SUMMARY
Actinobacteria are Gram-positive bacteria with high G+C DNA content that constitute one of the largest bacterial phyla, and they are ubiquitously distributed in both aquatic and terrestrial ecosystems. Many Actinobacteria have a mycelial lifestyle and undergo complex morphological differentiation. They also have an extensive secondary metabolism and produce about two-thirds of all naturally derived antibiotics in current clinical use, as well as many anticancer, anthelmintic, and antifungal compounds. Consequently, these bacteria are of major importance for biotechnology, medicine, and agriculture. Actinobacteria play diverse roles in their associations with various higher organisms, since their members have adopted different lifestyles, and the phylum includes pathogens (notably, species of Corynebacterium, Mycobacterium, Nocardia, Propionibacterium, and Tropheryma), soil inhabitants (e.g., Micromonospora and Streptomyces species), plant commensals (e.g., Frankia spp.), and gastrointestinal commensals (Bifidobacterium spp.). Actinobacteria also play an important role as symbionts and as pathogens in plant-associated microbial communities. This review presents an update on the biology of this important bacterial phylum.
INTRODUCTION
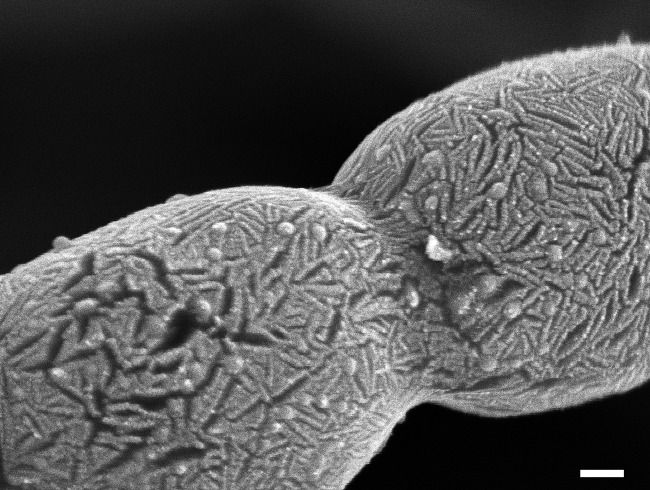
The phylum Actinobacteria is one of the largest taxonomic units among the major lineages currently recognized within the Bacteria domain (1). The actinobacterial genomes sequenced to date belong to organisms relevant to human and veterinary medicine, biotechnology, and ecology, and their observed genomic heterogeneity is assumed to reflect their biodiversity (2). The majority of the Actinobacteria are free-living organisms that are widely distributed in both terrestrial and aquatic (including marine) ecosystems (3). Actinobacteria are Gram-positive filamentous bacteria with a high guanine-plus-cytosine (G+C) content in their genomes. They grow by a combination of tip extension and branching of the hyphae. This is what gave them their name, which derives from the Greek words for ray (aktis or aktin) and fungi (mukēs). Traditionally, actinomycetes were considered transitional forms between fungi and bacteria. Indeed, like filamentous fungi, many Actinobacteria produce a mycelium, and many of these mycelial actinomycetes reproduce by sporulation. However, the comparison to fungi is only superficial: like all bacteria, actinomycetes' cells are thin with a chromosome that is organized in a prokaryotic nucleoid and a peptidoglycan cell wall; furthermore, the cells are susceptible to antibacterial agents (Fig. 1). Physiologically and ecologically, most Actinobacteria are aerobic, but there are exceptions. Further, they can be heterotrophic or chemoautotrophic, but most are chemoheterotrophic and able to use a wide variety of nutritional sources, including various complex polysaccharides (4, 5). Actinobacteria may be inhabitants of soil or aquatic environments (e.g., Streptomyces, Micromonospora, Rhodococcus, and Salinispora species), plant symbionts (e.g., Frankia spp.), plant or animal pathogens (e.g., Corynebacterium, Mycobacterium, or Nocardia species), or gastrointestinal commensals (e.g., Bifidobacterium spp.).
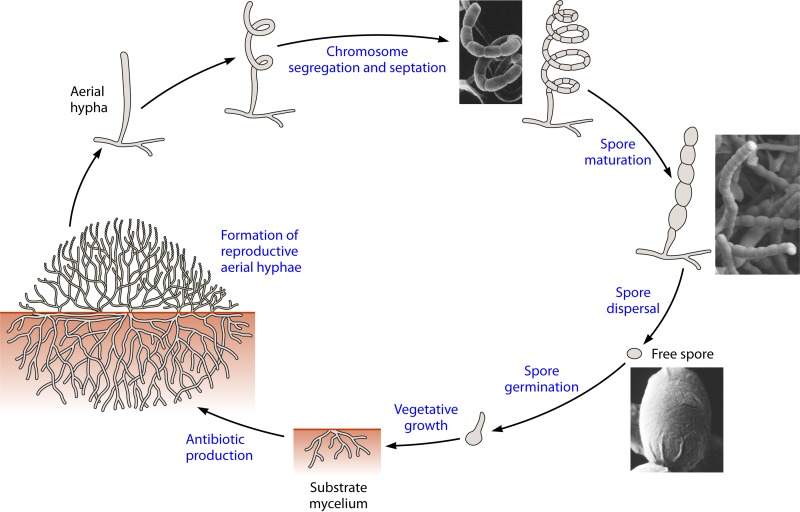
FIG 1.
Schematic representation of the life cycle of sporulating actinomycetes.
BIOLOGY OF ACTINOBACTERIA
Most of the Actinobacteria (the streptomycetes in particular) are saprophytic, soil-dwelling organisms that spend the majority of their life cycles as semidormant spores, especially under nutrient-limited conditions (6). However, the phylum has adapted to a wide range of ecological environments: actinomycetes are also present in soils, fresh and salt water, and the air. They are more abundant in soils than other media, especially in alkaline soils and soils rich in organic matter, where they constitute an important part of the microbial population. Actinobacteria can be found both on the soil surface and at depths of more than 2 m below ground (7).
The population density of Actinobacteria depends on their habitat and the prevailing climate conditions. They are typically present at densities on the order of 106 to 109 cells per gram of soil (7); soil populations are dominated by the genus Streptomyces, which accounts for over 95% of the Actinomycetales strains isolated from soil (8). Other factors, such as temperature, pH, and soil moisture, also influence the growth of Actinobacteria. Like other soil bacteria, Actinobacteria are mostly mesophilic, with optimal growth at temperatures between 25 and 30°C. However, thermophilic Actinobacteria can grow at temperatures ranging from 50 to 60°C (9). Vegetative growth of Actinobacteria in the soil is favored by low humidity, especially when the spores are submerged in water. In dry soils where the moisture tension is greater, growth is very limited and may be halted. Most Actinobacteria grow in soils with a neutral pH. They grow best at a pH between 6 and 9, with maximum growth around neutrality. However, a few strains of Streptomyces have been isolated from acidic soils (pH 3.5) (10). The first study on the effect of climate on the distribution of Actinobacteria was done by Hiltner and Strömer (11), who showed that these bacteria account for 20% of the microbial flora of the soil in spring and more than 30% in the autumn because of the large amounts of crop residues available at this time of year. However, during the winter, frost reduces their relative abundance to only 13%.
Taxonomy of Actinobacteria
Actinobacteria represent one of the largest taxonomic units among the 18 major lineages currently recognized within the Bacteria domain, including 5 subclasses, 6 orders, and 14 suborders (1). The genera of this phylum exhibit enormous diversity in terms of their morphology, physiology, and metabolic capabilities. The taxonomy of Actinobacteria has evolved significantly over time with the accumulation of knowledge. The order Actinomycetales, established by Buchanan in 1917 (12), belongs to this group of prokaryotic organisms.
The phylum Actinobacteria is delineated on the basis of its branching position in 16S rRNA gene trees. However, rRNA sequences do not discriminate well between closely related species or even genera, which can create ambiguity. For instance, the taxonomic status of the genus Kitasatospora (13) within the family Streptomycetaceae has been disputed for many years (1, 14, 15), although a recent detailed genetic analysis provided strong evidence that it should be regarded as a separate genus (16). A similar close relationship exists between Micromonospora, Verrucosispora, and Salinispora. Additional genetic markers have therefore been used to discriminate between closely related genera, including rpoB and, most recently, ssgB, which is particularly useful for discriminating between closely related genera (17). Moreover, the massive recent increase in the availability of genome sequence information has provided detailed insights into genome evolution and made it possible to identify genes specific to organisms at the level of genera and family (18).
An updated taxonomy of the phylum Actinobacteria that is based on 16S rRNA trees was recently reported (1). That update eliminated the taxonomic ranks of subclasses and suborders, elevating the former subclasses and suborders to the ranks of classes and orders, respectively (19). The phylum is thus divided into six classes: Actinobacteria, Acidimicrobiia, Coriobacteriia, Nitriliruptoria, Rubrobacteria, and Thermoleophilia.
The class Actinobacteria contains 16 orders, including both of the previously proposed orders, Actinomycetales and Bifidobacteriales (20). The order Actinomycetales is now restricted to the members of the family Actinomycetaceae, and the other suborders that were previously part of this order are now designated distinct orders (19). Consequently, 43 of the 53 families within the phylum Actinobacteria are assigned to a single class, Actinobacteria, whereas the other five classes together contain only 10 families (21).
Morphological classification.
The main characteristics used to delineate the taxonomy of Actinobacteria at the genus and species levels are microscopic morphology and chemotaxonomy. The latter of these characteristics primarily relates to the composition of the cell wall and the whole-cell sugar distribution, although phospholipid composition and menaquinone type may also be considered for fine-tuning purposes (22).
Mycelial fragmentation can be regarded as a special form of vegetative reproduction. However, the Actinobacteria with primarily mycelial lifestyles usually reproduce by forming asexual spores. Actinobacteria exhibit a wide variety of morphologies, differing mainly with respect to the presence or absence of a substrate mycelium or aerial mycelium, the color of the mycelium, the production of diffusible melanoid pigments, and the structure and appearance of their spores (Fig. 1).
(i) Mycelial morphology.
Except for Sporichthya sp., which produces aerial hyphae that are initiated upright on the surface of the medium by holdfasts, Actinobacteria form a substrate mycelium in both submerged and solid-grown cultures. However, on solid surfaces, many differentiate to form aerial hyphae, whose main purpose is to produce reproductive spores (23, 24). The substrate mycelium develops from outgrowth of a germinating spore. The branching substrate mycelium is often monopodial, but in some rare cases, Actinobacteria, such as Thermoactinomyces, exhibit dichotomous branching (25). On the other hand, members of the Micromonosporaceae family produce an extensive substrate mycelium with an absent or rudimentary aerial mycelium.
Actinobacteria exhibit a wide variety of morphologies, including coccoid (Micrococcus) and rod-coccoid (Arthrobacter), as well as fragmenting hyphal forms (Nocardia spp.) and also forms with permanent and highly differentiated branched mycelia (e.g., Streptomyces spp., Frankia) (26). Rhodococci form elongated filaments on the substrate and do not produce a true mycelium (27), while corynebacteria do not produce mycelia at all. However, as in other Actinobacteria, the filaments grow at the apex instead of by lateral wall extension (28, 29). Actinobacteria belonging to the genus Oerskovia are characterized by the formation of branched substrate hyphae that break up into flagellated motile elements (30). Further, mycobacteria and rhodococci do not usually form aerial hyphae, although some exceptions exist (31).
(ii) Spore chain morphology.
Spores are extremely important in the taxonomy of Actinobacteria (32). The initial steps of sporulation in several oligosporic Actinobacteria can be regarded as budding processes, because they satisfy the main criteria used to define budding in other bacteria (Fig. 2). Spores may be formed on the substrate and/or the aerial mycelium as single cells or in chains of different lengths. In other cases, spores may be harbored in special vesicles (sporangia) and endowed with flagella.
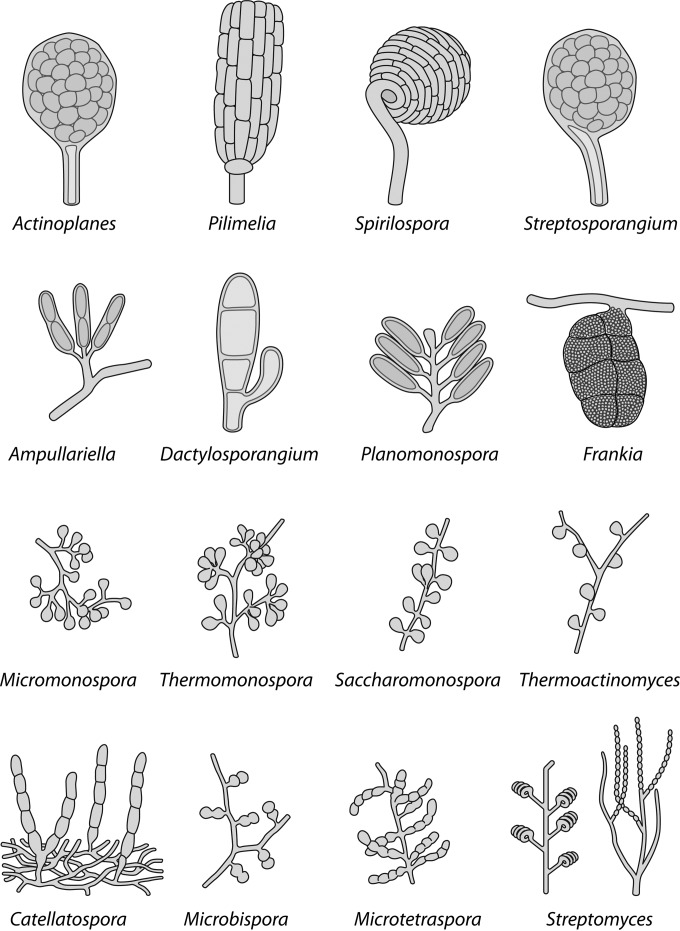
FIG 2.
Schematic drawings of the different types of spore chains produced by actinomycetes.
Thus, in the genera Micromonospora, Micropolyspora, and Thermoactinomycètes, spore formation occurs directly on the substrate mycelium (33), whereas in Streptomyces the spores grow out from the aerial mycelium. The Actinoplanes and Actinosynnema groups are characterized by motile spores, while Thermoactinomyces has unique heat-resistant endospores (33). Some other Actinobacteria genera have sclerotia (Chainia), synnemas (Actinosynnema), vesicles that contain spores (Frankia), or vesicles that are devoid of spores (Intrasporangium). Other genera, such as Actinoplanes, Ampulariella, Planomonospora, Planobispora, Dactylosporangium, and Streptosporangium, are classified based on their sporangial morphology. Figure 2 illustrates the different types of spores that can be found in actinomycetal genera. Finally, the morphology of the spores themselves can also be used to characterize species: they may have smooth, warty, spiny, hairy, or rugose surfaces (34).
(iii) Spore chain length.
The number of spores per spore chain varies widely from genus to genus. The genera Micromonospora, Salinispora, Thermomonospora, Saccharomonospora, and Promicromonospora produce isolated spores, while Microbispora produces spores in longitudinal pairs. Members of the genera Actinomadura, Saccharopolyspora, Sporicthya, and some Nocardia spp. have short spore chains, while members of the genera Streptomyces, Nocardioides, Kitasatospora, Streptoverticillium, and some Nocardia spp. produce very long chains of up to 100 spores. In contrast, Frankia species produce sporangia, which are essentially bags of spores. Streptomycetes' spore chains can be classified as being straight to flexuous (Rectus-Flexibilis), open loops (Relinaculam-Apertum), open or closed spirals (spira), or verticillate (35).
(iv) Melanoid pigments.
Melanins are polymers with diverse molecular structures that typically appear black or brown and are formed by the oxidative polymerization of phenolic and indolic compounds. They are produced by a broad range of organisms, ranging from bacteria to humans. Actinobacteria have long been known to produce pigments, which may be red, yellow, orange, pink, brownish, distinct brown, greenish brown, blue, or black, depending on the strain, the medium used, and the age of the culture (4).
Generally referred to as melanins, or melanoid pigments, these brown-black metabolic polymers are important not only because of their usefulness in taxonomic studies but also because of their similarity to soil humic substances (36, 37). Melanins are not essential for the organisms' growth and development, but they play a crucial role in improving their survival and competitiveness.
Chemotaxonomic classification.
Chemotaxonomy is the use of the distribution of chemical components to group organisms according to the similarities of their cellular chemistries (38, 39). The most commonly used chemical components in such systematics are cell wall amino acids, lipids, proteins, menaquinones, muramic acid types, sugars, and the base composition of DNA (40, 41). Chemotaxonomic classification and identification can also be performed on the basis of information derived from whole-organism chemical fingerprinting techniques. Below, we discuss chemotaxonomic markers that have been reported to be of particular value for the classification and identification of actinomycetes (1).
Analysis of the cell wall composition of Actinobacteria is taxonomically valuable because it differs between suborders (42). In particular, information on the chemical architecture of the peptidoglycan in the cell wall is valuable for classifying actinomycetes because it facilitates discrimination between groups of Actinobacteria above the genus level. Multiple discriminatory characteristics relating to the structure and composition of their peptidoglycans have been identified (43), including the identity of the amino acid in position 3 of the tetrapeptide side chain, the presence or absence of glycine in interpeptide bridges, and the peptidoglycan's sugar content (43). The presence or absence of specific optical isomers of the chiral nonproteinogenic amino acid 2,6-diaminopimelic acid (DAP) is another chemotaxonomically important characteristic of the cell walls of Gram-positive bacteria: the peptidoglycan of Actinobacteria may contain either ll- or dl-(meso)-DAP, depending on the genus. By considering DAP isomerism and the presence/absence of other amino acids and (amino)sugars, Lechevalier and Lechevalier (44) identified nine distinct actinobacterial cell wall chemotypes (Table 1). However, it is important to realize that while DAP analysis and other chemotaxonomic methods are extremely important in the taxonomy of Actinobacteria, diverse groups share the same DAP profile. For example, the genera Streptomyces, Streptoverticillium, Arachnia, and Nocardioides share the same chemotype (chemotype I), even though their different morphologies indicate that they belong to different families. Therefore, when assessing the phenotypic diversity of Actinobacteria, DAP profiling should be used in combination with other phenotypic or genotypic criteria (45). To this end, a system for classifying Actinobacteria based on both morphological and chemical characteristics has been proposed (4).
TABLE 1.
Different types of cell wall components in Actinomycetesa
| Cell wall type | Major parietal constituent(s) | Genera |
|---|---|---|
| I | ll-DAP, glycine, no sugar | Arachnia, Nocardioides, Pimelobacter, Streptomyces |
| II | meso-DAP, glycine, arabinose, xylose | Actinomyces, Actinoplanes, Ampulariella, Catellatosporia, Dactylosporangium, Glycomyces, Micromonospora, Pilimelia |
| III | meso-DAP, madurose (3-O-methyl-d-galactose) | Actinocorallia, Actinomadura, Dermatophylus, Frankia, Geodermatophilus, Kitasatospora, Maduromycetes, Microbispora, Microtetraspora, Nonomuraea, Planobispora, Planomonospora, Planotetraspora, some Frankia spp., Spirillosporia, Streptosporangium, Thermoactinomyces, Thermomonospora |
| IV | meso-DAP, arabinose, galactose | Micropolyspora, Nocardioforms |
| V | Deprived of DAP; possesses lysine and ornithine | Actinomyces |
| VI | Deprived of DAP; variable presence of aspartic acid, galactose | Arcanobacterium, Actinomyces, Microbacterium, Oerskovia, Promicromonospora |
| VII | Deprived of DAP; diaminobutyric acid, glycine, with lysine variable | Agromyces, Clavibacter |
| VIII | Deprived of DAP; ornithine | Aureobacterium, Curtobacterium, Cellulomonas |
Cellular fatty acid patterns are also very useful chemotaxonomic indicators for the identification of specific Actinobacteria genera (46). Bacterial fatty acids range in chain length from two (C2) to over 90 (C90) carbon atoms, but only those in the range of C10 to C24 are of particular taxonomic value (47). Three major types of fatty acid profiles have been identified in Actinobacteria (46).
Several types of isoprenoid quinones have been characterized in bacteria (48), of which menaquinones are most commonly found in actinomycete cell envelopes (46–49). Menaquinone analysis has provided valuable information for the classification of Actinomadura, Microtetraspora, and Streptomyces strains (46, 50–52). In addition, cyclic menaquinones are characteristic of members of the genus Nocardia (53, 54), while fully saturated cyclic menaquinones have been reported for Pyrobaculum organotrophum (54).
Different types of phospholipids are discontinuously distributed in actinomycetes' cytoplasmic membranes, providing useful information for the classification and identification of actinomycete genera (41, 55). Actinobacteria have been classified into five phospholipid groups based on semiquantitative analyses of major phospholipid markers found in whole-organism extracts (56–58). This classification system was used in the identification of Aeromicrobium (59) and Dietzia (60). Importantly, it has been reported that members of the same Actinobacteria genus have the same phospholipid type.
Finally, sugar composition analysis is also important in chemotaxonomy. At the suprageneric level, neutral sugars (the major constituents of actinomycete cell envelopes) are useful taxonomic markers (Table 2). On the basis of the discontinuous distribution of major diagnostic sugars, Actinomycetes can be divided into five groups. Group A comprises those species whose cell walls contain arabinose and galactose; group B cell walls contain madurose (3-O-methyl-d-galactose); group C consists of those with no diagnostic sugars; group D cell walls contain arabinose and xylose; group E cell walls contain galactose and rhamnose (22, 61). In addition, the presence of 3′-O-methyl-rhamnose in Catellatospora (62) and of tyvelose in Agromyces (63) has been valuable for the classification of some actinomycete taxa.
TABLE 2.
Taxonomic markers used as characteristics to differentiate the genera of Actinomycetes
| Amino acid present | Sugar(s) | Morphological characteristics | Genus |
|---|---|---|---|
| No diaminopimelic acid | Xylose, madurose | Only substrate mycelium, breaks into motile elements | Oerskovia |
| Sterile aerial mycelium, breaks into nonmotile elements | Promicromonospora | ||
| Sporangia with motile spores | Actinoplanes | ||
| Short chains of conidia on aerial mycelium | Actinomadura | ||
| l-Diaminopimelic acid | Xylose, madurose | Both aerial and substrate mycelia that break up into rods and coccoid elements | Nocardioides |
| Only substrate mycelium, bearing terminal or subterminal vesicles | Intrasporangium | ||
| Aerial mycelium with long chains of spores | Streptomyces, Kitasatospora | ||
| Sclerotia | Streptomyces | ||
| Very short chains of large conidia on the vegetative and aerial mycelia | Streptomyces | ||
| Whorls of small chains of spores | Streptoverticillium | ||
| No aerial mycelium, sporangia on the vegetative mycelium | Kineosporia | ||
| meso-Diaminopimelic acid | Xylose, arabinose | Conidia isolated on the vegetative mycelium | Micromonospora |
| No sporangia, short chains of conidia | Cattellatospora | ||
| Chains of conidia on the aerial mycelium | Glycomyces | ||
| Dactyloid oligosporic sporangia, motile spores | Dactylosporangium | ||
| Sporangia with spherical and motile spores formed on the surfaced of colonies | Actinoplanes | ||
| Sporangia with rod-shaped spores, motility via polar flagella | Ampullariella | ||
| Sporangia with lateral flagellated spores | Pilimelia | ||
| Multilocular sporangia, spores are nonmotile | Frankia | ||
| Madurose | Short chains of conidia on the aerial mycelium | Actinomadura | |
| Chains of conidia with spores | Microbispora | ||
| Chains of conidia with 2 to 6 spores | Microtetraspora | ||
| Sporangia with 2 motile spores | Planobispora | ||
| Sporangia with 1 motile spore | Planomonospora | ||
| Mycelium with spherical sporangia containing many rod-shaped, motile spores | Spirillospora | ||
| Fructose | Multilocular sporangia | Frankia | |
| Sporangia with motile spores | Actinoplanes | ||
| Rhamnose, galactose | Both substrate and aerial mycelia that break into nonmotile elements | Saccharothrix | |
| Rhamnose, galactose, mannose | Same as Streptomyces | Streptoalloteichus | |
| Galactose | Same as Streptomyces | Kitasatospora | |
| Arabinose, galactose | Presence of nocardiomycolic acid (NMA) in whole cells; both substrate and aerial mycelia fragment into rods and coccoid elements | Nocardia | |
| Presence of NMA; rods and extensively branched substrate mycelium that fragments into irregular rods and cocci | Rhodococcus | ||
| Presence of NMA; straight to slightly curved rods occur singly, in pairs, or in masses; cells are nonmotile, non-spore forming, and do not produce aerial hyphae | Tsukamurella | ||
| Presence of NMA; paired spores borne in longitudinal pairs on vegetative hyphae; aerial mycelium is sparse | Actinobispora | ||
| No NMA, spores are long, cylindrical on aerial mycelium, formed by budding | Pseudonocardia | ||
| No NMA; long chains of conidia on aerial mycelium | Saccharomonospora | ||
| No NMA; aerial mycelium bearing long chains of conidia; halophilic | Actinopolyspora | ||
| No NMA; substrate mycelium tends to break into nonmotile elements; aerial hyphae may form and may also segment | Amycolata, Amycolatopsis | ||
| No NMA; aerial mycelium bearing curled hyphae embedded in amorphous matrix | Kibdelosporangium | ||
| No NMA; both aerial and substrate mycelia bearing long chains of motile spores | Aktinokineospora | ||
| No NMA; aerial mycelium tends to fragment into rods and cocci, short chains of spores | Pseudoamycolata | ||
| Spores formed are not heat resistant | Thermomonospora | ||
| Long chains of spores on aerial mycelium | Nocardiopsis | ||
| Columnar hyphal structures called synnemata bearing chains of conidia capable of forming flagella | Actinosynnema | ||
| Multilocular sporangia containing motile spores | Geodermatophilus |
Molecular Classification
More recently, the morphological and chemical classification of actinomycetes have been challenged by molecular taxonomic data, much of which were obtained thanks to the rapid advancement of genome sequencing. Notably, some organisms that were inappropriately placed in certain taxonomic groups have recently been reclassified on the basis of molecular analyses (20). A recent example is the final definition of Kitasatospora as a separate genus within the Streptomycetaceae (17); genome sequencing resolved a long-running debate about this group's relationship with the genus Streptomyces and conclusively demonstrated that it is in fact a separate genus (15, 16, 64, 65).
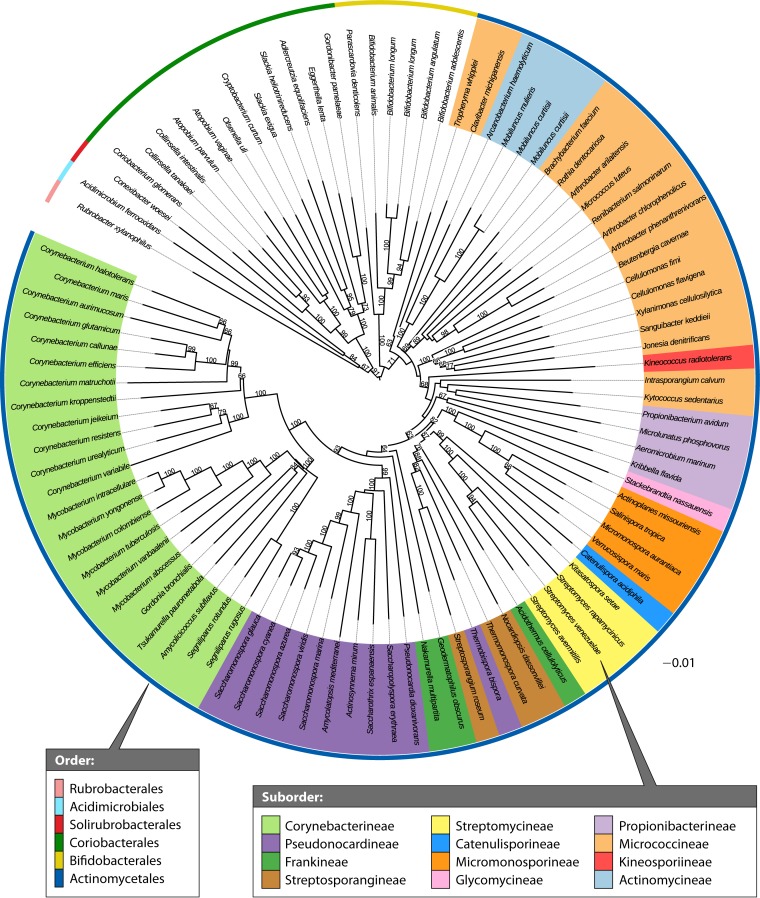
At present, a new species cannot be claimed without genetic analysis based on sequencing the 16S rRNA gene and DNA-DNA hybridization, and even genome sequencing is becoming routine. Molecular and chemical composition criteria have been used to group the order Actinomycetales into 14 suborders: Actinomycineae, Actinopolysporineae, Catenulisporineae, Corynebacterineae, Frankineae, Glycomycineae, Jiangellineae, Kineosporineae, Micrococcineae, Micromonosporineae, Propionibacterineae, Pseudonocardineae, Streptomycineae, and Streptosporangineae (66). Moreover, sequencing of 16S rRNA genes has led to the recognition of 39 families and 130 genera (Fig. 3). All groups previously assigned to the taxonomic rank of “order” were recovered as being strictly monophyletic based on these molecular and chemical criteria, but some paraphyletic groups were found within the rank “suborder.” This might be because the classification was mainly based on 16S rRNA gene trees, which were generated without bootstrap support and may thus include misleading results. The features of some of these genera are summarized below.
FIG 3.
A genome-based phylogenetic tree based on 97 genome sequences of the phylum Actinobacteria. Type strain genome projects were selected as previously described (676), provided that they yielded at most 25 contigs. Phylogenetic reconstruction, including the assessment of branch support, was done using amino acid sequences according to the methods described by Meier-Kolthoff et al. (677, 678). The tree was visualized by using ITOL (679). Branch support values below 60% are not shown, but the tree generally reveals high support throughout.
The genus Tropheryma.
The most-studied member of the genus Tropheryma is T. whipplei, the causative agent of Whipple's disease, which is characterized by intestinal malabsorption leading to cachexia and death. T. whipplei isolates are typically found in human intracellular niches, such as inside intestinal macrophages and circulating monocytes (67, 68). It has a condensed genome of only 925,938 bp, with a G+C content of only 46% (69, 70), whereas other actinomycete genomes have much larger genomes (up to 10 MBp) and higher G+C contents. T. whipplei has a tropism for myeloid cells, particularly macrophages, although it can be found in various cell types. Further, genome sequencing revealed a lack of key biosynthetic pathways and a lower capacity for energy metabolism. Its small genome and lack of metabolic capabilities suggest that T. whipplei has a host-restricted lifestyle (69). Recent findings have shown that T. whipplei survives phagocyte killing and replicates in macrophages by interfering with innate immune activation (71).
The genus Propionibacterium.
The genus Propionibacterium includes various species belonging to the human cutaneous propionibacteria, including P. acnes, P. avidum, P. granulosum, P. innocuum, and P. propionibacterium. Propionibacterium acnes is a non-spore-forming, anaerobic, pleomorphic rod whose end products of fermentation include propionic acid. The bacterium is omnipresent on human skin, predominantly within sebaceous follicles, where it is generally a harmless commensal. Nonetheless, P. acnes may be an opportunistic pathogen (72). Indeed, the bacterium has been isolated from sites of infection and inflammation in patients suffering from acne and other diverse conditions, including corneal ulcers, synovitis, hyperostosis, endocarditis, pulmonary angitis, and endophthalmitis (73, 74). Recently, Campisano et al. (75) reported a unique example of horizontal interkingdom transfer of P. acnes to the domesticated grapevine, Vitis vinifera L.
The genus Micromonospora.
Micromonospora species are widely distributed in nature, living in different environments. They have long been known as a significant source of secondary metabolites for medicine, and it was recently demonstrated that Micromonospora species may also influence plant growth and development (76); Micromonospora strains have been identified as natural endophytes of legume nodules, although the precise nature and mechanism of their effects on plant development and productivity are currently unclear. While the genus exhibits considerable physiological and biochemical diversity, Micromonospora constitutes a well-defined group in terms of morphology, phylogeny, and chemotaxonomy. Its colonies can be a variety of colors, including white, orange, rose, or brown. However, species of the genus Micromonospora are not always easy to differentiate on the basis of morphology alone. Consequently, phylogenies and species identifications are now more commonly derived by analyzing the sequence of the 16S rRNA gene or gyrB (the gene encoding DNA topoisomerase). The genus Micromonospora consists primarily of soil actinobacteria, which account for 32 of its species, according to the latest version of Bergey's manual (77), although 50 soil actinobacteria in this genus have been validly described as of the time of writing. Most of these species were isolated from alkaline or neutral soils and to a lesser extent from aquatic environments. The spore population of M. echinospora is known to be heterogeneous with respect to its heat response characteristics, suggesting that routine heat activation could be utilized to eliminate the natural variability that exists within populations of this species and its relatives (78). Further, analysis of the genome of M. lupini Lupac 08 revealed a diverse array of genes that may help the bacterium to survive in the soil or in plant tissues. However, despite having many genes that encode putative plant material-degrading enzymes, this bacterium is not regarded as a plant pathogen (79). In addition, genome comparisons showed that M. lupini Lupac 08 is metabolically closely related to Frankia sp. strains ACN14a, CcI3, and EAN1pec. These results suggest that the Micromonospora genus has undergone a previously unidentified process of adaptation from a purely terrestrial to a facultative endophytic lifestyle.
The genus has also been reported to produce a large number of antibiotics (80) and is second only to Streptomyces in this respect, synthesizing up to 500 different molecules with various properties (77). Micromonospora species can produce hydrolytic enzymes, which allows them to play an active role in the degradation of organic matter in their natural habitats. Marine Micromonospora species have recently been reviewed with respect to their broad distribution and their potential use as probiotics (76, 81). Like other endophytic actinobacteria, Micromonospora can suppress a number of pathogens both in vitro and in planta by activating key genes in the systemic acquired resistance (SAR) or jasmonate/ethylene (JA/ET) pathways (76). Unfortunately, there have been few genomic studies on Micromonospora species, and there is a lack of tools for their genetic analysis despite their acknowledged capacity for secondary metabolite production (76).
The genus Salinispora.
Salinispora belongs to the Micromonosporaceae and is the first Actinobacteria genus known to require seawater for growth (82). The genus is widely distributed in tropical and subtropical marine sediments (83) and includes three distinct but closely related clades corresponding to the species S. arenicola, S. pacifica, and S. tropica. Like their terrestrial actinomycete counterparts, Salinispora spp. produce numerous secondary metabolites with diverse potential pharmaceutical applications. For instance, salinosporamide A, isolated from S. tropica, is currently in phase 1 clinical trials in patients with multiple myeloma, lymphomas, leukemia, and solid tumors (84).
Although the three currently known species of Salinispora cooccur at six widely separated and distinct locations (82), only strains of S. tropica isolated from the Caribbean produce the potent anticancer compound salinosporamide A (85). In addition to its production of various secondary metabolites, this genus has attracted major interest for the novel phenomenon of species-specific secondary metabolite production (86, 87). Although it is clear that many of the genes for secondary metabolite production in the Salinispora genome were acquired via horizontal gene transfer, the ecological and evolutionary significance of these mechanisms remain unclear (86).
The genus Mycobacterium.
The relatively simple morphology of mycobacteria partly explains why it is sometimes overlooked when considering criteria for classifying actinomycetes (88, 89). With the genera Corynebacterium and Nocardia, Mycobacterium forms a monophyletic taxon within the Actinobacteria, the so-called CMN group (90). This group shares an unusual waxy cell envelope that contains mycolic acids, meaning these bacteria are unusual in being acid fast and alcohol fast. The mycobacterial cell wall contains various polysaccharide polymers, including arabinogalactan, lipomannan, lipoarabinomannan, and phosphatidylinositol mannosides (91, 92). Representatives of the genus Mycobacterium have been the subjects of three major 16S rRNA sequencing studies (93–95). Mycobacteria are generally free-living saprophytes (96), and they are the causative agents of a broad spectrum of human diseases. Mycobacterial diseases are very often associated with immunocompromised patients, especially those with AIDS. In addition, M. bovis and M. tuberculosis, isolated initially from infected animals, are most likely obligate parasites of humans (97). Both species can survive within macrophages and cause pulmonary disease, although organs other than lungs may be affected. M. leprae, which causes leprosy, lives in Schwann cells and macrophages; infection with this species results in a chronic granulomatous disease of the skin and peripheral nerves (98). Interestingly, the pathogenic M. ulcerans, which is the third most common causative agent of mycobacterial disease, has also been isolated as a soil inhabitant in symbiosis with roots of certain plants living in tropical rain forests and similar environments (99, 100). Mycobacterium marinum was initially identified as a causative organism of tuberculosis in fish in 1926 (101) and was subsequently shown to also cause skin disease in humans (102). M. marinum is a nontuberculosis mycobacterium that is a causative agent of human skin infections acquired through aquatic sources. Most cases of M. marinum infection are reported to have occurred after exposure to contaminated aquarium water or contact with fish and shellfish (103).
The genus Nocardia.
The genus Nocardia is a ubiquitous group of environmental bacteria that is most widely known as the causative agent of opportunistic infection in immunocompromised hosts. It forms a distinct clade that is associated with the genus Rhodococcus. Both the Nocardia and Rhodococcus genera belong to the family Nocardiaceae, which is a suborder of the “aerobic actinomycetes.” Nocardia species are ubiquitous soilborne aerobic actinomycetes, with more than 80 different species identified, of which at least 33 are pathogenic (104). Nocardia infections are mainly induced through inhalation or percutaneous inoculation from environmental sources (105), but nosocomial transmission has also been reported. The pathogen can spread to the brain, kidneys, joints, bones, soft tissues, and eyes, causing disseminated nocardiosis in humans and animals (106). Although Nocardia species are rare, they now account for 1 to 2% of all reported brain abscesses. However, the mortality rate for brain abscesses associated with Nocardia infection is substantially higher (31%) than that for brain abscesses in general (<10%) (107).
Moreover, Nocardia species produce industrially important bioactive molecules, such as antibiotics and enzymes (108, 109). Within the Nocardia clade, two sublines distinguishable by nucleotide differences in helix 37-1 are recognized; one consists of Nocardia asteroides and allied taxa, while the second consists of Nocardia otitidiscaviarum and related species. N. asteroides, the causal agent for most clinical human nocardial infections, was reorganized into multiple species on the basis of drug susceptibility patterns: Nocardia abscessus, the Nocardia brevicatena-Nocardia paucivorans complex, the Nocardia nova complex, the Nocardia transvalensis complex, Nocardia farcinica, and N. asteroides (104). Recently, Nocardia cyriacigeorgica was differentiated from N. asteroides (110).
