Abstract
TRIM22, a tripartite-motif (TRIM) protein, is upregulated upon interferon alpha (IFNα) administration to hepatitis C virus (HCV)-infected patients. However, the physiological role of TRIM22 upregulation remains unclear. Here, we describe a potential antiviral function of TRIM22's targeting of the HCV NS5A protein. NS5A is important for HCV replication and for resistance to IFNα therapy. During the first 24 h following the initiation of IFNα treatment, upregulation of TRIM22 in the peripheral blood mononuclear cells (PBMCs) of HCV patients correlated with a decrease in viral titer. This phenomenon was confirmed in the hepatocyte-derived cell line Huh-7, which is highly permissive for HCV infection. TRIM22 over-expression inhibited HCV replication, and Small interfering RNA (siRNA)-mediated knockdown of TRIM22 diminished IFNα-induced anti-HCV function. Furthermore, we determined that TRIM22 ubiquitinates NS5A in a concentration-dependent manner. In summary, our results suggest that TRIM22 upregulation is associated with HCV decline during IFNα treatment and plays an important role in controlling HCV replication in vitro.
Keywords: HCV, IFNα, NS5A, TRIM22, ubiquitin
Introduction
Hepatitis C virus (HCV) is a global problem, and according to the World Health Organization, 150 million people are currently infected.1 HCV is an enveloped, positive-sense single-stranded RNA virus of the family Flaviviridae.2 The viral RNA is translated as a precursor polyprotein, which is then spliced into 10 separate proteins.3 The three structural proteins are E1, E2 and core, while p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B are the non-structural proteins.3 HCV replication mainly occurs in hepatocytes. HCV infection is frequently not detected during the acute phase of infection, and most patients become chronically infected.4 Some chronic HCV patients may further develop liver cirrhosis or even liver cancer.4
Interferon alpha (IFNα) plus ribavirin is the standard therapy for HCV patients in China.5 IFNα is a broad-spectrum anti-viral molecule and plays an important role in the anti-viral innate immune response.6 Endogenous IFNα is induced by viruses through pattern recognition receptors, such as Toll-like receptors, RIG-I and MDA5.7,8 IFNα elicits anti-viral activity by binding to Heterodimer of interferon alpha receptor1(IFNAR1) and interferon alpha receptor 2 (IFNAR2) and inducing Janus-activated kinase 1 and tyrosine kinase 2, which in turn phosphorylate and thereby activate signal transducer and activator of transcription proteins 1 and 2 (STAT1 and STAT2).9 IFN-regulatory factor 9 forms a complex with the STAT1/2 complex in the cell nucleus, leading to the upregulation of interferon-stimulated genes (ISGs), which are involved in biological processes such as lipid metabolism, apoptosis, inflammation and protein degradation.9 Some ISGs affect the translation of viral proteins, such as protein kinase R.9 IFNα may also decrease viral RNA stability by inducing 2′,5′-oligoadenylate synthetase during HCV treatment.9
The tripartite motif (TRIM) family of proteins is defined by the highly conserved RBCC signature domain, which contains a RING finger domain, one or two B-Box domains, and a coiled-coil domain.10,11 The RING finger domain contains a specialized zinc finger and functions as a ubiquitin protein ligase, either alone or as a part of a multi-subunit E3 protein complex. The associated B-Box and coiled-coil domains are involved in protein interactions and homo/heterodimerization. TRIM proteins are involved in a variety of cellular functions, including apoptosis, transcription, differentiation and regulation of cell cycle progression.10,11 In addition, some TRIM proteins exhibit anti-viral activity and may be involved in innate immunity.10,11 Many TRIM proteins have recently emerged as IFN-induced proteins involved in various cellular processes, including cell proliferation and antiviral activities. TRIM22, also known as Staf-50, is a natural antiviral effector of HIV-1,12 HBV13 and influenza.14 The dependence of the anti-viral function of TRIM22 on its ubiquitination function varies depending on the virus.10,11 For example, TRIM22 inhibits influenza by targeting its nucleoprotein, but also inhibits the activity of the HBV core promoter not through ubiquitination.13
We previously determined that TRIM22 is highly upregulated in peripheral blood mononuclear cell (PBMC) samples from HCV patients treated with IFNα, as well as in Huh-7 cells treated with IFNα.13 The function of TRIM22 was further verified by examining its effects on the HCV Huh-7-Con-1 replicon system or JFH1 virus. Here, the mechanism of TRIM22 is explored.
Materials and methods
Subjects and study protocol
The study was approved by the ethics committee of the 1st Hospital of Jilin University and was registered with ClinicalTrials.gov, ID NCT01434212. All enrolled subjects were hepatitis B surface antigen-negative, hepatitis B surface antigen antibody-negative and HIV antibody-negative, with no serological or histological evidence of a cause of liver disease other than HCV. Treatment consisted of standard IFNα (Recombinant Human IFNα-2b; Kaiyinyisheng Inc., Beijing, China, FDA Approval number: S20030032) plus ribavirin (Weilake, FDA Approval number: H10940157) for 48 weeks. Five million units of IFNα-2b were administered subcutaneously every other day at the clinic throughout treatment. Ribavirin was dosed at 15 mg/kg/day in two divided doses. Serum HCV RNA levels were measured using the COBAS AmpliPrep/COBAS TaqMan assay (Roche Molecular Diagnostics, Pleasanton, CA, USA) with a lower limit of quantification of 15 IU/ml. Measurements were obtained at the following time points after IFNα treatment initiation: 0 h, 12 h, 24 h, 37 h and 43 h. PBMCs were separated using GE Percoll reagent (Product Code: 17-0891-01) following the manufacturer's instructions.
