Abstract
Bacteria use a wide variety of methyl-accepting chemotaxis proteins (MCPs) to mediate their attraction to or repulsion from different chemical signals in their environment. The bioluminescent marine bacterium Vibrio fischeri is the monospecific symbiont of the Hawaiian bobtail squid, Euprymna scolopes, and encodes a large repertoire of MCPs that are hypothesized to be used during different parts of its complex, multistage lifestyle. Here, we report the initial characterization of two such MCPs from V. fischeri that are responsible for mediating migration toward short- and medium-chain aliphatic (or fatty) acids. These receptors appear to be distributed among only members of the family Vibrionaceae and are likely descended from a receptor that has been lost by the majority of the members of this family. While chemotaxis greatly enhances the efficiency of host colonization by V. fischeri, fatty acids do not appear to be used as a chemical cue during this stage of the symbiosis. This study presents an example of straight-chain fatty acid chemoattraction and contributes to the growing body of characterized MCP-ligand interactions.
INTRODUCTION
Flagellar motility in bacteria is a complex, tightly regulated process. Through alternating cycles of swimming and tumbling, a bacterium is able to navigate its environment, either to move away from unfavorable conditions or to migrate toward a more favorable habitat (e.g., toward nutrient sources) (1, 2). This directed motility is termed chemotaxis and is mediated by the CheAY two-component signal transduction pathway. These proteins alter the relative time for which the flagellum rotates counterclockwise (CCW) or clockwise (CW) and, in turn, how much time the bacterium spends swimming or tumbling, respectively. This change in rotational direction is determined by the phosphorylation state of CheY, which is mediated primarily by membrane-bound receptors known as methyl-accepting chemotaxis proteins (MCPs). MCPs are ligand-binding sensory proteins and are generally embedded in the inner membrane with their ligand-binding domain (LBD) extended into the periplasm and their signaling domains present in the cytosol. Upon ligand binding, the MCP undergoes a conformational change that allows methylation of the signaling domain, thereby permitting a sensory signal to be transmitted across the inner membrane.
Chemotaxis and MCPs have been most thoroughly studied in Escherichia coli, which encodes three to five distinct MCPs shown to be involved in the sensing of compounds such as amino acids, peptides, sugars, and electron acceptors (3–8). However, as the number of sequenced bacterial genomes grows, it has become apparent that many other bacteria contain much larger repertoires of MCPs (1). For example, the genome of Pseudomonas aeruginosa encodes 24 MCPs, that of Vibrio cholerae encodes 44, and that of Magnetospirillum magnetotacticum encodes the largest known set of 61 MCPs. Despite these large numbers of receptors, the number of known ligands is surprisingly small. Previous work has shown little correlation between genome size and the number of MCPs, but the major contributing factor appears to be the complexity of the bacterium's lifestyle. Vibrio fischeri, the mutualistic symbiont of the Hawaiian bobtail squid, Euprymna scolopes, has 43 predicted MCPs in its genome and a life cycle that alternates between planktonic and symbiotic states (9, 10). Previous work has shown that chemotaxis is important to the establishment of this symbiosis, though the specific receptors involved are as yet unknown (11–13). To date, only one of these MCPs has been well characterized, the amino acid sensor VfcA; nevertheless, chemoattraction to several other compounds has also been observed (9, 14). In this study, we have investigated whether there may be redundancy among this large pool of MCPs and identified a pair of paralogous receptors that appear to function in the sensing of short- and medium-chain fatty acids.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All of the strains and plasmids used in this study are listed in Table 1. All of the V. fischeri strains are derived of strain ES114 (15), and the E. coli strain used for cloning was DH5α λpir (16–18). The sequences of the primers used in this study are listed in Table 2. Deletion mutants and fluorescently labeled strains were constructed as described previously (17–20). V. fischeri cells were grown in Luria-Bertani salt (LBS) broth (10 g/liter tryptone, 5 g/liter yeast extract, 20 g/liter NaCl) or seawater-based tryptone (SWT) broth (5 g/liter tryptone, 3 g/liter yeast extract, 0.3% glycerol, 700 ml Instant Ocean [IO; Aquarium Systems, Mentor, OH, USA] at 33 to 35 ppt, 300 ml of water) as indicated, at 28°C with shaking. E. coli cells were grown in Luria-Bertani broth (10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter NaCl) at 37°C with shaking. Antibiotics were used at the following concentrations when appropriate: erythromycin at 5 mg/ml for V. fischeri and 150 mg/ml for E. coli and chloramphenicol at 2.5 mg/ml for V. fischeri and 25 mg/ml for E. coli.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| V. fischeri strains | ||
| MJM1100 | ES114, sequenced wild-type strain | 15 |
| MB08701 | ES114 cheA::Tnerm | 9 |
| KCN43 | ES114 ΔvfcB ΔvfcB2 | This study |
| KCN44 | ES114 ΔvfcB ΔvfcB2/pVS102 | This study |
| KCN45 | ES114 ΔvfcB ΔvfcB2/pVS208 | This study |
| KCN54 | ES114 ΔvfcB ΔvfcB2/pCAB103A | This study |
| KCN60 | ES114 ΔvfcB ΔvfcB2/pKN57 | This study |
| E. coli DH5α λpir | Cloning vector | 16 |
| Plasmids | ||
| pEVS104 | Conjugative helper plasmid, Kanr | 17 |
| pKV363 | Suicide vector for chromosomal deletions in V. fischeri | 20 |
| pKN42 | pKV363::ΔVF_A0448ΔVF_A0447 | This study |
| pVSV105 | pES213-based complementation vector, Cmr | 18 |
| pCAB103A | pVSV105::vfcB | This study |
| pKN57 | pVSV105::vfcB2 | This study |
| pVSV102 | GFP marker plasmid, Kanr | 18 |
| pVSV208 | RFP marker plasmid, Cmr | 18 |
TABLE 2.
