Significance
Embryonic fibroblasts have long been considered a relatively homogeneous cell type, and many studies have been conducted on fibroblasts of varying provenance. The results of the present study indicate that embryonic fibroblasts are highly heterogeneous and exhibit anatomic and developmental variation that has both practical consequences for the interpretation of experimental results and theoretical implications for the nature and organization of metazoan organisms. Studies using embryonic fibroblasts to explore the consequences of altered alleles should take into account the possibility that fibroblast composition may be altered by allele expression.
Keywords: embryo, fibroblast, lineage, heterogeneity, phenotype
Abstract
Analysis of embryonic fibroblasts from GFP reporter mice indicates that the fibroblast cell type harbors a large collection of developmentally and phenotypically heterogeneous subtypes. Some of these cells exhibit multipotency, whereas others do not. Multiparameter flow cytometry analysis shows that a large number of distinct populations of fibroblast-like cells can be found in cultures initiated from different embryonic organs, and cells sorted according to their surface phenotype typically retain their characteristics on continued propagation in culture. Similarly, surface phenotypes of individual cloned fibroblast-like cells exhibit significant variation. The fibroblast cell class appears to contain a very large number of denumerable subtypes.
Fibroblasts are ubiquitous mesenchymal cells that are thought to play important roles in wound repair and healing, and that serve as reservoirs of multipotent progenitors capable of repopulating depleted cell compartments. The overgrowth of fibroblasts and elaboration of their secreted matrix products contributes to a variety of human fibrotic diseases (1). In the setting of tumor biology, the contributions of fibroblasts to tumor stroma have been recognized as important factors in the evolution of neoplastic tissue (2–6).
Embryos are a rich source of fibroblasts, and murine embryonic fibroblasts (MEFs) are frequently prepared to study the physiological consequences of selective gene ablations. MEFs are typically prepared by trypsinization and seeding of embryos into cell culture medium after removal of the head, tail, limbs, and internal organs. The cultures that result after a few passages are often considered largely homogeneous populations.
MEF cultures are widely believed to proliferate in response to mitogenic factors in serum that arise from the platelet degranulation resulting from coagulation of whole blood. Among the growth factors that have been identified from platelet releasates are TGF-β1, PDFG-β, angiopoietin 1, ECM protein 1, and vascular endothelial growth factor (VEGF)-C (7). Several of these growth factors are thought to play important roles in proliferation of tumor stromal fibroblasts (6).
Previous studies have characterized a line of transgenic mice expressing green fluorescent protein (GFP) under the control of a fragment from the promoter region of VEGF-A. GFP-positive cells seen to infiltrate tumors implanted in the transgenic mice were identified as fibroblasts, and MEFs isolated from transgenic mouse embryos were found to give rise to both GFP-positive and GFP-negative cells (8). MEF cultures consist of highly heterogeneous populations of distinct cell types. In the present study, we examined the origins of MEF heterogeneity in greater detail.
Results
Fibroblast-Like Populations from VEGF-GFP Transgenic Mice.
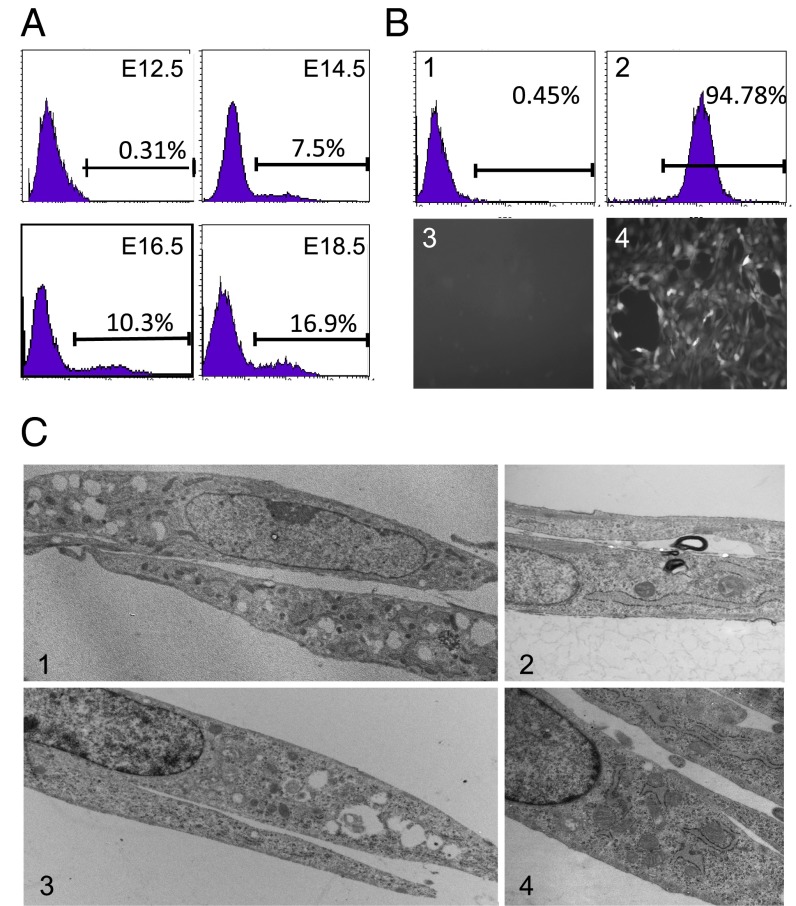
Cells derived from tumor stroma or from primary MEF cultures of VEGF-GFP transgenic mice contain both GFP-positive and GFP-negative cells resembling fibroblasts (8). Fibroblast-like cells isolated from VEGF-GFP mouse embryos of various ages ranging from embryo (postcoital) day E12.5 to E18.5 are morphologically indistinguishable by light or electron microscopy, but exhibit variable GFP expression. Populations from earlier stages, days E12.5 and E14.5, exhibit lower frequencies of GFP-positive cells compared with cultures prepared from day E16.5 and E18.5 embryos (Fig. 1A). GFP-positive cells typically replicate more rapidly than GFP-negative cells under standard culture conditions, leading to an increase in their prevalence with increasing culture passage (Fig. 1B).
Fig. 1.
