Abstract
Nanopore technology employs a nanoscale hole in an insulating membrane to stochastically sense with high throughput individual biomolecules in solution. The generality of the nanopore detection principle and the ease of single-molecule detection suggest many potential applications of nanopores in biotechnology. Recent progress has been made with nanopore fabrication and sophistication, as well as with applications in DNA/protein mapping, biomolecular structure analysis, protein detection, and DNA sequencing. In addition, concepts for DNA sequencing devices have been suggested, and computational efforts have been made. The state of the nanopore field is maturing and given the right type of nanopore and operating conditions, nearly every application could revolutionize medicine in terms of speed, cost, and quality. In this review, we summarize progress in nanopores for biotechnological applications over the past 2–3 years.
Introduction
Nanopores operate on a basic principle: a nanoscale hole is made between two electrolytic fluid chambers with an impermeable membrane between them. When voltage is applied to the chambers a steady-state ion current across the pore develops. Transient changes in the ion flux across the pore can result from occupation of a macromolecule in the pore, and therefore, monitoring the current across the pore enables molecular sensing. Addition of a sample of charged biomolecules (e.g., DNA and proteins) to one of the fluid chambers results in stochastic entry and exit of biomolecules from the pore, which produces a series of discrete fluctuations in the ion current signal. These current fluctuations communicate many properties of the sample, including the biomolecular size, concentration, and structure. By manipulating the dimensions of the pore, its surface characteristics, the applied voltage, and the solution conditions, one can tailor a nanopore for sensing different types of biomolecules. Since nanopore sensing does not require biomolecular modification, labeling, or surface immobilization, the technique is useful for detecting a broad range of molecules and complexes. However, nanopores of various dimensions in different materials are needed, for these purposes, and this review highlights, a combination of molecular biology techniques and advances in nanotechnology which have spawned a broad range of nanopore types for these biotechnological applications.
Various technical improvements have been made that allow for improved nanopore resolution and stability, and novel techniques and new forms of nanopores have been introduced to expand the utility of the technology. Furthermore, great strides have been made in the technology in the realistic hope that the nanopore will be able to work as a low cost, high efficiency DNA sequencing device.
Improvements to nanopore technology
Originating from experiments on lipid embedded α-hemolysin protein channels, over the past decade a wide range of nanopore types have been developed and explored. Most recently, new forms of biomimetic nanopores have been developed, including protein pores using brush-like filaments [1] and biomimetic nuclear pore complexes [2,3•]. Lateral electrodes formed across apertures have enabled electronic detection of biomolecules during their transport through the nanopore [4–6]. Ultrathin nanopores fabricated using plasma thinning [7] and ion-beam sculpting [8•,9] have also been developed. Alternative detection using electrostatic and field-effect phenomena has been demonstrated by coupling a scanning probe microscope [10] and Si-nanowire transistors [11•] to nanopores.
A promising new development for making various nanopore shapes involves using DNA origami structures as a scaffolding structure. Nanoscale DNA structures are obtained by combining a long single-stranded DNA with synthetic DNA staples, which mold the strand into a predetermined shape. Positioning onto a larger solid-state pore is then performed using a similar docking approach previously used for α-hemolysin docking [8•]. Biomolecule translocations have been measured through a conical origami-pore [12] and a nano-plate with a pore designed through it [13••]. This origami approach to nanopores will surely yield more chemically and biologically specific nanopore structures with near atomic precision.
Graphene, a thin and transparent lattice of sp2 carbon atoms, has also become a prime target as a membrane material for ultrathin nanopores [14–16]. The electronic properties of graphene, its robustness, atomic thickness, and ion impermeability, make it an intriguing material for nanopore-based electronic sequencing of DNA molecules. New improvements in the formation of suspended graphene flake membranes [17] and nanopore fabrication in self-aligned carbon electrodes [18] have recently been reported, which facilitate the integration of carbon nanostructure into nanopore technology.
Automated capturing of molecules into pores allows for interrogating a molecule’s structure and dynamics. Studies have used this technology to investigate and model the local diffusive effects of beads near a pore as groundwork for future biological studies [19]. Studies of ion transport through metallic pores, such as pores in gold surfaces, can be a way of creating species-selective pore systems that are of interest for detection of biomolecular systems [20] (Figure 1).
