Abstract
Specialized pro-resolving lipid mediators (SPMs) constitute a recently recognized class of bioactive molecules which promote the resolution of inflammation. We recently reported that the SPMs resolvin D1 (RvD1) and 17-hydroxydocosahexaenoic acid (17-HDHA) promote the differentiation of IgG-secreting B cells and enhance antibody-mediated immune responses. However, there is an important knowledge gap regarding whether or not SPMs regulate human B-cell IgE production, which is the key effector in diseases such as asthma and allergy. Therefore we investigated whether a panel of diverse SPMs influences B-cell IgE production. An important finding was that 17-HDHA and RvD1 inhibit IgE production by human B cells and suppress the differentiation of naïve B cells into IgE-secreting cells by specifically blocking epsilon germline transcription (εGLT). This effect is specific to human IgE, as the SPMs do not inhibit production of IgM and IgG and did not suppress other IL-4-upregulated genes. 17-HDHA and RvD1 act by stabilizing the transcriptional repressor Bcl-6, which competes with STAT6 for binding at the εGLT promoter. Overall, these new findings demonstrate that certain SPMs inhibit the differentiation of IgE-producing B cells, without being broadly immune-suppressive, representing a novel class of potential therapeutics for IgE-driven diseases such as asthma and allergy.
Keywords: pro-resolving mediator, inflammation resolution, human, B cell, IgE
Introduction
Acute inflammation is a protective response triggered by trauma, pathogens, toxins, and other tissue insults, which is initiated within minutes of recognition of “danger” signals by activation of the innate immune system. Resolution of inflammation is a dynamic and active process that regulates many cellular interactions in affected tissues to restore homeostasis [1–3]. Failure to re-establish homeostasis due to insufficient resolution can contribute to chronic inflammatory conditions, such as asthma [2, 3]. Recently, endogenous specialized pro-resolving lipid mediators (SPMs) were identified as important drivers of resolution of inflammation [3–6]. SPMs are derived from dietary polyunsaturated fatty acids, such as omega-3 and omega-6, and are classified into four families: lipoxins (LXs), resolvins (RvDs, RvEs), protectins (PDs) and maresins (MaRs) [7]. Many intermediates are involved in their biosynthetic pathway, such as 17-HDHA, which is a precursor of RvDs [1, 6]. Each lipid mediator has a unique chemical structure, which determines their specific bioaction on immune cells [8].
Recently, our lab has shown that the SPMs, 17-HDHA and RvD1, directly regulate human B-cell functions by promoting plasma cell differentiation and increasing IgM and IgG antibody production, and these may serve as a new class of adjuvants [9]. However, an important issue is whether or not SPMs affect human B-cell IgE antibody production.
IgE is an antibody produced by B cells that is responsible for the onset and maintenance of allergic diseases, including asthma [10, 11]. The vast majority of individuals with allergic asthma have elevated serum IgE levels, which is a reliable parameter that tracts with uncontrolled and severe asthma [12]. IgE is produced from B cells stimulated by cytokines and co-stimulatory signals from Th2 cells, in response to specific allergens. Antigen cross-linking of IgE bound to Fc receptors on mast cells or basophils triggers cascades of pro-inflammatory immune reactions presumably to clear pathogens such as parasites [13]. However, when this process is not terminated acutely or with repeated exposure to allergens, it can lead to chronic inflammation, causing tissue damage. Because IgE is a key player in allergic diseases, control of IgE levels, such as via anti-IgE antibodies, has emerged as an important therapeutic strategy [10, 11].
Given the important role of SPMs in promoting inflammation resolution, some studies have investigated the roles of SPMs in murine models of inflammation, including asthma [14–16]. Resolvin D1 (RvD1) reduces allergic airway inflammation by targeting eosinophils and pro-inflammatory mediators involved in Th2 signaling pathway, while resolvin E1 (RvE1) regulates the development of Th17 cells and IL-23 production [14, 15]. However, there is an important knowledge gap regarding the effects of SPMs on human B-cell IgE production, which has a central role in initiating allergic diseases. In this study, we asked whether SPMs can regulate human B-cell IgE production. We demonstrate that RvD1 and 17-HDHA specifically suppress IgE production in human B cells by inhibiting B-cell class switch to IgE. This is mediated by Bcl-6 (B cell lymphoma-6), the transcriptional repressor, which is known to control IgE production by competing with STAT6 (signal transducer and activator of transcription 6) for binding to the promoter region of epsilon germline transcript (εGLT). The important regulatory role of Bcl-6 was also shown in Bcl-6 knockout mice which exhibited extremely high serum IgE levels [17]. Therefore, these pro-resolving lipid mediators may represent a novel treatment pathway for allergic diseases with their unique actions on regulating human B-cell IgE production.
Results
17-HDHA and RvD1 decrease IgE production from human B cells
Several different IgE inducing cocktails have been reported, however, some modifiers such as IL-21 and CpG ODNs are reported to have different effects by different groups [18–22]. To investigate the effect of SPMs on IgE production, we first determined the optimal conditions for B-cell differentiation to IgE-producing cells in our hands. Human B cells were isolated from peripheral blood of healthy donors using CD19+ magnetic beads (purity>98%) and were cultured with IgE-inducing cocktails that are reported to mimic allergic responses and promote B-cell IgE production in vitro [18, 19]. CpG ODN2395 alone induced both IgE and IgM (Supporting Information Fig 1A). However, CpG ODN2395 alone did not induce epsilon germline transcription, which is required for class switch to IgE (Supporting Information Fig 1B). It has been reported that CpG ODN alone could stimulate antibody production from memory cells [23], and it is likely that there is a small proportion of IgE memory cells even in nominally healthy donors. IL-4 is required for IgE class switching ([24] and Supporting Information Fig 1B) and in our hands a cocktail of mCD40L, IL-4 and CpG ODN2395 induced the highest level of IgE production, the highest ratio of IgE to IgM, and the highest rate of εGLT transcription (Supporting Information Fig 1A, 1B). We also determined the optimal cell density and time course of antibody production (Supporting Information Fig 1C, 1D) and used these conditions for the remaining studies.
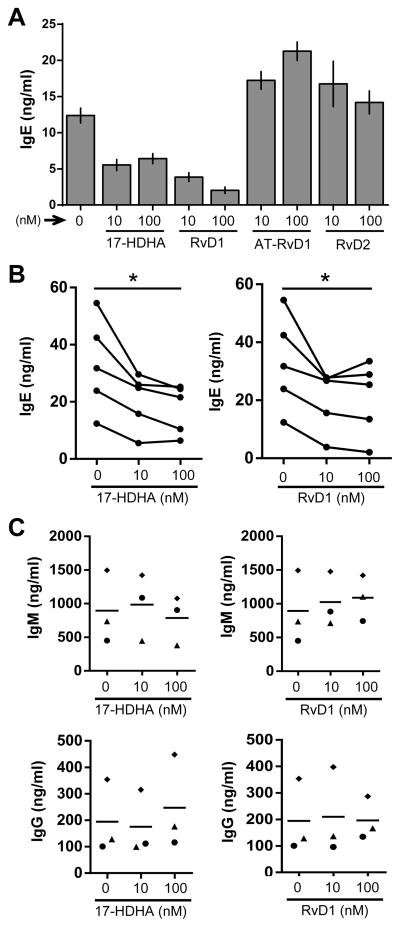
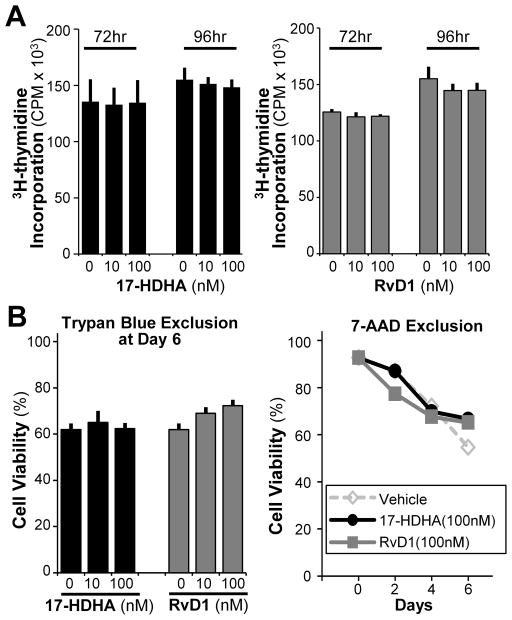
Next, the effects of certain SPMs on B-cell IgE production were analyzed by measuring IgE antibody levels using ELISA assay (Fig. 1A–B). 17-HDHA and RvD1 reduced IgE levels in a dose-dependent manner, unlike AT-RvD1 and RvD2 (Fig. 1A, Supporting Information Fig 2). Moreover, the inhibitory effects of 17-HDHA and RvD1 on B-cell IgE production were shown in multiple human donors (Fig 1B). To determine whether this effect was isotype-specific, we also measured production of IgM and IgG in an IgE-inducing environment. Neither IgM nor IgG levels were reduced by 17-HDHA or RvD1 (Fig 1C). To determine whether the decrease in IgE production was due to changes in cell proliferation or viability, thymidine incorporation assay and live/dead exclusion analysis were performed (Fig 2). There were no significant differences detected in proliferation or viability between SPM-treated and vehicle-treated B cells (Fig 2A–B). Therefore, we concluded that 17-HDHA and RvD1 specifically reduced human B-cell IgE production, without affecting cell proliferation or viability.