Primers used in this study
| Primer | Description | Sequence (5′−3′) |
|---|---|---|
| A0448_H1_Fwd | Amplification of VF_A0448 upstream homology arm | TCA GCC GTC GAC CCC AAT TTG AGT CTT CAT TTT TTA TCA |
| A0448_H1_Rev | Amplification of VF_A0448 upstream homology arm | TCA GCC GCA TGC TTG CAT TAA AGC CTC TTA AAT TTG AGA |
| A0447_H2_Fwd | Amplification of VF_A0447 downstream homology arm | TCA GCC GCA TGC TTC TTT AAA TTA TAG TTG TTC TCT TAA TGA AAG |
| A0447_H2_Rev | Amplification of VF_A0447 downstream homology arm | TCA GCC ACT AGT AAG ATG GAC GGT ATG TCG |
| A0448_Con_Fwd | Confirmation primer for ΔvfcB ΔvfcB2 clean deletion strain | GAA GCC GTA TAT AAT CAT TTA ATG CCC |
| A0447_Con_Rev | Confirmation primer for ΔvfcB ΔvfcB2 clean deletion strain | TAT GAA AAA GAC AGC ATT AGC CAT |
| A0448_compUS_Sph1 | Amplification of VF_A0448 for insertion into pVSV105 complementation vector | GGG CCC GCA TGC CTA ACA CAC AGG AAA CAG CTA TGC AAT TTT CAT TAA AAA ATA CG |
| A0448_compDS_Kpn1 | Amplification of VF_A0448 for insertion into pVSV105 complementation vector | GGG CCC GGT ACC GCA GCA AGA GAT TAA CCT TG |
| A0447_compUS_Sph1 | Amplification of VF_A0447 for insertion into pVSV105 complementation vector | GGG CCC GCA TGC CTA ACA CAC AGG AAA CAG CTA TGC AAT TTT CAC TTA AAA ATA CGT CT |
| A0447_compDS_Kpn1 | Amplification of VF_A0447 for insertion into pVSV105 complementation vector | GGG CCC GGT ACC GCG CTA AGG AGG GTG G |
Bioinformatic analysis.
Complete and draft genomes with VfcB BLASTP hits were retrieved from the NCBI genome database. Ortholog group prediction was determined with orthoMCL (21). The resulting 62 single-copy gene ortholog clusters were obtained and individually aligned at the protein level with mafft-linsi (22) and then back-translated with backtranseq (23) and realigned at the nucleotide level with mafft-FFT-NS-i. Individual alignments were trimmed for sites with >50% gaps with trimAl (24) and concatenated for phylogeny reconstruction with RAxML (25) by using the GTRGAMMA model and 1,000 bootstrap pseudoreplicates. For the genomes used to construct this tree, together with the VfcB homolog accession numbers for each genome, see Table S1 in the supplemental material. In addition, for a complete list of the 62 proteins used for the phylogeny reconstruction and the respective accession numbers, see Table S2 in the supplemental material.
To determine whether the distribution of VfcB homologs within members of the family Vibrionaceae is most likely due to horizontal gene transfer events and/or multiple gene losses within the family, a VfcB homolog phylogeny reconstruction was performed by the procedure described above. In this case, gene sequences were extracted from the genomes retrieved for the whole-genome phylogeny reconstruction (see Table S1 in the supplemental material). We used the codeML program of PAML (26) with the codon-aligned nucleotide sequences to infer the dN/dS ratio (ratio of nonsynonymous to synonymous evolutionary changes [or substitutions]) for each pair of sequences. In addition, the N- and C-terminal portions of the sequence were analyzed separately to compare differences between the amino acid substitution rates in these two regions of the protein.
Capillary chemotaxis assay.
Cultures from single bacterial colonies were grown overnight in a 28°C incubator shaker in LBS liquid medium, subsequently diluted 1:100 into SWT liquid medium, and grown until early log phase (optical density [OD] at 600 nm of ∼0.3 to 0.5). Two milliliters was harvested from the SWT broth culture at an OD of 0.3 by centrifugation (5 min at a relative centrifugal force of 850). Cells were resuspended in 2 ml of buffered artificial seawater (ASW; 100 mM MgSO4, 20 mM CaCl2, 20 mM KCl, 400 mM NaCl, 50 mM HEPES, pH 7.5) as previously described (9, 27). All chemoattractants were prepared in Nanopure (EMD Millipore, Billerica, MA, USA) water, which was added to 1.25× ASW to create a final 1 mM solution in 1× ASW. One-microliter capillary tubes (Drummond Scientific, Broomall, PA) previously sealed at one end were heated and then introduced into the microcentrifuge tubes containing the 1 mM chemoattractant solution. After rinsing of the capillary tubes with ASW, tubes were inserted into the culture resuspensions and incubated horizontally for 5 min at room temperature (∼24°C). The capillary tubes were removed and rinsed with ASW, and the contents were expelled into 150 μl of ASW. The tube contents were serially diluted with ASW and plated onto LBS for colony counting to determine total number of CFU that had entered the capillary.
Squid colonization assay.
Freshly hatched juvenile squid were exposed to a 1:1 ratio of the indicated green fluorescent protein (GFP)- or red fluorescent protein (RFP)-labeled strains of V. fischeri (∼1,500 total CFU/ml) for 3 h in filtered IO (FIO; 0.2-μm-pore-size filter). Squid were then transferred to individual vials containing 4 ml of fresh, bacterium-free FIO and stored for an additional 18 to 21 h. Colonized squid were anesthetized on ice and then surface sterilized by placement at −80°C. Individual squid were then homogenized and plated on LBS agar plates as described previously (28), and the number of colonies each strain present in the light organ population was calculated by counting the differently fluorescing colonies with a Leica MZFLIII fluorescent dissection scope. The relative competitive index (RCI) of the two cocolonizing strains was calculated as follows: RCI = log(CFU mutant/CFU wild type)/(inoculum CFU mutant/inoculum CFU wild type).
RESULTS
Predicted redundancy of V. fischeri MCPs.
Because V. fischeri encodes 43 predicted MCPs within its genome, we hypothesized that some of these MCPs might show redundant functionality. To address this theory, we performed a bioinformatic analysis of the N-terminal region of each MCP with the BLASTP search algorithm (29). The N-terminal region encompasses the predicted LBD of this protein family. The LBD for each MCP was defined as N terminal to either the aligned HAMP domain, when present, or, alternatively, the predicted periplasmic sequence based upon the presence of projected transmembrane helix domains. Each N-terminal domain was then used as a query against the V. fischeri ES114 genome, and MCPs that aligned with an e score of <1 × 10−9 were labeled as potentially having redundant or similar ligand-binding characteristics (Table 3).
TABLE 3.