Reagents and antibodies
Recombinant IFNα was purchased from eBioscience, San Diego, CA, USA. Anti-FLAG monoclonal antibody (M2) and anti-actin antibody were purchased from Sigma-Aldrich, St. Louis, MO, USA. Anti-HA monoclonal antibody was purchased from Covance. Rabbit anti-TRIM22 polyclonal antibody was purchased from Sigma (Cat. No. HPA003307).
Reverse transcription and real-time RT-PCR
Total RNA was obtained using TRIzol reagent (Life Technologies, Grand Island, NY, USA) according to the manufacturer's instructions. Total cellular RNA (2 µg) was reverse-transcribed with SuperScript II reverse transcriptase (Life Technologies), and 2 µl of the reverse-transcribed cDNA was used in real-time RT-PCR with the 7900HTFast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) and a traditional agarose gel assay. The primers used for TRIM22 detection were 5′-GGTTGAGGGGATCGTCAGTA-3′ (L) and 5′-TTGGAAACAGATTTTGGCTTC-3′ (R).
Plasmids
Sequences encoding TRIM22 were amplified by PCR from cDNA from Huh-7 cells challenged with IFNα for 12 h. TRIM22 and its truncation mutants were cloned into the pcDNA3.1-5′-FLAG vector. PCR amplification was performed in a Mastercycler (Eppendorf, Hamburg, Germany), and PCR products were separated on 1% agarose gels and visualized by ethidium bromide staining.
The sequences of the PCR primers were as follows:
TRIM22 forward: 5′-ATGGATTTCTCAGTAAAGGTA-3′
TRIM22 reverse: 5′-TCAGGAGCTCGGTGGGCACAC-3′
TRIM22A reverse: 5′-CCGCTCGAGtcaaatgtttggtgaccttggtgttcctgagac-3′
TRIM22B reverse: 5′-CCGCTCGAGtcacatcccactcagatctggtactcggaa-3′
TRIM22C forward: 5′-CCATCGATatggcaagttcttaaagagctgacagatgtccag-3′
TRIM22D forward: 5′- CCATCGATatgcataaacgaggtggtcaaggaatgtcagg-3′
NS5A forward: 5′-AATGAATTCatgtccggatcctggctccgcg-3′
NS5A reverse: 5′-ccgggtaccatgcagcacacggtggtatcgtcctc-3′
NS5A-D1 reverse: 5′-ccgggtaccattgatccccgtgccaagcgc-3′
NS5A-D12 reverse: 5′-ccgggtaccatttctgatatggtgctctcgctcagaccc-3′
NS5A-D23 forward: 5′- AATGAATTCatgcctccatctgaggcgagctcctca-3′
Virus, cell culture and transfection
HCV JFH1 virus was kindly provided by Takaji Wakita, National Institute of Infectious Diseases, Japan.15,16 CON1 replicon cells were provided by Ralf Bartenschlager, Department of Infectious Diseases, Molecular Virology, Heidelberg University, Heidelberg, Germany.17 JFH1 full genome Huh-7 cells were generated as previously described.18 HEK293T, Huh-7 or HCV replicon cells were cultured in DMEM (Life Technologies) supplemented with heat-inactivated fetal calf serum (10%), penicillin (100 U/ml) and streptomycin (100 µg/ml). The cells were grown at 37 °C in a humidified incubator with a 5% CO2 atmosphere.
HEK293T cells were seeded into 6-well plates or 24-well plates for 24 h. The cells were transiently transfected using Lipofectamine (Life Technologies) according to the manufacturer's instructions. The plasmid amounts were normalized by the addition of empty plasmid.
Immunoblotting and immunoprecipitation
For immunoblotting, cells were washed with ice-cold phosphate-buffered saline and lysed with ice-cold lysis buffer containing 1.0% (v/v) Nonidet P40, 20 mM Tris-HCl pH 8.0, 10% (v/v) glycerol, 150 mM NaCl, 1 mM NaF, 2 mM sodium orthovanadate, 1 mM sodium pyrophosphate and a protease inhibitor cocktail (Roche, Basel, Switzerland). After incubation on ice for 15 min, the supernatant was obtained by centrifuging the crude lysates at 12 000 g for 15 min to remove cell debris. Total protein was separated on a 10% SDS–polyacrylamide gel and transferred to a nitrocellulose membrane (Bio-Rad, Berkeley, CA, USA). For immune detection, membranes were washed with TBS-T (20 mM Tris, 500 mM NaCl, 0.1% Tween 20) and blocked in a 5% powdered-milk solution in TBS-T for 1 h. After washing with TBS-T, the membranes were separately probed with anti-HA (HA1.1) (Covance, Princeton, NJ, USA) (1∶1000), anti-FLAG M2 (1∶1000), anti-TRIM22 (1∶500) (Sigma) and anti-beta-actin (1∶1000) antibodies for 1 h. Secondary peroxidase-labeled anti-rabbit or anti-mouse antibodies were incubated for 1 h at room temperature. Anti-NS5A was raised in Dr Jin Zhong's lab. Protein detection was visualized by ECL according to the manufacturer's directions (Pierce, Waltham, MA, USA).
