Abstract
DNA mismatch repair (MMR) is one of the biological pathways, which plays a critical role in DNA homeostasis, primarily by repairing base-pair mismatches and insertion/deletion loops that occur during DNA replication. MMR also takes part in other metabolic pathways and regulates cell cycle arrest. Defects in MMR are associated with genomic instability, predisposition to certain types of cancers and resistance to certain therapeutic drugs. Moreover, genetic and epigenetic alterations in the MMR system demonstrate a significant relationship with human fertility and related treatments, which helps us to understand the etiology and susceptibility of human infertility. Alterations in the MMR system may also influence the health of offspring conceived by assisted reproductive technology in humans. However, further studies are needed to explore the specific mechanisms by which the MMR system may affect human infertility. This review addresses the physiological mechanisms of the MMR system and associations between alterations of the MMR system and human fertility and related treatments, and potential effects on the next generation.
Keywords: DNA mismatch repair, Infertility, Assisted reproductive technology
1. Introduction
Cellular DNA damage accumulates as a result of exposure to exogenous agents (biological physical or chemical) and endogenous sources including oxidative stress and errors associated with DNA processing. DNA damage if unrepaired creates the possibility of mutagenesis in somatic or germline cells which can alter normal function and result in diseases even in the next generation. Consequently there are multiple mechanisms to repair DNA damage and maintain the stability of the DNA sequence in all tissues and cells. One of them known as DNA mismatch repair (MMR) mainly repairs base-pair mismatches and insertion/deletion loops (IDLs) (Jascur and Boland 2006). MMR repairs DNA mismatches arising during replication thereby preventing deleterious mutations and maintaining genomic stability. The MMR system also plays a critical role in the meiotic process and gametogenesis. Defects in the MMR system are related to predisposition to certain types of cancer resistance to certain therapeutic drugs and infertility collectively known as DNA MMR defective abnormalities (Maduro et al. 2003; Li 2008).
Infertility is a common and complex condition affecting about 10%–15% of couples of reproductive age (Gnoth et al. 2005) and the causes of a considerable proportion of infertility cases remain unknown. Based on their important physiological functions and a growing amount of evidence MMR genes and proteins demonstrate an impact on human infertility even in the next generation conceived by assisted reproductive technology (ART). The molecular mechanisms of the DNA damage and repair in spermatogenesis have been reviewed recently (Gunes et al. 2015) focusing on the process of spermatogenesis origin of DNA damage and five types of DNA repair mechanisms. This review will concentrate on the relationship between DNA MMR alterations and human fertility fertility-related treatments and the potential influence on ART offspring
2. Components and mechanisms of the MMR system
First identified in Escherichia coli MMR is a biological pathway highly conserved throughout evolution to maintain genomic integrity. The bacterial MMR system is the best studied biochemically (Lahue et al. 1989; Modrich 1991) and yeast and mouse systems have provided valuable insights because of the power of genetic models (Kolodner and Marsischky 1999). In humans the MMR system is an excision and re-synthesis system that can be divided into three steps: recognition of the mismatch excision of the incorrect fragment and DNA re-synthesis (Table 1). The human MMR protein MutS which is a heterodimer composed of MutS homologues MSH2 and MSH6 (MutSα) or MSH2 and MSH3 (MutSβ) is an ATPase that plays a critical role in mismatch recognition and initiation of repair. MutSα preferentially recognizes base-pair mismatches and short IDLs while MutSβ recognizes larger IDLs. Then the DNA-MutS complex recruits MutLα a heterodimer of MutL homologues MLH1 and PMS2. Other potential proteins like exonuclease 1 (EXO1) DNA polymerase δ (Polδ) and its cofactors proliferation cell nuclear antigen (PCNA) and replication factor C (RFC) are recruited to accomplish the repair activity. Other members of the MMR system are yet to be found or confirmed such as DNA methyltransferase 1 (Dnmt1) (Guo et al. 2004; Kim et al. 2004).
Table 1.
Human MMR components and functions
| MMR components | Fuction |
| MutSα (MSH2-MSH6) MutSβ (MSH2-MSH3) | DNA mismatch recognition |
| MutLα (MLH1-PMS2) MutLβ (MLH1-PMS1) MutLγ (MLH1-MLH3) | Molecular matchmaker; recruitment of other related proteins |
| EXO1 | DNA mismatch excision |
| Polδ | DNA re-synthesis |
| PCNA | Initiation of DNA re-synthesis |
| RFC | Loading and unloading PCNA |
| RPA | Single strand DNA protection; termination of DNA excision |
| DNA ligase I | Nick ligation |
| Other members unidentified |
In addition to DNA MMR activity, MMR is also associated with a series of DNA damage signaling pathways. Plenty of articles have reported interactions between MMR and DNA damage regulators including MLH1 and ATM (Brown et al., 2003), MSH2 and ATR, Bcl-2 (Wang and Qin, 2003; Youn et al., 2005), MLH1, PMS2 and p53, p73 (Shimodaira et al., 2003; Chen and Sadowski, 2005), MLH1, PMS1, and PMS2 in cell cycle arrest (Stojic et al., 2005; Cannavo et al., 2007), all of which are involved in cell signaling/cycle arrest/apoptosis. Thus, the MMR system recognizes and repairs mismatches and eliminates severely damaged cells, preventing mutagenesis in the short term and tumorigenesis in the long term.
MMR proteins also take part in the meiotic process and are involved in gametogenesis. They have been implicated in somatic hypermutation, interstrand-crosslink repair, immunoglobulin class switching, trinucleotide repeat (TNR) expansion, and other processes. With so many proteins and regulators involved in the MMR system, it is not surprising that mistakes may occur and manifest themselves in different phenotypes. Loss of MMR function leads to failure to repair base-pair mismatches and IDLs, including short repetitive sequences known as microsatellites. Shortening or lengthening of microsatellites, referred to as microsatellite instability (MSI), is the hallmark of MMR system deficiency. The MSI status is commonly determined by five microsatellite markers (BAT25, BAT26, D2S123, D5S346, and D17S250).
The contribution of defective MMR to the development of human cancer has been recognized for more than two decades (Peltomaki, 2003). Alterations in MLH1, MSH2, MSH6, and PMS2 lead to the most common form of cancer, hereditary nonpolyposis colorectal cancer (HNPCC) or Lynch syndrome (LS). Loss of MMR function caused by either genetic or epigenetic variation of MMR genes is associated with various human cancers such as colorectal, endometrial, ovarian, cervical, breast, gastric, urological, skin, and other rare cancers (Watson and Lynch, 1994; Karamurzin et al., 2012). Defects in MMR can also trigger a multidrug resistance phenotype, resulting cellular resistance to certain alkylating, methylating, and platinating agents, antimetabolites, topoisomerase inhibitors, and DNA minor groove binders (Valentini et al., 2006) since the system is involved in DNA repair, cell cycle arrest, and many other metabolic pathways. In addition to its involvement in human cancer and drug resistance, the role of the MMR system in the meiotic process and gametogenesis should never be neglected. MMR alterations could manifest themselves in human infertility and infertility treatment. More research is needed to understand the origin and development of infertility.
