I. Introduction
The partitioning of the cytoplasm into functionally distinct compartments called organelles, delimited by lipidic membranes, is a hallmark of eukaryotic cells. By concentrating enzymes as well as substrates, each organelle is specialized for a set of reactions, for which compartmental conditions can be optimized. In addition to increasing the efficiency of individual and coupled reactions, compartmental organization also enables eukaryotic cell to simultaneously execute reactions that might be incompatible if pursued in a continuous cytoplasm. Membrane-bound compartments can also serve for dynamic storage, for example of calcium that can be rapidly transported from the lumen of an organelle via trans-membrane channels to the cytoplasm in response to signaling pathways. Additionally, molecules within the lumen of organelles can be exported to the cell exterior, which occurs upon fusion of the organellar and plasma membranes. Secretion via membrane fusion is the major mechanism of protein secretion from eukaryotic cells, a phenomenon of great physiological significance that allows cells to influence their environments in a multitude of ways, for both unicellular and multicellular organisms. The secretory apparatus consists of an array of morphologically and biochemically complex organelles, whose concerted activity is responsible for first translocating proteins out of the cytoplasm into the lumen of the secretory pathway, sorting those proteins to accommodate different modes of secretion, and subsequently releasing them into the environment. In many cases the proteins are covalently modified during this process. A second complex network of compartments, partially overlapping with the first, exists to receive and sort molecules that are internalized from the cell surface.
A major thrust of eukaryotic cell biology in the last 50 years has been to understand organelle activities and organization. Broadly speaking, a first aim has been to understand the basic mechanisms underlying organelle biogenesis and function, which are thought to be widely conserved among eukaryotes. Numerous insights have come from exploiting powerful approaches in budding yeast, in many cases providing initial identification of key components, or detailed mechanistic information, for steps that are conserved in mammalian cells. Generally, however, the pathways of membrane traffic in mammalian cells are more complex than in yeast, so a second broad aim has been to understand the additional features of specific mammalian cell types. For example, some polarized mammalian cells can maintain two or more parallel pathways of protein secretion, with each pathway directed toward a distinct cell surface(Weisz and Rodriguez-Boulan, 2009). These two lines of inquiry have also been explored in evolutionary terms. In particular, mechanisms that are conserved among eukaryotes are inferred to have been present in a shared eukaryotic ancestor, and therefore very ancient(Dacks and Field, 2007). In contrast, pathways and mechanisms that are specifically required for mammalian complexity can be inferred to have arisen as more recent adaptations that are consequently restricted to a subset of modern lineages.
When compared to pathways of membrane traffic in mammalian cells or fungi, the pathways in Tetrahymena are still relatively unexplored. Many historical studies, as reviewed by Frankel(Frankel, 2000), rely chiefly on morphological analysis. Published electron micrographs provide an excellent overview of many aspects of subcellular organization, and offer a strong starting point for molecular mechanistic studies. However, for many compartments there were and remain few if any identified molecular components. Such molecular markers are powerful tools for direct observations of compartments and their dynamics in living cells (e.g., by green fluorescent protein tagging). Secondly, in some cases molecular markers are essential for interpreting the compartments that are visualized. While some membrane structures are unmistakeable, such as the nuclear envelope, many others are highly pleiomorphic in other cells where they have been studied. Structures such as the Golgi, trans-Golgi network, and distinct classes of endosomes adopt different appearances depending on the cell type and cell activity and can therefore be difficult to identify without molecular markers. Fortunately, the resources available from the sequenced Tetrahymena genome should accelerate the development of such markers, in part by facilitating the identification of Tetrahymena homologs for markers established in other systems. The genomic data also facilitates proteomic approaches to identify the components of isolated organelles.
Although many details are lacking, we know that T. thermophila maintains a highly complex network of membrane trafficking pathways. For example, there is evidence for at least four distinct pathways of endocytic uptake. Cells form small endocytic vesicles at cortical invaginations called parasomal sacs(Nilsson and Van Deurs, 1983), while much larger vesicles (phagosomes) arise at the base of the oral apparatus(Nilsson, 1979). While it is clear that different mechanisms are involved in endocytosis from parasomal sacs vs the oral apparatus, neither of these pathways has been dissected in detail. A third pathway of endocytosis, which can be inferred from work in Paramecium, is coupled with the exocytosis of dense-core secretory vesicles and facilitates the rapid recovery of vesicle membranes (Hausmann and Allen, 1976). Fourth, endocytic membrane recovery also occurs upon phagosome fusion, at a cortical site called the cytoproct(Allen and Wolf, 1979). Similarly, Tetrahymena secrete proteins by at least 3 different routes: a pathway of rapid constitutive release of newly-synthesized proteins (for which the vesicular carriers have not been identified)(Bowman and Turkewitz, 2001; Madinger et al., 2010); regulated exocytosis from docked mucocysts(Turkewitz, 2004), and; release of hydrolytic enzymes via lysosome exocytosis(Kiy et al., 1993). There is also indirect evidence for cytoplasmic protein release via an exosome-like mechanism(Madinger et al., 2010). This list understates the complexity of the pathways of cell surface delivery, since for example there is also vesicle trafficking from the cytoproct (the site of phagosome exocytosis) to the oral apparatus(Allen and Fok, 1980; Bright et al., 2010). It is also not clear whether there is a single pathway of endocytosis from parasomal sacs or whether parallel pathways exist, as will be discussed below.
In our view, more detailed molecular studies of trafficking in T. thermophila could be significant for several reasons. While it may not be possible or even desirable to obtain a comprehensive understanding of the entire network of trafficking steps, there are individual pathways whose analysis could make major contributions to both of the aims outlined above. Unlike budding yeast whose successful evolutionary strategy was to become small and relatively simple, the ciliates, like animal cells, have undergone large expansions in gene families encoding key determinants of membrane trafficking, as the membrane trafficking pathways themselves grew increasingly complex(Bright et al., 2010; Eisen et al., 2006; Saito-Nakano et al., 2010). Because the adaptations to membrane traffic occurred independently in ciliates and animals, a comparison of these lineages offers one the chance to ask whether specific pathways were prone to expansion and adaptation. This question is also being asked more broadly, taking advantage of the wealth of sequenced genomes now available to ask whether the determinants of specific pathways have tended to expand in multiple lineages, to generate large gene subfamilies. For example, phylogenetic analysis of SNARE proteins in many lineages suggests that SNARE subfamilies associated with endocytosis have undergone more expansion than other subfamilies, suggesting that endocytosis has been a particularly rich substrate for innovations in membrane traffic(Kienle et al., 2009). These metagenomic studies can be complemented by more in-depth studies of individual organisms. Studying such questions in a specific complex non-animal lineage, such as higher plants or ciliates, allows one to confirm the phylogenetic predictions, i.e., test the underlying assumption that sequence comparison are reliable for assigning function. Secondly, such single-species studies are critical to understanding both how, and to what purpose, the genetic innovations have modified conserved pathways or generated new ones. In other words, how has selection acted on the organization and function of membrane trafficking pathways? Critically, the experimental tools available in T. thermophila already facilitate asking complex cell biological questions, and new approaches continue to be developed(Turkewitz et al., 2002). One such novel approach to studying membrane traffic in particular is discussed at the end of this chapter, based on the availability of extensive whole genome expression data.
Effective use of T. thermophila may come from exploiting unique features of its complex and unusual organization. For example, many sites of specific membrane trafficking steps at the plasma membrane are organized as precise arrays, allowing a microscopist to analyze multiple sites, simultaneously, at predictable locations. This aspect of ciliate organization has recently been brilliantly exploited to analyze basal bodies(Pearson and Winey, 2009). For membrane trafficking, such organized domains include sites of clathrin-mediated endocytosis and of regulated exocytosis(Allen, 1967; Elde et al., 2005; Satir et al., 1973). A second striking aspect of Tetrahymena is that there are structurally and functionally distinct variants of several organelles, maintained in the same cytoplasm. This is best known for the nucleus, where studies exploiting the differences between the macro- and micronucleus have made pivotal contributions to molecular biology(Pederson, 2010). Nuclear dimorphism in Tetrahymena has recently been exploited to analyze the role of nuclear pore components. Nuclear pores are selective gates that regulate traffic of cytosolic and membrane proteins into the nucleoplasm, and a major question in the field is how the components of nuclear pores act as gatekeepers, with much attention focusing on iterative motifs consisting of glycine-leucine-phenylalanine-glycine (GLFG) that are abundant in many proteins lining the pores (nucleoporins, or nups). In ciliates, the two functionally-distinct nuclei contain different sets of nucleoplasmic proteins, implying that nuclear pores in Mics and Macs are also distinct. Haraguchi and colleagues recently identified micronuclear- and macronuclear-specific versions of NUP98(Iwamoto et al., 2009). The repeats in the micronuclear (but not macronuclear) Nup98p were NIFN, rather than the canonical GLFG, and domain-swapping experiments provided evidence that the change in the Nup repeat motif has functional consequences for gatekeeping. This line of work therefore holds promise both to reveal mechanisms underlying nuclear dimorphism in Tetrahymena as well as providing a unique model system for dissecting features of nuclear pore selectivity.
Another example of organellar differentiation in Tetrahymena is that each cell contains both a “standard” endoplasmic reticulum (ER), including the nuclear envelope, but also distinct flattened cisternae called alveoli that tightly underlayer the plasma membraneto. While alveoli have been only glancingly studied in Tetrahymena, data from Paramecium make a strong case that alveoli function as a major store for mobilizable calcium, a classical activity of the ER, and also contain some ER proteins(Plattner et al., 1999; Stelly et al., 1995). ER subdomains in animal cells are recognized as an important aspect of the secretory pathway and of cellular signaling, and understanding the biogenesis and maintenance of the ER and alveoli in Tetrahymena may offer exceptional opportunities for illuminating mechanisms of protein and lipid sublocalization in this organelle.
Lastly, Tetrahymena, because of the strong experimental tools that have been developed, may be an excellent organism to appreciate “cell biodiversity”, namely the range of adaptations that have evolved in eukaryotes that are deeply divergent from animals. For example, the contractile vacuole is a multi-part organelle that collects water from the cytoplasm to pump it out of the cell, and is essential for osmotic homeostasis in fresh water organisms lacking cell walls. The remarkable properties of the ciliate contractile vacuole have been investigated in Paramecium, but virtually nothing is known about assembly or mechanism of action at the molecular level(Allen, 2000). Contractile vacuoles are also present in Amoebozoa and other distantly-related lineages, but whether these are homologous organelles to those in Ciliates (i.e., inherited from a common ancestor), or whether organelles as complex as contractile vacuoles have arisen multiple times, independently, is an open question. Pursuing such organelles in Tetrahymena, as they are also being studied in Dictyostelium, could help to provide a new perspective on the relative importance of inheritance vs innovation in the structures that animate modern eukaryotes.
II. Recent work on membrane traffic in Tetrahymena
Studies of membrane traffic in Tetrahymena up until the last decade have been authoritatively reviewed by Frankel, and we will therefore focus on work reported since that review(Frankel, 2000). At the end of each section we list additional papers reporting observations or reagents that may be of interest to those investigating membrane traffic.
A. Protein secretion
1. Constitutive secretion
Constitutive secretion refers to secretion of newly synthesized proteins in the absence of specific extracellular stimulation. Rapid secretion of newly-synthesized proteins in T. thermophila was visualized using pulse-chase biosynthetic labeling, where it was demonstrated that the proteins released via this pathway were different from those released via regulated exocytosis from mucocysts(Bowman and Turkewitz, 2001). It is likely that these proteins, after transport through the ER and Golgi, are transported from the trans-Golgi to the cell surface in vesicles or membrane tubules, but this process has not been directly visualized in Tetrahymena. Moreover, the site(s) of such protein release represents an interesting problem since most of the plasma membrane, with which vesicles must fuse to release their contents, is inaccessible from the cytoplasm because of the intervening alveoli(Allen, 1978).
A proteomic analysis of T. thermophila culture supernatants has recently supplied the first relatively comprehensive view of what this unicellular protest is releasing into its environment(Madinger et al., 2010). That list includes 207 proteins including many hydrolytic enzymes as well as novel proteins of unknown function, with the cohort of secreted proteins changing significantly depending on whether cells were incubated under nutritive vs starvation conditions. The authors also characterize protein secretion from a previously isolated Mendelian mutant, SB281, whose most striking defect is the failure to synthesize mucocysts. They showed that the constitutive proteins released in this mutant was different from wildtype cells, consistent with previous descriptive evidence(Bowman and Turkewitz, 2001). Interestingly, the authors find evidence that some proteins known to be released via mucocysts may also be released via constitutive exocytosis, and that some specific mucocyst proteins may be released differentially under growth vs starvation conditions. Following up on these observations may be interesting both from a mechanistic perspective and also to illuminate the role of mucocysts for Tetrahymena, which is not yet known.
Part of the interest in characterizing secretion comes from the question of whether Tetrahymena can be usefully engineered to produce and secrete heterologous proteins (Aldag et al., 2011; Hartmann et al., 2000).
