Summary
Exploring the molecular bases of intraspecific reproductive isolation captures the ongoing phenotypic consequences of genetic divergence and provides insights into the early onset of speciation. Recent species-wide surveys using natural populations of yeasts demonstrated that intrinsic post-zygotic reproductive isolation segregates readily within the same species, and revealed the multiplicity of the genetic mechanisms underlying such processes. These advances deepened our current understandings and opened further perspectives regarding the complete picture of molecular and evolutionary origins driving the onset intraspecific reproductive isolation in yeasts.
Main text
In all living organisms, genetic differences constantly emerge and accumulate, providing the raw material for phenotypic variation upon which natural selection operates. Natural populations of yeasts display considerable levels of genetic and phenotypic diversity, in part allowing them to conquer a wide range of ecological and geographical habitats (Liti et al. 2009, Schacherer et al. 2009, Liti and Schacherer 2011, Skelly et al. 2013, Almeida et al. 2014, Hirakawa et al. 2014, Fawcett et al. 2014, Sampathkumar and Drouin 2015, Ford et al. 2015, Jeffares et al. 2015). Nevertheless, as demonstrated by recent studies (Charron, et al. 2014, Hou, et al. 2014, Hou, et al. 2015), independently segregating genetic variants in diverging populations could occasionally cause them to be reproductively isolated, leading to reduced offspring viability upon hybridization. What kinds of variants are likely to cause such intraspecific reproductive isolation in yeasts, and what could be the molecular and evolutionary mechanisms involved?
In sexual populations, reproductive isolation acts as a gene flow barrier and is considered as a key step in the formation of new species (Coyne and Orr 2004). Among the mechanisms leading to reproductive isolation in yeasts, the role of chromosomal rearrangements such as large-scale reciprocal translocations has been well established within the Saccharomyces species complex (Fischer, et al. 2000). Several documented translocations were shown to explain the loss of offspring viability observed in hybrids between closely related species (Delneri, et al. 2003, Fischer, et al. 2000), and more recently, within populations of a same species (Charron, et al. 2014, Hou, et al. 2014). One extensively studied example involved a translocation between chromosome VIII and XVI in Saccharomyces cerevisiae, which conferred an advantageous trait of sulfite resistance and could be tightly linked to adaptation of wine making (Clowers, et al. 2015, Hou, et al. 2014, Perez-Ortin, et al. 2002). However, although such large chromosomal changes contribute to post-zygotic reproductive isolation at different temporal scales (within and between species), their distribution does not necessarily correlate with the severity of isolation and the degree of genetic divergence observed. For instance, between S. cerevisiae and its close relative S. paradoxus, most isolates have collinear genomes and yet are reproductively isolated. Consequently, the role of chromosomal rearrangements in speciation in yeasts remains indecisive, and other mechanisms must exist leading to the onset of reproductive isolation (Delneri, et al. 2003).
Besides large-scale genome changes, small sequence variations could also lead to intrinsic post-zygotic reproductive isolation. The idea was formulated almost eight decades ago by Theodosius Dobzhansky and Hermann Müller, whereby diverging populations, separated by geographical or ecological barriers, could accumulate independent mutations that cause negative epistatic interactions when brought together, leading to loss of hybrid fertility or viability. Over the years, examples of such Dobzhansky-Müller incompatibility have been found in species among various taxa (Coyne and Orr 2004) and more recently, within populations of the same species in model organisms such as Arabidopsis thaliana and Caenorhabditis elegans (Bikard, et al. 2009, Bomblies and Weigel 2010, Chae, et al. 2014, Seidel, et al. 2008). Curiously, the very existence of such genetic incompatibilities among yeasts species has long been a subject of debate, with few rare examples identified to date (Chou, et al. 2010, Heck, et al. 2006).
In fact, the concept of negative epistatic interactions is all but unfamiliar in yeast. The development of the synthetic genetic array (SGA) of deletion mutants in S. cerevisiae allowed for genome-wide profiling of the synthetic genetic interaction networks, providing the best understanding of the functional connections of genes at an organismal level in any species so far (Costanzo, et al. 2010). More generally, epistatic interactions also contribute to heritable phenotypic variations in yeast, explaining approximately 9% of the total phenotypic variance observed within a very large population of segregants from a specific cross in S. cerevisiae (Bloom, et al. 2013, Bloom, et al. 2015). Nevertheless, how epistasis takes part among genetic variations within natural populations was still not well understood, and whether it plays a role in the onset of reproductive isolation was unknown.
