Abstract
Classical swine fever (CSF) is a highly contagious systemic hemorrhagic viral disease of pigs. Wild boar plays a crucial role in the epidemiology of CSF. Between 2010 and 2014, samples were collected nationwide from 6,654 wild boars hunted in South Korea. Anti-CSF antibodies were identified in 0.59% (39 of 6,654) of the wild boar samples using a virus neutralization test and were primarily detected in wild boars living close to the demilitarized zone and the area of the Taebaek Mountains surroundings. The CSF virus (subgroup 2.1b) was isolated from two wild boars captured in a nearby border area. The criteria used to define high-risk areas for targeted CSF surveillance in South Korea should be further expanded to include other regions nationwide.
Keywords: border disease virus, bovine viral diarrhea virus, classical swine fever virus, wild boar
Classical swine fever virus (CSFV) possesses an enveloped, single-stranded, positive-sense RNA genome of approximately 12.3 kb [10]. It belongs to the genus Pestivirus within the family Flaviviridae, which also comprises bovine viral diarrhea virus (BVDV) [17] and border disease virus (BDV) [3] that infect cattle, sheep and pigs. Classical swine fever (CSF) causes a highly contagious disease among pigs that has a significant global economic impact on the pig industry. The CSFV strains can be categorized into groups 1, 2 and 3, each comprising three to four subgroups (1.1–1.4; 2.1–2.3; and 3.1–3.4) [9, 12, 13].
CSF, one of the most devastating diseases affecting the South Korean domestic pork industry, was first reported in 1947 [7], with occasional outbreaks occurring until 2013. The major approaches used to eradicate CSF in domestic pigs in Korea include the continuous use of vaccination programs, stamping out policies and strict quarantine measures during disease outbreaks. However, there is increasing concern that wild boars may act as an important reservoir for CSFV, which may then spill over in the domestic pig population. Sporadic outbreaks of CSF in domestic pigs throughout Europe have been linked to either indirect or direct contact with wild boar [8]. It has been estimated that 59% of primary outbreaks of CSF in Germany over a decade were caused by contact between wild boar and domestic pigs [2, 11]. In Germany, studies of previous outbreaks of CSFV in wild boar over extended periods of time indicate that young animals (less than one year old) were frequently infected, whereas older animals were rarely infected with CSFV [5, 6].
Surveillance and monitoring of CSF in the wild boar population in South Korea are essential to achieve a CSF disease free status according to the Terrestrial Animal Health Code (Chapter 15.2) of the Office International des Epizooties (OIE). In this study, the prevalence of CSFV-specific antibodies and antigens in wild boar over a 5-year period using the national surveillance program policy was examined.
To satisfy the OIE requirements for the surveillance of wild boar and feral pigs in CSF-free countries, wild boars were hunted in cooperation with the Korean Pork Producers Association and the Korean government from 2010. The objective of the wild boar hunting program was threefold: CSF surveillance, the culling of animals causing crop damage and controlling the wild boar population in restricted regions. In this study, blood and/or spleen and fecal samples were collected from 6,654 wild boars hunted in eight provinces in South Korea between November 2010 and December 2014.
To avoid false-positive results from possible cross-reactions to the related pestiviruses BVDV and BDV, neutralization tests were performed according to protocols described in the OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (7th Edition, 2012). Serum samples were collected from wild pigs and analyzed using a serum neutralization peroxidase-linked antibody assay. For cell staining, the monoclonal antibody 3B6 (Median Diagnostics, Chuncheon, South Korea) was used to detect the CSFV E2 protein, and the BA-2 monoclonal antibody (VMRD, Pullman, WA, U.S.A.) was used to detect BVDV subgroup 2. The neutralization test was evaluated by the cell pathogenic effect (CPE). The CSF reference strains used in the neutralization test were the LOM vaccine strain (subgroup 1.1; accession no. EU789580, GenBank), the wild boar YC11WB strain (subgroup 2.1; accession no. KC149990, GenBank) and the domestic pig YI strain (subgroup 3.2; accession no. AF521710, GenBank). The BVDV reference strains used were the 08GB44-1 strain (subgroup 1a; accession no. JQ418633, GenBank), the 08GB45-2 strain (subgroup 1b, accession no. JQ418634, GenBank) and the 08Q723 strain (subgroup 2a; accession no. JQ418635, GenBank). The BDV reference strain used was the Lyon2 strain from France, and the black goat fetal lung (BGFL) primary cells were used for the neutralization test.