For immunoprecipitation, total cell lysates were subjected to immunoprecipitation with 0.5 µg of mouse anti-HA (HA1.1) (Covance) monoclonal antibody or rabbit anti-TRIM22 polyclonal antibodies in 500 µl of 1% (v/v) lysis buffer. Binding between the antibody and antigen was allowed to occur at 4 °C for 2 h in suspension under constant rotation. Then, the protein A/G agarose suspension was added, and the mixture was incubated for 2 h at 4 °C with constant agitation. The immunoprecipitated complexes were washed three times with 1% IP buffer, and the proteins were eluted by adding 30 µl of 2% SDS–PAGE sample buffer, followed by boiling for 5 min. Sepharose beads were pelleted by centrifugation in a microfuge for 5 min. The supernatant containing proteins was separated by SDS-PAGE, followed by staining with mouse anti-FLAG M2 monoclonal antibody (Sigma).
siRNA
TRIM22 Stealth Select RNAi (Catalog# 1299003) was purchased from Life Technologies: short interfering negative control sequence 5′-UUCUCCGAACGUGUCACGUTT-3′, si1 5′-CACCAAACAUUCCGCAUAATT-3′, si2 5′-GGAUGCUGCAAGUUCUUAATT-3′. HEK293T cells or Huh-7 cells were transfected with the siRNA targeting TRIM22 or control siRNA (20 nM final concentration) using Lipofectamine 2000 (Life Technologies) or Lipofectamine LTX (Life Technologies) using the standard method described in the manufacturer's protocol. Empty vector was added to normalize the final plasmid amount. The degree of gene silencing was confirmed by RT-PCR or immunoblotting 24 h after transfection.
Statistical analysis
Normally distributed continuous variables were compared using t-tests. The Mann–Whitney test was used when a normal distribution was rejected (Shapiro test, P<0.05). In all cases, a P value ≤0.05 was considered significant. IBM SPSS V.19 software was used for statistical analysis.
Results
TRIM22 is induced in PBMCs from HCV patients by Type I IFNα treatment and is associated with a decrease in HCV titers
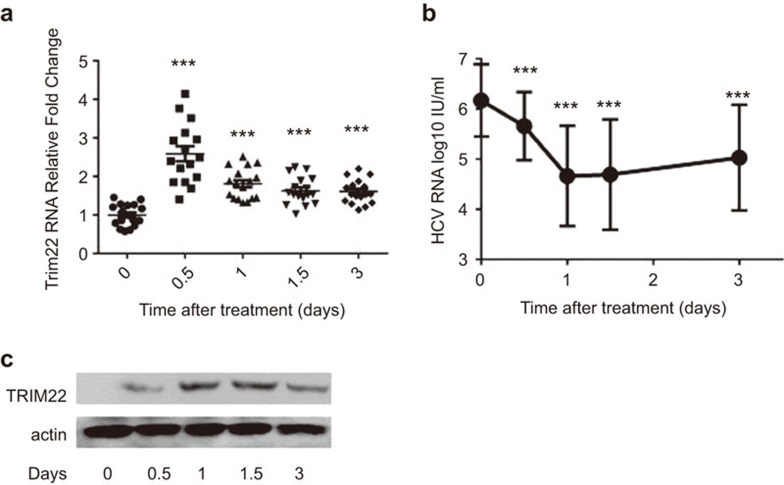
TRIM22 is upregulated by IFNα treatment. In this study, we evaluated whether this association occurred in HCV patients who were treated with IFNα. Patient PBMCs were collected after IFNα treatment, and TRIM22 expression was analyzed by both real-time PCR and immunoblotting. A significant increase in TRIM22 expression was observed in a real-time PCR assay 12 h after the initiation of IFNα treatment (Figure 1a). TRIM22 induction was also observed at the protein level by western blotting (Figure 1c). Concomitantly, the HCV virus titer in the blood decreased rapidly (Figure 1b). The change in early virus kinetics after IFNα administration suggests that TRIM22 is involved in IFNα-induced antiviral effects.
Figure 1.
TRIM22 is induced in PBMCs by Type I IFN treatment of HCV patients and is associated with a decrease in HCV levels. (a) The TRIM22 mRNA level was significantly increased in the PBMCs of HCV patients approximately 12 h after initiation of IFNα treatment, as measured by real-time PCR. Comparison of the HCV mRNA levels with the baseline HCV mRNA level at the remaining time points revealed significant differences. ***P<0.001 as determined by Student's t-test. (b) HCV viral titers (shown as the logarithm) in patients' blood decreased markedly, as measured by COBAS (as described in the section on ‘Materials and methods'). Comparison of the HCV titer with the baseline virus titration at other time points revealed significant differences. ***P<0.001, as determined by Student's t-test. (c) TRIM22 is induced in patient PBMC samples, as determined by western blotting (a representative patient result is shown). HCV, hepatitis C virus; IFN, interferon; PBMC, peripheral blood mononuclear cell; TRIM, tripartite motif.
TRIM22 is upregulated in Huh-7 cells treated with IFNα
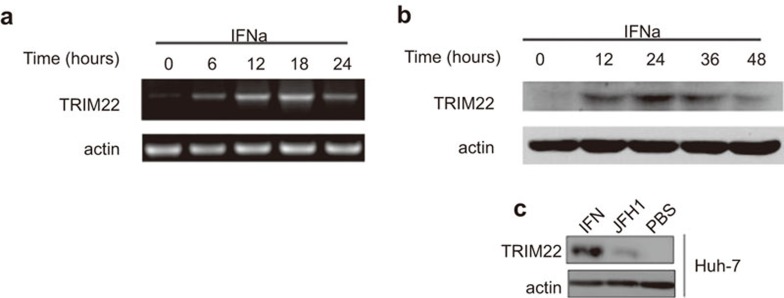
To determine whether IFNα is capable of inducing TRIM22, we examined TRIM22 expression in the Huh-7 human hepatocyte cell line after IFNα treatment. When Huh-7 cells were treated with IFNα for 6 h, TRIM22 mRNA expression increased, reaching a maximum at 18 h post-stimulation (Figure 2a). Accordingly, western blotting with a TRIM22-specific antibody revealed that TRIM22 protein levels increased significantly after 12 h of stimulation (Figure 2b). HCV infection triggered endogenous TRIM22 expression (Figure 2c), albeit at lower levels than that induced by direct IFNα stimulation. Taken together, these results demonstrate that TRIM22 is induced following IFNα stimulation of Huh-7 cells.
Figure 2.
