Abstract
A wealth of studies has demonstrated that resident microorganisms (microbiota) influence the pattern of nutrient allocation to animal protein and energy stores, but it is unclear how the effects of the microbiota interact with other determinants of animal nutrition, including animal genetic factors and diet. Here, we demonstrate that members of the gut microbiota in Drosophila melanogaster mediate the effect of certain animal genetic determinants on an important nutritional trait, triglyceride (lipid) content. Parallel analysis of the taxonomic composition of the associated bacterial community and host nutritional indices (glucose, glycogen, triglyceride, and protein contents) in multiple Drosophila genotypes revealed significant associations between the abundance of certain microbial taxa, especially Acetobacteraceae and Xanthamonadaceae, and host nutritional phenotype. By a genome-wide association study of Drosophila lines colonized with a defined microbiota, multiple host genes were statistically associated with the abundance of one bacterium, Acetobacter tropicalis. Experiments using mutant Drosophila validated the genetic association evidence and reveal that host genetic control of microbiota abundance affects the nutritional status of the flies. These data indicate that the abundance of the resident microbiota is influenced by host genotype, with consequent effects on nutrient allocation patterns, demonstrating that host genetic control of the microbiome contributes to the genotype-phenotype relationship of the animal host.
INTRODUCTION
The recognition that animals are routinely colonized by dense and often diverse communities of microorganisms is driving a major reassessment of fundamental aspects of animal biology (1). Notably, there is accumulating evidence that resident microorganisms influence the nutritional status of animals in multiple ways, including competition for ingested nutrients, providing supplementary nutrients (e.g., vitamins, short-chain fatty acids, essential amino acids), and by modulating the nutrient signaling circuits that regulate nutrient allocation (2–5). These discoveries demonstrate the inadequacy of traditional explanations of animal nutrition in terms of nutritional inputs (amount and composition of food ingested) and outputs (animal nutritional demand for activity, growth, reproduction, etc.) and highlight our ignorance of how microbial effects on animal nutrition interact with other factors, especially animal genotype (6–10).
The focus of this study is the impact of interactions between the gut microbiota and host genotype on animal nutrition. The nutritional consequence of the microbiota is known to vary with the abundance and composition of the microorganisms (7, 10–13). For example, comparison of nutritional phenotypes in Drosophila melanogaster raised with or without gut bacteria revealed that the microbiome can influence penetrance of host mutations (12). To the extent that the abundance and composition of the microbiota are determined by host genotype, the impact of animal genetic variation on nutrition may be mediated via effects on the microbiota, and host genotype-independent differences in the microbiota among individual animals may also make an appreciable contribution to the nutritional phenotype of animals (10). These issues are immediately relevant to the promise of microbial therapies and microbiologically informed dietary therapies for nutritional health (i.e., probiotics and prebiotics). The rational application of these therapies will require an understanding of how the effects of the microbiota and host genotype interact to shape animal nutrition.
The gut microbiota in the fruit fly Drosophila melanogaster is an excellent system to investigate the fundamentals of interactions between resident microorganisms and host genotype on animal nutrition. A nutritionally important component of the Drosophila microbiota is the gut-inhabiting bacteria, including members of the Acetobacteraceae (alphaproteobacteria), Lactobacillales, and gammaproteobacteria (14–16), which contribute to the B vitamin and protein nutrition of the host, and can reduce energy storage as triglyceride (TAG) and glycogen (9, 17–19). The Drosophila model is also supported by a wealth of genetic and genomic resources, including the Drosophila Genetic Reference Panel (DGRP) comprising multiple inbred lines with sequenced genomes used in this study (20, 21).
In this study, we investigate the relationship between the composition of the microbiota and nutritional phenotype of Drosophila, quantified as a set of nutritional indices (protein, TAG, glycogen, and glucose contents). Using the DGRP, we demonstrate that the composition of the microbiota varies in a Drosophila population and identify microbial species with previously unappreciated influence of host nutrition whose abundance correlates with different nutritional indices. We also identify candidate host genes that influence the abundance of one bacterium, Acetobacter tropicalis, with nutritional consequences. These results demonstrate the significance of host genetic control of the microbiota on animal nutrition.
MATERIALS AND METHODS
Drosophila stock cultures and manipulations.
The Drosophila melanogaster lines (see Table S2 in the supplemental material) were cultured at 25°C on a light-dark cycle (12-h light, 12-h dark). The Drosophila lines were fed a yeast-glucose diet (1 liter H2O, 100 g inactive brewer's yeast [catalog no. 903312; MP Biomedicals], 100 g glucose [catalog no. 158968; Sigma], 1.2% agar [catalog no. 66-103; Apex], 0.84% propionic acid, 0.08% phosphoric acid). Routine cultures were maintained on cooked, but not autoclaved, food, and experimental cultures were reared on sterile yeast-glucose diet prepared by autoclaving the diet, then aseptically adding acid preservatives, and transferring 7.5-ml aliquots into sterile 50-ml Falcon tubes. For experiments using Drosophila with unmanipulated microbiota (conventional Drosophila), eggs (<22 h old) were collected from grape juice agar plates, rinsed gently with double-distilled water (ddH2O), and transferred in groups of 30 to 50 to sterile diet. To prepare gnotobiotic flies (containing defined microbiota), eggs were collected as described above, then surface sterilized by two 2.5-min washes in 0.6% hypochlorite, rinsed three times with sterile water, and aseptically transferred to sterile diet in a biosafety cabinet. Bacterial inocula, comprising Acetobacter pomorum DmCS_004, Acetobacter tropicalis DmCS_006, Lactobacillus brevis DmCS_003, Lactobacillus fructivorans DmCS_002, and Lactobacillus plantarum DmCS_001 (22), were prepared from cells grown at 30°C to stationary-phase culture in mMRS medium (11), normalized to 5 × 107 total CFU ml−1, mixed in equal proportions, and added directly to the food surface in 50 μl within 3 h of egg transfer. All fly assays were performed on male flies, 5 to 8 days posteclosion, and 5 to 8 h into the daily light cycle.
Identification of microbiota community composition.