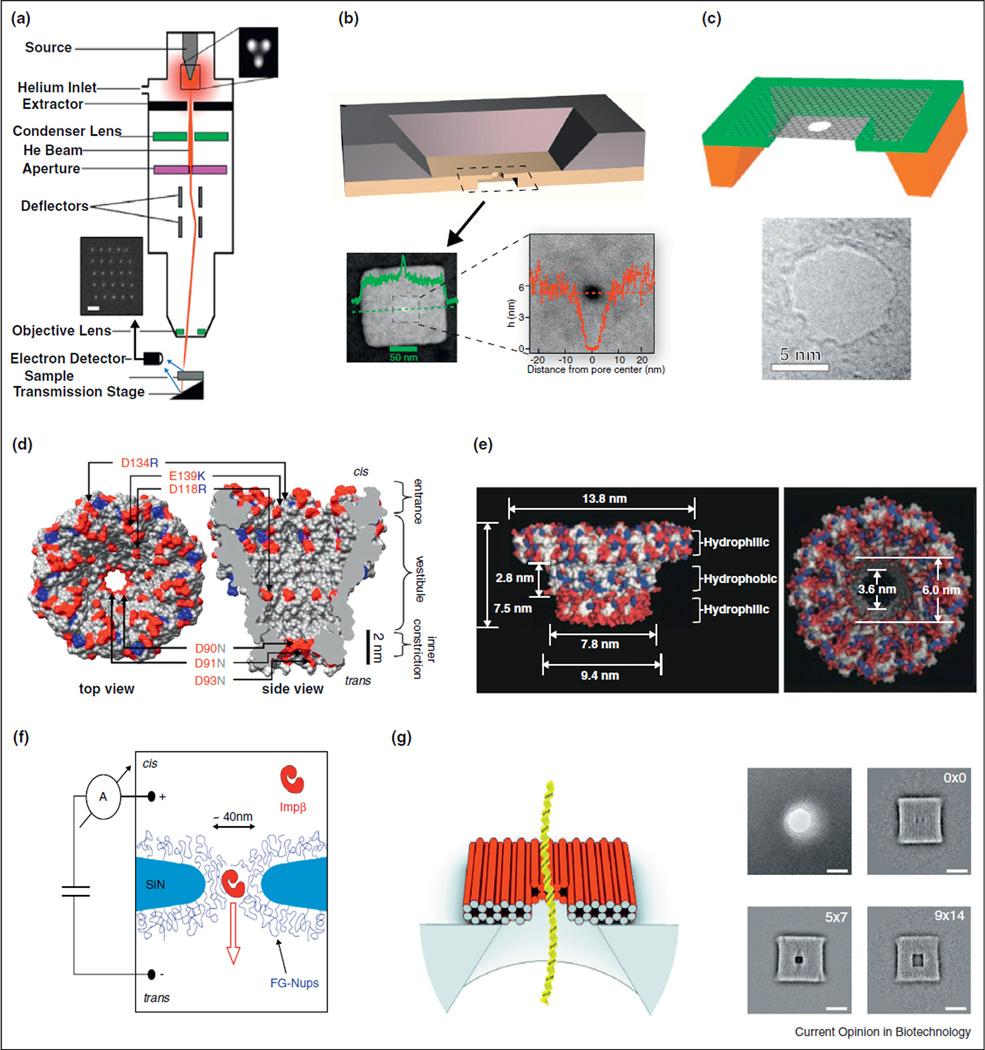
Figure 1.
(a) Nanopore manufacture using an ion beam [21••]. (b) Solid-State nanopore in an ultrathin silicon nitride membrane [7]. (c) Nanopore in a graphene layer [14]. (d) MspA biological pore from Mycobacterium smegmatis [22]. (e) Biological pore engineered from phi29 bacteriophage [23]. (f) Biomimetic nuclear pore complex [3•]. (g) DNA origami schematic and TEM images of DNA origami plates with different pore sizes [12].
