Abstract
Mammalian telomeres and subtelomeres are marked by heterochromatic epigenetic modifications, including repressive DNA methylation and histone methylation (e.g., H3K9me3 and H4K20me3). Loss of these epigenetic marks results in increased rates of telomere recombination and elongation. Other than these repressive epigenetic marks, telomeric and subtelomeric H3 and H4 are underacetylated. Yet, whether histone acetylation also regulates telomere length has not been directly addressed. We thought to test the effects of histone acetylation levels on telomere length using histone deacetylase (HDAC) inhibitor (sodium butyrate, NaB) that mediates histone hyperacetylation and histone acetyltransferase (HAT) inhibitor (C646) that mediates histone hypoacetylation. We show that histone hyperacetylation dramatically elongates telomeres in wild-type ES cells, and only slightly elongates telomeres in Terc−/− ES cells, suggesting that Terc is involved in histone acetylation-induced telomere elongation. In contrast, histone hypoacetylation shortens telomeres in both wild-type and Terc−/− ES cells. Additionally, histone hyperacetylation activates 2-cell (2C) specific genes including Zscan4, which is involved in telomere recombination and elongation, whereas histone hypoacetylation represses Zscan4 and 2C genes. These data suggest that histone acetylation levels affect the heterochromatic state at telomeres and subtelomeres, and regulate gene expression at subtelomeres, linking histone acetylation to telomere length maintenance.
Mammalian telomeres contain repetitive G-rich sequences and associated proteins at the ends of linear chromosomes (Blackburn, 2001). Telomeres protect chromosome ends and maintain chromosomal stability (Palm and de Lange, 2008). Telomere length maintenance is primarily achieved by telomerase that adds telomere repeats de novo during each cell division, counteracting telomere erosion (Chan and Blackburn, 2002). Telomere length also can be maintained by telomerase-independent mechanisms, including an alternative lengthening of telomeres (ALT) mechanism, based on homologous recombination between telomere repeats (Muntoni and Reddel, 2005).
Telomeres and subtelomeres are densely compacted with repressive DNA methylation and histone modifications, forming condensed heterochromatin structures (Blasco, 2007). Differential abundance of those epigenetic modifications at telomeres and subtelomeres contributes to the formation of a “closed” or “open” chromatin state, regulating telomere length, possibly through regulating the access of telomerase to telomeres or the ALT mechanism (Blasco, 2007). Mouse embryonic stem (ES) cells deficient for DNA methyltransferases Dnmt1 or Dnmt3a/3b exhibit reduced DNA methylation at subtelomere regions, increased telomere recombination as indicated by telomere sister-chromatid exchange (T-SCE), and elongated telomeres (Gonzalo et al., 2006). Repressive histones H3K9me3 and H4K20me3, as well as heterochromatin protein 1 isoforms, are also enriched at condensed heterochromatin regions (Blasco, 2007). H3K9me3 and H4K20me3 are detected at satellite, telomeres, and active long-terminal repeats, and can spread to proximal unique sequences (Mikkelsen et al., 2007). Mouse embryonic fibroblast (MEF) cells lacking Suv39h1 and Suv39h2 histone methyltransferases (HMTs), which govern methylation of heterochromatic H3K9me3, show abnormal telomere lengthening and increased T-SCE (Garcia-Cao et al., 2004), suggesting an essential role ofH3K9me3 in suppression of telomere length. Similarly, mouse ES and MEF cells deficient for Suv4-20h2 HMTs that is responsible for trimethylating H4K20 display abnormally elongated telomeres and increased T-SCE (Benetti et al., 2007). Furthermore, mouse MEF cells deficient for all three members of retinoblastoma gene family (RB1, RBL1 and RBL2) also exhibit decreased levels of H4K20me3 at telomeres and global reduction of DNA methylation, accompanied by aberrantly elongated telomeres (Gonzalo and Blasco, 2005). In addition, mammalian telomeres and subtelomeres are bound by low levels of acetylated H3 (AcH3) and H4 (AcH4) (Blasco, 2007; Wong, 2010). However, whether histone acetylation also participates in telomere length regulation in ES cells remains elusive.
ES cell cultures are a heterogeneous mixture of metastable cells with fluctuating activation of 2-cell embryo specific genes (2C-genes) and endogenous transposable element (TE) activities (Macfarlan et al., 2012; Torres-Padilla and Chambers, 2014), suggesting that ES cells in the 2C-state might resemble the totipotent zygotes/2C-stage embryos. In this regard, the 2C-state was postulated as a “super” state of ES cells (Surani and Tischler, 2012). Zscan4, specifically expressed in ES cells and 2-cell embryos (Falco et al., 2007; Zalzman et al., 2010), and highly enriched within 2C::tdTomato+ mouse ES cells (Macfarlan et al., 2012), can also faithfully represent the 2C-state of mouse ES cells. Zscan4 is only expressed in about 3–5% of ES cells at any given time, and Zscan4+ and Zscan4− ES cells can interconvert to each other and nearly all ES cells activate Zscan4 at least once during nine passages (Zalzman et al., 2010). Without intermittent activation of Zscan4, embryos delay preimlantation development and ES cells lose their ability to proliferate indefinitely (Falco et al., 2007; Zalzman et al., 2010), suggesting that the equilibrium between the 2C-state and canonical ES state is essential for properembryonic development. The major function of transient Zscan4 expression in ES cells is telomere lengthening by recombination involving T-SCE (Zalzman et al., 2010).
We find that histone acetylation positively regulates telomere length by Zscan4/2C-mediated telomere recombination as well as up-regulation of Terc.
Material and Methods
Mouse ES cells
ES cells were cultured as described (Dan et al., 2013; Dan et al., 2014). The ES cell culture medium consisted of knock-out DMEM (Gibco, Grand Island, NY) with 15% FBS (Hyclone, Grand Island, NY), 1000 U/ml mouse leukemia inhibitory factor (LIF) (ESGRO, Chemicon), 0.1 mM non-essential amino acids, 0.1 mM β-mercaptoethanol, 1 mM l-glutamine, and penicillin (100 U/ml) and streptomycin (100 µg/ml). The culture medium was changed daily and ES cells routinely passaged every 2 days. Histone deacetylase specific inhibitor sodium butyrate (NaB) (B5887, Sigma–Aldrich, Saint Louis, MO) was dissolved in water and histone acetyltransferase specific inhibitor C646 (SML0002, Sigma–Aldrich, Saint Louis, MO) dissolved in DMSO. NaB at 0.2 mM or C646 at 12.5 µM was added to ES culture medium during experiments.
