SUMMARY
Iron is an essential element for Vibrio spp., but the acquisition of iron is complicated by its tendency to form insoluble ferric complexes in nature and its association with high-affinity iron-binding proteins in the host. Vibrios occupy a variety of different niches, and each of these niches presents particular challenges for acquiring sufficient iron. Vibrio species have evolved a wide array of iron transport systems that allow the bacteria to compete for this essential element in each of its habitats. These systems include the secretion and uptake of high-affinity iron-binding compounds (siderophores) as well as transport systems for iron bound to host complexes. Transporters for ferric and ferrous iron not complexed to siderophores are also common to Vibrio species. Some of the genes encoding these systems show evidence of horizontal transmission, and the ability to acquire and incorporate additional iron transport systems may have allowed Vibrio species to more rapidly adapt to new environmental niches. While too little iron prevents growth of the bacteria, too much can be lethal. The appropriate balance is maintained in vibrios through complex regulatory networks involving transcriptional repressors and activators and small RNAs (sRNAs) that act posttranscriptionally. Examination of the number and variety of iron transport systems found in Vibrio spp. offers insights into how this group of bacteria has adapted to such a wide range of habitats.
INTRODUCTION
Vibrionaceae are inhabitants of estuarine and marine environments worldwide (1). These Gram-negative bacteria are motile, facultative anaerobes that can grow with a number of different carbon and nitrogen sources. Vibrios may be found as free-living, planktonic cells; in biofilms; or colonizing a variety of marine organisms. Their metabolic adaptability and ability to occupy a variety of niches likely reflect their evolutionary history, which shows evidence of frequent gene acquisition. Vibrio species genomes are organized into two chromosomes (2). The majority of the essential genes map to the larger chromosome. The smaller chromosome has a mosaic structure with pathogenicity islands and phage-like genes, indicating numerous horizontal gene transfer events.
The vast majority of the >80 Vibrio species identified thus far are harmless to humans and marine organisms. However, some members of the species are associated with devastating epidemics (Vibrio cholerae) or sporadic infections (Vibrio vulnificus and Vibrio parahaemolyticus, among others). Human infections may follow the consumption of contaminated food or water or exposure of wounds to contaminated water. Because of their effects on human health, pathogenic vibrios have been extensively studied and are the focus of much of this discussion.
Vibrio spp., like most other organisms, have an absolute requirement for iron (3). Iron occurs in both the ferric (Fe3+) and ferrous (Fe2+) states and can participate in electron transfer over a wide range of redox potentials. Iron is essential for the function of a number of crucial enzymes in the bacterial cell (3), including ribonucleotide reductase for the synthesis of DNA precursors, cytochromes for electron transport, and tricarboxylic acid (TCA) cycle enzymes for energy production. In these enzymes, the iron cofactor may be heme, an iron-sulfur cluster, or, less often, an iron atom. The amount of iron required by vibrios varies depending on the cell's physiology and metabolism, but concentrations in the medium in the range of 0.1 μM to 5.0 μM provide sufficient iron for optimal growth of bacteria under laboratory conditions (4, 5). V. cholerae cells growing in broth medium at pH 7 with aeration will accumulate ∼106 atoms of iron per cell (Carolyn Fisher, unpublished data), similar to amounts reported for Escherichia coli grown under similar conditions (6, 7).
Despite its abundance, iron is not readily available in most environments. The form and solubility of iron are strongly influenced by the Eh and pH of the environment and by the presence of anions that may form complexes with iron. Iron has low solubility in the presence of oxygen at neutral or slightly higher pH, where it is predominantly found as ferric hydroxides (8). The seawater environments that are home to the Vibrionaceae are slightly alkaline (pH 7.5 to 8.4), and the concentration of dissolved iron in the open ocean is usually in the nanomolar range but can be as low as 20 to 30 pM (9), well below the concentration required for most microorganisms. Iron is more abundant in coastal and estuarine areas due to runoff. The concentration of iron is lowest at the ocean surface (<0.2 nM) and is 4- to 5-fold higher at depths below 500 m. Some of the iron is released from oxygenated sediments on the continental margins and hydrothermal venting, but dust, particularly Saharan dust aerosol, is a major source of dissolved iron in the North Atlantic Ocean (10). In the ocean interior, remineralization from sinking particulate organic matter accounts for much of the soluble iron (10). The iron that is released at hydrothermal vents is converted to a bioavailable form by microbes present at the vents (11).
The low levels of available iron led Martin et al. (9) to suggest that iron is the growth-limiting factor in nutrient-rich ocean environments. Low iron levels not only reduce the rate of growth of bacteria but, in the case of vibrios, also affect their survival and ability to persist, particularly at the lower end of the ocean pH range (12). The addition of ferric oxide to water increased the survival of V. cholerae, even in the absence of nutrients (13). Vibrio spp. can enter a viable-but-nonculturable (VBNC) state under adverse conditions, such as low nutrient availability or cold (14, 15). Transcriptional analysis of genes upregulated by VBNC V. cholerae identified transport genes, including at least one iron transport system (16), further suggesting that iron acquisition is important for the survival of V. cholerae in the marine environment.
Although vibrios may be found as planktonic bacteria in ocean or estuarine waters, they are often found in biofilms (17), which can affect iron availability. Biofilms, in which the bacteria are embedded in an extracellular polysaccharide (EPS) matrix forming a complex three-dimensional structure, often form at solid-liquid or air-liquid interfaces. The effects of biofilm on iron availability are variable. Chelation of iron by negatively charged components of the EPS may increase the local concentration of iron. The amount of iron in Vibrio species EPS in nature is not known, but divalent cations are commonly found in biofilms (18). An analysis of pellicle biofilms isolated from mine drainage showed that iron accounted for ∼70% of the cations present in the EPS. However, the binding of iron to EPS components may reduce its availability to bacteria. Reduced diffusion within biofilms can also lead to iron limitation, and the localized anoxic zones that occur in biofilms (19) influence the oxidation state and solubility of iron within these areas. Under low-oxygen, low-Eh conditions, iron is more likely to be in the ferrous than in the ferric form. Several bacterial species, including Pseudomonas aeruginosa (20) and V. cholerae (21), have reduced biofilm formation in low-iron environments, suggesting that biofilms are not conducive to iron acquisition.
Many vibrios form species-specific associations with marine organisms. V. cholerae can be found in the mucilaginous sheath of the filamentous cyanobacterium Anabaena variabilis (22), and this association allows the bacteria to persist longer in some environments. Vibrio fischeri (23) and Vibrio logei (24) (reclassified as Aliivibrio spp. [25]) form symbiotic relationships with squid, inhabiting the squid light organ. Extensive studies of the V. fischeri-squid association have shown not only that the bacteria colonize and produce light within the squid but also that the vibrios are required for normal development of the light organ (23, 26). Other vibrios colonize copepods, fish, or human hosts, and these interactions may range from relatively neutral to lethal for the host.
Colonization of a host presents additional challenges for microbial iron acquisition. In mammals, the level of freely available iron is far below that required for bacterial multiplication (27). The majority of iron is intracellular as heme, in iron-sulfur clusters, or stored as ferritin. Extracellular iron is bound to high-affinity iron-binding proteins, transferrin in serum and lactoferrin in secretions. Heme or hemoglobin released from cells is bound to hemopexin or haptoglobin, respectively. These proteins normally are relatively unsaturated and inhibit microbial growth by limiting iron availability. In response to infection, the level of available iron is further restricted by a reduction in serum iron saturation. This active withholding of iron from bacterial pathogens is a form of nutritional immunity (28) that protects against infection. In some human disease states, such as hemochromatosis or malaria, iron saturation increases, allowing the pathogen to obtain iron more easily and establish a more severe infection. In particular, V. vulnificus infections are more common in patients with iron overload.
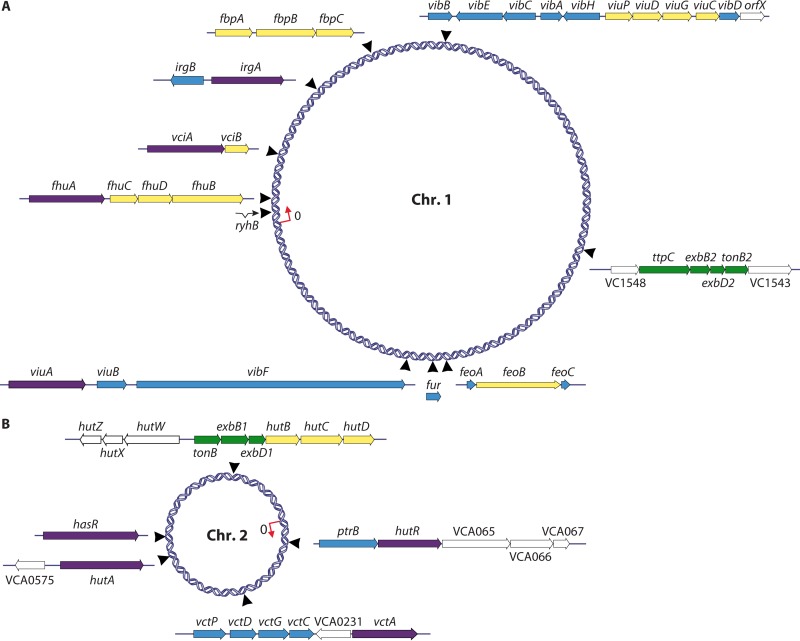
Vibrios, like other bacterial species, have evolved a variety of mechanisms to acquire iron in each environment that they inhabit. Some of these iron acquisition systems are closely related among all vibrios, reflecting their common ancestry. Other transport systems appear to have been acquired by horizontal transfer and may indicate selective advantages in specific niches occupied by those Vibrio species. Iron transport system genes are found on both chromosomes in vibrios and encode transporters for ferric or ferrous iron, proteins for the synthesis and transport of high-affinity iron chelators, and specific receptors for iron proteins and various chelated forms of iron. The iron transport systems are best defined in V. cholerae, in which >1% of the genome is devoted to iron transport systems (Fig. 1). These iron transport systems, the genes encoding them, and the regulatory systems that control iron uptake have been described in detail. Thus, V. cholerae serves as a model for understanding iron acquisition in vibrios. What is known for the other Vibrio species and the similarities with and differences from V. cholerae are described below.
FIG 1.
Map of V. cholerae chromosomes showing locations of known iron transport genes. Purple, outer membrane receptor genes; yellow, periplasmic binding protein and cytoplasmic membrane transporter genes; green, TonB system genes; blue, cytoplasmic protein genes. (Adapted from reference 31 [Fig. 1] with kind permission from Springer Science+Business Media.)
VIBRIO IRON TRANSPORT SYSTEMS
Transport of Iron Complexes
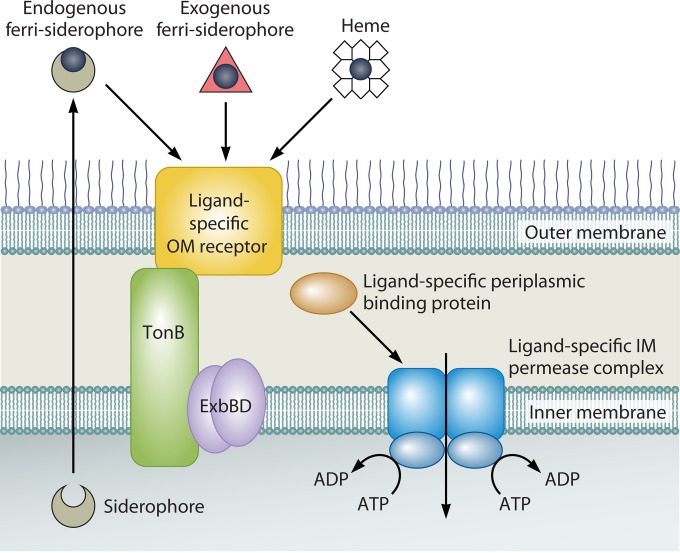
The majority of Vibrio iron transport systems recognize iron that is complexed to an iron chelator or carrier molecule (Fig. 2). The iron chelator may be synthesized by the bacteria or scavenged from other cells in the environment. Vibrio spp., like many other bacteria, produce one or more siderophores, small high-affinity ferric iron chelators (29). The siderophores are secreted into the environment or, less frequently, bound to the surface of the vibrio, and they serve as scavengers for iron. Once the siderophore binds iron, the ferrisiderophore complex is recognized by a specific receptor on the surface of the outer membrane (30, 31). Other iron complexes, including heme or siderophores made by other species (xenosiderophores), are also recognized by specific receptors. As described in more detail below, transport across the outer membrane via the receptor requires energy, which is supplied by the inner membrane protein TonB and its associated proteins ExbB and ExbD (32). Vibrios typically have more than one set of TonB/Exb proteins, and these proteins have specific as well as redundant functions (33). Once in the periplasm, the iron complex associates with a periplasmic binding protein (PBP) and is delivered to a cytoplasmic membrane permease for transport into the cytoplasm (34, 35). The periplasmic binding proteins and cytoplasmic permeases are less specific than the outer membrane receptors and recognize a broader class of complexes, usually additional members of the same class of siderophores. Once inside the bacterial cytoplasm, the iron is released from the siderophore by reduction of the iron and/or enzymatic cleavage of the siderophore, making the iron available for cellular needs (36, 37).
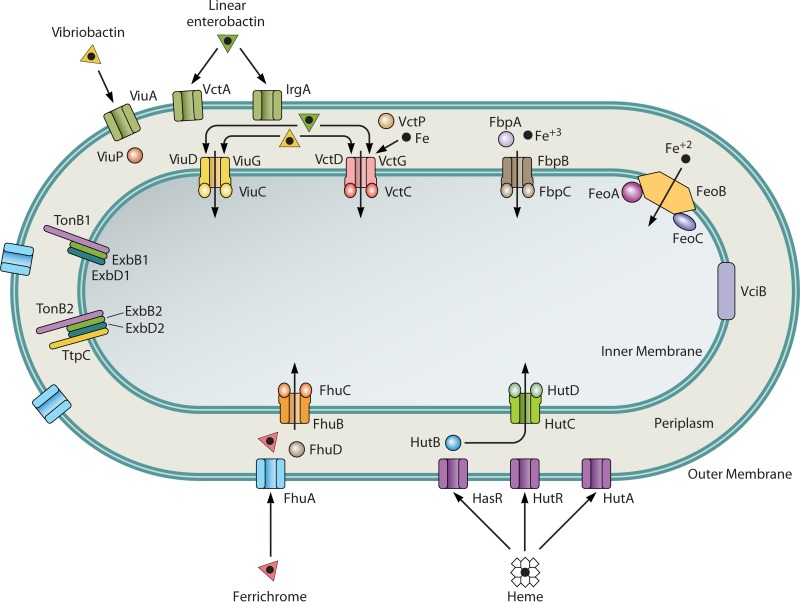
FIG 2.
V. cholerae iron transport systems. The cell envelope locations of components of the major iron transport systems and the compounds transported through each system are shown. (Adapted from reference 154 with permission of the publisher [copyright 2011 Blackwell Publishing Ltd.].)
Vibrio siderophores.
Each of the Vibrio species analyzed thus far produces one or more siderophores when iron is limiting. These molecules often have catechols or hydroxamates as part of the iron-binding moieties, although other structures have been identified. In some species, such as the squid symbiont V. fischeri, the structure of the siderophore is unknown (38), but a number of vibrio siderophores have been isolated and characterized, and there is considerable diversity in their structure and chemical properties. As described in more detail in the discussion of individual siderophores below, this diversity is likely a reflection of vibrios evolving to grow in different environments and to produce siderophores that can outcompete those produced by other members of the community when iron is scarce.
(i) Catechol siderophores.
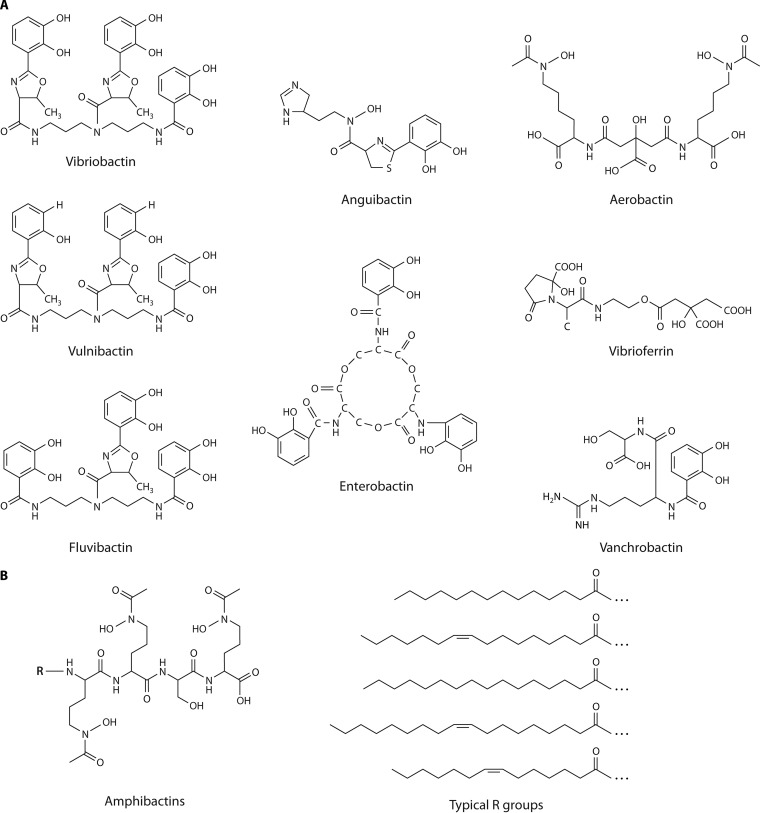
Catechol siderophores, which have 2,3-dihydroxybenzoic acid (DHBA) as the iron-chelating moiety, are common among Vibrio spp. Catechols and their biosynthesis were first described in Enterobacteriaceae, and little variation in catechol structures has been noted for this group of bacteria. Escherichia coli (39), Salmonella (40), and most Shigella spp. (41, 42) produce enterobactin, a cyclic trimer of 2,3-dihydroxybenzoylserine (Fig. 3A). The notable example of variation in catechol structure in this group is the salmochelins produced by some pathogenic Enterobacteriaceae, in which the 2,3-dihydroxybenzoylserine is glucosylated (43). In contrast, vibrios have a much more varied repertoire of catechols. The vibrio catechols are generally linear structures that differ in their backbones and in the numbers and types of amino acids linked to the catechols.
