical/clinical studies of SIRS/sepsis. Two of them are chemokines Cxcl1 and Cxcl10. Chemokines and their receptors are expressed in immune and endothelial cells, and play a fundamental role in recruiting immune cells into the inflammatory site [39, 40]. Therefore, the reduced expression of Cxcl1 and Cxcl10 observed in casp1−/− mice could have contributed to a better outcome against LPS-induced lethality, due to decreased immune cell recruitment.
We also investigated the expression of the GTPases Tgtp and Gbp2, another two genes that were differentially expressed between casp1−/− and wt mice. These two interferon-induced GTPases are expressed in immune cells, but their roles in SIRS/sepsis have not been clearly ascertained [41-43]. Overall, guanylate binding proteins have been characterized as downstream effectors of the interferon signaling pathway [44]. They play essential roles in fighting intracellular pathogens [44]. A recent study, showed that intracellular LPS from Gram-negative bacteria activated guanylate binding proteins, which in turn participated in a programmed cell death pathway termed pyroptosis [45]. Since we employed a large dose of LPS, it is likely that increased cytosolic accumulation of endotoxin could activate guanylate binding protein-induced pyroptosis. Thus, their decreased expression in casp1−/− mice could result in decreased inflammation, and consequently in improved survival after being exposed to a lethal dose of endotoxin. However, further studies need to be performed to elucidate the roles of GBP2 and TGTP during SIRS.
Finally, ADAMTS1, an enzyme that belongs to the family of the metalloproteases, was also differentially expressed between the two genotypes. ADAMTS1 is necessary for normal growth, female fertility, and kidney, adrenal and female reproductive organ structure and function [46]. Recently, it was shown that LPS dramatically increased the synthesis of ADAMTS1 within the splenic lymph and interstitial fluid to a concentration that was much higher than that of plasma, suggesting that the spleen is a major source of ADAMTS1, and that this metalloprotease is involved in the endotoxin-induced disease process [47]. Within the brain, metalloproteases play a key role in controlling blood brain barrier permeability during LPS-induced inflammation, and ADAMTS1 may have a role in regulating neural plasticity, as null female mice have a selective decline of synaptic protein levels in the frontal cortex [48].
There are apparent discrepancies between our current and previous studies. In our previous study, the transcriptional differences between both genotypes appeared to occur specifically within the brain, and not in the periphery, since both wt and casp1−/− showed similar LPS-induced increases in their expression level within the spleen [12]. In this study, we found that only the hippocampal transcriptional expression of two out of the eight genes, Nos2 and Ptgs2, showed similar differences to those previously reported in the whole brain of both genotypes [12]. These differences could be due to the fact that we previously employed a lower dose and less inflammatory serotype of LPS (e.g. Escherichia Coli 055:B5, 25 mg/kg) than the one employed at present [12]. The previous dose and serotype of LPS was shown to cause a slower progression towards death in wt mice (100% mortality after 5 days of injection) [49], than the LPS dose and serotype reported here (100% mortality within approximately 1.25 days) [11]. Furthermore, in our previous studies we employed whole brains [12], whereas here we used hippocampi. Moreover, we studied a small group of mice at 12h post-LPS injection, and this could have lowered the statistical power to detect differences at that particular time-point.
It was recently shown that casp1-targeted mice carry a 129 ES cell-derived mutation in the casp11 gene, which appeared to be involved, at least in part, in their resilience to LPS-induced lethality [50]. The synergistic key role of two of the downstream effectors controlled by casp1, namely IL-18 and IL-1ß, during LPS-induced lethality was recently shown [51]. The survival rate of casp1/11−/− was very similar to that of IL-1ß/IL-18-deficient mice after receiving a LD100 of LPS of the same serotype as the one used in the present studies [51]. The latter results clearly highlight the relevance of the casp1-orchestrated pathway during LPS-induced SIRS. However, when casp1/11−/− and IL-1ß/IL-18-deficient mice were subjected to other immunological challenges such as the cecal ligation and puncture (CLP) procedure, or the administration of a lethal dose of tumor necrosis alpha (TNF-α), casp1/11−/− displayed a lower survival rate than IL-1ß/IL-18-deficient mice, indicating that targeting downstream effectors could be more efficacious than inhibiting the upstream activators CASP1 or CASP11[51].
