Abstract
With recent technological advances at synchrotrons [Graber et al., J. Synchrotron Radiat. 18, 658–670 (2011)], it is feasible to rapidly collect time-resolved crystallographic data at multiple temperature settings [Schmidt et al., Acta Crystallogr. D 69, 2534–2542 (2013)], from which barriers of activation can be extracted. With the advent of fourth generation X-ray sources, new opportunities emerge to investigate structure and dynamics of biological macromolecules in real time [M. Schmidt, Adv. Condens. Matter Phys. 2013, 1–10] in crystals and potentially from single molecules in random orientation in solution [Poon et al., Adv. Condens. Matter Phys. 2013, 750371]. Kinetic data from time-resolved experiments on short time-scales must be interpreted in terms of chemical kinetics [Steinfeld et al., Chemical Kinetics and Dynamics, 2nd ed. (Prentience Hall, 1985)] and tied to existing time-resolved experiments on longer time-scales [Schmidt et al., Acta Crystallogr. D 69, 2534–2542 (2013); Jung et al., Nat. Chem. 5, 212–220 (2013)]. With this article, we will review and outline steps that are required to routinely determine the energetics of reactions in biomolecules in crystal and solution with newest X-ray sources. In eight sections, we aim to describe concepts and experimental details that may help to inspire new approaches to collect and interpret these data.
I. CONFORMATIONAL ENTROPY AND ENTHALPY
Determination of the energetics of a chemical reaction is synonymous with the determination of the free energy surface on which that reaction proceeds. The energy of an instantaneous structure of a single protein molecule can be considered as a point on a hypersurface called configurational energy surface with dimension 3N, with N the number of atoms in a molecule plus their weakly interacting solvent molecules. Each point on the surface corresponds to a different 3-dimensional arrangement of the atoms. The conformational free energy, G (free energy in short, Eq. (1), is an ensemble property. If the molecules distribute approximately in an even potential , which is assumed for simplicity, the configurational energy of the mean structure represents the energy minimum, which is the conformational enthalpy. The difference between the enthalpy of formation of the macromolecule and the conformational enthalpy is that the conformational enthalpy does not consider covalent bonds that hold the primary structure together and rather considers non-covalent interactions that are responsible for the tertiary and quaternary structure formation. G also contains the conformational entropy S and the temperature T
| (1) |
A single configuration on the configurational energy surface is not sufficient to determine the conformational entropy. The absolute conformational entropy is given by10
| (2) |
where S may be given either in units of the Boltzmann constant kB [1.381 × 10−23 J K−1] or the gas constant R [8.314 J mol−1 K−1]. is the probability density to find a molecule with configuration on the energy surface. A sufficiently large area on that surface must be sampled to evaluate the integral (Eq. (2)), which can be challenging.10 Methods have been developed to estimate the absolute conformational entropy from theoretical calculations such as molecular dynamics simulation10,11 or from experiment: Conformational entropy has been estimated from crystallographic B-factors assuming that the entire mean square deviation is caused by dynamic fluctuations,12,13 from nuclear magnetic resonance order parameters,14 or from incoherent neutron scattering15 that provide dynamic mean square deviations 〈x2〉dyn. For small proteins, the absolute conformational entropy is between 10 000 J mol−1 K−1 (Refs. 10 and 16) and 60 000 J mol−1 K−1,12 for larger proteins it can be more than 100 kJ mol−1 K−1, which is enormous. The absolute free energy of a state, therefore, consists of the conformational enthalpy and the conformational entropy as a measure of structural variability.
II. ENZYMATIC REACTION PATHWAYS
Proteins as one group of biological macromolecules may catalyze chemical reactions that take place in the active center of these molecules. Proteins are flexible and change their shape or conformation during the course of a reaction. Along the reaction, a protein molecule migrates on certain, energetically allowed pathways through conformational space (Fig. 1). These pathways are pre-determined by the structure of the protein and are therefore sometimes referred to as the reaction coordinate or the enzymatic reaction pathway.17,18 The migration along these pathways is decisively determined by the local configurational energies and the number of microstates accessible at each point on the hypersurface. Both, conformational entropies and enthalpies play major roles in the description of these processes.19
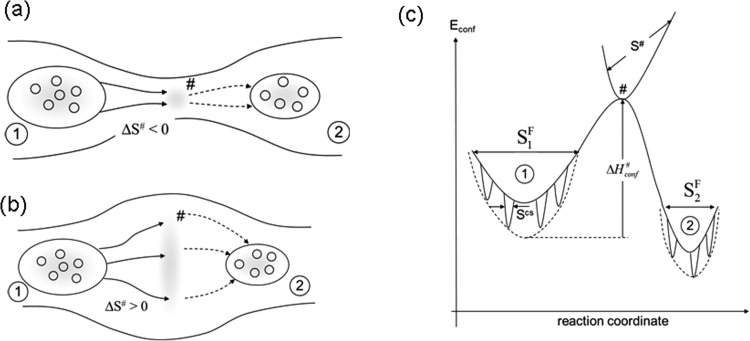
FIG. 1.
Reaction coordinates of a general reaction (schematic). (a) and (b) two dimensional projection of the conformational hypersurface, iso-energy lines are schematically shown. Migration pathways of molecules through configurational space shown as arrows. (a) Transition state # has low entropy; reaction proceeds through a narrow path, ΔS# is negative. (b) Similar to (a), transition state # has a larger entropy, the molecules are more flexible at the transition state, ΔS# is positive. (c) One dimensional projection of the configurational energy hypersurface with energy Econf along the reaction coordinate. S1F: conformational entropy of state 1 in the flexible state, S2F: conformational entropy of state 2 in the flexible state, Scs: conformational entropy of conformational substates. Here, S1F > S2F > Scs and reaction entropy ΔS is negative. S#: entropy of transition state. ΔH#conf: change of conformational enthalpy from state 1 to the transitions state.
Protein folding is only one form of reaction that proceeds through energy landscapes. Conformational free energy differences between folded and unfolded states of a typical protein can be determined from melting curves20 or even estimated from chain lengths.21 Conformational enthalpy differences, ΔH, are on the order of 500 kJ/mol for a medium sized protein, conformational entropy differences, ΔS, are around 1000 J mol−1 K−1.20,22 This means that at physiological temperatures a few hundred kJ/mol of energy has to be used to break non-covalent bonds of a small protein to go from the folded to the unfolded state, and simultaneously a similar amount of energy is available through the gain of conformational entropy.23 The typical free energy difference ΔG (Eq. (3)) of folding is consequently small on the order of 30–50 kJ mol−1 in favor of the folded state
| (3) |
III. SHORT LIVED (TRANSIENT) STATES
A transient state is a synonym for an intermediate state that is shortly (transiently) populated by reacting molecules. Transient states are energy minima (holes) in configurational space, which are subsequently occupied by reacting molecules. The conformational enthalpy can be considered as the depth of the minimum and the entropy is equivalent to the width. A transient state represents an ensemble of molecules, all with approximately the same structure. Emphasis lies here on approximately since the structures of individual molecules are slightly different and exhibit deviations form a mean structure in the same way as is the case for stable equilibrium states. Molecules will voluntarily migrate from state to state when the difference of free energy between the states, ΔG, is negative. The reaction is exergonic. Endergonic reactions are made possible in biology by coupling them to hydrolysis of energy rich compounds such as ATP or GTP.
The free energy depends on the temperature (Eq. (1)). Equation (3) shows that the direction of a reaction cannot simply be changed by changing the temperature unless there is an entropy difference ΔS. Structural flexibility and, connected to this, the conformational entropy is dependent on the environment. When, for example, the solvent in which the protein is embedded becomes more mobile, the protein may exhibit a larger phase space and its conformational entropy increases. Populations of molecules previously in equilibrium may then change with temperature. A descriptive example is again the situation when the protein melts. The denatured state above the melting temperature Tm exhibits less interactions between the atoms and is disordered. Accordingly, it has a more positive conformational enthalpy (=less bonds) and also a larger conformational entropy compared to the folded state below Tm. At a temperature above Tm, the reaction proceeds towards the melted state, below Tm the protein folds. There are a number of experimental techniques that can contribute to the assessment of free energy surfaces in biological macromolecules by either determining enthalpies, entropies or both. The most frequently used techniques are based on spectroscopic methods such as UV/Vis absorption spectroscopy,24,25 Moessbauer spectroscopy,4,5,13,26 and nuclear magnetic resonance.14 However, also scattering methods, such as transient grating light scattering,27 incoherent Neutron scattering,15 and X-ray diffraction,4,28 are used to extract thermodynamic properties of biological macromolecules to determine free energy surfaces.
