Abstract
Feline morbillivirus (FmoPV) is a new virus species and its detection is important, since correlation has been reported between FmoPV virus infection and tubulointerstitial nephritis in cats. Here, we report a real-time reverse transcription (RT)-PCR system that can detect the FmoPV L-gene sequence with more than 10-time higher sensitivity than a conventional PCR system, resulting in detection of less than 10 copies of the template DNA. The total FmoPV positive rate of urine samples from veterinary clinics and hospitals in Japan was 15.1% (25/166) using this system. This study demonstrates usefulness of the real-time RT-PCR system for detection of FmoPV for cat urine samples.
Keywords: detection, feline, paramyxovirus, PCR
Feline morbillivirus (FmoPV) is a new virus species which was first discovered in 2012 from specimens of stray cats in Hong Kong. Phylogenetic analyses of the virus genome sequences demonstrated that FmoPV is a distinct virus species from other morbilliviruses [5]. Discovery of FmoPV was particularly important, since a case-control study suggested correlation between the virus infection and tubulointerstitial nephritis in cats [5], in which about 30% of cats older than 15-year-old suffer from renal failures [1]. FmoPV was also detected in Japan, and Japanese strains were clearly clustered together with Hong Kong strains in the phylogenetic analyses [2,3,4]. Potential recombination between Japanese and Hong Kong lineages was reported based on the genome sequence of one of the strains detected in Japan [3]. Most reports about FmoPV used PCR amplification of virus-specific nucleic acids to detect FmoPV. However, conventional PCR assays use gel electrophoresis for detection of the target products which is neither quantitative nor convenient for analysis of a large number of samples. In this report, we developed a real-time PCR assay for detection of FmoPV, which was quantitative and applicable to a larger number of samples. The assay was used for cat urine samples and proved to be useful for detection of FmoPV in these samples.
Urine samples were collected from cats at veterinary clinics at three different areas in Japan (Hokkaido, Yamaguchi and Ehime prefectures). Viral RNA was extracted from the urine samples using Trizol® LS (Life Technologies, Carlsbad, CA, U.S.A.). For conventional RT-PCR, the extracted RNA was reverse transcribed with random primers (PrimeScript™ RT-PCR Kit, Takara, Otsu, Japan), and the 401 bp L-gene fragment was PCR amplified with a set of primers (forward, CCAAATCATGCATCTGCTGT; reverse, GCGAACAATCGACCTACCTC), which was separated on an agarose gel and stained with ethidium bromide. Real-time RT-PCR was performed with Applied Biosystems 7300 Real-Time PCR System (Life Technologies) with 96-well plate format. The PCR reaction mixture was made using One Step PrimeScript™ RT-PCR Kit (Takara) following the manufacturer’s instruction, in which each well contained 20 µl of the reaction mixture, containing Taq polymerase, reverse transcriptase, buffer, ROX reference dye, PCR primers (0.5 µM each final) and hybridization probe (0.25 µM final). Primers and hybridization probe were designed based on the FmoPV L-gene sequences obtained from 6 Japanese strains [2], and their specific sequences are as follows: forward primer, TACAATGTACCTTCGGGACAAG; reverse primer, ATCTTCTAGAGCTTGTAGGTAATCC; probe, AGTGAGTGGGATTCCGTTTATGCGAAAGA, which produce 121 bp PCR amplicon. The hybridization probe was labeled with 6-FAM at 5′-end, and fluorescence quenchers, IBFQ and ZEN at 3′-end and an internal site, respectively (Medical and Biological Laboratories, Nagoya, Japan). RT-PCR was performed at 42°C for 5 min, 95°C for 10 sec, followed by 40 cycles of 95°C for 10 sec and 60°C for 31 sec. The assay was carried out in duplicate for each sample and was regarded as positive when both reactions gave quantitative values. For making a standard curve, a 401-bp L-gene fragment containing the sequences of the primers and the probe used for the real-time RT-PCR assay was cloned into a TA-plasmid (Mighty TA-cloning kit, Takara). The plasmid DNA was purified (FastGene Plasmid Mini Kit, Nippon genetics, Tokyo, Japan), and the copy number concentration was calculated based on the DNA concentration and the molecular mass of the plasmid DNA.