Predicted homologous classes of V. fischeri MCPsa
| Class | MCP 1 | MCP 2 | MCP 3 | MCP 4 | MCP 5 | Median e score | Predicted ligand |
|---|---|---|---|---|---|---|---|
| I | VF_0698 | VF_1092 | 1.50E-16 | Nitric oxide | |||
| II | VF_A0246 | VF_A0300 | VF_A0302 | 1.04E-13 | Unknown | ||
| III | VF_0777b | VF_1369 | VF_A0107 | VF_A0389 | VF_A0865 | 1.50E-19 | Amino acids |
| IV | VF_1117 | VF_1652 | VF_A0859 | VF_A1084 | 5.00E-21 | Unknown | |
| V | VF_0827 | VF_2161 | 3.50E-19 | Unknown | |||
| VI | VF_1091 | VF_A0528 | VF_A1072 | 1.50E-67 | Unknown | ||
| VII | VF_A0169 | VF_A0170 | 1.04E-85 | Unknown | |||
| VIII | VF_A0447c | VF_A0448d | 3.20E-74 | See text | |||
| IX | VF_A0527 | VF_A1073 | 2.20E-11 | Unknown | |||
| X | VF_A0092 | VF_A1071 | 2.50E-39 | None (aerotaxis) | |||
| Unique | VF_0872, VF_1618, VF_A0481 | VF_0987, VF_1778, VF_A0677 | VF_1133, VF_1789, VF_A0891 | VF_1138, VF_2042, VF_A1069 | VF_1503, VF_2236 | VF_1504, VF_A0325 | Unknown |
V. fischeri contains 43 predicted MCPs, and 27 of these are distributed into 10 classes that contain putative homologous LBDs. Sixteen MCPs were identified that showed no homology to other LBDs and had no predicted ligands. These genes were VF_0872, VF_0987, VF_1133, VF_1138, VF_1503, VF_1504, VF_1618, VF_1778, VF_1789, VF_2042, VF_2236, VF_A0325, VF_A0481, VF_A0677, VF_A0891, and VF_A1069. The 10 classes contain up to five MCPs each. Some of these classes have predicted ligands based on homologs in other species. The previously characterized amino acid-sensing MCP VfcA (class III) has four predicted paralogs, while the fatty acid-sensing MCPs described here, VF_A0447 and VF_A0448 (class VIII), show similarity only to each other.
Encoded by vfcA.
Encoded by vfcB2.
Encoded by vfcB.
On the basis of this criterion, many of these MCPs are candidates for having (i) a recent shared ancestry and/or (ii) similar functionality. Of the 43 MCPs investigated, only 16 appear to be unique within the genome, while 27 can be segregated into 10 putative classes. The number of members of each of these classes ranges from two to five MCPs and includes four predicted paralogs to the previously characterized VfcA amino acid receptor (9). The results of this grouping are not fully consistent with a previously reported Pfam domain structure analysis (9). However, such an outcome is not unexpected on the basis of differences in both the assumptions used to define protein domains and the algorithms employed. We believe that the BLASTP alignment analysis reported here provides a more clearly resolved picture of the possible paralogs present in V. fischeri and that recognition of these paralog classes will help prevent confounding results in future MCP characterization studies.
VfcB and VfcB2 mediate fatty acid chemotaxis.
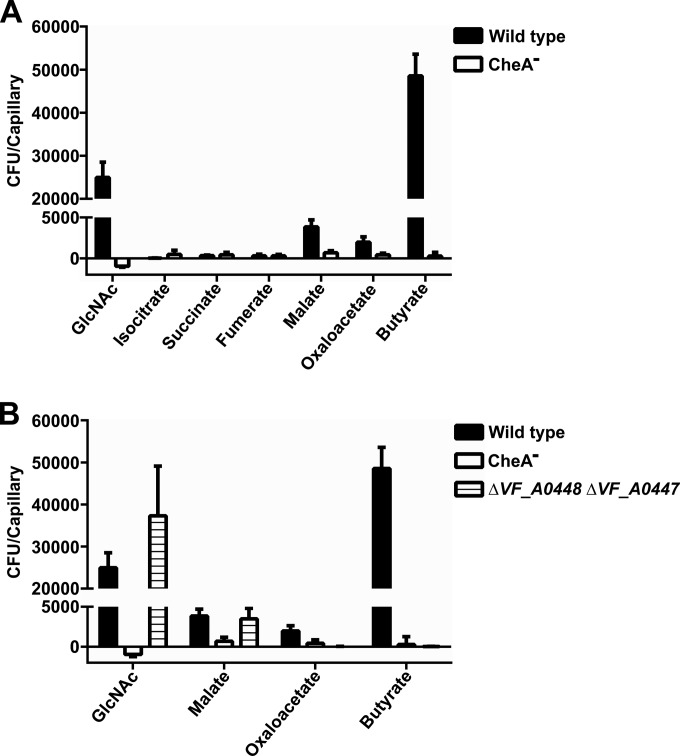
Previous work with Pseudomonas putida McpS and Comamonas testosteroni Mcp2201 identified MCPs responsible for the sensing of trichloroacetic acid (TCA) cycle intermediates (30, 31). We asked whether MCPs from V. fischeri might exhibit similar behavior and looked for chemoattraction to several of the compounds reported as attractants for P. putida (Fig. 1A). Of the compounds tested, we noted only weak attraction to malate and oxaloacetate for V. fischeri, both of which were reported as strong attractants of P. putida. However, we observed a strong attraction to butyrate, a relatively weak attractant of P. putida. Initial screening of MCP mutants previously generated in our lab revealed that a transposon mutant with VF_A0448 interrupted showed a complete loss of attraction to butyrate but only a partial reduction in attraction to malate and oxaloacetate (data not shown).
FIG 1.
Chemoattraction to metabolite ligands. V. fischeri strains were tested for chemoattraction to several TCA cycle intermediates (1 mM) previously described in the characterization of McpS from P. putida. Results are the mean ± the standard error of the mean of at least three independent experiments. (A) V. fischeri showed significant attraction only to malate, oxaloacetate, and butyrate. N-Acetylglucosamine (GlcNAc) is a known but structurally unrelated chemoattractant of V. fischeri, and a CheA mutant serves as a negative control. (B) Deletion of the VF_A0448 and VF_A0447 genes eliminated detectable chemoattraction to butyrate and oxaloacetate but did not impact attraction to either malate or GlcNAc.