With the advent of sequencing technology, there has been a renewed interest in the understanding of intraspecific diversity in various yeast species in the last decade (Friedrich, et al. 2015, Liti, et al. 2009, Schacherer, et al. 2009, Strope, et al. 2015). These advances provided valuable tools to systematically evaluate the onset of intraspecific reproductive isolation across the whole species diversity, to the end of precise characterization of the possible mechanisms involved to a molecular resolution. With these goals in mind, we have performed a first comprehensive effort characterizing intraspecific reproductive isolation using a large number of diverse natural populations of S. cerevisiae (Hou, et al. 2014). We have identified chromosomal rearrangements segregating in diverse populations and acting as the major mechanism leading to reduced offspring viabilities observed in 16% of the crosses. In parallel, a study within S. paradoxus populations reached similar conclusion (Charron, et al. 2014). By contrast, no evident case of Dobzhansky-Müller incompatibility was found in both species. Are yeasts just prone to this kind of phenomenon, or were we just looked at it wrong?
As a matter of fact, all studies up until now were performed under laboratory conditions, which involves crossing various isolates and then estimate the offspring viability on a rich permissive media that optimize yeast growth. Considering the vast ecological range and the diverse environmental stresses (Ho and Gasch 2015) that natural populations of yeasts encounter in nature, our view of reproductive isolation cases restricted to laboratory conditions might be over simplified. Indeed, when taking into account of different environmental factors, negative epistasis involving incompatible genic interactions were much more common than previously considered in the yeast S. cerevisiae (Hou, et al. 2015).
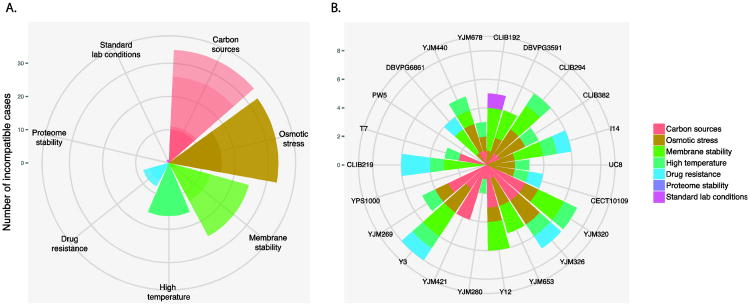
To assess the impact of environmental factors in the onset of reproductive isolation within S. cerevisiae, we performed a systematic survey using a population of 27 natural isolates originated from soil, tree barks, immuno-compromised patients and various fermentations across different continents. All selected isolates were previously shown to yield high offspring viability on the rich media when crossed with the reference strain S288c (Hou, et al. 2014). Interestingly, these highly compatible crosses could be sometimes incompatible on other conditions. In fact, we evaluated the offspring viabilities of these crosses in the presence of 20 culture conditions including different carbon sources, chemical stress and temperatures, and found over 24% (117/481) of the cases showing different degrees of offspring loss ranging from 1% to 62% (Figure 1). Within all identified cases, negative epistasis were found in conditions related to various stress types (Figure 1A), and were randomly distributed among different isolates, regardless of their origin or the level of sequence divergence between the parental pair (Figure 1B). These results clearly indicated that relying on standard laboratory media was simply biased (Figure 1A), and that negative epistasis readily segregates within yeast natural populations and contributes to reproductive isolation at the intraspecific scale.
Figure 1. Negative epistasis segregates within populations of S. cerevisiae related to various environmental conditions.
A. Distribution of epistasis cases according to various stress types. Shades of colors represent different conditions tested that belong to the same category. A total of 117 cases are categorized.
B. Distribution of epistasis cases according to isolates crossed with S288c. Colors correspond to type of stress as indicated. Isolates are organized clockwise according to the level of sequence divergence compared to S288c, with CLIB192 being most closely related with a divergence of 0.11%.
