Abstract
Background
In models extensively used in studies of aging and extended lifespan, such as C. elegans and Drosophila, adult senescence is regulated by gene networks that are likely to be similar to ones that underlie lifespan extension during dormancy. These include the evolutionarily conserved insulin/IGF, TOR and germ line-signaling pathways. Dormancy, also known as dauer stage in the larval worm or adult diapause in the fly, is triggered by adverse environmental conditions, and results in drastically extended lifespan with negligible senescence. It is furthermore characterized by increased stress resistance and somatic maintenance, developmental arrest and reallocated energy resources. In the fly Drosophila melanogaster adult reproductive diapause is additionally manifested in arrested ovary development, improved immune defense and altered metabolism. However, the molecular mechanisms behind this adaptive lifespan extension are not well understood.
Results
A genome wide analysis of transcript changes in diapausing D. melanogaster revealed a differential regulation of more than 4600 genes. Gene ontology (GO) and KEGG pathway analysis reveal that many of these genes are part of signaling pathways that regulate metabolism, stress responses, detoxification, immunity, protein synthesis and processes during aging. More specifically, gene readouts and detailed mapping of the pathways indicate downregulation of insulin-IGF (IIS), target of rapamycin (TOR) and MAP kinase signaling, whereas Toll-dependent immune signaling, Jun-N-terminal kinase (JNK) and Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathways are upregulated during diapause. Furthermore, we detected transcriptional regulation of a large number of genes specifically associated with aging and longevity.
Conclusions
We find that many affected genes and signal pathways are shared between dormancy, aging and lifespan extension, including IIS, TOR, JAK/STAT and JNK. A substantial fraction of the genes affected by diapause have also been found to alter their expression in response to starvation and cold exposure in D. melanogaster, and the pathways overlap those reported in GO analysis of other invertebrates in dormancy or even hibernating mammals. Our study, thus, shows that D. melanogaster is a genetically tractable model for dormancy in other organisms and effects of dormancy on aging and lifespan.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-2383-1) contains supplementary material, which is available to authorized users.
Background
Aging and adult senescence in the fly Drosophila and the worm Caenorhabditis elegans are regulated by gene networks that may overlap with ones that underlie lifespan extension during dormancy in these organisms [1–7]. Among the regulatory networks likely to be shared are the evolutionarily conserved insulin/IGF, TOR and germ line-signaling pathways [1–3, 8]. This hypothesis is supported by the phenotype characteristic of dormancy, which includes developmental arrest, increased stress resistance and somatic maintenance, reallocated energy resources, accompanied by a drastically extended lifespan with negligible senescence [8–10]. Dormancy, in insects also known as diapause, is an adaptive shift in life history that can be triggered by particularly adverse environmental challenges [8, 11–15].
Diapause appears to be part of a spectrum of general and partially conserved stress syndromes in animals, with a physiological shift from a reproduction mode to extended survival [9, 14, 16]. In insects, this dormancy, with its accompanying suite of co-adapted traits, is preprogrammed and can occur at different stages of the life cycle, but almost always in a single specific stage in each species [12]. The fruitfly Drosophila melanogaster can enter a reproductive diapause in the adult stage that is facultative, shallow, and rapidly reversible [9, 17–20]. In the laboratory diapause is induced by exposing female flies to low temperature and short photoperiod soon after the adults have eclosed, and is characterized by attenuated production of vitellogenic eggs, increased nutrient stores and diminished senescence [9, 10, 20]. Since the molecular mechanisms behind dormancy and its links to slowed aging are not well understood in any organism, we embarked on an analysis of genes involved in diapause of the genetic model insect D. melanogaster.
In a recent paper we reported in detail the dynamic phenotypic changes associated with reproductive diapause in D. melanogaster [20]. Diapausing flies display reduced food intake, increased stores of carbohydrates and lipids, activated immune genes, altered expression of genes related to insulin- and glucagon-like signaling, display low mortality and overall a drastically extended lifespan [20]. It was furthermore shown that flies with loss of function mutations in insulin-like peptide genes dilp2-3 and dilp5 are more prone to enter diapause than wild type flies [20]. This is in agreement with several reports on the role of insulin signaling in insect diapause and other forms of stress responses [9, 21–27]. Insulin/IGF signaling (IIS) is also known to be critical in regulation of fecundity, metabolism, stress resistance and longevity [28–32], all of which are affected by the quiescence during diapause [8, 10, 27]. Thus, to further investigate the role of IIS and other signaling pathways in slowed aging during diapause, we performed a genome-wide transcriptome analysis of flies kept for three weeks in diapause. The resultant data set indicates a broad effect of diapause on gene regulation, with over 4500 genes showing at least twofold up- or downregulation, compared to flies kept at normal conditions. Our analysis reveals that a large fraction of the genes associated with aging and longevity in Drosophila [2, 3, 5, 33] are affected by diapause. Furthermore, we show that several relevant signaling pathways associated with increased stress tolerance and extended lifespan were differentially regulated at the transcript level. These include the IIS and target of rapamycin (TOR) pathways, as well as Toll, JAK/STAT, JNK and MAP kinase signaling, and there are also effects on peptide hormone, juvenile hormone and ecdysone signaling components. Importantly, we identified a large number of altered genes that are in common with other genome-wide searches for adaptive changes in life history traits in D. melanogaster, for instance cold resistance and starvation responses, indicating the modular usage of stress signaling pathways. Our study clearly shows that D. melanogaster is a powerful organism for modeling genetic regulation of dormancy and slowed aging across a range of species.
Results and discussion
Experimental design
In a previous study of D. melanogaster we showed that vitellogenesis is at its most reduced level after 3 weeks in diapause-inducing conditions at 11 °C and short photoperiod (10 L:14D) [20]. These conditions were also established as optimal for reproductive diapause by Saunders et al. [18], and we, thus, did not test the effects of low temperature only. More than 90 % of the virgin female flies (Canton S strain) exhibit previtellogenic ovaries after this period of induction. This state of ovary maturation is commonly used as a marker of reproductive diapause [9, 18]. Additional physiological traits also display a strong phenotype after three weeks [20]. Hence, in our present genome-wide transcription study we compared flies (Canton S) kept for 3 weeks in diapause conditions (3wD) with flies kept in non-diapause conditions (25 °C, 12 L:12D). Since senescence appears reduced during diapause [9, 20], we chose two time points for control flies to determine how aging of flies at 25 °C would influence gene expression. Thus, we collected sibling flies that were three weeks (3wN) and one week old (1wN) as controls.
Bioinformatics analysis
Samples of virgin female flies were collected in four biological replicates and purified RNA was hybridized to Affymetrix GeneChip Drosophila Genome 2.0 arrays. A heat map of the 100 most variable probes (Additional file 1: Figure S1) indicates that the majority of the differences detected in our microarrays are due to transcriptome changes in diapause samples, whereas the age of our control flies (1wN or 3wN) only has a minor effect. Principle Component Analysis, (PCA; Additional file 2: Figure S2), revealed distinct transcriptional profiles for diapause samples versus controls, but failed to distinguish between 1wN and 3wN transcriptional profiles. Thus, for further bioinformatics analysis we focused on comparing our diapause samples primarily with 1wN control flies. The transcriptome data for all three comparisons can be found in the Additional file 3: Dataset S1.
We identified 4624 differentially expressed transcripts (absolute fold change ≥ 2, q < 0.05), of which 2412 were upregulated and 2212 were downregulated in diapausing females. The Venn diagram in Fig. 1a shows that almost 80 % of these was also differentially regulated in comparison with the 3wN control. As verified by functional classification based on Gene Ontology (GO) terms (Fig. 1b) the diapause-regulated genes are mostly involved in metabolic processes (44 %) and cellular processes (21 %), which includes genes mainly associated with cell communication and the cell cycle. Significant changes were also observed in sets of genes regulating developmental processes (6 %), such as cell death and morphogenesis, as well as neurological system processes (5 %), and response to stimulus (5 %), where stimuli can be stress, abiotic or other external stimuli, pheromones, immune or toxic agents. About 4 % of the genes represent immune genes, and only a small subset of the genes (2 %) is involved in reproduction and gamete generation.
Fig. 1.
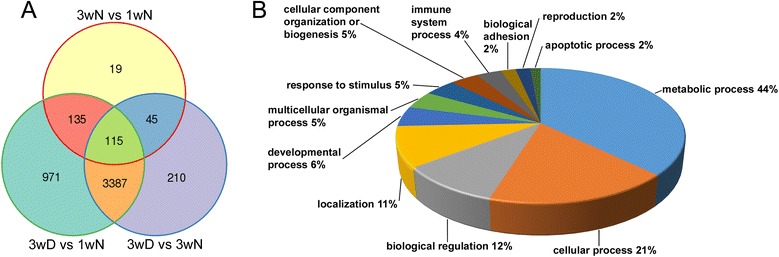
Diagrams summarizing differentially expressed transcripts under diapause and control conditions, and their functional classification based on gene ontology (GO) terms. a Out of 4624 differentially expressed transcripts, 3387 are detected in flies diapausing for 3 weeks (3wD) compared to both 1 (1wN) and 3 (3wN) week controls. Very few transcripts (19) differ due to the age difference between control flies. The transcripts shown display at least an absolute two-fold change and q < 0.05. b The functional classification of GO terms reveals that the majority of the affected genes are involved in metabolic (44 %) and cellular processes (21 %)
Among the top 25 most upregulated genes there is an enrichment of genes predominantly expressed in midgut and fat body (Fig. 2). In contrast the majority of the 25 most downregulated genes display enriched expression in the ovaries (Fig. 2). This apparent tissue distribution of the most strongly altered genes is in line with morphogenetic changes occurring in diapausing flies, as described earlier [20].
Fig. 2.
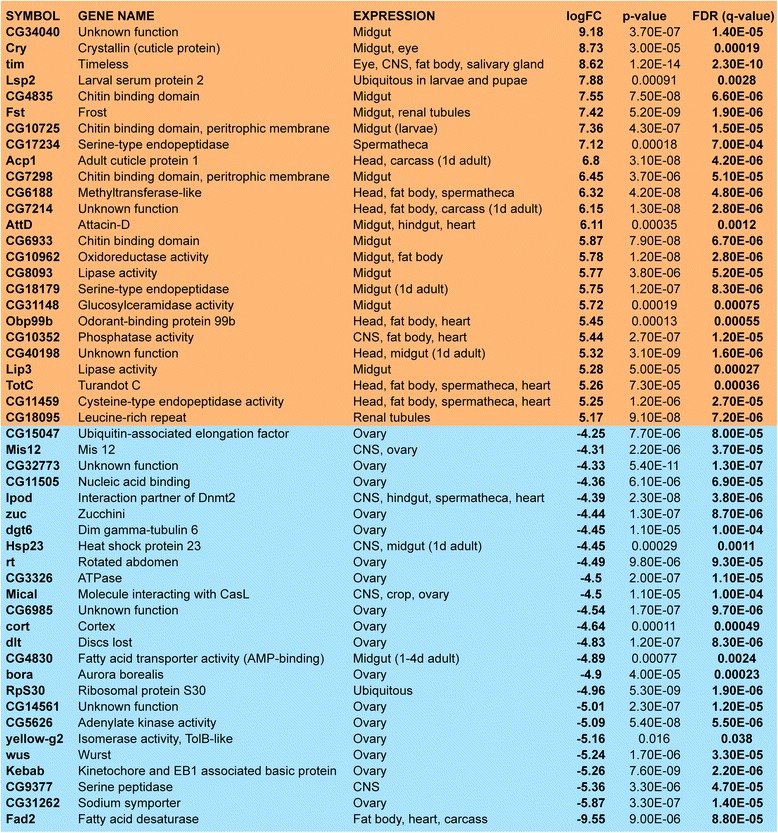
The 25 most up- and downregulated transcripts in diapausing flies. Flies diapausing for three weeks (3wD) were compared to one week old flies (1wN) kept at normal conditions. Upregulated transcripts are predominantly associated with the intestinal structures, fat body, spermatheca and heart, while the most downregulated ones are expressed in the ovaries intestinal structures and fat body (only Fad2). The changes are given as logarithmic fold change (logFC). For a complete list of altered gene expression see Additional file 3: Dataset S1
Gene set enrichment analysis (GSEA) of KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways (Additional file 4: Table S1) confirms the functional GO classification analysis and adds additional information. The altered transcripts belong to 126 different KEGG pathways. From those GSEA identified 48 to be significantly enriched in our study. Of these, 30 KEGG pathways are mostly enriched for upregulated genes and 18 for downregulated genes (Additional file 4: Table S1). As in the GO classification, we detected strong enrichment of upregulated genes associated with different metabolic pathways (mostly basic energetic and storage metabolism), but also drug metabolism and detoxification with cytochrome P450. We also found enhanced Extracellular Matrix (ECM) receptor interactions, many genes involved in the lysosome pathway, and enrichment of genes involved in circadian rhythm. GSEA identified a number of significantly downregulated genes involved in DNA replication, homologous recombination, nucleotide excision repair, transcription, translation and protein processing (Additional file 4: Table S1). This is likely due to general downregulation or silencing of cell division and protein synthesis during dormancy and low cellular activity at low temperature.
However, from GSEA alone we cannot determine how expression of specific genes influences signaling in a particular pathway, since the genes can represent both inhibitors and positive regulators. To circumvent this we implemented Signal pathway impact analysis (SPIA) [34]. We used the KEGG pathway database for search with SPIA and found 4 pathways that are significantly influenced (Additional file 5: Table S2); all of them were previously identified in the GSEA. The SPIA confirmed that ECM receptor interactions are activated during diapause. Furthermore suppression of the circadian rhythm pathway in diapause was seen. In this context we curiosly found an increased expression of the clock genes timeless (tim), period (per), shaggy (sgg), and vrille (vri), which are among the top 100 most strongly induced genes in our study, and slightly (twofold) down-regulated cycle (Cyc) (Additional file 3: Dataset S1). It should be noted that clock genes are also expressed outside of the bona fide clock neurons of the brain, and since our analysis is performed on whole animals we cannot exclude that transcript changes are in other tissues. The KEGG pathways with ID numbers 3460 and 4914 are defined only on the basis of sequence similarity of Drosophila genes with those of humans and other mammals, and therefore their relevance in Drosophila is uncertain. However, since KEGG number 4914 (Progesterone-mediated oocyte maturation) partially overlaps with insulin and MAPK signaling pathways and was identified by SPIA to be significantly downregulated, we decided to analyze selected pathways closer by manual annotation.
Confirmation of altered gene expression by qPCR
We chose 12 genes from the microarray for confirmation by qPCR. Analysis of RNA extracted from flies kept at 3wN and 3wD showed that the qPCR data for 11 of these are in agreement with the changes of transcript levels seen in the microarray (Fig. 3). One gene, the Vm32E (Vitelline membrane 32E) gene, exhibits a large decrease in the microarray, but low statistical significance. With qPCR we confirmed the variability in expression of Vm32E in 3wD samples, which explains the incongruence of statistical results (Fig. 3). The variability in expression of this gene may reflect that most, but not all, of the Canton S females suppress vitellogenesis in diapause; we always observed a small percentage of escapers [20]. Two of the genes tested (fatty acid desaturase, Fad2, and timeless, tim) are among the top three differentially regulated transcripts in the microarray, and this is confirmed by qPCR. For tim, microarray probes differentiate between subsets of the 11 known isoforms of the corresponding mRNA. Microarray data indicated that either one or both of the isoforms N and O were strongly upregulated during diapause, while other isoforms were not. Data from qPCR corroborates this pattern, with strong upregulation of N and/or O, and little change for other isoforms. Some gene transcripts had been assayed earlier from 1wN and 3wD flies [20] and the data are in agreement with the present microarray results. These (see Fig. 4) are genes encoding peptide hormones (dilps2, 3, 5, 6, and Akh), and metabolic read-out genes (4ebp, tobi, bmm and pepck).
Fig. 3.