Figure 1. 17-HDHA and RvD1 decrease human B-cell IgE production.
Human CD19+ B cells purified from healthy donors were pretreated with 17-hydroxydocosahexaenoic acid (17-HDHA), resolvin D1 (RvD1), aspirin-triggered resolvin D1 (AT-RvD1) or resolvin D2 (RvD2) at the indicated concentrations, followed by stimulation with the IgE-inducing cocktail. SPM treatments were repeated daily, and antibody levels in the supernatants were measured by ELISA on day 6. (A) Different SPMs were added to human B cells and culture supernatant IgE levels were measured. Data are shown as mean±SEM of triplicates from one representative donor of 3 donors. (B) 17-HDHA and RvD1 were added to B cells from five different donors and culture supernatant IgE levels were measured. (C) The effects of 17-HDHA and RvD1 on IgM or IgG antibody production were evaluated. Each symbol represents an individual donor and bars represent means. Data analyzed by repeated measures ANOVA with Tukey’s post test, *P<0.05.
Figure 2. 17-HDHA and RvD1 do not affect B-cell proliferation or viability.
B cells were pretreated with the SPMs, stimulated with the IgE-inducing cocktail, and collected at different time points (0–6 days) to measure cell viability. (A) B-cell proliferation was measured using [3H]-thymidine incorporation at 72 or 96 hours post-stimulation with 17-HDHA (left) and RvD1 (right). (B) B-cell viability was measured using two different staining methods; Trypan blue (left) and 7-AAD exclusion (right). Data shown are mean±SEM of triplicates for one representative donor of 3 donors tested and analyzed by ANOVA with Tukey’s post test.
17-HDHA and RvD1 decrease the number of IgE-secreting B cells in culture
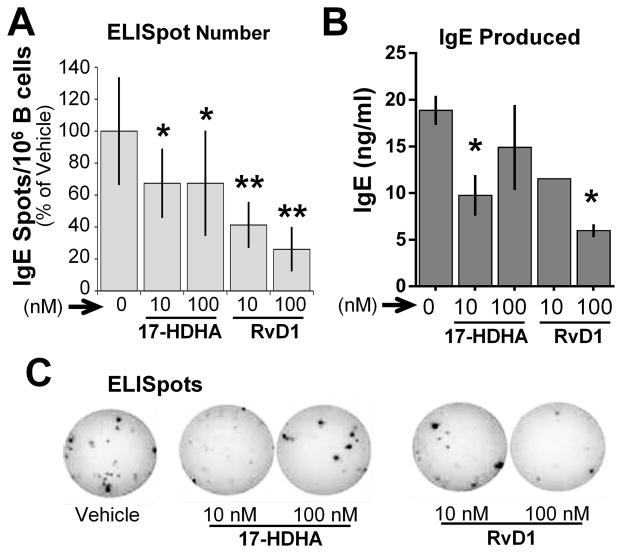
The 17-HDHA and RvD1-mediated decrease of IgE antibody could be due to reduced IgE produced per cell or to fewer IgE-secreting B cells. To address this question, we measured the number of IgE-secreting B cells using the ELISpot assay. B cells from healthy donors were treated with SPMs plus an IgE-inducing cocktail for 6 days, then transferred to ELISpot plates for 24 hours. 17-HDHA and RvD1 reduced the number of IgE-secreting B cells, and the total amount of IgE produced (Fig 3).
Figure 3. 17-HDHA and RvD1 decrease the number of IgE-secreting B cells in culture.
CD19+ B cells isolated from healthy donors were pretreated with vehicle or SPMs (17-HDHA, RvD1) followed by stimulation with the IgE-inducing cocktail. On day 6, the cells were transferred to anti-IgE-coated ELISpot plates and further cultured overnight. (A) The number of IgE-producing B cells, and (B) IgE antibody levels were measured in the day 6 culture supernatants by ELISA. Data shown are mean±SEM for one representative donor of 3 donors tested. *P≤0.05 by one-way ANOVA with Tukey’s post test. (C) Representative photograph of ELISpots from a donor with and without SPM treatment.
RvD1 inhibits B-cell class switching to IgE by suppressing εGLT transcription
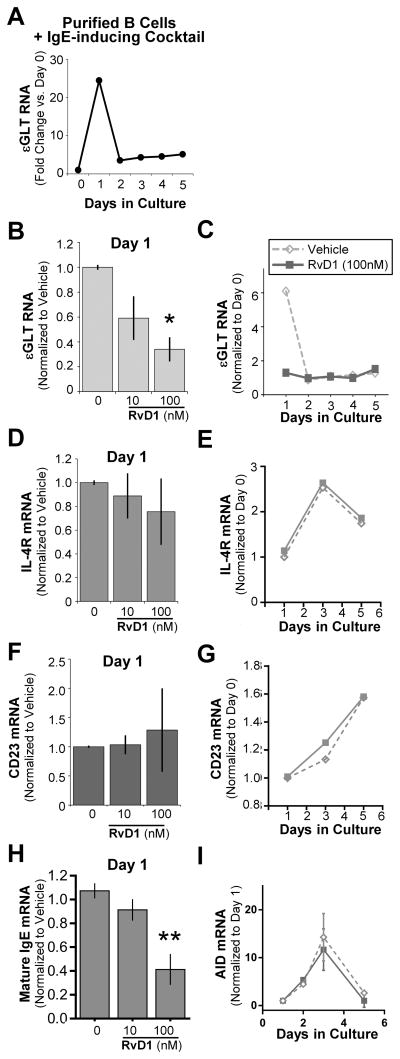
There are two main processes that naïve B cells undergo to become IgE-producing cells; class switching to IgE and differentiation to plasma cells [25]. To investigate the mechanism(s) by which SPMs reduce the number of IgE-secreting cells, we focused on the effects of RvD1 on B-cell class switching. Epsilon germline transcription is an essential process that precedes Ab class switching, and is known to be a reliable indicator of class switching [26, 27]. We measured the kinetics of epsilon germline transcript (εGLT) RNA expression in B cells stimulated with an IgE-inducing cocktail up to day 5. εGLT RNA levels rapidly peaked at day 1, then reduced to near-basal levels through day 5 (Fig. 4A). Therefore, we decided to measure the effects of RvD1 at day 1, when εGLT expression is the highest. Purified B cells were pretreated with RvD1, followed by stimulation, and εGLT RNA levels were determined. RvD1 reduced εGLT RNA levels in a dose-dependent manner (Fig. 4B). However, at later time points (day 2–5), there were no changes in εGLT RNA levels between vehicle-treated and RvD1-treated B cells (Fig. 4C). Activation of the IL-4 signaling pathway induces transcription of a subset of genes including εGLT [28]. To determine whether or not RvD1 acts on εGLT via the IL-4 pathway, we measured changes in two other IL-4-inducible genes, IL-4R and CD23. Surprisingly, there were no significant changes in the mRNA levels of either IL-4R or CD23 with RvD1 treatment at day 1 (Fig. 4D, F), and even at later time points up to day 6 (Fig. 4E, G). While εGLT transcription precedes the class switch recombination (CSR) process, mature IgE mRNA transcription occurs after the CSR. Therefore, we have also measured the changes in mature IgE mRNA levels in RvD1-treated human B cells, a direct indicator of class switching. As expected, RvD1 reduced IgE mRNA levels in a concentration-dependent manner (Fig. 4F). Due to its important role in catalyzing antibody class switching to IgG, IgA or IgE, we measured activation-induced cytidine deaminase (AID) mRNA levels. B cells were pretreated with RvD1 (100nM) or vehicle, and then stimulated with the IgE-inducing cocktail as described above. AID mRNA levels peaked at day 3 post-stimulation, but RvD1 treatment did not affect the AID mRNA levels (Fig. 4G).
