Abstract
The horseshoe crab Tachypleus tridentatus is a unique marine species and a potential model for marine invertebrate. Limited genomic and transcriptional data are currently available to understand the molecular mechanisms underlying the embryonic development of T. tridentatus. Here, we reported for the first time the de novo transcriptome assembly for T. tridentatus at embryonic developmental stage using Illumina RNA-seq platform. Approximate 38 million reads were obtained and further assembled into 133,212 unigenes. Sequence homology analysis against public databases revealed that 33,796 unigenes could be annotated with gene descriptions. Of the annotated unigenes, we identified a number of key components of several conserved metazoan signaling pathways (Hedgehog, Wnt, TGF-beta and Notch pathways) and other important regulatory genes involved in embryonic development. Targeted searching of Pax family genes which play critical roles in the formation of tissue and organ during embryonic development identified a complete set of Pax family genes. Moreover, the full length T. tridentatus Pax1/9a (TtPax1/9a) and Pax1/9b (TtPax1/9b) cDNA sequences were determined based on the transcriptome, demonstrating the immediate application of our database. Using quantitative real time PCR, we analyzed the expression patterns of TtPax1/9a and TtPax1/9b in different tissues of horseshoe crab. Taking advantage of Drosophila model, we further found that TtPax1/9b, but not TtPax1/9a, can partly rescue the Drosophila homolog Poxm dysfunction-caused lethality at the larval stage. Our study provides the embryonic transcriptome of T. tridentatus which could be immediately used for gene discovery and characterization, functional genomics studies in T. tridentatus. This transcriptome database will also facilitate the investigations of molecular mechanisms underlying embryonic development of T. tridentatus and other marine arthropods as well.
Introduction
Emergence of new model organisms plays increasingly critical roles in embryogenesis and evolutionary developmental study. Until now, the most well-known model organism in arthropods is the fruit fly Drosophila melanogaster (belonging to Class Insecta), which is widely used in studies of genetics and embryogenesis [1,2]. The genome of water flea Daphnia pulex (belonging to Class Crustacea) was also published recently, which facilitates the study of cellular response to environmental challenges [3]. Nevertheless, due to lack of genomic information, understanding of developmental and molecular mechanisms of most arthropods is still lagged far behind that of the vertebrates, which consequentially hinders the appearance of new model organisms within arthropods. Fortunately, high throughput de novo transcriptome assembly has proven to be a valuable technology to obtain sequence information and expression level of large-scale target genes involved in a particular biological process without any prior knowledge of reference genome [4–7]. In fact, this technology has been applied to analyze transcriptomes from a variety of species in metazoan [8–12].
Horseshoe crabs (Family Limulidae, Order Xiphosura, Class Merostomata), which are an extremely ancient marine group, have emerged as a valuable laboratory animal model in developmental study of marine invertebrate for decades [13–16]. They inhabit the areas around the shallow coastal seas and breed on intertidal shores. Even though horseshoe crabs show some common features of crustaceans (crab-like shell and claws), they are more closely related to arachnids, such as spiders and scorpions [17,18]. To date, only four extant horseshoe crab species have been discovered in two distinctly separate regions of the world, viz. Tachypleus tridentatus, Tachypleus gigas, Carcinoscorpius rotundicauda (mainly found in Southeast Asia) and Limulus polyphemus (only found in western Atlantic coast of North America). T. tridentatus was once very common along the southeast coast of Mainland China. Unfortunately, its population has been declining for several decades due to excessive hunting by humans and environmental pollution. Fossil evidence revealed that the earliest horseshoe crab lived during the late Ordovician period, around 445 million years ago [19]. More strikingly, the horseshoe crab remains unchanged in overall morphology for over 200 million years, and therefore is considered as a “living fossil” [20]. Besides their importance for the evolutionary studies and preservation of ecological diversity, horseshoe crabs also serve as a multiple-use animal resource. For example, the Limulus Amebocyte Lysate test, which is widely applied in the detection and quantification of bacterial endotoxins, is based on an aqueous extract of amebocytes from horseshoe crab [21]. Moreover, it is believed that horseshoe crabs are essential for the maintenance of the ecology of estuarine and coastal communities [22].
It is reported that the horseshoe crabs take about 10–15 years to reach sexual maturity from fertilized eggs and more than ten molts occur during this period [23], which makes it difficult to record the whole growth process of horseshoe crab in natural habitat without interruption. Growth observation of horseshoe crabs from larvae to adulthood in laboratory also turned out to be a failure [24]. On the other hand, over the recent decades, the horseshoe crab has emerged as an experimental model for studying marine invertebrate embryology, structure-function relationship of the visual system and nervous system [15,25]. Morphological changes during early embryonic development of two horseshoe crab species, including T. tridentatus and L. polyphemus, have been studied [14,23]. According to Sekiguchi’s classification [23], the embryonic development of horseshoe crab could be roughly divided into the 21 stages, mainly including cleavage, blastula, gastrula, appearance of germ disc, formation of appendages and finally hatch-out to the first instar “trilobite larvae”. During hatch-out stage, the embryo grows remarkably and several organs further develop. For instance, the central eye becomes discernable as a brownish spot. The appendages of the prosoma are further expanded [23]. All these changes enable the horseshoe crab to survive in the new challenging circumstance without the protection of chorion.
Although the study of horseshoe crab morphological changes during embryonic development has been described, the detailed molecular mechanism underlying this process remains unknown, which is largely due to the lack of genomic information. In the present study, we employed high-throughput Illumina Solexa sequencing and gene annotation to characterize the transcriptome of T. tridentatus embryo at the hatch-out stage. We reported for the first time a comprehensive analysis of large-scale gene expression profile during T. tridentatus embryonic development, which could be immediately used for further gene discovery and functional genomics study. Our data indicated that the major signaling pathways and key regulatory factors involved in embryonic development were highly conserved between T. tridentatus and other metazoans, especially D. melanogaster. Molecular cloning and functional study of T. tridentatus Pax1/9a and Pax1/9b were also investigated in our study. Therefore, the transcriptome analysis reported here have important applications to the understanding of molecular mechanisms underlying T. tridentatus embryonic development.
Materials and Methods
Horseshoe crab maintenance and breeding
Adult T. tridentatus were maintained in a 3 m×1 m×2 m tank containing natural seawater (temperature 25°C, salinity 30 ppt) and fed with bivalves. Fertilized eggs were obtained by natural spawning and cultured in the laboratory with standard procedures [26]. The staging of embryos was according to Sekiguchi’s developmental tables [23]. The embryos at Stage 21 (the hatch-out stage) were collected for high throughput transcriptome sequencing.
