Abstract
Background
Less than 20 % of familial breast cancer patients who undergo genetic testing for BRCA1 and BRCA2 carry a pathogenic mutation in one of these two genes. The GENESIS (GENE SISter) study was designed to identify new breast cancer susceptibility genes in women attending cancer genetics clinics and with no BRCA1/2 mutation.
Methods
The study involved the French national network of family cancer clinics. It was based on enrichment in genetic factors of the recruited population through case selection relying on familial criteria, but also on the consideration of environmental factors and endophenotypes like mammary density or tumor characteristics to assess potential genetic heterogeneity. One of the initial aims of GENESIS was to recruit affected sibpairs. Siblings were eligible when index cases and at least one affected sister were diagnosed with infiltrating mammary or ductal adenocarcinoma, with no BRCA1/2 mutation. In addition, unrelated controls and unaffected sisters were recruited. The enrolment of patients, their relatives and their controls, the collection of the clinical, epidemiological, familial and biological data were centralized by a coordinating center.
Results
Inclusion of participants started in February 2007 and ended in December 2013. A total of 1721 index cases, 826 affected sisters, 599 unaffected sisters and 1419 controls were included. 98 % of participants completed the epidemiological questionnaire, 97 % provided a blood sample, and 76 % were able to provide mammograms. Index cases were on average 59 years old at inclusion, were born in 1950, and were 49.7 years of age at breast cancer diagnosis. The mean age at diagnosis of affected sisters was slightly higher (51.4 years). The representativeness of the control group was verified.
Conclusions
The size of the study, the availability of biological specimens and the clinical data collection together with the detailed and complete epidemiological questionnaire make this a unique national resource for investigation of the missing heritability of breast cancer, by taking into account environmental and life style factors and stratifying data on endophenotypes to decrease genetic heterogeneity.
Background
Less than 20 % of women affected by breast cancer (BC) and qualified for BRCA1/2 testing are carrying a deleterious (or pathological) mutation in one of these genes [1]. Mutations in other genes causing familial syndromes in which BC incidence is highly increased (TP53, PTEN, STK11 and CDH1) are estimated to cause 5 % of the familial forms of BC and an additional 5 % is accounted for by moderate penetrance genes (i.e. associated with an odds ratio (OR) below 3), such as ATM, CHEK2, and the Fanconi anemia pathway genes (BRIP1, PALB2, RAD51C, RAD51D and XRCC2). Therefore, the majority of the familial forms of BC remain unexplained. Schematically, the studies performed to elucidate the missing heritability have either been on high-risk populations using mainly linkage analysis approaches to detect “major” genes or on the general population using association studies to detect “more” common genetic “variations”. Linkage analyses failed to identify new “major” loci [2] while genome-wide association studies performed on large case-control studies of BC have identified about 100 common BC susceptibility loci (single nucleotide polymorphisms or SNPs) to date (e.g. [3–13]). However, the effect sizes detected by these large-scale studies were small, for the vast majority, the associated OR rarely being greater than 1.20, and altogether may account for only 14 % of the missing heritability [14].
There is likely a genetic heterogeneity, with different types of predisposing situations observed among women at risk. These genetic “sub-entities” resulting from the combination of several factors may be associated with particular characteristics of the individual or of the tumor. For example, several SNPs identified by genome-wide association studies were shown to be associated with the estrogen receptor status of the breast tumor both in the general population [3, 4, 15–17] and the population of BRCA2 mutation carriers [18–21].
Our proposal was to set up a study to investigate the missing heritability of BC in a high-risk population with unrelated controls for conducting association studies. The novelty of the GENESIS (GENE SISters) study is the recruitment of a study population enriched in susceptibility factors by case selection based on familial criteria, with consideration of environmental factors. Potential genetic heterogeneity was accounted for by stratifying the study sample on proxy such as particular individual epidemiological or clinical characteristics (mammary density, for example), or tumor characteristics.
The GENESIS study is an integrative genetic epidemiological project based on the involvement of all French family cancer clinic consultants who belong to the “Groupe Génétique et Cancer” (GGC) of Unicancer, the centralized enrolment of patients and collection of their clinical, epidemiological, familial and biological data by a coordinating center (CC) at the Institut Curie (Paris, France). Here we describe the design and logistics of the study and the available data. We also discuss the participation rates, the prevalence of the BC cases, and the representativeness of the participants and of the population of controls.