2. Regulated secretion
In animal cells, proteins can be released in response to extracellular stimuli both from small vesicles such as synaptic vesicles, or from larger dense-core secretory vesicles, also called secretory granules. These two types of vesicles arise via distinct biosynthetic pathways and serve a wide range of physiological roles in different tissues(Gumbiner and Kelly, 1982). Ciliates synthesize vesicles containing dense-cores, which because of their distinct appearance are prominent features in many cytological studies(Rosati and Modeo, 2003). In T. thermophila these vesicles are called mucocysts, and they have been explored as a system for understanding biosynthetic mechanisms that are still rather poorly understood in animals, but which appear to differ from canonical mechanisms involved in vesicle formation(Tooze et al., 2001; Turkewitz, 2004). Mucocysts contain two major families of soluble (i.e., not membrane-bound) proteins, which are coordinately expressed(Rahaman et al., 2009). The first, whose corresponding genes were called GRL for granule lattice, are required to form a protein crystal that comprises the bulk of the dense core(Cowan et al., 2005). A subset of the GRL-encoded proteins may however play roles that are less structural than regulatory, since gene knockout did not affect the appearance of the dense core but reduced mucocyst accumulation(Cowan et al., 2005). The GRLs have been identified both biochemically but also by forward genetics, using an unbiased screen based on antisense ribosomes(Cowan et al., 2005). The Grl proteins are synthesized as proproteins, and endoproteolytic cleavage is closely connected with assembly of the dense core(Verbsky and Turkewitz, 1998). This assembly appears to take place in a post-Golgi vesicular compartment and intermediates can be visualized by EM(Bowman et al., 2005a). A link between proprotein processing and dense core assembly is established in mammalian systems, and the similarity in ciliates is intriguing(Creemers et al., 1998). In addition, biochemical and genetic experiments demonstrated that granule assembly intermediates in Tetrahymena form in the endoplasmic reticulum(Cowan et al., 2005).
A second family of granule proteins in T. thermophila is defined by a common C-terminal β/γ crystalline domain, the remainder of the proteins consisting of a variable number of repeats of several different domains(Bowman et al., 2005b). Two members of this family have been investigated and neither is proteolytically processed(Bowman et al., 2005a; Haddad et al., 2002). In addition, none of the genes in this family that have been disrupted is essential for core assembly, but the double disruption of two related genes subtly changed the properties of the mucocyst core following its exocytic release, so apparently the proteins in this family are playing distinct roles from the Grls(Rahaman et al., 2009). Only one protein in the non-Grl family has been localized and was found, remarkably, to be highly concentrated at the end of the secretory granule where it docks at the plasma membrane, prior to exocytosis(Bowman et al., 2005a). The protein, called Grt1p for Granule tip, fails to polarize in two Mendelian mutants that are defective in a late stage of mucocyst assembly as well as mucocyst docking(Bowman et al., 2005a). One possibility is that Grt1p and other proteins in that family can interact with specific proteins in the mucocyst membrane and thereby organize membrane zones with specific activities, such as docking and exocytic fusion. Interestingly, some electron micrographs of mucocysts appear to show that, in addition to the crystalline core, there are other components that are more closely associated with the membrane(Williams and Luft, 1968).
The mechanisms enabling accurate sorting to mucocysts of the Grl and β/γ crystallin-containing protein families are not yet known. One model, based on work in animal cells, is that proteins destined for dense core vesicles have a predisposition to co-aggregate in a late Golgi compartment(Chanat and Huttner, 1991). However, neither genetic nor biochemical experiments has demonstrated any interaction between the two families of proteins in T. thermophila dense core vesicles(Rahaman et al., 2009). These are suggestive rather than conclusive results, since the system is sufficiently complex so that physiologically important interactions may have escaped detection, for example due to functional redundancy. In addition, biochemical interactions may be highly sensitive to the ionic conditions within specific compartments of the secretory pathway, about which very little in known in ciliates.
A third demonstrated pathway of secretion in T. thermophila is that of secretory lysosomes, and is reviewed in Frankel(Frankel, 2000). A pathway in Tetrahymena that has not been investigated at the molecular level is secretion from the contractile vacuole upon its cyclic fusion at plasma membrane pores.
Other studies of interest in T. thermophila:
Identification of PGP1, an HSP70 homolog, of the GRP170 subfamily, whose product localizes to the endoplasmic reticulum and is shown to be a glycyosylated, glycylated protein. PGP1 is induced on cell stress but also essential for vegetative growth(Xie et al., 2007). The C-terminal peptide KQTDL functions as an ER-retention signal. KDEL and related sequences have previously been shown to act as functional ER retention signals in T. thermophila(Cowan et al., 2005).
A novel gene, CDA13, encodes a predicted transmembrane protein which may reside in a post-Golgi compartment of the secretory pathway(Zweifel et al., 2009).
Identification of DRP6, a highly divergent dynamin-related protein that localizes to the macronuclear envelope and a vesicular ER-like compartment, and which is essential for nuclear remodeling during conjugation(Rahaman et al., 2008).
Analysis, including localization and functional studies, of the nucleoporins and karyopherins involved in nuclear import(Malone et al., 2008).
Analysis of the expression of Ser antigens, the best-known cell surface proteins in this system(Doerder and Gerber, 2000). Work on antigenic variation in ciliates has recently been reviewed(Simon and Schmidt, 2007).
Analysis of the carbohydrate structure of secretory proteins(Becker and Rusing, 2003).
Analysis of extracellular cysteine proteases(Herrmann et al., 2006).
B. Endocytosis
In mammalian cells, endocytosis is a critical pathway for the uptake of extracellular macromolecules, modulation of signaling pathways, and turnover of membrane proteins. A number of different endocytic pathways exist in animals, the best characterized of which involves assembly of the protein clathrin, which interacts with the heterotetrameric adaptor protein AP-2 during endocytic vesicle formation. The scission of the vesicle membrane from the plasma membrane, releasing it into the cytoplasm, involves a GTPase of the dynamin family, and actin is involved at multiple steps in the process(Kirchhausen, 2009).
In Tetrahymena, the alveoli limit contact between the cytoplasm and plasma membrane. One interruption in the alveoli are indentations called parasomal sacs that are found proximal to each ciliary basal body, and these are sites of endocytosis as shown by EM studies of cationized ferritin uptake(Nilsson and Van Deurs, 1983). Endocytosis at parasomal sacs has more recently been confirmed in living T. thermophila using a styryl dye, FM1–43, which had been shown in other systems to be a useful marker for endocytic vesicles(Cousin and Robinson, 1999). Tetrahymena incubated with FM dyes first show fluorescence in small puncta that form an array near the cell cortex, as would be expected for endocytic vesicles that have just undergone scission at parasomal sacs(Elde et al., 2005). Thereafter the puncta are highly mobile, and within minutes appear to collect toward the cell posterior. The appearance of the posterior fluorescent structures, which can on the basis of the FM1-43 labeling be classified as endosomes, suggests that the initial endocytic vesicles have undergone fusion events to create larger, heterogeneous structures. The FM1-43 accumulation in these posterior endosomes persists for at least tens of minutes. A related dye, FM4-64 has also been shown to accumulate after long labeling periods in moderate-sized vesicles at some distance from the cortex, located throughout the cell(Zweifel et al., 2009).
The FM1-43 uptake assay facilitated analysis of the protein requirements for endocytosis. Using GFP-tagging the authors demonstrated that clathrin was localized to parasomal sacs, and involved in endocytosis since induced expression of a truncated clathrin heavy chain, which acts as a dominant negative form, suppressed FM1–43 uptake(Elde et al., 2005). Similarly, GFP-tagging was used to localize four AP complexes, and only AP-2 was found to localize to parasomal sacs. A third similarity with animal cells was a requirement for dynamin in endocytosis. T. thermophila was found to encode 8 members of the dynamin family, called DRP1-8 for dynamin-related proteins, a remarkably large number for a unicellular organism. Drp1p and Drp2p were found to localize to parasomal sacs (Elde et al., 2005)(Rahaman and Turkewitz, unpublished) and the endocytic activity of Drp1p, an essential gene, was demonstrated by several genetic approaches. A 28 amino acid stretch of Drp1p was sufficient, when exchanged with the same region of a different Drp family member, to redirect the chimeric protein to sites of endocytosis. Surprisingly, actin did not appear to be required for endocytosis, judging by results with pharmacological actin inhibitors. If this result is correct, Tetrahymena may be unique in having evolved actin-independent endocytic mechanisms. However, the experimental results could not rule out the possibility that divergent actin isoforms, which are insensitive to the drugs used, are involved in endocytosis. Other interesting possibilities are discussed below, as well as additional endocytic markers identified in a screen of Rab GTPases.
It is worth noting that neither the expression of the dominant negative alleles of clathrin or DRP1, nor disruption of the endogenous DRP1 gene, led to a complete block in FM1-43 uptake(Elde et al., 2005). The appearance of the residual FM accumulation in these strains suggested that the signal was due to vesicle formation at parasomal sacs. These results raise the question of whether Tetrahymena also has a clathrin- and DRP1-independent pathway of endocytosis. To investigate this it will be important to analyze the function of Drp2p, which also localizes to parasomal sacs (Rahaman and Turkewitz, unpublished).
Additional papers of interest:
Analysis of a novel predicted membrane protein, Cda12p, which localizes to a putative endocytic compartment. The knockdown (via antisense ribosomes) phenotype includes defects in cytokinesis and some aspect of endosome formation. During cytokinesis as well as in mating cells, the protein localizes in regions where there is likely to be active membrane remodeling. This provides a hint about how remodeling of endosomal compartments might underlie structural changes at specific stages in the life cycle(Zweifel et al., 2009).
C. Phagocytosis and phagosome maturation
T. thermophila is magnificently adapted for bactivory, sweeping small particles into the base of the oral apparatus where they are ingested via formation of large food vacuoles called phagosomes(Frankel, 2000). The digestion of phagosome contents takes place via a series of remodeling steps, collectively termed maturation, in which the nascent phagosome fuses with vesicles that deliver acidification machinery as well as hydrolytic enzymes, while other components are selectively removed/recycled via vesicle budding(Stuart and Ezekowitz, 2005). Remodelling of the phagosome membrane by cytosolic factors is also likely to be important, as has been shown for mammalian cells(Huynh et al., 2007). Compartment maturation is an important theme in membrane traffic, and phagosomes are a particularly attractive pathway for detailed analysis because phagosomes are large and easily labeled by loading specific cargo (e.g., fluorescent bacteria, or bacteria-sized latex beads).
Phagosome formation and maturation in Tetrahymena are actin-dependent processes. First, there is a clear requirement for dynamic actin during phagosome formation, which has been demonstrated using pharmacological inhibition of actin dynamics but also on disruption of genes encoding the major actin gene, and the actin-assembly cofactor, profilin(Wilkes and Otto, 2003; Williams et al., 2006). Actin filaments may also be indirectly involved in phagosome maturation, based on reports that the movement of phagosomes from the oral apparatus in the cell anterior, to the cytoproct at the cell posterior, involves an actin-based myosin motor encoded by MYO1(Hosein et al., 2005). In addition, the microtubule-based dynein motor DYH1 has also been implicated in phagosome formation(Lee et al., 1999).
The dynamin Drp1p, involved in clathrin-mediated endocytosis at parasomal sacs, is also required for phagocytosis. Cells expressing a dominant-negative DRP1 allele (K51E) or in which the level of wildtype gene expression is reduced by methods described above, showed no discernable phagocytic uptake of particles (e.g., India ink particles) from the medium (N. Elde and A.P. Turkewitz, unpublished)(N.Elde, PhD thesis). This phenotype was distinct from that of cells treated with actin inhibitors. The actin-inhibited cells showed no accumulation of ink particles in cytoplasmic phagosomes, but nearly all cells accumulated ink in a large vesicle that formed at, but failed to detach from, the base of the oral apparatus. DRP1 mutant cells failed to accumulate even this single frustrated phagosome, suggesting that Drp1p acts upstream of actin during phagosome formation. A potential hint of the function of Drp1p in this pathway is that GFP-tagged Drp1p labels puncta along the so-called deep fiber, a cytoskeletal filament that extends from the base of the oral apparatus that appears to act as a vesicle highway. The deep fiber and nearby structures also appear to be sites of localization of calmodulin and a number of calmodulin-binding proteins, and pharmacological inhibition of calmodulin activity and calcium-based signaling block phagosome formation(Gonda et al., 2000; Moya and Jacobs, 2006).
The sequencing of the T. thermophila genome made it possible to easily identify homologs to many proteins previously studied in other systems, facilitating many of the studies cited above(Eisen et al., 2006). A second important consequence was facilitation of proteomic studies, since the predicted T. thermophila proteome could be used to identify proteins in isolated organelle fractions using mass spectrometry data. This has been very fruitfully applied to phagosomes, which were highly purified by taking advantage of the aforementioned ability to identify, and change the fractionating properties of, phagosomes that had taken up polystyrene beads(Jacobs et al., 2006). Proteins associated with the purified phagosomes were then analyzed by mass spectrometry, resulting in the identification of 73 genes. This extensive list allowed the authors to gauge the similarity of phagosomes between T. thermophila and other organisms, since 28 of the proteins had been associated with phagocytosis in other organisms. In addition, the authors choose four candidate genes from the survey and, by expressing these as GFP-tagged copies, demonstrated that three of these were phagosome-associated. Taken together, these results suggest that many mechanisms are conserved between the phagosome pathways in multiple lineages. Since many of the conserved proteins including several associated with human disease have functions that are not well understood, T. thermophila may offer an attractive system for addressing questions about the mammalian phagosome pathway.
Additional papers of interest:
-
i
A study showing that Pseudopterosin A, a marine natural product, inhibits phagocytosis, with pharmacological evidence arguing for a G protein-coupled receptor mechanism of action involving a calcium-dependent step(Moya and Jacobs, 2006).
-
ii
A study showing that degradation of the old macronucleus during conjugation has features of an unusual autophagy(Akematsu et al., 2010). Additional insightful studies from the same group focus on the role of mitochondria and mitochondrial signaling factors during macronuclear breakdown(Akematsu and Endoh, 2010).
-
ii
Mass spectrometric analysis of the mitochondrial proteome(Smith et al., 2007b), the conjugation junction(Cole et al., 2008), and the ciliome(Smith et al., 2005).
-
iii
A particularly elegant study identifying a basal body proteome, including extensive ultrastructural analysis(Kilburn et al., 2007).
-
iv
Studies on the effect of passage through the phagosome on bacterial conjugation and infectivity(Klobutcher et al., 2006; Matsuo et al., 2010).
-
v
The important role of cytoskeletal-based motor proteins in membrane traffic is well established. Most work on motor proteins in T. thermophila has focused on ciliary beat, but informatics-based surveys indicate that a large number of cytosolic proteins remain to be explored(Sugita et al., 2011; Wilkes et al., 2008).