Figure 4. RvD1 inhibits B-cell class switch to IgE by suppressing εGLT transcription.
(A) Purified CD19+ B cells were stimulated with the IgE-inducing cocktail and cells were collected each day until day 5. RNA levels of εGLT were measured by RT-qPCR and normalized to 18S rRNA. εGLT fold changes are shown as RNA levels normalized to the basal RNA level expressed at day 0. (B–G) Purified B cells pretreated with RvD1 followed by stimulation with the IgE-inducing cocktail were collected at day 1 and mRNA levels of (B) εGLT, (D) IL-4R, and (F) CD23 were quantified using RT-qPCR (left). (C, E, G) The kinetics of (C) εGLT, (E) IL-4R, and (G) CD23 mRNA levels in B cells treated with RvD1 (100 nM) are shown from a representative donor. (H) Mature IgE mRNA levels in RvD1 preteated B cells were measured at day 1. (I) AID mRNA levels in B cells pretreated with RvD1 (100 nM) or vehicle, followed by stimulation with the IgE-inducing cocktail were measured at different time points using RT-qPCR. Data shown are mean±SEM for 3 donors.*P≤0.05 by one-way repeated measures ANOVA with Tukey’s post test.
RvD1 prevents STAT6 binding to the εGLT promoter region
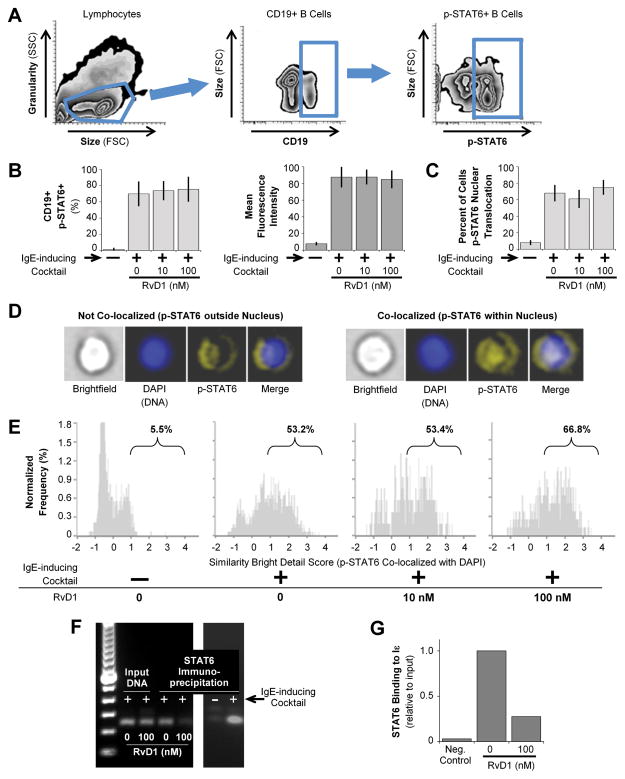
Once the IL-4 signaling pathway is initiated, STAT6 acts as a master transcription factor that translocates to the nucleus and promotes transcription of εGLT and other IL-4-inducible genes [25]. Therefore, we next assessed STAT6 phosphorylation and nuclear localization in B cells treated with RvD1. First, we performed flow cytometry analysis for intracellular phospho-STAT6 (p-STAT6) expression. PBMCs from healthy donors were pretreated with RvD1, followed by stimulation for 60 minutes. Figure 5A shows the gating strategy used to measure p-STAT6 in CD19+ B cells. B cells started to express p-STAT6 as soon as 5 minutes post-stimulation (data not shown), peaking at 60 minutes. There was no significant effect of RvD1 on the percentage of B cells expressing p-STAT6 or on the fluorescence intensity of p-STAT6 (Fig. 5B). Next, to measure nuclear translocation of p-STAT6, we used ImageStream analysis to determine co-localization of p-STAT6 and the nuclear stain DAPI using the Similarity Bright Detail Score algorithm (Fig 5C–E). As expected, B-cell activation led to a large increase in the percentage of cells with p-STAT6 nuclear localization (Fig. 5D, E). However, RvD1 treatment did not affect the percentage of B cells with pSTAT6 translocated to the nucleus (Fig. 5C), showing no significant effect of RvD1 on STAT6 phosphorylation or nuclear translocation. With no significant changes in STAT6 activation and nuclear translocation in RvD1-treated B cells, we hypothesized that RvD1 may prevent STAT6 binding to the εGLT promoter region and hence reduce εGLT transcription. To test this, we performed a Chromatin ImmunoPrecepitation (ChIP) assay on PBMCs isolated from healthy human donors pretreated with RvD1 or vehicle, and then stimulated with the IgE-inducing cocktail for 4 hours. Anti-STAT6 antibody was added to enrich for STAT6-bound DNA. Primers specific for the εGLT promoter region that included the STAT6 binding site were used for quantitative PCR analysis of the precipitated DNA. STAT6 binding efficiency to the εGLT promoter region was quantified relative to the input DNA which was extracted prior to ChIP (Fig. 5G). In Figure 5F and 5G, we show that in B cells stimulated with the IgE-inducing cocktail, STAT6 efficiently binds to the εGLT promoter region compared to the unstimulated cells. However, with the RvD1 (100nM) pretreatment, the association of STAT6 on the εGLT promoter region was greatly reduced in B cells stimulated with the IgE-inducing cocktail compared to the vehicle. This supports the idea that RvD1 prevents STAT6 binding to the εGLT promoter region without affecting the the phosphorylation and translocation of STAT6.
Figure 5. RvD1 does not affect STAT6 phosphorylation or nuclear translocation.
PBMCs from healthy donors were pretreated with RvD1, followed by stimulation with the IgE-inducing cocktail for 60 minutes. Cells were stained for surface CD19 and intracellular p-STAT6. (A) Gating strategy to identify p-STAT6 expression in B cells. (B) The percentage of B cells expressing p-STAT6 (right) and mean fluorescence intensity (left) for p-STAT6 staining were determined by flow cytometry. Results shown are mean±SEM for 3 donors. (C–E) Purified CD19+ B cells were pretreated with RvD1, and then stimulated with the IgE-inducing cocktail for 60 minutes. Co-localization of p-STAT6 and DAPI staining was determined by ImageStream analysis (C) The percentage of B cells with pSTAT6 nuclear translocalization is shown as mean±SEM for 3 donors. (D) Representative images and (E) ImageStream analysis showing the Similarity Bright Detail Score (SBDS) for DAPI and p-STAT6 in one representative donor of 3 tested are shown. A SBDS score<1 indicates no significant overlap of stained regions. Labels indicate percentage of cells with SBDS>1. (F, G) PBMCs isolated from healthy human donors were pretreated with 100nM of RvD1, followed by stimulation with the IgE-inducing cocktail for 4 h. ChIP was carried out using anti-STAT6 antibody followed by qPCR analysis. (F) PCR products obtained with primers specific to the εGLT promoter region. Two images are from different gels. (G) Quantification of STAT6 binding to the εGLT promoter region relative to input DNA. Data shown are from one human donor, representative of two donors.
