Significance
Life in aerobic environments inevitably generates reactive oxygen species (ROS), which damage proteins and other cellular components, often irreversibly. Cells must degrade these potentially harmful, damaged macromolecules. A bacterial ROS sensor, the transcriptional repressor PerR, is itself controlled by irreversible oxidation of its metal-binding site. We found that active PerR is stable, but that this oxidation marks PerR for degradation by the protease LonA. We identified a LonA-binding site in PerR, which is exposed in structures of unliganded or oxidized PerR, but buried in the liganded active form. Therefore, we propose that an oxidation-induced and activity-coupled conformational change in PerR triggers its degradation. Similar allostery may explain how site-specific and mild oxidation can commit proteins to degradation.
Keywords: oxidative damage, proteolysis, degradation signals, Lon
Abstract
Peroxide operon regulator (PerR) is a broadly conserved hydrogen peroxide sensor in bacteria, and oxidation of PerR at its regulatory metal-binding site is considered irreversible. Here, we tested whether this oxidation specifically targets PerR for proteolysis. We find that oxidizing conditions stimulate PerR degradation in vivo, and LonA is the principal AAA+ (ATPases associated with diverse cellular activities) protease that degrades PerR. Degradation of PerR by LonA is recapitulated in vitro, and biochemical dissection of this degradation reveals that the presence of regulatory metal and PerR-binding DNA dramatically extends the half-life of the protein. We identified a LonA-recognition site critical for oxidation-controlled PerR turnover. Key residues for LonA-interaction are exposed to solvent in PerR lacking metal, but are buried in the metal-bound form. Furthermore, one residue critical for Lon recognition is also essential for specific DNA-binding by PerR, thus explaining how both the metal and DNA ligands prevent PerR degradation. This ligand-controlled allosteric mechanism for protease recognition provides a compelling explanation for how the oxidation-induced conformational change in PerR triggers degradation. Interestingly, the critical residues recognized by LonA and exposed by oxidation do not function as a degron, because they are not sufficient to convert a nonsubstrate protein into a LonA substrate. Rather, these residues are a conformation-discriminator sequence, which must work together with other residues in PerR to evoke efficient degradation. This mechanism provides a useful example of how other proteins with only mild or localized oxidative damage can be targeted for degradation without the need for extensive oxidation-dependent protein denaturation.
In aerobic respiratory organisms, reactive oxygen species (ROS) oxidize proteins (1), which can lead to loss of proper protein function. Accumulation of oxidatively damaged proteins threatens cellular homeostasis. To avoid and ameliorate recovery from oxidative damage, cells encode systems to protect biological molecules and repair oxidized proteins. Expression of these protection and repair genes is often controlled by oxidation-sensitive transcription regulators, and most of these regulators sense intracellular oxidative stress via their own oxidation. For example, the activity of a bacterial H2O2-responsive transcriptional regulator OxyR is controlled by reversible oxidation of two specific cysteines to form a disulfide bond (2, 3). Oxidized OxyR is in the active conformation and promotes expression of target genes involved in H2O2 detoxification, such as catalases. Once the intracellular levels of H2O2 decrease to subtoxic levels, the oxidized Cys residues are chemically reduced by glutathione to regenerate the sensor state of OxyR (2).
Not all oxidative stress sensors, however, are reversibly oxidized and reduced. A second class of H2O2 sensors, the peroxide operon regulator (PerR) family of transcriptional repressors, is inactivated by oxidation to trigger transcription of the oxidative response genes. The oxidative-stress-sensing mechanism of PerR is best characterized in Bacillus subtilis (4). PerR is a transcriptional repressor of genes involved in peroxide resistance. PerR is active when iron or manganese is bound at its sensor metal-binding site. As intracellular H2O2 levels rise, metal-catalyzed oxidation (MCO) of specific histidine residues (H37 or H91) in PerR generates 2-oxo-histidine adducts at these critical sensor site residues. Upon this site-specific oxidation, PerR releases its bound iron (or manganese) and adopts a more open conformation that does not tightly bind its operator sites. In contrast to the reversible disulfide bonding of the OxyR sensor cysteines, the 2-oxo-histidine modification in PerR is proposed to be irreversible (4). Therefore, once oxidized, PerR is expected to permanently lose its activity as a repressor. Because of this irreversible oxidization, PerR family members are characterized as “suicide regulators,” wherein each protein molecule can carry out only one cycle of ROS-controlled transcriptional regulation.
Although the ultimate fate of oxidized PerR has been unknown, it is likely that this protein will be degraded to prevent its accumulation in the oxidized form. Here, we show that oxidized PerR is a substrate of LonA (Lon was named to describe the long shape cells adopt in the absence of Lon-directed protein degradation), a widely conserved AAA+ (ATPases associated with diverse cellular activities) protease. We also report that the protein conformation stabilized by the bound regulatory metal is a key feature in protecting the active repressor form of PerR from degradation. Identification of sequences crucial for PerR recognition by LonA, together with the previously described crystal structures of PerR in the oxidized, metal-bound, and apo forms (5–7), lead us to propose an attractive model for how a metal-stabilized conformation protects the active form of PerR from proteolysis. Furthermore, many bacteria encode PerR homologs as ROS-stress sensors. Conserved features of PerR homologs suggest that selective degradation of oxidized sensors triggered by conformational changes may be common in a broad group of bacteria.