3. MMR alterations and infertility
Infertility is a worldwide problem that occurs in 10%–15% of couples. Male factors account for about half of all infertility cases. However, the etiology of male infertility remains unknown in about half of all cases. Such cases are classified as idiopathic. This is particularly relevant to men with non-obstructive azoospermia or severe oligospermia. The human testis is composed mainly of three types of cells: spermatogenic cells to produce spermatids; Sertoli cells to protect, support, and supply spermatogenic cells, making sure that spermatids are well produced; and Leydig cells to secrete androgen. Male infertility could be an outcome of genetic, epigenetic, endocrine, or environmental changes that lead to aberrant sperm production or function. Substantial evidence has indicated that several members of the MMR family participate in the meiotic recombination process and gametogenesis (Kunkel and Erie, 2005; Iyer et al., 2006; Jiricny, 2006). A defective MMR system has been shown to be related to spermatogenetic failure and spermatic dysfunction in some infertile individuals and animal models (Mukherjee et al., 2010). Later, we will discuss recent progress in understanding the genetic and epigenetic variation in the MMR system associated with male infertility.
Animal knockout models have shown the importance of MMR genes in both male and female infertility. First discovered in 1995, mice with defective PMS2 exhibit male infertility due to failure of chromosomal synapsis (Baker et al., 1995). Disruption of other genes, including MLH1, MLH3, and EXO1 (Baker et al., 1996; Lipkin et al., 2002; Wei et al., 2003), also leads to phenotypes of infertility (Table 2). However, there has been little progress in understanding links between the MMR system and human male infertility. Maduro et al. (2003) proposed that MSI and defective MMR proteins like MLH1 and MSH2 are presented in both testicular and peripheral blood cells in some azoospermic men, predominantly in Sertoli cell-only patients. This indicates that defects in MMR may underlie some forms of male infertility such as Sertoli cell-only, hypospermatogenesis type, and maturation arrest. Recently, Sertoli cells have been recognized as having several common metabolic features analogous to those of cancer cells and could be used as a good model for exploring new perspectives of the Warburg effect (Oliveira et al., 2015). Sertoli cells may play a significant role in obesity-induced male infertility (Martins et al., 2015). The above studies signify important linkages between MMR alterations and human infertility and Sertoli cells, which deserve further detailed study.
Table 2.
MMR genes involved in mouse infertility models
| Gene | Phenotype |
| MSH4 | Infertile: failure of spermatogonial maturation beyond zygonema |
| MSH5 | Infertile: incomplete and non-homologous chromosomal pairing |
| MLH1 | Infertile: failure of crossing over |
| PMS2 | Male infertile: disruption of normal chromosomal synapsis |
| MLH3 | Infertile |
| EXO1 | Infertile |
MMR proteins participate in the meiotic recombination process in yeast and mammals. Among these proteins there are two MutS homologues (MSH4, MSH5) and three MutL homologues (MLH1, MLH3, PMS2) (Surtees et al., 2004; Her et al., 2007). Terribas et al. (2010) studied 13 infertile patients each with one of two types of spermatogenic arrest: maturation arrest or hypospermatogenesis. These two types showed significantly decreased MMR expression values (MLH1, MLH3, MSH4, MSH5) in testicular samples, except for PMS2, and the more serious type, the maturation arrest group, showed a greater reduction. Ferras et al. (2007) showed that the testicular MLH3 gene in 13 infertile patients with spermatogenic arrest contained four missense (T2896C, C2531T) and eight intronic (IVS9+66G/A) variants. Among these 13 patients, the combination of C2531T and IVS9+66G/A variants was identified only in patients with primary spermatogenic arrest, indicating that the presence of two simultaneous MLH3 variants might be a cause of the arrest. MLH3 is certainly involved in the mammalian meiotic process, but its role in MMR during DNA replication is controversial (Lipkin et al., 2000; Wu et al., 2001; Hienonen et al., 2003; Liu et al., 2003; Korhonen et al., 2007). Cytological studies in mice have shown that MLH3 is found in zygotene, combines with recombination nodules in early pachytene, and associates with MLH1 from mid-pachytene to the early diplonema stages (Lipkin et al., 2002; Santucci-Darmanin et al., 2002; Marcon and Moens, 2003). Also, both male and female MLH3 −/− mice manifested infertility (Lipkin et al., 2002). Therefore, human MLH3 variants might interfere in the meiotic process resulting in male infertility.
Ferras et al. (2012) found that one patient with two MLH3 mutations, the combination of C2531T and IVS9+66G/A, showed overexpression of MLH3 and MLH1. Since MLH3 and PMS2 share the same interaction domain on MLH1, it is possible that the quantitative balance of the MLH1-binding partners (MLH3 and PMS2) plays a crucial role in specifying different outcomes during meiosis (Kondo et al., 2001; Korhonen et al., 2007). Accordingly, mutation-related overexpression of MLH3 and MLH1 may result in a predominance of MLH1-MLH3 complexes and a relative reduction in MLH1-PMS2 complexes. Consequently, meiotic failure might occur as MLH1-PMS2 complexes ought to replace MLH1-MLH3 complexes during diplonema. However, due to the relatively small sample sizes in the above studies, further evaluation of the correlations and mechanisms linking MMR defects and male infertility is required.
Recently, numerous sequencing analyses of single nucleotide polymorphisms (SNPs) in candidate genes have helped to clarify the etiology and susceptibility of both male and female infertility (Kang et al., 2014; Ni et al., 2014). Xu et al. (2010) identified two SNPs, MSH5 (C85T, Pro29Ser) and MLH3 (C2531T, Pro844Leu) associated with male infertility, especially in non-obstructive azoospermia or severe oligozoospermia. The allele carriers MSH5 Pro29Ser and MLH3 Pro844Leu demonstrated a 2.89-fold and 2.25-fold increased risk of azoospermia or oligozoospermia, respectively. However, two major problems have limited the value of medical sequencing studies. First, the actual rates of these reported SNPs are quite low and could explain only a small percentage of idiopathic male infertility. Growing evidence suggests that common genetic variations are rarely responsible for the whole disease phenotype (Frazer et al., 2009; Manolio et al., 2009). The “common disease, common variant” model is not suitable for most complex diseases, including male infertility. Further studies have suggested that risk or causative genetic variants for most cases of idiopathic male infertility might be related to the identification of rare polymorphisms and copy number variants (CNVs). Second, most reports of SNPs associated with male infertility have either not been followed up with validation studies, or often failed validation in follow-up studies.