RvD1 enhances the expression of Bcl-6 in primary human B cells and in malignant B cell lymphoma
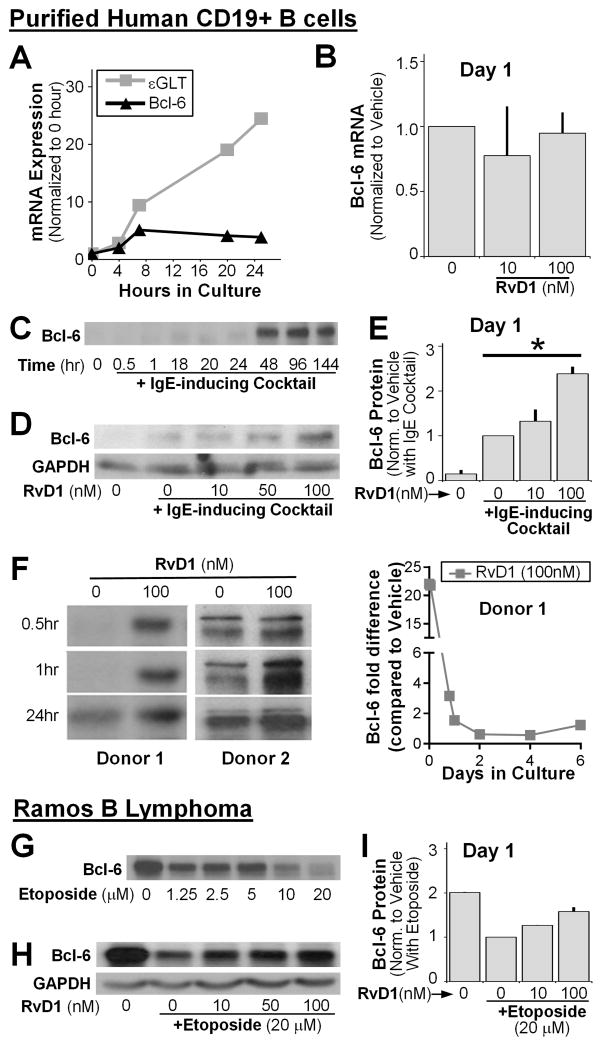
We next investigated how RvD1 could reduce STAT6 binding affinity to the εGLT promoter region without affecting the transcription of other IL4-inducible genes such as IL-4R and CD23. Bcl-6 is a transcriptional repressor that controls B-cell IgE production by antagonizing STAT6 activity on a limited subset of IL-4-responsive genes [29]. Therefore, we investigated the role of Bcl-6 in RvD1-mediated suppression of IgE production. We measured the kinetics of Bcl-6 mRNA expression in B cells stimulated with an IgE-inducing cocktail up to 24 hours. A small increase in Bcl-6 mRNA levels at 24 hours post-stimulation was observed, in contrast to the rapid induction of εGLT RNA expression (Fig. 6A). To evaluate the effect of RvD1 on Bcl-6 mRNA expression, cells were pretreated with RvD1 and stimulated with the IgE-inducing cocktail for 24 hours. There were no significant changes observed in Bcl-6 mRNA level between RvD-treated and vehicle-treated B cells (Fig. 6B). Bcl-6 expression could be regulated at the translational and post-translational level rather than by transcription [30], so we next measured the effect of RvD1 on Bcl-6 protein expression. Bcl-6 protein expression levels were measured in B cells stimulated with an IgE-inducing cocktail at different time points (0–6 days). Bcl-6 expression was barely detectable at early time points (<24 hours), and remarkably enhanced at day 2 (Fig. 6C), consistent with the rapid decline in εGLT RNA levels seen at the same time (Fig. 4A). To evaluate the effects of RvD1 on Bcl-6 expression at the time point when εGLT RNA level peaked, B cells were treated with RvD1, and then stimulated for 24 hours. RvD1 enhanced Bcl-6 protein expression in a dose-dependent manner, consistent with suppression of εGLT transcription (Fig. 6D–E) [29]. The effects of RvD1 to enhance Bcl-6 protein levels were even stronger at earlier time points (< 24hr) when the basal Bcl-6 levels were barely detectable (Fig. 6F). However, at later time points (2–6 days), RvD1 did not affect Bcl-6 expression which already reached the level to suppress the transcription of εGLT (data not shown). To further confirm the effects of RvD1 on Bcl-6, we tested RvD1 in a lymphoma B cell line. The Ramos cell line is a human Burkitt’s lymphoma that constitutively expresses high levels of Bcl-6 protein [31]. Treating cells with the DNA damaging agent, etoposide, induced Bcl-6 protein degradation (Fig. 6G). Importantly, RvD1 protected Bcl-6 from etoposide-induced degradation (Fig. 6H, I).
Figure 6. RvD1 enhances Bcl-6 expression in primary human B cells and the Ramos B lymphoma cell line.
(A–F) Purified human CD19+ B cells were pretreated with RvD1 or left untreated, followed by stimulation with the IgE-inducing cocktail and cell lysates were collected. (A) The kinetics of Bcl-6 mRNA levels and εGLT RNA levels were measured using RT-qPCR. (B) Changes in Bcl-6 levels in RvD1-treated B cells at day 1 are shown. (C) Kinetic of Bcl-6 protein expression levels in B cells stimulated with an IgE-inducing cocktail was measured by western blot. (D) B cells pretreated with RvD1 (10, 50, 100nM), and then stimulated with the IgE-inducing cocktail, were collected at day 1 and Bcl-6 protein levels were measured by western blot. GAPDH was used as a control. (E) Western blot densitometry analysis for 3 donors (mean±SEM), samples normalized to vehicle control (no treatment). (F) Western blot images of Bcl-6 at earlier time points (<24) in B cells treated with RvD1 (100 nM) from two different human donors are shown. Fold differences in Bcl-6 protein levels in RvD1 (100 nM) treated B cells compared to vehicle from one representative donor were determined by densitometry analysis. (G) The Ramos cell line was treated with different concentrations of etoposide for 24 hours, and Bcl-6 expression levels were measured by western blot. (H) Bcl-6 protein levels were measured in the Ramos cell line pretreated with RvD1 in three different concentrations, followed by stimulation with 20μM of etoposide using western blot. (I) Densitometry analysis for two separate experiments (mean±SEM). *P≤0.05 by one-way repeated measures ANOVA with Tukey’s post test.
Discussion
Here, we show that the pro-resolving lipid mediators, RvD1 and 17-HDHA, reduce human B-cell IgE production by suppressing class switching. SPMs have shown great utility in promoting resolution in preclinical animal models of inflammatory diseases, including allergic airway inflammation, cigarette smoke exposure, and peritonitis (11, 12, 24). Different SPMs appear to act via different mechanisms in a cell type-specific and disease-specific manner, promoting the resolution of inflammation. For example, in a mouse model of allergic airway inflammation, RvD1 reduces inflammation by inhibiting excessive eosinophil infiltration and Th2 cytokine production, whereas RvE1 inhibits IL-23 and Th17 cells (11, 12). Here, we show for the first time, that the SPMs directly regulate B-cell IgE production, a major step forward in promoting the potential use of SPMs as novel treatments for IgE-mediated diseases.