D. Rab GTPases as markers for membrane traffic
Rabs are small GTPases that act as key determinants of compartmental specificity by recruiting, when in their GTP-bound, membrane-tethered state, a wide range of effectors(Segev, 2001). Rabs exist as products of large gene families in which each family member associates with one or a small number of cellular compartments. Thus the number of Rabs expressed in a cell likely reflects the complexity of membrane trafficking pathways in that cell(Stenmark and Olkkonen, 2001). There are 12 Rabs in S. cerevisiae, compared with 63 in humans(Pereira-Leal and Seabra, 2001).
Two groups have recently characterized the Rabs in T. thermophila, which were identified via homology searches in the macronuclear database based on the fact that Rabs can be distinguished from other related small GTPases based on a number of conserved motifs. Bright et al. characterized Rabs by combining phylogenetic and expression analysis with localization data, the last based on GFP-tagging the large majority of the Rab family members(Bright et al., 2010). Phylogenetic and expression analysis of the Rab superfamily was also reported by Numata and colleagues(Saito-Nakano et al., 2010). Where they overlap, the results of the two groups are largely similar, the most important difference being that different criteria were used to assign orthology, an issue discussed below(Turkewitz and Bright, in press). In addition, the Numata group took the valuable step of experimentally verifying the sequences predicted by genome annotation.
The T. thermophila genome encodes 63 Rabs, a number greater than the 33 in Drosophila melanogaster or 29 in C. elegans. There are an additional 25 Rab-like proteins, which differ from Rabs in lacking identifiable C-terminal prenylation motifs(Saito-Nakano et al., 2010). A first question was how many of these Rabs are expressed concurrently. In animal cells, many Rab isoforms are expressed preferentially in particular tissues, so that the total number of isoforms reflects the range of adaptations of membrane traffic for distinct tissues(Zhang et al., 2007). For unicellular organisms that possess large Rab families, one possibility is that subsets are expressed under different conditions, i.e., that the large number reflects adaptations of membrane traffic for different environments or life stages. For Tetrahymena, this question was answered by mining a public database in which all transcripts were measured in cultures sampled at a variety of physiologically-relevant states(Miao et al., 2009). A small number of Rabs showed strikingly stage-specific expression (e.g., undetectable expression under growth conditions, and high expression upon starvation or during mating)(Bright et al., 2010; Saito-Nakano et al., 2010). However, the large majority of Rabs were co-expressed, and many at very high levels, in growing cells. The large number of co-expressed Rabs suggests that T. thermophila maintains a membrane trafficking network that is roughly as elaborate as that of mammalian cells.
To investigate the functions of the large set of Rabs, Bright et al. expressed each in T. thermophila as an N-terminal GFP fusion, and took advantage of a novel thermally-controlled gel to immobilize the normally fast-swimming cells to capture time-lapse movies showing the dynamics of the GFP-Rab-labeled structures(Jeong et al., 2007). Given the paucity of molecular markers for many pathways in Ciliates, the individual Rabs may become important tools since they localize to a wide range of cellular structures, many of which could be tentatively identified even at the level of light microscopy. For example, Rabs associated with endocytosis were identified by using FM4–64 as an endocytic tracer, while another set of Rabs could be assigned to phagocytosis-related structures (i.e., the oral apparatus, phagosomes, or the cytoproct) by labeling phagosomes with fluorescent bacteria or India ink. Rabs associated with unique large structures, such as the contractile vacuole, could be assigned in the absence of any other compartmental marker. Movies showing the dynamic behavior of many of the GFP-Rab-labeled structures can be viewed at http://tetrahymenacell.uchicago.edu.
Since the Rabs are GFP-tagged and can be viewed in living cells, they offer the possibility of studying membrane dynamics in these cells. For example, phagocytosis in Tetrahymena and in human macrophages have many similarities but one difference is that, in the former, egestion of undigested contents in fully matured phagosomes occurs at a unique site on the plasma membrane called the cytoproct(Allen and Wolf, 1979). Several Rabs were found to associate only with phagosomes that were positioned right at the cytoproct, and time-lapse movies showed that egestion resulted in transient transfer of those Rabs to the PM and subsequent retrieval(Bright et al., 2010). These “terminal Rabs” may be activated by proteins (e.g., Rab-GEFs, or GTP-exchange factors) that are present at the cytoproct itself, so that some stages in phagosome maturation are influenced by cortical determinants. Another Rab is uniquely associated with what appear to be elongated vesicles being transported, primarily toward the cell anterior, along cytoplasmic microtubules that extend from the cytoproct region toward the oral apparatus. These vesicles may underlie the recycling of phagosome membrane components following retrieval of the phagosome membrane at the cytoproct, an actin-dependent process(Allen and Fok, 1980; Sugita et al., 2009).
II. Studies on membrane lipids in Tetrahymena
While the discussion above has focused on the role of proteins in membrane traffic, it is also increasingly appreciated that lipids are not merely passive structural elements in cells but also key determinants. We therefore review recent work in Tetrahymena on three different aspects of lipids.
A. Phosphoinositides
Lipid-anchored phosphoinositols (which are called phosphatidyl inositols, or PtdIns) can serve as important determinants of compartmental identity and are therefore key elements of membrane traffic. Their activity depends upon the fact that the inositol ring, when phosphorylated in specific combinations at the 3, 4 and/or 5 positions, can be recognized by large numbers of proteins, which can thereby be activated or recruited. In metazoa, this has important consequences for cytosolic signaling, membrane trafficking, nuclear events, cytoskeleton integrity, permeability and transport(Di Paolo and De Camilli, 2006).
Because of their ease of culture, including the possibility of precisely defined growth medium, Tetrahymena were used in a large number of classical studies on lipid metabolism. More recently, several groups have focused on phosphoinositides in Tetrahymena species. Work in T. vorax, T. pyriformis and thermophila was reviewed by Ryals in 2009, focusing primarily on biochemical aspects(Ryals, 2009). Our goal here is to view these data from a genetic/molecular perspective, focusing on T. thermophila and exploiting information that can be gathered using, for example, the genomic and gene expression databases of Tetrahymena thermophila (TGD and TGED, respectively).
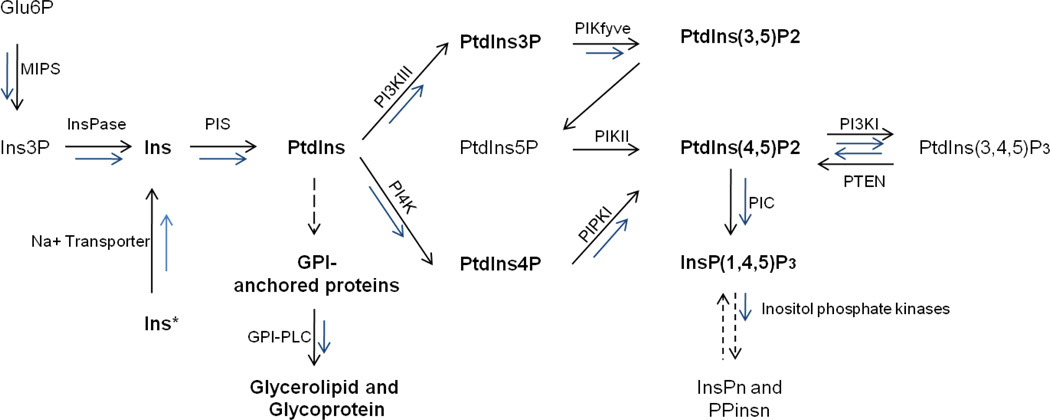
The metabolic pathways that generate inositol derivatives are schematized in Figure 1. This figure, adapted from Michell(Michell, 2008), shows the steps for which T. thermophila enzymes have been identified or can be inferred, as described below in the text.
Figure 1.
To begin, cells can take up inositol from the environment or synthesize it from D-glucose-6-phosphate. Synthesis involves two enzymes: myo-inositol-3-phosphate synthase (MIPS), which catalyzes the cyclization of D-glucose-6-phosphate to D-myo-inositol-3-phosphate (Ins3P)(Michell, 2008), and inositolmonophosphatase (InsPase), which dephosphorylates Ins3P. While neither activity has been reported in T. thermophila, there are clear homologs encoded in the genome. TTHERM_00519810 shows a high homology (E<1.0 e−120) with MIPS from plants (A. thaliana), animals (H. sapiens), fungi (S. cerevisiae), amobozoa (D. discoideum) and kinetoplastids (T. cruzi). The second enzyme, TTHERM_00318610, contains a highly conserved inositol monophosphate domain (PFAM PF00459). Both enzymes are expressed in growing, starved, and conjugating cell cultures (TGED)(Miao et al., 2009).
Environmental inositol import in Tetahymena vorax involves a sodium-dependent mechanism with features similar to those in other organisms(Ryals and Kersting, 1999). The genes are also likely to be similar, and homology searches identify a strong set of candidates (i.e., TTHERM_00852790, TTHERM_01080450 and TTHERM_00473200) that are related to other eukaryotic Na+/myo-inositol symporters. Inositol exists as a number of stereoisomers, the most common of which is myo-inositol, but other isomers are also present in cells including scyllo-, neo-, epi-D-chiro- and muco-inositols. In T. vorax, non-myo-inositols have also been detected, with evidence that these can be taken up from the medium(Kersting et al., 2003; Kersting and Ryals, 2004; Ryals and Kersting, 1999). Conversion between inositol isomers may also occur via inositol epimerases(Sun et al., 2002), though no conclusive genetic or biochemical data have been published in Tetrahymena to date. An open question is whether the free inositols in Tetrahymena have a function independent of their role as biosynthetic precursors. In other organisms, the presence of these organic solutes, together with other polyalcohols, has been linked to a cytoprotective response against environmental stress. For example, in mammalian kidney cells, inositols play a role in osmoregulation, while in plants and archaea they play a role in the stabilization of cellular proteins(Yancey, 2005).
Inositols are a substrate for PtdIns synthesis, carried out by phosphatidylinositol synthase (PIS) starting with inositol and cytidine diphosphate-diacylglycerol (CDP-DAG). This reaction has been characterized in T. vorax microsomes, where the substrates included both myo- and non-myo-inositol isomers(Riggs et al., 2007). T. thermophila has a single PIS gene (TTHERM_00678350) whose expression profile is similar to the enzymes involved in the synthesis of inositol (MIPS and InsPase). Its high homology (E value between 1.0 e−20 and 1.0e−35) with PIS of species belonging to different eukaryotic supergroups supports its broad conservation, as previously reported(Michell, 2008).
Starting from PtdIns, a set of kinases and phosphatases are involved in the synthesis of phosphatidylinositol phosphates (PtdInsPs, also called phosphoinositides), generating seven possible species (see Figure 1). The species actually present have been determined by HNMR (see below)(Leondaritis and Galanopoulou, 2000). PtdIns in T. pyriformis strain W, which makes up approximately 4% of total phospholipids, is found exclusively as diacylphospholipid(Pieringer and Conner, 1979), whereas other phospholipids (phosphatidylcholine (PC) and aminoethylphosphonoglyceride (AEPL)) are predominantly found as alkylacyl lipids(Leondaritis and Galanopoulou, 2000). Other differences between PtdIns and PC/PE in T. pyriformis include high myristic acid content, fully saturated acyl chains and the absence of C18 fatty acid. PC and PE may have similar fatty acid content because they derive from a shared biosynthetic pathway, whereas PtdIns synthesis depends on a distinct phosphatidic acid pool for the generation of CDP- DAG(Leondaritis and Galanopoulou, 2000).
Work by Galanopoulou and colleagues on phosphoinositides in Tetrahymena pyriformis and thermophila identified PtdIns(3)P, PtdIns(4)P, PtdIns(3,5)P2 and PtdIns(4,5)P2, but not PtdIns(5)P, PtdIns(3,4)P2 or PtdIns(3,4,5)P3(Deli et al., 2008; Leondaritis et al., 2005). One major source of interest in phosphoinositides in mammalian cells comes from their role as compartment-specific determinants, based on the ability of individual phosphoinositide species to recruit proteins with the corresponding phosphoinositide-binding domains. As described earlier in this chapter, the pathways of membrane trafficking in ciliates depend on a network of Rab GTPase determinants as extensive as that in animals, so an interesting question is whether phosphoinositide isomers provide a second set of determinants in ciliates as they do in animals. However, we still lack information about localization of the identified phosphoinositide species in Tetrahymena. One important question is whether the individual phosphoinositides are concentrated in specific compartments or act in specific pathways. In many eukaryotes PtdIns(3)P and PtdIns(3,5)P2 are determinants for endocytic traffic, and PtdIns(4)P has been implicated in maintaining Golgi structure and function (Di Paolo and De Camilli, 2006). A hint regarding phosphoinositide function in T. thermophila comes from studies using wortmannin, a specific inhibitor of phosphoinositide 3-kinases (PI3Ks), which reduces the levels of PtdIns(3)P and PtdIns(3,5)P2 but not D4-phosphoinositides. In T. thermophila, wortmannin treatment led to increased secretion of lysosomal enzymes(Kovacs and Pallinger, 2003; Leondaritis et al., 2005). Interestingly, this enhancement was absent or reduced in two mutant strains that are deficient, respectively, in a late stage of secretion from lysosomes (MS-1), and in phagocytosis (A2). Unfortunately, neither the precise cell biological nor genetic deficiencies are known in these mutants, but the fact that the mutations attenuate the effect of wortmannin may indicate, as suggested by the authors, that an InsPtd phosphorylated at the 3-position primarily functions in phagosomes/phagolysosomes A complicating factor is that the MS-1 mutant shows elevated levels of PtdIns(4)P, which could be a direct or indirect effect of the mutation(Deli et al., 2008). Wortmannin treatment also significantly inhibited phagocytic activity in T. pyriformis, possibly due to inhibition of actin polymerization(Kovacs and Pallinger, 2003). Further studies, particularly if more fully characterized mutant strains become available, should reveal more details of pathway regulation by phosphoinositides.