To assess the microbiota composition of flies, total genomic DNA was extracted from five pooled male flies as described previously (23, 24). 16S rRNA amplicons of the V2 region were prepared by triplicate PCRs (23), using the 16S rRNA gene primers 27F (F stands for forward) (5′-AGAGTTTGATCMTGGCTCAG-3′) and 338R (R stands for reverse) (5′-TGCTGCCTCCCGTAGGAGT-3′), tagged with different molecular identifiers (MIDs) (see Table S3 in the supplemental material). Equal amounts of the triplicate products per sample were mixed, purified by Sephadex filtration, quantified by Quant-iT PicoGreen, and normalized to 109 molecules per μl. Emulsion PCR was conducted at 1.5 copies per bead using only “A” beads for unidirectional 454 GS-FLX pyrosequencing with standard titanium chemistry. Pyrosequencing flowgrams were analyzed by the procedure of Wong et al. (16) using QIIME 1.7.0 (25), except that reads from two half plates were separately demultiplexed. A total of 669,705 reads were obtained. Following quality filtering and removal of chimeras, the reads were clustered at 97% identity (see Data Set S1A in the supplemental material). After removal of reads representing Wolbachia sequences, all samples were rarefied to 501 reads, and single reads were discarded (Data Set S1B). Assignments to taxonomic ranks were performed in QIIME using the Greengenes database (26) (class, order, family, genus, species). Operational taxonomic units (OTUs) were binned based on QIIME taxonomic assignments during correlation analysis. Every OTU in the reagent-only controls had <10 reads, ensuring minimal contamination of experimental samples.
Assays of fly weight and nutritional indices.
Flies were lightly anesthetized on CO2, and males were sorted for downstream assays. For dry weight measurements, flies were flash-frozen on dry ice and desiccated over 7 days at 55°C prior to weighing. To quantify nutritional indices, triplicate samples of five flies each were homogenized in 125 μl TET buffer (10 mM Tris [pH 8], 1 mM EDTA, 0.1% Triton X-100) with 1.4-mm ceramic beads (MP Biomedicals) in a FastPrep-24 instrument (MP Biomedicals) for 30 s. Twenty microliters of homogenate was frozen at −80°C, and 40 μl was heat treated at 72°C for 15 min prior to freezing at −80°C. As described previously (12), commercial kits were used to assay protein content (catalog no. 500-0111; Bio-Rad) in directly frozen samples, and glucose and glycogen (catalog no. GAGO20-1KT; Sigma-Aldrich) and triglyceride (catalog no. TR0100-1KT; Sigma-Aldrich) content in the heat-treated samples. All nutritional indices were normalized to dry weight.
Bacterial culture and monoassociation experiments.
For total bacterial counts, pools of five flies were homogenized in 125 μl phosphate-buffered saline (PBS), and the homogenate was directly plated onto mMRS plates using a spiral plater (WASP-2 instrument; Microbiology International). To measure Lactobacillus abundance, a replicate plate from the same homogenate was plated onto mMRS medium and incubated in a CO2-flooded airtight container. To obtain selective growth of A. tropicalis from gnotobiotic flies containing both A. tropicalis and A. pomorum, the flies were homogenized in TET buffer, which disrupted growth of the A. pomorum cells. Bacterial colonies were read using a Protocol 3 colony counter (Microbiology International).
Microbiota abundance correlations with nutritional indices.
Spearman's rank correlations performed in R (27) identified significant correlations between nutritional indices and microbiota abundance (OTU abundance at every taxonomic level assigned in QIIME OTU table [class, order, family, genus, species, OTU [97%] with Bonferroni's correction]).
Genome-wide association.
The DGRP lines used for genome-wide association (GWA) (see Table S2A in the supplemental material) were raised with A. pomorum, A. tropicalis, L. brevis, L. fructivorans, and L. plantarum. Male flies were sorted and homogenized in TET buffer (as described above), and A. tropicalis load was quantified in three triplicate pools of five flies each. Dry weights of three pools of five flies were also collected, as described above. A. tropicalis was distinguished from A. pomorum by growth after homogenization in TET buffer (A. pomorum does not grow), and from Lactobacillus species by color (11). Each DGRP line was tested in one of five experimental blocks.
The GWA was conducted with custom R scripts and the nlme R package (28), with single nucleotide polymorphism (SNP) data from the Freeze 1.0 DGRP release. Log+1-transformed A. tropicalis CFU number per fly, measured in triplicate and expressed as a fraction of the total number of CFU, was the response variable, SNP identity was the fixed effect, and experimental block and DGRP genotype were random effects. Following established procedures (29), the most frequent (major) and second-most frequent (minor) SNP identities were retained at each locus; all other alleles were omitted from the analysis, and any SNP with the minor allele present in three or fewer lines was discarded. In total, 2,027,631 SNPs were tested, and significance values for SNP effect are reported. Genetic contributions to variance were calculated as the square of the standard deviation for the random effect of DGRP genotype in the model.
GWA validation.
Drosophila mutants (see Table S2B in the supplemental material) corresponding to genes containing or near GWA-identified SNPs and background stocks were obtained from the Bloomington Drosophila Stock Center and used directly in subsequent assays, following the experimental design of our previous work and others (12, 29–31). Gnotobiotic flies were raised with the five-species microbiota, as in the GWA, and A. tropicalis abundance and nutritional indices were determined. Data were collected in triplicate for each of three separate experiments, and significant differences between mutant and background lines were identified by a linear mixed model (28), with the vial nested within the experiment as a random effect. Data were normalized by log or square root (sqrt) transformation, whichever gave the highest significance value above 0.05 in a Shapiro test. Significance values were assigned using multcomp (32) (critical threshold P < 0.05). Mutant flies and their backgrounds were also raised with a three-species microbiota of L. brevis, L. fructivorans, and L. plantarum and analyzed exactly as described above, except that the experimental replicate was the only random effect used in the models.
RESULTS
Relationship between the microbiota and nutritional traits in the DGRP.