FIG 3.
Structures of representative vibrio siderophores. (A) Catechol and carboxylate siderophores. (B) Amphibactins.
(a) Vibriobactin.
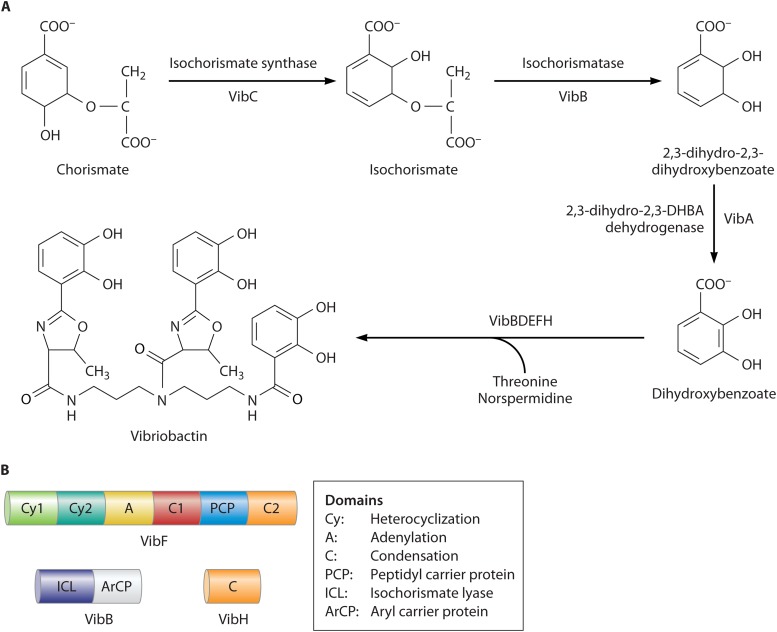
V. cholerae produces the catechol vibriobactin, which has three catechol groups attached to a norspermidine backbone (44) (Fig. 3A). The genes for norspermidine synthesis are not commonly found in bacteria but are widely distributed in Vibrio spp. (45). Two of the DHBA molecules are linked to the backbone through threonines that are cyclized, forming oxazoline rings. The vibriobactin biosynthesis mechanisms and pathway were determined by the Walsh group (46–49) (Fig. 4A). VibF, a nonribosomal peptide synthetase (NRPS), together with the vibriobactin synthase proteins VibE, VibH, and VibB assemble the siderophore from its precursors DHBA, threonine, and norspermidine, (46). DHBA is the product of VibA, VibB, and VibC (50), which have the same functions as their E. coli homologs EntA, EntB, and EntC (51). VibF acts as the scaffold on which vibriobactin synthesis is completed and has six domains organized in an assembly-line fashion (Fig. 4B). There are two cyclization domains, followed by an adenylation domain, a condensation domain, a peptidyl carrier protein (PCP) domain, and a second condensation domain (46). This assembly process on a NRPS with distinct, modular domains is characteristic of bacterial catechol siderophores and other secondary metabolites, including antibiotics, and has been described in detail for E. coli and other species (52–54). The domain structure of the NRPSs has facilitated the evolution of a large family of distinct siderophores with variations in the peptide backbones, chelating moieties, or other modifications. There is considerable homology among the NRPSs, but differences in the numbers of domains and in their substrates are evident. Duplications or substitutions of domains allow different amino acids or chelating groups to be incorporated into the final structure, resulting in a family of related, but distinct, siderophores produced by different Vibrio species.
FIG 4.
Vibriobactin biosynthesis. (A) Biosynthetic pathway. 2,3-Dihydroxybenzoate is synthesized from chorismate by the sequential activities of VibC, VibB, and VibA. VibB is a bifunctional enzyme that is also required for a later step in biosynthesis. VibB and VibF are modified by the attachment of phosphopantetheine arms to the aryl carrier protein domain of VibB and the peptidyl carrier domain of VibF (46). VibD, which is required for late steps in vibriobactin synthesis (263), is predicted to be the phosphopantetheine transferase based on homology and its ability to complement an E. coli entD mutation (50). VibE activates DHBA forming the acyl adenylate and transfers it to the free thiol of the phosphopantetheine (47). This DHB thioester is combined with norspermidine by the condensation domain protein VibH to produce N1-(2,3-dihydroxybenzoyl)norspermidine (47). VibF activates and covalently loads the PCP domain with l-threonine and heterocyclizes 2,3-dihydroxybenzoyl-VibB with l-Thr. The aryl oxazoline is then transferred to N1-(2,3-dihydroxybenzoyl)norspermidine. VibF catalyzes a second oxazoline acylation, yielding vibriobactin. (B) Domain structure of the vibriobactin biosynthesis enzymes VibF, VibB, and VibH. The NRPS VibF is the scaffold for assembly, and the indicated domains act in an assembly-line fashion to construct the final product.
The genes encoding vibriobactin synthesis are found in two separate clusters on the large chromosome (Fig. 1). The genes for the synthesis of the catechol moiety, vibABC, are linked to genes for subsequent steps in biosynthesis, vibD, vibE, and vibH, and to genes for catechol transport, viuPDGC (50). The NRPS VibF is encoded by a second vibriobactin gene cluster on the large chromosome linked to genes for vibriobactin transport, viuA, and utilization, viuB (55) (Fig. 1). It is not uncommon in vibrios for the core catechol genes to be found as one module and one or more genes encoding the species-specific modifications occurring elsewhere in the genome as a second module.
(b) Fluvibactin and vulnibactin.
Vibrio fluvialis and Vibrio vulnificus produce catechol siderophores, fluvibactin and vulnibactin, that are structurally related to vibriobactin (56, 57) (Fig. 3A). Like vibriobactin, these siderophores have a linear, norspermidine backbone with the iron-binding moieties linked directly or through threonine to the backbone. In fluvibactin, however, only one of the three DHBA moieties is linked to norspermidine by forming an oxazoline ring with threonine, and the other two are linked directly to norspermidine. Vulnibactin represents another variation on the theme: it has a norspermidine backbone with one catechol linked directly to the backbone, but it differs in having two salicylate residues, rather than DHBA, forming the oxazoline rings with threonine (56) (Fig. 3A). A mutant of V. vulnificus defective in salicylate biosynthesis was rescued for growth in low iron by the addition of DHBA, suggesting that V. vulnificus may also be able to produce a siderophore having DHBA instead of salicylate (58). The genes encoding vulnibactin have not been fully characterized, but the identification of venB (isochorismate lyase) (59) and two AMP ligase genes (58) within a cluster of homologs of known NRPSs suggests that a region on chromosome 2 of V. vulnificus (VV2_0828 to VV2_0844) encodes vulnibactin biosynthesis.
(c) Anguibactin, a mixed catechol and hydroxamate siderophore.
The V. anguillarum O1 serotype, a fish pathogen, also produces a catechol siderophore, but its structure is distinct in containing both catechol and hydroxamate groups as part of the iron-binding residues (60) (Fig. 3A). This siderophore, anguibactin, is synthesized from DHBA, which provides the catechol group; N-hydroxyhistamine, which contributes the N-hydroxyl group characteristic of hydroxamate siderophores; and l-cysteine (61–63).
Most of the genes required for anguibactin synthesis are carried on a 65-kb plasmid, but there is some redundancy with chromosomal genes. Genes for DHBA synthesis (angABC) and activation (angE) are chromosomal (64), but the NRPS domain proteins are encoded on the plasmid. The plasmid has homologs of the angB, angC, and angE genes, but there is not an angA equivalent among the plasmid-borne genes. Some of the plasmid-borne genes are flanked by insertion sequences, forming a composite transposon-like structure (65, 66). This suggests that V. anguillarum acquired the anguibactin iron transport system by horizontal transmission. Homologs of the plasmid genes are present in the chromosome of Vibrio harveyi, which also produces anguibactin, further supporting horizontal transfer of the genes (67).
V. anguillarum O1 strains that lack the anguibactin plasmid, as well as strains of a number of other V. anguillarum serotypes, synthesize vanchrobactin (Fig. 3A), a catechol siderophore distinct from anguibactin (68–70). Vanchrobactin is a linear dipeptide derived from arginine and serine bound to DHBA (71), indicating some similarity to the enteric siderophore enterobactin (69).
(ii) Carboxylate siderophore.
Vibrio parahaemolyticus produces a very hydrophilic carboxylate siderophore that is unrelated to other characterized Vibrio siderophores. This compound, given the trivial name vibrioferrin, is composed of alanine, citric acid, ethanolamine, and 2-ketoglutarate (72) (Fig. 3A). Vibrioferrin has a lower affinity for iron than do the Vibrio catechol siderophores. In addition, it is more sensitive to photolysis than other photoreactive siderophores, resulting in the release of iron even under low-light conditions (73). Although the instability and low iron affinity of vibrioferrin would appear to be disadvantageous, studies of another Gram-negative marine bacterium, Marinobacter, offer insights into a unique role for vibrioferrin. Vibrioferrin has been isolated from alga-associated subclades of Marinobacter, and it was proposed that its low affinity and photoreactivity are evolutionary adaptations to the bacterium-alga mutualism (74). The phototactic dinoflagellates form aggregates with Marinobacter at the sea surface, where sunlight promotes photolysis of the vibrioferrin secreted by the bacteria. This results in a local increase in the concentration of uncomplexed ferric iron that can be used by both Marinobacter and the dinoflagellates. The iron allows the algae to increase the amount of fixed carbon, some of which is made available to the bacteria (74, 75). V. parahaemolyticus, like Marinobacter, can associate with marine algae (76), and it is possible that mutualistic interactions involving vibrioferrin occur with V. parahaemolyticus as well.
(iii) Hydroxamate siderophores.
Although many of the siderophores produced by vibrios are catechols, hydroxamate compounds have also been found in some species. Aerobactin, a hydroxamate siderophore found in many enteric bacteria, including E. coli, Shigella, and Salmonella, is also produced by some Vibrio species. Vibrio mimicus (77), Vibrio hollisae (78), and a planktonic marine Vibrio sp. (79) all produce this siderophore. Interestingly, the genes for aerobactin synthesis and transport (iucABCD and iutA) in V. mimicus and V. hollisae have the same arrangement as that in the enteric species, and the proteins show considerable identity to the corresponding E. coli proteins (78). In the Enterobacteriaceae, the genes are found on plasmids (80, 81) and in pathogenicity islands (82–85). Thus, it is likely that these genes have spread among the various bacterial strains by horizontal transmission. A strain of Vibrio harveyi isolated from the open ocean of the Gulf of Mannar produced a dihydroxamate siderophore (86). However, its complete structure has not been determined. V. vulnificus also produces a hydroxamate siderophore (87) in addition to its catechol vulnibactin, but the exact structure of the hydroxamate is not known.
(iv) Amphiphilic siderophores.
Many marine bacteria have been found to produce amphiphilic siderophores, which have a hydrophobic tail attached to the hydrophilic siderophore backbone. These siderophores are often produced by the bacteria as a suite of related compounds with a conserved ferric iron-binding peptide or citrate-based head group and one or two fatty acids (88–90). The fatty acid appendages may vary in length and the degree of saturation and hydroxylation. Depending on the peptidic head group and length of the acyl chain, these siderophores may be predominantly cell associated or more water soluble and secreted into the environment (90). Both types of siderophores, termed amphibactins, have been isolated from marine Vibrio species. Vibrio species strain HC0601C5 produces a hydrophilic suite of amphibactins that are primarily secreted into the medium (90), while Vibrio sp. strain R-10 produces amphibactins that are cell associated (89) (Fig. 3B). The amphibactins have a hydroxamate-containing head group composed of a serine and three ornithine residues with acyl groups ranging from C12 to C18. The moanachelins, produced by Vibrio sp. strain Nt1, are related to the amphibactins but differ in the head group, in which the serine is replaced by glycine or alanine (91). V. harveyi produces an amphiphilic variant of enterobactin, the catechol siderophore produced by most Enterobacteriaceae (92). V. harveyi has a cluster of genes homologous to the E. coli ent genes with a long-chain fatty acid coenzyme A (CoA) ligase gene located nearby. The structure of the siderophore is similar to that of Ent but has an acyl-l-serine in addition to the three dihydroxybenzoyl-serines that make the cyclic backbone of Ent. Like the other amphiphilic siderophores produced by Vibrio spp., amphi-enterobactin is produced as a family of siderophores differing in the acyl chain (92). The production of a collection of these compounds by a Vibrio species may result in the less-hydrophobic siderophores being released into the environment, while the more hydrophobic members of the suite would remain cell associated. It was proposed that this would create a gradient of iron chelators extending from the bacterium and would allow the iron to be passed from the secreted siderophores to those at the surface (89). How the iron transits from the membrane-associated siderophore to the interior of the cell is not known, but hydrolysis of the fatty acids could release the membrane-bound form of the siderophore for its transport into the cell (93). An amide hydrolase produced by a Marinobacter species strain in late log phase hydrolyzed the Marinobacter amphiphilic siderophores. In addition, when Marinobacter sp. and Halomonas aquamarina were cocultured, the Marinobacter enzyme hydrolyzed the suite of aquachelins produced by Halomonas, even though these aquachelins have a different peptidic head group (93). Thus, enzymatic modifications of secreted or cell surface-associated siderophores may occur in the aquatic environment, where a variety of bacterial species coexist.
Siderophore receptors.
The fully assembled siderophore is secreted from the cell or localized to the outer surface, where it binds ferric iron from the extracellular environment. Binding results in a conformational change in the siderophore that in vibriobactin is evidenced by the change in color of the siderophore from colorless to deep purple when the siderophore is complexed to ferric iron (44). This iron-siderophore complex cannot diffuse back into the cell but requires a specific outer membrane receptor (37, 94–96) (Fig. 2). These receptors belong to the TonB-dependent receptor family and require TonB to energize transport. Each Vibrio species produces a receptor specific for its siderophore, and the receptor gene is usually linked to genes encoding the siderophore biosynthesis enzymes. V. cholerae transports vibriobactin via the outer membrane receptor ViuA (94). Similarly, PvuA is the vibrioferrin receptor in V. parahaemolyticus (97), VuuA is the vulnibactin receptor in V. vulnificus (98), and FvtA transports vanchrobactin in V. anguillarum (99). The siderophore receptors are typically localized to the outer membrane without the need for accessory factors. However, the V. anguillarum anguibactin receptor FatA (100, 101) requires O-antigen side-chain biosynthesis for outer membrane localization (102). The phenotype of the O-antigen mutants was similar to that of a fatA mutant. This indicates a novel role for O-antigen in the localization or stability of the siderophore receptor in V. anguillarum.
Xenosiderophore receptors.
Cheating, or at least taking advantage of the neighbors, is a common phenomenon in iron transport. Most bacteria express one or more receptors for siderophores that they do not themselves synthesize, allowing them to acquire iron bound to siderophores made by other species. Examples of the variety of siderophores transported by Vibrio spp. are shown in Table 1. Because vibrios live in polymicrobial habitats, there may be strong evolutionary pressure to acquire xenosiderophore transport systems. Iron is a limited resource, and the secreted siderophores that can deliver this resource represent a common good. A siderophore secreted by one species would promote the growth of other members of that species and simultaneously sequester iron from unrelated species that are unable to use the siderophore. Therefore, the production of species-specific siderophores represents a form of kin discrimination (103). Members of other species bypass this kin discrimination by the acquisition of receptors allowing the recognition and transport of the xenosiderophore. Vibrio spp. exhibit natural competence and readily acquire new genes by transformation (104, 105). Such horizontal gene transfer events appear to be responsible for many of the genes encoding xenosiderophore transporters.
TABLE 1.
Siderophore synthesis and use by selected Vibrio species
| Species and siderophore | Reference(s) |
|---|---|
| V. cholerae | |
| Endogenous siderophore | |
| Vibriobactin | 44, 46 |
| Xenosiderophores | |
| Linear enterobactin derivatives | 44, 106, 108 |
| Ferrichrome | 44, 107 |
| V. vulnificus | |
| Endogenous siderophores | |
| Vulnibactin | 56 |
| Uncharacterized hydroxamate | 87, 186 |
| Xenosiderophores | |
| Aerobactin | 257 |
| Deferoxamine B | 111, 112 |
| Vibriobactin | 117 |
| V. parahaemolyticus | |
| Endogenous siderophore | |
| Vibrioferrin | 72 |
| Xenosiderophores | |
| Enterobactin | 258, 259 |
| Aerobactin | 109 |
| Ferrichrome | 110 |
| Vibriobactin(?) | 117 |
| Fluvibactin(?) | 117 |
| V. anguillarum | |
| Endogenous siderophores | |
| Vanchrobactin (strains lacking plasmid pJM1) | 68–70 |
| Anguibactin (strains with plasmid pJM1) | 60 |
| Xenosiderophores | |
| Enterobactin | 260 |
| Rhodotorulic acid | 261 |
| Ferrichrome | 261 |
| Citrate | 262 |
V. cholerae has receptors for nonnative catechol siderophores (106) and for ferrichrome, a fungal hydroxamate siderophore (107). The V. cholerae catechol receptors VctA and IrgA recognize linear derivatives of enterobactin, which is produced by E. coli and other enteric bacteria, while fluvibactin can be transported by either of these receptors or by ViuA (108). V. parahaemolyticus can use aerobactin (109) and ferrichrome (110). Both V. vulnificus (111, 112) and V. furnissii (113) have a receptor, DesA, for the hydroxamate deferoxamine B. desA expression requires a positive transcription factor, DesR. This AraC-like regulator promotes the transcription of desA in the presence of deferoxamine B (113). Thus, the expression of this system is induced when the siderophore is present in the environment.