Methods
Eligible individuals
Index cases (and their affected sisters) were identified through the French family cancer clinics of the GGC (i.e. 42 centers) and were eligible when diagnosed with infiltrating mammary or ductal adenocarcinoma, were negative for BRCA1 and BRCA2 mutations, and had a sister with BC. The mutation screening strategy was similar for all the clinics. The full coding sequences and the exon-intron junctions of the BRCA1 and BRCA2 genes were screened for mutations, based on pre-screening (Denaturing High-Performance Liquid Chromatography (dHPLC), High-Resolution Melting (HRM) or Enhanced Mismatch Mutation Analysis (EMMA)) and sequencing. For a subset of the index cases; large rearrangements were screened by large cDNA sequencing, Multiplex Ligation-dependent Probe Amplification (MLPA), Quantitative Multiplex PCR of Short Fragments (QMPSF), Quantitative PCR (qPCR), Quantitative PCR High Resolution Melting (qPCR HRM), EMMA or dedicated array Comparative Genomic Hybridization (array CGH). Sisters with infiltrating mammary adenocarcinoma or in situ ductal carcinoma, regardless of their age at diagnosis, were eligible. If the index case had more than one affected sister, all were approached. Two types of controls were included: unrelated controls and unaffected sisters. The unrelated controls were selected among the unaffected friends and/or colleagues of the cases. The year of birth of controls was matched to that of the corresponding case (+/− 3 years). The parents, brothers and unaffected sisters were also contacted, when possible.
Geneticists of family cancer clinics identified index cases and invited them to participate in GENESIS by referring them to the CC. Each family cancer clinic invited index cases to participate in the study by letter (retrospective index cases with a molecular diagnosis performed between 2003 and 2007) or during consultations informing patients of their BRCA1/2 negative results (prospective index cases). The CC organized the inclusion of index cases and of their relatives and unrelated controls. The index case then sent a response coupon to the CC to obtain the complete study file including a detailed information letter and a consent form to be completed. Subjects were included in the study when they sent back their signed consent, with the possibility of a telephone contact with a member of the CC team and/or a genetic consultation for additional information. The index case contacted her sisters (affected and unaffected), parents and brothers, and unrelated unaffected friends or colleagues and gave them an information letter and response coupon. After their agreement, the CC sent them the study file including the detailed information letter and the consent form. Again, relatives and unrelated subjects were included in the study when they sent back their signed consent. The CC organized the collection of blood samples from the index case and other participants by sending them a prescription for blood sampling, a letter for the medical analysis laboratory or the nurse who took the blood sample, and appropriate prepaid packaging for dispatch of the samples directly to the biological resource center at the Centre Léon Bérard (Lyon, France).
The study was examined by the appropriate committees: ethics (CCP Ile-de-France III, 3 October 2006, agreement no. 2373) and by the data protection agency (CNIL, 22 May 2006, agreement no. 1170775), all of which approved the study.
Data collected
All patients (index cases and affected sisters) and controls completed a questionnaire on environmental, lifestyle and reproductive factors and family history of cancer. This self-report questionnaire contained detailed questions concerning demographic data, alcohol and tobacco consumption, pregnancies, breastfeeding, contraception, hormone replacement, physical activity, personal medical history and exposure to irradiation at work and for medical purposes and pedigree, with detailed information on medical history for each first- and second-degree relative.
Mammograms taken at the time of diagnosis or one to three years before diagnosis for the cases and most recent mammography for the controls and unaffected sisters at inclusion were collected. Craniocaudal and medio-lateral oblique views of both breasts were digitized. A VIDAR DiagnosticPro Advantage scanner, with a resolution of 570 dpi, was used to record the information required for quantitative calculations of mammary density. The images obtained were recorded and the identity of the participant was erased from images with Adobe® Photoshop® software. The incidence and date of the mammograms were recorded on the images.
Blood samples were collected from index cases, affected and unaffected sisters and controls. Viable lymphocytes from index cases were frozen if this had not already been done by the laboratory having carried out BRCA1/2 analyses. Part of each blood sample was frozen, while the rest was processed in order to obtain plasma, serum, and lymphocyte pellets, all of which were then frozen. DNA was subsequently extracted using the AutopureLS Instrument (Qiagen).
No systematic collection of tumor specimens has been performed. However, pathology reports and information of sample storage conditions and location for mammary tumors have been collected and are being coded and computerized. This information will facilitate access to the tumor samples for specific projects to come.
Study power
These data will be used to study the missing heritability of BC by taking into account environmental factors. The power of the study depends on the study design and strategy employed for detecting BC susceptibility alleles. For instance, SNP genotyping, mutation screening of candidate genes or whole exome sequencing may be undertaken in all subjects or specific subset of participants, since stratifying data on endophenotypes may help decreasing genetic heterogeneity. Interaction effects according to the genes under study may also be investigated. A “simple” power calculation showed that a genetic association with an amplitude of 3 (relative risk associated with a susceptibility genotype) sought by a candidate gene approach in a case-control study design can be identified with a power of 80 % (alpha = 0.05) for allelic frequencies greater than 0.5 % for a dominant inheritance and greater than 10 % for a recessive inheritance. The power will decrease with decreasing risk amplitude if the study eligibility criteria lead to an underrepresentation of high-risk families. A priori power calculations are challenging and will depend on the hypotheses and models used (single SNP analyses, gene pathway-based approaches, single environmental/lifestyle factor, exposure profiles with or without interaction…).
