Abstract
The rising interest in understanding the functions, regulation and maintenance of the epitranscriptome calls for robust and accurate analytical methods for the identification and quantification of post-transcriptionally modified nucleosides in RNA. Mono-methylations of cytidine and adenosine are common post-transcriptional modifications in RNA. Herein, we developed an LC-MS/MS/MS coupled with the stable isotope-dilution method for the sensitive and accurate quantifications of 5-methylcytidine (m5C), 2′-O-methylcytidine (Cm), N6-methyladenosine (m6A) and 2′-O-methyladenosine (Am) in RNA isolated from mammalian cells and tissues. Our results showed that the distributions of the four methylated nucleosides are tissue-specific. In addition, the 2′-O-methylated ribonucleosides (Cm and Am) are present at higher levels than the corresponding methylated nucleobase products (m5C and m6A) in total RNA isolated from mouse brain, pancreas and spleen, but not mouse heart. We also found that the levels of m5C, Cm and Am are significantly lower (by 6.5-43 fold) in mRNA than in total RNA isolated from HEK293T cells, whereas the level of m6A was slightly higher (by 1.6 fold) in mRNA than in total RNA. The availability of this analytical method, in combination with genetic manipulation, may facilitate the future discovery of proteins involved in the maintenance and regulation of these RNA modifications.
Introduction
More than 100 types of post-transcriptional modifications are known to exist in RNA and they play very important roles in the metabolic and regulatory processes of RNA. The biological functions of individual types of RNA modifications and their contributions to gene regulation remain largely unknown.1 Among these RNA modifications, mono-methylated cytidine and adenosine, including 5-methylcytidine (m5C), 2′-O-methylcytidine (Cm), N6-methyladenosine (m6A) and 2′-O-methyladenosine (Am) commonly occur in all RNA species.2
Previous investigations about the mono-methylated ribonucleosides have been mostly confined to transfer RNA (tRNA) and ribosomal RNA (rRNA), especially for cytidine modifications. It has been reported that m5C contributes to the stabilization of secondary structures, codon recognition, and aminoacylation of tRNA.3-5 In rRNA, m5C sites have been thought to regulate translational fidelity and tRNA recognition.6 Cm in tRNA was found to prevent the hydrolysis of the phosphodiester backbone,7 whereas the Cm located in the cap structure of mRNA inhibits its 5′→3′ degradation8 and distinguishes self from non-self RNA.9
Aside from their functions in tRNA and rRNA, recent studies suggested that nucleobase methylations in mRNA may also play a very important role in gene regulation. In this vein, transcriptome-wide mapping studies have revealed a widespread occurrence of m5C and m6A in messenger RNA (mRNA) and non-coding RNA.10-12 Sequencing data indicate that m6A is localized around stop codons and present in both 3′-untranslated regions (3′-UTRs) and long internal exons,10 whereas the m5C sites are enriched in untranslated regions and near Argonaute protein binding sites.12 Those studies suggested that m6A may modulate pre-mRNA splicing, mRNA stability, translation, turnover and nuclear export,13-15 whereas m5C may play a role in microRNA (miRNA)-mediated mRNA degradation and affect the interactions of long non-coding RNA with chromatin-associated protein complexes.12, 16
The recent identification of enzymes fostering RNA methylation and demethylation highlights the importance in furthering our current understanding of the role of RNA methylation in gene regulation. In this vein, FTO (Fat mass and obesity-associated protein) and ALKBH5 (Alkylated DNA repair protein alkB homolog 5) were found to be capable of demethylating m6A in mRNA.17, 18 Subsequently, human YTH domain family proteins (YTHDF1-3) were shown to bind to m6A and affect the stability of m6A-harboring mRNA.19 Furthermore, the heterodimeric METTL3-METTL14 (human methyltransferase-like 3 and 14) core-complex was observed to deposit m6A on mammalian nuclear RNAs.20 Apart from these regulatory proteins of m6A, the ten-eleven translocation family of Fe(II)- and 2-oxoglutarate-dependent dioxygenases 3 (Tet3) can induce the formation of 5-hydroxymethylcytidine from m5C in cellular RNA.21 Additionally, TRDMT1 (tRNA aspartic acid methyltransferase 1) and NSUN2 (NOP2/Sun domain family, member 2) have been identified to be the cytosine-5-methyltransferase in tRNA and mRNA.12, 22 Taken together, the identification and characterizations of proteins involved in the deposition, removal and recognition of m6A and m5C provide strong support for a reversible post-transcriptional modification pathway of RNA, which may constitute an important, yet under-appreciated mechanism of gene regulation.23, 24
To better exploit the mechanisms of RNA epigenetics, a robust analytical method is required for assessing the occurrence of these modifications in cellular RNA. Traditional methods for analyzing RNA modifications include 32P-labelling and two-dimensional thin-layer chromatography,25 dot-blot,18 and capillary electrophoresis coupled with laser-induced fluorescence (CE-LIF) detection.26 Apart from being tedious, semi-quantitative, and low-throughput, these methods require a large amount of RNA and are not compatible with the analysis of RNA species of low abundance. Recently, high-performance liquid chromatography coupled with a triple-quadrupole mass spectrometer, along with the use of external standards, was employed to measure m6A in mRNA 17, 18 and other RNA modifications in tRNA and small RNA, and femtomole level of sensitivity was achieved.27, 28 We reason that the application of stable isotope-labeled internal standards will offer unambiguous and accurate measurements of these post-transcriptionally modified nucleosides in cellular RNA species. Herein, we developed an LC-MS/MS/MS coupled with the stable isotope-dilution method to achieve sensitive, accurate and simultaneous quantifications of the global levels of the mono-methylated cytidine and adenosine in RNA. By using this method, we quantified the levels of m5C, Cm, m6A, and Am in total RNA isolated from cultured human cells and mammalian tissues, and in mRNA isolated from HEK293T human embryonic kidney cells.