Phenotypic comparisons of GFP-negative and GFP-positive cells. Fibroblasts from the VEGF-GFP mouse embryos at E12.5, E14.5, E16.5, and E18.5 were isolated and cultured as described in Materials and Methods. (A) Flow cytometry analyses of GFP-positive and -negative cells. (B) Fibroblasts from E12.5 embryos at passage 28 (1) or from E18.5 embryos at passage 30 (2) were analyzed cytometrically or by epifluorescence microscopy (3 and 4). (C) Representative electron micrographs of GFP-negative cells (1 and 2; E12.5 from VEGF-GFP embryos) and GFP-positive cells (3 and 4; E18.5 embryos) at 2,000× magnification. Both GFP-positive and GFP-negative cell types exhibited characteristics of fibroblast-like cells.
Following fluorescence-activated cell sorting of newly initiated cultures, GFP-positive and GFP-negative cells can be propagated as apparently homogeneous populations. The GFP-positive cells resemble cells from the immortalized fibroblast line NIH 3T3, whereas the GFP-negative cells retain the characteristics of the cells initially placed in culture and resemble the C3H10T1/2 cell line (SI Appendix, Fig. S1A). On electron microscopy (Fig. 1C), both cell types show typical characteristics of fibroblasts: fairly smooth plasma membrane and nuclear membrane surfaces with delicate cytoplasmic processes, abundant intracytoplasmic ribosomes and undilated or dilated rough endoplasmic reticulum, and occasional mitochondria and pinocytic vesicles. No tight gap junctions are apparent between cells. Analysis of cytokine expression showed that the GFP-positive cells expressed VEGF-A, whereas the GFP-negative cells did not, consistent with the structure of the transgene (SI Appendix, Fig. S1B). These data show that fibroblast populations can be heterogeneous with respect to expression in a way that is not immediately apparent, and raise the question of whether additional heterogeneity can be found.
Analysis of Fibroblast-Like Cells by Immunofluorescence.
In a flow cytometry survey, although the GFP-positive cells were relatively homogeneous, GFP-negative cells exhibited substantial heterogeneity. Most of the GFP-negative population did not express the cell surface proteins typical of committed hematopoietic, endothelial, or epithelial lineages (SI Appendix, Table S1). The pattern of expression also diverged from that reported for various compilations of mesenchymal stem cell (MSC) surface proteins (9–14).
GFP-negative cells variably expressed CD14, CD24, CD34, and podoplanin (PDPN; Pdpn); expressed little CD73 and CD105; and did not express CD133. In contrast, MSCs have not been reported to express CD14, CD24, CD34 or PDPN, but have been shown to express CD73, CD105, and CD133 (SI Appendix, Table S1). Sca-1 (Ly6a) has been found to be expressed by bone marrow-derived MSCs (15), and the cells in this study were also positive for this marker (SI Appendix, Table S1). Other markers of pluripotency, including Oct-4 (Pou5f1), Sox-2 (Sox2), Nanog (Nanog), Myc (Myc), nucleostemin (NST; Gnl3), nestin (Nes), Cox-2 (Cox2), Pax-7 (Pax7), Fra-1 (Fosl1), and Klf-4 (Klf4) were confirmed to be expressed by murine ES cells (SI Appendix, Fig. S2A), whereas NIH 3T3 fibroblasts did not express most of these markers (SI Appendix, Fig. S2B). Expression of NST and Fra-1 was observed in NIH 3T3 cells, in agreement with earlier findings (16, 17). GFP-negative cells were found to express nucleolar NST and nestin, along with limited quantities of nuclear Fra-1, but none of the other proteins characteristic of pluripotent cells (SI Appendix, Fig. S3A). GFP-positive cells did not differ greatly from the GFP-negative cells in this respect (SI Appendix, Fig. S3B). Nestin and NST expression was higher in GFP-negative cells, whereas Fra-1 expression was higher in GFP-positive cells. In addition, the GFP-negative cells did not express the embryonic stem cell markers CD31, SSEA1, SSEA3, and podocalyxin (Tra-1-81) (SI Appendix, Table S1).
Some classification of fibroblast subtypes has been achieved by examining the pattern of expression of contractile and intermediate filament proteins, such as alpha-smooth muscle actin (αSMA; Acta2), smooth muscle protein 22-α (sM22α; Tagln), desmin (Des), vimentin (Vim), and myosin heavy chain (smMHC; Myh11) in fibroblasts, myofibroblasts, and smooth muscle cells (18). Both GFP-positive and GFP-negative cells were positive for αSMA, and sM22α (smooth muscle or myofibroblast markers) and for vimentin and negative for smMHC, a marker of more mature muscle cells, and desmin, a smooth muscle cell protein not normally found in fibroblasts or myofibroblasts (SI Appendix, Fig. S4 A and B).
Two candidate reagents that have been used to identify fibroblasts are anti-FAPα, (Fap), which recognizes a member of the dipeptidyl peptidase family expressed by tumor stromal fibroblasts (19), and ER-TR7, an antibody raised against thymic stromal cells that recognizes an unidentified intracellular antigen (20, 21). GFP-positive cells express the ER-TR7 antigen and FAPα (SI Appendix, Fig. S4A), whereas GFP-negative cells express only FAPα (SI Appendix, Fig. S4B).
The results of the foregoing studies show that GFP-negative fibroblasts can be further divided into several subpopulations. The patterns of expression appear to be diverse and distinctive, suggesting either that multiple cell types are present or that an otherwise homogeneous population can display wide variation as a possible consequence of other influences, such as growth conditions or duration of culture.
Multiparameter Analysis Exposes a Highly Heterogeneous Population.