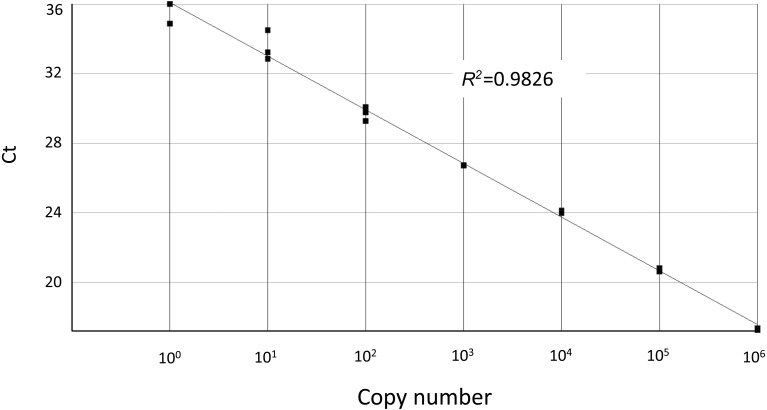
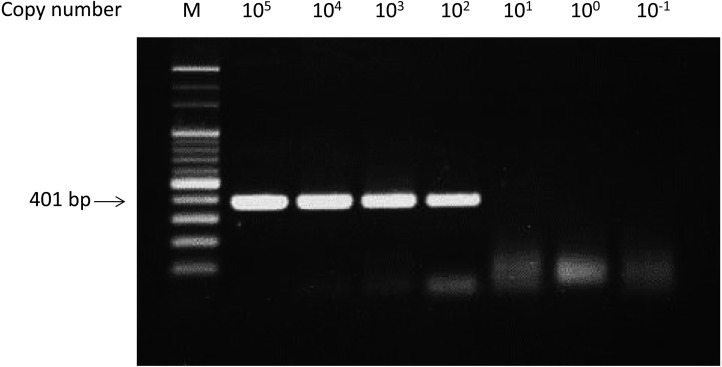
Since a part of the FmoPV L-gene (401 bp) was amplified by RT-PCR for detection of Japanese FmoPV strains in a previous study [2], primers and probe were designed in the same 401 bp region of the L-gene for a real-time RT-PCR detection system with the hybridization method. This portion of the DNA was also cloned into a plasmid vector to make a standard DNA for estimation of the assay sensitivity and calculation of copy numbers of the virus nucleic acid in the real-time RT-PCR assay. First, the sensitivity of the assay was examined by quantitation of serial dilutions of the standard DNA. As Fig. 1 demonstrates, the assay detected less than 10 copies of the standard DNA in the reaction mix, although the signal from close to one copy was not always consistent. Next, a conventional RT-PCR assay amplifying the same L-gene portion used for the real-time RT-PCR assay was performed with the same serial dilutions of the standard DNA to compare its sensitivity with that of the real-time PCR assay. The results demonstrated that the conventional RT-PCR could detect only up to 100 copies of the standard DNA (Fig. 2), which indicated the real-time RT-PCR had more than 10-time higher sensitivity in terms of copies of the standard DNA. Based on the results, the real-time PCR assay developed in this study appeared to be more sensitive than the conventional RT-PCR assay when FmoPV template sequences match completely with the primers and the probe used in the assay.
Fig. 1.
Standard curve of the real-time RT-PCR assay for FmoPV. Ten-fold dilutions of control plasmid DNA harboring the 401 bp L-gene fragment were assayed with the real-time RT-PCR assay. Log10 copy numbers and cycle threshold values (Ct) are plotted on the X-asis and the Y-axis, respectively. Each dot represents the result of triplicate of the reactions, and the coefficient of determination (R2) is shown.