In the last 2 decades, Nocardia infections have become regarded as an emerging disease among humans and domestic animals worldwide because of improved methods for pathogen isolation and molecular identification and a growing immunocompromised population (111). Nocardia species are recognized as opportunistic pathogens (112) and are known to compromise immune function. Moreover, they have been associated with organ and bone marrow transplants (113), long-term steroid use, connective tissue diseases, human immunodeficiency virus (HIV) infections, chronic obstructive pulmonary disease, alcoholism, cirrhosis, systemic vasculitis, ulcerative colitis, and renal failure (114).
In companion animals, Nocardia infections are usually reported as coinfections with immunosuppressive infectious diseases such as distemper in dogs and leukemia and immunodeficiency in cats (115).
The genus Corynebacterium.
The genus Corynebacterium was initially defined in 1896 to accommodate mainly pathogenic species exhibiting morphological similarity to the diphtheroid bacillus (116). Therefore, the genus comprised, for several decades, an extremely diverse collection of morphologically similar Gram-positive microorganisms, including nonpathogenic soil bacteria (117). Following chemotaxonomic studies and 16S rRNA sequence analysis, there are currently almost 70 recognized Corynebacterium species. Some well-known representatives include C. glutamicum, which (like the thermostable C. efficiens) is widely used in industry for the production of amino acids such as l-glutamic acid and l-lysine for human and animal nutrition, respectively (118). Several genome sequences of Corynebacterium species have been reported, including those of C. ulcerans (119), C. kutscheri (120), C. kroppenstedtii (121), and C. argentoratense (122), providing important new insights into the genomic architecture of the genus. A prophage, CGP3, that integrates into the genome of C. glutamicum and encodes an actin-like protein, AlpC, was recently described (123). CGP3 appears to be inactive in terms of cell lysis and virion production and is therefore referred to as a cryptic prophage, which likely became trapped in the genome in the course of evolution (123). This suggests that bacterial phages use an actin-based transport system similar to that found in vertebrate viruses, such as the herpesvirus. Among the known pathogenic members of Corynebacterium are C. diphtheria, which is a notorious strictly human-adapted species and the causative agent of the acute, communicable disease diphtheria, which is characterized by local growth of the bacterium in the pharynx along with the formation of an inflammatory pseudomembrane (124). The virulence factor in diphtheria is an exotoxin that targets host protein synthesis (125). Another important Corynebacterium pathogen is C. ulcerans, which is increasingly acknowledged as an emerging pathogen in various countries; infections with this species can mimic diphtheria because it harbors lysogenic-β-corynephages that carry the the diphtheria toxin (DT) gene, which is responsible for most of the systemic symptoms of diphtheria (126). C. ulcerans also induces clinical symptoms in the lower respiratory tract, including pneumonia (127) and pulmonary granulomatous nodules (128). However, its pathogenicity does not necessarily depend on the production of DT (129). A final important pathogen in this genus is C. jeikeium, which was initially isolated from human blood cultures and is associated with bacterial endocarditis contracted following cardiac surgery (130). It was subsequently shown to be a natural inhabitant of human skin and has been implicated in a variety of nosocomial infections (131).
The genus Gordonia.
Initially proposed by Tsukamura (132), this genus has been isolated from the sputum of patients with pulmonary disease and also from soil samples. There are currently 29 validly described species in this genus (1). Bacteria of this genus are aerobic and catalase positive, forming rods and cocci. The gordonae are widely distributed and are common in soil, but some strains have been linked with foams found in activated sludge at sewage treatment plants. Three species originally assigned to Rhodococcus, namely, R. bronchialis (132), R. rubropertinctus (133), and R. terrae (132), have more recently been reaffiliated to the genus Gordona as Gordona bronchialis (132), Gordona rubropertincta (133), and Gordona terrae (132). The original spelling Gordona (sic) was corrected to Gordonia by Stackebrandt et al. (134).
The genus Rhodococcus.
The genus Rhodococcus is a heterogeneous group of microorganisms whose members are more closely related to those of the genus Nocardia than to those of the genus Mycobacterium. Rhodococcus species include symbionts (Rhodococcus rhodnii) and pathogens to animals (e.g., R. equi), plants (Rhodococcus fascians), and humans (e.g., R. equi, R. rhodochrous, and R. erythropolis) (135). Rhodococcus equi is the Rhodococcus species that is most likely to act as a pulmonary pathogen in young horses and HIV-infected humans (136).
The Rhodococcus genus has had a long and confused taxonomic pedigree (137, 138). However, many of the early uncertainties have been resolved satisfactorily through the application of chemotaxonomic and phylogenetic character analyses. In the last edition of Bergey's Manual of Systematic Bacteriology, rhodococci were assigned to two aggregate groups based primarily on chemical and serological properties (21). Key diagnostic characteristics for rhodococci are the presence of tuberculostearic acid, mycolic acids with lengths of between 34 and 64 carbon atoms, and with the major menaquinone type being dihydrogenated menaquinones that possess eight isoprenoid units but which lack the cyclic element that is the characteristic motif of the Nocardia genus (135).
Rhodococci are aerobic, Gram-positive, catalase-positive, partially acid-fast, nonmotile actinomycetes that can grow as rods but also as extensively branched substrate hyphae. Some strains produce sparse, aerial hyphae that may be branched or form aerial synnemata, which consist of unbranched filaments that coalesce and project upwards (53). Rhodococci are very important organisms with remarkably catabolic versatility, because they carry genes encoding enzymes that can degrade an impressive array of xenobiotic and organic compounds (139). In addition to their bioremediation potential, they produce metabolites of industrial potential, such as carotenoids, biosurfactants, and bioflocculation agents (140). Some species, such as Rhodococcus rhodochrous, also synthesize commercially valuable products, such as acrylamide (135).
The nomenclature of Rhodococcus equi remains controversial. In a commentary on the nomenclature of this equine pathogen, Goodfellow et al. (141) noted that the taxon is regrettably left without a valid name, because Rhodococcus itself is an illegitimate name and, according to the nomenclature code, should not be used. “Prescottella equi” was suggested as a new name for the taxon that would provide nomenclatural stability; consequently, clinicians and scientists working on this taxon should adopt the name “P. equi.”
The genus Leifsonia.
Evtushenko et al. (142) introduced the genus Leifsonia to accommodate Gram-positive, non-spore-forming, irregular rod- or filament-shaped, motile, mesophilic, catalase-positive bacteria containing dl-2,4-diaminobutyric acid in their peptidoglycan layer. Currently, the genus harbors 12 species and two subspecies, with Leifsonia aquatica as the type species. Members of the genus Leifsonia have been isolated from different ecological niches, including plants (L. poae and L. xyli), soil (L. naganoensis and L. shinshuensis), distilled water (e.g., L. aquatica), Himalayan glaciers, and Antarctic ponds (L. rubra and L. aurea) (142–146).
Leifsonia xyli comprises two subspecies: L. xyli subsp. cynodontis, a pathogen that causes stunting in Bermuda grass (Cynodon dactylon), and L. xyli subsp. xyli (142). Information on the biology and pathogenicity of L. xyli subsp. xyli is limited. Like the gammaproteobacterium Xylella fastidiosa, L. xyli subsp. xyli belongs to a unique group of xylem-limited and fastidious bacterial pathogens and is the causative agent of ratoon stunting disease, the main sugarcane disease worldwide (147).
The genus Bifidobacterium.
Bifidobacteria, first isolated by Tissier (148), are the only family of bacteria in the order Bifidobacteriales. The Bifidobacteriaceae family contains the type genus Bifidobacterium (149), and members of the family Bifidobacteriaceae have different shapes, including curved, short, and bifurcated Y shapes. They were initially classified as Bacillus bifidus communis. The cells have no capsule and they are non-spore-forming, nonmotile, and nonfilamentous bacteria. The genus encompasses bacteria with health-promoting or probiotic properties, such as antimicrobial activity against pathogens that is mediated through the process of competitive exclusion (150), and also bile salt hydrolase activity, immune modulation, and the ability to adhere to mucus or the intestinal epithelium (151). For commercial exploitation, bifidobacterial strains are typically selected for fast growth, antibacterial activity, good adhesion properties, and utilization of prebiotic substrates (151). Among the many probiotic features that have been attributed to bifidobacteria are (i) the induction of immunoglobulin production, (ii) improvement of a food's nutritional value by assimilation of substrates not metabolized by the host, (iii) anticarcinogenic activity, and (iv) folic acid synthesis (152–154). Some bifidobacteria produce antimicrobials (155) and notably, also bacteriocins (156, 157).
The genus Gardnerella.
Classification for the genus Gardnerella is controversial: the genus has often been described as Gram variable but has a Gram-positive wall type (158). Gardnerella vaginalis is a facultative anaerobic bacterium and the only species of this genus belonging to the Bifidobacteriaceae family (159). G. vaginalis is strongly associated with bacterial vaginosis, a disease characterized by malodorous vaginal discharge, but it also occurs frequently in the vaginal microbiota of healthy individuals (160). G. vaginalis-associated vaginosis is a risk factor for poor obstetric and gynecologic outcomes, as well as the acquisition of some sexually transmitted diseases. In addition, clinical studies have demonstrated a relationship between G. vaginalis and preterm delivery (161). The issue of G. vaginalis commensalism is still ambiguous, as the vaginal bacterial community is dynamic and tends to change over the menstrual cycle, leading to a transient dominance of G. vaginalis even in healthy women (162).
The genus Streptomyces.
The various mycelial genera of Actinobacteria harbor some of the most complex known bacteria (163), such as Streptomyces, Thermobifida, and Frankia. Of the three genera, Streptomyces has received particular attention for three main reasons. First, streptomycetes are abundant and important in the soil, where they play major roles in the cycling of carbon trapped in insoluble organic debris, particularly from plants and fungi. This action is enabled by the production of diverse hydrolytic exoenzymes. Second, the genus exhibits a fairly wide phylogenetic spread (164). Third, streptomycetes are among Nature's most competent chemists and produce a stunning multitude and diversity of bioactive secondary metabolites; consequently, they are of great interest in medicine and industry (165). Streptomycetes are the only morphologically complex Actinobacteria whose development has been considered in detail. For more details on this genus, which serves as a model system for bacterial antibiotic production, see the section on “Physiology and Antibiotic Production of Streptomyces,” below.
The genus Frankia.
Frankia is the only nitrogen-fixing actinobacterium and can be distinguished by its ability to enter into symbiotic associations with diverse woody angiosperms known collectively as actinorhizal plants. The most notable plant genera in this group are Alnus, Casuarina, and Elaeaginus, and their symbiosis with Frankia enables them to grow well in nitrogen-poor soils (166, 167). Like Streptomyces, the DNA of Frankia has a particularly high G+C content of 72 to 73% (2). Frankia can form three different cell types, growing as mycelia or as multilocular sporangia. Under nitrogen-limited and aerobic conditions, Frankia develops so-called vesicles at the tips of hyphae or at the ends of short side hyphae (168). For a long time, Frankia spp. were believed to be the only bacteria within the Actinobacteria able to fix atmospheric nitrogen. However, Gtari et al. (169) recently reviewed the sparse physiological and biochemical studies conducted on Actinobacteria over the last 50 years and concluded that nitrogen fixation within this group is unlikely to be restricted to frankiae.
The genus Thermobifida.
The genus Thermobifida, established by Zhang et al. (170), was originally assigned to the highly heterogeneous genus Thermomonospora. A phylogenetic analysis based on 16S rRNA sequences prompted the reclassification of Thermobifida alba and Thermobifida fusca, which were previously classified as Thermomonospora species (33, 137). Later, Thermobifida cellulolytica was added to this genus (171). More recently, Thermobifida halotolerans sp. nov. was proposed as representative of a novel species of Thermobifida (172). Members of the genus Thermobifida are Gram-positive, non-acid-fast, chemo-organotrophic aerobic organisms that form an extensively branched substrate mycelium. Thermobifida species are moderately thermophilic, growing optimally at 55°C, and act as major degraders of plant cell walls in heated organic materials, such as compost heaps, rotting hay, manure piles, or mushroom growth medium.
PHYSIOLOGY AND ANTIBIOTIC PRODUCTION OF STREPTOMYCES
The Streptomyces Life Cycle
Streptomycetes play key roles in soil ecology because of their ability to scavenge nutrients and, in particular, to hydrolyze a wide range of polysaccharides (cellulose, chitin, xylan, and agar) and other natural macromolecules (173). The life cycle of the multicellular mycelial Streptomyces starts with the germination of a spore that grows out to form vegetative hyphae, after which a process of hyphal growth and branching results in an intricately branched vegetative mycelium (174). A prominent feature of the vegetative hyphae of Streptomyces is that they grow by tip extension (28). This in contrast to unicellular bacteria, like Bacillus subtilis and Escherichia coli, where cell elongation is achieved by incorporation of new cell wall material in the lateral wall (175). Exponential growth of the vegetative hyphae is achieved by a combination of tip growth and branching. The fact that cell division during vegetative growth does not lead to cell fission but rather to cross-walls that separate the hyphae into connected compartments (176) makes streptomycetes a rare example of a multicellular bacterium, with each compartment containing multiple copies of the chromosome (177, 178). The spacing of the vegetative cross-walls varies significantly, both between different Streptomyces species and within individual species between different growth conditions and mycelial ages.
Under adverse conditions, such as nutrient depletion, the vegetative mycelium differentiates to form erected sporogenic structures called aerial hyphae. This is also the moment in the life cycle when most antibiotics are produced (179, 180). Streptomyces and other filamentous microorganisms are sessile; when nutrient depletion occurs, the vegetative or substrate mycelium is autolytically degraded by a programmed cell death (PCD)-like mechanism to acquire the building blocks needed to erect a second mass of (aerial) mycelium (181–183). PCD results in the accumulation of amino acids, aminosugars, nucleotides, and lipids around the lysing substrate mycelium (184–186), which inevitably attract motile competing microbes in the habitat; it is logical to assume that antibiotics are produced at this time to protect the pool of nutrients. One well-studied system revolves around the PCD-responsive nutrient sensory regulator DasR, which controls early development and antibiotic production and responds to the accumulation of cell wall-derived N-acetylglucosamine (186, 187). The role of DasR as a regulator of antibiotic production is discussed in more detail in the section on controlling antibiotic production. A cascade of extracellular proteases and protease inhibitors also plays a well-established role in PCD and development in streptomycetes, as reviewed elsewhere (173, 188).
Two rounds of PCD occur during the Streptomyces life cycle (189). After spore germination, a compartmentalized mycelium grows out and then undergoes a first round of PCD that affects the material formed during early vegetative growth. This is then followed by a second round of PCD that is initiated during the onset of development (189). At this stage, the vegetative or substrate hyphae are lysed so as to provide nutrients for the next round of biomass formation, i.e., the growth of the aerial mycelium. The aerial hyphae give the colonies their characteristic fluffy appearance and eventually differentiate to form chains of unigenomic spores (23). Genes that are required for the formation of aerial hyphae are referred to as bld genes, in reference to the bald (“hairless”) phenotype of mutants lacking the fluffy aerial hyphae (190), while mutants whose development is blocked at a stage prior to sporulation are called whi (white), due to their failure to produce the gray spore pigment (174, 191).
Genes that are required for aerial growth or for sporulation were originally identified by screening for mutants after random mutagenesis by using UV irradiation or treatment with chemical mutagens, or by transposon-mediated mutagenesis, resulting in a collection of bld and whi mutants that were subsequently classified on the basis of their morphology (174, 190–195). Several new classes of developmental genes have been identified on the basis of physiological criteria, such as the acceleration of aerial mycelium formation in S. lividans (ram genes, for rapid aerial mycelium [196]), complementation of mutants of S. griseus with disturbed sporulation (ssgA-like genes, for sporulation of Streptomyces griseus [197]), or disruptions in sugar metabolism (186, 198, 199).
Most bld and whi genes that have been identified to date have a (predicted) regulatory function at the transcriptional or translational level, with many encoding predicted transcription factors. Some of the best-studied examples are bldD, a highly pleiotropic transcription factor that controls hundreds of development-related genes (200–202), the RNA polymerase σ factors bldN (203) and whiG (204, 205), which control early events during sporulation (although bldN is also strongly transcribed during aerial growth), and whiH, which controls the onset of sporulation-specific cell division (206, 207). There is also extensive control at the translational level. A wonderful example is bldA, which specifies a tRNA molecule responsible for the translation of the rare leucine codon UUA (208, 209). Deletion of bldA has a pleiotropic effect on gene expression in streptomycetes (210, 211). A major target of bldA-mediated translational control is bldH (adpA), which encodes an important global regulator of development and antibiotic production (212–215). Transcription of adpA is activated in response to the γ-butyrolactone A-factor in S. griseus and to the related molecule SCB1 in S. coelicolor (216–220). An interesting feedback loop exists whereby the translation of the adpA mRNA depends on BldA (221, 222), while AdpA in turn controls bldA transcription (223).
Recently, it was elegantly shown by the group of Mark Buttner that the activity of BldD, which represses many developmental genes during vegetative growth, is controlled posttranslationally by the signaling molecule cyclic-di-GMP (CDG) (224). Binding of tetrameric CDG to BldD brings together the DNA binding domains of the BldD dimer, thus enabling the protein to bind to its target sites (224). An example of metabolic control is presented by the pleiotropic nutrient sensory regulator DasR, which is essential for development and pleiotropically represses antibiotic production (see below). DNA binding by DasR is controlled by the binding of GlcNAc-related metabolites as ligands (186, 225). An overview of key developmental events and regulatory networks in streptomycetes is presented in Fig. 4. An extensive overview of the very complex and intriguing regulatory networks that control the onset of sporulation is beyond the scope of this review; we refer the reader to the excellent previously published reviews of this field for further information (23, 173, 188, 226, 227).
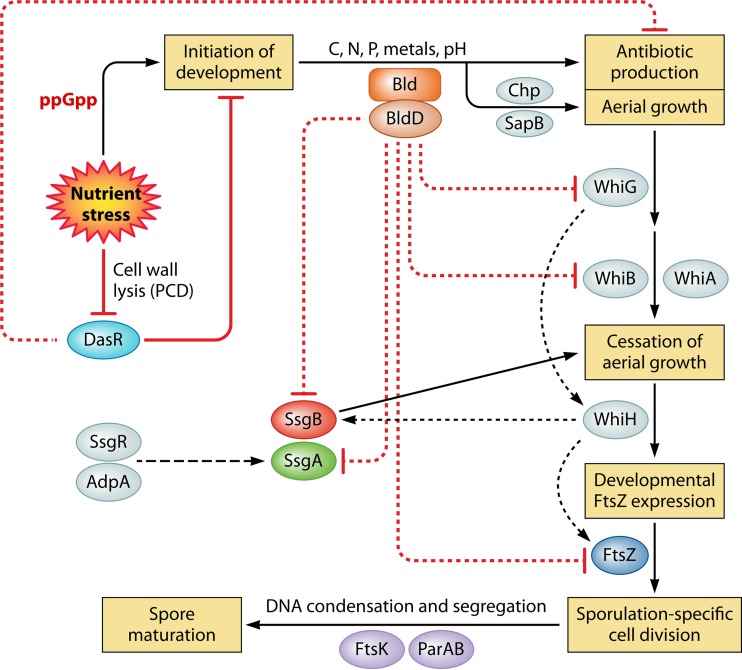
FIG 4.
Major events during development of Streptomyces. Nutrient stress is a major trigger of development, leading to the accumulation of ppGpp, resulting in cessation of early growth and repression of the nutrient sensory DasR protein by cell wall-derived metabolites following PCD of the substrate mycelium. Bld proteins and environmental signals control the procession toward aerial growth and antibiotic production. The developmental master regulator BldD (when bound to tetrameric cyclic-di-GMP) represses the transcription of genes for many key developmental regulatory proteins, including WhiB, WhiG, SsgA, and SsgB, as well as FtsZ. Chaplins and SapB provide a supportive hydrophobic layer to allow aerial hyphae to become erect and break through the moist soil surface. White proteins control aerial growth, whereby WhiAB and SsgB likely play a role in growth cessation. Eventually, FtsZ accumulates and localizes to septum sites in an SsgAB-dependent manner. Ladders of FtsZ are formed, which subsequently delimit the spore compartments. Chromosome condensation and segregation are followed by septum closure and spore maturation. The onset of antibiotic production typically correlates temporally to the transition from vegetative to aerial growth. Solid black arrows represent major transitions in development. Dark dotted lines indicate transcriptional control (arrows for activation, ovals for repression).
Environmental Control of Aerial Hypha Formation
In addition to being defective in aerial hypha formation, early developmental (bld) mutants also exhibit disrupted antibiotic production. This underlines the connection between development and secondary metabolism (see below). Most bld mutants fail to produce antibiotics, although some, in particular bldF, are antibiotic overproducers. By definition, all of the nonessential genes that are required for aerial hypha formation are bld genes. Extracellular complementation experiments where bld mutants were grown in close proximity to one another without physical contact suggested the existence of a hierarchical relationship between at least some of the bld genes (228–231). Aerial hypha formation could be restored from one bld mutant to another, which is consistent with the idea of a signaling cascade that generates a signal that ultimately leads to the onset of development. However, these experiments were almost exclusively performed on a single reference medium, namely, nutrient-rich R2YE agar plates with glucose, and many bld mutants have a conditional bald phenotype—in other words, they are able to produce at least some aerial hyphae and spores on minimal media with nonrepressive carbon sources, such as mannitol (192, 198, 227). A logical assumption is that this is the result of carbon catabolite repression (CCR), whereby favorable carbon sources such as glucose signal the presence of abundant food, thus favoring growth over development and antibiotic production (232, 233).
In streptomycetes, CCR largely depends on the glycolytic enzyme glucose kinase, and deletion of the glkA gene encoding glucose kinase therefore abolishes CCR (232, 234, 235). Suggestively, deleting glkA in bldA mutants of S. coelicolor restores their ability to sporulate on glucose-containing media (236). Conversely, mutants that lack the bldB gene (which encodes a small 99-amino-acid [aa] protein) are defective in CCR, although the mode of action of BldB is as yet unclear (192, 237). It should be noted that Glk-independent pathways of CCR that affect development and antibiotic production also exist, adding further complexity to the picture (238). Other bld genes relevant to sugar metabolism are ptsH, ptsI, and crr, which encode the global components HPr, enzyme I (EI), and enzyme IIA (EIIACrr), respectively, of the phospohoenolpyruvate (PEP)-dependent phosphotransferase system (PTS), which transports sugars such as N-acetylglucosamine and fructose in S. coelicolor (239, 240). Other examples are dasABC, which encodes a chitobiose sugar transporter (199, 241), and the pleiotropic sugar regulators atrA (242) and dasR (186, 187). Perhaps surprisingly, the nonsporulating phenotype of the das and pts transport mutants is independent of the carbon source and thus probably also of the transport activity (186, 199, 241). These examples highlight the important and complex connections between carbon utilization and development in streptomycetes.
Metals also play a key role in the onset of development. Streptomyces lividans requires a large amount of copper for proper aerial growth (243). This defect can be rescued by enhanced expression of the ram cluster (196), which ultimately leads to the production of the surfactant SapB (see the next section). Recently, Sébastien Rigali and colleagues showed that development can be restored to bldJ and bldK mutants by supplementing R2YE agar with iron (244). The bldK gene cluster encodes an oligopeptide transporter (228, 245), while the function of bldJ is unknown (228). Interestingly, mass spectrometric analysis revealed that all of the bld mutants that were tested showed either severely reduced (for bldA, bldJ, and ptsH mutants) or enhanced (for bldF, bldK, crr, and ptsI mutants) production of the iron-binding siderophore desferrioxamine (244). The same paper also mentioned unpublished data suggesting that deregulated desferrioxamine production occurs in bldB, bldC, bldD, dasA, and adpA mutants when grown on R2YE agar plates and may thus be a more general feature of many bld mutants (244). Further complications arise from the fact that both bldJ mutants and mutants lacking the citA gene for citrate synthase (which also fail to develop on R2YE under “standard” conditions), do successfully develop on R2YE when the medium is strongly buffered (246). Besides shedding more light on the nature of bld mutations, these experiments also show that environmental factors, such as metal availability, pH, and carbon and nitrogen sources, have profound effects on the onset of development, and so the composition of the medium should be considered very carefully when planning experiments to study development in streptomycetes.
Facilitating Aerial Growth: the Roles of Chaplins, Rodlins, and SapB
Aerial hyphae differ substantially from vegetative hyphae. One major difference is that aerial hyphae of wild-type cells typically do not branch extensively, are nearly twice as wide as vegetative hyphae, and undergo rapid growth. The aerial hyphae are also surrounded by a sheath that later becomes part of the spore coat (247–251). This sheath is hydrophobic on the air-facing side, allowing the aerial hyphae to break through the moist soil-air surface with the assistance of the turgor pressure generated by the hyphae (252, 253), similarly to what has been proposed for fungi (254). Chaplins are potent surfactants, reducing the surface tension from 72 to 24 mJ m−2 (247, 255). An important function of the sheath may also be to create a channel along the outer hyphal wall that can facilitate nutrient transport, as proposed by Keith Chater (173, 256). This argument suggests that nutrients and other metabolites might diffuse from the vegetative hyphae in the basal part of the colony up to the growing tips of the aerial hyphae (173, 256), which would be an attractive alternative to transport through hyphae and could potentially resolve the longstanding debate about how nutrients are transported efficiently across long distances over the cross-walls (188).
The sheath consists of a number of hydrophobic proteins, in particular chaplins and rodlins (230, 247, 249, 257–260). Together, these proteins form the so-called rodlet layer, which decorates the spores with a seemingly random pattern of small lines running in all directions; high-resolution electron microscopy shows that these lines consist of small protein assemblies (Fig. 5). Typical rodlet layer formation depends on the rodlin proteins RdlA and RdlB; deletion of the rdl genes instead results in decoration with fine lines consisting of the chaplins (248). S. coelicolor contains eight chaplins, three large ones (ChpABC) with two chaplin domains and a sortase domain and five smaller chaplins (ChpDEFGH) bearing a single chaplin domain (247, 249). The chaplins assemble on the hyphal surface into an amphipathic protein layer that consists of amyloid-like fibrils. Of the chaplins, the vegetatively expressed ChpC, ChpE, and ChpH proteins are sufficient for sporulation (261). ChpE and ChpH are secreted into the surrounding medium to reduce the surface tension so as to enable the hyphae to grow into the air (247, 255).
FIG 5.
Scanning electron micrograph of the surface layer of mature spores, revealing a distinctive rodlet layer. This layer consists of hydrophobic chaplin (Chp) and rodlin (Rdl) proteins. Bar, 100 nm.
Closer analysis of the ChpH protein showed that it has two amyloidogenic domains at its N and C termini, which are both required for aerial hypha formation, while only the C-terminal domain is required for assembly of the rodlet ultrastructure (262). In addition to the chaplins, there are two rodlin proteins that also contribute to the development of the sheath's rodlet ultrastructure and the spore surface (258). Suggestively, most of the genes for the chaplins and the two rodlin genes lie in close proximity on the genome (263). However, the rodlins are not required for sporulation, even though they are needed for the sheath's development into paired rodlet structures (258). It is clear that these hydrophobic structural proteins play key roles in the aerial development of streptomycetes, but the precise role of each of the individual components remains to be resolved.
In addition to ChpE and ChpH, the onset of aerial growth requires the extracellular accumulation of yet another hydrophobic surfactant, SapB (230, 264). SapB-type proteins are widespread in streptomycetes; well-studied examples include AmfS in S. griseus and SapT in S. tendae (265, 266). In S. coelicolor, SapB is encoded by the ramS gene in the ramCSAB gene cluster (196, 267), which is controlled by the orphan response regulatory gene ramR (268–270). In turn, at least in S. griseus, transcription of the amf operon, and thus of amfS, depends on AdpA and therefore ultimately also on BldA (215). RamS is produced as a 42-aa propeptide that is subsequently modified and exported in a way very similar to that for lantibiotics, although the way the propeptide is processed is yet unknown. During modification by RamC, four dehydroalanine residues and two lanthionine bridges are introduced (271). Thus, a highly modified 21-aa molecule of 2,027 Da is produced (271), with all the structural and genetic features of type II lantibiotics (259, 272). Despite the exciting insights that have been obtained into the biological role of SapB so far, the precise mechanism by which it controls the developmental growth of streptomycetes awaits further elucidation, as do the transcriptional and posttranscriptional control mechanisms underlying its biosynthesis (273).
As a final comment on this topic, it is important to note that even the extracellular addition of the fungal hydrophobin SC3 (obtained from the basidiomycete Schizophyllum commune) restores aerial growth to several bld mutants of S. coelicolor that are deficient in the production of chaplins and/or SapB (264, 265). This again underlines the importance of the extracellular accumulation of a hydrophobic layer for the early stages of aerial growth.
From Aerial Hyphae to Spores: Sporulation-Specific Cell Division and the Cytoskeleton
Like vegetative hyphae, aerial hyphae grow by tip extension. Once sufficient aerial biomass is generated, a signal is transmitted that results in growth cessation, followed by the onset of sporulation. The signal for growth cessation is not yet known but likely relates to the Whi regulatory proteins WhiA and WhiB, as well as the cell division activator SsgB. Mutations of whiA and whiB produce identical phenotypes, with hypercoiling and very long aerial hyphae that fail to initiate cell division (23, 174, 274), while ssgB mutations produce a large colony phenotype, forming an extremely large aerial biomass (275).
The landmark event in the onset of sporulation is the initiation of sporulation-specific cell division, which is notable because the process of cell division is completely different between vegetative and aerial hyphae. Wonderful movies of septum formation during early growth of the hyphae of S. coelicolor show how irregular the placement of septa is in vegetative hyphae, with cross-walls dividing the vegetative hyphae into multigenomic compartments (276). In contrast, during sporulation-specific cell division in aerial hyphae, many septa are formed almost simultaneously and in a highly symmetrical fashion, followed by the formation of spore compartments and cell fission, resulting in chains of spores that each contain a single copy of the chromosome (reviewed in references 277 and 278). Most bacteria divide by binary fission, whereby a single mother cell symmetrically divides into identical daughter cells. This process involves the formation of a cytokinetic ring structure, of which the scaffold is formed by the polymerization of thousands of copies of the tubulin homolog FtsZ at division sites (279–283). However, in streptomycetes, the long aerial hyphae differentiate into chains of spores after a uniquely coordinated cell division event. Distinctive ladders of FtsZ are thereby produced that consist of up to 100 septa, and this eventually leads to the production of chains of haploid spores (284–286). Sufficient accumulation of FtsZ is required to support sporulation, and developmental ftsZ transcription is largely dependent on the “early” whi regulatory genes whiA, whiB, whiG, whiH, whiI, and whiJ (287). Consistent with the notion that the control of ftsZ transcription may be a key event, at least in S. coelicolor, the nonsporulating phenotype of many of these early whi mutants could be overruled by constitutive expression of ftsZ during development (288). This also suggests that no other genes that are required for sporulation completely depend on these whi genes, at least not when FtsZ is overexpressed.
A unique feature of Streptomyces biology is that cell division is not required for growth and ftsZ null mutants are viable (289). While the ftsZ mutant also fails to make cross-walls, most of the other cell division mutants are only defective in sporulation-specific cell division (278, 290–293). This illustrates a major difference between vegetative and aerial cell division.
While sporulation-specific cell division is mechanistically very similar to that in bacteria that divide by binary fission, the way septum site localization is controlled is completely different, involving actinomycete-specific proteins (177, 277, 278). In unicellular bacteria, the positioning and timing of the formation of a septum involves the action of negative-control systems such as Min, which prevents Z-ring assembly at the cell poles (294, 295), and nucleoid occlusion, which prevents DNA damage by blocking Z-ring formation over nonsegregated chromosomes (296). The Z-ring is tethered to the membrane by dedicated anchoring proteins, such as FtsA and ZipA in Escherichia coli (297, 298). All of these systems are absent in streptomycetes. It is therefore unclear how streptomycetes avoid DNA damage in the multinucleoid hyphae. Elegant work on DNA partitioning revealed the important role of the ParAB proteins in DNA segregation during growth and development (299–301). FtsK helps to avoid “guillotining” of the DNA by pumping chromosomes into the spore compartments prior to septum closure, and ftsK mutants frequently generate spores with incomplete chromosomes (302–304). Other proteins that should be considered are SmeA and SffA, which play key roles in DNA translocation during sporulation (302), and also the DNA-packaging proteins HupS (305), sIHF (306, 307), Smc (308), and Dps (309).
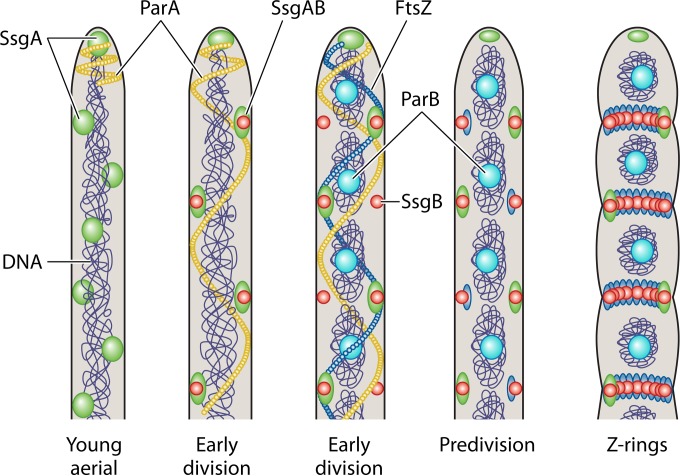
In terms of septum site localization, a key role is played by the SsgA-like proteins (SALPs), which only occur in sporulating actinobacteria (310, 311). SsgA activates sporulation-specific cell division (312, 313), and both ssgA and ssgB are required for sporulation (275, 314, 315). The symmetrical spacing of the many Z-rings is achieved by SsgB, which directly recruits FtsZ and also stimulates its polymerization (316). SsgB localizes to future division sites prior to and independent of FtsZ (316). Thus, cell division is positively controlled in streptomycetes (Fig. 6). The next obvious question is how SsgB itself is localized, especially given that it lacks a membrane domain. Another important cell division protein that controls sporulation-specific cell division in Streptomyces is CrgA, which affects sporulation-specific cell division by influencing Z-ring assembly (317). In contrast to the SALPs, CrgA also occurs in nonsporulating actinomycetes, and it interacts with FtsZ, FtsI, and FtsQ in Mycobacteria (318). The phenotypes of crgA null mutants and overexpressing strains suggest that CrgA affects both cell division and the cytoskeleton, although its precise mode of action is still unknown.
FIG 6.
Model for the control of sporulation-specific cell division in Streptomyces. When sporulation starts, SsgA localizes dynamically in young aerial hyphae, while SsgB and FtsZ are still diffuse at this stge. At this point, ParA is constrained to the hyphal tip. During early cell division, SsgA and SsgB colocalize temporarily at either side of the aerial hyphae, with ParA extending downward as filaments along the aerial hypha. ParB complexes are then formed over the uncondensed chromosomes, while FtsZ assembles in spiral-like filaments. Subsequently, FtsZ and SsgB colocalize and stay together until FtsZ disperses, whereby SsgB recruits FtsZ and stimulate its polymerization into protofilaments. The way the SsgB-FtsZ complex is tethered to the membrane in the absence of a membrane domain in either protein is unclear, but a likely role is played by the SepG protein (SCO2078 in S. coelicolor) encoded by a gene upstream of divIVA (L. Zhang, J. Willemse, D. Claessen, and G. P. van Wezel, unpublished data). Z-rings are then formed at the sporulation stage, followed by chromosome condensation and segregation and the production of sporulation septa. SsgA eventually marks the future germination sites. The figure was adapted from references 277 and 316.
Streptomycetes probably have a much more elaborate cytoskeleton than most other bacteria, which may be explained by their hyphal rather than planktonic growth (319). Besides the tubulin homolog FtsZ and the actin-like proteins MreB and Mbl (320, 321), a large number of proteins with coiled-coil structural elements occur in these bacteria, and evidence is accumulating regarding their important role in growth, cell shape, and morphogenesis (319, 322–324). The protein FilP forms intermediate filament-like structures that contribute to mechanical stress resistance (322). In addition, the Scy protein, encoded by a gene immediately adjacent to filP, apparently functions as a “molecular assembler” and sequesters DivIVA (323). DivIVA is essential for growth in streptomycetes and localizes to tips to drive apical growth, although the molecular mechanism of this process is still unclear (325, 326). In this way, Scy establishes growth nuclei for apical growth and branching. It also interacts with the chromosome-partitioning protein ParA (327) and the intermediate filament-like protein FilP, which in turn interacts with DivIVA. The apical assembly that drives tip growth was termed the tip organizing complex (TIPOC) by Gabriella Kelemen and colleagues (323, 328, 329). SsgA (330) and the polysaccharide synthase CslA (323, 328, 329) are other proteins that are part of this TIPOC. Clearly, we can only see the tip of the iceberg at present, and future discoveries will undoubtedly shed new light on these processes.
STREPTOMYCETES AS ANTIBIOTIC FACTORIES
Actinomycetes produce approximately two-thirds of all known antibiotics, the majority of which are produced by streptomycetes. Consequently, these microorganisms are very important in the fight against emerging multidrug-resistant pathogens (331–333). Streptomyces coelicolor is a model system for studying (the control of) antibiotic production. Scientists have marveled for decades at the ability of single streptomycetes species to produce a plethora of different antibiotic compounds. For example, those produced by S. coelicolor include actinorhodin (Act [334]), undecylprodigiosin (Red [335]), calcium-dependent antibiotic (CDA [336]), and methylenomycin (Mmy [337]), the latter of which is carried on a plasmid. However, when the genome sequence of S. coelicolor was published (263), it became apparent that this species' true potential as a producer of natural products had actually been underestimated: over 20 biosynthetic gene clusters for secondary metabolites were identified (338), including one that appears to be for the production of a cryptic polyketide antibiotic (Cpk [339]). It rapidly became apparent that such “concealment” of antibiotic-producing capabilities is the norm rather than the exception, with some streptomycetes harboring more than 50 different secondary metabolite gene clusters (340–343). It therefore appears that the potential of these organisms for novel drug production is much greater than originally anticipated. This has prompted extensive research in applied genomics into so-called cryptic, silent, or sleeping antibiotics (reviewed in references 344 to 347) and methods for activating their biosynthesis (348–352).
Correlation between Growth and Antibiotic Production
Programmed cell death and the DasR system.
The production of antibiotics (and other secondary metabolites) is temporally correlated to the onset of development in the Streptomyces life cycle (179, 180). This correlation may exist because of the need to defend the colony when it is undergoing PCD. Evidence supporting such a direct link between PCD and antibiotic production was provided by the observation that cell wall-derived N-acetylglucosamine (GlcNAc) acts as a signal for the onset of development and as a global elicitor molecule for antibiotic production (186, 187). In competitive soil habitats, the timing of development is crucial. However, it is not clear how colonies know when to initiate this process, which has such major consequences for the colony. As long as sufficient nutrients are available, growth should prevail over development, while during starvation, sporulation and subsequent spore dispersal are essential for the survival of the progeny. The signals that trigger such events should be unmistakable, and GlcNAc may serve this purpose well. In nature, GlcNAc can be obtained from hydrolysis of the abundant natural polymer chitin by the chitinolytic system, or from hydrolysis of microorganism cell walls. For bacteria, GlcNAc is a favorable C and N source and a major constituent of the cell wall peptidoglycan. Some 13 chitinases and chitosanases have been identified in S. coelicolor (353–355).