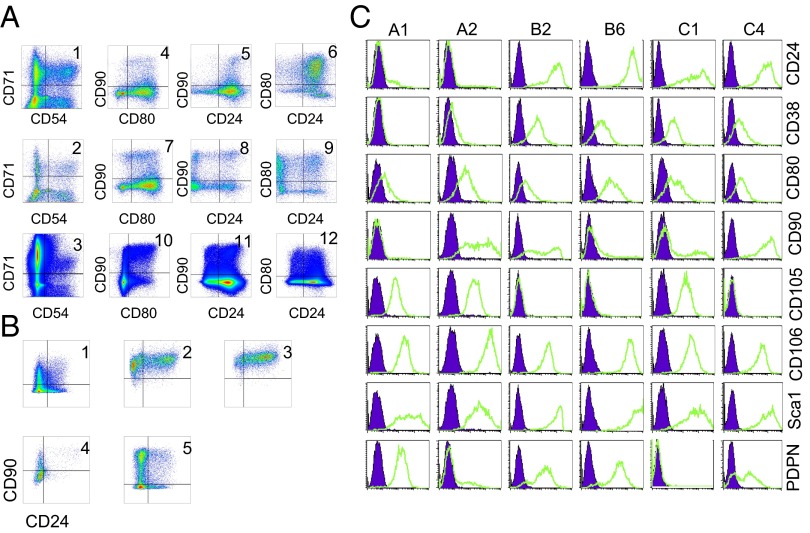
Because the heterogeneity observed for individual cell surface proteins in the GFP-negative population could reflect the diversity of a single population, each member of which could show variation, or could arise from the presence of multiple subpopulations with distinct patterns of expression, we conducted a multiparameter cytometry experiment. A culture that was homogeneous by morphology was divided by expression of CD73 and CD146 (two proteins that reliably demarcate subsets), and three resulting subpopulations were analyzed for CD54 and CD71 expression. Analysis of the latter showed at least four distinct expression patterns, corresponding to CD54 high and low and CD71 high and low (Fig. 2A, 1–3). Analysis of CD24, CD80, and CD90.1 within these subgroups revealed additional subpopulations (Fig. 2A, 4–6, 7–9, and 10–12).
Fig. 2.
Heterogeneity within fibroblast-like cells. (A) Subset analyses. First column (1–3), CD71 and CD54 subsets; subsequent columns, analysis of the CD71+ CD54− subset by CD24, CD80, and CD90. Top row (1, 4, 5, 6), CD146+ CD73+ population; middle row (2, 7, 8, 9), CD146− CD73+ population; bottom row (3, 10, 11, 12), CD146+ CD73− population. (B) Stability of surface phenotype. Cultures initiated from subpopulations were reanalyzed after propagation in culture for four to six passages. Top row (1–3), reanalysis of CD54− CD73− Sca+ CD80− cells that were originally CD24− CD90− (1), CD24− CD90+ (2), or CD24+ CD90+ (3). Bottom row (4 and 5), CD54− CD73− Sca+ CD80+ cells, originally CD24− CD90− (4) or CD24− CD90+ (5). (C) Heterogeneity of cell populations derived from isolated colonies. Individual colonies were picked and expanded, and the surface expression patterns of the progeny were analyzed by flow cytometry.
To identify whether the observed variation represented the multiple manifestations of a single cell type capable of facile conversion between phenotypes or the presence of a complex population of cells stably expressing distinct patterns of cell surface proteins, we subjected selected subpopulations to fluorescence-activated cell sorting and initiated cultures from the sorted cells. We then reanalyzed the cultures for their sort markers after four to six passages. Most of the expanded subpopulations retained their original surface phenotype, and in settings in which drift toward a less distinct expression pattern was observed, the pattern of drift was typically specific to the population observed (Fig. 2B, 1–5). After CD54−, CD73−, and Sca-1+ cells were sorted for CD24, CD80, and CD90.1 expression and propagated in culture, ∼60% of the CD24−, CD80−, CD90.1− cells and ∼86% of the CD24+, CD80−, CD90.1+ cells retained their initial pattern of expression (Fig. 2B, 1 and 3). These data suggest that the observed heterogeneity of fibroblast-like cells largely reflects the expression programs of phenotypically stable cell types.
Single Colonies Give Rise to Different Phenotypes.
If the cells of the MEF population stably express distinct cell surface phenotypes, then it should be possible to isolate clonally derived progeny that exhibit those phenotypes. To test this, we plated MEFs from day E18.5 embryos in medium containing 20% calf serum to yield visible colonies of various sizes at 6–8 wk in culture. Isolated colonies were individually picked and cultured in 96-well microplates. The serum percentage was gradually reduced to 10% as the clones expanded. Failures of expansion were common, and many of the colonies did not fill wells of a 96-well plate or subsequently did not fill wells of a 48-well plate, with some showing signs of senescence. Out of 227 colonies with normal morphology, 20 clones could be expanded (SI Appendix, Table S2). Each of the clones was analyzed by microscopy and cytometry for GFP status (SI Appendix, Fig. S5 A and B). Three GFP-positive cell populations were obtained, a proportion roughly consistent with the observed percentage of GFP-positive cells in day E18.5 cultures. Individual clones could acquire characteristics of fat, muscle, and bone following exposure to the appropriate media (SI Appendix, Fig. S6).
Consistent with the interpretation that the starting population was heterogeneous, surface phenotyping of cells derived from GFP-negative colonies revealed great variability (Fig. 2C and SI Appendix, Table S3). These data suggest that the phenotypic diversity observed in unfractionated populations could be retained by single cells and their progeny through many rounds of cellular division.
Single-Cell Transcriptome Analysis.