On the basis of this partial reduction, we asked whether VF_A0448 might have a paralogous receptor. According to our bioinformatic analysis, VF_A0448 had one predicted paralog: VF_A0447; the LBDs of these two proteins show 61.5% DNA identity and 52.6% amino acid identity (32). To determine whether VF_A0447 also contributes to the sensing of malate and oxaloacetate, we attempted to construct independent in-frame VF_A0447 and VF_A0448 deletion mutants. However, because of their close proximity and significant sequence similarity to other regions of the genome immediately upstream and downstream, we were only able to construct a deletion of the genes encoding both chemoreceptors. The resulting double-deletion mutant (ΔA0448 ΔA0447) showed no observable growth phenotype (see Fig. S1 in the supplemental material). We then assessed the ability of this double mutant to sense the compounds of interest and observed the elimination of any response to either butyrate or oxaloacetate; however, chemoattraction to malate was not significantly affected (Fig. 1B).
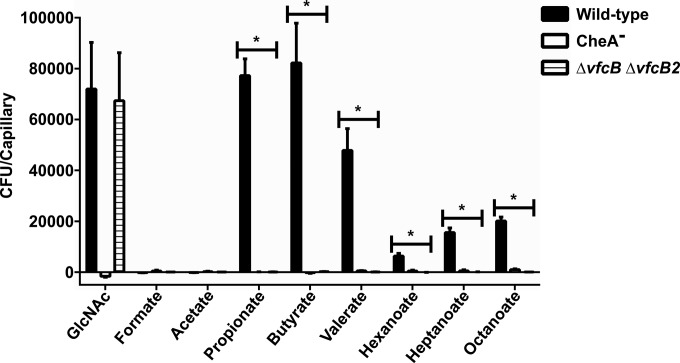
To determine the extent of ligand specificity of these two MCPs, we looked for chemoattraction to compounds structurally similar to butyrate. We assessed the ability of V. fischeri to move toward straight-chain fatty acids of various lengths, from formate to octanoate (Fig. 2; see Fig. S2 in the supplemental material). There was a strong attraction to fatty acids with carbon chain lengths of ≥3 but no evidence of chemoattraction to either formate or acetate. Maximum sensitivity to propionate, valerate, and butyrate was observed, with lower sensitivity to hexanoate, heptanoate, and octanoate. In all cases, the ΔA0448 ΔA0447 mutant showed a complete elimination of fatty acid attraction, with the exception that a low-level chemotactic response to propionate was observed at the highest concentration tested. The observed attraction to oxaloacetate was also eliminated within the double-mutant background. However, oxaloacetate is dissimilar in structure to the other ligands and is known to undergo decarboxylation to pyruvate, which is structurally similar to propionate, under aqueous conditions (33). Pyruvate was also found to act as an attractant specific to these receptors (see Fig. S3 in the supplemental material).
FIG 2.
VfcB and VfcB2 are MCP receptors for C3 to C8 fatty acids. V. fischeri showed strong attraction to fatty acids (1 mM) having a minimum backbone length of three carbons (C3). Propionate (C3), butyrate (C4), and valerate (C5) provided the strongest responses; attraction to hexanoate (C6), heptanoate (C7), and octanoate (C8) was less but significantly above the background (*, significantly different by t test with a false-discovery rate of 1%). Deletion of both the vfcB and vfcB2 genes completely eliminated chemoattraction to all of these compounds but GlcNAc. Results are the mean ± the standard error of the mean of at least three independent experiments.
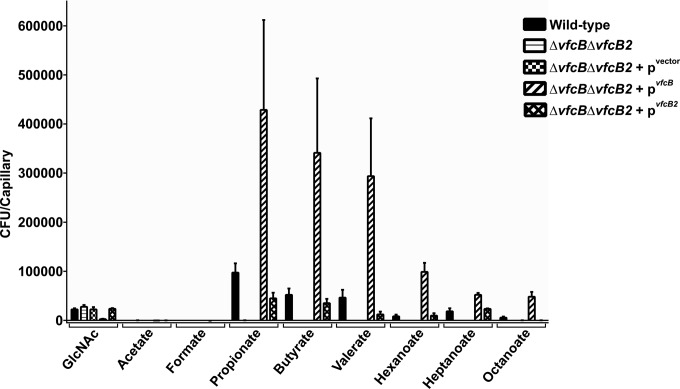
In lieu of the single mutants, we constructed complementation plasmids to individually express each receptor endogenously and asked whether they were able to restore fatty acid chemotaxis in the double-mutant background (Fig. 3; see Fig. S3 in the supplemental material). Complementation with the gene encoding either MCP successfully restored the chemotactic response to the short- and medium-chain-length fatty acids tested. On the basis of this similarity in ligand specificity, we have named VF_A0448 and VF_A0447 V. fischeri chemotaxis protein B (VfcB) and V. fischeri chemotaxis protein B2 (VfcB2), respectively. Interestingly, the two complemented strains did not show the same sensitivity to the different ligands and VfcB2 was unable to restore detectable chemotaxis to octanoate.
FIG 3.
Chemoattraction to fatty acids is restored by genetic complementation. The ΔvfcB ΔvfcB2 double mutant could be complemented by a plasmid endogenously expressing either VfcB (pvfcB) or VfcB2 (pvfcB2). For all of the fatty acids tested, the ΔvfcB and ΔvfcB2 mutations eliminated chemoattraction, and carriage of either vfcB or vfcB2 was able to restore responsiveness, with the exception that vfcB2 did not restore responsiveness to octanoate. A plasmid vector control (pvector) showed no restoration of chemoattraction to any of the ligands tested. Results are the mean ± the standard error of the mean of at least three independent experiments.
The VfcB-type receptor is found only within the family Vibrionaceae.
