Abstract
The interaction of crop pests with their natural enemies is a fundament to their control. Natural enemies of fungal pathogens of crops are poorly known relative to those of insect pests, despite the diversity of fungal pathogens and their economic importance. Currently, many regions across Latin America are experiencing unprecedented epidemics of coffee rust (Hemileia vastatrix). Identification of natural enemies of coffee rust could aid in developing management strategies or in pinpointing species that could be used for biocontrol. In the present study, we characterized fungal communities associated with coffee rust lesions by single-molecule DNA sequencing of fungal rRNA gene bar codes from leaf discs (≈28 mm2) containing rust lesions and control discs with no rust lesions. The leaf disc communities were hyperdiverse in terms of fungi, with up to 69 operational taxonomic units (putative species) per control disc, and the diversity was only slightly reduced in rust-infected discs, with up to 63 putative species. However, geography had a greater influence on the fungal community than whether the disc was infected by coffee rust. Through comparisons between control and rust-infected leaf discs, as well as taxonomic criteria, we identified 15 putative mycoparasitic fungi. These fungi are concentrated in the fungal family Cordycipitaceae and the order Tremellales. These data emphasize the complexity of diverse fungi of unknown ecological function within a leaf that might influence plant disease epidemics or lead to the development of species for biocontrol of fungal disease.
INTRODUCTION
Fungal biodiversity is closely linked to plant biodiversity, because a dominant number of fungal taxa are symbiotic with plants (1, 2). On one end of the symbiosis spectrum are plant pathogens, whose impacts are enormous in agricultural production, dramatically reducing yields or even eliminating production altogether, as well as in natural ecosystems, where they may facilitate plant biodiversity by Janzen-Connell effects, coevolutionary processes, or other mechanisms. At the other end of the spectrum are mutualists of plants, including beneficial mycorrhizae and endophytes that promote plant growth and facilitate stress tolerance (3, 4). Somewhere in the middle are the majority of plant-symbiotic fungi, with copious species detected within and on asymptomatic plant tissues. Most of these fungi have unclear roles with respect to plant health but comprise a massive and ubiquitous proportion of overall fungal biodiversity, especially in the lowland tropics, where every leaf sampled reveals the presence of endophytes by use of culture techniques (5, 6). Although some endophytes may have no direct effect on the host, what is becoming increasingly more appreciated is that endophytes can benefit the host through indirect actions, such as by protection from plant pests and pathogens (7, 8).
In natural ecosystems, cascading trophic interactions are tied to ecosystem stability and diversity (9). These trophic interactions involve numerous natural enemies of plant herbivores and pathogens. However, agricultural ecosystems provide scenarios in which pests and pathogens have fewer naturally existing enemies due to a loss of complexity of habitat. In situations where habitat complexity is built into the system, for example, in shade farming of coffee or cacao, the presence of natural enemies can reduce the need and cost for management by use of chemicals. Losses in crop yields due to pathogens are estimated to comprise 16% of potential yields (10), and the cost of preventing these losses is substantial. For example, approximately $2.8 billion is spent globally per year solely on chemical control of late blight of potato (11). On the other hand, fungi can also be used in the war against pests; such biocontrol agents include the insect pathogens Beauvaria bassiana and Nosema locustae. Only recently has attention been turned to the use of fungi as biocontrol agents of other fungi, in part because fungi occupying this niche are poorly known (12–14).
Among tropical crops, coffee ranks high in both its total value and its role in maintaining biodiversity when grown on a small scale and as an understory plant (i.e., shade coffee production) (15). In the last 2 years, coffee rust (Hemileia vastatrix) has emerged as a major disease in South and Central America, leading to reductions in annual production of over 10% (16) and threatening the livelihoods of hundreds of thousands of small-scale farmers in the region. Coffee rust has a history of devastation, having been discovered first in Sri Lanka (completely eliminating all production [17]) and later migrating to South America by 1970 and to Central America by 1976 (18). Only now is the disease becoming epidemic in the Americas, perhaps due to the potential for the fungus to overcome resistant plant varieties through evolution (19) and because of changes in farming practices that may increase disease transmission. The application of copper-based fungicides has been shown to be effective for treating the rust, but these chemicals are not readily available to the small-scale farmer and may have unintended consequences on potentially beneficial fungi. New approaches to combating rust are needed, and among these is the use of mycoparasites as biocontrol agents.
The best-known mycoparasite of H. vastatrix is the white halo fungus Lecanicillium lecanii (Cordycipitaceae), which is best known for being a parasite of scale insects, against which it is also a marketed product (Mycotal). In at least one field trial, the fungus showed high efficacy against coffee rust (20). Because of the complex trophic interactions of L. lecanii (21) and its relatively low growth rate in culture, it is unclear whether this fungus is the most virulent mycoparasitic natural enemy of coffee rust or the best candidate for development of a biocontrol strategy. The known diversity of mycoparasitic (or fungicolous, i.e., growing on other fungi) fungi on H. vastatrix is nine species (22). Additional studies to characterize the biotic community associated with coffee rust infection are needed to better understand the full spectrum of fungi, as well as other predators, involved in trophic interactions with the rust.
To begin documenting all of the fungi associated with coffee rust that may be mycoparasitic in nature, we developed a catalog of fungicolous fungi associated with H. vastatrix uredinia on coffee by using advanced DNA methods that allow high-throughput, unbiased molecular biodiversity discovery (23). We surveyed rust lesions from Puerto Rico and Mexico that had visible signs of coinfection with other fungi by using fungus-specific PCR and single-molecule DNA sequencing and then compared the data to those for leaf tissue lacking rust pustules. These data reveal strikingly diverse populations of fungi, confirm suspected mycoparasites, and identify putative novel mycoparasites that may be involved in controlling rust in more natural coffee ecosystems.
MATERIALS AND METHODS
Field sampling.
Sampling was conducted in the summer of 2013 on three coffee farms in the Soconusco region in Chiapas, Mexico (Tapachula municipality), on June 20, and on five coffee farms in the central mountain region of Puerto Rico (municipalities of Orocovis and Utuado), between July 16 and 23. Coffee plants were selected based on evident signs of rust lesions and visible potential coinfection with other fungi. Infected rust pustules were sampled by punching out a leaf disc (≈28 mm2), using a sterilized hole puncher. Sixteen rust-infected samples were obtained in Mexico, and 23 were obtained in Puerto Rico. In Puerto Rico, additional samples were taken from leaves that were rust infected, but from leaf regions that lacked rust pustules (control samples). Samples were stored in 95% ethanol (EtOH) in the field and transported back to the lab for DNA analysis. From these samples, 39 rust-infected samples and 7 control samples were subsampled for DNA analysis.