Results
Inclusion of participants started in February 2007 and ended in December 2013. Description of the GENESIS population is based on data available on 17 November 2014. Thus, the population may increase slightly when residual signed inform consents are received by the CC. Figure 1 shows the cumulative number of index cases over the time-course of inclusion, both for retrospective and prospective inclusions.
Fig. 1.
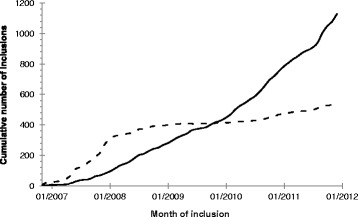
Cumulative number of index case inclusions over time. Legend:  Index cases with retrospective molecular diagnosis.
Index cases with retrospective molecular diagnosis.  Index cases with prospective molecular diagnosis
Index cases with prospective molecular diagnosis
2543 patients qualified according to the study criteria were listed and invited to participate by 26 centers. 1669 women sent back a reply coupon to the CC for complete information about the study and 1315 agreed to participate (i.e. 52 % of patients invited). 539 additional cases, invited by 16 centers without being listed beforehand and meeting the inclusion criteria returned a reply coupon and 406 of them agreed to participate and were therefore included. A total of 1721 patients were included, 1483 of whom had at least one living sister affected by BC and potentially able to participate in the study, and 238 of whom had no living affected sister. Since the participation rate was not very high, the representativeness of the included cases could be questioned. Birth year, age at diagnosis and year of diagnosis were compared in the eligible population (631 index cases) of the Institut Curie clinic where the information was available. The characteristics of eligible index cases and the characteristics of index cases included were similar, i.e. 1948, 50 years and 1998 on average for birth year, age at diagnosis and year of diagnosis, respectively. Thus, we can be confident that the population of index cases is representative of the targeted population.
98 % of index cases completed the epidemiological questionnaire and provided a blood sample; 68 % were able to provide mammograms. The information on mammograms was extracted from the epidemiological questionnaire. The collection of mammograms is still on-going since many are kept in the care centers where the women were treated for their cancer (cf. Table 1). Table 2 shows that all index cases included were on average 59 years old at inclusion (minimum (min): 31 years old; maximum (max): 90 years old), were born in 1950 (min: 1918; max: 1977) and were 49.7 years old at the BC diagnosis (min: 20 years old; max: 80 years old). The mean interval between the date of diagnosis and the date of inclusion was 9.3 years (min: 0 year; max: 48 years).
Table 1.
Available data in GENESIS per type of participant
| Type of participant | Signed consent form | Completed questionnaire | Blood sample | At least one mammogram performeda | At least one mammogram collected |
|---|---|---|---|---|---|
| N | N (%) | N (%) | N | N (%) | |
| Index case | 1721 | 1682 (98 %) | 1695 (98 %) | 1721 | 1169 (68 %) |
| Affected sister | 826 | 807 (98 %) | 805 (97 %) | 826 | 546 (66 %) |
| Unaffected sister | 599 | 592 (99 %) | 582 (97 %) | 589 | 493 (84 %) |
| Control | 1419 | 1411 (99 %) | 1360 (96 %) | 1322 | 1201 (91 %) |
| Total | 4565 | 4492 (98 %) | 4442 (97 %) | 4458 | 3409 (76 %) |
N, number; %, percentage, abased on questionnaire information for unaffected women
Table 2.
Characteristics of the GENESIS population according to the type of participant
| Type of participant | Year of birth | Age at breast cancer diagnosis (years) | Age at inclusion (years) | Interval between diagnosis and inclusion | ||||
|---|---|---|---|---|---|---|---|---|
| By class | ||||||||
| ≤4 years | 5-9 years | ≥10 years | ||||||
| N | mean (SD) [min, max] | mean (SD) [min, max] | mean (SD) [min, max] | mean (SD) | N (%) | N (%) | N (%) | |
| Index case | 1682 | 1950 (9.3) [1918, 1977] | 49.7 (9.3) [20, 80] | 59.0 (9.3) [31, 90] | 9.3 (7.4) | 559 (33 %) | 480 (29 %) | 643 (38 %) |
| Affected sister | 807 | 1949 (9.0) [1926, 1978] | 51.4 (8.7) [27, 77] | 59.5 (8.8) [30, 83] | 8.1 (6.4) | 297 (37 %) | 249 (31 %) | 258 (32 %) |
| Unaffected sister | 592 | 1952 (9.3) [1926, 1978] | 57.0 (9.2) [30, 84] | |||||
| Control | 1411 | 1953 (10.0) [1926, 1991] | 55.7 (9.9) [19, 83] | |||||
N, number; SD, standard deviation; min, minimum; max, maximum; %, percentage
This interval has an effect on the familial phenotype of the index cases. 32 % of the index case families had 3 or more cases of BC when the interval was greater than 10 years; this percentage decreases to 26 % when the interval was less than 10 years. This difference is mostly due to a longer survey of the families. When the follow-up of the family members was censured at the year of diagnosis of the index case, the percentage of index case families with 3 or more BC cases was 24 % when the interval was greater than 10 years and 23 % when it was less than 10 years. Thus, ascertainment by familial phenotype criteria appears constant over time.