The enzymes involved in phosphoinositide synthesis have also been investigated, at least by informatics approaches. Based on work in other eukaryotes, phosphoinositide-3-kinase activity can be encoded by enzymes belonging to three classes, all of which have a PI3K enzymatic core inhibited by wortmannin but which diverge in other structural features. Importantly, because the 3 classes use overlapping but not-identical substrates, they can generate different 3-phosphoinositides that consequently recruit or activate distinct effectors(Vanhaesebroeck et al., 2010). In the genome of T. thermophila, four putative PI3Ks have been identified, three of which bear structural features of group I enzymes (TtPI3K 1–3: TTHERM_00655270, TTHERM_00323020, TTHERM_00951960) and one of group III (TtPI3K III (TTHERM_00649380)(Leondaritis et al., 2005). Interestingly, the Class I enzymes appear to be absent in plants, fungi, and many other protozoa(Michell, 2008). All four T. thermophila genes are expressed in growing, starved, and conjugating cell cultures, but with different expression patterns suggesting non-redundant functions. The only published information on the roles of these putative PI3K genes comes from pharmacologic studies. Inhibition of PI3K activity by wortmannin, LY294002 and 3-methyladenine (which do not distinguish between the 3 groups of PI3Ks (Vanhaesebroeck et al., 2001; Wu et al., 2010)) blocked programmed nuclear degradation (PND) in conjugating T. thermophila leading to accumulation of additional micronuclei and macronuclei(Yakisich and Kapler, 2004). The authors proposed that a product of PI3K is associated with PND activation and the degradation of non-exchanged pronuclei and later the macronucleus, and suggested that the active species may be PtdIns(3,4,5)P3. Understanding the precise role of PI3K in PND is complicated by the uncertain specificity of the available inhibitors, but future work may be able to illuminate noted similarities between PND in Tetrahymena and autophagy in other organisms, a pathway in which different classes of kinases are known to act(Wu et al., 2010). It may also be useful to consider enzymes that potentially degrade or convert PI3K. We detected 4 genes encoding putative homologs of PTEN, a phosphatase responsible for degrading PtdIns(3,4,5)P3 to PtdIns(4,5)P2 (TTHERM_00538980, TTHERM_00313160, TTHERM_00421160 and TTHERM_00467300). Interestingly, the expression of three of these is maximal during the stages of conjugation (C6 and C8) where PND occurs.
Another way to interrogate the roles of phosphoinositides is to identify proteins with putative phosphoinositide-binding domains. Many such proteins appear to be encoded in the T. thermophila genome. We used a domain-based search, with the SMART database in Genomic Mode(Letunic et al., 2006), to identify 49 genes possessing a pleckstrin homology (PH) domain, 39 possessing a phox homology (PX) domain and seven possessing an FYVE domain. In other searches we uncovered additional elements of the T. thermophila phosphoinositide genetic toolkit. For example, PIKfyve, which produces PtdIns(3,5)P2 from PtdIns3P, is present in the genome of T. thermophila in a single copy (TTHERM_01005090). In addition, several putative homologs can be found for PI4K, which converts PtdIns to PtdIns(4)P, and PIPKI, which converts PtdIns(4)P to PtdIns(4,5)P2, (Figure 1). These suggestive hits, which will need to be confirmed using genetic and biochemical approaches, suggest that T. thermophila could be an excellent model system for understanding how phosphoinositides contribute to the organization of complex cells, and could be particularly useful for investigating the Class I PI3K, as noted above.
In addition to their role as membrane-linked determinants, PtdIns are used as substrates to generate soluble molecules acting as secondary messengers. In particular, metazoans activate phospholipase C (PLC) via G protein-coupled receptors (GPCR) to hydrolyze PtdIns(4,5)P2, forming the second messengers Ins(1,4,5)P3 and DAG, with downstream effects on Ca2+ mobilization and protein phosphorylation. Outside of the metazoa, there is little convincing evidence linking PtdIns(4,5)P2 to these functions(Michell, 2008). However, the picture is becoming clearer in ciliates, through the study of PLC in Tetrahymena and the Ca2+ release channels (CRC) in Paramecium. Though beyond the scope of this chapter focused on membrane traffic, both bacterial-like and eukaryotic PI-PLCs have been identified in T. thermophila and pyriformis and putative homologs in both classes are encoded in the T. thermophila genome(Leondaritis et al., 2011). The bacterial PI-PLCs, which are likely to have been acquired via lateral gene transfer, may function in hydrolysis of the ciliate GPI anchors and the degradation of phospholipids in the extracellular space, along with other phospholipases(Florin-Christensen et al., 1986).
B. Sterol metabolism
Sterols affect membrane fluidity and permeability(Ohvo-Rekila et al., 2002). In addition, they are essential components of the “lipid rafts” that have been characterized principally in animal cells, which are currently understood as membrane microdomains whose formation depends upon the affinity of sterols for sphingolipids. The partitioning of proteins in lipid rafts may be important for regulation of signal transduction pathways(Simons and Toomre, 2000). Sterols also serve as precursors of bile salts and steroid hormones in mammals, brassinosteroids in plants and fungi and ecdysteroids in arthropods.
Eukaryotic organisms can satisfy their sterol requirement by de novo synthesis in vertebrates (cholesterol), plants (stigmasterol, sitosterol, and campesterol), and fungi (ergosterol), or by obtaining them from food. Sterol auxotrophs include invertebrates (nematodes and arthropods), some ciliates (Paramecium tetraurelia), apicomplexans (Plasmodium falciparum) and some flagellated parasites (Giardia intestinalis and Trichomonas vaginalis). T. thermophila is unusual in this regard, having no detectable sterols in its membranes and, accordingly, no sterol requirement. Instead, it synthesizes tetrahymanol, a compound similar to hopanoids found in bacteria, which acts as a surrogate sterol. However, when sterols are added to the growth medium, tetrahymanol synthesis is suppressed and T. thermophila incorporates the exogenous sterol, either with or without modifications(Conner et al., 1968). In particular the ciliate desaturates sterols at positions C5(6), C7(8) and C22(23) and removes the C24 ethyl group in C29 sterols (phytosterols)(Mallory and Conner, 1971). By the activity of these three sterol desaturases (C-5, C-7 and C-22 sterol desaturases) and C-24 sterol deethylation, the ciliate modifies exogenous sterols and accumulates the tri-unsaturated products in its membrane.
C-22 sterol desaturases have been characterized in other eukaryotes. In T. thermophila, the C-7 and C-22 sterol-desaturating activities, found mainly in a microsomal fraction, require cytochrome b5 as shown by their inhibition with azide and cyanide(Nusblat et al., 2005; Valcarce et al., 2000). This cytochrome b5 dependence is not characteristic of the C22 desaturases of plants and fungi, which require cytochrome P450. The difference is underscored by the insensitivity of the ciliate C22 desaturase to azole, a compound that strongly inhibits the corresponding plant and fungal activities. Moreover, no clear orthologs can be found in the T. thermophila genome for known C-22 sterol desaturases(Morikawa et al., 2006). These observations suggest that the T. thermophila enzyme represents a new class of C-22 sterol desaturases.
The C-5 sterol desaturase present in most eukaryotic cells belongs to the fatty acid hydroxylase (FAH) superfamily of integral membrane proteins that bind an iron cofactor via a 3-histidine motif. The C-5 sterol desaturase in T. thermophila, DES5A, was identified by characterizing the phenotype resulting from deletion of a putative FAH gene(Nusblat et al., 2009). The deletion mutant, which was fully viable, showed strongly diminished C-5 sterol desaturase activity, while C-7(8) and C-22(23) desaturase activities were unaffected.
The gene involved in C-24 sterol deethylation, DES24, was similarly confirmed by the disruption of putative FAH genes(Tomazic et al., 2011), resulting in a strain unable to eliminate the C-24 ethyl group from different phytosterols, and probably defective at the first step in dealkylation. Interestingly, the mutant strain was highly sensitive to phytosterols in the culture media, showing defects in growth and morphology and altered tetrahymanol biosynthesis. This observation suggests that C29 sterols can impair the normal growth of Tetrahymena. While C-24 sterol deethylation activity has been characterized in other organisms including nematodes, arthropods and green algae, the Tetrahymena enzyme represents the first molecular characterization. However, DES24 clusters phylogenetically with bacterial FAH sequences of unknown function, with no obvious orthologs in other eukaryotes, and may therefore have been acquired by lateral transfer. A variety of other observations, including substrate specificity and inhibitor studies, are also consistent with the hypothesis that the mechanism of T. thermophila C-24 deethylation differs from that in other eukaryotes.
T. thermophila, which is exposed in its environment to phytoplankton, higher plants and algae, may have developed the ability to metabolize otherwise-harmful phytosterols upon acquisition of DES24 from bacteria. Interestingly, however, Paramecium tetraurelia does not have C-24 dealkylation activity(Conner et al., 1971) and requires phytosterols(Whitaker and Nelson, 1987). Overall, sterol metabolism in T. thermophila seems to be the evolutionary product of a fascinating combination of gene losses (e.g., typical eukaryotic genes involved in sterol biosynthesis) combined with acquisition of bacterial genes to allow for synthesis of unusual compounds, with potentially novel mechanisms of sterol modification. This evolutionary history may be illuminated by interrogating the genomes of other Tetrahymena species as these are sequenced. In addition, further studies of the sterol pathways in T. thermophila may yield more information about lipid diversity and function.
C. Role of lipids in membrane curvature
Membrane fusion occurs when two separate lipid membranes merge into a single continuous bilayer. It underlies all membrane traffic as well as other important intra- and inter-cellular phenomena. The propensity of lipid bilayers to fuse in vitro is sensitive to lipid composition. One potential factor is that different lipids prefer, from an energetic perspective, to form surfaces with specific curvatures, and curvature affects fusogenicity in experimental pure lipid systems. Cone-shaped lipid like phosphatidylethanolamine (PE) and diacylglycerol (DAG) induce negative spontaneous curvature whereas inverted cone–shaped lipids like lysophosphatidylcholine (LPC) can induce positive spontaneous curvature. On the other hand, cylindrical phosphatidylcholine (PC) forms an almost flat monolayer. However, real biological membranes are also rich in proteins, and assessing the relative contributions of proteins and lipids to membrane fusion is a long-standing challenge. One issue has been whether lipids can drive changes in membrane curvature or simply accommodate changes that are driven by proteins. An approach that has been pioneered in Tetrahymena is to examine the distribution of lipids with fine resolution in sub-cellular membranes of defined curvature.
T. thermophila cultures can be induced to undergo synchronous mating, during which they form conjugation junctions containing hundreds of fusion pores in a small, well-defined zone, though which micronuclei are exchanged between paired cells(Wolfe, 1982, 1985). During formation of this zone, local lipid composition could change due to either de novo lipid synthesis, known to be required during conjugation, or to lipid exchange between cellular membranes. Ewing and colleagues exploited synchronous conjugation in Tetrahymena, combined with secondary ion mass spectrometry (SIMS), to ask whether the lipids in the fusion zone were enriched in species predicted to favor positively curved membranes, and also depleted in lipids whose shapes would resist such curvature(Ostrowski et al., 2004). The SIMs technique allows for visualization of the spatial distribution of molecular species according to the mass/charge ion ratio(Murphy et al., 2009). (For a detailed description of the technique, see (Heien et al., 2010).) The results indicated that the mating junction has a lower concentration of phosphatidylcholine, relative to the cell body, as expected since PC tends to favor flat membranes. In contrast, the mating junction contained a higher concentration of 2-aminoethylphosphonolipid, a phosphonolipid analog of phosphatidylethanolamine whose cone-shape would favor highly curved membranes.
These results, important in showing a set of predicted deviations within the lipid composition of an in vivo fusogenic zone, could not address the question of whether such lipid domains were present prior to or following the formation of the fusion pores. If the former, the lipids could be acting as fusion determinants; if the latter, the change in lipid composition could be accommodating the curvature imposed by other mechanisms, e.g., proteins. In a second paper, the same group addressed this by studying pairs during a time course of Tetrahymena mating, once again exploiting the synchronicity that can be achieved in the laboratory, and using the stability of formed pairs as a proxy for whether a zone of fusion pores had formed, based on earlier EM studies(Kurczy et al., 2010). The results suggested that change in lipid composition of the mating cell junction occur after fusion pores have formed, supporting a model in which changes in lipid composition arise subsequent to structural changes that are imposed by proteins. These studies, although still based on correlations, represent a beautiful example of using advanced technology to build upon the wealth of classical studies in T. thermophila, and using unique features to address fundamental questions in cell biology. If fusion pore formation is driven by proteins that are selectively expressed during conjugation, the identification of such proteins and disruption of the corresponding genes could facilitate future studies on lipid composition in which the correlations established in the studies described above could be tested by direct manipulation.
Space limitations prevent us from discussing other interesting work in the lipid field, including studies on sphingolipids, phospholipase D, and endocannabinoids(Wang et al., 2001; Wang et al., 2002) (Anagnostopoulos et al., 2010).
III. Conservation vs. innovation
With the data outlined above, one can begin to ask questions about the extent of evolutionary innovation to generate complex pathways of membrane traffic. A view ensconced in many textbooks is that modern cells are overwhelmingly similar, their shared features and pathways reflecting the shared inheritance from a common ancestor. A potential problem with such a blanket conclusion is that it stems from cell biology studies that have historically been conducted on a narrow swath of eukaryotic diversity, with all animal and fungal “model organisms” belonging to a single lineage, the Opisthokonts(Parfrey et al., 2006). In this regard, cell biological studies in higher plants are of great value, since these constitute a more divergent evolutionary branch. The greater divergence means that gain-of-function mutations in membrane trafficking determinants may have arisen and been positively selected following the split from Opisthokonts. Similarly, Ciliates represent another deeply divergent eukaryotic branch. The question in such divergent lineages is not whether innovations occurred (since they must have), but how they may have shaped specific pathways. An exceptionally interesting example of radical innovation was illustrated in recent work showing that T. thermophila, and probably all Alveolates, has invented a novel means to operate the mitochondrial ATP synthase complex that was previously believed to be conserved in structure and mechanism throughout eukaryotes(Balabaskaran Nina et al., 2010).