RNA extraction and quality determination
Total RNA of T. tridentatus embryos was isolated by TRIzol (Invitrogen, Carlsbad, CA, USA) according to the standard protocol. The RNA samples were treated with DNase I (TaKaRa, Japan) for 4 h. RNA was quantified by measuring the absorbance at 260 nm using a NanoDrop spectrophotometer (Thermo Fisher Scientific Inc., San Jose, CA, USA). The purity of RNA was assessed by the ratio of the absorbance at 260 and 280 nm. The integrity of the RNA samples was examined with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
Cloning of full length T. tridentatus Pax1/9 cDNAs
To obtain the full length cDNAs of Pax1/9 genes, the BLAST search of human Pax1 and Pax9 genes were performed against the T. tridentatus transcriptome database, which resulted in two sequences with high homology. The 3’and 5’ ends were obtained by rapid amplification of cDNA ends (RACE) approaches using 3’-Full RACE Core Set with PrimeScript™ RTase and 5’-Full RACE Kit with TAP (TaKaRa, Japan) following the manufacturer’s instructions. Primers for 3’-RACE and 5’-RACE were listed in S1 Table. The PCR products were ligated into pMD-19T vector (TaKaARa, Japan) and transformed into the competent E. coli TOP10 cells. Positive clones with the expected-size inserts were determined by colony PCR and DNA sequencing.
cDNA library preparation, Illumina sequencing and sequence assembly
The cDNA library was prepared using the TruSeqTM RNA Sample Preparation Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Poly(A)-containing mRNA was purified by Oligo(dT) magnetic beads from 10 μg total RNA sample and fragmented using divalent cations. The cleaved RNA fragments were used for the first strand cDNA synthesis using reverse transcriptase and random primers, followed by second strand cDNA synthesis using DNA polymerase I and RNase H. After second strain cDNA synthesis, fragments were treated with end repair, A-base tailing and adapter ligation consecutively. The sample was further treated by gel size fractionation and PCR amplification to create final cDNA library. The cDNA library was sequenced on the Illumina Cluster Station and Illumina Genome Analyzer system according to the manufacturer’s instructions. The Trinity method was used for de novo assembly of Illumina reads of T. tridentatus embryos [27]. Briefly, the trinity using de Bruijn graph algorithm was run on the paired-end sequences with the fixed default k-mer size of 25, minimum contig length of 200 and paired fragment length of 500.
Functional annotation
All possible coding sequences were predicted by GetORF model of EMBOSS (http://emboss.sourceforge.net/apps/cvs/emboss/apps/getorf.html) with default parameters. The longest ORF was considered as the candidate coding sequence (CDS). The assembled unigenes were annotated based on sequence similarity by sequential BLAST searches against National Center for Biotechnology Information (NCBI) non-redundant protein database (NR) and nucleotide sequences database (NT), the Swiss-Prot protein database, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database, the Cluster of Orthologous Groups (COG) database and the Translated EMBL Nucleotide Sequence Database (TrEMBL). The Blast2GO software was used for blasting and assigning associated gene ontology (GO) terms describing biological processes, molecular functions and cellular components.
Phylogenetic analysis
Related sequences from homo sapiens and Drosophila melanogaster were included in the phylogenetic analysis (S2 Table). The DNA binding domains of Pax family proteins was aligned manually and Mega 3 [28] was used to generate the phylogenetic trees. The neighbor joining method with 1000 bootstrap replications was used to calculate each tree.
Expression of TtPax1/9a and TtPax1/9b in T. tridentatus tissues
To assess the tissue distribution of TtPax1/9a and TtPax1/9b transcripts, total RNA was extracted from intestine, liver, yellow connective tissue, heart, stomach, muscle and gill, respectively. First strand cDNA synthesized from 2 μg total RNA was used as templates for quantitative real time PCR using IQTM 5 Multicolor Real-time PCR Detection System (Bio-Rad, Richmond, CA, USA). The TtPax1/9a specific primers are Pax1/9a-Fw (5’-AGCCGTTTACCTGAATCGAC-3’) and Pax1/9a-Rv (5’-AAATATTGTGCACTTGCTGGA-3’). The TtPax1/9b specific primers are Pax1/9b-Fw (5’-CCAGTGTCCATGCCATTAAG-3’) and Pax1/9b-Rv (5’-GTTTGCGGTGACACTGTTCT-3’). The T. tridentatus GAPDH gene was used as the internal control. Specific primers for T. tridentatus GAPDH are GAPDH-Fw (5’-ATCATCAGCAATGCCTCTTG-3’) and GAPDH-Rv (5’-GCCTTAGAGCTTGGTCCATC-3’).
Generation of transgenic flies
D. melanogaster were cultured on standard medium with 12/12 light/dark cycle at 25°C. The poxm loss-of-function mutant fly PoxmR361 and transgenic line poxm8.4-Gal4 have been described previously [29]. Taking advantage of EcoR I and Xho I restriction sites, the full length cDNA of TtPax1/9a and TtPax1/9b were subclone into pUAST vector. The UAS-TtPax1/9a and UAS-TtPax1/9b transgenic fly lines were generated by germline transformation with co-injection of the helper plasmid Δ2–3 according to standard protocol [30]. The UAS-TtPax1/9a or UAS-TtPax1/9b transgenic fly lines were crossed with poxm8.4-Gal4 fly to obtain the TtPax1/9a- and UAS-TtPax1/9b-expressing flies under the control of poxm8.4 upstream region. The transgenic flies were imaged with Leica M205FA microscope (Leica, Wetzlar, German).
Results and Discussion
Sequencing and transcriptome assembly
In this study, 10 μg of total RNA isolated from the T. tridentatus embryos at hatch-out stage were used to prepare cDNA library for subsequent transcriptome sequencing using an Illumina HiSeq2000 sequencer. RNA was pooled from multiple T. tridentatus embryos to prepare one sample for sequencing. The summary of Illumima sequencing and annotation was shown in Table 1. Sequencing of cDNA library generated a total of 37,902,792 paired end reads. These data are available from the NCBI Short Read Archive under accession number SRR946952. Transcriptome assembly was completed using RNAseq de novo assembler Trinity (k-mer length = 25). 4,864,778 contigs were identified with the nucleotide composition of A, T, C and G being 30.98%, 29.67%, 20.22% and 19.12% respectively, which gives rise to an overall GC content of 39.34% in the whole transcriptome. The contigs were further assembled into 176,231 transcripts falling into 133,212 unigenes (> 200 bp) with a mean unigene length of 746 bp and an N50 of 568 bp (Table 1). The length distribution of transcripts and unigenes were shown in S1 Fig.
Table 1. Summary of T. tridentatus transcriptome sequencing, assembly and annotation.
| Stage | Data | |
|---|---|---|
| Sequencing | Sequencing reads | 37,902,792 |
| Assembly | Total transcripts | 176,231 |
| N50 length of transcripts (bp) | 1147 | |
| Mean length of transcripts (bp) | 713 | |
| Max length of transcripts (bp) | 11,337 | |
| Total unigenes | 133,212 | |
| N50 length of unigenes (bp) | 746 | |
| Mean length of unigenes (bp) | 568 | |
| Max length of unigenes (bp) | 11,337 | |
| Annotation | Total unigenes annotated | 33,796 |
| Total unigenes unannotated | 99,416 | |
| Unigenes annotated against NR database | 29,918 | |
| Unigenes annotated against NT database | 11,116 | |
| Unigenes annotated against Swiss-Prot database | 24,118 | |
| Unigenes annotated against TrEMBL database | 29,884 | |
| Unigenes annotated against GO database | 15,768 | |
| Unigenes annotated against COG database | 6,920 | |
| Unigenes annotated against KEGG database | 10,999 |
Unigene annotation was achieved by BLASTx and BLASTn searches against NR, NT, Swissprot, and TrEMBL with an e-value less than 1×10−5. In total, there were 33,796 unigenes showing hits in one or more databases (Table 1), among which the NR database gave rise to the most annotations (29,918 hits). However, a large proportion of the sequences did not show significant blast hit. This was probably due to the lack of characterization of genes from closely related species, as the length distribution and the coverage of the annotated and unannotated unigenes were in a similar pattern (S2 Fig). The assembled sequences generated in our study are available from the authors upon request.