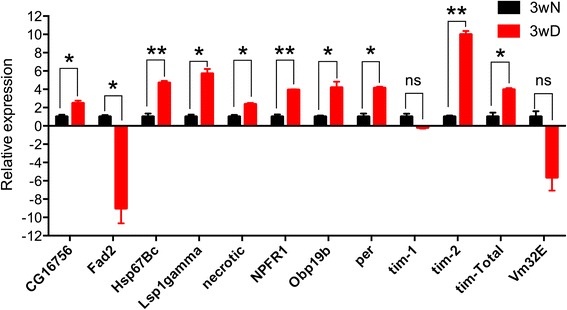
Quantitative real-time PCR confirms transcript changes in diapausing flies. Estimate of relative gene expression, comparing (ΔΔCt) three week old diapausing flies (3wD) with three week old flies kept under normal conditions (3wN). Directionality and relative magnitude of change matches microarray data in all cases, except Vm32E. We used Welch’s unpaired t-test with corrections for non-equal variances, comparing expression at 3wD and 3wN for each gene, ns, not significant; *, p < 0.05; **, p < 0.01. Error bars show standard error of the mean, n = 4. Acronyms used: FAD2 - Fatty Acid Desaturase 2, HS67Bc - Heat Shock 67 Bc, LSPgamma - Larval Serum Protein gamma, NeurpepRecF - Neuropeptide receptor F, OBP19b - Odorant Binding Protein 19 b, Per – Period, Tim-1 - Timeless, (targets all Timeless isoforms except N and O), Tim-2 - Timeless, (targets N and O isoforms of Timeless), Tim-Total - Timeless, (targets all Timeless isoforms), VME32e - Vitelline Membrane 32e
Fig. 4.
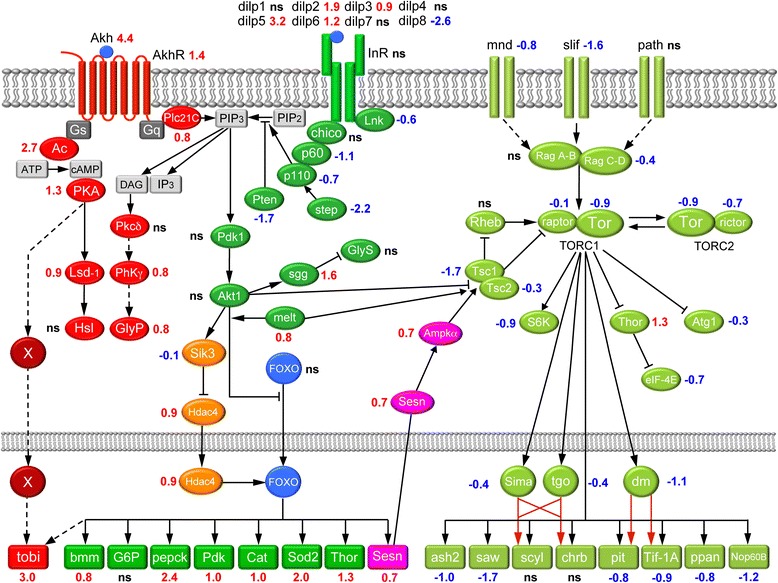
Altered gene expression in AKH, insulin/IGF- and TOR-signaling pathways in diapausing flies. The scheme displays a generalized assembly of relevant genes (regardless tissue-specificity) in the three signal pathways. Transcript levels (logarithmic fold change, LogFC) are given in red for upregulated, blue for downregulated and black for no significant change (ns; LogFC close to 0). The acronyms are listed in Additional file 6: Table S3 where also references and details of gene/protein functions are given. The AKH signaling is generally upregulated and the expression of the target gene tobi increased (X depicts an unidentified transcription factor). The Insulin/IGF signaling is generally downregulated and as a result several of the FOXO-regulated target genes are upregulated. The TOR-signaling is also downregulated, together with a number of its target genes. The AKH signaling scheme was compiled after [68, 171], the insulin/IGF signaling based on elements from [29, 36] and TOR signaling assembled after [35, 57]
Insulin/IGF, TOR and AKH signaling is altered during diapause
Drosophila responds to environmental challenges and maintains metabolic and energy homeostasis by evolutionarily conserved signaling pathways, such as the insulin/IGF signaling (IIS) and target of rapamycin (TOR) pathways [31, 35–38]. These signaling pathways display crosstalk in regulation of metabolism, homeostasis, growth, development and longevity [32, 35, 36]. Previous studies have indicated that the IIS pathway may be central in diapause regulation in flies and mosquitos [9, 21, 22, 24, 39], whereas, to our knowledge, there is no experimental data on TOR signaling (reviewed in [8, 23]).
To identify whether a pathway is generally up- or downregulated in diapausing flies we assembled the transcript data into their presumed functional context in signaling pathways. We particularly focused on readouts from specific transcriptional targets that are important in metabolism, stress responses, development and growth. Fig. 4 shows an assembly of transcript changes in the IIS, adipokinetic hormone (AKH) and TOR pathways based on the comparison of 3wD and 1wN flies. Note that due to the large number of genes discussed for these pathways and the ones in the following sections we have assembled a set of tables (Additional file 6: Table S3) where details are given for each gene, including relevant literature references.
Insulin/IGF signaling is downregulated
Based on transcriptional changes in several known IIS and FOXO target genes and the downregulation of several core IIS pathway components, our analysis suggests that IIS generally is downregulated during diapause (Fig. 4).
In diapausing flies we noted an up-regulation of dilp2, 3, 5 and 6, similar to a previous study [20], but a strong down-regulation of dilp8, a known coordinator of growth and development [40, 41]. We failed to observe any change in expression of the insulin receptor, InR, or the receptor substrate chico. However, Lnk (SH2B family of adaptor molecules; [42]), as well as the catalytic (Dp110) and adaptor (p60) subunits of PI3K, together with Steppke (step), an upstream regulator of PI3K [43] were down-regulated. Akt1 and Pdk1 (phosphatidylinositol-dependent kinase 1) expression was unaffected by diapause. Taken together, these data suggest attenuated IIS signaling during diapause, leading to reduced FOXO phosphorylation and thus increased FOXO translocation into the nucleus. This is supported by an increase in transcription of HDAC4, which may activate FOXO [44] in diapausing flies via increased deacetylation. The downregulation of IIS is further supported by the decreased expression of salt-inducible kinase 3 (Sik3), responsible for the phosphorylation and inactivation of HDAC4 [44]. Increased translocation of unphosphorylated HDAC4 into the nucleus could result in FOXO deacetylation and increased DNA-binding activity [44, 45]. The increased FOXO action is likely to increase activation of transcription of catabolic enzymes and other FOXO targets, including phosphoenolpyruvate carboxykinase (pepck) and brummer (bmm) TAG lipase [44], as observed in diapausing D. melanogaster. The enhanced transcription of bmm, the fly homolog of adipose triglyceride lipase (ATGL) [46], indicates an activation of lipolysis. Increased pepck transcription points towards an enhancement of both glyconeogenesis and glyceroneogenesis [47] and was previously recorded in diapausing larvae of the mosquito Wyeomyia smithii [48]. Furthermore, our data indicate an upregulation of pyruvate dehydrogenase kinase (Pdk), responsible for a re-direction of pyruvate into anaerobic oxidation [49]. Activation of gluconeogenesis and other anaerobic metabolic pathways under diapause is a universal mechanism found in different insects [8, 10].
Among other transcriptional targets of FOXO, we detected increased expression of the antioxidant enzymes, catalase (Cat) and manganese superoxide dismutase (Sod2) in diapausing flies. This supports an enhanced stress resistance during diapause, consistent with earlier findings in Drosophila [9, 15]. Moreover, diapause triggered an increase in Thor transcript, encoding the eukaryotic initiation factor 4 binding protein (4EBP), an inhibitor of translation [50] that might be responsible for the diminished protein synthesis in diapausing flies. Finally, we observed an activation of shaggy (sgg), a GSK homolog. GSK3 is responsible for glycogen synthase (GlyS) inhibition [51], suggesting diminished glycogen synthesis in diapausing flies. In summary, our data suggest that the IIS pathway is significantly downregulated during diapause and that altered gene transcription may lead to increases in lipolysis, glycogenolysis, gluconeogenesis, glyceroneogenesis and stress resistance, and results in lowered aerobic pyruvate oxidation and glycogen synthesis (see Additional file 7: Figure S3). Importantly, downregulated IIS is known to extend lifespan in Drosophila and C. elegans [32, 52–54].
TOR signaling is downregulated
The transcript changes in diapausing flies reveal effects on TOR pathway components responsible for ribosome and protein biosynthesis, including AMPK-mediated growth suppression, as well as hypoxia-responsive transcription, and suggest a general downregulation of TOR signaling. Nutrient dependent TOR signaling is involved in the regulation of growth and metabolism and interacts with IIS, also in aging [32, 33, 36, 55, 56].
The diapause-induced downregulation of the rapamycin-sensitive TOR partners raptor and Tor in the TORC1 complex could indicate diminished translation and ribosome biosynthesis, possibly via phosphorylation of downstream targets such as S6 kinase (S6K) and 4E-BP (Thor) [57]. Downregulation of Rag GTPases (RagC-D, but not RagA-B) excludes stimulation of the TORC1 complex in diapausing flies. Concomitantly, but independently, the TORC1 complex is negatively regulated by the tuberous sclerosis complex (TSC), which inactivate the GTPase Rheb [58]. In diapausing flies the expression of the TORC1 suppressors Tsc1 and Tsc2 was decreased, whereas the TORC1 activators, RagA-B, RagC-D and Rheb were either not changed or decreased as well (Fig. 4).
The TORC1 decrease noted during diapause may be mediated by another mechanism involving AMPK (AMP-activated protein kinase), which can phosphorylate TSC2, and lead to TORC1 inhibition [59]. A further inhibitory effect of AMPK on TORC1 through phosphorylation of Raptor [60] cannot be excluded, while there is an energy deficit during diapause that requires AMPK-mediated suppression of growth and biosynthetic processes [8]. Moreover, the inhibitory effect of AMPK on TOR signaling during diapause could be enhanced by elevated sestrin (Sesn) expression, since Sesn transcription in Drosophila is under FOXO control and leads to AMPK activation and in turn an enhancement of Tsc1/Tsc2 inhibition of TORC1 [61]. The latter mechanism may be general in diapause regulation and has been suggested earlier for flesh flies and apple maggot flies [39, 62].
We find that decreased TOR signaling in diapausing flies is accompanied by a downregulation of S6 kinase (S6K), which together with downregulated Pol I transcription factor Tif-IA and transcription factor diminutive (dm), a Drosophila Myc homolog, may explain the decreased ribosome biogenesis (see above and [57, 58]). The enhanced Thor expression in our study is accompanied by a decreased level of eIF-4E transcript, indicating a suppression of transcription [50] during diapause. An enhanced transcription of the Thor gene, via FOXO, and lack of inhibitory phosphorylation of 4EBP protein from suppressed TORC1, was previously found to provide an adaptive decrease in energy-consuming translation during nutrient restriction with decreased IIS and TOR signaling [36, 50]. The diminished biosynthetic processes in diapausing flies was accompanied by a lower expression of a number of TOR readout genes, the growth regulators (saw and ash2), as well as pitchoune (pit), peter pan (ppan) and Nop60B/minifly (Nop60B/mnf), indicating arrested differentiation and development together with lowered fertility [63]. In addition we observed downregulation of the main hypoxia-inducible transcription factors Similar (Sima) and Tango (tgo/Arnt). These findings suggest reduced hypoxia-responsive transcription during diapause. In summary, our analysis indicates a general downregulation of the TOR signaling cascade during diapause. Furthermore, activators of TOR signaling, including nutrient sensing through amino acid transporters, like slimfast (slif) [64], or Akt-mediated stimulation of the TORC1 complex [65], are repressed in diapausing flies.
AKH signaling is upregulated
Our data suggest that AKH signaling may be upregulated during diapause. In Drosophila and other insects AKH signaling mobilizes stored energy reserves [66–70]. AKH signaling furthermore plays a role under stressful conditions, to potentiate rapid production of energy for survival of the organism [68]. A recent study indicates that upregulated AKH signaling extends lifespan, although the mechanisms are yet to be determined [71]. We find upregulated expression of both neuropeptide (Akh) and AKH receptor (AkhR/GRHR) transcripts, and several components of the two possible second messenger cascades downstream the receptor (Fig. 4, Table 1). These pathways act via phospholipase C (PLC; Plc-21C) or adenylyl cyclase (Ac) [68, 70, 72, 73]. The activation of PLC via the AKH receptor may lead to stimulation of glycogenolysis during diapause (see [69]). Elevation of cAMP, via Ac, and activation of protein kinase A (Pka), may suggest enhanced lipolysis in diapausing flies (see [74]). Furthermore, diapausing flies display elevated expression of target of brain insulin (tobi), which was postulated to be under control of AKH signaling through an unknown transcription factor [75]. Thus, it seems that in diapausing flies AKH signaling enhances glycogenolysis, either via activation of glycogen phosphorylase (GlyP) or stimulation of tobi expression, or both pathways.
Table 1.
Neuropeptides, peptide hormones and receptors affected by diapause
| Neuropeptide and/or GPCRa | Expressionb | CG number | Log FC |
|---|---|---|---|
| Adipokinetic hormone (AKH) | Corpora cardiaca | CG1171 | 4.4 |
| AKH receptor (AKHR) | Fat body, spermatheca | CG11325 | 1.4 |
| Allatostatin A (Ast-A) | CNS, midgut | CG13633 | 1.9 |
| Allatostatin B (Ast-B or MIPc) | CNS | CG6456 | 1.1 |
| Allatostatin C (Ast-C) | CNS, midgut | CG14919 | 1.7 |
| CAPA receptor (capaR) | Renal tubules | CG14575 | 1.3 |
| CCHamide 2 | Midgut, fat body | CG14375 | 2.1 |
| Corazonin (CRZ) | CNS | CG3302 | 1.3 |
| CRZ receptor (CrzR) | Fat body, heart | CG10698 | 2.0 |
| DH31 receptor 1 (DH31–R1) | CNS, gut, renal tubules | CG32843 | 2.3 |
| Diuretic hormone 44 (DH44) | CNS | CG8348 | 3.4 |
| DH44 receptor 2 (DH44–R2) | CNS, gut, renal tubules | CG12370 | 2.5 |
| Insulin-like peptide 2 (DILP2) | Brain | CG8167 | 1.9 |
| DILP5 | Brain, ovary | CG33273 | 3.2 |
| DILP6 | Fat body | CG14049 | 1.2 |
| DILP8 | Ovary | CG14059 | −2.6 |
| DLGR1d | Hindgut, salivary gland | CG7665 | 3.1 |
| Dromyosuppressin (DMS) | Brain, heart | CG6440 | 1.5 |
| Drosulfakinin (DSK) | Brain | CG18090 | 1.4 |
| Ion transport peptide (ITP) | CNS, PNS | CG13586 | 1.2 |
| ITG (Apis-ITG-like) | CNS | CG8216 | 3.0 |
| Leucokinin (LK) | CNS | CG13480 | 2.1 |
| LK receptor (LK-R) | CNS, renal tubules | CG10626 | 1.3 |
| Neuropeptide F receptor (NPFR) | CNS, renal tubules | CG1147 | 2.4 |
| NPLP1e | CNS | CG3441 | 2.4 |
| NPLP3 | Head, carcass | CG13061 | 2.1 |
| NPLP4 | Fat body (larva), Eye | CG15361 | 1.3 |
| Orcokinin | CNS, gut | CG13565 | 1.6 |
| Proctolin | CNS | CG7105 | 2.2 |
| PTTHf | CNS, Renal tubules? | CG13687 | 1.1 |
| Short neuropeptide F (sNPF) | CNS | CG13968 | 1.5 |
| Tachykinin (DTK) | CNS, midgut | CG14734 | 2.5 |
| Torsog | Ovary | CG1389 | −3.3 |
Notes: The transcripts are sorted alphabetically (significantly >2 fold up- or downregulated; LogFC >1)
aAcronyms used are those for the proteins/peptides
bBased on FlyAtlas and/or modENCODE [116, 117], as well as research papers (summarized in [115]). Expression data are for adult flies, except NPLP4, where expression is most prominent in larvae
cMIP, myoinhibitory peptide
dLeucine-rich repeat-containing G protein-coupled receptor 1. Ligand is the dimeric GPA2/GPB5 protein
eNeuropeptide-like precursor 1
fProthoracicotropic hormone
gReceptor tyrosine kinase, can be activated by PTTH [132]
JAK/STAT signaling is affected in a tissue-specific manner
Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling coordinates important biological processes in Drosophila during development and in adult physiology, such as cell proliferation and growth [76], organ formation [77], stem cell maintenance [78, 79] and immune response [80, 81] important during aging. At the whole organism level we observed a decreased expression of the core components of the JAK/STAT signaling and some read-out genes in diapausing flies (Fig. 5). The downregulated genes in the signaling pathway include domeless (dome) encoding a cytokine-like receptor, and hopscotch (hop) encoding Drosophila Janus kinase (JAK), as well as a single Drosophila STAT gene Stat92E [76, 81, 82]. Further downregulated genes are shown in Fig. 5 and details on gene function are given in Additional file 6: Table S3. Notable exceptions are the upregulated Stat92E targets Turandot A (TotA), known to play important roles in stress tolerance and immune response [83, 84], vir-1 (virus-induced RNA-1), required for antiviral protection [85] and Tep2 (Thioester-containing protein 2) that is part of the humoral immune response [86, 87]. These findings suggest that diapause triggers a tissue-specific upregulation of JAK/STAT signaling.
