Abstract
Neurosensory hearing loss is a growing problem of super-aged societies. Cochlear implants can restore some hearing, but rebuilding a lost hearing organ would be superior. Research has discovered many cellular and molecular steps to develop a hearing organ but translating those insights into hearing organ restoration remains unclear. We cannot make various hair cell types and arrange them into their specific patterns surrounded by the right type of supporting cells in the right numbers. Our overview of the topologically highly organized and functionally diversified cellular mosaic of the mammalian hearing organ highlights what is known and unknown about its development. Following this analysis, we suggest critical steps to guide future attempts toward restoration of a functional OC. We argue that generating mutant mouse lines that mimic human pathology to fine-tune attempts toward long-term functional restoration are needed to go beyond the hope generated by restoring single hair cells.
Keywords: cochlea, organ of Corti, development, patterning, diffusible factors, lateral inhibition, Atoh1, expression regulation
“What I cannot create, I do not understand”
R. Feynman (http://archives.caltech.edu/pictures/1.10-29.jpg)
Introduction
Development progressively restricts alternative cell fates [1] via sophisticated regulatory loops within and between cells [2–4] to regulate gene expression levels in a given cell by morphogen gradients [5] that are fine-tuned by local cellular interactions [6–8]. Combined, these signals lead to gene expression profiles that specify cellular phenotype in the right position. Such processes are best understood in flies, where single continuous rows of cells can be defined out of homogenous precursors [9,10].
The vertebrate ear is among the few biological examples that has yielded molecular insights into progressive developmental commitment and cellular specification that transform a uniform epithelium into an organ of Corti (OC) [11,12]. The OC consists of rows of hair cells (HC) that are able to transduce sound related mechanical stimuli into ionic influx at the tip of interconnected stereocilia [13]. This ionic influx leads to a graded change of the HC potential and stimulation of postsynaptic neurons to conduct frequency-specific information to the brain [14,15]. The OC is the most sophisticated, two-dimensional cellular mosaic of the vertebrate body, consisting of a HC and supporting cell layer [16,17]. Such a two-dimensional structure can be easily imaged and yet, 130 years after the first description of its development [18], we cannot predict when we will be able to restore a lost OC, and thus hearing, to the millions of deaf people worldwide [19,20]. This problem is becoming more profound with the increasing longevity of the super-aged society despite significant efforts to generate new HCs or delay their loss through various means [21,22]. In the last 15 years, ear research has mostly focused on a few transcription factors known to regulate general HC differentiation such as Atoh1 [23–27]. Additional work aimed at revealing how HCs interact with their immediate neighbors through cell-cell interactions (Delta-Notch) but also with diffusible factors (Fgf’s) in a complex network [28–31] to generate a mosaic of cells organized into the functionally relevant cellular architecture of the mature OC (Fig. 1).
Figure 1.
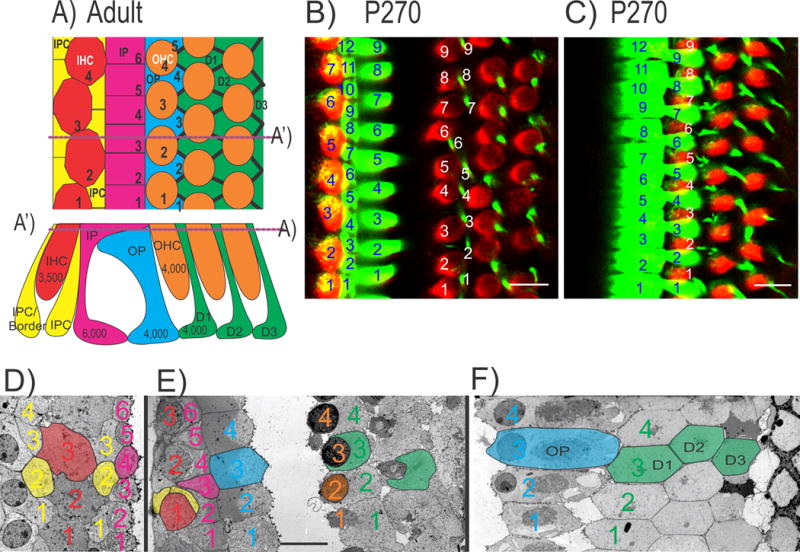
The diagram shows the adult mosaic of hair cells (HCs) and supporting cells of the organ of Corti (OC; a) with numbers of cell types inserted for the adult human cochlea. Dotted line in (a) indicates the section plane in (a’) and vice versa. The total number of inner hair cells (IHCs) to inner pillar (IP) cells is almost a 1:2 ratio whereas each of the three rows of outer hair cells (OHCs) to outer pillar (OP) and Deiters’ cells (D1, D2, D3) is a 1:1 ratio (a–f). The ratio of IP to OP cells (4:3) indicates numerical differences between the medial and lateral compartments possibly related to distinct quantitative regulatory mechanisms. The distribution of HCs and supporting cells in the mouse cochlea show a similar pattern revealed by immunohistochemistry of Myo7a and Tubulin at P270 (b, c). The ratio of IHCs to IP cells is nearly 2:3 but the ratio of OHCs to OP/Deiters’ cells is 1:1 with an IP to OP cells ratio of 4:3 (b, c). The lower row shows transmission electron microscopy (TEM) sections taken parallel to the reticular lamina of an adult guinea pig through the inner compartment (d), the inner and outer compartment (e) and only the outer compartment (f). Colored numbers identify HCs and supporting cells with the same color code as in the drawing in (a). Note that IP cells are most abundant (6 in d, e). There are four inner phalangeal cells (IPC, sometimes also referred to as border cells) medial (yellow 1–4 in d) and 3.5 inner phalangeal cells lateral to the 3.5 IHCs, suggesting a ratio of nearly 2:1 between IPCs and IHCs. IHCs are in broad contact with each other at this section plane but are only in limited contact with IP cells. Note that the outer compartment consists of a mosaic of 4 OP cells, 4 OHCs and 4 Deiters’ cells (g) per row. IP cells are the most numerous cells forming a single row with no obvious ratio to other cell types of the OC. Scale bar represents 10 μm in b–f. Modified after [56,88,116].
While great progress has been achieved, this review stresses gaps in our understanding that prevent translating developmental insights into restoring an OC. To paraphrase R. Feynman, we are not (yet) able to create an OC in vitro or to transform in vivo a non-mammalian vertebrate hearing organ into an OC. Restoring HCs in adult vestibular organs is a great step in the right direction [32] but the added complexity of the cellular assembly of the OC must be considered to restore a functional OC. We argue that the current inability to restore hearing is in part related to incomplete understanding of the molecular development of the cellular mosaic of the OC and intracellular signal pathways of HCs to generate OC specific cell types and distribution pattern, including how these intracellular signals are modified by cell-cell interactions and diffusible gradients.
The Organ of Corti has locally variable cell arrangements
The OC develops from a uniform otocyst epithelium into a mosaic of two types of HCs and up to five types of supporting cells [inner (IP) and outer pillar (OP), Deiters’, inner phalangeal (IPC), including border cells]. A checkerboard is obvious for outer hair cells (OHCs) and Deiters’ cells in every mammalian species (Fig. 1,2). However, IHCs always form a single row of broadly contacting cells adjacent to a single row of broadly contacting IP cells [16]. Limits of this patterning process are revealed in species with additional rows of randomly distributed HCs or widely interrupted distributions of IHCs [33].
Figure 2.
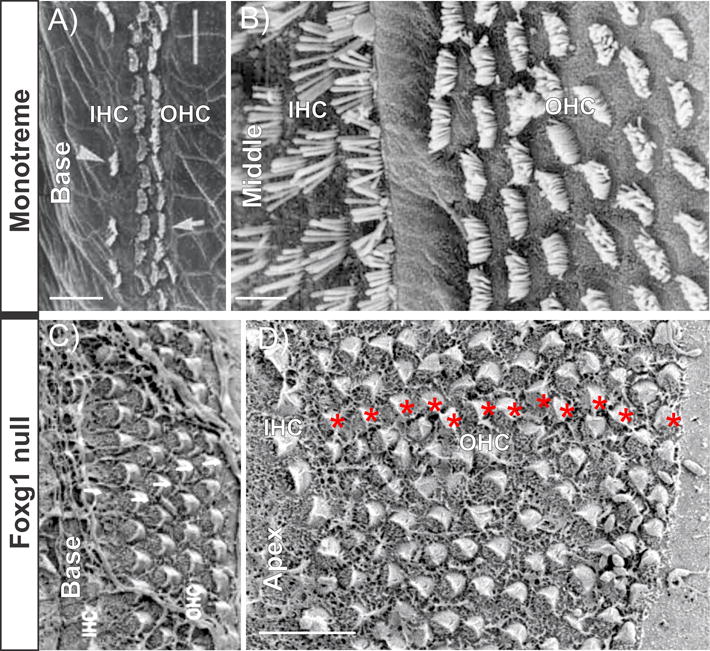
Scanning electron microscopy (SEM) images show the similarity of HC distribution in a monotreme (a, b) and Foxg1 null mouse cochlea (c, d). Monotreme cochlea has multiple rows of both IHCs and OHCs in the apex (b). However, the base of the monotreme cochlea has only one row of IHCs and two rows of OHCs (a). The Foxg1 null mouse cochlea has up to 12 rows of OHCs in the apex (red * in d) but HC distribution remains near normal in the base (c). Both monotreme and Foxg1 null mouse have a much shortened cochlea. Absence of convergent extension of the growing cochlear duct can affect the apical HCs without having much effect on the base whereas reduced proliferation always leads to shortened OC. Scale bar represents 10 μm. Modified after [33,37].