IV. EQUILIBRIUM DYNAMICS AND ITS RELATIONSHIP TO TRANSIENT STATE DYNAMICS
Dynamic properties of proteins at thermal equilibrium can be characterized by methods that allow for the dynamic autocorrelation function to be determined for a structural probe at a time dependent position in the molecule. c(τ) is the average of the position of the probe at time t dotted into the position of the same probe at time t + τ. Once c(τ) is determined accurately, the dynamic mean square displacement can be determined from it13 and from this the conformational entropy.12 Typically, c(τ) is measured by methods that probe an ensemble incoherently, such as inelastic, incoherent neutron scattering,29 or Moessbauer spectroscopy.13 Incoherent means that scattering or absorption of one molecule in the ensemble has no (or an irrelevant) phase relation to a similar event in another molecule in the ensemble. If the dynamics is governed by phonons, c(τ) oscillates. On the other hand, non-oscillatory fluctuations exist which can be described by structural relaxations. The conformation is disturbed by random forces, for example, by fluctuating solvent molecules in the sample, and the structure adjusts slowly. Relaxations can be described mathematically by overdamped oscillations. Then the c(τ) decays exponentially within a characteristic time τrelax which can be quite long if the system is strongly overdamped. The appearance of τrelax requires that the method measures over a sufficiently large time τexp that τrelax can be accurately determined.30 The reciprocity between τexp and energy automatically means that the method must have energy resolution. A relaxation with τrelax that is much longer than τexp must be considered static for the method. τexp is ∼140 ns for 57Fe Moessbauer spectroscopy with an energy resolution (=line width) Γ on the order of 5 neV. For inelastic, incoherent neutron scattering experiments, τexp is a few picoseconds with an energy resolution of μeV.29 Accordingly, different time scales are explored with different methods30 that provide different estimates how much of conformational space is sampled by individual molecules during the time τexp. The important outcome is the dynamic mean square displacement characteristic to the method. During the limited time τexp, the individual molecules may not be able to sample the entire accessible conformational space and the may be an inadequate measure of the absolute conformational entropy.
Frauenfelder and colleagues31–34 described how protein conformational substrates are hierarchically arranged within a conformation. Substrates represent a multitude of slightly different minima below the level of the top-tier conformation (Fig. 1), each minimum with a smaller width than the width of the hierarchically higher conformation. This means that each minimum represents an ensemble of molecules having restricted conformational entropy. Temperature dependent Moessbauer spectroscopic investigations can shine light on the nature of substrates, since these investigations are able to extract a dynamic mean square displacement, , through the Lamb-Moessbauer factor. The observed below a characteristic temperature of Tc ∼ 180 °C is small. The can be explained by a normal mode analysis performed on the atomic structure of the protein.26,35–37 The normal mode analysis provides an estimate for the conformational entropy Sconf.10 When the temperature increases, the molecules pass over to a more flexible state. This transition is known as the dynamic transition. Harmonic oscillations that are at the base of a normal mode analysis are not sufficient to explain the . Diffusive motions described by relaxations in restricted space dominate the mean square displacement.5 At elevated temperature, a protein molecule can access and explore a larger phase space by populating a multitude of protein substrates by means of relaxations in the flexible state. The difference of conformational entropy of such a transition from a narrow structural distribution in the substrate to the broader distribution in the flexible state is known for certain proteins (Table I).4,5 It is on the order of 100 J mol−1 K−1. At 300 K, the entropy contributes ∼30 kJ mol−1 to the conformational free energy. It helps to keep the molecules in a flexible state ready to perform their function. At low temperatures, the entropic contribution to the free energy is small. As a consequence, the molecules stay in the rigid state and are non-functional. The free energy change of the dynamic transition from the rigid to the flexible state is relatively small (on the order of 1–5 kJ/mol at 30 °C, see Table I), since entropic and enthalpic contributions to the dynamic transition almost cancel. So far, no obvious differences in the dynamic transition temperature between molecules in the crystal and in solution are observed.5,26,29 The dynamic transition has similarities to melting although Tc (∼180 K) is much smaller than Tm (∼350 K for photoactive yellow protein for example): Tc as well as Tm are points where ΔH and TΔS are equal. In both cases, entropy and enthalpy differences are positive when going from lower to higher temperatures. The difference is, however, that at temperatures above Tc diffusive motions in restricted space dominate the protein's 〈x2〉dyn, whereas above Tm the molecules lose their structural integrity altogether.
TABLE I.
Thermodynamic parameters ΔH, ΔS, and ΔG (at 300 K) of the dynamic transition in heme proteins. The free energy is negative at 300 K, which means that the molecular population shift from the rigid states (CS) to the flexible state.
A reaction scheme such as the one depicted in Fig. 1 must be augmented by protein conformational substrates (CS) which are the small, local minima within a state's potential. For a reaction to occur, the molecules lift out of the CS and either surmount the barrier of activation (see below) or explore other CS. At lower temperatures, the CS decisively determines the time-dependence of the reaction, which becomes non-exponential.31 ΔH# distributes along a range of energies. At temperatures higher than ∼200 K (Ref. 26) kinetic averaging, also called motional narrowing, restores exponential behavior.26,38,39
V. TIME-RESOLVED TECHNIQUES TO EXPLORE CONFORMATIONAL SPACE
If a reaction is initiated in biological macromolecules, its progress can be followed by time-resolved techniques. Typical examples are mixing of substrate and enzymes or photoinitiating a process in a photoresponsive protein using an intense Laser pulse. For enzymes the difference of free energy between the substrate and the product will drive the reaction. In photoinitiated processes, a large amount of energy is injected into the system by the Laser pulse that dissipates through the reaction. The energy injected is usually absorbed by a central chromophore whose temperature can rise by several hundreds of K.40 This energy is gradually released through different processes. A substantial amount of this energy is dissipated rapidly into the phonon modes of motion and results in a moderate adiabatic temperature change of the protein. For example, the activation of the photocycle in the photoactive yellow protein (PYP, see Fig. 2) requires an energy of 245 kJ/mol which is the energy of the blue photon that excites the central chromophore p-coumaric acid (PCA). With a molar mass of 14.7 kg/mol and assuming a heat capacitance of a typical protein being 5 kJ kg−1 K−1, the adjabatic temperature rise of a single PYP molecule would be , given that all the energy absorbed locally by the chromophore dissipates in the phonon modes of motion of the entire protein. However, in PYP, a substantial amount of the photon energy is used to twist the chromophore into a geometrically unfavorable configuration (Fig. 2(a)). The structural mechanism how this is achieved is subject to extensive theoretical and experimental work.6,9,41–48
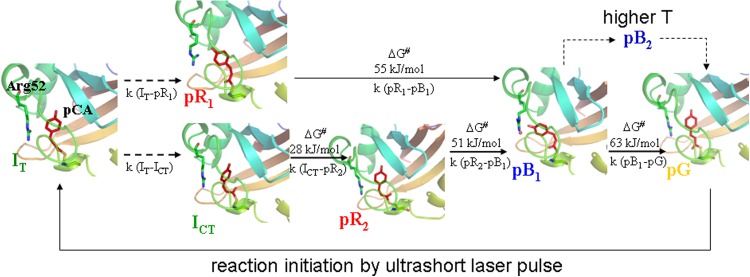
FIG. 2.
PYP photocycle from a time-resolved crystallographer's perspective. The early intermediate IT crosses over to pR1 and ICT in a hula-twist and a bicycle pedal motion, respectively. ICT decays to pR2. pR2 and pR1 both react to pB1 that may decay directly to the dark state. At higher temperatures pB2 accumulates in addition. Mechanism is simplified by omitting less important transition pathways; see Ref. 2 for a more comprehensive mechanism. Free energies of the barriers of activation are shown as determined from five-dimensional crystallography for several transitions.