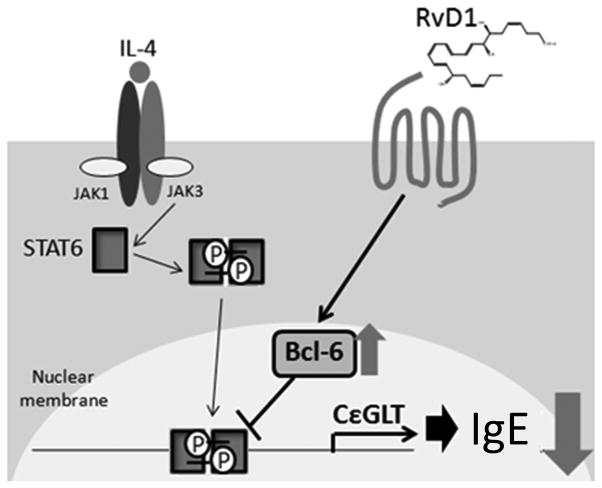
Our data show that the SPMs reduce human B-cell IgE production by reducing the number of IgE-secreting cells (Fig. 3). To determine the mechanism(s) of reduction in IgE producing cell number, we focused on B-cell Ab class switching. B-cell class switch is an essential process for the production of IgG, IgA and IgE antibody isotypes [32], which is catalyzed by lymphocyte-specific AID. However, due to the low number of IgE-producing B cells compared to IgG-producing cells in total B cell culture even with the specific stimulation, we were not able to see significant reduction in AID mRNA levels with RvD1 treatment. Class-switch to IgE is a tightly regulated process and is preceded by transcription of εGLT driven by the transcription factor STAT6. Although RvD1 blocked εGLT transcription, the effect was not directly mediated through STAT6, as it did not interfere with STAT6 phosphorylation, nuclear translocation or transcription of other STAT6 regulated genes (see Fig. 7 diagram). Most importantly, STAT6 binding activity on the εGLT promoter region was strongly reduced by RvD1 treatment, which led us to investigate other cofactors and mediators that could regulate STAT6 activity. Interestingly, a recent study reported that RvD1 enhanced IL-4 and STAT6 mediated transcriptional activation in a mouse microglial cell line [33], supporting a developing broad paradigm that the effects of SPMs are context and cell-type dependent.
Figure 7. RvD1 suppresses human B-cell class switching to IgE by stabilizing the expression of Bcl-6.
IL-4 activates the signaling pathway mediated by the transcription factor, STAT6, promoting the transcription of genes involved in B-cell Ab class switching to IgE such as εGLT, IL-4R and CD23. However, RvD1 suppresses εGLT transcription by stabilizing the expression of Bcl-6 in B cells. Bcl-6 is a transcription repressor that antagonizes the activity of STAT6 only in a limited subset of genes, with stronger activity on εGLT promoter region compared to IL-4R or CD23. Therefore, RvD1 decreases B-cell Ab class switch to IgE by specifically suppressing the transcription of εGLT.
Several other transcription factors regulate εGLT transcription including the positive regulators, NF-κB, AP-1, PU.1, C/EBP, bZip, and the inhibitory regulator, Bcl-6 [25]. Bcl-6 is a transcriptional repressor that competes with STAT6 for binding to the εGLT promoter region and therefore regulates IgE class switching and antibody production [17, 34]. For example, Bcl-6 knockout mice exhibited enhanced ability to produce IgE-secreting cells within multi-organ inflammatory disease and showed extremely high serum IgE levels [17]. Here, we investigated the effect of RvD1 on Bcl-6 using human peripheral blood B cells and the Ramos B cell lymphoma. RvD1 enhanced Bcl-6 protein levels in human B cells which in turn prevented STAT6 binding to the εGLT promoter region, reduced εGLT transcription and consequently B-cell class switching (Fig. 6 and 7). Moreover, RvD1 also rescued Bcl-6 protein levels in Ramos B lymphoma cells treated with etoposide. Etoposide is a DNA topoisomerase inhibitor that induces cell cycle arrest and apoptosis via promoting the degradation of Bcl-6 [31, 35]. RvD1 did not alter Bcl-6 mRNA levels but did prevent loss of Bcl-6 protein induced by etoposide, which suggests that RvD1 stabilizes the Bcl-6 protein. While it may also be that RvD1 increases Bcl-6 translation, furture studies will be needed to address this. In IgE-producing B cells, the affinity of Bcl-6 for different DNA promoter regions varies, with the εGLT promoter region being the highest [34]. Therefore, increased Bcl-6 protein levels in RvD1-treated B cells can impact εGLT transcription more than other STAT6-binding genes with lower Bcl-6 binding affinity. By regulating the level of Bcl-6, instead of the master transcription factor STAT6, RvD1 specifically suppresses IgE production without affecting other STAT6 regulated genes, a very provocative finding. Moreover, Bcl-6 is known to suppress excessive allergic reaction by regulating various immune cells, such as mast cells, Th2 cells and airway epithelial cells [36]. These findings serve to support the development of SPMs as novel therapeutics as they may have fewer unintended or off-target effects and may impact overall allergic immune responses.
In our study, RvD1 and 17-HDHA inhibited IgE class switching in B cells from healthy donors treated with an IgE-inducing cocktail, without adversely affecting IgM or IgG production. Interestingly, we previously reported that RvD1 and 17-HDHA increase IgM and IgG antibody production in B cells stimulated with a different polyclonal activator which mimics B-cell responses against pathogenic organisms [9]. Therefore, we suggest that the actions of SPMs in B cells depend on the microenvironment and the nature of the ligands used to stimulate the B-cell receptor. Bcl-6 does not play a role in IgG class switch, so the interaction of Bcl-6 and RvD1 may have different effects or no effect in B cells stimulated with an IgG-promoting stimulus [25]. Moreover, different stimulatory signals on B cells might induce different expression levels of SPM receptors, which would alter the ability of B cells to respond to SPMs. RvD1 is an agonist for two different receptors, GPR32 and ALX/FPR2, each of which has a different binding affinity for RvD1 and might activate different signaling pathways [1, 37]. Therefore, further studies on the expression of SPM receptors in an allergic response environment will provide additional insights into how B cells respond to SPMs.
Most asthma patients have high serum IgE levels and have circulating IgE-secreting cells. Therapeutic reduction of IgE levels has proven successful in severe asthma patients using neutralizing antibody for free IgE (e.g. omalizumab) [38]. Because IgE is a key player in allergic asthma, it represents a prime target for therapeutic intervention. For many years, the beneficial effects of omega-3 fatty acids, which are found in fish oil, have been suggested in various inflammatory diseases, including asthma. Observational studies have shown that omega-3 dietary intake improves the symptoms of asthma [39–41]. However, there has been no accepted mechanism due to lack of molecular evidence in vitro, the high concentrations needed to show an effect in vivo, and the randomly successful clinical trials that have tried to prove a benefit for omega-3 fatty acids. Therefore, SPMs such as 17-HDHA and RvD1 might be the “missing link” between the actions of omega-3 and inflammatory diseases.
Current asthma therapies mostly treat the symptoms without addressing the underlying causes of the disease. Although anti-IgE antibody therapy is used, it is very expensive, must be given intravenously, and it targets the endpoint of dysregulated antibody production, rather than the cause. Some treatments, including corticosteroids and 5-lipoxygenase inhibitors, may actively suppress production of pro-resolving mediators, while also inhibiting pro-inflammatory mediators. SPMs are endogenous natural products that specifically inhibit differentiation of IgE-producing B cells without being broadly immunosuppressive. Therefore, SPMs target one of the root causes of allergic diseases, IgE production, and represent an exciting and novel class of potential therapeutics for asthma, atopy and other allergic diseases.
Materials & Methods
B lymphocyte isolation
Human peripheral blood B cells from healthy donors were isolated as previously described [42]. Briefly, the buffy coat was separated and diluted in 1x PBS and the PBMCs were isolated using Ficoll-Paque (Amersham Biosciences, Piscataway, NJ) gradient centrifugation. B cells were then purified from the PBMCs using CD19 Dynabeads (Invitrogen, Carlsbad, CA). CD19 Dynabead-cell rosettes were disrupted using CD19 Detach beads (Invitrogen, Carlsbad, CA). Cells obtained by this method of isolation were >98% CD19+ as determined by flow cytometry. Where indicated, some experiments were performed on PBMCs. All donors gave informed written consent in accordance with the Declaration of Helsinki and the protocol was approved by the University of Rochester Research Subjects Review Board.