Some papers view the development of the OC as the development of a simple checkerboard [6,8,34] while others depict it as parallel rows of different cells [35]. Both views are true but for two distinctly patterned compartments of the OC: the outer compartment has three rows of OHC alternating with Deiters’ cells and 1 rows of OP cells [sometimes up to four rows [36]]. The inner compartment consists of IPCs (including Border cells) and one contiguous row of IHCs. Variations of rows of HCs and pillar cells can be experimentally generated in specific mutations (Fig. 2), such as Foxg1 null mutants [37]. Separating both compartments is a single row of IP cells adjacent to a row of OP cells (Fig. 1). How the single rows of IHCs, IP and OP cells are determined to form the highly unusual configuration of three contiguous and parallel rows of cells – not found in any other sensory organ of the vertebrate ear [24] – is unknown. This problem is further aggravated by the numerical differences in these two adjacent supporting cell types with a different numerical ratio of IP: OP as well as IHC: OHC (Fig. 1).
Given the numerical ratio of IHC to OHC (Fig. 1), different cell patterns (alternating or rows) and the various supporting cell types, there is no easy way to imagine the entire development of the OC to be uniformly guided by a single uniform lateral inhibition principle, even if more recent variations are taken into consideration [7,8,38]. Importantly, it is conceptually difficult to understand how IP and OP should be derived in their specific location based only on a lateral inhibition model that assumes uniformity of cell types prior to the selection process [6,8,34]. The fact that two supporting cells (IP, OP) are in broad contact with each other radially is difficult to reconcile with a ubiquitous Delta-Notch inhibition, even considering different compartmental effects of ligands [39] and differential distribution of downstream effectors such as Hes1/5 [40] and various functional variations of notch signaling [34,41].
The formation of the single row of IP cells in the OC is reminiscent of the single row of single-minded (sim) gene expressing cells in the developing fly embryo [9]. This single line of bHLH gene expressing cells in flies may come about through Delta-Notch interaction with Tgf [34], and defines two flanking compartments. For the OC it is conceivable that Delta/Notch cellular interactions may cooperate with the BMP4/Fgf gradients across the OC [11,42] to define the IP cells and other supporting cells in two distinctly patterned compartments. Given the known complexity of the Delta/Notch signaling pathway alone with different ligands, downstream effectors and the Notch receptor responding likely to ligands expressed at diverse cells differently [7,34,38,43] more work is needed to sort out how the cell-cell interactions via the Delta/Notch system cooperates with the various morphogen gradients identified in the developing OC [11,44] to define the position and differentiation of the molecular and anatomically distinct single row of IP cells. Clearly, sorting out this single line of special supporting cells requires as much consideration as formation of the single line of sim positive cells in flies has attracted over the last 30 years.
In summary, the formation of the IP as the dividing line between the inner and outer cochlear compartment is a crucial event in OC evolution [24] and a central problem of OC development that may also directly tie into the OC elongation [45]. Any model of OC development needs to explain how the odd number of contiguous IP cells comes about and how IP cells are placed exactly on the bony lip of Rosenthal’s canal to segregate the differently patterned inner and outer compartment (Fig. 1). Figuring out how this is achieved matters from a theoretical perspective of patterning analysis as a compromise between cellular interactions and diffusible factors [10]. On a practical level, numerical and topological correct formation of the IP cells is essential for OC function as shown in Fgfr3 mutant mice [46]. IP cells are truly unique also in their molecular development, expressing both proneural bHLH genes [47,48] and multiple Hes/Hey factors [40]. There is a long-term viability of IP cells in molecularly engineered cases of HC loss [49,50], which is also found in certain ablation studies of HCs [51]. Combined with the known variation in human cochlear ducts [52], these variations in OC prior to and, in particular, after HC loss makes a straightforward therapeutic approach to restore an OC extremely difficult because inter-individual differences need to be considered related to absence and presence of IP cells.
Evolving gene networks help differentiate OC specific cell types
Part 1 outlined the complexity of the OC with an emphasis on various support cells. The remaining part of this review will present a transcription factor centric perspective of OC development with a focus on HCs. We will highlight expression of the basic helix-loop-helix (bHLH) transcription factor Atoh1 in the developing OC, its interaction with other transcription factors, including factors of the same family, how it regulates intercellular signals (both diffusible and cell-cell interactions), and what might be needed to turn Atoh1 into a factor to regenerate different types of HCs. Given the prominence of Atoh1 for HC differentiation, attempts to generate new HCs in vitro or in vivo either express Atoh1 or upregulate Atoh1 through de-repression [21,22,53,54]. While progress has been made, no OC specific HCs that can function for many years have been generated. This is not unusual because as it is still not possible to generate a fully functional, lasting β-cell [55]. Given the added complexity of the OC [56], it will likely require additional levels of understanding of normal development to ensure the topographically and numerically correct placement of the two HC types. Like many factors that can restore embryonic features [4], Atoh1 is involved in carcinogenesis [57]. Restoring embryonic properties to improve regeneration bear a significant risk for tumor formation, thus far unknown for the OC.
Atoh1 belongs to an ancient family of bHLH transcription factors already found in bacteria plants and fungi. Diversification and formation of complex interactions of bHLH TFs appears to be the basis for animal cellular diversification [2]. The bHLH gene Atoh1 forms a clade with other bHLH genes that drive neuronal differentiation (Neurod1, Neurog1, Ascl1). These bHLH genes interact extensively with other bHLH genes [58] that are either needed as co-factors for DNA binding (Tcf’s/E-proteins), or inhibit DNA binding through absence of a basic domain (Id1-4). Other bHLH genes inhibit the transcription of proneural bHLH genes (e.g., Hes1 and Hey1) through heterodimerization with Tcf’s/E-proteins or direct suppression of DNA binding [58].
These transcription factors form a network of cross-regulation to fine tune expression of downstream genes. For example, da (daughterless; Tcf/E-proteins in mammals) are partners for DNA binding of proneural transcription factors (Fig. 3) such as the fly atonal (Atoh1 in mammals). Heterodimers bind to target genes and define the specific signaling [59]. Tcf/E proteins also have signal potential of their own as homodimers [60] like regulating the expression of emc (extra macrochaete; Id proteins in mammals) by suppressing emc expression in the neuronal lineage while promoting emc expression in the non-neural lineage [61]. This feedback-feedforward loop of bHLH genes (Fig. 3), combined with the specifically regulated expression of proneural genes such as atonal in flies, defines the regular mosaic of omatidia during fly eye development. A similar network of interacting TF’s is present in mammalian OC development with the added complexity of multiple interacting proneural TFs within cells (Fig. 3).
Figure 3.
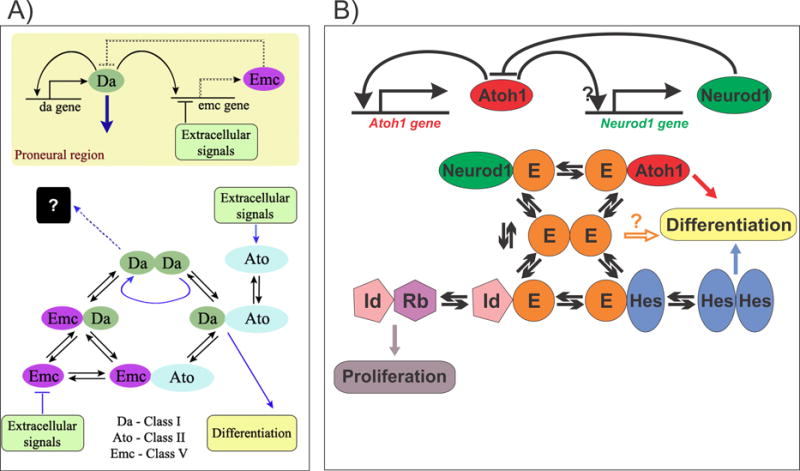
Interactions between bHLH genes are shown in the developing fly eye and mouse ear. In the fly (a), bHLH gene Ato (atonal) forms a heterodimer with Da (doughterless) and this binding preference promotes the differentiation by outcompeting the binding with Emc (extramacrochaete). Comparable data in mice (b) suggest that proneural transcription factors (such as Atoh1 and Neurod1) compete for E-proteins to form heterodimers for enhancer binding. In addition, the Hes/Hey factors and Id (inhibitor of DNA binding) proteins can form heterodimers with E-proteins, limiting availability of E-proteins for proneural heterodimerization. E-protein binding preferences and concentration will determine the differentiation of HCs/ neuron/ supporting cells or continuation of proliferation of prosensory precursors. Modified after [61,88,117].
