Abstract
The infection status of 15 viruses in 120 pigs aged about 6 months was investigated based on tonsil specimens collected from a slaughterhouse. Only 5 species of porcine parvoviruses and porcine circovirus type 2 (PCV2) were detected at high frequencies; 67% for porcine parvovirus (PPV) (PPV-Kr or -NADL2 as the new abbreviation), 58% for PPV2 (CnP-PARV4), 39% for PPV3 (P-PARV4), 33% for PPV4 (PPV4), 55% for PBo-likeV (PBoV7) and 80% for PCV2. A phylogenetic analysis of PPV3 suggested that Japanese PPV3s showed a slight variation, and possibly, there were farms harboring homogeneous or heterogeneous PPV3s. Statistical analyses indicated that the detection of PCV2 was significantly coincidental with each detection of PPV, PPV2 and PPV3, and PPV and PPV4 were also coincidentally detected. The concurrent infection with PCV2 and porcine parvoviruses in the subclinically infected pigs may resemble the infection status of pigs with the clinical manifestations of porcine circovirus associated disease which occurs in 3–5 months old pigs and is thought to be primarily caused by the PCV2 infection.
Keywords: coinfection, porcine circovirus associated disease, porcine circovirus type 2, porcine parvoviruses, prevalence
A number of new parvoviruses have been identified during the past 15 years and given various names, and thereby, the updated taxonomy of the family Parvoviridae was proposed in 2014 [6]. The classical porcine parvovirus (PPV), which was first identified in the 1960s [4] and now present worldwide, causes embryonic death, stillbirths and mummification when embryos or fetuses in seronegative dams are infected. The newly identified porcine parvoviruses have been detected in various areas of the world, but its relationship with any diseases remains unclear. PPV is thought to be one of the cofactors for porcine circovirus associated disease (PCVAD) whose main etiologic agent is porcine circovirus type 2 (PCV2) [1, 10, 13]. The PCV2 infection alone does not cause a clinical disease, but concurrent viral or bacterial infections may augment the severity of PCVAD possibly through stimulating the PCV2 replication or suppressing the PCV2 clearance by altered cytokine regulation [8, 9, 18].
During our screening for known viral genomes and newly identified porcine parvovirus genomes in specimens of apparently healthy pigs, we found and now report that the genomes of PCV2 and the classical and new porcine parvoviruses were coincidentally detected. The 5 porcine parvoviruses we studied include PPV [4], PPV2 [11], PPV3 [14], PPV4 [5] and porcine bocavirus-like virus (PBo-likeV) [2]. According to the proposed taxonomy of the family Parvoviridae [6], most of the virus names have been changed as indicated in Table 1. However, we use the previous abbreviations in this paper to avoid confusion.
Table 1. Prevalence of 15 virus genomes in 120 pigs.
| Virus | Abbreviation | Prevalence (%, n=120) |
|---|---|---|
| porcine parvovirus (porcine parvovirus)a) | PPV | 67 |
| porcine parvovirus 2 (porcine Cn virus)a) | PPV2 (CnP-PARV4)a) | 58 |
| porcine parvovirus 3 (porcine hokovirus)a) | PPV3 (P-PARV4)a) | 39 |
| porcine parvovirus 4 (porcine parvovirus 4)a) | PPV4 (PPV4)a) | 33 |
| porcine boca-like virus (porcine bocavirus 7)a) | PBo-likeV (PBoV7)a) | 55 |
| porcine circovirus 2 | PCV2 | 80 |
| suid herpesvirus 1 | SuHV1 | 0 |
| hepatitis E virus | HEV | 0 |
| swine influenza virus | SIV | 0 |
| porcine reproductive and respiratory syndrome virus | PRRSV | 0 |
| Japanese encephalitis virus | JEV | 0 |
| porcine epidemic diarrhea virus | PEDV | 0 |
| porcine rotavirus A | PoRV-A | 0 |
| transmissible gastroenteritis virus | TGEV | 0 |
| Getah virus | GETV | 0 |
MATERIALS AND METHODS
Sample collection and viral nucleic acid purification: Tonsil specimens from 120 pigs were collected from a slaughterhouse in 2010 when most of the pigs were probably not injected with the inactivated PCV2 vaccine in Japan. The pigs were about 6 months old and obtained from 22 farms with 1–10 samples per farm.