With regard to membrane traffic, one potential example of innovation generating a largely novel pathway in Tetrahymena, rather than simply tweaking a pre-existing pathway, is regulated secretion, that is, the synthesis of the dense-core secretory vesicles called mucocysts. A large number of protein components of mucocysts have been deduced using biochemical and genetic approaches, as well as by identifying T. thermophila homologs of proteins required for trichocyst exocytosis in Paramecium (reviewed in (Bowman et al., 2005b; Turkewitz, 2004))(A. Turkewitz, unpublished). While some of these proteins contain identifiable domains, e.g., β/γ crystallin domains, none of the proteins has an identifiable homolog outside of ciliates, with some possible weak exceptions in the related Apicomplexans. Thus all of the identified components of dense core vesicles in ciliates appear to reflect mutations that occurred after ciliates (or perhaps Alveolates) had branched from other organisms. Furthermore, it appears that the endoproteases responsible for processing of the dense core vesicle proproteins (GRL-encoded in Tetrahymena) are not related to the endoproteases that serve the homologous function in mammalian endocrine dense core granules (P. Romei and A. Turkewitz, unpublished). A tentative conclusion, based on these data, is that the striking functional similarities between the regulated secretory pathways in animals and ciliates primarily reflect independent innovation in the two lineages, shaped by similar selective pressures(Elde et al., 2007). However, such conclusions should be considered tentative. First, the genes that have been identified to date may reflect biases in the genetic and biochemical methods used. Secondly, there is no information yet available on the cellular machinery involved in mucocyst synthesis, so one possibility is that animals and ciliates have both adapted the same conserved biosynthetic machinery to create dense core vesicles, albeit from different ingredients.
The analysis of endocytosis from parasomal sacs revealed significant conservation between ciliates and animals in a clathrin-dependent pathway that also involved AP-2(Elde et al., 2005). Another apparent similarity is the endocytic involvement of dynamin in both lineages, but phylogenetic analysis revealed that this conservation has an innovative twist. In particular, the phylogenetic reconstruction argued that dynamin existed in a common ancestor of ciliates and animals, but that ancestral dynamin was unlikely to be involved in endocytosis(Elde et al., 2005). Instead, independent mutations in animals and ciliates subsequently led to the targeting of a dynamin paralog (i.e., a dynamin gene arising from a gene duplication within each lineage) to the endocytic pathway. An inference of this analysis was that the mechanism of targeting of the animal and ciliate dynamins could be different. While the ciliate mechanism is not yet known, the targeting motif identified in Drp1p does not resemble the known motif in the mammalian dynamins, consistent with the hypothesis(Elde et al., 2005). An interesting question is why dynamin was recruited for endocytosis in Tetrahymena, since this does not appear to have occurred in many other protist lineages. As discussed above, Tetrahymena may be very unusual in not using dynamic actin during endocytosis, and dynamin may be providing a function contributed by actin in animal cells.
A broader but shallower dataset to assess the relative contribution of innovation to membrane traffic exists in the Rab GTPase survey discussed above. Rabs are particularly well suited to addressing such questions because they determine compartmental identity, and must have co-evolved with their associated compartments(Pereira-Leal and Seabra, 2001). Consistent with this idea, the phylogenetic and functional comparison of yeast and human Rabs confirms that sequence-relatedness correlates with functional relatedness(Pereira-Leal, 2008). This indicates that many Rabs existed in the common ancestor of yeast and humans, associated with compartments that were retained in both the fungal and animal lineages.
Roughly ¼ of the T. thermophila Rabs, falling into 6 major branches, are conserved with homologs in distant lineages(Bright et al., 2010; Turkewitz and Bright, in press). Five of these six branches have previously been argued to represent “core Rabs”(Dacks and Field, 2007). The human Rabs in these branches are Rabs 4, 5, 7, 11 and 21 (all associated with stages of endocytosis), Rab 1 (associated with ER-to-Golgi traffic), and Rab 6 (associated with retrograde Golgi traffic). Tetrahymena appears to be missing Rabs in the three other identified core groups, corresponding to golgi-related (two clades), and regulated exocytic pathways. This may reflect lineage-restricted loss, but could be an artifact of failing to detect true homologs due to excessive sequence divergence. (Note that the absence of a Tetrahymena Rab in the regulated exocytic branch would be consistent with independent evolution of mucocysts and secretory vesicles in animals.) Two Tetrahymena Rabs fall into a robust branch with human Rab32 as well as RabE in Dictyostelium discoideum, suggesting that this group may be a 9th highly conserved Rab clade. Rab32 has been associated with several different organelles in mammals, including mitochondria and lysosome-related organelles(Tamura et al., 2009).
The fact that most conserved Tetrahymena Rabs belong to endocytic clades suggests that much of the endocytic machinery in Tetrahymena was inherited from an ancient eukaryotic ancestor, but this does not tell the whole story. Of the Rabs that were experimentally determined to be associated with endocytic compartments based on co-localization with FM4-64, roughly half belonged to conserved endocytic clades, while the rest were highly divergent (RabsD4, D5, D24, D27, D28, D35)(Bright et al., 2010). This suggests that a substantial part of the expansion within Rabs during Tetrahymena evolution was devoted to lineage-specific adaptations to endocytic pathways. A surprising observation was that four Rabs (Rabs4A, 4B, 11B, 31) that were assigned to the endocytic clade based on sequence, did not co-localize with FM4-64. This suggests that some Rabs may have retained the sequence signatures of conserved clades but have changed their compartmental localization. If such role-switching has indeed occurred, such unexpected plasticity in sequence-conserved Rabs would mean that inferring Rab function in divergent lineages simply based on sequence may sometimes be misleading. However, it is important to note that robust phylogenetic clustering of Tetrahymena Rabs required that the hypervariable C-terminal domains be excluded during the tree-building. Since these domains may contain targeting information, the role-switching may have been driven by specific C-terminal mutations that would be invisible to the phylogenetic methods used by both groups that have analyzed this important gene family to date.
Almost one third of the T. thermophila Rabs localized to phagosomes or structures associated with the phagocytic pathway, i.e., the oral apparatus and cytoproct(Bright et al., 2010). A similarly large number of Rabs have been associated with phagosomes in mammalian cells(Smith et al., 2007a). Given the results of the phagosome proteome project cited above (i.e., 28/73 Tetrahymena proteins were homologous to putative phagosome proteins in other systems), the expectation was that a large set of Tetrahymena and mammalian phagosomal Rabs would be mutually orthologous. However, only two of the phagosomal Rabs appeared orthologous(Bright et al., 2010). Moreover, these two Rabs are also associated with late endosomal compartments, so the orthology may reflect conservation in endocytic pathways that also intersect with the phagocytic pathway. In this regard, it will be important to learn what fraction of the phagosome proteome constituents are restricted to phagosomes. The Rab data, taken by itself, does not strongly support a common origin for the phagocytic pathways in ciliates and animals. However, it is also possible that the failure to detect orthology is due to the limitations of phylogenetic analysis for highly divergent lineages such as ciliates.
As implied by the discussion above, it is not always straightforward to generate robust phylogenetic trees using Tetrahymena gene sequences. An example of this can be seen in a comparison between two analyses of the myosin family in T. thermophila, in which the earlier survey underestimated the similarity of Tetrahymena myosins to those in other eukayotes(Sugita et al., 2011; Williams and Gavin, 2005). A very important issue in analyzing members of gene families is being able to distinguish orthologs (homologs that diverged in sequence following a speciation event, and which often retain the same function) vs paralogs (homologs that diverged within a species, and often diverged in function). While the two papers surveying Tetrahymena Rabs reach identical conclusions for many of the family members, they also differ in some cases. This is both because the datasets are slightly different and because different criteria were used to assign orthology. Some of the differences have been resolved, but researchers using phylogenetic tools to analyze Tetrahymena genes should be aware that some standard approaches may yield ambiguous or spurious results when applied to such divergent sequences, so collaboration with experts may be useful. In the relatively near future, some aspects of phylogenetic analysis should be simplified when additional Tetrahymena species genomes are sequenced.
Additional paper of interest.
Genetic and functional analysis of the septins, a family of 3 genes. The data indicate that the Tetrahymena septins are primarily involved in mitochondrial functions. Though septins are found throughout eukaryotes, such mitochondrial roles have previously been found only in mammals, suggesting independent recruitment for similar functions in mammals and ciliates(Wloga et al., 2008).
IV. Using expression data to elucidate pathways of membrane traffic
To better understand both mechanistic and evolutionary aspects of membrane traffic in Tetrahymena, it will be important to assemble a more complete parts list of the proteins associated with specific compartments and pathways. While significant progress has been made by simply pursuing Tetrahymena homologs of relevant animal and fungal proteins, as outlined above, this approach is clearly limited, in part because distinguishing orthologs from paralogs is not always possible. Secondly, the elucidation of membrane traffic is far from complete even in the best-studied systems like budding yeast, so limiting oneself to a homology-based approach would eliminate any contribution that Tetrahymena could make to identifying new factors in membrane traffic. One less biased approach would be to use biochemical approaches to identify Rab-interacting proteins starting with Rabs associated with specific compartments, which has been a powerful approach in mammalian cells(Christoforidis and Zerial, 2000).
A relatively novel approach that appears promising as a tool in Tetrahymena is based on a systems biology approach in mammalian cells, namely correlating the transcriptional profiles of genes involved in membrane trafficking factors. Balch and colleagues compiled microarray expression data from a large number of different tissues and cell lines, collected under a wide range of conditions, and calculated the degree of co-regulation between genes known to be involved in membrane trafficking(Gurkan et al., 2005). Based on the observed patterns of co-regulation, the authors proposed that membrane trafficking events are orchestrated by Rab-regulated protein “hubs” that are transcriptionally linked to the machinery involved in processes including coat formation, tethering and membrane fusion at those hubs. To test the significance of the observed co-regulation, the authors turned to the extensive biochemical and genetic data on membrane trafficking available in both mammalian and fungal systems. In some cases, these experimental data strongly supported the idea that co-regulated genes encoded products that were associated with the same hub. While the significance of many of their findings remains to be tested, the suggestion is that expression data, which are relatively simple to collect, could potentially be used to identify novel components of hubs, or to identify new hubs. Moreover, the data could also give hints regarding the conditions under which specific hubs are most physiologically significant.
In T. thermophila, whole-genome microarrays have been used for a variety of purposes, including to identify genes up-regulated during induced synthesis of mucocysts (L. Bright and A. Turkewitz, unpublished). A particularly rich dataset, referred to above, was collected by Gorovsky and colleagues by sampling Tetrahymena cultures under growing and starved conditions as well as during conjugation(Miao et al., 2009). The processed data are publicly available, curated by Miao and colleagues, in a format in which co-expressed genes can be identified based on Pearson correlation coefficients (http://tged.ihb.ac.cn/). More recently, this group has used additional approaches to recognize co-regulation within the dataset(Xiong et al., 2011).
We have begun to ask whether the Tetrahymena expression database can provide insights into the organization of membrane traffic in this organism. A hint that this might be true was mentioned earlier, namely that the genes encoding all known protein components of Tetrahymena mucocysts are co-regulated(Rahaman et al., 2009). However, this represents a very specific example and is limited to the proteins residing in the mucocyst lumen and membrane, rather than the still-unknown proteins involved in mucocyst biosynthesis. To pursue the larger questions, we have begun by asking whether proteins involved in membrane trafficking are co-regulated according to the Gorovsky expression database. Heterotetrameric adaptor complexes, such as AP2 discussed above, play highly conserved roles including the recruitment of coat proteins to membranes during vesicle budding. T. thermophila has 5 predicted AP µ subunits, including two µ subunits that fall into the AP1 cluster, and one each of AP2, AP3 and AP4, which would be expected to act at different sites and with different effectors. In the terms of Balch and colleagues, each AP complex represents a different hub. In Table I, we show the results of querying the database to identify genes that are coregulated with each of these µ subunits. In each case, we started with the APµ gene (here called APM, to conform to T. thermophila genetic nomenclature) as a “seed” and asked what genes in the dataset were most highly co-regulated, judging by the Pearson correlation coefficients. We then analyzed the hits using BLAST searches and gene ontology databases. We considered all hits with correlation coefficients ≥0.85, based on data presented on co-regulation in the whole genome dataset(Miao et al., 2009).
Table I.
Genes coregulated with the µ subunit genes of 5 heterotetrameric adaptors in T. thermophila. We list, in order of decreasing correlation coefficients, all coregulated genes with Pearson’s correlation coefficients ≥0.85 for each µ subunit (to a maximum of 20, for reasons of space). PCC = Pearson's Correlation Coefficient. Not included in these lists are coregulated genes for which no functions or conserved domains can be specified (denoted 'hypothetical protein' in NCBI). The number of such hypothetical genes with correlation coefficients equal or larger than those of the genes shown is as follows: for AMP1A, 14; for APM1B, 35; for APM2, 21; for APM4, 24; for APM5, 55.