This intracellular bHLH gene network (Fig. 3) is further increased in its complexity through the involvement of the Delta-Notch system of lateral inhibition that regulates the level of expression of Hes/Hey genes between cells that interact with several other bHLH genes [8,34,40]. In addition, the level of proneural bHLH factors further complexes the intracellular bHLH gene network. A crucial step in unfolding the interactions is the spatial upregulation of proneural bHLH genes to drive neuronal differentiation and of emc/Id in non-neural cells to retain them as supporting cells as demonstrated in flies [61]. These intracellular interactions (Fig. 3) are reinforced by the Delta-Notch mediated upregulation of Hes/Hey genes. Enhancers for bHLH gene expression are known [62] but what drives Atoh1 expression specifically in hair cell precursors is unknown.
Given that this model of progressive change of bHLH gene expression requires outside signal input to regulate various bHLH TFs in different cell types, one needs to define these inputs and how they themselves are regulated to result in localized activation of the downstream effector genes. Obviously, such signals must be tied into diffusible signals that can generate the pre-pattern necessary to regulate the cell-specific upregulation of proneural bHLH genes. In the fly retina, the hedgehog and wingless signal play this role in an iterative fashion [63]. Ear [64] and brain development follows a similar developmental principle in vertebrates [65], just using different subsets of the same gene families. How many equivalent steps are taken in brain and ear development remains to be seen beyond those genes that are already known to be essential for ear and brain development such as Foxg1 [37], Pax2/8 [66], Lmx1a [67], Foxi family members [68] and Gata3 [69].
In summary, Atoh1 belongs to an ancient family of genes that have undergone multiple rounds of duplication and diversification [70]. Throughout this process, the basic DNA binding domain has remained highly conserved [27,70] comparable to the evolution of the BAF complex [4]. Recent replacement experiments of Atoh7 with Neurod1 show identical function in the retinal ganglion cells [71]. In contrast, replacing Neurod1 with Atoh7 results in reprogramming of amacrine cells (inhibitory neurons) [72], indicating that the function of each gene is highly context dependent. In the ear, replacing Atoh1 with Neurog1 leads to very limited initiation of HC differentiation, implying a role for Atoh1 in stereocilia differentiation [73]. Finding a bHLH TF to substitute for Atoh1 to initiate HC development in postmitotic SCs without transforming adult SCs into HCs [54] could avoid depleting the functionally essential SCs of the OC.
Atoh1 is a gene for many cell types and developmental stages
Atoh1 was first identified in the cerebellum, regulating the most abundant cells of the mammalian brain [74] and later in HC differentiation of the ear [26]. These initial papers were followed by expression analysis in a number of other neuronal, sensory, and non-neuronal systems such as cochlear nuclei [75], Merkel cells [76], multiple other neurons of the proprioceptive pathway [77], and the Paneth cells in the intestine [78]. These data show three things:
Atoh1 is mainly associated with neurosensory but also with non-sensory development.
Atoh1 alone does not define a specific cell type.
Atoh1 function is context-dependent.
In the ear, other factors are needed to set the stage for Atoh1 action such as Sox2 [79], Six1/Eya1 [62], Pax2/8 [66] and Gata3 [69] and many other factors [35]. Context dependency of effects of a given gene is already commonplace in other developing systems where single genes can have inverse interactions in adjacent cells, depending on the context [80].
Beyond the expression of Atoh1 in different cell types in the developing mammalian embryo, Atoh1 is not specific to a given step in development. For example, in the cerebellum, Atoh1 is expressed in proliferating granule cell precursors [57]. In the developing intestine, Atoh1 defines the Paneth cells, which in turn define the stem cells that give rise to enteroendocrine cells [78]. As in the cerebellum [81], Atoh1 drives Neurod1 which in turn inhibits Atoh1 expression [82]. The OC lags behind in understanding the complexity of interactions needed for an in vitro recapitulation of normal development as is now possible with the ‘in vitro’ intestine [82]. Recent work strongly supports the notion of context for Atoh1 function in HC development, showing that level of Atoh1 expression depends on upstream genes that regulate downstream genes, such as Pou4f3 [62], needed for effective in vitro HC development [83] but also for long term maintenance of HCs in vivo [84,85].
Atoh1 expression levels fine tune OC development
Atoh1, like most other gene expression profiles that have been analyzed [86] will presumably follow sigmoidal upregulations (Fig. 4). PCR data show that Atoh1 expression starts in the ear at E10.5 [47]. PCR lags crucial topological information thus precluding to define the cell types. Several techniques that allow quantification of local mRNA in cells have been developed [87] and are needed to investigate cell based quantitative expression profiling in the ear. Tissue based data are less sensitive and lag behind PCR thus showing delayed expression reflecting the sensitivity relative to PCR.
Figure 4.
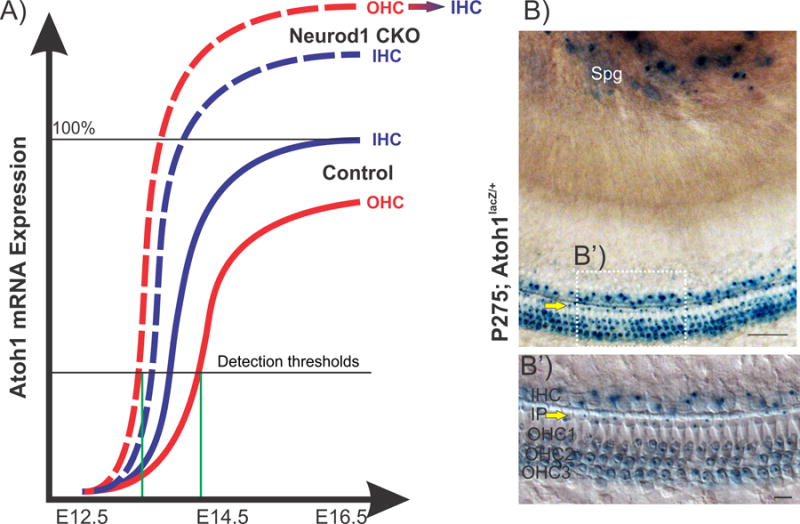
This hypothetical diagram correlates semi-quantitative mRNA expression of Atoh1 at different time points in mouse cochlear development in two HC types (a). In control (solid lines), Atoh1 is first expressed in IHCs followed by OHCs. But in the Neurod1 conditional null (CKO, Pax2-Cre; Neurod1f/f) mouse (dashed lines), this expression pattern is reversed and might be responsible for the transdifferentiation of some OHCs into ectopic IHCs in this mutant. Blue lines represent expression in IHCs and red lines for expression in OHCs. Green lines indicate the detection threshold of Atoh1 expression in OHCs, which is reached much earlier in Neurod1 CKO than in control. Atoh1-LacZ expression (b) in P275 mice shows reaction product in inner pillar cells (IP, yellow arrows) and in all HCs. Spg, spiral ganglion. Images modified after [25,47,88,89]. Scale bars represent 100 um.
Many markers exist for Atoh1 such as LacZ knockin, eGFP transgenes, in situ probes and monoclonal and polyclonal antibodies (Fig. 4, 5). All data agree that Atoh1 expression starts in the cochlea around embryonic day 13.5 in the upper middle turn, progressing toward the base and the apex [47,88]. In situ hybridization data show a downregulation after birth (Fig. 5) in a base to apex progression [89,90]. q-PCR data are needed to confirm the steepness of the Atoh1 downregulation [91], also suggested by immunohistochemistry [48]. Atoh1-LacZ reveals expression 9 months after birth in HC and IP cells (Fig. 4) [47], implying very low expression difficult to detect by current techniques. Limited expression of Atoh1 would be consistent with HC loss in Atoh1 hypomorphic mice [92] and rescue of HC damage with Atoh1 overexpression [93]. Inconsistencies of expression data go beyond the temporal expression in HCs. For example, expression of Atoh1 in IP cells and neurons [47] were ignored until recently confirmed [48,94] during early development.