The procedures for the viral DNA and RNA isolation were previously described [23]. Briefly, the tonsil homogenates were prepared using a Micro Smash machine (Tomy Seiko, Tokyo, Japan), and after centrifugation at 15,000 g for 15 min, aliquots of the supernatant were stored at −80°C. The viral DNA and RNA were isolated by a DNA/RNA purification machine, Magtration System 6GC (Precision System Science, Chiba, Japan) and a solution kit, GC series Magtration-MagaZorb RNA Common Kit (Precision System Science). The isolated nucleic acids were reverse-transcribed by Superscript II reverse transcriptase and primers of random hexamers according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, U.S.A.) and used as templates for the various PCRs detecting the genomes of both the DNA and RNA viruses. For the validation of reverse transcription-PCR assay, control RNA and PCR primers of the kit were used in each experiment.
Detection of viral genome by PCR: Two multiplex PCRs for 3 DNA viruses (porcine circovirus type 2 (PCV2), suid herpesvirus 1 and porcine parvovirus (PPV)) and 6 RNA viruses (porcine reproductive and respiratory syndrome virus (PRRSV), Japanese encephalitis virus, porcine rotavirus A (PoRV-A), porcine epidemic diarrhea virus (PEDV), transmissible gastroenteritis virus (TGEV) and Getah virus) were performed in separate tubes according to the published method [17]. Other PCR primer pairs included; NP1200 and NP1529 for swine influenza virus [15], HE5-1 and HE5-4m for hepatitis E virus [26], Q1 F and Q2 R for porcine parvovirus 2 [11], PPV3 F and PPV3 R for porcine parvovirus 3 [25], PPV4 F and PPV4 R for porcine parvovirus 4 [25], and SbocaF and SbocaR for PBo-likeV [32].
Viral genomes were amplified by PCR using Quick Taq HS DyeMix (Toyobo, Osaka, Japan) including Taq polymerase. The PCR consisted of an initial enzyme activation step at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec, extension at 72°C for 30 sec and a final extension at 72°C for 7 min.
Phylogenetic analysis: For the phylogenetic analysis of PPV3, the 622 bp of the VP gene were examined after amplifying the 713 bp fragment of the VP region (nucleotide positions from 3,359 to 4,072 of the stain AB916464) with the PCR primers PPV3 P7F (3′-GGGGCACTCATTTCTCTGAT-5′) and PPV3 P7R (3′-CTGGCCTTTTCCACTTAGGA-5′) [25] and sequencing with both primers and the two internal primers PPV3 7F2 (3′- GGAGAATAATGTTCTTCCTC-5′) and PPV3 7R2 (3′-TCGTACTCATCAAGCAGCTG-5′).
The sequence data and phylogenetic tree were compiled and analyzed using MEGA 5.1 [27] and Genetyx (Genetyx Co., Tokyo, Japan). The phylogenetic trees were generated by the maximum likelihood method.
The partial sequences of the PCR products of PPV3 have been deposited in DDBJ under accession numbers from LC011459 to LC011478.
Statistical analysis: Chi-square tests were used to evaluate the statistical significance of co-isolation of two viral genomes among PPV, PPV2, PPV3, PPV4, PBo-likeV and PCV2 by dividing two categories, PCR-positive and PCR-negative individuals for each virus. P values of <0.05 were considered statistically significant.
RESULTS
Prevalence of porcine parvoviruses and porcine circovirus 2 in 120 Japanese pigs: We previously analyzed the prevalence of the PPV2 genomes in the tonsil specimens from 120 pigs [23]. With the same specimens, we extended such a screening for 14 other viral genomes as listed in Table 1. Five of the 14 viral genomes were detected; four were members of the family Parvoviridae and another one was PCV2. The prevalences were 67% for PPV, 39% for PPV3, 33% for PPV4, 55% for PBo-likeV and 80% for PCV2 (Table 1), in addition to 58% for PPV2 [23]. Multiple viral genomes were detected from the individual pigs, and thereby, as for the 5 examined porcine parvoviruses, 3%, 23%, 53%, 78% and 93% of the pigs were positive for more than 5, 4, 3, 2 and 1 virus (es), respectively. Only 9 pigs of various farms were negative for the 5 parvovirus DNAs, and 4 of the 9 pigs were negative for PCV2 DNA. Among the 8 farms with larger sample numbers (8 to 10 samples per farm), 7 farms were positive for all 5 parvoviruses, and one farm was negative (0/10) for only one (PPV4) of the 5 parvoviruses. The results suggested that a high proportion of the pigs in most farms were co-infected with the five parvoviruses and PCV2.