The TTHERM IDs of genes that appear more than once in the Table, i.e, that appear to be coregulated with more than one adaptor, are emboldened.
| APM1A query (TTHERM_01108620) | APM2 query (TTHERM_00577030) | ||||
|---|---|---|---|---|---|
| Coregulated gene name (or putative) | Ttherm ID | PCC | Coregulated gene name (or putative) | Ttherm ID | PCC |
| Drp1, dynamin-related protein 1 | _00486790 | 0.9434 | Fimbrin-like | _00136510 | 0.9148 |
| Drp2, dynamin-related protein 2 | _00188910 | 0.9433 | Succinate dehydrogenase/fumarate reductase | _00241700 | 0.9073 |
| Adaptor-related protein complex 4, epsilon 1 | _00316010 | 0.9339 | Proteasome A-type and B-type family | _00487110 | 0.9072 |
| SNARE; syntaxin 5-2 | _00558250 | 0.9298 | Mov34/MPN/PAD-1 family | _00049450 | 0.9066 |
| Rab6C GTPase | _00079900 | 0.9042 | Proteasome A-type and B-type family | _00147560 | 0.9044 |
| N-terminal YjeF-domain | _00149620 | 0.9021 | Phosphoglycerate mutase 1 family | _00641240 | 0.9071 |
| RabD4 GTPase | _00825210 | 0.8965 | Ubiquinol-cytochrome c reductase | _00295080 | 0.8969 |
| Ubiquitin carboxyl-terminal hydrolase family | _00077370 | 0.8861 | Actin family | _01020680 | 0.8962 |
| ARF5 GTPase | _00735240 | 0.8846 | Adaptor-related protein complex 1, beta 1 | _00219370 | 0.8959 |
| RCC1-domain protein | _00586630 | 0.8793 | Protein phosphatase 2C–containing | _00446510 | 0.8955 |
| Endonuclease/exonuclease/phosphatase family | _00316140 | 0.8783 | Coatomer protein complex, subunit beta 1 | _00488350 | 0.8937 |
| Protein kinase domain-containing | _00128930 | 0.8760 | V-type ATPase, D subunit | _00821870 | 0.8936 |
| Rab GGTB geranylgeranyltransferase | _00630460 | 0.8739 | EF hand-containing | _00059190 | 0.8917 |
| Yip1 domain family (GDI displacement factor) | _00193890 | 0.8737 | Proteasome A-type and B-type family | _00316530 | 0.8912 |
| HECT domain and RCC1-like domain | _00586640 | 0.8721 | Cytochrome C1 family | _00918500 | 0.8908 |
| NSF (N-ethylmaleimide-sensitive factor) | _00039210 | 0.8718 | DnaJ domain-containing | _00195890 | 0.8888 |
| von Willebrand factor type A domain-containing | _00809300 | 0.8698 | ATP synthase F1, delta | _00684790 | 0.8886 |
| TRAF-type family | _00666410 | 0.8695 | Ribosomal protein L13 | _01207660 | 0.8884 |
| DHHC zinc finger domain-containing | _00581830 | 0.8683 | Proteasome A-type and B-type family | _00043880 | 0.8834 |
| Adaptor-related protein complex 1, gamma 1 | _00715750 | 0.8673 | IQ calmodulin-binding motif family | _01194640 | 0.8822 |
| APM1B query (TTHERM_00455300) | APM4 query (TTHERM_00545860) | ||||
| Coregulated gene name (or putative) | Ttherm ID | PCC | Coregulated gene name (or putative) | Ttherm ID | PCC |
| Phosphatidylinositol glycan, class Q (PIGQ) | _00945240 | 0.9392 | Oxidoreductase family | _00334360 | 0.9517 |
| RabD40 GTPase | _01129680 | 0.9346 | ARP2/3 complex 20 kDa subunit (ARPC4) | _00857810 | 0.9397 |
| Hydrolase, NUDIX family | _01084270 | 0.9214 | Amidase family | _00361450 | 0.9383 |
| TB2/DP1, HVA22 family | _00270350 | 0.9184 | Coatomer zeta coat | _00052600 | 0.9378 |
| Adaptor-related protein complex 4, beta-1 | _00225780 | 0.9141 | EF hand-containing | _01055480 | 0.9364 |
| NSF (N-ethylmaleimide-sensitive factor) | _00039210 | 0.9078 | F-actin capping protein, beta subunit-containing | _00784430 | 0.9339 |
| SNARE; syntaxin 5-2 | _00558250 | 0.8983 | Coatomer epsilon subunit family | _00459320 | 0.9304 |
| HIT domain-containing | _00378790 | 0.8978 | Actin family | _01020680 | 0.9299 |
| N-terminal YjeF-domain | _00149620 | 0.8977 | Fimbrin-like 71 K | _00136510 | 0.9291 |
| Oxidoreductase | _00334360 | 0.8941 | ADP-ribosylation factor | _00340150 | 0.9286 |
| Eukaryotic phosphomannomutase family | _00059380 | 0.8860 | Signal peptidase I family | _01093560 | 0.9283 |
| Protein kinase domain-containing | _00715960 | 0.8851 | Peptidyl-prolyl cis-trans isomerase | _00548130 | 0.9279 |
| Adaptor-related protein complex 4, sigma 1 | _01227730 | 0.8843 | Peptidyl-prolyl cis-trans isomerase | _00532620 | 0.9239 |
| Adaptor-related protein complex 1, gamma 1 | _00287890 | 0.8783 | Phosphatidylserine decarboxylase family | _00859340 | 0.9221 |
| RabX23 Rab-like protein; GTPase | _00298510 | 0.8676 | HCaRG | _00059120 | 0.9205 |
| Adaptor-related protein complex 4, mu 1 | _00545860 | 0.8604 | Coatomer protein complex, gamma 2 | _00444320 | 0.9199 |
| Hydrolase, NUDIX family | _00113100 | 0.8536 | Coatomer alpha | _01345820 | 0.9196 |
| ADP-ribosylation factor | _00448910 | 0.8528 | Endomembrane protein 70-containing | _00723110 | 0.9187 |
| EF Tu C-terminal domain-containing | _01234330 | 0.8505 | Roadblock/LC7 domain-containing | _00348650 | 0.9180 |
| EF hand-containing | _00497600 | 0.9177 | |||
| APM3 query (TTHERM_00572100) | |||||
| Coregulated gene name (or putative) | Ttherm ID | PCC | |||
| Adaptor-related protein complex 3, beta 1 | _00703490 | 0.9581 | |||
| Protein kinase domain-containing | _01015890 | 0.9467 | |||
| VPS10 domain (sortilin 2) | _00410210 | 0.9428 | |||
| Protein kinase domain-containing | _01052880 | 0.9378 | |||
| VPS10 domain (sortilin 4) | _00313130 | 0.9350 | |||
| MAC/Perforin domain-containing | _01141420 | 0.9231 | |||
| Rab4B GTPase | _01097960 | 0.9190 | |||
| Thioredoxin (TRX) family | _00664030 | 0.9175 | |||
| Golgin; GRIP domain-containing | _01143870 | 0.9168 | |||
| Eukaryotic aspartyl protease family | _00440510 | 0.9156 | |||
| SNARE; synaptobrevin 3 | _00152090 | 0.9116 | |||
| Amidase family | _00361390 | 0.9086 | |||
| Taurine catabolism dioxygenase TauD, TfdA family | _00823760 | 0.8995 | |||
| Rab21A GTPase | _00540110 | 0.8758 | |||
| HAD-superfamily hydrolase, subfamily IIA | _01093670 | 0.8722 | |||
| Histidine acid phosphatase family | _00649220 | 0.8666 | |||
| Protein kinase domain-containing | _01164090 | 0.8638 | |||
| Calpain family cysteine protease | _00196650 | 0.8618 | |||
| Mitochondrial carrier protein | _00637710 | 0.8609 |
Many of the results are intriguing. For example, all of the top co-regulated genes with the AP1 µ subunit encode other genes involved in membrane traffic, including two dynamin-family proteins and the ε subunit of the AP4 adaptor. For a second example, the genes co-regulated with APM3 include multiple sortilin homologs, which are receptors known to be sorted in an AP3-dependent mechanism in other organisms, while genes co-regulated with APM4 include multiple coatamer subunits and a collection of actin-binding proteins. Interestingly, while the majority of coregulated genes are non-overlapping between the 5 lists, a small number of genes (whose TTHERM IDs are emboldened in the Table) appear to be co-regulated with more than one of the AP µ genes. One of these is a SNARE, a protein involved in membrane fusion, while a second is NSF, a complex required for SNARE disassembly. While the significance of the hits has not been tested, these preliminary results suggest that exploiting Tetrahymena expression databases may be a rich new vein to explore for cell biologists interested in membrane traffic.
Acknowledgments
We thank Joe Briguglio for insightful comments on the manuscript and, and Mimi Pixque for support. Work in APT’s laboratory is supported by NSF MCB0422011 and NIH GM077607. ADN was supported by the International funding program of the National Scientific and Technical Research Council (CONICET) of Argentina. LJB was supported by Genetics and Regulation Training Grant T32 GM007197. APT is a member of the scientific advisory board of TetraGenetics, Inc.
References
- Akematsu T, Endoh H. Role of apoptosis-inducing factor (AIF) in programmed nuclear death during conjugation in Tetrahymena thermophila. BMC Cell Biol. 2010;11:13. doi: 10.1186/1471-2121-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akematsu T, Pearlman RE, Endoh H. Gigantic macroautophagy in programmed nuclear death of Tetrahymena thermophila. Autophagy. 2010;6:901–911. doi: 10.4161/auto.6.7.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldag I, Bockau U, Rossdorf J, Laarmann S, Raaben W, Herrmann L, Weide T, Hartmann MW. Expression, secretion and surface display of a human alkaline phosphatase by the ciliate Tetrahymena thermophila. BMC Biotechnol. 2011;11:11. doi: 10.1186/1472-6750-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RD. Fine structure, reconstruction and possible functions of components of the cortex of Tetrahymena pyriformis. J Protozool. 1967;14:553–565. doi: 10.1111/j.1550-7408.1967.tb02042.x. [DOI] [PubMed] [Google Scholar]
- Allen RD. Membranes of ciliates: ultrastructure, biochemistry and fusion, Membrane Fusion. Amsterdam: Elsevier/North-Holland Biomed. Press; 1978. pp. 657–763. [Google Scholar]
- Allen RD. The contractile vacuole and its membrane dynamics. Bioessays. 2000;22:1035–1042. doi: 10.1002/1521-1878(200011)22:11<1035::AID-BIES10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Allen RD, Fok AK. Membrane recycling and endocytosis in Paramecium confirmed by horseradish peroxidase pulse-chase studies. J Cell Sci. 1980;45:131–145. doi: 10.1242/jcs.45.1.131. [DOI] [PubMed] [Google Scholar]
- Allen RD, Wolf RW. Membrane recycling at the cytoproct of Tetrahymena. J Cell Sci. 1979;35:217–227. doi: 10.1242/jcs.35.1.217. [DOI] [PubMed] [Google Scholar]
- Anagnostopoulos D, Rakiec C, Wood J, Pandarinathan L, Zvonok N, Makriyannis A, Siafaka-Kapadai A. Identification of endocannabinoids and related N-acylethanolamines in tetrahymena. A new class of compounds for Tetrahymena. Protist. 2010;161:452–465. doi: 10.1016/j.protis.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Balabaskaran Nina P, Dudkina NV, Kane LA, van Eyk JE, Boekema EJ, Mather MW, Vaidya AB. Highly divergent mitochondrial ATP synthase complexes in Tetrahymena thermophila. PLoS Biol. 2010;8:e1000418. doi: 10.1371/journal.pbio.1000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B, Rusing M. Structure of N-glycosidic carbohydrates of secretory proteins of Tetrahymena thermophila. J Eukaryot Microbiol. 2003;50:235–239. doi: 10.1111/j.1550-7408.2003.tb00125.x. [DOI] [PubMed] [Google Scholar]
- Bowman GR, Elde NC, Morgan G, Winey M, Turkewitz AP. Core Formation and the Acquisition of Fusion Competence are Linked During Secretory Granule Maturation in Tetrahymena. Traffic. 2005a;6:303–323. doi: 10.1111/j.1600-0854.2005.00273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR, Smith DG, Michael Siu KW, Pearlman RE, Turkewitz AP. Genomic and proteomic evidence for a second family of dense core granule cargo proteins in Tetrahymena thermophila. J Eukaryot Microbiol. 2005b;52:291–297. doi: 10.1111/j.1550-7408.2005.00045.x. [DOI] [PubMed] [Google Scholar]
- Bowman GR, Turkewitz AP. Analysis of a mutant exhibiting conditional sorting to dense core secretory granules in Tetrahymena thermophila. Genetics. 2001;159:1605–1616. doi: 10.1093/genetics/159.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright LJ, Kambesis N, Nelson SB, Jeong B, Turkewitz AP. Comprehensive analysis reveals dynamic and evolutionary plasticity of Rab GTPases and membrane traffic in Tetrahymena thermophila. PLoS Genetics. 2010;6:e1001155. doi: 10.1371/journal.pgen.1001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanat E, Huttner WB. Milieu-induced, selective aggregation of regulated secretory proteins in the trans-Golgi network. J. Cell Biol. 1991;115:1505–1519. doi: 10.1083/jcb.115.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis S, Zerial M. Purification and identification of novel Rab effectors using affinity chromatography. Methods. 2000;20:403–410. doi: 10.1006/meth.2000.0953. [DOI] [PubMed] [Google Scholar]
- Cole ES, Anderson PC, Fulton RB, Majerus ME, Rooney MG, Savage JM, Chalker D, Honts J, Welch ME, Wentland AL, Zweifel E, Beussman DJ. A proteomics approach to cloning fenestrin from the nuclear exchange junction of tetrahymena. J Eukaryot Microbiol. 2008;55:245–256. doi: 10.1111/j.1550-7408.2008.00337.x. [DOI] [PubMed] [Google Scholar]
- Conner RL, Landrey JR, Burns CH, Mallory FB. Cholesterol inhibition of pentacyclic triterpenoid biosynthesis in Tetrahymena pyriformis. J Protozool. 1968;15:600–605. doi: 10.1111/j.1550-7408.1968.tb02178.x. [DOI] [PubMed] [Google Scholar]
- Conner RL, Landrey JR, Kaneshiro ES, Van Wagtendonk WJ. The metabolism of stigmasterol and cholesterol by Paramecium aurelia. Biochim Biophys Acta. 1971;239:312–319. doi: 10.1016/0005-2760(71)90176-7. [DOI] [PubMed] [Google Scholar]