Figure 5.
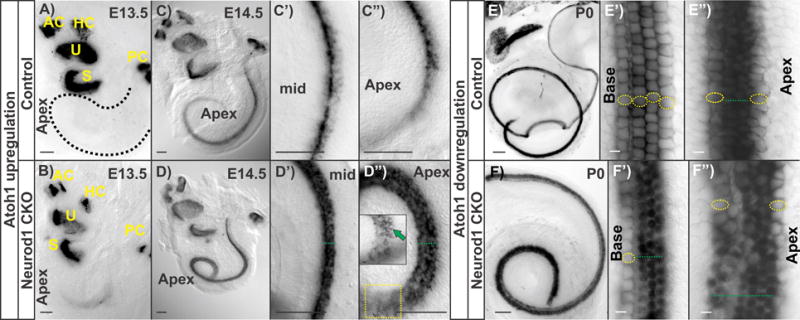
Atoh1 expression revealed by in situ hybridization in control and in Neurod1 CKO (Pax2-Cre; Neurod1f/f) in different stages is compared with the upregulation and downregulation pattern in different cochlear HCs. In control cochlea, Atoh1 is upregulated longitudinally in a base to apex progression and radially from IHCs to OHCs (a, c-c”). In the Neurod1 CKO, Atoh1 is prematurely expressed in the apex and the OHCs first (green arrow in insert in d”, b, d-d”). Downregulation of Atoh1 follows a base to apex progression where Atoh1 is reduced earlier in IHCs and outermost OHCs in the control P0 cochlea (e-e”). In addition to premature upregulation, Atoh1 expression persists longer in Neurod1 CKO compared to the control cochlea (f-f”). Yellow circles indicate the cells with downregulated Atoh1 expression and green dotted lines indicate the persistence of Atoh1 expression. Scale bar represents 100 μm. AC, anterior canal crista; HC, horizontal canal crista; PC, posterior canal crista; S, saccule; U, utricle. Modified after [25,89].
In summary, the existing data on Atoh1 expression agree on the overall onset and topology of expression but either disagree or do not report expression in non-HCs [88]. At present, the function of Atoh1 in developing IP cells [40] is merely speculative. Interestingly, IP cells are, with respect to co-expression of a proneural bHLH TF (Atoh1) [47] and Hes/Hey [40], comparable to the single row of sim expressing cells in flies [9]. It appears that Atoh1 expression is not sufficient to turn just any Atoh1 expressing supporting cell into a long term viable HC, but that it requires absence or presence of additional co-expressed factors, in particular Hes/Hey [40].
Another point of variable interpretation is how Atoh1 relates to HC specification/differentiation [12]. HC exit the cell cycle in the apex [95] before Atoh1 is expressed in the apex [47]. This expression suggests that Atoh1 acts as a HC differentiation factor to be upregulated in postmitotic cells that express proteins that keep precursors from re-entering the cell cycle. Eliminating such proteins that block HC from re-entering the cell cycle (Rb mutants, p27 mutants) results in extra hair cells [96,97]. Consistently, eliminating proteins that reduce proliferation also reduces HC formation including loss of the basal turn [98]. In summary, Atoh1 expression is insufficient to drive HC commitment, regardless of whether the expression is inside or outside the ear. Like expression of p27, Atoh1 expression appears to be a consequence of cell cycle exit and thus HC precursor determination instead of causing HC determination. This is important to recognize in the context of Atoh1 mis/over-expression [54,99] and Atoh1 de-repression [100], the two leading attempts to drive supporting cells to differentiate as HCs. If Atoh1 is expressed naturally in supporting cells such as in IP cells (Fig. 4), we need to understand why those cells are not converted into HCs by this expression [40] and what conditions can change that [35] to reliably use this for restoration of lost HCs.
Atoh1 interacts with other bHLH genes to regulate cell types
Atoh1 belongs to a family of transcription factors that multiplied and diversified the DNA binding domain to regulate different downstream genes. Following multiplication, the enhancer regions [70] were modified for differential expression in cell types or sequential regulation within a cell type [3]. This resulted in complicated cross-regulation of co-expressed family members that ensure transitions from a single original sensory cell to two types of neurosensory cells found in vertebrates: the neurons and the HCs [101].
Consistent with this evolutionary scenario is work on Neurog1 null mutants that showed not only absence of neurons [102] but also reduction of HCs, in particular in the saccule and the OC. Subsequent work showed expression of Atoh1-lacZ in some spiral ganglion neurons [47]. The data suggested that common sensory neuron-HC precursors exist [64], consistent with work showing that neurotrophin expression is in precursors of HCs and neurons [103,104]. Later findings confirmed that Neurog1 and Atoh1 interact in some common precursors of HCs and sensory neurons for certain vestibular sensory epithelia [105]. Further insights into this complexity of cross-regulation was revealed by manipulating Neurod1, a bHLH transcription factor that interacts with both Neurog1 and Atoh1 [89].
In the cerebellum, Atoh1 regulates Neurod1 expression, which negatively regulates Atoh1 [81]. A similar feedback loop exists in the ear where Neurod1 suppresses Atoh1 expression in neurons. In the absence of Neurod1, Atoh1 expression is maintained and neurons turn into intraganglionic HCs [25]. The effect of Neurod1 on HCs is more subtle, and lack of Neurod1 causes a premature expression of Atoh1 in the apex and in OHCs earlier than IHCs (Fig. 4, 5) [25]. These changes in expression translate into alterations in HC type, turning some OHCs into IHC-like cells with respect to the diameter of the stereocilia. Interestingly, deleting one copy of Atoh1 partially rescues the HC alteration phenotype in Neurod1 mutants, leading to fewer ectopic IHCs compared to the Neurod1 deletion alone (Fig. 6). This suggests an effect of Atoh1 haploinsufficiency not seen before in Atoh1, possibly unmasked through altered Atoh1 expression in the critical range for HC type determination (Fig. 4).
Figure 6.
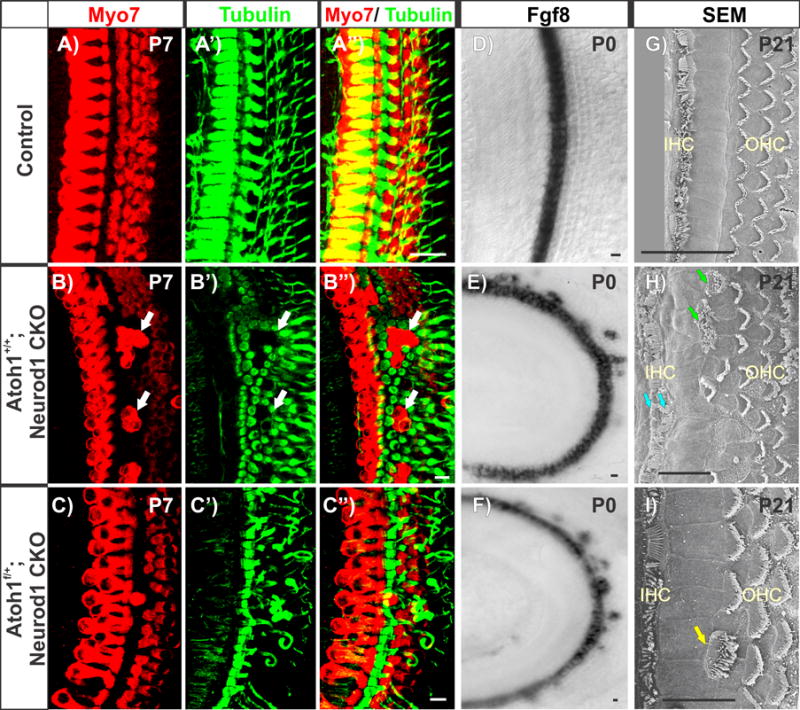
Immunohistochemistry of Myo7a and Tubulin in Neurod1 CKO mice shows absence of Neurod1 results in disorganized apical OC with multiple rows of IHCs, ectopic IHCs in the position of OHCs (white arrows in b-b”), and formation of extra pillar cells surrounding ectopic IHCs (b-b”). Deletion of one Atoh1 allele in the Neurod1 CKO mutant (Pax2-Cre; Neurod1f/f; Atoh1f/+) can partially correct the disorganization in the apex (c-c”) including correction of extra rows of pillar cells, which is consistent with HC type formation being dependent on the level of Atoh1 expression. However, this mutant occasionally eliminates the outermost row of OHCs (c, c”). Fgf8 is expressed in one row of IHCs in combined Atoh1 haploinsufficient and Neurod1 CKO mice instead of multiple rows as in Neurod1 CKO, but shows some misexpression in OHCs at P0 (e, f). This suggests that altered Atoh1 expression in the absence of Neurod1 (Fig. 5) can misexpress Fgf8 (e) and reduce Atoh1 expression level in this combined mutant mice (f). This may suggest spurious regulation of Atoh1 and Fgf8 bu Neurod1 or other unknown factors. Note that simple Atoh1 heterozygous mice do not have any phenotype. The scanning electron microscopy (SEM) images show two rows of IHCs (cyan arrows in h) and ectopic IHCs with thick stereocilia in the position of OHCs (green arrows in h) in P21 Neurod1 CKO mouse cochleae (h). Atoh1 haploinsufficient and Neurod1 CKO mice reveal dramatic correction of the OC disorganization with occasional ectopic IHCs (yellow arrow in i). These data suggest a critical dosage effect of Atoh1 in HC specific stereocilia thickness. Scale bar represents 10 μm except 20 μm in g. Modified after [25].