Not surprisingly, some bacteria have evolved mechanisms to outwit the cheaters and scavengers. This can be seen in studies of polymicrobial communities in which some bacteria produce siderophores that not only cannot be used by competitors but also inhibit the competition. For example, the addition of the siderophore produced by a potential fish probiont, Pseudomonas fluorescens strain AH2, to a growing culture of V. anguillarum caused immediate growth arrest (114). Growth was restored by the addition of iron, suggesting that the P. fluorescens siderophore outcompetes V. anguillarum for iron, resulting in iron starvation. Further analysis of the V. anguillarum response to the antagonistic siderophore showed the induction of rpoS, encoding the alternative sigma factor associated with the stationary phase and the stress response. Thus, P. fluorescens may protect fish from infection with V. anguillarum by producing an antagonistic siderophore that withholds iron and induces stationary-phase-like growth arrest in the vibrios. Similarly, Shewanella algae can inhibit V. alginolyticus. V. alginolyticus has at least a dozen genes encoding putative siderophore receptors (115) and can use ferrichrome (116) and siderophores secreted by V. cholerae, V. fluvialis, and V. parahaemolyticus, in addition to its own siderophore (117). However, avaroferrin, a cyclic dihydroxamate siderophore secreted by S. algae isolated from the same red seaweed as V. alginolyticus, inhibited V. alginolyticus. Swarming of V. alginolyticus was inhibited at low concentrations of the siderophore, and this inhibition was greater than that observed with other xenosiderophores (115). Structural analysis of avaroferrin indicates that it is a chimera of two known siderophores, bisucaberin and putrebactin. The chimeric nature of the siderophore is reflected in the genetics of avaroferrin biosynthesis genes. The biosynthesis genes have homologs in the Vibrio salmonicida (bisucaberin) and Shewanella putrefaciens (putrebactin) siderophore biosynthesis gene clusters, and Böttcher and Clardy (115) point out that the modular nature of siderophore biosynthesis genes may favor recombination and the evolution of new siderophores. In this case, the chimeric siderophore can be used by the species that synthesizes it but is inhibitory to other bacteria, at least until they acquire the genes necessary for the use of avaroferrin. Thus, the evolutionary arms race for iron continues.
Heme receptors.
In addition to the siderophore receptors, vibrios have one or more receptors for the transport of heme and can use heme for growth in low-iron environments. These receptors are also members of the TonB-dependent transporter family and have homology to the siderophore receptors. V. cholerae has three heme receptors, designated HutA, HutR, and HasR (118–120). HutA and HutR share significant homology, while HasR more closely resembles the HasR hemophore-dependent heme transporters in Pseudomonas aeruginosa and Serratia marcescens (120). The hemophore-dependent receptors in P. aeruginosa and S. marcescens bind heme complexed to a heme carrier protein, the hemophore (121), but no hemophores have been identified in V. cholerae or other Vibrio species. Each of these receptors allows growth in medium containing heme as the sole iron source. Heme is transported into the cell as an intact iron-porphyrin complex and supports the growth of a heme biosynthesis mutant (118). It is not known whether the heme is used by V. cholerae solely as an intact heme complex or if it is broken down following transport to release the iron. A candidate for a heme oxygenase-like enzyme to degrade heme is the cytoplasmic protein HutZ, which is needed for the efficient use of heme as an iron source in V. cholerae (122). An analysis of HutZ in vitro indicated that it has a high affinity for heme and degrades it via the same intermediates as heme oxygenase (123). Thus, HutZ may serve to release iron from heme following transport.
Heme receptors in other species include HupA (124), HvtA (125), and HupO in V. fluvialis (126); HuvA in V. anguillarum (127, 128); and MhuA in V. mimicus (129). HvtA has 68% similarity and 51% identity to V. cholerae HutR, and the operon organization is the same in the two species, indicating a close genetic relationship (120, 125). Although V. cholerae has only a limited ability to use hemoglobin as an iron source, other Vibrio species, including V. parahaemolyticus, V. fluvialis (126), V. alginolyticus (130), V. anguillarum (131), and V. vulnificus (132, 133), have the ability to use hemoglobin efficiently. This is dependent on specific receptors that bind hemoglobin or proteases that can release the heme from heme proteins. In V. vulnificus, for example, both HupA and HvtA can transport hemin, but only HupA permits the use of hemoglobin (125). V. vulnificus can use hemoglobin even when it is bound to the hemoglobin-binding protein haptoglobin (133, 134). Hemoglobin can also be bound by albumin. The ability of V. vulnificus to obtain heme from the hemoglobin-albumin complex was associated with the production of an extracellular protease that liberated heme from the proteins (134).
TonB Systems in Vibrio Iron Transport
The transport of siderophores and heme across the outer membrane of Vibrio requires energy. This is problematic since there is no ATP or other potential energy source available to transporters in the outer membrane. Vibrio spp. and other Gram-negative bacteria solve this problem by coupling outer membrane receptors for iron sources to the cytoplasmic membrane proton motive force. The TonB-ExbB-ExbD complex that is embedded in the cytoplasmic membrane of Gram-negative bacteria transduces the energy across the periplasm to the outer membrane receptors (32) (Fig. 5). Interestingly, the vibrios have more than one set of TonB/Exb proteins. V. cholerae (33, 120, 135), V. anguillarum (136), and V. alginolyticus (116) have two TonB systems, and V. vulnificus (137) has three. These systems differ in their recognition of specific receptors and in the environments in which they function, as discussed below. In addition to the Exb proteins, some Vibrio TonB systems also include an additional factor, TtpC (138–140). All of the Vibrio tonB2 gene clusters, as well as some tonB3 operons, include a ttpC gene (139). TtpC is a cytoplasmic membrane protein that is predicted to have three membrane-spanning domains with its C terminus in the cytosol and its N terminus located within the periplasm (140). TtpC is part of the TonB2-ExbB2-ExbD2 complex and is required for the transport of heme or siderophores that use the cognate outer membrane receptors (140).
FIG 5.
General scheme for TonB-dependent iron transport systems. The iron complex is recognized by a specific receptor in the outer membrane (OM). Vibrio spp. have receptors for their own endogenously synthesized siderophores and for exogenous siderophores produced by siblings or by members of other species (xenosiderophores). Transport across the outer membrane is dependent on energy provided by the TonB-ExbB-ExbD complex. Following transport, the siderophore or heme is bound to a periplasmic binding protein and is delivered to the ATP-dependent inner membrane (IM) permease.
The first TonB system characterized in Vibrio, the TonB1-ExbB1-ExbD1 system, is encoded on the smaller of the two chromosomes of V. cholerae, and the genes are in an operon containing genes for the use of heme as an iron source (33) (Fig. 1). A second system, the TonB2-ExbB2-ExbD2 system, maps to the larger chromosome (33) (Fig. 1). Analysis of the TonB1 and TonB2 systems indicates that they have overlapping functions but are not fully redundant. Vibriobactin and ferrichrome are transported efficiently using either TonB1 or TonB2 (33, 135). Either TonB system also allowed heme transport via HutA and HutR, although transport was more efficient in strains carrying the tonB1 system genes (120). In contrast, heme uptake via HasR was TonB2 dependent (120). The TonB2 system is required for catechol transport via the VctA and IrgA receptors in V. cholerae (106), and tonB2, but not tonB1, can complement E. coli tonB mutations for transport of the catechol siderophore enterobactin (33). It was shown through characterization of chimeric TonB molecules, as well as analyses of TonB1 point mutants, that the specificity of TonB1 for outer membrane receptors in V. cholerae resides within the carboxy terminus of TonB1 (141). Conversely, the ability of a receptor to be energized by TonB1 is determined by a single amino acid residue in the N-terminal domain of these receptors (141).
TonB1 interacts with a restricted group of V. cholerae receptors but has the ability to function under environmental conditions, i.e., growth with high salt levels, that do not allow TonB2 to support transport through some receptors (135). TonB1 is 38 amino acids longer than TonB2, and most of this extension maps to the proline-rich region that spans the periplasm. Under conditions of increased osmolarity, such as those of seawater, which increase the periplasmic space (142), the longer TonB1 system could span the periplasm to interact with the receptors. The shorter TonB2 system does not function with a subset of the outer membrane receptors, including the heme receptors, in medium with increased salt concentrations. Deletion of 35 amino acids of the periplasmic domain region of TonB1 did not affect its ability to function at low osmolarity but eliminated its interaction with the heme receptors at higher osmolarity (135). Thus, TonB1 may specifically be required for transport through a subset of TonB-dependent receptors when V. cholerae is in the marine environment, while TonB2 is required for the use of some catechol xenosiderophores.
V. alginolyticus also has two TonB systems that have distinct functions. While both the TonB1 and TonB2 systems supported the use of vibrioferrin and the xenosiderophore ferrichrome, TonB1 was specifically required for heme use (116).
Transport from the Periplasm to the Cytoplasm
Following transport across the outer membrane, the siderophore or heme is bound by a periplasmic binding protein (PBP) and handed off to an ATP-binding cassette (ABC) transporter for transit across the cytoplasmic membrane. The periplasmic and cytoplasmic membrane proteins recognize specific ligands but tend to be less discriminating than the outer membrane receptors. Thus, vibriobactin is recognized by the outer membrane receptor ViuA (94) and not by the catechol receptors VctA and IrgA (106), but once in the periplasm, either of the catechol PBP and ABC transporters VctPDGC and ViuPDGC can transport the siderophore into the cell (106). Similarly, enterobactin derivatives are not recognized by ViuA but can be transported by both the Vct and Viu periplasmic and cytoplasmic membrane transport systems (96).
The transport of hydroxamate siderophores has been characterized in less detail than catechol transport in the Vibrionaceae. Hydroxamate siderophores also have specific outer membrane receptors but often share periplasmic binding proteins and cytoplasmic transporters (143). In V. parahaemolyticus, ferrichrome and aerobactin have distinct receptors, but FhuBCD transports both of these hydroxamates through the periplasm and across the cytoplasmic membrane (110).
Once in the cytoplasm, iron must be removed from the siderophore for use by the cell. This can be accomplished by the reduction of ferric iron to ferrous iron, for which the siderophores have low affinity, or by enzymatic cleavage of the siderophore. The use of vibriobactin-bound iron requires ViuB (37), and based on its homology to the E. coli siderophore-interacting protein YqjH (144), it is likely to act as a ferric reductase and to release iron from vibriobactin. Although E. coli uses an esterase, Fes, to break down enterobactin to release iron (36), no homolog of Fes is found in V. cholerae. Because V. cholerae uses linear rather than cyclic catechols, it may not be necessary to degrade the siderophore to release the iron (108). Other Vibrio spp., however, have Fes homologs, including V. parahaemolyticus (75% amino acid sequence identity), V. alginolyticus (37% identity), and V. anguillarum (36% identity). The V. anguillarum Fes homolog VabH has been partially characterized and was shown to be required for the use of the V. anguillarum siderophore vanchrobactin (145). Vanchrobactin is also a linear catechol but resembles enterobactin in that it contains serine linked to the catechol.
Transport of Unchelated Iron: Ferrous and Ferric Iron Transporters
Although siderophores have extremely high affinities for iron and can promote growth under many conditions of limited iron availability, they must not be sufficient for iron acquisition under all conditions, since all the vibrios tested have been found to have additional systems. This suggests that there has been selection for iron transport systems that can provide iron under conditions where the siderophores do not meet the cell's need for iron. In environments where only ferrous iron is available, for example, the ferric iron-binding siderophores would not efficiently transport iron. Transporters for both ferric iron and ferrous iron that is not complexed to a carrier have been identified in Vibrio spp.
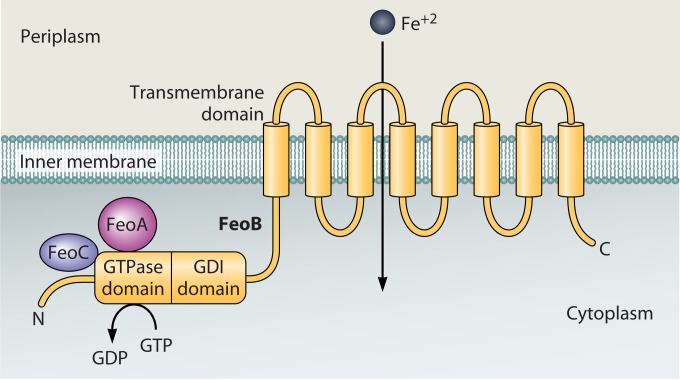
FeoABC, a ferrous iron transporter, is found in all the Vibrio species analyzed (31, 146). This system is likely the most ancient bacterial iron transport system, since homologs of the feo genes are widely distributed among bacterial species, and prior to the evolution of oxygen in the Earth's atmosphere, Fe2+ would have been the predominant iron species. The Feo transporter was first described in E. coli (147) and consists of FeoB, an 85-kDa cytoplasmic membrane protein, and two small (∼8.5-kDa) cytoplasmic proteins, FeoA and FeoC (Fig. 6). FeoB is the most highly conserved of these proteins in the vibrios, while FeoC is the least conserved (146). In V. cholerae, the feoABC genes form an operon, and all 3 genes are required for Feo-mediated iron transport (146, 148). Although this system has not been fully characterized, it is thought that FeoB forms the ferrous iron transport channel in the cytoplasmic membrane and that FeoA and FeoC serve accessory functions. FeoB is embedded in the cytoplasmic membrane and has a cytoplasmic, amino-terminal extension with homology to eukaryotic G proteins. The GTPase activity is required for transport, but its precise function has not been established (149). Similarly, the roles of FeoA and FeoC are poorly understood. Interactions between V. cholerae FeoB and FeoC have been shown (146). In Salmonella, FeoC has been shown to stabilize FeoB by providing protection against protease degradation (150), and it may play a similar role in the Vibrio Feo transporter. FeoA has also been shown to interact with FeoB in Salmonella (151), but how FeoA functions in ferrous iron transport is unknown. Another question that remains to be answered is how ferrous iron gains access to the periplasm for transport by Feo. No ferrous iron transporter has been identified in the outer membrane. The ferrous iron may diffuse through porins or other relatively nonspecific channels, consistent with the lack of a requirement for TonB for Feo-mediated iron transport (152).
FIG 6.
The ferrous iron transporter Feo. FeoB is localized to the cytoplasmic membrane. The accessory proteins FeoA and FeoC likely associate with the N-terminal cytoplasmic domain of FeoB. GDI, guanosine nucleotide dissociation inhibitor.
In V. cholerae, ferrous iron transport is facilitated by a cytoplasmic membrane protein designated VciB (152). The transport of ferrous iron and growth of bacteria are increased when VciB is present in addition to a ferrous iron transport system, including V. cholerae Feo, E. coli Feo, or Shigella flexneri Sit (152). The lack of specificity for V. cholerae Feo suggests that VciB does not recognize and interact with the ferrous iron transporter but may serve a more general function, such as in the reduction of ferric iron to ferrous iron for subsequent transport. Unlike most of the V. cholerae iron transporters, which have homologs in the other Vibrio species, VciB has few relatives in the genome sequence databases. VciB is found in all the sequenced V. cholerae genomes and may play a unique role in V. cholerae iron metabolism. Although it does not have homology to VciB, a ferric reductase has been identified in V. vulnificus (153), and it may play a similar role as VciB in making iron more available for transport via Feo.
V. cholerae also has a transporter for ferric iron, FbpABC, an ABC-type transporter in the cytoplasmic membrane (148). Based on homology with other ABC transporters, it was proposed that ferric iron bound to FbpA, the periplasmic binding protein component of the system, is delivered to FbpBC in the cytoplasmic membrane, which uses ATP hydrolysis to drive the transport of iron into the cytoplasm. As noted for the ferrous iron ligand for the Feo system, ferric iron transport by Fbp is TonB independent (148), and no specific outer membrane receptor has been identified. FbpABC is widely conserved in the Vibrionaceae and appears to represent a common mode of ferric iron acquisition in this family.
A third system for the transport of iron not complexed to a siderophore has been identified in V. cholerae. VctP, which is the periplasmic binding protein component of the Vct system, one of two V. cholerae transporters for catechol siderophores, also recognizes iron in the absence of the catechol. These two transport functions can be separated genetically. A tyrosine-to-phenylalanine substitution had no effect on ferri-catechol transport but eliminated the transport of iron in the absence of the siderophore (154). Thus, the Vct system can transport iron in either the presence or absence of the catechol siderophore.
IRON ACQUISITION WITHIN THE HOST
Vibrios are often found in association with other organisms. These associations present both opportunities for the bacteria to take advantage of new iron sources and hazards of outcompetition by host iron-binding proteins.
In humans, most of the iron is complexed to heme, primarily in red blood cell hemoglobin but also in heme proteins in other cells. Ferritin and Fe-S proteins account for much of the remainder of intracellular iron. Potential sources of iron in extracellular fluids are host iron-binding proteins, transferrin in the circulation and lactoferrin in secretions. However, these proteins are usually relatively unsaturated with iron, reducing the level of available iron in serum and secretions to a level below that needed to support microbial growth (155–159). Furthermore, lactoferrin has been found to have bactericidal activity against V. cholerae (160, 161). Lactoferrin can be cleaved by proteases to yield the antimicrobial peptide lactoferricin, which is active against a variety of Gram-negative pathogens (162), and it is possible that this accounts for the bactericidal effect against V. cholerae. In addition to the effects of iron-binding proteins, the vertebrate hormone hepcidin helps starve invading pathogens for iron by regulating the absorption of iron and its distribution within the body, reducing available iron in response to infection (158, 163, 164). This withholding of iron by host proteins and the active sequestration of iron in mammals in response to infection have been termed nutritional immunity (165, 166). Conditions that result in increased available iron in humans are associated with higher rates of infection and more severe infections. Hemochromatosis is associated with a variety of severe infections (167), including V. vulnificus (168) and non-O1 V. cholerae (169) infections. V. vulnificus, in particular, is associated with infections in people with iron overload (168, 170–172), and this can be reproduced in an animal model (173). It has been suggested that the sensitivity of hemochromatosis patients to fatal V. vulnificus infections is due, in part, to a failure to produce sufficient hepcidin (174). Studies using a hepcidin-deficient mouse model demonstrated that reduced hepcidin levels were associated with increased bacteremia, while administration of hepcidin agonists prevented death from V. vulnificus infection in these mice (175). The abrogation of this iron-withholding nutritional immunity favors growth of the pathogen.