Ji et al. (2012) conducted a prospective case-control study of 1292 idiopathic male infertility patients and 480 fertile controls in a Chinese population. The idiopathic infertility patients were divided into two subgroups: 524 patients with non-obstructive azoospermia or oligozoospermia and 768 with a normal sperm count. Twenty-one tagging SNPs in five MMR genes (MLH1, MLH3, PMS2, MSH4, and MSH5) were examined by the sequence detection system and the SNPstream 12-plex platform. The results showed that the genotype frequencies of three SNPs, MLH1 (rs4647269), PMS2 (rs1059060, Ser775Asn), and MSH5 (rs2075789, Pro29Ser), were significantly increased (by from 6% to 17%) in patients with azoospermia or oligozoospermia. Therefore, the presence of these SNPs seems to be a risk factor for the development of azoospermia or oligozoospermia. Another SNP in PMS2 rs1059060 appeared to contribute to the risk of male infertility in patients with a normal sperm count. Guerrette et al. (1999) localized the MLH1-PMS2 interaction region to amino acids 506–675 of MLH1 and 675–850 of PMS2. This study provided evidence that the PMS2 Ser775Asn variant attenuates the binding of MLH1 and PMS2 detected by fluorescence resonance energy transfer (FRET) and co-immunoprecipitation assay, further supporting the potential interaction between MLH1-PMS2 and MLH1-MLH3 discussed above. The MSH5 Pro29Ser polymorphism is located within the MSH4-MSH5 interacting domain and leads to a weakened formation of the MSH4-MSH5 heterocomplex (Yi et al., 2005). This was supported by the study of Xu et al. (2010). Gene knockout of MSH4 or MSH5 results in infertility in mice since they are unable to resolve meiotic chromosomal crossovers (de Vries et al., 1999; Edelmann et al., 1999; Kneitz et al., 2000). Consequently, MSH5 Pro29Ser alteration is related to a significantly increased risk of male infertility. However, the detailed molecular mechanisms are unknown.
Compared with the current understanding of the MMR system in male infertility, its potential involvement in female infertility has received much less attention. A case-control study including 41 women with premature ovarian failure and 39 controls suggested that MSH5 Pro29Ser polymorphism might be an explanation for premature ovarian failure (Mandon-Pepin et al., 2008), indicating that MMR gene mutation is likely to be related to female infertility. Pashaiefar et al. (2013) suggested that MLH3 C2531T polymorphism can be associated with the risk of unexplained fertility in Iranian women. Perry et al. (2014) showed that gene mutation-related low expression of MSH6 due to one SNP in MSH6 (rs1800932) is associated with earlier menopause, suggesting that MMR is a key process in determining female reproductive lifespan and could be a crucial therapeutic target for female infertility.
With an increased incidence and a younger age of patients, certain forms of MMR-related genital system cancers which require surgical treatment may contribute to female “structural” infertility. For instance, endometrial cancer has an intimate connection with genetic and epigenetic variation of the MMR system. Studies demonstrated that aberrant methylation of MLH1 is detected in about 40% of endometrial cancer cases and is considered to be an important process in the early stage of endometrial carcinogenesis (Muraki et al., 2009). Conservative surgical and hormonal therapy is currently practical in clinical treatment. This kind of therapy ensures that a considerable proportion of women with endometrial cancer successfully achieve pregnancies resulting in live births with or without ART (Park et al., 2013). However, some other problems still exist. Although current studies have demonstrated that there is no definitive evidence of a significant association between MMR status and survival in endometrial cancer (Diaz-Padilla et al., 2013), deficient MMR is associated with a higher risk of high-grade endometrial cancer and worse clinical outcomes in women aged 40 years or younger (Garg et al., 2009; Shih et al., 2011). Also, MMR status appears to be linked to body mass index (BMI) and endometrial cancer (Win et al., 2011; Joehlin-Price et al., 2014), indicating a possible interactive system between MMR and some types of metabolic diseases and cancers. Therefore, cautious and comprehensive assessment should be carried out before, during, and after conservative therapy in young women who have a strong desire to preserve fertility, to maintain the safety and health of both the mother and offspring.
Apart from the genetic components involved in human infertility, a growing body of evidence clearly indicates the importance of other factors such as epigenetic variation (Dada et al., 2012; Richardson et al., 2014). Whether epigenetic variation in the MMR system may be etiologic of infertility is unknown, but the fast developing field of epigenetics may be yet another area related to infertility and should be considered together with genetic factors.
4. MMR alterations and ART
With the application of ART including in vitro fertilization (IVF), intracytoplasmic sperm injection (ICSI), and preimplantation genetic diagnosis (PGD), it is now practical to bypass the natural barriers to produce offspring. Interestingly, Terribas et al. (2010) considered the expression of testicular MSH4 could be useful as a surrogate marker for intratesticular elongated spermatids in patients with non-obstructive azoospermia and may be useful in predicting the viability of assisted reproduction. This approach needs further evaluation for sperm quality selection. Zheng et al. (2005) showed that in vitro culture could lead to dysregulation of many genes and possibly affect the expression of MSH2, which may affect the viability of Rhesus monkey embryos. This raises the possibility of the same possible biological activity in humans and implies that the MMR system might play a role in the ART process and affect the outcome. ART offspring are associated with various risks such as preterm delivery, low birth weight, congenital abnormality, and genomic imprinting syndromes (Maher et al., 2003; Kurinczuk et al., 2004; de Ligt et al., 2012). However, it is difficult to establish whether these risks are related to the technology itself or the inherited genetic background. Whether the incidence of congenital abnormalities increases following ART is controversial (Halliday et al., 2010; Simon et al., 2010; Woldringh et al., 2010).
ART has been shown to have a definite relationship with an increased number of genetic and epigenetic alterations (Kochanski et al., 2013). ART may create de novo mutations through diverse chromosomal and molecular mechanisms. It enables the transmission of pre-existing mutations which are related to infertility and the natural process of early pregnancy termination. A Y-chromosome microdeletion is found in about 1 in 4000 in the general population and is responsible for male infertility. Feng et al. (2008) proposed that ART offspring may show a rising risk of a gene static mutation, azoospermia factor (AZF) microdeletion, even when their fathers have a normal spermatogenesis and genetic background. Bianchi et al. (2002) showed that Y chromosome MSI and AZF microdeletion testing in five cases of HNPCC strongly suggested a correlation between heterozygosity for MLH1 or other MMR gene mutations, Y chromosome instability, and AZF microdeletions. Thus, MMR gene mutations may play a role in the origin of AZF microdeletions. Given the fact of increased vertical transmission of Y-chromosome microdeletions in the offspring via ICSI (Serebrovska et al., 2006), the potential benefits and risks of adopting ART for those specific individuals who carry defective MMR genes should be considered.
Zheng et al. (2013) indicated that ART conceived babies display an increased incidence of gene dynamic mutation, also defined as TNR instability, which is related to more than 20 neuromuscular and neurodegenerative disorders including Huntington’s disease, myotonic dystrophy, and fragile X syndrome. Due to the specificity of TNR structure, the DNA damage repair system plays an important role in the formation of TNR alterations, except for its function in repairing de novo mutations (Fig. 1). As MMR protein expression can alter TNR in diverse ways by either enhancing or suppressing different human TNR types (Lin and Wilson, 2009; Halabi et al., 2012; Pluciennik et al., 2013), ART application should be considered more cautiously to prevent severe diseases.