In summary, we described tissue and time-dependent expression profiles of a set of genes regulated by casp-1, after LPS-induced SIRS. Our data suggest that three elements may contribute to casp1−/− mice resilience to SIRS-induced lethality: 1) a decreased inflammatory cascade due to a lower expression of casp1-orchestrated genes and a faster resolution of the pro-inflammatory process, 2) increased Adamts1 activation in the hippocampus, which could potentially increase neural plasticity, and 3) loss of gene-gene interaction in the hippocampus. Future studies need to confirm whether changes in CNS gene-gene interactions or ADAMTS1 biological activity are associated with the increased survival of casp1−/− mice after a lethal dose of LPS. Additionally, we will examine whether inhibitors of casp1 activity promote similar changes in genetic expression. The elucidation of these transcription pattern changes could provide a foundation for the development of molecular biomarkers and therapeutic targets against SIRS/sepsis.
Supplementary Material
Acknowledgments
This research was supported in part by a National Institutes of Health Grant K24RR017365 to MLW, and by institutional funds from the University of Miami and The Australian National University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Takala A, Nupponen I, Kylanpaa-Back ML, Repo H. Markers of inflammation in sepsis. Ann Med. 2002;34:614–623. doi: 10.1080/078538902321117841. [DOI] [PubMed] [Google Scholar]
- 2.Hotchkiss RSIE, Karl The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348:138–150. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 3.Wong ML, Xie B, Beatini N, Phu P, Marathe S, Johns A, Gold PW, Hirsch E, Williams KJ, Licinio J, Tabas I. Acute systemic inflammation up-regulates secretory sphingomyelinase in vivo: a possible link between inflammatory cytokines and atherogenesis. Proc Natl Acad Sci U S A. 2000;97:8681–8686. doi: 10.1073/pnas.150098097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mastronardi CA, Srivastava V, Yu WH, Les Dees W, McCann SM. Lipopolysaccharide-induced leptin synthesis and release are differentially controlled by alpha-melanocyte-stimulating hormone. Neuroimmunomodulation. 2005;12:182–188. doi: 10.1159/000084851. [DOI] [PubMed] [Google Scholar]
- 5.Mastronardi CA, Yu WH, McCann S. Lipopolysaccharide-induced tumor necrosis factor-alpha release is controlled by the central nervous system. Neuroimmunomodulation. 2001;9:148–156. doi: 10.1159/000049019. [DOI] [PubMed] [Google Scholar]
- 6.Mastronardi CA, Yu WH, Rettori V, McCann S. Lipopolysaccharide-induced leptin release is not mediated by nitric oxide, but is blocked by dexamethasone. Neuroimmunomodulation. 2000;8:91–97. doi: 10.1159/000026458. [DOI] [PubMed] [Google Scholar]
- 7.Wong ML, Bongiorno PB, Rettori V, McCann SM, Licinio J. Interleukin (IL) 1beta, IL-1 receptor antagonist, IL-10, and IL-13 gene expression in the central nervous system and anterior pituitary during systemic inflammation: pathophysiological implications. Proc Natl Acad Sci U S A. 1997;94:227–232. doi: 10.1073/pnas.94.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong M-L, Rettori V, Al-Shekhlee A, Bongiorno PB, Canteros G, McCann SM, Gold PW, Licinio J. Inducible nitric oxide synthase gene expression in the brain during systemic inflammation. Nature Med. 1996;2:581–584. doi: 10.1038/nm0596-581. [DOI] [PubMed] [Google Scholar]
