Significance
Repressor element 1-silencing transcription factor (REST) is a transcriptional repressor that regulates nervous system development. Normally expressed at low levels by mature neurons, REST is up-regulated in various brain pathologies. Using a light-sensitive domain from the oat plant (Avena sativa), we engineered novel optogenetic proteins that inhibit REST activity when illuminated by blue light, thus obtaining the spatial and temporal control of the transcription of REST target genes. This approach may have an impact in the development of new therapies for all the diseases in which REST is dysregulated, such as epilepsy, ischemia, and cancers of various origin, as such therapies may counteract the long-term changes in gene expression that take place in the context of the pathological brain.
Keywords: optogenetics, AsLOV2, REST/NRSF, gene transcription, molecular dynamics
Abstract
Optogenetics provides new ways to activate gene transcription; however, no attempts have been made as yet to modulate mammalian transcription factors. We report the light-mediated regulation of the repressor element 1 (RE1)-silencing transcription factor (REST), a master regulator of neural genes. To tune REST activity, we selected two protein domains that impair REST-DNA binding or recruitment of the cofactor mSin3a. Computational modeling guided the fusion of the inhibitory domains to the light-sensitive Avena sativa light–oxygen–voltage-sensing (LOV) 2-phototrophin 1 (AsLOV2). By expressing AsLOV2 chimeras in Neuro2a cells, we achieved light-dependent modulation of REST target genes that was associated with an improved neural differentiation. In primary neurons, light-mediated REST inhibition increased Na+-channel 1.2 and brain-derived neurotrophic factor transcription and boosted Na+ currents and neuronal firing. This optogenetic approach allows the coordinated expression of a cluster of genes impinging on neuronal activity, providing a tool for studying neuronal physiology and correcting gene expression changes taking place in brain diseases.
The development and maturation of the nervous system rely on the temporally and spatially precise modulation of gene expression, coordinated by transcriptional enhancers and repressors that bind specific sequences on gene promoters (1, 2). In this context, the key role of the repressor element 1 (RE1)-silencing transcription factor/neural restrictive silencer factor (REST/NRSF, henceforth referred to as REST) has been extensively reported (3, 4). REST binds to gene promoters containing the RE1 consensus site and mediates cell-specific gene repression (5) by recruiting the corepressors mSin3a (6) and CoREST (7) at its N- and C-terminal domains, respectively. Corepressors, in turn, recruit multiple chromatin remodeling factors that ultimately repress gene transcription by densely packing the genomic material. REST levels decrease during brain development and are relatively low in mature neurons (8); however, increased REST expression has been reported in a large array of brain pathologies. In this regard, whether the increase of REST is protective or deleterious for neural cells is still debated. A transient increase in REST expression was associated with homeostatic plasticity in vitro, resulting in the repression of Na+ channel 1.2 (Nav1.2) during sustained network hyperexcitation (9). Moreover, REST is induced by nonautonomous Wnt signaling in the aging brain, protecting neurons from oxidative stress and amyloid β-induced toxicity (10). On the other hand, increased REST levels have been associated with the onset of several brain diseases such as Huntington disease (HD) (11), epilepsy (12), stroke (13), and tumors of neural origin (14, 15). To interfere with REST dysregulation in neuronal pathologies, various molecular strategies have been developed with the purpose of restoring the correct levels of expression of REST target genes (16–18). Such approaches, however, suffer from a number of limitations, including their transient activity or the possibility to generate important side effects when used in long-term applications.
Optogenetics has the ability to exert an on-demand multimodal control of several cellular processes with appropriate time scales (19). Recently, an array of light-gated protein modules has provided the possibility to dissect the function of cellular networks, including signal transduction (20), translation (21), and transcription processes (22). Thus, to effectively and dynamically modulate REST activity, we engineered recombinant light-sensitive proteins able to modulate REST action, by using interfering domains fused to the photosensitive light–oxygen–voltage-sensing (LOV) 2 domain of Avena sativa phototrophin 1 (AsLOV2). AsLOV2 comprises an inner flavin-binding domain and a C-terminal α-helix (Jα) folded in the dark. On blue light (450–470 nm) stimulation, photon absorption leads to major conformational changes that result in the unwinding of the Jα helix and exposure of the C-terminal fused domain (23, 24).
In this work, we demonstrate the activity of two photo-switchable chimeras composed of AsLOV2 fused to the minimal REST-interacting sequence of the corepressor mSin3a (25) (AsLOV2-PAH1) or to the active domain of the REST inhibitor REST-interacting LIM domain protein (RILP) (26) (AsLOV2-RILP N313). The former chimera interferes with the recruitment of endogenous mSin3a by direct competition for its binding site with REST, whereas the latter directly competes with REST-DNA binding by binding the Zn-finger domains of the repressor factor. Both chimeras specifically inhibit REST activity on illumination, causing an increase in the transcription of REST target genes and enhanced firing activity in primary neurons. These probes potentially provide a way to finely tune the transcription of the large array of REST-controlled neuronal genes, possibly counteracting the long-term changes in gene expression taking place in brain diseases.
Results
Strategies to Inhibit REST Activity.
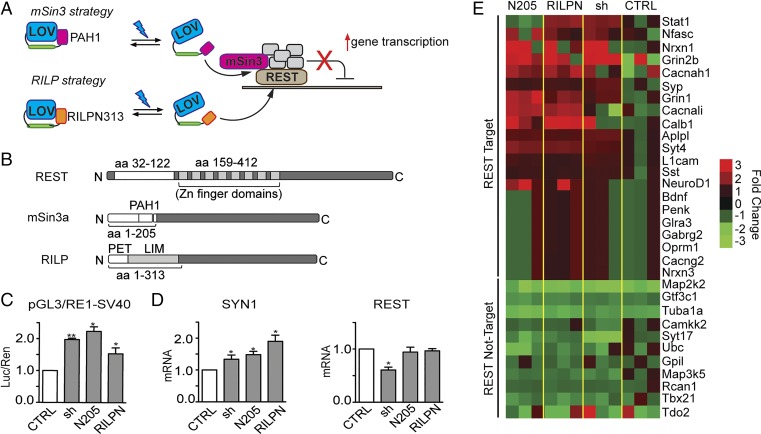
To modulate REST activity, we planned to engineer chimeric proteins formed by the light-sensitive AsLOV2 driving the effects of a REST-interfering domain. According to our model, AsLOV2 would sterically block the effector domain fused to its C terminus in the dark, whereas the unfolding of the Jα-helix upon illumination would cause the reversible exposure of the effector sequence. Such domains would bind REST, thus preventing the assembly of the inhibitory complex and consequently increasing the transcription of REST-target genes (Fig. 1A). To select REST-interfering molecules, we focused on domains able to alter two key events in REST activity, namely the recruitment of the cofactor mSin3a and the REST-RE1 site interaction. To impair mSin3a recruitment, we used the N-terminal portion of mSin3a (mSin3a-N205), which was identified as the minimal REST binding region of mSin3a (6) (Fig. 1B). To interfere with RE1 recognition, we chose REST-interacting LIM domain protein (RILP), an endogenous REST interactor that, by binding to the Zn-finger domains of the repressor, displaces it from the target chromatin (26, 27). We used the first N-terminal portion of RILP (RILP-N313), which contains the minimal REST binding domain (26) (Fig. 1B).
Fig. 1.
Strategies for REST inhibition. (A) Schematic cartoon of the inhibition strategy. mSin3a (Upper) or RILP (Lower) interfering domains are directly fused to AsLOV2. In the dark, AsLOV2 is in a closed conformation, masking the REST-binding sites. On blue light illumination (470 nm), AsLOV2 unfolds thus freeing the C-terminal domains to interact with REST, displacing the endogenous mSin3a or the entire REST complex from target DNA. This would result in the increased transcription of REST-target genes. (B) Schematic representation of REST, mSin3a, and RILP protein sequences. The interacting portions between REST and mSin3a/RILP are highlighted. PAH1, paired amphipathic helix 1; PET, Prickle Espinas Testin; LIM, Lin11, Isl-1, and Mec-3. (C) HeLa cells were transfected with a reporter vector in which the expression of the luciferase gene is modulated by the SV40 promoter fused to a RE1 cis-site (pGL3-RE1/SV40), in the absence (white bars) or presence (gray bars) of expression plasmids encoding for mSin3a N205 (N205), RILP N313 (RILPN), or shRNA against REST (sh), as indicated. Control samples (CTRL) were cotransfected with the empty vector corresponding to the effector plasmids. Luciferase activity was measured 48 h after transfection. Data were first normalized to the activity of the cotransfected TK-Renilla reporter vector and subsequently to the activity of the reporter gene alone, set to 1 (*P < 0.05; **P < 0.01; one-way ANOVA followed by the Tukey's multiple comparison test vs. control; n = 3 independent experiments). Luc/Ren, Luciferase/Renilla ratio. (D) SYN1 and REST mRNA levels were quantified by qRT-PCR in N2a cells 48 h after the transfection of expression plasmids encoding for mSin3a N205, RILP N313, or shRNA against REST, as indicated. RPS9, GAPDH, and HPRT1 were used as control housekeeping genes. (*P < 0.05 vs. control; one-way ANOVA followed by the Tukey's multiple comparison test vs. control; n = 3 independent experiments). (E) Heat map of various RE1-containing and noncontaining genes whose transcription was analyzed by the NanoString nCounter gene expression system in undifferentiated N2a cells 48 h after the transfection of expression plasmids encoding for mSin3a N205, RILP N313, shRNA against REST or an empty vector (CTRL). Values were normalized against five housekeeping genes (PPIA; Pgk1; Hdac3; GAPDH; and HPRT) and then reported as ratio of the control samples. The color represents the expression level of each gene (red for high expression, green for low expression). The analysis was performed using the nSolver Analysis Software 2.5. Primer sequences and numerical values are reported in Tables S1 and S2.
If these domains inhibit the binding of REST, their expression will result in an increased transcription of REST-target genes. To verify this hypothesis, we performed gene reporter assays using a construct in which luciferase expression was driven by the SV40 promoter fused to a single RE1 cis-site. Both RILP-N313 and mSin3a-N205 were able to decrease REST-mediated inhibition of the SV40 promoter, increasing luciferase transcription, whereas the same peptides tested on a non–RE1-containing reporter were totally ineffective (Fig. 1C and Fig. S1A). We then tested the effect of the domains on nuclear chromatin, by analyzing their ability to modulate the expression of the neuron-specific SYN1 gene, a well-known REST target (28), in Neuro2a (N2a) neuroblastoma cells. Indeed, a comparable increase in SYN1 gene transcription was apparent when cells were transfected with either mSin3a-N205 or RILP-N313 or silenced for REST with shRNAs (Fig. 1D). Interestingly, no change in REST mRNA levels was revealed under the same experimental conditions, confirming that expression of either mSin3a-N205 or RILP-N313 did not affect REST expression, but only REST activity on target genes (Fig. 1D). A more comprehensive analysis of several REST-target (n = 21) and nontarget (n = 11) genes was performed using the NanoString nCounter gene expression system, a technology able to capture and count individual mRNA transcripts (29). An increase in mRNA of REST-target genes, comparable to the effect of REST silencing, was observed in N2a expressing either mSin3A N205 or RILP N313, whereas no effect was detected on non–REST-target genes abundance (Fig. 1E and Tables S1 and S2). All together, these data confirm that the mSin3a-N205 and RILP-N313 interfering domains effectively decrease REST activity both on exogenous promoters and on endogenous REST-target genes.
Fig. S1.
(A) HeLa cells were transfected with the pGL3-SV40 reporter vector in the absence (white bars) or presence (gray bars) of expression plasmids encoding for mSin3a N205 or RILP N313. (B) HeLa cells were transfected with the pGL3-SV40 reporter vector in the absence (white bars) or presence (gray bars) of expression plasmids encoding for the closed (C) or open (O) forms of AsLOV2-PAH1b or RILP N313. Control samples were cotransfected with the empty vector corresponding to the effector plasmids. Luciferase activity was measured 48 h after transfection. Data were first normalized to the activity of the cotransfected TK-Renilla reporter vector and subsequently to the activity of the reporter gene alone, set to 1 (one-way ANOVA followed by the Tukey’s multiple comparison test; n = 3 independent experiments).
Table S1.