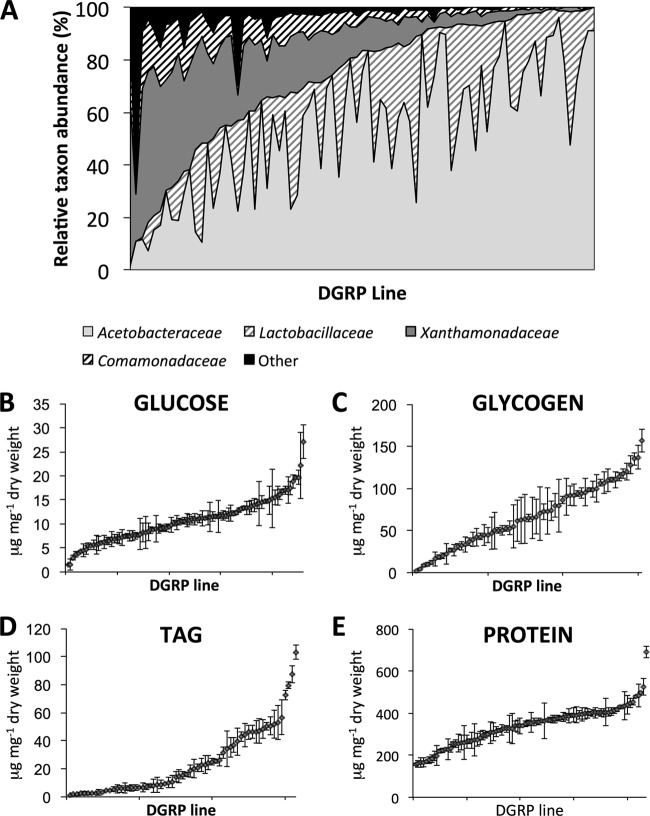
Pyrosequencing of 16S rRNA gene amplicons in 5- or 6-day-old male flies from 79 DGRP lines yielded 177 bacterial OTUs at 97% sequence identity (see Data Set S1A in the supplemental material). Consistent with the previously reported dominance of Acetobacteraceae and Lactobacillaceae in laboratory cultures of Drosophila (14–16, 23, 33–35), all the DGRP lines yielded reads assigned to Acetobacteraceae, and all but eight of the lines also bore Lactobacillaceae; these bacteria accounted for 54 and 19%, respectively, of the reads after removal of sequences assigned to Wolbachia and rarefaction (Fig. 1A; Data Set S1B). The Acetobacteraceae included representation of three genera, Acetobacter, Gluconobacter, and Komagataeibacter (Gluconacetobacter [36]), but all the Lactobacillaceae were members of one genus, Lactobacillus, of which 76% of the reads could be assigned to a single species, L. brevis. In addition, the Xanthomonadaceae and Comamonadaceae were highly prevalent (present in all but two and eight DGRP lines, respectively), representing 17% and 6% of the total reads. As found in previous analyses (14, 16), a core microbiota (i.e., OTUs present in all lines) was not detected at 97% sequence identity. The most prevalent OTU (OTU160 in Xanthomonadaceae; Data Set S1B) was present in 77 of 79 lines, but further analysis revealed nine sequence variants within this OTU, including two (OTU4 and OTU131 in Data Set S1C) with nearly mutually exclusive distributions (detected in 53 and 28 DGRP lines, respectively). These OTUs were both most similar to Stenotrophomonas species.
FIG 1.
Bacterial communities and phenotypic traits of 79 Drosophila lines from DGRP. (A) Microbiota composition was assessed by pyrosequencing with OTUs called at 97% sequence identity (see also Data Set S1 in the supplemental material). (B to E) Nutritional indices (in micrograms per milligram [dry weight]), with data represented as means ± standard errors of the means (SEMs) (error bars). In each panel, Drosophila lines are ordered by the sum of Acetobacter and Lactobacillus species (A) or by mean nutritional index value (B to E).
A parallel analysis of nutritional indices in the DGRP flies revealed wide between-line variation (Fig. 1B to D) that correlated with the relative abundance of associated microbes (Table 1; see Data Set S2 in the supplemental material). The results are congruent with published studies on the nutritional traits of Drosophila in monoassociation (11, 22, 37) or polyassociation (12) with different bacteria. A previous analysis of the same nutritional indices in an overlapping subset of the DGRP raised under axenic or five-species gnotobiotic conditions demonstrated contributions of both host genotype and presence of bacteria to variation in nutritional indices (12). The current analysis extends these published data by demonstrating that the microbial influence is sufficient to yield significant correlations between microbial abundance and nutritional traits identified even across genetically distinct hosts. Specifically, Acetobacteraceae of the genera Gluconobacter and Komagataeibacter, as well as the previously documented Acetobacter, are associated with reduced energy storage (11, 17, 18, 37), while a single Lactobacillus OTU is predicted to influence TAG content, and Xanthomonadaceae and Achromobacter (Betaproteobacteria) are associated with high glycogen content. In principle, the correlations between abundance of specific bacterial taxa and host nutritional traits can be attributed to bacterium-mediated modulation of Drosophila nutritional indices or the suitability of hosts with different nutritional phenotypes for different bacteria. A causal role of Acetobacteraceae is indicated by published demonstrations of significantly reduced energy storage indices in Drosophila monoassociated with these bacteria (11, 18, 19, 22, 37). Multiple attempts to culture Xanthomonadaceae from the DGRP were unsuccessful, although non-Xanthomonadaceae were readily isolated, and the causal basis of both the Xanthomonadaceae and Achromobacter on glycogen content remains to be investigated. In summary, correlations obtained in this study between microbial composition and nutritional traits in genetically distinct conventional Drosophila can identify taxa with causal effects on host nutrition demonstrated previously through monoassociation, and suggest putative roles for novel taxa in the Achromobacter and Xanthamonadaceae.
TABLE 1.
Correlation between Drosophila nutritional indices and bacterial taxa in DGRP lines
| Drosophila nutritional index and bacterial taxona | No. of lines | Spearman rank order correlation |
||
|---|---|---|---|---|
| rho | P value | Critical probability (no. of samples)b | ||
| TAG content | ||||
| Komagataeibacter | 10 | −0.42 | 0.003 | 0.0055 (9) |
| Acetobacteraceae | 49 | −0.43 | 0.002 | 0.006 (8) |
| Lactobacillus OTU7 | 21 | +0.44 | 0.002 | 0.002 (25) |
| Glucose content | ||||
| Komagataeibacter OTU5 | 14 | −0.37 | 0.002 | 0.0017 (29) |
| Komagataeibacter | 14 | −0.38 | 0.001 | 0.0055 (9) |
| Komagataeibacter OTU115 | 11 | −0.42 | <0.001 | 0.0017 (29) |
| Glycogen content | ||||
| Comamonas | 40 | +0.40 | 0.005 | 0.007 (7) |
| Comamonadaceae | 40 | +0.40 | 0.005 | 0.008 (6) |
| Achromobacter | 29 | +0.42 | 0.003 | 0.007 (7) |
| Alcaligenaceae | 29 | +0.42 | 0.003 | 0.007 (7) |
| Xanthomonadaceae OTU160 | 45 | +0.45 | 0.002 | 0.0022 (23) |
| Achromobacter OTU10 | 29 | +0.45 | 0.001 | 0.0022 (23) |
| Xanthomonadaceae | 45 | +0.45 | 0.001 | 0.008 (6) |
| Acetobacteraceae | 47 | −0.54 | <0.001 | 0.008 (6) |
The bacterial taxa in DGRP lines were family, genus, or OTU as indicated.