Fig. 5.
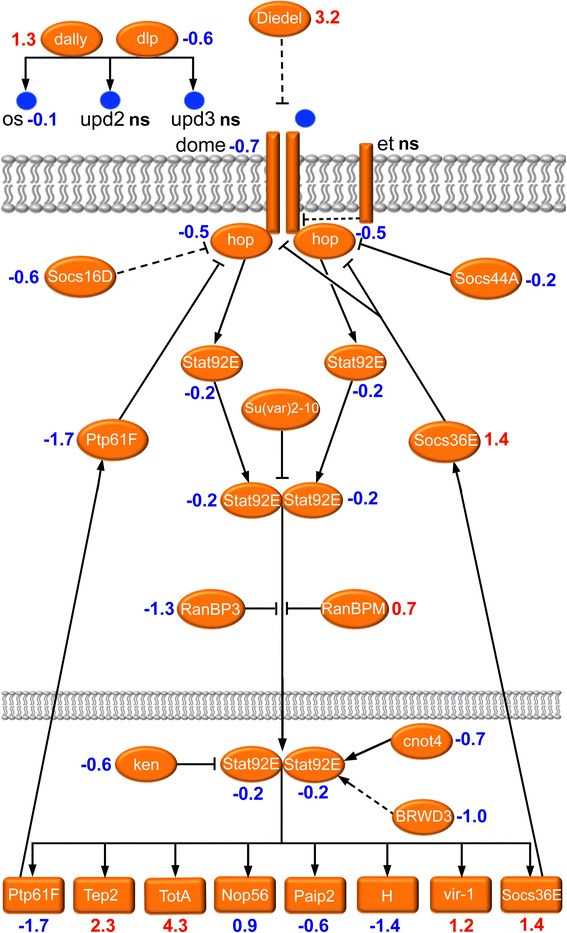
Altered gene expression in the JAK/STAT signaling pathway during diapause. This scheme displays a generalized assembly (regardless tissue specificity) of relevant genes in the JAK/STAT signal pathway. Transcript levels (logarithmic fold change, LogFC) are given in red for upregulated, blue for downregulated and black for no significant change (ns; LogFC close to 0). The acronyms are listed in Additional file 6: Table S3 where also references and details of gene/protein functions are given. At the whole organism level we observed a decreased expression of the core components of the JAK/STAT signaling and some read-out genes in diapausing flies. Some exceptions are seen in certain genes, involved in humoral immune response and stress resistance, which were upregulated (probably tissue-specific). The scheme is based partly on [172] and [80]. Further details are given in Additional file 6: Table S3 and text
Other genes used as readout of the JAK/STAT pathway, including dome [88] and Ptp61F [89] displayed decreased expressions, whereas Socs36E [90] and Turandot A (TotA) [83] expression was increased. Most JAK/STAT target genes with Stat92E binding sites [91], including growth regulator Nop56, Juvenile hormone epoxide hydrolase (Jheh2), zinc finger transcription factor ftz-f1, negative regulator of Notch pathway Hairless (H) and negative regulator of translation polyA-binding protein interacting protein 2 (Paip2) were downregulated.
The upregulation of TotA, vir-1 and Tep2 during diapause suggest a possible role of JAK/STAT in enhancing the stress and immune defense. This is supported by the upregulation of other Turandot genes, including TotA, TotC, TotM, and TotX (see Additional file 6: Table S3). Taken together, our findings indicate that the JAK/STAT pathway may regulate different target genes in a tissue-specific manner in diapausing flies. To test this hypothesis we kept flies expressing a 10xStat92E-GFP reporter [76] under diapause and normal conditions and screened for tissue-specific activation of JAK/STAT signaling. Strong GFP fluorescence was detected in specific thoracic regions, including thorax, legs, and ovaries of diapausing flies (Fig. 6a,b). The overall GFP expression increased significantly during diapause (Fig. 6c)
Fig. 6.
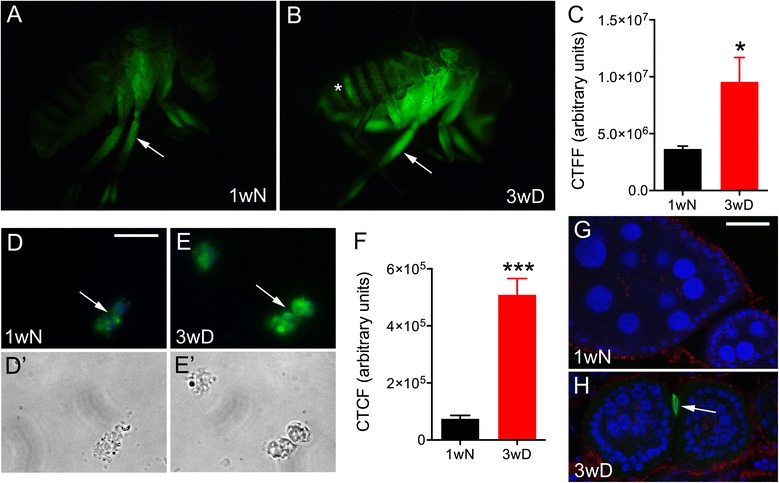
Tissue specific JAK/STAT signaling in diapausing flies. The Stat92E reporter 10xStat92E-GFP expression in the whole body is compared between control flies (1wN) (a) and flies in diapause conditions (3wD) (b) and shows an increased fluorescence in 3wD, as quantified in (c) with corrected total fly fluorescence (CTFF). The statistical significance was verified by Welch’s unpaired t-test, * p < 0.05, n = 6 (error bars show standard error of the mean). In (a) and (b) arrows point to hemocytes that accumulated in the legs of the fly, which largely contributes to the fluorescence signal, and the asterisk in B indicates GFP signal in the ovaries. d and e Hemocytes exhibit a strong accumulation of the 10xStat92E-GFP signal upon diapause, 3wD (e) compared to controls, 1wN (d). In these figures green label is 10xStat92E-GFP, and blue label hemocyte nuclei stained with DAPI. E´ and D´ The corresponding hemocytes displayed in bright field. The scale bar corresponds to 20 μm. f Corrected total cell fluorescence (CTCF) for diapause and control hemocytes is calculated as an average from n = 15, statistical significance verified by Welch’s unpaired t-test, ***, p < 0.001. Error bars show standard error of the mean. g and h Portions of ovaries from control (1wN) and diapausing (3wD) flies. In diapausing flies there is an increased expression of the 10xStat92E-GFP reporter (green) in polar cells of ovaries (arrow in h), which is not detectable in directly developing ovaries of control flies (g). Blue label, nuclei stained with DAPI; red label phalloidin-TRITC. Scale bars: D, E 50 μm, G, H 25 μm
The strong GFP signal of the 10xStat92E-GFP reporter in the whole fly body appears to derive mainly from hemocytes (Fig. 6d-f). Adult hemocytes are mostly sessile and tend to accumulate in legs and thorax close to the halteres [92]. This immune tissue specific regulation might explain why we detected predominantly stress-related and immune-response targets of JAK/STAT in our microarray. It has been shown that JAK/STAT-signaling regulates humoral defense factors, including several ones produced by hemocytes [81]. We noted an upregulation of Tep2, 3 and 4 (see Fig. 5), which resemble both the vertebrate complement factors and members of the α-macroglobulin family of protease inhibitors, and are predominantly expressed by hemocytes [81, 87]. A more detailed analysis of the ovaries revealed that the GFP signal is localized to specialized follicle cells (polar cells) and close to stem cells (Fig. 6g, h). It is known that JAK/STAT signaling is critically important for early differentiation of follicle cells and proper germ line cell encapsulation during Drosophila oogenesis [88, 93, 94], as well as subsequent reduction of polar cell number [95] and border cell migration which occurs during stage 9 [96]. Oogenesis in diapausing females is in most cases arrested at previtellogenic stages (stage 7 and earlier) [20], when strong JAK/STAT signaling in polar cells is active and necessary to keep egg chamber morphology stable.
Innate immunity: the Toll pathway is activated
Analysis of the array data confirmed our previous results [20] that Drosomycin as well as other Drosomycin-like genes exhibit increased expression in flies that have entered diapause. Drosomycin is considered a canonical target of Toll (Tl) signaling (see [97]). A summary diagram is shown in Fig. 7 and further details on immune genes are given in Additional file 6: Table S3. We found significant and strong upregulation for two additional targets of Tl signaling, namely Immune induced molecule 1 (IM1) and Immune induced molecule 2 (IM2) (see [98] and Fig. 7). A detailed pathway analysis revealed upregulation of genes for Peptidoglycan recognition proteins (PGRPs) and Gram-negative bacteria binding proteins (GNBPs), which bind to immune elicitors and activate an extracellular proteolytic cascade ultimately leading to proteolytic activation of Spätzle (encoded by spz) to its active form. Several extracellular members of the Tl-activating proteolytic cascade were upregulated upon diapause (see Fig. 7). Since both the serine protease Persephone (psh), which is part of the Spätzle-activating cascade [99], but also its inhibitor Necrotic (nec), were induced, activation of Tl signaling occurs most likely downstream of ModSP/Grass, a multifunctional member of the cascade. Despite a slight downregulation of Tl itself and its ligand Spätzle (encoded by spz), we could clearly show that the induction of both Drs and Drsl5 is regulated via Spätzle, since spz mutants failed to induce both peptides, while flies mutant in the transcription factor relish (rel), which is part of the IMD (Drosophila immune deficiency) pathway show no such difference (Fig. 8a and b). Downstream of Tl-signaling during diapause the combined upregulation of the transcription factor dorsal (dl) and the downregulation of the inhibitor cactus (cact), leads to increased transcription of dl (and possibly dif) target genes.
Fig. 7.
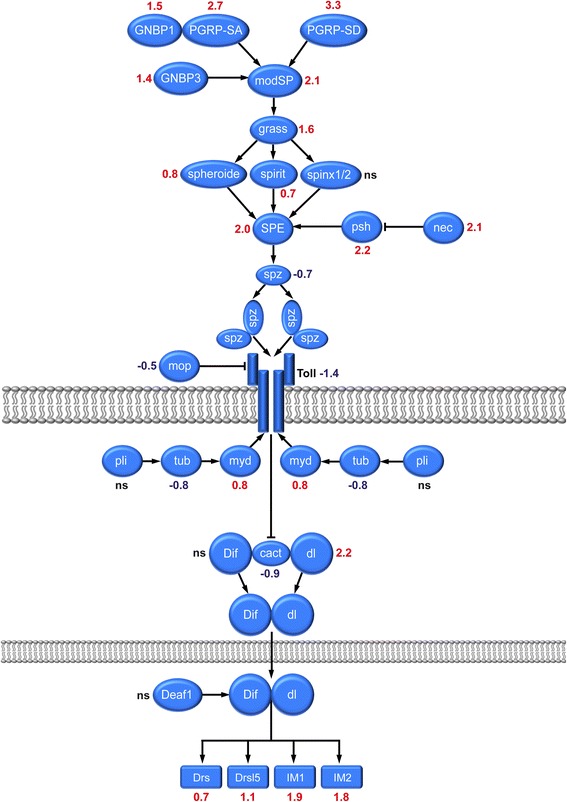
Altered gene expression in Toll (Tl) signaling pathway during diapause. The scheme displays a generalized assembly (regardless tissue specificity) of relevant genes in the Toll signal pathway. Transcript levels (logarithmic fold change, LogFC) are given in red for upregulated, blue for downregulated and black for no significant change (ns; LogFC close to 0). The acronyms are listed in Additional file 6: Table S3 where also references and details of gene/protein functions are given. The Toll pathway is activated in diapausing flies as seen from increases of target genes such as Drosomycin (Drs), Drosomycin-like 5 (Drsl5), IM1 and IM2. Also several extracellular members of the Toll-activating proteolytic cascade were upregulated. The signaling scheme is based on [97]
Fig. 8.
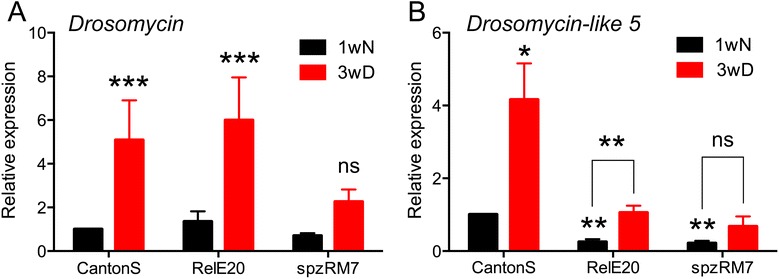
Drosomycin (Drs) and Drosomycin-like 5 (Drsl5) transcription during diapause is regulated by Spätzle and Toll signaling. Upregulation of Drosomycin (a) and Drosomycin-like 5 (b) is significant in the control genotype (Canton S), as well as in a mutant in a component of the Imd pathway, the transcription factor Relish (RelE20). Conversely transcription of Drs and Drsl5 was blocked in a mutant in the Toll ligand Spätzle (spzRM7). Significance compared to one-week-old Canton S flies (1wN) was verified by ANOVA, followed by Tukey test (ns, not significant; * p < 0.05; ** p < 0.01; *** p < 0.001). Error bars show standard error of the mean, n = 3
The ERK/MAPK pathway is downregulated
The extracellular signal-regulated kinase/mitogen-activated protein kinase (ERK/MAPK) signaling cascade regulates cell proliferation, differentiation and survival [100, 101] and we detected a downregulation of key components of this pathway. ERK/MAPK signaling can be triggered by ligands binding to receptor tyrosine kinases (RTKs) with subsequent Ras activation at the plasma membrane or via Ras induction from intracellular sites [101]. As seen in Additional file 8: Figure S4 downregulation of the whole MAPK signaling cascade in diapausing flies is supported by decreased expressions of all kinases, including MAPKKK/Raf1 (encoded by phl), MAPKK/MEK (encoded by Dsor1) and MAPK/ERK (encoded by rl). In addition we found diminished expressions of almost all positive regulators in the pathway, participating in Ras (βggt-I, Fnta, Hmgcr), Raf1 (ave, ksr, cnk, Cdc37, 14-3-3ε), MEK (gfzf) and ERK (mago, Fip1, Cdk12, eIF4AIII) activation [100]. Further details on specific genes are provided in Additional file 6: Table S3. At the receptor level we noted an activation of genes encoding the RTK Sevenless and its ligand Boss in the diapausing flies. Thus, there might be a role of Boss in regulation of carbohydrate and lipid metabolism during diapause, similar to the one found during larva to pupa transition [102]. Possibly downregulation of MAPK signaling is of general importance during diapause, while its activation is responsible for the initiation of development following diapause termination in several insect species [103–105].