NanoString nCounter code set design
| Gene name | Accession no. | REST-target | Position | Target sequence |
| REST-target genes | ||||
| Stat1 | NM_009283.3 | YES | 1591–1690 | ACGCTGGGAACAGAACTAATGAGGGGCCTCTCATTGTCACCGAAGAACTTCACTCTCTTAGCTTTGAAACCCAGTTGTGCCAGCCAGGCTTGGTGATTGA |
| Nfasc | NM_001160316.1 | YES | 4936–5035 | CTCTTCGTATGCCTTATACAGCTCGGATCTAACCCTGCGGTTCCCAGATCCCTAGCCCTATTCTGCAAGTCTTGATGCCTCCAGTGATGTGTGTCTCGGG |
| Nrxn1 | NM_177284.2 | YES | 721–820 | AGATGCCCTCACTTAAACATCATCGGATGCAAATGGATCAGTGATCGCTCTTGAGCCTCGGTGGCCCTCTTTTTCAGAACGTTGCCTCCAAAGTGTATCC |
| Grin2b | NM_008171.3 | YES | 6341–6440 | GGGAAAGCTCTTCGTATAAGGCTTTGTGAAAGAGCCATTACAGTAGGGTGAGAGAGGGGGATGTTTTTAGTCATTAACGGTAGGGTTAGTGAGAAAGGGG |
| Cacna1h | NM_021415.4 | YES | 3213–3312 | ATCGTCACTGTGTTTCAGATCTTGACACAGGAAGACTGGAATGTGGTTCTTTACAACGGCATGGCTTCCACCTCGTCCTGGGCTGCCCTTTACTTTGTGG |
| Syp | NM_009305.2 | YES | 733–832 | TGTTTGGCTTCCTGAACCTGGTGCTCTGGGTTGGCAACCTATGGTTCGTGTTCAAGGAGACAGGCTGGGCCGCCCCATTCATGCGCGCACCTCCAGGCGC |
| Grin1 | NM_008169.2 | YES | 493–592 | ACAGATGGCCCTGTCAGTGTGTGAGGACCTCATCTCTAGCCAGGTCTACGCTATCCTAGTTAGTCACCCGCCTACTCCCAACGACCACTTCACTCCCACC |
| Cacna1i | NM_001044308.2 | YES | 9301–9400 | TGCCCTGGCTTATTCCTGTCTTCGGCTCTGGTCCTGGCTTTCCCTCAGAGGAGGATGAACGAATCATGAGCAGTACTTACCTGTCTGCTTGTCCCTAATA |
| Calb1 | NM_009788.4 | YES | 344–443 | ATGGAAAAATAGGAATTGTAGAGTTGGCTCACGTCTTACCCACAGAAGAGAATTTCTTGCTGCTCTTTCGATGCCAGCAACTGAAGTCCTGCGAGGAATT |
| Aplp1 | NM_007467.3 | YES | 2301–2400 | CTGGGTCCCAGGTATGTATGTCACTCCCTGGAATTCACCATCCCACGTTTCTTCACTAACATCCCAATAAAGTCCTCTTTCCCACCCGGCCAAAAAAAAA |
| Syt4 | NM_009308.3 | YES | 1121–1220 | TCTGGTCTCTCTCTGTTATCAGTCCACTACAAACACGCTCACTGTGGTGGTCTTAAAAGCGCGGCACCTACCGAAATCTGATGTGTCTGGACTTTCAGAT |
| L1cam | NM_008478.3 | YES | 3561–3660 | TGAAGACTAATGGAACTGGCCCTGTGCGAGTTTCTACTACAGGTAGCTTTGCCTCCGAGGGCTGGTTCATCGCCTTTGTCAGCGCTATCATTCTCTTGCT |
| Sst | NM_009215.1 | YES | 47–146 | CCTGCGACTAGACTGACCCACCGCGCTCCAGCTTGGCTGCCTGAGGCAAGGAAGATGCTGTCCTGCCGTCTCCAGTGCGCCCTGGCTGCGCTCTGCATCG |
| Neurod1 | NM_010894.2 | YES | 565–664 | CCTGCGCTCAGGCAAAAGCCCTGATCTGGTCTCCTTCGTACAGACGCTCTGCAAAGGTTTGTCCCAGCCCACTACCAATTTGGTCGCCGGCTGCCTGCAG |
| Bdnf | NM_007540.4 | YES | 3261–3360 | AGTCCCGTCTGTACTTTACCCTTTGGGGTTAGAAGTCAAGTTGGAAGCCTGAATGAATGGACCCAATGAGAACTAGTGTTAAGCCCATTTCCCTAGTCAG |
| Penk | NM_001002927.2 | YES | 1011–1110 | CGCTTTGCTGAGTCTCTGCCCTCCGATGAAGAAGGCGAAAATTACTCGAAAGAAGTTCCTGAGATAGAGAAAAGATACGGGGGCTTTATGCGGTTCTGAA |
| Glra3 | NM_080438.2 | YES | 757–856 | GGACTTGCGGTACTGCACTAAACACTACAATACAGGAAAGTTTACATGCATAGAAGTGCGATTCCATCTTGAGCGTCAGATGGGCTATTACTTGATCCAG |
| Gabrg2 | NM_177408.5 | YES | 1614–1713 | TTAGAACAAGAGCTGTTACACTGAGCAAGATACCTTTGAGCAACAGCAATGAAAACAGTGGAAGCTGGGAGGGTTTAAAGTGGCATTATCAGTCTTTGAC |
| Oprm1 | NM_001039652.1 | YES | 1197–1296 | GATTCCAGAAACCACTTTCCAGACTGTTTCCTGGCACTTCTGCATTGCCTTGGGTTACACAAACAGCTGCCTGAACCCAGTTCTTTATGCGTTCCTGGAT |
| Cacng2 | NM_007583.2 | YES | 645–744 | GACGCGGACTACGAAGCTGACACCGCAGAGTATTTCCTCCGGGCCGTGAGGGCCTCGAGTATCTTCCCGATCCTGAGTGTGATCCTGCTTTTCATGGGTG |
| Nrxn3 | NM_001198587.1 | YES | 6111–6210 | CTCAAAATCTACCATGGCATTCACTCCATGTAGCAGGTTGTGGTGTCTCTAGAACCAATTGTTAGTTTCCTAATGCTTTGGTGAACCCATGTGGGATGAC |
| Non–REST-target and housekeeping genes | ||||
| Map2k2 | NM_023138.4 | NO | 2268–2367 | CACATTCCTAAATACTAGGAAGGCTGAGTCGGGAAAACGACAGGTTTTGGGCCACTGTGGGCTACCTAGTGAGAATGTCTTACATCATGGAAATGGTGCA |
| Gtf3c1 | NM_207239.1 | NO | 4587–4686 | ACCAACGGCATGCTAGACCAGCCTGATCATTTTTCTTTCAAGGACCTGGATAGCAGTGACCCTTCAAATGACCTGGTGGCATTTTCTTTGGACAGCCCTG |
| Tuba1a | NM_011653.2 | NO | 1379–1478 | GGGAGGAAGAAGGAGAGGAATACTAAATTAAATGTCACAAGGTGCTGCTTCCACAGGGATGTTTATTGTGTTCCAACACAGAAAGTTGTGGTCTGATCAG |
| Camkk2 | NM_145358.2 | NO | 1321–1420 | AAAGGCGTTGGATGTTTGGGCCATGGGTGTGACGCTGTACTGCTTTGTCTTTGGCCAGTGCCCTTTCATGGATGAACGAATCATGTGTTTGCACAGTAAG |
| Syt17 | NM_138649.1 | NO | 1391–1490 | AACTGGAAAACGCCAGCCTAGTATTCACAGTGTTCGGCCACAACATGAAAAGCAGCAATGACTTCATCGGGAGGATCGTCATCGGCCAGTATTCCTCCGG |
| Ubc | NM_019639.4 | NO | 22–121 | GCGGAGTCGCCCGAGGTCACAGCCCTGCCCTCCCACACAAAGCCCCTCAATCTCTGGACGCCACCGTGAAACAACTCCGTGAGAGAGACGATGCAGATCT |
| Gpi1 | NM_008155.3 | NO | 2676–2775 | CAGGGATGAACTCGATTATACCCTTCAAAAGACAGTTTTAGCAGGGTGTCATTACTGTCACATTCATATCCTGGCTATCAGTGAGTCCTTGTGAGACTGA |
| Map3k5 | NM_008580.4 | NO | 641–740 | CATCATCCTCTACTGCGATACTAATTCCGATTCACTCCAGTCCCTGAAGGAAATTATTTGCCAGAAGAATACTGTGTGCACCGGGAACTACACCTTCATC |
| Rcan1 | NM_001081549.1 | NO | 1061–1160 | ACACAAGGACACTGGGGACATCCTGAGAAAACTGATAGTTCTTGTAATTGCTCATTTCTAGGTTCTGTTTTTGGCAAGGACAGGTTGACTGGTGGCCCAG |
| Tbx21 | NM_019507.1 | NO | 626–725 | CACTAAGCAAGGACGGCGAATGTTCCCATTCCTGTCCTTCACCGTGGCTGGGCTGGAGCCCACAAGCCATTACAGGATGTTTGTGGATGTGGTCTTGGTG |
| Tdo2 | NM_019911.2 | NO | 496–595 | AGAGTCTACAGTTCCGGCTGCTGGAAAATAAGATTGGTGTTCTTCAGAGCTTGAGAGTCCCTTACAACAGGAAACACTATCGTGATAACTTTGGAGGAGA |
| Gapdh | NM_001001303.1 | HK | 891–990 | AGGTTGTCTCCTGCGACTTCAACAGCAACTCCCACTCTTCCACCTTCGATGCCGGGGCTGGCATTGCTCTCAATGACAACTTTGTCAAGCTCATTTCCTG |
| Hdac3 | NM_010411.2 | HK | 1026–1125 | GGCCATTAGTGAGGAACTTCCCTATAGTGAATACTTCGAGTACTTTGCCCCAGATTTCACACTCCATCCAGATGTCAGCACCCGCATCGAGAATCAGAAC |
| Hprt | NM_013556.2 | HK | 31–130 | TGCTGAGGCGGCGAGGGAGAGCGTTGGGCTTACCTCACTGCTTTCCGGAGCGGTAGCACCTCCTCCGCCGGCTTCCTCCTCAGACCGCTTTTTGCCGCGA |
| Pgk1 | NM_008828.2 | HK | 37–136 | CCGGCATTCTGCACGCTTCAAAAGCGCACGTCTGCCGCGCTGTTCTCCTCTTCCTCATCTCCGGGCCTTTCGACCTCACGGTGTTGCCAAAATGTCGCTT |
| Ppia | NM_008907.1 | HK | 391–490 | CCAAGACTGAATGGCTGGATGGCAAGCATGTGGTCTTTGGGAAGGTGAAAGAAGGCATGAACATTGTGGAAGCCATGGAGCGTTTTGGGTCCAGGAATGG |
Optimal sequences were designed on the chosen genes by Nanostring Technologies. The table reports the following for each gene: (i) the gene name; (ii) the accession number; (iii) whether it is a REST-target gene (YES/NO) or a housekeeping (HK) gene; the position (iv) and the sequence (v) of the targeted region.
Table S2.