Vector constructs
A putative Zscan4c promoter containing the 2570 bp upstream sequences from Zscan4c start codon (Zalzman et al., 2010) was amplified from mouse ES cell genomic DNA with TransStar Fastpfu polymerase (Transgene, Beijing, China) using the following primers: forward: AGAGATGCTTCTGCATCTGT; reverse: TGTGGTGACAATGGTGTGAAAG. The PCR product was inserted into pEGFP-1 vector at SalI/KpnI sites. The vector was designated as pEGFP-1-Zscan4. The 2570 full-length putative Zscan4c promoter was cut using SacI/SmaI enzymes from pEGFP-1-Zscan4 and inserted into pGL3-basic vector containing several base pairs from pEGFP-1 vector, and the luciferase vector named as pGL3-Zscan4.
Generation of pZscan4-EGFP cells
The pEGFP-1-Zscan4 vector was linearized by XhoI digestion and purified by PCR purification kit (Transgene, Beijing, China). Feeder-free J1 ES cells were transfected with 2 µg linearized vector using Lipofectamine 2000 (Invitrogen, Grand Island, NY) and selected with 400 µg/ml G418 (Invitrogen) for 2 weeks, and a clone with bright green fluorescence was picked and expanded for further experiments.
FACS analysis of Zscan4 positive ES cells
Zscan4-EGFP J1 ES cells (1 × 105 per well in 12-well plate) were cultured with or without 0.2 mM sodium butyrate (NaB) or 12.5 µM C646, or valproic acid (VPA, HDAC inhibitor) at various concentrations for 48 h, and analyzed for percentage of Zscan4-positive cells by fluorescence activated cell sorting using BD FACSAria.
Luciferase reporter assay
Feeder-free Zscan4-EGFP J1 ES cells (1 × 105 per well in 24-well plate) were transfected with 0.8 µg pGL3-Zscan4 vector and 8 ng pRL-SV40 vector (as control) using Lipofectamine 2000 (Invitrogen, Grand Island, NY) according to manufacturer’s instructions. Transfected J1 ES cells were lysed 24 h later with 1 × PLB (positive lysis buffer, Promega, Madison, WI), shaken for 15 min, and then centrifuged at 13,000 rpm for 10 min at 4°C. The supernatants were collected and analyzed for luciferase activity by dual luciferase reporter assay according to manufacturer’s instructions (E1910, Promega).
Western blot
Cells were washed twice in PBS, collected, lysed and boiled in SDS sample buffer at 99°Cfor 5 min. Equal amount of proteins of each cell extracts were resolved by 10–12% Bis-Tris SDS-PAGE and transferred to polyvinylidene difluoride membrane (PVDF, Millipore, Billerica, MA). Non-specific binding was blocked by incubation in 5% skim milk in TBST at room temperature for 1–2 h. Blots were then probed with various primary antibodies, AcH3 (06-599, Millipore, Billerica, MA), H3K9me3 (ab8898, Abcam, Cambridge, UK), Zscan4 (#5114, custom-made), Histone H3 (ab1791, Abcam, Cambridge, UK) and β-actin (P30002, Abmart, Shanghai, China) by overnight incubation at 4°C in 5% skim milk in TBST. Immunoreactive bands were then probed for 1–2 h at room temperature with the appropriate horseradish peroxidase-conjugated secondary anti-Rabbit IgG-HRP (GE Healthcare, NA934V, Piscataway, NJ). The protein bands were detected by Enhanced ECL Amersham™ prime Western blotting detection reagent (GE Healthcare, RPN2232).
Gene expression analysis by quantitative real-time PCR
Total RNA was isolated from cells using RNeasy mini kit (Qiagen, Hilden, Germany). Two micrograms of RNA were subjected to cDNA synthesis using M-MLV Reverse Transcriptase (Invitrogen). Real-time quantitative PCR reactions were set up with the FastStart Universal SYBR Green Master (ROX) (Roche, Basel, Switzerland) and run on the iCycler iQ5 2.0 Standard Edition Optical System (Bio-Rad, Hercules, CA) using primers (Table S1). β-actin served as the internal control.
Telomere quantitative fluorescence in situ hybridization (QFISH)
Telomere length and function (telomere integrity and chromosome stability) were estimated by QFISH. Cells were incubated with 0.5 µg/ml nocodazole for 1.5 h to enrich cells at metaphase. Chromosome spreads were made by a routine method. Metaphase-enriched cells were exposed to hypotonic treatment with 75 mM KCl solution, fixed with methanol:glacial acetic acid (3:1) and spread onto clean slides. Telomere FISH and quantification were performed as described previously (Poon et al., 1999), except for FITC-labeled(CCCTAA)peptide nucleic acid (PNA) probe used in this study. Telomeres were denatured at 80°C for 3 min and hybridized with telomere specific PNA probe (0.5 µg/ml) (Panagene, Korea). Chromosomes were counter-stained with 0.5 µg/ml DAPI. Fluorescence from chromosomes and telomeres was digitally imaged on a Zeiss microscope with FITC/DAPI filters, using AxioCam and AxioVision software 4.6. (Carl Zeiss, Oberkochen, Germany) For quantitative measurement of telomere length, telomere fluorescence intensity was integrated using the TFL-TELO program (gift kindly provided by P. Lansdorp, Terry Fox Laboratory, Vancouver, Canada).