TRIM22 is upregulated in Huh-7 cells treated with IFNα. (a, b) TRIM22 mRNA is upregulated in Huh-7 cells treated with IFNα (50 IU/ml). Time-series (time points are shown in the figure) samples were collected and prepared for RT-PCR or western blotting. The primers are described in the section on ‘Materials and methods'. (c) TRIM22 is induced by JFH-1 (MOI=0.1) 24 h after JFH-1 infection but at a lower level than induced by IFNα (50 IU/ml). HCV, hepatitis C virus; IFN, interferon; MOI, multiplicity of infection.
TRIM22 is an anti-HCV molecule
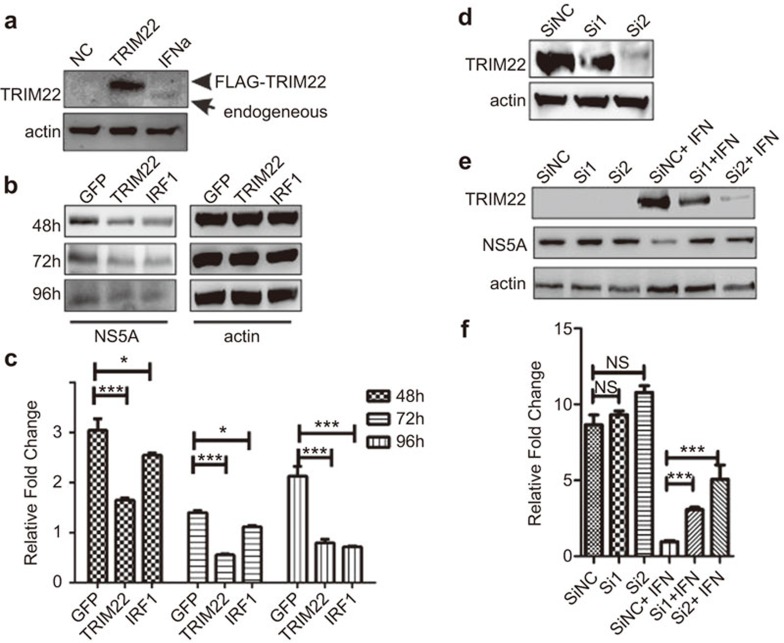
We next determined whether IFNα-induced TRIM22 can suppress HCV replication. The Huh-7 cell line was transfected with the Con1 replicon genome as described previously by Lohmann et al.17 After selection with G418, we obtained a cell line that expressed the deleted Con1 genome stably, Huh-7-Con1-rep. TRIM22 was over-expressed in Huh-7-Con1-rep cells (Figure 3a), leading to a decrease in HCV levels of approximately 50% compared to control cells 48 h following transfection and approximately 72% at 96 h (Figure 3c). Inhibition of HCV was also observed at the protein level by western blot assay (Figure 3b). To suppress TRIM22 expression, two siRNAs were designed, and their knockdown efficiency was evaluated by western blotting (Figure 3d). Cells were infected with the HCV JFH-1 virus (multiplicity of infection (MOI) of 0.1). When the infected cells were treated with control siRNA and IFNα in combination, HCV RNA was significantly decreased. By contrast, under the same conditions, si1 and si2 against TRIM22 at the same concentration as the control siRNA suppressed TRIM22 expression and subsequently inhibited IFNα treatment-mediated suppression of HCV at both the protein and RNA levels (Figure 3e and f). Thus, TRIM22 is an anti-HCV molecule that can negatively regulate HCV replication.
Figure 3.
TRIM22 has anti-viral activity against HCV. (a) Western blot analysis of TRIM22 expression with anti-TRIM22 antibody after transfection of cells with FLAG-TRIM22 (0.5 µg/well, 24-well plate). Endogenous TRIM22 is shown as a positive control in the lane labeled ‘IFNα' in Huh-7 cells that were treated with IFNα (50 IU/ml) for 24 h and collected for western blotting. (b, c) A marked decrease in the virus replicon levels was observed in TRIM22-over-expressing Huh-7-con1-replicon cells 48–96 h after infection. Huh-7-con1-replicon cells were transfected with GFP, TRIM22 and the anti-viral ISG and IRF1 plasmids. Samples were collected at 48 h, 72 h and 96 h after transfection. Real-time RT-PCR and western blotting were performed as described in the section on ‘Materials and methods'. (d) The knockdown efficiency of the siRNAs was determined by western blotting. Huh-7 cells were transfected with siRNA or TRIM22 (siRNA: 50 nmol per well/24-well plate, TRIM22: 100 ng per well/24-well plate), and cells were collected for western blotting 24 h after transfection. (e, f) Knockdown of TRIM22 diminished the anti-viral effects of IFNα. Huh-7 cells were transfected with siRNA (50 nmol per well/24-well plate). After 24 h, the transfected cells were infected with JFH1 (MOI=0.1). At 24 h after infection, the cells were treated with IFNα (50 IU/ml) for an additional 48 h. Samples were collected for real-time PCR assays and western blotting. *P<0.05 as determined by Student's t-test. ***P<0.001 as determined by Student's t-test. HCV, hepatitis C virus; IFN, interferon; IRF, IFN-regulatory factor; ISG, interferon-stimulated gene; PBMC, peripheral blood mononuclear cell; TRIM, tripartite motif.