- 9.Mastronardi CA, Yu WH, Srivastava VK, Dees WL, McCann SM. Lipopolysaccharide-induced leptin release is neurally controlled. Proc Natl Acad Sci U S A. 2001;98:14720–14725. doi: 10.1073/pnas.251543598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong ML, O'Kirwan F, Khan N, Hannestad J, Wu KH, Elashoff D, Lawson G, Gold PW, McCann SM, Licinio J. Identification, characterization, and gene expression profiling of endotoxin-induced myocarditis. Proc Natl Acad Sci U S A. 2003;100:14241–14246. doi: 10.1073/pnas.2336220100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li P, Allen H, Banerjee S, Franklin S, Herzog L, Johnston C, McDowell J, Paskind M, Rodman L, Salfeld J, et al. Mice deficient in IL-1 beta-converting enzyme are defective in production of mature IL-1 beta and resistant to endotoxic shock. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 12.Mastronardi C, Whelan F, Yildiz OA, Hannestad J, Elashoff D, McCann SM, Licinio J, Wong ML. Caspase 1 deficiency reduces inflammation-induced brain transcription. Proc Natl Acad Sci U S A. 2007;104:7205–7210. doi: 10.1073/pnas.0701366104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chesnokova V, Melmed S. Minireview: Neuro-immuno-endocrine modulation of the hypothalamic-pituitary-adrenal (HPA) axis by gp130 signaling molecules. Endocrinology. 2002;143:1571–1574. doi: 10.1210/endo.143.5.8861. [DOI] [PubMed] [Google Scholar]
- 14.Carnio EC, Moreto V, Giusti-Paiva A, Antunes-Rodrigues J. Neuro-immune-endocrine mechanisms during septic shock: role for nitric oxide in vasopressin and oxytocin release. Endocrine, metabolic & immune disorders drug targets. 2006;6:137–142. doi: 10.2174/187153006777442396. [DOI] [PubMed] [Google Scholar]
- 15.Chrousos GP. The hypothalamic-pituitary-adrenal axis and immune-mediated inflammation. The New England journal of medicine. 1995;332:1351–1362. doi: 10.1056/NEJM199505183322008. [DOI] [PubMed] [Google Scholar]
- 16.Tracey KJ. Physiology and immunology of the cholinergic antiinflammatory pathway. J Clin Invest. 2007;117:289–296. doi: 10.1172/JCI30555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Kloet ER, Vreugdenhil E, Oitzl MS, Joels M. Brain corticosteroid receptor balance in health and disease. Endocr Rev. 1998;19:269–301. doi: 10.1210/edrv.19.3.0331. [DOI] [PubMed] [Google Scholar]
- 18.Schobitz B, Sutanto W, Carey MP, Holsboer F, de Kloet ER. Endotoxin and interleukin 1 decrease the affinity of hippocampal mineralocorticoid (type I) receptor in parallel to activation of the hypothalamic-pituitary-adrenal axis. Neuroendocrinology. 1994;60:124–133. doi: 10.1159/000126742. [DOI] [PubMed] [Google Scholar]
- 19.Bener D, Wohlman A, Itzik A, Yirmiya R, Ben-Hur T, Weidenfeld J. Glucocorticoid resistance following herpes simplex-1 infection: role of hippocampal glucocorticoid receptors. Neuroendocrinology. 2007;85:207–215. doi: 10.1159/000102976. [DOI] [PubMed] [Google Scholar]
- 20.Yao JH, Ye SM, Burgess W, Zachary JF, Kelley KW, Johnson RW. Mice deficient in interleukin-1beta converting enzyme resist anorexia induced by central lipopolysaccharide. Am J Physiol. 1999;277:R1435–1443. doi: 10.1152/ajpregu.1999.277.5.R1435. [DOI] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 22.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 23.Mastronardi CA, Licinio J, Wong ML. Candidate biomarkers for systemic inflammatory response syndrome and inflammation: a pathway for novel translational therapeutics. Neuroimmunomodulation. 2010;17:359–368. doi: 10.1159/000292040. [DOI] [PubMed] [Google Scholar]
- 24.Thiemermann C, Vane J. Inhibition of nitric oxide synthesis reduces the hypotension induced by bacterial lipopolysaccharides in the rat in vivo. Eur J Pharmacol. 1990;182:591–595. doi: 10.1016/0014-2999(90)90062-b. [DOI] [PubMed] [Google Scholar]
- 25.Kilbourn RG, Gross SS, Jubran A, Adams J, Griffith OW, Levi R, Lodato RF. NG-methyl-L-arginine inhibits tumor necrosis factor-induced hypotension: implications for the involvement of nitric oxide. Proc Natl Acad Sci U S A. 1990;87:3629–3632. doi: 10.1073/pnas.87.9.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petros A, BennettP D. Vallance: Effect of nitric oxide synthase inhibitors on hypotension in patients with septic shock. Lancet. 1991;338:1557–1558. doi: 10.1016/0140-6736(91)92376-d. [DOI] [PubMed] [Google Scholar]