Functional annotation
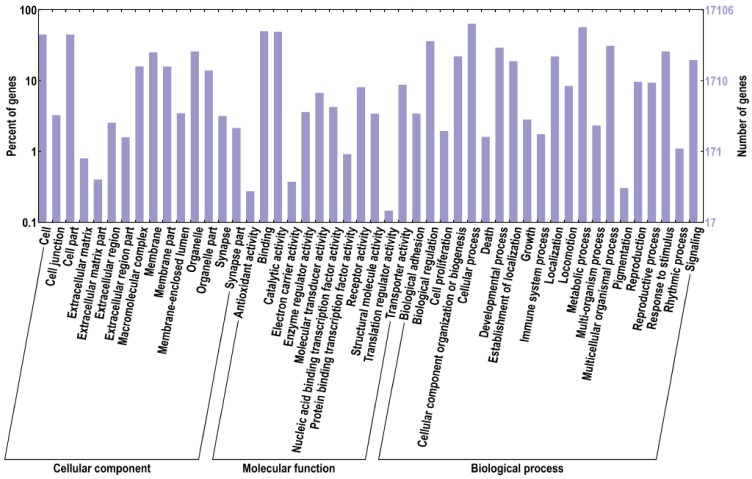
In order to predict the functions of these unigenes, all the sequences were analyzed according to gene ontology (GO) database and clusters of orthologous groups (COGs). 15,768 genes were successfully annotated with 95,305 GO terms and were separated into three categories: biological process, cellular component, and molecular function, which were further divided into 15, 12 and 21 functional groups, respectively (Fig 1). Among these GO terms, 49,724 unigene sequences were assigned to biological process, 27,948 to molecular function, and 17,363 to cellular component. Interestingly, the top 5 groups with most unigenes involved in the biological process were: cellular process, metabolic process, biological regulation, developmental process, and multicellular organismal process. While the cell part and the cell functional groups were dominant in the cellular component category, and the binding and the catalytic activity were dominant in the molecular function category. The integral to membrane (GO: 0016021) contained 1572 unigenes, which belonged to the cellular component category. It had the largest number of unigenes among all the groups. In addition, the ATP binding (GO: 0005524) containing 1478 unigenes and the oxidation-reduction process (GO: 0055114) containing 619 unigenes were the most represented GO terms for the molecular function and biological process categories, respectively. A high percentage of unigenes involved in the following functional groups should also be noted: cellular process (GO: 0009987) with 561unigenes, protein phosphorylation (GO: 0006468) with 596 unigenes and regulation of transcription, DNA-dependent (GO: 0006355) with 556 unigenes. We were interested in the embryonic development process of T. tridentatus, and thus investigated the unigenes with GO terms containing “embryo” or “development”. As a result, a total of 183 GO terms were found with unigene numbers ranging from 1 to 241. The top 20 terms with the most number of unigenes were listed in Table 2. This demonstrated that our transcriptome database contained a variety of unigenes related to the embryonic development, which provided an abundant resource for further investigation of regulatory mechanisms of T. tridentatus embryonic development.
Fig 1. GO distribution of the T. tridentatus unigenes.
Go ontology distribution of the T. tridentatus unigenes were derived using BLAST2GO. The X-axis represents three main GO categories: cellular component, molecular function and biological process, which further separated into 15, 12 and 21 functional groups, respectively. The Y-axis represents percentages and numbers of unigenes mapping to the given functional GO group.
Table 2. Top 20 GO terms with most unigenes involved in embryonic development.
| GO term | GO index | No. of unigenes |
|---|---|---|
| mushroom body development | 0016319 | 241 |
| peripheral nervous system development | 0007422 | 207 |
| mesoderm development | 0007498 | 173 |
| central nervous system development | 0007417 | 162 |
| multicellular organismal development | 0007275 | 153 |
| open tracheal system development | 0007424 | 146 |
| muscle organ development | 0007517 | 140 |
| embryo development ending in birth or egg hatching | 0009792 | 125 |
| compound eye development | 0048749 | 113 |
| neuromuscular junction development | 0007528 | 105 |
| embryonic development via the syncytial blastoderm | 0001700 | 86 |
| wing disc development | 0035220 | 86 |
| nervous system development | 0007399 | 85 |
| nematode larval development | 0002119 | 82 |
| organ development | 0048513 | 75 |
| chaeta development | 0022416 | 74 |
| gonad development | 0008406 | 69 |
| heart development | 0007507 | 68 |
| instar larval development | 0002168 | 65 |
| sensory organ development | 0007423 | 62 |
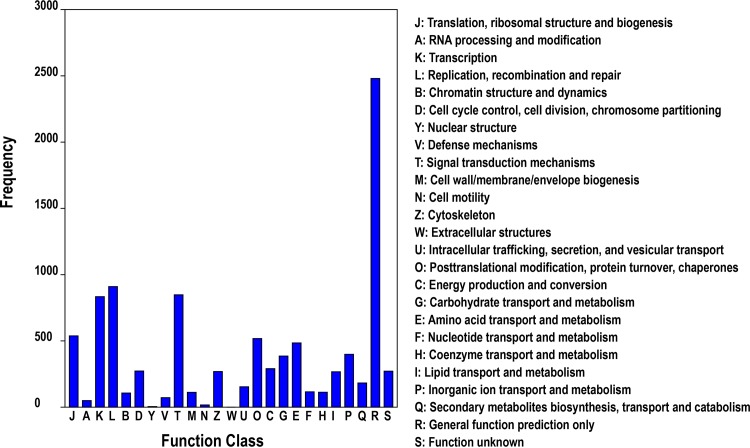
Meanwhile, COG annotation was used for phylogenetic classification of the putative proteins from the T. tridentatus transcriptome, in which a total of 6,920 proteins were assigned to 25 clusters (Fig 2). Among the COG library, the cluster of “General function prediction only” consisted of the largest number of unigenes. The other five largest categories were: “replication, recombination and repair” (14.47%), “signal transduction mechanisms” (13.42%), “transcription” (13.12%), “translation, ribosomal structure and biogenesis” (8.44%), and “posttranslational modification, protein turnover, chaperones” (8.24%), suggesting the specific responses and molecular mechanisms of T. tridentatus early development to some extent.
Fig 2. COG functional classification of the T. tridentatus transcriptome.
To investigate the biological pathways which are possibly involved in early embryonic development process of T. tridentatus, we further mapped the unigenes to the reference canonical pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. A total of 10,999 unigenes were assigned to 298 KEGG pathways. The pathways most represented by numbers of unigene sequences were “transcription factors” (865 unigenes), “chromosome” (802 unigenes), “ubiquitin system” (798 unigenes), “protein kinases” (784 unigenes), and “pathways in cancer” (518 unigenes).