TRIM22 interacts with HCV NS5A
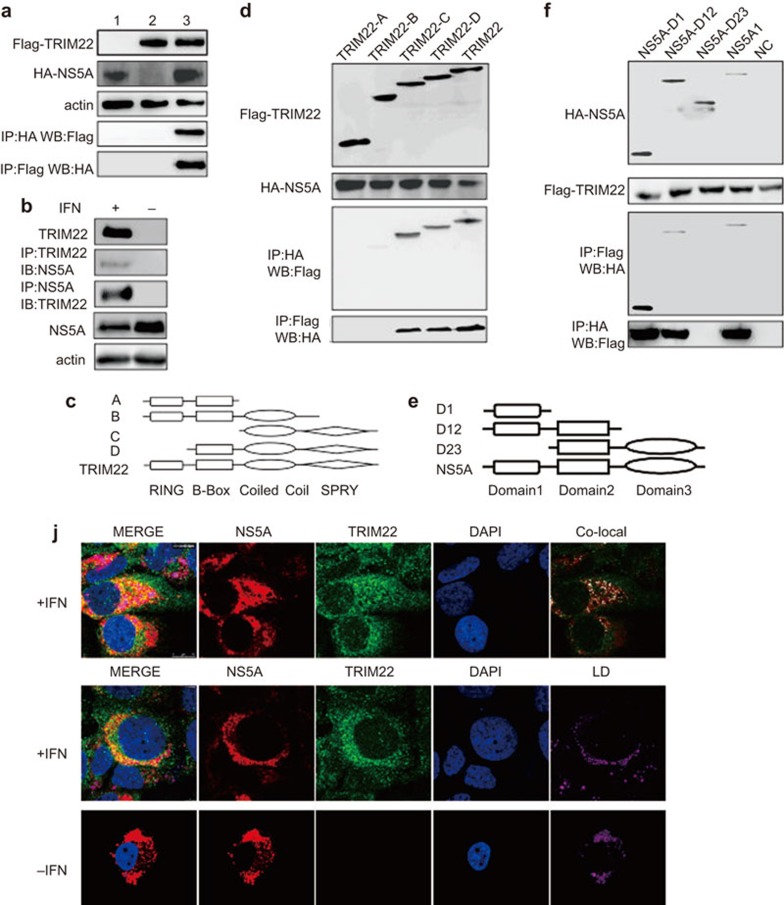
Because TRIM22 is an anti-HCV ISG, we determined whether TRIM22 directly interacts with the proteins encoded by HCV. First, we screened all HCV proteins by co-immunoprecipitation (co-IP) (Supplementary Figure 1), which revealed that TRIM22 specifically binds the NS5A protein (Figure 4a). Second, the endogenous TRIM22-NS5A interaction was observed (Figure 4b). We next identified the domain of TRIM22 that is essential for the interaction between TRIM22 and NS5A. Four truncations of TRIM22 lacking the SPRY and the coiled coil domains (A), the SPRY domain (B), the RING and B-box domains (C) or only the RING domain (D) were constructed to identify the domains required for the interaction between TRIM22 and NS5A (Figure 4c). TRIM22 mutants A and B (which lack the SPRY domain) were unable to interact with NS5A (Figure 4d), indicating that the SPRY domain is important for the interaction between TRIM22 and NS5A. This interaction suggests that the NS5A protein plays a role in the inhibition of HCV function by TRIM22. We also determined which domain of NS5A is essential for the interaction between TRIM22 and NS5A. NS5A contains three protein domains: Domain 1, Domain 2 and Domain 3. We created three truncations of NS5A, NS5A-D1 (Domain 1), NS5A-D12 (Domain 1 and 2) and NS5A-D23 (Domain 2 and 3) (Figure 4e). Only NS5A-D23 did not interact with TRIM22, indicating that domain 1 is essential for the interaction between NS5A and TRIM22 (Figure 4f). In addition, TRIM22 and NS5A colocalized in the cytoplasm of Huh-7 cells around lipid droplets (Figure 4j and Supplementary Figure 2).
Figure 4.
TRIM22 interacts with HCV NS5A. (a) Both FLAG-TRIM22 (1.5 µg/well, six-well plate) and HA-NS5A (1.5 µg/well, six-well plate) were expressed in 293T cells. For lane 1, cells were transfected with only HA-NS5A. For lane 2, cells were transfected with only FLAG-TRIM22. For lane 3, cells were transfected with both HA-NS5A and FLAG-TRIM22. Co-IP experiments were performed with anti-FLAG and anti-HA antibodies 24 h after transfection of cells with the plasmids. The results of the co-IP experiments revealed that TRIM22 interacts with HCV NS5A. (b) The endogenous interaction of TRIM22 and NS5A was evaluated. Twenty-four hours after adding IFNα (50 IU/ml) to the HCV Huh-7-Con-1 replicon, the cells were harvested and subjected to endogenous IP with anti-TRIM22 and anti-NS5A antibodies. (c) Schematic diagram of the four truncated forms of TRIM22: TRIM22A (lacks coiled-coil and SPRY domains), B (lacks SPRY domain), C (lacks RING and B-Box domains) and D (lacks RING domain). (d) The SPRY and coiled-coil domains are important for the interaction of TRIM22 with NS5A because only TRIM22 C and TRIM22 D interact with NS5A. All TRIM22 truncations (1.5 µg/well, six-well plate) and HA-NS5A (1.5 µg/well, six-well plate) were expressed in 293T cells. Co-IP experiments were performed using anti-FLAG and anti-HA antibodies 24 h after transfection of cells with the plasmids. (e) Schematic diagram of the three truncated forms of NS5A: NS5A-D1 (contains only domain 1), NS5A-D12 (contains domains 1 and 2) and NS5A-D23 (contains domains 2 and 3). (f) Domain 1 of NS5A is important for the interaction of TRIM22 with NS5A because only NS5A-D23 does not interact with NS5A. All HA-NS5A truncations (1.5 µg/well, six-well plate) and FLAG-TRIM22 (1.5 µg/well, six-well plate) were expressed in 293T cells. Co-IP experiments were performed using anti-FLAG and anti-HA antibodies 24 h after transfection of cell with the plasmids. (j) IFNα (50 IU/ml) treatment was initiated 24 h after Huh-7 cells were infected with the JFH-1 virus (MOI of 0.1). TRIM22 was colocalized with NS5A in the cytoplasm. Immunofluorescence was performed IFNα using anti-TRIM22 and anti-NS5A 12 h after IFNα treatment. LD staining was conducted with bodipy dye (Life Technologies D2228). The region of co-localization is indicated in white in the figure labeled ‘Co-local'. TRIM22 and NS5A were colocalized near the lipid droplet, which is reported to be important for HCV assembly. co-IP, co-immunoprecipitation; HCV, hepatitis C virus; IFN, interferon; MOI, multiplicity of infection; TRIM, tripartite motif.