DNA methods.
The discs were dried under vacuum to remove residual EtOH. DNAs were extracted from the 46 discs by use of Qiagen Plant minikits. DNA was diluted to approximately 1 to 5 ng/μl and amplified by PCR, using 10 μl of template with GoTaq Green PCR mix (Promega) in an Eppendorf Mastercycler Pro S thermocycler. We first confirmed the presence or absence of H. vastatrix on infected/control discs by using rRNA gene primers specific to the rust internal transcribed spacer (ITS) region. The forward primer was Hv-ITS-F (5′-CTGCGGCAATTTATTGCTTA), and the reverse primer was Hv-ITS-R (5′-AATGGCAAGCACCCAATATC). The PCR conditions were 94°C for 3 min followed by 25 cycles of 94°C for 1 min, 55°C for 30 s, and 72°C for 1 min, with a 7-min final extension at 72°C. Both control and infected-disc extracts were used to amplify the rRNA gene ITS region with fungus-specific primers ITS1-F and ITS4 (24). Primers were bar coded using 16-bp extensions on the ITS1-F primer for the purposes of multiplexing, using sequences provided by PacBio. PCR was conducted using the following temperature profile: 94°C for 3 min followed by 25 cycles of 94°C for 45 s, 53°C for 30 s, and 72°C for 2 min, with a 7-min final extension at 72°C. Amplification was checked by gel electrophoresis, and then the products were purified using a Qiagen PCR purification kit. DNA was quantified on a NanoDrop spectrophotometer. The 24 samples were then pooled by combining 40 ng of each purified sample in a final volume of 40 μl.
We also added to the pooled samples a mock community of six species of fungi to determine sequencing error rates. The mock community was a combination of DNAs extracted from pure cultures isolated from green coffee beans (Aspergillus niveoglaucus, Penicillium cf. citrinum, Sporobolomyces sp., Sporidiobolus ruineniae, Fusarium cf. lateritium, and Cystofilobasidium ferigula). DNAs were extracted from these cultures by using tissue removed from the surface of nutrient agar following the method of James et al. (25).
The pooled sample was then used to generate a P4-C2 library by using a DNA Template Prep 2.0 kit for sequencing on a single SMRT cell of a PacBio-RS II machine at the University of Michigan Sequencing Core. A single SMRT cell movie yielded 52,393 reads of insert, with a mean of 12.3 passes and 27.7 Mb of total data.
Data analysis.
The bash5tools.py script from the pbh5tools package (https://github.com/PacificBiosciences/pbh5tools) was used to extract the circular consensus sequencing (CCS) sequences with a minimum of 6 passes (n = 40,110). The data were demultiplexed and trimmed using the trim.seqs command in mothur v 1.32.1 (26), with the following parameters: qaverage = 71, checkorient = t, maxambig = 2, maxhomop = 20, bdiffs = 1, pdiffs = 1, and minlength = 300. We detected chimeras by using the uchime algorithm implemented in mothur. Operational taxonomic units (OTUs) were constructed by evaluating all pairwise distances between unique sequences, and then clustering using the average neighbor method with a 0.03 distance was used for all analyses. Representative OTU sequences were extracted for taxonomic classification using a recent Unite (27) database (sh_refs_qiime_ver6_dynamic_10.09.2014). The program ITSx (28) was used to divide the representative sequences into ITS1 and ITS2 spacer regions, and the full-length sequence as well as the ITS1 and ITS2 regions was separately classified using the classify.seqs command in mothur with the Unite database.
After quality control (see the next section), community analyses were performed in mothur v.1.32.1 (26) and R v. 3.1.2 (29). The mothur sub.sample command was used to generate subsamples from each sample with the same number of sequences. Rarefaction curves for each sample were generated using the mothur rarefaction.single command. The mothur merge.groups and venn commands were used to combine samples in each group and to visualize overlap in the number of OTUs. Measures of alpha diversity (Chao1 richness estimator and inverse Simpson diversity index) averaged across 1,000 subsamples of 143 sequences from each sample were calculated using the mothur summary.single command. Kruskal-Wallis tests were used in R to compare the numbers of OTUs, the Chao1 estimator values, and the inverse Simpson indexes among samples from the three groups: Mexico infected (Mex), Puerto Rico infected (PR), and Puerto Rico control (PR_C). Kruskal-Wallis tests were used due to heteroscedasticity in the response variables across groups.
To examine beta diversity among our samples, a distance matrix was generated by calculating Yue and Clayton's theta value, a measure of community dissimilarity, via the mothur dist.shared command. Principle coordinate analysis was performed using the resulting matrix via the mothur pcoa command, and samples were plotted along the first two pcoa axes in R. Analysis of molecular variance (AMOVA) was used to examine differences among samples from the three groups via the mothur amova command. Finally, differences between the PR and PR_C groups in the relative abundance of each OTU were examined using the mothur metastats command.
Quality control of PacBio CCS data.
Because PacBio sequencing technology is relatively new and has not been used widely for rRNA gene sequencing, we analyzed the quality of the data in a number of ways. The 40,110 6-pass consensus sequences were first demultiplexed in mothur. Of these, 24,273 were successfully demultiplexed, with the failures being due to poor sequencing quality near the bar code or primers. We examined the impact of adjusting the average read Q score on the recovery of CCS sequences and on the sequence differences among the dominant species in the mock community (Aspergillus niveoglaucus). These data revealed that the 6-pass CCS sequences had a very low mean error rate, with mean distances ranging from 0.005 to 0.001 between sequences from A. niveoglaucus (across the range of average Q scores of 67 to 72 [denoted here as Q67 to Q72]). While the mean error was low, some putatively lower-quality sequences remained, as evidenced by pairwise distances above 0.03 at average Q scores of 70 or less (see Fig. S1 in the supplemental material). Using the entire set of 24 samples, we investigated the effects of quality filtering and distance used for clustering OTUs on the number of OTUs recovered, both with and without filtering OTUs found fewer than 5 times. The number of OTUs was sensitive to quality filtering at a clustering distance of 0.03, with values ranging from 776 to 1,405 (excluding Q72) (see Fig. S2A). However, this variation largely disappeared when only OTUs found 5 or more times were considered, with the number of OTUs ranging from 322 to 443 (see Fig. S2B). These data show that Q71 is a highly conservative quality filter for the data and is not very sensitive to the cutoff level for OTU clustering, especially considering only nonsingleton OTUs. For example, the difference between OTU distance cutoffs of 0.01 and 0.03 for Q71 is 342 versus 322. We therefore proceeded with our analysis by using reads with an average quality score of Q71, an average OTU distance of 0.03, and a minimum OTU count of 5 sequence reads.