Adjusted with Bonferroni's correction for number of comparisons at each taxonomic level (see Data Set S2 in the supplemental material).
Host genetic determinants of the abundance of the bacterium A. tropicalis.
We hypothesized that host genetic factors may contribute to the variation in the bacterial communities among the DGRP. As a test of this hypothesis, we took a discovery-based approach, using genome-wide association (GWA) to identify Drosophila mutations that influence microbial abundance. The test flies were colonized with a defined, five-species microbiota used previously (9, 11, 22) to ensure uniform access to bacterial species, and the 103 DGRP lines comprised 61 lines used for pyrosequencing (Fig. 1A) (18 of the pyrosequenced lines were unsuitable for gnotobiotic culture, and 42 lines were added to increase sampling of genetic variation). We focused on the abundance of one bacterial species, A. tropicalis, which displayed high between-line variation in abundance, ranging from undetected (in three lines) to 20,000 CFU mg−1 fly (Fig. 2A). Overall, most of this variation (78%) could be attributed to host genotype. Then, as a surrogate for a functional screen for Drosophila genes that contribute to variation in A. tropicalis abundance, we conducted a genome-wide association of the abundance of A. tropicalis CFU with SNP identity. As in other Drosophila GWA studies (GWAS), P value distributions displayed bias (12, 38) (see Text S1, Fig. S1, and Fig. S2 in the supplemental material). Subsequent analysis was restricted to genes near SNPs with P values below a nominal threshold of 2 × 10−8. Seven SNPs associated with six unique genes fit these criteria (Table 2; all results shown in Data Set S3). Loss-of-function mutants derived by P- and Minos-elements or chemical mutagenesis were readily available for four of six genes: polychaetoid (pyd), paralytic (para), heartless (htl), and dunce (dnc) (Table S2B). The two other SNP-neighboring genes were Calnexin 14D (Cnx14D) and defensin (def), which have known functions in neural function and antimicrobial response, respectively.
FIG 2.
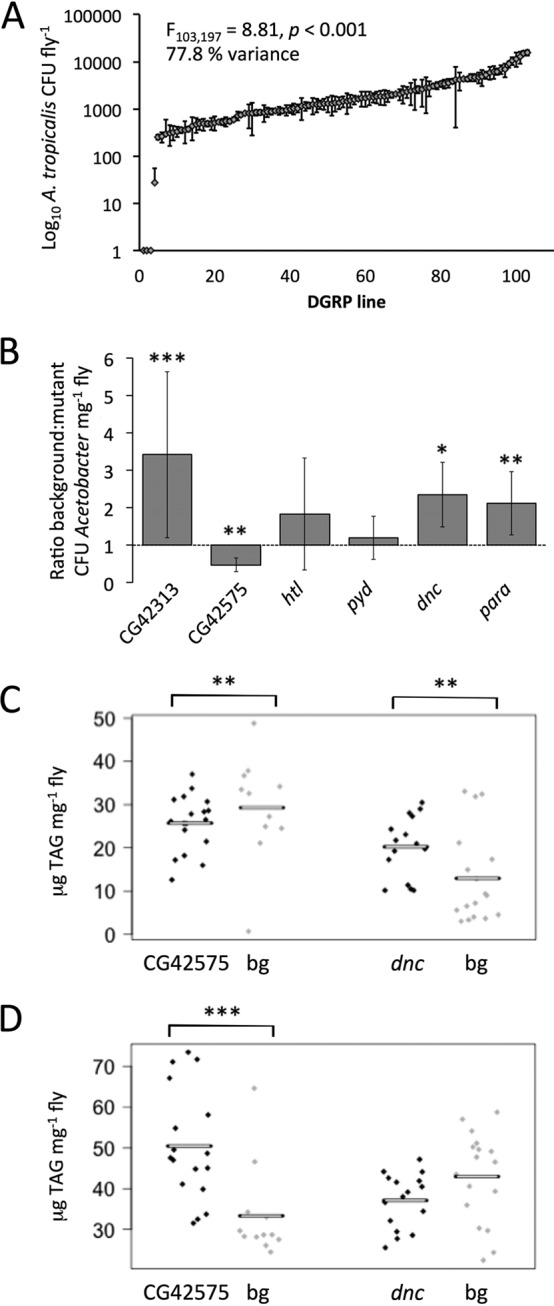
Host genetic control of the microbiota. (A) CFU counts per fly of A. tropicalis in five-species gnotobiotic DGRP lines (colonized with Acetobacter pomorum DmCS_004, A. tropicalis DmCS_006, Lactobacillus brevis DmCS_003, L. fructivorans DmCS_002, and L. plantarum DmCS_001), ranked by CFU abundance, and expressed as log10(number of CFUs +1). The contribution of genotype to the total variance (77.8%) was calculated as the square of the standard deviation when DGRP genotype was included as a random effect in the linear mixed model (LMM). (B) Validation of A. tropicalis abundance GWA. Differences between mutant and control lines each raised with the defined five-species microbiota were identified using a linear mixed model. Data are represented as means ± SEMs. (C) Mutants with altered microbiota composition have altered nutritional status. TAG content was measured in five-species gnotobiotic fly lines with mutations in genes that exert genetic control over the microbiota (CG42575, dnc) relative to background controls (bg). (D) Host genetic control of the microbiota mediates fly leanness. TAG content was measured in three-species (Lactobacillus brevis, L. fructivorans, and L. plantarum) gnotobiotic mutant fly lines (omission of Acetobacter species relieves host genetic control of Acetobacter-mediated TAG effects). In panels C and D, each symbol represents the value for an individual fly, and the horizontal bar shows the mean for the group. The data were evaluated by LMM and analysis of variance (ANOVA), with all statistical results shown in Table S1 in the supplemental material. Values that are significantly different are indicated by asterisks as follows: *, P < 0.05, ** P ≤ 0.01, *** P ≤ 0.001. bg, background.
TABLE 2.