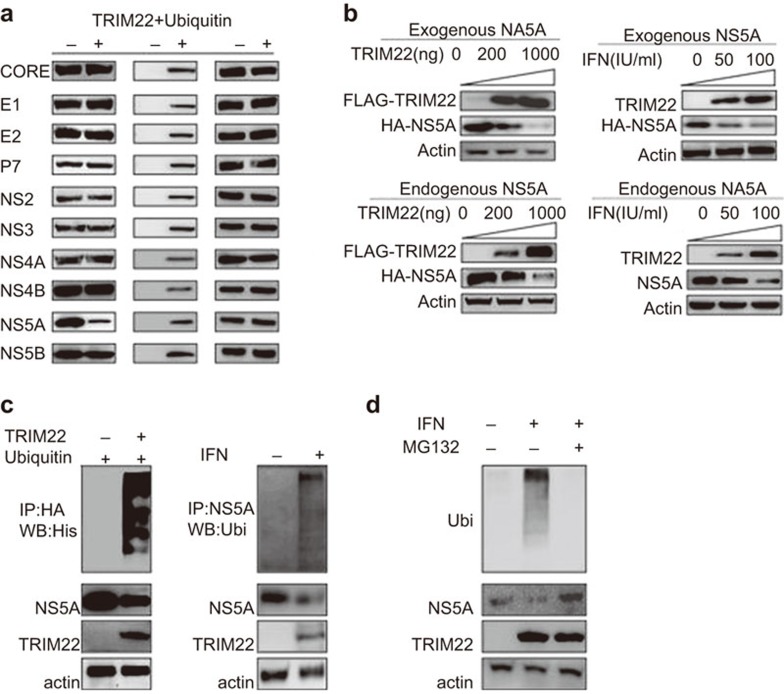
TRIM22 is an ubiquitin ligase for NS5A
TRIM family members feature a common E3 ubiquitin ligase domain; thus, TRIM22 may be a ubiquitin ligase for NS5A. TRIM22 efficiently ligated ubiquitin to NS5A (Figure 5c) and degraded NS5A in a concentration-dependent manner (Figure 5b). MG132 blocked the TRIM22-mediated degradation of NS5A (Figure 5d). TRIM22 does not degrade other HCV proteins, indicating that the degradation is specific for NS5A (Figure 5a). Based on the importance of NS5A in HCV replication and the results of our study, we conclude that TRIM22 is induced by IFNα and plays a role in negatively regulating HCV replication by ligating ubiquitin to NS5A.
Figure 5.
TRIM22 is an ubiquitin ligase of NS5A. (a) TRIM22 specifically degrades NS5A. 293T cells are transfected. HCV proteins are transfected together with TRIM22 and ubiquitin. Western blotting was performed 48 h after plasmid transfection. (b) Exogenous NS5A: Huh-7 cells transfected with HA-NS5A plasmid (100 ng/well in a 24-well plate) were transfected with FLAG-TRIM22 (upper left: 0, 200 and 1000 ng/well in a 24-well plate) and HIS-Ubiquitin (40 ng/well in a 24-well plate) plasmids or treated with IFNα (upper right: IFNα 0, 50 and 100 IU/ml triggered 24 h after HA-NS5A plasmid transfection). After 48 h, samples were collected for western blotting. TRIM22 degraded NS5A (100 ng/well in a 24-well plate) in a dose-dependent manner. Endogenous NS5A: Huh-7-Con-1 replicon cells were transfected with FLAG-TRIM22 (lower left: 0, 200 and 1000 ng/well in a 24-well plate) and HIS-Ubiquitin (40 ng/well in a 24-well plate) or treated with IFNα (lower right: 50 IU/ml). Samples were detected by western blotting 48 h after plasmid transfection or IFNα treatment. TRIM22 degraded endogenous NS5A (100 ng/well in a 24-well plate) in a dose-dependent manner. (c) Upper panel: FLAG-TRIM22 (1.5 µg/well, six-well plate), HA-NS5A (1.5 µg/well, six-well plate) and HIS-Ubiquitin (0.3 µg/well, six-well plate) were expressed in 293T cells. Immunoprecipitation of NS5A with anti-HA antibody and western blotting of ubiquitin with an anti-HIS antibody were performed 24 h after plasmid transfection. TRIM22 efficiently ligated ubiquitin to NS5A. Lower panel: endogenous ubiquitination of NS5A was detected. Huh-7-Con-1 replicon cells were treated with IFNα (50 IU/ml). Twenty-four hours after IFNα treatment, cells were harvested and immunoprecipitated with an anti-NS5A antibody. Then, western blotting was performed with an anti-ubiquitin antibody. (d) MG132 (20 nM) successfully blocked the endogenous ubiquitination of NS5A and NS5A degradation. HCV, hepatitis C virus; IFN, interferon; TRIM, tripartite motif.
Discussion
In the present study, upregulation of TRIM22 by IFNα was associated with a decrease in HCV titer in the blood of patients on the first day after IFNα administration. Because liver biopsy samples were not available, it is unclear whether TRIM22 was also induced in hepatocytes. Previous research13 has demonstrated that TRIM22 is upregulated in liver cell lines, consistent with its up-regulation in PBMCs. In the present study, upregulation of TRIM22 was also observed in Huh-7 cells after IFNα treatment.