JNK signaling is activated in diapause
The Jun-N-terminal Kinase (JNK) signaling pathway is an evolutionarily conserved stress-activated protein kinase pathway that is induced by a range of intrinsic and extrinsic stressors [106, 107]. JNK signaling is also involved in developmental and metabolic regulation, immune responses, as well as cell death and lifespan extension [106, 108]. In Drosophila JNK signaling involves a conserved core of kinases (JNKKK-JNKK-JNK) and a mitogen activated protein kinase (MAPK) pathway, and can be loosely classified into “canonical” and “non-canonical” signaling pathways [107]. Our transcriptome data indicate a moderate general activation of the “non-canonical” JNK signaling pathway in diapausing flies.
We assembled an overview of the JNK signaling cascade in D. melanogaster (Based on [106, 107]) with associated transcript changes induced by diapause (Fig. 9). Details on genes are provided in Additional file 6: Table S3. The “canonical” JNK pathway seems to be downregulated in diapausing flies, since we detect decreased expression of transcripts of the main upstream acting molecules, like non-receptor tyrosine kinases of the Src oncogene family (Src64B and Btk29A, but not Src42A) and the kinase shark, together with its adaptor protein Downstream of kinase (Dok). Further components of the “canonical” pathway were also downregulated, including the specific JNKKK Slipper (slpr), together with the kinase Misshapen (msn), as well as the main activator Rac1 (a small GTPase).
Fig. 9.
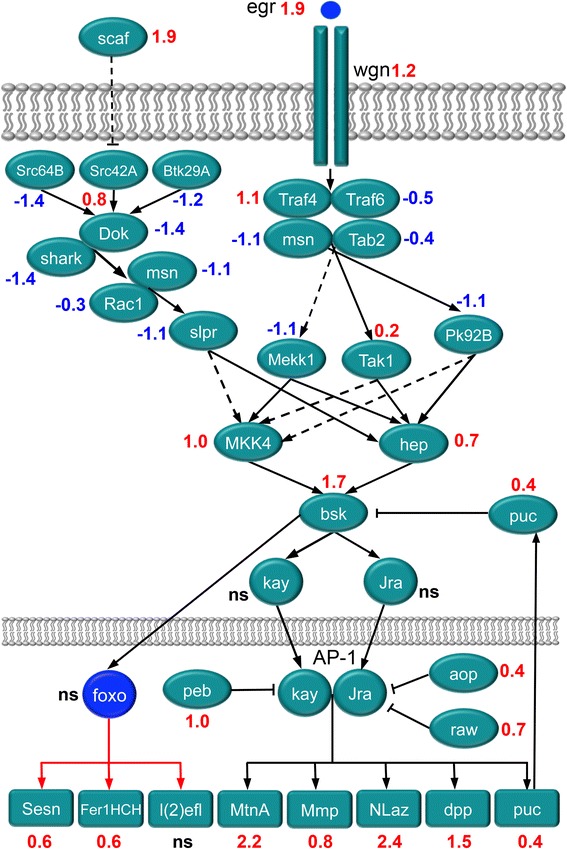
Altered gene expression in the JNK signaling pathway during diapause. This scheme displays a generalized assembly (regardless tissue specificity) of relevant genes in the JNK signal pathway. Transcript levels (logarithmic fold change, LogFC) are given in red for upregulated, blue for downregulated and black for no significant change (ns; LogFC close to 0). The acronyms are listed in Additional file 6: Table S3 where also references and details of gene/protein functions are given. At the whole organism level diapausing flies demonstrate a moderate activation of JNK signaling as judged from an elevated expression of some target genes. There is also enhanced expression of JNK-responsive genes, known to be under FOXO (IIS) transcriptional control (Sesn and Fer1HCH)
In the “non-canonical” JNK pathway a ligand, the tumor necrosis factor homolog Eiger (egr/TNF) [109] and a receptor, Wengen (wgn/TNFR) [110] have been identified, both of which were upregulated in diapausing flies (Fig. 9). The activation of “non-canonical” JNK signaling in diapause is further suggested by elevated transcriptional levels of the main target genes of the transcriptional complex AP-1, including a Drosophila member of transforming growth factor-β (TGF-β) family decapentaplegic (dpp) and puckered (puc), which encodes a JNK-specific phosphatase (Fig. 9). Previous studies of diapausing D. melanogaster actually revealed that upregulation of the JNK target dpp varies in Australian and North American clines [26, 111]. Among JNK signaling responsive genes, upregulated in diapausing flies, we also found the homolog of vertebrate profilin chickadee (chick), matrix metalloproteinase (Mmp1), free radical scavenger Ferritin 1 heavy chain (Fer1HCH), the antioxidants metallothionein A (MtnA) and Sestrin (Sesn) and the lipocalin Neural Lazarillo (NLaz).
Moderate activation of JNK signaling results in increased stress tolerance and extended lifespan [106], due to increased expression of cytoprotective JNK target genes in various tissues [112, 113]. This mechanism is balanced by an antagonism between JNK and IIS signaling pathways that regulate FOXO in opposite ways [108]. The same mechanism may be involved in Drosophila diapause leading to extended lifespan and increased stress tolerance [9, 15, 20]. Taken together our findings indicate a moderate general activation of “non-canonical” JNK signaling in diapausing D. melanogaster that could contribute to extended lifespan.
Neuropeptides and peptide hormone signaling are selectively affected
In Drosophila, as in other animals, neuroendocrine peptides are central to the regulation of development, reproduction, physiology and behavior [114, 115] and at least insulin-like peptides and AKH have been implicated in regulation of longevity [28, 32, 71]. Thus, we screened for diapause-induced changes in expression of genes encoding neuropeptide/peptide hormone precursors and their G-protein coupled receptors (GPCRs).
The transcripts of 23 neuroendocrine peptide precursors were significantly upregulated in diapausing flies (Table 1). The only precursor transcript that was significantly down-regulated was dilp8, proposed to be expressed in ovaries (FlyAtlas [116], ModENCODE [117]). For four of the peptide ligands also the corresponding GPCR transcripts were significantly upregulated: those of AKH, corazonin (CRZ) diuretic hormone 44 (DH44), and leucokinin (LK). Transcripts of five peptide GPCRs were upregulated, but with ligand levels unchanged: the DH31-receptor 1 (DH31-R1), the capability (CAPA) receptor (capaR), DLGR1 (ligand is the dimeric protein GPA2/GPB5) and the neuropeptide F receptor (NPFR). Four of the peptide hormone signaling pathways with increased transcripts (capaR, DH31-R1, DH44, LK) are known to regulate diuresis by action on the Malpighian tubules, and the NPFR is expressed in this tissue [118]. Furthermore, in mosquitos DLGR1 is involved in hindgut regulation of ion and water balance [119]. Some of the upregulated peptides modulate feeding [allatostatin A (AstA), CCHamide2, short neuropeptide F (sNPF), NPF, drosulfakinin (DSK)] and others (CRZ, sNPF, tachykinin) may regulate stress responses [115, 118, 120–122]. Finally, five of the peptides (AstA, CRZ, CCHamide2, sNPF, tachykinin) are known to act on the insulin producing cells of the brain and thus modulate IIS [122–124]. In summary, peptidergic signaling appears altered in pathways involved in regulation of feeding, carbohydrate and lipid metabolism, IIS, stress responses, diuretic functions and longevity.
Juvenile hormone, ecdysone and monoamine signaling is affected by diapause
Since monoamines, ecdysone (Ecd) and juvenile hormone (JH) have been implicated in diapause, reproduction and stress responses in several insects (see [8, 25, 27, 125–127]) we assembled transcript data for relevant genes in these signaling pathways (Table 2). Several genes associated with serotonin, dopamine and octopamine signaling displayed increased expression (Table 2). Whereas dopamine signaling may be confined to the CNS in Drosophila, serotonin and octopamine are also known to have peripheral targets [128].
Table 2.
Hormonal and monoamine signaling components affected by diapause
| Gene transcript | Role (main enrichmenta) | CG number | Log FC |
|---|---|---|---|
| Monoamine signaling | |||
| 5-HT2A (serotonin receptor 2) | Serotonin signaling (salivary gland, eye) | CG1056 | 2.2 |
| SerT (serotonin transporter) | Serotonin signaling (CNS, eye) | CG4545 | 1.4 |
| Ddc (Dopa decarboxylase) +2.82 CG10697 | Dopamine biosynthesis (CNS) | CG10697 | 2.8 |
| DAT (Dopamine transporter) +1.7 CG8380 + 1.35 CG3856 | Dopamine storage (CNS) | CG8380 | 1.7 |
| DopR2 (Dopamine receptor 2) +1.15 CG18741 | Dopamine signaling (CNS) | CG18741 | 1.1 |
| DopEcdR (Dopamine/Ecd receptor) +2.87 CG18314 | Dopamine ecdysone signaling (CNS) | CG18314 | 2.9 |
| Dat (Dopamine N acetyltransferase) +1.35 CG3318 | Dopamine inactivation (CNS, gut) | CG3318 | 1.4 |
| Tdc1 (Tyrosine decarboxylase 1) +1.56 CG30445 | Octopamine/tyramine biosynt. (gut) | CG30445 | 1.6 |
| Oamb (octopamine receptor) | Octopamine (OA) signaling (CNS) | CG3856 | 1.4 |
| Oct-TyrR (OA-Tyramine receptor) +1.84 CG7485 | Octopamine/tyramine signal. (CNS) | CG7485 | 1.8 |
| Ecdysone signaling | |||
| Spo (Spook) Cytochrome P450 | Ecdysone (Ecd) biosynthesis (ovary) | CG10594 | −1.6 |
| Eo (ecdysone oxidase) +2.44 CG9504 | Ecd metabolism (renal tubules; RT) | CG9504 | 2.4 |
| Usp (ultraspiracle) | Ecd receptor partner (ovary) | CG4380 | −1.3 |
| Cyp18a1 Cytochrome P450b | Ecd inducible (fat body, spermatheca) | CG6816 | 1.6 |
| Eip63F1 (Ecd induced protein 63 F1)c | Ecd inducible protein (CNS, gut, RT) | CG15855 | 2.0 |
| Eip75B (Ecd induced protein 75B)b + 1.1 CG8127) | Ecd inducible protein (ubiq.) | CG8127 | 1.1 |
| Eip74EF (Ecd induced prot. 74EF) +1.67 CG32180 | Ecd inducible proteind (brain, crop) | CG32180 | 1.7 |
| Ftz-f1 (ftz transcription factor 1) | Ecd and JHe inducible gene (ovary) | CG4059 | −1.9 |
| ImpL3 (ecd inducible gene L3) | Ecd inducible gene (midgut) | CG10160 | 2.4 |
| Br (Broad) | DNA-binding, Ecd response (CNS) | CG11491 | −1.5 |
| ecd (ecdysoneless) | Ecd biosynthesis ? (CNS, ovary) | CG5714 | −1.1 |
| Juvenile hormone signaling | |||
| Jhamt (JH acid methyltransferase) | JH biosynthesis (brain) | CG17330 | 1.8 |
| Jheh1 (JH epoxide hydrolase 1) | JH catabolic process (fat body, gut) | CG15101 | 1.3 |
| Jeheh3 (JH epoxide hydrolase 3) | JH catabolic process (gut) | CG15106 | 1.2 |
| Jhedup (JH esterase duplication) | Carboxylesterase activity (head) | CG8424 | 3.2 |
| Jhi1 (JH-inducible protein 1) | endoribonuclease activity (ubiq.) | CG3298 | −1.5 |
| jhi26 (JH-inducible protein 26) | Protein kinase-like (renal tubules) | CG3767 | 2.9 |
Notes: These transcripts are significantly affected more than two-fold (>LogFC 1)
aAdult expression (according to FlyAtlas)
bRead-out for increased ecdysone or 20-hydroxy ecdysone signaling during development [129, 130]
cEF-Hand 1, calcium-binding site
dmay be involved in autophagy
eJH, juvenile hormone
Ecd signaling may be upregulated after three weeks of diapause since the genes Eip75B (E75B) and Cyp18a1, often used as proxys for elevated Ecd titer [129, 130], display increased transcript levels (Table 2). Also the Ecd-induced genes, Eip74EF and ImpL3 as well as the Dopamine/Ecd receptor (DopEcdR) are upregulated. Prothoracicotropic hormone (PTTH), a peptide known to induce Ecd biosynthesis [131] is also significantly upregulated in diapausing flies (Table 1). However, genes encoding the biosynthetic enzymes Spook, ecdysoneless, the Ecd receptor partner Ultraspiracle, the DNA-binding Broad, and the Ecd-induced transcription factor Ftz-f1 are downregulated and Ecd oxidase upregulated during diapause. As shown in Table 2 several of the downregulated genes, including that of the PTTH receptor Torso [132] (see Table 1), are normally enriched in ovaries, suggesting a possible tissue specific response in adult flies.
In the JH signaling pathway genes in catabolic processes are upregulated, whereas one JH-inducible gene is upregulated (jhi1) and one is downregulated (jh26). Thus it is not clear from our transcript data how JH signaling is affected by three weeks of diapause at the organism level. In summary, it is likely that the dynamics of JH and Ecd signaling is most drastic during the initial phases of diapause and thus our data reflect a later, steady state transcriptional snapshot. Therefore, we may miss transcript changes reflecting critical transient hormone pulses or decreases, as well as feedback mechanisms.
Altered transcripts of numerous genes implicated in aging and lifespan extension in Drosophila
We compared our list of diapause induced transcript changes with a set of genes from the GenAge database [133, 134]. Out of 136 genes, which are known to influence longevity in D melanogaster, we found 100 to be altered also during dormancy (Table 3, Additional file 9: Table S4). Whereas genes with pro-longevity effects are mostly strongly upregulated, anti-longevity genes are downregulated as a response to dormancy. This is in line with the drastic extension of lifespan with reduced senescence shown in diapausing flies [20]. Performing a broader comparison, we revealed that about 28 % of the genes in our study (Fig. 10a, for details see Additional file 9: Table S4) have previously been recorded as part of a transcriptional response to aging [7, 135, 136], or were identified as candidate genes affecting longevity [33, 135]. According to the Panther GO overrepresentation test the majority of these shared genes do not have mapped GO terms, however some are involved in actomyosin organization and meiotic and mitotic division (Additional file 10: Figure S5).
Table 3.
Genes shared between 3 weeks diapause and aging in Drosophila
| Symbol | Gene name | 3wD (logFC) | Longevity effect |
|---|---|---|---|
| ry | rosy | 2.4 | pro |
| Nlaz | Neural Lazarillo | 2.38 | pro |
| to | takeout | 2.21 | pro |
| Zw | Zwischenferment | 2.18 | pro |
| elav | embryonic lethal abnormal vision | 1.83 | pro |
| bsk | basket | 1.69 | pro |
| GstS1 | Glutathione S transferase S1 | 1.68 | pro |
| Nf1 | Neurofibromin 1 | 1.63 | pro |
| CG1623 | hebe | 1.54 | pro |
| Mlp84B | Muscle LIM protein at 84B | 1.45 | pro |
| CG11546 | kermit | 1.43 | pro |
| CG8846 | Thor | 1.3 | pro |
| Ilp6 | Insulin-like peptide 6 | 1.17 | pro |
| cher | cheerio | 1.13 | pro |
| rut | rutabaga | 1.01 | pro |
| srl | spargel | −1.06 | pro |
| bam | bag of marbles | −1.18 | pro |
| Daxx | Daxx-like protein | −1.4 | pro |
| POSH | Plenty of SH3s | −1.42 | pro |
| CG5671 | Pten | −1.71 | pro |
| Mt2 | Methyltransferase 2 | −2.14 | pro |
| Hsp26 | Heat shock protein 26 | −2.89 | pro |
| Hsp27 | Heat shock protein 27 | −3.15 | pro |
| CG6284 | Sirt6 | −3.41 | pro |
| a-Man-Ia | alpha Mannosidase I | 2.01 | anti |
| Ilp2 | Insulin-like peptide 2 | 1.9 | anti |
| E(z) | Enhancer of zeste | −1 | anti |
| snz | snazarus | −1.2 | anti |
| Edem1 | ER degradation enhancer mannosidase alpha-like 1 | −1.35 | anti |
| kuk | kugelkern | −1.36 | anti |
| LBR | Lamin B receptor | −1.91 | anti |
| CG6824 | ovo | −1.98 | anti |
| esc | extra sexcombs | −2.44 | anti |
| Hsp22 | Hsp22 | −1.13 | both |
Notes: During diapause 34 genes out of 136 hits for aging genes in GenAge database were found to be significantly regulated (q < 0.05, |logFC| > 1). Comparison with the GenAge database [133, 134] http://genomics.senescence.info/genes/
Fig. 10.