Results
PerR Is Degraded Faster Under Oxidizing Conditions in Vivo.
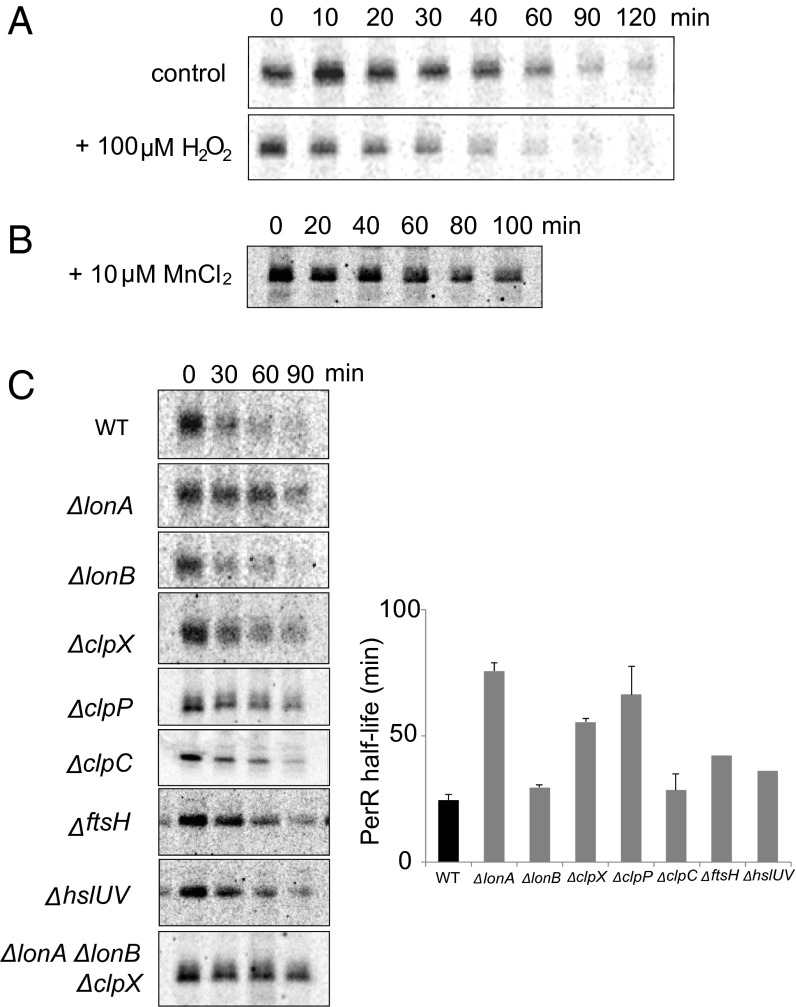
To understand the life cycle of the PerR regulator, we sought to compare the stability of unoxidized active PerR with that of irreversibly oxidized PerR. We measured the in vivo rate of PerR degradation in H2O2-treated cells (oxidizing conditions) and untreated cells by pulse-chase and immunoprecipitation (Fig. 1A). The half-life of PerR in the presence of H2O2 was estimated as 25 ± 4 min, whereas that measured in the untreated culture was 42 ± 3 min. These data reveal that PerR is degraded more rapidly in vivo under oxidizing conditions. However, because the extent of PerR oxidation in either growth condition has not been determined and the untreated cells were still exposed to oxygen, we cannot directly measure the relative stability of oxidized and nonoxidized PerR from these results.
Fig. 1.
In vivo degradation of PerR under oxidizing conditions and in AAA+ protease mutant strains. (A) The half-life of PerR was measured by pulse-chase experiment using [35S] methionine and anti-PerR sera under H2O2-treated (Lower) and nontreated (Upper) conditions. For the H2O2-treated sample, 100 µM H2O2 was added to the medium immediately after addition of the cold methionine. (B) Addition of manganese to the medium increased PerR half-life. This experiment was performed without H2O2 treatment. (C) PerR stability was determined in various AAA+ protease mutant strains in the presence of 100 µM H2O2. Left shows representative autoradiograms of pulse-chase degradation experiments with each mutant, and the bars in Right represent the average half-life of PerR in each mutant strain calculated from three independent experiments with the error bars representing the SEM, except the disruption mutants of ftsH and hslUV, which were not measured enough times to calculate errors accurately.
To gain insight into the contribution of direct PerR oxidation to its degradation under oxidizing conditions, we took advantage of the specific protective effect of manganese against PerR oxidation. PerR plays a cellular role by “monitoring” intracellular Fe and Mn levels. Under most conditions, PerR is principally an H2O2 sensor, because Fe, which is essential for its H2O2 response, binds more tightly than Mn to the protein’s sensor site. However, if cells are starved for both Fe and Mn, the sensor site may become unoccupied, and the PerR regulon would be induced. [PerR also has a structurally important zinc bound by a distinct site that is unaffected by oxidation under our conditions (8) (see below).]