We tested the possibility that the detections of these highly prevalent viruses were random or coincidental. The chi square analyses indicated that PCV2 was coincidentally detected with PPV (χ2=5.86, P<0.02), PPV2 (χ2=4.91, P<0.03) or PPV3 (χ2=4.23, P<0.04) and that PPV and PPV4 were also coincidentally detected (χ2=6.15, P<0.02) (Table 2).
Table 2. Chi square analysis for coincidental detection among genomes of 4 parvoviruses and PCV2.
| Relationship between two viruses | Number of pigs | χ2 value | P value | Significance | ||||
|---|---|---|---|---|---|---|---|---|
| +/+ | −/+ | +/− | −/− | |||||
| PPV / | PPV2 | 47 | 22 | 33 | 18 | 0.153 | 0.695 | |
| PPV3 | 33 | 14 | 47 | 26 | 0.437 | 0.508 | ||
| PPV4 | 32 | 7 | 48 | 33 | 6.154 | 0.013 | * a | |
| PBo-likeV | 46 | 20 | 34 | 20 | 0.606 | 0.436 | ||
| PCV2 | 69 | 27 | 11 | 13 | 5.859 | 0.015 | * | |
| PPV2 / | PPV3 | 28 | 19 | 41 | 32 | 0.136 | 0.712 | |
| PPV4 | 25 | 14 | 44 | 37 | 1.031 | 0.310 | ||
| PBo-likeV | 39 | 27 | 30 | 24 | 0.152 | 0.697 | ||
| PCV2 | 60 | 36 | 9 | 15 | 4.910 | 0.027 | * | |
| PPV3 / | PPV4 | 15 | 24 | 32 | 49 | 0.012 | 0.913 | |
| PBo-likeV | 28 | 38 | 19 | 35 | 0.653 | 0.419 | ||
| PCV2 | 42 | 54 | 5 | 19 | 4.232 | 0.040 | * | |
| PPV4 / | PBo-likeV | 26 | 40 | 13 | 41 | 3.177 | 0.075 | |
| PCV2 | 32 | 64 | 7 | 17 | 0.152 | 0.697 | ||
| PBo-likeV/ | PCV2 | 53 | 43 | 13 | 11 | 0.008 | 0.927 | |
a, *: significant (0.01<P<0.05). Others without asterisk mean not significant (P>0.05).
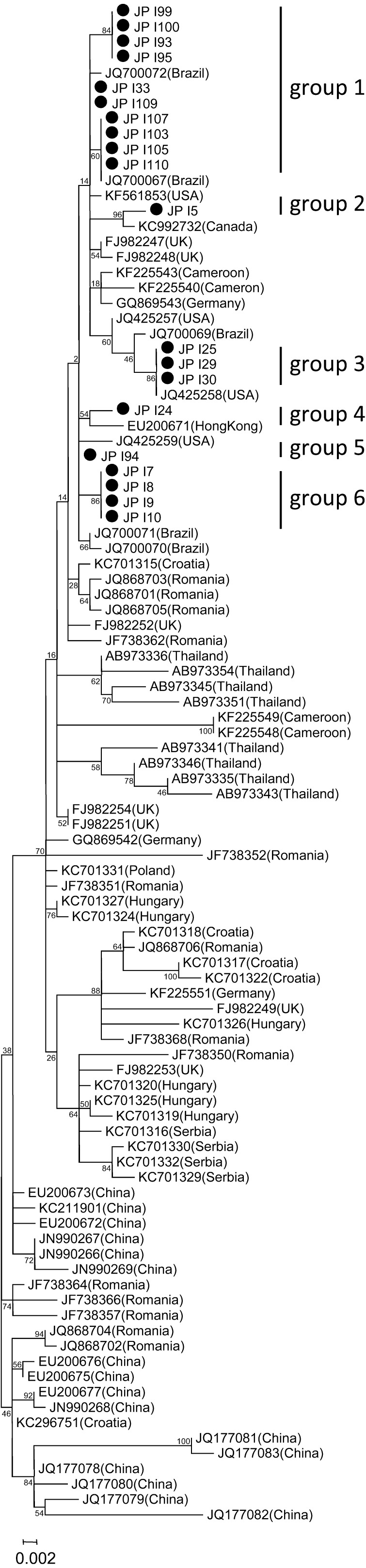
Nucleotide sequence diversity of PPV3: In order to know the genetic diversity of the Japanese PPV3s, a phylogenetic analysis based on the 622 bases of the VP gene was performed using 20 Japanese samples, 5 samples each from 4 farms and 87 reference sequences from around the world. The Japanese PPV3s were slightly diverged in the phylogenetic tree with 1.6% (10/622 bases) of the maximum nucleotide difference and closely related to the other PPV3s detected in Europe, North America, South America and Hong Kong (Fig. 1).