Signaling pathways and regulatory genes involved in the development of T. tridentatus
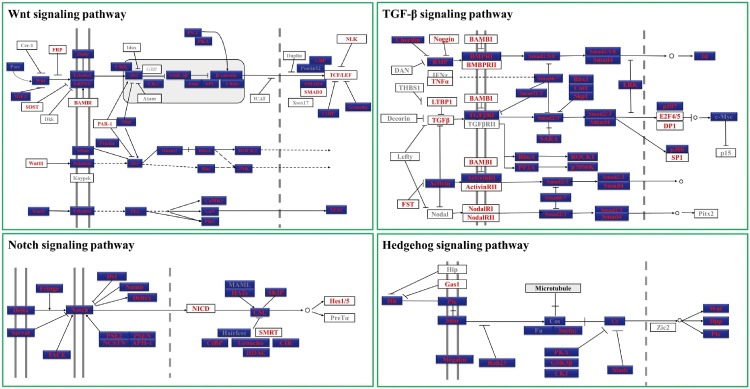
Formation of different cell types and patterns during development of multicellular organisms depends on a complex cascade of developmental decisions. An evolutionarily conserved network of signaling pathways and regulatory genes plays key roles during this process [31]. Analyzing these pathways and factors involved in the early embryonic development of T. tridentatus is of great importance for understanding of the developmental mechanisms of this ancient marine creature and may provide insights into the evolutional study of conserved signaling pathways and regulatory genes. In all the annotated unigenes of T. tridentatus transcriptome, we identified a number of key components of four major signaling pathways (including Hedgehog, Wnt, transforming growth factor-β and Notch pathways) conserved in metazoan. As shown in Fig 3, 98 possible homologues were identified from 126 key genes involved in these signaling pathways, representing 77.78% coverage of the total genes. The percentage of genes found in each pathway is 78.85% (Wnt), 77.27% (TGF-β), 77.78% (Hedgehog) and 87.5% (Notch) respectively. In case of D. melanogaster-specific pathways, the percentage was much higher, about 91.57% (76/83) homologues could be found in T. tridentatus transcriptome, indicating the higher similarity of T. tridentatus with D. melanogaster than other animal models in the aspect of signaling transduction pathways. It should also be noted that the sequence length of most gene fragments (> 200 bp) is sufficient for functional studies of these genes by modern molecular technology, such as real-time PCR quantification, in situ hybridization and antibody preparation. Moreover, with the sequence information of these gene fragments, it would be more efficient and reliable to obtain the full length of desired genes comparing with common degenerate PCR method. This is particularly helpful in case of gene discovery and functional study in the “non-model” organism horseshoe crab.
Fig 3. Major components from Conserved metazoan signaling pathways identified in T. tridentatus.
The results were generated by BLASTing the unigenes of T. tridentatus to known homologues with the e-value cutoff at 1e-10. Pathway schematics were adapted from KEGG pathway models (http://www.genome.jp/kegg/). Drosophila melanogaster-specific pathways are colored blue. Genes identified and not identified in T. tridentatus were marked with red font and grey font respectively.
Regulatory genes are also key players in the network governing embryonic development [32]. D. melanogaster is one of the best understood models of embryonic development, especially pattern formation process, and thus we analyzed the regulatory genes potentially involved in the T. tridentatus axis specification and patterning by using D. melanogaster as a reference model. The genetic control of axis specification and patterning in D. melanogaster requires a cascade of gene regulation events before the onset of blastoderm stage. The major genes involved in this developmental process of D. melanogaster can grouped into four categories, viz. maternal effect genes, gap genes, pair rule genes and segment polarity genes [33,34]. This developmental process starts with the diffusion of the maternal effect genes which are responsible for setting the anterior-posterior polarity of the embryo. The gap genes, which are among the first genes transcribed in the embryo, participate in the establishment of the sub-domain of body plan under the control of maternal gradients. The pair rule genes subsequently divide the embryo into periodic units, whereas the segment polarity genes activated by pair rule genes further establish the periodicity of the embryo by dividing it into 14 segment-wide units [35,36]. Key regulatory genes of each category used in our study were selected according to S.F. Gilbert’s and R.M. Twyman’s classification [37,38]. Among all the 53 genes listed in Table 3, we identified a total of 40 genes (75.47%) which had homologs in T. tridentatus, suggesting that functions of these genes conserved in T. tridentatus. It should be pointed out that the percentage of gene homologs identified in the maternal effect genes category was much lower (11/18, 61.11%) comparing to that of the other categories: gap genes (12/14, 85.71%), pair rule genes (7/8, 87.5%) and segment polarity genes (13/16, 81.25%). This is possibly because the mRNA products of the maternal gene homologs have been consumed at the hatch-out stage, the last stage of T. tridentatus embryonic development. In addition, we also failed to identify some well-studied gene homologs (including knirps, tailless, even-skipped, invected, fused and costal 2), which may be due to the extremely low expression of these gene homologs in our sample. Anyway, we identified the majority of the candidate regulatory genes that are potentially involved in the axis specification and patterning of T. tridentatus embryo. Further study is required to characterize the function of these putative genes.
Table 3. Representative regulatory genes involved in the early embryonic development of T. tridentatus.
| Name | Length (range) | No. of transcripts | Read number | Hit ID | target organism | E-value |
|---|---|---|---|---|---|---|
| Major maternal effect genes | ||||||
| bicoid | 215–717 | 7 | 84 | gb|EHJ73092.1| | Danaus plexippus | 3E-96 |
| hunchbacka | 281 | 2 | 3 | emb|CAK50841.1| | Glomeris marginata | 1E-13 |
| caudala | 286–1412 | 5 | 967 | ref|XP_002405030.1| | Ixodes scapularis | 5E-06 |
| staufen | 3588 | 1 | 3580 | gb|ELT94341.1| | Capitella teleta | 3E-141 |
| oskar | 214–4349 | 14 | 13341 | ref|XP_004932537.1| | Bombyx mori | 2E-34 |
| tudor | 480–1849 | 4 | 358 | gb|EKC34332.1| | Crassostrea gigas | 1E-11 |