Applications in biotechnology
DNA is by far the most common polymer studied using nanopores, beginning with the pioneering demonstration of DNA detection using lipid embedded α-hemolysin channels. Recently, solid-state nanopores have been used to detect differences in nucleosomal substructures [24] as well as key parts of RNA polymerase DNA transcription [25], opening up new opportunities in understanding chromatin structure and transcription research, respectively. Biological nanopores have been shown to detect the guanine rich G-quadruplex, which plays an important role in genomics and epigenetics [26]. Abasic sites can also be dynamically detected using nanopores by spiking the electrolyte solution with ionophores [27]. Protein translocation through pores assisted by high voltage, and discrimination among proteins using aptamer-modified pores, were demonstrated [28,29]. When made to translocate certain proteins will unzip, allowing for the translocation to become a measure of the unzipping kinetics. A variety of these proteins have been well studied in their unzipping behavior, allowing for the dynamic use of nanopores as a label free efficient force spectroscopy instrument [30,31••,32,33]. Important neurological transmitters have also been dynamically differentiated in real time with the hope that studies can be done on the brain’s chemical response to drugs in real time [34]. These capabilities are promising as the nanopore is label free, cheap and considerably quicker than most other techniques to investigate the same properties, but without a marked decrease in accuracy or ability (Figure 2).
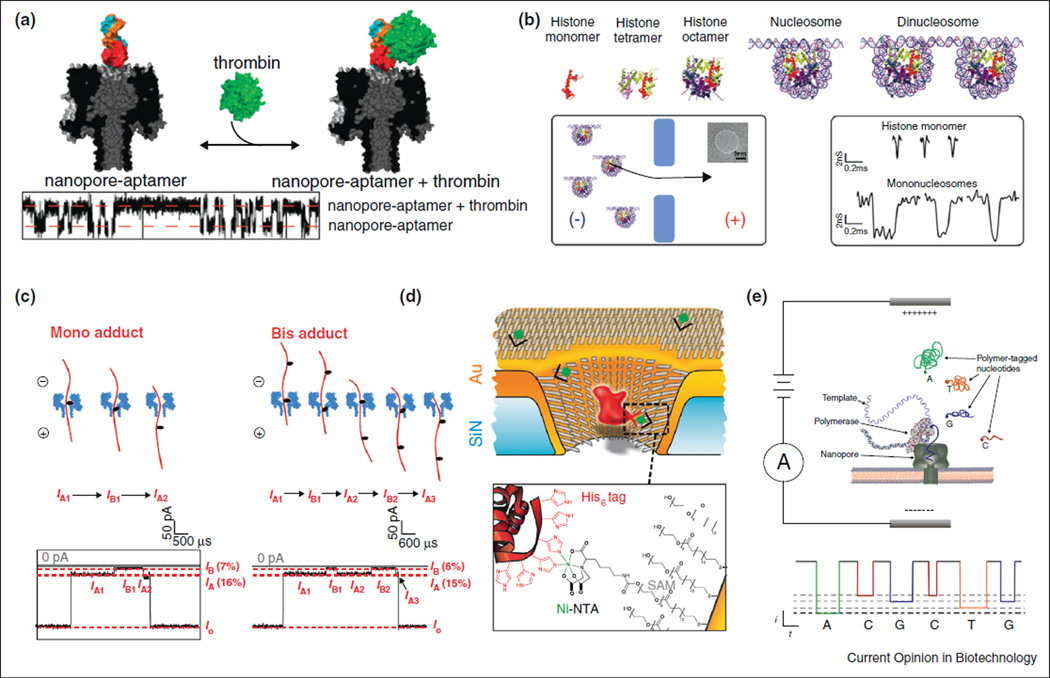
Figure 2.
(a) Thrombin detection using a biological nanopore modified with an apatmer [28]. (b) Detection of different nucleosomal structures [24]. (c) Detection of AP-18c6 DNA adduct sites [27]. (d) Pore coated with receptor sites used to measure ligand binding [35•]. (e) Current-based DNA sequencing using polymer-tagged nucleotides [36].
Other nanopore structures offer a variety of new studies and technologies in the biological realm. Large solid-state nanopores can be used to dynamically trap and release bacteria, presenting a faster and cheaper method of dynamic single-cell capture [37]. Using a lipopeptidecoated solid-state nanopore, the interactions of DNA with the membrane near a pore have been probed [38]. The development of ‘smart’ pores has advanced with proposed thermo-responsive polymers [39] as a means for creating devices that dynamically respond to temperature, and biological pores made of electrolyte brushes may be able to control salt conductance near nanopore openings [40].