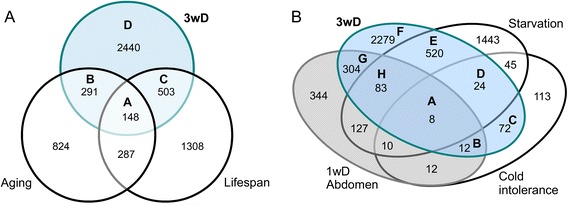
Venn diagrams comparing differentially regulated genes in genome-wide transcriptome studies of D. melanogaster. a Venn diagram comparing differentially regulated genes after 3 weeks of diapause (3wD, blue area; our study) with genes responding to aging [136] and a list of candidate genes affecting lifespan [135]. In Additional file 10: Figure S5 we show the results of Overrepresentation test of GO terms, performed in PANTHER for genes altered in dormancy in comparisons in groups A-D. b Comparisons with starvation, one-week diapause (abdomen) and cold tolerance. The blue area displays the number of differentially regulated genes after 3 weeks of diapause (3wD; our study). The grey area shows number of genes regulated in abdomens of flies after one week of diapause (1wD Abdomen) [140]. The other two sets contain genes regulated due to cold intolerance [141] and starvation [143]. In Additional file 11: Figure S6 we present a gene set enrichment analysis (GSEA) of Gene Ontology (GO) terms for comparisons among the four genome-wide transcription studies for each of the fields A-H in 10B
We find that this overlap with several aging studies is not only observed at the level of gene lists, but also in the occurrence of affected signaling pathways and GO terms. Using the Drosophila melanogaster genetic reference panel (DGRP) for a genome-wide analysis of single nucleotide polymorphisms (SNPs) influencing lifespan a number of genes were identified with roles in processes such as carbohydrate metabolism, TOR signaling, proteolysis, cell death and others [33]. Of the top ranked genes identified in that study we also identified TOR, slif, mipp2, Thor, sima, dm of the TOR pathway, and brummer (and Thor) of the IIS pathway, as well as the protease nephrilysin1 (Nep1) and the organic cation transporter (Orct), possibly downstream of S6K in TOR signaling (see Fig. 4 and Additional file 3: Dataset S1, Additional file 9: Table S4).
Increasing S-adenosylmethionine (SAM) synthesis by FOXO-dependent glycine N methyltransferase (Gnmt) extends the lifespan in Drosophila and thus overexpression of Gnmt increases longevity, cooperatively with dietary restriction and lowered IIS [137]. We see a 6.3 LogFC (increase) in Gnmt in three week diapausing flies (Additional file 3: Dataset S1, Additional file 9: Table S4). Another gene implicated in Drosophila lifespan extension is Tequila a multiple-domain serine protease known to be upregulated during infection [138]. These authors showed that knockdown of Tequila in insulin producing cells increases longevity, probably due to decreased systemic IIS. In our transcript data Tequila is significantly upregulated during diapause (1.7 LogFC).
Finally, it was noted that aging is associated with alterations of genes involved in responses to light and expressed in the eye [139], and we found a few genes associated with phototransduction significantly upregulated: NinaC (CG5125, 2.1 LogFC), InaE (CG33174, 1.6 LogFC), Rhodopsin 3 (CG10888, 1.2 LogFC) and Gβ5 (CG10763, 2.15 LogFC). Thus, apart from components of the IIS and TOR pathways, a number of single genes with important roles in extending lifespan have been implicated in various screens and are also seen in our gene list from diapausing flies.
Comparisons with other genome-wide investigations of dormancy and stress
Despite the high level of complexity of transcriptional changes in fly diapause, as reflected by the large number of affected genes in our analysis, there are certain commonalities with gene regulation found in other stress syndromes, including other forms of dormancy in animals. Especially comparisons at a gene ontology level show considerable conservation.
First, we compared our gene list with previously published genome-wide transcription studies in D. melanogaster, where relevant physiological phenotypes were tested. Not surprisingly, we found considerable overlap with transcripts identified in a study focusing on egg development during one week in diapause conditions [140]. Almost 50 % of the altered genes of that genome-wide transcription study were changed also in our analysis (Fig. 10b). According to the GO analysis, these genes are mostly involved in DNA replication and repair, chitin based cuticle attachment to epithelium, anatomical structure development, regulation of mitosis and maintenance of RNA localization (see Additional file 11: Figure S6, groups G and H). Note that the egg development study only analyzed the transcriptome of the abdomen, whereas our study covers transcriptional changes in the whole fly. Thus, our work includes additional genes expressed in the CNS and other tissues in the head and thorax, likely to be important in association with the diapause syndrome.
After three weeks in diapause at 11 °C the flies seem well adapted to cold conditions. Thus we compared our data to a genome-wide analysis of transcriptional changes in a cold-sensitive fly strain compared to its cold resistant siblings [141]. We find that 40 % of the genes are shared between these studies (Fig. 10b). The majority of the genes are regulated in opposite directions compared to our 3wD flies. The GO terms common for the shared genes are response to heat, immune response (covers part of the Toll pathway and stress response pathway JAK/STAT) and response to oxidative stress (see Additional file 11: Figure S6, group C). In this context, the gene frost (CG9434) is among the most upregulated in our study (LogFC 7.4; Fig. 2) and is known to be essential for cold tolerance [142].
During diapause, food intake of flies is strongly reduced [20]. Indeed, a comparison with a study of starved flies [143] reveals about 30 % shared genes and GO terms, such as inhibition of cell division, changes in biosynthesis, carbohydrate catabolism (for GO term enrichment analysis see Additional file 11: Figure S6, groups D and E) and downregulation of TOR pathway response genes (e.g. ash2). A comparison with abdominal genes after 1wD [140] only highlights 10 % shared genes (Fig. 10b). Starvation usually results in many pathologies, eventually leading to increased mortality, whereas during diapause flies still feed at a reduced rate, aging is slowed down and flies display extended lifespan [20]. Possibly the adult diapause is more similar to food restriction [144]. Indeed, a comparison to transcriptome changes after food restriction show 50 % of the transcripts we identified in our analysis (Additional file 12: Dataset S2). However, most genes exhibit much higher fold change in diapause than upon food restriction. The shared genes are mostly connected to cell cycle and gene expression (Additional file 13: Dataset S3). Surprisingly, 120 genes were found to be regulated in opposite direction; these mostly comprise carbohydrate metabolism, as verified by GO term enrichment analysis (Additional file 13: Dataset S3), and a comparison of GSEA of KEGG pathways between ours and the food restriction study (compare Additional file 4: Table S1 with [145]). Whereas in diapause we found many upregulated genes in starch, sucrose, galactose metabolism, as well as pentose cycle, those pathways are mostly downregulated upon food restriction. Another discrepancy we found is in expression of cytochromes p450, which are highly upregulated upon diapause.
Next, we compared our transcriptome analysis to studies on D. melanogaster where the focus was on mapping genetic variation that could be implicated in diapause, revealing many interesting similarities. As already mentioned, the importance of insulin-regulated genes such as dp110 for diapause has been found also in genetic studies (e.g. [24]), and coach potato, previously identified as a candidate gene for climatic adaptations in D. melanogaster [146] was differentially expressed in our transcriptome. Tauber et al. [147] provided evidence that a mutation in the clock gene timeless has spread in Europe over the last 10 000 years, and suggested a link to an enhancement of diapause (see also [148]). Notably, timeless was one of the most strongly differentially regulated genes in our study. Finally, a genome-wide sequencing study [26] comparing populations along the North American east coast found that many of the most strongly differentiated genes along the cline of increased diapause intensity northwards are involved in functional pathways indicated also by our transcriptome study, i.e. insulin/TOR, JAK/STAT, immunity and circadian rhythm pathways. In all, this concordance between studies of different types suggests that the same genes are targeted by genetic adaptation and transcriptional responses, and indicates that an understanding of the pathways involved in diapause is emerging.
What about other species? Only a limited number of transcriptome studies on reproductive diapause in other insect species are available so far, and these usually identify only a small number of significantly changed genes. In a comparison with a targeted mini-array performed on Drosophila montana, that live in temperate regions and has a photoperiodically regulated reproductive diapause [149], we found three genes out of 17 to be regulated in the same manner in D. melanogaster (Additional file 14: Dataset S4). These include upregulated couch potato, and downregulated CG7650 (encoding a phosducin-like protein) and Hsp70/Hsp90 organizing protein homolog. Transcriptional changes in Hsp70 and Hsp90 during diapause have been described for several insect species [10] and these molecular chaperons may be involved in increased resistance to stress. In our analysis of D. melanogaster four small heat shock proteins (Hsp22, 23, 26 and 27) were found downregulated (Additional file 3: Dataset S1). Three of these are normally enriched in the ovaries. In the Asian temperate Drosophila species D. triauraria Drosomycin and Drosomycin-like genes were found to be strongly upregulated during diapause, whereas other immune response genes like Drosocin and Defensin were not [150]. This is in line with our findings, that Drosomycin and Drosomycin-like genes are upregulated via the Toll-pathway, whereas the imd pathway is not influenced upon diapause. Another study in D. triauraria reveled allelic variations in timeless and cryptochrome genes, which influence the incidence of diapause [151]. This might be related to our findings that timeless isoforms are regulated differentially upon diapause in D. melanogaster.
More distantly in dipteran insects, we made a comparison with candidate genes of diapausing mosquitos, Culex pipiens, revealed from suppressive subtractive hybridization [152]. We identified 5 out of 40 genes that are in common with our study (Additional file 14: Dataset S4). These include the extracellular matrix component Multiplexin, CG34227 (encoding a secreted protein with unknown function), Mediator complex subunit 14 (encoding an RNA polymerase II complex subunit) and two heat shock proteins (Hsp23 and Hsp27), which are downregulated in diapausing mosquitos, but upregulated in diapausing fruitflies. A variable Heat shock protein expression (species specific or transcript specific), has been described previously for many insect species, and is known to vary among species with reproductive diapause, from flies to mosquitoes and beetles [10].
More general comparisons of KEGG or GO analyses performed on insects displaying different forms of diapause (including other developmental stages), and also the dormant dauer state in C. elegans reveal that similar signal pathways are affected, such as IIS, TOR, stress responses, carbohydrate and lipid metabolism [62, 153–156]. However, so far limited transcriptional similarities have been detected across species, and it has even been suggested that there may be many transcriptional strategies for producing similar dormancy phenotypes [62]. Possibly, the same genetic and physiological modules are involved across taxa, but the details of how they interconnect may differ, leading to unexpected variation in the expression of specific genes when extrapolating between species.
Conclusions
The drastically extended lifespan and diminished senescence associated with adult diapause is a likely to be caused by multiple factors that increase resistance to the deleterious effects of stress and aging. Indeed, we found that a substantial portion of the genes and signal pathways affected by D. melanogaster diapause is shared with the ones revealed in studies of aging and extended lifespan, and also include pathways regulating development, detoxification, as well as stress and immune responses [2, 4, 7, 135, 136].
Diapause induces a massive alteration of gene expression in D. melanogaster with more than 4500 differentially regulated genes. Gene Ontology, GSEA and KEGG analysis provide clues to processes that are affected by diapause. These include metabolic processes (energetic and storage metabolism), cell communication, developmental processes, neuronal function, reproduction, lysosome pathway, transcription, translation, protein processing, clock system, drug metabolism and detoxification. Our manual annotation of transcripts in signaling pathways reveals downregulation of IIS, TOR and MAPK signaling and upregulation of JNK and Toll immune signaling, as well as tissue specific upregulation of JAK/STAT signaling in hemocytes. All these pathways are likely involved in the diapause phenotype with reduced food intake, diminished metabolism, increased nutrient stores, arrested vitellogenesis, and increased stress resistance. Several of the pathways have also been implicated in extended lifespan and diminished senescence [2, 7, 32, 33, 135, 136]. Furthermore, we find that many genes whose expressions are affected in diapausing flies are shared with ones found in D. melanogaster exposed to restricted diet or cold [141, 144]. A portion of the transcript changes probably reflects the transition from reproduction to survival mode, characteristic of adult diapause. Thus, experimentally induced diapause is a suitable means to study regulation of processes underlying aging related phenomena.
Comparisons with other insects that display adult reproductive diapause such as D. montana and the mosquito Culex pipiens, where limited transcriptome analyses have been performed [149, 152], revealed some overlap, though restricted by the small number of transcripts assessed in these studies. However, general comparisons of transcript changes in components of signal pathways by KEGG or GO analyses suggest similarities between reproductive diapause in Drosophila and dormancy in other insects and C. elegans, including IIS and TOR signaling, stress responses and metabolism, as well as energy storage [62, 153–156]. This comparison can be extended to mammals, where the hibernation phenotype involves altered metabolism and TOR signaling [157, 158]. Taken together our data provide an organism-wide and detailed analysis of transcriptional changes induced by diapause, and shows the power of D. melanogaster as a model to analyze genetics of dormancy and its effects on lifespan and aging.
Methods
Drosophila strains and diapause induction
Drosophila melanogaster of the Canton S strain, obtained from the Bloomington Drosophila Stock Center (BDSC, Bloomington, IN) was used in the microarray analysis. For analysis of transcriptional regulation of immune response genes during diapause we also tested RelE20 [159] and spzrm7 [160] mutants kindly provided by Bruno Lemaitre (Lausanne, Switzerland). Furthermore, for monitoring activity in the JAK/STAT pathway we used w1118; 10xStat92E-GFP [76] reporter flies from BDSC. For diapause induction we followed the protocol described in our previous study [20].
RNA isolation and sampling
As our experimental group for microarray analysis we used Canton S female virgin flies kept in diapause conditions (11 °C, short photoperiod 10 L:14D) for 3 weeks (3wD). Flies were placed under diapause conditions 4-6 h after adult eclosion. Control virgin female flies (1wN) were kept for either 1 week in normal conditions (25 °C, 12 L:12D), or as a sibling control we used flies kept for 3 weeks in normal conditions (3wN). All flies were fed standard Drosophila food. Total RNA from whole flies was extracted using Trizol reagent (Invitrogen) according to manufacturer’s protocol and subsequently cleaned with NucleoSpin RNA II kit (Macherey Nagel). 15 flies were used for each biological replicate. Quality and concentration of the RNA were measured with a NanoDrop 2000 spectrophotometer (Thermo Scientific). RNA integrity was analyzed in an Agilent 2100 Bioanalyzer. We included only samples with an intact RNA profile. The same RNA isolation procedure was also used for preparation of samples for quantitative real-time PCR (qPCR).
Expression profiling
The Affymetrix GeneChip® Drosophila Genome 2.0 Array System was used for microarray analysis following the standard protocol [100 ng RNA was amplified with GeneChip 3 ‘ IVT Express Kit (Affymetrix) and 10 μg of labeled cRNA was hybridized to the chip according to the manufacturer’s instructions].