Of the 5095 sisters and controls invited by the index cases and identified through the list of invitations sent back by the index case, 2518 women agreed to participate, i.e. 685 sisters with BC (57 %), 541 unaffected sisters (54 %) and 1292 controls (45 %). An extra 141 affected sisters, 58 unaffected sisters and 127 controls, not previously listed in the index cases’ invitation list, agreed to participate. Thus a total of 826 affected sisters, 599 unaffected sisters and 1419 controls were included.
98 % of the affected sisters and 99 % of the unaffected sisters and controls completed the epidemiological questionnaire, and 97 % of the sisters and 96 % of the controls provided a blood sample. 66 % of the affected sisters provided mammograms, a percentage similar to that of the index cases, again because many of the mammograms are kept in the care centers where the sisters with BC were treated for cancer. The percentage was higher for the unaffected sisters and the controls: 84 % and 91 %, respectively (cf. Table 1). The mean ages at diagnosis and at inclusion of the affected sisters were slightly higher than those of the index cases, i.e. 51.4 and 59.5 years, respectively, with therefore a slight decrease in the interval between diagnosis and inclusion (8.1 years vs 9.3 years). On average, the unaffected sisters were two years younger than the index cases and the controls three years younger (cf. Table 2).
Our target of recruiting 1000 sibpairs, based on power calculations, was not reached. Among the 1483 index cases with at least one sister alive and with BC, 696 had at least one affected sister included (47 %). A total of 788 affected sisters agreed to participate. The size of the sibships with participating affected sisters varied from 2 to 5 (cf. Table 3). Of the 696 sibships with at least two affected sisters included, 678 completed the epidemiological questionnaires, 669 provided blood samples and 362 provided mammograms. The absolute mean difference in the ages at diagnosis within sibpairs was 5.7 years (Standard Deviation (SD): 8.2 years).
Table 3.
Available data on sibships with affected sisters
| Number of sisters with breast cancer per sibship | Number of sibships with | |||
|---|---|---|---|---|
| Signed consent forms | Completed questionnaires | Blood samples | Mammograms collected | |
| at least 2a | 696 | 678 | 669 | 362 |
| exactly 2a | 616 | 600 | 592 | 324 |
| exactly 3a | 69 | 67 | 67 | 34 |
| exactly 4a | 10 | 10 | 9 | 4 |
| exactly 5a | 1 | 1 | 1 | 0 |
aWhether the women are index cases or affected sisters
Since the controls were supposed to be either a friend or a colleague of an index case, the representativeness of this group is questionable in terms of family history of cancer. Indeed, friends or colleagues may have participated because they have a “strong” family history of cancer. This bias might lead to an underestimate of the effect of genetic factors. Therefore, we analyzed the pedigree of the 1411 unrelated controls by comparing the cancer incidences in these pedigrees (first degree of relationship) to the national incidences using SAS 9.3 software (SAS Institute, Cary, NC) and the estimated national cancer incidences from 1975 to 2005 [22]. We observed a slight increased incidence of cancer within control families for all sites (Standardized Incidence Ratio (SIR): 1.11 95%CI: 1.02–1.22), for breast cancer (SIR: 1.21 95%CI: 1.04–1.39) and for ovarian cancer (SIR: 1.16 95%CI: 0.73–1.76) and this should be taken into account in the further analyses.
Discussion
The GENESIS study is a unique national resource to study the missing heritability of BC. This is a large study including over 6000 participants, and the biological, clinical and epidemiological data collected are detailed and complete. The rate of agreement to participate was moderate for each participant category, around 50 %. However, the constraint of asking the index cases to contact the other participants impaired assessment of the true agreement rates as for these people we relied on a list completed by the index cases. Indeed, people not listed beforehand were included (more than 11 % of the inclusions). Even though we have full confidence in the representativeness of the population of the included index cases and controls, it may be more effective in future studies to contact relatives and other participants directly.
Even though part of the study was prospective, most index cases and their affected sisters were prevalent cases (only 5.5 % were included in the calendar year of their BC diagnosis or in the following year). This has to be taken into account when seeking for BC susceptibility genes in order to avoid false conclusion. For instance, identified genes could be in fact involved in survival; conversely, one could miss BC susceptibility genes that are also involved in poor prognosis. Hence, for future studies using this resource, a comparison between pseudo-incident cases (interval between diagnosis and interview less than 5 years, for example) and prevalent cases will be useful.