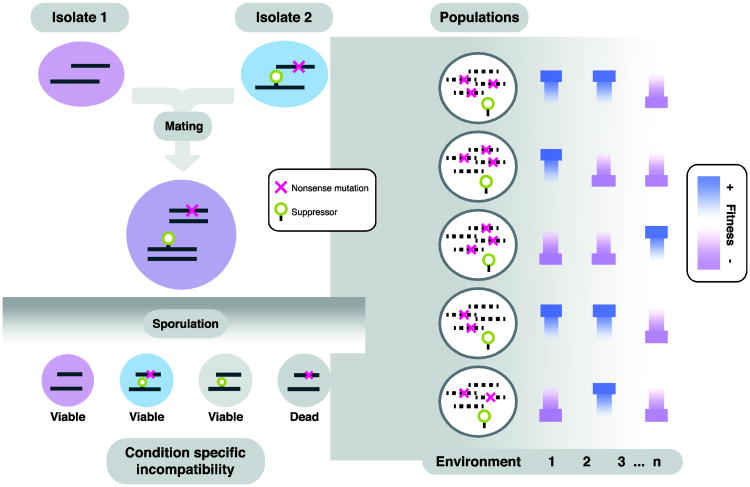
Using a combination of classical genetic analysis of lethal phenotypic segregation in the offspring and high-throughput genomic mapping strategy, we further identified and characterized the first example of two loci Dobzhansky-Müller incompatibility in yeast related to respiratory conditions, such as the non fermentable carbon source glycerol. In this case, a clinical isolate YJM421 was incompatible when crossed with the reference S288c, yielding only 75% of offspring that are able to grow on glycerol (Figure 2). The rest of the offspring were unable to respire because of a specific incompatible allelic combination between two genes: COX15, a mitochondrial inner membrane cargo protein that is essential for respiration, and SUP7, a tRNA nonsense suppressor. In fact, the incompatible isolate YJM421 has a nonsense mutation in the COX15 gene, and in the same time a nonsense suppressor SUP7 to compensate the effect of the mutated COX15. However, as the reference strain S288c does not possess the suppressor mutation, 25% of the offspring obtained by mating these two strains will inherit only the non-functional COX15 and not the suppressor, therefore become incapable to grow on respiratory conditions (Figure 2).
Figure 2. The molecular basis of the first example of two loci Dobzhansky-Müller incompatibility related to respiration in S. cerevisiae.
Crossing between isolate carrying allelic combination of a nonsense mutation and a tRNA nonsense suppressor with isolate without this combination. After meiosis, 25% of the offspring inheriting only the nonsense mutation but not the suppressor were non-viable on conditions with non-fermentable carbon sources, as the nonsense mutation was essential for respiration. Nevertheless, when the nonsense suppressor was introduced other genetic background, it could confer to gain or loss of fitness in different environmental conditions, possibly indicating an adaptive role of certain incompatible alleles in complex ecological contexts.
It is now clear that in addition to chromosomal rearrangements, negative epistasis could also lead to the onset of reproductive isolation within yeast natural populations in a condition-specific manner. Nonetheless, it is interesting to note that even though the frequency of potential incompatibilities is relatively high (117/483), most of them were not shared among different isolates, suggesting a rather unique genetic origin for different cases (Figure 1). Taking for example the identified Dobzhansky-Müller case, the allelic combination of cox15stop and SUP7 were only found the clinical isolate YJM421, making this isolate universally incompatible with more than 100 natural isolates of S. cerevisiae that do not possess this combination. What could be the force that drives the onset and the maintenance of this particular configuration?
As it turned out, alleles causing hybrid incompatibility may sometimes offer fitness advantages under stress conditions. When the suppressor mutation SUP7 in YJM421 was ectopically expressed in other isolates, it conferred to diverse fitness effects. Some isolates displayed significant gain of fitness in some conditions in the presence of the suppressor, and others displayed the exact opposite (Figure 2). This observation suggests that carrying the suppressor might be advantageous in certain environmental conditions, thus balancing the effect of potential offspring loss upon hybridization.
Overall, it is evident that even at the intraspecific scale, mechanisms leading to the onset of reproductive isolation are multiple. Nevertheless, our understanding of how intraspecific genetic diversity impacts the onset of reproductive isolation is still incomplete. More particularly, studies in S. cerevisiae were still biased as all crosses were performed between natural isolates and the laboratory reference strain S288c. Would the conclusions be somewhat different in unbiased pairwise crosses? In addition to epistasis within the nuclear genome, is there a role of interactions between the mitochondrial and nuclear genome to the onset of reproductive isolation (Paliwal, et al. 2014)? What is the relative importance of chromosomal rearrangements vs. negative epistasis? Are these events random or related to selection? And finally, what should we expect to see in other yeast species, when the observed genetic diversity is usually higher?
Exploring the mechanisms leading to the onset of reproductive isolation stems from the fundamental interest of understanding biodiversity. Recent systematic surveys within yeasts have revealed the multiplicity regarding the molecular and evolutionary origin of intraspecific reproductive isolation, yet much is still left to explore. Whether these mechanisms would eventually lead to speciation is unclear, yet it is worth noting that potential barriers to gene flow could segregate contemporaneously within natural populations of a single species of yeast.