Viral RNA was extracted from 6,654 wild boar samples using a QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA, U.S.A.) in accordance to the manufacturer’s instructions. The extracted RNA was amplified using one-step RT-PCR (Qiagen) and primers specific to the E2 gene as previously described [12]. The amplified PCR products (190 bp) were cloned into the pGEM-T Vector System IITM (Promega, Madison, WI, U.S.A.), and the cloned genes were sequenced with T7 and SP6 sequencing primers using an ABI Prism® 3730xi DNA Sequencer at the Macrogen Institute (Macrogen, Seoul, South Korea)
The nucleotide sequences of partial E2 genes (190 bp) derived from 106 CSFV isolates in various countries were obtained from the NCBI GenBank database (Fig. 1). Sequence alignment was performed using the ClustalX 1.83 software [18]. Multiple alignment files saved by ClustalX 1.83 in the Clustal format (*.aln) were converted to the MEGA format (*.meg) using the MEGA version 6.06 software [16]. Phylogenetic distances were performed by the neighbor-joining (NJ) method and calculated using 1,000 bootstrap replications.
Fig. 1.
Phylogenetic tree based on the E2 fragment (190 bp) of 106 classical swine fever viruses isolated in various countries. Nucleotide sequences of partial E2 genes derived from CSFV isolates were obtained from the NCBI GenBank database. The phylogenetic tree was constructed using the neighbor-joining method in the MEGA 6.06 program and calculated using 1,000 bootstrap replications for each nucleotide sequence. The scale bar indicates the genetic distance (expressed as substitutions per 100 bases). The triangle mark represents strains isolated from wild boars in Europe. The upper gray box represents CSFV strains isolated from wild boars in South Korea. The middle gray box represents subgroup 2.1b strains isolated since 2002 in South Korea. The lower gray box represents subgroup 3.2 strains isolated prior to 2000. The sequence of signs respectively corresponds to the strain, country of origin, year of isolation and GenBank accession number.
The strains YC11WB and PC11WB were identified in 6,654 wild boar samples by CSFV RT-PCR, and the viruses were successfully isolated using porcine kidney cells (PK-15). The isolated strains were obtained from two wild boars captured in the regions Yoenchen and Pochen bordering North Korea in 2011 (Fig. 2). Complete genome sequences were available for the YC11WB and PC11WB strains [4].
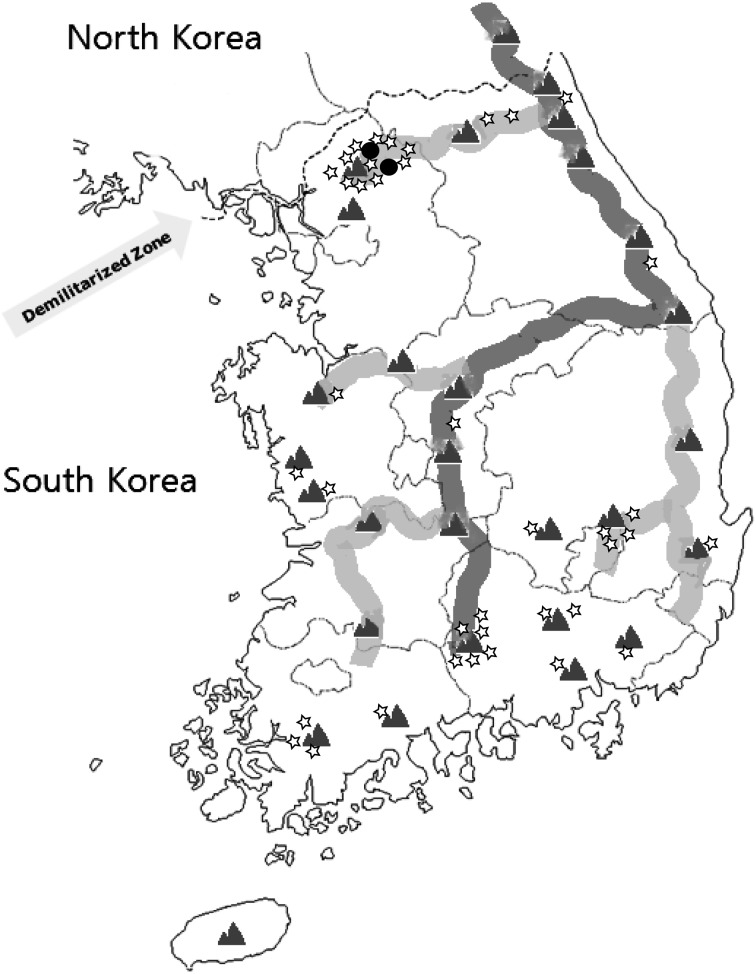
Fig. 2.