From the initial 24,273 demultiplexed reads, we removed 11,989 CCS sequences due to overall low quality, another 53 comprising a single sample of low yield, and 689 from the mock community, leaving 11,542 sequences for analysis. Chimeras (n = 19) were removed and the data clustered into 770 OTUs. We eliminated OTUs found fewer than 5 times, leaving 318 high-confidence OTUs. We then used three criteria to further detect remaining chimeras: sequences that were found in only one sample, had a BLAST match to GenBank of <97%, and showed clear (>80% similarity and >50% coverage) BLAST matches to different taxonomic orders when classified separately for ITS1 and ITS2. Five additional OTUs were removed, leaving a final number of 313 OTUs.
Nucleotide sequence accession numbers.
All 313 unique, high-confidence OTUs were submitted to GenBank, using a single representative of each OTU, under accession numbers KT328605 to KT328917.
RESULTS
We successfully PCR amplified the ribosomal ITS region specific to H. vastatrix for 34 of the 39 lesion samples but none of the 7 controls. Rust was not amplified using the universal fungus-specific primers ITS1-F and ITS4 under the thermocycling conditions we utilized. The number of lesions to be analyzed by community profiling was then restricted to those in which we successfully detected H. vastatrix and reduced to a number that would maximize the number of reads per sample produced from a single SMRT cell. We proceeded forward with PacBio sequencing of amplicons from 24 samples, including the 7 controls and the mock community. After sequencing, one control sample was removed due to low sequence recovery.
Community analyses.
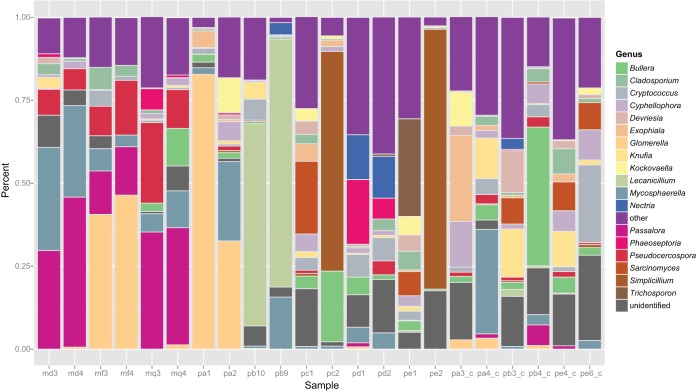
The 313 OTUs from the leaf samples showed a typical rank abundance distribution, with a few dominant taxa found in more than 10 samples and a large number of taxa found in low abundance (Table 1; see Table S1 in the supplemental material). The majority of sequences were in Ascomycota classes Dothideomycetes and Sordariomycetes. Samples were typically dominated by two or three genera with high prevalences as well as containing a number of rarer genera (Fig. 1).
TABLE 1.
Top 20 OTUs by abundancea
| OTU | OTU ID | Class | Order | No. of sequences | No. of samples |
|---|---|---|---|---|---|
| 1 | Glomerella cingulata (46) | Sordariomycetes | Incertae sedis | 1,284 | 10 |
| 2 | Passalora sp. CBS 113378 (49) | Dothideomycetes | Capnodiales | 1,004 | 10 |
| 3 | Mycosphaerella sp. AA_2012 (53) | Dothideomycetes | Capnodiales | 468 | 10 |
| 4 | Pseudocercospora norchiensis (100) | Dothideomycetes | Capnodiales | 458 | 14 |
| 5 | Lecanicillium sp. (30) | Sordariomycetes | Hypocreales | 439 | 6 |
| 6 | Simplicillium lanosoniveum (94) | Sordariomycetes | Hypocreales | 340 | 5 |
| 7 | Lecanicillium fusisporum (90) | Sordariomycetes | Hypocreales | 327 | 1 |
| 8 | Mycosphaerella sp. (100) | Dothideomycetes | Capnodiales | 270 | 12 |
| 9 | Bullera sp. VY_86 (26) | Tremellomycetes | Tremellales | 255 | 7 |
| 10 | Phaeoseptoria sp. FF_2011 (100) | Dothideomycetes | Pleosporales | 176 | 9 |
| 11 | Cladosporium ramotenellum (49) | Dothideomycetes | Capnodiales | 174 | 17 |
| 12 | Nectria aurantiaca (15) | Sordariomycetes | Incertae sedis | 147 | 7 |
| 13 | Bullera sp. VY_86 (79) | Tremellomycetes | Tremellales | 131 | 9 |
| 14 | Trichosporon laibachii (100) | Tremellomycetes | Trichosporonales | 124 | 4 |
| 15 | Sarcinomyces sp. SL_2011 (24) | Eurotiomycetes | Incertae sedis | 112 | 4 |
| 16 | Exophiala eucalyptorum (23) | Eurotiomycetes | Chaetothyriales | 103 | 1 |
| 17 | Mycosphaerella yunnanensis (95) | Dothideomycetes | Capnodiales | 95 | 4 |
| 18 | Cyphellophora eugeniae (66) | Eurotiomycetes | Chaetothyriales | 95 | 4 |
| 19 | Knufia perforans (42) | Incertae sedis | Incertae sedis | 92 | 1 |
| 20 | Kockovaella schimae (52) | Tremellomycetes | Tremellales | 87 | 2 |
Samples were classified using classify.seqs in mothur with the sh_refs_qiime_ver6_dynamic_10.09.2014 database. The OTU ID shows the tentative species assignment found, with the bootstrap confidence interval shown in parentheses. Class and order are assignments from the mothur classify.seqs algorithm. The number of sequences shows the total number of sequences of the OTU across all samples. The number of samples shows the number of samples from which the OTU was recovered.
FIG 1.
Relative abundances of fungal genera across 22 samples. Samples beginning with “m” are from Mexico (rust infected), samples beginning with “p” are from Puerto Rico (rust infected), and samples ending with “_c” are control, uninfected leaves from Puerto Rico.