| vasa | 257–3575 | 2 | 4032 | gb|ACT35657.1| | Haliotis asinina | 2E-180 |
| pumilio | 274–322 | 2 | 5 | ref|XP_002601469.1| | Branchiostoma floridae | 1E-45 |
| torso | 256–344 | 5 | 22 | ref|XP_002408935.1| | Ixodes scapularis | 2E-10 |
| trunk | 236–2925 | 13 | 754 | gb|EFX70037.1| | Daphnia pulex | 3E-03 |
| rolled | 442–858 | 2 | 58876 | gb|ADV40083.1| | Latrodectus hesperus | 6E-59 |
| Maternal effect genes not found in T. tridentatus transcriptome: exuperantia, swallow, nanos, valios, torsolike, nasrat and polehole. | ||||||
| Major gap genes | ||||||
| hunchbacka | 281 | 2 | 3 | emb|CAK50841.1| | Glomeris marginata | 1E-13 |
| krüppel | 369–4091 | 3 | 1306 | ref|XP_002435294.1| | Ixodes scapularis | 4E-09 |
| giant | 468–1490 | 4 | 210 | ref|XP_002429035.1| | Pediculus humanus corporis | 5E-43 |
| caudala | 286–1412 | 5 | 967 | ref|XP_002405030.1| | Ixodes scapularis | 5E-06 |
| huckebein | 321 | 1 | 322 | ref|NP_524221.1| | Drosophila melanogaster | 9E-27 |
| buttonhead | 388 | 1 | 7 | ref|XP_004375586.1| | Trichechus manatus latirostris | 5E-59 |
| cap'n'collar | 522–3345 | 4 | 1471 | gb|EFA11557.1| | Tribolium castaneum | 5E-46 |
| knot | 652–787 | 2 | 118 | gb|EEZ99300.1| | Tribolium castaneum | 7E-108 |
| crocodile | 289–588 | 2 | 28 | emb|CAK50838.1| | Glomeris marginata | 3E-47 |
| empty spiracles | 209–1313 | 2 | 142 | ref|NP_001107793.1| | Tribolium castaneum | 3E-47 |
| orthodenticle | 226–975 | 5 | 83 | gb|AAU85255.1| | Euscorpius flavicaudis | 2E-05 |
| sloppy pairedb | 336 | 1 | 2 | ref|NP_001071091.1| | Tribolium castaneum | 1E-62 |
| Gap genes not found in T. tridentatus transcriptome: knirps and tailless. | ||||||
| Major pair rule genes | ||||||
| fushi tarazu | 240–714 | 3 | 45 | ref|XP_002406407.1| | Ixodes scapularis | 6E-17 |
| hairy | 242–2184 | 8 | 355 | gb|EFA13687.1| | Tribolium castaneum | 1E-29 |
| runt | 499–2102 | 3 | 696 | emb|CAB89493.1| | Cupiennius salei | 7E-75 |
| odd-paired | 1488 | 1 | 72 | ref|NP_001158430.1| | Saccoglossus kowalevskii | 7E-169 |
| odd-skipped | 691–2683 | 4 | 1333 | ref|XP_002399401.1| | Ixodes scapularis | 3E-79 |
| paired | 363 | 1 | 11 | gb|ACT79975.1| | Nasonia vitripennis | 5E-62 |
| sloppy pairedb | 336 | 1 | 2 | ref|NP_001071091.1| | Tribolium castaneum | 1E-62 |
| Pair rule genes not found in T. tridentatus transcriptome: even-skipped. | ||||||
| Major segment polarity genes | ||||||
| engrailed | 565–1432 | 2 | 154 | gb|AAB40144.1| | Branchiostoma floridae | 3E-48 |
| hedgehog | 224–569 | 2 | 15 | emb|CAE83646.1| | Glomeris marginata | 3E-16 |
| patched | 680–3166 | 2 | 550 | dbj|BAI83404.1| | Parasteatoda tepidariorum | 5E-103 |
| smoothened | 1739 | 1 | 170 | dbj|BAI83345.1| | Parasteatoda tepidariorum | 3E-48 |
| pangolin | 473–3616 | 3 | 711 | gb|EKC28444.1| | Crassostrea gigas | 4E-75 |
| decapentaplegic | 390–2164 | 6 | 280 | gb|AEK81570.1| | Ptychodera flava | 3E-45 |
| wingless | 1711 | 1 | 135 | ref|XP_002403230.1| | Ixodes scapularis | 6E-14 |
| cubitus interruptus | 405–1012 | 3 | 99 | ref|XP_002435743.1| | Ixodes scapularis | 6E-43 |
| frizzled | 290–1569 | 8 | 292 | ref|XP_002433979.1| | Ixodes scapularis | 6E-39 |
| Disheveled-1 | 706–1987 | 2 | 254 | ref|XP_002403641.1| | Ixodes scapularis | 1E-07 |
| shaggy | 526 | 1 | 15 | gb|EFN78376.1| | Harpegnathos saltator | 1E-08 |
| armadillo | 350–657 | 5 | 75 | gb|EKC38770.1| | Crassostrea gigas | 1E-73 |
| gooseberry | 771 | 1 | 23 | gb|EHJ65894.1| | Danaus plexippus | 2E-103 |
| Segment polarity genes not found in T. tridentatus transcriptome: invected, fused and costal 2. | ||||||
a gene grouped in both maternal effect genes and gap genes
b gene grouped in both gap genes and segment polarity genes
Case study: Detailed analysis of Pax family genes
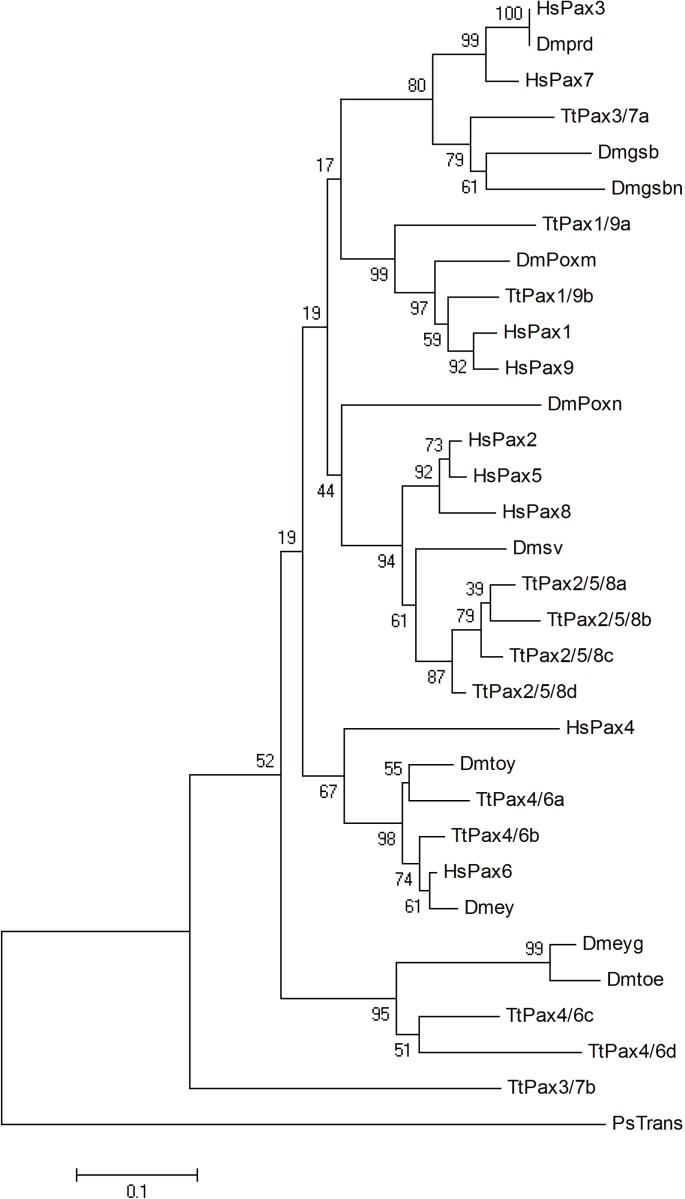
In addition to transcriptome analysis of conserved signaling pathway components and regulatory genes, we also tried to identify genes known to be important for embryogenesis based on our transcriptome data. Here, we focused specifically on the Pax family genes. Pax genes, which are grouped into 4 subfamilies (Pax1/9, Pax2/5/8, Pax3/7 and Pax4/6), encode a group of transcription factors that have been conserved through millions of years of evolution and play roles in early development [39–41]. According to the homology within the highly conserved paired domain, the putative horseshoe crab Pax genes were identified. A complete set of Pax family gene homologs was found in T. tridentatus. Phylogenetic tree was constructed by aligning the conserved paired domain with neighbor joining method using Pseudomonas transposase as outgroup (Fig 4). Genes belonging to Pax4/6 subfamily branched out at the base of the tree. Genes from Pax4/6 subfamily were divided into two clades. One included Pax4/6 genes from Homo sapiens (HsPax4 and HsPax6), D. melanogaster (Dmtoy and Dmey) and T. tridentatus (TtPax4/6a and TtPax4/6b). The second clade was composed of two sister groups with one containing the genes Dmeyg and Dmtoe from D. melanogaster, and the other consisting of TtPax4/6c and TtPaX4/6d from T. tridentatus. Interestingly, all the Dmeyg, Dmtoe, TtPax4/6a and TtPaX4/6b had a truncated paired domain. It is possible that the truncated paired domain already existed in the common ancestor of horseshoe crab and Drosophila. After split into two lineages, specific gene duplications occurred independently in horseshoe crab and Drosophila. The second clade includes Pax2/5/8 subfamily genes. All members of TtPax2/5/8 genes were gathered closely, indicating that relatively recent duplications appear to have occurred in horseshoe crabs. The third clade was composed of Pax1/9 subfamily genes. TtPax1/9a branched out at the base against all other genes in this clade, whereas TtPax1/9b gathered with the vertebrate Pax1 and Pax9 genes. The last clade was constituted of Pax3/7 subfamily. The horseshoe crab TtPax3/7a was clustered with Drosophila Dmgsb and Dmgsbn genes.