Not surprisingly, many bacteria that infect humans or other vertebrates have evolved mechanisms to circumvent nutritional immunity and acquire iron bound to host proteins. These mechanisms include the expression of bacterial genes encoding iron acquisition systems in the host environment. Evidence that iron transport genes are expressed during infection has been obtained by using animal models or experimental infections of humans. Lombardo et al. (176) used in vivo expression technology (IVET) to determine the V. cholerae genes expressed during infection of human volunteers or in an infant mouse model. fhuC, encoding the ATPase of the hydroxamate transport system, was identified in both infected humans and the mouse model. Transcriptome sequencing (RNA-seq) analysis of V. cholerae isolated from the intestines of mice showed that a number of iron transport genes were highly induced in mouse (177). Furthermore, analysis of vibrios isolated directly from infected patients provided evidence for iron transport gene expression during naturally occurring infections. Genes for vulnibactin synthesis and the ferric vulnibactin receptor were detected in V. vulnificus obtained from the tissue of a patient with a severe soft tissue infection (178). Similarly, transcriptional profiling of V. cholerae obtained from stool of cholera patients showed the expression of the vct and fbp iron transport genes (179), and Fbp was also detected in the proteome of V. cholerae obtained from human stool (180). In contrast, neither V. cholerae (177) nor V. parahaemolyticus (181) showed significant expression of iron transport genes when isolated from the intestine of experimentally infected infant rabbits. This suggests that rabbit intestine is not as iron limited as human or mouse intestine and may not be the best model for studying iron transport in vivo.
Because multiple iron transport genes and proteins may be expressed in the host, it is not clear which of these systems are required for iron acquisition and colonization. Several studies have used mutational analysis to assess the roles of specific iron transporters in acquiring iron from the host by infecting vibrios. The heme uptake gene cluster in V. fischeri contributes to symbiotic colonization of the squid light organ (182). The heme uptake genes were expressed during colonization of the squid light organ, and a mutant containing a deletion of the heme transport genes had reduced fitness in colonization. In contrast, heme transport was not essential for V. cholerae colonization in a mouse model of infection (120).
In the vertebrate host, transferrin is one of the iron-binding proteins that can limit bacterial growth. Some vibrios have the ability to acquire iron from transferrin, either directly or through their siderophores. Transferrin receptors, which bind host transferrin at the cell surface, where the iron is removed for transport, have been described for several groups of pathogens, including Neisseria spp. In the case of Neisseria gonorrhoeae, the transferrin receptor is required for human infection (183). Until recently, transferrin receptors had not been identified in Vibrio species. The lack of identification of transferrin-binding proteins on the surface of vibrios may be a result of the specificity of the receptors for host transferrin. A recent analysis of V. vulnificus biotype 2 strains, which cause disease in eels as well as humans, identified a receptor for eel, but not human, transferrin (184). This receptor, Vep20, has homology (∼30% identity) to the Neisseria receptor for human transferrin, TbpA (185). Vep20 is required for full virulence and is plasmid encoded (184). Plasmids carrying the vep20 gene were also found in other fish-pathogenic vibrios, including V. harveyi, suggesting horizontal transmission of this iron acquisition gene.
Among those vibrios that lack transferrin receptors, the secreted siderophore may be able to capture iron from transferrin. V. vulnificus type 2 (186), V. parahaemolyticus (187), and V. damsela (188) use their siderophores for removal of iron from human transferrin. Okujo et al. (189) showed that V. vulnificus produced an extracellular protease that cleaved transferrin and lactoferrin, making the iron more accessible to the bacteria. However, subsequent studies by Shin et al. (190) and Kim et al. (191) demonstrated that the metalloprotease VvpE was not required for iron uptake from holotransferrin; the siderophore vulnibactin allowed V. vulnificus to use transferrin iron irrespective of the presence of the protease.
While many siderophores are able to remove iron from transferrin in vitro, this does not ensure that they can acquire transferrin iron in vivo. In human, siderocalin (lipocalin 2) can bind catechol siderophores and remove them from the circulation (192–194). Not to be outwitted, bacteria have evolved siderophore modifications that defeat the antisiderophore properties of siderocalin. For example, some enteric pathogens glucosylate enterobactin, making it unable to be bound by siderocalin (43). It was originally reported that the V. cholerae siderophore vibriobactin was also a stealth siderophore, able to escape siderocalin binding because of its weaker negative charge and phenolate-oxazoline coordination mode (195). A subsequent study by Allred et al., however, showed that siderocalin shifts the coordination mode of vibriobactin from the phenolate-oxazoline mode to the distinct catecholate mode, allowing high-affinity binding by siderocalin (196).
Vibriobactin synthesis was not required for infection or fluid accumulation in an infant mouse model of infection (197). Similarly, a mutant defective in heme transport had no effect in this model (120), although a mutant defective in both the siderophore and heme iron uptake systems showed some reduction in colonization (198). A mutant defective in both the TonB1 and TonB2 systems also had reduced fitness in the infant mouse model (135), but testing of mutants defective in each of the known iron transport systems has not revealed any single system that is essential for V. cholerae colonization in the mouse model (148, 152, 198). Because V. cholerae has so many iron transport systems, there may be compensation for the loss of any one system. It is also likely that subtle losses in fitness cannot be detected in short-term animal experiments. In nature, even a small reduction in fitness due to the loss of an iron transporter may result in a loss of the mutant from the population. V. cholerae would not be expected to retain multiple iron transport systems if each one did not provide some advantage in a particular environment.
IRON AND REGULATION OF GENE EXPRESSION
Regulation of Iron Transport Systems
Although iron is essential, too much iron is lethal. Intracellular iron that is not bound to proteins or chelated can act as a catalyst for the Haber-Weiss reaction and participate in Fenton reactions, leading to the generation of toxic hydroxyl radicals (199, 200). Hydroxyl radicals damage DNA, unsaturated lipids, and proteins, resulting in increased mutations and cell damage or death (201, 202). Thus, any excess iron is sequestered in the cell, and the bacteria respond to excess iron by rapidly repressing iron uptake systems. There is a coordination of iron transport with the metabolic activities of the cells and tight regulation of iron transport to meet, but not exceed, the demand for iron.
The major regulator of iron transport in Gram-negative bacteria is Fur. Fur is an iron-binding transcriptional regulator that was first identified in E. coli (200). It forms a homodimer, and in the presence of iron, the iron-Fur complex binds to specific sites on the DNA. These sites, termed Fur boxes, are typically located within the −10 and −35 regions of the promoters of iron-regulated genes, and the association of Fur with sites at this location blocks transcription.
All Vibrio spp. analyzed thus far have a Fur homolog. V. cholerae fur was identified by Litwin et al. (203), and a fur mutant was found to derepress the expression of two genes involved in iron transport, irgA and viuA, suggesting that it functioned much like Fur in E. coli (204). Crystallization of the V. cholerae Fur protein provided evidence for two metal-binding sites per monomer: a regulatory site where iron or other divalent metal cations can bind and an auxiliary zinc-binding site (205). No binding to DNA occurred when the metals were removed from the protein. There was no evidence that metal binding required any of the conserved cysteines in the protein (205). The Fur protein is relatively abundant for a transcriptional regulator; there are ∼2,500 copies per cell in exponentially growing V. cholerae bacteria, increasing to ∼7,500 copies in stationary-phase cells (206). Although Fur is relatively abundant, its levels are modulated in response to iron and the metabolic status of the cell. There is moderate negative regulation of fur by Fe-Fur in E. coli (207), while studies of V. vulnificus have shown positive regulation of fur by Fur in the absence of iron (208, 209). These types of autoregulation may control the ratios of Fur to Fe-Fur in the cell to limit the formation of aberrantly active dimers between iron-bound and iron-free Fur. fur is also positively regulated by cyclic AMP receptor protein (Crp) (207), and this may help couple cell metabolism to iron uptake.
Transcriptional analysis of Fur regulation in V. cholerae revealed positive as well as negative effects on transcription, although regulation was predominantly negative (210). Fur repressed the genes involved in iron acquisition when the cells were grown under iron-replete conditions but positively affected the expression of ompT, encoding a major porin protein (210, 211). For most of the negatively regulated genes, a putative Fur box is found in the −10 and −35 regions, but the binding site for Fur in the ompT promoter is upstream, and increased expression may be due to Fur competing with a negative regulator at this binding site.
A more recent analysis of transcriptional regulation of Fur discriminated between direct and indirect effects. Davies et al. (212) used chromatin immunoprecipitation sequencing (ChIP-seq) to identify the Fur-binding sites in the V. cholerae genome. This study verified a number of the sites predicted by genetic and transcriptional analyses and revealed new sites, including several small RNAs (sRNAs). Based on their analysis, a revised 21-bp Fur box consensus sequence was defined, and the site of the Fur box relative to the transcriptional start site was found to be variable.
There is also evidence for strain differences in the regulation of iron transporters. There are two distinct biotypes of V. cholerae, classical and El Tor, which differ in virulence, environmental persistence, hemolysin production, and other phenotypes. The El Tor biotype was found to have higher expression levels of iron transport genes than classical V. cholerae (213), and El Tor and non-O1 strains were noted to produce larger amounts of vibriobactin and to have greater resistance to chemical iron chelators than classical strains (5). Whether this is related to differences in Fur or other regulation is unknown.
Fur has been shown to play similar roles in V. vulnificus (214–216), V. anguillarum (217, 218), and V. salmonicida (219). In each of these vibrios, Fur negatively regulates the expression of genes for iron acquisition systems, thereby reducing the chance of toxicity due to iron overload.
Many of the vibrios secrete hemolysins that may indirectly aid in iron acquisition by lysing erythrocytes or other host cells, releasing internal heme and iron. Fur negatively regulates the expression of hemolysins in V. cholerae (220), V. parahaemolyticus (221), and V. vulnificus (215). V. vulnificus produces a protease that aids in the acquisition of heme from heme-albumin (222). This proteolytic activity is not regulated by iron but is induced by heme or hemoglobin.
Although Fur acts as a global regulator to repress iron transport systems, there are a number of examples of additional regulators that affect individual iron transport systems. In V. anguillarum, anguibactin transport is positively regulated by AngR and Taf (223–225). Taf is a transcriptional activator (226), while AngR is a bifunctional protein required for the synthesis of anguibactin as well as for the regulation of its transport proteins (225, 227). Other examples of positive regulation are the activation of irgA, encoding a V. cholerae enterobactin receptor, and hupR, encoding the V. vulnificus heme receptor, both of which are divergently transcribed from positive regulators. IrgB activates the transcription of irgA (228, 229), and HupR induces hupA (230). Both IrgB and HupR are members of the LysR family of positive transcriptional activators (228, 230). A number of other Vibrio iron transport genes, including vctA, have known or putative regulators that are divergently transcribed. In each of the systems with positive regulation, the genes for the activators, as well as the iron transporters, are negatively regulated by Fur. This arrangement amplifies the response to low-iron environments and allows a much greater fold change in expression as a function of the iron concentration than would Fur regulation alone.
Iron and Regulatory RNAs
Transcriptional analyses of iron and Fur regulation have shown that the concentration of iron and the presence or absence of Fur have widespread effects on the cell. Not only is iron transport tightly controlled by Fur, but changes in superoxide dismutase (SodB), TCA cycle enzymes, biofilm formation, and a variety of other phenotypes are influenced by Fur. While the effects on iron transport genes are direct, many of the other genes are indirectly regulated by Fur via RyhB, an sRNA. RyhB, first described in E. coli, repressed the synthesis of the iron-containing superoxide dismutase, encoded by sodB, and aconitase, encoded by acnB, among others, by controlling mRNA stability and translation (231, 232). RyhB can bind to the transcripts of these genes in the presence of Hfq and direct their degradation by RNase E (233). The expression of ryhB is negatively regulated by Fur (232). Thus, Fur can positively affect gene expression by reducing the levels of the repressor RyhB. RyhB provides a mechanism for linking cellular metabolism to iron availability and shutting down iron-greedy enzymes and iron storage proteins when iron is less plentiful (234). The complexity of the regulation and level of control of Fur, RyhB, and their targets is further illustrated by the fact that RyhB influences the levels of Fur. RyhB downregulates the expression of Fur in E. coli and is predicted to regulate fur in Vibrio spp. (235). There is complementarity between RyhB and the translation initiation region of the V. cholerae fur gene that may result in the downregulation of fur mRNA.
In V. cholerae, RyhB plays a role similar to that of the E. coli homolog, linking iron acquisition and overall metabolism. Analysis of the RyhB regulon in V. cholerae showed that RyhB influenced the synthesis of superoxide dismutase, TCA cycle enzymes, and proteins containing heme or iron-sulfur clusters (21, 236). However, V. cholerae RyhB is considerably larger than its E. coli homolog (>200 nucleotides, compared to 90 for E. coli) and has a larger regulon, influencing the expression of genes involved in motility, chemotaxis, and biofilm formation. A mutant with a deletion in ryhB had reduced chemotactic motility and was unable to form wild-type biofilms in low-iron medium (21). The addition of iron restored biofilm formation in the V. cholerae ryhB mutant, but excess iron did not enhance biofilm formation in the wild-type strain (21). This suggests that the ryhB mutant is iron stressed in biofilms, and the inability of the ryhB mutant to properly maintain iron homeostasis likely contributes to the defects in motility and biofilm formation.
The role of RyhB in iron regulation is even more complex in V. parahaemolyticus, where RyhB positively regulates the production of the siderophore vibrioferrin (237). RyhB, with the aid of Hfq, can directly bind to and stabilize the 5′ untranslated region (UTR) of pvsO, a polycistronic message for vibrioferrin biosynthesis, leading to increased vibrioferrin synthesis.
In V. anguillarum, another RNA is involved in the regulation of siderophore transport. An antisense RNA, RNAα, specifically controls the expression of the transport genes fatA and fatB by binding to the fatA and fatB mRNAs and reducing their translation (238, 239). Fur is required for RNAα synthesis and controls transcription initiation, independent of the iron status of the cell. It is not known how Fur regulates gene expression in the absence of iron. However, iron-independent Fur regulation of gene expression was noted in an analysis of the Fur and iron regulons in V. cholerae (210). Iron plays a role in RNAα expression, but the effect of iron is posttranscriptional: RNAα is stabilized in cells grown in the presence of iron (238). The effect of RNAα in addition to Fur allows tighter repression of FatA and FatB synthesis in the presence of iron.
Other Environmental Sensors and Regulators for Iron Acquisition
In addition to iron, there are a variety of other environmental factors that influence the expression of iron transport genes, including temperature, oxygen, carbon sources, and quorum-sensing molecules. These factors, summarized in Table 2, may correlate with the form of iron available and serve as indirect signals for specific iron transport systems. Higher oxygen levels are associated with the presence of ferric, rather than ferrous, iron. Thus, the presence of oxygen may signal that it would be advantageous to express ferric iron transporters, while anoxic environments may increase the synthesis of ferrous iron transporters (240). Similarly, higher temperatures signal a mammalian host environment and could favor the expression of transporters that acquire iron from host proteins. Expression of the cell's entire repertoire of iron transporters under all conditions would be metabolically costly and could lead to the import of toxic levels of iron. Thus, additional layers of regulation would allow the cell to optimize iron acquisition and growth by tuning the expression of the transport genes to the most likely available iron source.
TABLE 2.
Regulation of iron transport by signals in addition to iron availabilitya
| Environment | Most likely iron source(s)b | Environmental signalsc | Possible Vibrio regulator(s) | Effect(s) on Vibrio iron transporters |
|---|---|---|---|---|
| Marine surface waters | Fe3+ compounds | O2 present | Fnr,d ArcABd | ↑ siderophore |
| Neutral or alkaline pH | PepAe | ↓ Feo | ||
| Sediments, solid surfaces, or biofilms | Fe2+ compounds | Low O2 concn, reduced diffusion, and crowding in biofilm | Fnr,d ArcABd | ↑ Feo |
| LitR,f quorum sensingf | ↓ siderophore, ↓ heme transporters | |||
| Host | Heme and transferrin in blood and tissues, lactoferrin in secretions, and Fe2+ iron complexes in intestine | Temperature | H-NSd | ↑ heme transporters |
| Specific C sources | Crpf | ↑ TonB | ||
| Specific N sources in squid | GlnDf | ↑ siderophore | ||
| Low O2 concn in intestine | Fnr,d ArcABd | ↓ siderophore, ↑ Feo | ||
| Bile or antimicrobial peptides in intestine | Cpxf | ↑ heme transporters, ↑ siderophore |
Shown is a summary of the effects of environmental conditions on the regulation of iron transport systems. Conditions that have been shown to affect specific iron transporters are discussed in the text. Arrows indicate a positive (up arrow) or negative (down arrow) effect on expression of the iron transport genes.
Iron sources present in each environment are predicted based on the known distributions of iron complexes.
Environmental signals are predicted based on conditions known to exist in each environment.
Regulator that affects iron transport in other species and for which homologs are present in Vibrio spp.
Regulator that is present in Vibrio spp. and is known to regulate genes in response to the stated conditions but that has not been shown to regulate iron acquisition genes.
Regulator that is known to affect Vibrio iron transport.
Regulation in response to signals found within the host.
The sources of iron available to vibrios in the host vary depending on whether the bacteria are found on body surfaces or in tissues or blood. Heme is the most abundant iron source in animal hosts, and linking the expression of heme or hemoglobin receptors to host conditions allows the bacteria to take advantage of this iron source. In V. vulnificus, the heme receptor gene hupA is regulated by temperature: there is a significant increase in the level of transcription at 40°C compared to that at 30°C (241). HupA is required for maximal virulence in mice, and linking its expression to higher temperatures allows its expression in the mammalian host but not in the generally cooler, external environments. In contrast, V. salmonicida, which causes cold-water vibriosis in Atlantic salmon, produces siderophores only at lower temperatures (242). Siderophores and iron-regulated outer membrane transport proteins are synthesized at 10°C, the water temperature at which hemorrhagic disease occurs, but not at temperatures of 15°C or higher. The mechanism by which temperature affects the expression of Vibrio iron transport genes has not been determined, but the nucleoid-associated protein H-NS is a possible mediator. H-NS is found in Vibrio and is known to regulate gene expression as a function of temperature in other species.
Other signals in the host include the presence of specific carbon and nitrogen sources; however, these and other signals, such as O2 concentration and pH, are not limited to the host environment but may be found in other habitats. It is the ability of the vibrios to integrate multiple environmental signals to regulate iron transport that allows them to more precisely determine their location and possible iron sources.
Effects of metabolism on expression of iron transport genes.
