Abstract
The present work details the in vitro interactions between Lactobacillus pentosus and yeast strains isolated from table olive processing to form mixed biofilms. Among the different pairs assayed, the strongest biofilms were obtained from L. pentosus and Candida boidinii strain cocultures. However, biofilm formation was inhibited in the presence of d-(+)-mannose. In addition, biofilm formation by C. boidinii monoculture was stimulated in the absence of cell-cell contact with L. pentosus. Scanning electron microscopy revealed that a sort of “sticky” material formed by the yeasts contributed to substrate adherence. Hence, the data obtained in this work suggest that yeast-lactobacilli biofilms may be favored by the presence of a specific mate of yeast and L. pentosus, and that more than one mechanism might be implicated in the biofilm formation. This knowledge will help in the design of appropriate mixed starter cultures of L. pentosus-yeast species pairs that are able to improve the quality and safety of Spanish-style green table olive processing.
INTRODUCTION
Until recently, microbiologists had studied microorganisms as pure cultures of nonaggregated planktonic cells. However, it is now well accepted that in nature, microorganisms can exist within microbial communities named biofilms (1). Biofilms are defined as sessile microbial communities that are attached to a surface or each other, surrounded by a matrix of exopolysaccharide material (EPS) and extracellular DNA (eDNA) produced and released by the same microorganisms. According to Costerton et al. (2), the biofilm life cycle is part of a dominant survival strategy in natural environments.
It is known that monocultural biofilms are rarely found in nature. Instead, they are composed of mixed species of bacteria and eukaryotes, which are thought to be more stable than monospecies films (3, 4). This behavior is also the case with microbial communities in food environments (5–10). In the particular case of olive table fermentations, the presence of polymicrobial communities composed of yeasts and lactic acid bacteria (LAB) attached to both biotic (skin of the olives) and abiotic (inner fermenter walls) surfaces has been reported (11–14). The predominant LAB adhered to the surface of this fruit belong mainly to the species Lactobacillus pentosus, although a variety of yeast species coexist in the same biofilms during fermentation. Candida boidinii, Debaryomyces etchelsii, Issatchenkia occidentalis, Pichia membranifaciens, Saccharomyces cerevisiae, and Wickerhamomyces anomalus are among the most prominent yeast species found in this fermented fruit (15).
Apart from contributing to the sensory attributes of the final product and other advantageous activities, it has been recognized that yeasts have a beneficial role in the olive fermentation ecosystem by promoting the growth of L. pentosus through the production of essential B vitamins (15–18). In the last few years, various authors have described the biofilm formation process with mixed species of L. pentosus and yeasts in natural olive fermentations (11, 12, 15, 19). These microbial communities attach not only to the skins of the fruits but also to the surface of fermentation vessels and other surfaces available in the fermentation environment, which act as a natural reservoir of inocula for other fermentations.
In this work, we studied the in vitro ability of specific pairs of L. pentosus and yeast isolates from olive fermentations to produce mixed biofilms. Also, using different methods, we assayed the possible interactions of L. pentosus and yeasts leading to biofilm formation by yeasts in a monoculture. A polystyrene surface was used as a model for studying these interactions. Scanning electron microscopy (SEM) was applied to visualize such interactions between lactobacilli and yeasts. The knowledge obtained may aid in the design of the appropriated mixed starter cultures of yeasts and L. pentosus strains able to improve the Spanish-style green olive fermentation process.
MATERIALS AND METHODS
Microorganisms and culture conditions.
The six strains of L. pentosus used in the present study (Lp2, Lp43, Lp209, Lp13, Lp57, and Lp13B4) were all previously isolated from cover brines or from biofilms of Spanish-style green table olive fermentations. They were isolated from different processing plants in the south of Spain and propagated in de Man, Rogosa, and Sharpe (MRS) agar medium (Oxoid Unipath Ltd., Basingstoke, Hampshire, England) supplemented with 0.02% (wt/vol) sodium azide. The yeast species C. boidinii, D. etchellsii, I. occidentalis, S. cerevisiae, and W. anomalus chosen for the study were also isolated from diverse table olive fermentations. They were propagated in a yeast mold (YM) agar medium (Difco; Becton and Dickinson Co., Sparks, MD, USA) supplemented with 100 mg/ml oxytetracycline as the selective agent for the yeasts.
Biofilm formation.
The ability of L. pentosus and/or yeasts to form a biofilm under in vitro conditions was assayed as previously described (20, 21), with minor modifications. A polystyrene surface was used as a model to study biofilm formation.
To assay biofilm formation in monocultures, L. pentosus and yeast strains were grown overnight at 30°C in MRS broth and YM broth, respectively. Next, the cells were diluted 1:40 in fresh medium, and 200 μl of the bacterial or yeast suspension was grown in sterile 96-well Nunclon Delta surface microtiter plates (Nunc A/S, Denmark). For biofilm formation in coculture, L. pentosus or yeast cultures were grown as described above, and after dilution in fresh medium, 100 μl of each L. pentosus strain and 100 μl of the yeast species suspension were inoculated together in sterile 96-well microtiter plates. After inoculation, both mono- and cocultures were incubated for 7 days at 30°C, and then the biomass in biofilm was quantified, as described below.