Statistical analysis of array data
Analysis was performed in four replicates (except for 1wN controls where 3 replicates were used). Data were preprocessed in Partek Genomic Suit (Partek). The transcription profiles were background corrected using the GCRMA method, quantile normalized and variance stabilized using base-2 logarithmic transformation. Analysis of variance yielded transcripts differentially expressed between analyzed samples (within LIMMA [161]); Storey’s q values [162] were used to select significantly differentially transcribed genes, q < 0.05. The transcription data are MIAME compliant and deposited in the ArrayExpress database (accession E-MTAB-3546).
Statistical analyses were performed in R (http://www.Rproject.org) and within Bioconductor [163] with additional database searches in AmiGO 2 [164]. Functional classification of Gene Ontology (GO) terms [165] was performed in PANTHER [166] with default settings. Differentially expressed genes were selected for gene set enrichment analysis (GSEA). We performed GSEA on genes that mapped to KEGG pathways [167] using the Fisher test and the approach of Tian et al. [168]. For GSEA genes with q < 0.05 and |log FC| ≥ 0.4 were considered differentially expressed. To identify significantly perturbed pathways, we performed SPIA analysis [34] on KEGG pathways.
Quantitative Real-Time PCR analysis
Quantitative Real-Time PCR (qPCR) experiments were performed on aliquots of RNA extractions performed for microarray tests (see above) as controls of gene expression. Life Technologies SuperScript III Reverse Transcriptase (Life Technologies) was used for cDNA synthesis. Primers were designed using Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/), except for two cases, where previously designed primers were used: for Actin 88 F [20], and for primers targeting all timeless isoforms except N and O [169]. Primers are shown in Additional file 15: Table S5. Levels of mRNA were estimated using SensiFAST SYBR Hi-ROX (Bioline Reagents, London, UK) on a StepOnePlus Real-Time PCR System (Applied Biosystems/Thermo Fisher Scientific). Samples were compared using the ΔΔCt method, with Actin 88 F as reference gene, using the StepOne software v 2.3 (Applied Biosystems). For measurement of transcripts of Toll response genes we followed the protocol for immune genes described in our previous study [20]. We used TaqMan Gene Expression Assays (Applied Biosystems) for following genes: Drosomycin (Dm01822006_s1), Drosomycin-like 5 (Dm02332286_g1) and internal control gene RpL32 (Dm02151827_g1). Statistical analysis of data was performed in Prism 6 (Graphpad Software Inc, San Diego, CA).
Microscopy and 10xStat92E-GFP intensity measurements
A Leica MZ FLIII fluorescence stereomicroscope associated with a Panasonic DMC-G2 camera was used to visualize whole flies anesthetized with CO2. Confocal images of ovaries were produced in a Zeiss LSM 780 microscope. The following dyes were used: DAPI (1:1000 dilution, Sigma-Aldrich), and Phalloidin-TRITC (1:1000 dilution, Sigma-Aldrich). The samples were mounted in Fluoromount-G (SouthernBiotech). To retrieve the hemocytes, flies were anesthetized, preinjected with small amount of Schneider’s medium (Sigma-Aldrich) containing the anti-coagulant phenylthiourea (PTU) and incubated on ice for 5 min. Thereafter cells were collected on a glass slide with 25 μl of Schneider’s medium with PTU and incubated in a wet chamber for 30 min to allow them to adhere. For fixation 4 % paraformaldehyde in PBS was used for 10 min. After staining with DAPI (1:1000 dilution, Sigma-Aldrich), samples were mounted in Fluoromount-G (SouthernBiotech) and observed under a Zeiss Axioplan 2 microscope coupled to a Hamamatsu ORCA-ER camera (C4742-95). The ImageJ program was used to measure the intensity of GFP. Fluorescence values were recalculated to Corrected total fluorescence (CTF) according to [170].
Availability of supporting data
Gene microarray data have been deposited at ArrayExpress, accession number E-MTAB-3546.
Acknowledgements
We thank Dr Bruno Lemaitre (Lausanne, Switzerland) and the Bloomington Drosophila Stock Center (BDSC, Bloomington, IN) for providing fly strains. The authors are grateful to the service laboratory at IMG, and especially to Martina Chmelikova for technical assistance. We also gratefully acknowledge the help of Anna Šenovská with immune genes experiments. Stina Höglund and the Imaging Facility at Stockholm University (IFSU) are acknowledged for maintenance of the confocal microscopes.
Additional files
Heat map of the top 100 most variable genes in our microarray study. Columns depicted: 3wD (1_3wD-4_3wD) = samples from flies kept for 3 weeks in diapause conditions (11 °C, short photoperiod 10 L:14D); 1wN (1_1wN-3_1wN) = control flies kept in normal conditions (25 °C, 12 L:12D) for 1 week, and 3wN (1-3wN-4_3wN) = sibling controls, 3 week old flies kept in normal conditions (25 °C, 12 L:12D). Each column represents an independent sample. The color key represents the level of regulation (red is up- and blue downregulation). Dark intensities indicate the most up- and down- regulated genes, respectively. The largest subset of most variable genes is upregulated in diapause (3wD) samples and downregulated in 1wN and 3wN controls. However there is also a smaller number of genes, which are downregulated in diapause, clearly upregulated in 3wN control, but less upregulated in 1wN controls. (TIF 1817 kb)
Principal component analysis (PCA) plot. The Y-axis represent the biggest variability in our samples. Our samples fall into two groups. One represents all our samples from diapause conditions (3wD, above line), the second all our control samples (1wN and 3wN under line). Whereas the PCA clearly grouped all 3wD samples (orange subset), it failed to distinguish between 1wN control samples (green subset) and 3wN sibling control (blue subset). (TIF 553 kb)
Gene list of significantly regulated probes and reciprocal comparison. Transcripts that are significantly altered more than two-fold (|log2FC| ≥ 1, q ≤ 0.05) are shown. 3wDvs1wN represent transcripts regulated upon 3 weeks of diapause compared to 1 week control flies, 3wDvs3wN represent genes regulated in diapause samples with comparison to 3 week old sibling controls, 3wNvs1wN represent genes influenced by aging in 3 week old control flies compared to 1 week old control flies. Significantly inhibited transcripts are highlighted in blue, significantly activated in orange. Links to Flybase, Wikigene, Genecard, NCBI, ENSEMBL and ExPASy databases are included. (XLSX 2131 kb)
GSEA of KEGG pathway analysis. (A) The set of KEGG pathways enriched for upregulated genes encompass mostly carbohydrate metabolic pathways, drug metabolism and circadian rhythm pathway. (B) The set of KEGG pathways enriched for downregulated genes covers mostly RNA and DNA metabolism and protein degradation. Genes with |log2FC| ≥ 0.04 and q ≤ 0.05 were considered significantly regulated. Pathways highlighted in italics were defined only on the basis of experiments performed in mammalian systems. (PDF 45 kb)
SPIA of KEGG pathways. Significantly inhibited pathways are highlighted in blue and significantly activated ones in orange. Genes with |log2FC| ≥ 0.1 and q ≤ 0.05 were considered significantly regulated. Pathways highlighted in italics were defined only on the basis of experiments performed in mammalian systems. (PDF 42 kb)
Detailed data on genes in seven signal pathways. Description of genes shown in Figs. 4, 5, 7, 9 and Additional file 8: Figure S4 and discussed in the corresponding sections of the paper. References are also given for these genes. Note that the genes are listed as they are shown in the figures (from top to bottom in the pathways). (PDF 328 kb)
Read-outs from AKH-IIS-TOR pathways. Summary of likely effects of altered signaling in these pathways, based on transcript changes in read-out genes. Based on Fig. 4. (TIF 3492 kb)
Altered gene expression in the MAPK signaling pathway during diapause. This scheme displays a generalized assembly (regardless tissue specificity) of relevant genes in the MAPK signal pathway. Transcript levels (logarithmic fold change, LogFC) are given in red for upregulated, blue for downregulated and black for no significant change (ns; LogFC close to 0). The acronyms are listed in Additional file 6: Table S3 where also references and details of gene/protein functions are given. Decreased expression of all MAP kinases (Ras85D, phl, Dsor1, rl) and most positive regulators support a general downregulation of the whole MAPK signaling cascade in diapausing flies. Only one read-out gene was downregulated (dm, diminutive a Drosophila Myc). (TIF 1437 kb)
Genes affected by 3 weeks diapause compared to ones implicated in aging and increased longevity. Microarray study of aging response genes [136], longevity mapping study [33], transcriptome changes in mated and non-mated aging females [7] and candidate study for lifespan genes [135]. Significantly inhibited transcripts are highlighted in blue, significantly activated in orange. (XLSX 77 kb)
Results of overrepresentation test of GO biological process terms for comparisons between our genome-wide transcription study of D. melanogaster dormancy and aging studies. 3wD, three weeks diapause (whole animals) our study; Aging, aging response genes [136] and candidate lifespan extending genes [135]. Comparison based on genes in the Venn diagram in Fig. 10a. Term enrichment analysis of Groups A – D are shown separately. (PDF 405 kb)
Term enrichment analysis of GO biological process terms for comparisons among four genome-wide transcription studies of D. melanogaster. 3wD, three weeks diapause (whole animals) our study; 1wD, abdomens sampled after one week diapause [140]; cold intolerance [141] and starvation [143]. Comparison based on genes in the Venn diagram in Fig. 10b. Term analysis of Groups A – H are shown separately. (PDF 1267 kb)
List of genes shared between diapause and response to restricted diet in D. melanogaster. We compared our study of three weeks diapause (3wD) to that of an analysis of effects of restricted diet (RF) [145]. Significantly inhibited transcripts are highlighted in blue, significantly activated in orange (for the RF study, genes with q ≤ 0.05 were considered significantly regulated, as was used in original study). Links to Flybase and Genecard databases are included. The Venn diagram summarizes the numbers of significantly regulated genes and how they overlap. (XLSX 173 kb)
Gene set enrichment analysis (GSEA) of GO terms comparing diapause and response to restricted diet in D. melanogaster. We compare our study of three weeks diapause (3wD) to that of an analysis of effects of restricted diet (RF) [145]. (A) GSEA for upregulated transcripts identified mostly terms connected to cell cycle and gene expression. (B) Transcripts mostly from carbohydrate metabolism are regulated in opposite way in 3wD and RF. (C) For downregulated transcripts no significant enriched GO terms (ns) were identified. (XLSX 11 kb)
Comparisons of diapause response genes from our study of D. melanogaster with other transcription studies on reproductive diapause. We compared our study to a D. montana candidate gene microarray study [149] and Culex pipiens diapause response genes [152]. Significantly inhibited transcripts are highlighted in blue, significantly activated in orange. Links to Flybase are included at genes shared with D. melanogaster. (XLSX 15 kb)
Primers used for quantitative real-time PCR. (PDF 55 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LK, OIK, UT and DRN designed the study. LK, OIK, JB, and HS performed the experiments. LK, OIK, JB, HS, SN, UT and DRN analyzed the data. LK, OIK, and DRN wrote the manuscript. DRN supervised the study. All authors read, edited and approved the final manuscript. SN obtained funding.
Contributor Information
Lucie Kučerová, Email: lucie.kucerova@su.se.
Olga I. Kubrak, Email: olga.kubrak@zoologi.su.se
Jonas M. Bengtsson, Email: jonas.bengtsson@zoologi.su.se
Hynek Strnad, Email: strnad@img.cas.cz.
Sören Nylin, Email: soren.nylin@zoologi.su.se.
Ulrich Theopold, Email: uli.theopold@su.se.
