Summary
A suspected case of sexual transmission from a male survivor of Ebola virus disease (EVD) to his female partner (the patient in this report) occurred in Liberia in March 2015. Ebola virus (EBOV) genomes assembled from blood samples from the patient and a semen sample from the survivor were consistent with direct transmission. The genomes shared three substitutions that were absent from all other Western African EBOV sequences and that were distinct from the last documented transmission chain in Liberia before this case. Combined with epidemiologic data, the genomic analysis provides evidence of sexual transmission of EBOV and evidence of the persistence of infective EBOV in semen for 179 days or more after the onset of EVD. (Funded by the Defense Threat Reduction Agency and others.)
In December 2013, EBOV emerged in Guinea and quickly spread to several neighboring countries, resulting in the largest recorded outbreak of EVD in history.1 On September 3, 2015, Liberia was declared to be free from EVD for the second time, and although new cases were still being reported in Guinea and Sierra Leone as of September 9, 2015, weekly numbers were just a fraction of those reported during the peak of the outbreak.1 As the EVD outbreak in western Africa wanes, the affected countries must transition from controlling an EVD epidemic to addressing the needs of an unprecedented number of survivors of EVD who often have substantial medical sequelae.2
EBOV is detectable in the bloodstream only during acute illness, but the virus may persist for longer periods of time within immune-privileged sites. For instance, among convalescent patients, EBOV RNA has been detected in breast milk up to 15 days after the onset of the disease, in vaginal secretions up to 33 days after onset, in ocular aqueous humor up to 98 days after onset, and in semen up to 101 days after onset.3–5 In addition, EBOV has been cultured from semen samples that were collected 40, 61, and 82 days after disease onset when EBOV was cleared from the blood.3,4,6
This long-term persistence may provide an opportunity for the transmission of EBOV from survivors even after the official end of an outbreak, which is currently defined by the World Health Organization (WHO) as 42 days after the last direct contact with a patient or burial (i.e., two incubation periods after blood samples from the last patient with confirmed disease have tested negative twice for the virus). Because Marburg virus, a distant relative of EBOV, has been sexually transmitted at least once,7 sexual transmission of EBOV is thought to be plausible. Consequently, the WHO and the Centers for Disease Control and Prevention advised survivors of EVD to abstain from sexual intercourse or to use condoms during sexual relations for at least 3 months after the onset of EVD. (Note that the epidemiologic investigation of this case8 has resulted in a change to this recommendation9.) However, evidence of the sexual transmission of EBOV has thus far been limited to scarce and inconclusive data.10
Case Report
History and Epidemiology
On March 20, 2015, a 44-year-old woman from Montserrado County, Liberia, was confirmed to have EVD. Blood samples from the patient were confirmed to be positive for EBOV RNA by quantitative reverse-transcriptase–polymerasechain-reaction (RT-PCR) assay at the Eternal Love Winning Africa (ELWA) Laboratory in Paynesville, Liberia. The patient died on March 27, 2015.
The case investigation did not reveal an immediate source of infection, such as contact with patients with acute EVD. However, the patient reported that on March 7, 2015, she had had unprotected vaginal intercourse with a male Liberian survivor of EVD.8 Subsequent to the patient’s EVD diagnosis, 192 contacts were identified,11 all of whom were free from clinical signs.
The survivor also lived in Montserrado County. Several members of his family had had EVD, beginning in late August 2014. The survivor’s older brother, who presented with clinical signs of EVD on August 22, died during the night on September 5–6, 2014, and was confirmed to be positive for EBOV RNA by means of a postmortem quantitative RT-PCR assay. The survivor is thought to have had symptoms of EVD beginning on September 9, 2014, which is the estimated triage date,8 and he was admitted to the nearby Island Clinic Ebola Virus Disease Treatment Unit on September 23.