At the same time, to visualize biofilm formation in cocultures using scanning electron microscopy (SEM), 2 ml of each L. pentosus strain and yeast species was added to 48 ml of YM broth in 50-ml Falcon tubes in which glass slides were placed. After cultivation for 7 days at 30°C, the slides were removed and processed for analysis by SEM, as described below.
Biofilm formation without physical contact between L. pentosus and yeasts.
To investigate whether L. pentosus-yeast contact was necessary for biofilm formation, a coculture method was used. The device consisted of a two-compartment system in which each well of a 24-well polystyrene plate (Cellstar; Greiner Bio-One North America, Inc., Monroe, NC, USA) was separated into two compartments of 0.4-μm-pore-size ThinCert-TC insert membranes (Greiner Bio-One North America, Inc.). Two hundred microliters of an overnight culture in the YM broth of C. boidinii was inoculated into the lower compartment (24-well plate) and filled with fresh YM broth up to 2 ml. Next, 200 μl of the selected lactobacillus (previously incubated overnight in MRS, centrifuged, and with pellets diluted 1:40 in YM broth) was added to the upper compartment. Simultaneously, monocultures of the L. pentosus strains or yeasts were used as controls. After incubation for 72 h at 30°C, the biofilms in the lower compartments were quantified.
To investigate the possible presence of any signaling between the lactobacilli and yeast, the L. pentosus strains were grown overnight in MRS broth at 30°C. Next, the cultures were centrifuged (at 15,000 × g and 4°C for 5 min), and the supernatants were filter sterilized through a 0.22-μm-pore-size filter (Millipore Iberica, Madrid, Spain). One hundred microliters of these cell-free supernatants was added to wells in a polystyrene plate together with 100 μl of an overnight culture of C. boidinii, which was then filled to 2 ml with a fresh YM broth medium. After incubation for 72 h at 30°C, the samples were removed with a plastic spatula, spread onto the surface of a glass slide, and then processed for SEM visualization, as described below. Wells to which uninoculated fresh MRS broth was added were used as controls. They were treated as described above.
Aggregation assay.
To study the ability of the L. pentosus strains to coaggregate with yeast cells, the protocol for S. cerevisiae described previously (22) was used, with minor modifications. d-(+)-Mannose was used as an inhibitory substance of lactobacillus-yeast aggregation, C. boidinii was assayed instead of S. cerevisiae, and coaggregation was examined by SEM.
PCR.
In an attempt to identify a candidate gene potentially involved in mannose adhesion of the L. pentosus strains, a PCR assay was carried out. Based on the genomic sequence of L. pentosus IG1 (23), we designed locus-specific primers of the LPENT_00013 gene. This gene encodes a protein that is 78% identical to a mannose-specific adhesin (Msa) in Lactobacillus plantarum WCFS1 (22). The primers used were LPENT_00013for (5′-CTACTCATGGCTAATAGTAATAAACCACTCAGTG-3′) and LPENT_00013rev (3′-ATGACTTATTCACCGATTCAAAAGGCAATCATAAATTATA-5′). DNA from the L. pentosus strains was extracted as previously described (11). DNA from L. pentosus IG1 was supplied by J. L. Ruiz-Barba (Instituto de la Grasa, Consejo Superior de Investigaciones Científicas, Seville, Spain). The PCR was accomplished according to a previously described protocol (24).
Quantification of biofilms.
For the biomass quantification of biofilms, the supernatants in the wells were aseptically removed, the wells were gently washed twice with sterile phosphate-buffered saline (PBS), and then the plates were dried in an inverted position. Two hundred microliters of 0.8% crystal violet (CV) solution was added to each well and allowed to stain the cells for 15 to 30 min at room temperature. The wells were rinsed again with PBS, and the CV in each well was solubilized in 100 μl of a mixture of ethanol and acetone (80:20 [vol/vol]). Next, the optical density at 595 nm (OD595) was measured in an iMark microplate reader (Bio-Rad, Hercules, CA, USA). The monocultures and cocultures were classified for their biofilm-forming abilities, as described previously (21).
Scanning electron microscopy.
The method of Kubota et al. (25) adapted by Domínguez-Manzano et al. (11) was used. Briefly, this method includes fixation of the biofilms with glutaraldehyde, followed by dehydration through graded ethanol steps and a final treatment in tert-butyl alcohol. The samples were coated with gold and observed using a Jeol JSM-6460LV scanning electron microscope (Jeol USA, Inc., Peabody, MA) at the Centro de Investigación, Tecnología, e Innovación de la Universidad de Sevilla (CITIUS, Seville, Spain).
Statistical data analysis.