Fig. 1.
The phylogenetic tree was constructed, based on the 622 bases of the PPV3 VP gene, with the 20 Japanese PPV3s and 87 PPV3s currently deposited in the data bank. For the Japanese sequences, the 6 tentative sequence groups (Sequence groups 1–6) were defined by phylogenetic branch and % nucleotide difference, i.e.,<0.3% (2/622) within each sequence group. The relationship between the farm and the sequence group of the detected PPV3 sequences is indicated in Table 3.
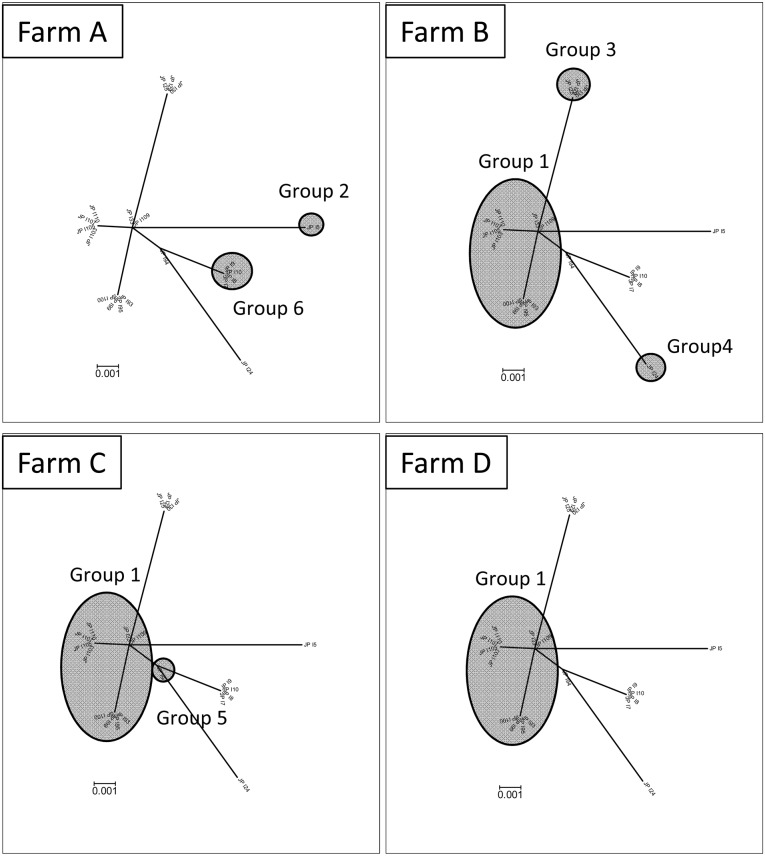
To characterize the variation in the nucleotide sequence among the farms or within a farm, the 20 Japanese PPV3 sequences were tentatively separated into 6 sequence groups based on thier phylogenetic branch and % nucleotide difference (Figs. 1, 2 and Table 3). The sequence group 1 was a major one to which 10 of the 20 sequences belonged. Farms C and D appeared homogeneous, having 4 and 5 sequences of the sequence group 1, respectively, while farms A and B had relatively heterogeneous PPV3 sequences. Farm A had the sequence groups 2 and 6 with a 1.3% nucleotide difference (8/622 bases), and farm B had the sequence groups 1, 3 and 4 with 0.6–1.4% nucleotide differences (4–9/622 bases) (Table 3).
Fig. 2.
The phylogenetic tree was constructed with the 20 Japanese PPV3s detected from the 4 farms based on the 622 bases of the PPV3 VP gene. The 6 sequence groups were tentatively defined by the phylogenetic branch and % nucleotide difference, i.e., <0.5% (3/622) within each sequence group. The 6 sequence groups detected from the 4 pig farms are indicated. The relationship among the sequence data, the sequence group and the farm is indicated in Table 3.