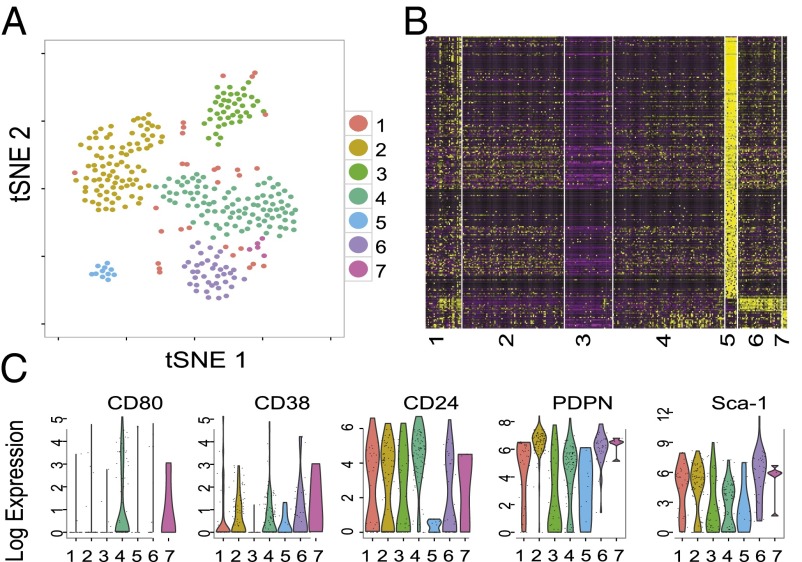
Single-cell transcriptome data were collected from 136 unfractionated cells of an E12.5 embryo, 86 fibroblasts from E18.5 embryo skeletal muscle, and 88 cells from a sorted and cultured MEF population (CD80−, Sca-1−, CD24−, CD38−, CD90−, CD107a−). Analysis of principal components yielded 10 statistically significant components that could be reduced to two dimensions using t-distributed stochastic neighbor embedding (tSNE) (22–24). Density clustering with differential expression analysis gave seven transcriptionally distinct clusters ranging in size from 5 to 96 cells (Fig. 3A). Populations 2–7 are classified as distinct, whereas population 1 is composed of cells dissimilar to one another and the cells of populations 2–7. Populations 2 and 4 are the largest, and although they are classified as single populations computationally, elements of diversity that do not rise to the level of statistical significance are clearly present (Fig. 3B). Of the markers used for flow cytometry analysis, only five (CD80, CD38, CD24, PDPN, and Sca-1) were found to be expressed at levels adequate for analysis, and for each the expression levels were widely distributed, consistent with the observed cytometric heterogeneity (Fig. 3C). The genes shown in the heat map are listed in SI Appendix, Table S4 and a more comprehensive list of the highly variable genes is presented in SI Appendix, Table S5. These experiments show that profound differences in transcriptional profile can be detected among single fibroblast cells, even when a single organ type serves as the fibroblast source.
Fig. 3.
Single-cell analyses identify transcriptionally distinct clusters. (A) A tSNE plot of seven distinct populations. Cell populations range from the very distinct and small (e.g., population 5) to the larger populations with potential further subdivisions (e.g., population 2). (B) Heat map of gene expression by markers that best differentiate the seven cell populations. (C) Violin density plots in the seven cell populations for CD80, CD38, CD24, PDPN, and Sca-1. The ordinate is the gene expression level measured in counts per million mapped reads. Each dot represents an expression value for one cell. The width is a smoothed probability density of the expression values.
Organotypic Expression of Fibroblast Surface Phenotypes.
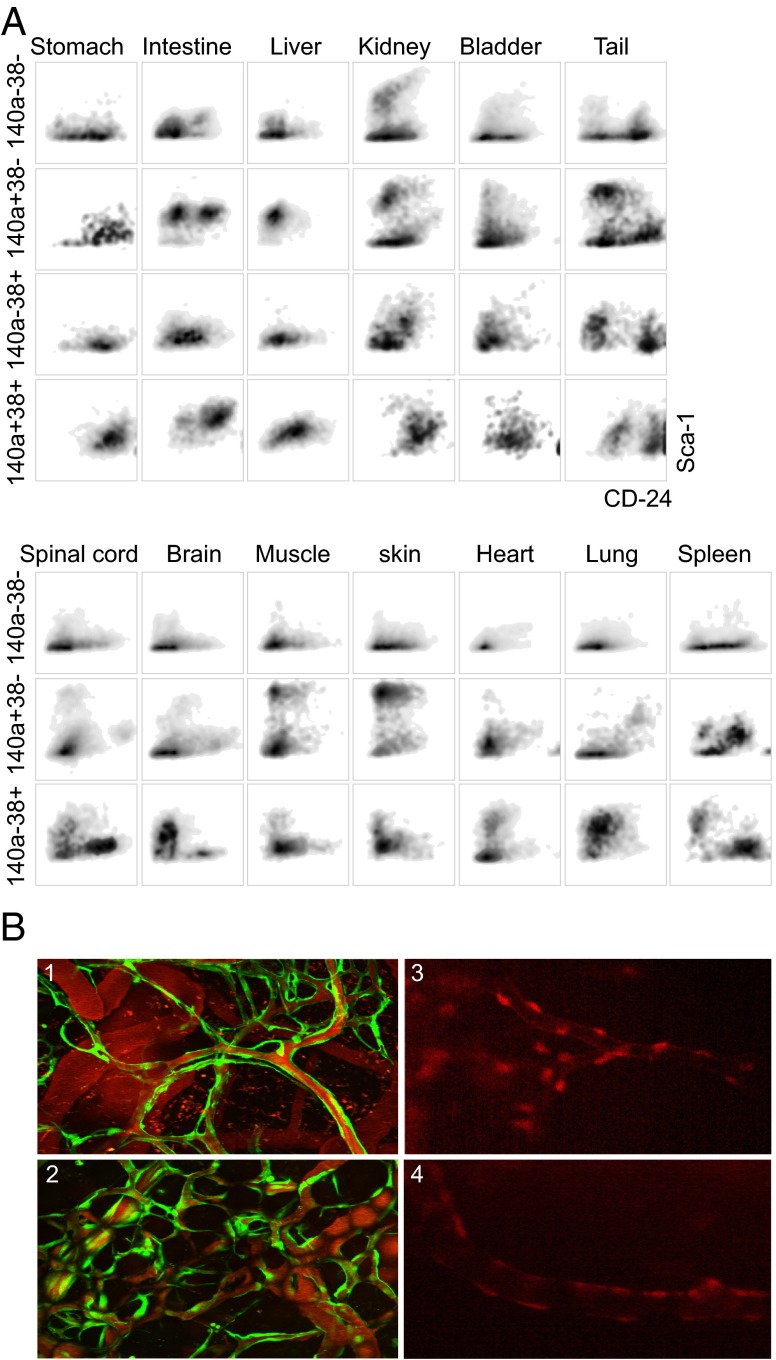
To determine whether embryonic location contributes to the diversity of fibroblast subtypes, we isolated fibroblasts from multiple embryonic organs. Cells from each organ were subjected to multiparameter flow cytometry without being subjected to culture conditions. FAPα was used to gate a population that could be considered distinct from cells of neuroepithelial origin. Cells expressing FAPα were present in all organs, albeit to varying degrees (SI Appendix, Fig. S7). Subpopulations characterized by differential expression of CD38 and CD140a were further analyzed for CD24 and Sca-1 expression, which revealed a variety of distinguishable fibroblast subtypes (Fig. 4A and SI Appendix, Fig. S9). Organ-specific patterns of embryonic fibroblast subsets were readily detected.