An analysis of variance (ANOVA) was performed by means of the factorial ANOVA module of Statistica software version 7.0 (StatSoft Inc., Tulsa, OK, USA), using time (7 levels, at 0, 1, 2, 3, 4, 5, and 6 days) and type of microorganism culture (41 levels, with mono- and cocultures of C. boidinii, I. occidentalis, W. anomalus, S. cerevisiae, and D. etchelsii with L. pentosus Lp2, Lp13, Lp43, Lp57, Lp209, and Lp13B4) as categorical predictor variables. The dependent variables introduced for the analysis came from the biomass of biofilm, as measured by the OD595. To check for significant differences among treatments and to form homogeneous groups, a post hoc comparison test was applied using the Scheffé test, which is considered to be one of the most conservative post hoc tests (26). A second advantage of the Scheffé test is that it can also be used with unequal sample sizes. In this way, when statistical significance is obtained in an ANOVA (P ≤ 0.05), we can reject the null hypothesis of no differences between the means and accept the alternative hypothesis, that is, that there are at least significant differences between two means. All experiments were repeated at least 6 times, resulting in a total of 1,722 data points (7 time levels × 41 culture levels × 6 replicates).
RESULTS
Biofilm formation by L. pentosus and yeast strains in mono- and cocultures.
Based on the adherence values of Enterococcus faecalis strains to a surface of polystyrene (21), all the L. pentosus strains or yeasts tested in monocultures and most of L. pentosus-yeast combinations could be considered non-biofilm forming, because their OD595 was ≤1.0 in all cases (Fig. 1). Only W. anomalus-Lp2 and W. anomalus-Lp43 mixed cultures were considered weak biofilm-forming combinations, because they presented adherence values ranging from 1.0 to 2.0. On the contrary, C. boidinii always formed a medium-strength biofilm regardless of the L. pentosus strain, because all these cocultures showed the highest biofilm mass levels, with significant differences (P ≤ 0.05) with respect to all the L. pentosus-yeast pairs tested (OD595 between 2.0 and 3.0).
FIG 1.
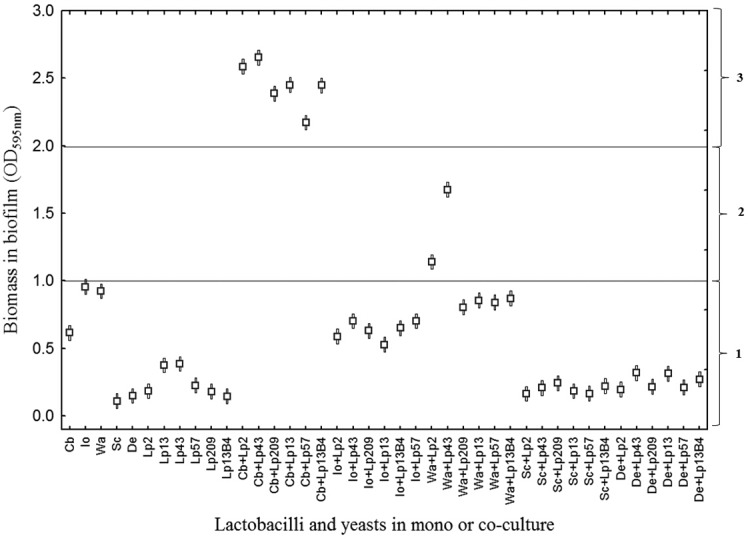
ANOVA graphical representation of the biomass of biofilm (measured as OD595) obtained for the different mono- and cocultures between C. boidinii (Cb), I. occidentalis (Io), W. anomalus (Wa), S. cerevisiae (Sc), and D. etchelsii (De) with L. pentosus Lp2, Lp13, Lp43, Lp57, Lp209, and Lp13B4. Horizontal sections: 1, non-biofilm forming; 2, weak biofilms; 3, medium biofilms.
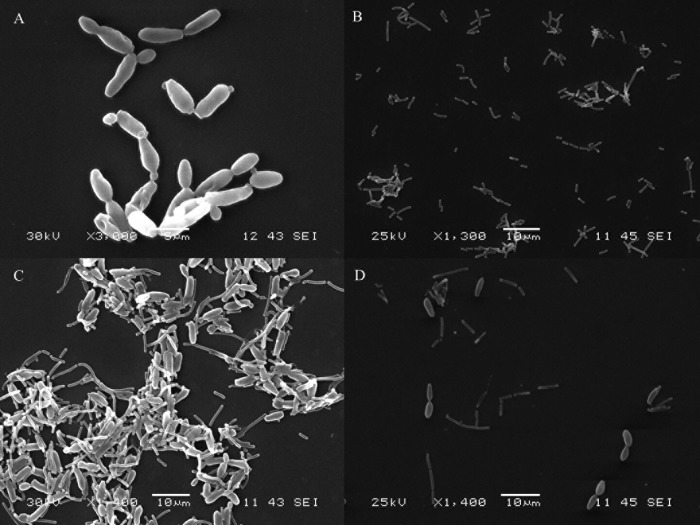
Biofilm formation in the cocultures was visualized by SEM. The images showed that none of the pairs of L. pentosus and yeast species assayed in this work were able to form biofilms, except the combination of L. pentosus and C. boidinii mentioned above. As an example, the biofilm formed between L. pentosus Lp13 and C. boidinii is shown in Fig. 2. In monocultures, the cells of L. pentosus Lp13 or C. boidinii appeared to disperse without any apparent linkage among them (Fig. 2A and B), whereas in cocultures, the cells of both microorganisms appeared by forming aggregates (Fig. 2C). However, when d-(+)-mannose was added to the cultures of C. boidinii prior to the addition of the L. pentosus strains, no coaggregation among lactobacilli and the yeast was observed (Fig. 2D). In addition to this, the LPENT_00013 gene in L. pentosus IG1 (23), which encodes a protein that is 78% identical to a mannose-specific adhesin (Msa) encoded by the lp_1229 gene in L. plantarum WCFS1 (26), was amplified (651-bp amplicon) in all L. pentosus strains that coaggregated with C. boidinii (Fig. 3).