- Cousin MA, Robinson PJ. Mechanisms of synaptic vesicle recycling illuminated by fluorescent dyes. J Neurochem. 1999;73:2227–2239. doi: 10.1046/j.1471-4159.1999.0732227.x. [DOI] [PubMed] [Google Scholar]
- Cowan AT, Bowman GR, Edwards KF, Emerson JJ, Turkewitz AP. Genetic, genomic, and functional analysis of the granule lattice proteins in Tetrahymena secretory granules. Mol Biol Cell. 2005;16:4046–4060. doi: 10.1091/mbc.E05-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creemers JW, Jackson RS, Hutton JC. Molecular and cellular regulation of prohormone processing. Semin Cell Dev Biol. 1998;9:3–10. doi: 10.1006/scdb.1997.0195. [DOI] [PubMed] [Google Scholar]
- Dacks JB, Field MC. Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J Cell Sci. 2007;120:2977–2985. doi: 10.1242/jcs.013250. [DOI] [PubMed] [Google Scholar]
- Deli D, Leondaritis G, Tiedtke A, Galanopoulou D. Deficiency in lysosomal enzyme secretion is associated with upregulation of phosphatidylinositol 4-phosphate in tetrahymena. J Eukaryot Microbiol. 2008;55:343–350. doi: 10.1111/j.1550-7408.2008.00329.x. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Doerder FP, Gerber CA. Molecular characterization of the SerL paralogs of Tetrahymena thermophila. Biochem Biophys Res Commun. 2000;278:621–626. doi: 10.1006/bbrc.2000.3857. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith RK, Jr, Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC, Turkewitz AP, Asai DJ, Wilkes DE, Wang Y, Cai H, Collins K, Stewart BA, Lee SR, Wilamowska K, Weinberg Z, Ruzzo WL, Wloga D, Gaertig J, Frankel J, Tsao CC, Gorovsky MA, Keeling PJ, Waller RF, Patron NJ, Cherry JM, Stover NA, Krieger CJ, del Toro C, Ryder HF, Williamson SC, Barbeau RA, Hamilton EP, Orias E. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde NC, Long M, Turkewitz AP. A role for convergent evolution in the secretory life of cells. Trends Cell Biol. 2007;17:157–164. doi: 10.1016/j.tcb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Elde NC, Morgan G, Winey M, Sperling L, Turkewitz AP. Elucidation of Clathrin-Mediated Endocytosis in Tetrahymena Reveals an Evolutionarily Convergent Recruitment of Dynamin. PLoS Genet. 2005;1:e52. doi: 10.1371/journal.pgen.0010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin-Christensen J, Florin-Christensen M, Knudsen J, Rasmussen L. Phospholipases and phosphonolipids in a ciliate: an attack and defense system? Trends Biochem Sci. 1986;11:354–355. [Google Scholar]
- Frankel J. Cell biology of Tetrahymena thermophila. Methods Cell Biol. 2000;6299432807:27–125. doi: 10.1016/s0091-679x(08)61528-9. [DOI] [PubMed] [Google Scholar]
- Gonda K, Komatsu M, Numata O. Calmodulin and Ca2+/calmodulin-binding proteins are involved in Tetrahymena thermophila phagocytosis. Cell Struct Funct. 2000;25:243–251. doi: 10.1247/csf.25.243. [DOI] [PubMed] [Google Scholar]
- Gumbiner B, Kelly RB. Two distinct intracellular pathways transport secretory and membrane glycoproteins to the surface of pituitary tumor cells. Cell. 1982;28:51–59. doi: 10.1016/0092-8674(82)90374-9. [DOI] [PubMed] [Google Scholar]
- Gurkan C, Lapp H, Alory C, Su AI, Hogenesch JB, Balch WE. Large-scale profiling of Rab GTPase trafficking networks: the membrome. Mol Biol Cell. 2005;16:3847–3864. doi: 10.1091/mbc.E05-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad A, Bowman GR, Turkewitz AP. A new class of cargo protein in Tetrahymena thermophila dense core secretory granules. Eukaryot Cell. 2002;1:583–593. doi: 10.1128/EC.1.4.583-593.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M, Guberman A, Florin-Christensen M, Tiedtke A. Screening for and characterization of phospholipase A1 hypersecretory mutants of Tetrahymena thermophila. Appl Microbiol Biotechnol. 2000;54:390–396. doi: 10.1007/s002530000405. [DOI] [PubMed] [Google Scholar]
- Hausmann K, Allen RD. Membrane behavior of exocytic vesicles. II. Fate of the trichocyst membranes in Paramecium after induced trichocyst discharge. J Cell Biol. 1976;69:313–326. doi: 10.1083/jcb.69.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heien ML, Piehowski PD, Winograd N, Ewing AG. Lipid detection, identification, and imaging single cells with SIMS. Methods Mol Biol. 2010;656:85–97. doi: 10.1007/978-1-60761-746-4_4. [DOI] [PubMed] [Google Scholar]
- Herrmann L, Erkelenz M, Aldag I, Tiedtke A, Hartmann MW. Biochemical and molecular characterisation of Tetrahymena thermophila extracellular cysteine proteases. BMC Microbiol. 2006;6:19. doi: 10.1186/1471-2180-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosein RE, Williams SA, Gavin RH. Directed motility of phagosomes in Tetrahymena thermophila requires actin and Myo1p, a novel unconventional myosin. Cell Motil Cytoskeleton. 2005;61:49–60. doi: 10.1002/cm.20065. [DOI] [PubMed] [Google Scholar]
- Huynh KK, Kay JG, Stow JL, Grinstein S. Fusion, fission, and secretion during phagocytosis. Physiology (Bethesda) 2007;22:366–372. doi: 10.1152/physiol.00028.2007. [DOI] [PubMed] [Google Scholar]
- Iwamoto M, Mori C, Kojidani T, Bunai F, Hori T, Fukagawa T, Hiraoka Y, Haraguchi T. Two distinct repeat sequences of Nup98 nucleoporins characterize dual nuclei in the binucleated ciliate tetrahymena. Curr Biol. 2009;19:843–847. doi: 10.1016/j.cub.2009.03.055. [DOI] [PubMed] [Google Scholar]
- Jacobs ME, DeSouza LV, Samaranayake H, Pearlman RE, Siu KW, Klobutcher LA. The Tetrahymena thermophila phagosome proteome. Eukaryot Cell. 2006;5:1990–2000. doi: 10.1128/EC.00195-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y, Joo MK, Sohn YS, Jeong B. Reverse thermal organogels. Adv Mater. 2007;19:3947–3950. [Google Scholar]
- Kersting MC, Boyette M, Massey JH, Ryals PE. Identification of the inositol isomers present in Tetrahymena. J Eukaryot Microbiol. 2003;50:164–168. doi: 10.1111/j.1550-7408.2003.tb00111.x. [DOI] [PubMed] [Google Scholar]
- Kersting MC, Ryals PE. Sodium-dependent transport of [3H](1D)chiro-inositol by Tetrahymena. J Eukaryot Microbiol. 2004;51:307–311. doi: 10.1111/j.1550-7408.2004.tb00571.x. [DOI] [PubMed] [Google Scholar]
- Kienle N, Kloepper TH, Fasshauer D. Differences in the SNARE evolution of fungi and metazoa. Biochem Soc Trans. 2009;37:787–791. doi: 10.1042/BST0370787. [DOI] [PubMed] [Google Scholar]
- Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Jr, Culver BP, Yates JR, 3rd, Winey M. New Tetrahymena basal body protein components identify basal body domain structure. J Cell Biol. 2007;178:905–912. doi: 10.1083/jcb.200703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T. Imaging endocytic clathrin structures in living cells. Trends Cell Biol. 2009;19:596–605. doi: 10.1016/j.tcb.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiy T, Vosskuhler C, Rasmussen L, Tiedtke A. Three pools of lysosomal enzymes in Tetrahymena thermophila. Exp Cell Res. 1993;205:286–292. doi: 10.1006/excr.1993.1088. [DOI] [PubMed] [Google Scholar]
- Klobutcher LA, Ragkousi K, Setlow P. The Bacillus subtilis spore coat provides “eat resistance” during phagocytic predation by the protozoan Tetrahymena thermophila. Proc Natl Acad Sci U S A. 2006;103:165–170. doi: 10.1073/pnas.0507121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs P, Pallinger E. Phosphatidylinositol 3-kinase-like activity in Tetrahymena. Effects of wortmannin and LY294002. Acta Protozool. 2003;42:277–285. [Google Scholar]
- Kurczy ME, Piehowski PD, Van Bell CT, Heien ML, Winograd N, Ewing AG. Mass spectrometry imaging of mating Tetrahymena show that changes in cell morphology regulate lipid domain formation. Proc Natl Acad Sci U S A. 2010;107:2751–2756. doi: 10.1073/pnas.0908101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Wisniewski JC, Dentler WL, Asai DJ. Gene knockouts reveal separate functions for two cytoplasmic dyneins in Tetrahymena thermophila. Mol Biol Cell. 1999;10:771–784. doi: 10.1091/mbc.10.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leondaritis G, Galanopoulou D. Characterization of inositol phospholipids and identification of a mastoparan-induced polyphosphoinositide response in Tetrahymena pyriformis. Lipids. 2000;35:525–532. doi: 10.1007/s11745-000-552-8. [DOI] [PubMed] [Google Scholar]
- Leondaritis G, Sarri T, Dafnis I, Efstathiou A, Galanopoulou D. Biochemical and genetic evidence for the presence of multiple phosphatidylinositol- and phosphatidylinositol 4,5-bisphosphate-specific phospholipases C in Tetrahymena. Eukaryot Cell. 2011;10:412–422. doi: 10.1128/EC.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leondaritis G, Tiedtke A, Galanopoulou D. D-3 phosphoinositides of the ciliate Tetrahymena: characterization and study of their regulatory role in lysosomal enzyme secretion. Biochim Biophys Acta. 2005;1745:330–341. doi: 10.1016/j.bbamcr.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madinger CL, Collins K, Fields LG, Taron CH, Benner JS. Constitutive secretion in Tetrahymena thermophila. Eukaryot Cell. 2010;9:674–681. doi: 10.1128/EC.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory FB, Conner RL. Dehydrogenation and dealkylation of various sterols by Tetrahymena pyriformis. Lipids. 1971;6:149–153. doi: 10.1007/BF02533028. [DOI] [PubMed] [Google Scholar]
- Malone CD, Falkowska KA, Li AY, Galanti SE, Kanuru RC, LaMont EG, Mazzarella KC, Micev AJ, Osman MM, Piotrowski NK, Suszko JW, Timm AC, Xu MM, Liu L, Chalker DL. Nucleus-specific importin alpha proteins and nucleoporins regulate protein import and nuclear division in the binucleate Tetrahymena thermophila. Eukaryot Cell. 2008;7:1487–1499. doi: 10.1128/EC.00193-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo J, Oguri S, Nakamura S, Hanawa T, Fukumoto T, Hayashi Y, Kawaguchi K, Mizutani Y, Yao T, Akizawa K, Suzuki H, Simizu C, Matsuno K, Kamiya S, Yamaguchi H. Ciliates rapidly enhance the frequency of conjugation between Escherichia coli strains through bacterial accumulation in vesicles. Res Microbiol. 2010;161:711–719. doi: 10.1016/j.resmic.2010.07.004. [DOI] [PubMed] [Google Scholar]
- Miao W, Xiong J, Bowen J, Wang W, Liu Y, Braguinets O, Grigull J, Pearlman RE, Orias E, Gorovsky MA. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS ONE. 2009;4:e4429. doi: 10.1371/journal.pone.0004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michell RH. Inositol derivatives: evolution and functions. Nat Rev Mol Cell Biol. 2008;9:151–161. doi: 10.1038/nrm2334. [DOI] [PubMed] [Google Scholar]
- Morikawa T, Mizutani M, Aoki N, Watanabe B, Saga H, Saito S, Oikawa A, Suzuki H, Sakurai N, Shibata D, Wadano A, Sakata K, Ohta D. Cytochrome P450 CYP710A encodes the sterol C-22 desaturase in Arabidopsis and tomato. Plant Cell. 2006;18:1008–1022. doi: 10.1105/tpc.105.036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya CE, Jacobs RS. Pseudopterosin A inhibits phagocytosis and alters intracellular calcium turnover in a pertussis toxin sensitive site in Tetrahymena thermophila. Comp Biochem Physiol C Toxicol Pharmacol. 2006;143:436–443. doi: 10.1016/j.cbpc.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Murphy RC, Hankin JA, Barkley RM. Imaging of lipid species by MALDI mass spectrometry. J Lipid Res. 2009;50(Suppl):S317–S322. doi: 10.1194/jlr.R800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson JR. Phagotrophy in Tetrahymena. In: Levandowsky M, Hutner SH, editors. Biochemistry and Physiology of Protozoa. New York: Academic Press; 1979. pp. 339–379. [Google Scholar]
- Nilsson JR, Van Deurs B. Coated pits and pinocytosis in Tetrahymena. J. Cell Sci. 1983;63:209–222. doi: 10.1242/jcs.63.1.209. [DOI] [PubMed] [Google Scholar]
- Nusblat AD, Munoz L, Valcarce GA, Nudel CB. Characterization and properties of cholesterol desaturases from the ciliate Tetrahymena thermophila. J Eukaryot Microbiol. 2005;52:61–67. doi: 10.1111/j.1550-7408.2005.3279rr.x. [DOI] [PubMed] [Google Scholar]
- Nusblat AD, Najle SR, Tomazic ML, Uttaro AD, Nudel CB. C-5(6) sterol desaturase from tetrahymena thermophila: Gene identification and knockout, sequence analysis, and comparison to other C-5(6) sterol desaturases. Eukaryot Cell. 2009;8:1287–1297. doi: 10.1128/EC.00057-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohvo-Rekila H, Ramstedt B, Leppimaki P, Slotte JP. Cholesterol interactions with phospholipids in membranes. Prog Lipid Res. 2002;41:66–97. doi: 10.1016/s0163-7827(01)00020-0. [DOI] [PubMed] [Google Scholar]