Advances in nanopores for DNA sequencing have also been made recently. By controlling single-stranded DNA transport through a graphene pore using a DNA polymerase as a ratchet, high precision readout of a nucleotide sequence can be obtained, as indicated by a recent molecular dynamics study [41]. The ability to selectively immobilize a DNA strand using streptavidin in an α-hemolysin pore allows for very high resolution nucleotide differentiation at different geometric locations inside the pore [42]. Slowing down translocation has been suggested using a p–n semiconductor junction, capable of dynamic voltage control during the translocation process [43]. Using sophisticated electronics engineering, a new CMOS-based amplifier has been realized that allows submicrosecond temporal current detection [44•]. High temporal resolution is desirable for monitoring structural information during the translocation process: since average transport speeds are below 1 µs per base in many nanopore experiments, submicrosecond-scale resolution can in principle probe base information during voltage-driven transport.
Theoretical works related to DNA sequencing have suggested several potential methods of overcoming the above mentioned speed limitations of ion current measurements. Simulations have shown that a pore in a graphene nanoribbon can exploit local current density on the edges of the pore, thereby producing a superior resolution to tunneling methods [45]. Conductance changes across a graphene nanoribbon placed perpendicular to a nanochannel has also been proposed as a translocation-based DNA sequencing device [46]. Another proposed approach involves embedding gold nanoparticles within the nanopore membrane [47].
Finally, various simulations have been performed to investigate the principle of electrical discrimination among different bases in a DNA strand. Brownian dynamics studies of the base-sequence dependence of the ion current have been performed [48], and spring-like modeling of the pulling force profile for different bases has been explored [41], although the latter approach fails to distinguish between cytosines and thymines. Simulations have also shown the ability to differentiate among base pairs in graphene pores, but potential complications include control over translocation speeds and base repeat errors [49••]. Finally, a hidden Markov model of translocation data has been suggested for decoding triplet bases in a DNA strand, which relieves the need for single-base resolution and allows contextual information to improve the error in nucleotide reading [50].
Conclusions
The nanopore field continues to deliver interesting new pore types and biotechnological ideas. Smart pore materials based on origami and other programmable materials can greatly aid in the development of reproducible pore structures, and in combination with new technology for synthetic pore supports, these hybrid devices should be able to perform extremely well in the detection of nearly any biomolecular species. Finally, nanopore-based DNA sequencing can revolutionize healthcare by allowing genomes to be sequenced at permissive costs and speeds. Advancement of these technologies to fruition in the years to come will undoubtedly influence personalized medicine and early screening/treatment of genetic diseases.
During the publication of our manuscript, a new paper by Langecker et al. has shown that DNA origami can be designed to function as an ion channel [51]. This paper clearly is a novel and exciting breakthrough in the nanopore field and is well-worth attention.
Acknowledgments
This work was supported partly by National Institutes of Health grant 1R01HG006321-01 (NHGRI).
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Pongprayoon P, Beckstein O, Sansom MSP. Biomimetic design of a brush-like nanopore: simulation studies. J Phys Chem B. 2012;116:462–468. doi: 10.1021/jp206754w. [DOI] [PubMed] [Google Scholar]