Table 3. Nucleotide sequence diversity of PPV3 within 4 farms.
| Farm | “sequence group” | Nucleotide difference | |||||
|---|---|---|---|---|---|---|---|
| 1a) | 2 | 3 | 4 | 5 | 6 | ||
| A | JP I5 | JP I7 | Sequence groups 2 vs 6: 1.3% (8/622) | ||||
| JP I8 | |||||||
| JP I9 | |||||||
| JP I10 | |||||||
| B | JP I33 | JP I25 | JP I24 | Sequence groups 1 vs 3: 0.8% (5/622) | |||
| JP I29 | Sequence groups 1 vs 4: 0.6% (4/622) | ||||||
| JP I30 | Sequence groups 3 vs 4: 1.4% (9/622) | ||||||
| C | JP I93 | JP I94 | Sequence groups 1 vs 5: 0.5% (3/622) | ||||
| JP I95 | |||||||
| JP I99 | |||||||
| JP I100 | |||||||
| D | valign="middle"JP I103 | ||||||
| JP I105 | |||||||
| JP I107 | |||||||
| JP I110 | |||||||
| (JP I109)b) | |||||||
PCR products were directly sequenced, and the sequence data of the 622 bases of the PPV3 VP gene were subjected to a phylogenetic analysis. The “sequence group” in this analysis was defined by the phylogenetic branch (Fig. 1) and % nucleotide difference. In this table, nucleotide sequences in a box were identical, except for the JP I109 sequence of the sequence group 1 in farm D. a), In the “sequence group 1”, the JP I33 sequence of farm B was identical to the JP I109 sequence of farm D, which are located at the center of the phylogenetic tree of Fig. 2. The 4 sequences of farm C (JP I93, JP I95, JP I99 and JP I100) were identical, and the 4 sequences of farm D (JP I103, JP I105, JP I107 and JP I110) were also identical. The sequence of JP I33 and JP I109 differed by 2 bases from the 4 identical sequences of farm C and by 1 base from the 4 identical sequences of farm D. The 4 sequences of farm C and the 4 sequences of farm D differed by 3 bases. b), the JP I109 sequence was slightly different (0.2% (1/622)) from the other 4 sequences at the same farm.
DISCUSSION
The present study, together with our previous study [23], examined the tonsil specimens of 120 apparently healthy pigs for the screening of 15 viruses which can infect pigs. Only the five porcine parvoviruses, i.e., PPV, PPV2, PPV3, PPV4 and PBo-likeV, and PCV2 were detected, and their prevalences were quite high, ranging from 33% to 80% (Table 1) [23]. The high prevalences of the classical PPV and PCV2 at the age of about 6 months are common in most pig-producing countries, whereas the prevalences of PPV3, PPV4 and PBo-likeV are the first observations in Japanese pigs.
The PPV3 DNA was detected in 39% of the 120 Japanese pigs (Table 1). Since the first identification of PPV3 [14], the prevalence has been reported in several countries and appears to widely vary from lower frequencies (6–20%) in Hungary [7], China [21], the U.S.A. [29] and Germany [25] to higher frequencies (44–73%) in Hong Kong [14], China [16] and Thailand [22].
The prevalence of the PPV4 genome was 33% in this study which is comparable to the prevalence (44%) in Thailand [22], but higher than those of several other countries, that is, 1% in China [12], 6% in Hungary [7], 7% in Germany [25] and 4% in the U.S.A. [30].
PBo-likeV [2], which was also called PBoV (PBoV-SX) [31] or PBoV1 [24, 33], is one of several porcine bocaviruses which have recently been discovered [28]. The PBo-likeV infection was initially supposed to be associated with respiratory tract diseases in pigs due to the remarkable difference in the prevalences between sick (39% (74/191)) and healthy (7% (3/41)) pigs [32]. The prevalence of PBo-likeV was 55% in our study (Table 1), in contrast to 18% in Thailand [22], 7% in China [32], 63% in different areas of China [24], 2% in Romania [7] and 13% in the wild boars of Romania [3].
Although the prevalences of PPV3, PPV4 and PBo-likeV show some variation among countries, the available data suggest that these newly identified parvoviruses have already spread worldwide.
The phylogenetic analysis of PPV3 suggested that, compared to the variation of 87 sequences deposited from around the world, the 20 Japanese sequences were less variable and belonged to limited branches (Fig. 1). In the 4 farms we analyzed, 2 farms appeared to have heterogeneous PPV3s (Table 3 and Fig. 2). Although the variations within the farms were not high, this raises the possibility that the observed variation within a farm resulted from multiple invasions of different strains rather than natural mutations within a farm after the invasion of one strain. The coexistence of different strains in a farm and coinfection of a pig with different strains must be risk factors for vaccine strategies and generation of a new recombinant virus strain.