- 27.Petros A, Lamb G, Leone A, Moncada S, Bennett D, Vallance P. Effects of a nitric oxide synthase inhibitor in humans with septic shock. Cardiovasc Res. 1994;28:34–39. doi: 10.1093/cvr/28.1.34. [DOI] [PubMed] [Google Scholar]
- 28.Avontuur JA, Tutein Nolthenius RP, van Bodegom JW, Bruining HA. Prolonged inhibition of nitric oxide synthesis in severe septic shock: a clinical study. Crit Care Med. 1998;26:660–667. doi: 10.1097/00003246-199804000-00012. [DOI] [PubMed] [Google Scholar]
- 29.Lopez A, Lorente JA, Steingrub J, Bakker J, McLuckie A, Willatts S, Brockway M, Anzueto A, Holzapfel L, Breen D, Silverman MS, Takala J, Donaldson J, Arneson C, Grove G, Grossman S, Grover R. Multiple-center, randomized, placebo-controlled, double-blind study of the nitric oxide synthase inhibitor 546C88: effect on survival in patients with septic shock. Crit Care Med. 2004;32:21–30. doi: 10.1097/01.CCM.0000105581.01815.C6. [DOI] [PubMed] [Google Scholar]
- 30.Kinasewitz GT, Privalle CT, Imm A, Steingrub JS, Malcynski JT, Balk RA, DeAngelo J. Multicenter, randomized, placebo-controlled study of the nitric oxide scavenger pyridoxalated hemoglobin polyoxyethylene in distributive shock. Crit Care Med. 2008;36:1999–2007. doi: 10.1097/CCM.0b013e31817bfe84. [DOI] [PubMed] [Google Scholar]
- 31.Aronoff DM. Cyclooxygenase inhibition in sepsis: is there life after death? Mediators of inflammation. 2012;2012:696897. doi: 10.1155/2012/696897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernard GR, Wheeler AP, Russell JA, Schein R, Summer WR, Steinberg KP, Fulkerson WJ, Wright PE, Christman BW, Dupont WD, Higgins SB, Swindell BB. The effects of ibuprofen on the physiology and survival of patients with sepsis. The Ibuprofen in Sepsis Study Group. The New England journal of medicine. 1997;336:912–918. doi: 10.1056/NEJM199703273361303. [DOI] [PubMed] [Google Scholar]
- 33.Haupt MT, Jastremski MS, Clemmer TP, Metz CA, Goris GB. Effect of ibuprofen in patients with severe sepsis: a randomized, double-blind, multicenter study. The Ibuprofen Study Group. Crit Care Med. 1991;19:1339–1347. doi: 10.1097/00003246-199111000-00006. [DOI] [PubMed] [Google Scholar]
- 34.Memis D, Karamanlioglu B, Turan A, Koyuncu O, Pamukcu Z. Effects of lornoxicam on the physiology of severe sepsis. Critical care. 2004;8:R474–482. doi: 10.1186/cc2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arons MM, Wheeler AP, Bernard GR, Christman BW, Russell JA, Schein R, Summer WR, Steinberg KP, Fulkerson W, Wright P, Dupont WD, Swindell BB. Effects of ibuprofen on the physiology and survival of hypothermic sepsis. Ibuprofen in Sepsis Study Group. Crit Care Med. 1999;27:699–707. doi: 10.1097/00003246-199904000-00020. [DOI] [PubMed] [Google Scholar]
- 36.Hirsch E, Irikura VM, Paul SM, Hirsh D. Functions of interleukin 1 receptor antagonist in gene knockout and overproducing mice. Proc Natl Acad Sci U S A. 1996;93:11008–11013. doi: 10.1073/pnas.93.20.11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Opal SM, Fisher CJ, Jr., Dhainaut JF, Vincent JL, Brase R, Lowry SF, Sadoff JC, Slotman GJ, Levy H, Balk RA, Shelly MP, Pribble JP, LaBrecque JF, Lookabaugh J, Donovan H, Dubin H, Baughman R, Norman J, DeMaria E, Matzel K, Abraham E, Seneff M. Confirmatory interleukin-1 receptor antagonist trial in severe sepsis: a phase III, randomized, double-blind, placebo-controlled, multicenter trial. The Interleukin-1 Receptor Antagonist Sepsis Investigator Group. Crit Care Med. 1997;25:1115–1124. doi: 10.1097/00003246-199707000-00010. [DOI] [PubMed] [Google Scholar]
- 38.Fisher CJ, Jr., Opal SM, Lowry SF, Sadoff JC, LaBrecque JF, Donovan HC, Lookabaugh JL, Lemke J, Pribble JP, Stromatt SC, et al. Role of interleukin-1 and the therapeutic potential of interleukin-1 receptor antagonist in sepsis. Circ Shock. 1994;44:1–8. [PubMed] [Google Scholar]