Immunofluorescence microscopy
Zscan4-EGFP J1 ES cells were washed twice in phosphate buffered saline (PBS), then fixed in freshly prepared 3.7% paraformaldehyde in PBS (pH 7.4) for 15 min on ice, permeabilized in 0.1% Triton X-100 (Sigma–Aldrich, Saint Louis, MO) in blocking solution (3% goat serum plus 0.5% BSA in PBS) for 30 min at room temperature, washed three times (each for 15 min), and left in blocking solution for 1 h. Cells were then incubated overnight at 4°C with primary antibodies against Oct4 (sc5279, Santa Cruz, Santa Cruz, CA), Nanog (ab80892, Abcam), Sox2 (AB5603, Millipore), washed three times (each for 15 min), and incubated for 1 h with secondary antibodies, Alexa Fluor 568 Goat anti-Rabbit (A-11011, Invitrogen, Grand Island, NY), or Texas red conjugated anti-mouse IgG (Vector, TI-2000, Burlingame, CA), diluted 1:200 with blocking solution. Samples were washed, and counterstained with 0.5 µg/ml Hoechst33342 (H1398, MP) in Vectashield mounting medium. Fluorescence was detected and imaged using a Zeiss fluorescence microscope (AxioImager Z1).
ChIP-qPCR analysis
ChIP and data analysis were performed as we described previously (Dan et al., 2014), using primary antibody to AcH3 (06-599, Millipore). Normal Rabbit immunoglobulin G (IgG) (Santa Cruz, sc-2027) served as control. Briefly, control and NaB-treated mouse J1 ES cells (48 h) were fixed in freshly prepared 1% paraformaldehyde for 10 min at room temperature. Cells were harvested, and their nuclei extracted, lysed, and sonicated. DNA fragments were then enriched by immunoprecipitation with AcH3 antibody (06-599, Millipore). The eluted protein:DNA complex was reverse-crosslinked at 65°C overnight. DNA was recovered with a MiniElute column (Qiagen) after Proteinase and RNase A treatment. The ChIP-enriched DNA was analyzed by real-time PCR using primers for Zscan4c loci (Table S2) and relative occupancy was normalized to pan-H3 levels.
Statistical analysis
Data were analyzed by ANOVA and means compared by Fisher’s protected least-significant difference (PLSD) using the StatView software from SAS Institute, Inc. (Cary, NC). Significant differences were defined as P < 0.05, 0.01, or lower.
Results
Histone acetylation regulates telomere length of mouse ES cells
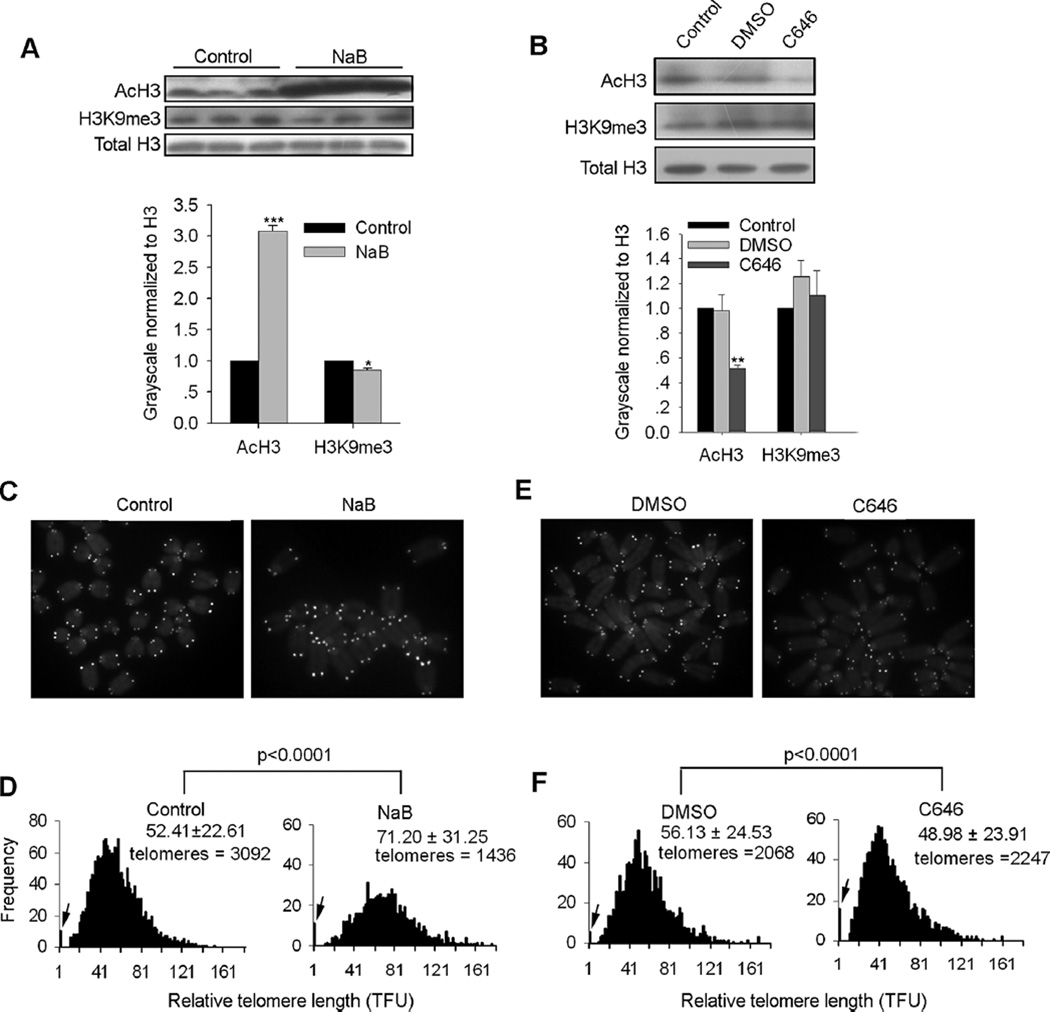
In order to address the effects of levels of histone acetylation on telomere length regulation, we used two commonly accepted small molecules, specific histone deacetylase (HDAC) inhibitor sodium butyrate (NaB) (Ware et al., 2009) and specific histone acetyltransferase (HAT) inhibitor C646 (Bowers et al., 2010). We confirmed that ES cells treated with NaB showed increased levels of acetylated H3 (AcH3) compared with controls by western blot analysis (Fig. 1A, upper and lower panels), whereas C646 treatment reduced levels of AcH3 compared with DMSO treatment as vehicle control (Fig. 1B, upper and lower panels). Next, we performed telomere quantitative fluorescence in situ hybridization (QFISH) (Poon et al., 1999) to assess the effects of NaB or C646 on telomere length. NaB markedly elongated telomeres of ES cells (Fig. 1C and D). In contrast, C646 shortened telomeres in ES cells compared with DMSO treated controls, and also increased telomere loss (Fig. 1E and F). NaB decreased levels of H3K9me3 in ES cells (Fig. 1A), butC646 did not influence levels of H3K9me3 (Fig. 1B) based on western blot and quantification estimated by Bio-Rad Quantity One software. Loss of H3K9me3 led to abnormal telomere lengthening (Garcia-Cao et al., 2004), suggesting that reduced levels of H3K9me3 may partly contribute to telomere elongation in NaB treated ES cells. Taken together, these data suggest that active epigenetic mark histone acetylation also regulates telomere length.