Quantitative RT-PCR testing for EBOV RNA in the survivor’s first blood sample on September 28 yielded ambiguous results. Repeated testing of this sample on September 29 yielded a negative result for EBOV. A subsequent test performed on October 3 (presumably from a second blood sample, although this information could not be confirmed owing to the absence of a sample record) was also negative. The survivor was discharged from the Ebola treatment unit on October 7 and reported no subsequent illness.
On September 20, clinical signs of EVD developed in the survivor’s former wife, who was estranged from the survivor. She was admitted to an ELWA Ebola treatment unit on September 24 and died the following day.
As a result of the case investigation into the patient’s illness, the survivor voluntarily provided a blood sample on March 23, 2015, and a semen sample on March 27, 2015 (199 days after the estimated onset of EVD and 175 days after the survivor’s blood tested negative for EBOV). The blood sample tested negative for EBOV RNA on quantitative RT-PCR assay, but the sample tested positive for EBOV glycoprotein-specific and nucleoprotein-specific IgG antibodies.8 The semen sample tested positive for EBOV RNA on quantitative RT-PCR assay, but attempts to culture virus were unsuccessful.
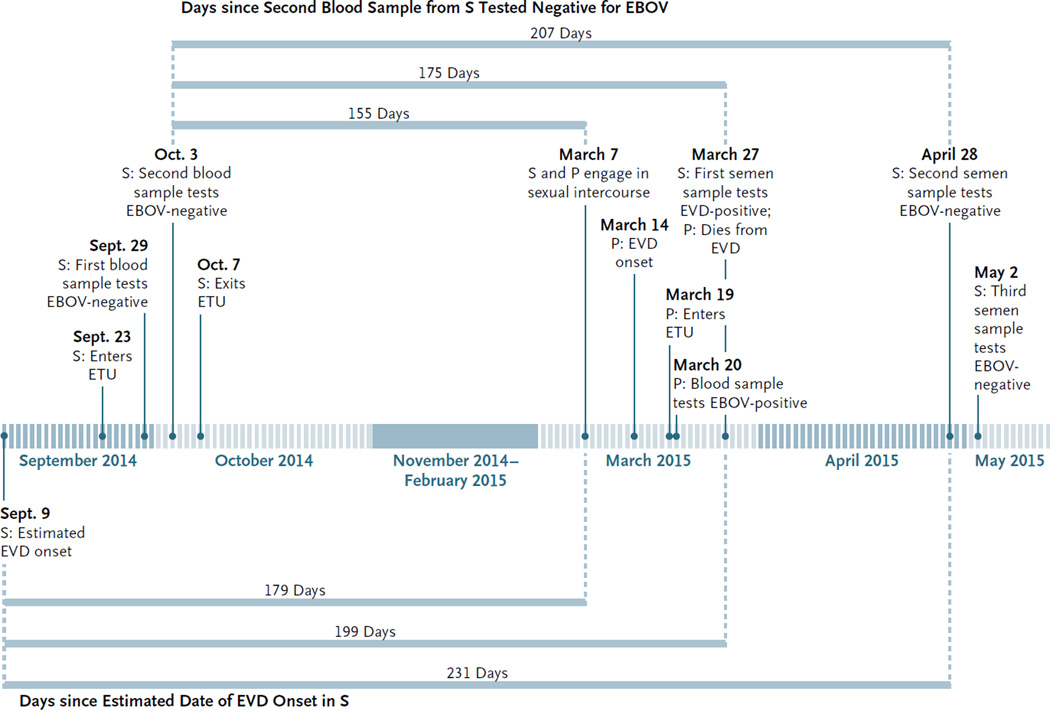
On April 28 (32 days later), the survivor provided a second semen sample for diagnostic testing at the Liberian National Public Health Reference Laboratory in Margibi County. No EBOV RNA was detected by the quantitative RT-PCR assay. A third semen sample, collected 3 days later, on May 1, also tested negative for EBOV on quantitative RT-PCR assay, which suggests that there was possible EBOV clearance from semen 231 days after the estimated onset of EVD and 207 days after the survivor’s blood tested negative. A timeline of the events is shown in Fig. 1.