PerR binds Fe at the sensor site ∼30-fold tighter than it binds Mn (8). When neither metal is limiting, the available Fe and Mn concentrations are similar, and >95% of the PerR is bound to Fe; however, when Mn is abundant and Fe is limiting, the PerR’s sensor metal binding site is principally occupied by Mn. Whereas the iron bound by PerR efficiently promotes MCO of one of the two sensor-site histidines (H37 or H91) to make it a good H2O2 sensor, manganese protects PerR from the oxidative damage by occupying the sensor site in place of iron, but not stimulating oxidation (4). Consistent with this protective effect of manganese on the PerR-sensor site, the half-life of PerR was substantially increased (>80 min) when manganese was present (Fig. 1B). This protection by manganese suggests that nonoxidized PerR, with metal bound in its sensor site, is rarely turned over.
LonA Deficiency Has the Largest Effect on PerR Degradation in Vivo.
B. subtilis has several AAA+ proteases, most of which are broadly conserved among bacteria. To identify specific AAA+ protease(s) that degrade oxidized PerR, we determined how PerR stability changed in strains carrying null alleles for the AAA+ protease genes (Fig. 1C). Although most of the strains with disruption mutations in one or more protease genes degraded PerR somewhat more slowly than wild type, the lonA disruption resulted in the largest increase in the half-life of PerR (76 ± 6 min), to a rate very similar to that measured under manganese-protective conditions. These data, together with the extended half-lives observed in some of the double- or triple-knockout strains (e.g., lonA−, lonB−, and clpX−; Fig. 1C) indicate that, although multiple AAA+ proteases influence PerR stability in vivo, LonA likely has the largest role. Thus, we focused on determining whether PerR is a direct substrate of LonA and whether this degradation is modulated by sensor-site oxidation.
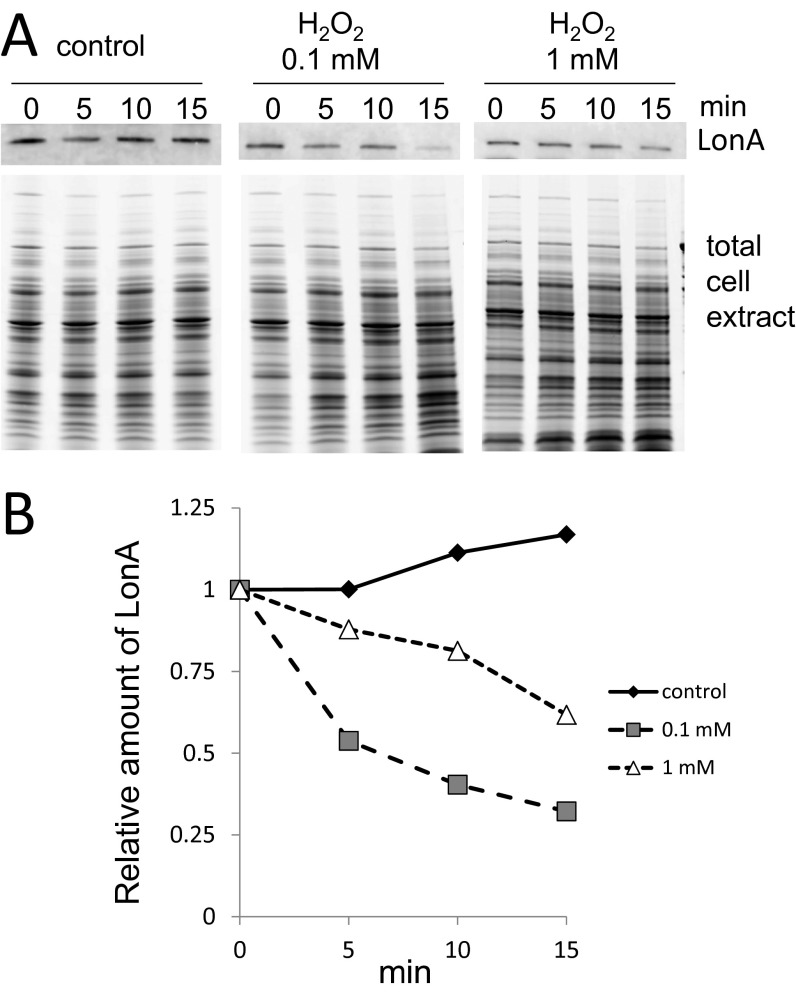
Because Lon protease is a stress-induced protein, we tested whether the oxidative stress conditions used for in vivo degradation up-regulated expression of LonA, because high concentrations of LonA could nonselectively increase protein degradation. Although a previous report indicated that 1 mM H2O2 substantially enhanced transcription of B. subtilis LonA (9), we did not observe any increase in LonA protein levels with 100 µM H2O2 (Fig. S1), where we observed enhanced degradation of PerR (Fig. 1). Thus, we sought to test whether selective proteolysis of oxidized PerR causes the increased degradation of this regulatory protein under oxidizing conditions.
Fig. S1.
LonA expression level in the presence of H2O2. (A) Time-based observation of LonA protein levels in the absence or presence of H2O2. A concentration of 100 µM (0.1 mM) or 1 mM H2O2 was added in the LB medium when the cell density was 0.6 at OD600. LonA protein level was detected by immunoblot. (B) Relative amount of LonA calculated from immunoblot signal intensity and the total protein content of cell extract in A.