The PCV2 genome was detected at a high frequency (80%) (Table 1) which is common worldwide. Interestingly, PCV2 was coincidentally detected along with PPV, PPV2 or PPV3, and PPV and PPV4 were also coincidentally detected (Table 2). These associations were weak, but statistically significant (0.01<P<0.05). Recently, similar associations were observed in pigs with PCVAD; the prevalences of the PPV and PPV2 DNAs were significantly higher in the PCVAD cases containing high amounts of PCV2 DNA than in the non-PCVAD cases, while, in contrast to our data, PPV3, PPV4 and PPV5 were not correlated with the amount of PCV2 [20]. The major difference between the two studies is that they analyzed the lungs of pigs with PCVAD probably aged 3–5 months while we used the tonsils of subclinical pigs aged about 6 months.
PCV2 is recognized as a causative agent of PCVAD. The clinical features of PCVAD or formerly called postweaning multisystemic wasting syndrome (PMWS) caused by PCV2 are systemic including enlargement of the lymph nodes, progressive loss of body weight or wasting combined with difficulty in breathing, diarrhea, pale skin and jaundice [9, 19]. The histopathologic changes in the affected lymphoid tissues are a severe lymphoid depletion, a diffuse infiltration of histiocytic cells and various inflammatory lesions. The pathogenesis of PCVAD or PCV2-induced diseases is complex, probably involving PCV2 infection and cofactors, such as other infections and altered cytokine or immune responses [8]. Particularly, the concurrent infection of PCV2-infected pigs by viruses (PPV, PRRSV, etc.), bacteria (Mycoplasma hyopneumoniae) or parasites may not be only a secondary infection after PCV2-induced depletion of lymphocytes, but could be important for the disease manifestation [18]. The experimental inoculation with PCV2 and PPV, but not PCV2 alone, could reproduce lesions similar to those of the field cases of PMWS [1, 10, 13]. The mechanism for the synergetic effect of coinfection was proposed that coinfection may promote the PCV2 infection by stimulating immune cells and providing target cells for the PCV2 replication or suppressing the PCV2 clearance by alteration of the cytokine production and profiles [1, 18].
The coincidental detections of PCV2 and PPVs in various combinations have been observed in both pigs with PCVAD at 3–5 months old and healthy pigs at about 6 months old. Therefore, the two stages may share a common mechanism for the proliferation of these viruses regardless of the presence or absence of PCVAD. Since circovirus and parvovirus are both DNA viruses, which require actively proliferating cells for efficient viral replication, lymphoproliferation or immunosuppression induced by infection with a virus could support the growth of other viruses.
Acknowledgments
We thank Dr. Masumi Kamiya and other members of Ibaraki Ken-sei Meat Inspection Office for their support to collect the pig samples. This study was supported in part by Grant-in-Aids for Scientific Research from the Ministry of Health, Labor and Welfare (H20-Shokuhin-Ippan-014).
REFERENCES
- 1.Allan G. M., Kennedy S., McNeilly F., Foster J. C., Ellis J. A., Krakowka S. J., Meehan B. M., Adair B. M.1999. Experimental reproduction of severe wasting disease by co-infection of pigs with porcine circovirus and porcine parvovirus. J. Comp. Pathol. 121: 1–11. doi: 10.1053/jcpa.1998.0295 [DOI] [PubMed] [Google Scholar]
- 2.Blomström A. L., Belák S., Fossum C., McKillen J., Allan G., Wallgren P., Berg M.2009. Detection of a novel porcine boca-like virus in the background of porcine circovirus type 2 induced postweaning multisystemic wasting syndrome. Virus Res. 146: 125–129. doi: 10.1016/j.virusres.2009.09.006 [DOI] [PubMed] [Google Scholar]