NanoString nCounter gene expression data
| Gene name | Accession no. | REST-target | mSin3 N205_1 | mSin3 N205_2 | mSin3 N205_3 | RILP N313_1 | RILP N313_2 | RILP N313_3 | shREST_1 | shREST_2 | shREST_3 | CTRL_1 | CTRL_2 | CTRL_3 |
| REST-target genes | ||||||||||||||
| Stat1 | NM_009283.3 | YES | −1.05 | −1.32 | −1.13 | 1.9 | 1.85 | 2.06 | 1.87 | 1.72 | 1.83 | 1.02 | −1.09 | 1.07 |
| Nfasc | NM_001160316.1 | YES | 2.08 | −1.13 | 2.31 | 1.75 | 1.27 | 1.04 | 1.84 | 1.64 | 1.06 | 1.25 | 1.06 | −1.32 |
| Nrxn1 | NM_177284.2 | YES | 3.78 | 2.71 | −1.07 | 8.92 | −1.07 | 1.62 | 4.75 | 5.89 | −1.07 | −1.07 | −1.03 | 1.09 |
| Grin2b | NM_008171.3 | YES | 2.56 | 4.79 | 1.99 | 5 | 2.35 | −1.1 | 2.66 | 2.74 | 5.08 | −1.9 | 3.17 | −1.67 |
| Cacna1h | NM_021415.4 | YES | 2.25 | 1.82 | 1.64 | 2.17 | 1.62 | 2.4 | 1.96 | −1.06 | 1.1 | −1.86 | −1.13 | 2.11 |
| Syp | NM_009305.2 | YES | 1.23 | 1.11 | 1.22 | 1.26 | 1.22 | 1.14 | 1.37 | 1.58 | 1.54 | −1.02 | 1.1 | −1.08 |
| Grin1 | NM_008169.2 | YES | 2.34 | 2.11 | 2.81 | 1.42 | 1.94 | 1.76 | 1.21 | 1.5 | 1.51 | −1.03 | −1.13 | 1.17 |
| Cacna1i | NM_001044308.2 | YES | 2.39 | 2.18 | 2.29 | 2.03 | 2.28 | 2.2 | 1.06 | −1.01 | −2.58 | 1.3 | −1.22 | −1.06 |
| Calb1 | NM_009788.4 | YES | 2.98 | 1.98 | 1.24 | 4.53 | 4.19 | 2.59 | 4.75 | −1.05 | −1.07 | −1.07 | −1.03 | 1.09 |
| Aplp1 | NM_007467.3 | YES | 1.44 | 1.42 | 1.46 | 1.37 | 1.31 | 1.38 | 1.54 | 1.59 | 1.56 | 1 | −1.01 | 1.01 |
| Syt4 | NM_009308.3 | YES | 1.9 | 1.72 | 1.72 | 1.67 | 1.7 | 1.64 | 1.48 | 1.39 | 1.34 | 1.01 | −1.04 | 1.02 |
| L1cam | NM_008478.3 | YES | 1.29 | 1.18 | 1.2 | 1.07 | 1.06 | 1.12 | 1.31 | 1.26 | 1.25 | 1 | −1.02 | 1.02 |
| Sst | NM_009215.1 | YES | 1.09 | 1.17 | 1.16 | 1.03 | 1.12 | 1.11 | 1.19 | 1.21 | 1.19 | 1.09 | −1.19 | 1.09 |
| Neurod1 | NM_010894.2 | YES | 1.98 | 3.8 | 1.37 | 1.08 | 6.49 | 1.44 | 1.15 | 1.22 | −1.02 | −1.07 | −1.03 | 1.09 |
| Bdnf | NM_007540.4 | YES | −1.07 | −1.07 | 1.37 | 1.08 | 1.02 | 1.44 | 1.15 | 1.22 | −1.02 | −1.07 | −1.03 | 1.09 |
| Penk | NM_001002927.2 | YES | −1.07 | −1.07 | 1.37 | 1.08 | 1.02 | 1.44 | 1.15 | 1.22 | −1.02 | −1.07 | −1.03 | 1.09 |
| Glra3 | NM_080438.2 | YES | −1.07 | −1.07 | 1.37 | 1.08 | 1.02 | 1.44 | 1.15 | 1.22 | −1.02 | −1.07 | −1.03 | 1.09 |
| Gabrg2 | NM_177408.5 | YES | −1.07 | −1.07 | 1.37 | 1.08 | 1.02 | 1.44 | 1.15 | 1.22 | −1.02 | −1.07 | −1.03 | 1.09 |
| Oprm1 | NM_001039652.1 | YES | −1.07 | −1.07 | 1.37 | 1.08 | 1.02 | 1.44 | 1.15 | 1.22 | −1.02 | −1.07 | −1.03 | 1.09 |
| Cacng2 | NM_007583.2 | YES | −1.07 | −1.07 | 1.37 | 1 | 1.02 | 1.44 | 1.15 | 1.22 | −1.02 | −1.07 | −1.03 | 1.09 |
| Nrxn3 | NM_001198587.1 | YES | −1.07 | −1.07 | 1.37 | 1 | 1.02 | 1.44 | 1.15 | 1.22 | −1.02 | −1.07 | −1.03 | 1.09 |
| Non–REST-target and housekeeping genes | ||||||||||||||
| Map2k2 | NM_023138.4 | NO | −1.48 | −2.34 | −1.75 | −1.84 | −1.9 | −1.63 | −1.87 | −2.03 | −2.05 | −1.86 | −1.6 | −1.93 |
| Gtf3c1 | NM_207239.1 | NO | −1.36 | −1.41 | −1.26 | −1.3 | −1.31 | −1.18 | −1.21 | −1.18 | −1.29 | −1.2 | −1.24 | −1.1 |
| Tuba1a | NM_011653.2 | NO | −1.63 | −1.82 | −1.7 | −1.49 | −1.59 | −1.49 | −1.28 | −1.43 | −1.38 | −1.44 | −1.46 | −1.49 |
| Camkk2 | NM_145358.2 | NO | −1.01 | −1.14 | −1.04 | −1.05 | −1.02 | 1 | −1.04 | −1.28 | −1.27 | 1.02 | −1.05 | 1.05 |
| Syt17 | NM_138649.1 | NO | −1.22 | −1.63 | −1.33 | −1.26 | −1.14 | −1.11 | −1.9 | −1.99 | −2.02 | 1.03 | −1 | −1.03 |
| Ubc | NM_019639.4 | NO | −1.74 | −1.76 | −1.25 | −1.08 | −1.33 | −1.15 | −1.21 | −1.51 | 1.09 | 1.14 | −1.31 | 1.15 |
| Gpi1 | NM_008155.3 | NO | −1.15 | 1.06 | −1.13 | −1.01 | −1.27 | −1.05 | 1.2 | −1.21 | −1.01 | −1.01 | 1.1 | −1.09 |
| Map3k5 | NM_008580.4 | NO | −1.22 | −1.12 | −1.15 | −1.15 | −1.09 | −1.25 | −1.29 | −1.16 | −1.39 | 1.03 | −1.09 | 1.06 |
| Rcan1 | NM_001081549.1 | NO | −1.07 | −1.04 | −1 | −1.11 | −1.06 | −1.07 | −1.03 | −1.04 | −1.11 | −1.08 | −1.05 | 1.14 |
| Tbx21 | NM_019507.1 | NO | −1.38 | −1.23 | −1.08 | −1.21 | −1.43 | −1.26 | −1.23 | −1.39 | −1.01 | −1.28 | 1.34 | −1.05 |
| Tdo2 | NM_019911.2 | NO | −1.13 | −1.65 | 1.34 | −1.65 | −1.65 | 1.86 | 2.87 | −1.27 | −1.58 | 2.26 | −1.59 | −1.42 |
| Gapdh | NM_001001303.1 | HK | ||||||||||||
| Hdac3 | NM_010411.2 | HK | ||||||||||||
| Hprt | NM_013556.2 | HK | ||||||||||||
| Pgk1 | NM_008828.2 | HK | ||||||||||||
| Ppia | NM_008907.1 | HK |
NanoString nCounter gene expression data. The table reports the following for each gene: (i) the gene name; (ii) the accession number; (iii) whether it is a REST-target gene (YES/NO) or a housekeeping (HK) gene; and (iv) the fold change of expression. Values were processed by using the nSolver Analysis Software Version 2.5 (nanoString Technologies). by removing the experimental background; normalizing to the HK genes; calculating the ratio with respect to the control samples; and visualizing the obtained data as a heat map (Fig. 1E).
Structural Predictions and Optimization of the AsLOV2-Based Competitor of the mSin3a/REST Complex.
To obtain the optogenetic control of mSin3a-N205 activity, we directly fused the C terminus of the complete AsLOV2 sequence to the N terminus of the interfering peptide (AsLOV2-N205). To test whether AsLOV2-N205 activity was conformation dependent, we cotransfected N2a cells with the RE1-SV40 reporter and the AsLOV2-N205 chimera containing two light-insensitive mutated forms of the LOV2 domain: (i) the C450A mutant mimicking the closed “dark” state (30) and (ii) the I539E mutant maintaining the unfolded “lit” state by a constitutive destabilization of the Jα helix (31). Although abundantly expressed, the constitutively open AsLOV2-N205 construct was not able to interfere with REST (Fig. S2), likely due to a persistent steric blockade by AsLOV2 despite the Jα-helix unfolding. We thus restricted the N205 sequence to its minimal REST-interacting portion, i.e., the paired amphipathic helix 1 (PAH1) domain of mSin3a, whose interaction with the N terminus of REST was previously characterized (25, 32, 33) (Fig. 2A).
Fig. S2.
(Left) N2a cells were transfected with a control plasmid (CTRL) or an effector vector encoding for AsLOV2-N205, as indicated. The expression of the construct (asterisk) in total cell lysate was analyzed by Western blot by using an anti-histidine tag antibody. (Right) HeLa cells were transfected with the pGL3-RE1/SV40 reporter vector in the absence (white bars) or presence (gray bars) of expression plasmids encoding for closed (C) or open (O) AsLOV2-N205 constructs, as indicated. Control samples were cotransfected with the empty vector corresponding to the effector plasmids. Luciferase activity was measured 48 h after transfection. Data were first normalized to the activity of the cotransfected TK-Renilla reporter vector and subsequently to the activity of the reporter gene alone, set to 1. Luc/Ren, luciferase/Renilla ratio (one-way ANOVA followed by the Tukey’s multiple comparison test; n = 3 independent experiments).
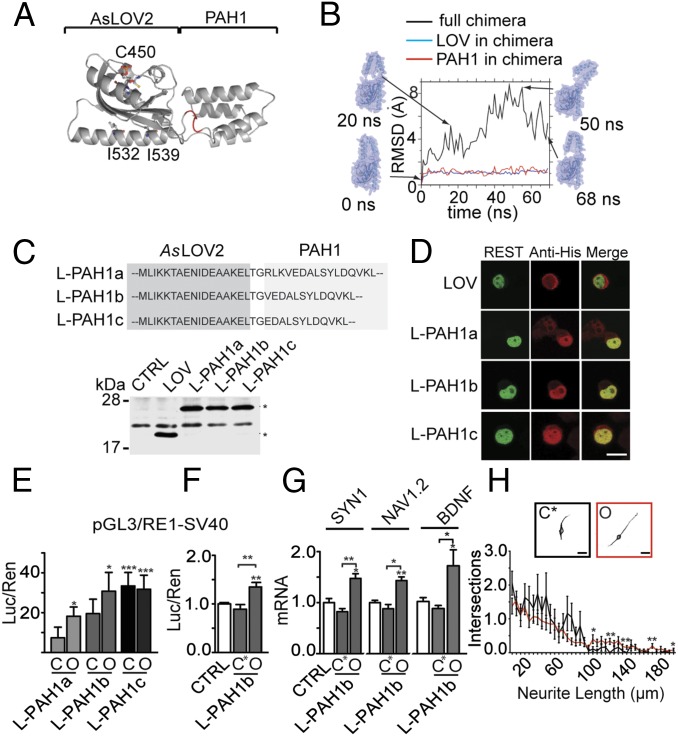
Fig. 2.
Building of the AsLOV2-PAH1 chimera. (A) Model of the AsLOV2-PAH1 chimera resulting from RosettaDock (35) and built using the LOV2 domain of A. sativa phototropin 1 (PDB ID code 2V1A) and the PAH1 domain of mSin3a (PDB ID code 2CZY). The backbone of AsLOV2 and PAH1 domains is represented as gray ribbons, whereas the LOV2 chromophore and the residues C450, I532, and I539 are represented as ball-and-stick. The linker amino acids are highlighted in red. (B) Conformational heterogeneity of the full chimera and its two domains along the MD simulation, calculated as RMSD from the starting structure. Large RMSD values for the full chimera correspond to highly distorted conformations generated because of the linker. Snapshots of conformations at different simulation times are also shown, with the molecular surface represented as transparent spheres. (C, Upper) Amino acid sequences of AsLOV2-PAH1 constructs, limited to the linker region. (C, Lower) N2a cells were transfected with a control plasmid (CTRL) or effector vectors encoding for AsLOV2 alone or AsLOV2-PAH1a, -b, or -c, as indicated. The expression of the constructs in total cellular lysates was analyzed by Western blot by using anti-histidine tag antibodies. Specific immunoreactive bands are indicated with asterisks. (D) Confocal images of N2a cells cotransfected with GFP-REST and either AsLOV2 alone or AsLOV2-PAH1a, -b, or -c, as indicated. N2a cells were processed for indirect immunofluorescence using anti-histidine tag antibodies (red) to detect AsLOV2 constructs or anti-EGFP (green) to detect GFP-REST. The overlay images (merge) reveal colocalization of REST and the AsLOV2-PAH1 constructs in the nuclear compartment. (Scale bar, 10 μm.) (E and F) HeLa cells were transfected with the pGL3-RE1/SV40 reporter vector in the absence (white bars) or presence (gray bars) of expression plasmids encoding for closed (C) or open (O) AsLOV2-PAH1a, AsLOV2-PAH1b, AsLOV2-PAH1c (E) or expression plasmids encoding for the closed double mutant (C450A-I532A) or open AsLOV2-PAH1b (F). Control samples were cotransfected with the empty vector corresponding to the effector plasmids. Luciferase activity was measured 48 h after transfection. Data were first normalized to the activity of the cotransfected TK-Renilla reporter vector and subsequently to the activity of the reporter gene alone, set to 1. (*P < 0.05; **P < 0.01 ***P < 0.001; one-way ANOVA followed by the Tukey’s multiple comparison test vs. control or the indicated group; n = 3 independent experiments). Luc/Ren, luciferase/Renilla ratio. (G) Undifferentiated N2a cells were transfected with plasmids encoding for the closed double mutant (C450A-I532A) or open variants of the LOV2-PAH1b chimera, as indicated. The endogenous mRNA levels of SYN1, NAV1.2, and BDNF genes were quantified via qRT-PCR. GAPDH and HPRT1 were used as control housekeeping genes (*P < 0.05; **P < 0.01; one-way ANOVA followed by the Tukey’s multiple comparison test vs. control or the indicated group; n = 3 independent experiments). (H) Sholl analysis of N2a cells transfected with expression plasmids encoding for the closed (C) or open (O) mutants of AsLOV2-PAH1b and differentiated with Retinoic Acid (*P < 0.05; **P < 0.01; Student t test; n = 3 independent experiments). Representative cells are shown in the Insets. (Scale bar, 20 μm.) CTRL, control; LOV, AsLOV2; L-PAH1, AsLOV2-PAH1; L-PAH1b C*, AsLOV2-PAH1b (C450A-I532A).