Experiment Section
Materials
All chemicals and enzymes, unless otherwise specified, were purchased from Sigma-Aldrich (St. Louis, MO) and New England Biolabs (Ipswich, WA). Erythro-9-(2-hydroxy-3-nonyl) adenine (EHNA) hydrochloride was purchased from Tocris Bioscience (Ellisville, MO). 15N3-cytidine-5′-triphosphate was obtained from Sigma-Aldrich (St. Louis, MO) and all other stable isotope-labeled nucleoside starting materials were from Cambridge Isotope Laboratories (Tewksbury, MA). Mouse tissues were obtained from 19-21 week old animals. The HEK293T embryonic kidney cells, MCF7 human breast cancer cells,HCT116 human colon cancer cells, HeLa human cervical cancer cells, WM-266-4 human melanoma cells, and cell culture reagents were purchased from ATCC (Manassas, VA).
Syntheses of Stable Isotope-labeled Ribonucleosides
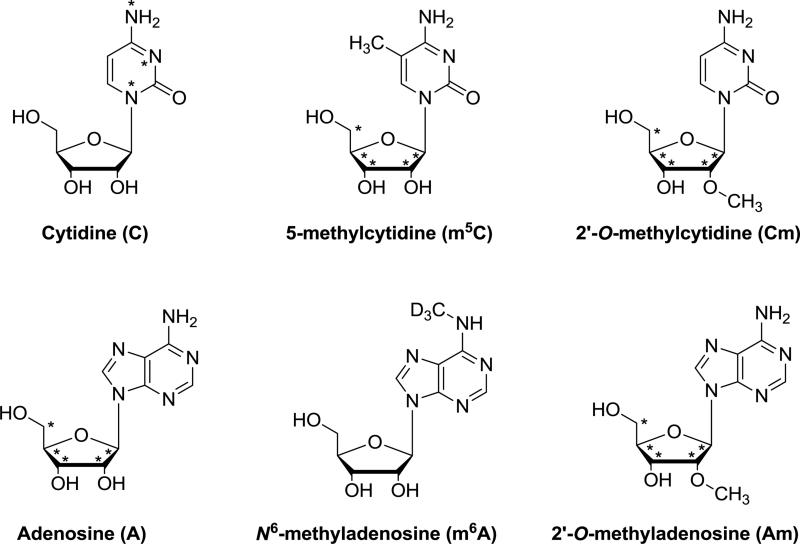
The stable isotope-labeled nucleosides employed in this study are shown in Scheme 1.
Scheme 1.
The chemical structures of the stable isotopic-labeled nucleosides. Asterisk (*) indicates the site of 15N or 13C labeling; D = deuterium.
15N3-cytidine
15N3-cytidine-5′-triphosphate was treated with alkaline phosphatase in 50 mM Tris-HCl buffer (pH 8.9) at 37°C for 2 hrs. The enzyme was removed by chloroform extraction and the aqueous layer was dried in a Speed-vac. The resulting 15N3-cytidine was purified by HPLC.