Carbon metabolism influences the expression of Vibrio iron transport genes. Crp binds upstream of the V. vulnificus hupA gene and activates its expression (241). Crp also induces the expression of tonB3 (137) and the aerobactin receptor gene iutA (243) in V. vulnificus. Crp and cAMP may couple growth on carbon sources other than glucose with the increased need for iron required for full metabolism of these substrates. While glycolysis does not require iron-containing enzymes, other pathways, such as the Entner-Doudoroff pathway and the TCA cycle, have enzymes that require iron for activity. Under low-iron conditions, E. coli shifts its metabolism to increase glycolysis and decrease the level of TCA cycle enzymes (244). It is likely that vibrios alter their metabolism in a similar way. In the absence of glucose, additional iron would be needed for carbon metabolism via pathways other than or in addition to glycolysis.
Another example of a link between metabolism and iron transport has been identified in V. fischeri, which couples nitrogen metabolism to iron acquisition. A mutation in glnD, which acts as a nitrogen sensor, not only caused a defect in nitrogen metabolism but also reduced siderophore synthesis by these bacteria (38). The glnD mutant was unable to persist in the squid light organ, but excess iron restored persistence to wild-type levels, indicating that the colonization defect was a result of the effect of the glnD mutation on iron acquisition. Thus, the presence of specific nitrogen sources in the squid light organ may serve as a signal for this host environment and increase the synthesis of the siderophore needed for colonization.
Quorum sensing regulation and iron transport.
Several studies have also linked quorum sensing to iron metabolism in Vibrio spp. Quorum sensing modulates a variety of vibrio activities, including biofilm formation and the expression of virulence factors. In V. vulnificus, quorum sensing repressed vvsA and vvsB, which encode a NRPS required for vulnibactin synthesis. The primary quorum sensing regulator, SmcR, binds upstream of the vvsAB transcription start site and overlaps the Fur-binding site. Under high-iron conditions, Fur repressed vvsA transcription, while in low-iron environments, SmcR regulated the expression of the siderophore biosynthesis genes (245). Using a luxS mutant defective in the synthesis of autoinducer 2 (AI-2), Kim and Shin observed an increase in the synthesis of the siderophore receptors for aerobactin (IutA) and heme (HupA), suggesting that they are also repressed by quorum sensing (246). Additional studies revealed a more complex connection between iron and quorum sensing (247). Fur bound at two sites upstream of the transcriptional start site of smcR, repressing the production of SmcR under high-iron conditions. However, the affinity of Fur at these sites was low enough that smcR was induced by quorum sensing at a high cell density, even under iron-rich conditions. In low-iron environments, Fur did not bind, and smcR expression was regulated solely by quorum sensing. This dual control by iron (via Fur) and quorum sensing (via SmcR) allows fine-tuning of iron acquisition. At a high cell density, higher levels of the siderophore in the environment, representing a common good coupled with the reduced need for iron as the cells enter stationary phase, could result in cells importing too much iron. Monitoring cell density as a secondary level of negative regulation provides tighter control over cellular iron levels. This cross talk between iron and quorum sensing regulation may be a common theme in vibrios: Septer et al. (248) showed that Fur represses LitR, a positive regulator of V. fischeri quorum sensing that is a homolog of V. vulnificus SmcR.
Membrane stress response and regulation of iron transport.
Another environmental sensor that plays a role in the regulation of Vibrio iron transport is the two-component Cpx pathway that responds to envelope stress (249). Iron starvation in V. cholerae led to Cpx activation, and overexpression of the response regulator gene cpxR induced the expression of a number of iron-regulated genes, including those for iron transport proteins. The Cpx response was inhibited by the addition of iron, suggesting that iron is a common component of Cpx-inducing signals in V. cholerae (249). Cpx may respond to the accumulation of potentially toxic apo-cofactors for iron-containing proteins in the membrane or periplasm, or it may be a response to the large number of membrane proteins induced by iron starvation. Additionally, the presence of bile and membrane-damaging antimicrobial peptides in the intestine could activate the Cpx response and signal the need for increased levels of iron transporters in the host environment. Thus, Cpx may aid in the response to membrane perturbations associated with iron stress and help coordinate cellular functions in response to iron starvation.
Growth on surfaces induces iron transport genes.
In V. parahaemolyticus, some iron transport proteins were induced by growth on a surface (250). A comparison of genes expressed in cells grown on solid medium to those expressed in cells grown in broth showed the induction of a large number of genes, including those encoding swarming motility proteins, virulence factors, and sensory enzymes involved in chemoreception and cyclic di-GMP (c-di-GMP) signaling. Iron-regulated genes, including those encoding the enterobactin and ferrichrome receptors, were also induced when the bacteria were grown on the surface of agar medium. This finding is consistent with data from previous studies showing that restricted diffusion of iron in agar media limits its availability to V. parahaemolyticus cells growing on surfaces, and iron is a key signal for swarmer cell differentiation (251). Differentiation in response to iron limitation requires Fur, but it is not known whether the effect of Fur is direct or indirect (L. McCarter, personal communication).
Iron Regulates Expression of Genes for Systems Other than Iron Metabolism
In addition to its role as an essential element in maintaining cell function, iron also serves as an environmental sensor for a wide range of bacterial activities. In particular, it has long been noted that environments deficient in iron trigger the synthesis of virulence factors such as diphtheria toxin (252) and Shiga toxin (253), suggesting that low iron levels are a signal that bacteria use to sense that they are within a vertebrate host. This iron regulation is dependent on Fur or Fur-like repressors (254–256), representing another example of virulence factors coopting basic cellular regulators of metabolism for control of virulence gene expression. In V. cholerae, Fur positively regulates genes located in the V. cholerae pathogenicity island, which encodes the toxin-coregulated pilus (TCP), a major virulence factor of V. cholerae, as well as genes within the mega-integron (210). Unlike the effects of Fur on iron transport systems, the regulation of pathogenicity island genes was independent of the iron concentration. Consistent with the regulation of TCP synthesis or assembly, a fur mutant exhibited very weak autoagglutination, independent of the iron concentration. Furthermore, the fur mutant had a strong phenotype in the infant mouse model of cholera. The fur mutant was highly attenuated and failed to compete effectively with the wild type for colonization of the mouse intestine (210).
CONCLUSIONS
Iron acquisition is critical to the survival of Vibrio spp. These bacteria inhabit a wide variety of habitats and encounter iron in different forms depending on the habitat. The available iron may be ferric or ferrous and may be in insoluble hydroxides or part of heme or iron sulfur clusters in host proteins. Vibrio species have evolved systems to acquire iron in all of these forms, and there is evidence that iron transport systems may have been acquired from other bacterial species by horizontal transmission. Thus, these bacteria are well adapted for survival in marine habitats and for rapid adaptation to new environments when they transition to a host or to polymicrobial communities. The selective pressures that shape the repertoire of systems found in any of the Vibrio spp. may be positive (ability to acquire iron in a new environment or to reduce competition with other bacteria) and negative (TonB-dependent receptors may be antigenic or may be receptors for bacteriophages). From the vibrio point of view, it is apparent that more is better when it comes to iron transport systems.
ACKNOWLEDGMENTS
We thank Carolyn Fisher and the members of the Payne laboratory for helpful discussions and editing of the manuscript.
This work was supported by NIH grant AI091957.
REFERENCES
- 1.Farmer JJI. 2006. The family Vibrionaceae, p 495–507. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (ed), The prokaryotes. Springer, New York, NY. [Google Scholar]
- 2.Trucksis M, Michalski J, Deng YK, Kaper JB. 1998. The Vibrio cholerae genome contains two unique circular chromosomes. Proc Natl Acad Sci U S A 95:14464–14469. doi: 10.1073/pnas.95.24.14464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crosa JH, Payne SM, Mey AR. 2004. Iron transport in bacteria. ASM Press, Washington, DC. [Google Scholar]
- 4.Ju C-H, Yeung PSM, Oesterling J, Seigerman DA, Boor KJ. 2006. Vibrio parahaemolyticus growth under low-iron conditions and survival under high-magnesium conditions. J Food Prot 69:1040–1045. [DOI] [PubMed] [Google Scholar]
- 5.Sigel SP, Payne SM. 1982. Effect of iron limitation on growth, siderophore production, and expression of outer membrane proteins of Vibrio cholerae. J Bacteriol 150:148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrews SC, Robinson AK, Rodríguez-Quiñones F. 2003. Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 7.Abdul-Tehrani H, Hudson AJ, Chang Y-S, Timms AR, Hawkins C, Williams JM, Harrison PM, Guest JR, Andrews SC. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J Bacteriol 181:1415–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hem JD. 1972. Chemical factors that influence the availability of iron and manganese in aqueous systems. Geol Soc Am Spec Pap 140:17–24. doi: 10.1130/SPE140-p17. [DOI] [Google Scholar]
- 9.Martin JH, Gordon RM, Fitzwater SE. 1991. Iron limitation? The case for iron. Limnol Oceanogr 36:1793–1802. doi: 10.4319/lo.1991.36.8.1793. [DOI] [Google Scholar]
- 10.Conway TM, John SG. 2014. Quantification of dissolved iron sources to the North Atlantic Ocean. Nature 511:212–215. doi: 10.1038/nature13482. [DOI] [PubMed] [Google Scholar]
- 11.Li M, Toner BM, Baker BJ, Breier JA, Sheik CS, Dick GJ. 2014. Microbial iron uptake as a mechanism for dispersing iron from deep-sea hydrothermal vents. Nat Commun 5:3192. doi: 10.1038/ncomms4192. [DOI] [PubMed] [Google Scholar]
- 12.Patel M, Isaäcson M, Gouws E. 1995. Effect of iron and pH on the survival of Vibrio cholerae in water. Trans R Soc Trop Med Hyg 89:175–177. doi: 10.1016/0035-9203(95)90484-0. [DOI] [PubMed] [Google Scholar]
- 13.Joseph S, Bhat KG. 2000. Effect of iron on the survival of Vibrio cholerae in water. Indian J Med Res 111:115–117. [PubMed] [Google Scholar]
- 14.Nilsson L, Oliver JD, Kjelleberg S. 1991. Resuscitation of Vibrio vulnificus from the viable but nonculturable state. J Bacteriol 173:5054–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu HS, Roberts N, Singleton FL, Attwell RW, Grimes DJ, Colwell RR. 1982. Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb Ecol 8:313–323. doi: 10.1007/BF02010671. [DOI] [PubMed] [Google Scholar]
- 16.Asakura H, Ishiwa A, Arakawa E, Makino S, Okada Y, Yamamoto S, Igimi S. 2007. Gene expression profile of Vibrio cholerae in the cold stress-induced viable but non-culturable state. Environ Microbiol 9:869–879. doi: 10.1111/j.1462-2920.2006.01206.x. [DOI] [PubMed] [Google Scholar]
- 17.Yildiz FH, Visick KL. 2009. Vibrio biofilms: so much the same yet so different. Trends Microbiol 17:109–118. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sutherland IW. 2001. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147:3–9. doi: 10.1099/00221287-147-1-3. [DOI] [PubMed] [Google Scholar]
- 19.Stewart PS. 2003. Diffusion in biofilms. J Bacteriol 185:1485–1491. doi: 10.1128/JB.185.5.1485-1491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh PK, Parsek MR, Greenberg EP, Welsh MJ. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- 21.Mey AR, Craig SA, Payne SM. 2005. Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect Immun 73:5706–5719. doi: 10.1128/IAI.73.9.5706-5719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Islam MS, Drasar BS, Bradley DJ. 1990. Long-term persistence of toxigenic Vibrio cholerae 01 in the mucilaginous sheath of a blue-green alga, Anabaena variabilis. J Trop Med Hyg 93:133–139. [PubMed] [Google Scholar]
- 23.Ruby EG, McFall-Ngai MJ. 1992. A squid that glows in the night: development of an animal-bacterial mutualism. J Bacteriol 174:4865–4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fidopiastis PM, von Boletzky S, Ruby EG. 1998. A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J Bacteriol 180:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urbanczyk H, Ast JC, Higgins MJ, Carson J, Dunlap PV. 2007. Reclassification of Vibrio fischeri, Vibrio logei, Vibrio salmonicida and Vibrio wodanis as Aliivibrio fischeri gen. nov., comb. nov., Aliivibrio logei comb. nov., Aliivibrio salmonicida comb. nov. and Aliivibrio wodanis comb. nov. Int J Syst Evol Microbiol 57:2823–2829. doi: 10.1099/ijs.0.65081-0. [DOI] [PubMed] [Google Scholar]
- 26.Montgomery MK, McFall-Ngai M. 1994. Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development 120:1719–1729. [DOI] [PubMed] [Google Scholar]
- 27.Cassat JE, Skaar EP. 2013. Iron in infection and immunity. Cell Host Microbe 13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinberg ED. 2009. Iron availability and infection. Biochim Biophys Acta 1790:600–605. doi: 10.1016/j.bbagen.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Neilands JB. 1981. Microbial iron compounds. Annu Rev Biochem 50:715–731. doi: 10.1146/annurev.bi.50.070181.003435. [DOI] [PubMed] [Google Scholar]
- 30.Neilands JB. 1982. Microbial envelope proteins related to iron. Annu Rev Microbiol 36:285–309. doi: 10.1146/annurev.mi.36.100182.001441. [DOI] [PubMed] [Google Scholar]
- 31.Wyckoff EE, Mey AR, Payne SM. 2007. Iron acquisition in Vibrio cholerae. Biometals 20:405–416. doi: 10.1007/s10534-006-9073-4. [DOI] [PubMed] [Google Scholar]
- 32.Postle K, Larsen RA. 2007. TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals 20:453–465. doi: 10.1007/s10534-006-9071-6. [DOI] [PubMed] [Google Scholar]
- 33.Occhino DA, Wyckoff EE, Henderson DP, Wrona TJ, Payne SM. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol Microbiol 29:1493–1507. [DOI] [PubMed] [Google Scholar]
- 34.Chu BCH, Vogel HJ. 2011. A structural and functional analysis of type III periplasmic and substrate binding proteins: their role in bacterial siderophore and heme transport. Biol Chem 392:39–52. doi: 10.1515/BC.2011.012. [DOI] [PubMed] [Google Scholar]
- 35.Braun V, Hantke K. 2011. Recent insights into iron import by bacteria. Curr Opin Chem Biol 15:328–334. doi: 10.1016/j.cbpa.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 36.Brickman TJ, McIntosh MA. 1992. Overexpression and purification of ferric enterobactin esterase from Escherichia coli. Demonstration of enzymatic hydrolysis of enterobactin and its iron complex J Biol Chem 267:12350–12355. [PubMed] [Google Scholar]
- 37.Butterton JR, Calderwood SB. 1994. Identification, cloning, and sequencing of a gene required for ferric vibriobactin utilization by Vibrio cholerae. J Bacteriol 176:5631–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graf J, Ruby EG. 2000. Novel effects of a transposon insertion in the Vibrio fischeri glnD gene: defects in iron uptake and symbiotic persistence in addition to nitrogen utilization. Mol Microbiol 37:168–179. doi: 10.1046/j.1365-2958.2000.01984.x. [DOI] [PubMed] [Google Scholar]
- 39.O'Brien IG, Gibson F. 1970. The structure of enterochelin and related 2,3-dihydroxy-N-benzoylserine conjugates from Escherichia coli. Biochim Biophys Acta 215:393–402. doi: 10.1016/0304-4165(70)90038-3. [DOI] [PubMed] [Google Scholar]
- 40.Pollack JR, Neilands JB. 1970. Enterobactin, an iron transport compound from Salmonella typhimurium. Biochem Biophys Res Commun 38:989–992. doi: 10.1016/0006-291X(70)90819-3. [DOI] [PubMed] [Google Scholar]
- 41.Payne SM, Niesel DW, Peixotto SS, Lawlor KM. 1983. Expression of hydroxamate and phenolate siderophores by Shigella flexneri. J Bacteriol 155:949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perry RD, San Clemente CL. 1979. Siderophore synthesis in Klebsiella pneumoniae and Shigella sonnei during iron deficiency. J Bacteriol 140:1129–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hantke K, Nicholson G, Rabsch W, Winkelmann G. 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc Natl Acad Sci U S A 100:3677–3682. doi: 10.1073/pnas.0737682100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Griffiths GL, Sigel SP, Payne SM, Neilands JB. 1984. Vibriobactin, a siderophore from Vibrio cholerae. J Biol Chem 259:383–385. [PubMed] [Google Scholar]
- 45.Yamamoto S, Shinoda S, Kawaguchi M, Wakamatsu K, Makita M. 1983. Polyamine distribution in Vibrionaceae: norspermidine as a general constituent of Vibrio species. Can J Microbiol 29:724–728. doi: 10.1139/m83-118. [DOI] [Google Scholar]