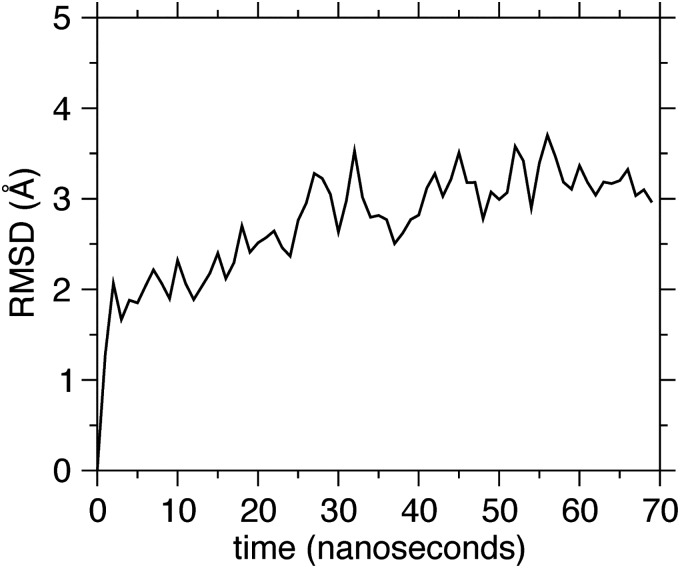
The PAH1 structure comprises a bundle of four parallel α-helices, with the REST-interacting residues situated at one side of the bundle, close to the N terminus (Fig. 2A). Thus, we anticipated that by fusing the AsLOV2 C-terminal helix to the N terminus of PAH1, the REST binding region would be occluded in the dark state, whereas the light-evoked unwinding of the Jα-helix would bring the domains apart and make the binding site accessible to REST. To test this hypothesis and obtain information on the full protein structure and conformations at atomic resolution, we built a computational model of the chimera, starting from the previously determined all-atom structures of the AsLOV2 (in the dark state) and PAH1 domains (33, 34). First, using the RosettaDock program (35), we searched the optimal relative orientation of the two domains in an associated complex, starting from arbitrary positions characterized by a small distance between the AsLOV2 C terminus and the PAH1 N terminus. Among the best scoring conformations, we picked the one that minimized the exposure of the REST binding domain in PAH1. A single protein chain was then created by connecting the two termini (Fig. 2A). This structure was then subjected to molecular dynamics (MD) simulations in solution to assess its stability and investigate its possible conformations. The two domains maintained their functional folded patterns (Fig. 2B); however, we observed a high degree of flexibility in the linker region connecting the AsLOV2-Jα helix to the effector PAH1 that might induce exposure of the PAH1-REST binding region even in the dark. To avoid this, we designed a chimera with a truncated form of the PAH1 domain in which three N-terminal residues were deleted from the linker and subjected it to MD simulations. Results showed that the conformation of the truncated chimera is more stable than the full length, with no exposure of the PAH1-REST binding surface in the dark state (Fig. S3). In conclusion, the computational analysis suggests a role of the linker length in the efficiency of the construct.
Fig. S3.
Conformational stability of the dark state structure of the AsLOV2-PAH1 chimera after deletion of three residues at the linker region, calculated along an MD simulation as RMSD from the starting structure.
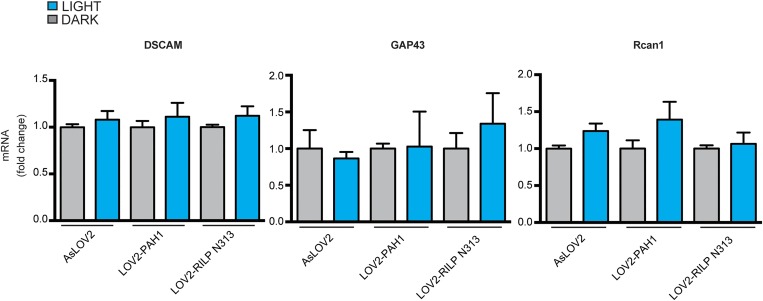
Building on these results, we engineered three chimeras with various linker lengths, obtained by progressively shortening the N terminus of the PAH1 domain (AsLOV2-PAH1a, -b, and -c; Fig. 2C, Upper). All of the constructs were abundantly expressed in N2a cells (Fig. 2C, Lower) in which they colocalized with REST in the nucleus, whereas AsLOV2 alone was strictly cytosolic (Fig. 2D). Remarkably, the chimeras were specifically targeted to the nucleus, i.e., to the appropriate cellular compartment to influence REST activity on chromatin, even in the absence of nuclear localization sequences. When tested in a gene reporter assay, the constitutively open PAH1a and PAH1b chimeras significantly inhibited REST activity on target RE1 elements, whereas the corresponding dark chimeras or the same probes tested on a RE1-null reporter were virtually ineffective (Fig. 2E and Fig. S1B). On the other hand, the PAH1c chimera, containing the shortest linker, inhibited REST activity also in the constitutively closed form, suggesting that PAH1 escaped the LOV2 steric inhibition (Fig. 2E). We chose the AsLOV2-PAH1b construct for all subsequent experiments, as it was the one showing the best effect in the open conformation. To further improve the stability of the chimera in the dark, we introduced the I532A point mutation in the AsLOV2 sequence, known to increase Jα-helix stability (36). The introduction of the second mutation completely abolished the residual activity of the dark construct on the exogenously expressed RE1-SV40 promoter (Fig. 2F). Similarly, when the AsLOV2-PAH1b constructs were expressed in N2a cells, the transcription of the endogenous SYN1, NAV1.2, and BDNF genes, which are well-known REST targets (9, 28), was significantly up-regulated only in cells containing the open chimera (Fig. 2G). On the contrary, the analysis of four additional genes not containing RE1 sites, namely APP, DSCAM, GAP43, and Rcan1, did not detect any difference (Fig. S4).
Fig. S4.
Undifferentiated N2a cells were transfected with plasmids encoding for the closed (C) or open (O) variants of the LOV2-PAH1b or LOV2-RILPN313 chimeras, as indicated. The endogenous mRNA levels of the reported genes were quantified by qRT-PCR. GAPDH and HPRT1 were used as control housekeeping genes (one-way ANOVA followed by the Tukey’s multiple comparison test; n = 3 independent experiments).
To study whether these transcriptional changes were accompanied by changes in the neuronal differentiation of N2a cells, we measured neurite length and arborization after transfection with the open and closed forms of AsLOV2-PAH1b. We previously found that differentiation of N2a cells induced by retinoic acid (RA) treatment is associated with loss of endogenous REST activity and increased expression of its target genes (28). Indeed, we found that neurite outgrowth was enhanced in differentiating N2a cells transfected with the open form of the AsLOV2-PAH1b with respect to the cells expressing the closed form of the chimera (Fig. 2H). Taken together, the above results indicate that the optimized chimeric protein AsLOV2-PAH1b effectively interferes with the formation of the REST/mSin3a complex in a conformation-dependent manner.
Expression and Validation of the AsLOV2-Based Inhibitor of REST-DNA Binding.
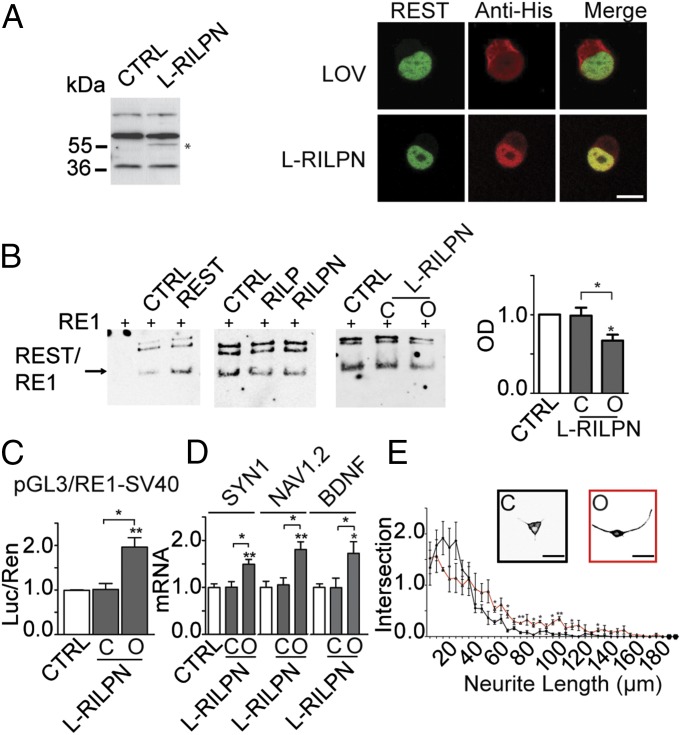
To achieve the light-driven modulation of REST-DNA binding, we used the REST binding domain of RILP. Because the atomic structure of the RILP protein has not been resolved, it was not possible to obtain a detailed structural model. Thus, we fused the C terminus of AsLOV2 in frame with the N terminus of the previously described RILP N313 domain, obtaining AsLOV2-RILP N313. The fusion protein was expressed in mammalian cells and colocalized with REST in the nuclear compartment, similar to what observed with the AsLOV2-PAH1 probe (Fig. 3A). To verify the ability of AsLOV2-RILP N313 to inhibit the binding of REST to specific DNA targets, we performed EMSAs on nuclear extracts from N2a cells, using a canonical RE1 sequence as probe, and chimeras containing either the C450A (dark, closed) (30) or the I539E (lit, open) (31) AsLOV2 mutant (Fig. 3B). In agreement with the results obtained with the exogenous RE1-SV40 promoter (Fig. 1C), REST-RE1 binding was reduced on transfection of both full-length RILP and RILP N313, demonstrating that the latter was as effective as the full-length protein in inhibiting the REST-DNA interaction. Interestingly, the intensity of the REST-RE1 specific band was not affected in cells transfected with the closed AsLOV2-RILP N313, whereas it was markedly reduced on expression of the open AsLOV2-RILP N313 chimera (Fig. 3B).
Fig. 3.
Engineering of AsLOV2-RILP N313. (A, Left) N2a cells were transfected with control (CTRL) or effector vectors encoding for AsLOV2-RILP N313 as indicated. The expression of the constructs in total cellular lysates was analyzed by Western blot by using an anti-histidine tag antibody. The specific immunoreactive band is indicated with an asterisk. (A, Right) Confocal images of N2a cells cotransfected with GFP-REST and either AsLOV2 alone or AsLOV2-RILP N313, as indicated. N2a cells were processed using anti-histidine tag antibody (red) to detect AsLOV2 constructs or anti-EGFP (green) to detect GFP-REST. The overlay images (merge) reveal colocalization of REST and the AsLOV2-RILP N313 construct in the nuclear compartment. (Scale bar, 10 μm.) (B, Left) gel shift assays of biotinylated RE1. The probe was incubated with nuclear extracts from undifferentiated N2a cells that had been previously transfected with the indicated constructs. RE1-containing bands were revealed by HRP-streptavidin followed by chemiluminescence. The REST-specific band is indicated with an arrow. (B, Right) quantification of the REST/RE1 complex is provided, data are expressed as fold change with respect to the CTRL sample, set to 1 (*P < 0.05; one-way ANOVA followed by the Tukey’s multiple comparison test vs. control or the indicated group; n = 4 independent experiments). (C) HeLa cells were transfected with the pGL3-RE1/SV40 reporter vector in the absence (white bars) or presence (gray bars) of expression plasmids encoding for the closed (C) or open (O) mutants of AsLOV2-RILP N313, as indicated. Control samples were cotransfected with the empty vector corresponding to the effector plasmids. Luciferase activity was measured 48 h after transfection. Data were first normalized to the activity of the cotransfected TK-Renilla reporter vector and subsequently to the activity of the reporter gene alone, set to 1. (*P < 0.05; **P < 0.01; one-way ANOVA followed by the Tukey’s multiple comparison test vs. control or the indicated group; n = 3 independent experiments). Luc/Ren, luciferase/Renilla ratio. (D) Undifferentiated N2a cells were transfected with plasmids encoding for the closed or open variants of the LOV2-RILP N313 chimera, as indicated. The endogenous mRNA levels of the SYN1, NAV1.2, and BDNF genes were quantified via qRT-PCR. GAPDH and HPRT1 were used as control housekeeping genes (*P < 0.05; **P < 0.01; one-way ANOVA followed by the Tukey’s multiple comparison test vs. control or the indicated group; n = 3 independent experiments). (E) Sholl analysis of N2a cells transfected with expression plasmids encoding for the closed (C) or open (O) mutants of AsLOV2-RILP N313 and differentiated with retinoic acid (*P < 0.05; **P < 0.01; Student t test; n = 3 independent experiments). Representative cells are shown in the Insets. (Scale bar, 20 μm.) CTRL, control; L-RILPN, AsLOV2-RILP N313.
To clarify whether the competition of the REST-RE1 binding was associated with an inhibition of REST activity, we performed gene reporter assays using the RE1-SV40 reporter construct and the AsLOV2-RILP N313 chimeras. Interestingly, the open AsLOV2-RILP N313 construct was able to increase luciferase transcription, whereas no effect was observed when the corresponding closed construct was used or when both probes were tested on a RE1-null reporter (Fig. 3C and Fig. S1B). Moreover, the AsLOV2-RILP N313 open mutant induced a significant increase in the transcription of the REST target genes SYN1, NAV1.2, and BDNF, whereas no change was observed by the closed chimera or on non–RE1-containing genes (Fig. 3D and Fig. S4). We also investigated whether the open or closed form of AsLOV2-RILP N313 affected neurite length and arborization in N2a cells. Similar to what found for AsLOV2-PAH1b, neurite outgrowth was enhanced in differentiating N2a cells transfected with the open form of the AsLOV2-RILP N313 with respect to the cells expressing the closed form of the chimera (Fig. 3E). Altogether, these data demonstrate that the AsLOV2-RILP N313 chimera allows a conformation-dependent inhibition of REST activity through its displacement from target DNA.
AsLOV2-PAH1 and AsLOV2-RILP N313 Inhibit the Formation of the REST Complex on Target Chromatin.