- 3.Cadar D., Cságola A., Lorincz M., Tombácz K., Kiss T., Spînu M., Tuboly T.2011. Genetic detection and analysis of porcine bocavirus type 1 (PoBoV1) in European wild boar (Sus scrofa). Virus Genes 43: 376–379. doi: 10.1007/s11262-011-0650-4 [DOI] [PubMed] [Google Scholar]
- 4.Cartwright S. F., Huck R. A.1967. Viruses isolated in association with herd infertility, abortions and stillbirths in pigs. Vet. Rec. 81: 196–197. [Google Scholar]
- 5.Cheung A. K., Wu G., Wang D., Bayles D. O., Lager K. M., Vincent A. L.2010. Identification and molecular cloning of a novel porcine parvovirus. Arch. Virol. 155: 801–806. doi: 10.1007/s00705-010-0646-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotmore S. F., Agbandje-McKenna M., Chiorini J. A., Mukha D. V., Pintel D. J., Qiu J., Soderlund-Venermo M., Tattersall P., Tijssen P., Gatherer D., Davison A. J.2014. The family Parvoviridae. Arch. Virol. 159: 1239–1247. doi: 10.1007/s00705-013-1914-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cságola A., Lőrincz M., Cadar D., Tombácz K., Biksi I., Tuboly T.2012. Detection, prevalence and analysis of emerging porcine parvovirus infections. Arch. Virol. 157: 1003–1010. doi: 10.1007/s00705-012-1257-3 [DOI] [PubMed] [Google Scholar]
- 8.Darwich L., Mateu E.2012. Immunology of porcine circovirus type 2 (PCV2). Virus Res. 164: 61–67. doi: 10.1016/j.virusres.2011.12.003 [DOI] [PubMed] [Google Scholar]
- 9.Ellis J.2014. Porcine circovirus: a historical perspective. Vet. Pathol. 51: 315–327. doi: 10.1177/0300985814521245 [DOI] [PubMed] [Google Scholar]
- 10.Ellis J., Krakowka S., Lairmore M., Haines D., Bratanich A., Clark E., Allan G., Konoby C., Hassard L., Meehan B., Martin K., Harding J., Kennedy S., McNeilly F.1999. Reproduction of lesions of postweaning multisystemic wasting syndrome in gnotobiotic piglets. J. Vet. Diagn. Invest. 11: 3–14. doi: 10.1177/104063879901100101 [DOI] [PubMed] [Google Scholar]
- 11.Hijikata M., Abe K., Win K. M., Shimizu Y. K., Keicho N., Yoshikura H.2001. Identification of new parvovirus DNA sequence in swine sera from Myanmar. Jpn. J. Infect. Dis. 54: 244–245. [PubMed] [Google Scholar]
- 12.Huang L., Zhai S. L., Cheung A. K., Zhang H. B., Long J. X., Yuan S. S.2010. Detection of a novel porcine parvovirus, PPV4, in Chinese swine herds. Virol. J. 7: 333. doi: 10.1186/1743-422X-7-333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J., Choi C., Chae C.2003. Pathogenesis of postweaning multisystemic wasting syndrome reproduced by co-infection with Korean isolates of porcine circovirus 2 and porcine parvovirus. J. Comp. Pathol. 128: 52–59. doi: 10.1053/jcpa.2002.0605 [DOI] [PubMed] [Google Scholar]
- 14.Lau S. K., Woo P. C., Tse H., Fu C. T., Au W. K., Chen X. C., Tsoi H. W., Tsang T. H., Chan J. S., Tsang D. N., Li K. S., Tse C. W., Ng T. K., Tsang O. T., Zheng B. J., Tam S., Chan K. H., Zhou B., Yuen K. Y.2008. Identification of novel porcine and bovine parvoviruses closely related to human parvovirus 4. J. Gen. Virol. 89: 1840–1848. doi: 10.1099/vir.0.2008/000380-0 [DOI] [PubMed] [Google Scholar]
- 15.Lee M. S., Chang P. C., Shien J. H., Cheng M. C., Shieh H. K.2001. Identification and subtyping of avian influenza viruses by reverse transcription-PCR. J. Virol. Methods 97: 13–22. doi: 10.1016/S0166-0934(01)00301-9 [DOI] [PubMed] [Google Scholar]
- 16.Li S., Wei Y., Liu J., Tang Q., Liu C.2013. Prevalence of porcine hokovirus and its co-infection with porcine circovirus 2 in China. Arch. Virol. 158: 1987–1991. doi: 10.1007/s00705-013-1690-y [DOI] [PubMed] [Google Scholar]
- 17.Ogawa H., Taira O., Hirai T., Takeuchi H., Nagao A., Ishikawa Y., Tuchiya K., Nunoya T., Ueda S.2009. Multiplex PCR and multiplex RT-PCR for inclusive detection of major swine DNA and RNA viruses in pigs with multiple infections. J. Virol. Methods 160: 210–214. doi: 10.1016/j.jviromet.2009.05.010 [DOI] [PubMed] [Google Scholar]