- 46.Keating TA, Marshall CG, Walsh CT. 2000. Reconstitution and characterization of the Vibrio cholerae vibriobactin synthetase from VibB, VibE, VibF, and VibH. Biochemistry 39:15522–15530. doi: 10.1021/bi0016523. [DOI] [PubMed] [Google Scholar]
- 47.Keating TA, Marshall CG, Walsh CT. 2000. Vibriobactin biosynthesis in Vibrio cholerae: VibH is an amide synthase homologous to nonribosomal peptide synthetase condensation domains. Biochemistry 39:15513–15521. doi: 10.1021/bi001651a. [DOI] [PubMed] [Google Scholar]
- 48.Marshall CG, Burkart MD, Keating TA, Walsh CT. 2001. Heterocycle formation in vibriobactin biosynthesis: alternative substrate utilization and identification of a condensed intermediate. Biochemistry 40:10655–10663. doi: 10.1021/bi010937s. [DOI] [PubMed] [Google Scholar]
- 49.Marshall CG, Hillson NJ, Walsh CT. 2002. Catalytic mapping of the vibriobactin biosynthetic enzyme VibF. Biochemistry 41:244–250. doi: 10.1021/bi011852u. [DOI] [PubMed] [Google Scholar]
- 50.Wyckoff EE, Stoebner JA, Reed KE, Payne SM. 1997. Cloning of a Vibrio cholerae vibriobactin gene cluster: identification of genes required for early steps in siderophore biosynthesis. J Bacteriol 179:7055–7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young IG, Langman L, Luke RK, Gibson F. 1971. Biosynthesis of the iron-transport compound enterochelin: mutants of Escherichia coli unable to synthesize 2,3-dihydroxybenzoate. J Bacteriol 106:51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crosa JH, Walsh CT. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev 66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mansson M, Gram L, Larsen TO. 2011. Production of bioactive secondary metabolites by marine vibrionaceae. Mar Drugs 9:1440–1468. doi: 10.3390/md9091440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koglin A, Walsh CT. 2009. Structural insights into nonribosomal peptide enzymatic assembly lines. Nat Prod Rep 26:987–1000. doi: 10.1039/b904543k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Butterton JR, Choi MH, Watnick PI, Carroll PA, Calderwood SB. 2000. Vibrio cholerae VibF is required for vibriobactin synthesis and is a member of the family of nonribosomal peptide synthetases. J Bacteriol 182:1731–1738. doi: 10.1128/JB.182.6.1731-1738.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okujo N, Saito M, Yamamoto S, Yoshida T, Miyoshi S, Shinoda S. 1994. Structure of vulnibactin, a new polyamine-containing siderophore from Vibrio vulnificus. Biometals 7:109–116. [DOI] [PubMed] [Google Scholar]
- 57.Yamamoto S, Okujo N, Fujita Y, Saito M, Yoshida T, Shinoda S. 1993. Structures of two polyamine-containing catecholate siderophores from Vibrio fluvialis. J Biochem 113:538–544. [DOI] [PubMed] [Google Scholar]
- 58.Tan W, Verma V, Jeong K, Kim SY, Jung C-H, Lee SE, Rhee JH. 2014. Molecular characterization of vulnibactin biosynthesis in Vibrio vulnificus indicates the existence of an alternative siderophore. Front Microbiol 5:1. doi: 10.3389/fmicb.2014.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Litwin CM, Rayback TW, Skinner J. 1996. Role of catechol siderophore synthesis in Vibrio vulnificus virulence. Infect Immun 64:2834–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Actis LA, Fish W, Crosa JH, Kellerman K, Ellenberger SR, Hauser FM, Sanders-Loehr J. 1986. Characterization of anguibactin, a novel siderophore from Vibrio anguillarum 775(pJM1). J Bacteriol 167:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Di Lorenzo M, Poppelaars S, Stork M, Nagasawa M, Tolmasky ME, Crosa JH. 2004. A nonribosomal peptide synthetase with a novel domain organization is essential for siderophore biosynthesis in Vibrio anguillarum. J Bacteriol 186:7327–7336. doi: 10.1128/JB.186.21.7327-7336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Di Lorenzo M, Stork M, Naka H, Tolmasky ME, Crosa JH. 2008. Tandem heterocyclization domains in a nonribosomal peptide synthetase essential for siderophore biosynthesis in Vibrio anguillarum. Biometals 21:635–648. doi: 10.1007/s10534-008-9149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tolmasky ME, Actis LA, Crosa JH. 1995. A histidine decarboxylase gene encoded by the Vibrio anguillarum plasmid pJM1 is essential for virulence: histamine is a precursor in the biosynthesis of anguibactin. Mol Microbiol 15:87–95. doi: 10.1111/j.1365-2958.1995.tb02223.x. [DOI] [PubMed] [Google Scholar]
- 64.Alice AF, López CS, Crosa JH. 2005. Plasmid- and chromosome-encoded redundant and specific functions are involved in biosynthesis of the siderophore anguibactin in Vibrio anguillarum 775: a case of chance and necessity? J Bacteriol 187:2209–2214. doi: 10.1128/JB.187.6.2209-2214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Di Lorenzo M, Stork M, Tolmasky ME, Actis LA, Farrell D, Welch TJ, Crosa LM, Wertheimer AM, Chen Q, Salinas P, Waldbeser L, Crosa JH. 2003. Complete sequence of virulence plasmid pJM1 from the marine fish pathogen Vibrio anguillarum strain 775. J Bacteriol 185:5822–5830. doi: 10.1128/JB.185.19.5822-5830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tolmasky ME, Crosa JH. 1995. Iron transport genes of the pJM1-mediated iron uptake system of Vibrio anguillarum are included in a transposonlike structure. Plasmid 33:180–190. doi: 10.1006/plas.1995.1019. [DOI] [PubMed] [Google Scholar]
- 67.Naka H, Actis LA, Crosa JH. 2013. The anguibactin biosynthesis and transport genes are encoded in the chromosome of Vibrio harveyi: a possible evolutionary origin for the pJM1 plasmid-encoded system of Vibrio anguillarum? Microbiologyopen 2:182–194. doi: 10.1002/mbo3.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Conchas RF, Lemos ML, Barja JL, Toranzo AE. 1991. Distribution of plasmid- and chromosome-mediated iron uptake systems in Vibrio anguillarum strains of different origins. Appl Environ Microbiol 57:2956–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lemos ML, Salinas P, Toranzo AE, Barja JL, Crosa JH. 1988. Chromosome-mediated iron uptake system in pathogenic strains of Vibrio anguillarum. J Bacteriol 170:1920–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lemos ML, Balado M, Osorio CR. 2010. Anguibactin- versus vanchrobactin-mediated iron uptake in Vibrio anguillarum: evolution and ecology of a fish pathogen. Environ Microbiol Rep 2:19–26. doi: 10.1111/j.1758-2229.2009.00103.x. [DOI] [PubMed] [Google Scholar]
- 71.Soengas RG, Anta C, Espada A, Paz V, Ares IR, Balado M, Rodríguez J, Lemos ML, Jiménez C. 2006. Structural characterization of vanchrobactin, a new catechol siderophore produced by the fish pathogen Vibrio anguillarum serotype O2. Tetrahedron Lett 47:7113–7116. doi: 10.1016/j.tetlet.2006.07.104. [DOI] [Google Scholar]
- 72.Yamamoto S, Okujo N, Yoshida T, Matsuura S, Shinoda S. 1994. Structure and iron transport activity of vibrioferrin, a new siderophore of Vibrio parahaemolyticus. J Biochem 115:868–874. [DOI] [PubMed] [Google Scholar]
- 73.Amin SA, Green DH, Küpper FC, Carrano CJ. 2009. Vibrioferrin, an unusual marine siderophore: iron binding, photochemistry, and biological implications. Inorg Chem 48:11451–11458. doi: 10.1021/ic9016883. [DOI] [PubMed] [Google Scholar]
- 74.Amin SA, Green DH, Hart MC, Küpper FC, Sunda WG, Carrano CJ. 2009. Photolysis of iron-siderophore chelates promotes bacterial-algal mutualism. Proc Natl Acad Sci U S A 106:17071–17076. doi: 10.1073/pnas.0905512106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kirchman DL, Meon B, Cottrell MT, Hutchins DA, Weeks D, Bruland KW. 2000. Carbon versus iron limitation of bacterial growth in the California upwelling regime. Limnol Oceanogr 45:1681–1688. doi: 10.4319/lo.2000.45.8.1681. [DOI] [Google Scholar]
- 76.Kumazawa NH, Fukuma N, Komoda Y. 1991. Attachment of Vibrio parahaemolyticus strains to estuarine algae. J Vet Med Sci 53:201–205. doi: 10.1292/jvms.53.201. [DOI] [PubMed] [Google Scholar]
- 77.Moon Y-H, Tanabe T, Funahashi T, Shiuchi K, Nakao H, Yamamoto S. 2004. Identification and characterization of two contiguous operons required for aerobactin transport and biosynthesis in Vibrio mimicus. Microbiol Immunol 48:389–398. doi: 10.1111/j.1348-0421.2004.tb03528.x. [DOI] [PubMed] [Google Scholar]
- 78.Suzuki K, Tanabe T, Moon Y-H, Funahashi T, Nakao H, Narimatsu S, Yamamoto S. 2006. Identification and transcriptional organization of aerobactin transport and biosynthesis cluster genes of Vibrio hollisae. Res Microbiol 157:730–740. doi: 10.1016/j.resmic.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 79.Haygood MG, Holt PD, Butler A. 1993. Aerobactin production by a planktonic marine Vibrio sp. Limnol Oceanogr 38:1091–1097. doi: 10.4319/lo.1993.38.5.1091. [DOI] [Google Scholar]
- 80.Colonna B, Nicoletti M, Visca P, Casalino M, Valenti P, Maimone F. 1985. Composite IS1 elements encoding hydroxamate-mediated iron uptake in FIme plasmids from epidemic Salmonella spp. J Bacteriol 162:307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.De Lorenzo V, Bindereif A, Paw BH, Neilands JB. 1986. Aerobactin biosynthesis and transport genes of plasmid ColV-K30 in Escherichia coli K-12. J Bacteriol 165:570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marolda CL, Valvano MA, Lawlor KM, Payne SM, Crosa JH. 1987. Flanking and internal regions of chromosomal genes mediating aerobactin iron uptake systems in enteroinvasive Escherichia coli and Shigella flexneri. J Gen Microbiol 133:2269–2278. [DOI] [PubMed] [Google Scholar]
- 83.Moss JE, Cardozo TJ, Zychlinsky A, Groisman EA. 1999. The selC-associated SHI-2 pathogenicity island of Shigella flexneri. Mol Microbiol 33:74–83. doi: 10.1046/j.1365-2958.1999.01449.x. [DOI] [PubMed] [Google Scholar]
- 84.Purdy GE, Payne SM. 2001. The SHI-3 iron transport island of Shigella boydii 0-1392 carries the genes for aerobactin synthesis and transport. J Bacteriol 183:4176–4182. doi: 10.1128/JB.183.14.4176-4182.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vokes SA, Reeves SA, Torres AG, Payne SM. 1999. The aerobactin iron transport system genes in Shigella flexneri are present within a pathogenicity island. Mol Microbiol 33:63–73. doi: 10.1046/j.1365-2958.1999.01448.x. [DOI] [PubMed] [Google Scholar]
- 86.Murugappan RM, Aravinth A, Karthikeyan M. 2011. Chemical and structural characterization of hydroxamate siderophore produced by marine Vibrio harveyi. J Ind Microbiol Biotechnol 38:265–273. doi: 10.1007/s10295-010-0769-7. [DOI] [PubMed] [Google Scholar]
- 87.Simpson LM, Oliver JD. 1983. Siderophore production by Vibrio vulnificus. Infect Immun 41:644–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martinez JS, Zhang GP, Holt PD, Jung HT, Carrano CJ, Haygood MG, Butler A. 2000. Self-assembling amphiphilic siderophores from marine bacteria. Science 287:1245–1247. doi: 10.1126/science.287.5456.1245. [DOI] [PubMed] [Google Scholar]
- 89.Martinez JS, Carter-Franklin JN, Mann EL, Martin JD, Haygood MG, Butler A. 2003. Structure and membrane affinity of a suite of amphiphilic siderophores produced by a marine bacterium. Proc Natl Acad Sci U S A 100:3754–3759. doi: 10.1073/pnas.0637444100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vraspir JM, Holt PD, Butler A. 2011. Identification of new members within suites of amphiphilic marine siderophores. Biometals 24:85–92. doi: 10.1007/s10534-010-9378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gauglitz JM, Butler A. 2013. Amino acid variability in the peptide composition of a suite of amphiphilic peptide siderophores from an open ocean Vibrio species. J Biol Inorg Chem 18:489–497. doi: 10.1007/s00775-013-0995-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zane HK, Naka H, Rosconi F, Sandy M, Haygood MG, Butler A. 2014. Biosynthesis of amphi-enterobactin siderophores by Vibrio harveyi BAA-1116: identification of a bifunctional nonribosomal peptide synthetase condensation domain. J Am Chem Soc 136:5615–5618. doi: 10.1021/ja5019942. [DOI] [PubMed] [Google Scholar]
- 93.Gauglitz JM, Iinishi A, Ito Y, Butler A. 2014. Microbial tailoring of acyl peptidic siderophores. Biochemistry 53:2624–2631. doi: 10.1021/bi500266x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Butterton JR, Stoebner JA, Payne SM, Calderwood SB. 1992. Cloning, sequencing, and transcriptional regulation of viuA, the gene encoding the ferric vibriobactin receptor of Vibrio cholerae. J Bacteriol 174:3729–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stoebner JA, Butterton JR, Calderwood SB, Payne SM. 1992. Identification of the vibriobactin receptor of Vibrio cholerae. J Bacteriol 174:3270–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wyckoff EE, Valle AM, Smith SL, Payne SM. 1999. A multifunctional ATP-binding cassette transporter system from Vibrio cholerae transports vibriobactin and enterobactin. J Bacteriol 181:7588–7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Funahashi T, Moriya K, Uemura S, Miyoshi S, Shinoda S, Narimatsu S, Yamamoto S. 2002. Identification and characterization of pvuA, a gene encoding the ferric vibrioferrin receptor protein in Vibrio parahaemolyticus. J Bacteriol 184:936–946. doi: 10.1128/jb.184.4.936-946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Webster AC, Litwin CM. 2000. Cloning and characterization of vuuA, a gene encoding the Vibrio vulnificus ferric vulnibactin receptor. Infect Immun 68:526–534. doi: 10.1128/IAI.68.2.526-534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Balado M, Osorio CR, Lemos ML. 2009. FvtA is the receptor for the siderophore vanchrobactin in Vibrio anguillarum: utility as a route of entry for vanchrobactin analogues. Appl Environ Microbiol 75:2775–2783. doi: 10.1128/AEM.02897-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Actis LA, Tolmasky ME, Farrell DH, Crosa JH. 1988. Genetic and molecular characterization of essential components of the Vibrio anguillarum plasmid-mediated iron-transport system. J Biol Chem 263:2853–2860. [PubMed] [Google Scholar]
- 101.Köster WL, Actis LA, Waldbeser LS, Tolmasky ME, Crosa JH. 1991. Molecular characterization of the iron transport system mediated by the pJM1 plasmid in Vibrio anguillarum 775. J Biol Chem 266:23829–23833. [PubMed] [Google Scholar]
- 102.Welch TJ, Crosa JH. 2005. Novel role of the lipopolysaccharide O1 side chain in ferric siderophore transport and virulence of Vibrio anguillarum. Infect Immun 73:5864–5872. doi: 10.1128/IAI.73.9.5864-5872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Strassmann JE, Gilbert OM, Queller DC. 2011. Kin discrimination and cooperation in microbes. Annu Rev Microbiol 65:349–367. doi: 10.1146/annurev.micro.112408.134109. [DOI] [PubMed] [Google Scholar]
- 104.Meibom KL, Blokesch M, Dolganov NA, Wu C-Y, Schoolnik GK. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 105.Sun Y, Bernardy EE, Hammer BK, Miyashiro T. 2013. Competence and natural transformation in vibrios. Mol Microbiol 89:583–595. doi: 10.1111/mmi.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mey AR, Wyckoff EE, Oglesby AG, Rab E, Taylor RK, Payne SM. 2002. Identification of the Vibrio cholerae enterobactin receptors VctA and IrgA: IrgA is not required for virulence. Infect Immun 70:3419–3426. doi: 10.1128/IAI.70.7.3419-3426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rogers MB, Sexton JA, DeCastro GJ, Calderwood SB. 2000. Identification of an operon required for ferrichrome iron utilization in Vibrio cholerae. J Bacteriol 182:2350–2353. doi: 10.1128/JB.182.8.2350-2353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wyckoff EE, Allred BE, Raymond KN, Payne SM. 2015. Catechol siderophore transport by Vibrio cholerae. J Bacteriol 197:2840–2849. doi: 10.1128/JB.00417-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Funahashi T, Tanabe T, Aso H, Nakao H, Fujii Y, Okamoto K, Narimatsu S, Yamamoto S. 2003. An iron-regulated gene required for utilization of aerobactin as an exogenous siderophore in Vibrio parahaemolyticus. Microbiology 149:1217–1225. doi: 10.1099/mic.0.26066-0. [DOI] [PubMed] [Google Scholar]
- 110.Funahashi T, Tanabe T, Shiuchi K, Nakao H, Yamamoto S. 2009. Identification and characterization of genes required for utilization of desferri-ferrichrome and aerobactin in Vibrio parahaemolyticus. Biol Pharm Bull 32:359–365. doi: 10.1248/bpb.32.359. [DOI] [PubMed] [Google Scholar]
- 111.Aso H, Miyoshi S, Nakao H, Okamoto K, Yamamoto S. 2002. Induction of an outer membrane protein of 78 kDa in Vibrio vulnificus cultured in the presence of desferrioxamine B under iron-limiting conditions. FEMS Microbiol Lett 212:65–70. doi: 10.1111/j.1574-6968.2002.tb11246.x. [DOI] [PubMed] [Google Scholar]
- 112.Kim C-M, Park Y-J, Shin S-H. 2007. A widespread deferoxamine-mediated iron-uptake system in Vibrio vulnificus. J Infect Dis 196:1537–1545. doi: 10.1086/523108. [DOI] [PubMed] [Google Scholar]
- 113.Tanabe T, Funahashi T, Miyamoto K, Tsujibo H, Yamamoto S. 2011. Identification of genes, desR and desA, required for utilization of desferrioxamine B as a xenosiderophore in Vibrio furnissii. Biol Pharm Bull 34:570–574. doi: 10.1248/bpb.34.570. [DOI] [PubMed] [Google Scholar]
- 114.Holmstrøm K, Gram L. 2003. Elucidation of the Vibrio anguillarum genetic response to the potential fish probiont Pseudomonas fluorescens AH2, using RNA-arbitrarily primed PCR. J Bacteriol 185:831–842. doi: 10.1128/JB.185.3.831-842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Böttcher T, Clardy J. 2014. A chimeric siderophore halts swarming Vibrio. Angew Chem Int Ed Engl 53:3510–3513. doi: 10.1002/anie.201310729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang Q, Liu Q, Cao X, Yang M, Zhang Y. 2008. Characterization of two TonB systems in marine fish pathogen Vibrio alginolyticus: their roles in iron utilization and virulence. Arch Microbiol 190:595–603. doi: 10.1007/s00203-008-0407-1. [DOI] [PubMed] [Google Scholar]