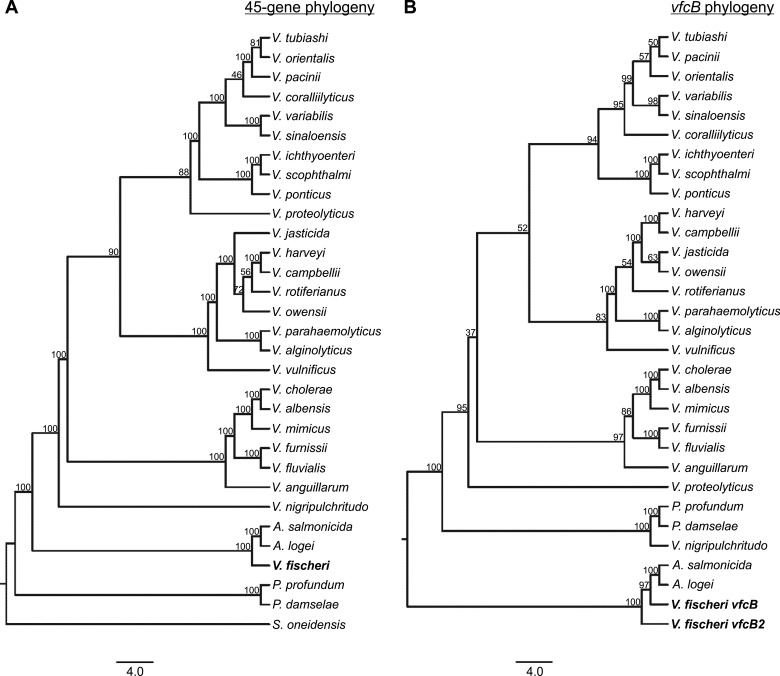
To determine the phylogenetic distribution of this receptor type, we used the N-terminal domain of VfcB as a query sequence and searched the NCBI nonredundant protein database with the BLASTP algorithm. To remain conservative in our identification of VfcB-type MCPs that might have similar functions, we used a cutoff of 75% query coverage and 40% sequence identity. Within these parameters, we were able to identify closely related MCPs within 30 different species (Fig. 4A). The species carrying these MCPs are strictly within the family Vibrionaceae. As there are currently 142 characterized species within the family Vibrionaceae (34), these putative fatty acid receptors are found in only approximately 20% of the species within this family. In addition, within two of these species (V. fischeri and Vibrio harveyi), the number of similar homologous receptors appears to vary between different strains. While the majority of bacterial species contain only one identifiable homolog, strains of V. fischeri carry up to three, and one strain of V. harveyi encoded two.
FIG 4.
Phylogeny of the VfcB MCP LBD. Thirty bacterial species encoded predicted receptor proteins homologous to VfcB, and all of these were within the family Vibrionaceae. (A) A phylogenetic tree of those species that encoded a predicted VfcB homologue was constructed on the basis of the relatedness of 62 core genes in their genomes. These species are broadly distributed throughout a complete phylogenetic tree of the family Vibrionaceae. S. oneidensis is included as an outgroup and does not contain a predicted VfcB homologue. (B) A second tree was constructed by using only the sequences of the predicted vfcB homologues from each species. Compared to the tree in panel A, the second tree shows a high degree of reconstruction of the terminal nodes, consistent with a single acquisition of this gene in a common ancestor of the family Vibrionaceae. The vfcB gene from V. fischeri clusters more closely to those of Aliivibrio salmonicida and Aliivibrio logei than to vfcB2, indicating that vfcB is most likely the ancestral copy. In both trees, bootstrap values from 1,000 pseudoreplicates are indicated.
As this receptor type was observed only in some of the members of the family Vibrionaceae, we next asked whether it was more likely that this gene had been (i) horizontally transferred repeatedly or (ii) lost by most of the members of this family over time. To address this question, we constructed a phylogenetic tree of the VfcB-containing species by using only the genes encoding putative VfcB homologs (Fig. 4B). The initial tree was constructed by using 62 genes from the most complete genomes available for each of the species indicated, and we found that the majority of the clustering was retained in the vfcB homolog tree. The most parsimonious explanation of this result is that these genes were originally derived from an early common ancestor within the family Vibrionaceae but that a large majority (80%) of the species had lost these receptors over time. Furthermore, calculating the dN/dS ratios of (i) the full sequences of VfcB2 and VfcB and the corresponding (ii) N-terminal and (iii) C-terminal regions yielded ratios of (i) 0.3373, (ii) 0.1130, and (iii) 0.0483. Because these values are all <1, both of these paralogs appear to be under negative selection pressure. However, the functional constraint appears to be less strong within the N-terminal LBD, for which the dN/dS ratio is more than twice that of the C-terminal domain. Combined with the closer clustering of vfcB to homologs from the other Aliivibrio clade species (Fig. 4B), we predict that vfcB, rather than vfcB2, is most closely related to the ancestral gene.
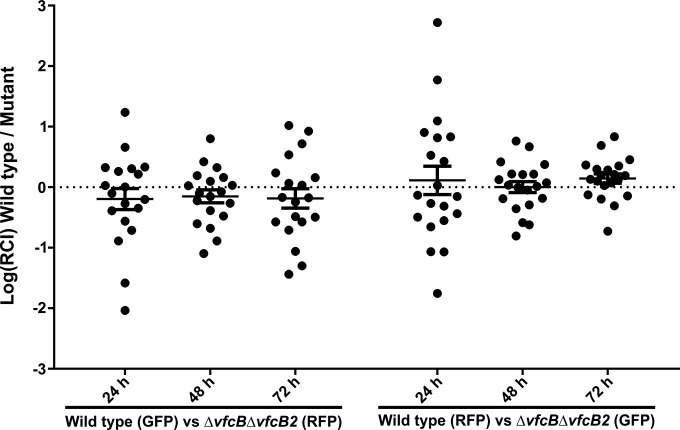
VfcB and VfcB2 are not critical to early host colonization.
V. fischeri is the monospecific symbiont of the Hawaiian bobtail squid, E. scolopes. Prior studies have shown that chemotaxis plays a significant role during the initial stages of host colonization (12), and thus, we asked whether fatty acids secreted from the host light organ might serve as a chemical cue for the bacteria during their migration into host tissues. However, we found no difference in the abilities of the wild-type strain and the ΔVfcB ΔVfcB2 mutant strain to initiate colonization of the host (Fig. 5). Other studies, most notably with a Δlux mutant, have shown that mutant backgrounds with no initiation phenotype can, instead, fail to persist in the host (35). Nevertheless, when we assessed the persistence of the ΔVfcB ΔVfcB2 mutant strain over the first 72 h postinoculation, we still found no phenotypic difference from the wild type. These results indicate that fatty acid chemotaxis is likely not involved during the early stages of host colonization in the squid-vibrio symbiosis.
FIG 5.