Fig. 2.
Detection of the 401 bp L-gene fragment with conventional PCR assay. Ten-fold dilutions of the control plasmid DNA were assayed with conventional PCR. Copy numbers were indicated above the gel. Lane M, 100 bp DNA ladder. Arrow shows the position of 401-bp L-gene DNA fragment.
By using this real-time RT-PCR system, we examined cat urine samples including 82 samples used in the previous study [2] for detection of FmoPV RNA. In the total number of 166 samples tested, 25 (15.1%) samples were considered as positives based on the assay. Six samples were positive based on the real-time RT-PCR system among the 82 samples tested in the previous study [2]. Copy numbers in the urine samples estimated with the real-time RT-PCR varied, between 10.85 and 26,620, and the mean value was 3,576 copies per ml.
The primer pairs and the probe used in the real-time RT-PCR assay developed in this study were designed using a consensus sequence based on the six Japanese FmoPV strains detected by the authors previously (2). Since then, some additional FmoPV strains were sequenced, and it appears that the assay system in this study may not be able to detect some FmoPV strains. Therefore, it is possible that the positive rates of FmoPV found in this study are lower than actual positive rates. Although it may be difficult to test all the FmoPV strains reported using the real-time RT-PCR in this study, it will be important to design other primers and probes based on the sequence information including the newer sequences available, and assay the samples tested in this study to find whether there are additional positive samples.
In conclusion, the sensitivity of the real-time RT PCR assay developed in this study exceeds conventional PCR amplification used in the previous studies and is applicable to large numbers of urine samples. This assay system is useful for diagnostic purposes for FmoPV infection and basic research of the virus, and can be used as a standard assay for detection and quantitation of the virus.
Acknowledgments
We would like to express sincere appreciation to all veterinary clinics and hospitals for providing cat urine samples.
REFERENCES
- 1.Bartges J. W.2012. Chronic kidney disease in dogs and cats. Vet. Clin. North Am. Small Anim. Pract. 42: 669–692, vi. doi: 10.1016/j.cvsm.2012.04.008 [DOI] [PubMed] [Google Scholar]
- 2.Furuya T., Sassa Y., Omatsu T., Nagai M., Fukushima R., Shibutani M., Yamaguchi T., Uematsu Y., Shirota K., Mizutani T.2014. Existence of feline morbillivirus infection in Japanese cat populations. Arch. Virol. 159: 371–373. doi: 10.1007/s00705-013-1813-5 [DOI] [PubMed] [Google Scholar]
- 3.Park E. S., Suzuki M., Kimura M., Maruyama K., Mizutani H., Saito R., Kubota N., Furuya T., Mizutani T., Imaoka K., Morikawa S.2014. Identification of a natural recombination in the F and H genes of feline morbillivirus. Virology 468–470: 524–531. doi: 10.1016/j.virol.2014.09.003 [DOI] [PubMed] [Google Scholar]
- 4.Sakaguchi S., Nakagawa S., Yoshikawa R., Kuwahara C., Hagiwara H., Asai K., Kawakami K., Yamamoto Y., Ogawa M., Miyazawa T.2014. Genetic diversity of feline morbilliviruses isolated in Japan. J. Gen. Virol. 95: 1464–1468. doi: 10.1099/vir.0.065029-0 [DOI] [PubMed] [Google Scholar]
- 5.Woo P. C., Lau S. K., Wong B. H., Fan R. Y., Wong A. Y., Zhang A. J., Wu Y., Choi G. K., Li K. S., Hui J., Wang M., Zheng B. J., Chan K. H., Yuen K. Y.2012. Feline morbillivirus, a previously undescribed paramyxovirus associated with tubulointerstitial nephritis in domestic cats. Proc. Natl. Acad. Sci. U.S.A. 109: 5435–5440. doi: 10.1073/pnas.1119972109 [DOI] [PMC free article] [PubMed] [Google Scholar]