Acknowledgments
The authors thank the Agence Nationale de la Recherche (ANR grant 2011-JSV6-004-01) and the National Institutes of Health (NIH grant R01 GM101091-01) for financial support. J.H. is supported in part by a grant from the Ministère de l'Enseignement Supérieur et de la Recherche and in part by a fellowship from the La Ligue contre le Cancer.
References
- Almeida P, Gonçalves C, Teixeira S, Libkind D, Bontrager M, Masneuf-Pomarède I, Albertin W, Durrens P, Sherman DJ, Marullo P, Hittinger CT, Gonçalves P, Sampaio JP. A Gondwanan imprint on global diversity and domestication of wine and cider yeast Saccharomyces uvarum. Nat Commun. 2014 Jun 2;5:4044. doi: 10.1038/ncomms5044. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D, Patel D, Le Mette C, Giorgi V, Camilleri C, Bennett MJ, Loudet O. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science. 2009;323:623–626. doi: 10.1126/science.1165917. [DOI] [PubMed] [Google Scholar]
- Bloom JS, Ehrenreich IM, Loo WT, Lite TL, Kruglyak L. Finding the sources of missing heritability in a yeast cross. Nature. 2013;494:234–237. doi: 10.1038/nature11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JS, Kotenko I, Sadhu M, Treusch S, Albert FW, Kruglyak L. The role of genetic interactions in yeast quantitative traits. 2015 doi: 10.1038/ncomms9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Weigel D. Arabidopsis and relatives as models for the study of genetic and genomic incompatibilities. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2010;365:1815–1823. doi: 10.1098/rstb.2009.0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae E, Bomblies K, Kim ST, Karelina D, Zaidem M, Ossowski S, Martin-Pizarro C, Laitinen RA, Rowan BA, Tenenboim H, Lechner S, Demar M, Habring-Muller A, Lanz C, Ratsch G, Weigel D. Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell. 2014;159:1341–1351. doi: 10.1016/j.cell.2014.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron G, Leducq JB, Landry CR. Chromosomal variation segregates within incipient species and correlates with reproductive isolation. Molecular ecology. 2014;23:4362–4372. doi: 10.1111/mec.12864. [DOI] [PubMed] [Google Scholar]
- Chou JY, Hung YS, Lin KH, Lee HY, Leu JY. Multiple molecular mechanisms cause reproductive isolation between three yeast species. PLoS biology. 2010;8:e1000432. doi: 10.1371/journal.pbio.1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowers KJ, Heilberger J, Piotrowski JS, Will JL, Gasch AP. Ecological and genetic barriers differentiate natural populations of Saccharomyces cerevisiae. Molecular biology and evolution. 2015 doi: 10.1093/molbev/msv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, Prinz J, St Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M, Deshpande R, Li Z, Lin ZY, Liang W, Marback M, Paw J, San Luis BJ, Shuteriqi E, Tong AH, van Dyk N, Wallace IM, Whitney JA, Weirauch MT, Zhong G, Zhu H, Houry WA, Brudno M, Ragibizadeh S, Papp B, Pal C, Roth FP, Giaever G, Nislow C, Troyanskaya OG, Bussey H, Bader GD, Gingras AC, Morris QD, Kim PM, Kaiser CA, Myers CL, Andrews BJ, Boone C. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA 2004 Speciation Sinauer Associates, Sunderland, Mass.