5-methyl-13C5-cytidine and 13C5-adenosine
Ribose-13C5-cytidine (5.0 mg, 0.020 mmol) was fully acetylated by treating with acetic anhydride (40 μL, 0.409 mmol) at 60°C for 3 hrs in 1-mL anhydrous pyridine. The resulting crude tetra-acetylated ribose-13C5-cytidine was dissolved in anhydrous acetonitrile (1 mL) in the presence of 5-methyl-N4-benzoylcytosine (7.4 mg, 0.032 mmol) or N6-benzoyladenine (9.7 mg, 0.040 mmol) and stirred at room temperature for 10 min. Bis-trimethylsilylacetamide (20 μL, 0.070 mmol) was subsequently added and the reaction mixture was heated to 70°C. After stirring at 70°C for 15 min, TMS-triflate (4 μL, 0.020 mmol) was added to the reaction flask and the reaction was refluxed at 70°C for 4 hrs. The solvent was removed, and the resulting crude mixture was dissolved in 2 M ammonia in methanol (4 mL) and stirred at 40°C for 24 hrs. Subsequently, 30% ammonium hydroxide (4 mL) was added to the reaction mixture and stirred at 40°C for 48 hrs. The resulting crude 5-methyl-13C5-cytidine and 13C5-adenosine were evaporated of solvent and purified by HPLC using a reverse-phase Alltima C18 column (5 μm in particle size, Grace Davison, Deerfield, IL). The purified 5-methyl-13C5-cytidine and 13C5-adenosine were confirmed by LC-MS and MS/MS analyses (Figure S1). A portion of the purified 13C5-adenosine was then used to synthesize 2′-O-methyl-13C5-adenosine.
2′-O-methyl-13C5-cytidine, 2′-O-methyl-13C5-adenosine and D3-N6-methyladenosine
2′-O-methyl-13C5-cytidine and 2′-O-methyl-13C5-adenosine were synthesized according to established procedures.29 6-Chloro-9-(β-D-ribofuranosyl)purine was synthesized following published procedures 30 and then reacted with D3-methylamine to yield D3-N6-methyladenosine.31
Isolation of total RNA and mRNA
Total RNA was isolated from mammalian cells and tissues using TRI Reagent® following the manufacturer's recommended procedures. The poly(A) messenger RNA (mRNA) was extracted using PolyATtract® mRNA Isolation Systems (Promega), immediately followed with the removal of rRNA contaminations by using RiboMinus Transcriptome Isolation Kit (Invitrogen). The mRNA concentrations were measured using UV spectrophotometry. The quality of mRNA was analyzed using an Agilent 2100 Bioanalyzer equipped with an RNA PicoChip, and the results showed that the isolated mRNA samples were free of rRNA contamination (Figure S2).
Digestion of RNA
To 500 ng of RNA were added 0.05 unit of nuclease P1, 0.125 nmol of EHNA, 0.0000625 unit of phosphodiesterase 2 and 1 μL solution containing 300 mM sodium acetate (pH 5.6) and 10 mM zinc chloride. The EHNA was added to minimize the potential deamination of adenosine. Doubly distilled water (ddH2O) was added to the reaction mixture to reach a final volume of 10 μL. The reaction mixture was incubated at 37°C for 4 hrs. To the resulting mixture were subsequently added 0.05 unit of alkaline phosphatase, 0.005 unit of phosphodiesterase 1, 1.5 μL of 0.5 M Tris-HCl buffer (pH 8.9) and ddH2O to reach a final volume of 15 μL. After digestion at 37°C for 2 hrs, the resulting digestion mixture was dried using a Speed-vac and the dried residue was reconstituted in 500 μL of ddH2O.
For the quantification of m5C and Cm in total RNA, to a 5-μL aliquot of the digestion mixture of total RNA (5 ng) were added 25.5 fmol of 5-methyl-13C5-cytidine, 19.6 fmol of 2′-O-methyl-13C5-cytidine and 3.3 pmol of 15N-labeled cytidine. For the quantification of m6A and Am in total RNA, to a 1-μL aliquot of total RNA (1 ng) were added 8.5 fmol D3-N6-methyladenosine, 6.9 fmol of 2′-O-methyl-13C5-adenosine and 1.6 pmol 13C5-labeled adenosine. For the quantification of m5C and Cm in mRNA, to a 10-μL aliquot of the digestion mixture of mRNA (10 ng) were added 5.1 fmol of 5-methyl-13C5-cytidine, 9.7 fmol of 2′-O-methyl-13C5-cytidine and 3.3 pmol of 15N-labeled cytidine. For the quantification of m6A and Am in mRNA, 8.5 fmol D3-N6-methyladenosine, 3.4 fmol of 2′-O-methyl-13C5-adenosine and 1.6 pmol 13C5-labeled adenosine were added to a 2-μL aliquot of the digestion mixture of mRNA. All enzymes used for RNA digestion were subsequently removed by chloroform extraction. The resulting aqueous layer was dried and reconstituted in ddH2O. For the total RNA samples, ¼ of the total sample was used for nLC-MS3 analysis, while for the mRNA, ½ of the total sample was subjected to nLC-MS3 analysis.