LonA Degrades both Nonoxidized and Oxidized PerR in Vitro.
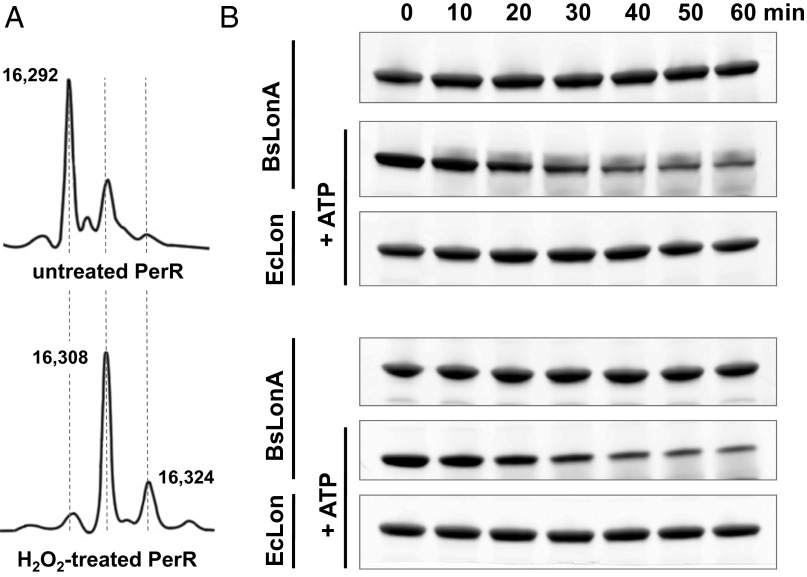
We produced an oxidized PerR sample containing 2-oxo-histidine at H37 and/or H91 in its metal-binding sensor site by treating cultures with 100 µM H2O2 directly before harvesting PerR-overexpressing Escherichia coli cells (4). We also produced an untreated PerR sample by the same expression and purification procedure, but omitting H2O2. Even without H2O2 treatment, however, the purified PerR sample contained considerable oxidized protein. As shown in Fig. 2A, in a typical preparation of non-H2O2-exposed PerR, 30–40% of the protein was oxidized at one of the sensor-site histidines; this level of contaminating oxidized PerR is similar to that observed previously (4).
Fig. 2.
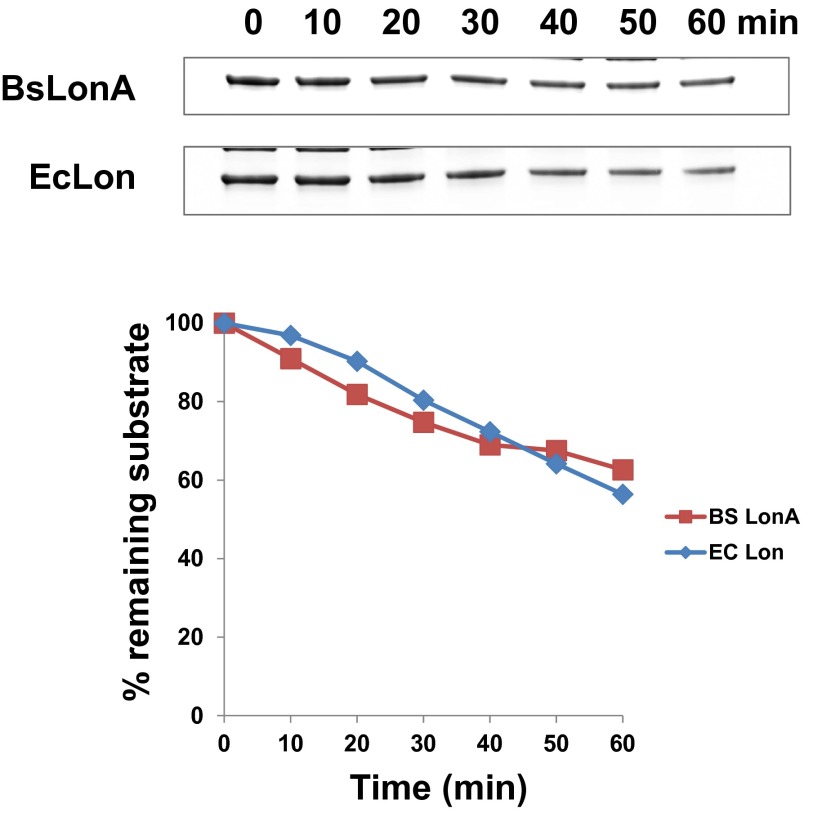
In vitro degradation of PerR by LonA. (A) Mass spectra of untreated (Upper) and H2O2-treated (Lower) PerR. The original molecular mass of the PerR monomer is 16,292 Da, and single oxidation increases it to 16,308 Da; this increase corresponds to the molecular mass of single oxygen (16 Da). All purification steps were performed in aerobic conditions. (B) Both untreated (Upper) and H2O2-treated (Lower) PerR are degraded by B. subtilis LonA in the presence of an ATP-regenerating system; however, PerR was not detectably degraded by E. coli Lon in vitro.