Fig. 1.
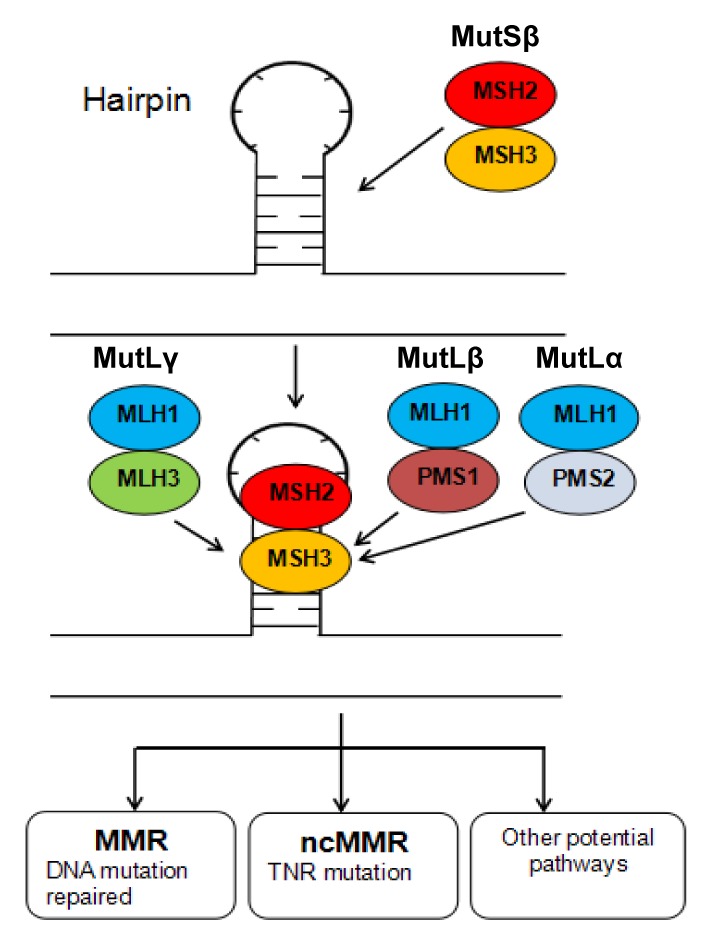
Proposed function model of MutS and MutL in relation to TNR instability
TNR-related hairpin structure occurring during DNA replication is initially recognized by the MutSβ complex, and then the MutL complex is recruited. Various pathways including canonical MMR and noncanonical MMR (ncMMR) may be engaged, leading to different outcomes of TNR instability: DNA damage repaired in time or otherwise, eventual TNR mutation
As for epigenetic variation, possible risks related to ART may be caused by either the use of incomplete reprogrammed sperm or in vitro embryo culture during a time of epigenetic reprogramming (Lucifero et al., 2004; Niemitz and Feinberg, 2004; Thompson and Williams, 2005). Geneticists have reported an increased incidence of selected epigenetic defects in ART offspring, such as Beckwith-Weidemann syndrome and Angelman syndrome (Maher et al., 2003; de Ligt et al., 2012). Although there is no evidence that the MMR system is involved in the epigenetic stability of ART offspring, we can assume that there might be an interaction since both MMR and epigenetic biological mechanisms are extremely active during gametogenesis and embryonic development. Our unpublished data show that ART procedures and the infertility background might affect MMR epigenetic modifications like DNA methylation. This needs further investigation.
Studies in mice and humans have reported that some genetic and epigenetic modifications caused by ART can be transmitted to the next or even to the F2 generation (van Montfoort et al., 2012; Wang et al., 2013). However, there have been no reports of transgenerational effects caused by MMR alterations. In consideration of its function in the parental generation, the inheritance of abnormal MMR gene status might affect aspects of an offspring’s health condition including predisposition to certain types of cancer, resistance to certain therapeutic drugs, and infertility. This area needs further study.
Given that the above, augmented genetic mutations seen in infertile individuals may raise the probability of these variants being inherited, conferring increased risk of infertility and then possible somatic diseases in later life in ART offspring. Research has shown that epigenetic mechanisms not only lead to inappropriate expression of the affected gene but may also expose hidden genetic variation, contributing to a predisposition to epigenetic instability in offspring conceived by ART (Sollars et al., 2003). Alternatively, the ART procedures and the genetic background may alter the genetic and epigenetic status of these offspring. More detailed information about infertility-related factors, gene mutations, DNA SNPs, other genetic and epigenetic forms of the MMR system, and correlated biomarkers should be found and ultimately applied clinically to provide better counseling and treatment for infertile couples considering IVF, ICSI, or even PGD, which aims to avoid inheriting defective genes. Also, long term follow-up studies of children born from ART are required to understand the possible effects on these offspring.
5. Conclusions
Apart from its primary role to correct replication-associated base-pair mismatches and IDLs, MMR also plays a crucial role in DNA recombination and is involved in gametogenesis. Infertility is an extremely complex disorder caused by various genetic, epigenetic, endocrine, and environmental factors. Although the etiology of human infertility is still largely unknown, the impact of genetic and epigenetic MMR system alterations on infertility is beyond debate. However, further studies are required to elucidate the molecular mechanisms of MMR genes and other additive risk factors in infertility. A better understanding of MMR alterations might be helpful in reducing DNA damage, developing gene targeted therapy, and providing diagnostic applications for ART.
The diverse functions of the MMR system in the human body, the applicability to humans of research based on transgenic, knock-out or knock-in mice models, and the great difficulty in obtaining and ethically using human gametes, especially oocytes and embryos, are huge obstacles to improving our understanding of MMR mechanisms in human infertility. More than thirty years of ART application and thirty-six years of observation of the first “in vitro” baby are still insufficient to thoroughly understand the potential changes and risks in later life resulting from infertility treatments. More work is urgently needed to investigate the interaction between MMR alterations and DNA integrity, human infertility, and ART security, to establish a better medical system with improved preventive, diagnostic, and therapeutic treatments of human infertility.
There is no doubt that ART will continue to be a major medical approach for infertile couples. Much work remains to be done to maximize the safety and reduce the risks of ART offspring. Long-term follow-up programs will yield significant results for fertility specialists providing ART, neonatologists working with ART offspring and geneticists counseling infertile parents. The efforts of global scientists, increasing maturity of the ART process, and the development of scientific technologies such as microarrays and single cell level sequencing, will lead to a better understanding of the cellular and molecular mechanisms of the MMR system involved in human infertility and related treatments. This will provide a better perspective to improve diagnostic capability, patient care, and ultimately the quality of the offspring.