Fig. 1.
Histone acetylation regulates telomere length in ES cells. (A) Western blot analysis of protein levels of AcH3 and H3K9me3 following treatment of sodium butyrate (NaB) in ES cells (upper panel). Relative protein quantity of AcH3 and H3K9me3 normalized to H3 (H3 was arbitrarily set as 1.0) following NaB treatment was estimated by Bio-Rad Quantity One software (lower panel) (three replicates). ES cells were treated with 2 mM NaB in ES medium for 48 h, and total cell lysate used for western blot analysis. (B) Western blot analysis of protein levels of AcH3 and H3K9me3 following treatment of C646 in ES cells (upper panel). Relative protein quantity of AcH3 and H3K9me3 normalized to H3 (H3 was arbitrarily set as 1.0) following C646 treatment was estimated by Bio-Rad Quantity One software (lower panel) (three replicates). ES cells were treated with 12.5 µM C646 in ES medium for 48 h, and total cell lysates used for western blot analysis. A, B, values indicate mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 compared to water controls, analyzed by ANOVA and PLSD analysis. (C, E) QFISH images of wild-type ES cells treated with NaB (C) or C646 (E), compared with control or DMSO vehicle controls, respectively. (D, F) Distribution histogram of relative telomere length of wild-type ES cells treated with NaB (D) or C646 (F), compared with water or DMSO served as controls, respectively, analyzed by telomere Q-FISH and the TFL-TELO software. Significant differences (P < 0.0001) were analyzed by ANOVA and PLSD analysis using Statview software compared to controls. TFU, arbitrary telomere fluorescence unit. Upper right hand corner, the average length ± SD. Arrows on Y-axis, frequency of telomere loss.
Telomerase activity is required in histone hyperacetylation-mediated telomere lengthening
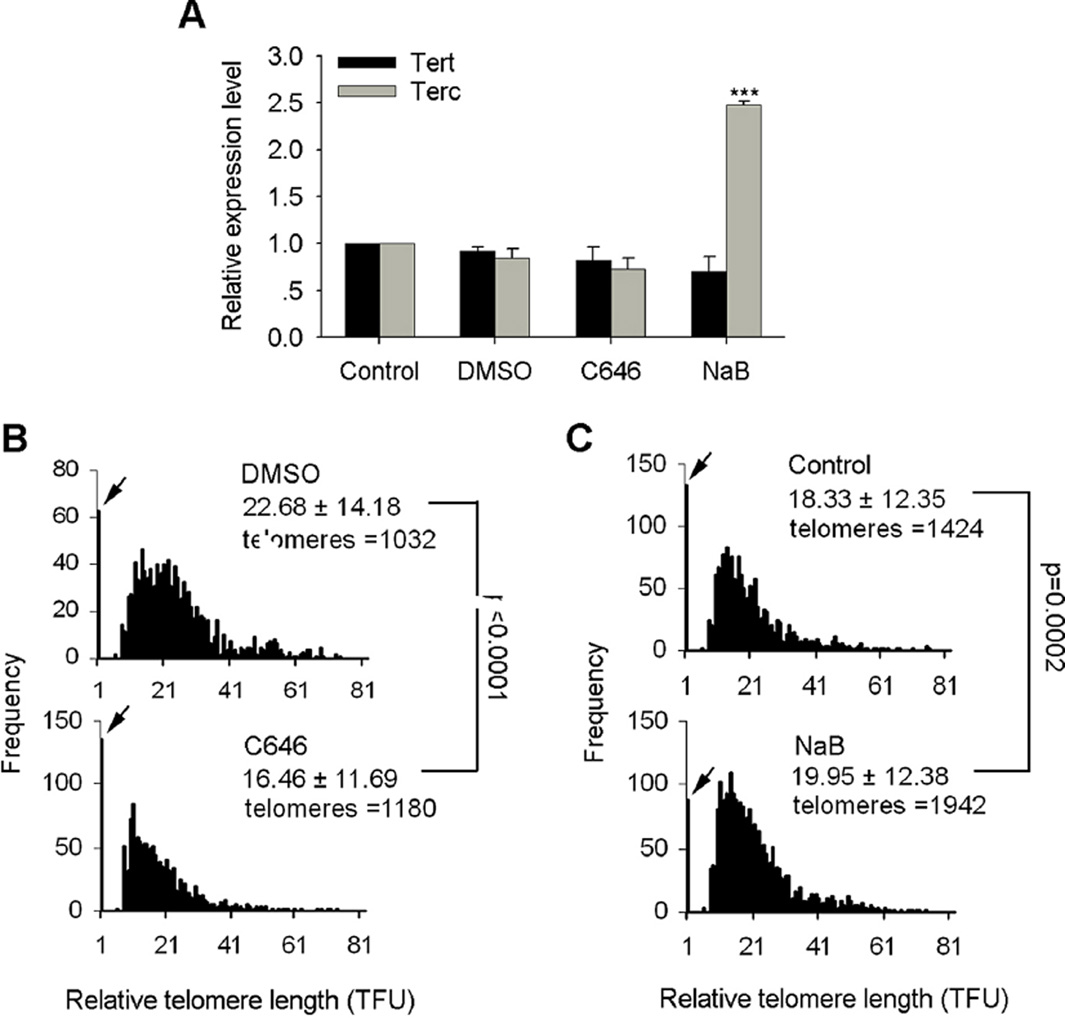
Mammalian telomeres are primarily maintained by telomerase, which is expressed only in a subset of stem cells, including ES cells. We found that levels of histone acetylation mediated by NaB or C646 regulate telomere length in wild-type (WT) ES cells (Fig. 1). It was unclear, however, whether NaB or C646 influences telomere length by affecting telomerase activity. Thus, we analyzed expression levels of telomerase subunits Tert and Terc following NaB and C646 treatments. Expression of Tert and Terc remained at similar levels between C646 treated ES cells and DMSO controls (Fig. 2A). Expression levels of Tert were also not changed in NaB-treated ES cells, but Terc expression was upregulated compared with controls (Fig. 2A), suggesting that increased telomerase expression induced by NaB also may contribute to telomere elongation. Next, we analyzed telomere length in Terc−/− ES cells treated with NaB and C646. Terc−/− ES cells treated with C646 showed noticeable telomere shortening compared with DMSO-treated ES cells (Fig. 2B). However, NaB still elongated telomeres in Terc−/− ES cells (Fig. 2C), although to less extent than WT cells, further supporting the involvement of telomerase activity in histone hyperacetylation-mediated telomere elongation.