LC-MS3 Analyses of m5C, Cm, m6A and Am
LC-MS3 measurements were conducted on an LTQ XL linear ion trap mass spectrometer equipped with a nanoelectrospray ionization source and coupled to an EASY-nLC II (Thermo Fisher Scientific, San Jose, CA, USA). The temperature for the ion transport tube of the mass spectrometer was maintained at 275°C. The instrument was operated in the positive-ion mode, with the spray, capillary, and tube lens voltages being 2.0 kV, 12 V, and 100 V, respectively. The sensitivities for detecting the four mono-methylated ribonucleosides were optimized by varying the normalized collision energy and activation Q of the LTQ mass spectrometer (Table S1 in the Supporting Information).
For the measurements of m5C and Cm, the samples were loaded onto a pre-column (150 μm × 70 mm) packed with porous graphitic carbon (PGC, 5 μm in particle size, Thermo Fisher Scientific). The samples were then eluted, at a flow rate of 2.5 μL/min, onto an in-house packed Zorbax SB-C18 column (75 μm × 250 mm, 5 μm beads, 100 Å in pore size, Agilent). A solution of 0.1% (v/v) formic acid in water (solution A) and a solution of 0.1% (v/v) formic acid in acetonitrile (solution B) were used as the mobile phases. The modified nucleosides were separated using a gradient of 0-5% B in 5 min, 5-15% B in 37 min, 15% B in 18 min, 15-90% B in 1 min, and finally at 90% B for 10 min. The flow rate was 300 nL/min.
The measurements of m6A and Am were conducted in a similar way except that a 150 μm × 50 mm pre-column and a 75 μm × 150 mm analytical column, packed with Magic AQ reversed-phase C18 resin (5 μm beads, 100 Å in pore size; Michrom BioResources, Auburn, CA, USA), were used. The modified nucleosides were separated using a gradient of 0-10% B in 50 min followed by 10% B for another10 min, and the flow rate was 300 nL/min.
Method Validation
The intra- and inter-day accuracy and precision were assessed by analyzing standard solutions of the methylated ribonucleosides at three different concentrations, following the Food and Drug Administration Guidance for Industry Bioanalytical Method Validation (http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm368107.pdf). At least 5 determinations were made for each replicate and at least 3 replicates were analyzed for each concentration of the standard solutions. The intra-day results were obtained from 5 determinations of one replicate within one day. The inter-day results were obtained from at least 15 determination of three replicate analyses conducted on 3 separate days. The mean accuracy was expressed as percent recovery and the precision was expressed as relative standard deviation (RSD). The stabilities of analytes present in total RNA were evaluated after three cycles of freeze (at - 20°C for 24 hrs) and thaw (at room temperature). The percent recovery was calculated by comparing the levels after the freeze-and-thaw cycles with those prior to freeze-and-thaw, and the relative standard deviations was calculated based on the recovery obtained from 3 aliquots of RNA samples.
Results
Nano-LC-MS/MS/MS analyses of m5C, Cm, m6A and Am
We set out to develop an nLC-MS/MS/MS in combination with the stable isotope-dilution method for the accurate assessment of the levels of m5C, Cm, m6A and Am in total RNA isolated from cultured cells and tissues. First, we examined the efficiencies of the pre-columns packed with various stationary phase materials including Zorbax SB-C18, Magic C18, Magic C18-AQ and porous graphitic carbon (PGC) in trapping the modified nucleosides. We found that m5C and Cm could be retained very well on a trapping column packed with PGC, but not on the other three types of packing materials. On the other hand, Magic C18-AQ displayed the most efficient trapping of m6A and Am.
We next tested the performance of the analytical columns packed with the four types of stationary phase materials. Our results showed that, even though m5C and Cm could be efficiently trapped on the PGC column, the use of PGC as the packing material for the analytical column yielded poor reproducibility and displayed severe issues with matrix interferences for the modified cytidines. On the other hand, the analytical columns packed with Magic C18 AQ or Zorbax SB-C18 exhibited excellent reproducibility and low matrix interferences when used with a slow gradient, with the Zorbax SB-C18 analytical column displaying slightly better performance. Therefore, we chose the combination of PGC trapping column with Zorbax SB-C18 analytical column for the analyses of m5C and Cm. On the other hand, we found that the use of Magic C18 AQ as the stationary phase material for both the trapping and analytical columns was the most suitable for measuring the two modified adenosine derivatives, i.e. m6A and Am.