FIG 2.
Visualization by scanning electron microscopy (SEM) of the biofilm formed between L. pentosus Lp13 and C. boidinii made on glass slides. (A) C. boidinii cells in monoculture. (B) L. pentosus Lp13 in monoculture. (C) C. boidinii and L. pentosus Lp13 in coculture. (D) C. boidinii and L. pentosus Lp13 in coculture in the presence of d-(+)-mannose.
FIG 3.
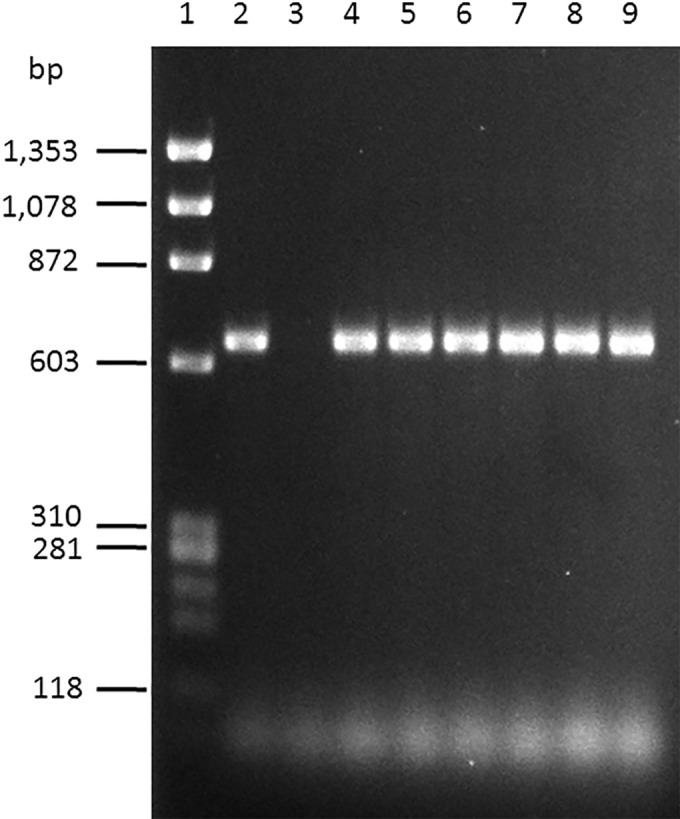
PCR with locus-specific primers of the total DNA of L. pentosus strains used in this study showing the 641-bp amplicon corresponding to the LPENT_00013 gene in L. pentosus IG1. Lane 1, ϕX174 DNA-HaeIII digestion (New England BioLabs, L'Hospitalet de Llobregat, Barcelona, Spain) was used as molecular weight marker; lane 2, L. pentosus IG1 as a positive control; lane 3, Escherichia coli DH5α as a negative control; lane 4, L. pentosus Lp2; lane 5, L. pentosus Lp13; lane 6, L. pentosus Lp43; lane 7, L. pentosus Lp57; lane 8, L. pentosus Lp209; lane 9, L. pentosus Lp13B4.
Ability of C. boidinii to form biofilms without physical contact with L. pentosus.
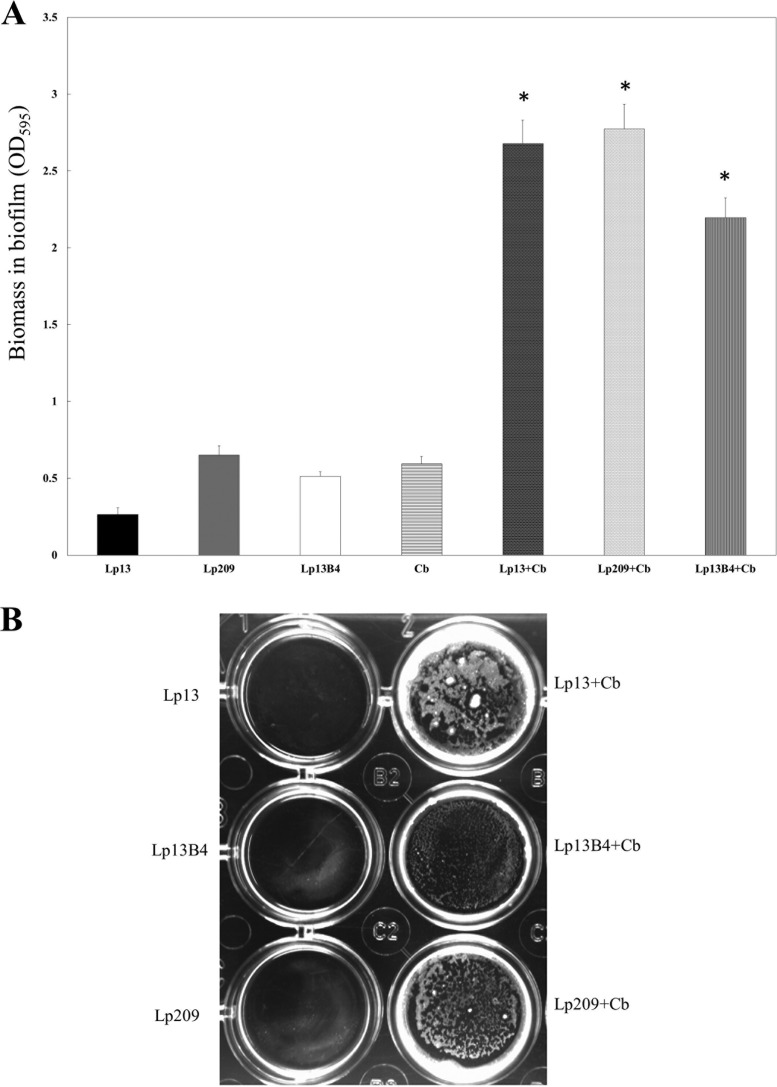
In an attempt to investigate the possible interactions between C. boidinii and the L. pentosus strains, as a prerequisite to forming mixed biofilms, a two-compartment system to prevent cell-cell contact during coculture was used. As can be seen in Fig. 4, neither the L. pentosus strains nor C. boidinii strains were able to form biofilms in monocultures (Fig. 4A and B), corroborating the results previously obtained (see Fig. 1). Surprisingly, C. boidinii, without a direct contact with cells of the six L. pentosus strains, was able to form as much biofilm mass as it was in the cocultures with the same strains (Fig. 1 and 2). A visualization of the attachment of C. boidinii to the surface of polystyrene plates after incubation in the two-compartment system is shown in Fig. 4B.
FIG 4.