Fig 4. Phylogenetic analysis of paired domains of T. tridentatus Pax family proteins.
Neighbour joining tree of the paired domains of Pax proteins with paired deletion option. Since TtPax4/6 has only partial paired domain available, the phylogeny of Pax4/6 subfamily was constructed with complete deletion option. A pseudomonas transposase sequence (AF169828) serves as outgroup. Numbers above branches are the percentage of the trees in which the topology appears. Paired domain sequences of Pax proteins were shown in S2 Table. Tt: Tachypleus tridentatus. Hs: homo sapiens. Dm: Drosophila melanogaster.
While the majority of horseshoe crab genes of the Pax family show obvious homology to their respective subfamily, the placement of TtPax3/7b in the phylogenetic tree is ambiguous. It occupied the most basal position of the tree without obvious orthologs. The homeodomain sequence of TtPax3/7b was blasted against the GenBank database. Sequence comparison result showed that all the genes highly similar (over 78%) with TtPax3/7b homeodomain belonged to the Pax3/7 subfamily. Therefore, it is possible that TtPax3/7b genes belong to the Pax3/7 subfamily, but have been evolving at a rate that obscures its orthology.
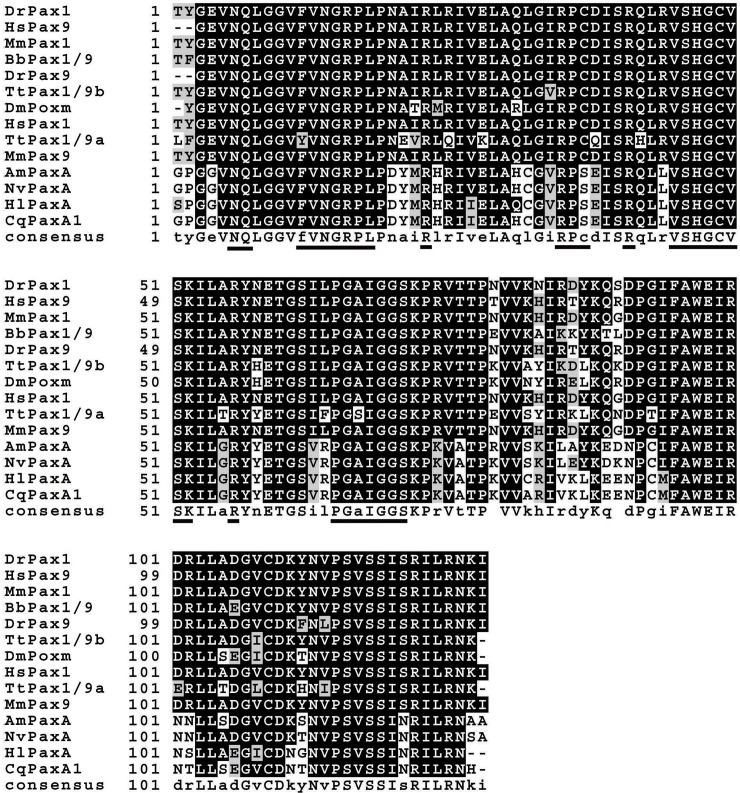
To further test the reliability of our database in gene discovery, the full length cDNAs encoding TtPax1/9a and TtPax1/9b proteins were cloned based on the partial sequences obtained from transcriptome assembly. The complete TtPax1/9a cDNA contained 258 bp of 5’ untranslated region (UTR), 681 bp of open reading frame (ORF) and 150 bp of 3’ UTR. The TtPax1/9a ORF can be translated into a polypeptide of 226 amino acids (aa) with a predicted molecular weight (MW) of 25.4 KDa. A conserved paired domain of 127 aa was included in the predicted protein sequence (S3 Fig). The full length TtPax1/9b cDNA contains 1893 bp, while the ORF is of 912 bp encoding 303 aa with a predicted MW of 33.2 KDa. A conserved paired domain with 127 aa in length is also present in the TtPax1/9b protein sequence (S4 Fig). A multiple alignment of the paired domain sequences of Pax1/9 was performed in T. tridentatus and other representative metazoan species. As shown in Fig 5, the Pax1 and Pax9 showed high similarity among all the species examined. Interestingly, although both T. tridentatus and D. melanogaster belong to the Arthropoda, TtPax1/9a and TtPax1/9b showed higher protein identity to zebrafish (Danio renio), human (Homo sapiens) and mouse (Mus musculus) which belong to Chordata, comparing to D. melanogaster. Whereas, polypeptide identities of Pax paired domain between T. tridentatus and the Coelenterata species, including Acropora millepora, Chrysaora quinquecirrha, Hydra littoralis and Nematostella vectensis, were relatively low. In summary, our data demonstrated the immediate application of the transcriptome data for gene discovery in the “non-model” organism T. tridentatus.
Fig 5. Multiple alignment of paired domain of Pax1/9 proteins from representative metazoan.
The polypeptides were aligned using ClustalX 2.1 program and Boxshade online software (http://www.ch.embnet.org/software/BOX_form.html) were employed to highlight conserved amino acids. Conserved DNA binding motifs were underlined. The sequences used in the alignment are as follows: AmPaxA: Acropora millepora PaxA [GenBank: AAC15713.2], BbPax1/9: Branchiostoma belcheri Pax1/9 [GenBank: ABK54274.1], CqPaxA1: Chrysaora quinquecirrha PaxA1 [GenBank: AAB58292.1], DmPoxm: Drosophila melanogaster Poxm [GenBank: ABE69189.1], HlPaxA: Hydra littoralis PaxA [GenBank: AAB58290.1], DrPax1: Danio rerio Pax1 [GenBank: NP_001074061.1], DrPax9: Danio rerio Pax9 [GenBank: NP_571373.1], HsPax1: Homo sapiens Pax1 [GenBank: NP_006183.2], HsPax9: Homo sapiens Pax9 [GenBank: NP_006185.1], MmPax1: Mus musculus Pax1 [GenBank: AAK01146.1], MmPax9: Mus musculus Pax9 [GenBank: NP_035171.1] and NvPaxA: Nematostella vectensis PaxA [GenBank: AAW29066.1].