The number of OTUs per sample ranged from 13 to 69 OTUs after quality control and after removing low-abundance OTUs (<5 sequences). Sample pb3_c, a control sample, had the most OTUs, and sample pe1 had the most OTUs for a rust-infected sample, with 62 OTUs (Table 2). The sample with the smallest number of sequences was pd2, with 143 sequences. To normalize the data for further analyses, a subsample of 143 sequences was generated from each sample. Rarefaction curves showed that most of the samples had not reached saturated sequencing at that level of sampling (data not shown). The rarefaction curves averaged across samples for the three sample groups (Mex, PR, and PR_C) showed that the control samples tended to have higher diversity (Fig. 2).
TABLE 2.
Diversity indices for sampled leaf discsa
| Disc | Treatment | No. of sequences | No. of OTUsb | S.obs | InvS | InvS_lci | InvS_hci | Chao1 | Chao1_lci | Chao1_hci |
|---|---|---|---|---|---|---|---|---|---|---|
| md3 | Mex | 360 | 27 | 20.66 | 5.10 | 4.18 | 6.52 | 27.56 | 22.20 | 52.35 |
| md4 | Mex | 485 | 26 | 19.04 | 3.54 | 2.94 | 4.44 | 25.10 | 20.37 | 47.37 |
| mf3 | Mex | 643 | 28 | 19.37 | 5.00 | 3.94 | 6.85 | 26.70 | 20.97 | 54.34 |
| mf4 | Mex | 485 | 24 | 16.04 | 3.76 | 3.07 | 4.87 | 22.70 | 17.43 | 49.53 |
| mq3 | Mex | 564 | 41 | 24.23 | 5.18 | 4.18 | 6.80 | 37.85 | 27.87 | 76.04 |
| mq4 | Mex | 830 | 37 | 24.14 | 6.09 | 4.78 | 8.39 | 33.82 | 26.56 | 63.56 |
| pa1 | PR | 704 | 24 | 12.44 | 1.46 | 1.28 | 1.71 | 19.56 | 13.94 | 47.76 |
| pa2 | PR | 518 | 45 | 30.20 | 5.74 | 4.59 | 7.65 | 45.55 | 34.88 | 80.97 |
| pb10 | PR | 539 | 38 | 22.28 | 2.65 | 2.14 | 3.48 | 35.32 | 25.69 | 73.17 |
| pb9 | PR | 566 | 18 | 9.17 | 1.73 | 1.50 | 2.06 | 15.64 | 10.45 | 43.92 |
| pc1 | PR | 397 | 48 | 39.29 | 23.32 | 18.39 | 31.91 | 47.12 | 41.41 | 68.69 |
| pc2 | PR | 464 | 22 | 12.66 | 2.12 | 1.81 | 2.56 | 21.35 | 14.58 | 53.86 |
| pd1 | PR | 570 | 62 | 43.39 | 14.81 | 11.02 | 22.62 | 59.32 | 48.73 | 91.23 |
| pd2 | PR | 143 | 33 | 33.00 | 23.18 | 18.54 | 30.91 | 35.63 | 33.47 | 47.67 |
| pe1 | PR | 411 | 63 | 43.86 | 10.23 | 7.30 | 17.09 | 62.76 | 50.33 | 99.32 |
| pe2 | PR | 221 | 13 | 11.06 | 5.08 | 4.38 | 6.05 | 15.40 | 11.80 | 37.79 |
| pa3_c | PR_C | 398 | 29 | 22.79 | 7.89 | 6.38 | 10.33 | 28.19 | 23.93 | 49.26 |
| pa4_c | PR_C | 394 | 37 | 31.22 | 12.70 | 9.42 | 19.51 | 35.17 | 32.04 | 51.02 |
| pb3_c | PR_C | 847 | 69 | 49.50 | 27.03 | 20.64 | 39.26 | 67.03 | 55.72 | 99.20 |
| pb4_c | PR_C | 354 | 32 | 25.06 | 5.30 | 4.00 | 7.83 | 32.25 | 26.66 | 58.37 |
| pe4_c | PR_C | 289 | 55 | 46.22 | 37.36 | 30.30 | 48.77 | 56.64 | 49.32 | 81.55 |
| pe6_c | PR_C | 342 | 43 | 35.10 | 12.86 | 9.79 | 18.76 | 42.45 | 37.03 | 63.68 |
Mex, Mexico infected; PR, Puerto Rico infected; PR_C, Puerto Rico control; S.obs, average number of OTUs present in subsamples of 143 sequences; InvS, inverse Simpson diversity index, with lower (lci) and upper (hci) 95% confidence intervals; Chao1, Chao1 richness estimator, with lower (lci) and upper (hci) 95% confidence intervals.
Ignoring those occurring fewer than five times per sample.
FIG 2.
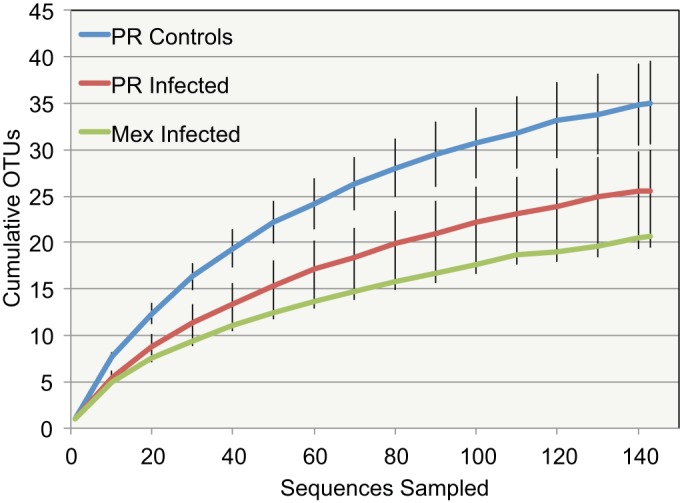
OTU accumulation curves averaged for infected samples from Mexico (Mex infected) and infected (PR infected) and control (PR controls) samples from Puerto Rico after rarefaction to 143 sequences per sample. Error bars show standard errors.