Biomass in biofilms (measured as OD595) between L. pentosus and C. boidinii strains. Biofilm formation was assayed in a coculture method consisting of a two-compartment system in which L. pentosus strains and the yeast were separated by a membrane (for more details, refer to Materials and Methods). (A) The ordinate axis shows the absorption of crystal violet extracted from crystal violet-stained biofilms. The mean values and standard deviations are shown. (B) Picture of the lower compartments showing biofilm formation by L. pentosus and C. boidinii strains. (A and B) Lp13, L. pentosus Lp13 in monoculture; Lp209, L. pentosus Lp209 in monoculture; Lp13B4, L. pentosus Lp13B4 in monoculture; Cb, C. boidinii in monoculture; Lp13+Cb, L. pentosus Lp13 and C. boidinii in coculture; Lp209+Cb, L. pentosus Lp209 and C. boidinii in coculture; Lp13B4+Cb, L. pentosus 13B4 and C. boidinii in coculture. *, values are statistically significant (P ≤ 0.05).
In addition to this, when cultures of C. boidinii were incubated in the presence of cell-free supernatants from every L. pentosus strain, individual yeast cells appeared to adhere to each other and the substrate by a sort of sticky material joining the cells and then appeared to form microcolony-like structures (Fig. 5A). In contrast, no connection by a joining sticky material between isolated cells was noticed, nor were microcolony-like structures observed when uninoculated fresh MRS was added to the C. boidinii cultures as a control (Fig. 5B).
FIG 5.
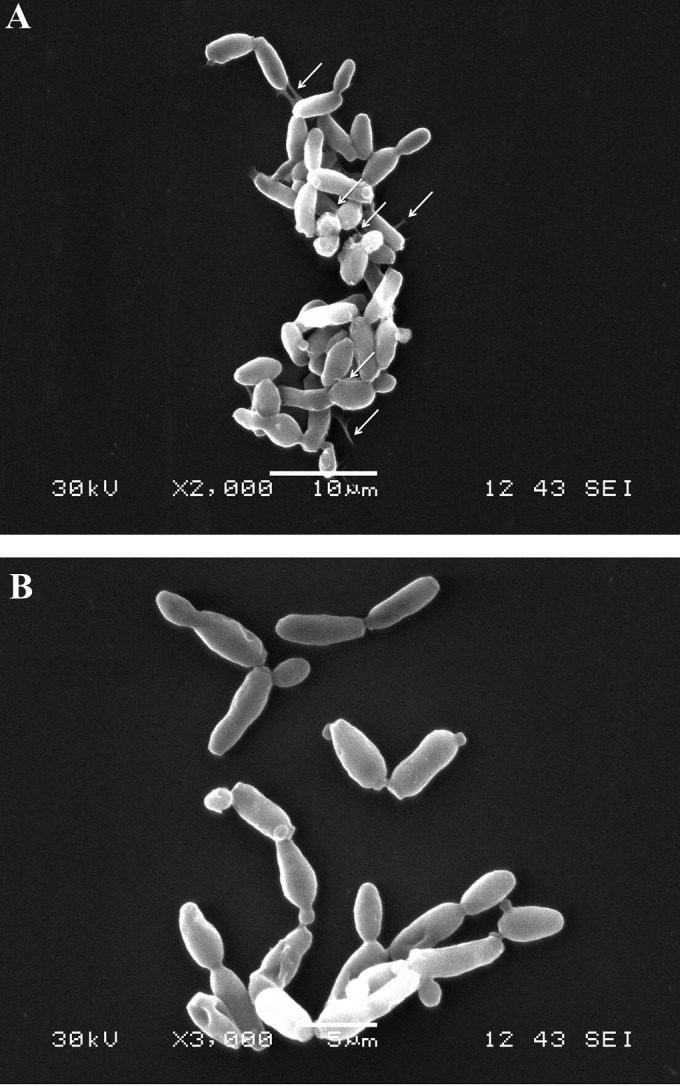
Visualization by scanning electron microscopy (SEM) of C. boidinii cells after coculture with cell-free supernatant from L. pentosus Lp13B4, as described in Materials and Methods. (A) C. boidinii culture in cell-free supernatant from L. pentosus Lp13B4. (B) C. boidinii culture in fresh MRS broth. In panel A, the solid white arrows indicate junctions among yeasts and to the glass surface.