- 2.Jovanovic-Talisman T, Tetenbaum-Novatt J, McKenney AS, Zilman A, Peters R, Rout MP, Chait BT. Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature. 2009;457:1023–1027. doi: 10.1038/nature07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kowalczyk SW, Kapinos L, Blosser TR, Magalhaes T, van Nies P, Lim RYH, Dekker C. Single-molecule transport across an individual biomimetic nuclear pore complex. Nat Nanotechnol. 2011;6:433–438. doi: 10.1038/nnano.2011.88. Modification of single solid-state pores with proteins, which demonstrate selective transport of one protein over others
- 4.Tsutsui M, Taniguchi M, Yokota K, Kawai T. Identifying single nucleotides by tunnelling current. Nat Nanotechnol. 2010;5:286–290. doi: 10.1038/nnano.2010.42. [DOI] [PubMed] [Google Scholar]
- 5.Jiang ZJ, Mihovilovic M, Chan J, Stein D. Fabrication of nanopores with embedded annular electrodes and transverse carbon nanotube electrodes. J Phys Condens Matter. 2010;22 doi: 10.1088/0953-8984/22/45/454114. [DOI] [PubMed] [Google Scholar]
- 6.Ivanov AP, Instuli E, McGilvery CM, Baldwin G, McComb DW, Albrecht T, Edel JB. DNA Tunneling detector embedded in a nanopore. Nano Lett. 2011;11:279–285. doi: 10.1021/nl103873a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wanunu M, Dadosh T, Ray V, Jin JM, McReynolds L, Drndic M. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nat Nanotechnol. 2010;5:807–814. doi: 10.1038/nnano.2010.202. [DOI] [PubMed] [Google Scholar]
- 8. Hall AR, Scott A, Rotem D, Mehta KK, Bayley H, Dekker C. Hybrid pore formation by directed insertion of alpha-haemolysin into solid-state nanopores. Nat Nanotechnol. 2010;5:874–877. doi: 10.1038/nnano.2010.237. Insertion of α-hemolysin channels into solid-state nanopores in order to form a precise, protein-based channel within a robust synthetic membrane
- 9.Kuan AT, Golovchenko JA. Nanometer-thin solid-state nanopores by cold ion beam sculpting. Appl Phys Lett. 2012;100 doi: 10.1063/1.4719679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyun C, Rollings R, Li JL. Probing access resistance of solidstate nanopores with a scanning-probe microscope tip. Small. 2012;8:385–392. doi: 10.1002/smll.201101337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xie P, Xiong QH, Fang Y, Qing Q, Lieber CM. Local electrical potential detection of DNA by nanowire-nanopore sensors. Nat Nanotechnol. 2012;7:119–125. doi: 10.1038/nnano.2011.217. Electron-beam formation of a nanopore at the site of a silicon nanowire, which affords single-molecule sensing by high-bandwidth measurements of the nanowire current
- 12.Bell NAW, Engst CR, Ablay M, Divitini G, Ducati C, Liedl T, Keyser UF. DNA origami nanopores. Nano Lett. 2012;12:512–517. doi: 10.1021/nl204098n. [DOI] [PubMed] [Google Scholar]
- 13. Wei RS, Martin TG, Rant U, Dietz H. DNA Origami gatekeepers for solid-state nanopores. Angew Chem Int Ed. 2012;51:4864–4867. doi: 10.1002/anie.201200688. Design of DNA-based plate that has a pore through its center. The pore size and chemical characteristics were controlled by choice of the appropriate DNA staples, which allowed for chemically modified pores with near-atomic precision
- 14.Garaj S, Hubbard W, Reina A, Kong J, Branton D, Golovchenko JA. Graphene as a subnanometre trans-electrode membrane. Nature. 2010;467:190–193. doi: 10.1038/nature09379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merchant CA, Healy K, Wanunu M, Ray V, Peterman N, Bartel J, Fischbein MD, Venta K, Luo ZT, Johnson ATC, et al. DNA translocation through graphene nanopores. Nano Lett. 2010;10:2915–2921. doi: 10.1021/nl101046t. [DOI] [PubMed] [Google Scholar]
- 16.Schneider GF, Kowalczyk SW, Calado VE, Pandraud G, Zandbergen HW, Vandersypen LMK, Dekker C. DNA translocation through graphene nanopores. Nano Lett. 2010;10:3163–3167. doi: 10.1021/nl102069z. [DOI] [PubMed] [Google Scholar]
- 17.Liu S, Zhao Q, Xu J, Yan K, Peng HL, Yang FH, You LP, Yu DP. Fast and controllable fabrication of suspended graphene nanopore devices. Nanotechnology. 2012;23 doi: 10.1088/0957-4484/23/8/085301. [DOI] [PubMed] [Google Scholar]
- 18.Spinney PS, Collins SD, Howitt DG, Smith RL. Fabrication and characterization of a solid-state nanopore with self-aligned carbon nanoelectrodes for molecular detection. Nanotechnology. 2012;23 doi: 10.1088/0957-4484/23/13/135501. [DOI] [PubMed] [Google Scholar]