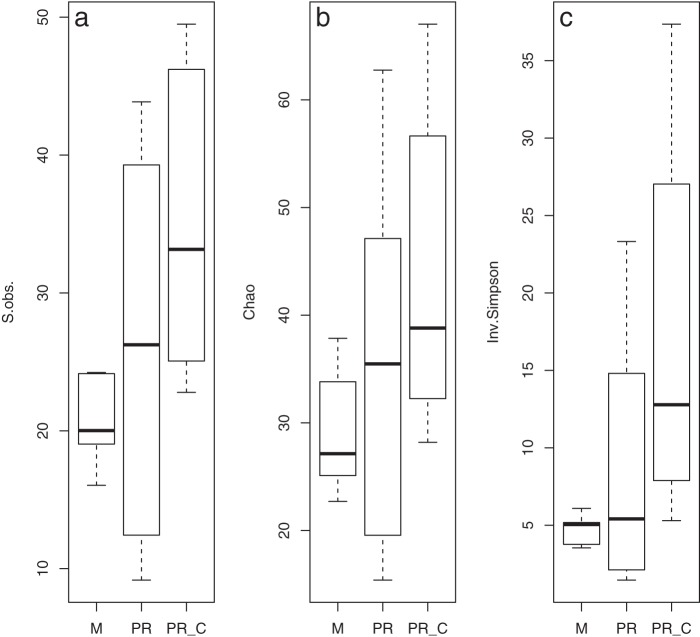
The number of OTUs in each subsample and the Chao1 and inverse Simpson index values (including confidence intervals) are reported in Table 2. Control samples tended to have more OTUs than samples from infected leaves at either site. After rarefaction, among the controls, sample pb3_c had the largest number of OTUs (n = 50), and among the rust-infected samples pe1 had the largest number of OTUs (n = 44). However, the number of OTUs observed and the Chao1 richness estimator did not differ significantly between the three groups (Fig. 3a and b) (by the Kruskal-Wallis test, for the number of OTUs observed, χ2 = 5.10, df = 2, and P = 0.078; and for Chao1, χ2 = 2.58, df = 2, and P = 0.28). The inverse Simpson diversity index differed marginally among groups (Fig. 3c) (by the Kruskal-Wallis test, χ2 = 5.88, df = 2, and P = 0.053).
FIG 3.
Box plots for the number of OTUs observed (S.obs.) (a), the Chao1 richness estimator (b), and the inverse Simpson diversity index (Inv.Simpson) (c), calculated for an average of 1,000 subsamples of 143 sequences from each sample. Samples are grouped into Mexico infected (M), Puerto Rico infected (PR), and control (PR_C) groups.
The overlap among OTUs in each sample group is shown in Fig. 4, revealing that more OTUs were shared between the two Puerto Rican samples (control and rust infected) than with the Mexican rust-infected samples. All samples were plotted along the first two coordinates of a principle coordinate analysis, which explained 16.35% and 14.56% of the variation in the fungal community composition of samples (Fig. 5). The plot shows that the Mexican samples cluster separately from both the PR and PR_C samples. The two PR samples that are closest to the Mexican samples in the ordination shared the presence of the most abundant OTU, Glomerella cingulata (see Table S1 in the supplemental material). AMOVA results further suggested that the fungal community composition of the Mexican samples differed significantly from that of the Puerto Rican samples and that the compositions of the control and infected samples from Puerto Rico did not differ from each other (Table 3).
FIG 4.
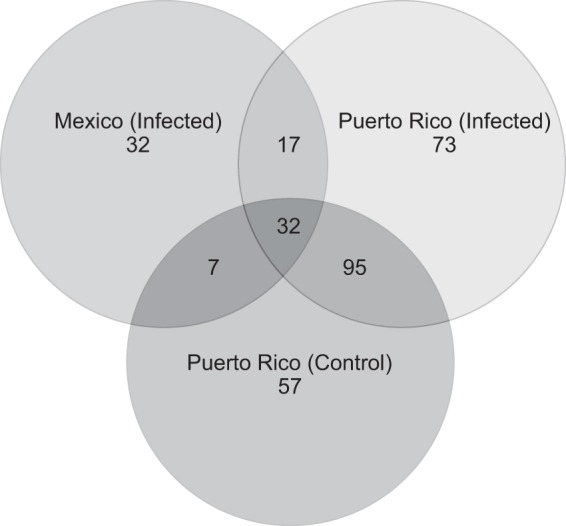
Venn diagram showing the overlaps of OTUs for samples from Mexico (infected), Puerto Rico (control), and Puerto Rico (infected).
FIG 5.
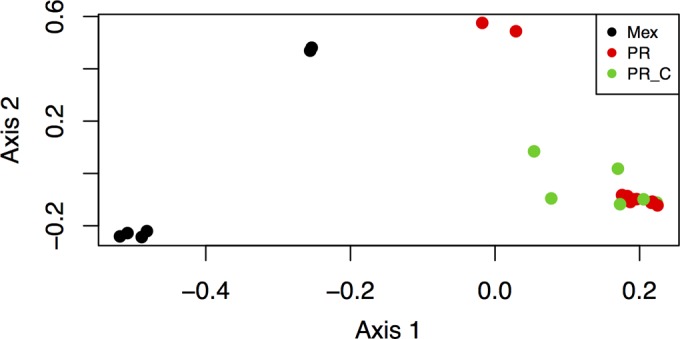
Plot of samples along the first two principle coordinates of a principle coordinate analysis. Samples are grouped into Mexico infected (black), Puerto Rico infected (red), and control (green) groups.
TABLE 3.
AMOVA results for analyses of community composition including all three groups of samples (3-way comparison) and all pairwise comparisons
| Comparison | F value | df (among, within) | P value |
|---|---|---|---|
| 3-way comparison | 2.34 | 2, 19 | <0.001 |
| Paired (Mex-PR) | 3.47 | 1, 14 | <0.001 |
| Paired (Mex-PR_C) | 3.41 | 1, 10 | 0.003 |
| Paired (PR-PR_C) | 0.92 | 1, 14 | 0.66 |
The results of the metastats analysis of a random subsample of 168 sequences (the smallest number of sequences in a sample when including rare OTUs) suggest that 32 OTUs differed significantly between PR and PR_C (the top 20 based on P values are shown in Table S2 in the supplemental material). However, taking multiple comparisons into account by using the false-discovery rate (see the q values in Table S2), it is difficult to say with confidence that any particular OTU differed significantly between the samples. On the other hand, a listing of the most abundant (or across samples) OTUs primarily found in the infected samples shows that a number of these samples are from genera known to be mycoparasites, e.g., Lecanicillium and Simplicillium (Table 1). Moreover, there were a number of additional low-abundance OTUs that are putative mycoparasites also detected in this study, and the complete list of putative mycoparasites is given in Table S3.