To determine whether PerR is a direct substrate of LonA, we reconstituted degradation in vitro. In support of our original hypothesis, LonA degraded H2O2-treated PerR. Furthermore, LonA also degraded the untreated PerR lacking either Fe or Mn at the sensor site with similar kinetics (Fig. 2B). We refer to untreated PerR that is not bound to either Fe or Mn as “apo,” although the structural Zn molecules remained bound (see below). Similar in vitro degradation rates for the oxidized and apo form of PerR was perhaps not surprising, considering that in the absence of metal ligand at the sensor site, nonoxidized apo-PerR and histidine-oxidized PerR adopt very similar conformations in crystal structures (7).
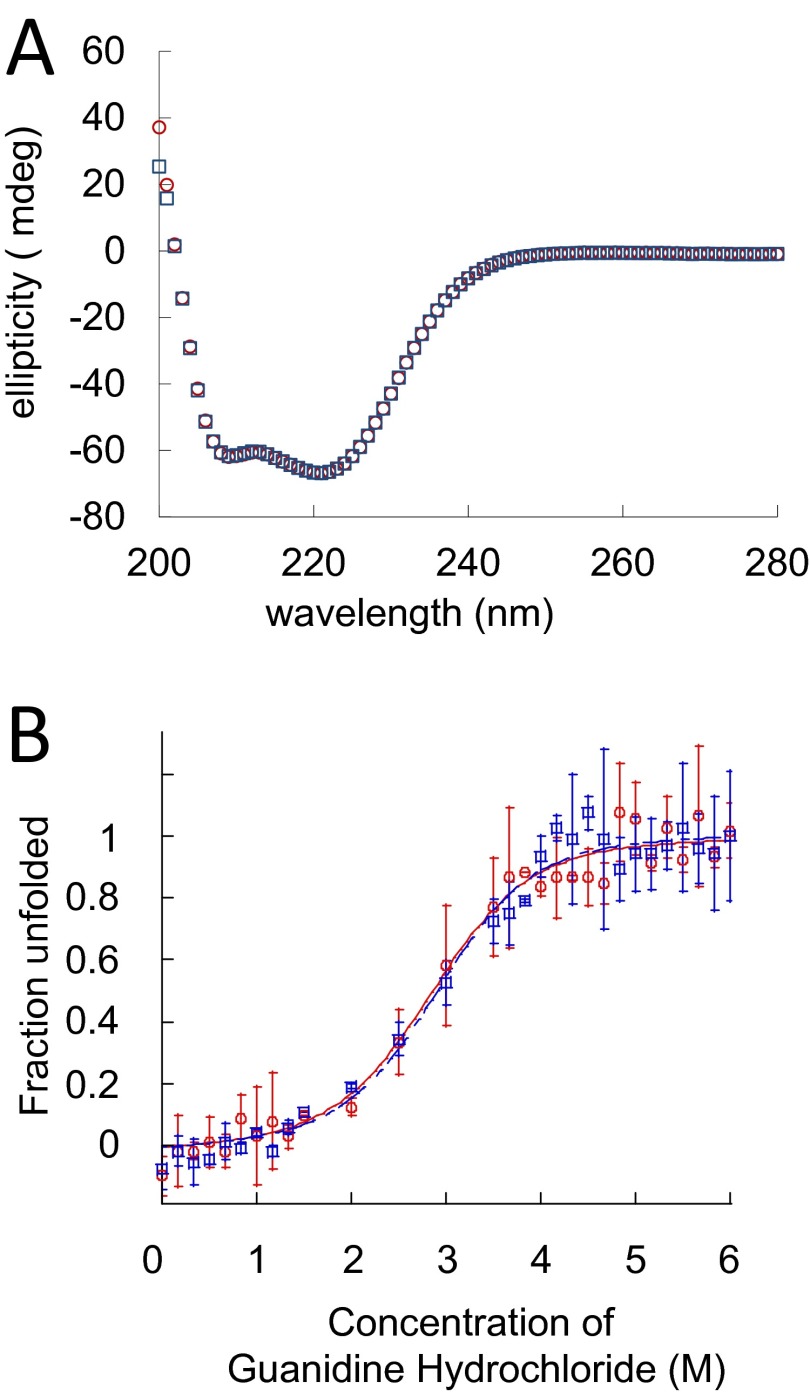
We also found that H2O2-treatment in our experiment does not reduce PerR’s intrinsic stability. Protein stability was determined by circular dichroism (CD) spectrum; both H2O2-treated and apo PerR (untreated) were unfolded by using 0–6 M concentrations of guanidine hydrochloride (Gu⋅HCl). The two CD spectra (Fig. S2A) and denaturation profiles (Fig. S2B) were virtually indistinguishable. These data strongly indicate that histidine oxidation does not significantly affect the intrinsic thermodynamic stability of PerR. Furthermore, the results demonstrate that our oxidation conditions do not disrupt binding of PerR’s structural zinc molecules, because loss of zinc would certainly affect the protein’s secondary structure and stability.
Fig. S2.