DISCUSSION
In this survey, we studied the ability of every coculture among six L. pentosus strains and five yeast species, all of them isolated from Spanish-style green olive fermentations, to form in vitro mixed-species biofilms. Based on the adherence values of E. faecalis strains to a surface of polystyrene (21), except for the L. pentosus and C. boidinii pairs and some L. pentosus and W. anomalus pairs, no stable in vitro biofilms were noticed for other dual cultures.
In vitro, biofilm formation between LAB and yeast species isolated from many foods has been reported (20, 27–29). In the natural environment of Spanish-style green olive fermentation, mixed-species biofilms between yeasts and lactobacilli in the skins of fruits have also been found (11, 12, 14, 15, 19). Therefore, one would expect that yeasts and lactobacilli isolated from olive fermentations in vitro are also able to form stable biofilms when cocultured. On the contrary, the data shown here suggest that mixed-species biofilm formation in vitro is an uncommon feature and might eventually be found to be related to the yeast strain. It may be due to the different environmental conditions that occur in the natural ecosystem provided by the olive fermentations, particularly on the olive surface, which may be quite different from those found in the synthetic media, e.g., pH and salt stresses. Interestingly, the architecture of the in vitro mixed biofilms between C. boidinii and L. pentosus resembles that of mixed-microbial communities attached to abiotic surfaces during Spanish-style green olive fermentation (11, 12). Thus, SEM micrographs showed that yeast and bacterial cells in coculture appear as mixed biofilms but with no extracellular matrix surrounding them.
The coaggregative interaction between LAB and yeast species has extensively been reported in the last few years (10, 20, 22, 30, 31). As mentioned by Fukurawa et al. (28), cell-cell adhesion (coaggregation) and cell-surface adhesion might be controlled by different mechanisms in lactobacilli, as reported in L. plantarum-S. cerevisiae mixed-species biofilm formation. Coaggregation between these two microorganisms needed cell-cell contact, and it has been shown that the interaction between proteins on the L. plantarum ML11-11 surface and the surface mannan of the yeast contribute significantly to mixed-species biofilm formation (31). The same was demonstrated for S. cerevisiae-Lactococcus lactis IL1403, for which the cytosolic proteins, including the heat shock proteins DnaK and GroEL displayed on the cell wall of the L. lactis strain, cause its adherence to the yeast (32). In the case of L. plantarum and Lactobacillus fermentum, the msa gene encodes the mannose-specific adhesin (Msa). This protein, which displays known carbohydrate-binding domains, has been implicated in cell-cell interactions (22, 33). In our case, the presence of d-(+)-mannose in the cultures of C. boidinii before the addition of the L. pentosus strains prevented coaggregation among them. Also, the LPENT_00013 gene in L. pentosus IG1 (23), which encodes a protein that is 78% identical to a mannose-specific adhesin (Msa) encoded by the lp_1229 gene in L. plantarum WCFS1 (22), was amplified in all six L. pentosus strains. It might indicate that a mannose-specific adhesin is implicated in the capacity of the L. pentosus strains to coaggregate with C. boidinii.
Kawarai et al. (20) reported that addition of cell-free supernatants from Lactobacillus casei subsp. rhamnosus IFO3831 induces biofilm formation by S. cerevisiae Kyokai-10 in monocultures. Apparently, an active substance of a molecular mass between 3 and 5 kDa present in the bacterial supernatant induces many protrusions visible on the yeast surface. Similar bud scars were observed in the cell surface of the Kyokai-10 strain in coculture biofilms with strain IFO3831. The authors concluded that this active substance induces S. cerevisiae to form a biofilm in the absence of the lactobacilli. In our case, C. boidinii without a direct contact with L. pentosus strains was able to form as much biofilm mass as that in the coculture with the same strains, but no visible changes in the morphology of the cell surface of the yeast were noticed (data not shown). Interestingly, it seems that for biofilm formation between LAB and yeasts isolated from food environments, cell-cell contact is required, except for the cases reported by Kawarai et al. (20) and the data provided by this study.
As the proper fermentation of Spanish-style table olives relies on the development of L. pentosus-yeast species biofilms in the skins of fruits, a deeper knowledge about the lactobacillus-yeast interactions will serve us in the design of further mixed starter cultures to improve the quality and safety of fermented Spanish-style green olives.