Genes with associated SNPs that had P values < 10−9 in GWAS or were tested for an effect on microbiota composition
| Gene name | FlyBase IDa | P value | SNP rank(s)b | Mean CFU of A. tropicalis fly−1c |
No. of fly lines with the following allele type: |
Function | Human homologd |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| Major | Minor | Major | Minor | No. of homologs | Function | |||||
| Polychaetoid | FBgn 0262614 | 1.3e−10 | 1,5,9 | 2,839 | 406 | 81 | 8 | PDZ domain-containing | 3 | Tight junction proteins 1 to 3 |
| Paralytic | FBgn 0264255 | 3.1e−10 | 2 | 2,713 | 332 | 87 | 6 | Sodium channel | 9 | Sodium channels |
| Calnexin 14D | FBgn 0264077 | 3.1e−10 | 2 | 2,713 | 332 | 87 | 6 | Ca2+ binding | 4 | Including Ca2+ binding |
| Defensin | FBgn 0010385 | 8.6e−10 | 3 | 2,649 | 146 | 88 | 4 | Antimicrobial peptide | NA | NA |
| Heartless | FBgn 0010389 | 1.0e−09 | 4 | 2,572 | 150 | 85 | 5 | Fibroblast growth factor receptor (FGFR 1) | 4 | Including FGFR 1 to 4 |
| Dunce | FBgn 0000479 | 3.1e−09 | 6 | 2,868 | 379 | 81 | 11 | cAMP phosphodiesterase (PDE) | 4 | PDE4 (A to D) |
| CG42575 | FBgn 0260795 | 1.5e−07 | 17 | 2,568 | 157 | 84 | 4 | Unknown | 2 | Phosphate transporters |
| CG42313 | FBgn 0259213 | 2.7e−07 | 26 | 2,708 | 1,010 | 80 | 10 | Unknown | NA | NA |
FlyBase ID, FlyBase identifier.
Rank 1 has the lowest probability.
A. tropicalis with the major or minor allele.
NA, not available.
To validate genes that affect A. tropicalis abundance, we used Drosophila lines bearing mutations in the four genes in the preceding paragraph and, to identify roles for genes with uncharacterized functions, two genes of unknown function with the lowest P values for which mutants were readily available (CG42575 and CG42313 [Table 2]). As in previous experiments, eggs from each Drosophila line were raised under axenic or five-bacterial-species gnotobiotic conditions. Four of the six mutations, including mutations in the well-characterized dnc and para genes, had significant effects on A. tropicalis abundance relative to background controls (Fig. 2B; statistics in Table S1A in the supplemental material). dnc is a classical learning gene in Drosophila (39), encoding a cyclic AMP (cAMP) phosphodiesterase (40), and has homology to human genes associated with disease (e.g., schizophrenia [41]). para encodes a sodium channel that, when disrupted, leads to paralytic phenotypes at elevated temperatures (42) or to olfactory dysfunction (43), and its human homologs are associated with epilepsy (44). In summary, these findings identify specific host genetic factors with homology to human genes that influence the abundance of associated microorganisms in Drosophila.
Relationship between D. melanogaster genotype and A. tropicalis-dependent nutrition.
Our demonstration that host nutritional phenotype, first, varies with microbiota composition (Table 1) and, second, is associated with certain host genes (Table 2 and Fig. 2B) suggests that host genetic control of microbiota abundance may contribute to the effect of host genotype on nutritional indices. To test this hypothesis, our analysis focused on TAG, following the published demonstration of negative correlations between Acetobacter abundance and TAG content in one Drosophila line, Canton S (11). The first experiments colonized mutant flies and background control flies with five-species associations of Acetobacter and Lactobacillus species. As predicted, the dnc mutant (with reduced A. tropicalis load) displayed significantly elevated TAG levels, and the CG42575 mutant (with increased A. tropicalis load) had reduced TAG levels (Fig. 2C; statistics in Table S1B in the supplemental material). The TAG contents of para and CG42313 mutants were not significantly altered compared to background (P > 0.05; data not shown), perhaps because of genotype-specific effects in these mutants, and were omitted from subsequent analysis.
As a test that the interactions between host genotype and TAG content are mediated by Acetobacter, the mutant flies were reared with the three Lactobacillus species from the five-species microbiota used in the previous experiment. These associations were predicted not to replicate the effect of Acetobacter on TAG content, since Lactobacillus species do not reduce fly TAG content relative to axenic flies (11) (Table 1). Consistent with the expectation, the TAG content was not significantly affected by dnc mutation in Lactobacillus-colonized flies (Fig. 2D; statistics in Table S1C in the supplemental material) and was significantly increased by a Lactobacillus-only microbiota in the CG42575 mutant, the reverse of the effect in Acetobacter-colonized flies. These data demonstrate that the effects of the dnc and CG42575 mutations on host TAG content are congruent with their effects on the abundance of TAG-reducing Acetobacter and abolished in flies lacking Acetobacter. Taken together, the most parsimonious explanation is that some host genetic factors do not influence the metabolic determinants of TAG content directly but by their effect on the abundance of associated microorganisms.
DISCUSSION
This study investigated microbiota effects on the relationship between host genotype and phenotype, with respect to Drosophila nutritional traits. It extends previous research demonstrating strong statistical associations between genotype and multiple nutritional indices in the DGRP (45) to reveal that members of the microbiota are correlated with certain host nutritional indices. Multiple lines of evidence indicate that the bacteria are the causal basis of these correlations, at least with respect to the Acetobacteraceae: flies bearing Acetobacteraceae, but not Lactobacillus, display the predicted reduction in TAG and glycogen contents (18, 37; this study), likely driven by bacterially mediated competition for dietary carbohydrate (37) and stimulation of insulin signaling in the Drosophila host (18). Variation in the contributions of distinct bacterial taxa to different traits (Table 1; see Data Set S2 in the supplemental material) is consistent with previous demonstrations of strain-specific effects in Drosophila that are not limited to high-level taxonomic classifications (19, 22, 37).
Correlations between microbiota composition and host lipid levels have been obtained previously for mouse mutants (Toll-like receptor 5 [TLR5], MyD88, and NOD2), with the microbiota identified as a causal factor by reproducing the deleterious metabolic phenotype via transplantation of gut microbiota from mutant to wild-type mice (46–48). Our study of Drosophila shows that the interactive effects of host genotype and microbiota on nutritional phenotype are not unique to mice (or mammals), and may be general to animals. Furthermore, the ease with which associations with different defined bacterial communities can be constructed in Drosophila enabled us to identify empirically the critical bacterial taxa. As our study illustrates, relatively small changes in the abundance of certain microbial taxa can have significant impacts on host phenotype, emphasizing the importance of attention to both microbial identity and abundance (e.g., reference 49).