DISCUSSION
We recovered a surprisingly high level of fungal diversity from extraordinarily small samples of coffee leaf material (≈28 mm2). As many as 44 fungal OTUs could be recovered from as few as 143 sequences for a given rust-infected sample. This is further remarkable because the samples were visibly dominated by coffee rust biomass. Our data corroborate the hyperdiversity of fungal endophytes and epiphytes known from tropical ecosystems (5, 30, 31) and clearly demonstrate the compact and intermingled nature of fungal leaf-inhabiting communities. “Hyperdiversity” implies a higher-than-average diversity relative to those of other types of communities. The communities associated with coffee leaves are considered hyperdiverse because they revealed over 300 fungal OTUs after sampling of an area much smaller than that of an average single leaf (≈6 cm2), and many more if OTUs represented by only a single occurrence are considered (770 OTUs). Studies that have investigated foliar fungi at a very fine spatial scale have similarly shown that the extent of fungal mycelium within and on individual leaves may be quite reduced (6, 32). Our surveys revealed diverse fungi from nearly every ecological guild, e.g., saprotrophs, endophytes, plant pathogens, mycoparasites, etc. (see Table S1 in the supplemental material for a full listing of OTU assignments). The majority of the fungi in these ecosystems are not well characterized with respect to their ecological role, or their role may depend on environmental context, and thus they all should be considered potentially part of the disease epidemiology of coffee rust.
Our strategy of sequencing the fungal communities of visibly colonized rust lesions identified at least 15 likely mycoparasites, either because they were significantly associated with the lesions compared to controls or because they are phylogenetically related to suspected mycoparasitic species (see Table S3 in the supplemental material). Previous surveys of mycoparasites on coffee rust relied on opportunistic surveys that used culturing and morphological identification to identify six species (22). The absence of DNA sequence data from previous studies of coffee rust mycoparasites and the shifting classification of the simple anamorphic fungi identified make it hard to know whether we recovered the same species as those in earlier studies. The prior studies were also performed before advances in DNA methods made it straightforward to match species across studies through bar codes and to reveal cryptic species within morphological species. The need for such an approach is obvious, for example, considering that we detected four distinct species of Simplicillium in this study, and these species may have different levels of virulence on H. vastatrix. Moreover, the 15 mycoparasitic “species” identified here are likely an underestimate because of the lack of resolution provided by the ITS rRNA gene marker locus that we used. As an example, the most common OTU in our sample, Colletotrichum gloeosporioides/Glomerella cingulata, is known to be a name applied to a number of closely related species that are poorly separated using ITS sequences (33). A similar phenomenon of multiple species within what we are calling a single OTU is likely to exist within other taxa identified in this study.
The statistical approach (see Table S2 in the supplemental material) was less powerful for identifying mycoparasites than phylogenetic approaches, which we believe is due to the high diversity and stochastic composition of parasitized rust lesions. In fact, most of the known mycoparasites were not detected by this method, e.g., the Lecanicillium and Simplicillium OTUs that dominated samples pb9, pb10, pc2, and pe2 (Fig. 1). These high-abundance mycoparasites are the best candidates for species specifically parasitizing the rust. Lecanicillium lecanii is already well known as an attacker of coffee rust pustules as well as a pathogen of insects (34–36). The fact that one fungus could reduce two coffee pests may be a bonus for farmers, but this species may not be the ideal rust biocontrol agent, and its role in suppression of coffee rust involves a number of complex, context-dependent ecological interactions (21). One of our OTUs, OTU 5, has its closest match to Cordyceps confragosa (95% identity), a known teleomorph of L. lecanii. Other OTUs match other Lecanicillium spp. and Simplicillium spp., which all have similar verticillium-like morphologies (37). Given that we identified a number of distinct anamorphic Cordycipitaceae species on rust pustules within a small geographical region, it is clear that the taxonomy and virulence properties of these species will need to be resolved before assuming that all “white halo” infections of scales and coffee rust are caused by the same species. Moreover, additional studies need to be performed to determine whether the same fungus attacks both coffee rust and coffee scale insects, though the spatial association of local coffee scale insect epidemics caused by Lecanicillium is associated with a reduced local abundance of coffee rust in subsequent years (38).
Geography was shown to be a greater determinant of fungal community structure than infection status (Fig. 5). Multiple factors, such as management (coffee varieties, use of fungicides, or use of shade trees), climatic conditions (higher precipitation at the Mexico site than at the Puerto Rico site), biogeography (historical movement of coffee plants), and the background ecological community surrounding the farms, could be causative, as these factors dramatically affect fungal communities. Unfortunately, the absence of control samples from Mexico and other samples across management practices and geographies partially limits our ability to infer the mechanism by which leaf fungal communities differ. An intensive culture-based survey of coffee endophytes similarly reported little overlap of OTUs across countries (39). The culture-based studies have all shown that Colletotrichum is perhaps the most common endophyte taxon in coffee (39–41). In the present study, a species (or set of closely related species) of Colletotrichum was also the most common OTU, but surprisingly, when it was the dominant taxon of a leaf disc, the disc was rust infected. In general, however, the overlap between the surveys of coffee endophytes and the fungi we identified is quite small. Only 24 of the 257 OTUs found by Vega et al. (39) were also detected in our survey. Our results also differed in taxonomic spectrum from culture-based methods in having a higher diversity of taxa from Dothideomycetes and Basidiomycota. One important factor is that our methods sampled a large proportion of the community, including epiphytes, endophytes, and transient spores that may have been attached to the coffee leaves.
Culturing and inoculation studies should now be used to test whether the candidate mycoparasitic fungi we identified here are actual rust mycoparasites. We are particularly interested in testing the idea that Colletotrichum/Glomerella and the abundant Capnodiales OTUs behave as mycoparasites, though their typical roles are considered endophytic or plant parasitic. The most abundant OTU (Glomerella cingulata = Colletotrichum gloeosporioides) was found at high abundance on rust lesions in both Mexico and Puerto Rico. C. gloeosporioides is a very common endophyte of multiple tropical tree species, and it has been shown to behave as a suppressor of fungal diseases of cacao in inoculation trials (7, 12). In this scenario, positive effects on the host from the C. gloeosporioides inoculation may be the result of direct negative effects on fungal pathogens of coffee.