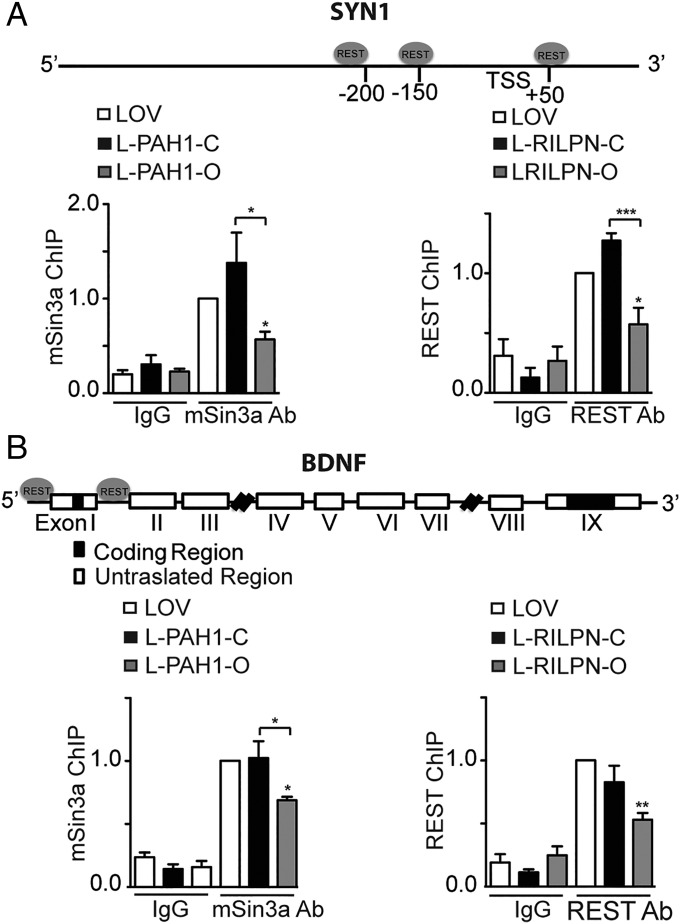
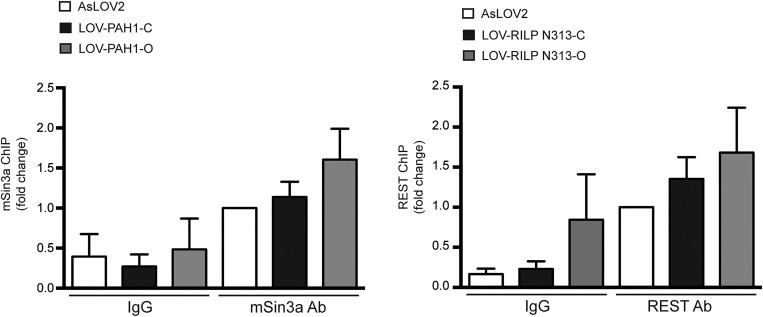
To characterize the molecular mechanisms by which AsLOV2-PAH1b (henceforth AsLOV2-PAH1) and AsLOV2-RILP N313 affect the activity of the REST complex on target chromatin, we measured the occupancy of the RE1 locus on the SYN1 (28) and BDNF (37) promoters by REST and mSin3a after transfection of either chimera in the open or closed form. Chromatin from transfected N2a cells was immunoprecipitated with mSin3a- or REST-specific antibodies, and the respective occupancy of mSin3a and REST at the SYN1 (Fig. 4A) or BDNF (Fig. 4B) promoters was evaluated by quantitative PCR (qPCR). Results indicate that the occupancy of mSin3a and REST was significantly reduced in cells containing the open construct of AsLOV2-PAH1 and AsLOV2-RILP N313, respectively, whereas no significant effects were observed in the presence of the closed chimeras (Fig. 4). As a control, we performed ChIP on a non-RE1 promoter, such as the one of the GAPDH gene. As expected, no differences in promoter pull-down were observed with both AsLOV2-PAH1 and AsLOV2-RILP N313 constructs (Fig. S5). Thus, AsLOV2-PAH1 and AsLOV2-RILP N313 interfere with the formation of the multiprotein REST complex in a conformation-specific way.
Fig. 4.
AsLOV2-PAH1 and AsLOV2-RILP N313 impair the formation of the REST complex on target chromatin. N2a cells were transfected with the indicated expression vectors and then subjected to chromatin immunoprecipitation of the SYN1 (A) or BDNF (B) promoter regions using 2 μg anti-mSin3a, anti-REST, or anti-rabbit IgG antibodies, as indicated. mSin3a or REST binding was normalized against the input DNA value and subsequently normalized to the binding in the control (AsLOV2) sample, set to 1 (*P < 0.05; **P < 0.01; one-way ANOVA followed by the Tukey’s multiple comparison test vs. control or the indicated group; n = 3 independent experiments). LOV, AsLOV2; L-PAH1, AsLOV2-PAH1; L-RILPN, AsLOV2-RILP N313.
Fig. S5.
N2a cells were transfected with the indicated expression vectors and then subjected to chromatin immunoprecipitation of the GAPDH promoter regions using 2 μg anti-mSin3a, anti-REST, or anti-rabbit IgG antibodies, as indicated. mSin3a or REST binding was normalized to the input DNA value and subsequently normalized to the binding in the control (AsLOV2) sample, set to 1 (one-way ANOVA followed by the Tukey’s multiple comparison test; n = 3 independent experiments).
Light-Driven Modulation of REST Activity.
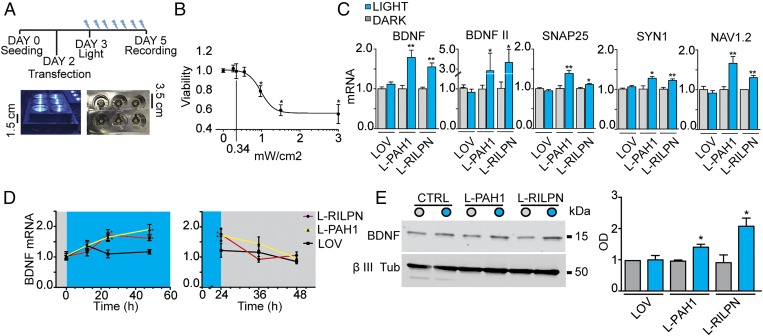
To evaluate the activity of the WT AsLOV2 constructs in response to specific patterns of illumination, N2a cells were transfected with the light-sensitive AsLOV2-PAH1 or AsLOV2-RILP N313 vectors. The transcription profile of the REST-regulated genes BDNF, SYN1, SNAP25, and NAV1.2 was measured after blue light illumination using a custom-made apparatus powered by an Arduino-driven array of blue LEDs (emission peak: 470 nm; Fig. 5A). Considering that AsLOV2 is known to return to the closed conformation in a timescale of tens of seconds (38–40), we tested several light/dark duty cycles by varying the relative duration of the dark vs. light period and selected for our experiments the best performing protocol, which used constant dark/light pulses (with 1-s pulses at 0.5 Hz). In a first series of experiments, we monitored cell viability by varying light intensity, keeping constant the light/dark duty cycle (Fig. 5B). When N2a cells were exposed to 470-nm LED illumination of increasing intensity for 48 h, we reported an optimal viability up to 0.55-mW/cm2 irradiation power, whereas higher intensities induced a progressively increasing percentage of cell death. Thus, to minimize phototoxic damage, a light power of 0.34 mW/cm2 was used in all subsequent experiments.
Fig. 5.
Photostimulation of AsLOV2-PAH1 and AsLOV2-RILP N313 constructs. (A) Illustration of the illumination protocol and LED stimulation apparatus. (Upper) Cells were seeded at day 0 and transfected at day 2. The illumination protocol was started 1 d after transfection, for 48 h. (Lower) Cells are 1.5 cm above a 470-nm light emitting LED. (B) Effect of light exposure on N2a cells viability. Cells were subjected to 470-nm light illumination (0.5 Hz) at the indicated intensities, and viability was measured after 48 h of stimulation (*P < 0.05; one-way ANOVA followed by the Tukey’s multiple comparison test vs. control; n = 3 independent experiments). (C) Undifferentiated N2a cells were transfected with the indicated constructs and then subjected to 470-nm light illumination (0.5 Hz) or kept in the dark, as indicated. After 48 h, the mRNA levels of the indicated genes were quantified via qRT-PCR. GAPDH and HPRT1 were used as control housekeeping genes (*P < 0.05; **P < 0.01; Student t test; n = 3 independent experiments). (D) Time-course of light-dependent BDNF up-regulation and postillumination recovery (*P < 0.05; Student t test vs. respective dark condition; n = 3 independent experiments). (E) Western blot analysis of BDNF in undifferentiated N2a cells transfected with the indicated constructs. A representative experiment is shown on the left, whereas the quantification is shown on the right (*P < 0.05; Student t test vs. the respective dark conditions; n = 3 independent experiments). LOV, AsLOV2; L-PAH1, AsLOV2-PAH1; L-RILPN, AsLOV2-RILP N313.
We then transfected undifferentiated N2a cells with either AsLOV2-PAH1 or AsLOV2-RILP N313. After 24 h, cells were transferred to the illumination apparatus and subjected to the light stimulation protocol. A significant increase in the transcription of the BDNF, BDNFII, SYN1, SNAP25, and NAV1.2 mRNAs was observed on illumination in cells expressing either chimera. No increase in gene transcription was observed in parallel samples kept in the dark or in control cells transfected with AsLOV2 alone and exposed to the same illumination pattern (Fig. 5C). The transcription of non-REST target genes DSCAM, GAP43, and Rcan1 did not change under all conditions tested (Fig. S6), confirming that gene up-regulation was specifically driven by light acting on the light-sensitive constructs at the level of RE1-containing genes. Although the increase in transcription of the tested REST target genes varied in magnitude, the two chimeras had comparable effects on transcription, indicating a final common pathway of REST inhibition.
Fig. S6.
Undifferentiated N2a cells were transfected with the indicated constructs and then subjected to 470-nm illumination (0.5 Hz) or kept in the dark, as indicated. After 48 h, the mRNA levels of the indicated genes were quantified via qRT-PCR. GAPDH and HPRT1 were used as control housekeeping genes (one-way ANOVA followed by the Tukey’s multiple comparison test; n = 3 independent experiments).
Next, we wanted to define the time course of the light-induced transcriptional changes from the beginning of the light stimulation protocol up to a maximum time of 48 h, using BDNF as a target gene. We found that, with both AsLOV2 chimeras, BDNF expression levels significantly increased already after 12 h of illumination, reached a plateau of ∼1.5- to 2-fold increase after 24 h (Fig. 5D, Left), and returned to baseline levels in ∼12 h after switching off the light (Fig. 5D, Right). To evaluate whether the observed changes in mRNA levels were followed by a parallel change in protein levels, BDNF protein was measured in cell extracts by Western blotting after 48 h of light stimulation. As expected, a significant increase in BDNF protein was observed only in N2a cells expressing the active probes and exposed to light (Fig. 5E). Taken together, these data demonstrate that both AsLOV2-PAH1 and AsLOV2-RILP N313 effectively act as light-modulated inhibitors of REST activity on target genes, resulting in increased levels of transcription and translation.
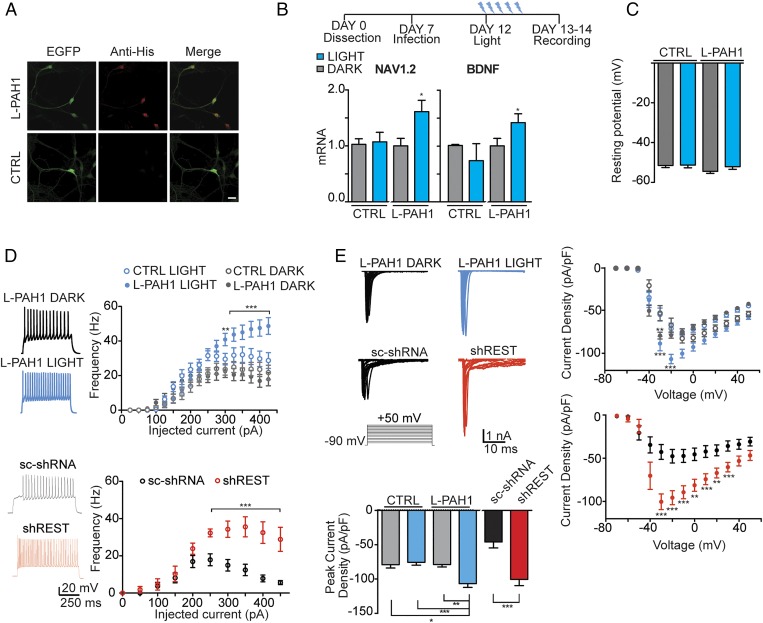
Transduction of AsLOV2-PAH1 in Primary Neurons Increases Intrinsic Excitability.
We previously reported that REST activity can influence neuronal firing through the modulation of NAV1.2 gene transcription (9). To study the effects of the light-driven modulation of REST in primary neurons, we engineered a bidirectional lentiviral vector expressing AsLOV2-PAH1 and EGFP. Once transduced into primary neurons, the probe showed a clear enrichment in the nuclear compartment, similar to what observed in transfected N2a cells (Fig. 6A, see also Figs. 2D and 3A). Primary cortical neurons were infected at 7 d in vitro (DIV) and then subjected to 470-nm light stimulation at 12 DIV for 24 h. Starting from the illumination parameters used for N2a cells, we optimized our protocol for primary neurons, by changing both the illumination intensity and the duty cycle. We found that the optimal conditions for primary neurons, which maximized cell viability while inducing appreciable changes in gene transcription, were 0.34 mW/cm2; 250-ms pulses; 0.25 Hz. The analysis of gene transcription reported a significant increase in both NAV1.2 and BDNF mRNAs in AsLOV2-PAH1 infected neurons exposed to light compared with the same neurons kept in the dark or to neurons infected with a control vector expressing only EGFP (Fig. 6B), indicating that the lentiviral probe was effective in inhibiting REST activity in primary neurons.