- 39.Puneet P, Moochhala S, Bhatia M. Chemokines in acute respiratory distress syndrome. American journal of physiology. Lung cellular and molecular physiology. 2005;288:L3–15. doi: 10.1152/ajplung.00405.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oynebraten I, Bakke O, Brandtzaeg P, Johansen FE, Haraldsen G. Rapid chemokine secretion from endothelial cells originates from 2 distinct compartments. Blood. 2004;104:314–320. doi: 10.1182/blood-2003-08-2891. [DOI] [PubMed] [Google Scholar]
- 41.Carlow DA, Teh SJ, Teh HS. Specific antiviral activity demonstrated by TGTP, a member of a new family of interferon-induced GTPases. J Immunol. 1998;161:2348–2355. [PubMed] [Google Scholar]
- 42.Carlow DA, Marth J, Clark-Lewis I, Teh HS. Isolation of a gene encoding a developmentally regulated T cell-specific protein with a guanine nucleotide triphosphate-binding motif. Journal of Immunology. 1995;154:1724–1734. [PubMed] [Google Scholar]
- 43.Lafuse WP, Brown D, Castle L, Zwilling BS. Cloning and characterization of a novel cDNA that is IFN-gamma-induced in mouse peritoneal macrophages and encodes a putative GTP-binding protein. J Leukoc Biol. 1995;57:477–483. doi: 10.1002/jlb.57.3.477. [DOI] [PubMed] [Google Scholar]
- 44.MacMicking JD. IFN-inducible GTPases and immunity to intracellular pathogens. Trends in Immunology. 2004;25:601–609. doi: 10.1016/j.it.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 45.Pilla DM, Hagar JA, Haldar AK, Mason AK, Degrandi D, Pfeffer K, Ernst RK, Yamamoto M, Miao EA, Coers J. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc Natl Acad Sci U S A. 2014;111:6046–6051. doi: 10.1073/pnas.1321700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shindo T, Kurihara H, Kuno K, Yokoyama H, Wada T, Kurihara Y, Imai T, Wang Y, Ogata M, Nishimatsu H, Moriyama N, Oh-hashi Y, Morita H, Ishikawa T, Nagai R, Yazaki Y, Matsushima K. ADAMTS-1: a metalloproteinase-disintegrin essential for normal growth, fertility, and organ morphology and function. J Clin Invest. 2000;105:1345–1352. doi: 10.1172/JCI8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oveland E, Karlsen TV, Haslene-Hox H, Semaeva E, Janaczyk B, Tenstad O, Wiig H. Proteomic evaluation of inflammatory proteins in rat spleen interstitial fluid and lymph during LPS-induced systemic inflammation reveals increased levels of ADAMST1. J Proteome Res. 2012;11:5338–5349. doi: 10.1021/pr3005666. [DOI] [PubMed] [Google Scholar]
- 48.Howell MD, Torres-Collado AX, Iruela-Arispe ML, Gottschall PE. Selective decline of synaptic protein levels in the frontal cortex of female mice deficient in the extracellular metalloproteinase ADAMTS1. PLoS One. 2012;7:e47226. doi: 10.1371/journal.pone.0047226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JH, Jung HS, Giang PM, Jin X, Lee S, Son PT, Lee D, Hong YS, Lee K, Lee JJ. Blockade of nuclear factor-kappaB signaling pathway and anti-inflammatory activity of cardamomin, a chalcone analog from Alpinia conchigera. J Pharmacol Exp Ther. 2006;316:271–278. doi: 10.1124/jpet.105.092486. [DOI] [PubMed] [Google Scholar]
- 50.Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, Zhang J, Lee WP, Roose-Girma M, Dixit VM. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 51.Vanden Berghe T, Demon D, Bogaert P, Vandendriessche B, Goethals A, Depuydt B, Vuylsteke M, Roelandt R, Van Wonterghem E, Vandenbroecke J, Choi SM, Meyer E, Krautwald S, Declercq W, Takahashi N, Cauwels A, Vandenabeele P. Simultaneous targeting of IL-1 and IL-18 is required for protection against inflammatory and septic shock. Am J Respir Crit Care Med. 2014;189:282–291. doi: 10.1164/rccm.201308-1535OC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.