Figure 1. Clinical Timelines for the Patient and the Survivor, from September 2014 through May 2015.
Shown are key dates regarding the Ebola virus disease (EVD) presentation, diagnostic tests, and outcomes for the survivor (S) and the patient (P). Horizontal bars estimate the number of days of persistence of the Ebola virus (EBOV) since the date of disease onset and since the date of clearance from blood. ETU denotes Ebola treatment unit.
Specific consent was obtained from the survivor. For all other samples collected for testing during the EVD outbreak, informed consent was not obtained because this work was conducted at the Liberian Institute for Biomedical Research (LIBR) as part of the EVD response and EBOV surveillance. With the consent of the National Incident Management System of the Ebola Virus Disease Outbreak and the Liberian Ministry of Health and Social Welfare, the work was supervised by the LIBR institutional review board. All the information obtained from the participants was anonymized for this report. The WHO Liberia Country Office team coordinated field epidemiologic investigations and support to the survivor and the patient.
Molecular Investigation
As part of the investigation into the source of the patient’s EBOV infection, the following samples were examined: whole blood from the patient was tested on March 20 and 21, 2015 (two samples); whole blood from the survivor’s older brother was tested on September 9, 2014; whole blood from the survivor’s former wife was tested on September 24, 2014; and semen from the survivor was tested on March 27, 2015. Viral RNA that was potentially present in all five samples was initially sequenced on an Illumina MiSeq at the LIBR with the use of methods that have been described previously.12
Nearly complete EBOV genome sequences (97.4 to 99.7% coverage) were assembled from the samples obtained from the patient, the survivor’s older brother, and the survivor’s former wife. No EBOV sequences were obtained from the survivor’s semen sample with the use of this sequencing method. Therefore, we enriched the semen sample for EBOV genomic RNA using the TruSeq RNA Access kit (Illumina) with custom capture probes designed against EBOV, along with other modifications (see the Supplementary Appendix, available with the full text of this article at NEJM.org). The semen sample was processed and sequenced separately to avoid contamination. With the combined data from four independent enrichment libraries, 85.1% genome coverage was achieved. A minimum of 3× sequencing depth was required to determine a genome position. However, the enrichment process resulted in a large number of duplicate reads. Therefore, the duplicate-adjusted sequencing depth for the semen sample was less than 3× in some positions (Table 1). The assembled genomes are available at GenBank under accession numbers KT587343, KT587344, KT587346, and KT587345.
Table 1.
Distinct Ebola Virus Genome Substitutions in the Patient, the Survivor, and the Survivor’s Older Brother.*
| Position† | Reference | Alternative | Samples with Alternative |
Survivor- Corrected Depth‡ |
Nature of Substitution§ |
|---|---|---|---|---|---|
| 4,107 | G | A | P, S | 1 | VP35, V327I |
| 8,592 | A | T | P, S | 1 | VP30, synonymous |
| 16,636 | G | A | P, S | 5 | L, G1686S |
| 4,384 | A | C | P, S, SB | 3 | Noncoding |
| 12,996 | C | A | P, S, SB | 1 | L, synonymous |
| 18,399 | AAAAAA | AAAAAAA | P, S, SB | 2 | Noncoding |
| 11,263 | C | T | S | 1 | Noncoding |
The GenBank accession numbers for the tested genomes are as follows: for the patient (P), the number is KT587343, for the survivor (S), the number is KT587344, and for the survivor’s older brother (SB), the number is KT587346. L denotes RNA-dependent RNA polymerase, and VP viral protein.
Positions were relative to the reference genome Ebola virus/H.sapiens-wt/GIN/2014/Makona-C15 (GenBank accession number, KJ660346.2).
The number indicates the depth at each position from the survivor after correction for duplicates resulting from polymerase- chain-reaction amplification.
The gene abbreviation is provided for substitutions within coding regions, followed by a description of the amino acid change for substitutions that are nonsynonymous.