PYP features a quintessential photocycle, with a number of intermediates on time-scales from femtoseconds49,50 to seconds (Fig. 2). The photocycle is active and can be investigated both in crystal and in solution. PYP and its photocycle were discovered in the 90th of the last century.51 Since then, a large number of publication report extensive details of the photocycle determined with methods with increasingly better time resolution. Initial time-resolved investigations were all performed using time-resolved spectroscopy in solution.52–56 After the structure of PYP has been determined,57 time-resolved crystallographic techniques were used to investigate the photocycle. First, the decay of a photostationary state produced by a long blue laser pulse (λ = 496.5 nm, 200 ms) was investigated on the millisecond time-scale with a single map collected with a 10 ms shutter opening from 2 to 12 ms.58 That revealed the structure of the intermediate we know now as pB. With the advent of a global analysis based on the singular value decomposition (SVD),59 the entire photocycle can be kinetically characterized using a comprehensive time-series of time-resolved X-ray data.1,47,48,60 Recently, even the picosecond time-scale has been covered by two different publications.6,9 Here, the results from Jung et al.9 are referred to, since in the other study crystals were grown in unusual conditions which might have changed to outcome. The structures of the photocycle intermediates IT, ICT, pR1 (formerly pRE46Q), pR2 (formerly Icw), pB1 and pB2 are now known (see Fig. 2) together with a plausible chemical, kinetic mechanism. In solution time-resolutions as good as 8 fs are reached.61,62 However, with spectroscopy, structural information is sparse.63 There are comparative investigations on crystal and solution on the μs to ms time scales.64,65 The former study investigated the decay of a photostationary state at several temperatures to gain information on barriers of activation of that decay. In the latter study, single laser pulses were used to initiate the PYP photocycle and data were collected at a single temperature (15 °C). Results were comparatively analyzed on a time range from 1 μs to 150 ms by kinetic target analysis.50 Both studies found kinetic differences between crystal and solution. The most striking difference is that the photocycle lasts about a factor 5 longer in solution than in the crystal. Obviously, barriers of activation in solution and in the crystal may be very different.
VI. BARRIERS OF ACTIVATION
Barriers of activation represent saddle points on the conformational energy surface that connect two adjacent states (Fig. 1). They decisively determine the magnitude of the microscopic rate coefficients k of a reaction. The transition state theory66 accounts for the magnitude of k according to
| (4) |
where Na is the Avogadro number, h is the Planck constant, R is the gas constant, ΔG# is the free energy difference from a state to the transition state, and ΔH# and ΔS# are the enthalpic and entropic contributions, respectively. The saddle point that represents the transition state in Fig. 1 is denoted by a double dagger (#). Similar to transient states a transition state can be considered to have its own phase space with its own conformational entropy S#. Transition states are notoriously difficult to characterize because their occupations are extremely low during a reaction. If, for example ΔG# is 50 kJ/mol (see, e.g., the transition from pR to pB in Fig. 2), the probability to find a molecule on top of the barrier of activation is only . Such an occupation is not measurable in any time-resolved experiment. However, transient states before and after the barrier can be characterized and the properties of the barrier deduced from this. Since the entropy change ΔS# is included in Eq. (4), one can estimate the number of microstates ΔQ that are accessible at the transition state # compared to those in the transient states that flank the barrier by
| (5) |
where R is the gas constant and ΔS# is measured in J mol−1 K−1. ΔQ might be smaller or larger depending whether ΔS# is negative (number is smaller, Fig. 1(a)) and positive (number is larger, Fig. 1(b)). A larger number as in Fig. 1(b) will accelerate a reaction, since the transition state offers a large number of microstates that can be reached through a multitude of pathways. This makes the reaction more probable. If, however, a reaction is confined to a narrow pathway, the entropy difference is negative. The reaction slows down because it is less probable that the molecules thread through a narrow, confined path (Fig. 1(a)). It is important to notice that barriers of activation have to be determined for true chemical rate coefficients. Apparent rates Λ that are observable in a kinetic experiment are linear combinations of the true rate coefficients.8 The temperature dependence of the Λ can be fit with the Van't Hoff Arrhenius equation to determine an energy of activation Ea and a prefactor ν, which can be understood as the number of attempts to surmount the barrier Ea. The Λ are fingerprints of a reaction, since they are, as mentioned, linear combination of k whose temperature dependences might be very different.
The appearance of diffusive motions in restricted space (see above) suggests that Kramer's theory67 instead of the mentioned transition state theory should be used to describe the temperature dependence of the rate coefficients.68 In Kramer's equation,67 the prefactor in Eq. (4) is parameterized differently. Pre-factors from Kramer's equation are much smaller with correspondingly longer time-scales.26,68 Parak et al.26 modified Kramer's equation to contain the factor α0, which is proportional to , as the prefactor (Eq. (6)). α0 can be obtained in a Moessbauer experiments from the linewidth of the broad Lorentzian that underlies the Moessbauer spectrum provided all molecules have left the conformational substrates and are in the flexible state
| (6) |
For α-helical as well as for β-sheet small proteins, such as myoglobin and nitrophorin, respectively, α0 is known to be ∼40 mm/s derived from 57Fe Moessbauer spectra,4 which corresponds to about 2000 neV that must be used in Eq. (6). From this a prefactor of ∼5 × 108 1/s is obtained. in Eq. (4) is 6 × 1012 1/s at 300 K. The difference in fitted values of ΔS#/R between Kramer's theory and the transition state theory is on the order of ln(104) = 9. The ΔS# itself deviates by ∼70 J mol−1 K−1, which is relatively large. This has to be used as a caveat and trends rather than absolute values must be considered. The small prefactor in Kramer's equation also implies that transitions faster than about 2 ns should reveal their diffusive nature and might deviate from simple exponential behavior. There are indeed time-resolved crystallographic and time-resolved spectroscopic photoflash experiments on myoglobin at room temperature where the initial, fast kinetic phase is non-exponential in Refs. 69–71. Detailed experiments that structurally probe the fast picosecond time-regime are necessary for these small proteins. Relaxations and barrier crossings last even longer in larger proteins,68 and the existing theories might need to be extended to describe their kinetics.72 However then, a molecular movie comes into reach that experimentally determines a trajectory through conformational space from an ensemble of reacting molecules.
VII. TRANSIENT STATE KINETIC EXPERIMENTS ON PYP TO DETERMINE BARRIERS OF ACTIVATION IN THE PHOTOCYCLE
Attempts to determine of barriers of activation in the PYP photocycle are sparse. All attempts so far used the transition state equation (Eq. (4)) or the Van't Hoff Arrhenius equation to fit the temperature dependence of the processes in the photocycle. Van Brederode et al., 199625 report barriers including entropy and enthalpy differences for the pR to pB and the pB to pG transition (Table II), and Ng et al.65 report energies of activation for the decay of a photostationary state to pG determined from single crystal and solution spectroscopy (see also Table II). Recently, a time-resolved crystallographic experiment was performed by collecting comprehensive time-series from 2 ns to seconds at 14 different temperatures from −40 °C to +70 °C.2 Global analysis with SVD and subsequent kinetic target analysis (posterior analysis) was used to extract apparent rates Λi for i = 1–3 processes as well as true rate coefficients of the mechanism shown in Fig. 2. Since the X-ray data were collected at 14 temperature settings, the temperature dependence of the rates and true rate coefficients could be used to determine activation energies, as well as entropy and enthalpy difference to transition states for six different rate coefficients. Since five variables, time, temperature, and space are involved, this method is called five-dimensional crystallography.73 The following picture emerges: Spectroscopically, in crystals, the transition of pB to pG is biphasic.2,64 Also the decay of a photostationary state to pG in crystals is biphasic with energies of activation of 23 kJ/mol and 48 kJ/mol for a fast and a slow phase respectively.65 With five-dimensional crystallography, only the slow phase is observed at T < 50 °C with Ea = 49.6 kJ/mol.2 pB1 is the dominant species.58 In solution the decay of the photostationary state is slower with Ea being about 6 kJ/mol larger than in the crystal.65 This trend can also be observed when the reaction is initiated by a single laser pulse. In solution, the pB to pG barrier ( ) is about 6 kJ/mol larger than that obtained in crystals from five-dimensional crystallography (see Table II). The pR to pB transition on the other hand is similar in the crystal and in solution. In the crystal, there are two pR species (pR1 and pR2). Barrier heights for the pR to pB transitions are = 54 kJ/mol and = 51 kJ/mol, respectively. Since the latter (pR2) is the species that is most populated with an approximate pR2 to pR1 ratio of 3:1, it dominates the transition. In solution is also 51 kJ/mol. Up to the pR relaxation the photocycles in crystal and solution show similar energetics. The energetics of even earlier intermediates such as ICT is only available to date from five-dimensional crystallography. The free energy of the ICT to pR2 transition is 28 kJ/mol (Fig. 2) which accounts for the much faster decay of ICT. So far, a consistent picture emerges, with the energetics in crystal and solution being similar except for the pB to pG transition. Apart from the difference of 6 kJ/mol between the barrier in the crystal and solution, significant differences emerge when entropy and enthalpy differences are inspected for the final pB to pG transition. In solution is only 9 kJ/mol. If the entropy would not play a role ( # ∼ 0), at 283 K (10 °C) pR would decay within 8 ps. However, is −196 J mol−1 K−1.25 As a result, at 283 K the rate coefficient for the pB decay is 7.5 s−1 with a characteristic time of 134 ms. It is the entropy of the transition state which decisively slows down the reaction. Since the entropy difference is negative, the transition state is much more ordered than the pB state itself. In solution, PYP refolds from a disordered structure with high conformational entropy through a well ordered transition state with a lower conformational entropy. In the crystal, the situation is different. There, is positive (42 J mol−1 K−1). The transition state is more disordered than pB, which helps to accelerate the reaction. From the crystallographic data, the reason for this becomes clear. In pB, the chromophore has swung out to the solvent and is bound to Arg52 and to one or two water molecules by hydrogen bonds. For reisomerization, it has to swing back into the chromophore pocket. First the hydrogen bonds have to be broken, and some rotation about the double bond is necessary. This requires substantial energy of about 75 kJ/mol. The protein then provides an enlarged, relaxed chromophore pocket so that this reisomerization is facilitated.