Fig. 6.
AsLOV2-PAH1 inhibits REST activity in primary neurons. (A) Confocal images of primary cortical neurons infected with a bidirectional lentiviral vector expressing either EGFP (CTRL) or AsLOV2-PAH1/EGFP. Neurons were stained using anti-Histidine tag antibodies (red) to detect the AsLOV2 construct, and anti-EGFP (green) antibodies to detect EGFP. (Scale bar, 10 μm.) (B, Upper) Schematic representation of the time course of the experiment: neurons were infected at day 7, the illumination protocol was started 5 d after infection (day 12) for 24 h. (B, Lower) qRT-PCR for NAV1.2 and BDNF mRNA in EGFP (CTRL) or AsLOV2-PAH1–expressing neurons kept under dark (gray bars) or lit (blue bars) conditions for 24 h. GAPDH and HPRT1 were used as control housekeeping genes (*P < 0.05; one-way ANOVA followed by the Tukey’s multiple comparison test vs. the respective dark control; n = 3 independent experiments). (C) Resting potential of cortical neurons infected with CTRL and AsLOV2-PAH1 and kept for 24 h under either dark or lit conditions. No significant differences were observed among the various conditions. (D, Upper) Plot of the mean (±SEM) firing frequency vs. injected current (F/I curve) in cortical neurons infected with either AsLOV2-PAH1 (closed symbols) or EGFP alone (CTRL, open symbols) and subjected to either 24-h light (blue symbols) or 24-h dark (gray symbols) (AsLOV2-PAH1/dark, n = 22; AsLOV2-PAH1/light, n = 21; EGFP/dark, n = 19; EGFP/light, n = 20; from three independent experiments). Representative current-clamp recordings of spike trains evoked by injection of 225 pA for 500 ms in neurons infected with AsLOV2-PAH1 and kept for 24 h under dark (black) or lit (blue) conditions are shown on the left. (D, Lower) The same experiment was performed in neurons infected with either a scrambled shRNA or shRNA against REST (n= 11 for each condition). (E) Mean current–voltage (I/V) relationship expressed as current density in cortical neurons infected with AsLOV2-PAH1, EGFP alone, scrambled shRNA, or shRNA against REST. Experimental conditions were as described in D (n= 30 for each condition). The stimulation protocol and representative INa+ traces obtained in the cell-attached configuration at various voltages are shown on the Upper Left. Current density was calculated by normalizing the Na+ current amplitude to the cell capacitance and plotted as means (±SEM) vs. the applied voltage. The mean (±SEM) peak Na+ current density evoked at −20 mV in the indicated groups is shown in the bar plot on the Lower Left. In D and E, *P < 0.05; **P < 0.01; ***P < 0.001; two-way ANOVA followed by the Bonferroni’s multiple comparison test. LOV, AsLOV2; L-PAH1, AsLOV2-PAH1; L-RILPN, AsLOV2-RILP N313; shREST, shRNA for REST; sc-shRNA, scrambled shRNA.
Next, we analyzed whether the optogenetic suppression of REST activity was associated with changes in intrinsic neural excitability. The resting potential of transduced neurons was not affected by the expression of the chimera and/or the light stimulation protocol (Fig. 6C). Then, transduced neurons, held at a membrane potential of –70 mV in whole-cell current-clamp mode were subjected to current injection of 500-ms duration and stepwise increasing amplitude. Interestingly, neurons expressing AsLOV2-PAH1 and subjected to light stimulation displayed a strongly increased firing frequency compared with the same transduced neurons kept in the dark or to parallel cultures infected with the control vector (Fig. 6D), in the absence of significant changes in the passive membrane properties or in action potential shape (Table S3). We then analyzed the Na+ current density to verify whether the increased NAV1.2 transcription was associated with an increased amount of functional channels exposed on the neuronal membrane. The plot of the current-voltage relationship in cortical neurons expressing AsLOV2-PAH1 and subjected to light stimulation revealed that the changes in NAV1.2 transcription were paralleled by a significant increase in the maximum Na+ current density with respect to neurons expressing AsLOV2-PAH1 and kept in the dark or to control neurons expressing EGFP only (Fig. 6E). Interestingly, the increase in the firing frequency (Fig. 6D) and in the Na+ current density (Fig. 6E) obtained by optogenetic inhibition of REST activity with either probe was fully comparable with that obtained by silencing REST with a lentiviral vector expressing a REST shRNA (9). These observations demonstrate that the AsLOV2-PAH1 probe is able to influence REST activity in primary neurons by inducing light-evoked transcriptional regulation with functional consequences on neuronal activity.
Table S3.
Electrophysiological properties of LOV2-PAH1–transduced primary neurons
| Cell parameter | LOV2-PAH1 dark | LOV2-PAH1 light |
| n | 21 | 21 |
| Maximum firing rate (Hz) | 18.57 ± 0.98* | 48.67 ± 4.86 |
| Current threshold (pA) | 152.4 ± 14.01 | 179.8 ± 11.52 |
| Membrane capacitance (pF) | 49.02 ± 4.56 | 50.05 ± 4.00 |
| Membrane resistance (MΩ) | 259.5 ± 23.32 | 210.4 ± 11.47 |
| Voltage threshold (mV) | −17.20 ± 2.31 | −18.31 ± 1.93 |
| AP amplitude (mV) | 49.51 ± 3.21 | 53.79 ± 2.37 |
| AHP (mV) | −21.58 ± 2.17 | −20.74 ± 1.59 |
| AP width 50% (ms) | 1.195 ± 0.061 | 1.171 ± 0.059 |
| Rising slope (V/s) | 22.71 ± 2.04 | 28.92 ± 2.66 |
| Repolarizing slope (V/s) | −9.490 ± 0.61 | −10.09 ± 0.57 |
Electrophysiological parameters describing the passive membrane properties and the shape of the first elicited action potential measured for neurons transduced with LOV2-PAH1 and kept under either dark or lit conditions for 24 h. The analysis refers to the experiments and data reported in Fig. 6.
P < 0.001; unpaired Student t test.
Discussion
In this work, we describe an optogenetic approach aimed at modulating gene transcription in a dynamic and reversible way, by acting on the activity of the master transcriptional repressor REST. We engineered chimerical proteins based on the blue light absorbing AsLOV2 domain and exploited the inhibitory potential of small protein domains belonging to the endogenous REST interactors mSin3a and RILP/PRICKLE1. Following this strategy, we were able to target two key events in REST physiology: (i) the assembly of the repressor complex on RE1 sites and (ii) REST binding to DNA. Optogenetic activation of our LOV-based opto-probes selectively induced transcription of REST target genes in neuroblastoma cells and in primary neurons. By applying specific illumination protocols, we were able to drive and modulate important physiological processes, such as the neural differentiation of N2a cells and the firing properties of cortical neurons.
Due to its crucial role in several neuropathologies (10, 11, 13, 14, 41, 42), various molecular strategies have been developed to target REST activity, including decoy oligodeoxynucleotides, interfering peptides, and stable expression of dominant-negative/constitutively active forms of REST (14, 16, 17). These approaches have been applied to a number of pathological models, including HD and medulloblastoma, encountering various levels of success. However, all these approaches are burdened by intrinsic limitations such as the rapid degradation of the probes or the constitutive and nontunable activation of REST-target genes, which may lead to unwanted side effects. The optogenetic tools described here are endowed with several features that circumvent these limitations, such as (i) their effect is reversible and can be finely modulated by specific illumination protocols; (ii) they can be used for long-term applications, being genetically encoded; and (iii) they counteract REST dysregulation without affecting REST expression.
Recently, various genetically encoded tools have been proposed to remotely control gene activity using light. Such tools were able to regulate the transcription of individual endogenous genes, such as the Vivid LOV (43) or the TALE-associated CRY2-CIB1-VP64 tool (22), or to modulate the activity of exogenous DNA (44). These mechanisms can effectively target the transcription of single genes. However, many cellular events are strictly dependent on the simultaneous and coordinated changes in the transcription of gene clusters, resulting from an increased or reduced activity of one or more transcriptional regulators. In this respect, our strategy is, to our knowledge, the first attempt to directly modulate an endogenous transcription factor, thus achieving the coordinated regulation of a cluster of neuron-specific genes.
The ability to regulate the activity of transcription factors represents an essential step for a better understanding of the molecular and epigenetic mechanisms that constitute the basis of cell physiology, and a promising approach in the treatment of pathologies linked to dysregulation of specific gene clusters. Epigenetic modifications recently emerged as a crucial event to rapidly adapt the neuronal transcriptional response to developmental and environmental hints (45). In this context, REST plays a central role in the determination of the neuronal fate (46), as well as in the modulation of neuronal activity and plasticity (9). The role of REST in the onset of pathologies is complex, acting under some circumstances as an oncogene and under other conditions as a promoter of insult-induced neuronal death (brain ischemia, HD) (11, 13, 41) or dysfunction (epilepsy) (42). Thus, a fundamental objective of next investigations would be to express the REST-modulating probes in vivo in experimental models of brain pathologies associated with REST overexpression or hyperactivity. To render the REST opto-probes useful for long-term experiments in vivo, we are currently engineering “self-activatable” optogenetic tools coupled to chemiluminescent probes that would emit light only on the injection of specific substrates, thus avoiding the implant of an optical fiber (47).
In conclusion, this work describes a previously unexplored optogenetic strategy to control the activity of an endogenous transcription factor, thus achieving the coordinated regulation of a large gene cluster. This approach allowed us to modulate fundamental physiological processes such as the neural differentiation of neuroblastoma cells and the intrinsic excitability of primary neurons and opens the possibility to devise novel therapeutic strategies for brain diseases based on the optogenetic control of the neuronal epigenome.
Materials and Methods
Materials.
All biochemical reagents and drugs were from Sigma-Aldrich, unless otherwise specified. Tissue culture reagents and media were from Gibco-Invitrogen (Life Technologies) or Sigma-Aldrich. The complete list of primary and secondary antibodies used throughout the paper is available in the SI Materials and Methods.
Mammalian Cell Culture and Light Stimulation Experiments.
Murine Neuro2a (N2a) neuroblastoma cells and human HeLa cells were cultured in DMEM (#11965-092) supplemented with 10% (vol/vol) FBS, glutamine (2 mM), and antibiotics, in a humidified 5% CO2 atmosphere at 37 °C. For light stimulation experiments, 750,000 cells were plated in 35-mm dishes, and the day after were transfected with 2.5 μg of the indicated vectors. Twenty-four hours after transfection, cells were subjected to light stimulation. Stimulation parameters were as follows: 0.34 mW/cm2, 470 nm, and 50% duty cycle (1-s light pulses) for the indicated time. After the stimulation period, RNA was extracted. For the control dark points, cells were maintained in the incubator wrapped in aluminum foil, to ensure complete dark conditions.
Sholl Analysis.
Undifferentiated N2a cells were transfected with the indicated constructs and subjected to RA treatment as previously described (28). Cells were fixed after 2 d and stained for β III tubulin and histidine. Images were acquired using an upright Leica TCS SP5 AOBS TANDEM confocal microscope equipped with a 20×/0.50 W UVI objective. At least 20 cells were analyzed for each condition, from three independent cell culture preparations. Sholl analysis was performed by using the Sholl plugin of ImageJ (starting radius 10 μm, radius step size 5 μm, ending radius 200 μm).
Protein extraction and Western blotting were performed following standard biochemical procedures. Details are provided in SI Materials and Methods.
Oligonucleotides and EMSA.
The RE1 cis-site sequence (5′-GTCGGATTCAGCACCACGGACAGCGCCCCT-3′) was chemically synthesized (Sigma-Aldrich). Fragments were end-labeled with biotin using the biotin 3′ end labeling kit (#89818; Pierce Biotech.). For each gel-shift reaction, a total of 20 nM biotin-labeled probe was dissolved in binding buffer [100 mM Hepes, pH 7.2, 400 mM KCl, 30 mM MgCl2, 10 mM DTT, 50% (vol/vol) glycerol] with 500 ng nuclear extracts and 100 ng poly(dI-dC). The reaction mixture was incubated for 25 min at room temperature and then resolved on a nondenaturing 6% (vol/vol) polyacrylamide gel. The signal of the biotin-labeled DNA was detected by using the Light Shift chemiluminescent EMSA kit (#20148; Pierce Biotech.).
ChIP.