Interestingly, under poor nutritional conditions, supplementing with GlcNAc accelerates both the onset of development and antibiotic production, suggesting that under these conditions GlcNAc signals nutrient stress, resulting in accelerated development. Conversely, in rich media, higher concentrations of GlcNAc block development and antibiotic production, thus inducing a response typical of vegetative growth (187). The different growth conditions of minimal and rich media likely resemble conditions of feast or famine in the natural environment (i.e., the soil), with GlcNAc acting as an important signaling molecule that would typically be derived from chitin in nutrient-rich soil during feast periods or from the Streptomyces cell walls during PCD (famine), respectively. The secret of this dual signaling role appears to lie in the nature of the sugar transporters. Monomeric GlcNAc enters the cell via the NagE2 permease (356), which is part of the PEP-dependent phosphotransferase system (PTS) (357, 358), while chitobiose (dimeric GlcNAc), which is the subunit of chitin, enters via the ABC transporters DasABC or NgcEFG (241, 353, 359). Subsequently, internalized GlcNAc is converted by the enzymes NagA and NagB to glucosamine-6-phosphate (GlcN-6-P) (360), a central metabolite that can then enter glycolysis (as fructose-6P) or the pathway toward peptidoglycan synthesis.
GlcNAc-derived GlcN-6-P acts as an allosteric effector of the GntR family regulator DasR (186), a global regulator that controls the GlcNAc regulon (186, 360, 361), and also the production of antibiotics (187) and siderophores (362). GlcNAc-dependent nutritional signaling is most likely mediated through changes in the intracellular level of GlcN-6-P, which binds as a ligand to the GntR family regulator DasR, leading to derepression of DasR-mediated control of antibiotic production (187). As shown by genome-wide transcription and ChIP-on-chip analysis, all pathway-specific activator genes for antibiotic biosynthetic gene clusters are controlled by DasR (363). Thus, antibiotic biosynthesis and secretion is induced by adding GlcNAc to minimal medium with a poor carbon source. As mentioned above, these conditions activated the cpk gene cluster (187), which encodes genes that for the cryptic polyketide Cpk and was more recently established as coelimycin P1 (364). This suggests that similar conditions could be used to activate other cryptic antibiotic gene clusters which are expressed poorly (or not at all) under normal growth conditions.
It was recently shown that “hostile” interactions between streptomycetes such as antibiosis and suppression of production by competitors primarily occur under nutrient-limiting conditions; conversely, under nutrient-rich conditions, social interactions are favored (365). These observations suggest that antibiotics are indeed used as weapons in nature. This concept is in contrast with studies suggesting that antibiotics act as signals for intercellular communication (366–368). The latter hypothesis is based on the rationale that the secreted antibiotics may not reach sufficiently high concentrations to cause appreciable growth inhibition and the observation that subinhibitory concentrations of antibiotics induce responses such as biofilm formation (369) or virulence (370) that may benefit the target cells (371). In terms of concentrations in the soil, the same argument can be made for GlcNAc, which only activates antibiotic production at millimolar concentrations on agar plates. Presumably, small molecules reach higher local concentrations, for instance, in close proximity to a producing colony or, in the case of natural products, by binding to dead plant or animal material. In this context, it is worth remembering that cellulose is a preferred column material for the purification of natural products. Information on the interactions between microbes is not just of ecological and evolutionary importance, since it could also be useful in drug discovery efforts. Indeed, data on the interactions between microbes could provide key clues concerning the activation of cryptic biosynthetic pathways (372–376).
Stringent control.
As discussed, nutrient deprivation and the resulting growth cessation are the primary triggers for the onset of antibiotic production. Starvation results in the depletion of amino acids and hence uncharged tRNAs, which then occupy the ribosomal A-site. This in turn induces the production of the small molecules guanosine tetraphosphate and pentaphosphate (377). However, (p)ppGpp, or “magic spot,” as it was originally known, is produced in response to basically all processes that relate to changes in nutrient availability and might affect growth, adaptation, secondary metabolism, survival, persistence, cell division, motility, biofilm formation, development, competence, and virulence (378). The synthesis of (p)ppGpp from GTP and ATP can be carried out by either RelA, which is ribosome associated and activated by the binding of uncharged tRNAs to the ribosome, or by SpoT, which produces (p)ppGpp in response to nutrient starvation (378). The ribosomal protein L11, encoded by rplI (relC), activates RelA and thus ppGpp synthesis (379). Deletion of the relA gene suppresses antibiotic production, underlining the important role of the stringent response in controlling antibiotic production (380). Indeed, under nitrogen-limiting conditions ppGpp causes a dramatic switch in the physiology of streptomycetes, activating the expression of genes involved in morphological and chemical differentiation, including the production of CDA and actinorhodin, and at the same time repressing genes involved in normal growth (381). The exact mechanism by which ppGpp acts upon such a wide range of genes remains to be elucidated. However, the response to fatty acid starvation involves the direct binding of the unacylated (“uncharged”) fatty acid acyl carrier protein (ACP) to SpoT (382), and it has been suggested (180) that perhaps the ACPs of polyketide synthases help to control the stringent response in a similar fashion, providing a possible explanation of the relationship between the stringent response and secondary metabolism.
Morphological control.
A complex relationship exists between morphology and antibiotic production. Streptomycetes display a great many different morphologies in submerged cultures, ranging from fragmented growth to dense clumps, and this can have a major influence on their production levels (24). For example, in Saccharopolyspora erythraea, erythromycin production is favored by clumps with a minimum size of around 90 μm in diameter (383). The connection between mycelial morphology and production is further exemplified by avermectin production by S. avermitilis, which is favored by small dense pellets (384), and by S. coelicolor, in which forced fragmentation by overexpression of the cell division activator protein SsgA abolishes actinorhodin production. However, it is dangerous to generalize, as the same strain shows a 20- to 50-fold increase in undecylprodigiosin production in a fermentor (310, 313, 315), and chloramphenicol production by Streptomyces venezuelae is not hampered by the extremely fragmented growth of the producing organism (385, 386). It was previously suggested that antibiotics that are produced during exponential growth may benefit from fragmented growth, while those produced during the transition or stationary phases are produced much more efficiently by clumps (236).
More insights into the genetic factors that control mycelial growth should allow scientists to significantly improve the yield of natural product formation by actinomycetes. A novel set of genes was recently discovered in Streptomyces that control pellet growth, called mat, for mycelial aggregation (387). The genes were discovered 30 years ago via reverse engineering of a strain of S. lividans that was selected for fragmented growth in a chemostat (388). Deletion of the mat genes (SCO2962 and SCO2963 in S. coelicolor) prevented pellet formation and increased both growth rate and enzyme production in S. lividans (387). The mat genes are probably responsible for the production of a secreted polysaccharide, which presumably glues the hyphae together and thus promotes pellet aggregation.
In silico models have been developed to better understand mycelial growth (389–391), although many were primarily based on physiological and nutritional parameters. Helped by the strong increase in computing power in the modern era, new models were developed recently (392, 393). In particular, a three-dimensional model was developed that includes parameters such as hyphal growth, branching, fragmentation, cross-wall formation, and collision detection, as well as oxygen diffusion (392). For the rational design of actinomycetes as production hosts, it is imperative that we better understand how morphology correlates with production. For example, when and especially where are natural products (and enzymes) secreted? Secretion at apical sites would imply that fragmented growth is favorable because it increases the number of hyphal tips per length unit, as opposed to when production primarily takes place inside mycelial clumps. Interestingly, production of enzymes through the twin arginine translocation (Tat) exporter occurs closely behind the hyphal tips in S. coelicolor (16, 394); in line with this concept, Tat substrates are secreted more efficiently in fragmenting strains of S. coelicolor and S. lividans (313). An extensive review of the industrial implications of the correlation between growth and natural product formation is beyond the scope of this review, so we refer the interested reader elsewhere (24, 395–398).
From global control to the activation of specific gene clusters.
The global regulatory networks ultimately relay information toward the individual biosynthetic gene clusters, acting at the level of pathway-specific control. Transcription of antibiotic biosynthetic gene clusters typically depends on a pathway-specific activator gene inside the cluster. Many of these are controlled in a growth phase-dependent manner, with the SARP family regulators (399) being the best-known examples. Particularly well-known members of this family are ActII-ORF4 and RedD, which activate the production of the pigmented antibiotics actinorhodin and undecylprodigiosin, respectively, in S. coelicolor, and StrR for production of the aminoglycoside streptomycin in S. griseus (400–402). As exemplified by the actinorhodin biosynthetic activator gene actII-ORF4, transcription of which is controlled by over 15 different regulatory proteins, the timing and accumulation of a pathway-specific activator may be very complex (236, 395). However, once activated, there appears to be little additional control downstream, as long as the necessary precursors are present. Indeed, when redD, the pathway-specific activator gene for production of prodigionines in S. coelicolor, is placed under the control of another regulatory element, the result is that control of prodigionin production becomes dictated by the regulatory network controlling that element (275, 314, 315). In other words, placing redD under the control of the promoter for the global nitrogen regulator (glnR) or the sporulation-specific sigma factor (sigF) ensures that production is controlled by nitrogen or produced in aerial hyphae, respectively. This implies that there may be few genetic limitations regarding the production of natural products in time and space once an appropriate activator is expressed. From a production point of view this is an advantage, because restrictions due to growth phase-related control mechanisms can be dealt with by changing the regulatory element. This is particularly important for heterologous expression of biosynthetic gene clusters that are being uncovered at a high rate in actinomycetes in the era of genome sequencing. In particular, the combination of synthetic biology approaches with expression in optimized heterologous Streptomyces production platforms is a promising development (403–408).
ACTINOBACTERIA AS SOURCES OF NATURAL PRODUCTS
Actinobacteria as Sources of Antibiotics
Actinobacteria are of great importance in the field of biotechnology, as producers of a plethora of bioactive secondary metabolites with extensive industrial, medical, and agricultural applications (Table 3 provides examples and corresonding references). In particular, Actinobacteria produce the majority of the naturally occurring antibiotics. The first antibiotics discovered in Actinobacteria were actinomycin from a culture of Streptomyces antibioticus in 1940 (409), streptothricin from Streptomyces lavendulae in 1942 (410), and streptomycin from Streptomyces griseus in 1944 (411), all of which were discovered by Waksman and colleagues. Streptomycetes have been the major source of clinical antibiotics and are responsible for over 80% of all antibiotics of actinobacterial origin (333). That actinomycin, streptomycin, and streptothricin were the first to be found is not surprising, as these molecules occur at much higher frequencies than many other antibiotics. For example, streptothricin is found in some 10% of all streptomycetes isolated randomly from soil and streptomycin is found in 1% and actinomycin in 0.1%, while conversely, erythromycin and vancomycin are found in around 10−5 soil isolates, and daptomycin is found only at a frequency of around 10−7 (412). Major classes of clinical antibiotics produced by actinomycetes are the following: aminoglycosides (neomycin, kanamycin, streptomycin (413–415), angucyclines (auricin; also, antitumor agents like landomycin and moromycin (416), ansamycins (rifamycin, geldanamycin) (417), anthracyclines (primarily antitumor agents, e.g., daunorubicin) (418, 419), β-lactams (cephamycins) (420) and also the important β-lactamase inhibitor clavulanic acid (421, 422), chloramphenicol (423), glutarimides (cycloheximide) (424), glycopeptides (vancomycin, teichoplanin) (425, 426), lipopeptides (daptomycin) (427), lantibiotics (mersacidin, actagardine) (272), macrolides (clarythromycin, erythromycin, tylosin, clarithromycin) (428, 429), oxazolidinones (cycloserine) (430), streptogramins (streptogramin) (431), and tetracyclines (432). The producing capacity of individual actinomycetes can also vary enormously. Some Streptomyces species produce a single antibiotic, while others produce a range of different compounds and compound classes.
TABLE 3.
Examples of bioactive molecules produced by Actinobacteria genera and their activities
| Type of compound and producing species | Bioactive agent(s) | Source or reference |
|---|---|---|
| Antibacterial agent producers | ||
| Verrucosispora spp. | Abyssomycin | 603 |
| Streptomyces anulatus | Actinomycins | 409 |
| Streptomyces canus | Amphomycin | 604 |
| Micromonospora spp. | Anthracyclin | 605 |
| Streptomyces cattley | Antibiotics and fluorometabolites | 606 |
| Streptomyces canus | Aspartocins | 607 |
| Streptomyces avermitilis | Avermectin | 608 |
| Streptomyces venezuelae | Chloramphenicol | 609 |
| Micromonospora spp. | Clostomicins | 609 |
| Streptomyces griseus | Cycloheximide | 620 |
| Streptomyces orchidaceus | Cycloserine | 610 |
| Streptomyces roseosporus | Daptomycin | 611 |
| Saccharopolyspora erythraea | Erythromycin (Ilotycin) | 612 |
| Micromonospora purpurea | Gentamicin | 613 |
| Streptomyces hygroscopicus | Hygromycin | 614 |
| Streptomyces kanamyceticus | Kanamycin | 615 |
| Streptomyces kitasoensis | Leucomycin | 616 |
| Streptomyces lincolnensis | Lincomycin | 617 |
| Marinispora spp. | Marinomycin | 618 |
| Streptomyces fradiae | Neomycins | 619 |
| Micromonospora spp. | Netamicin | 80 |
| Streptomyces niveus | Novobiocin | 620 |
| Streptomyces antibioticus | Oleandomycin | 621 |
| Streptomyces rimosus | Oxytetracycline | 622 |
| Streptomyces spp. | Pristinamycin | 623 |
| Streptomyces lindensis | Retamycin | 624 |
| Streptomyces mediterranei | Rifamycin | 625 |
| Nocardia lurida | Ristocetin | 621 |
| Streptomyces ambofaciens | Spiramycin | 626 |
| Streptomyces virginiae | Staphylomycin | 627 |
| Streptomyces endus | Stendomycin | 628 |
| Streptomyces lydicus | Streptolydigin | 629 |
| Streptomyces griseus | Streptomycin | 411 |
| Streptomyces lavendulae | Streptothricin | 410 |
| Streptomyces aureofaciens | Tetracycline | 630 |
| Micromonospora spp. | Thiocoraline | 631 |
| Amycolatopsis orientalis | Vancomycin | 632 |
| Antifungal agent producers | ||
| Streptomyces anulatus | Actinomycins | 603 |
| Streptomyces nodosus | Amphotericin B | 633 |
| Streptomyces griseochromogenes | Blasticidin | 634 |
| Streptomyces griseus | Candicidin | 635 |
| Streptomyces spp. | Carboxamycin | 636 |
| Streptomyces venezuelae | Chloramphenicol | 637 |
| Streptomyces padanus | Fungichromin | 638 |
| Streptomyces galbus | Galbonolides | 675 |
| Streptomyces violaceusniger YCED-9 | Guanidylfungin | 564 |
| Streptomyces venezuelae | Jadomycin | 639 |
| Streptomyces kasugaensis | Kasugamycin | 452 |
| Streptomyces spp. | Kitamycin | 640 |
| Streptomyces natalensis | Natamycin | 641 |
| Streptomyces tendae | Nikkomycin | 642 |
| Streptomyces diastatochromogenes | Oligomycin | 643 |
| Streptomyces humidus | Phenylacetate | 644 |
| Streptomyces cacaoi | Polyoxin B | 453 |
| Streptomyces canus | Resistomycin | 645 |
| Streptomyces lavendulae | Streptothricin | 410 |
| Streptomyces canus | Tetracenomycin | 645 |
| Nocardia transvalensis | Transvalencin | 646 |
| Streptomyces hygroscopicus | Validamycin | 647 |
| Bioherbicide/biopesticide producers | ||
| Actinomadura spp. | 2,4-Dihydro-4-(β-d-ribofuranosyl)-1, 2, 4 (3H)-triazol-3-one (herbicide) | 648 |
| Streptomyces hygroscopicus | Herbimycin | 649 |
| Streptomyces avermitilis | Ivermectin (derivative of avermectin) | 650 |
| Streptomyces prasinus | Prasinons | 651 |
| Saccharopolyspora spinosa | Spinosad (neurotoxic insecticides) | 652 |
| Antiparasitic agent producers | ||
| Streptomyces avermitilis | Avermectins | 608 |
| Streptomyces coelicolor | Prodiginine | 653 |
| Streptomyces bottropensis | Trioxacarcin | 654 |
| Antiviral agent producers | ||
| Streptomyces antibioticus | 9-β-d-Arabinofuranosyladénine | 655 |
| Streptomyces hygroscopicus | Hygromycin | 614 |
| Streptomyces spp. | Panosialins | 656 |
| Hypercholesterolemia agent producer | ||
| Streptomyces hygroscopicus | Rapamycin | 674 |
| Antitumor agent producers | ||
| Micromonospora spp. | Anthraquinones | 657 |
| Nocardia asteroides | Asterobactine | 658 |
| Streptomyces spp. | Borrelidine | 659 |
| Micromonospora spp. | Diazepinomicin | 660 |
| Actinomadura spp. | IB-00208 | 661 |
| Micromonospora spp. | LL-E33288 complex | 76 |
| Micromonospora spp. | Lomaiviticins | 76 |
| Micromonospora spp. | Lupinacidins | 657 |
| Thermoactinomyces spp. | Mechercharmycin | 662 |
| Marinospora spp. | Marinomycin | 618 |
| Salinispora tropica | Salinosporamide | 621 |
| Streptomyces peucetius | Doxorubicin (adriamycin) | 664 |
| Streptomyces peucetius | Daunorubicin (daunomycin) | 665 |
| Micromonospora spp. | Tetrocarcin | 76 |
| Micromonospora spp. | Thiocoraline | 666 |
| Immunostimulatory agent producers | ||
| Nocardia rubra | Rubratin | 667 |
| Streptomyces olivoreticuli | Bestatin | 668 |
| Kitasatospora kifunense | FR-900494 | 669 |
| Immunosuppressive agent producers | ||
| Nocardia brasiliensis | Brasilicardin | 670 |
| Streptomyces filipinensis | Hygromycin | 671 |
| Streptomyces filipinensis | Pentalenolactone | 671 |
| Therapeutic enzyme (antitumor) producers | ||
| Streptomyces spp. | l-Asparaginase | 672 |
| Streptomyces olivochromogenes | l-Glutaminase | 673 |
Besides antibiotics, Actinobacteria also produce a wide variety of other secondary metabolites with activity as herbicides (54), antifungals, antitumor or immunosuppressant drugs, and anthelmintic agents (433). Examples are given below.
Actinobacteria as Sources of Insecticides
Macrotetrolides are active against mites, insects (434–436), coccidia (437), and helminths (438), and they also show immunosuppressive effects (439). They are produced by a variety of Streptomyces species (for a review, see Jizba et al. [434]). However, with regard to the composition of the macrotetrolide complex, only S. aureus S-3466 (440), which produces a mixture of tetranactin (the most active member of the compound group) with dinactin and trinactin (435, 441), has been utilized for commercial purposes (442). Tetranactin, a cyclic antibiotic produced by Streptomyces aureus with a molecular structure related to cyclosporine, is used as emulsion against carmine mites of fruits and tea. A true success story in terms of anthelmintics is ivermectin (443), which is a dehydro derivative of avermectin produced by Streptomyces avermitilis. After its appearance in the late 1970s, ivermectin was the world's first endectocide, which at the time was a completely novel class of antiparasitic agents, with strong and broad-spectrum activity against both internal and external nematodes and arthropods. Recently, the Nobel Prize for Physiology or Medicine 2015 was awarded to Satoshi Omura and William C. Campbell for their discovery of avermectin, jointly with Youyou Tu for the discovery of the antimalarial drug artemisinin.
Actinobacteria as Sources of Bioherbicide and Bioinsecticide Agents
Mildiomycin, an antifungal metabolite isolated from cultures of Streptoverticillium rimofaciens Niida, is strongly active against several powdery mildews on various crops (444) and inhibits fungal protein biosynthesis (445). The primary sites of action of these antibiotics are at locations where chitin synthesis occurs in the cell wall, there is cation leakage from mitochondria, inositol biosynthesis is occurring, or sites of protein and DNA synthesis. The compounds mentioned above are a few examples of agroactive compounds isolated from Actinobacteria. Validamycin A was commercialized by Takeda for the control of pathogens in rice and other plants and as a tool for damping off diseases in vegetable seedlings. On the other hand, some secreted metabolites are cytotoxic and can include chemical structures such as macrolides, α-pyrones, lactones, indoles, terpenes, and quinones (446). For instance, resistomycin, a quinone-related antibiotic, has a unique structure and exhibits bactericidal and vasoconstrictive activity based on the inhibition of RNA and protein synthesis (447, 448).
The genome sequences of important Actinobacteria species reported to date indicate that as much as 90% of the chemical potential of these organisms remains undiscovered and that the biosynthetic machinery encoded by many of these genetic loci may be activatable under laboratory conditions (449). The predictive models of Watve et al. (450) suggested that over 150,000 bioactive metabolites from members of the genus Streptomyces alone within this order are still waiting to be discovered (328, 451). Molecular techniques such as combinatorial biosynthesis may lead to the discovery of drugs that cannot be found naturally and of biosynthetic components that can be interchanged and modified to produce bioactive products with unique properties.
Actinobacteria as Sources of Antifungal Agents
Kasugamycin is a bactericidal and fungicidal metabolite secreted by Streptomyces kasugaensis (452) that acts as an inhibitor of protein biosynthesis in microorganisms but not in mammals. The systemically active kasugamycin was marketed to control rice blast (Pyricularia oryzae cavara) and bacterial Pseudomonas diseases in several crops. In 1965, Isono et al. (453) isolated the first members of a new class of natural fungicides, polyoxins B and D, from metabolites of Streptomyces cacaoi var. asoensis. These substances act by interfering with fungal cell wall synthesis by inhibiting chitin synthase (454). Polyoxin B is applied against a number of fungal pathogens in fruits, vegetables, and ornamentals, while polyoxin D is used to control the causative agent of rice sheath blight, Rhizoctonia solani (455).
In 1968, the validamycin family was detected by researchers at Takeda Chemical Industries in a greenhouse assay for the treatment of sheath blight disease in rice plants caused by the fungus Rhizoctonia solani. Validamycin A, the major and most active component of the complex, was isolated from Streptomyces hygroscopicus var. limoneus. Within the fungal cell, validamycin is converted to validoxylamine A, a particularly strong inhibitor of trehalose that suppresses the breakdown of intracellular trehalose (456). Trehalose is well known as a storage carbohydrate, and trehalase plays an essential role in the transport of glucose in insects and fungi (457). This mode of action gives validamycin A a favorable biological selectivity, since vertebrates do not depend on the hydrolysis of the disaccharide trehalose for their metabolism (457).
INTERACTIONS BETWEEN ACTINOBACTERIA AND OTHER ORGANISMS
Interactions between Actinobacteria and Invertebrates
Insect-bacterium symbioses are widespread in the environment (458), and antibiotic-producing bacterial symbionts are often recruited to protect the host and/or their resources (459, 460). Many insects (e.g., ants, termites, gall midges, and beetles) have developed a specific association with their microbial communities. These interactions are diverse, ranging from antagonism and commensalism to mutualism, and from obligate to facultative (461).
Interaction with ants.
Microbial communities of many groups of insects have been widely studied (462), and particularly complex associations have been documented between gut bacteria and insects (463, 464). Attine ants have evolved a mutualism with Actinobacteria that produce antibiotics that the ants use as weedkillers to keep their fungal gardens free of other microbes (460, 465). For instance, the ants (genera Atta and Acromyrmex) cut leaves and then masticate them into a fine biomass that is fed to the symbiotic fungus (Leucoagaricus gongylophorus) which, in turn, provides lipid- and carbohydrate-rich hyphae known as gongylidia that will be used by the ants (466).
The ants rely on a similar mutualistic association with members of the Actinobacteria (genus Pseudonocardia) that produce antibiotics that help suppress Escovopsis, which has a devastating effect on the fungus gardens of leaf-cutting ants in the absence of the bacterium (467–470), by significantly reducing the colony fitness or even inducing colony death (471). Other Actinobacteria genera may play a similar role (472, 473). The presence and maintenance of Streptomyces bacteria seems to be of prime importance, as the bacteria appear to be the primary defense against Escovopsis, which ants possess (467, 474).
Interactions with beetles.
Multiple bacterial genera of the Gammaproteobacteria and Actinobacteria classes were found in the larvae, pupae, and adult guts of the bark beetle Dendroctonus rhizophagus. The class of Actinobacteria was represented by Ponticoccus gilvus and Kocuria marina, both of which can degrade carboxymethylcellulose in vitro. Neither Actinobacteria species has ever been reported in other bark beetles, suggesting that these bacteria could be involved in the degradation of cellulosic substrates such as pine bark and phloem, enabling them to serve as a carbon source (475). This postulate is supported by the presence of cellulose-degrading bacteria in the gut of insects that feed on woody tree tissues, such as wood-boring beetles, including Saperda vestita and Agrilus planipennis (476, 477).
Interactions with protozoans.
Mycobacterium ulcerans is responsible for a necrotizing cutaneous infection called Buruli ulcer, which has been reported in more than 30 countries worldwide, mainly in tropical and subtropical climates. However, M. ulcerans can probably not live freely due to its natural fragility, slow-growth development, and inability to withstand exposure to direct sunlight. Further, M. ulcerans is sensitive to several antibiotics, such as streptomycin and rifampin. M. ulcerans therefore rarely occurs as a free-living microorganism, even though Streptomyces griseus and Amycolatopsis rifamycinica, producers of streptomycin and rifampin, thrive under such conditions (478, 479).
To survive, Mycobacterium has adapted to a more protected niche by utilizing free-living amoebae (FLA) as carriers. In their protozoan hosts, “hidden” mycobacteria might find easier opportunities to infect vertebrate end hosts, multiplying within protozoans to escape immune reactions (480). This ability to persist within amoebae has been widely documented (481–483). The internalization of infectious agents inside other parasites represents an evolutionary strategy for survival that may sometimes enhance pathogenesis or transmissibility (480).
Interactions between Actinobacteria and Vertebrates
Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, and Proteobacteria dominate the gut microbiota, demonstrating a similar overall composition at the phylum level in various gastrointestinal tract locations, including human gastric fluid, intraoral niches (484, 485), throat (486), distal esophagus (487), stomach mucosa (486), and feces (488). Further, Actinobacteria are prominent among the identified microbiota of the oral cavity but are significantly less abundant in the lower gastrointestinal and genital tracts.
Children with diabetes reportedly exhibit substantially lower numbers of Actinobacteria and Firmicutes compared to healthy children. Further, within the Actinobacteria, the number of Bifidobacterium was significantly lower in children with diabetes (489).
Metagenomic studies of mucosal and fecal samples retrieved from healthy subjects demonstrated the presence of six dominant phylogenetic phyla, including Actinobacteria (490). The role of bifidobacteria in gut ecology illustrates the importance of Actinomyces and other Actinobacteria (491). The phyla Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, and Proteobacteria constitute more than 99% of all gut microbiota in dogs and cats. Compared with the case in humans, which harbor 1018 bacterial cells, Bifidobacteria is less abundant in cats and dogs (492, 493).
Although the aerobic Actinobacteria are infrequently encountered in clinical practice, they are important potential causes of serious human and animal infections. These bacteria have emerged as unusual but important potential human pathogens that cause significant disease, affecting not only immunocompetent hosts but also severely immunocompromised patients. McNeil and Brown (494) reviewed the data on medically important aerobic Actinobacteria and epidemic diseases that these organisms cause in animals and humans. For instance, the genus Nocardia comprises several species that are known to be unusual causes of a wide spectrum of clinical diseases in both humans and animals, infecting cattle, horses, dogs, and swine (494). While the majority of nocardial infections have been attributed to Nocardia asteroides, other pathogenic Nocardia species that have been reported include N. brasiliensis, N. otitidiscaviarum, and N. transvalensis. In a taxonomic revision of the N. asteroides taxon, two new species, N. farcinica and Nocardia nova, were separated (495).
Reports of human infection with Rhodococcus spp. have been rare (494). The disease they cause can have a variable clinical presentation depending upon the host's underlying immune status and possibly upon the site of inoculation and the virulence of the infecting microorganism. Consequently, in severely immunocompromised patients, primary pulmonary Rhodococcus equi infections (pneumonia and lung abscesses) have been reported most frequently (494).
Only a few invasive humans infections, such as mycetoma caused by Streptomyces spp., have been documented to date (496). They can, however, be caused by S. somaliensis and S. sudanensis (497, 498). The majority of invasive Streptomyces infections are associated with bacteremia and lung infections, namely, pneumonia, abscess, and pneumonitis (499). Streptomycetes are infrequent pathogens, although S. somaliensis and S. sudanensis can cause infections.
As for Rhodococcus, most of the infected patients had some underlying immunosuppressive condition, such as HIV infection, cancer, systemic lupus erythematosus (SLE), Crohn's disease, etc. Moreover, S. pelletieri, S. griseus, S. lanatus, and S. albus have been isolated from various patients with lung pathology (496, 500–502). Other opportunistic pathogenic Actinobacteria include Amycolata autotrophica, A. orientalis, Micromonospora spp., N. dassonvillei, and Oerskovia spp. (494).
Interactions between Actinobacteria and Plants
As stated previously, actinomycetes are abundantly present in soils and represent a high proportion of the microbial flora of the rhizosphere (503). Actinobacteria thus unsurprisingly play diverse roles in plant-associated microbial communities. Some genera are viewed predominantly as soil saprophytes with crucial roles in nutrient cycling, while others are endophytes, beneficial symbionts, or even pathogens of plants.
Plant-Actinobacteria deleterious interactions.
(i) Actinobacteria as plant pathogens.
The successful infection of a plant host is a complex multistep process, requiring the pathogen to sense the presence of a suitable host, penetrate and colonize the plant tissue, and survive in the presence of host defense mechanisms. In comparison to other bacteria, actinomycetes play a relatively minor role in plant diseases. However, they represent major pathogens of certain crops in particular areas, and under special conditions affect the quality and the quantity of agricultural products. This may result in huge agricultural losses, especially of potatoes but also of other root crops, such as beet, carrot, parsnip, radish, sweet potato, and turnip (504, 505).
Recent progress in molecular genetics and the understanding of the genomics of plant pathogenicity has been made for the Actinobacterial genera Clavibacter, Streptomyces, Leifsonia, and Rhodococcus. Further, plant-pathogenic Actinobacteria in the genera Streptomyces and Rhodococcus have very wide host ranges, including economically important crops and model plants. For instance, in the genus Streptomyces, plant pathogenicity is rare, with only a dozen or so species possessing this trait of the more than 900 species described. Nevertheless, such species have a significant impact on agricultural economies throughout the world due to their ability to cause important crop diseases, such as potato common scab, which is characterized by lesions that form on the potato tuber surface. The most well-known phytopathogenic Actinobacteria are Streptomyces scabiei, S. acidiscabies, and S. turgidiscabies, which induce devastating scab diseases on a broad spectrum of plants. Streptomyces scabiei, the most ancient of these pathogens, is found worldwide, whereas S. turgidiscabies and S. acidiscabies are emergent pathogens that were first described in Japan and the northeastern United States, respectively (506–508).
S. scabiei and S. turgidiscabies both cause “common” scab in potatoes (Solanum tuberosum); the disease is characterized by gray spores borne in spiral chains and that produce melanin (509). Another disease, the “acid scab,” is associated with S. acidiscabies (510). The typical and acid scab strains differ with respect to pigmentation, spore chain morphology, raffinose utilization, and tolerance of low pH. In addition, S. acidiscabies differs from S. scabiei in phenotype and ecology by having flexuous spore chains, a growth medium-dependent spore mass color ranging from white to salmon-pink, a red or yellow pH-sensitive diffusible pigment, and no melanin.
Russet scab is commonly restricted to nature of the potato's skin and affects the quality of the crop. This disease caused by soilborne streptomycetes different from S. scabies has been reported in Europe and the United States since the beginning of the century (511). The disease is divided into two types. The first type is American russet scab, caused by the genus Streptomyces and species different from S. scabiei (which forms a pigmented mycelium, flexuous spore chains, and no melanin) and from S. acidiscabies (mass spore color, inability to grow at pH 4.5). The second type is European russet (or netted) scab, which is apparently distinct from the American variant (512) with respect to cultivar susceptibility, root attack, and optimum soil temperature.
No specific taxonomic investigations have been carried out on S. ipomoeae, the causal agent of the sweet potato soil rot disease characterized by dwarfed plants with little or no growth and minor discolored leaves, with many plants dying before the end of the season. The organism apparently persists for long periods, even in the absence of the host plant.
In the 1980s, Actinobacteria were reported to plug the xylem vessels of silver, sugar, and Norway maples, leading to early decay and dieback of the tree branches (513, 514). A variety of streptomycetes of different species (S. parvus, S. sparsogenes, Streptomyces sp.) were isolated from the plugs. The isolates were capable of growing within the tree vessels and in vitro in the presence of several phenols. Although sugar maples in the northeastern United States are routinely tapped to collect maple sap for conversion to maple syrup, the mode of penetration of the Actinobacteria into the host is not known. Similarly, a lignocellulose-degrading streptomycete (S. flavovirens) was found to decompose the intact cell walls of the phloem of Douglas firs, and hyphae were found in the cavities deriving from the destruction of the walls of the parenchyma and sclereids (515).
Rhodococcus fascians was first isolated and identified in 1930 as the causal agent of sweet pea fasciations (516). Since then, the symptoms caused by R. fascians in diverse plant species have been described (reviewed by Goethals et al. [517]). The bacterium infects both monocot and dicot hosts, many of which are economically important (518). Extensive epiphytic growth precedes intercellular invasion through stomata. R. fascians causes various effects in its hosts, including leaf deformation and formation of witches' broom, fasciations, and leafy galls (517, 519). The symptoms are caused by the hyperinduction of shoots through activation of dormant axillary meristems and de novo meristem formation, probably as the result of elaborate manipulation of host hormone balances and pathogen-derived auxins and cytokinins (520, 521). In contrast to Streptomyces and Rhodococcus, plant-pathogenic species in the Actinobacteria genera Clavibacter and Leifsonia are host specific at the species or subspecies level.
Clavibacter michiganensis is another aerobic nonsporulating Gram-positive plant-pathogenic member of the Actinobacteria and is currently the only known species within the genus Clavibacter. C. michiganensis is composed of a number of host-specific subspecies, all of which colonize the xylem. Currently, C. michiganensis is represented by five subspecies: C. michiganensis subsp. insidiosus, C. michiganensis subsp. michiganensis, C. michiganensis subsp. nebraskensis, C. michiganensis subsp. sepedonicus, and C. michiganensis subsp. tesselarius.
C. michiganensis subsp. michiganensis provokes bacterial canker formation in tomatoes, a serious emerging disease of tomatoes wherever tomato plants are grown. This disease has caused substantial economic losses worldwide (522). Another important plant pathogen is C. michiganesis subsp. sepedonicus, which causes a disease in potatoes known as ring rot, due to the way it rots the vascular tissue inside potato tubers. C. michiganensis subsp. nebraskensis causes wilt and blight in maize. C. michiganensis subsp. tesselarius induces leaf freckles and leaf spots in wheat. Wilting and stunting in alfalfa (Medicago sativa) are induced by C. michiganensis subsp. insidiosus (523). The severity of these diseases and the difficulty of controlling the spread of the corresponding pathogens has resulted in the pathogens being classified as quarantine organisms under the European Union Plant Health Legislation, as well as the laws of many other countries.
The genus Leifsonia includes xylem-limited, fastidious, bacterial pathogens. The best-known of these pathogens is L. xyli subsp. xyli, the causative agent of a systemic disease called ratoon stunting of sugarcane. Plant growth inhibition, the hallmark of this disease, may be due to a putative fatty acid desaturase that modifies the carotenoid biosynthesis pathway to produce abscisic acid, a growth inhibitor (524).
(ii) Traits of pathogenicity.
The mechanisms by which a pathogen can induce the development of lesions on the host are still not well known, although recent advances in this field have provided some ideas. An important feature in the interaction of a pathogen and its host is the establishment of an equilibrium that allows both partners to survive in nature. If no resistant or tolerant plant cultivars are available, pathogen infection will inevitably lead to disease outbreaks. Despite the economic importance of plant-pathogenic Streptomyces species, very little is known about the molecular mechanisms used by these organisms to sense the presence of a suitable plant host, colonize the host's tissues, and resist its defense mechanisms.
The virulence factors of C. michiganensis subsp. michiganensis seem to be extracellular enzymes, particularly proteases. Dreier et al. (525) reported the participation of three proteases (Pat-1, ChpC, and ChpG) in the pathogen's initial interactions with the host plant, but their exact target remains unknown. Several potential virulence genes are clustered in the chp-tomA region of the bacterial genome, which may be a pathogenicity island and crucial for successful colonization. It was recently shown that the virulence of Clavibacter michiganensis subsp. michiganensis toward tomato plants can be modulated by three different mechanisms: loss of plasmids accompanied by the loss of the pathogenicity factors celA and pat-1, resulting in reduced virulence or even nonvirulence in a plasmid-free derivative; transfer of plasmids to plasmid-free C. michiganensis subsp. michiganensis derivatives, which restores full virulence; and loss of the pathogenicity island due to stress-activated recA-dependent recombination events, which leads to a low-titer colonizer that may carry the plasmids and the pathogenicity factors necessary for effective colonization but which can be considered a nonvirulent endophyte (526).
Ammonium assimilation and nitrogen control in M. tuberculosis have been studied intensively, with a particular focus on glutamine synthetase I (GSI), an enzyme whose extracellular release was identified as a potentially important determinant of pathogenicity (527).
Phytotoxin production is commonly involved in the pathogenicity of Streptomyces. Some early work on potato scab disease demonstrated that darkening of tuber cells during pathogen colonization was a response to the action of a toxin or enzyme secreted by the scab organism. A key virulence determinant in scab-causing streptomycetes is a family of phytotoxic secondary metabolites called thaxtomins, of which thaxtomin A is the most abundant (528). Thaxtomin production is usually positively correlated with pathogenicity. Further, thaxtomin induces a variety of phenotypic changes in the plant host, including cell hypertrophy, root and shoot stunting, tissue necrosis, alterations in plant Ca2+ and H+ ion influxes, inhibition of cellulose synthesis, programmed cell death, and production of the antimicrobial plant phytoalexin scopoletin (529–533). In addition, thaxtomin helps S. scabiei, S. turgidiscabies, and S. acidiscabies penetrate plant cell walls by inhibiting cellulose biosynthesis (reviewed by Loria et al. [505]), which presumably enables the bacterium to secrete proteins onto the host cell membrane. An additional virulence determinant that has been described in plant-pathogenic streptomycetes is a secreted necrogenic protein called Nec1. Thaxtomin and Nec1 were the first virulence determinants to be identified and are thought to contribute to tissue penetration and suppression of plant defense responses, respectively. Thaxtomins are cyclic dipeptides (2,5-diketopiperazines) derived from l-phenylalanine and l-tryptophan, and they contain a 4-nitroindole moiety that is essential for their phytotoxicity (528). The production of thaxtomin A was positively correlated with disease severity (534). Mutants of S. scabiei and S. acidiscabies that were deficient in thaxtomin A biosynthesis did not cause symptoms on potato tubers, establishing the thaxtomins as important pathogenicity determinants (535, 536). Eleven family members have been isolated and characterized and are distinguished by the presence or absence of N-methyl and/or hydroxyl groups. Thaxtomin A, the predominant family member produced by S. scabiei, S. turgidiscabies, and S. acidiscabies, is required for scab disease development (536–538). The S. scabiei coronafacic acid-like biosynthetic cluster contributes to host-pathogen interactions, as demonstrated recently by Bignell et al. (504). In addition, an extracellular esterase from S. scabiei has been characterized, sequenced, and identified as a potential virulence factor whose activity may be regulated by the availability of zinc (539).