The four assembled EBOV genomes were compared with all publicly available sequences (796 genomes, including 56 from cases in Liberia12–19) from the outbreak in Western Africa (Makona variant).20 The results were consistent with sexual transmission of EBOV from the survivor to the patient. First, the EBOV genome from the patient grouped phylogenetically with other genomes obtained from Liberian patients and was distinct from sequences from patients in Guinea, Sierra Leone, and Mali (Fig. 2). Thus, it is unlikely that the patient was infected owing to an undocumented reintroduction of EBOV to Liberia from a neighboring country with ongoing transmission.
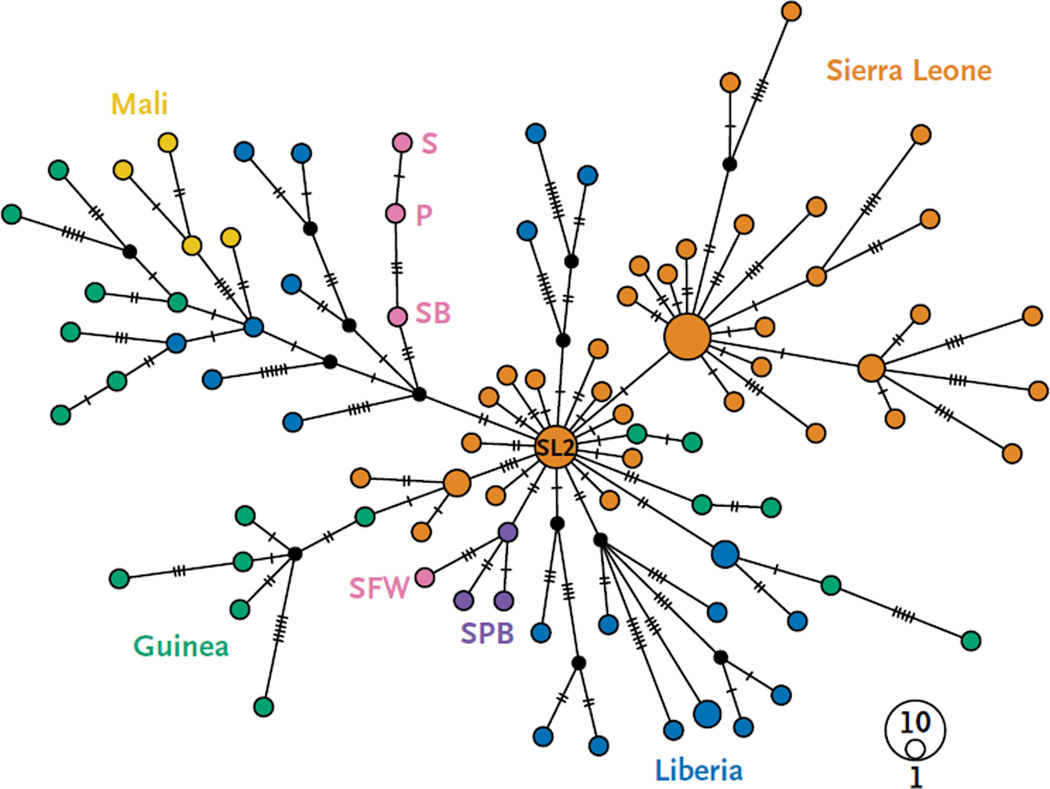
Figure 2. Median-Joining Haplotype Network.