A total of 10 × 106 N2a cells were incubated 10 min in 10 mL DMEM + 1% formaldehyde, and then 1 mL of 1.25 M glycine was added for 5 min. After, cells were washed twice with cold PBS and collected by centrifugation. The pellet was suspended in 300 μL swelling buffer [25 mM Hepes, pH 7.8, 1.5 mM MgCl2, 10 mM KCl, 0.1% octylphenoxypolyethoxyethanol (IGEPAL), 1 mM DTT, 0.5% PMSF, protease inhibitor mixture], incubated 10 min on ice, briefly vortexed, and centrifuged 5 min at 2,000 × g at 4 °C. The supernatant was removed, and the nuclear pellet was suspended in 200 μL Nuclei Lysis Buffer (50 mM Tris⋅HCl, pH 8.0, 10 mM EDTA, 1% SDS, protease inhibitor mixture) and incubated 10 min on ice. DNA was fragmented by five cycles of sonication using a Branson Sonifier (Power 1) and centrifuged 15 min at 10,000 × g, at 4 °C. The formaldehyde-fixed DNA-protein complex (100 μL) was added to 900 μL of immunoprecipitation dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris⋅HCl, pH 8.0, 167 mM NaCl, 1 mM PMSF, protease inhibitor mixture) and incubated with 2 μg specific antibody ChIPAb+ REST, anti-mSin3A (K-20), or anti-rabbit IgG antibodies (#PP54B; Millipore) for 2 h at 4 °C. Antibodies were previously bound to protein A beads (Dynabeads Protein A; Life Technologies) by 2-h incubation at 4 °C on a rotating wheel. DNA from a 100-μL aliquot from all experimental conditions was de–cross-linked and extracted as described below, to be used as input. After immunoprecipitation, samples were washed three times with 1 mL RIPA buffer (10 mM Tris⋅HCl, pH 7.5, 1 mM EDTA, 140 mM NaCl, 0.5 mM EGTA, 1% Triton X-100, 0.1% SDS, 0.1% Na-deoxycolate) and two times with 1 mL TE buffer (10 mM Tris⋅HCl, pH 8.0, 10 mM EDTA). Washed beads were pelleted, and 40 μL of 10% (vol/vol) Chelex-100 (#142-1253; Bio-Rad) was added. After a brief vortexing, samples were boiled for 10 min and then cooled to room temperature. One microliter Proteinase K (20 mg/mL) was added, and samples were incubated at 55 °C, 30 min on a thermomixer under constant shaking. Samples were then boiled 10 min and pelleted. Supernatant, containing de–cross-linked DNA, was transferred in clean tubes. Five microliters purified DNA was used for qPCR. The efficiency of ChIP was quantified on a standard curve prepared using various concentrations (from 100 to 5 ng) of genomic DNA. The amount of precipitated DNA was determined as relative to 1% input chromatin as (amount of ChIP DNA/amount of input DNA) × 100. The complete list of ChIP primers is available in Dataset S1.
Plasmids and Transfections.
Details about the plasmids used and relative cloning strategies are available in SI Materials and Methods. Reporter and expression vectors were transiently cotransfected into cultured cells using Lipofectamine 2000 (Life Technologies) following standard transfection procedures. Control samples were cotransfected with the empty vector corresponding to the effector plasmids. pRL-TK-SV40 control plasmid was used as internal control. Luciferase activity was assayed after 48 h using the Dual-Luciferase reporter assay system (#E1910; Promega).
AsLOV2 Mutagenesis.
To obtain AsLOV2 C450A and I532E mutants, 50 μg WT pcDNA3.1His/AsLOV2-PAH1 or pcDNA3.1His/AsLOV2-RILP N313 vectors were PCR amplified using Pfu DNA polymerase (#M7745; Promega). The complete list of primers is available in Dataset S1. PCR conditions were as follows: 95 °C, 5 min; 95 °C, 30 s; 55 °C, 30 s; 72 °C, 13 min; 30 cycles for pcDNA3.1His/AsLOV2-PAH1 mutants and 95 °C, 5 min; 95 °C, 30 s; 55 °C, 30 s; 72 °C, 14 min; 30 cycles for pcDNA3.1His/AsLOV2-RILPshort mutants. PCR products were digested using the DpnI enzyme (Promega) and transformed into DH5α cells. Positive colonies were verified by DNA sequencing.
RNA Preparation and qRT-PCR.
Total cellular RNA was extracted using TRIzol (Life Technologies), and isolated RNA was subjected to DNase I (Promega) treatment. cDNA was synthesized starting from 0.5 μg treated RNA according to the High-Capacity cDNA Reverse Transcription Kit manual (#4368814; Applied Biosystems) and used for qRT-PCR. The complete list of qRT-PCR primers is available in Dataset S1. For the Nanostring analysis of REST target and nontarget genes, fluorescently labeled probes were designed and synthetized by Nanostring Technologies (Table S1). One hundred nanograms total RNA per sample, prepared as described above, was processed in the Center for Genomic Science, Istituto Italiano di Tecnologia, Milano (Italy), following standard procedures. Data were analyzed by using the nSolver Analysis Software Version 2.5.
Immunofluorescence and Confocal Microscopy.
Cells were fixed with 4% (wt/vol) paraformaldehyde and 20% (wt/vol) sucrose in PBS for 15 min at room temperature and permeabilized with 0.1%. Triton X-100 in PBS for 5 min at room temperature. Samples were blocked for 30 min in immunofluorescence buffer [2% (wt/vol) BSA, 10% (vol/vol) goat serum in PBS]. Primary and secondary antibodies were diluted in immunofluorescence buffer and incubated for 45 min at room temperature. Coverslips were mounted using ProLong antifade (#P36931; Life Technologies) and imaged by confocal microscopy. Confocal fluorescent images were obtained using a Leica SP5 confocal scan with a 40× objective and analyzed with the Leica LAS AF software.
Photostimulation Hardware.
In vitro light stimulation experiments were performed using a custom built LED photostimulation device, which was fabricated with a precision-machined aluminum mounting plate and a 5-W high efficiency Blue LED array, with peak emission at 470 nm. LED intensity was regulated by a separate, 12 channels low-noise, linear power driver; on-off operation (and duty cycle) was modulated by an ARDUINO 2000 microcontroller (the system can accept every TTL or digital positive signal to enable channels output or can be used in stand-alone mode). Radiation output was measured from a distance of 1.5 cm above the array using a Thorlabs PM100 power meter and an S121b circular sensor probe. The complete Arduino code is available in Dataset S1.
Electrophysiological Recordings.
Primary cortical cultures were prepared from mouse C57BL/6J (E17–E18) embryos as previously described (48). All experiments were carried out in accordance with the guidelines established by the European Communities Council (Directive 2010/63/EU of 22 September 2010) and were approved by the Italian Ministry of Health. Lentivirus production and infection were performed following standard procedures (details are provided in SI Materials and Methods). Neurons were infected with lentiviral vectors encoding GFP, AsLOV2-PAH1b/GFP, scrambled/mCherry or REST shRNA/mCherry (9) and subjected to 24-h light stimulation or kept in the dark. One day after the end of the stimulation protocol, excitability and Na+ current density were studied by patch-clamp recordings in current-clamp and voltage-clamp configurations. All experiments were performed on transduced neurons identified by green or red fluorescence using a Multiclamp 700B amplifier (Axon Instruments, Molecular Devices) and an upright bx51WI microscope (Olympus) equipped with Nomarski optics. Current-clamp recordings were performed at a holding potential of −70 mV, and action potential firing was induced by injecting current steps of 25 pA lasting 500 ms. Sodium currents were obtained by stepping from a holding potential of −90 to −70 to +50 mV with 10-mV steps. Details about recordings and experimental conditions are provided in SI Materials and Methods.
Computational Methods.
An all-atom model of the chimera was built starting from the available high-resolution structures of the AsLOV2 (dark state) and PAH1 domains [Protein Data Bank (PDB) ID codes 2V1A and 2RMR] (33, 34). An optimal relative orientation for the two domains was determined using the RosettaDock software (35). The program generates several conformations of the encounter complex using random orientations and selects those with low energy values. We used as starting pose a conformation where the AsLOV2 C terminus was close to the N terminus of PAH1. Among the low energy conformations obtained, we selected those minimizing the exposed PAH1 binding domain. A single protein chain was then created by connecting the two termini with a chemical bond. The chimeric system, amounting to about 90.000 atoms, was then solvated in water box, neutralized with counter ions, and studied by MD simulations. Details about the simulation input parameters and methods are provided in SI Materials and Methods.
Statistical Analysis.
Results are expressed as means ± SEM throughout. Data were analyzed by either the unpaired Student t test or one-way ANOVA followed by multiple comparison tests, as indicated in the text.
SI Materials and Methods
List of Antibodies.
Horseradish peroxidase (HRP)-associated secondary antibodies for Western blot analysis were stabilized goat anti-mouse IgG (H+L), peroxidase conjugated (#32430; Thermo Scientific) and stabilized goat anti-rabbit IgG (H+L), peroxidase conjugated (#32460; Thermo Scientific). The following primary antibodies were used: polyclonal ChIPAb+ REST (#17–641; Millipore), polyclonal anti-GAPDH (#SAB3500247; Sigma-Aldrich), monoclonal anti-His-tag (HIS.H8, #sc-57598; Santa Cruz Biotechnology), rabbit anti-GFP (#A-11122; Life Technologies), polyclonal anti-mSin3A (K-20) (#sc-994; Santa Cruz Biotechnology), and polyclonal anti-BDNF (H-117) (#sc-20981; Santa Cruz Biotechnology).
Protein Extraction.
Total protein lysates were obtained from cells lysed on ice for 20 min in RIPA buffer [20 mM Tris⋅HCl, pH 7.4; 1% Triton X-100; 10% (vol/vol) glycerol; 150 mM NaCl; 1% PMSF; protease inhibitor mixture: Complete protease inhibitor mixture tablets; 04693116001; Roche Applied Science]. The final protein concentration was quantified by using the BCA protein assay kit (#23225; Pierce Biotech.). Nuclear extracts were prepared as follows: the pellet from 5 × 106 cells was resuspended in 400 μL cytoplasmic buffer (10 mM Hepes, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, protease inhibitor mixture) and incubated 20 min on ice with constant shaking. After the addition of 25 μL of 10% (vol/vol) IGEPAL, the solution was briefly vortexed and centrifuged 2 min at 8,000 × g at 4 °C. The supernatant containing the cytoplasmic extracts was collected, the pelleted nuclei were suspended in 50 μL nuclear buffer [20 mM Hepes, pH 7.9, 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 25% (vol/vol) glycerol, protease inhibitor mixture] and incubated 20 min on ice with constant shaking. Nuclei were centrifuged 25 min at 10,000 × g at 4 °C, and the supernatant (nuclear extract) was collected. The final protein concentration was quantified by using the Bradford protein assay (#23200; Pierce Biotech.).
Western Blotting.
Proteins were separated using precast 10% NuPAGE Novex Bis-Tris Gels (Life Technologies) and transferred to a nitrocellulose membrane. After incubation with primary antibodies, membranes were incubated with HRP-conjugated secondary antibodies and revealed by autoradiography using the SuperSignal West Pico Chemiluminescent Substrate (#34077; Thermo Scientific).
List of Plasmids and Cloning Strategies.
Expression vectors encoding for REST and shREST were a kind gift from J. Meldolesi, Fondazione San Raffaele, Milano, Italy; pCS2+MTmSin3AN205 was a kind gift from T. Kouzarides, Gurdon Institute, Cambridge, UK; and pCS2+PRIKLE1 was a kind gift from A. Bassuk, University of Iowa, Ames, IA. pcDNA3.1His-AsLOV2 was cloned as follows: the AsLOV2 sequence was amplified from a bacterial plasmid encoding for AsLOV2 (gift from T. R. Sosnick, University of Chicago, Chicago) and inserted in the pcDNA3.1V5/His vector (Life Technologies) between the HindIII and AgeI sites. pcDNA3.1His/AsLOV2-PAH1 and pcDNA3.1His/AsLOV2-RILPshort were obtained as follows: the PAH1 sequence was amplified from pCS2+MTmSin3AN205 and cloned in pcDNA3.1His-AsLOV2 at the AgeI site; the RILP N313 sequence was amplified from pCS2+PRIKLE1 and cloned in pcDNA3.1His-AsLOV2 at the AgeI site. pGFP-REST was cloned as follows: mouse REST cDNA was amplified with modified primers and inserted in pPAGFP-C1 between the EcoRI and BamHI sites. pGL3-RE1/NRSE was a kind gift from K. Nakashima, Nara Institute of Science and Technology, Nara, Japan. All constructs were verified by DNA sequencing.
Lentivirus Production and Infection Procedures.
The lentiviral pCCL.sin.cPPT.PGK.GFP.WPRE bidirectional expression vector and packaging plasmids were a kind gift from L. Naldini, TIGET, San Raffaele Sci. Institute, Milan, Italy. For the coordinated expression of AsLOV2-PAH1b and GFP or of scrambled/REST shRNA (described in ref. 9) and mCherry, the low-affinity nerve growth factor (NGF) receptor in the MA1 construct (49) was replaced with REST cDNA, leading to GFP expression from the miniCMV promoter and REST expression from the phosphoglycerate kinase (PGK) promoter. Third-generation lentiviruses were produced by transient four-plasmid cotransfection into HEK293T cells using the calcium phosphate transfection method. Supernatants were collected, passed through a 0.45-µm filter, and purified by ultracentrifugation as previously described. Viral vectors were titrated at concentrations ranging from 1 × 108 to 5 × 109 transducing units (TU)/mL and used at a multiplicity of infection (MOI) of 1–10. The efficiency of infection was estimated to range between 70% and 90% by counting neurons expressing GFP protein with respect to the total number of cells stained with DAPI. Primary cortical neurons were infected at 7 DIV. After 24 h, half of the medium was replaced with fresh medium. Experiments were performed 5–7 d after infection (between 12 and 14 DIV).
Electrophysiological Recordings.