Fig. 2.
Histone acetylation regulates telomere length in Terc−/− deficient ES cells. (A) QPCR experiment showed relative expression levels of Terc and Tert following NaB and C646 treatment in ES cells. Terc was upregulated in NaB treated ES cells compared to control ES cells (n = 4 replicates for each). Values indicate mean ± SEM. ***P < 0.001, compared to controls, analyzed by ANOVA and PLSD analysis using Statview software. (B, C) Distribution histogram of relative telomere length of Terc−/− ES cells (F19, first generation telomerase deficient (G1) Terc−/− ES cells) treated with C646 (B) or NaB (C) compared with DMSO or water controls, respectively, analyzed by telomere Q-FIS H and the TFL-TELO software. TFU, arbitrary telomere fluorescence unit. Upper right hand corner, the average length ± SD. Arrows on Y-axis, frequency of telomere loss. Significant differences were analyzed by ANOVA and PLSD analysis using Statview software.
Histone hyperacetylation activates Zscan4 and 2C-specific genes in ES cells
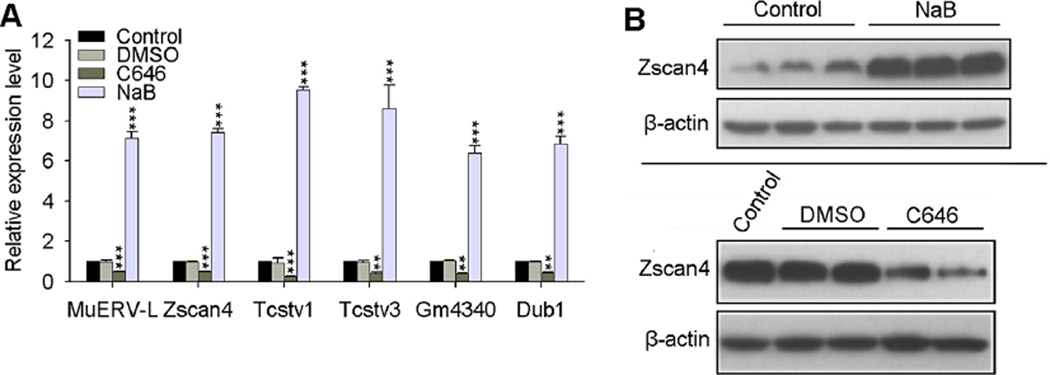
Telomeres lengthen rapidly in one- to two-cell stage embryos (Liu et al., 2007), and also may be driven by Zscan4-mediated telomere recombination or T-SCE (Zalzman et al., 2010). Notably, both mouse 2C::tomato+ mouse ES cells and 2C embryos show increased levels of AcH3 and AcH4 compared with 2C::tomato− mouse ES cells and oocytes, respectively (Wiekowski et al., 1997; Macfarlan et al., 2012), suggesting that Zscan4/2C also could be responsible for histone hyperacetylation-mediated telomere recombination and lengthening. In this regard, we tested whether Zscan4 expression can be regulated by histone acetylation levels. Zscan4, as well as several other 2C specific genes, including MuERV-L, Tcstv1/3, Gm4340, Dub1, all were regulated by the levels of histone acetylation following NaB and C646 treatment as analyzed by qPCR (Fig. 3A). Western blot analysis confirmed that Zscan4 protein was increased following NaB treatment, and decreased by C646 treatment (Fig. 3B). Zscan4 expression levels correlate with telomere length and knockdown of Zscan4 in ES cells shortens telomeres (Zalzman et al., 2010; Wang et al., 2012; Dan et al., 2014; Yin et al., 2014), while overexpression of Zscan4 alone in ES cells or coupled with Yamanaka factors during iPS induction elongates telomeres (Zalzman et al., 2010; Jiang et al., 2013). These data suggest that Zscan4-driven telomere recombination might be responsible for telomere elongation, regulated by the levels of histone acetylation in mouse ES cells.
Fig. 3.
Histone acetylation regulates expression of Zscan4 and other 2C genes. (A) QPCR experiment showed that histone acetylation regulates 2-cell specific genes in ES cells treated with NaB and C646 (n = 3 replicates for each). Values indicate mean ± SEM. **P < 0.01, ***P < 0.001 compared to water controls, analyzed by ANOVA and PLSD analysis. (B) Western blot experiment showed that NaB treatment in ES cells increased the protein level of Zscan4 (upper panel) (three replicates), while C646 treatment decreased the protein level of Zscan4 (lower panel) (two replicates).