Protein conformation and stability of apo- and H2O2-treated PerR. (A) CD spectra of apo (untreated) and H2O2-treated PerR at a concentration of 30 µM, 30 °C, in 20 mM Tris⋅HCl (pH 8.0) and 100 mM NaCl. (B) Apo (red circle) and H2O2-treated (blue square) PerR were denatured by Gu⋅HCl. Fraction of unfolded protein was measured by emitted fluorescence signal intensity when the sample was excited by 275-nm UV. The plotted signal intensity values are the average of three independent sets of experiments with calculated errors of ±1 SD. Fraction of unfolded protein (Fu) was calculated from the equation Fu = (Y − Yu)/(Yn − Yu), where Y is the observed signal intensity at each denaturant concentration, Yu is the observed signal intensity with the fully denatured protein, and Yn is the signal intensity of native protein.
Interestingly, in contrast to the B. subtilis enzyme, E. coli Lon did not detectably degrade either apo or treated PerR (Fig. 2B). This difference in degradation selectivity was evident despite the fact that E. coli Lon and B. subtilis LonA are very similar (56% identity and 74% similarity), and control experiments with a different substrate confirmed that E. coli Lon was active under our experimental conditions (Fig. S3). This unexpected species-specific degradation of PerR highlights that Lon enzymes can have differently evolved modes of substrate recognition and play alternative physiological roles in different species.
Fig. S3.
E. coli Lon is active to degrade a model substrate. The I27 domain of human titin (a common small model protein used in unfolding and degradation experiments) tagged with 20 amino acids of SulA C-terminal region (a Lon degron) (titin-sulA20) was used as a positive control substrate (10, 23) to confirm that both B. subtilis LonA and E. coli Lon are active under the same conditions used for PerR degradation in Fig. 2. Both Lon orthologs degraded the control substrate with a similar efficiency.
Ligand-Dependent Conformational Change in PerR Inhibits Degradation.
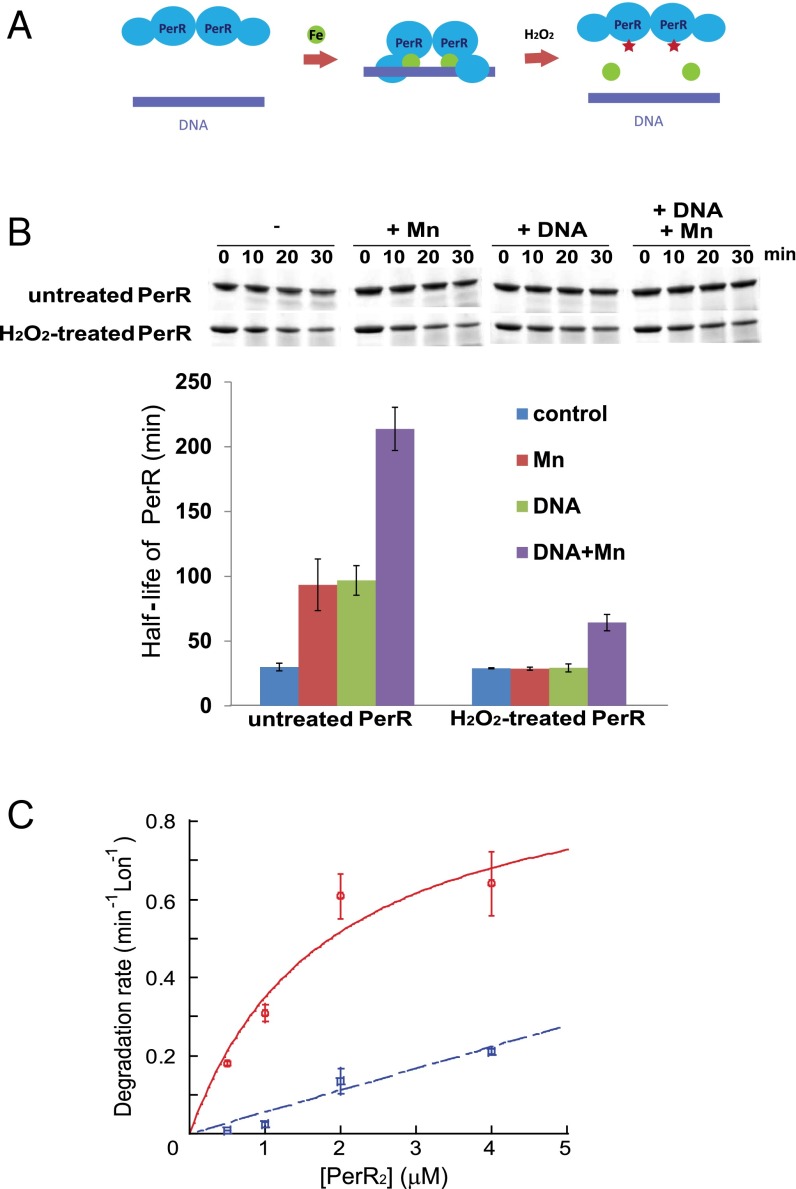
The PerR transcription regulator binds its operator sites in the promoter regions of genes it regulates, specifically in the presence of a regulatory metal (iron or manganese) (Fig. 3A). Metal binding to the sensor site promotes a change in PerR conformation from an extended to a more compact form that binds promoter DNA (5, 6). The metal-binding-promoted change of PerR to the compact conformation is explained by the fact that H37 and H91, both of which participate directly as metal binding, are located in the two different domains of PerR; thus, these residues can help bring the two PerR domains in each monomer in close proximity and stabilize the compact form.