- Ostrowski SG, Van Bell CT, Winograd N, Ewing AG. Mass spectrometric imaging of highly curved membranes during Tetrahymena mating. Science. 2004;305:71–73. doi: 10.1126/science.1099791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Barbero E, Lasser E, Dunthorn M, Bhattacharya D, Patterson DJ, Katz LA. Evaluating Support for the Current Classification of Eukaryotic Diversity. PLoS Genet. 2006;2:e220. doi: 10.1371/journal.pgen.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Winey M. Basal body assembly in ciliates: the power of numbers. Traffic. 2009;10:461–471. doi: 10.1111/j.1600-0854.2009.00885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. An olympian protozoan. Nucleus. 2010;1:2–3. doi: 10.4161/nucl.1.1.10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Leal JB. The Ypt/Rab family and the evolution of trafficking in fungi. Traffic. 2008;9:27–38. doi: 10.1111/j.1600-0854.2007.00667.x. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB, Seabra MC. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- Pieringer J, Conner RL. Positional distribution of fatty acids in the glycerophospholipids of Tetrahymena pyriformis. J Lipid Res. 1979;20:363–370. [PubMed] [Google Scholar]
- Plattner H, Flotenmeyer M, Kissmehl R, Pavlovic N, Hauser K, Momayezi M, Braun N, Tack J, Bachmann L. Microdomain arrangement of the SERCA-type Ca2+ pump (Ca2+-ATPase) in subplasmalemmal calcium stores of paramecium cells. J Histochem Cytochem. 1999;47:841–854. doi: 10.1177/002215549904700701. [DOI] [PubMed] [Google Scholar]
- Rahaman A, Elde NC, Turkewitz AP. A dynamin-related protein required for nuclear remodeling in Tetrahymena. Curr Biol. 2008;18:1227–1233. doi: 10.1016/j.cub.2008.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahaman A, Miao W, Turkewitz AP. Independent transport and sorting of functionally distinct protein families in Tetrahymena dense core secretory granules. Eukaryot Cell. 2009 doi: 10.1128/EC.00151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs BM, Lansley TA, Ryals PE. Phosphatidylinositol synthase of Tetrahymena: inositol isomers as substrates in phosphatidylinositol biosynthesis and headgroup exchange reactions. J Eukaryot Microbiol. 2007;54:119–124. doi: 10.1111/j.1550-7408.2007.00242.x. [DOI] [PubMed] [Google Scholar]
- Rosati G, Modeo L. Extrusomes in ciliates: diversification, distribution, and phylogenetic implications. J Eukaryot Microbiol. 2003;50:383–402. doi: 10.1111/j.1550-7408.2003.tb00260.x. [DOI] [PubMed] [Google Scholar]
- Ryals PE. Inositols and phosphoinositides in Tetrahymena. Acta Protozool. 2009;48:191–202. [Google Scholar]
- Ryals PE, Kersting MC. Sodium-dependent uptake of [3H]scyllo-inositol by Tetrahymena: incorporation into phosphatidylinositol, phosphatidylinositol-linked glycans, and polyphosphoinositols. Arch Biochem Biophys. 1999;366:261–266. doi: 10.1006/abbi.1999.1211. [DOI] [PubMed] [Google Scholar]
- Saito-Nakano Y, Nakahara T, Nakano K, Nozaki T, Numata O. Marked amplification and diversification of products of ras genes from rat brain, Rab GTPases, in the ciliates Tetrahymena thermophila and Paramecium tetraurelia. J Eukaryot Microbiol. 2010;57:389–399. doi: 10.1111/j.1550-7408.2010.00503.x. [DOI] [PubMed] [Google Scholar]
- Satir BH, Schooley C, Satir P. Membrane fusion in a model system. Mucocyst secretion in Tetrahymena. J. Cell Biol. 1973;56:153–176. doi: 10.1083/jcb.56.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N. Ypt/rab gtpases: regulators of protein trafficking. Sci STKE. 2001;2001:RE11. doi: 10.1126/stke.2001.100.re11. [DOI] [PubMed] [Google Scholar]
- Simon MC, Schmidt HJ. Antigenic variation in ciliates: antigen structure, function, expression. J Eukaryot Microbiol. 2007;54:1–7. doi: 10.1111/j.1550-7408.2006.00226.x. [DOI] [PubMed] [Google Scholar]
- Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- Smith AC, Heo WD, Braun V, Jiang X, Macrae C, Casanova JE, Scidmore MA, Grinstein S, Meyer T, Brumell JH. A network of Rab GTPases controls phagosome maturation and is modulated by Salmonella enterica serovar Typhimurium. J Cell Biol. 2007a;176:263–268. doi: 10.1083/jcb.200611056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DG, Gawryluk RM, Spencer DF, Pearlman RE, Siu KW, Gray MW. Exploring the mitochondrial proteome of the ciliate protozoon Tetrahymena thermophila: direct analysis by tandem mass spectrometry. J Mol Biol. 2007b;374:837–863. doi: 10.1016/j.jmb.2007.09.051. [DOI] [PubMed] [Google Scholar]
- Smith JC, Northey JG, Garg J, Pearlman RE, Siu KW. Robust method for proteome analysis by MS/MS using an entire translated genome: demonstration on the ciliome of Tetrahymena thermophila. J Proteome Res. 2005;4:909–919. doi: 10.1021/pr050013h. [DOI] [PubMed] [Google Scholar]
- Stelly N, Halpern S, Nicolas G, Fragu P, Adoutte A. Direct visualization of a vast cortical calcium compartment in Paramecium by secondary ion mass spectrometry (SIMS) microscopy: possible involvement in exocytosis. J. Cell Sci. 1995;108:1895–1909. doi: 10.1242/jcs.108.5.1895. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Olkkonen VM. The Rab GTPase family. Genome Biol. 2001;2:REVIEWS3007. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart LM, Ezekowitz RA. Phagocytosis: elegant complexity. Immunity. 2005;22:539–550. doi: 10.1016/j.immuni.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Sugita M, Iwataki Y, Nakano K, Numata O. Unique sequences and predicted functions of myosins in Tetrahymena thermophila. Gene. 2011 doi: 10.1016/j.gene.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Sugita M, Nakano K, Sato M, Toyooka K, Numata O. The roles of actin cytoskeleton and microtubules for membrane recycling of a food vacuole in Tetrahymena thermophila. Cell Motil Cytoskeleton. 2009;66:371–377. doi: 10.1002/cm.20374. [DOI] [PubMed] [Google Scholar]
- Sun TH, Heimark DB, Nguygen T, Nadler JL, Larner J. Both myo-inositol to chiro-inositol epimerase activities and chiro-inositol to myo-inositol ratios are decreased in tissues of GK type 2 diabetic rats compared to Wistar controls. Biochem Biophys Res Commun. 2002;293:1092–1098. doi: 10.1016/S0006-291X(02)00313-3. [DOI] [PubMed] [Google Scholar]
- Tamura K, Ohbayashi N, Maruta Y, Kanno E, Itoh T, Fukuda M. Varp is a novel Rab32/38-binding protein that regulates Tyrp1 trafficking in melanocytes. Mol Biol Cell. 2009;20:2900–2908. doi: 10.1091/mbc.E08-12-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomazic ML, Najle SR, Nusblat AD, Uttaro AD, Nudel CB. A novel sterol desaturase-like protein promoting dealkylation of phytosterols in Tetrahymena thermophila. Eukaryot Cell. 2011;10:423–434. doi: 10.1128/EC.00259-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze SA, Martens GJ, Huttner WB. Secretory granule biogenesis: rafting to the SNARE. Trends Cell Biol. 2001;11:116–122. doi: 10.1016/s0962-8924(00)01907-3. [DOI] [PubMed] [Google Scholar]
- Turkewitz AP. Out with a bang! Tetrahymena as a model system to study secretory granule biogenesis. Traffic. 2004;5:63–68. doi: 10.1046/j.1600-0854.2003.00155.x. [DOI] [PubMed] [Google Scholar]
- Turkewitz AP, Bright LJ. A Rab-based view of membrane traffic in the ciliate Tetrahymena thermophila. Small GTPases. 2:1–5. doi: 10.4161/sgtp.2.4.16706. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkewitz AP, Orias E, Kapler G. Functional genomics: the coming of age for Tetrahymena thermophila. Trends Genet. 2002;18:35–40. doi: 10.1016/s0168-9525(01)02560-4. [DOI] [PubMed] [Google Scholar]
- Valcarce G, Florin-Christensen J, Nudel C. Isolation of a delta7-cholesterol desaturase from Tetrahymena thermophila. Appl Microbiol Biotechnol. 2000;53:591–595. doi: 10.1007/s002530051661. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol. 2010;11:329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD. Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- Verbsky JW, Turkewitz AP. Proteolytic processing and Ca2+-binding activity of dense-core vesicle polypeptides in Tetrahymena. Mol. Biol. Cell. 1998;9:497–511. doi: 10.1091/mbc.9.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Banno Y, Nakashima S, Nozawa Y. Enzymatic characterization of phospholipase D of protozoan Tetrahymena cells. J Eukaryot Microbiol. 2001;48:194–201. doi: 10.1111/j.1550-7408.2001.tb00303.x. [DOI] [PubMed] [Google Scholar]
- Wang S, Banno Y, Nozawa Y. Two forms of membrane-bound sphingosine kinase in Tetrahymena and activity changes during growth and the cell cycle. J Eukaryot Microbiol. 2002;49:305–311. doi: 10.1111/j.1550-7408.2002.tb00374.x. [DOI] [PubMed] [Google Scholar]
- Weisz OA, Rodriguez-Boulan E. Apical trafficking in epithelial cells: signals, clusters and motors. J Cell Sci. 2009;122:4253–4266. doi: 10.1242/jcs.032615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker BD, Nelson DL. Growth support and metabolism of phytosterols in Paramecium tetraurelia. Lipids. 1987;22:386–396. doi: 10.1007/BF02537266. [DOI] [PubMed] [Google Scholar]
- Wilkes DE, Otto JJ. Profilin functions in cytokinesis, nuclear positioning, and stomatogenesis in Tetrahymena thermophila. J Eukaryot Microbiol. 2003;50:252–262. doi: 10.1111/j.1550-7408.2003.tb00130.x. [DOI] [PubMed] [Google Scholar]
- Wilkes DE, Watson HE, Mitchell DR, Asai DJ. Twenty-five dyneins in Tetrahymena: A re-examination of the multidynein hypothesis. Cell Motil Cytoskeleton. 2008;65:342–351. doi: 10.1002/cm.20264. [DOI] [PubMed] [Google Scholar]
- Williams NE, Luft JH. Use of a nitrogen mustard derivative in fixation for electron microscopy and observations of the ultrastructure of Tetrahymena. J Ultrastruct Res. 1968;25:271–292. doi: 10.1016/s0022-5320(68)80074-7. [DOI] [PubMed] [Google Scholar]
- Williams NE, Tsao CC, Bowen J, Hehman GL, Williams RJ, Frankel J. The actin gene ACT1 is required for phagocytosis, motility, and cell separation of Tetrahymena thermophila. Eukaryot Cell. 2006;5:555–567. doi: 10.1128/EC.5.3.555-567.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SA, Gavin RH. Myosin genes in Tetrahymena. Cell Motil Cytoskeleton. 2005;61:237–243. doi: 10.1002/cm.20078. [DOI] [PubMed] [Google Scholar]
- Wloga D, Strzyzewska-Jowko I, Gaertig J, Jerka-Dziadosz M. Septins stabilize mitochondria in Tetrahymena thermophila. Eukaryot Cell. 2008;7:1373–1386. doi: 10.1128/EC.00085-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe J. The conjugation junction of Tetrahymena: its structure and development. J Morphology. 1982;172:159–178. doi: 10.1002/jmor.1051720204. [DOI] [PubMed] [Google Scholar]
- Wolfe J. Cytoskeletal reorganization and plasma membrane fusion in conjugating Tetrahymena. J Cell Sci. 1985;73:69–85. doi: 10.1242/jcs.73.1.69. [DOI] [PubMed] [Google Scholar]
- Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, Ong CN, Codogno P, Shen HM. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 2010;285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R, Clark KM, Gorovsky MA. Endoplasmic reticulum retention signal-dependent glycylation of the Hsp70/Grp170-related Pgp1p in Tetrahymena. Eukaryot Cell. 2007;6:388–397. doi: 10.1128/EC.00366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Yuan D, Fillingham JS, Garg J, Lu X, Chang Y, Liu Y, Fu C, Pearlman RE, Miao W. Gene network landscape of the ciliate Tetrahymena thermophila. PLoS ONE. 2011;6:e20124. doi: 10.1371/journal.pone.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakisich JS, Kapler GM. The effect of phosphoinositide 3-kinase inhibitors on programmed nuclear degradation in Tetrahymena and fate of surviving nuclei. Cell Death Differ. 2004;11:1146–1149. doi: 10.1038/sj.cdd.4401473. [DOI] [PubMed] [Google Scholar]
- Yancey PH. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J Exp Biol. 2005;208:2819–2830. doi: 10.1242/jeb.01730. [DOI] [PubMed] [Google Scholar]
- Zhang J, Schulze KL, Hiesinger PR, Suyama K, Wang S, Fish M, Acar M, Hoskins RA, Bellen HJ, Scott MP. Thirty-one flavors of Drosophila rab proteins. Genetics. 2007;176:1307–1322. doi: 10.1534/genetics.106.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweifel E, Smith J, Romero D, Giddings TH, Jr, Winey M, Honts J, Dahlseid J, Schneider B, Cole ES. Nested genes CDA12 and CDA13 encode proteins associated with membrane trafficking in the ciliate Tetrahymena thermophila. Eukaryot Cell. 2009;8:899–912. doi: 10.1128/EC.00342-08. [DOI] [PMC free article] [PubMed] [Google Scholar]