Tissue distribution of Pax1/9a and Pax1/9b
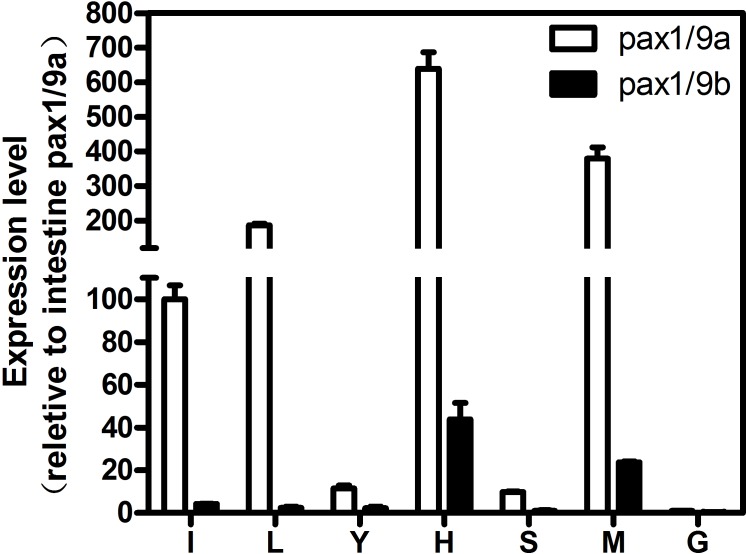
Expression patterns of TtPax1/9a and TtPax1/9b genes in different tissues of horseshoe crab were analyzed by quantitative RT-PCR. The highest expression level of TtPax1/9a was found in the heart (Fig 6). TtPax1/9a was also abundantly expressed in the muscle, liver and intestine. On the other hand, the expression level of TtPax1/9a in the stomach, yellow connective tissue and gill was significantly lower. Expression pattern of TtPax1/9b is similar to that of TtPax1/9a, albeit at much lower levels.
Fig 6. Tissue distribution of TtPax1/9a and TtPax1/9b determined by quantative real-time PCR.
The T. tridentatus GAPDH transcript was used as internal standard. I, intestine; L: liver; Y, yellow connective tissue; H, heart; S: stomach; M, muscle; G, gill.
TtPax1/9b partly rescues Poxm dysfunction-caused larval lethality of Drosophila
Due to lack of genetic manipulation tools for the gene function study in T. tridentatus, we further explore the function of TtPax1/9a and TtPax1/9b genes by taking advantage of Drosophila model. Drosophila Pax gene Pox meso (Poxm) plays a crucial role in the early development. Our previous study showed that the loss-of-function mutant in poxm, PoxmR361, causes the Drosophila death [29]. According to our protein sequence alignment data, TtPax1/9a and TtPax1/9b genes are homologs of the Drosophila poxm gene. Therefore in this experiment, we investigated whether the TtPax1/9a and TtPax1/9b genes rescue PoxmR361-caused embryonic lethality of Drosophila. The TtPax1/9a and TtPax1/9b genes were expressed in the Drosophila under the control of the 8.4 kb poxm upstream region using GAL4/UAS system [29]. The PoxmR361 mutant alone (Fig 7A) and the PoxmR361 mutant with TtPax1/9a gene expression (Fig 7B) resulted in the Drosophila death at the larval stage. Surprisingly, The PoxmR361 mutant in the presence of TtPax1/9b survived at the pupal stage (Fig 7C). These results imply that the TtPax1/9b can at least partly rescue Poxm dysfunction-caused larval lethality of Drosophila.
Fig 7. Rescue study of PoxmR361 mutant by the expression of TtPax1/9a and TtPax1/9b genes.
(a) PoxmR361 mutant. (b) PoxmR361 mutant with TtPax1/9a gene expression. (c) PoxmR361 mutant with TtPax1/9b gene expression. Anterior is to the left.
Conclusions
In this study, we, for the first time, performed de novo transcriptome sequencing of T. tridentatus embryos in the absence of a reference genome using Illumnia Solexa platform. Of 133,212 unigenes obtained, 33,796 were annotated by BLAST with NR, NT, Swiss-Prot, GO, COG and KEGG databases. We further identified a number of candidate genes potentially involved in embryonic development of T. tridentatus, shedding light on future study on the characterization and function of genes of interest and detailed molecular mechanisms underlying embryonic development of T. tridentatus. Moreover, the cloning and phylogenetic analysis of Pax family genes were performed, demonstrating that the transcriptome sequencing is a fast and reliable technology for high-throughput gene discovery in “non-model” organisms and for evolutionary developmental biology study as well. Rescue study further indicated that TtPax1/9b gene is functionally relative to Drosophila Poxm gene.
Supporting Information
All Illumina transcripts and unigenes with length over 200 bp were analyzed.
(TIF)
Transcripts were annotated using Blast2GO software. The Burrows-Wheeler Aligner (BWA) program was used for reads mapping. The color bar indicates log10 transformed count values.
(TIF)
Numbers on the left indicate numbers of nucleotides or amino acids. Boxing indicates the conserved paired domain and * stands for putative stop codon. The polyadenylation signal is underlined.
(TIF)
Numbers on the left indicate numbers of nucleotides or amino acids. Boxing indicates the conserved paired domain and * stands for putative stop codon.
(TIF)
(DOCX)
Tt: Tachypleus tridentatus, Hs: homo sapiens, Dm: Drosophila melanogaster.
(DOCX)
Acknowledgments
We thank Biomarker Technology Company (Beijing, China) for technical support. We are also grateful to Prof. Yi Tao from Xiamen University and Prof. Chaohong Weng from Jimei University for their thoughtful discussion and manuscript revision.
Data Availability
The transcriptome sequencing data is available from the NCBI Short Read Archive database (accession number SRR946952).
Funding Statement
This work was supported by marine public welfare grant 201005022 (http://www.soa.gov.cn/) to JC, COMRA research grant DY125-15-T-03 (http://www.comra.org/) to JC and grants from the National Natural Science Foundation of China 31171366 and 31271567 (http://www.nsfc.gov.cn/) to JC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, Butler R, et al. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res. 2009;37: D583–587. 10.1093/nar/gkn857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias AM. Drosophila melanogaster and the development of biology in the 20th century. Methods Mol Biol. 2008;420: 1–25. 10.1007/978-1-59745-583-1_1 [DOI] [PubMed] [Google Scholar]
- 3.Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011;331: 555–561. 10.1126/science.1197761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cahais V, Gayral P, Tsagkogeorga G, Melo-Ferreira J, Ballenghien M, Weinert L, et al. Reference-free transcriptome assembly in non-model animals from next-generation sequencing data. Mol Ecol Resour. 2012;12(5): 834–845. 10.1111/j.1755-0998.2012.03148.x [DOI] [PubMed] [Google Scholar]
- 5.Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011;12(10): 671–682. 10.1038/nrg3068 [DOI] [PubMed] [Google Scholar]
- 6.Surget-Groba Y, Montoya-Burgos JI. Optimization of de novo transcriptome assembly from next-generation sequencing data. Genome Res. 2010;20(10): 1432–1440. 10.1101/gr.103846.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7): 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Micallef G, Bicherdike R, Reiff C, Fernandes JM, Bowman AS, Martin SA. Exploring the transcriptome of Atlantic Salmon (Salmo salar) skin, a mjor defense organ. Mar Biotechno (NY). 2012;14(5): 559–569. [DOI] [PubMed] [Google Scholar]