Histone acetylation regulates Zscan4 promoter activity
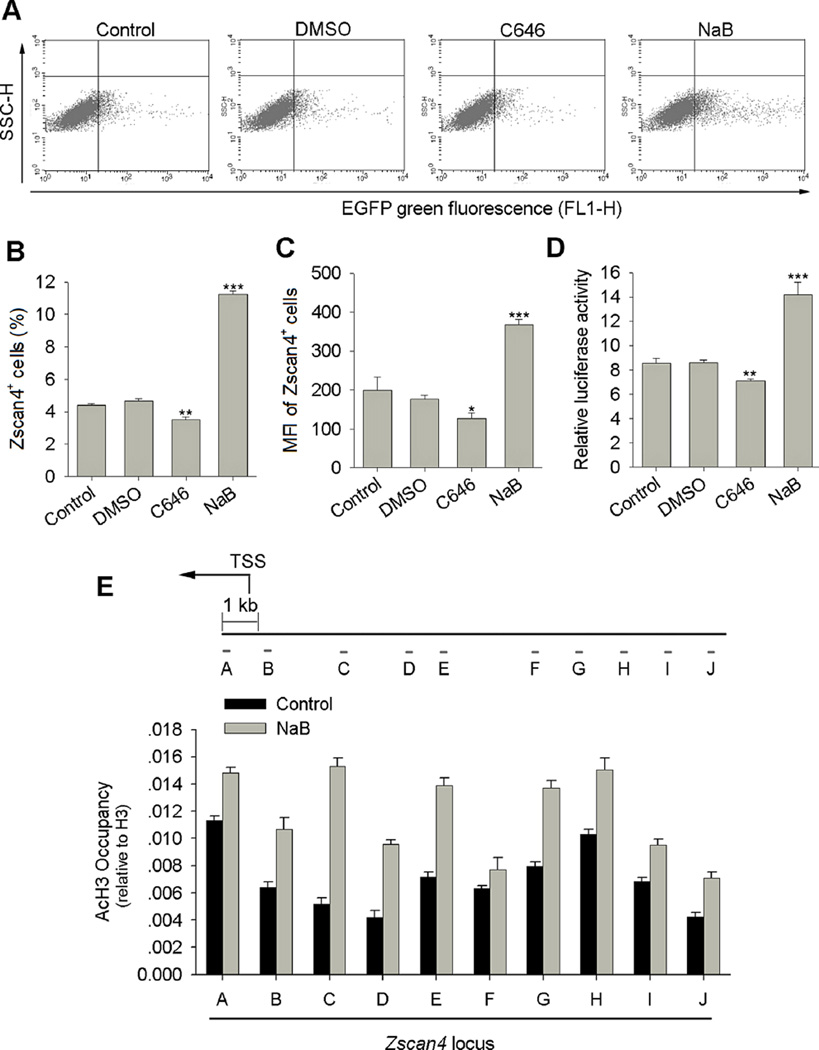
Next, we examined whether the Zscan4+/2C state of mouse ES cell culture was regulated by histone acetylation levels using a Zscan4c promoter-driven EGFP (Zscan4-EGFP) reporter ES cell line that has been shown to faithfully recapitulate the endogenous Zscan4 expression (Zalzman et al., 2010; Dan et al., 2013; Dan et al., 2014). Flow cytometry analysis confirmed that about 3–5% ES cells expressed Zscan4 at any given time (Fig. 4A and B), consistent with a previous report (Zalzman et al., 2010). Remarkably, histone hyperacetylation increased the percentage of Zscan4 positive ES cells by nearly threefold, reaching almost 12% of the ES cell population. Mean fluorescence intensity (MFI) of Zscan4+ ES cells also increased following NaB treatment of ES cells compared with controls (Fig. 4A–C). In contrast, histone hypoacetylation reduced the percentage of Zscan4+ cells and MFI of Zscan4+ cells following C646 treatment compared with DMSO controls (Fig. 4A–C). Valproic acid (VPA), another HDAC inhibitor, also increased the percentage of Zscan4+ ES cells and the MFI of Zscan4+ cells (Supplementary Fig. S1). Moreover, the increase in percentage of Zscan4+ cells was VPA concentration dependent (Supplementary Fig. S1A and B). Consistently, Zscan4 promoter activity analyzed by dual luciferase assay increased in NaB treated ES cells, but decreased in C646 treated ES cells compared with controls (Fig. 4D). The 2C state ES cells lack Oct4, Nanog, Sox2 core pluripotent protein labeling despite minimal changes in their mRNA levels (Macfarlan et al., 2012), indicating post-transcriptional regulation of these three core factors. Independently, we also confirmed the involvement of post-transcriptional regulation of Oct4, Nanog and Sox2 in Zscan4+/2C state ES cells (Supplementary Fig. S2). Collectively, levels of histone acetylation regulate Zscan4+/2C state of mouse ES cells and Zscan4 promoter activity.
Fig. 4.
Histone acetylation regulates Zscan4 promoter activity and 2C state of mouse ES cells. (A) Representative flow cytometry showing the percentage of Zscan4+ cells following NaB and C646 treatments for 48 h in Zscan4-EGFP J1 ES cells. Each one representative plot from four independent replicates is shown. (B) Percentage of Zscan4+ ES cells increases about three fold following NaB treatment, but decreases following C646 treatment. (C) Mean fluorescence intensity (MFI) of Zscan4+ cells increases remarkably following NaB treatment, but decreases following C646 treatment. B, C, four independent replicates; Values indicate mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 analyzed by ANOVA and PLSD analysis compared to water controls. (D) Zscan4c promoter activity increases following NaB treatment, but decreases following C646 treatment, as determined by dual-luciferase reporter assay (n = 4 replicates for each). Values indicate mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 analyzed by ANOVA and PLSD analysis compared to water controls. (E) ChIP-qPCR assay of AcH3 binding at proximal Zscan4 promoter in control and NaB treated ES cells (10 primer pairs 9 kb upstream of the Zscan4c proximal promoter were selected, shown as primer pairs A to J) (n = 3 replicates for each). Values indicate mean ± SD. TSS: Transcription Start Site.
Histone acetylation regulates Zscan4 expression by binding to Zscan4 promoter
Further analysis of the genes activated by histone hyperacetylation and repressed by histone hypoacetylation in Figure 3A identified that some of the genes, including Zscan4, Tcstv1, and Tcstv3, are located at subtelomeric regions, and Zscan4 gene clusters located at subtelomeric regions of chromosome 7. We hypothesized that histone hyperacetylation at Zscan4 promoter located at subtelomeric regions may induce a less condensed chromatin state and thus activation of Zscan4 expression. We performed chromatin immunoprecipitation (ChIP)-qPCR in ES cells using AcH3 antibody and 10 primer pairs 9 kb upstream of Zscan4c translational initiation code. Indeed, the AcH3 occupancy at Zscan4 proximal promoter regions increased following NaB treatment (Fig. 4E), supporting the notion that Zscan4 expression can be positively regulated by histone hyperacetylation.