The alteration of Atoh1 levels and progression is currently the only known cause of IHC versus OHC differentiation (Fig. 6), and may include the regulation of Fgf8 expression levels [25], the earliest marker for IHCs [30]. Consistent with the proposed important functional role of Fgf8 in the OC development [11], such misregulation of Fgf8 affects surrounding supporting cell development. Furthermore, Fgf8 is absent when Atoh1 is transiently expressed [90] or when Atoh1 is replaced by another bHLH transcription factor such as Neurog1 [25]. Given Fgf8’s role as a signaling center, for example in brain development [106], resolving the interplay of level of Atoh1 expression and signaling to regulate Fgf8 in OHCs could provide important clues to regenerate an OC.
The function of Atoh1 in HC differentiation [26] implied a potential mechanism to restore lost HCs. Atoh1 has the ability to turn some cells of the ear into HCs [54,99]. Unfortunately, those transformed cells have only a limited viability. Atoh1 function even in the ear will be context dependent, as demonstrated by the obvious expression of Atoh1 in many IP cells (Fig. 4) and sensory neurons [47] that require additional manipulations to be turned into HCs [25,54,89]. What the context is that allows IHC or OHC development, in part through altered Atoh1 expression profiles or signaling, remains to be shown.
Atoh1 loss causes HC loss
Already the initial paper on Atoh1 null mutants showed persisting expression of Atoh1-LacZ in ‘supporting cells’ [26]. Supporting cells and HCs share a common lineage [107] and this may indicate either that the common precursors expresses Atoh1 that persists in mutants or expression of Atoh1 in supporting cells is regulated independently. Atoh1-lacZ positive cells are reduced to a single line of cells in null mutants [108]. Expression data of genes flanking the OC medially (Fgf10 [109]) and laterally (BMP4 [110]) show that the OC is reduced to a single row of cells, possibly representing the IP cells [42]. If this interpretation is correct, it would imply that the expression regulation of Atoh1 in IP cells [47,48] becomes unmasked in the absence of HC differentiation, much like the Atoh1 expression in neurons becomes unmasked in neurons after Neurod1 loss [89]. Retaining IP cells could also explain the normal length of the OC in the absence of HC differentiation [108] because IP cells may drive normal extension of the cochlear duct [45]. How numbers of IP cells are regulated remains unclear but truncating precursor proliferation reduces OC length [37,98]. Specific markers for undifferentiated IP cells need to be investigated in Atoh1 null mice and mutant mice with a shortened OC.
A follow-up study on the effect of delayed loss of Atoh1 using the Atoh1 enhancer associated cre expression [47] showed stochastic effects of incomplete HC differentiation paired with progressive loss of the OC [90]. Even transient expression of Atoh1 suffices to rescue HCs for several weeks, indicating that long term viability of HC depends on length and level of expression of Atoh1, a conclusion also reached in an Atoh1 hypomorph mouse [92]. Knowing how long Atoh1 (or equivalent bHLH gene) expression is needed to differentiate long-term viable HCs remains unclear. Such treatment should avoid to convert supporting cells as is possible during a perinatal phase [54] and under certain conditions later [100].
Can developmental insights be used for in situ regeneration?
Transdifferentiating supporting cells into HCs or generating HCs from pluripotent stem cells has advanced since the important role of Atoh1 was first described in the ear of null mutants [26]. Generating vestibular HC-like differentiation from precursor cells [111] to form simple mosaic driven by Delta-Notch as found in vestibular sensory epithelia [32] may soon be possible. However, one would need to generate distinct cochlea HC types, surrounded by distinct supporting cell types in the right numbers and in the right position and pattern to achieve lasting restoration of the OC. Strategies need to go beyond HC replacement in avian sensory organs or in the mammalian vestibular organs in which simple conversion of supporting cells through the lateral inhibition via the Delta-Notch system suffices to establish a cellular pattern satisfactory for near normal function. While incomplete OC restoration immediately after HC loss may bring some benefit [100], this may not be the case after further deterioration of the OC due to prolonged HC loss [50,51].
After long-term HC loss the OC requires additional manipulations to be fully restored, possibly from a flat epithelium [112], with the orderly pattern of HC/supporting cell mosaic with their respective specializations that are unique to the mammalian cochlea [24,36]. An essential feature of the mammalian OC [24] is two adjacent and continuous rows of supporting cells, the IP and OP cells [56]. Re-patterning of Fgf8 expression from a diffuse distribution in nearly all HCs to an enhanced expression only in IHCs [25] may in part be responsible for this patterning of the mammalian OC. Verification of this suggestion requires forced expression of Fgf8 in a single row of HCs in vestibular organs to confirm that altered expression pattern of Fgf8 can indeed change the outcome of the cellular organization [11], cooperating with the Delta-Notch system of lateral inhibition [40] to generate a row of contiguous IP cells. How that can be achieved in a flat epithelium with little to no molecular markers to start this process remains unclear.
Beyond figuring out how to generate the cellular distribution in the inner compartment of the OC and the single row of IP cells (Fig. 1), one would need to know exactly what remains in an OC of variable genetic predisposition and variable progressive loss of cellular integrity. The ‘flat epithelium’ is certainly one possibility [112], but variable retention of IP cells [50,51] will be encountered in human hearing loss. Reconstitution of the OC would require an adjustable strategy that takes the local variation of the OC deterioration into account. Such a strategy could start with mapping precisely the residual hearing capacity currently used with short cochlear implants [113]. Even more desirable would be increased imaging resolution to identify not only residual cell types and their distribution, but have precise measurements of the apparently very variable cochlear duct [52] and hearing loss-related cellular loss [51]. These techniques could show the full scale of human OC variability at the time of attempted reconstitution. Adjusting the simple approaches currently at hand to the variable situations found in a given person requires new mouse models that go beyond current models. Simply wiping out all HCs rarely mimics the human condition and models are needed to present better genetically engineered simulation of human cochlea suffering from age related hearing loss. Newer models [50] can be employed to fine-tune attempts to reconstitute a functional OC from a mixed anatomy of variable loss of HCs and supporting cells.
Conclusion and outlook
We review OC and HC differentiation in the context of OC functional organization and the complexity of transcription factor interactions during ear development. While expressing Atoh1 alone or with other factors to restore HCs will continue to be valuable, parallel work needs to establish conditions to restore the various cell types. Understanding the function of the cellular organization of the OC has progressed greatly since its discovery by Alfonso Corti [114], but numerical ratios of various cellular constituents remain unclear, thus making it difficult to predict the outcome of partial restoration attempts. The evolution of the mammalian OC seemingly blocked all postnatal proliferation to maintain the functional integrity of this unique cellular assembly. Such alterations now pose a road block to restoring the OC through induced proliferation beyond early postnatal stages [20]. In the sense of Feynman, we propose that restoring fully functional OCs from partially formed OCs of mouse mutants, or from mouse OCs with incomplete or progressive loss of HCs, or through transformation of non-mammalian hearing organs are to be completed before attempts to restore human OC are initiated. A good first step toward OC restoration would be to repopulate a de-cellularized OC to show functional recovery as in limb regeneration [115] including differentiation of the various cell types in the right position. Alternatively, transforming the basilar papilla of a chicken into a mammalian-like OC would be proof that the basic molecular machinery leading to the very different cellular organization of the mammalian OC development is understood. Attempts to regenerate HCs in vestibular organs or non-mammalian hearing organs prove only that HC can be restored in a simple cellular mosaic; they are not evidence that OC restoration is resolved. Such proof is needed before initiating the translation of partially understood attempts to restore the OC in humans.
Acknowledgments
This work was supported by NIH (P30 DC 010362, R03 DC013655 to IJ), NASA Base Program and the OVPR, University of Iowa. We express our thanks to Dr. M Holley for allowing us to use high resolution images from Zetes et al., (2012) to make the point of cellular ratios. We thank two reviewers for their critical insights that helped shaped this review.