- 9.Zhao X, Wang Q, Jiao Y, Huang R, Deng Y, Wang H, et al. Identification of genes potentially related to biomineralization and immunity by transcriptome analysis of pearl sac in pearl oyster Pinctada martensii. Mar Biotechnol (NY). 2012;14(6): 730–739. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Jiang J, Chen S, Qi X, Peng H, Li P, et al. Next-generation sequencing of the Chrysanthemum nankingense (Asteraceae) transcriptome permits large-scale unigene assembly and SSR marker discovery. PLoS One. 2013;8(4): e62293 10.1371/journal.pone.0062293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vidotto M, Grapputo A, Boscari E, Barbisan F, Coppe A, Grandi G, et al. Transcriptome sequencing and de novo annotation of the critically endangered Adriatic sturgeon. BMC Genomics. 2013;14(1): 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng XL, Liu M, Jiang KY, Wang BJ, Tian X, Sun SJ, et al. De Novo Characterization of Japanese Scallop Mizuhopecten yessoensis Transcriptome and Analysis of Its Gene Expression following Cadmium Exposure. PLoS One. 2013;8(5): e64485 10.1371/journal.pone.0064485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugita H, Sekiguchi K. Horseshoe crab developmental studies II. Physiological adaptation of horseshoe crab embryos to the environment during embryonic development. Prog Clin Biol Res. 1982;81: 75–82. [PubMed] [Google Scholar]
- 14.Sekiguchi K, Yamamichi Y, Costlow JD. Horseshoe crab developmental studies I. Normal embryonic development of Limulus polyphemus compared with Tachypleus tridentatus. Prog Clin Biol Res. 1982;81: 53–73. [PubMed] [Google Scholar]
- 15.Harzsch S, Vilpoux K, Blackburn DC, Platchetzki D, Brown NL, Melzer R, et al. Evolution of arthropod visual systems: development of the eyes and central visual pathways in the horseshoe crab Limulus polyphemus Linnaeus, 1758 (Chelicerata, Xiphosura). Dev Dyn. 2006;235 (10): 2641–2655. [DOI] [PubMed] [Google Scholar]
- 16.Mittmann B. Early neurogenesis in the horseshoe crab Limulus polyphemus and its implication for arthropod relationships. Biol Bull. 2002;203(2): 221–222. [DOI] [PubMed] [Google Scholar]
- 17.Walls EA, Berkson J, Smith SA. The horseshoe crab, Limulus polyphemus: 200 million years of existence, 100 years of study. Rev Fish Sci. 2002; 10(1): 39–73. [Google Scholar]
- 18.Kamaruzzaman BY, Akbar JB, Zaleha K, Jalal KCA. Molecular Phylogeny of Horseshoe Crab. Asian Journal of Biotechnology. 2011;3: 302–309. [Google Scholar]
- 19.Rudkin DM, Young GA, Nowlan GS. The oldest horseshoe crab: a new xiphosurid from Late Ordovician Konservat-Lagerstätten deposits, Manitoba, Canada. Palaeontology. 2008;51(1): 1–9. [Google Scholar]
- 20.Fisher DC. The Xiphosurida: archetypes of bradytely? In: Eldredge N, Stanley SM editors. Living Fossils. New York: Springer; 1984. pp. 196–213. [Google Scholar]
- 21.Cooper JF, Pearson SM. Detection of endotoxin in biological products by the limulus test. Dev Biol Stand. 1997;34: 7–13. [PubMed] [Google Scholar]
- 22.Botton ML. The Ecological Importance of Horseshoe Crabs in Estuarine and Coastal Communities: A Review and Speculative Summary In: Tanacredi JT, Botton ML, Smith DR, editors. Biology and Conservation of Horseshoe Crabs. New York: Springer; 2009. pp. 45–64. [Google Scholar]
- 23.Sekiguchi K. Biology of horseshoe crabs Portland: International Specialized Book Service Incorporated; 1988. [Google Scholar]
- 24.Sekiguchi K, Seshimo H, Sugita H. Post-embryonic development of the horseshoe crab. Biological Bulletin. 1988;174(3): 337–345. [Google Scholar]
- 25.Mittmann B, Scholtz G. Development of the nervous system in the "head" of Limulus polyphemus (Chelicerata: Xiphosura): morphological evidence for a correspondence between the segments of the chelicerae and of the (first) antennae of Mandibulata. Dev Genes Evol. 2003;213(1): 9–17. [DOI] [PubMed] [Google Scholar]
- 26.Smith SA, Berkson J. Laboratory culture and maintenance of the horseshoe crab (Limulus polyphemus). Lab Anim (NY). 2005;34(7): 27–34. [DOI] [PubMed] [Google Scholar]
- 27.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7): 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5(2): 150–163. [DOI] [PubMed] [Google Scholar]
- 29.Duan H, Zhang C, Chen J, Sink H, Frei E, Noll M. A key role of Pox meso in somatic myogenesis of Drosophila. Development. 2007;134: 3985–3997. [DOI] [PubMed] [Google Scholar]
- 30.Del Valle Rodriguez A, Didiano D, Desplan C. Power tools for gene expression and clonal analysis in Drosophila. Nat Methods. 2011;9(1): 47–55. 10.1038/nmeth.1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pires-daSilva A, Sommer RJ. The evolution of signalling pathways in animal development. Nat Rev Genet. 2003;4(1): 39–49. [DOI] [PubMed] [Google Scholar]
- 32.Davidson EH, Levine MS. Properties of developmental gene regulatory networks. Proc Natl Acad Sci U S A. 2008;105(51): 20063–20066. 10.1073/pnas.0806007105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaeger J, Surkova S, Blagov M, Janssens H, Kosman D, Kozlov KN, et al. Dynamic control of positional information in the early Drosophila embryo. Nature. 2004;430(6997): 368–371. [DOI] [PubMed] [Google Scholar]
- 34.Wunderlich Z, DePace AH. Modeling transcriptional networks in Drosophila development at multiple scales. Curr Opin Genet Dev. 2011;21(6): 711–718. 10.1016/j.gde.2011.07.005 [DOI] [PubMed] [Google Scholar]
- 35.Jaeger J. The gap gene network. Cell Mol Life Sci. 2011;68(2): 243–274. 10.1007/s00018-010-0536-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroeder MD, Pearce M, Fak J, Fan H, Unnerstall U, Emberly E, et al. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2(9): E271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilbert SF. Developmental biology 8th ed. Oxford, UK: Sinauer Associates Inc; 2006. [Google Scholar]
- 38.Twyman R. BIOS Instant Notes in Developmental Biology Oxford, UK: Taylor & Francis Group; 2001. [Google Scholar]
- 39.Blake JA, Ziman MR. Pax genes: regulators of lineage specification and progenitor cell maintenance. Development. 2014;141(4): 737–751. 10.1242/dev.091785 [DOI] [PubMed] [Google Scholar]
- 40.Lang D, Powell SK, Plummer RS, Young KF, Ruggeri BA. PAX gene: role in development, pathophysiology, and cancer. Biochem Parmacol. 2007;73(1): 1–14. [DOI] [PubMed] [Google Scholar]
- 41.Noll M. Evolution and role of Pax genes. Curr Opin Genet Dev. 1993;3(4): 595–605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All Illumina transcripts and unigenes with length over 200 bp were analyzed.
(TIF)
Transcripts were annotated using Blast2GO software. The Burrows-Wheeler Aligner (BWA) program was used for reads mapping. The color bar indicates log10 transformed count values.
(TIF)
Numbers on the left indicate numbers of nucleotides or amino acids. Boxing indicates the conserved paired domain and * stands for putative stop codon. The polyadenylation signal is underlined.
(TIF)
Numbers on the left indicate numbers of nucleotides or amino acids. Boxing indicates the conserved paired domain and * stands for putative stop codon.
(TIF)
(DOCX)
Tt: Tachypleus tridentatus, Hs: homo sapiens, Dm: Drosophila melanogaster.
(DOCX)
Data Availability Statement
The transcriptome sequencing data is available from the NCBI Short Read Archive database (accession number SRR946952).