Discussion
Our data suggest that histone acetylation affects the heterochromatic state at telomeres and subtelomeres, and regulates the expression of nearby genes, including Zscan4.
Heterochromatin is found near centromeres, telomeres, and subtelomeres. Telomeres and subtelomeres are densely compacted with repressive DNA methylation and histone modifications (e.g., H3K9me3 and H4K20me3) that are important negative regulators of mammalian telomere length (Blasco, 2007; Wong, 2010). Moreover, these repressive DNA methylation and histone modifications at subtelomeric regions inhibit expression of nearby genes, recognized as telomere position effects (TPE) (Baur et al., 2001; Koering et al., 2002). Interestingly, we have previously demonstrated that reduction of DNA methylation or levels of H3K9me3 in mouse ES cells results in upregulation of Zscan4 and telomere elongation (Dan et al., 2013; Dan et al., 2014), providing evidence supporting the conclusion that TPE is influenced by repressive epigenetic marks, and suggesting that Zscan4 links reduced levels of repressive epigenetic marks, telomere recombination and telomere elongation. Interestingly, besides Zscan4, other 2C-specific genes regulated by endogenous retrovirus elements (Macfarlan et al., 2012) are also located at subtelomeric or centromeric regions enriched with repeat sequences. Our data show that histone hyperacetylation activates Zscan4+/2C-state in mouse ES cells likely by facilitating formation of a less condensed heterochromatin state at subtelomeres, which in turn de-represses subtelomeric genes, including Zscan4 and Tcstv1/3. In addition, NaB treatment results in upregulation of Terc expression. Zscan4-mediated telomere recombination and increased accessibility of telomerase to telomeres together may be responsible for notable telomere elongation following NaB-mediated histone hyperacetylation.
Consistently, chemical-induced histone acetylation can promote reprogramming to pluripotency as well as telomere elongation. The 2C genes are not properly activated in some cloned embryos (Suzuki et al., 2006), but cloned embryos treated with HDAC inhibitor or deficient for lysine-specific demethylase Lsd1/Kdm1a show enhanced reprogramming efficiency (Li et al., 2009; Shao et al., 2009). Both NaB-mediated hyperacetylation employed in the present study and Lsd1/Kdm1a deficiency in mouse ES cells (Macfarlan et al., 2011) derepress 2C-specific genes. Macfarlan et al. (2012) suggests that overexpression of one or several 2C stage specific genes or inhibition of other 2C gene repressors could be useful strategies to facilitate reprogramming when combined with Yamanaka factors. Indeed, forced expression of Zscan4 combined with Yamanaka factors enhances efficiency of iPS reprogramming from mouse fibroblasts (Hirata et al., 2012; Jiang et al., 2013), and improves the quality of iPS cells tested by tetraploid complementation (Jiang et al., 2013). Moreover, HDAC inhibitors VPA and butyrate can greatly enhance iPS cell reprogramming efficiency (Huangfu et al., 2008; Liang et al., 2010; Mali et al., 2010). Notably, trichostatin A (TSA), another HDAC inhibitor, facilitates telomere elongation in cloned embryos and pigs by somatic cell nuclear transfer (Kong et al., 2014), and also telomere lengthening in the resultant nuclear transfer ES cells generated from telomerase haplo-insufficient cells (Sung et al., 2014). It is possible that histone hyperacetylation by histone deacetylase inhibitors may have contributed to enhanced iPS reprogramming efficiency as well as telomere elongation and maintenance of pluripotent stem cells.
Supplementary Material
Acknowledgments
We thank Dr. Peter Lansdorp for the TFL-TELO software, Maja Okuka for the help of QFISH.
Contract grant sponsor: National Natural Science Foundation of China;
Contract grant number: 31271587.
Contract grant sponsor: Program of International S&T Cooperation;
Contract grant number: 2014DFA30450.
Contract grant sponsor: PCSIRT;
Contract grant number: IRT13023.
Footnotes
The authors declare that they have no competing interests.
Author Contributions: J.D., J.Y., and Y.L. performed all experiments and data analysis. J.D. designed the experiments and wrote the manuscript; A.X. designed and advised the experiments; L.L. designed and advised the experiments, and revised manuscripts.