VfcB and VfcB2 do not have a significant role in early host colonization. Strains carrying either a GFP or an RFP marker were used to coinoculate aposymbiotic juvenile E. scolopes. At each time indicated, the ratio of the two strains colonizing the organ was determined. The experiment was repeated with the markers switched between the strains to control for possible differential effects on colonization by the carriage of either fluorescent protein marker. No significant phenotypic difference from the wild type was observed in strains deficient in fatty acid chemoattraction, even up to 3 days postcolonization, indicating that the ΔvfcB ΔvfcB2 mutant strain is able to establish a robust and persistent symbiosis.
DISCUSSION
We have presented an initial characterization of a pair of MCPs from V. fischeri that are responsible for sensing and mediating chemoattraction to straight-chain fatty acids. Other studies have characterized MCPs responsible for attraction to acetate and propionate (36) or butyrate (30) but not for chemotaxis to the entire set of C3 through C8 fatty acid ligands reported here. This work serves to introduce a previously undescribed class of chemoreceptors, represented by VfcB and VfcB2 in V. fischeri, a species with a wide array of putative receptors. The LBDs of these two MCPs are noteworthy, as they exhibited no similarity to previously characterized domains either by Pfam analysis or by structural predictions with phyre2 (9, 37, 38). Prior studies have shown chemoattraction of V. fischeri to nucleotides, amino acids, and chitin-associated sugars, though to date, only amino acid sensing has been associated with a specific MCP (VfcA) (9, 14). We also investigated both the distribution and possible ecological role of these receptors in the context of the squid-vibrio symbiosis model system, thereby illustrating the ways in which bacteria with complex environmental lifestyles use chemotaxis under different conditions.
While examining the response of a ΔVfcB ΔVfcB2 double mutant of V. fischeri to organic acids, we discovered that wild-type cells are attracted to short- and medium-chain-length fatty acids with backbone lengths of C3 to C8. The most robust attraction was to one of three common fermentation products, i.e., propionate, valerate, and butyrate. Fatty acids longer than C8 were not assayed because of their poor solubility in aqueous solution. To confirm that both VfcB and VfcB2 were participating in the chemotactic response, we complemented each MCP independently on a constitutively expressing plasmid. Expressing either MCP restored responsiveness to the fatty acids tested in the double-mutant background, albeit to different levels. Furthermore, the VfcB2 receptor was unable to restore a detectable response to octanoate. It is possible that this difference is simply a result of different expression levels of the two MCPs when each is encoded on a plasmid. However, on the basis of the sequence differences and the high dN/dS ratio of these two receptors, we believe the more parsimonious explanation is that VfcB2 is undergoing genetic drift, perhaps reducing its sensitivity to fatty acids. While we attempted to purify the LBDs of these receptors for in vitro studies, they were both difficult to overexpress in E. coli and purify at a sufficient concentration; in addition, having purified them, we were unable to observe any evidence of specific binding by using isothermal titration calorimetry under a variety of conditions. It is possible that the relevant domain of these proteins requires either a V. fischeri-specific chaperone or a membrane integration step for proper folding. A second possibility is that these receptors require a secondary binding partner for ligand recognition, as was described by Hegde et al. (39) for the E. coli Tsr receptor's sensing of the AI-2 quorum-signaling molecule, or as occurs with chemotaxis toward ribose and galactose in E. coli (8). Further studies are needed to clarify the underlying reason for this difficulty.
In a previous Pfam analysis of the MCP repertoire of V. fischeri, the classification of these receptors by using recognized domain structures illustrated a strong likelihood of redundancy among these proteins (9). However, only a few of the large number of LBDs predicted among bacterial species have been functionally characterized. Thus, many of the V. fischeri MCPs grouped together as unknowns. In the present study, we attempted to disambiguate these receptors by comparing them against each other by using the BLASTP search algorithm and again found a high degree of apparent relatedness and, perhaps, redundancy among the 43 predicted MCPs in V. fischeri (Table 3). Overall, the BLASTP analysis predicted a greater number of redundant “classes” of MCPs than previously identified (9), with each class containing fewer individual receptors predicted to sense similar ligands. As additional MCP functional domains become better understood, both types of analysis will benefit from further refinement. As presented in this work, the BLASTP type analysis was able to predict the similar ligand-binding profiles of VfcB and VfcB2; however, this prediction does not rule out the possibility that other, unrelated, MCPs with similar functions will be found in the future, even within V. fischeri. It is also important to note that this method of predicting functional redundancy must be validated experimentally. While the amino acid sequence analysis used in this study was able to predict functional similarity between the VfcB and VfcB2 receptors, readers should be cautious about assuming that an amino acid sequence is sufficient for predicting similar functions. Some chemoreceptors (e.g., four-helix bundle receptors) may show a high degree of sequence similarity while mediating attraction to distinct sets of ligands.
Chemotaxis and motility play an integral role in initiating the colonization of the best-understood environmental niche of V. fischeri, the light organ of its eukaryotic host, E. scolopes (9, 11–13). These two partners form a monospecific mutualistic symbiosis wherein the squid provides its symbiont with nutrients and V. fischeri produces bioluminescence to provide counterillumination for the host (10). At each host generation, the symbionts are recruited from the bacterioplankton in part by chemotactic cues to host-derived chitobiose (12, 40). As the association matures, symbionts in the adult squid's light organ are directly incorporated into their cell membrane's long-chain (i.e., C20:4, C22:6) fatty acids provided by the host (41). Similarly, another study by Giles et al. showed that V. cholerae also incorporates into its cell membrane exogenous long-chain fatty acids derived either from the infected host or from marine sediment (42). In light of the importance of chemotaxis and fatty acids to the initiation and persistence of the symbiosis, we asked whether fatty acids presented by the host might be sensed by VfcB and VfcB2 as a chemotactic cue during the early stages of symbiotic colonization (Fig. 5). However, we found no evidence that a mutant deficient in the fatty acid-sensing MCPs had any colonization phenotype, at least during the first 72 h postcolonization. Thus, while it is unlikely that fatty acids are used to attract bacterial cells to the host light organ, these receptors might still play a role later in the symbiosis or there may be other MCPs that sense longer-chain fatty acids. Unfortunately, the currently available transcriptional data sets were unable to provide us with significant evidence as to their regulation.
Because there is evidence of a VfcB homolog in V. cholerae and other Vibrio species (Fig. 4A), we hypothesize that this class of receptors may have evolved to allow certain members of the family Vibrionaceae to find within their free-living or host-associated environments sources of fatty acids that are subsequently used in either anabolic or catabolic metabolism. While lipids are clearly enriched in animal tissues, the concentrations of long-chain fatty acids and neutral lipids in the ocean are quite low (43). Nevertheless, even in such a nutrient-poor environment, there is likely to be a selective advantage to sensing and migrating toward physiologically useful compounds such as fatty acids.