This network was constructed from a full genome alignment of 100 clinical sequences of the Ebola virus Makona variant, including those assembled from blood samples obtained from the patient (P), the survivor’s older brother (SB), and the survivor’s former wife (SFW), from a semen sample from the survivor (S), and from 96 additional genomes chosen from the 796 genomes that were analyzed to be representative of samples collected in Guinea, Liberia, Mali, and Sierra Leone. For visual clarity, the network was limited to 100 genomes. The GenBank accession numbers for the tested genomes are as follows: for P, the number is KT587343, for S, the number is KT587344, for SB, the number is KT587346, and for SFW, the number is KT587345. Each colored vertex represents a sampled viral haplotype. The vertex size is proportional to the number of sampled sequences. Genomes sequenced in this study are shown in pink. Purple vertexes (SPB) indicate samples from the last known cluster of EVD cases in Liberia before the infection of the patient discussed in this report. Other colors indicate the respective countries of origin. Edges are not drawn to scale; hatch marks indicate the number of substitutions along each edge. The vertex SL2 represents the ancestral haplotype that is thought to have been introduced into Liberia in the spring of 2014.12,15
Second, before the confirmation of EVD in the patient, the last known cluster of EVD cases in Liberia (December 29, 2014, to February 19, 2015) was linked to a single index case from a village near Saint Paul River Bridge, and the three sequenced EBOV genomes from this cluster (LIBR0993, LIBR1195, and LIBR1413) grouped together in an evolutionary lineage (SPB in Fig. 2) that was unrelated to the EBOV genome from the patient.21 Therefore, the infection in this patient is unlikely to have originated from this cluster of EVD cases.
Finally, the EBOV genomes from the patient and the survivor differed in only one position (11,263) across 15,808 nucleotides, a finding that is consistent with direct EBOV transmission (Fig. 2 and Table 1). Notably, EBOV genomes from the patient and the survivor shared eight substitutions relative to the ancestral haplotype (SL2)15 that is thought to have been originally introduced into Liberia,12 and three of these substitutions have thus far been seen only in the viruses that infected the patient and the survivor (Fig. 2 and Table 1). Although only 1× sequencing depth was obtained for several of these positions (after correction for duplicate reads), the detection in the survivor’s semen sample of every substitution that distinguished the patient’s EBOV sequence from the ancestral SL2 haplotype is indicative of a close epidemiologic link. The EBOV genome obtained from the survivor’s older brother shared five of eight substitutions with the EBOV genomes of the patient and the survivor, which suggests involvement of the survivor’s older brother in the same transmission chain (Fig. 2 and Table 1). The EBOV genome from the survivor’s former wife, however, was distinct (Fig. 2).
The EBOV genomes from the patient and the survivor differed at a single position (11,263), thus representing an additional substitution in the survivor’s EBOV genome relative to all other genomes assembled. After duplicates from PCR amplification were controlled for, this position had only 1× coverage depth. Given the low level of sequencing depth, this apparent substitution may simply represent a low-frequency allele in the survivor’s EBOV population or even a sequencing artifact. Alternatively, it could represent a shift in allele frequencies of EBOV subpopulations within the survivor during the 20 days that passed between the date of sexual intercourse and potential transmission (March 7, 2015) and the date of semen collection (March 27, 2015). Nevertheless, the nearly identical EBOV genomes place the survivor and the patient in the same transmission chain, and case tracing confirms contact by vaginal intercourse.
Discussion
Studies involving survivors from previous EVD outbreaks have indicated the possibility of sexual transmission owing to the presence of EBOV RNA in semen and vaginal secretions. However, our understanding of EBOV persistence in these bodily fluids is restricted to the examination of 13 samples (12 semen samples and 1 vaginal-secretion sample), each with a limited temporal span.3,4,22 Using a combination of genomic and epidemiologic data, we found that at least one case of EVD in the ongoing Liberian outbreak probably resulted from sexual transmission through unprotected vaginal intercourse. Although we cannot exclude the possibility of EBOV transmission from sources that were not sampled, contact tracing failed to uncover any other connections of the patient to possible or confirmed EVD cases. Furthermore, the EBOV genomes assembled from the survivor and the patient shared three substitutions that were not present in 796 EBOV genomes from western Africa. Together, these data provide evidence of human-to-human EBOV transmission through sexual contact.