TABLE II.
Thermodynamic parameters obtained from solution and crystal for barriers of activation for the pR to pB and the pB to pG transition by using Eq. (4). The activation energies (from the slopes of Arrhenius plots) of the decay of a photostationary state and for the pB to pR decay from single pulse experiments are also available.
| Solution a | Crystal b | ||
|---|---|---|---|
| pR‐pB | pR1-pB1 | pR2-pB1 | |
| ΔH# [kJ/mol] | 66 | 50.0 | 48.1 |
| ΔS# [J mol−1 K−1] | 51 | −15 | −10.2 |
| ΔG# [kJ/mol] at 300 K | 50.7 | 54.5 | 51.2 |
| pB-pG | pB1-pG | ||
| ΔH# [kJ/mol] | 9.2 | 75.2 | |
| ΔS# [J mol−1 K−1] | −196 | 41.9 | |
| ΔG# [kJ/mol] at 300 K | 69 | 63 | |
Compared to the enormous conformational entropy of the entire protein (see above), entropy differences to the transition states are small. Protein structures may have been evolved that relatively small local entropy changes control catalytic reactions. It is interesting to note that although kinetic differences between crystal and solution are relatively large, free energies of activation differ by only a few kJ/mol. It would be highly desirable to determine the solution structure and follow its decay with time-resolved methods capable of determining structural changes.
VIII. TIME-RESOLVED STRUCTURES FROM SOLUTION
With the advent of fourth generation X-ray sources such as the Linac Coherent Light Source (LCLS) in Stanford, the determination of protein structures from solution have come within reach.7,74,75 The coherent X-ray beam from these machines can be focused so much that the beamsize matches the size of a typical protein such as hemoglobin. The photon density becomes so large that a substantial number of photons can be scattered from a single molecule. Beamsizes as small as 100 nm are already achieved at the LCLS. Attempts to reduce the beam size further down are under way. In Fig. 3(c)), a beamsize of 10 nm is assumed into which 1012 photons are focused, so the photon density is about 1010 photons/nm2. The total scattering cross section of a PYP molecule is about 5000 × 10−10 nm2. Accordingly, a total number of 5000 photons would be scattered per single PYP. However, it is exceptionally difficult to hit a single PYP molecules with such a small X-ray beam (Fig. 3(a)). One way out is to prepare an ensemble in a larger liquid jet (Fig. 3(b)) and accept that more than one PYP molecule scatters. The volume of the shaded area V in Fig. 3(c) is 2.4 × 10−20 L. 40 mg/ml PYP corresponds to 16.4 × 1020 molecules per Liter. Accordingly, V contains about 40 molecules of PYP that would diffract coherently. The question is, can the diffraction volume of a single PYP molecule be retrieved from the coherent scattering of this small ensemble? The answer is yes, subject to certain conditions. A recent approach7,75 used pairwise correlations C2 = between intensities at q, I(q), and intensities I(q′,Δϕ) measured at q' an angular distance Δϕ apart in the diffraction pattern (Fig. 4(b)). It can be shown75,76 that the pairwise intensity correlation of an ensemble is the same as the pairwise intensity correlation of a single molecule. Each data set may contain millions of diffraction pattern such as the one shown in Fig. 4(b). The angular correlations found in each individual diffraction pattern are averaged over all diffraction patterns. Since only correlations are used, an additional phase problem emerges that can be solved for a number of scenarios. (i) High symmetry imposes strong constraints, so that a unique solution can be found.76–79 (ii) The existence of a reference structure in a time-resolved experiment allows structure determinations to be made independent of any symmetry of the structure.7,75 Then two data sets are collected, a reference data set with the molecules in the initial (dark) state and a time-dependent data set, where a reaction is initiated by a laser for example. Two sets of pairwise correlations C2 = can be extracted from the two data sets. Young's fringes and other cross terms between different molecules that emerge from coherent illumination (Fig. 4(b)) average out to a flat background, because the relative distances and orientations of the different molecules are uncorrelated in solution.75 The mathematics of this analysis is outlined in detail in two papers.7,75 The C2 can be related to the square of spherical harmonic expansion coefficients Il,m of the diffraction volume.80 The relevant quantity is , where m sums over the magnetic quantum number of the spherical harmonic expansion coefficients. The can be determined directly from C27,81 collected separately a time t after reaction initiation, and in the dark as a reference (ref) without reaction initiation, respectively. If the structural changes are not too large the difference of the two sets of Bl(q,q′) is simply . The are then related in a very specific manner (Eq. (7)) to the difference electron density75
| (7) |
Elements of matrix M are real numbers and are derived exclusively from known information provided by the dark state structure. δBl(q,q′) is a measured quantity, and is the difference electron density relative to the reference structure. can be retrieved directly from the δBs by inverting matrix M. A result from a simulation75 is shown in Fig. 4(c). were calculated from the dark state PYP model. Time-resolved were simulated by displacing the chromophore and a distant phenylalanine a significant amount. δρ was determined from the as described (Eq. (7)). Difference electron density δρ lights up in the frame of reference of the dark structure at the correct positions where the changes are made. This enables the difference electron density to be superimposed on the dark structure with standard crystallographic display software as in Fig. 4(c). With this method it might be possible for the first time to probe structures in solution at near atomic resolution and with ultra-fast time-resolution. For the PYP, it may become possible to directly observe the structural differences to the dark state in solution and compare those to known structural differences observed in the crystal.2,9,48 A kinetic analysis would be as straightforward as for time-resolved crystallographic data using the same SVD-based approaches. Solution structures determined by nuclear magnetic resonance82,83 as well as crystal structures9,48 may be used as reference structures to construct matrix M (Eq. (7)). The X-ray photon densities available at the free electron lasers should be sufficient to extract the correlations to high resolution even in the presence of multiple particles.84,85 By investigating the photocycle in terms of transient solution structures, it might well be that the structures of the transient states match those in the crystal, they might also be largely different, or a situation in between emerges. Results would shine light on why PYP behaves differently64 in crystal and solution, which would also apply to other proteins and enzymes. Since the method averages over a large number of molecules present in millions of snapshots, it can be expected that the method provides a mean square deviation from which the conformational entropy can be estimated.12 Moreover, by varying the temperature barriers of activation could be determined. Both conformational entropy and barriers can finally be compared to those obtained from crystals.