Fig. 4.
Diversity of FAPα-positive uncultured fibroblasts and multipotency of GFP-negative cells. (A) Heterogeneity among fibroblast-like cells from embryonic organs. Isolated cells were not cultured before analysis. Fibroblasts from several organs obtained from E18.5 embryos were analyzed using antibodies recognizing FAPα, CD140a, CD38, Sca-1, and CD24. FAPα-positive cells were selected for flow cytometry studies. (B) Two-photon micrographs of mouse cranial windows. (1 and 2) Windows were coimplanted with unlabeled GFP-negative fibroblasts and GFP-labeled HUVECs. Newly formed vessels were visualized with i.v.-injected tetramethylrhodamine dextran and HUVECs encircling them are in green, on days 39 (1) and 109 (2). (3 and 4) Windows were coimplanted with dsRED- labeled GFP-negative fibroblasts and unlabeled HUVECs at day 60, leading to dsRED-labeled cells surrounding the newly formed vessels.
We also initiated fibroblast cultures from 13 embryonic organs. Most cultures were almost entirely FAPα-positive (SI Appendix, Fig. S8), showing that FAPα can be expressed by fibroblasts in situ or in culture. A significant proportion of the fibroblast-like cells cultured from heart did not express FAPα, however. The cultured fibroblasts from various organs also displayed significant diversity (SI Appendix, Fig. S9).
GFP-Negative Cells Display Multipotency, But GFP-Positive Cells Do Not.
GFP-negative cells isolated from day E12.5 and E18.5 embryos were exposed to culture conditions that promote in vitro differentiation. Adipogenic differentiation was marked by an accumulation of intracellular fat droplets visualized by oil red O staining after an induction period of 2 wk (SI Appendix, Fig. S10A, 1). Quantitative PCR (qPCR) of cDNA prepared from differentiated cultures demonstrated an induction of adipocyte-specific genes (SI Appendix, Fig. S10 B, 6 and C, 6). No change in transcripts encoding caldesmon (taken to be a protein exhibiting invariant expression) was observed.
Osteogenic differentiation was evaluated by cytochemical staining with alizarin red for calcium deposition or alcian blue to visualize proteoglycan deposition, and for alkaline phosphatase (SI Appendix, Fig. S10A, 2–4). Osteogenic transcription patterns were revealed by qPCR evaluating the expression of osteoblast-specific markers (SI Appendix, Fig. S10 B, 5 and C, 5).
Myogenic differentiation was evaluated morphologically, by syncytium formation observed after 2–3 wk of culture. Confocal immunofluorescence microscopy showed greatly increased smooth muscle myosin heavy chain (Myh11) immunoreactivity compared with cells propagated under conventional conditions (SI Appendix, Figs. S4B and S10A, 9). qPCR demonstrated increased expression of muscle cell markers. (SI Appendix, Fig. S10 B, 4 and C, 4).
GFP-positive cells did not differentiate into adipogenic or osteogenic cells, as assessed by cytochemical staining (SI Appendix, Fig. S10A). In addition, qPCR did not reveal any significant shifts in transcript abundance characteristic of the respective lineages (SI Appendix, Fig. S10 B, 1–3 and C, 1–3). Expression of CD44, CD80, CD120a, and CD121 was reduced after induction of differentiation in all cases, whereas the densities of CD24, CD34, and CD106 were reduced in some, but not all, conditions (SI Appendix, Fig. S11). Expression of CD29, CD90.1, CD140a, Sca-1, and PDPN were little affected, and a CD36 population appeared in cultures of cells exposed to adipogenic conditions.
GFP-Negative Cells Can Form Pericyte-Like Structures in Vivo and Contract Collagen in Vitro.
Pericytes, cells that girdle the abluminal side of endothelial cells in the perivascular space, may be the most consequential cell type in the microvascular complications of diabetes (25, 26). The potential of the cell population to differentiate into pericytes has been explored in a mouse cranial window model that permits study of the microvasculature and associated cell types in vivo by intravital microscopy (27). Here, unlabeled GFP-negative cells and GFP-labeled human umbilical vein endothelial cells (HUVECs) were coimplanted in mouse cranial windows (28). At various intervals, the windows were examined by intravital microscopy to observe the ability of HUVECs to form complex microvessels. Newly formed microvessels were observed with green HUVECs (Fig. 4B, 1 and 2). Next, GFP-negative cells were labeled with dsRED (SI Appendix, Fig. S12A) and coimplanted with unlabeled HUVECs. Red fluorescent cells were observed around newly formed vessels (Fig. 4B, 3 and 4), suggesting the differentiation of the implanted cells into pericyte-like structures.
To further assess the multifunctional capacity of the GFP-negative cells in vitro, we performed collagen gel contraction assays. When cultured in a collagen type I gel in serum-free media for 1–2 d, GFP-negative cells reduced the diameter of the gel to <50% of the original size (SI Appendix, Fig. S12B). NIH 3T3 fibroblasts and GFP-positive cells did not produce significant contraction of the collagen gel. Using a matrigel culture support and serum-free medium, GFP-negative cells (SI Appendix, Fig. S12C, 1) and 10T1/2 cells adopted a smooth muscle morphology after only 1–3 d in culture (SI Appendix, Fig. S12C, 2). In comparison, GFP-positive cells at passage 6 or 34 did not show smooth muscle morphology (SI Appendix, Fig. S12C, 3 and 4). After detachment from the matrigel and immunofluorescence analysis for smooth muscle-specific caldesmon, nearly all cells were immunoreactive (SI Appendix, Fig. S12D, 1–4). These results demonstrate that fibroblast-like cells from embryos have the capacity to mature into cell types similar to those of pericytes.