Footnotes
Project supported by the National Basic Research Programs (973) of China (Nos. 2012CB944901 and 2014CB943302), the National Natural Science Foundation of China (Nos. 81200475, 81370760, and 81571500), and the Zhejiang Provincial Natural Science Foundation of China (No. LZ13H040001)
Compliance with ethics guidelines: Min-hao HU, Shu-yuan LIU, Ning WANG, Yan WU, and Fan JIN declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Baker SM, Bronner CE, Zhang L, et al. Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell. 1995;82(2):309–319. doi: 10.1016/0092-8674(95)90318-6. (Available from: http://dx.doi.org/10.1016/0092-8674(95)90318-6) [DOI] [PubMed] [Google Scholar]
- 2.Baker SM, Plug AW, Prolla TA, et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996;13(3):336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 3.Bianchi NO, Richard SM, Peltomaki P, et al. Mosaic AZF deletions and susceptibility to testicular tumors. Mutat Res. 2002;503(1-2):51–62. doi: 10.1016/s0027-5107(02)00072-6. (Available from: http://dx.doi.org/10.1016/S0027-5107(02)00072-6) [DOI] [PubMed] [Google Scholar]
- 4.Brown KD, Rathi A, Kamath R, et al. The mismatch repair system is required for S-phase checkpoint activation. Nat Genet. 2003;33(1):80–84. doi: 10.1038/ng1052. (Available from: http://dx.doi.org/10.1038/ng1052) [DOI] [PubMed] [Google Scholar]
- 5.Cannavo E, Gerrits B, Marra G, et al. Characterization of the interactome of the human MutL homologues MLH1, PMS1, and PMS2. J Biol Chem. 2007;282(5):2976–2986. doi: 10.1074/jbc.M609989200. (Available from: http://dx.doi.org/10.1074/jbc.M609989200) [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Sadowski I. Identification of the mismatch repair genes PMS2 and MLH1 as p53 target genes by using serial analysis of binding elements. PNAS. 2005;102(13):4813–4818. doi: 10.1073/pnas.0407069102. (Available from: http://dx.doi.org/10.1073/pnas.0407069102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dada R, Kumar M, Jesudasan R, et al. Epigenetics and its role in male infertility. J Assist Reprod Genet. 2012;29(3):213–223. doi: 10.1007/s10815-012-9715-0. (Available from: http://dx.doi.org/10.1007/s10815-012-9715-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Ligt J, Willemsen MH, van Bon BW, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367(20):1921–1929. doi: 10.1056/NEJMoa1206524. (Available from: http://dx.doi.org/10.1056/NEJMoa1206524) [DOI] [PubMed] [Google Scholar]
- 9.de Vries SS, Baart EB, Dekker M, et al. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 1999;13(5):523–531. doi: 10.1101/gad.13.5.523. (Available from: http://dx.doi.org/10.1101/gad.13.5.523) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaz-Padilla I, Romero N, Amir E, et al. Mismatch repair status and clinical outcome in endometrial cancer: a systematic review and meta-analysis. Crit Rev Oncol Hematol. 2013;88(1):154–167. doi: 10.1016/j.critrevonc.2013.03.002. (Available from: http://dx.doi.org/10.1016/j.critrevonc.2013.03.002) [DOI] [PubMed] [Google Scholar]
- 11.Edelmann W, Cohen PE, Kneitz B, et al. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat Genet. 1999;21(1):123–127. doi: 10.1038/5075. (Available from: http://dx.doi.org/10.1038/5075) [DOI] [PubMed] [Google Scholar]
- 12.Feng C, Wang L, Dong M, et al. Assisted reproductive technology may increase clinical mutation detection in male offspring. Fertil Steril. 2008;90(1):92–96. doi: 10.1016/j.fertnstert.2007.06.004. (Available from: http://dx.doi.org/10.1016/j.fertnstert.2007.06.004) [DOI] [PubMed] [Google Scholar]
- 13.Ferras C, Zhou XL, Sousa M, et al. DNA mismatch repair gene hMLH3 variants in meiotic arrest. Fertil Steril. 2007;88(6):1681–1684. doi: 10.1016/j.fertnstert.2007.01.063. (Available from: http://dx.doi.org/10.1016/j.fertnstert.2007.01.063) [DOI] [PubMed] [Google Scholar]
- 14.Ferras C, Fernandes S, Silva J, et al. Expression analysis of MLH3, MLH1, and MSH4 in maturation arrest. Reprod Sci. 2012;19(6):587–596. doi: 10.1177/1933719111428521. (Available from: http://dx.doi.org/10.1177/1933719111428521) [DOI] [PubMed] [Google Scholar]
- 15.Frazer KA, Murray SS, Schork NJ, et al. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10(4):241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 16.Garg K, Shih K, Barakat R, et al. Endometrial carcinomas in women aged 40 years and younger: tumors associated with loss of DNA mismatch repair proteins comprise a distinct clinicopathologic subset. Am J Surg Pathol. 2009;33(12):1869–1877. doi: 10.1097/PAS.0b013e3181bc9866. (Available from: http://dx.doi.org/10.1097/PAS.0b013e3181bc9866) [DOI] [PubMed] [Google Scholar]
- 17.Gnoth C, Godehardt E, Frank-Herrmann P, et al. Definition and prevalence of subfertility and infertility. Hum Reprod. 2005;20(5):1144–1147. doi: 10.1093/humrep/deh870. (Available from: http://dx.doi.org/10.1093/humrep/deh870) [DOI] [PubMed] [Google Scholar]
- 18.Guerrette S, Acharya S, Fishel R. The interaction of the human MutL homologues in hereditary nonpolyposis colon cancer. J Biol Chem. 1999;274(10):6336–6341. doi: 10.1074/jbc.274.10.6336. (Available from: http://dx.doi.org/10.1074/jbc.274.10.6336) [DOI] [PubMed] [Google Scholar]
- 19.Gunes S, Al-Sadaan M, Agarwal A. Spermatogenesis, DNA damage and DNA repair mechanisms in male infertility. Reprod BioMed Online. 2015;31(3):309–319. doi: 10.1016/j.rbmo.2015.06.010. (Available from: http://dx.doi.org/10.1016/j.rbmo.2015.06.010) [DOI] [PubMed] [Google Scholar]
- 20.Guo G, Wang W, Bradley A. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature. 2004;429(6994):891–895. doi: 10.1038/nature02653. (Available from: http://dx.doi.org/10.1038/nature02653) [DOI] [PubMed] [Google Scholar]
- 21.Halabi A, Ditch S, Wang J, et al. DNA mismatch repair complex MutSβ promotes GAA·TTC repeat expansion in human cells. J Biol Chem. 2012;287(35):29958–29967. doi: 10.1074/jbc.M112.356758. (Available from: http://dx.doi.org/10.1074/jbc.M112.356758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halliday JL, Ukoumunne OC, Baker HW, et al. Increased risk of blastogenesis birth defects, arising in the first 4 weeks of pregnancy, after assisted reproductive technologies. Hum Reprod. 2010;25(1):59–65. doi: 10.1093/humrep/dep364. (Available from: http://dx.doi.org/10.1093/humrep/dep364) [DOI] [PubMed] [Google Scholar]