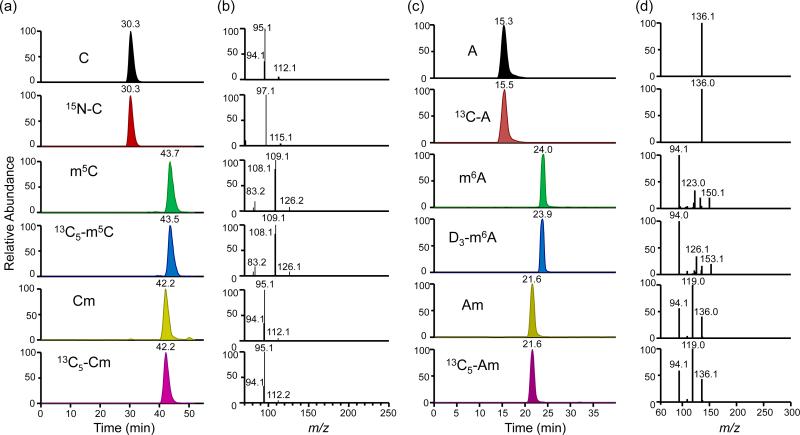
Upon collisional activation, the [M+H]+ ions of the four unlabeled methylated ribonucleosides readily eliminate the ribose moiety to yield the protonated ions of the nucleobase portions (i.e. m/z 126, 112, 150 and 136 for m5C, Cm, m6A and Am, respectively). Further collisional activation of the ions of m/z 126 and 112 leads to the facile losses of NH3 and H2O, yielding the fragment ions of m/z 109 and 108 in the MS3 for m5C, and the fragment ions of m/z 95 and 94 in the MS3 for Cm (Figure 1, b). On the other hand, collisional activation of the ion of m/z 150 results in the loss of C2H4N2 and HCN to yield product ions of m/z 94 and 123 in the MS3 of m6A. Additionally, further collisional activation of the ions of m/z 136 of Am gives rise to the elimination of C2H4N2 and NH3, yielding ions of m/z of 94 and 119, respectively, in the MS3 (Figure 1, d). The fragment ions of m/z 108, 95, 94 and 94 observed in the MS3 of these modified nucleosides were chosen for the quantification of the levels of m5C, Cm, m6A and Am, respectively (see representative SICs in Figure 1a & c). The nearly identical elution time and similar MS3 spectra for the analytes and their stable isotope-labeled counterparts, permit for the unambiguous identification and reliable quantification of the four modified ribonucleosides in the digestion mixture of total RNA. Calibration curves for the quantifications of rC, m5C, Cm, rA, m6A, and Am are shown in Figures S3-S4.
Figure 1.
Representative LC-MS/MS/MS results for the quantifications of m5C, Cm, m6A and Am in mouse brain. Shown are the selected-ion chromatograms for monitoring the indicated transitions for the analytes and the stable isotope-labeled standards (a & c), and the corresponding MS/MS/MS for the analytes and internal standards (b & d).
We next examined the limits of detection (LOD) and limits of quantification (LOQ) for the methylated nucleosides, which are defined as the amounts of analytes that give rise to signal-to-noise ratios of 3 and 10, respectively. Our results showed that low attomole levels of LOD and LOQ could be obtained for all the methylated ribonucleosides (Table S1).
We also assessed the intra- and inter-day accuracy and precision by analyzing three different concentrations of standard solutions of the methylated ribonucleosides. As shown in Tables S2, the method provides excellent accuracy and precision for measuring the methylated nucleosides. The percent recoveries for the four methylated ribonucleosides range from 89.6% to 105%, and the relative standard deviations for all the analytes were within 10%, with the exception of the measurement for 1.25 nM of m5C (12%, Table S2). We further investigated the stabilities of analytes present in total RNA after three cycles of freeze (at - 20°C for 24 hrs) and thaw (to room temperature). Our results showed that the analytes are reasonably stable under freezing/thaw conditions, as reflected by the observed percent recovery of 81-122% (Table S3).