Several bacterial and fungal genomes have been reported to encode proteins with distant homology to plant expansins (540). These proteins are required for virulence, have C-terminal expansin-like domains, and are found in many plant-associated microorganisms, including the phytopathogenic Actinobacteria organisms Clavibacter michiganensis subsp. sepedonicus (526, 541). The S. scabiei expansin-like proteins exhibit 66% homology with each other at the amino acid level and are also closely related to putative expansin-like proteins from two nonpathogenic Streptomyces species. It is tempting to speculate that SCAB44951 might be important for plant-microbe interactions, because it can mimic specific plant PR-1 proteins and thus manipulate plant defense responses during infection. Interestingly, the PR-1-type protein identified in S. scabiei is part of the pathogenome and is conserved in S. ipomoeae, further supporting a potential role for this protein in Streptomyces-plant interactions.
Propionibacterium acnes produces abundant porphyrins, which might contribute to skin damage (542). The interaction of porphyrins with oxygen is thought to contribute to keratinocyte damage and consequently to have implications regarding the pathogenesis of progressive macular hypomelanosis (543).
Concanamycins are produced by several Streptomyces species, including S. diastatochromogenes (544), S. neyagawaensis (545), S. graminofaciens (546), and S. scabiei (547, 548). For instance, concanamycins A and B, produced by S. scabiei, exhibit phytotoxic activity (549) and have been proposed (but not proven) to be virulence determinants in that organism. It should be noted that the other concanamycin family members are not produced in S. acidiscabies or S. turgidiscabies (548–550).
Plant-Actinobacteria beneficial interactions.
Actinobacteria are microorganisms capable of colonizing the rhizosphere through their antagonistic and competitive characteristics concerning other soil microorganisms (503). Like other beneficial microorganisms, Actinobacteria can affect plant growth in two general ways, either directly or indirectly. Indirect promotion occurs when they prevent the harmful effects of one or more deleterious microorganisms. This is chiefly done through biocontrol or antagonism toward soil plant pathogens. Specifically, colonization or the biosynthesis of antibiotics (551) and other secondary metabolites can prevent pathogen invasion and establishment. Direct promotion of plant growth occurs when the plant is supplied with a compound that is synthesized by the bacteria, or when the latter otherwise facilitates plant uptake of soil nutrients. Possible contributions of this sort include nitrogen fixation, siderophore synthesis, phytohormone synthesis, and solubilization of minerals to make them available for plant uptake and use (552).
(i) Actinobacteria as biological control agents.
The Actinobacteria are widely recognized for their potential in biocontrol (553–555) because they are important producers of bioactive compounds (556). Over the past 50 years, there have been many studies on the mechanisms by which Actinobacteria might inhibit pathogens in soil, including antibiosis, nutrient competition, production of degradative enzymes, nitrous oxide production, and quorum quenching (557–559). Their adaptability to different environments in the rhizosphere makes them a strong competitor. Some are known for their production of siderophores, which can chelate iron, depriving other organisms of this important micronutrient (560, 561). Siderophore production by S. griseorubiginouse is effective in the fight against Fusarium wilt of banana caused by Fusarium oxysporum f. sp. cubenese (561). Actinobacteria have also been reported to secrete enzymes that degrade the mycelial cell walls of fungal parasites, as described in several studies (556, 562–565). Several chitinolytic enzymes have been identified in some species of Actinobacteria, including S. antibioticus (566), S. aureofaciens (566), S. lividens (567), S. plicatus (568), S. halsteii AJ-7 (569), and S. lydicus WYEC108 (570). A biofungicide containing Streptomyces lydicus WYEC 108 was approved by AG (Natural Industries Inc., TX, USA) and registered in 2004 as Actinovate soluble (EPA registration number 73314-1). The product, which is completely water soluble, is a biofungicide that effectively protects against and controls many common foliar and soilborne diseases.
Actinobacteria are also widely known for their ability to produce antibiotics that allow them to inhibit plant pathogens (556, 571, 572). Trejo-Estrada et al. (564) have shown a correlation between the production of antibiotics in soil Actinobacteria and effectiveness in the fight against plant pathogens. For example, Streptomyces violaceusniger YCED9 produces three antifungal compounds, including nigrecine, geltanamycine, and guanidylfingine, that fight against plant pathogens (564). Similarly, several antibiotics produced by Actinobacteria are currently used in biological control (Table 3).
Millard (573) reported that green manures, or crops grown specifically for biomass to be incorporated into soil, could reduce the infection of potatoes by pathogenic Streptomyces scabiei. Later, in 1926, Sanford (574) noted that Actinomyces scabiei was “very sensitive to the secreted products of many molds and bacteria, some of which prevent its growth,” and suggested that green manures favored the antagonistic bacteria that inhibited the pathogen. Subsequently, Millard and Taylor (575) showed that soil inoculation with a saprophytic (nonpathogenic) Actinomycete isolate could significantly reduce both disease (potato scab) and pathogen populations, concluding that the saprophytic inoculated strain outcompetes the pathogen in soil, thereby reducing plant disease.
Antibiotic-mediated inhibition of pathogens is generally the primary focus in efforts to suppress plant diseases. However, the diversity of secondary metabolites produced by Streptomyces and other species also offers great potential for suppressing fungal, bacterial, oomycete, and nematode pathogens.
(ii) Actinobacteria as plant growth-promoting rhizobacteria.
In attempts to develop commercial biocontrol and plant growth-promoting products using rhizobacteria, it is important to recognize the specific challenges they present. To begin with, the interaction between plant growth-promoting rhizobacteria (PGPR) species and their plant symbionts appears to be specific, even within a crop or cultivar (552, 576). While a rhizobacterium screened for growth promotion may reveal positive effects on one crop, it may have no effect or even retard the growth of another (577, 578). Although rhizobacteria may present unique challenges to our attempts to harness their beneficial attributes, the prospects for improving agriculture by using PGPR for biocontrol seem to be excellent. The first step toward exploiting PGPR species to enhance plant growth will be to better understand the systems that enable them to act as efficient plant growth enhancers.
Since the 1990s, several nitrogen-fixing Actinobacteria have been recognized and found to be associated with plants. Corynebacterium sp. AN1 isolated from the plant forest phyllosphere reduces acetylene and can be regarded as a substitute for nitrogenous fertilizer as a means of promoting maize growth (579). Pseudonocardia dioxanivorans CB1190 isolates, which can grow on 1,4-dioxane as their sole source of carbon and energy, have also been shown to fix dinitrogen (580). Despite the well-documented history of Streptomyces in biocontrol and preliminary evidence of their capacity to enhance plant growth (581), the potential of Streptomyces species as PGPR has not been widely studied. This is surprising because streptomycetes, which generally account for an abundant percentage of the soil microflora, are particularly effective colonizers of plant root systems and are able to endure unfavorable growth conditions by forming spores (582).
Merriman et al. (583) reported the use of Streptomyces griseus (Krainsky) Waksman and Henrici isolates to treat the seeds of barley, oat, wheat, and carrot, in order to increase their growth. The isolate was originally selected for the biological control of the pathogen Rhizoctonia solani. Though the S. griseus isolate did increase the average grain yield, dry foliage weight, tiller number, and advanced head emergence for both wheat and oat relative to controls, the differences were not statistically significant. However, the isolate was more successful as a seed treatment for carrots. Marketable yields were increased over controls by 17% and 15% in two separate field trials. In addition, both trials also provided an improved yield of large- and very-large-grade carrots relative to controls (583). Nearly 20 years later, El-Abyad et al. (584) described the use of three Streptomyces spp. in the control of bacterial, Fusarium, and Verticillium wilts, early blight, and bacterial canker of tomato. In addition, tomato growth was significantly improved by the use of the antagonistic Streptomyces spp. for a seed coating. An increased availability of growth regulators produced by the inoculum was the reason proposed for the improvement in tomato growth, although this has not yet been formally tested (584). While studies conducted by El-Abyad et al. (584) and Merriman et al. (583) reported plant growth enhancement to be a function of the magnitude of inoculation with Streptomyces, but the behavior of the inocula under gnotobiotic conditions and the possible mechanisms of streptomycete-mediated growth promotion should be investigated further.
(iii) Actinobacteria as symbionts.
Streptomyces spp. constitute protective mutualistic symbioses in which the host feeds and protects the bacteria and in return the bacteria provide antibiotics to protect the host, or their resources, from pathogens (585). Other genera of Actinobacteria, namely, Frankia and Micromonospora, form mutualistic symbioses with higher organisms via nitrogen-fixing actinonodules in trees and shrubs (586, 587).
For a long time, diazotrophy in the Actinobacteria was thought to be limited to the genus Frankia. However, molecular studies have increased the number of known nifH-containing Actinobacteria beyond Frankia spp. (587–591). The discovery of these actinobaceria has stimulated further discussion and inquiries on the origin and emergence of diazotrophy among Actinobacteria. Nitrogen-fixing Actinobacteria in the genus Frankia live as soil saprophytes and as endophytic symbionts in over 200 plant species (592). The genus Frankia has a special significance as the nitrogen-fixing partner in a symbiosis with certain nonleguminous plants, most notably of the genera Alnus, Casuarina, and Elaeaginus, permitting these plants to grow well in nitrogen-poor soils. Some species of higher plants are symbiotic, forming endophytic associations with actinomycetes to achieve actinorhizal nitrogen fixation (168). The genus Frankia establishes a symbiotic relationship with many flowering plants. The best-known example is the alder (Alnus), where these Actinobacteria are found in the roots, in nodules where nitrogen gas is allowed to reach the nitrogenase (168).
(iv) Actinobacteria as endophytes.
Endophytic Actinobacteria have been isolated from a wide variety of plants. The most frequently observed species belong to the genera Microbispora, Nocardia, Micromonospora, and Streptomyces, the most abundant (566, 593). Unlike pathogenic streptomycetes, endophytic species persist inside the plant host for long periods of time without causing observable disorder symptoms and lack known virulence determinants shared with phytopathogenic Streptomyces spp. (503). Endophytic streptomycetes may improve the growth of their plant host by producing auxins that promote root growth and development (594, 595). Moreover, endophytic colonization of Pisum sativum with the endophyte S. lydicus improves the frequency of root nodulation by Rhizobium spp., causing enhanced iron and molybdenum assimilation and vigorous plant growth (596).
(v) Actinobacteria as elicitors of plant defense.
In addition to direct toxic effects on other microbes, nitrous oxide production by Streptomyces has been suggested to activate plant defenses, improving the plant's protection against pathogens (597). Recently, Mahmoudi et al. (559) reported that streptomycetes can degrade the signaling compounds that coordinate the expression of genes required for pathogenicity in Pectobacterium carotovorum, suggesting a further mechanism for disease suppression. In addition, the production of chitinases or plant growth-promoting compounds has been reported to contribute to disease suppression by some Streptomyces isolates (598–601).
CONCLUSIONS AND FUTURE PERSPECTIVES
In this review, we have provided a comprehensive overview of the current knowledge concerning the biology of the phylum Actinobacteria and applications of its members in medicine, agriculture, and industry. Distributed in both terrestrial and aquatic ecosystems, its members play a crucial role in the recycling of refractory biomaterials. The diversity of this phylum is large and includes many beneficial but also some pathogenic species. Mycobacterium tuberculosis is carried by 2 billion people in the world and is the causative agent of tuberculosis, killing several million people every year. There are also the scab-causing streptomycetes, which have a broad host range, infect plants, and are known for their ability to cause necrotic scab-like lesions on economically important root and tuber crops, such as potato. On the other hand, the Actinobacteria have numerous clear potential benefits for humans as sources of novel antibiotics, antifungals, anticancer agents, and other secondary metabolites that might be used in medicine or to improve plant growth and resistance to diseases. Actinobacteria are also very promising for biocontrol of pests and as plant growth promoters. With the rapid developments in the fields of genomics, synthetic biology, and ecology and the strong requirement for new antimicrobial compounds to combat antimicrobial resistance, the biology of the Actinobacteria is a highly dynamic research field and we expect to see many new advances in this field in the years to come.
ACKNOWLEDGMENTS
We are grateful to Dennis Claessen for discussions.
This work is supported by the research program “Assessing and reducing environmental risks from plant protection products” funded by the French Ministries in charge of Ecology and Agriculture.
Biographies

Essaid Ait Barka is a professor of Plant Physiology at the University of Reims. He studied Plant Biology at the Cady Ayad University in Marrakesh (1988) and got his Ph.D. from Reims University (1993), on the plant reaction to low temperatures stress. As a postdoc at Laval University (Canada) and Penn State University (USA), he worked as research professor at NSAC (Nova Scotia, Canada). He is interested in the interactions between plants and pathogenic/nonpathogenic microorganisms, and his current research is directed toward basic and applied aspects of using beneficial microorganisms as microbial inoculants to promote plant growth and provide biological resistance against plant biotic and abiotic stresses. His investigations aim to understand the molecular mechanisms of cross talk between plant defense signal transduction pathways and beneficial microorganisms. He has many partnerships with the private and public sectors and is coinventor of two patents in the field of biocontrol.

Christophe Clément is a Professor at the University of Reims, France. He received his Ph.D. in Plant Physiology from the University of Reims, France, in 1994. He obtained the head position of the lab in 2001. His research is focused on the physio-molecular analysis of plant responses to both biotic and abiotic stresses.

Gilles P. van Wezel is a Professor of Molecular Biotechnology, at the Institute of Biology at Leiden University. He studied biochemistry at the Free University in Amsterdam (1987) and performed his Ph.D. investigation in the group of Leendert Bosch in Leiden (1994) on the control of translational genes in Streptomyces. As a postdoc (EU Human Capital Mobility) in the laboratory of Mervyn Bibb at the John Innes Centre in Norwich (United Kingdom), he worked on the control of carbon metabolism in Streptomyces, and the link between sugar utilization and antibiotic production is still a major theme in his research. His current research focuses on the systems biology of growth, cell division, and antibiotic production of Streptomyces. A major application is the activation of silent biosynthetic gene clusters and elucidation of novel molecules with bioactivity against multidrug-resistant pathogens.
REFERENCES
- 1.Ludwig W, Euzéby J, Schumann P, Buss HJ, Trujillo ME, Kämpfer P, Whiteman WB. 2012. Road map of the phylum Actinobacteria, p 1–28. In Goodfellow M, Kämpfer P, Busse HJ, Trujillo ME, Suzuki KI, Ludwig W, Whitman WB (ed), Bergey's manual of systematic bacteriology, vol 5 Springer-Verlag, New York, NY. [Google Scholar]
- 2.Ventura M, Canchaya C, Fitzgerald GF, Gupta RS, van Sinderen D. 2007. Genomics as a means to understand bacterial phylogeny and ecological adaptation: the case of bifidobacteria. Antonie Van Leeuwenhoek 91:351–372. doi: 10.1007/s10482-006-9122-6. [DOI] [PubMed] [Google Scholar]
- 3.Macagnan D, Romeiro RDS, de Souza JT, Pomella AWV. 2006. Isolation of actinomycetes and endospore-forming bacteria from the cacao pod surface and their antagonistic activity against the witches' broom and black pod pathogens. Phytoparasitica 3:122–132. doi: 10.1007/BF02981312. [DOI] [Google Scholar]
- 4.Lechevalier HA, Lechevalier MP. 1965. Classification des actinomycètes aérobies basée sur leur morphologie et leur composition chimique. Ann Inst Pasteur 108:662–673. (In French.) [PubMed] [Google Scholar]
- 5.Zimmerman W. 1980. Degradation of lignin by bacteria. J Biotechnol 13:199–130. [Google Scholar]
- 6.Mayfield CI, Williams ST, Ruddick SM, Hatfield HL. 1972. Studies on the ecology of actinomycetes in soil. IV. Observations on the form and growth of streptomycetes in soil. Soil Biol Biochem 4:79–91. [Google Scholar]
- 7.Goodfellow M, Williams ST. 1983. Ecology of actinomycetes. Annu Rev Microbiol 37:189–216. doi: 10.1146/annurev.mi.37.100183.001201. [DOI] [PubMed] [Google Scholar]
- 8.Williams ST, Vickers JC. 1988. Detection of actinomycetes in the natural environment: problems and perspectives, p 165–270. In Okami Y, Beppu T, Ogawara H (ed), Biology of actinomycetes. Japan Scientific Societies Press, Tokyo, Japan. [Google Scholar]
- 9.Edwards C. 1993. Isolation properties and potential applications of thermophilic actinomycetes. Appl Biochem Biotechnol 42:161–179. doi: 10.1007/BF02788050. [DOI] [Google Scholar]
- 10.Kim SB, Lonsdale J, Seong C-N, Goodfellow M. 2003. Streptacidiphilus gen. nov., acidophilic actinomycetes with wall chemotype I and emendation of the family Streptomycetaceae (Waksman and Henrici (1943)AL) emend. Rainey et al. 1997. Antonie Van Leeuwenhoek 83:107–116. doi: 10.1023/A:1023397724023. [DOI] [PubMed] [Google Scholar]
- 11.Hiltner L, Störmer K. 1903. Studien über die Bakterienflora des Ackerbodens, mit besonderer Berücksichtigung ihres Verhaltens nach einer Behandlung mit Schwefelkohlenstoff und nach Brache. Arb Biol Reichsanst Land-Forstwirtsch 3:443–545. (In German.) [Google Scholar]
- 12.Buchanan RE. 1917. Studies in the nomenclature and classification of the bacteria. II. The primary subdivisions of the Schizomycetes. J Bacteriol 2:155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Omura S, Takahashi Y, Iwai Y, Tanaka H. 1982. Kitasatosporia, a new genus of the order Actinomycetales. J Antibiot 35:1013–1019. doi: 10.7164/antibiotics.35.1013. [DOI] [PubMed] [Google Scholar]
- 14.Wellington EM, Stackebrandt E, Sanders D, Wolstrup J, Jorgensen NO. 1992. Taxonomic status of Kitasatosporia, and proposed unification with Streptomyces on the basis of phenotypic and 16S rRNA analysis and emendation of Streptomyces Waksman and Henrici 1943, 339AL. Int J Syst Bacteriol 42:156–160. doi: 10.1099/00207713-42-1-156. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Wang Y, Ruan J. 1997. A proposal to revive the genus Kitasatospora (Omura, Takahashi, Iwai, and Tanaka 1982). Int J Syst Bacteriol 47:1048–1054. doi: 10.1099/00207713-47-4-1048. [DOI] [PubMed] [Google Scholar]
- 16.Girard G, Willemse J, Zhu H, Claessen D, Bukarasam K, Goodfellow M, van Wezel GP. 2014. Analysis of novel kitasatosporae reveals significant evolutionary changes in conserved developmental genes between Kitasatospora and Streptomyces. Antonie Van Leeuwenhoek 106:365–380. doi: 10.1007/s10482-014-0209-1. [DOI] [PubMed] [Google Scholar]
- 17.Girard G, Traag BA, Sangal V, Mascini N, Hoskisson PA, Goodfellow M, van Wezel GP. 2013. A novel taxonomic marker that discriminates between morphologically complex actinomycetes. Open Biol 3:130073. doi: 10.1098/rsob.130073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirby R. 2011. Chromosome diversity and similarity within the Actinomycetales. FEMS Microbiol Lett 319:1–10. doi: 10.1111/j.1574-6968.2011.02242.x. [DOI] [PubMed] [Google Scholar]
- 19.Gao B, Gupta RS. 2012. Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol Mol Biol Rev 76:66–112. doi: 10.1128/MMBR.05011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhi XY, Li WJ, Stackebrandt E. 2009. An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int J Syst Evol Microbiol 59:589–608. doi: 10.1099/ijs.0.65780-0. [DOI] [PubMed] [Google Scholar]
- 21.Goodfellow M, Kämpfer P, Busse HJ, Trujillo ME, Suzuki K, Ludwig W, Whitman WB (ed). 2012. Bergey's manual of systematic bacteriology, vol 5 The Actinobacteria, part A and B. Springer, New York, NY. [Google Scholar]
- 22.Labeda D. 1987. Actinomycete taxonomy: generic characterization. Dev Ind Microbiol 28:115–121. [Google Scholar]
- 23.Flärdh K, Buttner MJ. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol 7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- 24.van Dissel D, Claessen D, van Wezel GP. 2014. Morphogenesis of Streptomyces in submerged cultures. Adv Appl Microbiol 89:1–45. doi: 10.1016/B978-0-12-800259-9.00001-9. [DOI] [PubMed] [Google Scholar]
- 25.Kalakoutskii LV, Agre NS. 1976. Comparative aspects of development and differentiation in actinomycetes. Bacteriol Rev 40:469–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atlas R. 1997. Principles of microbiology. WCB McGraw-Hill, New York, NY. [Google Scholar]
- 27.Locci R, Schaal KP. 1980. Apical growth in facultative anaerobic actinomycetes as determined by immunofluorescent labeling. Zentralbl Bakteriol A 246:112–118. [PubMed] [Google Scholar]
- 28.Flärdh K. 2003. Growth polarity and cell division in Streptomyces. Curr Opin Microbiol 6:564–571. doi: 10.1016/j.mib.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 29.Letek M, Ordonez E, Vaquera J, Margolin W, Flärdh K, Mateos LM, Gil JA. 2008. DivIVA is required for polar growth in the MreB-lacking rod-shaped actinomycete Corynebacterium glutamicum. J Bacteriol 190:3283–3292. doi: 10.1128/JB.01934-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prauser H, Lechevalier MP, Lechevalier H. 1970. Description of Oerskovia gen. n. to harbor Orskov's motile Nocardia. Appl Microbiol 19:534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ochi K. 1995. Phylogenetic analysis of mycolic acid-containing wall-chemotype IV actinomycetes and allied taxa by partial sequencing of ribosomal protein AT-L30. Int J Syst Bacteriol 45:653–660. doi: 10.1099/00207713-45-4-653. [DOI] [PubMed] [Google Scholar]
- 32.Locci R, Sharples G. 1984. Morphology, p 165–199. In Goodfellow M, Mordarski M, Williams ST (ed), The biology of Actinomycetes. Academic Press, London, United Kingdom. [Google Scholar]
- 33.Cross T, Goodfellow M. 1973. Taxonomy and classification of the actinomycetes. Soc Appl Bacteriol Symp Ser 2:11–112. [PubMed] [Google Scholar]
- 34.Dietz A, Mathews J. 1971. Classification of Streptomyces spore surfaces into five groups. Appl Microbiol 21:527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pridham TG, Hesseltine CW, Benedict RG. 1958. A guide for the classification of streptomycetes according to selected groups; placement of strains in morphological sections. Appl Microbiol 6:52–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dastager S, Dayanand LWJA, Tang SK, Tian XP, Zhi XY, Xu LH, Jiang C. 2006. Separation, identification and analysis of pigment (melanin) production in Streptomyces. Afr J Biotechnol 5:1131–1134. [Google Scholar]
- 37.Manivasagan P, Venkatesan J, Sivakumar K, Kim SK. 2013. Marine actinobacterial metabolites: current status and future perspectives. Microbiol Res 168:311–332. doi: 10.1016/j.micres.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 38.Goodfellow M, Minnikin DE. 1985. Chemical methods in bacterial systematics. Academic Press, London, United Kingdom. [Google Scholar]
- 39.O'Donnell AG. 1988. Recognition of novel actinomycetes, p 69–88. In Goodfellow MM, Williams ST, Mordarski M (ed), Actinomycetes in biotechnology. Academic Press, London, United Kingdom. [Google Scholar]
- 40.Goodfellow M, O'Donnell AG. 1989. Search and discovery of industrially-significant actinomycetes, p 343–383. In Baumberg S, Hunter IS, Rhodes PM (ed), Microbial products: new approach. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 41.Williams ST, Goodfellow M, Alderson G. 1989. Genus Streptomyces Waksman and Henrici 1943, 339AL, p 2452–2492. In Williams ST, Sharpe ME, Holt JG (ed), Bergey's manual of systematic bacteriology, 1st ed, vol 4 Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 42.Berd D. 1973. Laboratory identification of clinically important aerobic actinomycetes. J Appl Microbiol 25:665–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willey JM, Sherwood LM, Woolverton CJ. 2010. Prescott's microbiology, 7th ed McGraw-Hill, New York, NY. [Google Scholar]
- 44.Lechevalier MP, Lechevalier HA. 1980. The chemotaxonomy of actinomycetes, p 225–292. In Dietz A, Thayer DW (ed), Actinomycetes taxonomy, vol A6 Virginia Society of Industrial Microbiology, Arlington, VA. [Google Scholar]
- 45.Bouizgarne B, Ait Ben Aoumar A. 2014. Diversity of plant associated Actinobacteria, p 41–100. In Maheshwari DK. (ed), Bacterial diversity in sustainable agriculture. Springer International, Heidelberg, Germany. [Google Scholar]
- 46.Kroppenstedt R. 1985. Fatty acid and menaquinone analysis of actinomycetes and related organisms, p 173–199. Chemical methods in bacterial systematics. SAB Technical Series 20, Academic Press, London, United Kingdom. [Google Scholar]
- 47.Suzuki K, Goodfellow M, O'Donnell AG. 1993. Cell envelopes and classification, p 195–250. In Goodfellow M, O'Donnell AG (ed), Handbook of new bacterial systematics. Academic Press, London, United Kingdom. [Google Scholar]
- 48.Collins MD, Goodfellow M, Minnikin DE, Alderson G. 1985. Menaquinone composition of mycolic acid-containing actinomycetes and some sporoactinomycetes. J Appl Bacteriol 58:77–86. doi: 10.1111/j.1365-2672.1985.tb01431.x. [DOI] [PubMed] [Google Scholar]
- 49.Collins MD. 1994. Isoprenoid quinones, p 265–309. In Goodfellow M, O'Donnell AG (ed), Chemical methods in prokaryotic systematics. Wiley, Chichester, United Kingdom. [Google Scholar]
- 50.Collins MD, Smida J, Dorsch M, Stackebrandt E. 1988. Tsukamurella gen. nov., harboring Corynebacterium paurometabolum and Rhodococcus aurantiacus. Int J Syst Bacteriol 38:385–391. doi: 10.1099/00207713-38-4-385. [DOI] [Google Scholar]
- 51.Kroppenstedt RM, Stackebrandt E, Goodfellow M. 1990. Taxonomic revision of the actinomycete genera Actinomadura and Microtetraspora. Syst Appl Microbiol 13:148–160. doi: 10.1016/S0723-2020(11)80162-1. [DOI] [Google Scholar]
- 52.Yamada Y, Aoki K, Tahara Y. 1982. The structure of hexa-hydrogenated isoprenoid sidechain menaquinone with nine isoprene units isolated from Actinomadura madurae. J Gen Appl Microbiol 28:321–329. doi: 10.2323/jgam.28.321. [DOI] [Google Scholar]
- 53.Goodfellow M. 1992. The family Nocardiaceae, p 1188–1212. In Balows A, Truper HG, Dworkin M, Harder W, Scheifer K-H (ed), The prokaryotes. Springer, New York, NY. [Google Scholar]
- 54.Tindall BJ, Kampfer P, Euzeby JP, Oren A. 2006. Valid publication of names of prokaryotes according to the rules of nomenclature: past history and current practice. Int J Syst Evol Microbiol 56:2715–2720. doi: 10.1099/ijs.0.64780-0. [DOI] [PubMed] [Google Scholar]
- 55.Goodfellow M. 1989. The genus Rhodococcus Zopf 1891, p 2362–2371. In Williams ST, Sharpe ME, Holt JG (ed), Bergey's manual of systematic bacteriology, 1st ed, vol 4 Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 56.Lechevalier MP. 1977. Lipids in bacterial taxonomy: a taxonomist's view. Crit Rev Microbiol 5:109–210. doi: 10.3109/10408417709102311. [DOI] [PubMed] [Google Scholar]
- 57.Lechevalier MP, De Bievre C, Lechevalier HA. 1977. Chemotaxonomy of aerobic actinomycetes: phospholipid composition. Biochem Syst Ecol 5:249–260. doi: 10.1016/0305-1978(77)90021-7. [DOI] [Google Scholar]
- 58.Lechevalier MP, Stern AE, Lechevalier HA. 1981. Phospholipids in the taxonomy of actinomycetes, p 111–116. In Schaal KP, Pulverer G (ed), Actinomycetes. Gustav Fischer Verlag, Stuttgart, Germany. [Google Scholar]
- 59.Yokota A, Tamura T. 1994. Transfer of Nocardioides fastidiosa Collins and Stackebrandt 1989 to the genus Aeromicrobium as Aeromicrobium fastidiosum comb. nov. Int J Syst Bacteriol 44:608–611. doi: 10.1099/00207713-44-4-608. [DOI] [Google Scholar]
- 60.Rainey FA, Klatte S, Kroppenstedt RM, Stackebrandt E. 1995. Dietzia, a new genus including Dietzia maris comb. nov., formerly Rhodococcus maris. Int J Syst Bacteriol 45:32–36. doi: 10.1099/00207713-45-1-32. [DOI] [PubMed] [Google Scholar]
- 61.Lechevalier MP, Lechevalier H. 1970. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol 20:435–443. doi: 10.1099/00207713-20-4-435. [DOI] [Google Scholar]
- 62.Asano K, Masunaga I, Kawamoto I. 1989. Catellatospora matsumotoense sp. nov. and C. tsunoense sp. nov., actinomycetes found in woodland soils. Int J Syst Bacteriol 39:309–313. doi: 10.1099/00207713-39-3-309. [DOI] [Google Scholar]
- 63.Maltsev II, Kalinovskii AI, Zgurskaya HI, Evtushenko LI. 1992. Tyvelose in Agromyces cell walls. Syst Appl Microbiol 15:187–189. doi: 10.1016/S0723-2020(11)80090-1. [DOI] [Google Scholar]
- 64.Ichikawa N, Oguchi A, Ikeda H, Ishikawa J, Kitani S, Watanabe Y, Nakamura S, Katano Y, Kishi E, Sasagawa M, Ankai A, Fukui S, Hashimoto Y, Kamata S, Otoguro M, Tanikawa S, Nihira T, Horinouchi S, Ohnishi Y, Hayakawa M, Kuzuyama T, Arisawa A, Nomoto F, Miura H, Takahashi Y, Fujita N. 2010. Genome sequence of Kitasatospora setae NBRC 14216T: an evolutionary snapshot of the family Streptomycetaceae. DNA Res 17:393–406. doi: 10.1093/dnares/dsq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim BJ, Kim CJ, Chun J, Koh YH, Lee SH, Hyun JW, Cha CY, Kook YH. 2004. Phylogenetic analysis of the genera Streptomyces and Kitasatospora based on partial RNA polymerase beta-subunit gene (rpoB) sequences. Int J Syst Evol Microbiol 54:593–598. doi: 10.1099/ijs.0.02941-0. [DOI] [PubMed] [Google Scholar]
- 66.Euzéby JP. 1997. List of bacterial names with standing in nomenclature: a folder available on the Internet. Int J Syst Bacteriol 47:590–592. doi: 10.1099/00207713-47-2-590. [DOI] [PubMed] [Google Scholar]
- 67.Raoult D, La Scola B, Lecocq P, Lepidi H, Fournier PE. 2001. Culture and immunological detection of Tropheryma whippelii from the duodenum of a patient with Whipple disease. JAMA 285:1039–1043. doi: 10.1001/jama.285.8.1039. [DOI] [PubMed] [Google Scholar]
- 68.Raoult D, Lepidi H, Harle JR. 2001. Tropheryma whipplei circulating in blood monocytes. N Engl J Med 345:548. doi: 10.1056/NEJM200108163450716. [DOI] [PubMed] [Google Scholar]
- 69.Bentley SD, Maiwald M, Murphy LD, Pallen MJ, Yeats CA, Dover LG, Norbertczak HT, Besra GS, Quail MA, Harris DE, von Herbay A, Goble A, Rutter S, Squares R, Squares S, Barrell BG, Parkhill J, Relman DA. 2003. Sequencing and analysis of the genome of the Whipple's disease bacterium Tropheryma whipplei. Lancet 361:637–644. doi: 10.1016/S0140-6736(03)12597-4. [DOI] [PubMed] [Google Scholar]
- 70.Raoult D, Ogata H, Audic S, Robert C, Suhre K, Drancourt M, Claverie JM. 2003. Tropheryma whipplei Twist: a human pathogenic Actinobacteria with a reduced genome. Genome Res 13:1800–1809. doi: 10.1101/gr.1474603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Desnues B, Al Moussawi K, Fenollar F. 2010. New insights into Whipple's disease and Tropheryma whipplei infections. Microbes Infect 12:1102–1110. doi: 10.1016/j.micinf.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 72.Ingham E. 1999. The immunology of Propionibacterium acnes and acne. Curr Opin Infect Dis 12:191–197. doi: 10.1097/00001432-199906000-00006. [DOI] [PubMed] [Google Scholar]
- 73.Csukas Z, Banizs B, Rozgonyi F. 2004. Studies on the cytotoxic effects of Propionibacterium acnes strains isolated from cornea. Microb Pathog 36:171–174. doi: 10.1016/j.micpath.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 74.Jakab E, Zbinden R, Gubler J, Ruef C, Von Graevenitz A, Krause M. 1996. Severe infections caused by Propionibacterium acnes: an underestimated pathogen in late postoperative infections. Yale J Biol Med 69:477–482. [PMC free article] [PubMed] [Google Scholar]
- 75.Campisano A, Ometto L, Compant S, Pancher M, Antonielli L, Yousaf S, Varotto C, Anfora G, Pertot I, Sessitsch A, Rota-Stabelli O. 2014. Interkingdom transfer of the acne-causing agent, Propionibacterium acnes, from human to grapevine. Mol Biol Evol 31:1059–1065. doi: 10.1093/molbev/msu075. [DOI] [PubMed] [Google Scholar]
- 76.Hirsch AM, Valdés M. 2009. Micromonospora: an important microbe for biomedicine and potentially for biocontrol and biofuels. Soil Biol Biochem 42:536–542. doi: 10.1016/j.soilbio.2009.11.023. [DOI] [Google Scholar]
- 77.Genilloud O. 2012. Genus I. Micromonospora, p 1039–1057. In Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB (ed), Bergey's manual of systematic bacteriology: the Actinobacteria, 2nd ed, vol 5 Springer-Verlag, New York, NY. [Google Scholar]
- 78.Hoskisson PA, Hobbs G, Sharples GP. 2000. Response of Micromonospora echinospora (NCIMB 12744) spores to heat treatment with evidence of a heat activation phenomenon. Lett Appl Microbiol 30:114–117. doi: 10.1046/j.1472-765x.2000.00680.x. [DOI] [PubMed] [Google Scholar]
- 79.Trujillo ME, Bacigalupe R, Pujic P, Igarashi Y, Benito P, Riesco R, Medigue C, Normand P. 2014. Genome features of the endophytic actinobacterium Micromonospora lupini strain Lupac 08: on the process of adaptation to an endophytic life style? PLoS One 9:e108522. doi: 10.1371/journal.pone.0108522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berdy J. 2005. Bioactive microbial metabolites. J Antibiot 58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 81.Das S, Ward LR, Burke C. 2008. Prospects of using marine actinobacteria as probiotics in aquaculture. Appl Microbiol Biotechnol 81:419–429. doi: 10.1007/s00253-008-1731-8. [DOI] [PubMed] [Google Scholar]