Immediately relevant to the increasing evidence for microbial impacts on host nutritional phenotypes is the basis of the variation in microbiota composition. Multiple studies have shown that the microbiome in Drosophila and other animals, including mammals, is variable. Processes contributing to this variation include stochastic variation, such that each individual host samples only a subset of the total compatible microorganisms (43), positive and antagonistic interactions among microbial taxa that may be mediated directly (e.g., metabolic cross-feeding, toxin production) and indirectly via the host immune system (11, 50–54), and environmental factors, including diet (9, 14, 55–60). Other studies are revealing significant associations of certain microbial taxa in the mammalian gut microbiota with both host genotype (10, 61–63) and host phylogeny (64, 65). The GWA of A. tropicalis abundance in Drosophila in this study confirms the importance of host genetic factors as determinants of the microbiota and identifies candidate host genes contributing to this variation. We note that the percentage of variance in microbial community composition attributed to host genotype in this GWAS analysis is high (Fig. 2A), and this is likely a consequence of all fly lines being inoculated with the same set of microbes at the same starting density. Host genotype likely accounts for a lower proportion of the variation in conventional flies in laboratory culture and wild populations, where the gut microbiota community composition can be influenced by the availability of bacteria in the external environment (14–16). Full understanding of the determinants of gut microbiota composition will require integration of these multiple genetic, physiological, and ecological factors.
Strongly represented among the genes identified from the GWAS and associated validation in this study are genes with annotated roles in neural function or preferential expression in neural tissues. This is intriguing given the growing evidence linking the microbiota with neural function and behavior in mammals (66–68). Although further research is required to establish the mechanistic basis of the relationship between neural genes and microbiota in Drosophila, variation in neural function may, for example, alter the feeding response, and hence the amount of food bearing Acetobacter that the flies ingest (demonstrated, e.g., by reference 69), or the suitability of the host environment for the colonizing Acetobacter. Also of great interest is the relationship between the microbiota effects and other phenotypic consequences of variation in host genes. For example, to what extent is the role of the functional dnc gene in sustaining Acetobacter populations causally linked to its contribution to multiple other phenotypic traits, including associative learning (39, 70), life span (71), reproduction (71, 72), courtship (73), circadian rhythm (74), and locomotion (72)? The ready availability of mutants and natural variants in Drosophila, together with methods to manipulate the microbial complement of the flies, provides the opportunity to dissect whether and how the different phenotypic traits are linked.
In conclusion, this study reveals host genotype-specific effects on microbiota composition as a causal determinant of animal phenotype. The general relevance of the results on the Drosophila model system is indicated by both broad parallels with data obtained in microbiome studies of mammals, including humans (75), and the strong representation of genes with mammalian homologs among the genes associated with microbiota phenotypes (Table 2). These results indicate that the fundamentals of animal-microbiota interactions may be evolutionarily conserved and driven by a common molecular processes.
ACKNOWLEDGMENTS
We thank L. Donahue, D. Duneau, S. Franzenburg, B. Gustin, S. Hermann, X. Jing, B. Lazzaro, D. Sannino, R. Schwenke, S. Short, R. Unckless, A. Wong, and S. Westmiller for technical support and J. Ayroles, A. G. Clark, B. Lazzaro, and R. Unckless for analytical support.
This study received financial support from NIH grant 1R01GM095372 to A.E.D. and Ruth L. Kirschstein NRSA postdoctoral fellowship 1F32GM099374-01 to P.D.N. Drosophila stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03301-15.
REFERENCES
- 1.McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Loso T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A 110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duca FA, Lam TK. 2014. Gut microbiota, nutrient sensing and energy balance. Diabetes Obes Metab 16(Suppl 1):S68–S76. doi: 10.1111/dom.12340. [DOI] [PubMed] [Google Scholar]
- 3.Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, Qiu Z, Maher L, Redinbo MR, Phillips RS, Fleet JC, Kortagere S, Mukherjee P, Fasano A, Le Ven J, Nicholson JK, Dumas ME, Khanna KM, Mani S. 2014. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flint HJ, Scott KP, Louis P, Duncan SH. 2012. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol 9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 5.Huang J-H, Douglas AE. 2015. Consumption of dietary sugar by gut bacteria determines Drosophila lipid content. Biol Lett 11:20150469. doi: 10.1098/rsbl.2015.0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manor O, Levy R, Borenstein E. 2014. Mapping the inner workings of the microbiome: genomic- and metagenomic-based study of metabolism and metabolic interactions in the human microbiome. Cell Metab 20:742–752. doi: 10.1016/j.cmet.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. 2013. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, Liu J, Houpt E, Li JV, Holmes E, Nicholson J, Knights D, Ursell LK, Knight R, Gordon JI. 2013. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong AC, Dobson AJ, Douglas AE. 2014. Gut microbiota dictates the metabolic response of Drosophila to diet. J Exp Biol 217:1894–1901. doi: 10.1242/jeb.101725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE. 2014. Human genetics shape the gut microbiome. Cell 159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newell PD, Douglas AE. 2014. Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in Drosophila melanogaster. Appl Environ Microbiol 80:788–796. doi: 10.1128/AEM.02742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobson AJ, Chaston JM, Newell PD, Donahue L, Hermann SL, Sannino DR, Westmiller S, Wong AC, Clark AG, Lazzaro BP, Douglas AE. 2015. Host genetic determinants of microbiota-dependent nutrition revealed by genome-wide analysis of Drosophila melanogaster. Nat Commun 6:6312. doi: 10.1038/ncomms7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandler JA, Lang JM, Bhatnagar S, Eisen JA, Kopp A. 2011. Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLoS Genet 7:e1002272. doi: 10.1371/journal.pgen.1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staubach F, Baines JF, Kunzel S, Bik EM, Petrov DA. 2013. Host species and environmental effects on bacterial communities associated with Drosophila in the laboratory and in the natural environment. PLoS One 8:e70749. doi: 10.1371/journal.pone.0070749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong AC, Chaston JM, Douglas AE. 2013. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J 7:1922–1932. doi: 10.1038/ismej.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ridley EV, Wong AC, Westmiller S, Douglas AE. 2012. Impact of the resident microbiota on the nutritional phenotype of Drosophila melanogaster. PLoS One 7:e36765. doi: 10.1371/journal.pone.0036765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, Lee WJ. 2011. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- 19.Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. 2011. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab 14:403–414. doi: 10.1016/j.cmet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 20.Mackay TF, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, Richardson MF, Anholt RR, Barron M, Bess C, Blankenburg KP, Carbone MA, Castellano D, Chaboub L, Duncan L, Harris Z, Javaid M, Jayaseelan JC, Jhangiani SN, Jordan KW, Lara F, Lawrence F, Lee SL, Librado P, Linheiro RS, Lyman RF, Mackey AJ, Munidasa M, Muzny DM, Nazareth L, Newsham I, Perales L, Pu LL, Qu C, Ramia M, Reid JG, Rollmann SM, Rozas J, Saada N, Turlapati L, Worley KC, Wu YQ, Yamamoto A, Zhu Y, Bergman CM, Thornton KR, et al. . 2012. The Drosophila melanogaster genetic reference panel. Nature 482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang W, Massouras A, Inoue Y, Peiffer J, Ramia M, Tarone AM, Turlapati L, Zichner T, Zhu D, Lyman RF, Magwire MM, Blankenburg K, Carbone MA, Chang K, Ellis LL, Fernandez S, Han Y, Highnam G, Hjelmen CE, Jack JR, Javaid M, Jayaseelan J, Kalra D, Lee S, Lewis L, Munidasa M, Ongeri F, Patel S, Perales L, Perez A, Pu L, Rollmann SM, Ruth R, Saada N, Warner C, Williams A, Wu YQ, Yamamoto A, Zhang Y, Zhu Y, Anholt RR, Korbel JO, Mittelman D, Muzny DM, Gibbs RA, Barbadilla A, Johnston JS, Stone EA, Richards S, Deplancke B, Mackay TF. 2014. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res 24:1193–1208. doi: 10.1101/gr.171546.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newell PD, Chaston JM, Wang Y, Winans NJ, Sannino DR, Wong ACN, Dobson AJ, Kagle J, Douglas AE. 2014. In vivo function and comparative genomic analyses of the Drosophila gut microbiota identify candidate symbiosis factors. Front Microbiol 5:576. doi: 10.3389/fmicb.2014.00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong CN, Ng P, Douglas AE. 2011. Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ Microbiol 13:1889–1900. doi: 10.1111/j.1462-2920.2011.02511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cenis JL, Perez P, Fereres A. 1993. Identification of aphid (Homoptera, Aphididae) species and clones by random amplified polymorphic DNA. Ann Entomol Soc Am 86:545–550. doi: 10.1093/aesa/86.5.545. [DOI] [Google Scholar]
- 25.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R Core Team. 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 28.Pinheiro J, Bates D, DebRoy S, Deepayan S, R Core Team . 2014. nlme: linear and nonlinear mixed effects models.
- 29.Ayroles JF, Carbone MA, Stone EA, Jordan KW, Lyman RF, Magwire MM, Rollmann SM, Duncan LH, Lawrence F, Anholt RR, Mackay TF. 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet 41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sundset MA, Edwards JE, Cheng YF, Senosiain RS, Fraile MN, Northwood KS, Praesteng KE, Glad T, Mathiesen SD, Wright AD. 2009. Molecular diversity of the rumen microbiome of Norwegian reindeer on natural summer pasture. Microb Ecol 57:335–348. doi: 10.1007/s00248-008-9414-7. [DOI] [PubMed] [Google Scholar]
- 31.Harbison ST, Carbone MA, Ayroles JF, Stone EA, Lyman RF, Mackay TF. 2009. Co-regulated transcriptional networks contribute to natural genetic variation in Drosophila sleep. Nat Genet 41:371–375. doi: 10.1038/ng.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hothorn T, Bretz F, Westfall P. 2008. Simultaneous inference in general parametric models. Biom J 50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 33.Brummel T, Ching A, Seroude L, Simon AF, Benzer S. 2004. Drosophila lifespan enhancement by exogenous bacteria. Proc Natl Acad Sci U S A 101:12974–12979. doi: 10.1073/pnas.0405207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cox CR, Gilmore MS. 2007. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect Immun 75:1565–1576. doi: 10.1128/IAI.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corby-Harris V, Pontaroli AC, Shimkets LJ, Bennetzen JL, Habel KE, Promislow DE. 2007. Geographical distribution and diversity of bacteria associated with natural populations of Drosophila melanogaster. Appl Environ Microbiol 73:3470–3479. doi: 10.1128/AEM.02120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamada Y, Yukphan P, Vu HTL, Muramatsu Y, Ochaikul D, Nakagawa Y. 2012. Subdivision of the genus Gluconacetobacter Yamada, Hoshino and Ishikawa 1998: the proposal of Komagatabacter gen. nov., for strains accomodated to the Gluconacetobacter xylinus group in the α-Proteobacteria. Ann Microbiol 62:849–859. doi: 10.1007/s13213-011-0288-4. [DOI] [Google Scholar]
- 37.Chaston JM, Newell PD, Douglas AE. 2014. Metagenome-wide association of microbial determinants of host phenotype in Drosophila melanogaster. mBio 5:e01631-14. doi: 10.1128/mBio.01631-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unckless RL, Rottschaefer SM, Lazzaro BP. 2015. The complex contributions of genetics and nutrition to immunity in Drosophila melanogaster. PLoS Genet 11:e1005030. doi: 10.1371/journal.pgen.1005030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis RL. 1996. Physiology and biochemistry of Drosophila learning mutants. Physiol Rev 76:299–317. [DOI] [PubMed] [Google Scholar]
- 40.Davis RL, Kiger JA Jr. 1981. Dunce mutants of Drosophila melanogaster: mutants defective in the cyclic AMP phosphodiesterase enzyme system. J Cell Biol 90:101–107. doi: 10.1083/jcb.90.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, Malloy MP, Chubb JE, Huston E, Baillie GS, Thomson PA, Hill EV, Brandon NJ, Rain JC, Camargo LM, Whiting PJ, Houslay MD, Blackwood DH, Muir WJ, Porteous DJ. 2005. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science 310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- 42.Grigliatti T, Suzuki DT, Williamson R. 1972. Temperature-sensitive mutation in Drosophila melanogaster. X. Developmental analysis of the paralytic mutation, para ts. Dev Biol 28:352–371. [DOI] [PubMed] [Google Scholar]