Footnotes
The authors declare that they have no conflict of interest.
References
- 1.Stergachis AB, Neph S, Reynolds A, Humbert R, et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fritzsch B, Jahan I, Pan N, Elliott KL. Evolving gene regulatory networks into cellular networks guiding adaptive behavior: an outline how single cells could have evolved into a centralized neurosensory system. Cell and tissue research. 2015;359:295–313. doi: 10.1007/s00441-014-2043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner A. The molecular origins of evolutionary innovations. Trends in genetics : TIG. 2011;27:397–410. doi: 10.1016/j.tig.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Science Advances. 2015;1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meinhardt H. Pattern Formation in Morphogenesis. Springer; 2013. From Hydra to Vertebrates: Models for the Transition from Radial-to Bilateral-Symmetric Body Plans; pp. 207–24. [Google Scholar]
- 6.Sprinzak D, Lakhanpal A, LeBon L, Garcia-Ojalvo J, et al. Mutual inactivation of Notch receptors and ligands facilitates developmental patterning. PLoS computational biology. 2011;7:e1002069. doi: 10.1371/journal.pcbi.1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamoto S, Schulze KL, Bellen HJ. Notch Signaling. Springer; 2014. Introduction to Notch Signaling; pp. 1–14. [DOI] [PubMed] [Google Scholar]
- 8.Petrovic J, Formosa-Jordan P, Luna-Escalante JC, Abelló G, et al. Ligand-dependent Notch signaling strength orchestrates lateral induction and lateral inhibition in the developing inner ear. Development. 2014;141:2313–24. doi: 10.1242/dev.108100. [DOI] [PubMed] [Google Scholar]
- 9.Morel V, Schweisguth F. Repression by suppressor of hairless and activation by Notch are required to define a single row of single-minded expressing cells in the Drosophila embryo. Genes Dev. 2000;14:377–88. [PMC free article] [PubMed] [Google Scholar]
- 10.Ozdemir A, Ma L, White KP, Stathopoulos A. Su(H)-mediated repression positions gene boundaries along the dorsal-ventral axis of Drosophila embryos. Dev Cell. 2014;31:100–13. doi: 10.1016/j.devcel.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groves AK, Fekete DM. Shaping sound in space: the regulation of inner ear patterning. Development. 2012;139:245–57. doi: 10.1242/dev.067074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fritzsch B, Pan N, Jahan I, Elliott KL. Inner ear development: building a spiral ganglion and an organ of Corti out of unspecified ectoderm. Cell Tissue Res. 2015;368:in press. doi: 10.1007/s00441-014-2031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powers RJ, Kulason S, Atilgan E, Brownell WE, et al. The Local Forces Acting on the Mechanotransduction Channel in Hair Cell Stereocilia. Biophysical journal. 2014;106:2519–28. doi: 10.1016/j.bpj.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hudspeth A. Integrating the active process of hair cells with cochlear function. Nature Reviews Neuroscience. 2014;15:600–14. doi: 10.1038/nrn3786. [DOI] [PubMed] [Google Scholar]
- 15.Muniak MA, Rivas A, Montey KL, May BJ, et al. 3D model of frequency representation in the cochlear nucleus of the CBA/J mouse. Journal of Comparative Neurology. 2013;521:1510–32. doi: 10.1002/cne.23238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Held H. Untersuchungen über den feineren Bau des Ohrlabyrinthes der Wirbeltier. BG Teubner; 1902. [Google Scholar]
- 17.Raphael Y, Altschuler RA. Structure and innervation of the cochlea. Brain research bulletin. 2003;60:397–422. doi: 10.1016/s0361-9230(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 18.Retzius G. Das Gehörorgan der Wirbelthiere: morphologisch-histologische Studien. 2, Das Gehörorgan der Reptilien, der Vögel und der Säugethiere. Samson & Wallin in Commission; 1884. [Google Scholar]
- 19.Yamasoba T, Lin FR, Someya S, Kashio A, et al. Current concepts in age-related hearing loss: epidemiology and mechanistic pathways. Hearing research. 2013;303:30–8. doi: 10.1016/j.heares.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zine A, Löwenheim H, Fritzsch B. Adult Stem Cells. Springer; 2014. Toward translating molecular ear development to generate hair cells from stem cells; pp. 111–61. [Google Scholar]
- 21.Müller U, Barr-Gillespie PG. New treatment options for hearing loss. Nature Reviews Drug Discovery. 2015 doi: 10.1038/nrd4533. [DOI] [PubMed] [Google Scholar]
- 22.Géléoc GS, Holt JR. Sound strategies for hearing restoration. Science. 2014;344:1241062. doi: 10.1126/science.1241062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulvaney J, Dabdoub A. Atoh1, an essential transcription factor in neurogenesis and intestinal and inner ear development: function, regulation, and context dependency. J Assoc Res Otolaryngol. 2012;13:281–93. doi: 10.1007/s10162-012-0317-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chonko KT, Jahan I, Stone J, Wright MC, et al. Atoh1 directs hair cell differentiation and survival in the late embryonic mouse inner ear. Dev Biol. 2013;381:401–10. doi: 10.1016/j.ydbio.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jahan I, Pan N, Kersigo J, Fritzsch B. Beyond generalized hair cells: molecular cues for hair cell types. Hear Res. 2013;297:30–41. doi: 10.1016/j.heares.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bermingham NA, Hassan BA, Price SD, Vollrath MA, et al. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–41. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- 27.Cai T, Jen H-I, Kang H, Klisch TJ, et al. Characterization of the Transcriptome of Nascent Hair Cells and Identification of Direct Targets of the Atoh1 Transcription Factor. The Journal of Neuroscience. 2015;35:5870–83. doi: 10.1523/JNEUROSCI.5083-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Wu Y, Zhao F, Wu Y, et al. Fgf-Signaling-Dependent Sox9a and Atoh1a Regulate Otic Neural Development in Zebrafish. The Journal of Neuroscience. 2015;35:234–44. doi: 10.1523/JNEUROSCI.3353-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raft S, Groves AK. Segregating neural and mechanosensory fates in the developing ear: patterning, signaling, and transcriptional control. Cell and tissue research. 2015;359:315–32. doi: 10.1007/s00441-014-1917-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pirvola U, Spencer-Dene B, Xing-Qun L, Kettunen P, et al. FGF/FGFR-2 (IIIb) signaling is essential for inner ear morphogenesis. The Journal of Neuroscience. 2000;20:6125–34. doi: 10.1523/JNEUROSCI.20-16-06125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huh S-H, Warchol ME, Ornitz DM. Cochlear progenitor number is controlled through mesenchymal FGF receptor signaling. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang T, Chai R, Kim GS, Pham N, et al. Lgr5+ cells regenerate hair cells via proliferation and direct transdifferentiation in damaged neonatal mouse utricle. Nature communications. 2015;6 doi: 10.1038/ncomms7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ladhams A, Pickles J. Morphology of the monotreme organ of Corti and macula lagena. Journal of Comparative Neurology. 1996;366:335–47. doi: 10.1002/(SICI)1096-9861(19960304)366:2<335::AID-CNE11>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 34.Benito-Gonzalez A, Doetzlhofer A. Hey1 and hey2 control the spatial and temporal pattern of mammalian auditory hair cell differentiation downstream of hedgehog signaling. The Journal of Neuroscience. 2014;34:12865–76. doi: 10.1523/JNEUROSCI.1494-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waldhaus J, Durruthy-Durruthy R, Heller S. Quantitative High-Resolution Cellular Map of the Organ of Corti. Cell reports. 2015 doi: 10.1016/j.celrep.2015.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vater M, Kössl M. Comparative aspects of cochlear functional organization in mammals. Hearing research. 2011;273:89–99. doi: 10.1016/j.heares.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 37.Pauley S, Lai E, Fritzsch B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev Dyn. 2006;235:2470–82. doi: 10.1002/dvdy.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmer WH, Jia D, Deng W-M. Cis-interactions between Notch and its ligands block ligand-independent Notch activity. eLife. 2015;3:e04415. doi: 10.7554/eLife.04415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan W, Jin Y, Stanger B, Kiernan AE. Notch signaling is required for the generation of hair cells and supporting cells in the mammalian inner ear. Proc Natl Acad Sci U S A. 2010;107:15798–803. doi: 10.1073/pnas.1003089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doetzlhofer A, Basch ML, Ohyama T, Gessler M, et al. Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev Cell. 2009;16:58–69. doi: 10.1016/j.devcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janody F, Treisman JE. Requirements for mediator complex subunits distinguish three classes of notch target genes at the Drosophila wing margin. Dev Dyn. 2011;240:2051–9. doi: 10.1002/dvdy.22705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan N, Jahan I, Kersigo J, Kopecky B, et al. Conditional deletion of Atoh1 using Pax2-Cre results in viable mice without differentiated cochlear hair cells that have lost most of the organ of Corti. Hearing Research. 