- 23.Her C, Zhao N, Wu X, et al. MutS homologues hMSH4 and hMSH5: diverse functional implications in humans. Front Biosci. 2007;12(1):905–911. doi: 10.2741/2112. (Available from: http://dx.doi.org/10.2741/2112) [DOI] [PubMed] [Google Scholar]
- 24.Hienonen T, Laiho P, Salovaara R, et al. Little evidence for involvement of MLH3 in colorectal cancer predisposition. Int J Cancer. 2003;106(2):292–296. doi: 10.1002/ijc.11218. (Available from: http://dx.doi.org/10.1002/ijc.11218) [DOI] [PubMed] [Google Scholar]
- 25.Iyer RR, Pluciennik A, Burdett V, et al. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106(2):302–323. doi: 10.1021/cr0404794. (Available from: http://dx.doi.org/10.1021/cr0404794) [DOI] [PubMed] [Google Scholar]
- 26.Jascur T, Boland CR. Structure and function of the components of the human DNA mismatch repair system. Int J Cancer. 2006;119(9):2030–2035. doi: 10.1002/ijc.22023. (Available from: http://dx.doi.org/10.1002/ijc.22023) [DOI] [PubMed] [Google Scholar]
- 27.Ji G, Long Y, Zhou Y, et al. Common variants in mismatch repair genes associated with increased risk of sperm DNA damage and male infertility. BMC Med. 2012;10(1):49. doi: 10.1186/1741-7015-10-49. (Available from: http://dx.doi.org/10.1186/1741-7015-10-49) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7(5):335–346. doi: 10.1038/nrm1907. (Available from: http://dx.doi.org/10.1038/nrm1907) [DOI] [PubMed] [Google Scholar]
- 29.Joehlin-Price AS, Perrino CM, Stephens J, et al. Mismatch repair protein expression in 1049 endometrial carcinomas, associations with body mass index, and other clinicopathologic variables. Gynecol Oncol. 2014;133(1):43–47. doi: 10.1016/j.ygyno.2014.01.017. (Available from: http://dx.doi.org/10.1016/j.ygyno.2014.01.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang S, Li Y, Li B, et al. Genetic variation of the E-cadherin gene is associated with primary infertility in patients with ovarian endometriosis. Fertil Steril. 2014;102(4):1149–1154. doi: 10.1016/j.fertnstert.2014.07.005. (Available from: http://dx.doi.org/10.1016/j.fertnstert.2014.07.005) [DOI] [PubMed] [Google Scholar]
- 31.Karamurzin Y, Zeng Z, Stadler ZK, et al. Unusual DNA mismatch repair-deficient tumors in Lynch syndrome: a report of new cases and review of the literature. Hum Pathol. 2012;43(10):1677–1687. doi: 10.1016/j.humpath.2011.12.012. (Available from: http://dx.doi.org/10.1016/j.humpath.2011.12.012) [DOI] [PubMed] [Google Scholar]
- 32.Kim M, Trinh BN, Long TI, et al. Dnmt1 deficiency leads to enhanced microsatellite instability in mouse embryonic stem cells. Nucleic Acids Res. 2004;32(19):5742–5749. doi: 10.1093/nar/gkh912. (Available from: http://dx.doi.org/10.1093/nar/gkh912) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kneitz B, Cohen PE, Avdievich E, et al. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 2000;14(9):1085–1097. (Available from: http://dx.doi.org/10.1101/gad.14.9.1085) [PMC free article] [PubMed] [Google Scholar]
- 34.Kochanski A, Merritt TA, Gadzinowski J, et al. The impact of assisted reproductive technologies on the genome and epigenome of the newborn. J Neonatal Perinatal Med. 2013;6(2):101–108. doi: 10.3233/NPM-1366812. (Available from: http://dx.doi.org/10.3233/NPM-1366812) [DOI] [PubMed] [Google Scholar]
- 35.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9(1):89–96. doi: 10.1016/s0959-437x(99)80013-6. (Available from: http://dx.doi.org/10.1016/S0959-437X(99)80013-6) [DOI] [PubMed] [Google Scholar]
- 36.Kondo E, Horii A, Fukushige S. The interacting domains of three MutL heterodimers in man: hMLH1 interacts with 36 homologous amino acid residues within hMLH3, hPMS1 and hPMS2. Nucleic Acids Res. 2001;29(8):1695–1702. doi: 10.1093/nar/29.8.1695. (Available from: http://dx.doi.org/10.1093/nar/29.8.1695) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korhonen MK, Raevaara TE, Lohi H, et al. Conditional nuclear localization of hMLH3 suggests a minor activity in mismatch repair and supports its role as a low-risk gene in HNPCC. Oncol Rep. 2007;17(2):351–354. (Available from: http://dx.doi.org/10.3892/or.17.2.351) [PubMed] [Google Scholar]
- 38.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74(1):681–710. doi: 10.1146/annurev.biochem.74.082803.133243. (Available from: http://dx.doi.org/10.1146/) [DOI] [PubMed] [Google Scholar]
- 39.Kurinczuk JJ, Hansen M, Bower C. The risk of birth defects in children born after assisted reproductive technologies. Curr Opin Obstet Gynecol. 2004;16(3):201–209. doi: 10.1097/00001703-200406000-00002. (Available from: http://dx.doi.org/10.1097/00001703-200406000-00002) [DOI] [PubMed] [Google Scholar]
- 40.Lahue RS, Au KG, Modrich P. DNA mismatch correction in a defined system. Science. 1989;245(4914):160–164. doi: 10.1126/science.2665076. (Available from: http://dx.doi.org/10.1126/science.2665076) [DOI] [PubMed] [Google Scholar]
- 41.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18(1):85–98. doi: 10.1038/cr.2007.115. (Available from: http://dx.doi.org/10.1038/cr.2007.115) [DOI] [PubMed] [Google Scholar]
- 42.Lin Y, Wilson JH. Diverse effects of individual mismatch repair components on transcription-induced CAG repeat instability in human cells. DNA Repair (Amst) 2009;8(8):878–885. doi: 10.1016/j.dnarep.2009.04.024. (Available from: http://dx.doi.org/10.1016/j.dnarep.2009.04.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lipkin SM, Wang V, Jacoby R, et al. MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nat Genet. 2000;24(1):27–35. doi: 10.1038/71643. (Available from: http://dx.doi.org/10.1038/71643) [DOI] [PubMed] [Google Scholar]
- 44.Lipkin SM, Moens PB, Wang V, et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet. 2002;31(4):385–390. doi: 10.1038/ng931. (Available from: http://dx.doi.org/10.1038/ng931) [DOI] [PubMed] [Google Scholar]