The unrelated controls were matched to the corresponding index cases on the basis of the year of birth (+/− 3 years) to simplify the index cases’ task of inviting controls. However, because the large majority of index cases were included prospectively, their invited controls had survived without BC years after the index cases’ year of diagnosis. This may therefore result in bias toward the alternative even after censuring controls at the index cases’ age at diagnosis. To avoid such bias, depending on the question under study, the controls could be matched either to the index case’s year of birth or age at BC diagnosis. The latter option should be avoided when a cohort effect is observed for the variables under study.
The eligibility criteria of GENESIS did not exclude patients carrying a mutation in “clinically actionable” susceptibility genes other than BRCA1 and BRCA2. In France, PALB2 testing was introduced in the clinical practices in July 2015; therefore PALB2 status in GENESIS index cases was not available at the time of inclusion. Subsequently, a case-control study performed in French BC families and including the first 40 % of the recruited GENESIS index cases has shown that PALB2 truncating mutation are found in 0.36 % of the familial cases [23]. Whole-exome and whole-genome sequencing projects are also ongoing on a subset of the GENESIS population, as well as targeted sequencing of a panel of high- to moderate-risk genes for more than 2000 GENESIS participants. Genes under investigation includes, among others, ATM, CHEK2, RAD51 paralogs and those included in panels used by the French diagnosis laboratories for research purpose (Lesueur et al. personal communication). Subjects carrying a mutation in one of these genes will be therefore identifiable for subsequent studies.
The low number of large families for which nearly all first-degree relatives have been recruited might be a limitation for powerful co-segregation analyses. However, in order to validate new potential BC susceptibility genes, it will be possible to use additional large sample set thanks to the GGC families’ recruitment or through participation to other international high-risk family studies.
Conclusions
The identification of new BC susceptibility genes will clearly have implications for the management of women at risk. It will enable adaptation of the follow-up of these women according to risk assessments based on new tests. If the risk is considered high, early and regular MRI-based screening could be offered.
The identification of new genes should improve our understanding of the origin of a proportion of BC sporadic cases and should make it possible to optimize the management of these cases’ relatives.
Finally, an understanding of the biological functions of these genes and their interactions with environmental factors may reveal new possibilities for the prevention and treatment of BC, the incidence of which is continuing to increase in Western populations.
The GENESIS study will be an asset for ongoing molecular studies aiming at identifying new BC susceptibility genes, and such studies using this resource have already started (e.g. [23–25]). GENESIS resource also contributes to international consortia like COMPLEXO (COMPLexity of EXOme) [26].
Special dedication
This article is dedicated to the memory of Olga M. Sinilnikova who died prematurely on June 30, 2014. Olga participated decisively in structuring research around hereditary predisposition to BC and in leading the GENESIS study.
Acknowledgments
Financial support for GENESIS was provided by the Ligue Nationale contre le Cancer (3 grants: PRE05/DSL, PRE07/DSL, PRE11/NA), the French National Institute of Cancer (INCa) and the comprehensive cancer center SiRIC, (Site de Recherche Intégrée sur le Cancer: Grant INCa-DGOS-4654).