ACKNOWLEDGMENTS
The Spanish Government supported this work through MICIIN projects AGL2009-08016/ALI, AGL2010-15529/ALI, and AGL2013-48300-R/ALI (project OliFilm) and the Junta de Andalucía (through financial support to project P11-AGR-7051). F.N.A.-L. was the recipient of a Ramón y Cajal postdoctoral research contract (Spanish Government), while Á.L.-R. and J.D.-M. were the beneficiaries of an FPI grant from the MICIIN and of a JAE-predoctoral grant from the CSIC, respectively.
REFERENCES
- 1.Stoodley P, Sauer K, Davies DG, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu Rev Microbiol 56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 2.Costerton JW, Geesey GG, Cheng KJ. 1978. How bacteria stick. Sci Am 238:86–95. doi: 10.1038/scientificamerican0178-86. [DOI] [PubMed] [Google Scholar]
- 3.Al-Bakri AG, Gilbert P, Allison DG. 2004. Immigration and emigration of Burkholderia cepacia and Pseudomonas aeruginosa between and within mixed biofilm communities. J Appl Microbiol 96:455–463. doi: 10.1111/j.1365-2672.2004.02201.x. [DOI] [PubMed] [Google Scholar]
- 4.Allison DG, McBain AJ, Gilbert P. 2000. Biofilms: problems of control, p 309–327. In Allison DG, Gilbert P, Lappin-Scott HM, Wilson M (ed), community structure and co-operation in biofilms. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 5.Campanaro S, Treu L, Vendramin V, Bovo B, Giacomini A, Corich V. 2014. Metagenomic analysis of the microbial community in fermented grape marc reveals that Lactobacillus fabifermentans is one of the dominant species: insights into its genome structure. Appl Microbiol Biotechnol 98:6015–6037. doi: 10.1007/s00253-014-5795-3. [DOI] [PubMed] [Google Scholar]
- 6.Ercolini D, Hill PJ, Dodd CER. 2003. Bacterial community structure and location in Stilton cheese. Appl Environ Microbiol 69:3540–3548. doi: 10.1128/AEM.69.6.3540-3548.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escalante A, Giles-Gómez M, Hernández G, Córdova-Aguilar MS, López-Munguía A, Gosset G, Bolívar F. 2008. Analysis of bacterial community during the fermentation of pulque, a traditional Mexican alcoholic beverage, using a polyphasic approach. Int J Food Microbiol 124:126–134. doi: 10.1016/j.ijfoodmicro.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Lortal S, Licitra G, Valence F. 2014. Wooden tools: reservoirs of microbial biodiversity in traditional cheesemaking. Microbiol Spectr 2:CM-0008-2012. doi: 10.1128/microbiolspec.CM-0008-2012. [DOI] [PubMed] [Google Scholar]
- 9.Mei J, Guo Q, Wu Y, Li Y. 2014. Microbial diversity of a Camembert-type cheese using freeze-dried Tibetan kefir coculture as starter culture by culture-dependent and culture-independent methods. PLoS One 9:e111648. doi: 10.1371/journal.pone.0111648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turpin W, Humblot C, Noordine M-L, Thomas M, Guyot JP. 2012. Lactobacillaceae and cell adhesion: genomic and functional screening. PLoS One 7:e38034. doi: 10.1371/journal.pone.0038034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domínguez-Manzano J, León-Romero Á, Olmo-Ruiz C, Bautista-Gallego J, Arroyo-López FN, Garrido-Fernández A, Jiménez-Díaz R. 2012. Biofilm formation on abiotic and biotic surfaces during Spanish style green table olive fermentation. Int J Food Microbiol 157:230–238. doi: 10.1016/j.ijfoodmicro.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 12.Domínguez-Manzano J. 2013. Estudio de las relaciones entre los microorganismos presentes en las fermentaciones de la aceituna de mesa. Ph.D. thesis. University of Seville, Seville, Spain: http://fondosdigitales.us.es/media/thesis/2323/I_T-PROV39.pdf. [Google Scholar]
- 13.Grounta A, Panagou EZ. 2014. Mono and dual species biofilm formation between Lactobacillus pentosus and Pichia membranifaciens on the surface of black olives under different sterile brine conditions. Ann Microbiol 64:1757–1767. doi: 10.1007/s13213-014-0820-4. [DOI] [Google Scholar]
- 14.Grounta A, Doulgeraki AI, Panagou EZ. 2015. Quantification and characterization of microbial biofilm community attached on the surface of fermentation vessels used in green table olive processing. Int J Food Microbiol 203:41–48. doi: 10.1016/j.ijfoodmicro.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Arroyo-López FN, Romero-Gil V, Bautista-Gallego J, Rodríguez-Gómez F, Jiménez-Díaz R, García-García P, Querol A, Garrido-Fernández A. 2012. Yeasts in table olive processing: desirable or spoilage microorganisms? Int J Food Microbiol 160:42–49. doi: 10.1016/j.ijfoodmicro.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Arroyo-López FN, Querol A, Bautista-Gallego J, Garrido-Fernández A. 2008. Role of yeasts in table olive production. Int J Food Microbiol 128:189–196. doi: 10.1016/j.ijfoodmicro.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 17.Ruiz-Barba JL, Jimenez-Diaz R. 1994. Vitamin and amino acid requirements of Lactobacillus plantarum strains isolated from green olive fermentations. J Appl Bacteriol 76:350–355. doi: 10.1111/j.1365-2672.1994.tb01639.x. [DOI] [PubMed] [Google Scholar]