VfcB and VfcB2 have a limited phylogenetic distribution, with possible homologous receptors restricted to the family Vibrionaceae (Fig. 4A). Furthermore, even within the family Vibrionaceae, this MCP class was present in only 30 of the 142 described species and almost all of these contained only a single ortholog. The congruence between a VfcB phylogenetic tree and a cladogram based on 62 other genes shared within the family Vibrionaceae (Fig. 4B) suggests that (i) this receptor arose once in a common ancestor and was subsequently lost in most modern species, and (ii) the ecological lifestyles of those species that have retained this class of MCPs benefit from fatty acid chemoattraction. The calculated dN/dS ratio of the encoded domains of VfcB and VfcB2 also indicates that both copies are under purifying selective pressure in V. fischeri and that relaxation of its function may have resulted in the DNA sequence variation found among the vfcB family members. It will be interesting to see whether this variation in the N-terminal region of the LBD might correspond to selection for a new or modified class of compounds. In addition, when considering the full-length protein, there is a difference in the frequency at which synonymous and nonsynonymous substitutions occur on both regions, perhaps because of asynchronous selective pressures. This difference suggests one explanation for why, following the presumed vfcB/vfcB2 duplication event, there is no indication that one copy is becoming a pseudogene. Because multiple copies are found almost exclusively within V. fischeri strains (two in strain ES114 and three in strain MJ11 [44]), it will be interesting to see whether this trend continues to hold true as further genomic sequences of this species become available. If it does, we wonder why V. fischeri retains multiple copies of a fatty acid chemoreceptor and whether they are differentially regulated as V. fischeri moves between its planktonic and symbiotic niches (45). Further characterization of the putative homologs from other members of the family Vibrionaceae will also identify the range of fatty acids (or other compounds) these bacteria can sense as chemoattractants.
This work presents the initial characterization of a novel pair of chemoreceptors that we have demonstrated are responsible for sensing and mediating chemoattraction to short- and medium-chain fatty acids. These also represent the second and third characterized MCPs from V. fischeri, out of its total repertoire of 43. As the genomes of environmental bacteria typically have similarly large repertoires of MCPs, the approach we present here for bioinformatically sorting them will provide a way for others to avoid confounding results from the presence of functionally similar proteins. In addition, future studies using V. fischeri will both help expand the current body of knowledge regarding MCP ligand specificity and validate and refine our ability to predict homologs among receptor families.
ACKNOWLEDGMENTS
We thank Eva Ziegelhoffer, John Parkinson, and the Ruby and McFall-Ngai laboratories for contributive discussions.
This work was funded by NIH grants RR12294/OD11024 (to E.G.R. and Margaret McFall-Ngai) and AI050661 (to Margaret McFall-Ngai). K.N. was supported by a Ruth L. Kirschstein National Research Service award from the NIGMS (F32GM112214).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02856-15.
REFERENCES
- 1.Krell T, Lacal J, Munoz-Martinez F, Reyes-Darias JA, Cadirci BH, García-Fontana C, Ramos JL. 2011. Diversity at its best: bacterial taxis. Environ Microbiol 13:1115–1124. doi: 10.1111/j.1462-2920.2010.02383.x. [DOI] [PubMed] [Google Scholar]
- 2.Sourjik V, Wingreen NS. 2012. Responding to chemical gradients: bacterial chemotaxis. Curr Opin Cell Biol 24:262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parkinson JS, Hazelbauer GL, Falke JJ. 2015. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol 23:257–266. doi: 10.1016/j.tim.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bibikov SI, Biran R, Rudd KE, Parkinson JS. 1997. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol 179:4075–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hazelbauer GL, Mesibov RE, Adler J. 1969. Escherichia coli mutants defective in chemotaxis toward specific chemicals. Proc Natl Acad Sci U S A 64:1300–1307. doi: 10.1073/pnas.64.4.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Springer MS, Goy MF, Adler J. 1977. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci U S A 74:3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manson MD, Blank V, Brade G, Higgins CF. 1986. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature 321:253–256. doi: 10.1038/321253a0. [DOI] [PubMed] [Google Scholar]
- 8.Kondoh H, Ball CB, Adler J. 1979. Identification of a methyl-accepting chemotaxis protein for the ribose and galactose chemoreceptors of Escherichia coli. Proc Natl Acad Sci U S A 76:260–264. doi: 10.1073/pnas.76.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennan CA, DeLoney-Marino CR, Mandel MJ. 2013. Chemoreceptor VfcA mediates amino acid chemotaxis in Vibrio fischeri. Appl Environ Microbiol 79:1889–1896. doi: 10.1128/AEM.03794-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nyholm SV, McFall-Ngai MJ. 2004. The winnowing: establishing the squid-vibrio symbiosis. Nat Rev Microbiol 2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 11.Brennan CA, Mandel MJ, Gyllborg MC, Thomasgard KA, Ruby EG. 2013. Genetic determinants of swimming motility in the squid light-organ symbiont Vibrio fischeri. Microbiologyopen 2:576–594. doi: 10.1002/mbo3.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandel MJ, Schaefer AL, Brennan CA, Heath-Heckman EA, Deloney-Marino CR, McFall-Ngai MJ, Ruby EG. 2012. Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Appl Environ Microbiol 78:4620–4626. doi: 10.1128/AEM.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deloney-Marino CR, Visick KL. 2012. Role for cheR of Vibrio fischeri in the Vibrio-squid symbiosis. Can J Microbiol 58:29–38. doi: 10.1139/w11-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLoney-Marino CR, Wolfe AJ, Visick KL. 2003. Chemoattraction of Vibrio fischeri to serine, nucleosides, and N-acetylneuraminic acid, a component of squid light-organ mucus. Appl Environ Microbiol 69:7527–7530. doi: 10.1128/AEM.69.12.7527-7530.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boettcher KJ, Ruby EG. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol 172:3701–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 17.Stabb EV, Ruby EG. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol 358:413–426. doi: 10.1016/S0076-6879(02)58106-4. [DOI] [PubMed] [Google Scholar]