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
Literature Cited
- Baur JA, Zou Y, Shay JW, Wright WE. Telomere position effect in human cells. Science. 2001;292:2075–2077. doi: 10.1126/science.1062329. [DOI] [PubMed] [Google Scholar]
- Benetti R, Gonzalo S, Jaco I, Schotta G, Klatt P, Jenuwein T, Blasco MA. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J Cell Biol. 2007;178:925–936. doi: 10.1083/jcb.200703081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, Larocca C, Saldanha SA, Abagyan R, Sun Y, Meyers DJ, Marmorstein R, Mahadevan LC, Alani RM, Cole PA. Virtual ligand screening of the p300/CBP histone acetyltransferase: Identification of a selective small molecule inhibitor. Chem Biol. 2010;17:471–482. doi: 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Blackburn EH. New ways not to make ends meet: Telomerase, DNA damage proteins and heterochromatin. Oncogene. 2002;21:553–563. doi: 10.1038/sj.onc.1205082. [DOI] [PubMed] [Google Scholar]
- Dan J, Li M, Yang J, Li J, Okuka M, Ye X, Liu L. Roles for Tbx3 in regulation of two-cell state and telomere elongation in mouse ES cells. Sci Rep. 2013;3:3492. doi: 10.1038/srep03492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J, Liu Y, Liu N, Chiourea M, Okuka M, Wu T, Ye X, Mou C, Wang L, Yin Y, Yuan J, Zuo B, Wang F, Li Z, Pan X, Yin Z, Chen L, Keefe DL, Gagos S, Xiao A, Liu L. Rif1 maintains telomere length homeostasis of ESCs by mediating heterochromatin silencing. Dev Cell. 2014;29:7–19. doi: 10.1016/j.devcel.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MS. Zscan4: A novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol. 2007;307:539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Blasco MA. Role of Rb family in the epigenetic definition of chromatin. Cell Cycle. 2005;4:752–755. doi: 10.4161/cc.4.6.1720. [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- Hirata T, Amano T, Nakatake Y, Amano M, Piao Y, Hoang HG, Ko MS. Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells. Sci Rep. 2012;2:208. doi: 10.1038/srep00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Jiang J, Lv W, Ye X, Wang L, Zhang M, Yang H, Okuka M, Zhou C, Zhang X, Liu L, Li J. Zscan4 promotes genomic stability during reprogramming and dramatically improves the quality of iPS cells as demonstrated by tetraploid complementation. Cell Res. 2013;23:92–106. doi: 10.1038/cr.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koering CE, Pollice A, Zibella MP, Bauwens S, Puisieux A, Brunori M, Brun C, Martins L, Sabatier L, Pulitzer JF, Gilson E. Human telomeric position effect is determined by chromosomal context and telomeric chromatin integrity. EMBO Rep. 2002;3:1055–1061. doi: 10.1093/embo-reports/kvf215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Ji G, Xie B, Li J, Mao J, Wang J, Liu S, Liu L, Liu Z. Telomere elongation facilitated by trichostatin a in cloned embryos and pigs by somatic cell nuclear transfer. Stem Cell Rev. 2014;10:399–407. doi: 10.1007/s12015-014-9499-y. [DOI] [PubMed] [Google Scholar]
- Li W, Zhou H, Abujarour R, Zhu S, Young Joo J, Lin T, Hao E, Scholer HR, Hayek A, Ding S. Generation of human-induced pluripotent stem cells in the absence of exogenous Sox2. Stem Cells. 2009;27:2992–3000. doi: 10.1002/stem.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Taranova O, Xia K, Zhang Y. Butyrate promotes induced pluripotent stem cell generation. J Biol Chem. 2010;285:25516–25521. doi: 10.1074/jbc.M110.142059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Bailey SM, Okuka M, Munoz P, Li C, Zhou L, Wu C, Czerwiec E, Sandler L, Seyfang A, Blasco MA, Keefe DL. Telomere lengthening early in development. Nat Cell Biol. 2007;9:1436–1441. doi: 10.1038/ncb1664. [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J, Andrews SE, Franco L, Rosenfeld MG, Ren B, Pfaff SL. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011;25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, Brodsky RA, Ohm JE, Yu W, Baylin SB, Yusa K, Bradley A, Meyers DJ, Mukherjee C, Cole PA, Cheng L. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28:713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntoni A, Reddel RR. The first molecular details of ALT in human tumor cells. Hum Mol Genet. 2005;14:R191–R196. doi: 10.1093/hmg/ddi266. [DOI] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Poon SS, Martens UM, Ward RK, Lansdorp PM. Telomere length measurements using digital fluorescence microscopy. Cytometry. 1999;36:267–278. doi: 10.1002/(sici)1097-0320(19990801)36:4<267::aid-cyto1>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Shao GB, Ding HM, Gao WL, Li SH, Wu CF, Xu YX, Liu HL. Effect of trychostatin A treatment on gene expression in cloned mouse embryos. Theriogenology. 2009;71:1245–1252. doi: 10.1016/j.theriogenology.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Sung LY, Chang WF, Zhang Q, Liu CC, Liou JY, Chang CC, Ou-Yang H, Guo R, Fu H, Cheng WT, Ding ST, Chen CM, Okuka M, Keefe DL, Chen YE, Liu L, Xu J. Telomere elongation and naive pluripotent stem cells achieved from telomerase haplo-insufficient cells by somatic cell nuclear transfer. Cell Rep. 2014;9:1603–1609. doi: 10.1016/j.celrep.2014.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani A, Tischler J. Stem cells: A sporadic super state. Nature. 2012;487:43–45. doi: 10.1038/487043a. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Minami N, Kono T, Imai H. Zygotically activated genes are suppressed in mouse nuclear transferred embryos. Cloning Stem Cells. 2006;8:295–304. doi: 10.1089/clo.2006.8.295. [DOI] [PubMed] [Google Scholar]
- Torres-Padilla ME, Chambers I. Transcription factor heterogeneity in pluripotent stem cells: A stochastic advantage. Development. 2014;141:2173–2181. doi: 10.1242/dev.102624. [DOI] [PubMed] [Google Scholar]
- Wang F, Yin Y, Ye X, Liu K, Zhu H, Wang L, Chiourea M, Okuka M, Ji G, Dan J, Zuo B, Li M, Zhang Q, Liu N, Chen L, Pan X, Gagos S, Keefe DL, Liu L. Molecular insights into the heterogeneity of telomere reprogramming in induced pluripotent stem cells. Cell Res. 2012;22:757–768. doi: 10.1038/cr.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CB, Wang L, Mecham BH, Shen L, Nelson AM, Bar M, Lamba DA, Dauphin DS, Buckingham B, Askari B, Lim R, Tewari M, Gartler SM, Issa JP, Pavlidis P, Duan Z, Blau CA. Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell Stem Cell. 2009;4:359–369. doi: 10.1016/j.stem.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiekowski M, Miranda M, Nothias JY, DePamphilis ML. Changes in histone synthesis and modification at the beginning of mouse development correlate with the establishment of chromatin mediated repression of transcription. J Cell Sci. 1997;110:1147–1158. doi: 10.1242/jcs.110.10.1147. [DOI] [PubMed] [Google Scholar]
- Wong LH. Epigenetic regulation of telomere chromatin integrity in pluripotent embryonic stem cells. Epigenomics. 2010;2:639–655. doi: 10.2217/epi.10.49. [DOI] [PubMed] [Google Scholar]
- Yin Y, Liu N, Ye X, Guo R, Hao J, Wang F, Liu L. Telomere elongation in parthenogenetic stem cells. Protein Cell. 2014;5:8–11. doi: 10.1007/s13238-013-0006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, Wersto RP, Ko MS. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.