- 82.Jensen PR, Mafnas C. 2006. Biogeography of the marine actinomycete Salinispora. Environ Microbiol 8:1881–1888. doi: 10.1111/j.1462-2920.2006.01093.x. [DOI] [PubMed] [Google Scholar]
- 83.Maldonado LA, Stach JE, Pathom-aree W, Ward AC, Bull AT, Goodfellow M. 2005. Diversity of cultivable actinobacteria in geographically widespread marine sediments. Antonie Van Leeuwenhoek 87:11–18. doi: 10.1007/s10482-004-6525-0. [DOI] [PubMed] [Google Scholar]
- 84.Fenical W, Jensen PR, Palladino MA, Lam KS, Lloyd GK, Potts BC. 2009. Discovery and development of the anticancer agent salinosporamide A (NPI-0052). Bioorg Med Chem 17:2175–2180. doi: 10.1016/j.bmc.2008.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jensen PR, Gontang E, Mafnas C, Mincer TJ, Fenical W. 2005. Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ Microbiol 7:1039–1048. doi: 10.1111/j.1462-2920.2005.00785.x. [DOI] [PubMed] [Google Scholar]
- 86.Bose U, Hewavitharana AK, Vidgen ME, Ng YK, Shaw PN, Fuerst JA, Hodson MP. 2014. Discovering the recondite secondary metabolome spectrum of Salinispora species: a study of inter-species diversity. PLoS One 9:e91488. doi: 10.1371/journal.pone.0091488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jensen PR, Williams PG, Oh DC, Zeigler L, Fenical W. 2007. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl Environ Microbiol 73:1146–1152. doi: 10.1128/AEM.01891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Waksman SA. 1961. The Actinomycetes, classification, identification and description of genera and species, vol 2 Williams & Wilkins Company, Baltimore, MD. [Google Scholar]
- 89.Waksman SA. 1967. The actinomycetes: a summary of current knowledge. Roland Press Company, New York, NY. [Google Scholar]
- 90.Embley T, Stackebrandt E. 1994. The molecular phylogency and systematics of the actinomycetes. Annu Rev Microbiol 48:257–289. doi: 10.1146/annurev.mi.48.100194.001353. [DOI] [PubMed] [Google Scholar]
- 91.Chatterjee D, Hunter W, McNeil M, Brennan PJ. 1992. Lipoarabinomannan: multiglycosylated form of the mycobacterial mannosylphosphatidylinositols. J Biol Chem 267:6228–6233. [PubMed] [Google Scholar]
- 92.Daffe M, Brennan PJ, McNeil M. 1990. Predominant structural features of the cell wall arabinogalactan of Mycobacterium tuberculosis as revealed through characterization of oligoglycosyl alditol fragments by gas chromatography/mass spectrometry and by 1H and 13C NMR analyses. J Biol Chem 265:6734–6743. [PubMed] [Google Scholar]
- 93.Pitulle C, Dorsch M, Kazda J, Wolters J, Stackebrandt E. 1992. Phylogeny of rapidly growing members of the genus Mycobacterium. Int J Syst Bacteriol 42:337–343. doi: 10.1099/00207713-42-3-337. [DOI] [PubMed] [Google Scholar]
- 94.Rogall T, Wolters J, Flohr T, Bottger EC. 1990. Towards a phylogeny and definition of species at the molecular level within the genus Mycobacterium. Int J Syst Bacteriol 40:323–330. doi: 10.1099/00207713-40-4-323. [DOI] [PubMed] [Google Scholar]
- 95.Stahl DA, Urbance JW. 1990. The division between fast- and slow-growing species corresponds to natural relationships among the Mycobacteria. J Bacteriol 172:116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Falkinham JO., III 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin Microbiol Rev 9:177–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Collins CH. 2000. The bovine tubercle Bacillus. Br J Biomed Sci 57:234–240. [PubMed] [Google Scholar]
- 98.Jacobson RR, Krahenbuhl JL. 1999. Leprosy. Lancet 353:655–660. doi: 10.1016/S0140-6736(98)06322-3. [DOI] [PubMed] [Google Scholar]
- 99.Hayman J. 1991. Postulated epidemiology of Mycobacterium ulcerans infection. Int J Epidemiol 20:1093–1098. doi: 10.1093/ije/20.4.1093. [DOI] [PubMed] [Google Scholar]
- 100.van der Werf TS, van der Graaf WT, Tappero JW, Asiedu K. 1999. Mycobacterium ulcerans infection. Lancet 354:1013–1018. doi: 10.1016/S0140-6736(99)01156-3. [DOI] [PubMed] [Google Scholar]
- 101.Aronson JD. 1926. Spontaneous tuberculosis in salt water fish. J Infect Dis 39:315–320. doi: 10.1093/infdis/39.4.315. [DOI] [Google Scholar]
- 102.Norden A, Linell F. 1951. A new type of pathogenic Mycobacterium. Nature 168:826. doi: 10.1038/168826a0. [DOI] [PubMed] [Google Scholar]
- 103.Patel SS, Tavana ML, Boger MS, Win SS, Rimawi BH. 2014. Necrotizing soft tissue infection occurring after exposure to Mycobacterium marinum. Case Rep Infect Dis 2014:702613. doi: 10.1155/2014/702613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brown-Elliott BA, Brown JM, Conville PS, Wallace RJ Jr. 2006. Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy. Clin Microbiol Rev 19:259–282. doi: 10.1128/CMR.19.2.259-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Abreu C, Rocha-Pereira N, Sarmento A, Magro F. 2015. Nocardia infections among immunomodulated inflammatory bowel disease patients: a review. World J Gastroenterol 21:6491–6498. doi: 10.3748/wjg.v21.i21.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Simpson RS, Read RC. 2014. Nocardiosis and actinomycosis. Medicine 42:23–25. doi: 10.1016/j.mpmed.2013.10.012. [DOI] [Google Scholar]
- 107.Yorke RF, Rouah E. 2003. Nocardiosis with brain abscess due to an unusual species, Nocardia transvalensis. Arch Pathol Lab Med 127:224–226. [DOI] [PubMed] [Google Scholar]
- 108.Coco WM, Levinson WE, Crist MJ, Hektor HJ, Darzins A, Pienkos PT, Squires CH, Monticello DJ. 2001. DNA shuffling method for generating highly recombined genes and evolved enzymes. Nat Biotechnol 19:354–359. doi: 10.1038/86744. [DOI] [PubMed] [Google Scholar]
- 109.Shigemori H, Komaki H, Yazawa K, Mikami Y, Nemoto A, Tanaka Y, Sasaki T, In Y, Ishida T, Kobayashi J. 1998. Brasilicardin A. A novel tricyclic metabolite with potent immunosuppressive activity from actinomycete Nocardia brasiliensis. J Org Chem 63:6900–6904. [DOI] [PubMed] [Google Scholar]
- 110.Schlaberg R, Huard RC, Della-Latta P. 2008. Nocardia cyriacigeorgica, an emerging pathogen in the United States. J Clin Microbiol 46:265–273. doi: 10.1128/JCM.00937-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sorrell TC, Mitchell DH, Iredell JR, Chen SC-A. 2009. Nocardia species, p 3199–3207. In Mandell GL, Bennett JE, Dolin R (ed), Principles and practice of infectious diseases, 7th ed Churchill Livingstone, Philadelphia, PA. [Google Scholar]
- 112.Mandell G, Douglas R, Bennett J, Dolin R (ed). 2005. Principles and practice of infectious diseases, 6th ed Elsevier/Churchill Livingstone, New York, NY. [Google Scholar]
- 113.Wiesmayr S, Stelzmueller I, Tabarelli W, Bargehr D, Graziadei I, Freund M, Ladurner R, Steurer W, Geltner C, Mark W, Margreiter R, Bonatti H. 2005. Nocardiosis following solid organ transplantation: a single-centre experience. Transpl Int 18:1048–1053. doi: 10.1111/j.1432-2277.2005.00177.x. [DOI] [PubMed] [Google Scholar]
- 114.Matulionyte R, Rohner P, Uckay I, Lew D, Garbino J. 2004. Secular trends of nocardia infection over 15 years in a tertiary care hospital. J Clin Pathol 57:807–812. doi: 10.1136/jcp.2004.016923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Greene CE. 2006. Actinomycosis and nocardiosis. Elsevier, Philadelphia, PA. [Google Scholar]
- 116.Barksdale L. 1970. Corynebacterium diphtheriae and its relatives. Bacteriol Rev 34:378–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Collins MD, Cummins CS. 1986. Genus Corynebacterium. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 118.Leuchtenberger W, Huthmacher K, Drauz K. 2005. Biotechnological production of amino acids and derivatives: current status and prospects. Appl Microbiol Biotechnol 69:1–8. doi: 10.1007/s00253-005-0155-y. [DOI] [PubMed] [Google Scholar]
- 119.Domingo MC, Fournier E, Masse C, Charest H, Bernard K, Cote JC, Tremblay C. 2015. Draft genome sequences of two toxigenic Corynebacterium ulcerans strains. Genome Announc 3(3):e00699-15. doi: 10.1128/genomeA.00699-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ruckert C, Albersmeier A, Winkler A, Tauch A. 2015. Complete genome sequence of Corynebacterium kutscheri DSM 20755, a corynebacterial type strain with remarkably low G+C content of chromosomal DNA. Genome Announce 3(3):e00571-15. doi: 10.1128/genomeA.00571-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fernandez-Natal MI, Soriano F, Ariza-Miguel J, Marrodan-Ciordia T, Acedo A, Hernandez M, Tauch A, Rodriguez-Lazaro D. 2015. Draft genome sequences of Corynebacterium kroppenstedtii CNM633/14 and CNM632/14, multidrug-resistant and antibiotic-sensitive isolates from nodules of granulomatous mastitis patients. Genome Announc 3(3):e00525-15. doi: 10.1128/genomeA.00525-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fernandez-Natal MI, Soriano F, Acedo A, Hernandez M, Tauch A, Rodriguez-Lazaro D. 2015. Draft genome sequences of the two unrelated macrolide-resistant Corynebacterium argentoratense strains CNM 463/05 and CNM 601/08, isolated from patients in the University Hospital of Leon, Spain. Genome Announc 3(4):e00765-15. doi: 10.1128/genomeA.00765-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Donovan C, Heyer A, Pfeifer E, Polen T, Wittmann A, Kramer R, Frunzke J, Bramkamp M. 2015. A prophage-encoded actin-like protein required for efficient viral DNA replication in bacteria. Nucleic Acids Res 43:5002–5016. doi: 10.1093/nar/gkv374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hadfield TL, McEvoy P, Polotsky Y, Tzinserling VA, Yakovlev AA. 2000. The pathology of diphtheria. J Infect Dis 181:S116–S120. doi: 10.1086/315551. [DOI] [PubMed] [Google Scholar]
- 125.Holmes RK. 2000. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J Infect Dis 181(Suppl 1):S156–S167. doi: 10.1086/315554. [DOI] [PubMed] [Google Scholar]
- 126.Mattos-Guaraldi AL, Sampaio JL, Santos CS, Pimenta FP, Pereira GA, Pacheco LG, Miyoshi A, Azevedo V, Moreira LO, Gutierrez FL, Costa JL, Costa-Filho R, Damasco PV, Camello TC, Hirata R Jr. 2008. First detection of Corynebacterium ulcerans producing a diphtheria-like toxin in a case of human with pulmonary infection in the Rio de Janeiro metropolitan area, Brazil. Mem Inst Oswaldo Cruz 103:396–400. doi: 10.1590/S0074-02762008000400014. [DOI] [PubMed] [Google Scholar]
- 127.Hatanaka A, Tsunoda A, Okamoto M, Ooe K, Nakamura A, Miyakoshi M, Komiya T, Takahashi M. 2003. Corynebacterium ulcerans diphtheria in Japan. Emerg Infect Dis 9:752–753. doi: 10.3201/eid0906.020645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dessau RB, Brandt-Christensen M, Jensen OJ, Tonnesen P. 1995. Pulmonary nodules due to Corynebacterium ulcerans. Eur Respir J 8:651–653. [PubMed] [Google Scholar]
- 129.Sing A, Bierschenk S, Heesemann J. 2005. Classical diphtheria caused by Corynebacterium ulcerans in Germany: amino acid sequence differences between diphtheria toxins from Corynebacterium diphtheriae and C. ulcerans. Clin Infect Dis 40:325–326. doi: 10.1086/426687. [DOI] [PubMed] [Google Scholar]
- 130.Jackman PJH, Pitcher DG, Pelczynska S, Borman P. 1987. Classification of Corynebacteria associated with endocarditis (group JK) as Corynebacterium jeikeium sp. nov. Syst Appl Microbiol 9:83–90. doi: 10.1016/S0723-2020(87)80060-7. [DOI] [Google Scholar]
- 131.Funke G, von Graevenitz A, Clarridge JE III, Bernard KA. 1997. Clinical microbiology of coryneform bacteria. Clin Microbiol Rev 10:125–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tsukamura M. 1971. Proposal of a new genus, Gordona, for slightly acid-fast organisms occurring in sputa of patients with pulmonary disease and in soil. J Gen Microbiol 68:15–26. doi: 10.1099/00221287-68-1-15. [DOI] [PubMed] [Google Scholar]
- 133.Hefferan M. 1904. A comparative and experimental study of bacilli producing red pigment. Zentralbl Bakteriol Parasitenkd Abt II 73:74–96. [Google Scholar]
- 134.Stackebrandt E, Sproer C, Rainey FA, Burghardt J, Pauker O, Hippe H. 1997. Phylogenetic analysis of the genus Desulfotomaculum: evidence for the misclassification of Desulfotomaculum guttoideum and description of Desulfotomaculum orientis as Desulfosporosinus orientis gen. nov., comb. nov. Int J Syst Bacteriol 47:1134–1139. doi: 10.1099/00207713-47-4-1134. [DOI] [PubMed] [Google Scholar]
- 135.Bell KS, Philp JC, Aw DW, Christofi N. 1998. The genus Rhodococcus. J Appl Microbiol 85:195–210. doi: 10.1046/j.1365-2672.1998.00525.x. [DOI] [PubMed] [Google Scholar]
- 136.Prescott JF. 1991. Rhodococcus equi: an animal and human pathogen. Clin Microbiol Rev 4:20–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Goodfellow M, Cross T. 1984. Classification, p 7–164. In Goodfellow M, Mordarski M, Williams ST (ed), The biology of the actinomycetes. Academic Press, London, United Kingdom. [Google Scholar]
- 138.Goodfellow M, Lechevalier MP. 1989. Genus Nocardia, p 235–236. In Williams ST, Sharpe ME, Holt JG (ed), Bergey's manual of systematic bacteriology, vol 4 Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 139.Larkin MJ, Kulakov LA, Allen CC. 2005. Biodegradation and Rhodococcus: masters of catabolic versatility. Curr Opin Biotechnol 16:282–290. doi: 10.1016/j.copbio.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 140.Jones AL, Goodfellow M. 2012. Genus IV. Rhodococcus (Zopf 1891) emend. Goodfellow, Alderson and Chun 1998a, p 437–463. In Whitman WB, Goodfellow M, Kämpfer P, Busse HJ, Trujillo ME, Ludwig W (ed), Bergey's manual of systematic bacteriology, vol 5 The Actinobacteria, part A. Springer, New York, NY. [Google Scholar]
- 141.Goodfellow M, Sangal V, Jones AL, Sutcliffe IC. 2015. Charting stormy waters: a commentary on the nomenclature of the equine pathogen variously named Prescottella equi, Rhodococcus equi and Rhodococcus hoagii. Equine Vet J 47:508–509. doi: 10.1111/evj.12399. [DOI] [PubMed] [Google Scholar]
- 142.Evtushenko LI, Dorofeeva LV, Subbotin SA, Cole JR, Tiedje JM. 2000. Leifsonia poae gen. nov., sp. nov., isolated from nematode galls on Poa annua, and reclassification of “Corynebacterium aquaticum” Leifson 1962 as Leifsonia aquatica (ex Leifson 1962) gen. nov., nom. rev., comb. nov. and Clavibacter xyli Davis et al. 1984 with two subspecies as Leifsonia xyli (Davis et al. 1984 gen. nov., comb. nov.). Int J Syst Evol Microbiol 50:371–380. doi: 10.1099/00207713-50-1-371. [DOI] [PubMed] [Google Scholar]
- 143.Dastager SG, Lee JC, Ju YJ, Park DJ, Kim CJ. 2008. Leifsonia bigeumensis sp. nov., isolated from soil on Bigeum Island, Korea. Int J Syst Evol Microbiol 58:1935–1938. doi: 10.1099/ijs.0.65572-0. [DOI] [PubMed] [Google Scholar]
- 144.Dastager SG, Lee JC, Ju YJ, Park DJ, Kim CJ. 2009. Leifsonia kribbensis sp. nov., isolated from soil. Int J Syst Evol Microbiol 59:18–21. doi: 10.1099/ijs.0.001925-0. [DOI] [PubMed] [Google Scholar]
- 145.Hao DC, Ge GB, Yang L. 2008. Bacterial diversity of Taxus rhizosphere: culture-independent and culture-dependent approaches. FEMS Microbiol Lett 284:204–212. doi: 10.1111/j.1574-6968.2008.01201.x. [DOI] [PubMed] [Google Scholar]
- 146.Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl Environ Microbiol 68:2391–2396. doi: 10.1128/AEM.68.5.2391-2396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sutcliffe IC, Hutchings MI. 2007. Putative lipoproteins identified by bioinformatic genome analysis of Leifsonia xyli ssp. xyli, the causative agent of sugarcane ratoon stunting disease. Mol Plant Pathol 8:121–128. doi: 10.1111/j.1364-3703.2006.00377.x. [DOI] [PubMed] [Google Scholar]
- 148.Tissier H. 1900. Recherches sur la flore intestinale normale et pathologique du nourrisson (état normale et pathologique). G. Carre et C. Naud, Paris, France. [Google Scholar]
- 149.Orla-Jensen S. 1924. La classification des bactéries lactiques. Lait 4:468–474. (In French.) [Google Scholar]
- 150.Lievin V, Peiffer I, Hudault S, Rochat F, Brassart D, Neeser JR, Servin AL. 2000. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut 47:646–652. doi: 10.1136/gut.47.5.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ouwehand AC, Salminen S, Isolauri E. 2002. Probiotics: an overview of beneficial effects. Antonie Van Leeuwenhoek 82:279–289. doi: 10.1023/A:1020620607611. [DOI] [PubMed] [Google Scholar]
- 152.Bevilacqua L, Ovidi M, Di Mattia E, Trovatelli LD, Canganella F. 2003. Screening of Bifidobacterium strains isolated from human faeces for antagonistic activities against potentially bacterial pathogens. Microbiol Res 158:179–185. doi: 10.1078/0944-5013-00192. [DOI] [PubMed] [Google Scholar]
- 153.Cheikhyoussef A, Pogori N, Chen H, Tian F, Chen W, Tang J, Zhang H. 2009. Antimicrobial activity and partial characterization of bacteriocin-like inhibitory substances (BLIS) produced by Bifidobacterium infantis BCRC 14602. Food Control 20:553–559. doi: 10.1016/j.foodcont.2008.08.003. [DOI] [Google Scholar]
- 154.Collado MC, Gonzalez A, Gonzalez R, Hernandez M, Ferrus MA, Sanz Y. 2005. Antimicrobial peptides are among the antagonistic metabolites produced by Bifidobacterium against Helicobacter pylori. Int J Antimicrob Agents 25:385–391. doi: 10.1016/j.ijantimicag.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 155.Cheikhyoussef A, Pogori N, Chen H, Zhao J, Tang J, Chen W, Zhang H. 2009. Comparison of three different methods for the isolation of bacteriocin-like inhibitory substances from Bifidobacterium infantis BCRC 14602. J Rapid Methods Automat Microbiol 17:182–194. doi: 10.1111/j.1745-4581.2009.00167.x. [DOI] [Google Scholar]
- 156.Cheikhyoussef A, Cheikhyoussef N, Chen HZJ, Tang J, Zhang H. 2010. Bifidin. I. A new bacteriocin produced by Bifidobacterium infantis BCRC 14602: purification and partial amino acid sequence. Food Control 21:746–753. doi: 10.1016/j.food.cont.2009.11.003. [DOI] [Google Scholar]
- 157.von Ah U. 2006. Identification of Bifidobacterium thermophilum RBL67 isolated from baby faeces and partial purification of its bacteriocin. Swiss Federal Institute of Technology, Zurich, Switzerland. [Google Scholar]
- 158.Harper J, Davis G. 1982. Cell wall analysis of Gardnerella vaginalis (Haemophilus vaginalis). Int J Syst Bacteriol 32:48–50. doi: 10.1099/00207713-32-1-48. [DOI] [Google Scholar]
- 159.Greenwood J, Pickett M. 1980. Transfer of Haemophilus vaginalis Gardner and Dukes to a new genus, Gardnerella: G. vaginalis (Gardner and Dukes) comb nov. Int J Syst Bacteriol 30:170–178. doi: 10.1099/00207713-30-1-170. [DOI] [Google Scholar]
- 160.Kim TK, Thomas SM, Ho M, Sharma S, Reich CI, Frank JA, Yeater KM, Biggs DR, Nakamura N, Stumpf R. 2009. Heterogeneity of vaginal microbial communities within individuals. J Clin Microbiol 47:1181–1189. doi: 10.1128/JCM.00854-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Menard JP, Mazouni C, Salem-Cherif I, Fenollar F, Raoult D, Boubli L, Gamerre M, Bretelle F. 2010. High vaginal concentrations of Atopobium vaginae and Gardnerella vaginalis in women undergoing preterm labor. Obstet Gynecol 115:134–140. doi: 10.1097/AOG.0b013e3181c391d7. [DOI] [PubMed] [Google Scholar]
- 162.Lamont RF, Sobel JD, Akins RA, Hassan SS, Chaiworapongsa T, Kusanovic JP, Romero R. 2011. The vaginal microbiome: new information about genital tract flora using molecular based techniques. BJOG 118:533–549. doi: 10.1111/j.1471-0528.2010.02840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Miyadoh S. 1997. Atlas of actinomycetes. Asakura Publishing Co, Tokyo, Japan. [Google Scholar]
- 164.Aderem A. 2005. Systems biology: its practice and challenges. Cell 121:511–513. doi: 10.1016/j.cell.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 165.Hopwood DA. 2007. Streptomyces in nature and medicine: the antibiotic makers. Oxford University Press, New York, NY. [Google Scholar]
- 166.Benson DR, Silvester WB. 1993. Biology of Frankia strains, actinomycete symbionts of actinorhizal plants. Microbiol Rev 57:293–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Perrine-Walker F, Gherbi H, Imanishi L, Hocher V, Ghodhbane-Gtari F, Lavenus J, Benabdoun FM, Nambiar-Veeti M, Svistoonoff S, Laplaze L. 2011. Symbiotic signaling in actinorhizal symbioses. Curr Protein Peptide Sci 12:156–164. doi: 10.2174/138920311795684896. [DOI] [PubMed] [Google Scholar]
- 168.Pawlowski K, Demchenko KN. 2012. The diversity of actinorhizal symbiosis. Protoplasma 249:967–979. doi: 10.1007/s00709-012-0388-4. [DOI] [PubMed] [Google Scholar]
- 169.Gtari M, Ghodhbane-Gtari F, Nouioui I, Beauchemin N, Tisa LS. 2012. Phylogenetic perspectives of nitrogen-fixing actinobacteria. Arch Microbiol 194:3–11. doi: 10.1007/s00203-011-0733-6. [DOI] [PubMed] [Google Scholar]
- 170.Zhang Z, Wang Y, Ruan J. 1998. Reclassification of Thermomonospora and Microtetraspora. Int J Syst Bacteriol 48:411–422. doi: 10.1099/00207713-48-2-411. [DOI] [PubMed] [Google Scholar]
- 171.Kukolya J, Nagy I, Laday M, Toth E, Oravecz O, Marialigeti K, Hornok L. 2002. Thermobifida cellulolytica sp. nov., a novel lignocellulose-decomposing actinomycete. Int J Syst Evol Microbiol 52:1193–1199. doi: 10.1099/00207713-52-4-1193. [DOI] [PubMed] [Google Scholar]
- 172.Yang LL, Tang SK, Zhang YQ, Zhi XY, Wang D, Xu LH, Li WJ. 2008. Thermobifida halotolerans sp. nov., isolated from a salt mine sample, and emended description of the genus Thermobifida. Int J Syst Evol Microbiol 58:1821–1825. doi: 10.1099/ijs.0.65732-0. [DOI] [PubMed] [Google Scholar]
- 173.Chater KF, Biro S, Lee KJ, Palmer T, Schrempf H. 2010. The complex extracellular biology of Streptomyces. FEMS Microbiol Rev 34:171–198. doi: 10.1111/j.1574-6976.2009.00206.x. [DOI] [PubMed] [Google Scholar]
- 174.Chater KF. 1972. A morphological and genetic mapping study of white colony mutants of Streptomyces coelicolor. J Gen Microbiol 72:9–28. doi: 10.1099/00221287-72-1-9. [DOI] [PubMed] [Google Scholar]
- 175.Donachie WD. 1993. The cell cycle of Escherichia coli. Annu Rev Microbiol 47:199–230. doi: 10.1146/annurev.mi.47.100193.001215. [DOI] [PubMed] [Google Scholar]
- 176.Wildermuth H, Hopwood DA. 1970. Septation during sporulation in Streptomyces coelicolor. J Gen Microbiol 60:51–59. doi: 10.1099/00221287-60-1-51. [DOI] [PubMed] [Google Scholar]
- 177.Claessen D, Rozen DE, Kuipers OP, Sogaard-Andersen L, van Wezel GP. 2014. Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat Rev Microbiol 12:115–124. doi: 10.1038/nrmicro3178. [DOI] [PubMed] [Google Scholar]
- 178.Elliot MA, Buttner MJ, Nodwell JR. 2008. Multicellular development in Streptomyces, p 419–438. In Whitworth DE. (ed), Myxobacteria: multicellularity and differentiation. ASM Press, Washington, DC. [Google Scholar]
- 179.Bibb MJ. 2005. Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol 8:208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 180.van Wezel GP, McDowall KJ. 2011. The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat Prod Rep 28:1311–1333. doi: 10.1039/c1np00003a. [DOI] [PubMed] [Google Scholar]
- 181.Méndez C, Braña AF, Manzanal MB, Hardisson C. 1985. Role of substrate mycelium in colony development in Streptomyces. Can J Microbiol 31:446–450. doi: 10.1139/m85-083. [DOI] [PubMed] [Google Scholar]
- 182.Miguélez EM, Hardisson C, Manzanal MB. 1999. Hyphal death during colony development in Streptomyces antibioticus: morphological evidence for the existence of a process of cell deletion in a multicellular prokaryote. J Cell Biol 145:515–525. doi: 10.1083/jcb.145.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wildermuth H. 1970. Development and organization of the aerial mycelium in Streptomyces coelicolor. J Gen Microbiol 60:43–50. doi: 10.1099/00221287-60-1-43. [DOI] [PubMed] [Google Scholar]
- 184.Fernández M, Sánchez J. 2002. Nuclease activities and cell death processes associated with the development of surface cultures of Streptomyce antibioticus. Microbiology 148:405–412. doi: 10.1099/00221287-148-2-405. [DOI] [PubMed] [Google Scholar]
- 185.Kim IS, Kang SG, Lee KJ. 1995. Physiological importance of trypsin like protease during morphological differentiation of Streptomyces spp. J Microbiol 33:315–321. [Google Scholar]
- 186.Rigali S, Nothaft H, Noens EE, Schlicht M, Colson S, Muller M, Joris B, Koerten HK, Hopwood DA, Titgemeyer F, van Wezel GP. 2006. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol Microbiol 61:1237–1251. doi: 10.1111/j.1365-2958.2006.05319.x. [DOI] [PubMed] [Google Scholar]
- 187.Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, Hopwood DA, van Wezel GP. 2008. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep 9:670–675. doi: 10.1038/embor.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chater KF. 2011. Differentiation in Streptomyces: the properties and programming of diverse cell-types, p 43–86. In Dyson P. (ed), Streptomyces: molecular biology and biotechnology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 189.Manteca A, Ye J, Sanchez J, Jensen ON. 2011. Phosphoproteome analysis of Streptomyces development reveals extensive protein phosphorylation accompanying bacterial differentiation. J Proteome Res 10:5481–5492. doi: 10.1021/pr200762y. [DOI] [PubMed] [Google Scholar]
- 190.Merrick MJ. 1976. A morphological and genetic mapping study of bald colony mutants of Streptomyces coelicolor. J Gen Microbiol 96:299–315. doi: 10.1099/00221287-96-2-299. [DOI] [PubMed] [Google Scholar]
- 191.Hopwood DA, Wildermuth H, Palmer HM. 1970. Mutants of Streptomyces coelicolor defective in sporulation. J Gen Microbiol 61:397–408. doi: 10.1099/00221287-61-3-397. [DOI] [PubMed] [Google Scholar]
- 192.Champness WC. 1988. New loci required for Streptomyces coelicolor morphological and physiological differentiation. J Bacteriol 170:1168–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gehring AM, Nodwell JR, Beverley SM, Losick R. 2000. Genomewide insertional mutagenesis in Streptomyces coelicolor reveals additional genes involved in morphological differentiation. Proc Natl Acad Sci U S A 97:9642–9647. doi: 10.1073/pnas.170059797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hopwood DA. 1999. Forty years of genetics with Streptomyces: from in vivo through in vitro to in silico. Microbiology 145:2183–2202. doi: 10.1099/00221287-145-9-2183. [DOI] [PubMed] [Google Scholar]
- 195.Ryding NJ, Bibb MJ, Molle V, Findlay KC, Chater KF, Buttner MJ. 1999. New sporulation loci in Streptomyces coelicolor A3(2). J Bacteriol 181:5419–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ma H, Kendall K. 1994. Cloning and analysis of a gene cluster from Streptomyces coelicolor that causes accelerated aerial mycelium formation in Streptomyces lividans. J Bacteriol 176:3800–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kawamoto S, Ensign JC. 1995. Isolation of mutants of Streptomyces griseus that sporulate in nutrient rich media: cloning of DNA fragments that suppress the mutations. Actinomycetologica 9:124–135. doi: 10.3209/saj.9_124. [DOI] [Google Scholar]
- 198.Pope MK, Green BD, Westpheling J. 1996. The bld mutants of Streptomyces coelicolor are defective in the regulation of carbon utilization, morphogenesis and cell–cell signalling. Mol Microbiol 19:747–756. doi: 10.1046/j.1365-2958.1996.414933.x. [DOI] [PubMed] [Google Scholar]
- 199.Seo JW, Ohnishi Y, Hirata A, Horinouchi S. 2002. ATP-binding cassette transport system involved in regulation of morphological differentiation in response to glucose in Streptomyces griseus. J Bacteriol 184:91–103. doi: 10.1128/JB.184.1.91-103.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.den Hengst CD, Tran NT, Bibb MJ, Chandra G, Leskiw BK, Buttner MJ. 2010. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol Microbiol 78:361–379. doi: 10.1111/j.1365-2958.2010.07338.x. [DOI] [PubMed] [Google Scholar]
- 201.Elliot MA, Bibb MJ, Buttner MJ, Leskiw BK. 2001. BldD is a direct regulator of key developmental genes in Streptomyces coelicolor A3(2). Mol Microbiol 40:257–269. doi: 10.1046/j.1365-2958.2001.02387.x. [DOI] [PubMed] [Google Scholar]
- 202.Elliot MA, Locke TR, Galibois CM, Leskiw BK. 2003. BldD from Streptomyces coelicolor is a non-essential global regulator that binds its own promoter as a dimer. FEMS Microbiol Lett 225:35–40. doi: 10.1016/S0378-1097(03)00474-9. [DOI] [PubMed] [Google Scholar]
- 203.Bibb MJ, Molle V, Buttner MJ. 2000. sigma(BldN), an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor A3(2). J Bacteriol 182:4606–4616. doi: 10.1128/JB.182.16.4606-4616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chater KF, Bruton CJ, Plaskitt KA, Buttner MJ, Mendez C, Helmann JD. 1989. The developmental fate of S. coelicolor hyphae depends upon a gene product homologous with the motility sigma factor of B. subtilis. Cell 59:133–143. doi: 10.1016/0092-8674(89)90876-3. [DOI] [PubMed] [Google Scholar]
- 205.Méndez C, Chater KF. 1987. Cloning of whiG, a gene critical for sporulation of Streptomyces coelicolor A3(2). J Bacteriol 169:5715–5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Persson J, Chater KF, Flärdh K. 2013. Molecular and cytological analysis of the expression of Streptomyces sporulation regulatory gene whiH. FEMS Microbiol Lett 341:96–105. doi: 10.1111/1574-6968.12099. [DOI] [PubMed] [Google Scholar]
- 207.Ryding NJ, Kelemen GH, Whatling CA, Flärdh K, Buttner MJ, Chater KF. 1998. A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2). Mol Microbiol 29:343–357. doi: 10.1046/j.1365-2958.1998.00939.x. [DOI] [PubMed] [Google Scholar]
- 208.Lawlor EJ, Baylis HA, Chater KF. 1987. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev 1:1305–1310. doi: 10.1101/gad.1.10.1305. [DOI] [PubMed] [Google Scholar]
- 209.Leskiw BK, Mah R, Lawlor EJ, Chater KF. 1993. Accumulation of bldA-specified tRNA is temporally regulated in Streptomyces coelicolor A3(2). J Bacteriol 175:1995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hesketh A, Bucca G, Laing E, Flett F, Hotchkiss G, Smith CP, Chater KF. 2007. New pleiotropic effects of eliminating a rare tRNA from Streptomyces coelicolor, revealed by combined proteomic and transcriptomic analysis of liquid cultures. BMC Genomics 8:261. doi: 10.1186/1471-2164-8-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kim DW, Chater K, Lee KJ, Hesketh A. 2005. Changes in the extracellular proteome caused by the absence of the bldA gene product, a developmentally significant tRNA, reveal a new target for the pleiotropic regulator AdpA in Streptomyces coelicolor. J Bacteriol 187:2957–2966. doi: 10.1128/JB.187.9.2957-2966.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Guyet A, Benaroudj N, Proux C, Gominet M, Coppee JY, Mazodier P. 2014. Identified members of the Streptomyces lividans AdpA regulon involved in differentiation and secondary metabolism. BMC Microbiol 14:81. doi: 10.1186/1471-2180-14-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Higo A, Hara H, Horinouchi S, Ohnishi Y. 2012. Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Res 19:259–273. doi: 10.1093/dnares/dss010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ohnishi Y, Yamazaki H, Kato JY, Tomono A, Horinouchi S. 2005. AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci Biotechnol Biochem 69:431–439. doi: 10.1271/bbb.69.431. [DOI] [PubMed] [Google Scholar]
- 215.Yamazaki H, Tomono A, Ohnishi Y, Horinouchi S. 2004. DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. Mol Microbiol 53:555–572. doi: 10.1111/j.1365-2958.2004.04153.x. [DOI] [PubMed] [Google Scholar]
- 216.Horinouchi S. 2002. A microbial hormone, A-factor, as a master switch for morphological differentiation and secondary metabolism in Streptomyces griseus. Front Biosci 7:d2045–d2057. doi: 10.2741/horinouc. [DOI] [PubMed] [Google Scholar]
- 217.Horinouchi S, Beppu T. 1994. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol Microbiol 12:859–864. doi: 10.1111/j.1365-2958.1994.tb01073.x. [DOI] [PubMed] [Google Scholar]
- 218.Khoklov AS, Tovarova II, Borisova N, Pliner SA, Schevchenko LA, Kornitskaya NS, Ivkina NS, Rapoport IA. 1967. A-faktor, obespecchivaiushchii biosintez streptomitsina mutantnym shtammom Actinomyces streptomycini. Dokl Akad Nauk SSSR 177:232–235. (In Russian.) [PubMed] [Google Scholar]
- 219.Ohnishi Y, Kameyama S, Onaka H, Horinouchi S. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol Microbiol 34:102–111. doi: 10.1046/j.1365-2958.1999.01579.x. [DOI] [PubMed] [Google Scholar]
- 220.Takano E, Chakraburtty R, Nihira T, Yamada Y, Bibb MJ. 2001. A complex role for the gamma-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol Microbiol 41:1015–1028. doi: 10.1046/j.1365-2958.2001.02562.x. [DOI] [PubMed] [Google Scholar]
- 221.Nguyen KT, Tenor J, Stettler H, Nguyen LT, Nguyen LD, Thompson CJ. 2003. Colonial differentiation in Streptomyces coelicolor depends on translation of a specific codon within the adpA gene. J Bacteriol 185:7291–7296. doi: 10.1128/JB.185.24.7291-7296.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Takano E, Tao M, Long F, Bibb MJ, Wang L, Li W, Buttner MJ, Deng ZX, Chater KF. 2003. A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of Streptomyces coelicolor. Mol Microbiol 50:475–486. doi: 10.1046/j.1365-2958.2003.03728.x. [DOI] [PubMed] [Google Scholar]
- 223.Higo A, Horinouchi S, Ohnishi Y. 2011. Strict regulation of morphological differentiation and secondary metabolism by a positive feedback loop between two global regulators AdpA and BldA in Streptomyces griseus. Mol Microbiol 81:1607–1622. doi: 10.1111/j.1365-2958.2011.07795.x. [DOI] [PubMed] [Google Scholar]
- 224.Tschowri N, Schumacher MA, Schlimpert S, Chinnam NB, Findlay KC, Brennan RG, Buttner MJ. 2014. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158:1136–1147. doi: 10.1016/j.cell.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Tenconi E, Urem M, Swiatek-Polatynska MA, Titgemeyer F, Muller YA, van Wezel GP, Rigali S. 2015. Multiple allosteric effectors control the affinity of DasR for its target sites. Biochem Biophys Res Commun 464:324–329. doi: 10.1016/j.bbrc.2015.06.152. [DOI] [PubMed] [Google Scholar]
- 226.Chater KF. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr Opin Microbiol 4:667–673. doi: 10.1016/S1369-5274(01)00267-3. [DOI] [PubMed] [Google Scholar]
- 227.Kelemen GH, Buttner MJ. 1998. Initiation of aerial mycelium formation in Streptomyces. Curr Opin Microbiol 1:656–662. doi: 10.1016/S1369-5274(98)80111-2. [DOI] [PubMed] [Google Scholar]
- 228.Nodwell JR, McGovern K, Losick R. 1996. An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol Microbiol 22:881–893. doi: 10.1046/j.1365-2958.1996.01540.x. [DOI] [PubMed] [Google Scholar]
- 229.Nodwell JR, Yang M, Kuo D, Losick R. 1999. Extracellular complementation and the identification of additional genes involved in aerial mycelium formation in Streptomyces coelicolor. Genetics 151:569–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Willey J, Santamaria R, Guijarro J, Geistlich M, Losick R. 1991. Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by S. coelicolor. Cell 65:641–650. doi: 10.1016/0092-8674(91)90096-H. [DOI] [PubMed] [Google Scholar]
- 231.Willey J, Schwedock J, Losick R. 1993. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes Dev 7:895–903. doi: 10.1101/gad.7.5.895. [DOI] [PubMed] [Google Scholar]