We wish to thank the genetic epidemiology platform (the PIGE, Plateforme d’Investigation en Génétique et Epidémiologie: S. Eon-Marchais, M. Marcou, D. Le Gal, L. Toulemonde, J. Beauvallet, N. Mebirouk, E. Cavaciuti, A. Fescia), the biological resource center (C. Verny-Pierre, L. Barjhoux, V. Sornin) and all the GENESIS collaborating cancer clinics (Clinique Sainte Catherine, Avignon: H. Dreyfus; Hôpital Saint Jacques, Besançon: M-A. Collonge-Rame; Institut Bergonié, Bordeaux: M. Longy, A. Floquet, E. Barouk-Simonet; CHU, Brest: S. Audebert; Centre François Baclesse, Caen: P. Berthet; Hôpital Dieu, Chambéry: S. Fert-Ferrer; Centre Jean Perrin, Clermont-Ferrand: Y-J. Bignon; Hôpital Pasteur, Colmar: J-M. Limacher; Hôpital d’Enfants CHU – Centre Georges François Leclerc, Dijon: L. Faivre; CHU, Fort de France: O. Bera; CHU Albert Michallon, Grenoble: D. Leroux; Hôpital Flaubert, Le Havre: V. Layet; Centre Oscar Lambret, Lille: P. Vennin†, C. Adenis; Hôpital Jeanne de Flandre, Lille: S. Lejeune-Dumoulin, S. Manouvier-Hanu; CHRU Dupuytren, Limoges: L. Venat-Bouvet; Centre Léon Bérard, Lyon: C. Lasset, V. Bonadona; Hôpital Edouard Herriot, Lyon: S. Giraud; Institut Paoli-Calmettes, Marseille: F. Eisinger, L. Huiart; Centre Val d’Aurelle – Paul Lamarque, Montpellier: I. Coupier; CHU Arnaud de Villeneuve, Montpellier: I. Coupier, P. Pujol; Centre René Gauducheau, Nantes: C. Delnatte; Centre Catherine de Sienne, Nantes: A. Lortholary; Centre Antoine Lacassagne, Nice: M. Frenay, V. Mari; Hôpital Caremeau, Nîmes: J. Chiesa; Réseau Oncogénétique Poitou Charente, Niort: P. Gesta; Institut Curie, Paris: D. Stoppa-Lyonnet, M. Gauthier-Villars, B. Buecher, A. de Pauw, C. Abadie, M. Belotti; Hôpital Saint-Louis, Paris: O. Cohen-Haguenauer; Centre Viggo-Petersen, Paris: F. Cornélis; Hôpital Tenon, Paris: A. Fajac; GH Pitié Salpétrière et Hôpital Beaujon, Paris: C. Colas, F. Soubrier, P. Hammel, A. Fajac; Institut Jean Godinot, Reims: C. Penet, T.D. Nguyen; Polyclinique Courlancy, Reims: L. Demange†, C. Penet; Centre Eugène Marquis, Rennes: C. Dugast; Centre Henri Becquerel, Rouen: A. Chevrier, T. Frebourg, J. Tinat, I. Tennevet, A. Rossi; Hôpital René Huguenin/Institut Curie, Saint Cloud: C. Noguès, L. Demange†, E. Mouret-Fourme; CHU, Saint-Etienne: F. Prieur; Centre Paul Strauss, Strasbourg: J-P. Fricker, H. Schuster; Hôpital Civil, Strasbourg: O. Caron, C. Maugard; Institut Claudius Regaud, Toulouse: L. Gladieff, V. Feillel; Hôpital Bretonneau, Tours: I. Mortemousque; Centre Alexis Vautrin, Vandoeuvre-les-Nancy: E. Luporsi; Hôpital de Bravois, Vandoeuvre-les-Nancy: P. Jonveaux; Gustave Roussy, Villejuif: A. Chompret†, O. Caron).
†: deceased prematurely
Abbreviations
- BC
Breast cancer
- CC
Coordinating Center
- GGC
Groupe Génétique et Cancer
- min
Minimum
- max
Maximum
- OR
Odds ratio
- SD
Standard deviation
- SIR
Standardized incidence ratio
- SNP
Single nucleotide polymorphism
- dHPLC
Denaturing high-performance liquid chromatography
- HRM
High-resolution melting
- EMMA
Enhanced mismatch mutation analysis
- MLPA
Multiplex ligation-dependent probe amplification
- QMPSF
Quantitative multiplex PCR of short fragments
- qPCR
Quantitative PCR
- qPCR HRM
Quantitative PCR High Resolution Melting
- array CGH
Array comparative genomic hybridization
Footnotes
Marie-Gabrielle Dondon Séverine Eon-Marchais Dominique Stoppa-Lyonnet and Nadine Andrieu contributed equally to this work.
Olga M. Sinilnikova and Liliane Demange, Deceased.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
OMS, DSL and NA led the GENESIS study and contributed to the protocol, design and search for funding. NA and SEM were responsible for coordination of the study. DSL, MB, OC, MGV, IC, BB, AL, CDu, PG, JPF, CN, LF, EL, PB, CDa, VB, CMM, PP, CL, ML, YJB, CA, LVB, LD, HD, MF, LG, IM, SAB, FS, SG, SLD, AC, JML, JC, AFa, AFl, FE, JT, CC, SFF, CP, TF, MACR, EBS, VL, DL, OC, FP, EMF, FC, PJ, OB are geneticists at family cancer clinics who made a major contribution to the invitation of index cases. SEM, MM, LT, JB, DLG, NM and EC were responsible for inclusion of participants, data collection and entry, and for organization of blood sample collection and the scanning of mammograms. OMS, FD, LB, CVP, VS, FL and SM were responsible for blood sample treatment and storage. AT provided expertise for mammograms digitalization and quantitative calculations of mammary density. MGD was responsible for data checking and preparation and conducted for the statistical analyses. NA supervised the analyses and wrote the report. All authors revised the report critically and gave final approval of the version to be published.
Contributor Information
Marie-Gabrielle Dondon, Email: mariegabrielle.dondon@curie.fr.
Séverine Eon-Marchais, Email: severine.eonmarchais@curie.fr.
Francesca Damiola, Email: francesca.damiola@lyon.unicancer.fr.
Laure Barjhoux, Email: laure.barjhoux@lyon.unicancer.fr.
Morgane Marcou, Email: morgane.marcou@apr.aphp.fr.
Carole Verny-Pierre, Email: carole.verny@lyon.unicancer.fr.
Valérie Sornin, Email: valerie.sornin@lyon.unicancer.fr.
Lucie Toulemonde, Email: lucietoulemonde@hotmail.com.