- 19.Lan WJ, White HS. Diffusional motion of a particle translocating through a nanopore. ACS Nano. 2012;6:1757–1765. doi: 10.1021/nn2047636. [DOI] [PubMed] [Google Scholar]
- 20.Makra I, Jagerszki G, Bitter I, Gyurcsanyi RE. Nernst-Planck/Poisson model for the potential response of permselective gold nanopores. Electrochim Acta. 2012;73:70–77. [Google Scholar]
- 21. Yang JJ, Ferranti DC, Stern LA, Sanford CA, Huang J, Ren Z, Qin LC, Hall AR. Rapid and precise scanning helium ion microscope milling of solid-state nanopores for biomolecule detection. Nanotechnology. 2011;22 doi: 10.1088/0957-4484/22/28/285310. A promising approach for fabricating nanopores is demonstrated using a focused beam of He ions. Contrary to heavier ions, the He beam does not contaminate the surface of the nanopore and can directly form small pores (<10 nm)
- 22.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Single-molecule DNA detection with an engineered MspA protein nanopore. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20647–20652. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wendell D, Jing P, Geng J, Subramaniam V, Lee TJ, Montemagno C, Guo PX. Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores. Nat Nanotechnol. 2009;4:765–772. doi: 10.1038/nnano.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soni GV, Dekker C. Detection of nucleosomal substructures using solid-state nanopores. Nano Lett. 2012;12:3180–3186. doi: 10.1021/nl301163m. [DOI] [PubMed] [Google Scholar]
- 25.Raillon C, Cousin P, Traversi F, Garcia-Cordero E, Hernandez N, Radenovic A. Nanopore detection of single molecule RNAP-DNA transcription complex. Nano Lett. 2012;12:1157–1164. doi: 10.1021/nl3002827. [DOI] [PubMed] [Google Scholar]
- 26.Shim J, Gu LQ. Single-molecule investigation of G-quadruplex using a nanopore sensor. Methods. 2012;57:40–46. doi: 10.1016/j.ymeth.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.An N, Fleming AM, White HS, Burrows CJ. Crown ether–electrolyte interactions permit nanopore detection of individual DNA abasic sites in single molecules. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11504–11509. doi: 10.1073/pnas.1201669109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rotem D, Jayasinghe L, Salichou M, Bayley H. Protein detection by nanopores equipped with aptamers. J Am Chem Soc. 2012;134:2781–2787. doi: 10.1021/ja2105653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cressiot B, Oukhaled A, Patriarche G, Pastoriza-Gallego M, Betton JM, Auvray L, Muthukumar M, Bacri L, Pelta J. Protein transport through a narrow solid-state nanopore at high voltage: experiments and theory. ACS Nano. 2012;6:6236–6243. doi: 10.1021/nn301672g. [DOI] [PubMed] [Google Scholar]
- 30.Pastoriza-Gallego M, Rabah L, Gibrat G, Thiebot B, Goot FG, Auvray L, Betton JM, Pelta J. Dynamics of unfolded protein transport through an aerolysin pore. J Am Chem Soc. 2011;133:2923–2931. doi: 10.1021/ja1073245. [DOI] [PubMed] [Google Scholar]
- 31. Schink S, Renner S, Alim K, Arnaut V, Simmel FC, Gerland U. Quantitative analysis of the nanopore translocation dynamics of simple structured polynucleotides. Biophys J. 2012;102:85–95. doi: 10.1016/j.bpj.2011.11.4011. Stability of hairpins of different sequences was quantitatively measured by their reverse passage through nanopores, and the data converged well with theory
- 32.Jin Q, Fleming AM, Burrows CJ, White HS. Unzipping kinetics of duplex DNA containing oxidized lesions in an alpha-hemolysin nanopore. J Am Chem Soc. 2012;134:11006–11011. doi: 10.1021/ja304169n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merstorf C, Cressiot B, Pastoriza-Gallego M, Oukhaled A, Betton JM, Auvray L, Pelta J. Wild type, Mutant protein unfolding and phase transition detected by single-nanopore recording. ACS Chem Biol. 2012;7:652–658. doi: 10.1021/cb2004737. [DOI] [PubMed] [Google Scholar]
- 34.Boersma AJ, Brain KL, Bayley H. Real-time stochastic detection of multiple neurotransmitters with a protein nanopore. ACS Nano. 2012;6:5304–5308. doi: 10.1021/nn301125y. [DOI] [PubMed] [Google Scholar]