Patch electrodes, fabricated from thick borosilicate glasses, were pulled to a final resistance of 4–6 MΩ. Recordings with leak current <100 pA and series resistance >20 MΩ were discarded. All recordings were acquired at 50 kHz. Neuron excitability was performed in Tyrode extracellular solution in which d-(−)-2-amino-5-phosphonopentanoic acid (d-AP5; 50 µM), 6-cyano-7 nitroquinoxaline-2,3-dione (CNQX; 10 µM), bicuculline methiodide (30 µM), and (2S)-3-[[(1S)-1-(3,4-dichlorophenyl)ethyl]amino-2-hydroxypropyl](phenylmethyl)phosphinic acid hydrochloride (CGP; 5 µM) were added to block NMDA, non-NMDA, GABAA, and GABAB receptors, respectively. The internal solution (K-gluconate) was composed of (in mM) 126 K gluconate, 4 NaCl, 1 MgSO4, 0.02 CaCl2, 0.1 BAPTA, 15 glucose, 5 Hepes, 3 ATP, and 0.1 GTP, pH 7.3. In voltage-clamp experiments, sodium currents were dissected using an extracellular solution containing 130 mM NaCl, 2 mM CaCl2, 2 mM MgSO4, 10 mM glucose, 5 mM tetraethylammonium chloride (TEA-Cl), and 10 mM Hepes/NaOH (pH 7.4). The patch pipette was filled with 130 mM CsCl2, 10 mM NaCl, 20 mM TEA-Cl, 10 mM EGTA, 2 mM MgCl2, 4 mM Mg-ATP, and 10 mM Hepes/CsOH (pH 7.4). All of the parameters were analyzed using the pClamp (Molecular Devices) and Prism (GraphPad Software) software.
Computational Methods.
A proper size for the simulation box corresponding to a pressure of 1 atm and temperature of 300 K was determined in the constant pressure and temperature (NPT) ensemble using the Nosé-Hoover Langevin piston method for 500 ps (50). The simulation ensemble was then switched to constant volume and temperature (NVT) for the rest of the simulation by keeping the temperature stationary around 300 K using Langevin dynamics. Two simulation runs were performed, one for the full chimera and one for the truncated linker, each lasting 70 ns. We used the MD program nanoscale molecular dynamics (NAMD), the chemistry at Harvard macromolecular mechanics (CHARMM27) force field with the cross-term energy map (CMAP) correction for protein backbone energetics (51), and transferable intermolecular potential, three-point (TIP3P) model for water molecules. Parameters for the flavin mononucleotide (FMN) chromophore were taken from ref. 52. Periodic boundary conditions were used to replicate the system and remove box surface effects. Short-range nonbonded interactions were cut off at 12 Å, whereas long-range electrostatic interactions were computed using the particle mesh Ewald method. Chemical bonds connecting hydrogen atoms to heavy atoms were kept fixed using the SHAKE algorithm (53). An integration time step of 1 fs was used to ensure stability of the dynamical algorithm.
Supplementary Material
Acknowledgments
We thank the following scientists for kindly providing us with the constructs used in our study: Dr. T. R. Sosnick (University of Chicago) for LOV2; Dr. J. Meldolesi (Fondazione San Raffaele) for REST and shREST constructs; Dr. A. Bassuk (University of Iowa) for PRICKLE1; Dr. T. Kouzarides (Gurdon Institute) for mSin3A; Dr. Luigi Naldini (Tiget) for help in lentiviral production protocols; Dr. S. Bianchi (Center for Genomic Science, Istituto Italiano di Tecnologia) for invaluable technical help in the Nanostring screening; A. L. Benfenati for help in programming the Arduino software; and Drs. M. Nanni, A. Mehilli, and C. Chiabrera for help with dissection of primary neurons. This work was supported by the Citizens United for Research in Epilepsy Innovator Award (to F.B.) and the European Union Integrating Project DESIRE (Development and Epilepsy–Strategies for Innovative Research to improve diagnosis, prevention and treatment in children with difficult to treat Epilepsy; FP7-HEALTH-2013-INNOVATION-1 Grant 602531 to F.B.). We acknowledge the IIT Platform “CompuNet” for CPU time and a PRACE (Partnership for Advanced Computing in Europe) infrastructure Award DECI-8 (to L.M.) allowing the access to resource Monte Rosa at Swiss National Supercomputing Centre.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507355112/-/DCSupplemental.
References
- 1.Bale TL, et al. Early life programming and neurodevelopmental disorders. Biol Psychiatry. 2010;68(4):314–319. doi: 10.1016/j.biopsych.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sweatt JD. The emerging field of neuroepigenetics. Neuron. 2013;80(3):624–632. doi: 10.1016/j.neuron.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121(4):645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 4.Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20(2):136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- 5.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): A coordinate repressor of multiple neuron-specific genes. Science. 1995;267(5202):1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 6.Grimes JA, et al. The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J Biol Chem. 2000;275(13):9461–9467. doi: 10.1074/jbc.275.13.9461. [DOI] [PubMed] [Google Scholar]
- 7.Abrajano JJ, et al. REST and CoREST modulate neuronal subtype specification, maturation and maintenance. PLoS One. 2009;4(12):e7936. doi: 10.1371/journal.pone.0007936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodenas-Ruano A, Chávez AE, Cossio MJ, Castillo PE, Zukin RS. REST-dependent epigenetic remodeling promotes the developmental switch in synaptic NMDA receptors. Nat Neurosci. 2012;15(10):1382–1390. doi: 10.1038/nn.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pozzi D, et al. REST/NRSF-mediated intrinsic homeostasis protects neuronal networks from hyperexcitability. EMBO J. 2013;32(22):2994–3007. doi: 10.1038/emboj.2013.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu T, et al. REST and stress resistance in ageing and Alzheimer’s disease. Nature. 2014;507(7493):448–454. doi: 10.1038/nature13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuccato C, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35(1):76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 12.Palm K, Belluardo N, Metsis M, Timmusk T. Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J Neurosci. 1998;18(4):1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calderone A, et al. Ischemic insults derepress the gene silencer REST in neurons destined to die. J Neurosci. 2003;23(6):2112–2121. doi: 10.1523/JNEUROSCI.23-06-02112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawinger P, et al. The neuronal repressor REST/NRSF is an essential regulator in medulloblastoma cells. Nat Med. 2000;6(7):826–831. doi: 10.1038/77565. [DOI] [PubMed] [Google Scholar]
- 15.Immaneni A, et al. REST-VP16 activates multiple neuronal differentiation genes in human NT2 cells. Nucleic Acids Res. 2000;28(17):3403–3410. doi: 10.1093/nar/28.17.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soldati C, Bithell A, Conforti P, Cattaneo E, Buckley NJ. Rescue of gene expression by modified REST decoy oligonucleotides in a cellular model of Huntington’s disease. J Neurochem. 2011;116(3):415–425. doi: 10.1111/j.1471-4159.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- 17.Conforti P, et al. In vivo delivery of DN:REST improves transcriptional changes of REST-regulated genes in HD mice. Gene Ther. 2013;20(6):678–685. doi: 10.1038/gt.2012.84. [DOI] [PubMed] [Google Scholar]
- 18.Conforti P, et al. Binding of the repressor complex REST-mSIN3b by small molecules restores neuronal gene transcription in Huntington’s disease models. J Neurochem. 2013;127(1):22–35. doi: 10.1111/jnc.12348. [DOI] [PubMed] [Google Scholar]
- 19.Zhang F, et al. Optogenetic interrogation of neural circuits: Technology for probing mammalian brain structures. Nat Protoc. 2010;5(3):439–456. doi: 10.1038/nprot.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautier A, et al. How to control proteins with light in living systems. Nat Chem Biol. 2014;10(7):533–541. doi: 10.1038/nchembio.1534. [DOI] [PubMed] [Google Scholar]
- 21.Cao J, Arha M, Sudrik C, Schaffer DV, Kane RS. Bidirectional regulation of mRNA translation in mammalian cells by using PUF domains. Angew Chem Int Ed Engl. 2014;53(19):4900–4904. doi: 10.1002/anie.201402095. [DOI] [PubMed] [Google Scholar]
- 22.Konermann S, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500(7463):472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu YI, et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461(7260):104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strickland D, Moffat K, Sosnick TR. Light-activated DNA binding in a designed allosteric protein. Proc Natl Acad Sci USA. 2008;105(31):10709–10714. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roopra A, et al. Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3-histone deacetylase complex. Mol Cell Biol. 2000;20(6):2147–2157. doi: 10.1128/mcb.20.6.2147-2157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimojo M, Hersh LB. Characterization of the REST/NRSF-interacting LIM domain protein (RILP): Localization and interaction with REST/NRSF. J Neurochem. 2006;96(4):1130–1138. doi: 10.1111/j.1471-4159.2005.03608.x. [DOI] [PubMed] [Google Scholar]
- 27.Bassuk AG, et al. A homozygous mutation in human PRICKLE1 causes an autosomal-recessive progressive myoclonus epilepsy-ataxia syndrome. Am J Hum Genet. 2008;83(5):572–581. doi: 10.1016/j.ajhg.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paonessa F, Latifi S, Scarongella H, Cesca F, Benfenati F. Specificity protein 1 (Sp1)-dependent activation of the synapsin I gene (SYN1) is modulated by RE1-silencing transcription factor (REST) and 5′-cytosine-phosphoguanine (CpG) methylation. J Biol Chem. 2013;288(5):3227–3239. doi: 10.1074/jbc.M112.399782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 30.Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39(31):9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 31.Harper SM, Christie JM, Gardner KH. Disruption of the LOV-Jalpha helix interaction activates phototropin kinase activity. Biochemistry. 2004;43(51):16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 32.Nomura M, Uda-Tochio H, Murai K, Mori N, Nishimura Y. The neural repressor NRSF/REST binds the PAH1 domain of the Sin3 corepressor by using its distinct short hydrophobic helix. J Mol Biol. 2005;354(4):903–915. doi: 10.1016/j.jmb.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 33.Sahu SC, et al. Conserved themes in target recognition by the PAH1 and PAH2 domains of the Sin3 transcriptional corepressor. J Mol Biol. 2008;375(5):1444–1456. doi: 10.1016/j.jmb.2007.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halavaty AS, Moffat K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry. 2007;46(49):14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 35.Chaudhury S, et al. Benchmarking and analysis of protein docking performance in Rosetta v3.2. PLoS One. 2011;6(8):e22477. doi: 10.1371/journal.pone.0022477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strickland D, et al. Rationally improving LOV domain-based photoswitches. Nat Methods. 2010;7(8):623–626. doi: 10.1038/nmeth.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hara D, et al. Remote control of activity-dependent BDNF gene promoter-I transcription mediated by REST/NRSF. Biochem Biophys Res Commun. 2009;384(4):506–511. doi: 10.1016/j.bbrc.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 38.Zayner JP, Sosnick TR. Factors that control the chemistry of the LOV domain photocycle. PLoS One. 2014;9(1):e87074. doi: 10.1371/journal.pone.0087074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo H, Kottke T, Hegemann P, Dick B. The phot LOV2 domain and its interaction with LOV1. Biophys J. 2005;89(1):402–412. doi: 10.1529/biophysj.104.058230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freddolino PL, Dittrich M, Schulten K. Dynamic switching mechanisms in LOV1 and LOV2 domains of plant phototropins. Biophys J. 2006;91(10):3630–3639. doi: 10.1529/biophysj.106.088609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noh KM, et al. Repressor element-1 silencing transcription factor (REST)-dependent epigenetic remodeling is critical to ischemia-induced neuronal death. Proc Natl Acad Sci USA. 2012;109(16):E962–E971. doi: 10.1073/pnas.1121568109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McClelland S, et al. The transcription factor NRSF contributes to epileptogenesis by selective repression of a subset of target genes. eLife. 2014;3:e01267. doi: 10.7554/eLife.01267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods. 2012;9(3):266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 44.Motta-Mena LB, et al. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat Chem Biol. 2014;10(3):196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riccio A. Dynamic epigenetic regulation in neurons: Enzymes, stimuli and signaling pathways. Nat Neurosci. 2010;13(11):1330–1337. doi: 10.1038/nn.2671. [DOI] [PubMed] [Google Scholar]
- 46.Mandel G, et al. Repressor element 1 silencing transcription factor (REST) controls radial migration and temporal neuronal specification during neocortical development. Proc Natl Acad Sci USA. 2011;108(40):16789–16794. doi: 10.1073/pnas.1113486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prescher JA, Contag CH. Guided by the light: Visualizing biomolecular processes in living animals with bioluminescence. Curr Opin Chem Biol. 2010;14(1):80–89. doi: 10.1016/j.cbpa.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 48.Cesca F, et al. Kidins220/ARMS mediates the integration of the neurotrophin and VEGF pathways in the vascular and nervous systems. Cell Death Differ. 2012;19(2):194–208. doi: 10.1038/cdd.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amendola M, Venneri MA, Biffi A, Vigna E, Naldini L. Coordinate dual-gene transgenesis by lentiviral vectors carrying synthetic bidirectional promoters. Nat Biotechnol. 2005;23(1):108–116. doi: 10.1038/nbt1049. [DOI] [PubMed] [Google Scholar]
- 50.Martyna GJ, Tobias DJ, Klein ML. Constant-pressure molecular-dynamics algorithms. J Chem Phys. 1994;101(5):4177–4189. [Google Scholar]
- 51.Mackerell AD, Jr, Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25(11):1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 52.Freddolino PL, Gardner KH, Schulten K. Signaling mechanisms of LOV domains: New insights from molecular dynamics studies. Photochem Photobiol Sci. 2013;12(7):1158–1170. doi: 10.1039/c3pp25400c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-integration of Cartesian equations of motion of a system with constraints: Molecular dynamics of N-alkanes. J Comput Phys. 1977;23(3):327–341. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.