- 18.Ruiz-Barba JL, Jiménez-Díaz R. 1995. Availability of essential B-group vitamins to Lactobacillus plantarum in green olive fermentation brines. Appl Environ Microbiol 61:1294–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nychas G-JE, Panagou EZ, Parker ML, Waldron KW, Tassou CC. 2002. Microbial colonization of naturally black olives during fermentation and associated biochemical activities in the cover brine. Lett Appl Microbiol 34:173–177. doi: 10.1046/j.1472-765x.2002.01077.x. [DOI] [PubMed] [Google Scholar]
- 20.Kawarai T, Furukawa S, Ogihara H, Yamasaki M. 2007. Mixed-species biofilm formation by lactic acid bacteria and rice wine yeasts. Appl Environ Microbiol 73:4673–4676. doi: 10.1128/AEM.02891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M, Amorena B, Leiva J, Penadés JR, Lasa I. 2001. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol 67:4538–4545. doi: 10.1128/AEM.67.10.4538-4545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pretzer G, Snel J, Molenaar D, Wiersma A, Bron PA, Lambert J, de Vos WM, van der Meer R, Smits MA, Kleerebezem M. 2005. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J Bacteriol 187:6128–6136. doi: 10.1128/JB.187.17.6128-6136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maldonado-Barragán A, Caballero-Guerrero B, Lucena-Padrós H, Ruiz-Barba JL. 2011. Genome sequence of Lactobacillus pentosus IG1, a strain isolated from Spanish-style green olive fermentations. J Bacteriol 193:5605. doi: 10.1128/JB.05736-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.León-Romero AM. 2014. Selección de cepas de Lactobacillus pentosus con propiedades potencialmente probióticas, aisladas de fermentaciones de aceitunas. Estudio de sus propiedades tecnológicas. Ph.D. thesis. University of Seville, Seville, Spain. [Google Scholar]
- 25.Kubota H, Senda S, Nomura N, Tokuda H, Uchiyama H. 2008. Biofilm formation by lactic acid bacteria and resistance to environmental stress. J Biosci Bioeng 106:381–386. doi: 10.1263/jbb.106.381. [DOI] [PubMed] [Google Scholar]
- 26.Winer BJ. 1962. Statistical principles in experimental design. McGraw-Hill, New York, NY. [Google Scholar]
- 27.Fukurawa S, Yoshida K, Ogihara H, Yamasaki M, Morinaga Y. 2010. Mixed-species biofilm formation by direct cell-cell contact between brewing yeasts and lactic acid bacteria. Biosci Biotechnol Biochem 74:2316–2319. doi: 10.1271/bbb.100350. [DOI] [PubMed] [Google Scholar]
- 28.Fukurawa S, Nojima N, Yoshida K, Hirayama S, Ogihara H, Morinaga Y. 2011. The importance of inter-species cell-cell co-aggregation between Lactobacillus plantarum ML11-11 and Saccharomyces cerevisiae BY4741 in mixed-species biofilm formation. Biosci Biotechnol Biochem 75:1430–1434. doi: 10.1271/bbb.100817. [DOI] [PubMed] [Google Scholar]
- 29.Jahid IK, Sang-Do H. 2014. The paradox of mixed-species biofilms in the context of food safety. Compr Rev Food Sci Food Saf 13:990–1011. doi: 10.1111/1541-4337.12087. [DOI] [Google Scholar]
- 30.Fukurawa S, Nojima N, Nozaka S, Hirayama S, Satoh A, Ogihara H, Morinaga Y. 2012. Mutants of Lactobacillus plantarum ML11-11 deficient in coaggregation with yeast exhibited reduced activities of mixed-species biofilm formation. Biosci Biotechnol Biochem 76:326–330. doi: 10.1271/bbb.110714. [DOI] [PubMed] [Google Scholar]
- 31.Hirayama S, Furukawa S, Ogihara H, Morinaga Y. 2012. Yeast mannan structure necessary for co-aggregation with Lactobacillus plantarum ML11-11. Biochem Biophys Res Commun 419:652–655. doi: 10.1016/j.bbrc.2012.02.068. [DOI] [PubMed] [Google Scholar]
- 32.Katakura Y, Sano R, Hashimoto T, Ninomiya K, Shioya S. 2010. Lactic acid bacteria display on the cell surface cytosolic proteins that recognize yeast mannan. Appl Microbiol Biotechnol 86:319–326. doi: 10.1007/s00253-009-2295-y. [DOI] [PubMed] [Google Scholar]
- 33.Turner MS, Hafner LM, Walsh T, Giffard PM. 2003. Peptide surface display and secretion using two LPXTG-containing surface proteins from Lactobacillus fermentum BR11. Appl Environ Microbiol 69:5855–5863. doi: 10.1128/AEM.69.10.5855-5863.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]