- Delneri D, Colson I, Grammenoudi S, Roberts IN, Louis EJ, Oliver SG. Engineering evolution to study speciation in yeasts. Nature. 2003;422:68–72. doi: 10.1038/nature01418. [DOI] [PubMed] [Google Scholar]
- Fawcett JA, Iida T, Takuno S, Sugino RP, Kado T, Kugou K, Mura S, Kobayashi T, Ohta K, Nakayama J, Innan H. Population genomics of the fission yeast Schizosaccharomyces pombe. PLoS One. 2014;9:e104241. doi: 10.1371/journal.pone.0104241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G, James SA, Roberts IN, Oliver SG, Louis EJ. Chromosomal evolution in Saccharomyces. Nature. 2000;405:451–454. doi: 10.1038/35013058. [DOI] [PubMed] [Google Scholar]
- Ford CB, Funt JM, Abbey D, Issi L, Guiducci C, Martinez DA, Delorey T, Li BY, White TC, Cuomo C, Rao RP, Berman J, Thompson DA, Regev A. The evolution of drug resistance in clinical isolates of Candida albicans. Elife. 2015;4:e00662. doi: 10.7554/eLife.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich A, Jung P, Reisser C, Fischer G, Schacherer J. Population genomics reveals chromosome-scale heterogeneous evolution in a protoploid yeast. Molecular biology and evolution. 2015;32:184–192. doi: 10.1093/molbev/msu295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck JA, Argueso JL, Gemici Z, Reeves RG, Bernard A, Aquadro CF, Alani E. Negative epistasis between natural variants of the Saccharomyces cerevisiae MLH1 and PMS1 genes results in a defect in mismatch repair. P Natl Acad Sci USA. 2006;103:3256–3261. doi: 10.1073/Pnas.0510998103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa MP, Martinez DA, Sakthikumar S, Anderson MZ, Berlin A, Gujja S, Zeng Q, Zisson E, Wang JM, Greenberg JM, Berman J, Bennett RJ, Cuomo CA. Genetic and phenotypic intra-species variation in Candida albicans. Genome Research. 2014;225:413–25. doi: 10.1101/gr.174623.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, Gasch Exploiting the yeast stress-activated signaling network to inform on stress biology and disease signaling. Curr Genet. 2015 doi: 10.1007/s00294-015-0491-0. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Friedrich A, de Montigny J, Schacherer J. Chromosomal rearrangements as a major mechanism in the onset of reproductive isolation in Saccharomyces cerevisiae. Current biology: CB. 2014;24:1153–1159. doi: 10.1016/j.cub.2014.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Friedrich A, Gounot JS, Schacherer J. Comprehensive survey of condition-specific reproductive isolation reveals genetic incompatibility in yeast. Nature communications. 2015;6:7214. doi: 10.1038/ncomms8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffares DC, Rallis C, Rieux A, Speed D, Převorovský M, Mourier T, Marsellach FX, Iqbal Z, Lau W, Cheng TM, Pracana R, Mülleder M, Lawson JL, Chessel A, Bala S, Hellenthal G, O'Fallon B, Keane T, Simpson JT, Bischof L, Tomiczek B, Bitton DA, Sideri T, Codlin S, Hellberg JE, van Trigt L, Jeffery L, Li JJ, Atkinson S, Thodberg M, Febrer M, McLay K, Drou N, Brown W, Hayles J, Carazo Salas RE, Ralser M, Maniatis N, Balding DJ, Balloux F, Durbin R, Bähler J. The genomic and phenotypic diversity of Schizosaccharomyces pombe. Nature Genetics. 2015;47:235–41. doi: 10.1038/ng.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, Tsai IJ, Bergman CM, Bensasson D, O'Kelly MJ, van Oudenaarden A, Barton DB, Bailes E, Nguyen AN, Jones M, Quail MA, Goodhead I, Sims S, Smith F, Blomberg A, Durbin R, Louis EJ. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Schacherer J. The rise of yeast population genomics. Comptes rendus biologies. 2011;334:612–619. doi: 10.1016/j.crvi.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Paliwal S, Fiumera AC, Fiumera HL. Mitochondrial-nuclear epistasis contributes to phenotypic variation and coadaptation in natural isolates of Saccharomyces cerevisiae. Genetics. 2014;198:1251–1265. doi: 10.1534/genetics.114.168575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Ortin JE, Querol A, Puig S, Barrio E. Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome research. 2002;12:1533–1539. doi: 10.1101/gr.436602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar, Drouin Purifying selection against gene conversions between the polyamine transport (TPO) genes of Saccharomyces species. Current Genetics. 2015;61:67–72. doi: 10.1007/s00294-014-0445-y. 2015. [DOI] [PubMed] [Google Scholar]
- Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458:342–345. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel HS, Rockman MV, Kruglyak L. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science. 2008;319:589–594. doi: 10.1126/science.1151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelly DA, Merrihew GE, Riffle M, Connelly CF, Kerr EO, Johansson M, Jaschob D, Graczyk B, Shulman NJ, Wakefield J, Cooper SJ, Fields S, Noble WS, Muller EG, Davis TN, Dunham MJ, Maccoss MJ, Akey JM. Integrative phenomics reveals insight into the structure of phenotypic diversity in budding yeast. Genome Research. 2013;23:1496–504. doi: 10.1101/gr.155762.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strope PK, Skelly DA, Kozmin SG, Mahadevan G, Stone EA, Magwene PM, Dietrich FS, McCusker JH. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome research. 2015;25:762–774. doi: 10.1101/gr.185538.114. [DOI] [PMC free article] [PubMed] [Google Scholar]