Juana Beauvallet, Email: juana.beauvallet@curie.fr.
Dorothée Le Gal, Email: dorothee.legal@curie.fr.
Noura Mebirouk, Email: noura.mebirouk@curie.fr.
Muriel Belotti, Email: muriel.belotti@curie.fr.
Olivier Caron, Email: olivier.caron@gustaveroussy.fr.
Marion Gauthier-Villars, Email: marion.gauthier-villars@curie.fr.
Isabelle Coupier, Email: i-coupier@chu-montpellier.fr.
Bruno Buecher, Email: bruno.buecher@curie.fr.
Alain Lortholary, Email: lortholary.alain@catherinedesienne.fr.
Catherine Dugast, Email: c.dugast@rennes.unicancer.fr.
Paul Gesta, Email: paul.gesta@ch-niort.fr.
Jean-Pierre Fricker, Email: jfricker@strasbourg.unicancer.fr.
Catherine Noguès, Email: catherine.nogues@curie.fr.
Laurence Faivre, Email: laurence.faivre@chu-dijon.fr.
Elisabeth Luporsi, Email: e.luporsi@nancy.unicancer.fr.
Pascaline Berthet, Email: p.berthet@baclesse.fr.
Capucine Delnatte, Email: capucine.delnatte@ico.unicancer.fr.
Valérie Bonadona, Email: valerie.bonadona@lyon.unicancer.fr.
Christine M. Maugard, Email: christine.maugard@chru-strasbourg.fr, Email: christinemaugard@gmail.com
Pascal Pujol, Email: p-pujol@chu-montpellier.fr.
Christine Lasset, Email: christine.lasset@lyon.unicancer.fr.
Michel Longy, Email: m.longy@bordeaux.unicancer.fr.
Yves-Jean Bignon, Email: yves-jean.bignon@cjp.fr.
Claude Adenis, Email: cl-adenis@o-lambret.fr.
Laurence Venat-Bouvet, Email: laurence.venat@chu-limoges.fr.
Hélène Dreyfus, Email: h.dreyfus@isc84.org.
Marc Frenay, Email: marc.frenay@nice.unicancer.fr.
Laurence Gladieff, Email: gladieff.laurence@iuct-oncopole.fr.
Isabelle Mortemousque, Email: i.mortemousque@chu-tours.fr.
Séverine Audebert-Bellanger, Email: severine.audebert@chu-brest.fr.
Florent Soubrier, Email: florent.soubrier@psl.aphp.fr.
Sophie Giraud, Email: sophie.giraud@chu-lyon.fr.
Sophie Lejeune-Dumoulin, Email: sophie.lejeune@chru-lille.fr.
Annie Chevrier, Email: annie.chevrier@chu-rouen.fr.
Jean-Marc Limacher, Email: limacher@transgene.fr.
Jean Chiesa, Email: jean.chiesa@chu-nimes.fr.
Anne Fajac, Email: anne.fajac@tnn.aphp.fr.
Anne Floquet, Email: a.floquet@bordeaux.unicancer.fr.
François Eisinger, Email: eisingerf@ipc.unicancer.fr.
Julie Tinat, Email: julie.tinat@chu-rouen.fr.
Chrystelle Colas, Email: chrystelle.colas@psl.aphp.fr.
Sandra Fert-Ferrer, Email: sandra.fertferrer@ch-chambery.fr.
Clotilde Penet, Email: cpenet@iccreims.fr.
Thierry Frebourg, Email: thierry.frebourg@chu-rouen.fr.
Marie-Agnès Collonge-Rame, Email: macollongerame@chu-besancon.fr.
Emmanuelle Barouk-Simonet, Email: e.barouksimonet@bordeaux.unicancer.fr.
Valérie Layet, Email: vlayet@ch-havre.fr.
Dominique Leroux, Email: dleroux@chu-grenoble.fr.
Odile Cohen-Haguenauer, Email: odile.cohen-haguenauer@sls.aphp.fr.
Fabienne Prieur, Email: fabienne.prieur@chu-st-etienne.fr.
Emmanuelle Mouret-Fourme, Email: emmanuelle.fourme@curie.fr.
François Cornélis, Email: fcornelis@chu-clermontferrand.fr.
Philippe Jonveaux, Email: p.jonveaux@chu-nancy.fr.
Odile Bera, Email: odile.bera@chu-fortdefrance.fr.
Eve Cavaciuti, Email: eve.cavaciuti@curie.fr.
Anne Tardivon, Email: anne.tardivon@curie.fr.
Fabienne Lesueur, Email: fabienne.lesueur@curie.fr.
Sylvie Mazoyer, Email: sylvie.mazoyer@lyon.unicancer.fr.
Dominique Stoppa-Lyonnet, Email: dominique.stoppa-lyonnet@curie.fr.
Nadine Andrieu, Email: nadine.andrieu@curie.fr.