There is still no consensus on the localization of TRIM22.19 When 293T or COS7 cells were transfected with human TRIM22 tagged with GFP, TRIM22 was localized in the cytoplasm.19,20,21 By contrast, when HepG2 cells were transfected with myc-tagged TRIM22, TRIM22 was localized in the nucleus.19,22,23 Moreover, in the serum-starved U2OS cell line, TRIM22 was localized in both the nucleus and cytoplasm.19,24,25 Gao and colleagues observed that TRIM22 was mainly localized in the nucleus of HepG2 cells when it suppresses HBV survival.13 The discrepancy in localization compared to our present study may be due to the different cell lines we used (Huh-7 cell lines).
The mechanism by which TRIM22 suppresses HCV replication is of substantial interest. Studies of changes in HCV kinetics during IFNα therapy have demonstrated that the dramatic decrease in viral titer on the first day of IFNα therapy is mainly caused by the inhibition of HCV replication.26 All HCV proteins were screened for interaction with TRIM22, resulting in the sole identification of NS5A. NS5A is a viral factor that helps HCV establish a niche in the host cell by turning off the IFNα signaling pathway.27 The sequence of a certain NS5A region, called the interferon sensitivity-determining region,28 is highly associated with the final outcome of IFNα therapy. Furthermore, experimental evidence suggests that the function of NS5A includes abrogation of STAT-1 translocation,29 suppression of STAT-1 phosphorylation 30 and repression of the protein kinase R protein kinase.31,32,33 Because of its important function in controlling HCV replication, many molecules have been designed to target NS5A, some of which have exhibited promising treatment outcomes in clinical research.34,35 Other potential mechanisms of the anti-HCV effects of TRIM22 should be evaluated in future research.
We performed experiments demonstrating that a single mutation (K to R) of NS5A can all be ubiquitinated, which implies that multiple ubiquitination sites might appear in NS5A (Supplementary Figure 3). At present, it is unclear which lysine in NS5A is ubiquitinated by TRIM22. The sequence of the HCV virus is constantly changing in the host.36 Therefore, a large HCV mutation repertoire exists in every HCV patient.37 To uncover the clinical association between the ubiquitinated sites and the resistance to IFNα, next generation sequencing should be used to determine the mutation status of NS5A at a population level because HCV has a highly dynamic mutational profile.37 Different time points should also be taken into consideration, and the association between the mutational profile and therapy should then be assessed. We will address those questions in a future study.
In this study, we observed that TRIM22 is induced after IFNα treatment and functions to degrade NS5A and, in turn, suppress HCV replication. Overall, our study indicates that TRIM22 has a novel anti-HCV effect during IFNα therapy in chronically HCV-infected patients and that this effect is mediated by the ubiquitination of NS5A.
Authors' contributions
BS, JN and JZ proposed and managed the project. CY designed the research project, performed most of the experiments and wrote the manuscript. XZ performed most of the experiments involving the HCV virus. DS performed the molecular cloning. LY performed the microarray and analyzed the results. SX and CC assisted with molecular cloning and immunoprecipitation experiments. YP and YG recruited the cohort and led the clinical research. MW, XS, HS and JL collected the biological samples and follow-up information from the patients. YG performed the HCV RNA titration assay. XC was responsible for ethical approval.
Acknowledgments
We thank R Bartenschlager for providing the CON1 replicon and T Wakita for providing the JFH1 virus. We thank Wanyin Tao, Qiang Ding, Yu Xiang and Yongfen Xu for experimental support. We thank Dahui Zhao, Shuai Yang and all of the other nurses who helped collect patient blood. We thank Shijie Sun and Jizheng He from Roche Diagnostics, China, for support with the COBAS technique. We thank Li Li and Wenjing Xuan for ordering and preparing the experimental reagents. This work was supported by a grant from the National 973 Key Project (2013CB530504), the National 863 Key Project (2012AA020103), National Science and Technology Major Projects (2013ZX10004-101-005, 2012ZX10002-007-003 and 2013ZX10004-003-003), grants from the National Natural Science Foundation of China (31030029, 31230024 and 81361120409) and by the CAS/SAFEA International Partnership Program for Creative Research Teams.
The authors of this study have no conflicts to disclose.
Footnotes
Supplementary Information accompanies the paper on Cellular & Molecular Immunology's website. (http://www.nature.com/cmi).