Quantification of m5C, Cm, m6A and Am in total RNA isolated from mammalian tissues
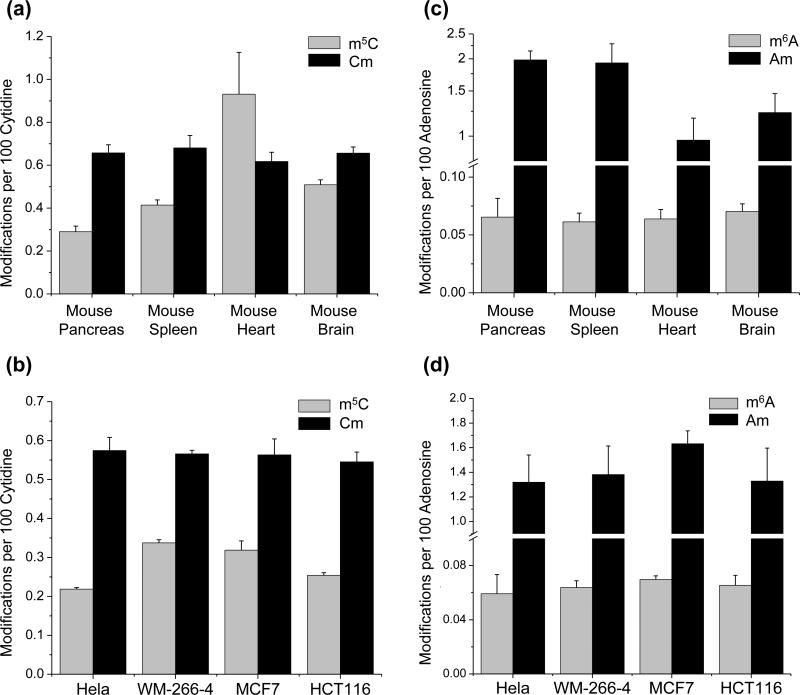
We first assessed the levels of the four methylated ribonucleosides in total RNA isolated from different mouse tissues. Our results showed that the levels of m5C were 0.29, 0.41, 0.93, and 0.51 modifications per 100 cytidines in the total RNA isolated from mouse pancreas, spleen, heart, and brain tissues, respectively, while the corresponding levels of Cm were 0.66, 0.68, 0.62, and 0.66 modifications per 100 cytidines, respectively (Figure 2a). In addition, the levels of Am (at 1.98, 1.93, 0.97 and 1.24 modifications per 100 adenosines, respectively) were significantly higher than those of m6A (at 0.065, 0.061, 0.064 and 0.070 modifications per 100 adenosines, respectively. Figure 2c). Furthermore, the levels of m5C are significantly higher in the heart than in other three types of mouse tissues. However, the levels of Cm and Am in RNA from the mouse heart are lower than those measured in RNA from the other three types of mouse tissues. Together, these results suggest that the distributions of these methylated ribonucleosides are tissue-specific.
Figure 2.
Quantification results for the levels of m5C and Cm (a), m6A and Am (c) in mouse tissues (n≥3). The tissue types include mouse pancreas, spleen, heart, and brain. Quantification results for the levels of m5C and Cm (b), m6A and Am (d) in cultured human cancer cells (n=3). The data represent the means and standard deviations of results from at least three separate mouse tissues or 3 individual RNA samples extracted from cultured human cells.
Quantification of m5C, Cm, m6A and Am in total RNA of cultured human cells
To evaluate if the levels of m5C, Cm, m6A and Am vary among different cancer cells, we isolated total RNA from four different human cancer cell lines, digested them with enzymes and subjected the resulting digestion mixtures to LC-MS3 analyses. Our results showed that the levels of m5C were 0.22, 0.34, 0.32, and 0.25 modifications per 100 cytidines in total RNA isolated from HeLa, WM-266-4, MCF7, and HCT116 cells, respectively, whereas the levels of Cm were consistently higher, at 0.57, 0.57, 0.56, and 0.55 modifications per 100 cytidines, respectively (Figure 2b). Additionally, the levels of m6A in total RNA isolated from these four cell lines were 0.059, 0.064, 0.070 and 0.065 modifications per 100 adenosines, respectively, whereas the levels of Am were 1.32, 1.38, 1.63 and 1.33 modifications per 100 adenosines, respectively (Figure 2d). Thus, these results indicate that the levels of Am and Cm are significantly higher in all human cancer cell lines compared to their respective mono-methylated nucleobase modifications (m6A and m5C). Additionally, these results indicate that the levels of m6A, Am, and Cm are similar among the human cancer cell lines, while the levels of m5C appear to be cell line-dependent. Finally, we measured the levels of these four mono-methylated nucleosides in the total RNA isolated from HEK293T human embryonic kidney cells (Figure 3, a). Our results showed that the levels of m6A, Am, and Cm in HEK293T cells are not significantly different from what we found for the cancer cells except the level of Am between HEK293T and MCF7 cells (p = 0.01). Intriguingly, the level of m5C in total RNA of HEK293T cells (0.28 modifications per 100 cytidines) is slightly higher than those found for HeLa and HCT116 cells, but slightly lower than those observed for WM-266-4 and MCF-7 cells. Taken together, these results suggest that the m5C levels in total RNA are cell line-dependent, whereas the levels of m6A, Am, and Cm are similar in cultured cancer cells and HEK293T cells.