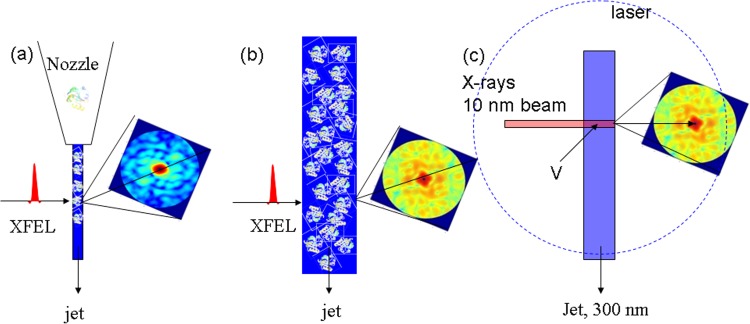
FIG. 3.
Geometry to determined time-resolved structures from solution. (a) Optimal setup: serial single molecule diffraction. Jet diameter is small. Due to jet and X-ray beam instabilities the hit rate is very small, if not non-existent. (b) More realistic scenario: Jet diameter is larger. The coherent XFEL beam interacts with an ensemble. (c) Geometry already in reach: X-ray beam is focused to 10 nm to increase the photon area density. Jet is large to allow for instabilities. With a PYP concentration of 40 mg/ml, the number of molecules in the intersecting volume V would be ∼40.
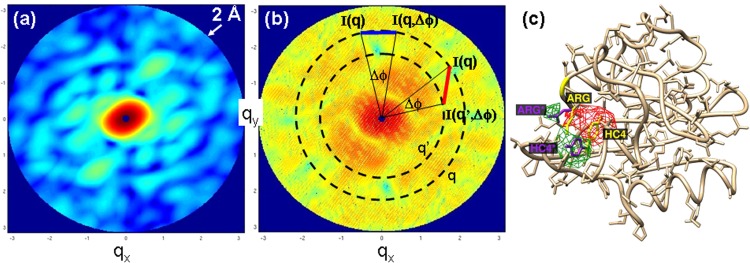
FIG. 4.
Structures from solution by analysis of the average angular correlations. (a) Intensity distribution from four molecules of random orientation and average distance of 150 Å from each other overlaid incoherently; is 2π with the scattering vector; resolution at the edge: 2 Å. (b) Coherent diffraction from an ensemble of four molecules of random orientation and average distance of 150 Å. Young's fringes can be easily identified. Blue bar: I(q) and I(q, Δϕ) on same resolution ring; red bar I(q) and I(q′, Δϕ) on different resolution rings. (c) Difference electron density recovered from simulated δBl(q,q′), courtesy of Kanupriya Pande. Position of the chromophore head as well as a Arg52 changed relative to the reference structure. Red: negative difference electron density, green: positive difference electron density.
IX. COMPARATIVE STRUCTURE BASED ENZYMOLOGY IN CRYSTAL AND SOLUTION
The European free electron laser for hard X-rays is designed to have X-ray pulse repetition rates of about 30 kHz.86 With this rate, 1 million diffraction patterns can be collected in 330 s, and 10 million patterns in an hour. This makes it feasible to collect comprehensive time-series from the start of a reaction to the very end in less than an 8 h shift. All sorts of enzymatic reactions including those occurring in the largest complexes such as the ribosome87–90 or other molecular machines that are prime drug targets91 could be investigated by this approach. To initiate an enzymatic reaction, the enzyme must be mixed with substrate and the mixture injected in the ultra-shortly pulsed X-ray beam. Diffusion times can then be as short as a few microseconds,3,92 and the reaction can be swiftly initiated. Alternatively, small micron-sized or nanosized crystals can be injected after mixing with substrate. Diffusion times are acceptable and in the millisecond to microsecond time range when micro- and nanocrystals are used3 respectively. With these two mix-and-inject approaches, structure based enzymology and drug design become feasible. The structures of the transient states can be determined rapidly and free energy landscapes characterized routinely.
ACKNOWLEDGMENTS
We acknowledge contributions by Kanupriya Pande. M.S. was supported by NSF-0952643 (Career). M.S. and D.K.S. were supported by Grant Nos. NSF-1158138 and NSF-STC 1231306.
REFERENCES
- 1. Graber T., Anderson S., Brewer H., Chen Y. S., Cho H. S., Dashdorj N., Henning R. W., Kosheleva I., Macha G., Meron M., Pahl R., Ren Z., Ruan S., Schotte F., Srajer V., Viccaro P. J., Westferro F., Anfinrud P., and Moffat K., “BioCARS: A synchrotron resource for time-resolved X-ray science,” J. Synchrotron Radiat. 18, 658–670 (2011). 10.1107/S0909049511009423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schmidt M., Srajer V., Henning R., Ihee H., Purwar N., Tenboer J., and Tripathi S., “Protein energy landscapes determined by five-dimensional crystallography,” Acta Crystallogr. D 69, 2534–2542 (2013). 10.1107/S0907444913025997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schmidt M., “Mix and inject, reaction initiation by diffusion for time-resolved macromolecular crystallography,” Adv. Condens. Matter Phys. 2013, 1–10. 10.1155/2013/167276 [DOI] [Google Scholar]
- 4. Schmidt M., Achterhold K., Prusakov V., and Parak F. G., “Protein dynamics of a beta-sheet protein,” Eur. Biophys. J. 38, 687–700 (2009). 10.1007/s00249-009-0427-z [DOI] [PubMed] [Google Scholar]
- 5. Parak F., Knapp E. W., and Kucheida D.,“Protein dynamics. Mossbauer spectroscopy on deoxymyoglobin crystals,” J. Mol. Biol. 161, 177–194 (1982). 10.1016/0022-2836(82)90285-6 [DOI] [PubMed] [Google Scholar]
- 6. Schotte F., Cho H. S., Kaila V. R., Kamikubo H., Dashdorj N., Henry E. R., Graber T. J., Henning R., Wulff M., Hummer G., Kataoka M., and Anfinrud P. A., “Watching a signaling protein function in real time via 100-ps time-resolved Laue crystallography,” Proc. Natl. Acad. Sci. U. S. A. 109, 19256–19261 (2012). 10.1073/pnas.1210938109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Poon H. C., Schmidt M., and Saldin D. K., “Extraction of fast changes in the structure of a disordered ensemble of photoexcited biomolecules,” Adv. Condens. Matter Phys. 2013, 750371. 10.1155/2013/750371 [DOI] [Google Scholar]
- 8. Steinfeld J. I., Francisco J. S., and Hase W. L., Chemical Kinetics and Dynamics, 2nd ed. (Prentience Hall, 1985). [Google Scholar]
- 9. Jung Y. O., Lee J. H., Kim J., Schmidt M., Moffat K., Srajer V., and Ihee H., “Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography,” Nat. Chem. 5, 212–220 (2013). 10.1038/nchem.1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hensen U., Lange O. F., and Grubmuller H., “Estimating absolute configurational entropies of macromolecules: The minimally coupled subspace approach,” PloS one 5, e9179 (2010). 10.1371/journal.pone.0009179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schlitter J., “Estimation of absolute and relative entropies of macromolecules using the covariance-matrix,” Chem. Phys. Lett. 215, 617–621 (1993). 10.1016/0009-2614(93)89366-P [DOI] [Google Scholar]
- 12. Polyansky A. A., Kuzmanic A., Hlevnjak M., and Zagrovic B., “On the contribution of linear correlations to quasi-harmonic conformational entropy in proteins,” J. Chem. Theo. Comput. 8, 3820–3829 (2012). 10.1021/ct300082q [DOI] [PubMed] [Google Scholar]
- 13. Parak F. and Knapp E. W., “A consistent picture of protein dynamics,” Proc. Natl. Acad. Sci. U. S. A. 81, 7088–7092 (1984). 10.1073/pnas.81.22.7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Akke M., Bruschweiler R., and Palmer A. G., “Nmr order parameters and free-energy - an analytical approach and its application to cooperative ca2+ binding by calbindin-D(9k),” J. Am. Chem. Soc. 115, 9832–9833 (1993). 10.1021/ja00074a073 [DOI] [Google Scholar]
- 15. Fitter J., “A measure of conformational entropy change during thermal protein unfolding using neutron spectroscopy,” Biophys. J. 84, 3924–3930 (2003). 10.1016/S0006-3495(03)75120-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karplus M., Ichiye T., and Pettitt B. M., “Configurational entropy of native proteins,” Biophys. J. 52, 1083–1085 (1987). 10.1016/S0006-3495(87)83303-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stroud R. M. and Finer-Moore J. S., “Conformational dynamics along an enzymatic reaction pathway: Thymidylate synthase,” "the movie”, Biochemistry 42, 239–247 (2003). 10.1021/bi020598i [DOI] [PubMed] [Google Scholar]