Fig. 3.
Binding of regulatory metal inhibits degradation of PerR by LonA. (A) Model for the PerR conformational changes induced/stabilized by regulatory metal-binding and oxidation by iron and H2O2. The green circles represent iron, and the red stars represent oxidized histidine residues at the sensor site. (B) In vitro degradation rates of untreated and H2O2-treated PerR by LonA were measured in the presence or absence of 100 µM MnCl2 and a 25-bp fragment of the mrgA promoter DNA, which carries a PerR-binding sequence (4). Upper is the representative figure of degradation reactions, and Lower presents the averaged PerR half-life from three independent experiments with ±1 SEM. (C) Manganese decreases productive interactions between PerR and LonA. Initial degradation rates for PerR by LonA at different concentrations of PerR in the absence (red circle, solid line) and presence (blue square, dashed line) of 100 µM MnCl2. PerR concentration values on the x axis are for the PerR dimer. Degradation rates of the PerR proteins were measured in the presence of 0.3 µM Lon monomer and 4 mM ATP. Plotted data represent the average value from three independent experiments with ±1 SEM and give a concentration for the half-maximal degradation (appKM) of apo-PerR by LonA of ∼1.9 ± 1.1 µM. The degradation rates were all normalized by dividing the observed rate by the concentration of Lon in the reaction (thus the rates shown are a hybrid between an initial velocity and a kcat value), and the rates are in units of PerR degraded per min, per Lon monomer.
Based on the hypothesis that protein conformation and/or DNA binding affect protease accessibility and degradation, we tested whether the presence of a binding metal and/or the PerR operator DNA sequence inhibits degradation of either oxidized or nonoxidized PerR (Fig. 3B). When manganese or iron binds in the PerR sensor site, the metal is assumed to promote the same conformational change (although the high-resolution structure of only the Mn-bound form is known) (5); however, the Mn-loaded protein is not subject to MCO (4). With untreated PerR, the presence of both DNA and manganese together had the largest inhibitory effect on PerR degradation (increasing the half-life approximately eightfold to a t1/2 > 200 min). Both operator DNA and manganese contributed to this increase, each having a threefold to fourfold stabilizing effect (Fig. 3B). In contrast, the degradation rate of H2O2-treated PerR, which is defective in binding Fe or Mn, was unaffected by either manganese or DNA and only slightly stabilized when both manganese and DNA were present (Fig. 3B). Control experiments with other substrates confirmed that neither the Mn nor DNA significantly influenced LonA activity (Fig. S4). Thus, we conclude that the slow degradation of PerR in the presence of these ligands is due to a specific effect on PerR as a substrate. These results indicate that the conformational change in PerR promoted by sensor-site bound metal and DNA binding is sufficient to protect the protein from degradation by LonA.
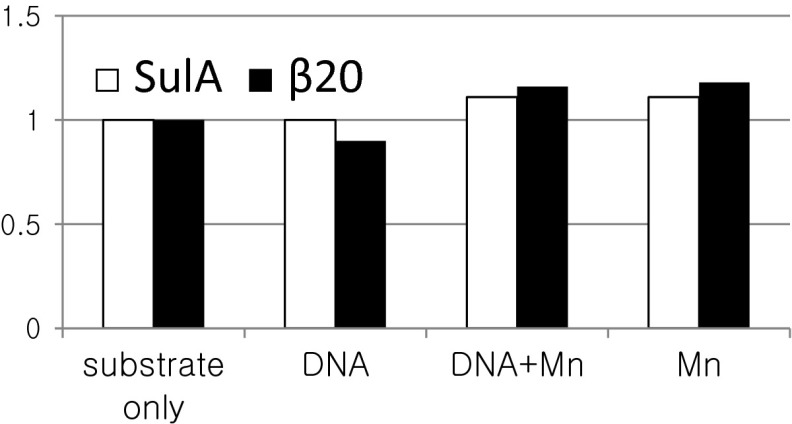
Fig. S4.
Test for a Mn2+ or DNA effect on LonA activity. Titin-I27 domain tagged at its C terminus with 20 amino acids from the SulA C terminus (open bar) or a specific internal 20 amino acids from beta-galactosidase (filled bar); both proteins are previously identified Lon substrates (10, 23) and were used to test whether the presence of Mn2+ or DNA nonspecifically affects LonA degradation activity. Degradation was monitored under the same conditions as in Fig. 3B. No significant difference was observed.
To gain insight into the step(s) of proteolysis affected by the metal-stabilized conformation of PerR, we determined the rate of degradation of apo-PerR and Mn-bound PerR (both untreated by H2O2) as a function of substrate concentration. The apparent KM for apo-PerR degradation by LonA was ∼1.9 ± 1.1 µM (using PerR dimer concentrations). In contrast, in the presence of Mn, PerR was extremely resistant to LonA. Although the degradation rates were too slow to be fitted accurately, it is clear that the apparent KM value of the productive PerR⋅LonA interaction was severely affected (Fig. 3C). These data strongly support the conclusion that LonA recognizes the “open” conformation of PerR much better than the compact conformation that is stabilized by metal and operator DNA-binding. To probe the mechanistic basis of this selective degradation, we investigated specific LonA-interacting sequences in PerR.