- 45.Liu HX, Zhou XL, Liu T, et al. The role of hMLH3 in familial colorectal cancer. Cancer Res. 2003;63(8):1894–1899. [PubMed] [Google Scholar]
- 46.Lucifero D, Mann MR, Bartolomei MS, et al. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum Mol Genet. 2004;13(8):839–849. doi: 10.1093/hmg/ddh104. (Available from: http://dx.doi.org/10.1093/hmg/ddh104) [DOI] [PubMed] [Google Scholar]
- 47.Maduro MR, Casella R, Kim E, et al. Microsatellite instability and defects in mismatch repair proteins: a new aetiology for Sertoli cell-only syndrome. Mol Hum Reprod. 2003;9(2):61–68. doi: 10.1093/molehr/gag013. (Available from: http://dx.doi.org/10.1093/molehr/gag013) [DOI] [PubMed] [Google Scholar]
- 48.Maher ER, Afnan M, Barratt CL. Epigenetic risks related to assisted reproductive technologies: epigenetics, imprinting, ART and icebergs? Hum Reprod. 2003;18(12):2508–2511. doi: 10.1093/humrep/deg486. (Available from: http://dx.doi.org/10.1093/humrep/deg486) [DOI] [PubMed] [Google Scholar]
- 49.Mandon-Pepin B, Touraine P, Kuttenn F, et al. Genetic investigation of four meiotic genes in women with premature ovarian failure. Eur J Endocrinol. 2008;158(1):107–115. doi: 10.1530/EJE-07-0400. (Available from: http://dx.doi.org/10.1530/EJE-07-0400) [DOI] [PubMed] [Google Scholar]
- 50.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. (Available from: http://dx.doi.org/10.1038/nature08494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marcon E, Moens P. MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics. 2003;165(4):2283–2287. doi: 10.1093/genetics/165.4.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martins AD, Moreira AC, Sa R, et al. Leptin modulates human Sertoli cells acetate production and glycolytic profile: a novel mechanism of obesity-induced male infertility. >Biochim Biophys Acta. 2015;1852(9):1824–1832. doi: 10.1016/j.bbadis.2015.06.005. (Available from: http://dx.doi.org/10.1016/j.bbadis.2015.06.005) [DOI] [PubMed] [Google Scholar]
- 53.Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25(1):229–253. doi: 10.1146/annurev.ge.25.120191.001305. (Available from: http://dx.doi.org/10.1146/annurev.ge.25.120191.001305) [DOI] [PubMed] [Google Scholar]
- 54.Mukherjee S, Ridgeway AD, Lamb DJ. DNA mismatch repair and infertility. Curr Opin Urol. 2010;20(6):525–532. doi: 10.1097/MOU.0b013e32833f1c21. (Available from: http://dx.doi.org/10.1097/MOU.0b013e32833f1c21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muraki Y, Banno K, Yanokura M, et al. Epigenetic DNA hypermethylation: clinical applications in endometrial cancer (Review) Oncol Rep. 2009;22(5):967–972. doi: 10.3892/or_00000523. (Available from: http://dx.doi.org/10.3892/or_00000523) [DOI] [PubMed] [Google Scholar]
- 56.Ni B, Ma H, Lin Y, et al. Genetic variants in Ser-Arg protein-coding genes are associated with the risk of nonobstructive azoospermia in Chinese men. Fertil Steril. 2014;101(6):1711–1717. doi: 10.1016/j.fertnstert.2014.02.033. (Available from: http://dx.doi.org/10.1016/j.fertnstert.2014.02.033) [DOI] [PubMed] [Google Scholar]
- 57.Niemitz EL, Feinberg AP. Epigenetics and assisted reproductive technology: a call for investigation. Am J Hum Genet. 2004;74(4):599–609. doi: 10.1086/382897. (Available from: http://dx.doi.org/10.1086/382897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oliveira PF, Martins AD, Moreira AC, et al. The Warburg effect revisited–lesson from the Sertoli cell. Med Res Rev. 2015;35(1):126–151. doi: 10.1002/med.21325. (Available from: http://dx.doi.org/10.1002/med.21325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park JY, Seong SJ, Kim TJ, et al. Pregnancy outcomes after fertility-sparing management in young women with early endometrial cancer. Obstet Gynecol. 2013;121(1):136–142. doi: 10.1097/aog.0b013e31827a0643. (Available from: http://dx.doi.org/10.1097/AOG.0b013e31827a0643) [DOI] [PubMed] [Google Scholar]
- 60.Pashaiefar H, Sheikhha MH, Kalantar SM, et al. Analysis of MLH3 C2531T polymorphism in Iranian women with unexplained infertility. Iran J Reprod Med. 2013;11(1):19–24. [PMC free article] [PubMed] [Google Scholar]
- 61.Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol. 2003;21(6):1174–1179. doi: 10.1200/JCO.2003.04.060. (Available from: http://dx.doi.org/10.1200/JCO.2003.04.060) [DOI] [PubMed] [Google Scholar]
- 62.Perry JR, Hsu YH, Chasman DI, et al. DNA mismatch repair gene MSH6 implicated in determining age at natural menopause. Hum Mol Genet. 2014;23(9):2490–2497. doi: 10.1093/hmg/ddt620. (Available from: http://dx.doi.org/10.1093/hmg/ddt620) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pluciennik A, Burdett V, Baitinger C, et al. Extrahelical (CAG)/(CTG) triplet repeat elements support proliferating cell nuclear antigen loading and MutLα endonuclease activation. PNAS. 2013;110(30):12277–12282. doi: 10.1073/pnas.1311325110. (Available from: http://dx.doi.org/10.1073/pnas.1311325110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richardson ME, Bleiziffer A, Tuttelmann F, et al. Epigenetic regulation of the RHOX homeobox gene cluster and its association with human male infertility. Hum Mol Genet. 2014;23(1):12–23. doi: 10.1093/hmg/ddt392. (Available from: http://dx.doi.org/10.1093/hmg/ddt392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Santucci-Darmanin S, Neyton S, Lespinasse F, et al. The DNA mismatch-repair MLH3 protein interacts with MSH4 in meiotic cells, supporting a role for this MutL homolog in mammalian meiotic recombination. Hum Mol Genet. 2002;11(15):1697–1706. doi: 10.1093/hmg/11.15.1697. (Available from: http://dx.doi.org/10.1093/hmg/11.15.1697) [DOI] [PubMed] [Google Scholar]
- 66.Serebrovska ZA, Serebrovskaya TV, Pyle RL, et al. Transmission of male infertility and intracytoplasmic sperm injection (mini-review) Fiziol Zh. 2006;52(3):110–118. [PubMed] [Google Scholar]
- 67.Shih KK, Garg K, Levine DA, et al. Clinicopathologic significance of DNA mismatch repair protein defects and endometrial cancer in women 40 years of age and younger. Gynecol Oncol. 2011;123(1):88–94. doi: 10.1016/j.ygyno.2011.06.005. (Available from: http://dx.doi.org/10.1016/j.ygyno.2011.06.005) [DOI] [PubMed] [Google Scholar]