- 18. Schlichting I., Berendzen J., Chu K., Stock A. M., Maves S. A., Benson D. E., Sweet B. M., Ringe D., Petsko G. A., and Sligar S. G., “The catalytic pathway of cytochrome P450cam at atomic resolution,” Science 287, 1615–1622 (2000). 10.1126/science.287.5458.1615 [DOI] [PubMed] [Google Scholar]
- 19. Daniel R. M., Dunn R. V., Finney J. L., and Smith J. C., “The role of dynamics in enzyme activity,” Annu. Rev. Biophys. Biomol. Struct. 32, 69–92 (2003). 10.1146/annurev.biophys.32.110601.142445 [DOI] [PubMed] [Google Scholar]
- 20. Pace C. N., Shirley B. A., McNutt M., and Gajiwala K., “Forces contributing to the conformational stability of proteins,” Faseb. J. 10, 75–83 (1996). [DOI] [PubMed] [Google Scholar]
- 21. Ghosh K. and Dill K. A., “Computing protein stabilities from their chain lengths,” Proc. Natl. Acad. Sci. U. S. A. 106, 10649–10654 (2009). 10.1073/pnas.0903995106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robertson A. D. and Murphy K. P., “Protein structure and the energetics of protein stability,” Chem. Rev. 97, 1251–1267 (1997). 10.1021/cr960383c [DOI] [PubMed] [Google Scholar]
- 23. Li D. W., Showalter S. A., and Bruschweiler R., “Entropy localization in proteins,” J. Phys. Chem. B 114, 16036–16044 (2010). 10.1021/jp109908u [DOI] [PubMed] [Google Scholar]
- 24. van Brederode M. E., Gensch T., Hoff W. D., Hellingwerf K. J., and Braslavsky S. E., “Photoinduced volume change and energy storage associated with the early transformations of the photoactive yellow protein from Ectothiorhodospira halophila,” Biophys. J. 68, 1101–1109 (1995). 10.1016/S0006-3495(95)80284-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Van Brederode M. E., Hoff W. D., Van Stokkum I. H., Groot M. L., and Hellingwerf K. J., “Protein folding thermodynamics applied to the photocycle of the photoactive yellow protein,” Biophys. J. 71, 365–380 (1996). 10.1016/S0006-3495(96)79234-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parak F. G., Achterhold K., Croci S., and Schmidt M., “A physical picture of protein dynamics and conformational changes,” J. Biol. Phys. 33, 371–387 (2007). 10.1007/s10867-008-9102-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takeshita K., Imamoto Y., Kataoka M., Tokunaga F., and Terazima M., “Themodynamic and transport properties of intermediate states of the photocyclic reaction of photoactive yellow protein,” Biochemistry 41, 3037–3048 (2002). 10.1021/bi0110600 [DOI] [PubMed] [Google Scholar]
- 28. Parak F., Achterhold K., Keppler C., vonBurck U., Potzel W., SchindelMann P., Knapp E. W., Melchers B., Ruffer R., Chumakov A. I., and Baron A. Q. R., “The myoglobin phonon-spectrum obtained from X-ray scattering analyzed by Mossbauer effect,” Prog. Biophys. Mol. Bio. 65, Pa322–Pa322 (1996). [Google Scholar]
- 29. Doster W., Cusack S., and Petry W., “Dynamical transition of myoglobin revealed by inelastic neutron-scattering,” Nature 337, 754–756 (1989). 10.1038/337754a0 [DOI] [PubMed] [Google Scholar]
- 30. Parak F. G., Achterhold K., Schmidt M., Prusakov V., and Croci S., “Protein dynamics on different timescales,” J. Non-Cryst. Solids 352, 4371–4378 (2006). 10.1016/j.jnoncrysol.2006.01.106 [DOI] [Google Scholar]
- 31. Austin R. H., Beeson K. W., Eisenstein L., Frauenfelder H., and Gunsalus I. C., “Dynamics of ligand binding to myoglobin,” Biochemistry 14, 5355–5373 (1975). 10.1021/bi00695a021 [DOI] [PubMed] [Google Scholar]
- 32. Frauenfelder H., Chen G., Berendzen J., Fenimore P. W., Jansson H., McMahon B. H., Stroe I. R., Swenson J., and Young R. D., “A unified model of protein dynamics,” Proc. Natl. Acad. Sci. U. S. A. 106, 5129–5134 (2009). 10.1073/pnas.0900336106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Young R. D., Frauenfelder H., and Fenimore P. W., “Mossbauer effect in proteins,” Phys. Rev. Lett. 107, 158102 (2011). 10.1103/PhysRevLett.107.158102 [DOI] [PubMed] [Google Scholar]
- 34. Frauenfelder H., Parak F., and Young R. D., “Conformational substrates in proteins,” Ann. Rev. Biophys. Biophys. Chem. 17, 451–479 (1988). 10.1146/annurev.bb.17.060188.002315 [DOI] [PubMed] [Google Scholar]
- 35. Achterhold K., Keppler C., vanBurck U., Potzel W., Schindelmann P., Knapp E. W., Melchers B., Chumakov A. I., Baron A. Q. R., Ruffer R., and Parak F., “Temperature dependent inelastic X-ray scattering of synchrotron radiation on myoglobin analyzed by the Mossbauer effect,” Eur. Biophys. J. Biophys. 25, 43–46 (1996). 10.1007/s002490050015 [DOI] [Google Scholar]
- 36. Melchers B., Knapp E. W., Parak F., Cordone L., Cupane A., and Leone M., “Structural fluctuations of myoglobin from normal-modes, Mossbauer, Raman, and absorption spectroscopy,” Biophys. J. 70, 2092–2099 (1996). 10.1016/S0006-3495(96)79775-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chong S. H., Joti Y., Kidera A., Go N., Ostermann A., Gassmann A., and Parak F., “Dynamical transition of myoglobin in a crystal: comparative studies of X–ray crystallography and Mossbauer spectroscopy,” Eur. Biophys. J. 30, 319–329 (2001). 10.1007/s002490100152 [DOI] [PubMed] [Google Scholar]
- 38. Tetreau C., Mouawad L., Murail S., Duchambon P., Blouquit Y., and Lavalette D., “Disentangling ligand migration and heme pocket relaxation in cytochrome P450cam,” Biophys. J. 88, 1250–1263 (2005). 10.1529/biophysj.104.050104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lavalette D., Tetreau C., and Mouawad L., “Ligand migration and escape pathways in haem proteins,” Biochem. Soc. Trans. 34, 975–978 (2006). 10.1042/BST0340975 [DOI] [PubMed] [Google Scholar]
- 40. Martin J. L., Migus A., Poyart C., Lecarpentier Y., Astier R., and Antonetti A., “Femtosecond photolysis of CO-ligated protoheme and hemoproteins: Appearance of deoxy species with a 350-fsec time constant,” Proc. Natl. Acad. Sci. U. S. A. 80, 173–177 (1983). 10.1073/pnas.80.1.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Groenhof G., Bouxin-Cademartory M., Hess B., De Visser S. P., Berendsen H. J., Olivucci M., Mark A. E., and Robb M. A., “Photoactivation of the photoactive yellow protein: why photon absorption triggers a trans-to-cis Isomerization of the chromophore in the protein,” J. Am. Chem. Soc. 126, 4228–4233 (2004). 10.1021/ja039557f [DOI] [PubMed] [Google Scholar]
- 42. Groenhof G., Lensink M. F., Berendsen H. J., and Mark A. E., “Signal transduction in the photoactive yellow protein. II. Proton transfer initiates conformational changes,” Proteins 48, 212–219 (2002). 10.1002/prot.10135 [DOI] [PubMed] [Google Scholar]