- 232.Kwakman JH, Postma PW. 1994. Glucose kinase has a regulatory role in carbon catabolite repression in Streptomyces coelicolor. J Bacteriol 176:2694–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sanchez S, Chavez A, Forero A, Garcia-Huante Y, Romero A, Sanchez M, Rocha D, Sanchez B, Avalos M, Guzman-Trampe S, Rodriguez-Sanoja R, Langley E, Ruiz B. 2010. Carbon source regulation of antibiotic production. J Antibiot 63:442–459. doi: 10.1038/ja.2010.78. [DOI] [PubMed] [Google Scholar]
- 234.Angell S, Lewis CG, Buttner MJ, Bibb MJ. 1994. Glucose repression in Streptomyces coelicolor A3(2): a likely regulatory role for glucose kinase. Mol Gen Genet 244:135–143. [DOI] [PubMed] [Google Scholar]
- 235.van Wezel GP, Konig M, Mahr K, Nothaft H, Thomae AW, Bibb M, Titgemeyer F. 2007. A new piece of an old jigsaw: glucose kinase is activated posttranslationally in a glucose transport-dependent manner in Streptomyces coelicolor A3(2). J Mol Microbiol Biotechnol 12:67–74. doi: 10.1159/000096461. [DOI] [PubMed] [Google Scholar]
- 236.van Wezel GP, McKenzie NL, Nodwell JR. 2009. Applying the genetics of secondary metabolism in model actinomycetes to the discovery of new antibiotics. Methods Enzymol 458:117–141. doi: 10.1016/S0076-6879(09)04805-8. [DOI] [PubMed] [Google Scholar]
- 237.Pope MK, Green B, Westpheling J. 1998. The bldB gene encodes a small protein required for morphogenesis, antibiotic production, and catabolite control in Streptomyces coelicolor. J Bacteriol 180:1556–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Gubbens J, Janus M, Florea BI, Overkleeft HS, van Wezel GP. 2012. Identification of glucose kinase dependent and independent pathways for carbon control of primary metabolism, development and antibiotic production in Streptomyces coelicolor by quantitative proteomics. Mol Microbiol 86:1490–1507. doi: 10.1111/mmi.12072. [DOI] [PubMed] [Google Scholar]
- 239.Nothaft H, Dresel D, Willimek A, Mahr K, Niederweis M, Titgemeyer F. 2003. The phosphotransferase system of Streptomyces coelicolor is biased for N-acetylglucosamine metabolism. J Bacteriol 185:7019–7023. doi: 10.1128/JB.185.23.7019-7023.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Parche S, Nothaft H, Kamionka A, Titgemeyer F. 2000. Sugar uptake and utilisation in Streptomyces coelicolor: a PTS view to the genome. Antonie Van Leeuwenhoek 78:243–251. doi: 10.1023/A:1010274317363. [DOI] [PubMed] [Google Scholar]
- 241.Colson S, van Wezel GP, Craig M, Noens EE, Nothaft H, Mommaas AM, Titgemeyer F, Joris B, Rigali S. 2008. The chitobiose-binding protein, DasA, acts as a link between chitin utilization and morphogenesis in Streptomyces coelicolor. Microbiology 154:373–382. doi: 10.1099/mic.0.2007/011940-0. [DOI] [PubMed] [Google Scholar]
- 242.Uguru GC, Stephens KE, Stead JA, Towle JE, Baumberg S, McDowall KJ. 2005. Transcriptional activation of the pathway-specific regulator of the actinorhodin biosynthetic genes in Streptomyces coelicolor. Mol Microbiol 58:131–150. doi: 10.1111/j.1365-2958.2005.04817.x. [DOI] [PubMed] [Google Scholar]
- 243.Keijser BJ, van Wezel GP, Canters GW, Kieser T, Vijgenboom E. 2000. The ram-dependence of Streptomyces lividans differentiation is bypassed by copper. J Mol Microbiol Biotechnol 2:565–574. [PubMed] [Google Scholar]
- 244.Lambert S, Traxler MF, Craig M, Maciejewska M, Ongena M, van Wezel GP, Kolter R, Rigali S. 2014. Altered desferrioxamine-mediated iron utilization is a common trait of bald mutants of Streptomyces coelicolor. Metallomics 6:1390–1399. doi: 10.1039/C4MT00068D. [DOI] [PubMed] [Google Scholar]
- 245.Nodwell JR, Losick R. 1998. Purification of an extracellular signaling molecule involved in production of aerial mycelium by Streptomyces coelicolor. J Bacteriol 180:1334–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Viollier PH, Minas W, Dale GE, Folcher M, Thompson CJ. 2001. Role of acid metabolism in Streptomyces coelicolor morphological differentiation and antibiotic biosynthesis. J Bacteriol 183:3184–3192. doi: 10.1128/JB.183.10.3184-3192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Claessen D, Rink R, de Jong W, Siebring J, de Vreugd P, Boersma FG, Dijkhuizen L, Wösten HA. 2003. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev 17:1714–1726. doi: 10.1101/gad.264303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Claessen D, Stokroos I, Deelstra HJ, Penninga NA, Bormann C, Salas JA, Dijkhuizen L, Wosten HA. 2004. The formation of the rodlet layer of streptomycetes is the result of the interplay between rodlins and chaplins. Mol Microbiol 53:433–443. doi: 10.1111/j.1365-2958.2004.04143.x. [DOI] [PubMed] [Google Scholar]
- 249.Elliot MA, Karoonuthaisiri N, Huang J, Bibb MJ, Cohen SN, Kao CM, Buttner MJ. 2003. The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev 17:1727–1740. doi: 10.1101/gad.264403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Glauert AM, Hopwood DA. 1961. The fine structure of Streptomyces violaceoruber (S. coelicolor). III. The walls of the mycelium and spores. J Biophys Biochem Cytol 10:505–516. doi: 10.1083/jcb.10.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wildermuth H, Wehrli E, Horne RW. 1971. The surface structure of spores and aerial mycelium in Streptomyces coelicolor. J Ultrastruct Res 35:168–180. doi: 10.1016/S0022-5320(71)80149-1. [DOI] [PubMed] [Google Scholar]
- 252.Claessen D, de Jong W, Dijkhuizen L, Wösten HA. 2006. Regulation of Streptomyces development: reach for the sky! Trends Microbiol 14:313–319. doi: 10.1016/j.tim.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 253.Wösten HA, Willey JM. 2000. Surface-active proteins enable microbial aerial hyphae to grow into the air. Microbiology 146:767–773. doi: 10.1099/00221287-146-4-767. [DOI] [PubMed] [Google Scholar]
- 254.Wösten HA, van Wetter MA, Lugones LG, van der Mei HC, Busscher HJ, Wessels JG. 1999. How a fungus escapes the water to grow into the air. Curr Biol 9:85–88. doi: 10.1016/S0960-9822(99)80019-0. [DOI] [PubMed] [Google Scholar]
- 255.Sawyer EB, Claessen D, Haas M, Hurgobin B, Gras SL. 2011. The assembly of individual chaplin peptides from Streptomyces coelicolor into functional amyloid fibrils. PLoS One 6:e18839. doi: 10.1371/journal.pone.0018839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Chater KF, Chandra G. 2006. The evolution of development in Streptomyces analysed by genome comparisons. FEMS Microbiol Rev 30:651–672. doi: 10.1111/j.1574-6976.2006.00033.x. [DOI] [PubMed] [Google Scholar]
- 257.Capstick DS, Willey JM, Buttner MJ, Elliot MA. 2007. SapB and the chaplins: connections between morphogenetic proteins in Streptomyces coelicolor. Mol Microbiol 64:602–613. doi: 10.1111/j.1365-2958.2007.05674.x. [DOI] [PubMed] [Google Scholar]
- 258.Claessen D, Wösten HA, van Keulen G, Faber OG, Alves AM, Meijer WG, Dijkhuizen L. 2002. Two novel homologous proteins of Streptomyces coelicolor and Streptomyces lividans are involved in the formation of the rodlet layer and mediate attachment to a hydrophobic surface. Mol Microbiol 44:1483–1492. doi: 10.1046/j.1365-2958.2002.02980.x. [DOI] [PubMed] [Google Scholar]
- 259.Willey JM, Gaskell AA. 2011. Morphogenetic signaling molecules of the streptomycetes. Chem Rev 111:174–187. doi: 10.1021/cr1000404. [DOI] [PubMed] [Google Scholar]
- 260.Willey JM, Willems A, Kodani S, Nodwell JR. 2006. Morphogenetic surfactants and their role in the formation of aerial hyphae in Streptomyces coelicolor. Mol Microbiol 59:731–742. doi: 10.1111/j.1365-2958.2005.05018.x. [DOI] [PubMed] [Google Scholar]
- 261.Di Berardo C, Capstick DS, Bibb MJ, Findlay KC, Buttner MJ, Elliot MA. 2008. Function and redundancy of the chaplin cell surface proteins in aerial hypha formation, rodlet assembly, and viability in Streptomyces coelicolor. J Bacteriol 190:5879–5889. doi: 10.1128/JB.00685-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Capstick DS, Jomaa A, Hanke C, Ortega J, Elliot MA. 2011. Dual amyloid domains promote differential functioning of the chaplin proteins during Streptomyces aerial morphogenesis. Proc Natl Acad Sci U S A 108:9821–9826. doi: 10.1073/pnas.1018715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 264.Tillotson RD, Wosten HA, Richter M, Willey JM. 1998. A surface active protein involved in aerial hyphae formation in the filamentous fungus Schizophillum commune restores the capacity of a bald mutant of the filamentous bacterium Streptomyces coelicolor to erect aerial structures. Mol Microbiol 30:595–602. doi: 10.1046/j.1365-2958.1998.01093.x. [DOI] [PubMed] [Google Scholar]
- 265.Kodani S, Lodato MA, Durrant MC, Picart F, Willey JM. 2005. SapT, a lanthionine-containing peptide involved in aerial hyphae formation in the streptomycetes. Mol Microbiol 58:1368–1380. doi: 10.1111/j.1365-2958.2005.04921.x. [DOI] [PubMed] [Google Scholar]
- 266.Ueda K, Oinuma K, Ikeda G, Hosono K, Ohnishi Y, Horinouchi S, Beppu T. 2002. AmfS, an extracellular peptidic morphogen in Streptomyces griseus. J Bacteriol 184:1488–1492. doi: 10.1128/JB.184.5.1488-1492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Ueda K, Hsheh CW, Tosaki T, Shinkawa H, Beppu T, Horinouchi S. 1998. Characterization of an A-factor-responsive repressor for amfR essential for onset of aerial mycelium formation in Streptomyces griseus. J Bacteriol 180:5085–5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Keijser BJ, van Wezel GP, Canters GW, Vijgenboom E. 2002. Developmental regulation of the Streptomyces lividans ram genes: involvement of RamR in regulation of the ramCSAB operon. J Bacteriol 184:4420–4429. doi: 10.1128/JB.184.16.4420-4429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Nguyen KT, Willey JM, Nguyen LD, Nguyen LT, Viollier PH, Thompson CJ. 2002. A central regulator of morphological differentiation in the multicellular bacterium Streptomyces coelicolor. Mol Microbiol 46:1223–1238. doi: 10.1046/j.1365-2958.2002.03255.x. [DOI] [PubMed] [Google Scholar]
- 270.O'Connor TJ, Nodwell JR. 2005. Pivotal roles for the receiver domain in the mechanism of action of the response regulator RamR of Streptomyces coelicolor. J Mol Biol 351:1030–1047. doi: 10.1016/j.jmb.2005.06.053. [DOI] [PubMed] [Google Scholar]
- 271.Kodani S, Hudson ME, Durrant MC, Buttner MJ, Nodwell JR, Willey JM. 2004. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc Natl Acad Sci U S A 101:11448–11453. doi: 10.1073/pnas.0404220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Willey JM, van der Donk WA. 2007. Lantibiotics: peptides of diverse structure and function. Annu Rev Microbiol 61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- 273.Gaskell AA, Giovinazzo JA, Fonte V, Willey JM. 2012. Multi-tier regulation of the streptomycete morphogenetic peptide SapB. Mol Microbiol 84:501–515. doi: 10.1111/j.1365-2958.2012.08041.x. [DOI] [PubMed] [Google Scholar]
- 274.Kormanec J, Sevcikova B, Sprusansky O, Benada O, Kofronova O, Novakova R, Rezuchova B, Potuckova L, Homerova D. 1998. The Streptomyces aureofaciens homologue of the whiB gene is essential for sporulation; its expression correlates with the developmental stage. Folia Microbiol (Praha) 43:605–612. doi: 10.1007/BF02816376. [DOI] [PubMed] [Google Scholar]
- 275.Keijser BJ, Noens EE, Kraal B, Koerten HK, van Wezel GP. 2003. The Streptomyces coelicolor ssgB gene is required for early stages of sporulation. FEMS Microbiol Lett 225:59–67. doi: 10.1016/S0378-1097(03)00481-6. [DOI] [PubMed] [Google Scholar]
- 276.Jyothikumar V, Tilley EJ, Wali R, Herron PR. 2008. Time-lapse microscopy of Streptomyces coelicolor growth and sporulation. Appl Environ Microbiol 74:6774–6781. doi: 10.1128/AEM.01233-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Jakimowicz D, van Wezel GP. 2012. Cell division and DNA segregation in Streptomyces: how to build a septum in the middle of nowhere? Mol Microbiol 85:393–404. doi: 10.1111/j.1365-2958.2012.08107.x. [DOI] [PubMed] [Google Scholar]
- 278.McCormick JR. 2009. Cell division is dispensable but not irrelevant in Streptomyces. Curr Opin Biotechnol 12:689–698. doi: 10.1016/j.mib.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 279.Bi EF, Lutkenhaus J. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 280.Lowe J, Amos LA. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 281.Lutkenhaus J, Addinall SG. 1997. Bacterial cell division and the Z ring. Annu Rev Biochem 66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 282.Margolin W. 2005. FtsZ and the division of prokaryotic cells and organelles. Nature Rev Mol Cell Biol 6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Romberg L, Levin PA. 2003. Assembly dynamics of the bacterial cell division protein FTSZ: poised at the edge of stability. Annu Rev Microbiol 57:125–154. doi: 10.1146/annurev.micro.57.012903.074300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Grantcharova N, Lustig U, Flärdh K. 2005. Dynamics of FtsZ assembly during sporulation in Streptomyces coelicolor A3(2). J Bacteriol 187:3227–3237. doi: 10.1128/JB.187.9.3227-3237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Schwedock J, Mccormick JR, Angert ER, Nodwell JR, Losick R. 1997. Assembly of the cell division protein FtsZ into ladder like structures in the aerial hyphae of Streptomyces coelicolor. Mol Microbiol 25:847–858. doi: 10.1111/j.1365-2958.1997.mmi507.x. [DOI] [PubMed] [Google Scholar]
- 286.Willemse J, van Wezel GP. 2009. Imaging of Streptomyces coelicolor A3(2) with reduced autofluorescence reveals a novel stage of FtsZ localization. PLoS One 4:e4242. doi: 10.1371/journal.pone.0004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Flärdh K, Leibovitz E, Buttner MJ, Chater KF. 2000. Generation of a non-sporulating strain of Streptomyces coelicolor A3(2) by the manipulation of a developmentally controlled ftsZ promoter. Mol Microbiol 38:737–749. doi: 10.1046/j.1365-2958.2000.02177.x. [DOI] [PubMed] [Google Scholar]
- 288.Willemse J, Mommaas AM, van Wezel GP. 2012. Constitutive expression of ftsZ overrides the whi developmental genes to initiate sporulation of Streptomyces coelicolor. Antonie Van Leeuwenhoek 101:619–632. doi: 10.1007/s10482-011-9678-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.McCormick JR, Su EP, Driks A, Losick R. 1994. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol Microbiol 14:243–254. doi: 10.1111/j.1365-2958.1994.tb01285.x. [DOI] [PubMed] [Google Scholar]
- 290.Bennett JA, Aimino RM, McCormick JR. 2007. Streptomyces coelicolor genes ftsL and divIC play a role in cell division but are dispensable for colony formation. J Bacteriol 189:8982–8992. doi: 10.1128/JB.01303-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Bennett JA, Yarnall J, Cadwallader AB, Kuennen R, Bidey P, Stadelmaier B, McCormick JR. 2009. Medium-dependent phenotypes of Streptomyces coelicolor with mutations in ftsI or ftsW. J Bacteriol 191:661–664. doi: 10.1128/JB.01048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.McCormick JR, Losick R. 1996. Cell division gene ftsQ is required for efficient sporulation but not growth and viability in Streptomyces coelicolor A3(2). J Bacteriol 178:5295–5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Mistry BV, Del Sol R, Wright C, Findlay K, Dyson P. 2008. FtsW is a dispensable cell division protein required for Z-ring stabilization during sporulation septation in Streptomyces coelicolor. J Bacteriol 190:5555–5566. doi: 10.1128/JB.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev 12:3419–3430. doi: 10.1101/gad.12.21.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Raskin DM, de Boer PA. 1997. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell 91:685–694. doi: 10.1016/S0092-8674(00)80455-9. [DOI] [PubMed] [Google Scholar]
- 296.Wu LJ, Errington J. 2012. Nucleoid occlusion and bacterial cell division. Nat Rev Microbiol 10:8–12. doi: 10.1038/nmicro2671. [DOI] [PubMed] [Google Scholar]
- 297.Begg K, Nikolaichik Y, Crossland N, Donachie WD. 1998. Roles of FtsA and FtsZ in activation of division sites. J Bacteriol 180:881–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Hale CA, de Boer PA. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88:175–185. doi: 10.1016/S0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 299.Jakimowicz D, Gust B, Zakrzewska-Czerwinska J, Chater KF. 2005. Developmental-stage-specific assembly of ParB complexes in Streptomyces coelicolor hyphae. J Bacteriol 187:3572–3580. doi: 10.1128/JB.187.10.3572-3580.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Jakimowicz D, Zydek P, Kois A, Zakrzewska-Czerwinska J, Chater KF. 2007. Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae. Mol Microbiol 65:625–641. doi: 10.1111/j.1365-2958.2007.05815.x. [DOI] [PubMed] [Google Scholar]
- 301.Kim HJ, Calcutt MJ, Schmidt FJ, Chater KF. 2000. Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-linked parAB locus. J Bacteriol 182:1313–1320. doi: 10.1128/JB.182.5.1313-1320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Ausmees N, Wahlstedt H, Bagchi S, Elliot MA, Buttner MJ, Flärdh K. 2007. SmeA, a small membrane protein with multiple functions in Streptomyces sporulation including targeting of a SpoIIIE/FtsK-like protein to cell division septa. Mol Microbiol 65:1458–1473. doi: 10.1111/j.1365-2958.2007.05877.x. [DOI] [PubMed] [Google Scholar]
- 303.Dedrick RM, Wildschutte H, McCormick JR. 2009. Genetic interactions of smc, ftsK, and parB genes in Streptomyces coelicolor and their developmental genome segregation phenotypes. J Bacteriol 191:320–332. doi: 10.1128/JB.00858-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Wang L, Vining LC. 2003. Control of growth, secondary metabolism and sporulation in Streptomyces venezuelae ISP5230 by jadW(1), a member of the afsA family of gamma-butyrolactone regulatory genes. Microbiology 149:1991–2004. doi: 10.1099/mic.0.26209-0. [DOI] [PubMed] [Google Scholar]
- 305.Salerno P, Larsson J, Bucca G, Laing E, Smith CP, Flärdh K. 2009. One of the two genes encoding nucleoid-associated HU proteins in Streptomyces coelicolor is developmentally regulated and specifically involved in spore maturation. J Bacteriol 191:6489–6500. doi: 10.1128/JB.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Swiercz JP, Nanji T, Gloyd M, Guarne A, Elliot MA. 2013. A novel nucleoid-associated protein specific to the actinobacteria. Nucleic Acids Res 41:4171–4184. doi: 10.1093/nar/gkt095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Yang YH, Song E, Willemse J, Park SH, Kim WS, Kim EJ, Lee BR, Kim JN, van Wezel GP, Kim BG. 2012. A novel function of Streptomyces integration host factor (sIHF) in the control of antibiotic production and sporulation in Streptomyces coelicolor. Antonie Van Leeuwenhoek 101:479–492. doi: 10.1007/s10482-011-9657-z. [DOI] [PubMed] [Google Scholar]
- 308.Kois A, Swiatek M, Jakimowicz D, Zakrzewska-Czerwinska J. 2009. SMC protein-dependent chromosome condensation during aerial hyphal development in Streptomyces. J Bacteriol 191:310–319. doi: 10.1128/JB.00513-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Facey PD, Hitchings MD, Saavedra-Garcia P, Fernandez-Martinez L, Dyson PJ, Del Sol R. 2009. Streptomyces coelicolor Dps-like proteins: differential dual roles in response to stress during vegetative growth and in nucleoid condensation during reproductive cell division. Mol Microbiol 73:1186–1202. doi: 10.1111/j.1365-2958.2009.06848.x. [DOI] [PubMed] [Google Scholar]
- 310.Traag BA, van Wezel GP. 2008. The SsgA-like proteins in actinomycetes: small proteins up to a big task. Antonie Van Leeuwenhoek 94:85–97. doi: 10.1007/s10482-008-9225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Xu Q, Traag BA, Willemse J, McMullan D, Miller MD, Elsliger MA, Abdubek P, Astakhova T, Axelrod HL, Bakolitsa C, Carlton D, Chen C, Chiu HJ, Chruszcz M, Clayton T, Das D, Deller MC, Duan L, Ellrott K, Ernst D, Farr CL, Feuerhelm J, Grant JC, Grzechnik A, Grzechnik SK, Han GW, Jaroszewski L, Jin KK, Klock HE, Knuth MW, Kozbial P, Krishna SS, Kumar A, Marciano D, Minor W, Mommaas AM, Morse AT, Nigoghossian E, Nopakun A, Okach L, Oommachen S, Paulsen J, Puckett C, Reyes R, Rife CL, Sefcovic N, Tien HJ, Trame CB, van den Bedem H, Wang S, Weekes D, Hodgson KO, Wooley J, Deacon AM, Godzik A, Lesley SA, Wilson I, van Wezel GP. 2009. Structural and functional characterizations of SsgB, a conserved activator of developmental cell division in morphologically complex actinomycetes. J Biol Chem 284:25268–25279. doi: 10.1074/jbc.M109.018564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Kawamoto S, Watanabe H, Hesketh A, Ensign JC, Ochi K. 1997. Expression analysis of the ssgA gene product, associated with sporulation and cell division in Streptomyces griseus. Microbiology 143:1077–1086. doi: 10.1099/00221287-143-4-1077. [DOI] [PubMed] [Google Scholar]
- 313.van Wezel GP, Krabben P, Traag BA, Keijser BJ, Kerste R, Vijgenboom E, Heijnen JJ, Kraal B. 2006. Unlocking Streptomyces spp. for use as sustainable industrial production platforms by morphological engineering. Appl Environ Microbiol 72:5283–5288. doi: 10.1128/AEM.00808-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Jiang H, Kendrick KE. 2000. Characterization of ssfR and ssgA, two genes involved in sporulation of Streptomyces griseus. J Bacteriol 182:5521–5529. doi: 10.1128/JB.182.19.5521-5529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.van Wezel GP, J van der Meulen Kawamoto S, Luiten RG, Koerten HK, Kraal B. 2000. ssgA is essential for sporulation of Streptomyces coelicolor A3(2) and affects hyphal development by stimulating septum formation. J Bacteriol 182:5653–5662. doi: 10.1128/JB.182.20.5653-5662.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. 2011. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev 25:89–99. doi: 10.1101/gad.600211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Del Sol R, Mullins JG, Grantcharova N, Flärdh K, Dyson P. 2006. Influence of CrgA on assembly of the cell division protein FtsZ during development of Streptomyces coelicolor. J Bacteriol 188:1540–1550. doi: 10.1128/JB.188.4.1540-1550.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Plocinski P, Ziolkiewicz M, Kiran M, Vadrevu SI, Nguyen HB, Hugonnet J, Veckerle C, Arthur M, Dziadek J, Cross TA, Madiraju M, Rajagopalan M. 2011. Characterization of CrgA, a new partner of the Mycobacterium tuberculosis peptidoglycan polymerization complexes. J Bacteriol 193:3246–3256. doi: 10.1128/JB.00188-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Celler K, Koning RI, Koster AJ, van Wezel GP. 2013. Multidimensional view of the bacterial cytoskeleton. J Bacteriol 195:1627–1636. doi: 10.1128/JB.02194-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Heichlinger A, Ammelburg M, Kleinschnitz EM, Latus A, Maldener I, Flärdh K, Wohlleben W, Muth G. 2011. The MreB-like protein Mbl of Streptomyces coelicolor A3(2) depends on MreB for proper localization and contributes to spore wall synthesis. J Bacteriol 193:1533–1542. doi: 10.1128/JB.01100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Mazza P, Noens EE, Schirner K, Grantcharova N, Mommaas AM, Koerten HK, Muth G, Flärdh K, van Wezel GP, Wohlleben W. 2006. MreB of Streptomyces coelicolor is not essential for vegetative growth but is required for the integrity of aerial hyphae and spores. Mol Microbiol 60:838–852. doi: 10.1111/j.1365-2958.2006.05134.x. [DOI] [PubMed] [Google Scholar]
- 322.Bagchi S, Tomenius H, Belova LM, Ausmees N. 2008. Intermediate filament-like proteins in bacteria and a cytoskeletal function in Streptomyces. Mol Microbiol 70:1037–1050. doi: 10.1111/j.1365-2958.2008.06473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Holmes NA, Walshaw J, Leggett RM, Thibessard A, Dalton KA, Gillespie MD, Hemmings AM, Gust B, Kelemen GH. 2013. Coiled-coil protein Scy is a key component of a multiprotein assembly controlling polarized growth in Streptomyces. Proc Natl Acad Sci U S A 110:E397–E406. doi: 10.1073/pnas.1210657110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Walshaw J, Gillespie MD, Kelemen GH. 2010. A novel coiled-coil repeat variant in a class of bacterial cytoskeletal proteins. J Struct Biol 170:202–215. doi: 10.1016/j.jsb.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 325.Flärdh K. 2010. Cell polarity and the control of apical growth in Streptomyces. Curr Opin Microbiol 13:758–765. doi: 10.1016/j.mib.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 326.Flärdh K. 2003. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol Microbiol 49:1523–1536. doi: 10.1046/j.1365-2958.2003.03660.x. [DOI] [PubMed] [Google Scholar]
- 327.Ditkowski B, Holmes N, Rydzak J, Donczew M, Bezulska M, Ginda K, Kedzierski P, Zakrzewska-Czerwinska J, Kelemen GH, Jakimowicz D. 2013. Dynamic interplay of ParA with the polarity protein, Scy, coordinates the growth with chromosome segregation in Streptomyces coelicolor. Open Biol 3:130006. doi: 10.1098/rsob.130006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Baltz RH. 2005. Antibiotic discovery from actinomycetes: will a renaissance follow the decline and fall? SIM News 55:186–196. [Google Scholar]
- 329.Xu H, Chater KF, Deng Z, Tao M. 2008. A cellulose synthase-like protein involved in hyphal tip growth and morphological differentiation in Streptomyces. J Bacteriol 190:4971–4978. doi: 10.1128/JB.01849-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Noens EE, Mersinias V, Willemse J, Traag BA, Laing E, Chater KF, Smith CP, Koerten HK, van Wezel GP. 2007. Loss of the controlled localization of growth stage-specific cell-wall synthesis pleiotropically affects developmental gene expression in an ssgA mutant of Streptomyces coelicolor. Mol Microbiol 64:1244–1259. doi: 10.1111/j.1365-2958.2007.05732.x. [DOI] [PubMed] [Google Scholar]
- 331.Bennett JW. 1998. Mycotechnology: the role of fungi in biotechnology. J Biotechnol 66:101–107. doi: 10.1016/S0168-1656(98)00133-3. [DOI] [PubMed] [Google Scholar]
- 332.Hopwood DA, Chater KF, Bibb MJ. 1995. Genetics of antibiotic production in Streptomyces coelicolor A3(2), a model streptomycete. Biotechnology 28:65–102. [DOI] [PubMed] [Google Scholar]
- 333.Iliĉ S, Konstantinoviĉ S, Todoroviĉ Z, Laziĉ M, Veljkoviĉ V, Jokoviĉ N, Radovanoviĉ B. 2007. Characterization and antimicrobial activity of the bioactive metabolites in streptomycete isolates. Mikrobiologiia 76:480–487. (In Russian.) [PubMed] [Google Scholar]
- 334.Rudd BA, Hopwood DA. 1979. Genetics of actinorhodin biosynthesis by Streptomyces coelicolor A3(2). J Gen Microbiol 114:35–43. doi: 10.1099/00221287-114-1-35. [DOI] [PubMed] [Google Scholar]
- 335.Feitelson JS, Malpartida F, Hopwood DA. 1985. Genetic and biochemical characterization of the red gene cluster of Streptomyces coelicolor A3(2). J Gen Microbiol 131:2431–2441. [DOI] [PubMed] [Google Scholar]
- 336.Hopwood DA, Wright HM. 1983. CDA is a new chromosomally-determined antibiotic from Streptomyces coelicolor A3(2). J Gen Microbiol 129:3575–3579. [DOI] [PubMed] [Google Scholar]
- 337.Wright LF, Hopwood DA. 1976. Identification of the antibiotic determined by the SCP1 plasmid of Streptomyces coelicolor A3(2). J Gen Microbiol 95:96–106. doi: 10.1099/00221287-95-1-96. [DOI] [PubMed] [Google Scholar]
- 338.Challis GL, Hopwood DA. 2003. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc Natl Acad Sci U S A 100(Suppl 2):S14555–S14561. doi: 10.1073/pnas.1934677100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Pawlik K, Kotowska M, Chater KF, Kuczek K, Takano E. 2007. A cryptic type I polyketide synthase (cpk) gene cluster in Streptomyces coelicolor A3(2). Arch Microbiol 187:87–99. doi: 10.1007/s00203-006-0176-7. [DOI] [PubMed] [Google Scholar]
- 340.Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol 21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 341.Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S. 2008. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J Bacteriol 190:4050–4060. doi: 10.1128/JB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF. 2007. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotechnol 25:447–453. doi: 10.1038/nbt1297. [DOI] [PubMed] [Google Scholar]
- 343.Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. 2007. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci U S A 104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Gross H. 2009. Genomic mining: a concept for the discovery of new bioactive natural products. Curr Opin Drug Discov Dev 12:207–219. [PubMed] [Google Scholar]
- 345.Medema MH, Breitling R, Bovenberg R, Takano E. 2011. Exploiting plug-and-play synthetic biology for drug discovery and production in microorganisms. Nat Rev Microbiol 9:131–137. doi: 10.1038/nrmicro2478. [DOI] [PubMed] [Google Scholar]
- 346.Nett M, Ikeda H, Moore BS. 2009. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep 26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Zerikly M, Challis GL. 2009. Strategies for the discovery of new natural products by genome mining. Chembiochem 10:625–633. doi: 10.1002/cbic.200800389. [DOI] [PubMed] [Google Scholar]
- 348.Baltz RH. 2008. Renaissance in antibacterial discovery from actinomycetes. Curr Opin Pharmacol 8:557–563. doi: 10.1016/j.coph.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 349.Ochi K, Tanaka Y, Tojo S. 2014. Activating the expression of bacterial cryptic genes by rpoB mutations in RNA polymerase or by rare earth elements. J Ind Microbiol Biotechnol 41:403–414. doi: 10.1007/s10295-013-1349-4. [DOI] [PubMed] [Google Scholar]
- 350.Rutledge PJ, Challis GL. 2015. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol 13:509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 351.Yoon V, Nodwell JR. 2014. Activating secondary metabolism with stress and chemicals. J Ind Microbiol Biotechnol 41:415–424. doi: 10.1007/s10295-013-1387-y. [DOI] [PubMed] [Google Scholar]
- 352.Zhu H, Sandiford SK, van Wezel GP. 2014. Triggers and cues that activate antibiotic production by actinomycetes. J Ind Microbiol Biotechnol 41:371–386. doi: 10.1007/s10295-013-1309-z. [DOI] [PubMed] [Google Scholar]
- 353.Colson S, Stephan J, Hertrich T, Saito A, van Wezel GP, Titgemeyer F, Rigali S. 2007. Conserved cis-acting elements upstream of genes composing the chitinolytic system of streptomycetes are DasR-responsive elements. J Mol Microbiol Biotechnol 12:60–66. doi: 10.1159/000096460. [DOI] [PubMed] [Google Scholar]
- 354.Delic I, Robbins P, Westpheling J. 1992. Direct repeat sequences are implicated in the regulation of two Streptomyces chitinase promoters that are subject to carbon catabolite control. Proc Natl Acad Sci U S A 89:1885–1889. doi: 10.1073/pnas.89.5.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Saito A, Fujii T, Miyashita K. 2003. Distribution and evolution of chitinase genes in Streptomyces species: involvement of gene-duplication and domain-deletion. Antonie Van Leeuwenhoek 84:7–15. doi: 10.1023/A:1024463113606. [DOI] [PubMed] [Google Scholar]
- 356.Nothaft H, Rigali S, Boomsma B, Swiatek M, McDowall KJ, van Wezel GP, Titgemeyer F. 2010. The permease gene nagE2 is the key to N-acetylglucosamine sensing and utilization in Streptomyces coelicolor and is subject to multi-level control. Mol Microbiol 75:1133–1144. doi: 10.1111/j.1365-2958.2009.07020.x. [DOI] [PubMed] [Google Scholar]
- 357.Postma PW, Lengeler JW, Jacobson GR. 1993. Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol Rev 57:543–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Titgemeyer F, Walkenhorst J, Reizer J, Stuiver MH, Cui X, Saier MH Jr. 1995. Identification and characterization of phosphoenolpyruvate:fructose phosphotransferase systems in three Streptomyces species. Microbiology 141:51–58. doi: 10.1099/00221287-141-1-51. [DOI] [PubMed] [Google Scholar]
- 359.Saito A, Shinya T, Miyamoto K, Yokoyama T, Kaku H, Minami E, Shibuya N, Tsujibo H, Nagata Y, Ando A, Fujii T, Miyashita K. 2007. The dasABC gene cluster, adjacent to dasR, encodes a novel ABC transporter for the uptake of N,N′-diacetylchitobiose in Streptomyces coelicolor A3(2). Appl Environ Microbiol 73:3000–3008. doi: 10.1128/AEM.02612-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Swiatek MA, Tenconi E, Rigali S, van Wezel GP. 2012. Functional analysis of the N-acetylglucosamine metabolic genes of Streptomyces coelicolor and role in control of development and antibiotic production. J Bacteriol 194:1136–1144. doi: 10.1128/JB.06370-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Nazari B, Kobayashi M, Saito A, Hassaninasab A, Miyashita K, Fujii T. 2013. Chitin-induced gene expression in secondary metabolic pathways of Streptomyces coelicolor A3(2) grown in soil. Appl Environ Microbiol 79:707–713. doi: 10.1128/AEM.02217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Craig M, Lambert S, Jourdan S, Tenconi E, Colson S, Maciejewska M, Ongena M, Martin JF, van Wezel G, Rigali S. 2012. Unsuspected control of siderophore production by N-acetylglucosamine in streptomycetes. Environ Microbiol Rep 4:512–521. doi: 10.1111/j.1758-2229.2012.00354.x. [DOI] [PubMed] [Google Scholar]
- 363.Świątek-Połatyńska MA, Bucca G, Laing E, Gubbens J, Titgemeyer F, Smith CP, Rigali S, van Wezel GP. 2015. Genome-wide analysis of in vivo binding of the master regulator DasR in Streptomyces coelicolor identifies novel non-canonical targets. PLoS One 10:e0122479. doi: 10.1371/journal.pone.0122479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Gomez-Escribano JP, Song L, Fox DJ, Yeo V, Bibb MJ, Challis GL. 2012. Structure and biosynthesis of the unusual polyketide alkaloid coelimycin P1, a metabolic product of the cpk gene cluster of Streptomyces coelicolor M145. Chem Sci 3:2716–2720. doi: 10.1039/c2sc20410j. [DOI] [Google Scholar]
- 365.Abrudan MI, Smakman F, Grimbergen AJ, Westhoff S, Miller EL, van Wezel GP, Rozen DE. 2015. Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc Natl Acad Sci U S A 112:11054–11059. doi: 10.1073/pnas.1504076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Davies J, Spiegelman GB, Yim G. 2006. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol 9:445–453. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 367.Linares JF, Gustafsson I, Baquero F, Martinez JL. 2006. Antibiotics as intermicrobial signaling agents instead of weapons. Proc Natl Acad Sci U S A 103:19484–19489. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Romero D, Traxler MF, Lopez D, Kolter R. 2011. Antibiotics as signal molecules. Chem Rev 111:5492–5505. doi: 10.1021/cr2000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 370.Stevens DL, Ma Y, Salmi DB, McIndoo E, Wallace RJ, Bryant AE. 2007. Impact of antibiotics on expression of virulence-associated exotoxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus. J Infect Dis 195:202–211. doi: 10.1086/510396. [DOI] [PubMed] [Google Scholar]
- 371.Fajardo A, Martinez JL. 2008. Antibiotics as signals that trigger specific bacterial responses. Curr Opin Microbiol 11:161–167. doi: 10.1016/j.mib.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 372.Angell S, Bench BJ, Williams H, Watanabe CM. 2006. Pyocyanin isolated from a marine microbial population: synergistic production between two distinct bacterial species and mode of action. Chem Biol 13:1349–1359. doi: 10.1016/j.chembiol.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 373.Konig CC, Scherlach K, Schroeckh V, Horn F, Nietzsche S, Brakhage AA, Hertweck C. 2013. Bacterium induces cryptic meroterpenoid pathway in the pathogenic fungus Aspergillus fumigatus. Chembiochem 14:938–942. doi: 10.1002/cbic.201300070. [DOI] [PubMed] [Google Scholar]