The analysis of the semen sample obtained from the survivor presented additional challenges beyond those encountered with whole-blood and oral-swab samples that were obtained from other patients with EVD. The high cycle-threshold values on the quantitative RT-PCR assay that were observed in the semen sample suggest a low viral load,8 which makes it difficult to obtain enough sequencing coverage with unbiased amplification of RNA. Therefore, we implemented a new target-enrichment strategy to obtain sufficient coverage. This approach resulted in nearly complete coverage of the EBOV genome, which was necessary given the low number of substitutions that discriminate distinct transmission chains within the current (2013–2015) EVD outbreak.15,19
The frequency of EBOV persistence among survivors is unknown, and available information suggests that sexual transmission is a relatively rare event. Nonetheless, persistent infections, in combination with unprotected sexual intercourse, could lead to flare-ups of EVD at close-to-random locations. We found that viral nucleic acids in the semen from a survivor of EVD persisted for at least 199 days after the estimated onset of EVD (175 days after the clearance from blood), which is more than four times as long as the WHO-defined waiting period for declaring a country to be free from EVD. Although the semen sample contained no detectable infectious EBOV, the assembly of a nearly complete genome suggested the possible presence of infectious particles. In addition, from the evidence of the sexual transmission between the patient and the survivor, we can infer that infectious EBOV was present in the survivor at least 179 days after the onset of disease (155 days after the clearance from blood). Larger and more systematic surveys of survivors are needed in order to determine the prevalence and risk of EBOV persistence in semen and other immunologically privileged sites.
Supplementary Material
Acknowledgments
Supported by the Defense Threat Reduction Agency, the Global Biosurveillance Technology Initiative, Global Emerging Infections Surveillance, and Illumina, by a contract (HHSN272200700016I) between Battelle Memorial Institute and the U.S. National Institute of Allergy and Infectious Diseases (NIAID), and by the Intramural Research Program, NIAID, National Institutes of Health.
We thank the survivor and his partner for willingly providing samples and information for the investigation; the personnel that provided infrastructure and personnel for general Ebola virus disease control in Liberia: Mark M. Bailey, Alissa L. Byrne, Gary W. Carter, Melissa A. Dugan, Joanna Fishback, Alex Hail, and Michelle A. Jefferson; Anthony R. Jones, Brian J. Kearney, Dave A. Norwood, Susana Padilla, Matthew A. Voorhees, and Neal Woollen (U.S. Army Medical Research Institute of Infectious Diseases [USAMRIID]); Trenton Bushmaker, Darryl Falzarano, Heinz Feldmann, Friederike Feldmann, Robert Fischer, Allison Groseth, Thomas Hoenen, Seth Judson, Andrea Marzi, Kristin McNally, Joseph Prescott, Kyle Rosenke, and Brandi Williamson (Rocky Mountain Laboratories, NIAID, NIH); Richard Bennett, Laura Bollinger, Michael R. Holbrook, Peter Jahrling, Krisztina Janosko, Joshua Johnson, Jason Kindrachuk, and Jonathan Marchand (Integrated Research Facility [IRF], NIAID, NIH); Augustine Fallah, Josiah S. George, Charlesetta Kanneh, Aaron T. Momolu, and Vera Yatta Walker (Liberian National Reference Laboratory); Eric Patten (Liberian Ministry of Health); U.S. Ambassador Deborah R. Malac for support of the USAMRIID, the Centers for Disease Control and Prevention, and the NIH–NIAID–IRF–Frederick Ebola Response Team; Tabitha Austill for arranging the shipment of sequencing and other equipment and supplies to the Liberian Institute for Biomedical Research; and Sean Lovett and Brad Pfeffer for the deposition of sequence data in public repositories.