- 117.Andrus CR, Walter M, Crosa JH, Payne SM. 1983. Synthesis of siderophores by pathogenic Vibrio species. Curr Microbiol 9:209–214. doi: 10.1007/BF01567583. [DOI] [Google Scholar]
- 118.Henderson DP, Payne SM. 1993. Cloning and characterization of the Vibrio cholerae genes encoding the utilization of iron from haemin and haemoglobin. Mol Microbiol 7:461–469. doi: 10.1111/j.1365-2958.1993.tb01137.x. [DOI] [PubMed] [Google Scholar]
- 119.Henderson DP, Payne SM. 1994. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J Bacteriol 176:3269–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mey AR, Payne SM. 2001. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol Microbiol 42:835–849. [DOI] [PubMed] [Google Scholar]
- 121.Cescau S, Cwerman H, Létoffé S, Delepelaire P, Wandersman C, Biville F. 2007. Heme acquisition by hemophores. Biometals 20:603–613. doi: 10.1007/s10534-006-9050-y. [DOI] [PubMed] [Google Scholar]
- 122.Wyckoff EE, Schmitt M, Wilks A, Payne SM. 2004. HutZ is required for efficient heme utilization in Vibrio cholerae. J Bacteriol 186:4142–4151. doi: 10.1128/JB.186.13.4142-4151.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Uchida T, Sekine Y, Matsui T, Ikeda-Saito M, Ishimori K. 2012. A heme degradation enzyme, HutZ, from Vibrio cholerae. Chem Commun (Camb) 48:6741–6743. doi: 10.1039/c2cc31147j. [DOI] [PubMed] [Google Scholar]
- 124.Litwin CM, Byrne BL. 1998. Cloning and characterization of an outer membrane protein of Vibrio vulnificus required for heme utilization: regulation of expression and determination of the gene sequence. Infect Immun 66:3134–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Datta S, Crosa JH. 2012. Identification and characterization of a novel outer membrane protein receptor required for hemin utilization in Vibrio vulnificus. Biometals 25:275–283. doi: 10.1007/s10534-011-9501-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ahn S-H, Han J-H, Lee J-H, Park K-J, Kong I-S. 2005. Identification of an iron-regulated hemin-binding outer membrane protein, HupO, in Vibrio fluvialis: effects on hemolytic activity and the oxidative stress response. Infect Immun 73:722–729. doi: 10.1128/IAI.73.2.722-729.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mazoy R, Lemos ML. 1996. Identification of heme-binding proteins in the cell membranes of Vibrio anguillarum. FEMS Microbiol Lett 135:265–270. doi: 10.1111/j.1574-6968.1996.tb07999.x. [DOI] [PubMed] [Google Scholar]
- 128.Mazoy R, Osorio CR, Toranzo AE, Lemos ML. 2003. Isolation of mutants of Vibrio anguillarum defective in haeme utilisation and cloning of huvA, a gene coding for an outer membrane protein involved in the use of haeme as iron source. Arch Microbiol 179:329–338. [DOI] [PubMed] [Google Scholar]
- 129.Tanabe T, Funahashi T, Moon Y-H, Tamai E, Yamamoto S. 2010. Identification and characterization of a Vibrio mimicus gene encoding the heme/hemoglobin receptor. Microbiol Immunol 54:606–617. doi: 10.1111/j.1348-0421.2010.00256.x. [DOI] [PubMed] [Google Scholar]
- 130.O'Malley SM, Mouton SL, Occhino DA, Deanda MT, Rashidi JR, Fuson KL, Rashidi CE, Mora MY, Payne SM, Henderson DP. 1999. Comparison of the heme iron utilization systems of pathogenic vibrios. J Bacteriol 181:3594–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mazoy R, Lemos DML. 1991. Iron-binding proteins and heme compounds as iron sources for Vibrio anguillarum. Curr Microbiol 23:221–226. doi: 10.1007/BF02092282. [DOI] [Google Scholar]
- 132.Fouz B, Mazoy R, Vázquez F, Lemos ML, Amaro C. 1997. Isolation of a hemin and hemoglobin binding outer membrane protein of Vibrio vulnificus biotype 2 (serogroup E). FEMS Microbiol Lett 156:187–191. doi: 10.1111/j.1574-6968.1997.tb12725.x. [DOI] [PubMed] [Google Scholar]
- 133.Zakaria-Meehan Z, Massad G, Simpson LM, Travis JC, Oliver JD. 1988. Ability of Vibrio vulnificus to obtain iron from hemoglobin-haptoglobin complexes. Infect Immun 56:275–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nishina Y, Miyoshi S, Nagase A, Shinoda S. 1992. Significant role of an exocellular protease in utilization of heme by Vibrio vulnificus. Infect Immun 60:2128–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Seliger SS, Mey AR, Valle AM, Payne SM. 2001. The two TonB systems of Vibrio cholerae: redundant and specific functions. Mol Microbiol 39:801–812. doi: 10.1046/j.1365-2958.2001.02273.x. [DOI] [PubMed] [Google Scholar]
- 136.Stork M, Di Lorenzo M, Mouriño S, Osorio CR, Lemos ML, Crosa JH. 2004. Two tonB systems function in iron transport in Vibrio anguillarum, but only one is essential for virulence. Infect Immun 72:7326–7329. doi: 10.1128/IAI.72.12.7326-7329.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Alice AF, Crosa JH. 2012. The TonB3 system in the human pathogen Vibrio vulnificus is under the control of the global regulators Lrp and cyclic AMP receptor protein. J Bacteriol 194:1897–1911. doi: 10.1128/JB.06614-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kuehl CJ, Crosa JH. 2009. Molecular and genetic characterization of the TonB2-cluster TtpC protein in pathogenic vibrios. Biometals 22:109–115. doi: 10.1007/s10534-008-9194-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kustusch RJ, Kuehl CJ, Crosa JH. 2012. The ttpC gene is contained in two of three TonB systems in the human pathogen Vibrio vulnificus, but only one is active in iron transport and virulence. J Bacteriol 194:3250–3259. doi: 10.1128/JB.00155-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Stork M, Otto BR, Crosa JH. 2007. A novel protein, TtpC, is a required component of the TonB2 complex for specific iron transport in the pathogens Vibrio anguillarum and Vibrio cholerae. J Bacteriol 189:1803–1815. doi: 10.1128/JB.00451-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mey AR, Payne SM. 2003. Analysis of residues determining specificity of Vibrio cholerae TonB1 for its receptors. J Bacteriol 185:1195–1207. doi: 10.1128/JB.185.4.1195-1207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Stock JB, Rauch B, Roseman S. 1977. Periplasmic space in Salmonella typhimurium and Escherichia coli. J Biol Chem 252:7850–7861. [PubMed] [Google Scholar]
- 143.Braun V, Hantke K, Köster W. 1998. Bacterial iron transport: mechanisms, genetics, and regulation. Met Ions Biol Syst 35:67–145. [PubMed] [Google Scholar]
- 144.Miethke M, Hou J, Marahiel MA. 2011. The siderophore-interacting protein YqjH acts as a ferric reductase in different iron assimilation pathways of Escherichia coli. Biochemistry 50:10951–10964. doi: 10.1021/bi201517h. [DOI] [PubMed] [Google Scholar]
- 145.Balado M, Osorio CR, Lemos ML. 2006. A gene cluster involved in the biosynthesis of vanchrobactin, a chromosome-encoded siderophore produced by Vibrio anguillarum. Microbiology 152:3517–3528. doi: 10.1099/mic.0.29298-0. [DOI] [PubMed] [Google Scholar]
- 146.Weaver EA, Wyckoff EE, Mey AR, Morrison R, Payne SM. 2013. FeoA and FeoC are essential components of the Vibrio cholerae ferrous iron uptake system, and FeoC interacts with FeoB. J Bacteriol 195:4826–4835. doi: 10.1128/JB.00738-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kammler M, Schon C, Hantke K. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J Bacteriol 175:6212–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wyckoff EE, Mey AR, Leimbach A, Fisher CF, Payne SM. 2006. Characterization of ferric and ferrous iron transport systems in Vibrio cholerae. J Bacteriol 188:6515–6523. doi: 10.1128/JB.00626-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Marlovits TC, Haase W, Herrmann C, Aller SG, Unger VM. 2002. The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc Natl Acad Sci U S A 99:16243–16248. doi: 10.1073/pnas.242338299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kim H, Lee H, Shin D. 2013. The FeoC protein leads to high cellular levels of the Fe(II) transporter FeoB by preventing FtsH protease regulation of FeoB in Salmonella enterica. J Bacteriol 195:3364–3370. doi: 10.1128/JB.00343-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kim H, Lee H, Shin D. 2012. The FeoA protein is necessary for the FeoB transporter to import ferrous iron. Biochem Biophys Res Commun 423:733–738. doi: 10.1016/j.bbrc.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 152.Mey AR, Wyckoff EE, Hoover LA, Fisher CR, Payne SM. 2008. Vibrio cholerae VciB promotes iron uptake via ferrous iron transporters. J Bacteriol 190:5953–5962. doi: 10.1128/JB.00569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mazoy R, Lopez EM, Fouz B, Amaro C, Lemos ML. 1999. Ferric-reductase activities in Vibrio vulnificus biotypes 1 and 2. FEMS Microbiol Lett 172:205–211. doi: 10.1111/j.1574-6968.1999.tb13470.x. [DOI] [PubMed] [Google Scholar]
- 154.Wyckoff EE, Payne SM. 2011. The Vibrio cholerae VctPDGC system transports catechol siderophores and a siderophore-free iron ligand. Mol Microbiol 81:1446–1458. doi: 10.1111/j.1365-2958.2011.07775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nairz M, Schroll A, Sonnweber T, Weiss G. 2010. The struggle for iron—a metal at the host-pathogen interface. Cell Microbiol 12:1691–1702. doi: 10.1111/j.1462-5822.2010.01529.x. [DOI] [PubMed] [Google Scholar]
- 156.Nairz M, Haschka D, Demetz E, Weiss G. 2014. Iron at the interface of immunity and infection. Front Pharmacol 5:152. doi: 10.3389/fphar.2014.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bullen JJ, Rogers HJ, Spalding PB, Ward CG. 2006. Natural resistance, iron and infection: a challenge for clinical medicine. J Med Microbiol 55:251–258. doi: 10.1099/jmm.0.46386-0. [DOI] [PubMed] [Google Scholar]
- 158.Johnson EE, Wessling-Resnick M. 2012. Iron metabolism and the innate immune response to infection. Microbes Infect 14:207–216. doi: 10.1016/j.micinf.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ratledge C. 2007. Iron metabolism and infection. Food Nutr Bull 28:S515–S523. [DOI] [PubMed] [Google Scholar]
- 160.Arnold RR, Cole MF, McGhee JR. 1977. A bactericidal effect for human lactoferrin. Science 197:263–265. doi: 10.1126/science.327545. [DOI] [PubMed] [Google Scholar]
- 161.Arnold RR, Brewer M, Gauthier JJ. 1980. Bactericidal activity of human lactoferrin: sensitivity of a variety of microorganisms. Infect Immun 28:893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gifford JL, Hunter HN, Vogel HJ. 2005. Lactoferricin: a lactoferrin-derived peptide with antimicrobial, antiviral, antitumor and immunological properties. Cell Mol Life Sci 62:2588–2598. doi: 10.1007/s00018-005-5373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Drakesmith H, Prentice AM. 2012. Hepcidin and the iron-infection axis. Science 338:768–772. doi: 10.1126/science.1224577. [DOI] [PubMed] [Google Scholar]
- 164.Yang C-G, Liu S-S, Sun B, Wang X-L, Wang N, Chen S-L. 2013. Iron-metabolic function and potential antibacterial role of hepcidin and its correlated genes (ferroportin 1 and transferrin receptor) in turbot (Scophthalmus maximus). Fish Shellfish Immunol 34:744–755. doi: 10.1016/j.fsi.2012.11.049. [DOI] [PubMed] [Google Scholar]
- 165.Weinberg ED. 1977. Infection and iron metabolism. Am J Clin Nutr 30:1485–1490. [DOI] [PubMed] [Google Scholar]
- 166.Weinberg ED. 2000. Microbial pathogens with impaired ability to acquire host iron. Biometals 13:85–89. doi: 10.1023/A:1009293500209. [DOI] [PubMed] [Google Scholar]
- 167.Khan FA, Fisher MA, Khakoo RA. 2007. Association of hemochromatosis with infectious diseases: expanding spectrum. Int J Infect Dis 11:482–487. doi: 10.1016/j.ijid.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 168.Gerhard GS, Levin KA, Price Goldstein J, Wojnar MM, Chorney MJ, Belchis DA. 2001. Vibrio vulnificus septicemia in a patient with the hemochromatosis HFE C282Y mutation. Arch Pathol Lab Med 125:1107–1109. [DOI] [PubMed] [Google Scholar]
- 169.Fernández JM, Serrano M, De Arriba JJ, Sánchez MV, Escribano E, Ferreras P. 2000. Bacteremic cellulitis caused by non-01, non-0139 Vibrio cholerae: report of a case in a patient with hemochromatosis. Diagn Microbiol Infect Dis 37:77–80. doi: 10.1016/S0732-8893(99)00153-4. [DOI] [PubMed] [Google Scholar]
- 170.Amaro C, Biosca EG, Fouz B, Toranzo AE, Garay E. 1994. Role of iron, capsule, and toxins in the pathogenicity of Vibrio vulnificus biotype 2 for mice. Infect Immun 62:759–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Brennt CE, Wright AC, Dutta SK, Morris JG. 1991. Growth of Vibrio vulnificus in serum from alcoholics: association with high transferrin iron saturation. J Infect Dis 164:1030–1032. doi: 10.1093/infdis/164.5.1030. [DOI] [PubMed] [Google Scholar]
- 172.Bullen JJ, Spalding PB, Ward CG, Gutteridge JM. 1991. Hemochromatosis, iron and septicemia caused by Vibrio vulnificus. Arch Intern Med 151:1606–1609. doi: 10.1001/archinte.1991.00400080096018. [DOI] [PubMed] [Google Scholar]
- 173.Starks AM, Schoeb TR, Tamplin ML, Parveen S, Doyle TJ, Bomeisl PE, Escudero GM, Gulig PA. 2000. Pathogenesis of infection by clinical and environmental strains of Vibrio vulnificus in iron-dextran-treated mice. Infect Immun 68:5785–5793. doi: 10.1128/IAI.68.10.5785-5793.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ashrafian H. 2003. Hepcidin: the missing link between hemochromatosis and infections. Infect Immun 71:6693–6700. doi: 10.1128/IAI.71.12.6693-6700.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Arezes J, Jung G, Gabayan V, Valore E, Ruchala P, Gulig PA, Ganz T, Nemeth E, Bulut Y. 2015. Hepcidin-induced hypoferremia is a critical host defense mechanism against the siderophilic bacterium Vibrio vulnificus. Cell Host Microbe 17:47–57. doi: 10.1016/j.chom.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lombardo M-J, Michalski J, Martinez-Wilson H, Morin C, Hilton T, Osorio CG, Nataro JP, Tacket CO, Camilli A, Kaper JB. 2007. An in vivo expression technology screen for Vibrio cholerae genes expressed in human volunteers. Proc Natl Acad Sci U S A 104:18229–18234. doi: 10.1073/pnas.0705636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mandlik A, Livny J, Robins WP, Ritchie JM, Mekalanos JJ, Waldor MK. 2011. RNA-seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 10:165–174. doi: 10.1016/j.chom.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bisharat N, Bronstein M, Korner M, Schnitzer T, Koton Y. 2013. Transcriptome profiling analysis of Vibrio vulnificus during human infection. Microbiology 159:1878–1887. doi: 10.1099/mic.0.067900-0. [DOI] [PubMed] [Google Scholar]
- 179.Merrell DS, Butler SM, Qadri F, Dolganov NA, Alam A, Cohen MB, Calderwood SB, Schoolnik GK, Camilli A. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417:642–645. doi: 10.1038/nature00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.LaRocque RC, Krastins B, Harris JB, Lebrun LM, Parker KC, Chase M, Ryan ET, Qadri F, Sarracino D, Calderwood SB. 2008. Proteomic analysis of Vibrio cholerae in human stool. Infect Immun 76:4145–4151. doi: 10.1128/IAI.00585-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Livny J, Zhou X, Mandlik A, Hubbard T, Davis BM, Waldor MK. 2014. Comparative RNA-Seq based dissection of the regulatory networks and environmental stimuli underlying Vibrio parahaemolyticus gene expression during infection. Nucleic Acids Res 42:12212–12223. doi: 10.1093/nar/gku891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Septer AN, Wang Y, Ruby EG, Stabb EV, Dunn AK. 2011. The haem-uptake gene cluster in Vibrio fischeri is regulated by Fur and contributes to symbiotic colonization. Environ Microbiol 13:2855–2864. doi: 10.1111/j.1462-2920.2011.02558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cornelissen CN, Kelley M, Hobbs MM, Anderson JE, Cannon JG, Cohen MS, Sparling PF. 1998. The transferrin receptor expressed by gonococcal strain FA1090 is required for the experimental infection of human male volunteers. Mol Microbiol 27:611–616. doi: 10.1046/j.1365-2958.1998.00710.x. [DOI] [PubMed] [Google Scholar]
- 184.Pajuelo D, Lee C-T, Roig F, Hor L-I, Amaro C. 2015. Novel host-specific iron acquisition system in the zoonotic pathogen Vibrio vulnificus. Environ Microbiol 17:2076–2089. doi: 10.1111/1462-2920.12782. [DOI] [PubMed] [Google Scholar]
- 185.Pajuelo D, Lee C-T, Roig FJ, Lemos ML, Hor L-I, Amaro C. 2014. Host-nonspecific iron acquisition systems and virulence in the zoonotic serovar of Vibrio vulnificus. Infect Immun 82:731–744. doi: 10.1128/IAI.01117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Biosca EG, Fouz B, Alcaide E, Amaro C. 1996. Siderophore-mediated iron acquisition mechanisms in Vibrio vulnificus biotype 2. Appl Environ Microbiol 62:928–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yamamoto S, Okujo N, Matsuura S, Fujiwara I, Fujita Y, Shinoda S. 1994. Siderophore-mediated utilization of iron bound to transferrin by Vibrio parahaemolyticus. Microbiol Immunol 38:687–693. doi: 10.1111/j.1348-0421.1994.tb01843.x. [DOI] [PubMed] [Google Scholar]
- 188.Fouz B, Biosca EG, Amaro C. 1997. High affinity iron-uptake systems in Vibrio damsela: role in the acquisition of iron from transferrin. J Appl Microbiol 82:157–167. doi: 10.1111/j.1365-2672.1997.tb02846.x. [DOI] [PubMed] [Google Scholar]
- 189.Okujo N, Akiyama T, Miyoshi S, Shinoda S, Yamamoto S. 1996. Involvement of vulnibactin and exocellular protease in utilization of transferrin- and lactoferrin-bound iron by Vibrio vulnificus. Microbiol Immunol 40:595–598. doi: 10.1111/j.1348-0421.1996.tb01114.x. [DOI] [PubMed] [Google Scholar]