2011;275:66–80. doi: 10.1016/j.heares.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiernan AE. Seminars in cell & developmental biology. Elsevier; 2013. Notch signaling during cell fate determination in the inner ear; pp. 470–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohyama T, Basch ML, Mishina Y, Lyons KM, et al. BMP signaling is necessary for patterning the sensory and nonsensory regions of the developing mammalian cochlea. The Journal of Neuroscience. 2010;30:15044–51. doi: 10.1523/JNEUROSCI.3547-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto N, Okano T, Ma X, Adelstein RS, et al. Myosin II regulates extension, growth and patterning in the mammalian cochlear duct. Development. 2009;136:1977–86. doi: 10.1242/dev.030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puligilla C, Feng F, Ishikawa K, Bertuzzi S, et al. Disruption of fibroblast growth factor receptor 3 signaling results in defects in cellular differentiation, neuronal patterning, and hearing impairment. Developmental Dynamics. 2007;236:1905–17. doi: 10.1002/dvdy.21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matei V, Pauley S, Kaing S, Rowitch D, et al. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev Dyn. 2005;234:633–50. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Driver EC, Sillers L, Coate TM, Rose MF, et al. The Atoh1-lineage gives rise to hair cells and supporting cells within the mammalian cochlea. Dev Biol. 2013;376:86–98. doi: 10.1016/j.ydbio.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pauley S, Kopecky B, Beisel K, Soukup G, et al. Stem cells and molecular strategies to restore hearing. Panminerva medica. 2008;50:41. [PMC free article] [PubMed] [Google Scholar]
- 50.Kersigo J, Fritzsch B. Inner ear hair cells deteriorate in mice engineered to have no or diminished innervation. Frontiers in Aging Neuroscience. 2015;7 doi: 10.3389/fnagi.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor RR, Jagger DJ, Forge A. Defining the cellular environment in the organ of Corti following extensive hair cell loss: a basis for future sensory cell replacement in the Cochlea. PLoS One. 2012;7:e30577. doi: 10.1371/journal.pone.0030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Avci E, Nauwelaers T, Lenarz T, Hamacher V, et al. Variations in microanatomy of the human cochlea. Journal of Comparative Neurology. 2014;522:3245–61. doi: 10.1002/cne.23594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cox BC, Chai R, Lenoir A, Liu Z, et al. Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development. 2014;141:816–29. doi: 10.1242/dev.103036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Z, Fang J, Dearman J, Zhang L, et al. In vivo generation of immature inner hair cells in neonatal mouse cochleae by ectopic atoh1 expression. PloS one. 2014;9:e89377. doi: 10.1371/journal.pone.0089377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raikwar SP, Kim E-M, Sivitz WI, Allamargot C, et al. Human iPS Cell-Derived Insulin Producing Cells Form Vascularized Organoids under the Kidney Capsules of Diabetic Mice. PLoS On e. 2015;10:e0116582. doi: 10.1371/journal.pone.0116582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zetes DE, Tolomeo JA, Holley MC. Structure and mechanics of supporting cells in the guinea pig organ of Corti. PLoS One. 2012;7:e49338. doi: 10.1371/journal.pone.0049338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Flora A, Klisch TJ, Schuster G, Zoghbi HY. Deletion of Atoh1 disrupts Sonic Hedgehog signaling in the developing cerebellum and prevents medulloblastoma. Science. 2009;326:1424–7. doi: 10.1126/science.1181453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skinner MK, Rawls A, Wilson-Rawls J, Roalson EH. Basic helix-loop-helix transcription factor gene family phylogenetics and nomenclature. Differentiation; research in biological diversity. 2010;80:1. doi: 10.1016/j.diff.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flora A, Garcia JJ, Thaller C, Zoghbi HY. The E-protein Tcf4 interacts with Math1 to regulate differentiation of a specific subset of neuronal progenitors. Proc Natl Acad Sci U S A. 2007;104:15382–7. doi: 10.1073/pnas.0707456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huret JL. TCF3 (transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47)) 2012. [Google Scholar]
- 61.Bhattacharya A, Baker NE. A network of broadly expressed HLH genes regulates tissue-specific cell fates. Cell. 2011;147:881–92. doi: 10.1016/j.cell.2011.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahmed M, Wong EY, Sun J, Xu J, et al. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev Cell. 2012;22:377–90. doi: 10.1016/j.devcel.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spratford CM, Kumar JP. Extramacrochaetae imposes order on the Drosophila eye by refining the activity of the Hedgehog signaling gradient. Development. 2013;140:1994–2004. doi: 10.1242/dev.088963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morris JK, Maklad A, Hansen LA, Feng F, et al. A disorganized innervation of the inner ear persists in the absence of ErbB2. Brain research. 2006;1091:186–99. doi: 10.1016/j.brainres.2006.02.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moody SA, Klein SL, Karpinski BA, Maynard TM, et al. On becoming neural: what the embryo can tell us about differentiating neural stem cells. American journal of stem cells. 2013;2:74. [PMC free article] [PubMed] [Google Scholar]
- 66.Bouchard M, Busslinger M, Xu P, De Caprona D, et al. PAX2 and PAX8 cooperate in mouse inner ear morphogenesis and innervation. BMC Dev Biol. 2010;10:89. doi: 10.1186/1471-213X-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nichols DH, Pauley S, Jahan I, Beisel KW, et al. Lmx1a is required for segregation of sensory epithelia and normal ear histogenesis and morphogenesis. Cell Tissue Res. 2008;334:339–58. doi: 10.1007/s00441-008-0709-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Edlund RK, Birol O, Groves AK. The Role of Foxi Family Transcription Factors in the Development of the Ear and Jaw. Current Topics in Developmental Biology. 2015 doi: 10.1016/bs.ctdb.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duncan JS, Fritzsch B. Continued expression of GATA3 is necessary for cochlear neurosensory development. PLoS One. 2013;8:e62046. doi: 10.1371/journal.pone.0062046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fritzsch B, Eberl DF, Beisel KW. The role of bHLH genes in ear development and evolution: revisiting a 10-year-old hypothesis. Cell Mol Life Sci. 2010;67:3089–99. doi: 10.1007/s00018-010-0403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mao CA, Wang SW, Pan P, Klein WH. Rewiring the retinal ganglion cell gene regulatory network: Neurod1 promotes retinal ganglion cell fate in the absence of Math5. Development. 2008;135:3379–88. doi: 10.1242/dev.024612. [DOI] [PubMed] [Google Scholar]
- 72.Mao CA, Cho JH, Wang J, Gao Z, et al. Reprogramming amacrine and photoreceptor progenitors into retinal ganglion cells by replacing Neurod1 with Atoh7. Development. 2013;140:541–51. doi: 10.1242/dev.085886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jahan I, Pan N, Kersigo J, Calisto LE, et al. Expression of Neurog1 instead of Atoh1 can partially rescue organ of Corti cell survival. PLoS One. 2012;7:e30853. doi: 10.1371/journal.pone.0030853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, et al. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–72. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- 75.Maricich SM, Xia A, Mathes EL, Wang VY, et al. Atoh1-lineal neurons are required for hearing and for the survival of neurons in the spiral ganglion and brainstem accessory auditory nuclei. J Neurosci. 2009;29:11123–33. doi: 10.1523/JNEUROSCI.2232-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maricich SM, Wellnitz SA, Nelson AM, Lesniak DR, et al. Merkel cells are essential for light-touch responses. Science. 2009;324:1580–2. doi: 10.1126/science.1172890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bermingham NA, Hassan BA, Wang VY, Fernandez M, et al. Proprioceptor pathway development is dependent on Math1. Neuron. 2001;30:411–22. doi: 10.1016/s0896-6273(01)00305-1. [DOI] [PubMed] [Google Scholar]