- 68.Shimodaira H, Yoshioka-Yamashita A, Kolodner RD, et al. Interaction of mismatch repair protein PMS2 and the p53-related transcription factor p73 in apoptosis response to cisplatin. PNAS. 2003;100(5):2420–2425. doi: 10.1073/pnas.0438031100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simon L, Brunborg G, Stevenson M, et al. Clinical significance of sperm DNA damage in assisted reproduction outcome. Hum Reprod. 2010;25(7):1594–1608. doi: 10.1093/humrep/deq103. (Available from: http://dx.doi.org/10.1093/humrep/deq103) [DOI] [PubMed] [Google Scholar]
- 70.Sollars V, Lu X, Xiao L, et al. Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nat Genet. 2003;33(1):70–74. doi: 10.1038/ng1067. (Available from: http://dx.doi.org/10.1038/ng1067) [DOI] [PubMed] [Google Scholar]
- 71.Stojic L, Cejka P, Jiricny J. High doses of SN1 type methylating agents activate DNA damage signaling cascades that are largely independent of mismatch repair. Cell Cycle. 2005;4(3):473–477. doi: 10.4161/cc.4.3.1528. (Available from: http://dx.doi.org/10.4161/cc.4.3.1528) [DOI] [PubMed] [Google Scholar]
- 72.Surtees JA, Argueso JL, Alani E. Mismatch repair proteins:key regulators of genetic recombination. Cytogenet Genome Res. 2004;107(3-4):146–159. doi: 10.1159/000080593. (Available from: http://dx.doi.org/10.1159/000080593) [DOI] [PubMed] [Google Scholar]
- 73.Terribas E, Bonache S, Garcia-Arevalo M, et al. Changes in the expression profile of the meiosis-involved mismatch repair genes in impaired human spermatogenesis. J Androl. 2010;31(4):346–357. doi: 10.2164/jandrol.109.008805. (Available from: http://dx.doi.org/10.2164/jandrol.109.008805) [DOI] [PubMed] [Google Scholar]
- 74.Thompson JR, Williams CJ. Genomic imprinting and assisted reproductive technology: connections and potential risks. Semin Reprod Med. 2005;23(3):285–295. doi: 10.1055/s-2005-872457. (Available from: http://dx.doi.org/10.1055/s-2005-872457) [DOI] [PubMed] [Google Scholar]
- 75.Valentini AM, Armentano R, Pirrelli M, et al. Chemotherapeutic agents for colorectal cancer with a defective mismatch repair system: the state of the art. Cancer Treat Rev. 2006;32(8):607–618. doi: 10.1016/j.ctrv.2006.08.001. (Available from: http://dx.doi.org/10.1016/j.ctrv.2006.08.001) [DOI] [PubMed] [Google Scholar]
- 76.van Montfoort AP, Hanssen LL, de Sutter P, et al. Assisted reproduction treatment and epigenetic inheritance. Hum Reprod Update. 2012;18(2):171–197. doi: 10.1093/humupd/dmr047. (Available from: http://dx.doi.org/10.1093/humupd/dmr047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang LY, Wang N, Le F, et al. Persistence and intergenerational transmission of differentially expressed genes in the testes of intracytoplasmic sperm injection conceived mice. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2013;14(5):372–381. doi: 10.1631/jzus.B1200321. (Available from: http://dx.doi.org/10.1631/jzus.B1200321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y, Qin J. MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. PNAS. 2003;100(26):15387–15392. doi: 10.1073/pnas.2536810100. (Available from: http://dx.doi.org/10.1073/pnas.2536810100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watson P, Lynch HT. The tumor spectrum in HNPCC. Anticancer Res. 1994;14(4B):1635–1639. [PubMed] [Google Scholar]
- 80.Wei K, Clark AB, Wong E, et al. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17(5):603–614. doi: 10.1101/gad.1060603. (Available from: http://dx.doi.org/10.1101/gad.1060603) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Win AK, Dowty JG, Antill YC, et al. Body mass index in early adulthood and endometrial cancer risk for mismatch repair gene mutation carriers. Obstet Gynecol. 2011;117(4):899–905. doi: 10.1097/AOG.0b013e3182110ea3. (Available from: http://dx.doi.org/10.1097/AOG.0b013e3182110ea3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Woldringh GH, Besselink DE, Tillema AH, et al. Karyotyping, congenital anomalies and follow-up of children after intracytoplasmic sperm injection with non-ejaculated sperm: a systematic review. Hum Reprod Update. 2010;16(1):12–19. doi: 10.1093/humupd/dmp030. (Available from: http://dx.doi.org/10.1093/humupd/dmp030) [DOI] [PubMed] [Google Scholar]
- 83.Wu Y, Berends MJ, Sijmons RH, et al. A role for MLH3 in hereditary nonpolyposis colorectal cancer. Nat Genet. 2001;29(2):137–138. doi: 10.1038/ng1001-137. (Available from: http://dx.doi.org/10.1038/ng1001-137) [DOI] [PubMed] [Google Scholar]
- 84.Xu K, Lu T, Zhou H, et al. The role of MSH5 C85T and MLH3 C2531T polymorphisms in the risk of male infertility with azoospermia or severe oligozoospermia. Clin Chim Acta. 2010;411(1-2):49–52. doi: 10.1016/j.cca.2009.09.038. (Available from: http://dx.doi.org/10.1016/j.cca.2009.09.038) [DOI] [PubMed] [Google Scholar]
- 85.Yi W, Wu X, Lee TH, et al. Two variants of MutS homolog hMSH5: prevalence in humans and effects on protein interaction. Biochem Biophys Res Commun. 2005;332(2):524–532. doi: 10.1016/j.bbrc.2005.04.154. (Available from: http://dx.doi.org/10.1016/j.bbrc.2005.04.154) [DOI] [PubMed] [Google Scholar]
- 86.Youn CK, Cho HJ, Kim SH, et al. Bcl-2 expression suppresses mismatch repair activity through inhibition of E2F transcriptional activity. Nat Cell Biol. 2005;7(2):137–147. doi: 10.1038/ncb1215. (Available from: http://dx.doi.org/10.1038/ncb1215) [DOI] [PubMed] [Google Scholar]
- 87.Zheng P, Schramm RD, Latham KE. Developmental regulation and in vitro culture effects on expression of DNA repair and cell cycle checkpoint control genes in rhesus monkey oocytes and embryos. Biol Reprod. 2005;72(6):1359–1369. doi: 10.1095/biolreprod.104.039073. (Available from: http://dx.doi.org/10.1095/biolreprod.104.039073) [DOI] [PubMed] [Google Scholar]
- 88.Zheng YM, Li L, Zhou LM, et al. Alterations in the frequency of trinucleotide repeat dynamic mutations in offspring conceived through assisted reproductive technology. Hum Reprod. 2013;28(9):2570–2580. doi: 10.1093/humrep/det294. (Available from: http://dx.doi.org/10.1093/humrep/det294) [DOI] [PubMed] [Google Scholar]