- 43. Groenhof G., Lensink M. F., Berendsen H. J., Snijders J. G., and Mark A. E., “Signal transduction in the photoactive yellow protein. I. Photon absorption and the isomerization of the chromophore,” Proteins 48, 202–211 (2002). 10.1002/prot.10136 [DOI] [PubMed] [Google Scholar]
- 44. Groenhof G., Schafer L. V., Boggio-Pasqua M., Grubmuller H., and Robb M. A., “Arginine52 controls the photoisomerization process in photoactive yellow protein,” J. Am. Chem. Soc. 130, 3250–3251 (2008). 10.1021/ja078024u [DOI] [PubMed] [Google Scholar]
- 45. Anderson S., Crosson S., and Moffat K., “Short hydrogen bonds in photoactive yellow protein,” Acta Crystallogr D 60, 1008–1016 (2004). 10.1107/S090744490400616X [DOI] [PubMed] [Google Scholar]
- 46. Anderson S., Srajer V., Pahl R., Rajagopal S., Schotte F., Anfinrud P., Wulff M., and Moffat K., “Chromophore conformation and the evolution of tertiary structural changes in photoactive yellow protein,” Structure 12, 1039–1045 (2004). 10.1016/j.str.2004.04.008 [DOI] [PubMed] [Google Scholar]
- 47. Schmidt M., Pahl R., Srajer V., Anderson S., Ren Z., Ihee H., Rajagopal S., and Moffat K., “Protein kinetics: Structures of intermediates and reaction mechanism from time-resolved x-ray data,” Proc. Natl. Acad. Sci. U. S. A. 101, 4799–4804 (2004). 10.1073/pnas.0305983101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ihee H., Rajagopal S., Srajer V., Pahl R., Anderson S., Schmidt M., Schotte F., Anfinrud P. A., Wulff M., and Moffat K., “Visualizing reaction pathways in photoactive yellow protein from nanoseconds to seconds,” Proc. Natl. Acad. Sci. U. S. A. 102, 7145–7150 (2005). 10.1073/pnas.0409035102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lincoln C. N., Fitzpatrick A. E., and van Thor J. J., “Photoisomerisation quantum yield and non-linear cross-sections with femtosecond excitation of the photoactive yellow protein,” Phys. Chem. Chem. Phys. 14, 15752–15764 (2012). 10.1039/c2cp41718a [DOI] [PubMed] [Google Scholar]
- 50. van Stokkum I. H., Larsen D. S., and van Grondelle R., “Global and target analysis of time—resolved spectra,” Biochim. Biophys. Acta 1657, 82–104 (2004). 10.1016/j.bbabio.2004.04.011 [DOI] [PubMed] [Google Scholar]
- 51. Meyer T. E., “Isolation and characterization of soluble cytochromes, ferredoxins and other chromophoric proteins from the halophilic phototrophic bacterium Ectothiorhodospira halophila,” Biochim. Biophys. Acta 806, 175–183 (1985). 10.1016/0005-2728(85)90094-5 [DOI] [PubMed] [Google Scholar]
- 52. Meyer T. E., Tollin G., Causgrove T. P., Cheng P., and Blankenship R. E., “Picosecond decay kinetics and quantum yield of fluorescence of the photoactive yellow protein from the halophilic purple phototrophic bacterium, ectothiorhodospira-halophila,” Biophys. J. 59, 988–991 (1991). 10.1016/S0006-3495(91)82313-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hoff W. D., van Stokkum I. H., van Ramesdonk H. J., van Brederode M. E., Brouwer A. M., Fitch J. C., Meyer T. E., van Grondelle R., and Hellingwerf K. J., “Measurement and global analysis of the absorbance changes in the photocycle of the photoactive yellow protein from Ectothiorhodospira halophila,” Biophys. J. 67, 1691–1705 (1994). 10.1016/S0006-3495(94)80643-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ujj L., Devanathan S., Meyer T. E., Cusanovich M. A., Tollin G., and Atkinson G. H., “New photocycle intermediates in the photoactive yellow protein from Ectothiorhodospira halophila: picosecond transient absorption spectroscopy,” Biophys. J. 75, 406–412 (1998). 10.1016/S0006-3495(98)77525-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kort R., Vonk H., Xu X., Hoff W. D., Crielaard W., and Hellingwerf K. J., “Evidence for trans-cis isomerization of the p-coumaric acid chromophore as the photochemical basis of the photocycle of photoactive yellow protein,” Febs. Lett. 382, 73–78 (1996). 10.1016/0014-5793(96)00149-4 [DOI] [PubMed] [Google Scholar]
- 56. Xie A., Hoff W. D., Kroon A. R., and Hellingwerf K. J., “Glu46 donates a proton to the 4-hydroxycinnamate anion chromophore during the photocycle of photoactive yellow protein,” Biochemistry 35, 14671–14678 (1996). 10.1021/bi9623035 [DOI] [PubMed] [Google Scholar]
- 57. Borgstahl G. E., Williams D. R., and Getzoff E. D., “1.4 A structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore,” Biochemistry 34, 6278–6287 (1995). 10.1021/bi00019a004 [DOI] [PubMed] [Google Scholar]
- 58. Genick U. K., Borgstahl G. E., Ng K., Ren Z., Pradervand C., Burke P. M., Srajer V., Teng T. Y., Schildkamp W., McRee D. E., Moffat K., and Getzoff E. D., “Structure of a protein photocycle intermediate by millisecond time-resolved crystallography,” Science 275, 1471–1475 (1997). 10.1126/science.275.5305.1471 [DOI] [PubMed] [Google Scholar]
- 59. Schmidt M., Rajagopal S., Ren Z., and Moffat K., “Application of singular value decomposition to the analysis of time-resolved macromolecular x-ray data,” Biophys. J. 84, 2112–2129 (2003). 10.1016/S0006-3495(03)75018-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rajagopal S., Schmidt M., Anderson S., Ihee H., and Moffat K., “Analysis of experimental time-resolved crystallographic data by singular value decomposition,” Acta crystallogr. D 60, 860–871 (2004). 10.1107/S0907444904004160 [DOI] [PubMed] [Google Scholar]
- 61. Mendonca L., Hache F., Changenet-Barret P., Plaza P., Chosrowjan H., Taniguchi S., and Imamoto Y., “Ultrafast carbonyl motion of the photoactive yellow protein chromophore probed by femtosecond circular dichroism,” J. Am. Chem. Soc. 135, 14637–14643 (2013). 10.1021/ja404503q [DOI] [PubMed] [Google Scholar]
- 62. Liu J., Yabushita A., Taniguchi S., Chosrowjan H., Imamoto Y., Sueda K., Miyanaga N., and Kobayashi T., “Ultrafast time-resolved pump-probe spectroscopy of PYP by a sub-8 fs pulse laser at 400 nm,” J. Phys. Chem. B 117, 4818–4826 (2013). 10.1021/jp4001016 [DOI] [PubMed] [Google Scholar]
- 63. Xie A., Kelemen L., Hendriks J., White B. J., Hellingwerf K. J., and Hoff W. D., “Formation of a new buried charge drives a large-amplitude protein quake in photoreceptor activation,” Biochemistry 40, 1510–1517 (2001). 10.1021/bi002449a [DOI] [PubMed] [Google Scholar]
- 64. Yeremenko S., van Stokkum I. H., Moffat K., and Hellingwerf K. J., “Influence of the crystalline state on photoinduced dynamics of photoactive yellow protein studied by ultraviolet-visible transient absorption spectroscopy,” Biophys. J. 90, 4224–4235 (2006). 10.1529/biophysj.105.074765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ng K., Getzoff E. D., and Moffat K., “Optical studies of a bacterial photoreceptor protein, photoactive yellow protein, in single-crystals,” Biochemistry 34, 879–890 (1995). 10.1021/bi00003a022 [DOI] [PubMed] [Google Scholar]
- 66. Eyring H., “The activated complex in chemical reaction,” J. Chem. Phys. 3, 107–115 (1935). 10.1063/1.1749604 [DOI] [Google Scholar]