Reagents and culture conditions
Purified CD19+ B cells or PBMCs were cultured in RPMI 1640 (GIBCO/Invitrogen, Carlsbad, CA) supplemented with 5% fetal bovine serum, 2mM L-glutamine, 5×10−5M 2-mercaptoethanol, 10mM HEPES and 50μg/ml gentamicin. mCD40 ligand (CD154) is prepared using an insect membrane culture as previously described [43]. The final IgE-inducing cocktail that we used was composed of mCD40L (1:50 dilution), IL-4 (50ng/ml, R&D system, Minneapolis, MN) and CpG ODN2395 (0.5μg/ml, Invivogen, San Diego, CA). Resolvin D1 (RvD1, 7S,8R,17S-trihydroxy-4Z,9E,11E,13Z,15E,19Z-docosahexaenoic acid), resolvin D2 (RvD2, 7S,16R,17S-trihydroxy-4Z,8E,10Z,12E,14E,19Z-docosahexaenoic acid), lipoxin B4 (LXB4, 5S, 14R, 15S-trihydroxy-6E, 8Z, 10E, 12E-eicosatetraenoic acid), lipoxin A4 (LXA4, 5S, 6R, 15S-trihydroxy-7E, 9E, 11Z, 13E-eicosatetraenoic acid), maresin-1 (MaR-1, 7R,14S-dihydroxy-docosa-4Z,8E,10E,12Z,16Z,19Z-hexaenoic acid) and 17(R)-hydroxydocosahexaenoic acid (17-HDHA, 17R-hydroxy-4Z, 7Z, 10Z, 13Z, 15E, 19Z-docosahexaenoic acid) (Cayman Chemical Company, Ann Harbor, MI) were suspended in ethanol and supplemented in cell culture at nanomolar concentrations. Vehicle control was defined as 1x PBS with 0.03% ethanol by volume, equivalent to the highest concentration of SPM used. Cells were pretreated with either vehicle control or SPMs for 30 minutes, then were treated with an IgE-inducing cocktail or left unstimulated. Additional SPM treatments were added every 24 hours for the duration of the experiment. Etoposide (Sigma Aldrich, Saint Louis, MO) was dissolved in DMSO and added to cell culture in micromolar concentrations. Some experiments used the Ramos cell line, a human Burkitt’s lymphoma cell line (ATCC, Manassas, Virginia).
Enzyme-linked immunosorbent assays (ELISA) and IgE-specific ELISpot assay
Purified CD19+ B cells (5×105 cells/ml) were cultured in triplicate in 96-well round-bottom plates for 6 days. Antibodies in the supernatant were measured by ELISA as specified by the manufacturer (Bethyl Laboratories, Montgomery, TX). For ELISpot assay, plates (Millipore, Billerica, MA) were coated with mouse anti-human IgE antibodies (Cosmo Bio Co.) at 1:1000 dilution. The B cells were transferred to the ELISpot plate and incubated for a further 24 hours. IgE-secreting B cells were detected with alkaline phosphatase-conjugated mouse anti-human IgE antibody (Sigma Aldrich, Saint Louis, MO) at 1:1000 dilution. Plates were developed using Vector AP substrate kit III (Vector Laboratories, Burlingame, CA) and spots were counted using an ImmunoSpot Series 5 Analyzer (Cellular Technology, Shaker Heights, OH).
Cell proliferation assay
Purified CD19+ B cells were cultured in round-bottom plates (5×105 cells/ml) in triplicate, and treated with 17-HDHA or RvD1 and stimulated with an IgE-inducing cocktail. [3H]Thymidine (1μCi/well) was added 12 hours prior to harvest. Incorporation was measured with a Topcount Luminometer (PerkinElmer, Boston, MA).
Real-time PCR
Purified CD19+ B cells pretreated with SPMs and stimulated with an IgE-inducing cocktail were harvested at the indicated time points. Total RNA was extracted with a Qiagen RNAeasy mini kit (Valencia, CA) using 1×106 cells/sample. 200ng of RNA from each sample was reverse transcribed with Superscript III and random primers (Invitrogen, Carlsbad, CA). The cDNA was amplified in a real time PCR using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) and quantified with Bio-Rad icycler software. Amplification of 18S rRNA was used to normalize and quantify the relative gene expression. The primers used were as follows:
εGLT forward, 5′-CACATCCACAGGCACCAAAT-3′;
εGLT reverse, 5′-ATCACCGGCTCCGGGAAGTA-3′;
IL-4R forward, 5′-GACCTGGAGCAACCCGTATC-3′;
IL-4R reverse, 5′-CAGGTGGTGTTATAGCACTG-3′;
CD23 forward, 5′-CTGCTTAAACCTCTGTCTCTGAC-3′;
CD23 reverse, 5′-GCTTGGATTCTCCCGATGATG-3′;
Bcl6 forward, 5′-AGACCGTCCATACCGG-3′;
Bcl6 reverse, 5′-CGCAAGTGAAGTCGCA-3′;
18S rRNA forward, 5′-GTAACCCGTTGAACCCCATT-3′;
18S rRNA reverse, 5′-CCATCCAATCGGTAGTAGCG-3′;
Mature IgE forward, 5′-ACCCTGGTCACCGTCTCCTCAG-3′;
Mature IgE reverse, 5′-CAGAGTCACGGAGGTGGCATT-3′;
AID forward, 5′-CGTGACAGTGCTACATCC-3′;
AID reverse, 5′-TGCGGTCCTCACAGAAGTAG-3′.
Western blotting
Purified human B cells were lysed in RIPA buffer (150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 50mM Tris, 0.1% SDS, pH 8.0) with protease inhibitor cocktail (Sigma Aldrich, Saint Louis, MO). Protein concentration was determined using Bio-Rad DC protein assay kit (BioRad, Hercules CA). Precast SDS-PAGE gels (Pierce/Thermo Fisher Scientific, Rockford, IL) were loaded with 10–30 μg of protein and transferred to PVDF membranes (Millipore, Billerica, MA). Western blots were probed with rabbit anti-human Bcl-6 (Cell Signaling Technology, Beverly, MA) and mouse anti-human GAPDH (Calbiochem Chemicals, Gibbstown, NJ). HRP conjugated goat anti-mouse or goat anti-rabbit secondary antibodies (Jackson ImmunoResearch, West Grove, PA) were used to detect specific probed antibodies. Western blots were visualized by autoradiography after incubation with ECL (Perkin Elmer Life Sciences Inc., Boston, MA).
Flow cytometry analysis
Purified CD19+ B cells or PBMCs were pretreated with SPMs, followed by stimulation with an IgE-inducing cocktail for 5, 20, 30 and 60 minutes. Cells were fixed with 4% paraformaldehyde EM grade (Electron Microscopy Sciences, Hatfield, PA) at 37°C for 10 minutes and permeabilized with BD Phosflow™ Perm Buffer III (BD Bioscience) on ice for 30 minutes. Cells were stained with anti-CD19-FITC (BD Biosciences) and anti-STAT6 (pY641)-Alexafluor647 (BD Biosciences) antibodies and incubated on ice for 30 minutes. Cell staining was analyzed using a 12-color LSRII flow cytometer (BD Biosciences) and analyzed with FlowJo software version 7.6.5 (Tree Star, Ashland, OR).
ImageStream
CD19+ B cells isolated from healthy donors were fixed with 4% paraformaldehyde EM grade (Electron Microscopy Sciences, Hatfield, PA, USA) and permeabilized with BD Phosflow™ Perm Buffer III. Cells were stained with anti-STAT6 (pY641)-Alexafluor647 (BD Biosciences) overnight, and DAPI (Invitrogen, Carlsbad, CA) was added just before analysis. Samples were run on an ImageStream X (Amnis, Seattle, WA, USA), and analyzed using IDEAS software version 6 (Amnis).