- 78.Shroyer NF, Helmrath MA, Wang VY, Antalffy B, et al. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132:2478–88. doi: 10.1053/j.gastro.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 79.Kiernan AE, Pelling AL, Leung KK, Tang AS, et al. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–5. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- 80.Mastracci TL, Anderson KR, Papizan JB, Sussel L. Regulation of Neurod1 contributes to the lineage potential of Neurogenin3+ endocrine precursor cells in the pancreas. PLoS genetics. 2013;9:e1003278. doi: 10.1371/journal.pgen.1003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pan N, Jahan I, Lee JE, Fritzsch B. Defects in the cerebella of conditional Neurod1 null mice correlate with effective Tg(Atoh1-cre) recombination and granule cell requirements for Neurod1 for differentiation. Cell Tissue Res. 2009;337:407–28. doi: 10.1007/s00441-009-0826-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clevers H. The Intestinal Crypt, A Prototype Stem Cell Compartment. Cell. 2013;154:274–84. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 83.Costa A, Sanchez-Guardado L, Juniat S, Gale JE, et al. Generation of sensory hair cells by genetic programming with a combination of transcription factors. Development. 2015;142:1948–59. doi: 10.1242/dev.119149. [DOI] [PubMed] [Google Scholar]
- 84.Xiang M, Maklad A, Pirvola U, Fritzsch B. Brn3c null mutant mice show long-term, incomplete retention of some afferent inner ear innervation. BMC neuroscience. 2003;4:2. doi: 10.1186/1471-2202-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hertzano R, Montcouquiol M, Rashi-Elkeles S, Elkon R, et al. Transcription profiling of inner ears from Pou4f3ddl/ddl identifies Gfi1 as a target of the Pou4f3 deafness gene. Human molecular genetics. 2004;13:2143–53. doi: 10.1093/hmg/ddh218. [DOI] [PubMed] [Google Scholar]
- 86.Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev Cell. 2011;20:483–96. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen KH, Boettiger AN, Moffitt JR, Wang S, et al. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jahan I, Pan N, Fritzsch B. Opportunities and limits of the one gene approach: the ability of Atoh1 to differentiate and maintain hair cells depends on the molecular context. Name: Frontiers in Cellular Neuroscience. 2015;9:26. doi: 10.3389/fncel.2015.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jahan I, Pan N, Kersigo J, Fritzsch B. Neurod1 suppresses hair cell differentiation in ear ganglia and regulates hair cell subtype development in the cochlea. PLoS One. 2010;5:e11661. doi: 10.1371/journal.pone.0011661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pan N, Jahan I, Kersigo J, Duncan J, et al. A novel Atoh1 ’self-terminating’ mouse model reveals the necessity of proper Atoh1 expression level and duration for inner ear hair cell differentiation and viability. PLoS One. 2012;7:e30358. doi: 10.1371/journal.pone.0030358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu H, Pecka JL, Zhang Q, Soukup GA, et al. Characterization of transcriptomes of cochlear inner and outer hair cells. The Journal of Neuroscience. 2014;34:11085–95. doi: 10.1523/JNEUROSCI.1690-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sheykholeslami K, Thimmappa V, Nava C, Bai X, et al. A new mutation of the Atoh1 gene in mice with normal life span allows analysis of inner ear and cerebellar phenotype in aging. PLoS One. 2013;8:e79791. doi: 10.1371/journal.pone.0079791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang SM, Chen W, Guo WW, Jia S, et al. Regeneration of stereocilia of hair cells by forced Atoh1 expression in the adult mammalian cochlea. PLoS One. 2012;7:e46355. doi: 10.1371/journal.pone.0046355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang H, Xie X, Deng M, Chen X, et al. Generation and characterization of Atoh1-Cre knock-in mouse line. Genesis. 2010;48:407–13. doi: 10.1002/dvg.20633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ruben RJ. Development of the inner ear of the mouse: a radioautographic study of terminal mitoses. Acta Otolaryngol. 1967;220(Suppl):1–44. [PubMed] [Google Scholar]
- 96.Mantela J, Jiang Z, Ylikoski J, Fritzsch B, et al. The retinoblastoma gene pathway regulates the postmitotic state of hair cells of the mouse inner ear. Development. 2005;132:2377–88. doi: 10.1242/dev.01834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schimmang T, Pirvola U. Seminars in cell & developmental biology. Elsevier; 2013. Coupling the cell cycle to development and regeneration of the inner ear; pp. 507–13. [DOI] [PubMed] [Google Scholar]
- 98.Kopecky BJ, Jahan I, Fritzsch B. Correct timing of proliferation and differentiation is necessary for normal inner ear development and auditory hair cell viability. Developmental Dynamics. 2013;242:132–47. doi: 10.1002/dvdy.23910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kelly MC, Chang Q, Pan A, Lin X, et al. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. The Journal of Neuroscience. 2012;32:6699–710. doi: 10.1523/JNEUROSCI.5420-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mizutari K, Fujioka M, Hosoya M, Bramhall N, et al. Notch inhibition induces cochlear hair cell regeneration and recovery of hearing after acoustic trauma. Neuron. 2013;77:58–69. doi: 10.1016/j.neuron.2012.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sienknecht UJ, Koppl C, Fritzsch B. Evolution and development of hair cell polarity and efferent function in the inner ear. Brain Behav Evol. 2014;83:150–61. doi: 10.1159/000357752. [DOI] [PubMed] [Google Scholar]
- 102.Ma Q, Anderson DJ, Fritzsch B. Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. J Assoc Res Otolaryngol. 2000;1:129–43. doi: 10.1007/s101620010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fariñas I, Jones KR, Tessarollo L, Vigers AJ, et al. Spatial shaping of cochlear innervation by temporally regulated neurotrophin expression. The Journal of Neuroscience. 2001;21:6170–80. doi: 10.1523/JNEUROSCI.21-16-06170.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Durruthy-Durruthy R, Gottlieb A, Hartman BH, Waldhaus J, et al. Reconstruction of the mouse otocyst and early neuroblast lineage at single-cell resolution. Cell. 2014;157:964–78. doi: 10.1016/j.cell.2014.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Raft S, Koundakjian EJ, Quinones H, Jayasena CS, et al. Cross-regulation of Ngn1 and Math1 coordinates the production of neurons and sensory hair cells during inner ear development. Development. 2007;134:4405–15. doi: 10.1242/dev.009118. [DOI] [PubMed] [Google Scholar]
- 106.Lee S, Danielian PS, Fritzsch B, McMahon AP. Evidence that FGF8 signalling from the midbrain-hindbrain junction regulates growth and polarity in the developing midbrain. Development. 1997;124:959–69. doi: 10.1242/dev.124.5.959. [DOI] [PubMed] [Google Scholar]
- 107.Jiang H, Wang L, Beier KT, Cepko CL, et al. Lineage analysis of the late otocyst stage mouse inner ear by transuterine microinjection of a retroviral vector encoding alkaline phosphatase and an oligonucleotide library. PLoS One. 2013;8:e69314. doi: 10.1371/journal.pone.0069314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fritzsch B, Matei V, Nichols D, Bermingham N, et al. Atoh1 null mice show directed afferent fiber growth to undifferentiated ear sensory epithelia followed by incomplete fiber retention. Developmental dynamics. 2005;233:570–83. doi: 10.1002/dvdy.20370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pauley S, Wright TJ, Pirvola U, Ornitz D, et al. Expression and function of FGF10 in mammalian inner ear development. Developmental dynamics. 2003;227:203–15. doi: 10.1002/dvdy.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Morsli H, Choo D, Ryan A, Johnson R, et al. Development of the mouse inner ear and origin of its sensory organs. The Journal of neuroscience. 1998;18:3327–35. doi: 10.1523/JNEUROSCI.18-09-03327.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Koehler KR, Malone AK, Hashino E. Recapitulating Inner Ear Development with Pluripotent Stem Cells: Biology and Translation. Development of Auditory and Vestibular Systems. 2014:213. [Google Scholar]
- 112.Izumikawa M, Batts SA, Miyazawa T, Swiderski DL, et al. Response of the flat cochlear epithelium to forced expression of Atoh1. Hear Res. 2008;240:52–6. doi: 10.1016/j.heares.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Reiss LA, Turner CW, Karsten SA, Erenberg SR, et al. Consonant recognition as a function of the number of stimulation channels in the Hybrid short-electrode cochlear implanta) The Journal of the Acoustical Society of America. 2012;132:3406–17. doi: 10.1121/1.4757735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Corti A. Recherches sur l’organe de l’ouie des mammiferes 1851 [Google Scholar]
- 115.Jank BJ, Xiong L, Moser PT, Guyette JP, et al. Engineered composite tissue as a bioartificial limb graft. Biomaterials. 2015;61:246–56. doi: 10.1016/j.biomaterials.2015.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Slepecky NB. The cochlea. Springer; 1996. Structure of the mammalian cochlea; pp. 44–129. [Google Scholar]
- 117.Pan N, Kopecky B, Jahan I, Fritzsch B. Understanding the evolution and development of neurosensory transcription factors of the ear to enhance therapeutic translation. Cell Tissue Res. 2012;349:415–32. doi: 10.1007/s00441-012-1454-0. [DOI] [PMC free article] [PubMed] [Google Scholar]