- 18.Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol 72:802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Roux F, Binesse J, Saulnier D, Mazel D. 2007. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl Environ Microbiol 73:777–784. doi: 10.1128/AEM.02147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibata S, Visick KL. 2012. Sensor kinase RscS induces the production of antigenically distinct outer membrane vesicles that depend on the symbiosis polysaccharide locus in Vibrio fischeri. J Bacteriol 194:185–194. doi: 10.1128/JB.05926-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer S, Brunk BP, Chen F, Gao X, Harb OS, Iodice JB, Shanmugam D, Roos DS, Stoeckert CJ Jr. 2011. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics Chapter 6:Unit 6.12.11-19. doi: 10.1002/0471250953.bi0612s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 24.Capella-Gutiérrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu B, Yang Z. 2013. PAMLX: a graphical user interface for PAML. Mol Biol Evol 30:2723–2724. doi: 10.1093/molbev/mst179. [DOI] [PubMed] [Google Scholar]
- 27.Ruby EG, Nealson KH. 1976. Symbiotic association of Photobacterium fischeri with the marine luminous fish Monocentris japonica; a model of symbiosis based on bacterial studies. Biol Bull 151:574–586. doi: 10.2307/1540507. [DOI] [PubMed] [Google Scholar]
- 28.Naughton LM, Mandel MJ. 2012. Colonization of Euprymna scolopes squid by Vibrio fischeri. J Vis Exp 61:e3758. doi: 10.3791/3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Lacal J, Alfonso C, Liu X, Parales RE, Morel B, Conejero-Lara F, Rivas G, Duque E, Ramos JL, Krell T. 2010. Identification of a chemoreceptor for tricarboxylic acid cycle intermediates: differential chemotactic response towards receptor ligands. J Biol Chem 285:23126–23136. doi: 10.1074/jbc.M110.110403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni B, Huang Z, Fan Z, Jiang CY, Liu SJ. 2013. Comamonas testosteroni uses a chemoreceptor for tricarboxylic acid cycle intermediates to trigger chemotactic responses towards aromatic compounds. Mol Microbiol 90:813–823. doi: 10.1111/mmi.12400. [DOI] [PubMed] [Google Scholar]
- 32.Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S, Park YM, Buso N, Lopez R. 2015. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 43:W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfenden R, Lewis CA Jr, Yuan Y. 2011. Kinetic challenges facing oxalate, malonate, acetoacetate, and oxaloacetate decarboxylases. J Am Chem Soc 133:5683–5685. doi: 10.1021/ja111457h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawabe T, Ogura Y, Matsumura Y, Feng G, Amin AR, Mino S, Nakagawa S, Sawabe T, Kumar R, Fukui Y, Satomi M, Matsushima R, Thompson FL, Gomez-Gil B, Christen R, Maruyama F, Kurokawa K, Hayashi T. 2013. Updating the Vibrio clades defined by multilocus sequence phylogeny: proposal of eight new clades, and the description of Vibrio tritonius sp. nov. Front Microbiol 4:414. doi: 10.3389/fmicb.2013.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lupp C, Urbanowski M, Greenberg EP, Ruby EG. 2003. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol Microbiol 50:319–331. doi: 10.1046/j.1365-2958.2003.t01-1-03585.x. [DOI] [PubMed] [Google Scholar]
- 36.García V, Reyes-Darias JA, Martin-Mora D, Morel B, Matilla MA, Krell T. 2015. Identification of a chemoreceptor for C2 and C3 carboxylic acids. Appl Environ Microbiol 81:5449–5457. doi: 10.1128/AEM.01529-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonnhammer EL, Eddy SR, Durbin R. 1997. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28:405–420. doi:. [DOI] [PubMed] [Google Scholar]
- 38.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hegde M, Englert DL, Schrock S, Cohn WB, Vogt C, Wood TK, Manson MD, Jayaraman A. 2011. Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein. J Bacteriol 193:768–773. doi: 10.1128/JB.01196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kremer N, Philipp EE, Carpentier MC, Brennan CA, Kraemer L, Altura MA, Augustin R, Hasler R, Heath-Heckman EA, Peyer SM, Schwartzman J, Rader BA, Ruby EG, Rosenstiel P, McFall-Ngai MJ. 2013. Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe 14:183–194. doi: 10.1016/j.chom.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiasse RP, Schaefer AL, Koroleva I, Splinter-Bondurant S, Brown B, Manzella L, Snir E, Almabrazi H, Scheetz TE, Bonaldo Mde F, Casavant TL, Soares MB, Cronan JE, Reed JL, Ruby EG, McFall-Ngai MJ. 2010. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci U S A 107:2259–2264. doi: 10.1073/pnas.0909712107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giles DK, Hankins JV, Guan Z, Trent MS. 2011. Remodelling of the Vibrio cholerae membrane by incorporation of exogenous fatty acids from host and aquatic environments. Mol Microbiol 79:716–728. doi: 10.1111/j.1365-2958.2010.07476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wakeham SG, Hedges JI, Lee C, Peterson ML, Hernes PJ. 1997. Composition and transport of lipid biomarkers through the water column and surficial sediments of the equatorial Pacific Ocean. Deep Sea Res Part 2 Top Stud Oceanogr 44:2131–2162. [Google Scholar]
- 44.Mandel MJ, Stabb EV, Ruby EG. 2008. Comparative genomics-based investigation of resequencing targets in Vibrio fischeri: focus on point miscalls and artefactual expansions. BMC Genomics 9:138. doi: 10.1186/1471-2164-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wollenberg MS, Ruby EG. 2012. Phylogeny and fitness of Vibrio fischeri from the light organs of Euprymna scolopes in two Oahu, Hawaii populations. ISME J 6:352–362. doi: 10.1038/ismej.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wollenberg MS, Preheim SP, Polz MF, Ruby EG. 2012. Polyphyly of non-bioluminescent Vibrio fischeri sharing a lux-locus deletion. Environ Microbiol 14:655–668. doi: 10.1111/j.1462-2920.2011.02608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruby EG, Lee KH. 1998. The Vibrio fischeri-Euprymna scolopes light organ association: current ecological paradigms. Appl Environ Microbiol 64:805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dikow RB, Smith WL. 2013. Genome-level homology and phylogeny of Vibrionaceae (Gammaproteobacteria: Vibrionales) with three new complete genome sequences. BMC Microbiol 13:80. doi: 10.1186/1471-2180-13-80. [DOI] [PMC free article] [PubMed] [Google Scholar]