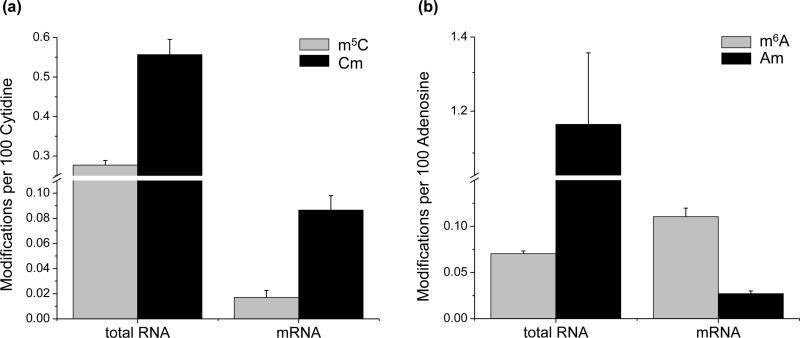
Figure 3.
Quantification results for the levels of m5C and Cm (a), m6A and Am (b) in total RNA and mRNA isolated from HEK 293T cells (n=4). The data represent the mean and standard deviation of measurement results for at least three separate total RNA and mRNA samples.
Quantification of m5C, Cm, m6A and Am in mRNA isolated from HEK293T cells
Lastly, we compared the global levels of these four mono-methylated ribonucleosides in mRNA isolated from HEK293T cells. Our results showed that the levels of m5C, Cm and Am in mRNA are lower than those in total RNA by16, 6.5, and 43 folds, respectively, whereas the level of m6A in mRNA is 1.6-fold higher than that in total RNA. The relatively large differences in the levels of m5C, Cm and Am between total RNA and mRNA are reasonable considering that mRNA constitutes only approximately 5% of the total cellular RNA. These results are also consistent with previous reports showing that m6A is the most abundant methylation product in mRNA.32-34 However, our measured level of m6A in mRNA from HEK293T cells (0.11 per 100 adenosines) is significantly lower than the previously reported level of m6A (~ 0.4 per 100 adenosine).18 This difference might be attributed to the methods through which the levels of m6A were quantified. Here, we employed stable isotope-labeled internal standards for the quantification, which offers unambiguous identification and accurate measurement of the levels of the analyte. On the other hand, external standards were utilized in the previous report,18 where the measured levels of the modified nucleosides could be potentially influenced by matrix effects. Our quantification results also showed that the levels of m5C, Cm, and Am in mRNA were 0.017, 0.086, and 0.024 modifications per 100 cytidines, respectively. These results demonstrate a higher level of Cm than m5C in both total RNA and mRNA of HEK293T cells. However, the relative level of m6A and Am displayed an opposite trend in total RNA and mRNA, with the level of m6A being higher than Am in mRNA, and lower than Am in total RNA.
Conclusions
In this study, we developed an LC-MS/MS/MS coupled with the stable isotope-dilution method to detect the levels of m5C, Cm, m6A and Am in total RNA isolated from cultured mammalian cells and tissues, as well as in mRNA isolated from HEK293T cells. This method has several advantages compared to previously reported methods. First, the measured levels of the analytes are not affected by alterations in sample matrices or LC-MS/MS/MS conditions because of the addition of stable isotope-labeled standards to the nucleoside mixture. Moreover, the analytes and the corresponding internal standards are analyzed simultaneously by LC-MS/MS/MS under identical conditions. Any variations in experimental conditions after enzymatic digestion and during LC-MS/MS/MS analysis do not affect the analytical accuracy. Second, our method allows for the unambiguous identification of each analyte. Both the analytes and their isotope-labeled standards co-elute and yield the same fragmentation patterns, offering unequivocal chemical specificity for analyte identification. Lastly, this method displays superior sensitivity. The limits of quantification for m5C, Cm, m6A and Am with our method were found to be 9±2, 7±2, 1.9±0.6 and 3.4±1.2 amol, respectively (Supplementary Table S1). A few μg of RNA was used in previously published methods, whereas nucleoside mixtures from digestion of only 0.5 ng of total RNA and less than 10 ng of mRNA were used for the analyses with this method.
Together, we developed a robust LC-MS3 coupled with the stable isotope-dilution method for the quantifications of m5C, Cm, m6A, and Am in total RNA isolated from mammalian tissues and cultured human cells. We were also able to accurately measure the levels of four mono-methylated ribonucleosides in mRNA isolated from HEK293T cells. To our knowledge, this is the first report about the global levels of m5C, Cm and Am in mRNA. It can be envisaged that this method can be generally applicable for examining the role of proteins involved in the deposition, removal and recognition of those RNA methylations. Although the emphasis of the present study was placed on cellular RNA, the method, owing to its excellent sensitivity, should also be applicable toward the analysis of these methylated ribonucleosides in mRNA isolated from animal tissues and in non-coding RNA.