- 374.Oh DC, Kauffman CA, Jensen PR, Fenical W. 2007. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J Nat Prod 70:515–520. doi: 10.1021/np060381f. [DOI] [PubMed] [Google Scholar]
- 375.Schroeckh V, Scherlach K, Nutzmann HW, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA. 2009. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci U S A 106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Wu C, Zacchetti B, Ram AF, van Wezel GP, Claessen D, Hae Choi Y. 2015. Expanding the chemical space for natural products by Aspergillus-Streptomyces co-cultivation and biotransformation. Sci Rep 5:10868. doi: 10.1038/srep10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Haseltine WA, Block R. 1973. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci U S A 70:1564–1568. doi: 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Potrykus K, Cashel M. 2008. (p) ppGpp: still magical? Annu Rev Microbiol 62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 379.Ochi K. 1990. A relaxed (rel) mutant of Streptomyces coelicolor A3(2) with a missing ribosomal protein lacks the ability to accumulate ppGpp, A-factor and prodigiosin. J Gen Microbiol 136:2405–2412. doi: 10.1099/00221287-136-12-2405. [DOI] [PubMed] [Google Scholar]
- 380.Chakraburtty R, Bibb M. 1997. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J Bacteriol 179:5854–5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Hesketh A, Chen WJ, Ryding J, Chang S, Bibb M. 2007. The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2). Genome Biol 8:R161. doi: 10.1186/gb-2007-8-8-r161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Battesti A, Bouveret E. 2006. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol Microbiol 62:1048–1063. doi: 10.1111/j.1365-2958.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- 383.Martin SM, Bushell ME. 1996. Effect of hyphal micromorphology on bioreactor performance of antibiotic-producing Saccharopolyspora erythraea cultures. Microbiology 142:1783–1788. doi: 10.1099/13500872-142-7-1783. [DOI] [Google Scholar]
- 384.Yin P, Wang YH, Zhang SL, Chu J, Zhuang YP, Wang ML, Zhou J. 2008. Isolation of soluble proteins from an industrial strain Streptomyces avermitilis in complex culture medium for two-dimensional gel electrophoresis. J Microbiol Methods 73:105–110. doi: 10.1016/j.mimet.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 385.Bewick MW, Williams ST, Veltkamp C. 1976. Growth and ultrastructure of Streptomyces venezuelae during chloramphenicol production. Microbios 16:191–199. [PubMed] [Google Scholar]
- 386.Glazebrook MA, Doull JL, Stuttard C, Vining LC. 1990. Sporulation of Streptomyces venezuelae in submerged cultures. J Gen Microbiol 136:581–588. doi: 10.1099/00221287-136-3-581. [DOI] [PubMed] [Google Scholar]
- 387.van Dissel D, Claessen D, Roth M, van Wezel GP. 2015. A novel locus for mycelial aggregation forms a gateway to improved Streptomyces cell factories. Microb Cell Fact 14:44. doi: 10.1186/s12934-015-0224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Roth M, Noack D, Geuther R. 1985. Maintenance of the recombinant plasmid pIJ2 in chemostat cultures of Streptomyces lividans 66 (pIJ2). J Basic Microbiol 25:265–271. doi: 10.1002/jobm.3620250407. [DOI] [Google Scholar]
- 389.Liu G, Xing M, Han Q. 2005. A population-based morphologically structured model for hyphal growth and product formation in streptomycin fermentation. World J Microbiol Biotechnol 21:1329–1338. doi: 10.1007/s11274-005-3648-z. [DOI] [Google Scholar]
- 390.Meyerhoff J, Tiller V, Bellgardt KH. 1995. Two mathematical models for the development of a single microbial pellet. Bioprocess Eng 12:305–313. doi: 10.1007/BF00369507. [DOI] [Google Scholar]
- 391.Tough AJ, Prosser JI. 1996. Experimental verification of a mathematical model for pelleted growth of Streptomyces coelicolor A3(2) in submerged batch culture. Microbiology 142:639–648. doi: 10.1099/13500872-142-3-639. [DOI] [PubMed] [Google Scholar]
- 392.Celler K, Picioreanu C, van Loosdrecht MC, van Wezel GP. 2012. Structured morphological modeling as a framework for rational strain design of Streptomyces species. Antonie Van Leeuwenhoek 102:409–423. doi: 10.1007/s10482-012-9760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Nieminen L, Webb S, Smith MC, Hoskisson PA. 2013. A flexible mathematical model platform for studying branching networks: experimentally validated using the model actinomycete, Streptomyces coelicolor. PLoS One 8:e54316. doi: 10.1371/journal.pone.0054316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Willemse J, Ruban-Osmialowska B, Widdick D, Celler K, Hutchings MI, van Wezel GP, Palmer T. 2012. Dynamic localization of Tat protein transport machinery components in Streptomyces coelicolor. J Bacteriol 194:6272–6281. doi: 10.1128/JB.01425-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Liu G, Chater KF, Chandra G, Niu G, Tan H. 2013. Molecular regulation of antibiotic biosynthesis in streptomyces. Microbiol Mol Biol Rev 77:112–143. doi: 10.1128/MMBR.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Olmos E, Mehmood N, Haj Husein L, Goergen JL, Fick M, Delaunay S. 2013. Effects of bioreactor hydrodynamics on the physiology of Streptomyces. Bioproc Biosyst Eng 36:259–272. doi: 10.1007/s00449-012-0794-1. [DOI] [PubMed] [Google Scholar]
- 397.Petkovic H, Cullum J, Hranueli D, Hunter IS, Peric-Concha N, Pigac J, Thamchaipenet A, Vujaklija D, Long PF. 2006. Genetics of Streptomyces rimosus, the oxytetracycline producer. Microbiol Mol Biol Rev 70:704–728. doi: 10.1128/MMBR.00004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Vanek Z, Janecek J. 1997. The physiology and biosynthesis of secondary metabolites. Acta Biol Hung 48:339–358. [PubMed] [Google Scholar]
- 399.Wietzorrek A, Bibb M. 1997. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol Microbiol 25:1181–1184. doi: 10.1046/j.1365-2958.1997.5421903.x. [DOI] [PubMed] [Google Scholar]
- 400.Gramajo HC, Takano E, Bibb MJ. 1993. Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolor A3(2) is transcriptionally regulated. Mol Microbiol 7:837–845. doi: 10.1111/j.1365-2958.1993.tb01174.x. [DOI] [PubMed] [Google Scholar]
- 401.Takano E, Gramajo HC, Strauch E, Andres N, White J, Bibb MJ. 1992. Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase-dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2). Mol Microbiol 6:2797–2804. doi: 10.1111/j.1365-2958.1992.tb01459.x. [DOI] [PubMed] [Google Scholar]
- 402.Tomono A, Tsai Y, Yamazaki H, Ohnishi Y, Horinouchi S. 2005. Transcriptional control by A-factor of strR, the pathway-specific transcriptional activator for streptomycin biosynthesis in Streptomyces griseus. J Bacteriol 187:5595–5604. doi: 10.1128/JB.187.16.5595-5604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Claesen J, Bibb M. 2010. Genome mining and genetic analysis of cypemycin biosynthesis reveal an unusual class of posttranslationally modified peptides. Proc Natl Acad Sci U S A 107:16297–16302. doi: 10.1073/pnas.1008608107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Flinspach K, Westrich L, Kaysser L, Siebenberg S, Gomez-Escribano JP, Bibb M, Gust B, Heide L. 2010. Heterologous expression of the biosynthetic gene clusters of coumermycin A(1), clorobiocin and caprazamycins in genetically modified Streptomyces coelicolor strains. Biopolymers 93:823–832. doi: 10.1002/bip.21493. [DOI] [PubMed] [Google Scholar]
- 405.Gverzdys TA. 2011. The development of protocols to engineer and screen Streptomyces in high throughput to test for the activation of cryptic clusters by the heterologous expression of pleiotropic regulators. M.S. dissertation. McMaster University, Hamilton, Ontario, Canada. [Google Scholar]
- 406.Komatsu M, Komatsu K, Koiwai H, Yamada Y, Kozone I, Izumikawa M, Hashimoto J, Takagi M, Omura S, Shin-ya K, Cane DE, Ikeda H. 2013. Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Synth Biol 2:384–396. doi: 10.1021/sb3001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H. 2010. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci U S A 107:2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Medema MH, Breitling R, Takano E. 2011. Synthetic biology in Streptomyces bacteria. Methods Enzymol 497:485–502. doi: 10.1016/B978-0-12-385075-1.00021-4. [DOI] [PubMed] [Google Scholar]
- 409.Waksman SA, Woodruff HB. 1940. Bacteriostatic and bactericidal substances produced by a soil actinomyces. Proc Soc Exp Biol Med 45:609. doi: 10.3181/00379727-45-11768. [DOI] [Google Scholar]
- 410.Waksman SA, Woodruff HB. 1942. Selective antibiotic action of various substances of microbial origin. J Bacteriol 44:373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Schatz A, Waksman SA. 1944. Effect of streptomycin and other antibiotic substances upon Mycobacterium tuberculosis and related organisms. Proc Soc Exp Biol Med 57:244–248. doi: 10.3181/00379727-57-14769. [DOI] [Google Scholar]
- 412.Baltz RH. 2007. Antimicrobials from actinomycetes: back to the future. Microbe 2:125–131. [Google Scholar]
- 413.Busscher GF, Rutjes FP, van Delft FL. 2005. 2-Deoxystreptamine: central scaffold of aminoglycoside antibiotics. Chem Rev 105:775–791. doi: 10.1021/cr0404085. [DOI] [PubMed] [Google Scholar]
- 414.Park SR, Park JW, Ban YH, Sohng JK, Yoon YJ. 2013. 2-Deoxystreptamine-containing aminoglycoside antibiotics: recent advances in the characterization and manipulation of their biosynthetic pathways. Nat Prod Rep 30:11–20. doi: 10.1039/C2NP20092A. [DOI] [PubMed] [Google Scholar]
- 415.Vakulenko SB, Mobashery S. 2003. Versatility of aminoglycosides and prospects for their future. Clin Microbiol Rev 16:430–450. doi: 10.1128/CMR.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Kharel MK, Pahari P, Shepherd MD, Tibrewal N, Nybo SE, Shaaban KA, Rohr J. 2012. Angucyclines: biosynthesis, mode-of-action, new natural products, and synthesis. Nat Prod Rep 29:264–325. doi: 10.1039/C1NP00068C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Kang Q, Shen Y, Bai L. 2012. Biosynthesis of 3,5-AHBA-derived natural products. Nat Prod Rep 29:243–263. doi: 10.1039/C2NP00019A. [DOI] [PubMed] [Google Scholar]
- 418.Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L. 2004. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev 56:185–229. doi: 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- 419.Nitiss JL. 2009. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer 9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Liras P. 1999. Biosynthesis and molecular genetics of cephamycins. Cephamycins produced by actinomycetes. Antonie Van Leeuwenhoek 75:109–124. [DOI] [PubMed] [Google Scholar]
- 421.Jensen SE, Paradkar AS. 1999. Biosynthesis and molecular genetics of clavulanic acid. Antonie Van Leeuwenhoek 75:125–133. doi: 10.1023/A:1001755724055. [DOI] [PubMed] [Google Scholar]
- 422.Saudagar PS, Survase SA, Singhal RS. 2008. Clavulanic acid: a review. Biotechnol Adv 26:335–351. doi: 10.1016/j.biotechadv.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 423.Vining LC, Stuttard C. 1994. Chloramphenicol, genetics and biochemistry of antibiotic production. Butterworth-Heinemann, Boston, MA. [Google Scholar]
- 424.Kominek L. 1975. Cycloheximide production by Streptomyces griseus: control mechanisms of cycloheximide biosynthesis. Antimicrob Agents Chem 7:856–860. doi: 10.1128/AAC.7.6.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Butler MS, Hansford KA, Blaskovich MA, Halai R, Cooper MA. 2014. Glycopeptide antibiotics: back to the future. J Antibiot 67:631–644. doi: 10.1038/ja.2014.111. [DOI] [PubMed] [Google Scholar]
- 426.Van Bambeke F. 2006. Glycopeptides and glycodepsipeptides in clinical development: a comparative review of their antibacterial spectrum, pharmacokinetics and clinical efficacy. Curr Opin Investig Drugs 7:740–749. [PubMed] [Google Scholar]
- 427.Baltz RH. 2010. Genomics and the ancient origins of the daptomycin biosynthetic gene cluster. J Antibiot 63:506–511. doi: 10.1038/ja.2010.82. [DOI] [PubMed] [Google Scholar]
- 428.Gaynor M, Mankin AS. 2003. Macrolide antibiotics: binding site, mechanism of action, resistance. Curr Top Med Chem 3:949–961. doi: 10.2174/1568026033452159. [DOI] [PubMed] [Google Scholar]
- 429.Poehlsgaard J, Douthwaite S. 2003. Macrolide antibiotic interaction and resistance on the bacterial ribosome. Curr Opin Investig Drugs 4:140–148. [PubMed] [Google Scholar]
- 430.Mulinos MG. 1955. Cycloserine: an antibiotic paradox. Antibiot Annu 3:131–135. [PubMed] [Google Scholar]
- 431.Johnston NJ, Mukhtar TA, Wright GD. 2002. Streptogramin antibiotics: mode of action and resistance. Curr Drug Targets 3:335–344. doi: 10.2174/1389450023347678. [DOI] [PubMed] [Google Scholar]
- 432.Okami Y, Hotta K. 1988. Search and discovery of new antibiotics, p 33–67. In Goodfellow M, Williams ST, Mordarski M (ed), Actinomycetes in biotechnology. Academic Press, San Diego, CA. [Google Scholar]
- 433.Behal V. 2000. Bioactive products from Streptomyces. Adv Appl Microbiol 47:113–157. doi: 10.1016/S0065-2164(00)47003-6. [DOI] [PubMed] [Google Scholar]
- 434.Jizba J, Sedmera P, Zima J, Beran M, Blumauerová M, Kandybin N, Samoukina G. 1991. Macrotetrolide antibiotics produced by Streptomyces globisporus. Folia Microbiol (Praha) 36:437–443. doi: 10.1007/BF02884062. [DOI] [PubMed] [Google Scholar]
- 435.Oishi H, Sugawa T, Okutomi T, Suzuki K, Hayashi T. 1970. Insecticidal activity of macrotetrolide antibiotics. J Antibiot 23:105–106. doi: 10.7164/antibiotics.23.105. [DOI] [PubMed] [Google Scholar]
- 436.Sagawa T, Hirano S, Takahashi H, Tanaka N, Oishi H. 1972. Tetranactin, a new miticidal antibiotic. 3. Miticidal and other biological properties. J Econ Entomol 65:372–375. [DOI] [PubMed] [Google Scholar]
- 437.Sakamoto K, Asano T, Mizuochi K, Sasaki K, Hasegawa K. 1978. Makrotetrolid-Antibiotikum zur Bekämpfung der Geflügelkokzidiose. Chem Abstr 89:152 732r German patent DE 28 02 455 C2. [Google Scholar]
- 438.Nippon Kayaku Co Ltd, Chugai Pharm Co Ltd. 1981. Polynactins as anthelmintics. Chem Abstr 95:108 866n Japan patent Kokai Tokkyo Koho 8157.714. [Google Scholar]
- 439.Shichi H, Tanouchi Y, Kamada Y. June 1989. Immunosuppressive agent. Chem Abstr 112:91 787g US patent 4,843,092. [Google Scholar]
- 440.Ando K, Oishi H, Hirano S, Okutomi T, Suzuki K. 1971. Tetranactin, a new miticidal antibiotic. I. Isolation, characterization and properties of tetranactin. J Antibiot 24:347–352. [DOI] [PubMed] [Google Scholar]
- 441.Ando K, Murakami Y, Nawata Y. 1971. Tetranactin, a new miticidal antibiotic. II. Structure of tetranactin. J Antibiot 24:418–422. [DOI] [PubMed] [Google Scholar]
- 442.Misato T. 1982. Present status and future prospects of agricultural antibiotics. J Pestic Sci 7:301–305. doi: 10.1584/jpestics.7.301. [DOI] [Google Scholar]
- 443.Omura S, Crump A. 2014. Ivermectin: panacea for resource-poor communities? Trends Parasitol 30:445–455. doi: 10.1016/j.pt.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 444.Harada S, Kishi T. 1978. Isolation and characterization of mildiomycin, a new nucleoside antibiotic. J Antibiot 31:519–524. doi: 10.7164/antibiotics.31.519. [DOI] [PubMed] [Google Scholar]
- 445.Feduchi E, Cosin M, Carrasco L. 1985. Mildiomycin: a nucleoside antibiotic that inhibits protein synthesis. J Antibiot 38:415–419. doi: 10.7164/antibiotics.38.415. [DOI] [PubMed] [Google Scholar]
- 446.Dharmaraj S. 2010. Marine Streptomyces as a novel source of bioactive substances. World J Microbiol Biotechnol 26:2123–2139. doi: 10.1007/s11274-010-0415-6. [DOI] [Google Scholar]
- 447.Arora SK. 1985. Molecular structure of heliomycin, an inhibitor of RNA synthesis. J Antibiot 38:113–115. doi: 10.7164/antibiotics.38.113. [DOI] [PubMed] [Google Scholar]
- 448.Barac T, Taghavi S, Borremans B, Provoost A, Oeyen L, Colpaert JV, Vangronsveld J, van der Lelie D. 2004. Engineered endophytic bacteria improve phytoremediation of water-soluble, volatile, organic pollutants. Nat Biotechnol 22:583–588. doi: 10.1038/nbt960. [DOI] [PubMed] [Google Scholar]
- 449.Wu MC, Law B, Wilkinson B, Micklefield J. 2012. Bioengineering natural product biosynthetic pathways for therapeutic applications. Curr Opin Biotechnol 23:931–940. doi: 10.1016/j.copbio.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 450.Watve MG, Tickoo R, Jog MM, Bhole BD. 2001. How many antibiotics are produced by the genus? Arch Microbiol 176:386–390. doi: 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- 451.Baltz RH. 2006. Marcel Faber Roundtable: is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration? J Ind Microbiol Biotechnol 33:507–513. doi: 10.1007/s10295-005-0077-9. [DOI] [PubMed] [Google Scholar]
- 452.Umezawa H, Okami Y, Hashimoto T, Suhara Y, Hamada M, Takeuchi T. 1965. A new antibiotic, kasugsmycin. J Antibiot 18:101–103. [PubMed] [Google Scholar]
- 453.Isono K, Nagatsu J, Kawashima Y, Suzuki S. 1965. Studies on polyoxins, antifungal antibiotics. Part I. Isolation and characterization of polyoxins A and B. Agric Biol Chem (Tokyo) 29:848–854. doi: 10.1080/00021369.1965.10858475. [DOI] [Google Scholar]
- 454.Endo A, Misato T. 1969. Polyoxin D, a competitive inhibitor of UDP-N-acetylglucosamine: chitin N-acetylglucosaminyltransferase in Neurospora crassa. Biochem Biophys Res Commun 37:718–722. doi: 10.1016/0006-291X(69)90870-5. [DOI] [PubMed] [Google Scholar]
- 455.British Crop Protection Council. 1994. The pesticide manual, 10th ed British Crop Protection Council, Alton, United Kingdom. [Google Scholar]
- 456.Kameda Y. 1987. Validoxylamines as trehalase inhibitors. J Antibiot 40:563–565. doi: 10.7164/antibiotics.40.563. [DOI] [PubMed] [Google Scholar]
- 457.Elbein AD, Pan YT, Pastuszak I, Carroll D. 2003. New insights on trehalose: a multifunctional molecule. Glycobiology 13:17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- 458.Moran NA. 2006. Symbiosis. Curr Biol 16:R866–R871. doi: 10.1016/j.cub.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 459.Seipke RF, Barke J, Ruiz-Gonzalez MX, Orivel J, Yu DW, Hutchings MI. 2012. Fungus-growing Allomerus ants are associated with antibiotic-producing actinobacteria. Antonie Van Leeuwenhoek 101:443–447. doi: 10.1007/s10482-011-9621-y. [DOI] [PubMed] [Google Scholar]
- 460.Seipke RF, Kaltenpoth M, Hutchings MI. 2012. Streptomyces as symbionts: an emerging and widespread theme? FEMS Microbiol Rev 36:862–876. doi: 10.1111/j.1574-6976.2011.00313.x. [DOI] [PubMed] [Google Scholar]
- 461.Leung TLF, Poulin R. 2008. Parasitism, commensalism, and mutualism: exploring the many shades of symbioses. Life Environ 58:107–115. [Google Scholar]
- 462.Brauman A, Dore J, Eggleton P, Bignell D, Breznak JA, Kane MD. 2001. Molecular phylogenetic profiling of prokaryotic communities in guts of termites with different feeding habits. FEMS Microbiol Ecol 35:27–36. doi: 10.1111/j.1574-6941.2001.tb00785.x. [DOI] [PubMed] [Google Scholar]
- 463.Breznak JA. 1982. Intestinal microbiota of termites and other xylophagous insects. Annu Rev Microbiol 36:323–343. doi: 10.1146/annurev.mi.36.100182.001543. [DOI] [PubMed] [Google Scholar]
- 464.Hongoh Y. 2011. Toward the functional analysis of uncultivable, symbiotic microorganisms in the termite gut. Cell Mol Life Sci 68:1311–1325. doi: 10.1007/s00018-011-0648-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Dejean A, Solano PJ, Ayroles J, Corbara B, Orivel J. 2005. Insect behaviour: arboreal ants build traps to capture prey. Nature 434:973. doi: 10.1038/434973a. [DOI] [PubMed] [Google Scholar]
- 466.Schultz TR, Brady SG. 2008. Major evolutionary transitions in ant agriculture. Proc Natl Acad Sci U S A 105:5435–5440. doi: 10.1073/pnas.0711024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Currie CR, Bot Anm , Boomsma JJ. 2003. Experimental evidence of a tripartite mutualism: bacteria protect ant fungal gardens from specialized parasites. Oikos 101:91–102. doi: 10.1034/j.1600-0706.2003.12036.x. [DOI] [Google Scholar]
- 468.Currie CR, Mueller UG, Malloch D. 1999. The agricultural pathology of ant fungus gardens. Proc Natl Acad Sci U S A 96:7998–8002. doi: 10.1073/pnas.96.14.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Currie CR, Wong B, Stuart AE, Schultz TR, Rehner SA, Mueller UG, Sung GH, Spatafora JW, Straus NA. 2003. Ancient tripartite coevolution in the attine ant-microbe symbiosis. Science 299:386–388. doi: 10.1126/science.1078155. [DOI] [PubMed] [Google Scholar]
- 470.Oh DC, Poulsen M, Currie CR, Clardy J. 2009. Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nat Chem Biol 5:391–393. doi: 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Currie CR. 2001. Prevalence and impact of a virulent parasite on a tripartite mutualism. Oecologia 128:99–106. doi: 10.1007/s004420100630. [DOI] [PubMed] [Google Scholar]
- 472.Barke J, Seipkel RF, Gruschow S, Heavens D, Drou N, Bibb MJ, Goss RJ, Yu DW, Hutchings MI. 2010. A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC Biol 8:109. doi: 10.1186/1741-7007-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Mueller UG, Dash D, Rabeling C, Rodrigues A. 2008. Coevolution between attine ants and actinomycete bacteria: a reevaluation. Evolution 62:2894–2912. doi: 10.1111/j.1558-5646.2008.00501.x. [DOI] [PubMed] [Google Scholar]
- 474.Currie CR, Scott JA, Summerbell RC, Malloch D. 1999. Fungus-growing ants use antibiotic producing bacteria to control garden parasites. Nature 398:701–704. doi: 10.1038/19519. [DOI] [Google Scholar]
- 475.Morales-Jimenez J, Vera-Ponce de Leon A, Garcia-Dominguez A, Martinez-Romero E, Zuniga G, Hernandez-Rodriguez C. 2013. Nitrogen-fixing and uricolytic bacteria associated with the gut of Dendroctonus rhizophagus and Dendroctonus valens (Curculionidae: Scolytinae). Microb Ecol 66:200–210. doi: 10.1007/s00248-013-0206-3. [DOI] [PubMed] [Google Scholar]
- 476.Hulcr J, Adams AS, Raffa K, Hofstetter RW, Klepzig KD, Currie CR. 2011. Presence and diversity of Streptomyces in Dendroctonus and sympatric bark beetle galleries across North America. Microb Ecol 61:759–768. doi: 10.1007/s00248-010-9797-0. [DOI] [PubMed] [Google Scholar]
- 477.Vasanthakumar A, Handelsman J, Schloss PD, Bauer LS, Raffa KF. 2008. Gut microbiota of an invasive subcortical beetle, Agrilus planipennis Fairmaire, across various life stages. Environ Entomol 37:1344–1353. doi: 10.1093/ee/37.5.1344. [DOI] [PubMed] [Google Scholar]
- 478.Miltner EC, Bermudez LE. 2000. Mycobacterium avium grown in Acanthamoeba castellanii is protected from the effects of antimicrobials. Antimicrob Agents Chemother 44:1990–1994. doi: 10.1128/AAC.44.7.1990-1994.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.van Ingen J, Boeree MJ, van Soolingen D, Iseman MD, Heifets LB, Daley CL. 2012. Are phylogenetic position, virulence, drug susceptibility and in vivo response to treatment in mycobacteria interrelated? Infect Genet Evol 12:832–837. doi: 10.1016/j.meegid.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 480.Greub G, Raoult D. 2004. Microorganisms resistant to free-living amoebae. Clin Microbiol Rev 17:413–433. doi: 10.1128/CMR.17.2.413-433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Amissah NA, Gryseels S, Tobias NJ, Ravadgar B, Suzuki M, Vandelannoote K, Durnez L, Leirs H, Stinear TP, Portaels F, Ablordey A, Eddyani M. 2014. Investigating the role of free-living amoebae as a reservoir for Mycobacterium ulcerans. PLoS Negl Trop Dis 8:e3148. doi: 10.1371/journal.pntd.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Gryseels S, Amissah D, Durnez L, Vandelannoote K, Leirs H, De Jonckheere J, Silva MT, Portaels F, Ablordey A, Eddyani M. 2012. Amoebae as potential environmental hosts for Mycobacterium ulcerans and other mycobacteria, but doubtful actors in Buruli ulcer epidemiology. PLoS Negl Trop Dis 6:e1764. doi: 10.1371/journal.pntd.0001764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Kennedy GM, Morisaki JH, Champion PA. 2012. Conserved mechanisms of Mycobacterium marinum pathogenesis within the environmental amoeba Acanthamoeba castellanii. Appl Environ Microbiol 78:2049–2052. doi: 10.1128/AEM.06965-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Crielaard W, Zaura E, Schuller AA, Huse SM, Montijn RC, Keijser BJ. 2011. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genomics 4:22. doi: 10.1186/1755-8794-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Zaura E, Keijser BJ, Huse SM, Crielaard W. 2009. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol 9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Pei Z, Bini EJ, Yang L, Zhou M, Francois F, Blaser MJ. 2004. Bacterial biota in the human distal esophagus. Proc Natl Acad Sci U S A 101:4250–4255. doi: 10.1073/pnas.0306398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, Soriguer F, Queipo-Ortuno MI. 2013. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med 11:46. doi: 10.1186/1741-7015-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Turroni F, Marchesi JR, Foroni E, Gueimonde M, Shanahan F, Margolles A, van Sinderen D, Ventura M. 2009. Microbiomic analysis of the bifidobacterial population in the human distal gut. ISME J 3:745–751. doi: 10.1038/ismej.2009.19. [DOI] [PubMed] [Google Scholar]
- 492.Inness V, McCartney A, Khoo C, Gross K, Gibson G. 2007. Molecular characterisation of the gut microflora of healthy and inflammatory bowel disease cats using fluorescence in situ hybridisation with special reference to Desulfovibrio spp. J Anim Physiol Anim Nutr 91:48–53. doi: 10.1111/j.1439-0396.2006.00640.x. [DOI] [PubMed] [Google Scholar]
- 493.Jia J, Frantz N, Khoo C, Gibson GR, Rastall RA, McCartney AL. 2010. Investigation of the faecal microbiota associated with canine chronic diarrhoea. FEMS Microbiol Ecol 71:304–312. doi: 10.1111/j.1574-6941.2009.00812.x. [DOI] [PubMed] [Google Scholar]
- 494.McNeil MM, Brown JM. 1994. The medically important aerobic actinomycetes: epidemiology and microbiology. Clin Microbiol Rev 7:357–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Tsukamura M. 1982. Numerical analysis of the taxonomy of Nocardiae and Rhodococci. Microbiol Immunol 26:1101–1119. doi: 10.1111/j.1348-0421.1982.tb00260.x. [DOI] [PubMed] [Google Scholar]
- 496.Kapadia M, Rolston KV, Han XY. 2007. Invasive Streptomyces infections: six cases and literature review. Am J Clin Pathol 127:619–624. doi: 10.1309/QJEBXP0BCGR54L15. [DOI] [PubMed] [Google Scholar]
- 497.Kirby R, Sangal V, Tucker NP, Zakrzewska-Czerwinska J, Wierzbicka K, Herron PR, Chu CJ, Chandra G, Fahal AH, Goodfellow M, Hoskisson PA. 2012. Draft genome sequence of the human pathogen Streptomyces somaliensis, a significant cause of actinomycetoma. J Bacteriol 194:3544–3545. doi: 10.1128/JB.00534-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Quintana ET, Wierzbicka K, Mackiewicz P, Osman A, Fahal AH, Hamid ME, Zakrzewska-Czerwinska J, Maldonado LA, Goodfellow M. 2008. Streptomyces sudanensis sp. nov., a new pathogen isolated from patients with actinomycetoma. Antonie Van Leeuwenhoek 93:305–313. doi: 10.1007/s10482-007-9205-z. [DOI] [PubMed] [Google Scholar]
- 499.Dunne EF, Burman WJ, Wilson ML. 1998. Streptomyces pneumonia in a patient with human immunodeficiency virus infection: case report and review of the literature on invasive Streptomyces infections. Clin Infect Dis 27:93–96. doi: 10.1086/514612. [DOI] [PubMed] [Google Scholar]
- 500.Datta P, Arora S, Jain R, Chander J, van de Sande W. 2012. Secondary peritonitis caused by Streptomyces viridis. J Clin Microbiol 50:1813–1814. doi: 10.1128/JCM.06045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Gugnani A, Unaogu I, Emeruwa C. 1993. Pulmonary infection due to Streptomyces griseus. J Commun Disord 25:38–40. [PubMed] [Google Scholar]
- 502.Kofteridis DP, Maraki S, Scoulica E, Tsioutis C, Maltezakis G, Gikas A. 2007. Streptomyces pneumonia in an immunocompetent patient: a case report and literature review. Diagn Microbiol Infect Dis 59:459–462. doi: 10.1016/j.diagmicrobio.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 503.Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. 2013. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 504.Bignell DR, Seipke RF, Huguet-Tapia JC, Chambers AH, Parry RJ, Loria R. 2010. Streptomyces scabies 87-22 contains a coronafacic acid-like biosynthetic cluster that contributes to plant-microbe interactions. Mol Plant Microbe Interact 23:161–175. doi: 10.1094/MPMI-23-2-0161. [DOI] [PubMed] [Google Scholar]
- 505.Loria R, Kers J, Joshi M. 2006. Evolution of plant pathogenicity in Streptomyces. Annu Rev Phytopathol 44:469–487. doi: 10.1146/annurev.phyto.44.032905.091147. [DOI] [PubMed] [Google Scholar]
- 506.Faucher E, Savard T, Bealieu C. 1992. Characterization of actinomycetes isolated from common scab lesions on potato tubers. Can J Plant Pathol 14:197–202. doi: 10.1080/07060669209500874. [DOI] [Google Scholar]
- 507.Kreuze JF, Suomalainen S, Paulin L, Valkonen JP. 1999. Phylogenetic analysis of 16S rRNA genes and PCR analysis of the nec1 gene from Streptomyces spp. causing common scab, pitted scab, and netted scab in Finland. Phytopathology 89:462–469. doi: 10.1094/PHYTO.1999.89.6.462. [DOI] [PubMed] [Google Scholar]
- 508.Wanner LA. 2006. A survey of genetic variation in Streptomyces isolates causing potato common scab in the United States. Phytopathology 96:1363–1371. doi: 10.1094/PHYTO-96-1363. [DOI] [PubMed] [Google Scholar]
- 509.Loria R, Bukhalid A, Fry B, King RR. 1997. Plant pathogenicity in the genus Streptomyces. Plant Dis 81:836–846. doi: 10.1094/PDIS.1997.81.8.836. [DOI] [PubMed] [Google Scholar]
- 510.Lambert DH, Loria R. 1989. Streptomyces scabies sp. nov., nom. rev. Int J Syst Bacteriol 39:387–392. doi: 10.1099/00207713-39-4-387. [DOI] [Google Scholar]
- 511.Harrison MD. 1962. Potato russet scab, its cause and factors affecting its development. Am J Potato Res 39:368–387. doi: 10.1007/BF02861618. [DOI] [Google Scholar]
- 512.Scholte K, Labruyere RE. 1985. Netted scab: a new name for an old disease in Europe. Potato Res 28:443–448. doi: 10.1007/BF02357520. [DOI] [Google Scholar]
- 513.Blanchette RA, Sutherland JB, Crawford DL. 1981. Actinomycetes in discolored wood of living silver maple. Can J Bot 59:1–7. doi: 10.1139/b81-001. [DOI] [Google Scholar]
- 514.Wallis G. 1983. Actinomycte and actinomycete-like vessel oclusions of sugar maple, Acer sacharum. M.S. dissertation. SUNY College of Environmental Science and Forestry, Syracuse, NY. [Google Scholar]
- 515.Sutherland JB, Blanchette RA, Crawford DL, Pometto AL. 1979. Breakdown of Douglas fir phloem by a lignocellulose-degrading Streptomyces. Curr Microbiol 2:123–126. doi: 10.1007/BF02603069. [DOI] [Google Scholar]
- 516.Tilford PE. 1936. Fasciation of sweet peas caused by Phytomonas fascians n. sp. J Agric Res 53:383–394. [Google Scholar]
- 517.Goethals K, Vereecke D, Jaziri M, Van Montagu M, Holsters M. 2001. Leafy gall formation by Rhodococcus fascians. Annu Rev Phytopathol 39:27–52. doi: 10.1146/annurev.phyto.39.1.27. [DOI] [PubMed] [Google Scholar]
- 518.Vereecke D, Burssens S, Simon-Mateo C, Inze D, Van Montagu M, Goethals K, Jaziri M. 2000. The Rhodococcus fascians-plant interaction: morphological traits and biotechnological applications. Planta 210:241–251. doi: 10.1007/PL00008131. [DOI] [PubMed] [Google Scholar]
- 519.Stes E, Francis I, Pertry I, Dolzblasz A, Depuydt S, Vereecke D. 2013. The leafy gall syndrome induced by Rhodococcus fascians. FEMS Microbiol Lett 342:187–194. doi: 10.1111/1574-6968.12119. [DOI] [PubMed] [Google Scholar]
- 520.de O Manes CL, Beeckman T, Ritsema T, Van Montagu M, Goethals K, Holsters M. 2004. Phenotypic alterations in Arabidopsis thaliana plants caused by Rhodococcus fascians infection. J Plant Res 117:139–145. doi: 10.1007/s10265-003-0138-y. [DOI] [PubMed] [Google Scholar]
- 521.Vandeputte O, Oden S, Mol A, Vereecke D, Goethals K, El Jaziri M, Prinsen E. 2005. Biosynthesis of auxin by the gram-positive phytopathogen Rhodococcus fascians is controlled by compounds specific to infected plant tissues. Appl Environ Microbiol 71:1169–1177. doi: 10.1128/AEM.71.3.1169-1177.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Flügel M, Becker A, Gartemann K-H, Eichenlaub R. 2012. Analysis of the interaction of Clavibacter michiganensis subsp. michiganensis with its host plant tomato by genome-wide expression profiling. J Biotechnol 160:42–54. doi: 10.1016/j.biotec.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 523.Eichenlaub R, Gartemann K-H. 2011. The Clavibacter michiganensis subspecies: molecular investigation of Gram-positive bacterial plant pathogens. Annu Rev Phytopathol 49:445–464. doi: 10.1146/annurev-phyto-072910-095258. [DOI] [PubMed] [Google Scholar]
- 524.Monteiro-Vitorello CB, Camargo LE, Van Sluys MA, Kitajima JP, Truffi D, do Amaral AM, Harakava R, de Oliveira JC, Wood D, de Oliveira MC, Miyaki C, Takita MA, da Silva AC, Furlan LR, Carraro DM, Camarotte G, Almeida NF Jr, Carrer H, Coutinho LL, El-Dorry HA, Ferro MI, Gagliardi PR, Giglioti E, Goldman MH, Goldman GH, Kimura ET, Ferro ES, Kuramae EE, Lemos EG, Lemos MV, Mauro SM, Machado MA, Marino CL, Menck CF, Nunes LR, Oliveira RC, Pereira GG, Siqueira W, de Souza AA, Tsai SM, Zanca AS, Simpson AJ, Brumbley SM, Setubal JC. 2004. The genome sequence of the gram-positive sugarcane pathogen Leifsonia xyli subsp. xyli. Mol Plant Microbe Interact 17:827–836. doi: 10.1094/MPMI.2004.17.8.827. [DOI] [PubMed] [Google Scholar]
- 525.Dreier J, Meletzus D, Eichenlaub R. 1997. Characterization of the plasmid encoded virulence region pat-1 of phytopathogenic Clavibacter michiganensis subsp. michiganensis. Mol Plant Microbe Interact 10:195–206. doi: 10.1094/MPMI.1997.10.2.195. [DOI] [PubMed] [Google Scholar]
- 526.Laine MJ, Haapalainen M, Wahlroos T, Kankare K, Nissinen R, Kassuwi S, Metzler MC. 2000. The cellulase encoded by the native plasmid of Clavibacter michiganensis ssp. sepedonicus plays a role in virulence and contains an expansin-like domain. Physiol Mol Plant P 57:221–233. doi: 10.1006/pmpp.2000.0301. [DOI] [Google Scholar]
- 527.Harth G, Horwitz MA. 1997. Expression and efficient export of enzymatically active Mycobacterium tuberculosis glutamine synthetase in Mycobacterium smegmatis and evidence that the information for export is contained within the protein. J Biol Chem 272:22728–22735. doi: 10.1074/jbc.272.36.22728. [DOI] [PubMed] [Google Scholar]
- 528.King RR, Calhoun LA. 2009. The thaxtomin phytotoxins: sources, synthesis, biosynthesis, biotransformation and biological activity. Phytochemistry 70:833–841. doi: 10.1016/j.phytochem.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 529.Duval I, Brochu V, Simard M, Beaulieu C, Beaudoin N. 2005. Thaxtomin A induces programmed cell death in Arabidopsis thaliana suspension-cultured cells. Planta 222:820–831. doi: 10.1007/s00425-005-0016-z. [DOI] [PubMed] [Google Scholar]
- 530.Errakhi R, Meimoun P, Lehner A, Vidal G, Briand J, Corbineau F, Rona J-P, Bouteau F. 2008. Anion channel activity is necessary to induce ethylene synthesis and programmed cell death in response to oxalic acid. J Exp Bot 59:3121–3129. doi: 10.1093/jxb/ern166. [DOI] [PubMed] [Google Scholar]
- 531.Fry BA, Loria R. 2002. Thaxtomin A: evidence for a plant cell wall target. Physiol Mol Plant Pathol 60:1–8. doi: 10.1006/pmpp.2001.0371. [DOI] [Google Scholar]