- 67. Hanggi P., Talkner P., and Borkovec M., “Reaction-rate theory—50 years after kramers,” Rev. Mod. Phys. 62, 251–341 (1990). 10.1103/RevModPhys.62.251 [DOI] [Google Scholar]
- 68. Chung H. S. and Eaton W. A., “Single-molecule fluorescence probes dynamics of barrier crossing,” Nature 502, 685 (2013). 10.1038/nature12649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bourgeois D., Vallone B., Arcovito A., Sciara G., Schotte F., Anfinrud P. A., and Brunori M., “Extended subnanosecond structural dynamics of myoglobin revealed by Laue crystallography,” Proc. Natl. Acad. Sci. U. S. A. 103, 4924–4929 (2006). 10.1073/pnas.0508880103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schmidt M., Nienhaus K., Pahl R., Krasselt A., Anderson S., Parak F., Nienhaus G. U., and Srajer V., “Ligand migration pathway and protein dynamics in myoglobin: A time-resolved crystallographic study on L29W MbCO,” Proc. Natl. Acad. Sci. U. S. A. 102, 11704–11709 (2005). 10.1073/pnas.0504932102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lim M. H., Jackson T. A., and Anfinrud P. A., “Nonexponential protein relaxation—Dynamics of conformational change in myoglobin,” Proc. Natl. Acad. Sci. U. S. A. 90, 5801–5804 (1993). 10.1073/pnas.90.12.5801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Moritsugu K. and Smith J. C., “Langevin model of the temperature and hydration dependence of protein vibrational dynamics,” J. Phys. Chem. B 109, 12182–12194 (2005). 10.1021/jp044272q [DOI] [PubMed] [Google Scholar]
- 73. Schmidt M., Graber T., Henning R., and Srajer V., “Five-dimensional crystallography,” Acta Crystallogr. Sect. A 66, 198–206 (2010). 10.1107/S0108767309054166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schwander P., Giannakis D., Yoon C. H., and Ourmazd A., “The symmetries of image formation by scattering. II. Applications,” Opt. Express 20, 12827–12849 (2012). 10.1364/OE.20.012827 [DOI] [PubMed] [Google Scholar]
- 75. Pande K., Schwander P., Schmidt M., and Saldin D. K., “Deducing fast electron density changes in randomly orientated uncrystallized biomolecules in a pump-probe experiment,” Philos. Trans. R. Soc. B (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Saldin D. K., Poon H. C., Shneerson V. L., Howells M., Chapman H. N., Kirian R. A., Schmidt K. E., and Spence J. C. H., “Beyond small-angle x-ray scattering: Exploiting angular correlations,” Phys. Rev. B 81, 174105 (2010). 10.1103/PhysRevB.81.174105 [DOI] [Google Scholar]
- 77. Liu H., Poon B. K., Saldin D. K., Spence J. C., and Zwart P. H., “Three-dimensional single-particle imaging using angular correlations from X-ray laser data,” Acta Crystallogr. Sect. A 69, 365–373 (2013). 10.1107/S0108767313006016 [DOI] [PubMed] [Google Scholar]
- 78. Poon H. C., Schwander P., Uddin M., and Saldin D. K., “Fiber diffraction without fibers,” Phys. Rev. Lett. 110, 265505 (2013). 10.1103/PhysRevLett.110.265505 [DOI] [PubMed] [Google Scholar]
- 79. Saldin D. K., Poon H. C., Schwander P., Uddin M., and Schmidt M., “Reconstructing an icosahedral virus from single-particle diffraction experiments,” Opt. Express 19, 17318–17335 (2011). 10.1364/OE.19.017318 [DOI] [PubMed] [Google Scholar]
- 80. Saldin D. K., Shneerson V. L., Fung R., and Ourmazd A., “Structure of isolated biomolecules obtained from ultrashort x-ray pulses: Exploiting the symmetry of random orientations,” J. Phys. Condens. Matter 21, 134014 (2009). 10.1088/0953-8984/21/13/134014 [DOI] [PubMed] [Google Scholar]
- 81. Fung R., Shneerson V., Saldin D. K., and Ourmazd A., “Structure from fleeting illumination of faint spinning objects in flight,” Nat. Phys. 5, 64–67 (2009). 10.1038/nphys1129 [DOI] [Google Scholar]
- 82. Rubinstenn G., Vuister G. W., Mulder F. A., Dux P. E., Boelens R., Hellingwerf K. J., and Kaptein R., “Structural and dynamic changes of photoactive yellow protein during its photocycle in solution,” Nat. Struct. Biol. 5, 568–570 (1998). 10.1038/823 [DOI] [PubMed] [Google Scholar]
- 83. Ramachandran P. L., Lovett J. E., Carl P. J., Cammarata M., Lee J. H., Jung Y. O., Ihee H., Timmel C. R., and van Thor J. J., “The short-lived signaling state of the photoactive yellow protein photoreceptor revealed by combined structural probes,” J. Am. Chem. Soc. 133, 9395–9404 (2011). 10.1021/ja200617t [DOI] [PubMed] [Google Scholar]
- 84. Kirian R. A., Schmidt K. E., Wang X., Doak R. B., and Spence J. C., “Signal, noise, and resolution in correlated fluctuations from snapshot small-angle x-ray scattering,” Phys. Rev. E 84, 011921 (2011). 10.1103/PhysRevE.84.011921 [DOI] [PubMed] [Google Scholar]
- 85. Kirian R. A., “Structure determination through correlated fluctuations in x-ray scattering,” J. Phys. B At. Mol. Opt. Phys. 45, 223001 (2012). 10.1088/0953-4075/45/22/223001 [DOI] [Google Scholar]
- 86. Porro M., Andricek L., Aschauer S., Bayer M., Becker J., Bombelli L., Castoldi A., De Vita G., Diehl I., Erdinger F., Facchinetti S., Fiorini C., Fischer P., Gerlach T., Graafsma H., Guazzoni C., Hansen K., Kalavakuru P., Klar H., Kugel A., Lechner P., Lemke M., Lutz G., Manghisoni M., Mezza D., Muntefering D., Pietsch U., Quartieri E., Randall M., Re V., Reckleben C., Sandow C., Soldat J., Struder L., Szymanski J., Weidenspointner G., and Wunderer C. B., “Development of the depfet sensor with signal compression: A large format x-ray imager with mega-frame readout capability for the European XFEL,” IEEE Trans. Nucl. Sci. 59, 3339–3351 (2012). 10.1109/TNS.2012.2217755 [DOI] [Google Scholar]
- 87. Nissen P., Hansen J., Ban N., Moore P. B., and Steitz T. A., “The structural basis of ribosome activity in peptide bond synthesis,” Science 289, 920–930 (2000). 10.1126/science.289.5481.920 [DOI] [PubMed] [Google Scholar]
- 88. Ban N., Nissen P., Hansen J., Moore P. B., and Steitz T. A., “The complete atomic structure of the large ribosomal subunit at 2.4 angstrom resolution,” Science 289, 905–920 (2000). 10.1126/science.289.5481.905 [DOI] [PubMed] [Google Scholar]
- 89. Harms J., Schluenzen F., Zarivach R., Bashan A., Gat S., Agmon I., Bartels H., Franceschi F., and Yonath A., “High resolution structure of the large ribosomal subunit from a mesophilic Eubacterium,” Cell 107, 679–688 (2001). 10.1016/S0092-8674(01)00546-3 [DOI] [PubMed] [Google Scholar]
- 90. Selmer M., Dunham C. M., Murphy F. V., Weixlbaumer A., Petry S., Kelley A. C., Weir J. R., and Ramakrishnan V., “Structure of the 70S ribosome complexed with mRNA and tRNA,” Science 313, 1935–1942 (2006). 10.1126/science.1131127 [DOI] [PubMed] [Google Scholar]
- 91. Yonath A., “Antibiotics targeting ribosomes: Resistance, selectivity, synergism, and cellular regulation,” Annu. Rev. Biochem. 74, 649–679 (2005). 10.1146/annurev.biochem.74.082803.133130 [DOI] [PubMed] [Google Scholar]
- 92. Adams S. R. and Tsien R. Y., “Controlling Cell Chemistry with Caged Compounds,” Annu. Rev. Physiol. 55, 755–784 (1993). 10.1146/annurev.ph.55.030193.003543 [DOI] [PubMed] [Google Scholar]