Chromatin immunoprecipitation (ChIP assay)
Human PBMCs isolated from healthy donors were stimulated for 4hr at density of 1×106 cells/ml with the IgE-inducing cocktail and then cross-linked with 1% formaldehyde for 10 min at room temperature. A total 2×107 cells per condition were used. Cross-linking was quenched by incubation with 0.125M glycine for 5 min, and cells were washed with PBS twice, resuspended in 1ml of ChIP lysis buffer (50mM Tris-HCl pH 8.0, 1% SDS, 5mM EDTA, 1X proteinase inhibitors Complete EDTA-free) and incubated on ice for 5 min. The lysates were sonicated on ice to shear DNA using a Misonix Sonicator 3000 at highest amplitude which is with 2×30 sec sonication, then another 1×20 sec sonication. DNA fragments of 100–500 bp were obtained. 10 μl of each sample were kept as input control and the remainder of samples were 5 times diluted with dilution buffer (1% Triton X-100, 2mM EDTA, 150mM NaCl, 20mM Tris-HCl pH8.0, 1X protease inhibitor Complete EDTA-free) and precleared with 50μl slurry of protein A-agarose beads (blocked by 0.2 mg/ml salmon sperm DNA in 1mg/ml of BSA) for 3hr on a rotating wheel at 4°C. Beads were pelleted in a benchtop centrifuge at 1000rpm for 1min at 4°C, precleared extract was transferred into new tube, and 2μg of anti-STAT6 (M20) (Santa Cruz biotechnology) or 1μg rabbit IgG were added, and samples were incubated rotating overnight at 4°C. 100μl of blocked protein A beads were added to the ChIP reactions and then incubated rotating for 2hr at 4°C. Supernatants were removed and beads were washed sequentially in Paro buffer I, II, III, listed below, and then 2 times with 1X TE buffer at RT. Paro buffer I: 0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris-HCl pH 8.1, 150mM NaCl, Paro buffer II: 0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris-HCl pH 8.1, 200mM NaCl, Paro buffer III: 0.25M LiCl, 1% NP-40, 1% deoxycholate, 1mM EDTA, 10mM Tris-HCl pH 8.1. Washes were 1ml each and there were 7–10min incubation in between paro washes. Elution was performed twice with 100ul of Elution buffer (1% SDS, 0.1M NaHCO3) and tap mixed every couple minutes. 6ul of 5mg/ml of proteinase K was added to the combined elutes and the input control samples, and then incubated overnight at 65°C. The next day, DNA was purified using Qiagen PCR purification kit (Valencia, CA) and q PCR was performed using primers for Iε germline promoter region, forward, 5′-CTAGAAAGAGGCCTACACCTG-3′; reverse, 5′-GCCAGACTGTTCTTATTCG-3′.
Statistical analysis
Each experiment was repeated with cells from at least three different human donors. Results are expressed as mean ± standard errors (SEM). Statistical analyses on normally distributed data were performed using repeated measure one-way analysis of variance (ANOVA) with Tukey’s posttest. Statistical analyses were performed using Prism version 6 (GraphPad, San Diego, CA).
Supplementary Material
Acknowledgments
We would like to thank the University of Rochester Medical Center Blood Bank for collecting blood samples, and Steven J. Pollock for assistance with formatting of the figures.
This research was supported in part by NIH grants T90DE021985 (University of Rochester Medical school), R21AI103690, T32HL066988 and the Mary Parkes Center for Asthma, Allergy and Pulmonary Care.
Abbreviation used
- SPM
Specialized pro-resolving mediator
- 17-HDHA
17-hydroxydocosahexaenoic acid
- RvD1
resolvin D1
- εGLT
epsilon germline transcript
Footnotes
Conflict of Interest
The authors declare no financial or commercial conflict of interest.
References
- 1.Serhan CN. The resolution of inflammation: the devil in the flask and in the details. FASEB J. 2011;25:1441–1448. doi: 10.1096/fj.11-0502ufm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serhan CN, Chiang N, Van Dyke TE. Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat Rev Immunol. 2008;8:349–361. doi: 10.1038/nri2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serhan CN. Resolution phase of inflammation: novel endogenous anti-inflammatory and proresolving lipid mediators and pathways. Annu Rev Immunol. 2007;25:101–137. doi: 10.1146/annurev.immunol.25.022106.141647. [DOI] [PubMed] [Google Scholar]
- 4.Serhan CN, Hamberg M, Samuelsson B. Lipoxins: novel series of biologically active compounds formed from arachidonic acid in human leukocytes. Proc Natl Acad Sci U S A. 1984;81:5335–5339. doi: 10.1073/pnas.81.17.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arita M, Bianchini F, Aliberti J, Sher A, Chiang N, Hong S, Yang R, et al. Stereochemical assignment, antiinflammatory properties, and receptor for the omega-3 lipid mediator resolvin E1. J Exp Med. 2005;201:713–722. doi: 10.1084/jem.20042031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serhan CN, Arita M, Hong S, Gotlinger K. Resolvins, docosatrienes, and neuroprotectins, novel omega-3-derived mediators, and their endogenous aspirin-triggered epimers. Lipids. 2004;39:1125–1132. doi: 10.1007/s11745-004-1339-7. [DOI] [PubMed] [Google Scholar]
- 7.Serhan CN, Yang R, Martinod K, Kasuga K, Pillai PS, Porter TF, Oh SF, et al. Maresins: novel macrophage mediators with potent antiinflammatory and proresolving actions. J Exp Med. 2009;206:15–23. doi: 10.1084/jem.20081880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serhan CN, Krishnamoorthy S, Recchiuti A, Chiang N. Novel anti-inflammatory--pro-resolving mediators and their receptors. Curr Top Med Chem. 2011;11:629–647. doi: 10.2174/1568026611109060629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramon S, Gao F, Serhan CN, Phipps RP. Specialized proresolving mediators enhance human B cell differentiation to antibody-secreting cells. J Immunol. 2012;189:1036–1042. doi: 10.4049/jimmunol.1103483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dullaers M, De Bruyne R, Ramadani F, Gould HJ, Gevaert P, Lambrecht BN. The who, where, and when of IgE in allergic airway disease. J Allergy Clin Immunol. 2012;129:635–645. doi: 10.1016/j.jaci.2011.10.029. [DOI] [PubMed] [Google Scholar]
- 11.Apter AJ. Advances in adult asthma diagnosis and treatment in 2012: potential therapeutics and gene-environment interactions. J Allergy Clin Immunol. 2013;131:47–54. doi: 10.1016/j.jaci.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 12.Davis LA. Omalizumab: a novel therapy for allergic asthma. Ann Pharmacother. 2004;38:1236–1242. doi: 10.1345/aph.1D626. [DOI] [PubMed] [Google Scholar]
- 13.Allen JE, Maizels RM. Diversity and dialogue in immunity to helminths. Nat Rev Immunol. 2011;11:375–388. doi: 10.1038/nri2992. [DOI] [PubMed] [Google Scholar]
- 14.Rogerio AP, Haworth O, Croze R, Oh SF, Uddin M, Carlo T, Pfeffer MA, et al. Resolvin D1 and aspirin-triggered resolvin D1 promote resolution of allergic airways responses. J Immunol. 2012;189:1983–1991. doi: 10.4049/jimmunol.1101665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haworth O, Cernadas M, Yang R, Serhan CN, Levy BD. Resolvin E1 regulates interleukin 23, interferon-gamma and lipoxin A4 to promote the resolution of allergic airway inflammation. Nat Immunol. 2008;9:873–879. doi: 10.1038/ni.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnamoorthy N, Burkett PR, Dalli J, Abdulnour RE, Colas R, Ramon S, Phipps RP, et al. Cutting Edge: Maresin-1 Engages Regulatory T Cells To Limit Type 2 Innate Lymphoid Cell Activation and Promote Resolution of Lung Inflammation. J Immunol. 2014 doi: 10.4049/jimmunol.1402534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris MB, Mostecki J, Rothman PB. Repression of an interleukin-4-responsive promoter requires cooperative BCL-6 function. J Biol Chem. 2005;280:13114–13121. doi: 10.1074/jbc.M412649200. [DOI] [PubMed] [Google Scholar]