Discussion
This study establishes that embryonic cells of the fibroblast class constitute a rich population of subtypes. From a transgenic reporter mouse line in which a VEGF promoter fragment drives the expression of GFP, cultures containing subpopulations of GFP-positive and GFP-negative cells with the typical characteristics of fibroblasts could be initiated. Within the GFP-negative cells, further subdivisions could be made based on the abundance of various cell surface proteins. Our analysis identified a large number of distinct subpopulations in cultures initiated from embryos. Cells with differing cell surface phenotypes were not easily distinguished from one another by morphology; for example, despite obvious differences in functionality, GFP-negative and GFP-positive fibroblasts were not distinguishable ultrastructurally.
GFP-negative fibroblasts displayed a well-spread morphology with prominent stress fibers and could be induced to differentiate into cell types of fat, muscle, and bone lineages. These features are also exhibited by the fibroblast-like cells identified as MSCs (29); however, the patterns of phenotypic expression identified here diverge from previously identified compilations of MSC phenotypes. MSCs from bone marrow have been purified by negative selection for antibodies against CD34, CD45, and CD14. In contrast, we found that GFP-negative cells expressed detectable levels of CD34 and CD14. GFP-negative cells show low levels of CD105 and CD73, in contrast to MSCs. MSCs have generally been shown to express Sca-1, PDGR-α (CD140a), NST, and CD133 (9, 13, 15, 30), whereas GFP-negative cells do not express CD133 and express variable levels of PDGR-α. GFP-negative cells express CD24, unlike preparations of MSCs. GFP-negative cells express PDPN, a marker suggested to be absent from MSCs (10, 11). The pattern of surface protein markers of the GFP-negative cells does not coincide with that of other known MSCs or the more primitive multipotent adult progenitor cells (14).
Although GFP-negative cells express some proteins found in ES cells, such as NST, nestin, and Fra-1, they do not express other stem cell markers, such as Sox-2, Klf-4, Oct-4, cMyc, CD31, SSEA-1, SSEA-3, and Tra-1-81. Both GFP-positive and GFP-negative cells express proteins found in contractile and intermediate filaments, such as αSMA, sM22α, and vimentin, but lack others, such as desmin and smMHC. GFP-positive cells express ER-TR7 and FAPα, whereas GFP-negative cells express only FAPα.
Following the identification of a small number of surface markers that could be used to subdivide the population by expression pattern, the fibroblast-like cells were found to be extremely heterogeneous, belying what appear to be relatively widespread assumptions about MEF uniformity. Multicolor flow cytometry revealed a complex population structure of subtypes with varying degrees of stability of the cell surface phenotype. Gating cells by CD73 and CD146 expression gave three subtypes that were further analyzed for CD54 and CD71 expression, producing 12 distinct patterns. Additional gating on CD24, CD80, and CD90.1 allowed further differentiation of subtypes. On expansion of sorted populations in culture, the most frequently observed behavior was retention of the expression pattern used to select the population.
Consistent with the observed retention of characteristics following culture of sorted subpopulations, cells from individual colonies showed unimodal abundance distributions and distinguishable characteristics, a finding inconsistent with the interpretation that the heterogeneous expression may be attributed to gene or gene network noise (31). Cells with differing surface phenotypes could be coerced into bone, muscle, and fat lineages, indicating that at least some of the cells exhibited true multipotency.
Single-cell expression profiling revealed intraorgan variation in cells captured from striated muscle. Multiparameter flow cytometric analysis of FAPα-positive cells from various organs showed that the diversity of fibroblast expression is exhibited by cells in vivo. Moreover, distinct patterns of expression were associated with different organs, indicating at least some form of organotypic gene expression. The use of FAPα as a marker was validated in part by the finding that a majority of fibroblasts growing in culture from various organ sources expressed FAPα.
Heterogeneity is an attribute of multiple cell types, including many sources of MSCs (32–36), hematopoietic progenitors (37, 38), adult cardiac side populations (39), and side populations of skeletal muscle (40). The extent to which this heterogeneity is transitory, representing gene or gene network noise, and to what extent it is stable, representing epigenetic subtypes, is not clear. For example, embryonic stem cell cultures show heterogeneity of expression of the transcription factors Nanog (41), Gata6, Rex1 (42), and Dppa3 (43), but the retention of pluripotency and the typically high fraction of clonal ES cell lines that can fully reconstitute all embryonic lineages support the view that the heterogeneity represents gene or gene network noise rather than variegated lineage-specific expression.
Fibroblast reprogramming has applications in drug discovery, disease modeling, tissue engineering, and regenerative medicine. The identification of extensive heterogeneity within fibroblast populations suggests that the developmental trajectories and possibly final states of the reprogrammed cells may vary considerably. Because reprogramming attempts often succeed for only a very small fraction of the cells targeted for fate reassignment, the possibility that the reprogrammed cells arise from a distinct subpopulation with unusual attributes should be considered.
From a practical perspective, the study of the phenotypic consequences of genetic changes is not easily carried out using fibroblasts unless specific steps are taken to guard against effects arising from inadvertent comparison of two cell lines of different fibroblast subtypes. One practice that can protect against this is the use of two sublines of a cloned fibroblast line from a null mouse, one subline consisting of the original (null) clone of fibroblast cells transduced with an empty retroviral vector and the other consisting of the null cell clone transduced with a retroviral vector that restores the missing gene product (44). If precautions of this type are not taken, there is a possibility that the variation observed between embryonic fibroblasts isolated from different mice could reflect the properties of different populations of fibroblasts, imperiling the conclusions reached.
Because the principal method for assigning subtypes in this study relied on cytometric classification of cell surface protein expression patterns, there is a reasonable likelihood that additional patterns will become apparent when less-biased methods are applied. At a minimum, the subclasses defined by single-cell transcriptome analysis do not obviously correspond to the subtypes defined by surface marker expression, and the identification of distinct patterns of expression in different tissues suggests that additional organ-specific characters may be present. Although a precise estimate of the number of fibroblast types present in vivo cannot be made, the possibility of tens to hundreds of subtypes seems plausible.