Appendix
The authors’ full names and academic degrees are as follows: Suzanne E. Mate, Ph.D., Jeffrey R. Kugelman, Ph.D., Tolbert G. Nyenswah, L.L.B., M.P.H., Jason T. Ladner, Ph.D., Michael R. Wiley, Ph.D., Thierry Cordier-Lassalle, M.B.A., D.E.S.S., Athalia Christie, M.I.A., Gary P. Schroth, Ph.D., Stephen M. Gross, Ph.D., Gloria J. Davies-Wayne, R.N., M.P.H., Shivam A. Shinde, M.B., B.S., Ratnesh Murugan, M.B., B.S., Sonpon B. Sieh, B.A., Moses Badio, M.Sc., Lawrence Fakoli, M.S., Fahn Taweh, B.S., Emmie de Wit, Ph.D., Neeltje van Doremalen, Ph.D., Vincent J. Munster, Ph.D., James Pettitt, M.S., Karla Prieto, M.S., Ben W. Humrighouse, M.S., M.P.H., Ute Ströher, Ph.D., Joseph W. DiClaro, Ph.D., Lisa E. Hensley, Ph.D., Randal J. Schoepp, Ph.D., David Safronetz, Ph.D., Joseph Fair, Ph.D., Jens H. Kuhn, M.D., Ph.D., David J. Blackley, Dr.P.H., A. Scott Laney, Ph.D., Desmond E. Williams, M.D., Ph.D., Terrence Lo, Dr.P.H., Alex Gasasira, M.B., Ch.B., M.P.H., Stuart T. Nichol, Ph.D., Pierre Formenty, D.V.M., M.P.H., Francis N. Kateh, M.D., Kevin M. De Cock, M.D., Fatorma Bolay, Ph.D., Mariano Sanchez-Lockhart, Ph.D., and Gustavo Palacios, Ph.D.
The authors’ affiliations are as follows: the Center for Genome Sciences (S.E.M., J.R.K., J.T.L., M.R.W., K.P., M.S.-L., G.P.) and Diagnostic Systems Division (R.J.S.), U.S. Army Medical Research Institute of Infectious Diseases, and the Division of Clinical Research, Integrated Research Facility at Fort Detrick, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH) (J.P., L.E.H., J.H.K.) — all in Frederick, MD; the Ministry of Health and Social Welfare (T.G.N., S.B.S., M.B., F.N.K.) and the World Health Organization (WHO) (G.J.D.-W., R.M.), Monrovia, and the Liberian Institute for Biomedical Research, Charlesville (L.F., F.T., F.B.) — all in Liberia; WHO, Geneva (T.C.-L., A.G., P.F.); the Centers for Disease Control and Prevention, Atlanta (A.C., B.W.H., U.S., D.J.B., A.S.L., D.E.W., T.L., S.T.N., K.M.D.C.); Illumina, San Diego, CA (G.P.S., S.M.G.); WHO, New Delhi, India (S.A.S); Rocky Mountain Laboratories, Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH, Hamilton, MT (E.W., N.D., V.J.M., D.S.); Naval Medical Research Unit 3, Cairo (J.W.D.); and the Foundation Mérieux, Washington, DC (J.F.).
Footnotes
The views expressed in this article are those of the authors and do not necessarily reflect the views or policies of the U.S. Department of Defense, the U.S. Department of Health and Human Services, the U.S. Department of the Army, or the institutions and companies affiliated with the authors.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Geneva: World Health Organization; 2015. Ebola situation reports. http://apps.who.int/ebola/ebola-situation-reports. [Google Scholar]
- 2.Clark DV, Kibuuka H, Millard M, et al. Long-term sequelae after Ebola virus disease in Bundibugyo, Uganda: a retrospective cohort study. Lancet Infect Dis. 2015;15:905–912. doi: 10.1016/S1473-3099(15)70152-0. [DOI] [PubMed] [Google Scholar]
- 3.Bausch DG, Towner JS, Dowell SF, et al. Assessment of the risk of Ebola virus transmission from bodily fluids and fomites. J Infect Dis. 2007;196(Suppl 2):S142–S147. doi: 10.1086/520545. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez LL, De Roo A, Guimard Y, et al. Persistence and genetic stability of Ebola virus during the outbreak in Kikwit, Democratic Republic of the Congo, 1995. J Infect Dis. 1999;179(Suppl 1):S170–S176. doi: 10.1086/514291. [DOI] [PubMed] [Google Scholar]