The diversity of fungi was marginally higher within and on uninfected leaf discs based on the inverse Simpson index, with a similar trend for the number of OTUs, but the differences are subtle. Lower diversity in infected leaves could be due to the competition of the rust fungus with endophytes, epiphytes, and other plant pathogens. On the other hand, the differences in community structure between infected and uninfected fungal communities in Puerto Rico as analyzed by AMOVA were not significant. This was unexpected, as the presence of rust fungus is expected to dramatically change the microenvironment experienced by other fungi. One possibility is that the control discs had background levels of rust fungi that were not as apparent as those in lesions. Although the rust fungus is not systemic (42), the rust may not have been readily apparent in leaf discs that comprised control samples, but nonetheless was there. However, our PCR assay for H. vastatrix should have reduced the likelihood of this occurring. Another possibility is that coffee rust may influence the rest of the fungal community at a larger scale than that addressed by our sampling here (e.g., at the scale of the individual plant or even the entire farm). In future studies, additional control samples from a farm entirely without rust or from a plant or leaf without rust should thus be informative.
This study demonstrates the power of next-generation sequencing for revealing hidden fungal diversity in complex samples. This method bypasses the labor-intensive step of culturing and identifying strains as well as avoiding the biases involved in culturing because of the slow-growing nature of some fungi. Similarly, endophyte communities of loblolly pine were also shown to be vastly different as determined by culture-based and culture-independent methods (43). On the other hand, PCR surveys also suffer from biases; for example, our PCR conditions were nonpermissive for Hemileia amplification, even though the majority of samples were heavily colonized by it. Ultimately, culture of the putative mycoparasitic fungi will be needed to better understand their effects on H. vastatrix and coffee. Lastly, the appreciation that endophytic fungi are beneficial to the host has been well considered. However, given that plant tissue is full of fungal hyphae, it needs to also be considered that some endophytes may actually be primarily symbiotic with other fungi and that the plant tissue is merely the environmental background within which these interactions take place.
Supplementary Material
ACKNOWLEDGMENTS
We thank Serena Zhao and Yuanying Su for assistance in preparing the samples for DNA sequencing. We also thank Edgardo and Annette Alvarado, Walter and Bernardo Peters, and the Edelman family for allowing us to work on their farms.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02639-15.
REFERENCES
- 1.Blackwell M. 2011. The Fungi: 1, 2, 3, 5.1 million species? Am J Bot 98:426–438. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- 2.Hawksworth DL. 1991. The fungal dimension of biodiversity—magnitude, significance, and conservation. Mycol Res 95:641–655. doi: 10.1016/S0953-7562(09)80810-1. [DOI] [Google Scholar]
- 3.Rodriguez RJ, White JF, Arnold AE, Redman RS. 2009. Fungal endophytes: diversity and functional roles. New Phytol 182:314–330. doi: 10.1111/j.1469-8137.2009.02773.x. [DOI] [PubMed] [Google Scholar]
- 4.van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller T, Wiemken A, Sanders IR. 1998. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396:69–72. doi: 10.1038/23932. [DOI] [Google Scholar]
- 5.Arnold AE, Maynard Z, Gilbert GS, Coley PD, Kursar TA. 2000. Are tropical fungal endophytes hyperdiverse? Ecol Lett 3:267–274. doi: 10.1046/j.1461-0248.2000.00159.x. [DOI] [Google Scholar]
- 6.Gamboa MA, Laureano S, Bayman P. 2002. Measuring diversity of endophytic fungi in leaf fragments: does size matter? Mycopathologia 156:41–45. doi: 10.1023/A:1021362217723. [DOI] [PubMed] [Google Scholar]
- 7.Arnold AE, Mejia LC, Kyllo D, Rojas EI, Maynard Z, Robbins N, Herre EA. 2003. Fungal endophytes limit pathogen damage in a tropical tree. Proc Natl Acad Sci U S A 100:15649–15654. doi: 10.1073/pnas.2533483100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee K, Pan JJ, May G. 2009. Endophytic Fusarium verticillioides reduces disease severity caused by Ustilago maydis on maize. FEMS Microbiol Lett 299:31–37. doi: 10.1111/j.1574-6968.2009.01719.x. [DOI] [PubMed] [Google Scholar]
- 9.McCann KS. 2000. The diversity-stability debate. Nature 405:228–233. doi: 10.1038/35012234. [DOI] [PubMed] [Google Scholar]
- 10.Oerke EC. 2006. Crop losses to pests. J Agric Sci 144:31–43. doi: 10.1017/S0021859605005708. [DOI] [Google Scholar]
- 11.Kromann P, Miethbauer T, Ortiz O, Forbes GA. 2014. Review of potato biotic constraints and experiences with integrated pest management interventions, p 245–268. In Pimentel D, Peshin R (ed), Integrated pest management. Springer, Dordrecht, Netherlands. [Google Scholar]
- 12.Mejia LC, Rojas EI, Maynard Z, Van Bael S, Arnold AE, Hebbar P, Samuels GJ, Robbins N, Herre EA. 2008. Endophytic fungi as biocontrol agents of Theobroma cacao pathogens. Biol Control 46:4–14. doi: 10.1016/j.biocontrol.2008.01.012. [DOI] [Google Scholar]
- 13.ten Hoopen GM, Rees R, Aisa P, Stirrup T, Krauss U. 2003. Population dynamics of epiphytic mycoparasites of the genera Clonostachys and Fusarium for the biocontrol of black pod (Phytophthora palmivora) and moniliasis (Moniliophthora roreri) on cocoa (Theobroma cacao). Mycol Res 107:587–596. doi: 10.1017/S095375620300772X. [DOI] [PubMed] [Google Scholar]
- 14.Viterbo A, Inbar J, Hadar Y, Chet I. 2007. Plant disease biocontrol and induced resistance via fungal mycoparasites, p 127–146. In Kubicek CP, Druzhinina IS (ed), Environmental and microbial relationships, vol 4 Springer, Berlin, Germany. [Google Scholar]