Dick R. Nässel, Phone: +46-8-164077, Email: dnassel@zoologi.su.se
References
- 1.Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408(6809):255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- 2.Pletcher SD, Macdonald SJ, Marguerie R, Certa U, Stearns SC, Goldstein DB, et al. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr Biol. 2002;12(9):712–723. doi: 10.1016/S0960-9822(02)00808-4. [DOI] [PubMed] [Google Scholar]
- 3.Partridge L, Alic N, Bjedov I, Piper MD. Ageing in Drosophila: the role of the insulin/Igf and TOR signalling network. Exp Gerontol. 2011;46(5):376–381. doi: 10.1016/j.exger.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou S, Meadows S, Sharp L, Jan LY, Jan YN. Genome-wide study of aging and oxidative stress response in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000;97(25):13726–13731. doi: 10.1073/pnas.260496697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin CS, Jan YN, et al. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat Genet. 2004;36(2):197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- 6.Lapierre LR, Hansen M. Lessons from C. elegans: signaling pathways for longevity. Trends Endocrinol Metabol. 2012;23(12):637–644. doi: 10.1016/j.tem.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou S, Mackay T, Anholt RR. Transcriptional and epigenetic responses to mating and aging in Drosophila melanogaster. BMC Genomics. 2014;15:927. doi: 10.1186/1471-2164-15-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hahn DA, Denlinger DL. Energetics of insect diapause. Ann Rev Entomol. 2011;56:103–121. doi: 10.1146/annurev-ento-112408-085436. [DOI] [PubMed] [Google Scholar]
- 9.Tatar M, Yin C. Slow aging during insect reproductive diapause: why butterflies, grasshoppers and flies are like worms. Exp Gerontol. 2001;36(4–6):723–738. doi: 10.1016/S0531-5565(00)00238-2. [DOI] [PubMed] [Google Scholar]
- 10.MacRae TH. Gene expression, metabolic regulation and stress tolerance during diapause. Cell Molec Life Sci. 2010;67(14):2405–2424. doi: 10.1007/s00018-010-0311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tauber MJ, Tauber CA, Masaki S. Seasonal Adaptations of Insects. New York: Oxford Univ Press; 1986. [Google Scholar]
- 12.Denlinger DL. Regulation of diapause. Ann Rev Entomol. 2002;47:93–122. doi: 10.1146/annurev.ento.47.091201.145137. [DOI] [PubMed] [Google Scholar]
- 13.Emerson KJ, Bradshaw WE, Holzapfel CM. Complications of complexity: integrating environmental, genetic and hormonal control of insect diapause. Trends Genet. 2009;25(5):217–225. doi: 10.1016/j.tig.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Flatt T, Amdam GV, Kirkwood TB, Omholt SW. Life-history evolution and the polyphenic regulation of somatic maintenance and survival. Q Rev Biol. 2013;88(3):185–218. doi: 10.1086/671484. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt PS, Paaby AB, Heschel MS. Genetic variance for diapause expression and associated life histories in Drosophila melanogaster. Evolution. 2005;59(12):2616–2625. doi: 10.1111/j.0014-3820.2005.tb00974.x. [DOI] [PubMed] [Google Scholar]
- 16.Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299(5611):1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- 17.Saunders DS. The circadian basis of ovarian diapause regulation in Drosophila melanogaster: is the period gene causally involved in photoperiodic time measurement? J Biol Rhythms. 1990;5(4):315–331. doi: 10.1177/074873049000500404. [DOI] [PubMed] [Google Scholar]
- 18.Saunders DS, Henrich VC, Gilbert LI. Induction of diapause in Drosophila melanogaster: photoperiodic regulation and the impact of arrhythmic clock mutations on time measurement. Proc Natl Acad Sci USA. 1989;86(10):3748–3752. doi: 10.1073/pnas.86.10.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saunders DS, Richard DS, Applebaum SW, Ma M, Gilbert LI. Photoperiodic diapause in Drosophila melanogaster involves a block to the juvenile hormone regulation of ovarian maturation. Gen Comp Endocrinol. 1990;79(2):174–184. doi: 10.1016/0016-6480(90)90102-R. [DOI] [PubMed] [Google Scholar]
- 20.Kubrak OI, Kucerova L, Theopold U, Nässel DR. The sleeping beauty: How reproductive diapause affects hormone signaling, metabolism, immune response and somatic maintenance in drosophila melanogaster. PLoS ONE. 2014;9(11) doi: 10.1371/journal.pone.0113051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sim C, Denlinger DL. Insulin signaling and FOXO regulate the overwintering diapause of the mosquito Culex pipiens. Proc Natl Acad Sci USA. 2008;105(18):6777–6781. doi: 10.1073/pnas.0802067105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sim C, Denlinger DL. A shut-down in expression of an insulin-like peptide, ILP-1, halts ovarian maturation during the overwintering diapause of the mosquito Culex pipiens. Insect Molec Biol. 2009;18(3):325–332. doi: 10.1111/j.1365-2583.2009.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sim C, Denlinger DL. Insulin signaling and the regulation of insect diapause. Front Physiol. 2013;4:189. doi: 10.3389/fphys.2013.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams KD, Busto M, Suster ML, So AK, Ben-Shahar Y, Leevers SJ, et al. Natural variation in Drosophila melanogaster diapause due to the insulin-regulated PI3-kinase. Proc Natl Acad Sci USA. 2006;103(43):15911–15915. doi: 10.1073/pnas.0604592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen MJ. What makes a fly enter diapause? Fly. 2007;1(6):307–310. doi: 10.4161/fly.5532. [DOI] [PubMed] [Google Scholar]
- 26.Fabian DK, Kapun M, Nolte V, Kofler R, Schmidt PS, Schlotterer C, et al. Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol Ecol. 2012;21(19):4748–4769. doi: 10.1111/j.1365-294X.2012.05731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schiesari L, Kyriacou CP, Costa R. The hormonal and circadian basis for insect photoperiodic timing. FEBS Lett. 2011;585(10):1450–1460. doi: 10.1016/j.febslet.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 28.Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A. 2005;102(8):3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antonova Y, Arik AJ, Moore W, Riehle MR, Brown MR. Insulin-like peptides: Structure, Signaling, and Function. In: Gilbert LI, editor. Insect Endocrinology. New York: Elsevier/Academic Press; 2012. pp. 63–92. [Google Scholar]
- 30.Broughton SJ, Slack C, Alic N, Metaxakis A, Bass TM, Driege Y, et al. DILP-producing median neurosecretory cells in the Drosophila brain mediate the response of lifespan to nutrition. Aging Cell. 2010;9(3):336–346. doi: 10.1111/j.1474-9726.2010.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giannakou ME, Partridge L. Role of insulin-like signalling in Drosophila lifespan. Trends Biochem Sci. 2007;32(4):180–188. doi: 10.1016/j.tibs.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 32.Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328(5976):321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivanov DK, Escott-Price V, Ziehm M, Magwire MM, Mackay TF, Partridge L, et al. Longevity GWAS Using the Drosophila Genetic Reference Panel. J Gerontol A Biol Sci Med Sci. 2015;70(12):1470–8. doi: 10.1093/gerona/glv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tarca AL, Draghici S, Khatri P, Hassan SS, Mittal P, Kim JS, et al. A novel signaling pathway impact analysis. Bioinformatics. 2009;25(1):75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teleman AA. Molecular mechanisms of metabolic regulation by insulin in Drosophila. Biochem J. 2010;425(1):13–26. doi: 10.1042/BJ20091181. [DOI] [PubMed] [Google Scholar]
- 36.Grewal SS. Insulin/TOR signaling in growth and homeostasis: a view from the fly world. Int J Biochem Cell Biol. 2009;41(5):1006–1010. doi: 10.1016/j.biocel.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 37.Garofalo RS. Genetic analysis of insulin signaling in Drosophila. Trends Endocrinol Metab. 2002;13(4):156–162. doi: 10.1016/S1043-2760(01)00548-3. [DOI] [PubMed] [Google Scholar]
- 38.Owusu-Ansah E, Perrimon N. Modeling metabolic homeostasis and nutrient sensing in Drosophila: implications for aging and metabolic diseases. Dis Model Mech. 2014;7(3):343–350. doi: 10.1242/dmm.012989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ragland GJ, Egan SP, Feder JL, Berlocher SH, Hahn DA. Developmental trajectories of gene expression reveal candidates for diapause termination: a key life-history transition in the apple maggot fly Rhagoletis pomonella. J Exp Biol. 2011;214(Pt 23):3948–3959. doi: 10.1242/jeb.061085. [DOI] [PubMed] [Google Scholar]
- 40.Garelli A, Gontijo AM, Miguela V, Caparros E, Dominguez M. Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science. 2012;336(6081):579–582. doi: 10.1126/science.1216735. [DOI] [PubMed] [Google Scholar]
- 41.Colombani J, Andersen DS, Leopold P. Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science. 2012;336(6081):582–585. doi: 10.1126/science.1216689. [DOI] [PubMed] [Google Scholar]
- 42.Werz C, Kohler K, Hafen E, Stocker H. The Drosophila SH2B family adaptor Lnk acts in parallel to chico in the insulin signaling pathway. PLoS genetics. 2009;5(8) doi: 10.1371/journal.pgen.1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuss B, Becker T, Zinke I, Hoch M. The cytohesin Steppke is essential for insulin signalling in Drosophila. Nature. 2006;444(7121):945–948. doi: 10.1038/nature05412. [DOI] [PubMed] [Google Scholar]
- 44.Wang B, Moya N, Niessen S, Hoover H, Mihaylova MM, Shaw RJ, et al. A hormone-dependent module regulating energy balance. Cell. 2011;145(4):596–606. doi: 10.1016/j.cell.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie Q, Chen J, Yuan Z. Post-translational regulation of FOXO. Acta Biochim Biophys Sin (Shanghai) 2012;44(11):897–901. doi: 10.1093/abbs/gms067. [DOI] [PubMed] [Google Scholar]
- 46.Grönke S, Mildner A, Fellert S, Tennagels N, Petry S, Muller G, et al. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metabol. 2005;1(5):323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Okamura T, Shimizu H, Nagao T, Ueda R, Ishii S. ATF-2 regulates fat metabolism in Drosophila. Mol Biol Cell. 2007;18(4):1519–1529. doi: 10.1091/mbc.E06-10-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emerson KJ, Bradshaw WE, Holzapfel CM. Microarrays reveal early transcriptional events during the termination of larval diapause in natural populations of the mosquito, Wyeomyia smithii. PLoS ONE. 2010;5(3) doi: 10.1371/journal.pone.0009574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Furuyama T, Kitayama K, Yamashita H, Mori N. Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. J Biochem. 2003;375(Pt 2):365–371. doi: 10.1042/bj20030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tettweiler G, Miron M, Jenkins M, Sonenberg N, Lasko PF. Starvation and oxidative stress resistance in Drosophila are mediated through the eIF4E-binding protein, d4E-BP. Gene Dev. 2005;19(16):1840–1843. doi: 10.1101/gad.1311805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frame S, Cohen P, Biondi RM. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Molecular cell. 2001;7(6):1321–1327. doi: 10.1016/S1097-2765(01)00253-2. [DOI] [PubMed] [Google Scholar]
- 52.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120(4):449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 53.Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292(5514):107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 54.Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292(5514):104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- 55.Luong N, Davies CR, Wessells RJ, Graham SM, King MT, Veech R, et al. Activated FOXO-mediated insulin resistance is blocked by reduction of TOR activity. Cell Metabol. 2006;4(2):133–142. doi: 10.1016/j.cmet.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 56.Oldham S, Hafen E. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 2003;13(2):79–85. doi: 10.1016/S0962-8924(02)00042-9. [DOI] [PubMed] [Google Scholar]
- 57.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 58.Grewal SS, Evans JR, Edgar BA. Drosophila TIF-IA is required for ribosome synthesis and cell growth and is regulated by the TOR pathway. J Cell Biol. 2007;179(6):1105–1113. doi: 10.1083/jcb.200709044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang YY, Neufeld TP. Autophagy takes flight in Drosophila. FEBS lett. 2010;584(7):1342–1349. doi: 10.1016/j.febslet.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7(2):167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 61.Dekanty A, Lavista-Llanos S, Irisarri M, Oldham S, Wappner P. The insulin-PI3K/TOR pathway induces a HIF-dependent transcriptional response in Drosophila by promoting nuclear localization of HIF-alpha/Sima. J Cell Sci. 2005;118(Pt 23):5431–5441. doi: 10.1242/jcs.02648. [DOI] [PubMed] [Google Scholar]
- 62.Ragland GJ, Denlinger DL, Hahn DA. Mechanisms of suspended animation are revealed by transcript profiling of diapause in the flesh fly. Proc Natl Acad Sci U S A. 2010;107(33):14909–14914. doi: 10.1073/pnas.1007075107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giordano E, Peluso I, Senger S, Furia M. minifly, a Drosophila gene required for ribosome biogenesis. J Cell Biol. 1999;144(6):1123–1133. doi: 10.1083/jcb.144.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114(6):739–749. doi: 10.1016/S0092-8674(03)00713-X. [DOI] [PubMed] [Google Scholar]
- 65.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. 2006;7(2):85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 66.Lee G, Park JH. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167(1):311–323. doi: 10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim SK, Rulifson EJ. Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature. 2004;431(7006):316–320. doi: 10.1038/nature02897. [DOI] [PubMed] [Google Scholar]
- 68.Bednarova A, Kodrik D, Krishnan N. Unique roles of glucagon and glucagon-like peptides: Parallels in understanding the functions of adipokinetic hormones in stress responses in insects. Comp Biochem Physiol A Mol Integr Physiol. 2013;164(1):91–100. doi: 10.1016/j.cbpa.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 69.Bharucha KN, Tarr P, Zipursky SL. A glucagon-like endocrine pathway in Drosophila modulates both lipid and carbohydrate homeostasis. J Exp Biol. 2008;211(Pt 19):3103–3110. doi: 10.1242/jeb.016451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baumbach J, Xu Y, Hehlert P, Kuhnlein RP. Galphaq, Ggamma1 and Plc21C control Drosophila body fat storage. J Genet Genomics. 2014;41(5):283–292. doi: 10.1016/j.jgg.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 71.Waterson MJ, Chung BY, Harvanek ZM, Ostojic I, Alcedo J, Pletcher SD. Water sensor pp k28 modulates Drosophila lifespan and physiology through AKH signaling. Proc Natl Acad Sci U S A. 2014;111(22):8137–8142. doi: 10.1073/pnas.1315461111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wicher D, Agricola HJ, Sohler S, Gundel M, Heinemann SH, Wollweber L, et al. Differential receptor activation by cockroach adipokinetic hormones produces differential effects on ion currents, neuronal activity, and locomotion. J Neurophysiol. 2006;95(4):2314–2325. doi: 10.1152/jn.01007.2005. [DOI] [PubMed] [Google Scholar]
- 73.Hauser F, Grimmelikhuijzen CJ. Evolution of the AKH/corazonin/ACP/GnRH receptor superfamily and their ligands in the Protostomia. Gen Comp Endocrinol. 2014;209C:35–49. doi: 10.1016/j.ygcen.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 74.Bi J, Xiang Y, Chen H, Liu Z, Gronke S, Kuhnlein RP, et al. Opposite and redundant roles of the two Drosophila perilipins in lipid mobilization. J Cell Sci. 2012;125(Pt 15):3568–3577. doi: 10.1242/jcs.101329. [DOI] [PubMed] [Google Scholar]
- 75.Buch S, Melcher C, Bauer M, Katzenberger J, Pankratz MJ. Opposing effects of dietary protein and sugar regulate a transcriptional target of Drosophila insulin-like peptide signaling. Cell Metab. 2008;7(4):321–332. doi: 10.1016/j.cmet.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 76.Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, et al. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns. 2007;7(3):323–331. doi: 10.1016/j.modgep.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 77.Luo H, Dearolf CR. The JAK/STAT pathway and Drosophila development. Bioessays. 2001;23(12):1138–1147. doi: 10.1002/bies.10016. [DOI] [PubMed] [Google Scholar]
- 78.Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294(5551):2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- 79.Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294(5551):2546–2549. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- 80.Myllymäki H, Rämet M. Jak/Stat Pathway in Drosophila Immunity. Scand J Immunol. 2014;79(6):377–385. doi: 10.1111/sji.12170. [DOI] [PubMed] [Google Scholar]
- 81.Agaisse H, Perrimon N. The roles of JAK/STAT signaling in Drosophila immune responses. Immunol Rev. 2004;198:72–82. doi: 10.1111/j.0105-2896.2004.0133.x. [DOI] [PubMed] [Google Scholar]
- 82.Arbouzova NI, Zeidler MP. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development. 2006;133(14):2605–2616. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- 83.Agaisse H, Petersen UM, Boutros M, Mathey-Prevot B, Perrimon N. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev Cell. 2003;5(3):441–450. doi: 10.1016/S1534-5807(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 84.Ekengren S, Tryselius Y, Dushay MS, Liu G, Steiner H, Hultmark D. A humoral stress response in Drosophila (vol 11, pg 714, 2001) Curr Biol. 2001;11(18):1479. doi: 10.1016/S0960-9822(01)00452-3. [DOI] [PubMed] [Google Scholar]
- 85.Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, et al. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol. 2005;6(9):946–953. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- 86.Lagueux M, Perrodou E, Levashina EA, Capovilla M, Hoffmann JA. Constitutive expression of a complement-like protein in toll and JAK gain-of-function mutants of Drosophila. Proc Natl Acad Sci U S A. 2000;97(21):11427–11432. doi: 10.1073/pnas.97.21.11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bou Aoun R, Hetru C, Troxler L, Doucet D, Ferrandon D, Matt N. Analysis of thioester-containing proteins during the innate immune response of Drosophila melanogaster. Journal of innate immunity. 2011;3(1):52–64. doi: 10.1159/000321554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ghiglione C, Devergne O, Georgenthum E, Carballes F, Medoni C, Cerezo D, et al. The Drosophila cytokine receptor Domeless controls border cell migration and epithelial polarization during oogenesis. Development. 2002;129(23):5437–5447. doi: 10.1242/dev.00116. [DOI] [PubMed] [Google Scholar]