Supplementary Information
References
- 1Slomski A. WHO issues guidelines on HCV amid drug cost controversy. JAMA 2014; 311: 2262–2263. [DOI] [PubMed] [Google Scholar]
- 2Chevaliez S, Pawlotsky JM. HCV genome and life cycle. In: Tan SL, editor. Hepatitis C Viruses: Genomes and Molecular Biology. Norfolk: Horizon Bioscience, 2006. [PubMed] [Google Scholar]
- 3Suzuki T, Aizaki H, Murakami K, Shoji I, Wakita T. Molecular biology of hepatitis C virus. J Gastroenterol 2007; 42: 411–423. [DOI] [PubMed] [Google Scholar]
- 4Levrero M. Viral hepatitis and liver cancer: the case of hepatitis C. Oncogene 2006; 25: 3834–3847. [DOI] [PubMed] [Google Scholar]
- 5Lai CL. Antiviral therapy for hepatitis B and C in Asians. J Gastroenterol Hepatol 1999; 14 Suppl: S19–S21. [DOI] [PubMed] [Google Scholar]
- 6Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 2011; 472: 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Prens EP, Kant M, van Dijk G, van der Wel LI, Mourits S, van der Fits L. IFN-alpha enhances poly-IC responses in human keratinocytes by inducing expression of cytosolic innate RNA receptors: relevance for psoriasis. J Invest Dermatol 2008; 128: 932–938. [DOI] [PubMed] [Google Scholar]
- 8Xagorari A, Chlichlia K. Toll-like receptors and viruses: induction of innate antiviral immune responses. Open Microbiol J 2008; 2: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Feld JJ, Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 2005; 436: 967–972. [DOI] [PubMed] [Google Scholar]
- 10Ozato K, Shin DM, Chang TH, Morse HC3rd. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol 2008; 8: 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol 2005; 3: 799–808. [DOI] [PubMed] [Google Scholar]
- 12Barr SD, Smiley JR, Bushman FD. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog 2008; 4: e1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Gao B, Duan Z, Xu W, Xiong S. Tripartite motif-containing 22 inhibits the activity of hepatitis B virus core promoter, which is dependent on nuclear-located RING domain. Hepatology 2009; 50: 424–433. [DOI] [PubMed] [Google Scholar]
- 14Di Pietro A, Kajaste-Rudnitski A, Oteiza A, Nicora L, Towers GJ, Mechti N et al. TRIM22 inhibits influenza A virus infection by targeting the viral nucleoprotein for degradation. J Virol 2013; 87: 4523–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 2005; 11: 791–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA 2005; 102: 9294–9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 1999; 285: 110–113. [DOI] [PubMed] [Google Scholar]
- 18He Y, Weng L, Li R, Li L, Toyoda T, Zhong J. The N-terminal helix alpha0 of hepatitis C virus NS3 protein dictates the subcellular localization and stability of NS3/NS4A complex. Virology 2012; 422: 214–223. [DOI] [PubMed] [Google Scholar]
- 19Hattlmann CJ, Kelly JN, Barr SD. TRIM22: a diverse and dynamic antiviral protein. Mol Biol Int 2012; 2012: 153415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Herr AM, Dressel R, Walter L. Different subcellular localisations of TRIM22 suggest species-specific function. Immunogenetics 2009; 61: 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L et al. The tripartite motif family identifies cell compartments. EMBO J 2001; 20: 2140–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Duan Z, Gao B, Xu W, Xiong S. Identification of TRIM22 as a RING finger E3 ubiquitin ligase. Biochem Biophys Res Commun 2008; 374: 502–506. [DOI] [PubMed] [Google Scholar]
- 23Yu S, Gao B, Duan Z, Xu W, Xiong S. Identification of tripartite motif-containing 22 (TRIM22) as a novel NF-kappaB activator. Biochem Biophys Res Commun 2011; 410: 247–251. [DOI] [PubMed] [Google Scholar]
- 24Petersson J, Lonnbro P, Herr AM, Morgelin M, Gullberg U, Drott K. The human IFN-inducible p53 target gene TRIM22 colocalizes with the centrosome independently of cell cycle phase. Exp Cell Res 2010; 316: 568–579. [DOI] [PubMed] [Google Scholar]
- 25Sivaramakrishnan G, Sun Y, Tan SK, Lin VC. Dynamic localization of tripartite motif-containing 22 in nuclear and nucleolar bodies. Exp Cell Res 2009; 315: 1521–1532. [DOI] [PubMed] [Google Scholar]
- 26Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 1998; 282: 103–107. [DOI] [PubMed] [Google Scholar]
- 27Qashqari H, Al-Mars A, Chaudhary A, Abuzenadah A, Damanhouri G, Alqahtani M et al. Understanding the molecular mechanism(s) of hepatitis C virus (HCV) induced interferon resistance. Infect Genet Evol 2013; 19: 113–119. [DOI] [PubMed] [Google Scholar]
- 28Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C et al. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med 1996; 334: 77–81. [DOI] [PubMed] [Google Scholar]
- 29Cao J, Zhou Y, Gong GZ. Effect of HCV NS5A on STAT1 phosphorylation and nuclear translocation induced by IFN alpha-2b. Zhonghua Gan Zang Bing Za Zhi 2006; 14: 894–897. Chinese. [PubMed] [Google Scholar]
- 30Gong GZ, Cao J, Jiang YF, Zhou Y, Liu B. Hepatitis C virus non-structural 5A abrogates signal transducer and activator of transcription-1 nuclear translocation induced by IFN-alpha through dephosphorylation. World J Gastroenterol 2007; 13: 4080–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Gale MJJr, Korth MJ, Katze MG. Repression of the PKR protein kinase by the hepatitis C virus NS5A protein: a potential mechanism of interferon resistance. Clin Diagn Virol 1998; 10: 157–162. [DOI] [PubMed] [Google Scholar]
- 32Gale MJr, Blakely CM, Kwieciszewski B, Tan SL, Dossett M, Tang NM et al. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol 1998; 18: 5208–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Gale MJJr, Korth MJ, Tang NM, Tan SL, Hopkins DA, Dever TE et al. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 1997; 230: 217–227. [DOI] [PubMed] [Google Scholar]
- 34Pereira AA, Jacobson IM. New and experimental therapies for HCV. Nat Rev Gastroenterol Hepatol 2009; 6: 403–411. [DOI] [PubMed] [Google Scholar]
- 35de Clercq E. The design of drugs for HIV and HCV. Nat Rev Drug Discov 2007; 6: 1001–1018. [DOI] [PubMed] [Google Scholar]
- 36Campo DS, Dimitrova Z, Yamasaki L, Skums P, Lau DT, Vaughan G et al. Next-generation sequencing reveals large connected networks of intra-host HCV variants. BMC Genomics 2014; 15 Suppl 5: S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37Beerenwinkel N, Gunthard HF, Roth V, Metzner KJ. Challenges and opportunities in estimating viral genetic diversity from next-generation sequencing data. Front Microbiol 2012; 3: 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.