- 5.Varkey JB, Shantha JG, Crozier I, et al. Persistence of Ebola virus in ocular fluid during convalescence. N Engl J Med. 2015;372:2423–2427. doi: 10.1056/NEJMoa1500306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emond RT, Evans B, Bowen ET, Lloyd G. A case of Ebola virus infection. Br Med J. 1977;2:541–544. doi: 10.1136/bmj.2.6086.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martini GA, Schmidt HA. Spermatogenic transmission of the “Marburg virus”: causes of “Marburg simian disease”. Klin Wochenschr. 1968;46:398–400. doi: 10.1007/BF01734141. (In German.) [DOI] [PubMed] [Google Scholar]
- 8.Possible sexual transmission of Ebola virus — Liberia, 2015. MMWR Morb Mortal Wkly Rep. 2015;64:479–481. [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization. Ebola virus disease: factsheet N 103. 2015 Aug; http://www.who.int/mediacentre/factsheets/fs103/en/
- 10.Rowe AK, Bertolli J, Khan AS, et al. Clinical, virologic, and immunologic follow- up of convalescent Ebola hemorrhagic fever patients and their household contacts, Kikwit, Democratic Republic of the Congo. J Infect Dis. 1999;179(Suppl 1):S28–S35. doi: 10.1086/514318. [DOI] [PubMed] [Google Scholar]
- 11.Cordier-Lassalle T. Geneva: World Health Organization; 2015. Summary of sector response model: a quality management system that led to Liberia’s success in fighting Ebola. http://www.who.int/csr/disease/ebola/sector-approach-liberia.pdf. [Google Scholar]
- 12.Kugelman JR, Wiley MR, Mate S, et al. Monitoring of Ebola virus Makona evolution through establishment of advanced genomic capability in Liberia. Emerg Infect Dis. 2015;21:1135–1143. doi: 10.3201/eid2107.150522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baize S, Pannetier D, Oestereich L, et al. Emergence of Zaire Ebola virus disease in Guinea. N Engl J Med. 2014;371:1418–1425. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- 14.Carroll MW, Matthews DA, Hiscox JA, et al. Temporal and spatial analysis of the 2014–2015 Ebola virus outbreak in West Africa. Nature. 2015;524:97–101. doi: 10.1038/nature14594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gire SK, Goba A, Andersen KG, et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345:1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoenen TS, Safronetz D, Groseth A, et al. Virology: mutation rate and genotype variation of Ebola virus from Mali case sequences. Science. 2015;348:117–119. doi: 10.1126/science.aaa5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park DJ, Dudas G, Wohl S, et al. Ebola virus epidemiology, transmission, and evolution during seven months in Sierra Leone. Cell. 2015;161:1516–1526. doi: 10.1016/j.cell.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simon-Loriere E, Faye O, Faye O, et al. Distinct lineages of Ebola virus in Guinea during the 2014 West African epidemic. Nature. 2015;524:102–204. doi: 10.1038/nature14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong YG, Shi WF, Liu D, et al. Genetic diversity and evolutionary dynamics of Ebola virus in Sierra Leone. Nature. 2015;524:93–96. doi: 10.1038/nature14490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhn JH, Andersen KG, Baize S, et al. Nomenclature- and database-compatible names for the two Ebola virus variants that emerged in Guinea and the Democratic Republic of the Congo in 2014. Viruses. 2014;6:4760–4799. doi: 10.3390/v6114760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nyenswah T, Fallah M, Sieh S, et al. Controlling the last known cluster of Ebola virus disease — Liberia, January–February 2015. MMWR Morb Mortal Wkly Rep. 2015;64:500–504. [PMC free article] [PubMed] [Google Scholar]
- 22.Richards GA, Murphy S, Jobson R, et al. Unexpected Ebola virus in a tertiary setting: clinical and epidemiologic aspects. Crit Care Med. 2000;28:240–244. doi: 10.1097/00003246-200001000-00041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.