Materials and Methods
Preparation of MEF Cell Cultures.
All animal studies were conducted following protocols approved by the Institutional Animal Care and Usage Committee of Massachusetts General Hospital. Embryos from VEGF-GFP transgenic mice were isolated between E12.5 and E18.5. After the heads, tails, limbs, and most of the internal organs were removed, the embryos were minced and typsinized for 20 min, and then seeded into T-75 cell culture dishes in 15 mL of complete MEF media (SI Appendix, Materials and Methods). The cells were split at 1:2–1:3 ratios when freshly confluent, passaged two or three times to obtain a morphologically homogenous culture, and then frozen or expanded for further studies.
Immunofluorescence.
For multicolor flow cytometry of fibroblasts from embryonic organs, extracted organs were minced into smaller pieces and digested using Liberase DL (Roche) and DNase I (Roche) in HBSS (SI Appendix, Materials and Methods) in a CO2 incubator at 37 °C for 20–30 min. Digested tissues were passed through a 100-µm mesh to obtain single-cell suspensions. Fc receptors were blocked, and cells were stained for flow cytometry analyses. Antibodies are listed in SI Appendix, Table S7.
Single-Cell Capture, mRNA Library Preparation, and Sequencing.
Cells were suspended at 250–500 cells/µL. The cell suspension and Fluidigm C1 suspension reagent were mixed at a 3:2 ratio. Then 20 µL of mix was loaded into a Fluidigm C1 integrated fluidic circuit system. RNA extraction and mRNA amplification were performed for single cells within the C1 system using the Clontech SMARTer system. cDNA products were prepared for sequencing using an Illumina Nextera XT-DNA sample preparation kit. The R package Seurat (45) was used to generate cell population clusters. Principal component analysis was performed on genes present in at least five cells with an expression value of one count per million mapped reads. Individual cells were projected based on their principal component scores onto a 2D plot using tSNE (24).
Mouse Cranial Window Preparation, Implantation, and Two-Photon Microscopy.
Mouse cranial windows were prepared as described previously (46). Tetramethylrhodamine dextran was used to visualize newly formed vessels.
More detailed information on the methods and reagents used in this study is provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Sylvie Roberge for assistance with the cranial window preparation, Peigen Huang for management of the mouse colony, and Gino Ferraro for comments on the manuscript. This research was supported by National Institutes of Health Grants P01CA080124, R35CA197743, and R01DK103879.
Footnotes
The authors declare a conflict of interest. R.K.J. has received consultant fees from Ophthotech, SPARC, SynDevRx, and XTuit; owns equity in Enlight, Ophthotech, SynDevRx, and XTuit; and serves on the Board of Directors of XTuit and the Board of Trustees of Tekla Healthcare Investors, Tekla Life Sciences Investors, Tekla Healthcare Opportunities Fund, and Tekla World Healthcare Fund. No reagents or funding from any these companies were used in this study.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522401112/-/DCSupplemental.
References
- 1.Schuppan D, Pinzani M. Anti-fibrotic therapy: Lost in translation? J Hepatol. 2012;56(Suppl 1):S66–S74. doi: 10.1016/S0168-8278(12)60008-7. [DOI] [PubMed] [Google Scholar]
- 2.Dvorak HF, Weaver VM, Tlsty TD, Bergers G. Tumor microenvironment and progression. J Surg Oncol. 2011;103(6):468–474. doi: 10.1002/jso.21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishii G, Ochiai A, Neri S. 2015. Phenotypic and functional heterogeneity of cancer-associated fibroblast within the tumor microenvironment. Adv Drug Deliv RevS0169-409X(15)00160-X.
- 4.Öhlund D, Elyada E, Tuveson D. Fibroblast heterogeneity in the cancer wound. J Exp Med. 2014;211(8):1503–1523. doi: 10.1084/jem.20140692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orimo A, Weinberg RA. Heterogeneity of stromal fibroblasts in tumors. Cancer Biol Ther. 2007;6(4):618–619. doi: 10.4161/cbt.6.4.4255. [DOI] [PubMed] [Google Scholar]
- 6.Pickup M, Novitskiy S, Moses HL. The roles of TGFβ in the tumour microenvironment. Nat Rev Cancer. 2013;13(11):788–799. doi: 10.1038/nrc3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wijten P, et al. High precision platelet releasate definition by quantitative reversed protein profiling: Brief report. Arterioscler Thromb Vasc Biol. 2013;33(7):1635–1638. doi: 10.1161/ATVBAHA.113.301147. [DOI] [PubMed] [Google Scholar]
- 8.Fukumura D, et al. Tumor induction of VEGF promoter activity in stromal cells. Cell. 1998;94(6):715–725. doi: 10.1016/s0092-8674(00)81731-6. [DOI] [PubMed] [Google Scholar]
- 9.Baddoo M, et al. Characterization of mesenchymal stem cells isolated from murine bone marrow by negative selection. J Cell Biochem. 2003;89(6):1235–1249. doi: 10.1002/jcb.10594. [DOI] [PubMed] [Google Scholar]
- 10.Conrad C, et al. Multipotent mesenchymal stem cells acquire a lymphendothelial phenotype and enhance lymphatic regeneration in vivo. Circulation. 2009;119(2):281–289. doi: 10.1161/CIRCULATIONAHA.108.793208. [DOI] [PubMed] [Google Scholar]
- 11.Imaizumi Y, Amano I, Tsuruga E, Kojima H, Sawa Y. Immunohistochemical examination for the distribution of podoplanin-expressing cells in developing mouse molar tooth germs. Acta Histochem Cytochem. 2010;43(5):115–121. doi: 10.1267/ahc.10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lv FJ, Tuan RS, Cheung KM, Leung VY. Concise review: The surface markers and identity of human mesenchymal stem cells. Stem Cells. 2014;32(6):1408–1419. doi: 10.1002/stem.1681. [DOI] [PubMed] [Google Scholar]
- 13.Tondreau T, et al. Mesenchymal stem cells derived from CD133-positive cells in mobilized peripheral blood and cord blood: Proliferation, Oct4 expression, and plasticity. Stem Cells. 2005;23(8):1105–1112. doi: 10.1634/stemcells.2004-0330. [DOI] [PubMed] [Google Scholar]