- 190.Shin S-H, Sun H-Y, Park R-Y, Kim C-M, Kim S-Y, Rhee J-H. 2005. Vibrio vulnificus metalloprotease VvpE has no direct effect on the iron-assimilation from human holotransferrin. FEMS Microbiol Lett 247:221–229. doi: 10.1016/j.femsle.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 191.Kim C-M, Park R-Y, Park J-H, Sun H-Y, Bai Y-H, Ryu P-Y, Kim S-Y, Rhee J-H, Shin S-H. 2006. Vibrio vulnificus vulnibactin, but not metalloprotease VvpE, is essentially required for iron-uptake from human holotransferrin. Biol Pharm Bull 29:911–918. doi: 10.1248/bpb.29.911. [DOI] [PubMed] [Google Scholar]
- 192.Abergel RJ, Clifton MC, Pizarro JC, Warner JA, Shuh DK, Strong RK, Raymond KN. 2008. The siderocalin/enterobactin interaction: a link between mammalian immunity and bacterial iron transport. J Am Chem Soc 130:11524–11534. doi: 10.1021/ja803524w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. 2004. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 194.Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. 2002. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell 10:1033–1043. doi: 10.1016/S1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 195.Li N, Zhang C, Li B, Liu X, Huang Y, Xu S, Gu L. 2012. Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response. J Biol Chem 287:8912–8919. doi: 10.1074/jbc.M111.316034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Allred BE, Correnti C, Clifton MC, Strong RK, Raymond KN. 2013. Siderocalin outwits the coordination chemistry of vibriobactin, a siderophore of Vibrio cholerae. ACS Chem Biol 8:1882–1887. doi: 10.1021/cb4002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sigel SP, Stoebner JA, Payne SM. 1985. Iron-vibriobactin transport system is not required for virulence of Vibrio cholerae. Infect Immun 47:360–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Henderson DP, Payne SM. 1994. Vibrio cholerae iron transport systems: roles of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect Immun 62:5120–5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Touati D. 2000. Iron and oxidative stress in bacteria. Arch Biochem Biophys 373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- 200.Kehrer JP. 2000. The Haber-Weiss reaction and mechanisms of toxicity. Toxicology 149:43–50. doi: 10.1016/S0300-483X(00)00231-6. [DOI] [PubMed] [Google Scholar]
- 201.Imlay JA, Linn S. 1988. DNA damage and oxygen radical toxicity. Science 240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 202.Iuchi S, Weiner L. 1996. Cellular and molecular physiology of Escherichia coli in the adaptation to aerobic environments. J Biochem 120:1055–1063. doi: 10.1093/oxfordjournals.jbchem.a021519. [DOI] [PubMed] [Google Scholar]
- 203.Litwin CM, Boyko SA, Calderwood SB. 1992. Cloning, sequencing, and transcriptional regulation of the Vibrio cholerae fur gene. J Bacteriol 174:1897–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Litwin CM, Calderwood SB. 1994. Analysis of the complexity of gene regulation by Fur in Vibrio cholerae. J Bacteriol 176:240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sheikh MA, Taylor GL. 2009. Crystal structure of the Vibrio cholerae ferric uptake regulator (Fur) reveals insights into metal co-ordination. Mol Microbiol 72:1208–1220. doi: 10.1111/j.1365-2958.2009.06718.x. [DOI] [PubMed] [Google Scholar]
- 206.Watnick PI, Eto T, Takahashi H, Calderwood SB. 1997. Purification of Vibrio cholerae Fur and estimation of its intracellular abundance by antibody sandwich enzyme-linked immunosorbent assay. J Bacteriol 179:243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.De Lorenzo V, Herrero M, Giovannini F, Neilands JB. 1988. Fur (ferric uptake regulation) protein and CAP (catabolite-activator protein) modulate transcription of fur gene in Escherichia coli. Eur J Biochem 173:537–546. doi: 10.1111/j.1432-1033.1988.tb14032.x. [DOI] [PubMed] [Google Scholar]
- 208.Lee H-J, Bang SH, Lee K-H, Park S-J. 2007. Positive regulation of fur gene expression via direct interaction of Fur in a pathogenic bacterium, Vibrio vulnificus. J Bacteriol 189:2629–2636. doi: 10.1128/JB.01791-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lee H-J, Park K-J, Lee AY, Park SG, Park BC, Lee K-H, Park S-J. 2003. Regulation of fur expression by RpoS and Fur in Vibrio vulnificus. J Bacteriol 185:5891–5896. doi: 10.1128/JB.185.19.5891-5896.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mey AR, Wyckoff EE, Kanukurthy V, Fisher CR, Payne SM. 2005. Iron and Fur regulation in Vibrio cholerae and the role of Fur in virulence. Infect Immun 73:8167–8178. doi: 10.1128/IAI.73.12.8167-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Craig SA, Carpenter CD, Mey AR, Wyckoff EE, Payne SM. 2011. Positive regulation of the Vibrio cholerae porin OmpT by iron and Fur. J Bacteriol 193:6505–6511. doi: 10.1128/JB.05681-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Davies BW, Bogard RW, Mekalanos JJ. 2011. Mapping the regulon of Vibrio cholerae ferric uptake regulator expands its known network of gene regulation. Proc Natl Acad Sci U S A 108:12467–12472. doi: 10.1073/pnas.1107894108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Beyhan S, Tischler AD, Camilli A, Yildiz FH. 2006. Differences in gene expression between the classical and El Tor biotypes of Vibrio cholerae O1. Infect Immun 74:3633–3642. doi: 10.1128/IAI.01750-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Alice AF, Naka H, Crosa JH. 2008. Global gene expression as a function of the iron status of the bacterial cell: influence of differentially expressed genes in the virulence of the human pathogen Vibrio vulnificus. Infect Immun 76:4019–4037. doi: 10.1128/IAI.00208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lee H-J, Kim J-A, Lee M-A, Park S-J, Lee K-H. 2013. Regulation of haemolysin (VvhA) production by ferric uptake regulator (Fur) in Vibrio vulnificus: repression of vvhA transcription by Fur and proteolysis of VvhA by Fur-repressive exoproteases. Mol Microbiol 88:813–826. doi: 10.1111/mmi.12224. [DOI] [PubMed] [Google Scholar]
- 216.Litwin CM, Calderwood SB. 1993. Cloning and genetic analysis of the Vibrio vulnificus fur gene and construction of a fur mutant by in vivo marker exchange. J Bacteriol 175:706–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Tolmasky ME, Wertheimer AM, Actis LA, Crosa JH. 1994. Characterization of the Vibrio anguillarum fur gene: role in regulation of expression of the FatA outer membrane protein and catechols. J Bacteriol 176:213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Zheleznova EE, Crosa JH, Brennan RG. 2000. Characterization of the DNA- and metal-binding properties of Vibrio anguillarum Fur reveals conservation of a structural Zn(2+) ion. J Bacteriol 182:6264–6267. doi: 10.1128/JB.182.21.6264-6267.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Pedersen HL, Ahmad R, Riise EK, Leiros H-KS, Hauglid S, Espelid S, Brandsdal BO, Leiros I, Willassen N-P, Haugen P. 2010. Experimental and computational characterization of the ferric uptake regulator from Aliivibrio salmonicida (Vibrio salmonicida). J Microbiol 48:174–183. doi: 10.1007/s12275-010-9199-5. [DOI] [PubMed] [Google Scholar]
- 220.Stoebner JA, Payne SM. 1988. Iron-regulated hemolysin production and utilization of heme and hemoglobin by Vibrio cholerae. Infect Immun 56:2891–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wong HC, Lee YS. 1994. Regulation of iron on bacterial growth and production of thermostable direct hemolysin by Vibrio parahaemolyticus in intraperitoneal infected mice. Microbiol Immunol 38:367–371. doi: 10.1111/j.1348-0421.1994.tb01792.x. [DOI] [PubMed] [Google Scholar]
- 222.Simpson LM, Oliver JD. 1993. Regulation of proteolytic activity of Vibrio vulnificus by iron-containing compounds. Microb Pathog 14:249–252. doi: 10.1006/mpat.1993.1024. [DOI] [PubMed] [Google Scholar]
- 223.Farrell DH, Mikesell P, Actis LA, Crosa JH. 1990. A regulatory gene, angR, of the iron uptake system of Vibrio anguillarum: similarity with phage P22 cro and regulation by iron. Gene 86:45–51. doi: 10.1016/0378-1119(90)90112-5. [DOI] [PubMed] [Google Scholar]
- 224.Salinas PC, Tolmasky ME, Crosa JH. 1989. Regulation of the iron uptake system in Vibrio anguillarum: evidence for a cooperative effect between two transcriptional activators. Proc Natl Acad Sci U S A 86:3529–3533. doi: 10.1073/pnas.86.10.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wertheimer AM, Verweij W, Chen Q, Crosa LM, Nagasawa M, Tolmasky ME, Actis LA, Crosa JH. 1999. Characterization of the angR gene of Vibrio anguillarum: essential role in virulence. Infect Immun 67:6496–6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tolmasky ME, Actis LA, Crosa JH. 1988. Genetic analysis of the iron uptake region of the Vibrio anguillarum plasmid pJM1: molecular cloning of genetic determinants encoding a novel trans activator of siderophore biosynthesis. J Bacteriol 170:1913–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Salinas PC, Crosa JH. 1995. Regulation of angR, a gene with regulatory and biosynthetic functions in the pJM1 plasmid-mediated iron uptake system of Vibrio anguillarum. Gene 160:17–23. doi: 10.1016/0378-1119(95)00213-P. [DOI] [PubMed] [Google Scholar]
- 228.Goldberg MB, Boyko SA, Calderwood SB. 1991. Positive transcriptional regulation of an iron-regulated virulence gene in Vibrio cholerae. Proc Natl Acad Sci U S A 88:1125–1129. doi: 10.1073/pnas.88.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Watnick PI, Butterton JR, Calderwood SB. 1998. The interaction of the Vibrio cholerae transcription factors, Fur and IrgB, with the overlapping promoters of two virulence genes, irgA and irgB. Gene 209:65–70. doi: 10.1016/S0378-1119(98)00018-3. [DOI] [PubMed] [Google Scholar]
- 230.Litwin CM, Quackenbush J. 2001. Characterization of a Vibrio vulnificus LysR homologue, HupR, which regulates expression of the haem uptake outer membrane protein, HupA. Microb Pathog 31:295–307. doi: 10.1006/mpat.2001.0472. [DOI] [PubMed] [Google Scholar]
- 231.Masse E, Vanderpool CK, Gottesman S. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol 187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Massé E, Gottesman S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci U S A 99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Masse E, Escorcia FE, Gottesman S. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Masse E, Majdalani N, Gottesman S. 2003. Regulatory roles for small RNAs in bacteria. Curr Opin Microbiol 6:120–124. doi: 10.1016/S1369-5274(03)00027-4. [DOI] [PubMed] [Google Scholar]
- 235.Večerek B, Moll I, Bläsi U. 2007. Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. EMBO J 26:965–975. doi: 10.1038/sj.emboj.7601553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Davis BM, Quinones M, Pratt J, Ding Y, Waldor MK. 2005. Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. J Bacteriol 187:4005–4014. doi: 10.1128/JB.187.12.4005-4014.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Tanabe T, Funahashi T, Nakao H, Maki J, Yamamoto S. 2013. The Vibrio parahaemolyticus small RNA RyhB promotes production of the siderophore vibrioferrin by stabilizing the polycistronic mRNA. J Bacteriol 195:3692–3703. doi: 10.1128/JB.00162-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Chen Q, Crosa JH. 1996. Antisense RNA, Fur, iron, and the regulation of iron transport genes in Vibrio anguillarum. J Biol Chem 271:18885–18891. doi: 10.1074/jbc.271.31.18885. [DOI] [PubMed] [Google Scholar]
- 239.Waldbeser LS, Chen Q, Crosa JH. 1995. Antisense RNA regulation of the fatB iron transport protein gene in Vibrio anguillarum. Mol Microbiol 17:747–756. doi: 10.1111/j.1365-2958.1995.mmi_17040747.x. [DOI] [PubMed] [Google Scholar]
- 240.Carpenter C, Payne SM. 2014. Regulation of iron transport systems in Enterobacteriaceae in response to oxygen and iron availability. J Inorg Biochem 133:110–117. doi: 10.1016/j.jinorgbio.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Oh MH, Lee SM, Lee DH, Choi SH. 2009. Regulation of the Vibrio vulnificus hupA gene by temperature alteration and cyclic AMP receptor protein and evaluation of its role in virulence. Infect Immun 77:1208–1215. doi: 10.1128/IAI.01006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Colquhoun DJ, Sørum H. 2001. Temperature dependent siderophore production in Vibrio salmonicida. Microb Pathog 31:213–219. doi: 10.1006/mpat.2001.0464. [DOI] [PubMed] [Google Scholar]
- 243.Kim C-M, Kim S-J, Shin S-H. 2012. Cyclic AMP-receptor protein activates aerobactin receptor IutA expression in Vibrio vulnificus. J Microbiol 50:320–325. doi: 10.1007/s12275-012-2056-y. [DOI] [PubMed] [Google Scholar]
- 244.Folsom JP, Parker AE, Carlson RP. 2014. Physiological and proteomic analysis of Escherichia coli iron-limited chemostat growth. J Bacteriol 196:2748–2761. doi: 10.1128/JB.01606-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Wen Y, Kim IH, Son J-S, Lee B-H, Kim K-S. 2012. Iron and quorum sensing coordinately regulate the expression of vulnibactin biosynthesis in Vibrio vulnificus. J Biol Chem 287:26727–26739. doi: 10.1074/jbc.M112.374165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Kim CM, Shin SH. 2011. Modulation of iron-uptake systems by a mutation of luxS encoding an autoinducer-2 synthase in Vibrio vulnificus. Biol Pharm Bull 34:632–637. doi: 10.1248/bpb.34.632. [DOI] [PubMed] [Google Scholar]
- 247.Kim IH, Wen Y, Son J-S, Lee K-H, Kim K-S. 2013. The Fur-iron complex modulates expression of the quorum-sensing master regulator, SmcR, to control expression of virulence factors in Vibrio vulnificus. Infect Immun 81:2888–2898. doi: 10.1128/IAI.00375-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Septer AN, Lyell NL, Stabb EV. 2013. The iron-dependent regulator Fur controls pheromone signaling systems and luminescence in the squid symbiont Vibrio fischeri ES114. Appl Environ Microbiol 79:1826–1834. doi: 10.1128/AEM.03079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Acosta N, Pukatzki S, Raivio TL. 2015. The Vibrio cholerae Cpx envelope stress response senses and mediates adaptation to low iron. J Bacteriol 197:262–276. doi: 10.1128/JB.01957-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Gode-Potratz CJ, Chodur DM, McCarter LL. 2010. Calcium and iron regulate swarming and type III secretion in Vibrio parahaemolyticus. J Bacteriol 192:6025–6038. doi: 10.1128/JB.00654-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.McCarter L, Silverman M. 1989. Iron regulation of swarmer cell differentiation of Vibrio parahaemolyticus. J Bacteriol 171:731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Pappenheimer AM, Johnson SJ. 1936. Studies in diphtheria toxin production. I. The effect of iron and copper. Br J Exp Pathol 17:335–341. [Google Scholar]
- 253.Dubos RJ, Geiger JW. 1946. Preparation and properties of Shiga toxin and toxoid. J Exp Med 84:143–156. doi: 10.1084/jem.84.2.143. [DOI] [PubMed] [Google Scholar]
- 254.Boyd J, Oza MN, Murphy JR. 1990. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc Natl Acad Sci U S A 87:5968–5972. doi: 10.1073/pnas.87.15.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Calderwood SB, Mekalanos JJ. 1987. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J Bacteriol 169:4759–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Schmitt MP, Twiddy EM, Holmes RK. 1992. Purification and characterization of the diphtheria toxin repressor. Proc Natl Acad Sci U S A 89:7576–7580. doi: 10.1073/pnas.89.16.7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Tanabe T, Naka A, Aso H, Nakao H, Narimatsu S, Inoue Y, Ono T, Yamamoto S. 2005. A novel aerobactin utilization cluster in Vibrio vulnificus with a gene involved in the transcription regulation of the iutA homologue. Microbiol Immunol 49:823–834. doi: 10.1111/j.1348-0421.2005.tb03671.x. [DOI] [PubMed] [Google Scholar]
- 258.Tanabe T, Funahashi T, Shiuchi K, Okajima N, Nakao H, Miyamoto K, Tsujibo H, Yamamoto S. 2012. Characterization of Vibrio parahaemolyticus genes encoding the systems for utilization of enterobactin as a xenosiderophore. Microbiology 158:2039–2049. doi: 10.1099/mic.0.059568-0. [DOI] [PubMed] [Google Scholar]
- 259.Tanabe T, Kato A, Shiuchi K, Miyamoto K, Tsujibo H, Maki J, Yamamoto S, Funahashi T. 2014. Regulation of the expression of the Vibrio parahaemolyticus peuA gene encoding an alternative ferric enterobactin receptor. PLoS One 9:e105749. doi: 10.1371/journal.pone.0105749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Naka H, Crosa JH. 2012. Identification and characterization of a novel outer membrane protein receptor FetA for ferric enterobactin transport in Vibrio anguillarum 775(pJM1). Biometals 25:125–133. doi: 10.1007/s10534-011-9488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Lemos ML, Osorio CR. 2010. Iron uptake in Vibrio and Aeromonas, p 117–141. In Cornelis P, Andrews SC (ed), Iron uptake and homeostasis in microorganisms. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 262.Mazoy R, Botana LM, Lemos ML. 1997. Iron uptake from ferric citrate by Vibrio anguillarum. FEMS Microbiol Lett 154:145–150. doi: 10.1111/j.1574-6968.1997.tb12636.x. [DOI] [Google Scholar]
- 263.Wyckoff EE, Smith SL, Payne SM. 2001. VibD and VibH are required for late steps in vibriobactin biosynthesis in Vibrio cholerae. J Bacteriol 183:1830–1834. doi: 10.1128/JB.183.5.1830-1834.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]