Distribution of wild boar samples confirmed as CSFV antigen or antibody positive in South Korea. The star symbol indicates the geographical distribution of wild boars positive for CSF antibody. The two black dots represent locations at which CSFVs were isolated from hunted wild boars. The dark gray line indicates the Taeback mountains, which are approximately 1,000 km in length and stretch across the Baekdu mountains in North Korea to the Jiri mountains in South Korea. The light gray line indicates secondary mountains that have split off from the Taebaek mountain range.
Phylogenetic analysis using the NJ method showed that the partial CSF E2 gene sequences were clustered into three genetically diverse groups (Fig. 1). Two strains isolated from Korean wild boar belonged to subgroup 2.1b, along with other strains identified in domestic pigs in South Korea since 2002. However, the two strains from Korean wild boar were more closely related to Chinese strains within subgroup 2.1b than to South Korean strains. Strains isolated from wild boars in Europe belong to group 2 and are further classified into the subgroups 2.2 (Italy) and 2.3 (Germany, Slovakia, Austria, the Czech Republic and France) (Fig. 1).
Comparative analysis of the complete genome sequences of the other CSFV revealed that the YC11WB and PC11WB strains showed 96.9% and 97.9% sequence homology, respectively, at the nucleotide level with strain ZJ0801 (accession no. FJ529205, GenBank), which was isolated in China in 2008 [4]. Further comparative analysis revealed low sequence homology of 94.5% and 95.2%, respectively, at the nucleotide level with strain SW03 strain, which had been isolated in South Korea in 2003 [4]. Since 2002, a switch has been observed in the CSFV strains in South Korean pig farms from group 3 to group 2 [1]. The source of inflow of CSFV in wild boars remains ambiguous, but may have occurred during the year 2000 or subsequently because of a switch to domestic pigs in 2002. Although two strains isolated from wild boar belonged to subgroup 2.1b, which also comprises strains isolated from domestic pigs in Korea, they may have been the result of an independent source because they showed low sequence homology to, and a greater genetic distance from strains isolated from domestic pigs belonging to this subgroup. Future studies are required to investigate the virulence level by examination of the challenge strain YC11WB in domestic pigs and wild boar.
Of 6,654 wild boar samples analyzed during the study period, 39 (0.59%) were positive for CSF antibodies (Supplemental Table 1). Although the 39 positive samples cross-reacted with BVDV and BDV to a certain extent, all reacted specifically with CSFV (Table 1). Samples positive for the three CSFV genotypes, type 1.1, type 2.1 and type 3.2, showed similar antibody levels (Table 1). Peak incidence of antibody positive cases collected from wild boars during the past 5 years was 12 in 2011 and 15 in 2012. Although an increasing number of wild boars have since then been captured, the incidence of positive antibody to CSFV has decreased to two in 2013 and five in 2014. Most of the positive samples were from hunted wild boars in two large geographical regions: the area close to the border with North Korea and the area of the Taeback mountains surroundings (Fig 2). Many of the positive samples were obtained from wild boars at the area surroundings of the Taeback mountains (average height, 800 m), which stretch approximately 1,000 km from the Baekdu mountain (height, 2,750 m) in North Korea to Jiri mountain (height, 1,916 m) in South Korea (Fig. 2). Therefore, the Taeback mountains geographically are easily able to migrate animals quite freely from north to south area.