- 14.Reyes M, et al. Origin of endothelial progenitors in human postnatal bone marrow. J Clin Invest. 2002;109(3):337–346. doi: 10.1172/JCI14327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houlihan DD, et al. Isolation of mouse mesenchymal stem cells on the basis of expression of Sca-1 and PDGFR-α. Nat Protoc. 2012;7(12):2103–2111. doi: 10.1038/nprot.2012.125. [DOI] [PubMed] [Google Scholar]
- 16.Gruda MC, Kovary K, Metz R, Bravo R. Regulation of Fra-1 and Fra-2 phosphorylation differs during the cell cycle of fibroblasts and phosphorylation in vitro by MAP kinase affects DNA binding activity. Oncogene. 1994;9(9):2537–2547. [PubMed] [Google Scholar]
- 17.Tsai RY, McKay RD. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16(23):2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abderrahim-Ferkoune A, et al. Transdifferentiation of preadipose cells into smooth muscle-like cells: Role of aortic carboxypeptidase-like protein. Exp Cell Res. 2004;293(2):219–228. doi: 10.1016/j.yexcr.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 19.Scanlan MJ, et al. Molecular cloning of fibroblast activation protein alpha, a member of the serine protease family selectively expressed in stromal fibroblasts of epithelial cancers. Proc Natl Acad Sci USA. 1994;91(12):5657–5661. doi: 10.1073/pnas.91.12.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Vliet E, Melis M, Van Ewijk W. Monoclonal antibodies to stromal cell types of the mouse thymus. Eur J Immunol. 1984;14(6):524–529. doi: 10.1002/eji.1830140608. [DOI] [PubMed] [Google Scholar]
- 21.Van Vliet E, Melis M, Foidart JM, Van Ewijk W. Reticular fibroblasts in peripheral lymphoid organs identified by a monoclonal antibody. J Histochem Cytochem. 1986;34(7):883–890. doi: 10.1177/34.7.3519751. [DOI] [PubMed] [Google Scholar]
- 22.Amir AD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31(6):545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 25.Arboleda-Velasquez JF, Valdez CN, Marko CK, D’Amore PA. From pathobiology to the targeting of pericytes for the treatment of diabetic retinopathy. Curr Diab Rep. 2015;15(2):573. doi: 10.1007/s11892-014-0573-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel-Hett S, D’Amore PA. Signal transduction in vasculogenesis and developmental angiogenesis. Int J Dev Biol. 2011;55(4-5):353–363. doi: 10.1387/ijdb.103213sp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan F, et al. Vascular permeability and microcirculation of gliomas and mammary carcinomas transplanted in rat and mouse cranial windows. Cancer Res. 1994;54(17):4564–4568. [PubMed] [Google Scholar]
- 28.Koike N, et al. Tissue engineering: Creation of long-lasting blood vessels. Nature. 2004;428(6979):138–139. doi: 10.1038/428138a. [DOI] [PubMed] [Google Scholar]
- 29.Pittenger MF, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 30.Kafienah W, Mistry S, Williams C, Hollander AP. Nucleostemin is a marker of proliferating stromal stem cells in adult human bone marrow. Stem Cells. 2006;24(4):1113–1120. doi: 10.1634/stemcells.2005-0416. [DOI] [PubMed] [Google Scholar]
- 31.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: From theories to phenotypes. Nat Rev Genet. 2005;6(6):451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 32.Ratajczak MZ, Kucia M, Majka M, Reca R, Ratajczak J. Heterogeneous populations of bone marrow stem cells: Are we spotting on the same cells from the different angles? Folia Histochem Cytobiol. 2004;42(3):139–146. [PubMed] [Google Scholar]
- 33.Okamoto T, et al. Clonal heterogeneity in differentiation potential of immortalized human mesenchymal stem cells. Biochem Biophys Res Commun. 2002;295(2):354–361. doi: 10.1016/s0006-291x(02)00661-7. [DOI] [PubMed] [Google Scholar]
- 34.Panepucci RA, et al. Comparison of gene expression of umbilical cord vein and bone marrow-derived mesenchymal stem cells. Stem Cells. 2004;22(7):1263–1278. doi: 10.1634/stemcells.2004-0024. [DOI] [PubMed] [Google Scholar]
- 35.Vogel W, et al. Heterogeneity among human bone marrow-derived mesenchymal stem cells and neural progenitor cells. Haematologica. 2003;88(2):126–133. [PubMed] [Google Scholar]
- 36.Colter DC, Sekiya I, Prockop DJ. Identification of a subpopulation of rapidly self-renewing and multipotential adult stem cells in colonies of human marrow stromal cells. Proc Natl Acad Sci USA. 2001;98(14):7841–7845. doi: 10.1073/pnas.141221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453(7194):544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morita Y, Ema H, Nakauchi H. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J Exp Med. 2010;207(6):1173–1182. doi: 10.1084/jem.20091318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamahara K, et al. Heterogeneic nature of adult cardiac side population cells. Biochem Biophys Res Commun. 2008;371(4):615–620. doi: 10.1016/j.bbrc.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 40.Uezumi A, et al. Functional heterogeneity of side population cells in skeletal muscle. Biochem Biophys Res Commun. 2006;341(3):864–873. doi: 10.1016/j.bbrc.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 41.Singh AM, Hamazaki T, Hankowski KE, Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25(10):2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- 42.Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135(5):909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- 43.Carter MG, et al. An in situ hybridization-based screen for heterogeneously expressed genes in mouse ES cells. Gene Expr Patterns. 2008;8(3):181–198. doi: 10.1016/j.gep.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou GL, Na SY, Niedra R, Seed B. Deficits in receptor-mediated endocytosis and recycling in cells from mice with Gpr107 locus disruption. J Cell Sci. 2014;127(Pt 18):3916–3927. doi: 10.1242/jcs.135269. [DOI] [PubMed] [Google Scholar]
- 45.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown E, Munn LL, Fukumura D, Jain RK. In vivo imaging of tumors. Cold Spring Harb Protoc. 2010 doi: 10.1101/pdb.prot5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.