Supplementary Material
Acknowledgement
The authors would like to thank the National Institutes of Health for supporting this research (R01 CA101864 and P01 AG043376). N.J.A. was supported by an NRSA Institutional Training Grant (T32 ES018827)
Footnotes
Supporting Information Available. Optimized instrumental parameters, LC-MS/MS data, method validation results, and calibration curves. This material is available free of charge via the Internet at http://pubs.acs.org.
References:
- 1.Dominissini D. Science. 2014;346:1192. doi: 10.1126/science.aaa1807. [DOI] [PubMed] [Google Scholar]
- 2.Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, Helm M, Bujnicki JM, Grosjean H. Nucleic Acids Res. 2013;41:D262–267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helm M. Nucleic Acids Res. 2006;34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motorin Y, Helm M. Biochemistry. 2010;49:4934–4944. doi: 10.1021/bi100408z. [DOI] [PubMed] [Google Scholar]
- 5.Squires JE, Preiss T. Epigenomics. 2010;2:709–715. doi: 10.2217/epi.10.47. [DOI] [PubMed] [Google Scholar]
- 6.Chow CS, Lamichhane TN, Mahto SK. ACS Chem Biol. 2007;2:610–619. doi: 10.1021/cb7001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawai G, Yamamoto Y, Kamimura T, Masegi T, Sekine M, Hata T, Iimori T, Watanabe T, Miyazawa T, Yokoyama S. Biochemistry. 1992;31:1040–1046. doi: 10.1021/bi00119a012. [DOI] [PubMed] [Google Scholar]
- 8.Cougot N, van Dijk E, Babajko S, Seraphin B. Trends Biochem Sci. 2004;29:436–444. doi: 10.1016/j.tibs.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Jr., Shi PY, Diamond MS. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 11.Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R. PLoS Genet. 2013;9:e1003602. doi: 10.1371/journal.pgen.1003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia G, Fu Y, He C. Trends Genet. 2013;29:108–115. doi: 10.1016/j.tig.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niu Y, Zhao X, Wu YS, Li MM, Wang XJ, Yang YG. Genomics Proteomics Bioinformatics. 2013;11:8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan T. Trends Biochem. Sci. 2013;38:204–209. doi: 10.1016/j.tibs.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amort T, Souliere MF, Wille A, Jia XY, Fiegl H, Worle H, Micura R, Lusser A. RNA Biol. 2013;10:1003–1008. doi: 10.4161/rna.24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RP, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, He C. Mol. Cell. 2014;56:5–12. doi: 10.1016/j.molcel.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, Dai Q, Chen W, He C. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S, Cai Q, Ji D, Jin SG, Niedernhofer LJ, Pfeifer GP, Xu GL, Wang Y. J Am Chem Soc. 2014;136:11582–11585. doi: 10.1021/ja505305z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 23.He C. Nat Chem Biol. 2010;6:863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, He C. Mol Cell. 2014;56:5–12. doi: 10.1016/j.molcel.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keith G. Biochimie. 1995;77:142–144. doi: 10.1016/0300-9084(96)88118-1. [DOI] [PubMed] [Google Scholar]
- 26.Cornelius MG, Schmeiser HH. Electrophoresis. 2007;28:3901–3907. doi: 10.1002/elps.200700127. [DOI] [PubMed] [Google Scholar]
- 27.Van Simaeys D, Turek D, Champanhac C, Vaizer J, Sefah K, Zhen J, Sutphen R, Tan W. Anal Chem. 2014;86:4521–4527. doi: 10.1021/ac500466x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan M, Wang Y, Hu Y, Feng Y, Dai C, Wu J, Wu D, Zhang F, Zhai Q. Anal Chem. 2013;85:12173–12181. doi: 10.1021/ac4036026. [DOI] [PubMed] [Google Scholar]
- 29.Yamauchi Kiyoshi, T. N., Kinoshita Masayoshi. J. Org. Chem. 1980;45:3865–3868. [Google Scholar]
- 30.Hiroki Kumamoto HH, Tanaka Hiromichi, Shindoh Satoru, Kato Keisuke, Miyasaka Tadashi, Endo Kazuki, Machida Haruhiko, Matsuda Akira. Nucleosides and Nucleotides. 1998;17:15–27. [Google Scholar]
- 31.Wang J, Wang Y. Nucleic Acids Res. 2010;38:6774–6784. doi: 10.1093/nar/gkq458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carroll SM, Narayan P, Rottman FM. Mol Cell Biol. 1990;10:4456–4465. doi: 10.1128/mcb.10.9.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desrosiers R, Friderici K, Rottman F. Proc Natl Acad Sci U S A. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narayan P, Rottman FM. Science. 1988;242:1159–1162. doi: 10.1126/science.3187541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.