- 18.Caven TH, Shelburne A, Sato J, Chan-Li Y, Becker S, Conrad DH. IL-21 dependent IgE production in human and mouse in vitro culture systems is cell density and cell division dependent and is augmented by IL-10. Cell Immunol. 2005;238:123–134. doi: 10.1016/j.cellimm.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Caven TH, Sturgill JL, Conrad DH. BCR ligation antagonizes the IL-21 enhancement of anti-CD40/IL-4 plasma cell differentiation and IgE production found in low density human B cell cultures. Cell Immunol. 2007;247:49–58. doi: 10.1016/j.cellimm.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harada M, Magara-Koyanagi K, Watarai H, Nagata Y, Ishii Y, Kojo S, Horiguchi S, et al. IL-21-induced Bepsilon cell apoptosis mediated by natural killer T cells suppresses IgE responses. J Exp Med. 2006;203:2929–2937. doi: 10.1084/jem.20062206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozcan E, Rauter I, Garibyan L, Dillon SR, Geha RS. Toll-like receptor 9, transmembrane activator and calcium-modulating cyclophilin ligand interactor, and CD40 synergize in causing B-cell activation. J Allergy Clin Immunol. 2011;128:601–609. e601–604. doi: 10.1016/j.jaci.2011.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gantner F, Hermann P, Nakashima K, Matsukawa S, Sakai K, Bacon KB. CD40-dependent and -independent activation of human tonsil B cells by CpG oligodeoxynucleotides. Eur J Immunol. 2003;33:1576–1585. doi: 10.1002/eji.200323444. [DOI] [PubMed] [Google Scholar]
- 23.Bernasconi NL, Traggiai E, Lanzavecchia A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science. 2002;298:2199–2202. doi: 10.1126/science.1076071. [DOI] [PubMed] [Google Scholar]
- 24.Pene J, Rousset F, Briere F, Chretien I, Paliard X, Banchereau J, Spits H, et al. IgE production by normal human B cells induced by alloreactive T cell clones is mediated by IL-4 and suppressed by IFN-gamma. J Immunol. 1988;141:1218–1224. [PubMed] [Google Scholar]
- 25.Hellman L. Regulation of IgE homeostasis, and the identification of potential targets for therapeutic intervention. Biomed Pharmacother. 2007;61:34–49. doi: 10.1016/j.biopha.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Jabara HH, Schneider LC, Shapira SK, Alfieri C, Moody CT, Kieff E, Geha RS, et al. Induction of germ-line and mature C epsilon transcripts in human B cells stimulated with rIL-4 and EBV. J Immunol. 1990;145:3468–3473. [PubMed] [Google Scholar]
- 27.Ichiki T, Takahashi W, Watanabe T. Regulation of the expression of human C epsilon germline transcript. Identification of a novel IL-4 responsive element. J Immunol. 1993;150:5408–5417. [PubMed] [Google Scholar]
- 28.Linehan LA, Warren WD, Thompson PA, Grusby MJ, Berton MT. STAT6 is required for IL-4-induced germline Ig gene transcription and switch recombination. J Immunol. 1998;161:302–310. [PubMed] [Google Scholar]
- 29.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 30.Cattoretti G, Chang CC, Cechova K, Zhang J, Ye BH, Falini B, Louie DC, et al. BCL-6 protein is expressed in germinal-center B cells. Blood. 1995;86:45–53. [PubMed] [Google Scholar]
- 31.Phan RT, Saito M, Kitagawa Y, Means AR, Dalla-Favera R. Genotoxic stress regulates expression of the proto-oncogene Bcl6 in germinal center B cells. Nat Immunol. 2007;8:1132–1139. doi: 10.1038/ni1508. [DOI] [PubMed] [Google Scholar]
- 32.Nacionales DC, Weinstein JS, Yan XJ, Albesiano E, Lee PY, Kelly-Scumpia KM, Lyons R, et al. B cell proliferation, somatic hypermutation, class switch recombination, and autoantibody production in ectopic lymphoid tissue in murine lupus. J Immunol. 2009;182:4226–4236. doi: 10.4049/jimmunol.0800771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L, Wu Y, Wang Y, Wu J, Song L, Xian W, Yuan S, et al. Resolvin D1 promotes the interleukin-4-induced alternative activation in BV-2 microglial cells. J Neuroinflammation. 2014;11:72. doi: 10.1186/1742-2094-11-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris MB, Chang CC, Berton MT, Danial NN, Zhang J, Kuehner D, Ye BH, et al. Transcriptional repression of Stat6-dependent interleukin-4-induced genes by BCL-6: specific regulation of iepsilon transcription and immunoglobulin E switching. Mol Cell Biol. 1999;19:7264–7275. doi: 10.1128/mcb.19.10.7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shao Y, Ling CC, Liu XQ. High concentrations of glucose suppress etoposide-induced cell death of B-cell lymphoma through BCL-6. Biochem Biophys Res Commun. 2014;450:227–233. doi: 10.1016/j.bbrc.2014.05.096. [DOI] [PubMed] [Google Scholar]
- 36.Arima M, Fukuda T, Tokuhisa T. Role of the Transcriptional Repressor BCL6 in Allergic Response and Inflammation. World Allergy Organ J. 2008;1:115–122. doi: 10.1097/WOX.0b013e31817dc522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norling LV, Dalli J, Flower RJ, Serhan CN, Perretti M. Resolvin D1 limits polymorphonuclear leukocyte recruitment to inflammatory loci: receptor-dependent actions. Arterioscler Thromb Vasc Biol. 2012;32:1970–1978. doi: 10.1161/ATVBAHA.112.249508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D’Amato G, Stanziola A, Sanduzzi A, Liccardi G, Salzillo A, Vitale C, Molino A, et al. Treating severe allergic asthma with anti-IgE monoclonal antibody (omalizumab): a review. Multidiscip Respir Med. 2014;9:23. doi: 10.1186/2049-6958-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D’Auria E, Miraglia Del Giudice M, Barberi S, Mandelli M, Verduci E, Leonardi S, Riva E, et al. Omega-3 fatty acids and asthma in children. Allergy Asthma Proc. 2014;35:233–240. doi: 10.2500/aap.2014.35.3736. [DOI] [PubMed] [Google Scholar]
- 40.Hodge L, Salome CM, Hughes JM, Liu-Brennan D, Rimmer J, Allman M, Pang D, et al. Effect of dietary intake of omega-3 and omega-6 fatty acids on severity of asthma in children. Eur Respir J. 1998;11:361–365. doi: 10.1183/09031936.98.11020361. [DOI] [PubMed] [Google Scholar]
- 41.Nagakura T, Matsuda S, Shichijyo K, Sugimoto H, Hata K. Dietary supplementation with fish oil rich in omega-3 polyunsaturated fatty acids in children with bronchial asthma. Eur Respir J. 2000;16:861–865. doi: 10.1183/09031936.00.16586100. [DOI] [PubMed] [Google Scholar]
- 42.Ryan EP, Pollock SJ, Murant TI, Bernstein SH, Felgar RE, Phipps RP. Activated human B lymphocytes express cyclooxygenase-2 and cyclooxygenase inhibitors attenuate antibody production. J Immunol. 2005;174:2619–2626. doi: 10.4049/jimmunol.174.5.2619. [DOI] [PubMed] [Google Scholar]
- 43.Kehry MR, Castle BE. Regulation of CD40 ligand expression and use of recombinant CD40 ligand for studying B cell growth and differentiation. Semin Immunol. 1994;6:287–294. doi: 10.1006/smim.1994.1037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.