Table 1. Cross-reactive neutralization antibody titers in wild boar in South Korea against classical swine fever virus, bovine viral diarrhea virus and border disease virus strains.
| Sample No. | Type | Classical swine fever virus | Bovine viral diarrhea virus | Border disease virus | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1.1 | 2.1 | 3.2 | 1a | 1b | 2a | 1 | ||||
| Strain | LOMa) | YC11b) | YIc) | 08GB44-1d) | 08GB45-2e) | 08Q723f) | Lyon2g) | |||
| 10K-52 | 256 | NTh) | NT | NT | NT | NT | NT | |||
| 10K-61 | 256 | NT | NT | NT | NT | NT | NT | |||
| 10K-94 | 256 | NT | NT | NT | NT | NT | NT | |||
| 10K-131 | 16 | NT | NT | NT | NT | NT | NT | |||
| 10K-190 | 128 | NT | NT | NT | NT | NT | NT | |||
| 11K-204 | 64 | NT | NT | NT | NT | NT | NT | |||
| 11K-344 | 1,024 | 1,024 | 1,024 | 32 | 32 | 16 | NT | |||
| 11K-392 | 512 | 1,024 | 1,024 | 16 | 8 | 16 | 16 | |||
| 11K-447 | 128 | 32 | 32 | 64 | 16 | 8 | 32 | |||
| 11K-506 | 256 | 2,048 | 1,024 | 16 | 8 | 16 | 8 | |||
| 11K-647 | 512 | 128 | 64 | 16 | 8 | 16 | 16 | |||
| 11K-691 | 64 | 16 | 4 | 16 | 4 | 8 | 4 | |||
| 11K-794 | 16 | 8 | 4 | 16 | 8 | 16 | 8 | |||
| 11K-967 | 64 | 16 | 8 | 8 | 16 | 8 | 8 | |||
| 11K-1216 | 16 | 8 | 8 | 2 | 2 | 8 | 2 | |||
| 11K-1276 | 32 | 16 | 16 | 8 | 4 | 8 | 2 | |||
| 11K-1352 | 32 | NT | NT | NT | NT | NT | NT | |||
| 12K-1395 | 256 | 1,024 | 512 | 64 | 8 | 16 | 16 | |||
| 12K-1453 | 64 | 16 | 8 | 16 | 8 | 2 | NT | |||
| 12K-1460 | 16 | 16 | 16 | 2 | 8 | 2 | NT | |||
| 12K-1495 | 16 | 16 | 8 | 4 | 4 | 2 | 2 | |||
| 12K-1507 | 256 | 16 | 64 | 32 | 16 | 64 | 8 | |||
| 12K-1663 | 256 | 32 | 32 | 32 | 16 | 8 | 16 | |||
| 12K-1747 | 256 | 1,024 | 2,048 | 64 | 16 | 8 | 16 | |||
| 12K-1763 | 1,024 | 1,024 | 1,024 | 64 | 32 | 64 | 32 | |||
| 12K-1879 | 16 | 32 | 16 | 8 | 8 | 4 | 4 | |||
| 12K-1881 | 256 | 128 | 64 | NT | NT | NT | NT | |||
| 12K-1924 | 512 | 512 | 512 | 64 | 32 | 128 | NT | |||
| 12K-2034 | 128 | 32 | 32 | 64 | 32 | 32 | 8 | |||
| 12K-2036 | 64 | 128 | 128 | 32 | 32 | 16 | 32 | |||
| 12K-2434 | 32 | 16 | 32 | 4 | 4 | 2 | 16 | |||
| 12K-2510 | 512 | 2,048 | 1,024 | 32 | 16 | 16 | 16 | |||
| 13K-3185 | 1,024 | 512 | 256 | 16 | 8 | 8 | 2 | |||
| 13K-3680 | 1,024 | 1,024 | 512 | 32 | 8 | 8 | 8 | |||
| 14K-5121 | 32 | NT | NT | NT | NT | NT | NT | |||
| 14K-5195 | 512 | NT | NT | NT | NT | NT | NT | |||
| 14K-5231 | 128 | NT | NT | NT | NT | NT | NT | |||
| 14K-5299 | 256 | NT | NT | NT | NT | NT | NT | |||
| 14K-5400 | 512 | NT | NT | NT | NT | NT | NT | |||
a) LOM, LOM vaccine strain (subgroup 1.1, accession no. EU789580); b) YC11, YC11WB strain (subgroup 2.1, accession no. KC149990) isolated from a wild boar; c) YI strain (subgroup 3.2, accession no. AF521710) isolated from a domestic pig; d) 08GB44-1, 08GB44-1 strain (subgroup 1a, accession no. JQ418633) isolated from a cow; e) 08GB45-2, 08GB45-2 strain (subgroup 1b, accession no. JQ418634) isolated from a cow; and f) 08Q723, 08Q723 strain (subgroup 2a, accession no. JQ418635) isolated from a cow. g) Lyon2, Lyon2 strain (group 1, accession no. DQ350165) was donated from CNEVA (Centre National d'Etudes Veterinaires et Alimentaires) in France; h) NT, Not Tested; no., number.