- 35. Wei RS, Gatterdam V, Wieneke R, Tampe R, Rant U. Stochastic sensing of proteins with receptor-modified solid-state nanopores. Nat Nanotechnol. 2012;7:257–263. doi: 10.1038/nnano.2012.24. Demonstration of gold-coated pores onto which self-assembled thiols were used to tether individual protein receptors, leading to single molecule measurements of protein binding/unbinding at the pore
- 36.Kumar S, Tao CJ, Chien MC, Hellner B, Balijepalli A, Robertson JWF, Li ZM, Russo JJ, Reiner JE, Kasianowicz JJ, Ju JY. PEG-labeled nucleotides and nanopore detection for single molecule DNA sequencing by synthesis. Scientific Reports. 2012;2 doi: 10.1038/srep00684. Article no. 684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo P, Hall EW, Schirhagl R, Mukaibo H, Martin CR, Zare RN. Microfluidic capture and release of bacteria in a conical nanopore array. Lab Chip. 2012;12:558–561. doi: 10.1039/c2lc21092d. [DOI] [PubMed] [Google Scholar]
- 38.Bessonov A, Takemoto JY, Simmel FC. Probing DNA–lipid membrane interactions with a lipopeptide nanopore. ACS Nano. 2012;6:3356–3363. doi: 10.1021/nn3003696. [DOI] [PubMed] [Google Scholar]
- 39.Zhou YH, Guo W, Cheng JS, Liu Y, Li JH, Jiang L. High-temperature gating of solid-state nanopores with thermo-responsive macromolecular nanoactuators in ionic liquids. Adv Mater. 2012;24:962. doi: 10.1002/adma.201104814. [DOI] [PubMed] [Google Scholar]
- 40.Yeh LH, Zhang M, Qian S, Hsu JP, Tseng S. Ion concentration polarization in polyelectrolyte-modified nanopores. J Phys Chem C. 2012;116:8672–8677. [Google Scholar]
- 41.Qiu H, Guo WL. Detecting ssDNA at single-nucleotide resolution by sub-2-nanometer pore in monoatomic graphene: A molecular dynamics study. Appl Phys Lett. 2012;100 [Google Scholar]
- 42.Franceschini L, Mikhailova E, Bayley H, Maglia G. Nucleobase recognition at alkaline pH and apparent pK(a) of single DNA bases immobilised within a biological nanopore. Chem Commun. 2012;48:1520–1522. doi: 10.1039/c1cc16124e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melnikov DV, Leburton JP, Gracheva ME. Slowing down and stretching DNA with an electrically tunable nanopore in a p–n semiconductor membrane. Nanotechnology. 2012;23 doi: 10.1088/0957-4484/23/25/255501. [DOI] [PubMed] [Google Scholar]
- 44. Rosenstein JK, Wanunu M, Merchant CA, Drndic M, Shepard KL. Integrated nanopore sensing platform with sub-microsecond temporal resolution. Nat Methods. 2012;9:487–U112. doi: 10.1038/nmeth.1932. The temporal resolution of nanopore measurements was improved by an order of magnitude over state-of-the-art by designing a new CMOS-based amplifier chip with on-board electrode
- 45.Saha KK, Drndic M, Nikolic BK. DNA base-specific modulation of microampere transverse edge currents through a metallic graphene nanoribbon with a nanopore. Nano Lett. 2012;12:50–55. doi: 10.1021/nl202870y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Min SK, Kim WY, Cho Y, Kim KS. Fast DNA sequencing with a graphene-based nanochannel device. Nat Nanotechnol. 2011;6:162–165. doi: 10.1038/nnano.2010.283. [DOI] [PubMed] [Google Scholar]
- 47.Pathak B, Lofas H, Prasongkit J, Grigoriev A, Ahuja R, Scheicher RH. Double-functionalized nanopore-embedded gold electrodes for rapid DNA sequencing. Appl Phys Lett. 2012;100 [Google Scholar]
- 48.Comer J, Aksimentiev A. Predicting the DNA sequence dependence of nanopore ion current using atomic-resolution brownian dynamics. J Phys Chem C. 2012;116:3376–3393. doi: 10.1021/jp210641j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wells DB, Belkin M, Comer J, Aksimentiev A. Assessing graphene nanopores for sequencing DNA. Nano Lett. 2012;12:4117–4123. doi: 10.1021/nl301655d. All-atom molecular dynamics were used to investigate the transport of DNA molecules through graphene nanopores, which reveals issues with DNA transport stochasticity and interactions with graphene/DNA bases
- 50.Timp W, Comer J, Aksimentiev A. DNA base-calling from a nanopore using a viterbi algorithm. Biophys J. 2012;102:L37–L39. doi: 10.1016/j.bpj.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langecker M, Arnaut V, Martin TG, List J, Renner S, Mayer M, Dietz H, Simmel FC. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science. 2012;338:932–936. doi: 10.1126/science.1225624. [DOI] [PMC free article] [PubMed] [Google Scholar]