- 532.Scheible WR, Fry B, Kochevenko A, Schindelasch D, Zimmerli L, Somerville S, Loria R, Somerville CR. 2003. An Arabidopsis mutant resistant to thaxtomin A, a cellulose synthesis inhibitor from Streptomyces species. Plant Cell 15:1781–1794. doi: 10.1105/tpc.013342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Tegg RS, Melian L, Wilson CR, Shabala S. 2005. Plant cell growth and ion flux responses to the streptomycete phytotoxin thaxtomin A: calcium and hydrogen flux patterns revealed by the non-invasive MIFE technique. Plant Cell Physiol 46:638–648. doi: 10.1093/pcp/pci069. [DOI] [PubMed] [Google Scholar]
- 534.Kinkel LL, Bowers JH, Shimizu K, Neeno-Eckwall EC, Schottel JL. 1998. Quantitative relationships among thaxtomin A production, potato scab severity, and fatty acid composition in Streptomyces. Can J Microbiol 44:768–776. [PubMed] [Google Scholar]
- 535.Goyer C, Vachon J, Beaulieu C. 1998. Pathogenicity of Streptomyces scabies mutants altered in thaxtomin A production. Phytopathology 88:442–445. doi: 10.1094/PHYTO.1998.88.5.442. [DOI] [PubMed] [Google Scholar]
- 536.Healy FG, Wach M, Krasnoff SB, Gibson DM, Loria R. 2000. The txtAB genes of the plant pathogen Streptomyces acidiscabies encode a peptide synthetase required for phytotoxin thaxtomin A production and pathogenicity. Mol Microbiol 38:794–804. doi: 10.1046/j.1365-2958.2000.02170.x. [DOI] [PubMed] [Google Scholar]
- 537.King RR, Lawrence CH, Clark MC, Calhoun LA. 1989. Isolation and characterization of phytotoxins associated with Streptomyces scabies. J Chem Soc Chem Commun 13:849–850. [Google Scholar]
- 538.Lawrence CH, Clark MC, King RR. 1990. Induction of common scab symptoms in aseptically cultured potato tubers by the vivotoxin, thaxtomin. Phytopathology 80:606–608. doi: 10.1094/Phyto-80-606. [DOI] [Google Scholar]
- 539.Raymer G, Willard JM, Schottel JL. 1990. Cloning, sequencing, and regulation of expression of an extracellular esterase gene from the plant pathogen Streptomyces scabies. J Bacteriol 172:7020–7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, McQueen-Mason SJ. 2002. Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol 128:854–864. doi: 10.1104/pp.010658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Stavrinides J, McCann HC, Guttman DS. 2008. Host-pathogen interplay and the evolution of bacterial effectors. Cell Microbiol 10:285–292. doi: 10.1111/j.1462-5822.2007.01078.x. [DOI] [PubMed] [Google Scholar]
- 542.Gribbon EM, Shoesmith J, Cunliffe W, Holland K. 1994. The microaerophily and photosensitivity of Propionibacterium acnes. J Appl Bacteriol 77:583–590. doi: 10.1111/j.1365-2672.1994.tb04405.x. [DOI] [PubMed] [Google Scholar]
- 543.Westerhof W, Relyveld GN, Kingswijk MM, de Man P, Menke HE. 2004. Propionibacterium acnes and the pathogenesis of progressive macular hypomelanosis. Arch Dermatol 140:210–214. doi: 10.1001/archderm.140.2.210. [DOI] [PubMed] [Google Scholar]
- 544.Kinashi H, Someno K, Sakaguchi K. 1984. Isolation and characterization of concanamycins A, B and C. J Antibiot 37:1333–1343. doi: 10.7164/antibiotics.37.1333. [DOI] [PubMed] [Google Scholar]
- 545.Haydock SF, Appleyard AN, Mironenko T, Lester J, Scott N, Leadlay PF. 2005. Organization of the biosynthetic gene cluster for the macrolide concanamycin A in Streptomyces neyagawaensis ATCC 27449. Microbiology 151:3161–3169. doi: 10.1099/mic.0.28194-0. [DOI] [PubMed] [Google Scholar]
- 546.Seki-Asano M, Okazaki T, Yamagishi M, Sakai N, Hanada K, Mizoue K. 1994. Isolation and characterization of new 18-membered macrolides FD-891 and FD-892. J Antibiot 47:1226–1233. doi: 10.7164/antibiotics.47.1226. [DOI] [PubMed] [Google Scholar]
- 547.Natsume M, Ryu R, Abe H. 1996. Production of phytotoxins, concanamycins A and B by Streptomyces spp. Ann Phytopathol Soc Jpn 62:411–413. doi: 10.3186/jjphytopath.62.411. [DOI] [Google Scholar]
- 548.Natsume M, Yamada A, Tashiro N, Abe H. 1998. Differential production of the phytotoxins thaxtomin A and concanamycins A and B by potato common scab-causing Streptomyces spp. Ann Phytopathol Soc Jpn 64:202–204. doi: 10.3186/jjphytopath.64.202. [DOI] [Google Scholar]
- 549.Natsume M, Komiya M, Koyanagi F, Tashiro N, Kawaide H, Abe H. 2005. Phytotoxin produced by Streptomyces sp. causing potato russet scab in Japan. J Gen Plant Pathol 71:364–369. doi: 10.1007/s10327-005-0211-6. [DOI] [Google Scholar]
- 550.Natsume M, Taki M, Tashiro N, Abe H. 2001. Phytotoxin production and aerial mycelium formation by Streptomyces scabies and S. acidiscabies in vitro. J Gen Plant Pathol 67:299–302. doi: 10.1007/PL00013035. [DOI] [Google Scholar]
- 551.Fenton A, Stephens P, Crowley J, O'Callaghan M, O'Gara F. 1992. Exploitation of gene(s) involved in 2,4-diacetylphloroglucinol biosynthesis to confer a new biocontrol capability to a Pseudomonas strain. Appl Environ Microbiol 58:3873–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Glick BR. 1995. The enhancement of plant growth by free-living bacteria. Can J Microbiol 41:109–117. doi: 10.1139/m95-015. [DOI] [Google Scholar]
- 553.Gomes R, Semedo L, Soares R, Alviano C, Linhares L, Coelho R. 2000. Chitinolytic activity of actinomycetes from a cerrado soil and their potential in biocontrol. Lett Appl Microbiol 30:146–150. doi: 10.1046/j.1472-765x.2000.00687.x. [DOI] [PubMed] [Google Scholar]
- 554.Kim BS, Moon SS, Hwang BK. 2000. Structure elucidation and antifungal activity of an anthracycline antibiotic, daunomycin, isolated from Actinomadura roseola. J Agric Food Chem 48:1875–1881. doi: 10.1021/jf990402u. [DOI] [PubMed] [Google Scholar]
- 555.Ouhdouch Y, Barakate M, Finace C. 2001. Actinomycetes from Maroccan habitats: screening for antifungal activites. Eur J Soil Biol 37:69–74. doi: 10.1016/S1164-5563(01)01069-X. [DOI] [Google Scholar]
- 556.El-Tarabily KA, Sivasithamparam K. 2006. Non-streptomycete actinomycetes as biocontrol agents of soil-borne fungal plant pathogens and as plant growth promoters. Soil Biol Biochem 38:1505–1520. doi: 10.1016/j.soilbio.2005.12.017. [DOI] [Google Scholar]
- 557.Cohen MF, Mazzola M. 2006. Resident bacteria, nitric oxide emission and particle size modulate the effect of Brassica napus seed meal on disease incited by Rhizoctonia solani and Pythium spp. Plant Soil 286:75–86. doi: 10.1007/s11104-006-9027-1. [DOI] [Google Scholar]
- 558.Doumbou CL, Salove MKH, Crawford DL, Beaulieu C. 2001. Actinomycetes, promising tools to control plant diseases and to promote plant growth. Phytoprotection 82:85–102. doi: 10.7202/706219ar. [DOI] [Google Scholar]
- 559.Mahmoudi E, Tabatabaei BES, Venturi V. 2011. Virulence attenuation of Pectobacterium carotovorum using N-acyl-homoserine lactone degrading bacteria isolated from potato rhizosphere. Plant Pathol J 27:242–248. doi: 10.5423/PPJ.2011.27.3.242. [DOI] [Google Scholar]
- 560.Cao L, Qiu Z, You J, Tan H, Zhou S. 2005. Isolation and characterization of endophytic streptomycete antagonists of Fusarium wilt pathogen from surface-sterilized banana roots. FEMS Microbiol Lett 247:147–152. doi: 10.1016/j.femsle.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 561.Getha K, Vikineswary S, Wong W, Seki T, Ward A, Goodfellow M. 2005. Evaluation of Streptomyces sp. strain g10 for suppression of Fusarium wilt and rhizosphere colonization in pot-grown banana plantlets. J Ind Microbiol Biotechnol 32:24–32. doi: 10.1007/s10295-004-0199-5. [DOI] [PubMed] [Google Scholar]
- 562.Errakhi R, Bouteau F, Lebrihi A, Barakate M. 2007. Evidences of biological control capacities of Streptomyces spp. against Sclerotium rolfsii responsible for damping-off disease in sugar beet (Beta vulgaris L.). World J Microbiol Biotechnol 23:1503–1509. doi: 10.1007/s11274-007-9394-7. [DOI] [Google Scholar]
- 563.Jain PK, Jain P. 2007. Isolation, characterization and antifungal activity of Streptomyces sampsonii GS 1322. Indian J Exp Biol 45:203–206. [PubMed] [Google Scholar]
- 564.Trejo-Estrada S, Paszczynski A, Crawford D. 1998. Antibiotics and enzymes produced by the biocontrol agent Streptomyces violaceusnuger YCED-9. J Ind Microbiol Biotechnol 21:81–90. doi: 10.1038/sj.jim.2900549. [DOI] [Google Scholar]
- 565.Xiao K, Samac DA, Kinkel LL. 2002. Biological control of Phytophthora root rots on alfalfa and soybean with Streptomyces. Biol Control 23:285–295. doi: 10.1006/bcon.2001.1015. [DOI] [Google Scholar]
- 566.Taechowisan T, Perberdy JF, Lumyong S. 2003. Isolation of endophytic actinomycetes from selected plants and their antifungal activity. World J Microbiol Biotechnol 19:381–385. doi: 10.1023/A:1023901107182. [DOI] [Google Scholar]
- 567.Miyashita K, Fujii T, Sawada Y. 1991. Molecular cloning and characterization of chitinase genes from Streptomyces lividans 66. J Gen Microbiol 137:2065–2072. doi: 10.1099/00221287-137-9-2065. [DOI] [Google Scholar]
- 568.Abd-Allah EF. 2001. Streptomyces plicatus as a model biocontrol agent. Folia Microbiol (Praha) 46:309–314. doi: 10.1007/BF02815619. [DOI] [PubMed] [Google Scholar]
- 569.Joo G-J. 2005. Production of an anti-fungal substance for biological control of Phytophthora capsici causing phytophthora blight in red-peppers by Streptomyces halstedii. Biotechnol Lett 27:201–205. doi: 10.1007/s10529-004-7879-0. [DOI] [PubMed] [Google Scholar]
- 570.Mahadevan B, Crawford DL. 1997. Properties of the chitinase of the antifungal biocontrol agent Streptomyces lydicus WYEC108. Enzyme Microb Technol 20:489–493. doi: 10.1016/S0141-0229(96)00175-5. [DOI] [Google Scholar]
- 571.Barakate M, Ouhdouch Y, Oufdou K, Beaulieu C. 2002. Characterization of rhizospheric soil strepomycetes from moroccan habitat and their antimicrobial activities. World J Microbiol Biotechnol 18:49–54. doi: 10.1023/A:1013966407890. [DOI] [Google Scholar]
- 572.Emmert EA, Handelsman J. 1999. Biocontrol of plant disease: a (Gram-) positive perspective. FEMS Microbiol Lett 171:1–9. doi: 10.1111/j.1574-6968.1999.tb13405.x. [DOI] [PubMed] [Google Scholar]
- 573.Millard WA. 1923. Common scab of potatoes. Ann Appl Biol 10:70–88. doi: 10.1111/j.1744-7348.1923.tb05654.x. [DOI] [Google Scholar]
- 574.Sanford GB. 1926. Some factors affecting the pathogenicity of Actinomyces scabies. Phytopathology 16:525–547. [Google Scholar]
- 575.Millard WA, Taylor CB. 1927. Antagonism of micro-organisms as the controlling factor in the inhibition of scab by green-manuring. Ann Appl Biol 14:202–216. doi: 10.1111/j.1744-7348.1927.tb07076.x. [DOI] [Google Scholar]
- 576.Kloepper JW. 1996. Host specificity in microbe-microbe interactions. Bioscience 46:406–409. [Google Scholar]
- 577.Gardner JM, Chandler JL, Feldman AW. 1984. Growth promotion and inhibition by antibiotic-producing fluorescent pseudomonads on citrus roots. Plant Soil 77:103–113. doi: 10.1007/BF02182816. [DOI] [Google Scholar]
- 578.O'Neill GA, Radley RA, Chanaway CP. 1992. Variable effects of emergence-promoting rhizobacteria on conifer seed growth under nursery conditions. Biol Fertil Soils 13:45–49. doi: 10.1007/BF00337237. [DOI] [Google Scholar]
- 579.Giri S, Pati B. 2004. A comparative study on phyllosphere nitrogen fixation by newly isolated Corynebacterium sp. & Flavobacterium sp. and their potentialities as biofertilizer. Acta Microbiol Immunol Hung 51:47–56. doi: 10.1556/AMicr.51.2004.1-2.3. [DOI] [PubMed] [Google Scholar]
- 580.Mahendra S, Alvarez-Cohen L. 2005. Pseudonocardia dioxanivoran sp. nov., a novel actinomycete that grows on 1,4-dioxane. Int J Syst Evol Microbiol 55:593–598. doi: 10.1099/ijs.0.63085-0. [DOI] [PubMed] [Google Scholar]
- 581.Aldesuquy HS, Mansour FA, Abo-Hamed SA. 1998. Effect of the culture filtrates of Streptomyces on growth and productivity of wheat plants. Folia Microbiol (Praha) 43:465–470. doi: 10.1007/BF02820792. [DOI] [Google Scholar]
- 582.Alexander M. 1977. Introduction to soil microbiology. Krieger Publishing Company, Malabar, FL. [Google Scholar]
- 583.Merriman PR, Price RD, Kollmorgen JF, Piggott T, Ridge EH. 1974. Effect of seed inoculation with Bacillus subtilis and Streptomyces griseus on the growth of cereals and carrots. Aust J Agric Res 25:219–226. doi: 10.1071/AR9740219. [DOI] [Google Scholar]
- 584.El-Abyad M, El-Sayed M, El-Shanshoury A, El-Sabbagh SM. 1993. Towards the biological control of fungal and bacterial diseases of tomato using antagonistic Streptomyces spp. Plant Soil 149:185–195. doi: 10.1007/BF00016608. [DOI] [Google Scholar]
- 585.Kaltenpoth M. 2009. Actinobacteria as mutualists: general healthcare for insects? Trends Microbiol 17:529–535. doi: 10.1016/j.tim.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 586.Kucho K, Hay AE, Normand P. 2010. The determinants of the actinorhizal symbiosis. Microbes Environ 25:241–252. doi: 10.1264/jsme2.ME10143. [DOI] [PubMed] [Google Scholar]
- 587.Trujillo ME, Alonso-Vega P, Rodriguez R, Carro L, Cerda E, Alonso P, Martinez-Molina E. 2010. The genus Micromonospora is widespread in legume root nodules: the example of Lupinus angustifolius. ISME J 4:1265–1281. doi: 10.1038/ismej.2010.55. [DOI] [PubMed] [Google Scholar]
- 588.Buckley DH, Huangyutitham V, Hsu SF, Nelson TA. 2007. Stable isotope probing with 15N2 reveals novel noncultivated diazotrophs in soil. Appl Environ Microbiol 73:3196–3204. doi: 10.1128/AEM.02610-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589.Gtari M, Brusetti L, Hassen A, Mora D, Daffonchio D, Boudabous A. 2007. Genetic diversity among Elaeagnus compatible Frankia strains and sympatric-related nitrogen-fixing actinobacteria revealed by nifH sequence analysis. Soil Biol Biochem 39:372–377. doi: 10.1016/j.soilbio.2006.07.005. [DOI] [Google Scholar]
- 590.Valdes M, Perez NO, Estrada-de Los Santos P, Caballero-Mellado J, Pena-Cabriales JJ, Normand P, Hirsch AM. 2005. Non-Frankia actinomycetes isolated from surface-sterilized roots of Casuarina equisetifolia fix nitrogen. Appl Environ Microbiol 71:460–466. doi: 10.1128/AEM.71.1.460-466.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Zakhia F, Jeder H, Willems A, Gillis M, Dreyfus B, de Lajudie P. 2006. Diverse bacteria associated with root nodules of spontaneous legumes in Tunisia and first report for nifH-like gene within the genera Microbacterium and Starkeya. Microb Ecol 51:375–393. doi: 10.1007/s00248-006-9025-0. [DOI] [PubMed] [Google Scholar]
- 592.Roy S, Khasa DP, Greer CW. 2007. Combining alders, frankiae, and mycorrhizae for the revegetation and remediation of contaminated ecosystems. Can J Bot 85:237–251. doi: 10.1139/B07-017. [DOI] [Google Scholar]
- 593.Sardi P, Saracchi M, Quaroni S, Petrolini B, Borgonovi GE, Merli S. 1992. Isolation of endophytic Streptomyces strains from surface-sterilized roots. Appl Environ Microbiol 58:2691–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Coombs JT, Michelsen P, Franco CMM. 2004. Evaluation of endophytic actinobacteria as antagonists of Gaeumannomyces graminis var. tritici in wheat. Biol Control 29:359–366. doi: 10.1016/j.biocontrol.2003.08.001. [DOI] [Google Scholar]
- 595.Overvoorde P, Fukaki H, Beeckman T. 2010. Auxin control of root development. Cold Spring Harb Perspect Biol 2:a001537. doi: 10.1101/cshperspect.a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596.Tokala RK, Strap JL, Jung CM, Crawford DL, Salove MH, Deobald LA, Bailey JF, Morra MJ. 2002. Novel plant-microbe rhizosphere interaction involving Streptomyces lydicus WYEC108 and the pea plant (Pisum sativum). Appl Environ Microbiol 68:2161–2171. doi: 10.1128/AEM.68.5.2161-2171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 597.Couillerot O, Loqman S, Toribio A, Hubert J, Gandner L, Nuzillard JM, Ouhdouch Y, Clement C, Barka EA, Renault JH. 2014. Purification of antibiotics from the biocontrol agent Streptomyces anulatus S37 by centrifugal partition chromatography. J Chromatogr B 944:30–34. doi: 10.1016/j.jchromb.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 598.Berg G, Marten P, Minkwitz A, Bruckner S. 2001. Efficient biological control of fungal plant diseases by Streptomyces sp. DSMZ 12424. J Plant Dis Prot 108:1–10. [Google Scholar]
- 599.El-Tarabily KA, Soliman MH, Nassar AH, Al-Hassani HA, Sivasithamparam K, McKenna F, Hardy GES. 2000. Biological control of Sclerotinia minor using a chitinolytic bacterium and actinomycetes. Plant Pathol 49:573–583. doi: 10.1046/j.1365-3059.2000.00494.x. [DOI] [Google Scholar]
- 600.Quecine MC, Araujo WL, Marcon J, Gai CS, Azevedo JL, Pizzirani-Kleiner AA. 2008. Chitinolytic activity of endophytic Streptomyces and potential for biocontrol. Lett Appl Microbiol 47:486–491. doi: 10.1111/j.1472-765X.2008.02428.x. [DOI] [PubMed] [Google Scholar]
- 601.Verma VC, Singh SK, Prakash S. 2011. Bio-control and plant growth promotion potential of siderophore producing endophytic Streptomyces from Azadirachta indica A Juss. J Basic Microbiol 51:550–556. doi: 10.1002/jobm.201000155. [DOI] [PubMed] [Google Scholar]
- 602.Murray RGE, Brenner DJ, Holt JG, Krieg NR, Mulder JW, Pfenning N, Sneath PH, Stoley JT, Williams ST (ed). 1989. Bergey's manual of systematic bacteriology, vol 4 Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 603.Bister B, Bischoff D, Strobele M, Riedlinger J, Reicke A, Wolter F, Bull AT, Zahner H, Fiedler HP, Sussmuth RD. 2004. Abyssomicin C: a polycyclic antibiotic from a marine Verrucosispora strain as an inhibitor of the p-aminobenzoic acid/tetrahydrofolate biosynthesis pathway. Angew Chem Int Ed Engl 43:2574–2576. doi: 10.1002/anie.200353160. [DOI] [PubMed] [Google Scholar]
- 604.Heinemann B, Kaplan MA, Muir RD, Hooper IR. 1953. Amphomycin, a new antibiotic. Antibiot Chemother 3:1239–1242. [PubMed] [Google Scholar]
- 605.Grein A, Merli S, Spalla C. 1980. New anthracycline glycosides from Micromonospora. I. Description of the producing strain. J Antibiot 33:1462–1467. [DOI] [PubMed] [Google Scholar]
- 606.Barbe V, Bouzon M, Mangenot S, Badet B, Poulain J, Segurens B, Vallenet D, Marliere P, Weissenbach J. 2011. Complete genome sequence of Streptomyces cattleya NRRL 8057, a producer of antibiotics and fluorometabolites. J Bacteriol 193:5055–5056. doi: 10.1128/JB.05583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Yang HJ, Huang XZ, Zhang ZL, Wang CX, Zhou J, Huang K, Zhou JM, Zheng W. 2014. Two novel amphomycin analogues from Streptomyces canus strain FIM-0916. Nat Prod Res 28:861–867. doi: 10.1080/14786419.2014.886210. [DOI] [PubMed] [Google Scholar]
- 608.Burg RW, Miller BM, Baker EE, Birnbaum J, Currie SA, Hartman R, Kong YL, Monaghan RL, Olson G, Putter I, Tunac JB, Wallick H, Stapley EO, Oiwa R, Omura S. 1979. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob Agents Chemother 15:361–367. doi: 10.1128/AAC.15.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609.Omura S, Imamura N, Oiwa R, Kuga H, Iwata R, Masuma R, Iwai Y. 1986. Clostomicins. New antibiotics produced by Micromonospora echinospora subsp. armeniaca subsp. nov. I. Production, isolation, and physico-chemical and biological properties. J Antibiot 39:1407–1412. [DOI] [PubMed] [Google Scholar]
- 610.Pendela M, Dragovic S, Bockx L, Hoogmartens J, Van Schepdael A, Adams E. 2008. Development of a liquid chromatographic method for the determination of related substances and assay of d-cycloserine. J Pharm Biomed Anal 47:807–811. doi: 10.1016/j.jpba.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 611.Mchenney MA, Hosted TJ, Dehoff BS, Rosteck PR Jr, Baltz RH. 1998. Molecular cloning and physical mapping of the daptomycin gene cluster from Streptomyces roseosporus. J Bacteriol 180:143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612.Mc Guire JM, Bunch RL, Anderson RC, Boaz HE, Flynn EH, Powell HM, Smith JW. 1952. Ilotycin, a new antibiotic. Antibiot Chemother (Northfield) 2:281–283. [PubMed] [Google Scholar]
- 613.Weinstein MJ, Luedemann GM, Oden EM, Wagman GH, Rosselet JP, Marquez JA, Coniglio CT, Charney W, Herzog HL, Black J. 1963. Gentamicin, a new antibiotic complex from Micromonospora. J Med Chem 6:463–464. doi: 10.1021/jm00340a034. [DOI] [PubMed] [Google Scholar]
- 614.Gonzalez A, Jimenez A, Vazquez D, Davies J, Schindler D. 1978. Studies on the mode of action of hygromycin B, an inhibitor of translocation in eukaryotes. Biochim Biophys Acta 521:459–469. [DOI] [PubMed] [Google Scholar]
- 615.Umezawa H, Ueda M, Maeda K, Yagishita K, Kondo S, Okami Y, Utahara R, Osato Y, Nitta K, Takeuchi T. 1957. Production and isolation of a new antibiotic: kanamycin. J Antibiot 10:181–188. [PubMed] [Google Scholar]
- 616.Hata T, Koga F, Kanamori H. 1953. Studies on leucomycin. II. Bacteriological properties of Streptomyces kitasatoensis. J Antibiot 6:109–112. [PubMed] [Google Scholar]
- 617.Mason DJ, Lewis C. 1964. Biological activity of the lincomycin-related antibiotics. Antimicrob Agents Chemother 10:7–12. [PubMed] [Google Scholar]
- 618.Kwon HC, Kauffman CA, Jensen PR, Fenical W. 2006. Marinomycins A-D, antitumor-antibiotics of a new structure class from a marine actinomycete of the recently discovered genus “Marinispora.” J Am Chem Soc 128:1622–1632. [DOI] [PubMed] [Google Scholar]
- 619.Dulmage HT. 1951. The production of neomycin by Stretomyces fradiae 3535. Ph.D. thesis. Rutgers University, New Brunswick, NJ. [Google Scholar]
- 620.Kominek LA. 1972. Biosynthesis of novobiocin by Streptomyces niveus. Antimicrob Agents Chemother 1:123–134. doi: 10.1128/AAC.1.2.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 621.Williams DH, Rajananda V, Williamson MP, Bojesen G. 1980. The vancomycin and ristocetin group of antibiotics, p 119–158. In Sammes PG. (ed), Topics in antibiotic chemistry, vol 5 Ellis Horwood, Chichester, United Kingdom. [Google Scholar]
- 622.Rhodes PM. 1984. The production of oxytetracycline in chemostat culture. Biotechnol Bioeng 26:382–385. doi: 10.1002/bit.260260415. [DOI] [PubMed] [Google Scholar]
- 623.Blanc V, Lagneaux D, Didier P, Gil P, Lacroix P, Crouzet J. 1995. Cloning and analysis of structural genes from Streptomyces pristinaespiralis encoding enzymes involved in the conversion of pristinamycin IIB to pristinamycin IIA (PIIA): PIIA synthase and NADH:riboflavin 5′-phosphate oxidoreductase. J Bacteriol 177:5206–5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624.Pamboukian CR, Facciotti MC. 2004. Production of antitumoral retamycin during fed-batch fermentations of Streptomyces olindensis. Appl Biochem Biotechnol 112:111–122. doi: 10.1385/ABAB:112:2:111. [DOI] [PubMed] [Google Scholar]
- 625.Margalith P, Beretta G. 1960. Rifomycin. XI. Taxonomic study on Streptomyces mediterranei nov. sp. Mycopathol Mycol Applic 13:321–330. [Google Scholar]
- 626.Pinnert-Sindico S. 1954. Une nouvelle espèce de Streptomyces productrice d'antibiotiques: Streptomyces ambofaciens n. sp. Ann Inst Pasteur 87:702–707. (In French.). [PubMed] [Google Scholar]
- 627.Yanagimoto M. 1983. Novel actions of inducer in staphylomycin production by Streptomyces virginiae. J Ferment Technol 61:443–448. [Google Scholar]
- 628.Thompson RQ, Hughes MS. 1963. Stendomycin: a new antifungal antibiotic. J Antibiot 16:187–194. [PubMed] [Google Scholar]
- 629.Li XB, Qiao B, Yuan YJ. 2006. Differential analysis of secondary metabolites by LC-MS following strain improvement of Streptomyces lydicus AS 4.2501. Biotechnol Appl Biochem 45:107–118. doi: 10.1042/BA20060042. [DOI] [PubMed] [Google Scholar]
- 630.Darken MA, Berenson H, Shirk RJ, Sjolander NO. 1960. Production of tetracycline by Streptomyces aureofaciens in synthetic media. Appl Microbiol 8:46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 631.Romero F, Espliego F, Perez Baz J, Garcia de Quesada T, Gravalos D, de la Calle F, Fernandez-Puentes JL. 1997. Thiocoraline, a new depsipeptide with antitumor activity produced by a marine Micromonospora. I. Taxonomy, fermentation, isolation, and biological activities. J Antibiot 50:734–737. [DOI] [PubMed] [Google Scholar]
- 632.Brigham RB, Pittenger RC. 1956. Streptomyces orientalis, n. sp., the source of vancomycin. Antibiot Chemother (Northfield) 6:642–647. [PubMed] [Google Scholar]
- 633.Linke HA, Mechlinski W, Schaffner CP. 1974. Production of amphotericin B-14C by Streptomyces nodosus fermentation, and preparation of the amphotericin B-14C-methyl-ester. J Antibiot 27:155–160. doi: 10.7164/antibiotics.27.155. [DOI] [PubMed] [Google Scholar]
- 634.Takeuchi S, Hirayama K, Ueda K, Sakai H, Yonehara H. 1958. Blasticidin S, a new antibiotic. J Antibiot 11:1–5. [PubMed] [Google Scholar]
- 635.Acker RF, Lechevalier H. 1954. Some nutritional requirements of Streptomyces griseus 3570 for growth and candicidin production. Appl Microbiol 2:152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 636.Hohmann C, Schneider K, Bruntner C, Irran E, Nicholson G, Bull AT, Jones AL, Brown R, Stach JE, Goodfellow M, Beil W, Kramer M, Imhoff JF, Sussmuth RD, Fiedler HP. 2009. Caboxamycin, a new antibiotic of the benzoxazole family produced by the deep-sea strain Streptomyces sp. NTK 937. J Antibiot 62:99–104. doi: 10.1038/ja.2008.24. [DOI] [PubMed] [Google Scholar]
- 637.Matsuoka M, Yagishita K, Umezawa H. 1953. Studies on the intermediate metabolism of chloramphenicol production. II. On the carbohydrate metabolism of Streptomyces venezuelae. Jpn J Med Sci Biol 6:161–169. [DOI] [PubMed] [Google Scholar]
- 638.Shih HD, Liu YC, Hsu FL, Mulabagal V, Dodda R, Huang JW. 2003. Fungichromin: a substance from Streptomyces padanus with inhibitory effects on Rhizoctonia solani. J Agric Food Chem 51:95–99. doi: 10.1021/jf025879b. [DOI] [PubMed] [Google Scholar]
- 639.Doull JL, Ayer SW, Singh AK, Thibault P. 1993. Production of a novel polyketide antibiotic, jadomycin B, by Streptomyces venezuelae following heat shock. J Antibiot 46:869–871. doi: 10.7164/antibiotics.46.869. [DOI] [PubMed] [Google Scholar]
- 640.Hayashi Ki, Nozaki H. 1999. Kitamycins, new antimycin antibiotics produced by Streptomyces sp. J Antibiot 52:325–328. doi: 10.7164/antibiotics.52.325. [DOI] [PubMed] [Google Scholar]
- 641.Struyk AP, Hoette I, Drost G, Waisvisz JM, van Eek T, Hoogerheide JC. 1958. Pimaricin, a new antifungal antibiotic, p 878–885. In Welch H, Marti-Ibanez F (ed), Antibiotics annual 1957-1958. Medical Encylopedia, Inc., New York, NY. [PubMed] [Google Scholar]
- 642.Bormann C, Huhn W, Zahner H, Rathmann R, Hahn H, Konig WA. 1985. Metabolic products of microorganisms. 228. New nikkomycins produced by mutants of Streptomyces tendae. J Antibiot 38:9–16. [DOI] [PubMed] [Google Scholar]
- 643.Smith RM, Peterson WH, McCoy E. 1954. Oligomycin, a new antifungal antibiotic. Antibiot Chemother (Northfield) 4:962–970. [PubMed] [Google Scholar]
- 644.Hwang BK, Lim SW, Kim BS, Lee J Y, Moon SS. 2001. Isolation and in vivo and in vitro antifungal activity of phenylacetic acid and sodium phenylacetate from Streptomyces humidus. Appl Environ Microbiol 67:3739–3745. doi: 10.1128/AEM.67.8.3739-3745.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 645.Zhang YL, Li S, Jiang DH, Kong LC, Zhang PH, Xu JD. 2013. Antifungal activities of metabolites produced by a termite-associated Streptomyces canus BYB02. J Agric Food Chem 61:1521–1524. doi: 10.1021/jf305210u. [DOI] [PubMed] [Google Scholar]
- 646.Hoshino Y, Mukai A, Yazawa K, Uno J, Ando A, Mikami Y, Fukai T, Ishikawa J, Yamaguchi K. 2004. Transvalencin A, a thiazolidine zinc complex antibiotic produced by a clinical isolate of Nocardia transvalensis. II. Structure elucidation. J Antibiot 57:803–807. doi: 10.7164/antibiotics.57.803. [DOI] [PubMed] [Google Scholar]
- 647.Iwasa T, Yamamoto H, Shibata M. 1970. Studies on validamycins, new antibiotics. I. Streptomyces hygroscopicus var. limoneus nov. var., validamycin-producing organism. J Antibiot 23:595–602. [PubMed] [Google Scholar]
- 648.Schmitzer PR, Graupner PR, Chapin EL, Fields SC, Gilbert JR, Gray JA, Peacock CL, Gerwick BC. 2000. Ribofuranosyl triazolone: a natural product herbicide with activity on adenylosuccinate synthetase following phosphorylation. J Nat Prod 63:777–781. doi: 10.1021/np990590i. [DOI] [PubMed] [Google Scholar]
- 649.Omura S, Iwai Y, Takahashi Y, Sadakane N, Nakagawa A, Oiwa H, Hasegawa Y, Ikai T. 1979. Herbimycin, a new antibiotic produced by a strain of Streptomyces. J Antibiot 32:255–261. doi: 10.7164/antibiotics.32.255. [DOI] [PubMed] [Google Scholar]
- 650.Omura S, Crump A. 2004. The life and times of ivermectin: a success story. Nat Rev Microbiol 2:984–989. doi: 10.1038/nrmicro1048. [DOI] [PubMed] [Google Scholar]
- 651.Box SJ, Cole M, Yeoman GH. 1973. Prasinons A and B: potent insecticides from Streptomyces prasinus. Appl Microbiol 26:699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 652.Waldron C, Matsushima P, Rosteck PR Jr, Broughton MC, Turner J, Madduri K, Crawford KP, Merlo DJ, Baltz RH. 2001. Cloning and analysis of the spinosad biosynthetic gene cluster of Saccharopolyspora spinosa. Chem Biol 8:487–499. doi: 10.1016/S1074-5521(01)00029-1. [DOI] [PubMed] [Google Scholar]
- 653.Cerdeno AM, Bibb MJ, Challis GL. 2001. Analysis of the prodiginine biosynthesis gene cluster of Streptomyces coelicolor A3(2): new mechanisms for chain initiation and termination in modular multienzymes. Chem Biol 8:817–829. doi: 10.1016/S1074-5521(01)00054-0. [DOI] [PubMed] [Google Scholar]
- 654.Tomita F, Tamaoki T, Morimoto M, Fujimoto K. 1981. Trioxacarcins, novel antitumor antibiotics. I. Producing organism, fermentation and biological activities. J Antibiot 34:1519–1524. [DOI] [PubMed] [Google Scholar]
- 655.Farmer PB, Suhadolnik RJ. 1972. Nucleoside antibiotics. Biosynthesis of arabinofuranosyladenine by Streptomyces antibioticus. Biochemistry 11:911–916. [DOI] [PubMed] [Google Scholar]
- 656.Aoyagi T, Yagisawa M, Kumagai M, Hamada M, Okami Y. 1971. An enzyme inhibitor, panosialin, produced by Streptomyces. I. Biological activity, isolation and characterization of panosialin. J Antibiot 24:860–869. [DOI] [PubMed] [Google Scholar]
- 657.Igarashi Y, Trujillo ME, Martinez-Molina E, Yanase S, Miyanaga S, Obata T, Sakurai H, Saiki I, Fujita T, Furumai T. 2007. Antitumor anthraquinones from an endophytic actinomycete Micromonospora lupini sp. nov. Bioorg Med Chem Lett 17:3702–3705. doi: 10.1016/j.bmcl.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 658.Nemoto A, Hoshino Y, Yazawa K, Ando A, Mikami Y, Komaki H, Tanaka Y, Grafe U. 2002. Asterobactin, a new siderophore group antibiotic from Nocardia asteroides. J Antibiot 55:593–597. doi: 10.7164/antibiotics.55.593. [DOI] [PubMed] [Google Scholar]
- 659.Vino S, Lokesh KR. 2008. Borrelidin: a promising anticancer agent from Streptomyces species. Adv Biotech 6:22–26. [Google Scholar]
- 660.Charan RD, Schlingmann G, Janso J, Bernan V, Feng X, Carter GT. 2004. Diazepinomicin, a new antimicrobial alkaloid from a marine Micromonospora sp. J Nat Prod 67:1431–1433. doi: 10.1021/np040042r. [DOI] [PubMed] [Google Scholar]
- 661.Malet-Cascon L, Romero F, Espliego-Vazquez F, Gravalos D, Fernandez-Puentes JL. 2003. IB-00208, a new cytotoxic polycyclic xanthone produced by a marine-derived Actinomadura. I. Isolation of the strain, taxonomy and biological activites. J Antibiot 56:219–225. doi: 10.7164/antibiotics.56.219. [DOI] [PubMed] [Google Scholar]
- 662.Kanoh K, Matsuo Y, Adachi K, Imagawa H, Nishizawa M, Shizuri Y. 2005. Mechercharmycins A and B, cytotoxic substances from marine-derived Thermoactinomyces sp. YM3-251. J Antibiot 58:289–292. doi: 10.1038/ja.2005.36. [DOI] [PubMed] [Google Scholar]
- 663.Williams PG, Buchanan GO, Feling RH, Kauffman CA, Jensen PR, Fenical W. 2005. New cytotoxic salinosporamides from the marine Actinomycete Salinispora tropica. J Org Chem 70:6196–6203. doi: 10.1021/jo050511+. [DOI] [PubMed] [Google Scholar]
- 664.Arcamone F, Cassinelli G, Fantini G, Grein A, Orezzi P, Pol C, Spalla C. 1969. Adriamycin, 14-hydroxydaunomycin, a new antitumor antibiotic from S. peucetius var. caesius. Biotechnol Bioeng 11:1101–1110. doi: 10.1002/bit.260110607. [DOI] [PubMed] [Google Scholar]
- 665.White RJ, Stroshane RM. 1984. Daunorubicin and adriamycin: properties, biosynthesis, and fermentation, p 569–594. In Vandamme EJ. (ed), Biotechnology of industrial antibiotics, vol 22 Marcel Dekker, Inc., New York, NY. [Google Scholar]
- 666.Perez Baz J, Canedo LM, Fernandez Puentes JL, Silva Elipe MV. 1997. Thiocoraline, a novel depsipeptide with antitumor activity produced by a marine Micromonospora. II. Physico-chemical properties and structure determination. J Antibiot 50:738–741. [DOI] [PubMed] [Google Scholar]
- 667.de Reijke TM, de Boer EC, Schamhart DH, Kurth KH. 1997. Immunostimulation in the urinary bladder by local application of Nocardia rubra cell wall skeleton preparation (Rubratin) for superficial bladder cancer immunotherapy: a phase I/II study. Urol Res 25:117–120. doi: 10.1007/BF01037926. [DOI] [PubMed] [Google Scholar]
- 668.Blomgren H, Strender LE, Edsmyr F. 1980. Bestatin treatment and the peripheral lymphocyte population in cancer patients. Recent Res Cancer 75:133–138. doi: 10.1007/978-3-642-81491-4_21. [DOI] [PubMed] [Google Scholar]
- 669.Iwami M, Nakayama O, Terano H, Kohsaka M, Aoki H, Imanaka H. 1987. A new immunomodulator, FR-900494: taxonomy, fermentation, isolation, and physico-chemical and biological characteristics. J Antibiot 40:612–622. doi: 10.7164/antibiotics.40.612. [DOI] [PubMed] [Google Scholar]
- 670.Komaki H, Nemoto A, Tanaka Y, Takagi H, Yazawa K, Mikami Y, Shigemori H, Kobayashi J, Ando A, Nagata Y. 1999. Brasilicardin A, a new terpenoid antibiotic from pathogenic Nocardia brasiliensis: fermentation, isolation and biological activity. J Antibiot 52:13–19. doi: 10.7164/antibiotics.52.13. [DOI] [PubMed] [Google Scholar]
- 671.Uyeda M, Mizukami M, Yokomizo K, Suzuki K. 2001. Pentalenolactone I and hygromycin A, immunosuppressants produced by Streptomyces filipinensis and Streptomyces hygroscopicus. Biosci Biotechnol Biochem 65:1252–1254. doi: 10.1271/bbb.65.1252. [DOI] [PubMed] [Google Scholar]
- 672.DeJong P. 1972. l-Asparaginase production by Streptomyces griseus. Appl Microbiol 23:1163–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 673.Balagurunatha R, Radhakrishnon M, Somasumdarom ST. 2010. l-Glutaminase producing Actinomycetes from marine sediments-selective isolation, semiquantitative assay and characterization of potential strain. Aust J Basic Appl Sci 4:698–705. [Google Scholar]
- 674.Chen Y, Krol J, Sterkin V, Fan W, Yan X, Huang W, Cino J, Julien C. 1999. New process control strategy used in as rapamycin fermentation. Process Biochem 34:383–389. doi: 10.1016/S0032-9592(98)00108-3. [DOI] [Google Scholar]
- 675.Fauth U, Zahner H, Muhlenfeld A, Achenbach H. 1986. Galbonolides A and B: two non-glycosidic antifungal macrolides. J Antibiot 39:1760–1764. doi: 10.7164/antibiotics.39.1760. [DOI] [PubMed] [Google Scholar]
- 676.Göker M, Klenk H. 2013. Phylogeny-driven target selection for large-scale genome-sequencing (and other) projects. Stand Genomic Sci 8:360–374. doi: 10.4056/sigs.3446951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 677.Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 678.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. 2013. Highly parallelized inference of large genome-based phylogenies. Concurrency Comput Pract Experience 26:1715–1729. doi: 10.1002/cpe.3112. [DOI] [Google Scholar]
- 679.Letunic I, Bork P. 2011. Interactive Tree of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]