The circulation of CSFV has been found to correspond with wild boar movements, which are in turn primarily correlated with forest conformation [15]. Maintenance of CSFV in wild boar populations is facilitated by increasing population density. High density wild boar populations usually contain relatively large numbers of young animals, which are more susceptible to infection [5, 14]. Recently, an emergency disease outbreak of CSF occurred in an unvaccinated pig farm in October, 2013 in the city of Sacheon in the Gyeungnam province. The CSFV strain detected was found to have a high level of similarity with the SW03 strain. To investigate the cause of the inflow of CSFV, hunters captured 28 wild boars from within a 20 km radius of the outbreak farm over a period of six months. No evidence for CSFV spillover events from wild boar to domestic pigs was found. Therefore, the cause of the CSF outbreak in 2013 was suspected to be the result of circulation of CSFV and hidden infections in pig farms.
Wild boars play a significant role in the transmission and spread of many diseases and act as reservoirs for many pathogens. Although the seroprevalence and detection of CSF antigens are quite low in wild boars in Korea, and the probability of virus spillover to domestic pigs is quite low, an epidemiological surveillance network should be established to continuously monitor the status of CSF in wild boar in the regions bordering North and South Korea and within the mountains surroundings of the Taebaek mountains range, if CSF is to be eradicated in South Korea. Infection of wild boar with CSFV from domestic pigs is possible via contact with infected feral pigs. This should be taken into account when establishing CSF monitoring programs.
Supplementary Material
Acknowledgments
This study was supported by a grant from the Animal and Plant Quarantine Agency (Project Code No. B-AD14-2012-14-03) and Ministry for Food, Agriculture, Forestry and Fisheries, South Korea.
REFERENCES
- 1.Cha S. H., Choi E. J., Park J. H., Yoon S. R., Kwon J. H., Yoon K. J., Song J. Y.2007. Phylogenetic characterization of classical swine fever viruses isolated in Korea between 1988 and 2003. Virus Res. 126: 256–261. doi: 10.1016/j.virusres.2007.01.017 [DOI] [PubMed] [Google Scholar]
- 2.Fritzemeier J., Teuffert J., Greiser-Wilke I., Staubach C., Schlüter H., Moennig V.2000. Epidemiology of classical swine fever in Germany in the 1990s. Vet. Microbiol. 77: 29–41. doi: 10.1016/S0378-1135(00)00254-6 [DOI] [PubMed] [Google Scholar]
- 3.Greiser-Wilke I., Blome S., Moennig V.2007. Diagnostic methods for detection of Classical swine fever virus—status quo and new developments. Vaccine 25: 5524–5530. doi: 10.1016/j.vaccine.2006.11.043 [DOI] [PubMed] [Google Scholar]
- 4.Jeoung H. Y., Lim J. A., Lim S. I., Kim J. J., Song J. Y., Hyun B. H., Kim Y. K., An D. J.2013. Complete genome sequences of classical Swine Fever virus strains isolated from wild boars in South Korea. Genome Announc. 1: e0014713. doi: 10.1128/genomeA.00147-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaden V., Steyer H., Schnabel J., Bruer W.2005. Classical swine fever (CSF) in wild boar: the role of the transplacental infection in the perpetuation of CSF. J. Vet. Med. B Infect. Dis. Vet. Public Health 52: 161–164. doi: 10.1111/j.1439-0450.2005.00838.x [DOI] [PubMed] [Google Scholar]