- 15.Perfecto I, Vandermeer J, Philpott SM. 2014. Complex ecological interactions in the coffee agroecosystem. Annu Rev Ecol Evol Syst 45:137–158. doi: 10.1146/annurev-ecolsys-120213-091923. [DOI] [Google Scholar]
- 16.Avelino J, Cristancho M, Georgiou S, Imbach P, Aguilar L, Bornemann G, Läderach P, Anzueto F, Hruska AJ, Morales C. 2015. The coffee rust crises in Colombia and Central America (2008–2013): impacts, plausible causes and proposed solutions. Food Sec 7:303–321. doi: 10.1007/s12571-015-0446-9. [DOI] [Google Scholar]
- 17.Ward MH. 1882. Researches on the life-history of Hemileia vastatrix, the fungus of the “coffee-leaf disease.” J Linnean Soc Lond Bot 19:299–335. [Google Scholar]
- 18.Webster J, Weber RWS. 2007. Introduction to fungi. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 19.Carvalho CR, Fernandes RC, Almeida Carvalho GM, Barreto RW, Evans HC. 2011. Cryptosexuality and the genetic diversity paradox in coffee rust, Hemileia vastatrix. PLoS One 6:e26387. doi: 10.1371/journal.pone.0026387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alarcón R, Carrión G. 1994. Uso de Verticillium lecanii en cafetales como control biológico de la roya del cafeto. Fitopatologia 29:82–85. [Google Scholar]
- 21.Vandermeer J, Perfecto I, Liere H. 2009. Evidence for hyperparasitism of coffee rust (Hemileia vastatrix) by the entomogenous fungus, Lecanicillium lecanii, through a complex ecological web. Plant Pathol 58:636–641. doi: 10.1111/j.1365-3059.2009.02067.x. [DOI] [Google Scholar]
- 22.Carrión G, Rico-Gray V. 2002. Mycoparasites on the coffee rust in Mexico. Fungal Divers 11:49–60. [Google Scholar]
- 23.Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. 2012. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 25.James TY, Stenlid J, Olson A, Johannesson H. 2008. Evolutionary significance of imbalanced nuclear ratios within heterokaryons of the basidiomycete fungus Heterobasidion parviporum. Evolution 62:2279–2296. doi: 10.1111/j.1558-5646.2008.00462.x. [DOI] [PubMed] [Google Scholar]
- 26.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Duenas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Luecking R, Martin MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Poldmaa K, Saag L, Saar I, Schuessler A, Scott J A, Senes C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson K-H. 2013. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 28.Bengtsson-Palme J, Ryberg M, Hartmann M, Branco S, Wang Z, Godhe A, De Wit P, Sanchez-Garcia M, Ebersberger I, de Sousa F, Amend AS, Jumpponen A, Unterseher M, Kristiansson E, Abarenkov K, Bertrand YJ K, Sanli K, Eriksson KM, Vik U, Veldre V, Nilsson RH. 2013. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol 4:914–919. [Google Scholar]
- 29.R Core Team. 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 30.Fröhlich J, Hyde KD. 1999. Biodiversity of palm fungi in the tropics: are global fungal diversity estimates realistic? Biodivers Conserv 8:977–1004. doi: 10.1023/A:1008895913857. [DOI] [Google Scholar]
- 31.Zimmerman NB, Vitousek PM. 2012. Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape. Proc Natl Acad Sci U S A 109:13022–13027. doi: 10.1073/pnas.1209872109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carroll G. 1995. Forest endophytes—pattern and process. Can J Bot 73:S1316–S1324. [Google Scholar]
- 33.Rojas EI, Rehner SA, Samuels GJ, Van Bael SA, Herre EA, Cannon P, Chen R, Pang JF, Wang RW, Zhang YP, Peng YQ, Sha T. 2010. Colletotrichum gloeosporioides s.l. associated with Theobroma cacao and other plants in Panama: multilocus phylogenies distinguish host-associated pathogens from asymptomatic endophytes. Mycologia 102:1318–1338. doi: 10.3852/09-244. [DOI] [PubMed] [Google Scholar]
- 34.Allen DJ. 1982. Verticillium lecanii on the bean rust fungus, Uromyces appendiculatus. Trans Br Mycol Soc 79:362–364. doi: 10.1016/S0007-1536(82)80132-0. [DOI] [Google Scholar]
- 35.Hall RA. 1984. Epizootic potential for aphids of different isolates of the fungus, Verticillium lecanii. Entomophaga 29:311–321. doi: 10.1007/BF02372119. [DOI] [Google Scholar]
- 36.McKenzie EHC, Hudson HJ. 1976. Mycoflora of rust-infected and non-infected plant material during decay. Trans Br Mycol Soc 66:223–238. doi: 10.1016/S0007-1536(76)80050-2. [DOI] [Google Scholar]
- 37.Zare R, Gams W. 2001. A revision of Verticillium section Prostrata. IV. The genera Lecanicillium and Simplicillium gen. nov. Nova Hedwigia 73:1–50. [Google Scholar]
- 38.Jackson D, Skillman J, Vandermeer J. 2012. Indirect biological control of the coffee leaf rust, Hemileia vastatrix, by the entomogenous fungus Lecanicillium lecanii in a complex coffee agroecosystem. Biol Control 61:89–97. doi: 10.1016/j.biocontrol.2012.01.004. [DOI] [Google Scholar]
- 39.Vega FE, Simpkins A, Aime MC, Posada F, Peterson SW, Rehner SA, Infante F, Castillo A, Arnold AE. 2010. Fungal endophyte diversity in coffee plants from Colombia, Hawai'i, Mexico and Puerto Rico. Fungal Ecol 3:122–138. doi: 10.1016/j.funeco.2009.07.002. [DOI] [Google Scholar]
- 40.Santamaria J, Bayman P. 2005. Fungal epiphytes and endophytes of coffee leaves (Coffea arabica). Microb Ecol 50:1–8. doi: 10.1007/s00248-004-0002-1. [DOI] [PubMed] [Google Scholar]
- 41.Saucedo-García A, Luisa Anaya A, Espinosa-García FJ, González MC. 2014. Diversity and communities of foliar endophytic fungi from different agroecosystems of Coffea arabica L. in two regions of Veracruz, Mexico. PLoS One 9:e98454. doi: 10.1371/journal.pone.0098454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward MH. 1882. On the morphology of Hemileia vastatrix Berk. and Br. (the fungus of the coffee disease of Ceylon). Q J Microscope Sci 22:1–11. [Google Scholar]
- 43.Arnold AE, Henk DA, Eells RL, Lutzoni F, Vilgalys R. 2007. Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia 99:185–206. doi: 10.3852/mycologia.99.2.185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.