References
- 1.Inca. Synthèse de l’activité d’oncogénétique 2012 - Consultations et laboratoires. BILONCOGEN13. 2014. Available: http://www.e-cancer.fr/Expertises-et-publications/Catalogue-des-publications/Synthese-de-l-activite-d-oncogenetique-2012-Consultations-et-laboratoires [Accessed 11 26 2014]. 2014.
- 2.Antoniou AC, Easton DF. Models of genetic susceptibility to breast cancer. Oncogene. 2006;25:5898–905. doi: 10.1038/sj.onc.1209879. [DOI] [PubMed] [Google Scholar]
- 3.Bojesen SE, Pooley KA, Johnatty SE, Beesley J, Michailidou K, Tyrer JP, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013;45:371–2. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–2. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–9. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 8.Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41:585–90. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41:579–84. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stacey SN, Manolescu A, Sulem P, Thorlacius S, Gudjonsson SA, Jonsson GF, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40:703–6. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 11.Zheng W, Long J, Gao YT, Li C, Zheng Y, Xiang YB, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–8. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–7. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghoussaini M, Fletcher O, Michailidou K, Turnbull C, Schmidt MK, Dicks E, et al. Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nat Genet. 2012;44:312–8. doi: 10.1038/ng.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melchor L, Benitez J. The complex genetic landscape of familial breast cancer. Hum Genet. 2013;132:845–63. doi: 10.1007/s00439-013-1299-y. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45:392. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siddiq A, Couch FJ, Chen GK, Lindström S, Eccles D, Millikan RC, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. 2012;21:5373–84. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevens KN, Fredericksen Z, Vachon CM, Wang X, Margolin S, Lindblom A, et al. 19p13.1 is a triple-negative-specific breast cancer susceptibility locus. Cancer Res. 2012;72:1795–803. doi: 10.1158/0008-5472.CAN-11-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaudet MM, Kuchenbaecker KB, Vijai J, Klein RJ, Kirchhoff T, McGuffog L, et al. Identification of a BRCA2-specific modifier locus at 6p24 related to breast cancer risk. PLoS Genet. 2013;9:e1003173. doi: 10.1371/journal.pgen.1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couch FJ, Wang X, McGuffog L, Lee A, Olswold C, Kuchenbaecker KB, et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 2013;9:e1003212. doi: 10.1371/journal.pgen.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antoniou AC, Kuchenbaecker KB, Soucy P, Beesley J, Chen X, McGuffog L, et al. Common variants at 12p11, 12q24, 9p21, 9q31.2 and in ZNF365 are associated with breast cancer risk for BRCA1 and/or BRCA2 mutation carriers. Breast Cancer Res. 2012;14:R33. doi: 10.1186/bcr3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulligan AM, Couch FJ, Barrowdale D, Domchek SM, Eccles D, Nevanlinna H, et al. Common breast cancer susceptibility alleles are associated with tumour subtypes in BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2. Breast Cancer Res. 2011;13:R110. doi: 10.1186/bcr3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Binder-Foucard F, Rasamimanana Cerf N, Belot A, Bossard N: Estimation nationale de l’incidence et de la mortalité par cancer en France entre 1980 et 2012. Étude à partir des registres des cancers du réseau Francim. Partie 1 – Tumeurs solides. Saint-Maurice (Fra): Institut de veille sanitaire; 2013. Available: http://www.invs.sante.fr/pmb/invs/(id)/PMB_11619. 2014.
- 23.Damiola F, Schultz I, Barjhoux L, Sornin V, Dondon MG, Eon-Marchais S, et al. Mutation analysis of PALB2 gene in French breast cancer families. Breast Cancer Res Treat. 2015;154(3):463–71. doi: 10.1007/s10549-015-3625-7. [DOI] [PubMed] [Google Scholar]
- 24.Blein S, Barjhoux L, GENESIS investigators. Damiola F, Dondon MG, Eon-Marchais S, et al. Targeted Sequencing of the Mitochondrial Genome of Women at High Risk of Breast Cancer without Detectable Mutations in BRCA1/2. PLoS One. 2015;10(9):e0136192. doi: 10.1371/journal.pone.0136192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterlongo P, Catucci I, Colombo M, Caleca L, Mucaki E, Bogliolo M, et al. FANCM c.5791C > T nonsense mutation (rs144567652) induces exon skipping, affects DNA repair activity and is a familial breast cancer risk factor. Hum Mol Genet. 2015;24(18):5345–55. doi: 10.1093/hmg/ddv251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.COMPLEXO. Southey MC, Park DJ, Nguyen-Dumont T, Campbell I, Thompson E, et al. COMPLEXO: identifying the missing heritability of breast cancer via next generation collaboration. Breast Cancer Res. 2013;15(3):402. doi: 10.1186/bcr3434. [DOI] [PMC free article] [PubMed] [Google Scholar]