- 6.Kern B., Depner K. R., Letz W., Rott M., Thalheim S., Nitschke B., Plagemann R., Liess B.1999. Incidence of classical swine fever (CSF) in wild boar in a densely populated area indicating CSF virus persistence as a mechanism for virus perpetuation. Zentralbl. Veterinarmed. B. 46: 63–67. [DOI] [PubMed] [Google Scholar]
- 7.Kim B., Song J. Y., Tark D. S., Lim S. I., Choi E. J., Kim J., Park C. K., Lee B. Y., Wee S. H., Bae Y. C., Lee O. S., Kwon J. H., Kang W. C., Kim T. Y., Kim J. H., Lee J. H., Kang M. I.2008. Feed contaminated with classical swine fever vaccine virus (LOM strain) can induce antibodies to the virus in pigs. Vet. Rec. 162: 12–17. doi: 10.1136/vr.162.1.12 [DOI] [PubMed] [Google Scholar]
- 8.Laddomada A.2000. Incidence and control of CSF in wild boar in Europe. Vet. Microbiol. 73: 121–130. doi: 10.1016/S0378-1135(00)00139-5 [DOI] [PubMed] [Google Scholar]
- 9.Lowings P., Ibata G., Needham J., Paton D.1996. Classical swine fever virus diversity and evolution. J. Gen. Virol. 77: 1311–1321. doi: 10.1099/0022-1317-77-6-1311 [DOI] [PubMed] [Google Scholar]
- 10.Meyers G., Rümenapf T., Thiel H. J.1989. Molecular cloning and nucleotide sequence of the genome of hog cholera virus. Virology 171: 555–567. doi: 10.1016/0042-6822(89)90625-9 [DOI] [PubMed] [Google Scholar]
- 11.Mintiens K., Verloo D., Venot E., Laevens H., Dufey J., Dewulf J., Boelaert F., Kerkhofs P., Koenen F.2005. Estimating the probability of freedom of classical swine fever virus of the East-Belgium wild-boar population. Prev. Vet. Med. 70: 211–222. doi: 10.1016/j.prevetmed.2005.03.008 [DOI] [PubMed] [Google Scholar]
- 12.Paton D. J., McGoldrick A., Greiser-Wilke I., Parchariyanon S., Song J. Y., Liou P. P., Stadejek T., Lowings J. P., Björklund H., Belák S.2000. Genetic typing of classical swine fever virus. Vet. Microbiol. 73: 137–157. doi: 10.1016/S0378-1135(00)00141-3 [DOI] [PubMed] [Google Scholar]
- 13.Postel A., Schmeiser S., Perera C. L., Rodríguez L. J., Frias-Lepoureau M. T., Becher P.2013. Classical swine fever virus isolates from Cuba form a new subgenotype 1.4. Vet. Microbiol. 161: 334–338. doi: 10.1016/j.vetmic.2012.07.045 [DOI] [PubMed] [Google Scholar]
- 14.Rossi S., Fromont E., Pontier D., Crucière C., Hars J., Barrat J., Pacholek X., Artois M.2005. Incidence and persistence of classical swine fever in free-ranging wild boar (Sus scrofa). Epidemiol. Infect. 133: 559–568. doi: 10.1017/S0950268804003553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon G., Le Dimna M., Le Potier M. F., Pol F.2013. Molecular tracing of classical swine fever viruses isolated from wild boars and pigs in France from 2002 to 2011. Vet. Microbiol. 166: 631–638. doi: 10.1016/j.vetmic.2013.06.032 [DOI] [PubMed] [Google Scholar]
- 16.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thiel H. J., Collett M. S., Gould E. A., Heinz F. X., Meyers G., Purcell R. H., Rice C. M., Houghton M.2005. Flaviviridae. In: Virus Taxonomy, the eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego. [Google Scholar]
- 18.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G.1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. doi: 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.