- 89.Baeg GH, Zhou R, Perrimon N. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev. 2005;19(16):1861–1870. doi: 10.1101/gad.1320705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Karsten P, Hader S, Zeidler MP. Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech Dev. 2002;117(1–2):343–346. doi: 10.1016/S0925-4773(02)00216-2. [DOI] [PubMed] [Google Scholar]
- 91.Wang HB, Chen X, He T, Zhou YN, Luo H. Evidence for Tissue-Specific JAK/STAT Target Genes in Drosophila Optic Lobe Development. Genetics. 2013;195(4):1291−+. doi: 10.1534/genetics.113.155945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lanot R, Zachary D, Holder F, Meister M. Postembryonic hematopoiesis in Drosophila. Dev Biol. 2001;230(2):243–257. doi: 10.1006/dbio.2000.0123. [DOI] [PubMed] [Google Scholar]
- 93.Baksa K, Parke T, Dobens LL, Dearolf CR. The Drosophila STAT protein, stat92E, regulates follicle cell differentiation during oogenesis. Dev Biol. 2002;243(1):166–175. doi: 10.1006/dbio.2001.0539. [DOI] [PubMed] [Google Scholar]
- 94.McGregor JR, Xi R, Harrison DA. JAK signaling is somatically required for follicle cell differentiation in Drosophila. Development. 2002;129(3):705–717. doi: 10.1242/dev.129.3.705. [DOI] [PubMed] [Google Scholar]
- 95.Silver DL, Montell DJ. Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell. 2001;107(7):831–841. doi: 10.1016/S0092-8674(01)00607-9. [DOI] [PubMed] [Google Scholar]
- 96.Borensztejn A, Boissoneau E, Fernandez G, Agnes F, Pret AM. JAK/STAT autocontrol of ligand-producing cell number through apoptosis. Development. 2013;140(1):195–204. doi: 10.1242/dev.079046. [DOI] [PubMed] [Google Scholar]
- 97.Valanne S, Wang JH, Ramet M. The Drosophila Toll signaling pathway. J Immunol. 2011;186(2):649–656. doi: 10.4049/jimmunol.1002302. [DOI] [PubMed] [Google Scholar]
- 98.De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002;21(11):2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ming M, Obata F, Kuranaga E, Miura M. Persephone/Spätzle pathogen sensors mediate the activation of Toll receptor signaling in response to endogenous danger signals in apoptosis-deficient Drosophila. J Biol Chem. 2014;289(11):7558–7568. doi: 10.1074/jbc.M113.543884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ashton-Beaucage D, Udell CM, Gendron P, Sahmi M, Lefrancois M, Baril C, et al. A functional screen reveals an extensive layer of transcriptional and splicing control underlying RAS/MAPK signaling in Drosophila. PLoS Biol. 2014;12(3) doi: 10.1371/journal.pbio.1001809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26(22):3113–3121. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- 102.Kohyama-Koganeya A, Kim YJ, Miura M, Hirabayashi Y. A Drosophila orphan G protein-coupled receptor BOSS functions as a glucose-responding receptor: loss of boss causes abnormal energy metabolism. Proc Natl Acad Sci U S A. 2008;105(40):15328–15333. doi: 10.1073/pnas.0807833105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fujiwara Y, Denlinger DL. High temperature and hexane break pupal diapause in the flesh fly, Sarcophaga crassipalpis, by activating ERK/MAPK. J Insect Physiol. 2007;53(12):1276–1282. doi: 10.1016/j.jinsphys.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 104.Kidokoro K, Iwata K, Takeda M, Fujiwara Y. Involvement of ERK/MAPK in regulation of diapause intensity in the false melon beetle, Atrachya menetriesi. J Insect Physiol. 2006;52(11–12):1189–1193. doi: 10.1016/j.jinsphys.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 105.Fujiwara Y, Shindome C, Takeda M, Shiomi K. The roles of ERK and P38 MAPK signaling cascades on embryonic diapause initiation and termination of the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2006;36(1):47–53. doi: 10.1016/j.ibmb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 106.Biteau B, Karpac J, Hwangbo D, Jasper H. Regulation of Drosophila lifespan by JNK signaling. Exp Gerontol. 2011;46(5):349–354. doi: 10.1016/j.exger.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rios-Barrera LD, Riesgo-Escovar JR. Regulating cell morphogenesis: the Drosophila Jun N-terminal kinase pathway. Genesis. 2013;51(3):147–162. doi: 10.1002/dvg.22354. [DOI] [PubMed] [Google Scholar]
- 108.Karpac J, Hull-Thompson J, Falleur M, Jasper H. JNK signaling in insulin-producing cells is required for adaptive responses to stress in Drosophila. Aging Cell. 2009;8(3):288–295. doi: 10.1111/j.1474-9726.2009.00476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Igaki T, Kanda H, Yamamoto-Goto Y, Kanuka H, Kuranaga E, Aigaki T, et al. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J. 2002;21(12):3009–3018. doi: 10.1093/emboj/cdf306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kauppila S, Maaty WS, Chen P, Tomar RS, Eby MT, Chapo J, et al. Eiger and its receptor, Wengen, comprise a TNF-like system in Drosophila. Oncogene. 2003;22(31):4860–4867. doi: 10.1038/sj.onc.1206715. [DOI] [PubMed] [Google Scholar]
- 111.Kolaczkowski B, Kern AD, Holloway AK, Begun DJ. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics. 2011;187(1):245–260. doi: 10.1534/genetics.110.123059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang MC, Bohmann D, Jasper H. JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Dev Cell. 2003;5(5):811–816. doi: 10.1016/S1534-5807(03)00323-X. [DOI] [PubMed] [Google Scholar]
- 113.Wang MC, Bohmann D, Jasper H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121(1):115–125. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 114.Taghert PH, Nitabach MN. Peptide neuromodulation in invertebrate model systems. Neuron. 2012;76(1):82–97. doi: 10.1016/j.neuron.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nässel DR, Winther ÅM. Drosophila neuropeptides in regulation of physiology and behavior. Progr Neurobiol. 2010;92(1):42–104. doi: 10.1016/j.pneurobio.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 116.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39(6):715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 117.Contrino S, Smith RN, Butano D, Carr A, Hu F, Lyne R, et al. modMine: flexible access to modENCODE data. Nucleic Acids Res. 2012;40(Database issue):D1082–1088. doi: 10.1093/nar/gkr921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dow JA. Insights into the Malpighian tubule from functional genomics. J Exp Biol. 2009;212(Pt 3):435–445. doi: 10.1242/jeb.024224. [DOI] [PubMed] [Google Scholar]
- 119.Paluzzi JP, Vanderveken M, O'Donnell MJ. The heterodimeric glycoprotein hormone, GPA2/GPB5, regulates ion transport across the hindgut of the adult mosquito, Aedes aegypti. PLoS ONE. 2014;9(1) doi: 10.1371/journal.pone.0086386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hergarden AC, Tayler TD, Anderson DJ. Allatostatin-A neurons inhibit feeding behavior in adult Drosophila. Proc Natl Acad Sci U S A. 2012;109(10):3967–3972. doi: 10.1073/pnas.1200778109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhao Y, Bretz CA, Hawksworth SA, Hirsh J, Johnson EC. Corazonin neurons function in sexually dimorphic circuitry that shape behavioral responses to stress in Drosophila. PLoS ONE. 2010;5(2) doi: 10.1371/journal.pone.0009141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sano H, Nakamura A, Texada MJ, Truman JW, Ishimoto H, Kamikouchi A, et al. The Nutrient-Responsive Hormone CCHamide-2 Controls Growth by Regulating Insulin-like Peptides in the Brain of Drosophila melanogaster. PLoS genetics. 2015;11(5) doi: 10.1371/journal.pgen.1005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nässel DR, Kubrak OI, Liu Y, Luo J, Lushchak OV. Factors that regulate insulin producing cells and their output in Drosophila. Front Physiol. 2013;4:252. doi: 10.3389/fphys.2013.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hentze JL, Carlsson MA, Kondo S, Nässel DR, Rewitz KF. The neuropeptide allatostatin A regulates metabolism and feeding decisions in Drosophila. Sci Rep. 2015;5:11680. doi: 10.1038/srep11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gruntenko NE, Wen D, Karpova EK, Adonyeva NV, Liu Y, He Q, et al. Altered juvenile hormone metabolism, reproduction and stress response in Drosophila adults with genetic ablation of the corpus allatum cells. Insect Biochem Mol Biol. 2010;40(12):891–897. doi: 10.1016/j.ibmb.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 126.Rauschenbach IY, Karpova EK, Adonyeva NV, Andreenkova OV, Faddeeva NV, Burdina EV, et al. Disruption of insulin signalling affects the neuroendocrine stress reaction in Drosophila females. J Exp Biol. 2014;217(Pt 20):3733–3741. doi: 10.1242/jeb.106815. [DOI] [PubMed] [Google Scholar]
- 127.Noguchi H, Hayakawa Y. Role of dopamine at the onset of pupal diapause in the cabbage armyworm Mamestra brassicae. FEBS lett. 1997;413(1):157–161. doi: 10.1016/S0014-5793(97)00848-X. [DOI] [PubMed] [Google Scholar]
- 128.Monastirioti M. Biogenic amine systems in the fruit fly Drosophila melanogaster. Microsc Res Tech. 1999;45(2):106–121. doi: 10.1002/(SICI)1097-0029(19990415)45:2<106::AID-JEMT5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 129.Colombani J, Bianchini L, Layalle S, Pondeville E, Dauphin-Villemant C, Antoniewski C, et al. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science. 2005;310(5748):667–670. doi: 10.1126/science.1119432. [DOI] [PubMed] [Google Scholar]
- 130.Rewitz KF, Yamanaka N, O'Connor MB. Steroid hormone inactivation is required during the juvenile-adult transition in Drosophila. Dev cell. 2010;19(6):895–902. doi: 10.1016/j.devcel.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McBrayer Z, Ono H, Shimell M, Parvy JP, Beckstead RB, Warren JT, et al. Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev Cell. 2007;13(6):857–871. doi: 10.1016/j.devcel.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rewitz KF, Yamanaka N, Gilbert LI, O'Connor MB. The insect neuropeptide PTTH activates receptor tyrosine kinase torso to initiate metamorphosis. Science. 2009;326(5958):1403–1405. doi: 10.1126/science.1176450. [DOI] [PubMed] [Google Scholar]
- 133.Tacutu R, Craig T, Budovsky A, Wuttke D, Lehmann G, Taranukha D, et al. Human Ageing Genomic Resources: integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 2013;41(Database issue):D1027–1033. doi: 10.1093/nar/gks1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.de Magalhaes JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25(7):875–881. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lai CQ, Parnell LD, Lyman RF, Ordovas JM, Mackay TF. Candidate genes affecting Drosophila life span identified by integrating microarray gene expression analysis and QTL mapping. Mech Ageing Dev. 2007;128(3):237–249. doi: 10.1016/j.mad.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 136.Carlson KA, Gardner K, Pashaj A, Carlson DJ, Yu F, Eudy JD, et al. Genome-wide gene expression in relation to Age in large laboratory cohorts of Drosophila melanogaster. Genet Res Int. 2015;2015:835624. doi: 10.1155/2015/835624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Obata F, Miura M. Enhancing S-adenosyl-methionine catabolism extends Drosophila lifespan. Nat Comm. 2015;6:8332. doi: 10.1038/ncomms9332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Huang CW, Wang HD, Bai H, Wu MS, Yen JH, Tatar M, et al. Tequila Regulates Insulin-Like Signaling and Extends Life Span in Drosophila melanogaster. J Gerontol A Biol Sci Med Sci. 2015;70(12):1461–9. doi: 10.1093/gerona/glv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim SN, Rhee JH, Song YH, Park DY, Hwang M, Lee SL, et al. Age-dependent changes of gene expression in the Drosophila head. Neurobiol Aging. 2005;26(7):1083–1091. doi: 10.1016/j.neurobiolaging.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 140.Baker DA, Russell S. Gene expression during Drosophila melanogaster egg development before and after reproductive diapause. BMC Genomics. 2009;10:242. doi: 10.1186/1471-2164-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vermeulen CJ, Sorensen P, Kirilova Gagalova K, Loeschcke V. Transcriptomic analysis of inbreeding depression in cold-sensitive Drosophila melanogaster shows upregulation of the immune response. J Evol Biol. 2013;26(9):1890–1902. doi: 10.1111/jeb.12183. [DOI] [PubMed] [Google Scholar]
- 142.Colinet H, Lee SF, Hoffmann A. Functional characterization of the Frost gene in Drosophila melanogaster: importance for recovery from chill coma. PLoS ONE. 2010;5(6) doi: 10.1371/journal.pone.0010925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bauer M, Katzenberger JD, Hamm AC, Bonaus M, Zinke I, Jaekel J, et al. Purine and folate metabolism as a potential target of sex-specific nutrient allocation in Drosophila and its implication for lifespan-reproduction tradeoff. Physiol Genom. 2006;25(3):393–404. doi: 10.1152/physiolgenomics.00009.2006. [DOI] [PubMed] [Google Scholar]
- 144.Whitaker R, Gil MP, Ding F, Tatar M, Helfand SL, Neretti N. Dietary switch reveals fast coordinated gene expression changes in Drosophila melanogaster. Aging. 2014;6(5):355–368. doi: 10.18632/aging.100662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ding F, Gil MP, Franklin M, Ferreira J, Tatar M, Helfand SL, et al. Transcriptional response to dietary restriction in Drosophila melanogaster. J Insect Physiol. 2014;69:101–106. doi: 10.1016/j.jinsphys.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schmidt PS, Zhu CT, Das J, Batavia M, Yang L, Eanes WF. An amino acid polymorphism in the couch potato gene forms the basis for climatic adaptation in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2008;105(42):16207–16211. doi: 10.1073/pnas.0805485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tauber E, Zordan M, Sandrelli F, Pegoraro M, Osterwalder N, Breda C, et al. Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science. 2007;316(5833):1895–1898. doi: 10.1126/science.1138412. [DOI] [PubMed] [Google Scholar]
- 148.Sandrelli F, Tauber E, Pegoraro M, Mazzotta G, Cisotto P, Landskron J, et al. A molecular basis for natural selection at the timeless locus in Drosophila melanogaster. Science. 2007;316(5833):1898–1900. doi: 10.1126/science.1138426. [DOI] [PubMed] [Google Scholar]
- 149.Kankare M, Salminen T, Laiho A, Vesala L, Hoikkala A. Changes in gene expression linked with adult reproductive diapause in a northern malt fly species: a candidate gene microarray study. BMC Ecol. 2010;10:3. doi: 10.1186/1472-6785-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Daibo S, Kimura MT, Goto SG. Upregulation of genes belonging to the drosomycin family in diapausing adults of Drosophila triauraria. Gene. 2001;278(1–2):177–184. doi: 10.1016/S0378-1119(01)00713-2. [DOI] [PubMed] [Google Scholar]
- 151.Yamada H, Yamamoto MT. Association between circadian clock genes and diapause incidence in Drosophila triauraria. PLoS One. 2011;6(12) doi: 10.1371/journal.pone.0027493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Robich RM, Rinehart JP, Kitchen LJ, Denlinger DL. Diapause-specific gene expression in the northern house mosquito, Culex pipiens L., identified by suppressive subtractive hybridization. J Insect Physiol. 2007;53(3):235–245. doi: 10.1016/j.jinsphys.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Honda Y, Tanaka M, Honda S. Modulation of longevity and diapause by redox regulation mechanisms under the insulin-like signaling control in Caenorhabditis elegans. Exp Gerontol. 2008;43(6):520–529. doi: 10.1016/j.exger.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 154.McElwee JJ, Schuster E, Blanc E, Thornton J, Gems D. Diapause-associated metabolic traits reiterated in long-lived daf-2 mutants in the nematode Caenorhabditis elegans. Mech Ageing Dev. 2006;127(5):458–472. doi: 10.1016/j.mad.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 155.Zhu H, Gegear RJ, Casselman A, Kanginakudru S, Reppert SM. Defining behavioral and molecular differences between summer and migratory monarch butterflies. BMC Biol. 2009;7:14. doi: 10.1186/1741-7007-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sim C, Denlinger DL. Transcription profiling and regulation of fat metabolism genes in diapausing adults of the mosquito Culex pipiens. Physiol Genom. 2009;39(3):202–209. doi: 10.1152/physiolgenomics.00095.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Storey KB, Heldmaier G, Rider MH. Mammalian hibernation: physiological, cell signaling and gene controls on metabolic rate depression. In: Lubzens E, Cerda J, Clark M, editors. Dormancy and resistance in harsh environments. Heidelberg: Springer; 2010. pp. 227–252. [Google Scholar]
- 158.Wu CW, Storey KB. Regulation of the mTOR signaling network in hibernating thirteen-lined ground squirrels. J Exp Biol. 2012;215(Pt 10):1720–1727. doi: 10.1242/jeb.066225. [DOI] [PubMed] [Google Scholar]
- 159.Hedengren M, Asling B, Dushay MS, Ando I, Ekengren S, Wihlborg M, et al. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol Cell. 1999;4(5):827–837. doi: 10.1016/S1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- 160.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86(6):973–983. doi: 10.1016/S0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 161.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 162.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25(2):288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8(8):1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tian L, Greenberg SA, Kong SW, Altschuler J, Kohane IS, Park PJ. Discovering statistically significant pathways in expression profiling studies. Proc Natl Acad Sci U S A. 2005;102(38):13544–13549. doi: 10.1073/pnas.0506577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kula-Eversole E, Nagoshi E, Shang Y, Rodriguez J, Allada R, Rosbash M. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc Natl Acad Sci U S A. 2010;107(30):13497–13502. doi: 10.1073/pnas.1002081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Burgess A, Vigneron S, Brioudes E, Labbe JC, Lorca T, Castro A. Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc Natl Acad Sci U S A. 2010;107(28):12564–12569. doi: 10.1073/pnas.0914191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gäde G, Auerswald L. Mode of action of neuropeptides from the adipokinetic hormone family. Gen Comp Endocrinol. 2003;132(1):10–20. doi: 10.1016/S0016-6480(03)00159-X. [DOI] [PubMed] [Google Scholar]
- 172.Zeidler MP, Bach EA, Perrimon N. The roles of the Drosophila JAK/STAT pathway. Oncogene. 2000;19(21):2598–2606. doi: 10.1038/sj.onc.1203482. [DOI] [PubMed] [Google Scholar]