- 18.Opriessnig T., Halbur P. G.2012. Concurrent infections are important for expression of porcine circovirus associated disease. Virus Res. 164: 20–32. doi: 10.1016/j.virusres.2011.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Opriessnig T., Langohr I.2013. Current state of knowledge on porcine circovirus type 2-associated lesions. Vet. Pathol. 50: 23–38. doi: 10.1177/0300985812450726 [DOI] [PubMed] [Google Scholar]
- 20.Opriessnig T., Xiao C. T., Gerber P. F., Halbur P. G.2014. Identification of recently described porcine parvoviruses in archived North American samples from 1996 and association with porcine circovirus associated disease. Vet. Microbiol. 173: 9–16. doi: 10.1016/j.vetmic.2014.06.024 [DOI] [PubMed] [Google Scholar]
- 21.Pan Y., Zeng Q., Zhu C., Hua X., Wang M., Pan K., Cui L.2012. Frequency and characterization of porcine hokovirus (PHoV) in domestic pigs in eastern China. Arch. Virol. 157: 1785–1788. doi: 10.1007/s00705-012-1350-7 [DOI] [PubMed] [Google Scholar]
- 22.Saekhow P., Ikeda H.2015. Prevalence and genomic characterization of porcine parvoviruses detected in Chiangmai area of Thailand in 2011. Microbiol. Immunol. 59: 82–88. doi: 10.1111/1348-0421.12218 [DOI] [PubMed] [Google Scholar]
- 23.Saekhow P., Mawatari T., Ikeda H.2014. Coexistence of multiple strains of porcine parvovirus 2 in pig farms. Microbiol. Immunol. 58: 382–387. doi: 10.1111/1348-0421.12159 [DOI] [PubMed] [Google Scholar]
- 24.Shan T., Lan D., Li L., Wang C., Cui L., Zhang W., Hua X., Zhu C., Zhao W., Delwart E.2011. Genomic characterization and high prevalence of bocaviruses in swine. PLoS ONE 6: e17292. doi: 10.1371/journal.pone.0017292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Streck A. F., Homeier T., Foerster T., Fischer S., Truyen U.2013. Analysis of porcine parvoviruses in tonsils and hearts from healthy pigs reveals high prevalence and genetic diversity in Germany. Arch. Virol. 158: 1173–1180. doi: 10.1007/s00705-013-1603-0 [DOI] [PubMed] [Google Scholar]
- 26.Takahashi K., Iwata K., Watanabe N., Hatahara T., Ohta Y., Baba K., Mishiro S.2001. Full-genome nucleotide sequence of a hepatitis E virus strain that may be indigenous to Japan. Virology 287: 9–12. doi: 10.1006/viro.2001.1017 [DOI] [PubMed] [Google Scholar]
- 27.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S.2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao C. T., Halbur P. G., Opriessnig T.2013. Molecular evolutionary genetic analysis of emerging parvoviruses identified in pigs. Infect. Genet. Evol. 16: 369–376. doi: 10.1016/j.meegid.2013.03.017 [DOI] [PubMed] [Google Scholar]
- 29.Xiao C. T., Giménez-Lirola L. G., Halbur P. G., Opriessnig T.2012. Increasing porcine PARV4 prevalence with pig age in the U.S. pig population. Vet. Microbiol. 160: 290–296. doi: 10.1016/j.vetmic.2012.05.038 [DOI] [PubMed] [Google Scholar]
- 30.Xiao C. T., Giménez-Lirola L. G., Jiang Y. H., Halbur P. G., Opriessnig T.2013. Characterization of a novel porcine parvovirus tentatively designated PPV5. PLoS ONE 8: e65312. doi: 10.1371/journal.pone.0065312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng S., Wang D., Fang L., Ma J., Song T., Zhang R., Chen H., Xiao S.2011. Complete coding sequences and phylogenetic analysis of porcine bocavirus. J. Gen. Virol. 92: 784–788. doi: 10.1099/vir.0.028340-0 [DOI] [PubMed] [Google Scholar]
- 32.Zhai S., Yue C., Wei Z., Long J., Ran D., Lin T., Deng Y., Huang L., Sun L., Zheng H., Gao F., Zheng H., Chen S., Yuan S.2010. High prevalence of a novel porcine bocavirus in weanling piglets with respiratory tract symptoms in China. Arch. Virol. 155: 1313–1317. doi: 10.1007/s00705-010-0698-9 [DOI] [PubMed] [Google Scholar]
- 33.Zhang H. B., Huang L., Liu Y. J., Lin T., Sun C. Q., Deng Y., Wei Z. Z., Cheung A. K., Long J. X., Yuan S. S.2011. Porcine bocaviruses: genetic analysis and prevalence in Chinese swine population. Epidemiol. Infect. 139: 1581–1586. doi: 10.1017/S0950268811000847 [DOI] [PubMed] [Google Scholar]