- 43.Lilly M, Kreber R, Ganetzky B, Carlson JR. 1994. Evidence that the Drosophila olfactory mutant smellblind defines a novel class of sodium channel mutation. Genetics 136:1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mulley JC, Scheffer IE, Petrou S, Dibbens LM, Berkovic SF, Harkin LA. 2005. SCN1A mutations and epilepsy. Hum Mutat 25:535–542. doi: 10.1002/humu.20178. [DOI] [PubMed] [Google Scholar]
- 45.Jumbo-Lucioni P, Ayroles JF, Chambers MM, Jordan KW, Leips J, Mackay TF, De Luca M. 2010. Systems genetics analysis of body weight and energy metabolism traits in Drosophila melanogaster. BMC Genomics 11:297. doi: 10.1186/1471-2164-11-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. 2010. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Everard A, Geurts L, Caesar R, Van Hul M, Matamoros S, Duparc T, Denis RG, Cochez P, Pierard F, Castel J, Bindels LB, Plovier H, Robine S, Muccioli GG, Renauld JC, Dumoutier L, Delzenne NM, Luquet S, Backhed F, Cani PD. 2014. Intestinal epithelial MyD88 is a sensor switching host metabolism towards obesity according to nutritional status. Nat Commun 5:5648. doi: 10.1038/ncomms6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denou E, Lolmede K, Garidou L, Pomie C, Chabo C, Lau TC, Fullerton MD, Nigro G, Zakaroff-Girard A, Luche E, Garret C, Serino M, Amar J, Courtney M, Cavallari JF, Henriksbo BD, Barra NG, Foley KP, McPhee JB, Duggan BM, O'Neill HM, Lee AJ, Sansonetti P, Ashkar AA, Khan WI, Surette MG, Bouloumie A, Steinberg GR, Burcelin R, Schertzer JD. 2015. Defective NOD2 peptidoglycan sensing promotes diet-induced inflammation, dysbiosis, and insulin resistance. EMBO Mol Med 7:259–274. doi: 10.15252/emmm.201404169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, et al. . 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 50.Lee KA, Kim SH, Kim EK, Ha EM, You H, Kim B, Kim MJ, Kwon Y, Ryu JH, Lee WJ. 2013. Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell 153:797–811. doi: 10.1016/j.cell.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 51.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. 2011. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. 2009. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Gene Dev 23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fischbach MA, Sonnenburg JL. 2011. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe 10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, Mercante JW, Alam A, Gates CL, Wu H, Swanson PA, Lambeth JD, Denning PW, Neish AS. 2013. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J 32:3017–3028. doi: 10.1038/emboj.2013.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O'Sullivan O, Fitzgerald GF, Deane J, O'Connor M, Harnedy N, O'Connor K, O'Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP, O'Toole PW. 2012. Gut microbiota composition correlates with diet and health in the elderly. Nature 488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 57.Faith JJ, McNulty NP, Rey FE, Gordon JI. 2011. Predicting a human gut microbiota's response to diet in gnotobiotic mice. Science 333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Degnan PH, Barry NA, Mok KC, Taga ME, Goodman AL. 2014. Human gut microbes use multiple transporters to distinguish vitamin B12 analogs and compete in the gut. Cell Host Microbe 15:47–57. doi: 10.1016/j.chom.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carmody RN, Gerber GK, Luevano JM Jr, Gatti DM, Somes L, Svenson KL, Turnbaugh PJ. 2015. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knights D, Silverberg MS, Weersma RK, Gevers D, Dijkstra G, Huang H, Tyler AD, van Sommeren S, Imhann F, Stempak JM, Huang H, Vangay P, Al-Ghalith GA, Russell C, Sauk J, Knight J, Daly MJ, Huttenhower C, Xavier RJ. 2014. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med 6:107. doi: 10.1186/s13073-014-0107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, Kachman SD, Moriyama EN, Walter J, Peterson DA, Pomp D. 2010. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A 107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leamy LJ, Kelly SA, Nietfeldt J, Legge RM, Ma F, Hua K, Sinha R, Peterson DA, Walter J, Benson AK, Pomp D. 2014. Host genetics and diet, but not immunoglobulin A expression, converge to shape compositional features of the gut microbiome in an advanced intercross population of mice. Genome Biol 15:552. doi: 10.1186/s13059-014-0552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ochman H, Worobey M, Kuo CH, Ndjango JB, Peeters M, Hahn BH, Hugenholtz P. 2010. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol 8:e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oh PL, Benson AK, Peterson DA, Patil PB, Moriyama EN, Roos S, Walter J. 2010. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J 4:377–387. doi: 10.1038/ismej.2009.123. [DOI] [PubMed] [Google Scholar]
- 66.Cryan JF, Dinan TG. 2012. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci 13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 67.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, Patterson PH, Mazmanian SK. 2013. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, Backhed F, Mithieux G. 2014. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 69.Broderick NA, Buchon N, Lemaitre B. 2014. Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. mBio 5:e01117-14. doi: 10.1128/mBio.01117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Byers D, Davis RL, Kiger JA Jr. 1981. Defect in cyclic AMP phosphodiesterase due to the dunce mutation of learning in Drosophila melanogaster. Nature 289:79–81. doi: 10.1038/289079a0. [DOI] [PubMed] [Google Scholar]
- 71.Bellen HJ, Kiger JA Jr. 1987. Sexual hyperactivity and reduced longevity of dunce females of Drosophila melanogaster. Genetics 115:153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sokolowski MB. 2001. Drosophila: genetics meets behaviour. Nat Rev Genet 2:879–890. doi: 10.1038/35098592. [DOI] [PubMed] [Google Scholar]
- 73.Greenspan RJ, Ferveur JF. 2000. Courtship in Drosophila. Annu Rev Genet 34:205–232. doi: 10.1146/annurev.genet.34.1.205. [DOI] [PubMed] [Google Scholar]
- 74.Stanewsky R. 2003. Genetic analysis of the circadian system in Drosophila melanogaster and mammals. J Neurobiol 54:111–147. doi: 10.1002/neu.10164. [DOI] [PubMed] [Google Scholar]
- 75.Erkosar B, Storelli G, Defaye A, Leulier F. 2013. Host-intestinal microbiota mutualism: “learning on the fly.” Cell Host Microbe 13:8–14. doi: 10.1016/j.chom.2012.12.004. [DOI] [PubMed] [Google Scholar]