Lon-Recognition Site(s) on PerR.
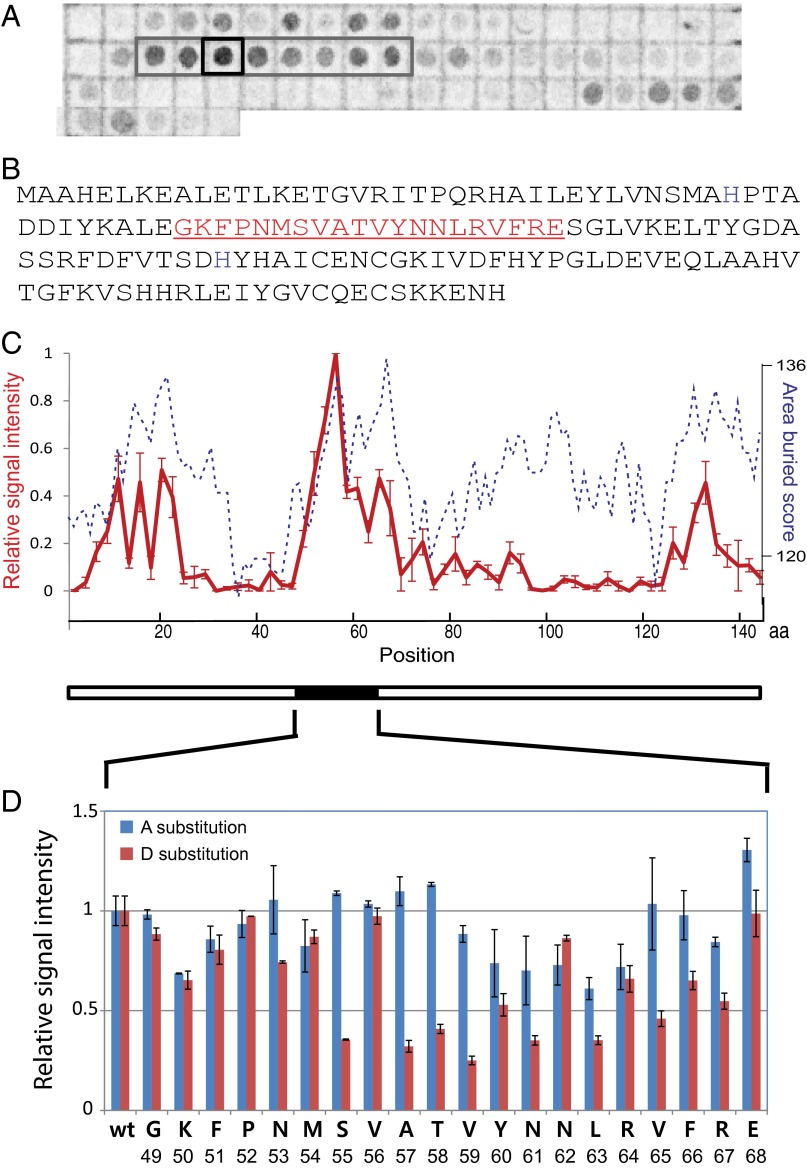
To identify regions of PerR that interact with LonA, we probed a peptide spot-blot array that displayed the entire PerR amino acid sequence with overlapping 20 amino acid peptides with 35S-labeled LonA. Spots with high radioactive signals were considered to contain peptides carrying candidate Lon-interacting sequences (Fig. 4A).
Fig. 4.
A region of the PerR polypeptide strongly interacts with LonA. (A) Membranes displaying spots of specific peptides from PerR were used to determine LonA-interacting region(s). Spots are composed of 20-amino-acid peptides from across the entire PerR sequence, and peptides were incubated with radioactive (S35-Met labeled) LonA. The gray box indicates a region of high signal intensity, and the black square indicates the spot with the strongest signal. The blot presented here is representative of three independent experiments. (B) Amino acid sequence of PerR. The peptide sequence of the spot that showed the strongest interaction signal with LonA (the black square in A) is indicated in red. (C) Relative interaction affinities of Lon to each PerR peptide fragment presented in A (red solid line, left y axis, scaled as the relative radioactive signal intensities of spots with ±1 SEM) was compared with calculated surface-burial scores of corresponding peptides (blue dashed line, right y axis; Materials and Methods). The x axis displays amino acid positions in PerR sequence. (D) Relative intensity of radioactive signals of Ala (blue bars) or Asp (red bars) substitution in individual positions. Data are plotted as average values of three independent experiments with ±1 SEM.
Peptides with high surface-burial scores have been shown to be well correlated with Lon degrons (10). We also observed that the peptide regions from PerR that interacted well with LonA all had relatively high surface-burial scores (Fig. 4C) (see also Materials and Methods). However, throughout the sequence of PerR, there were many other regions that had a high surface burial score but were not efficient LonA binding sites. Thus, the LonA binding and surface-burial scores of the PerR peptides were not highly correlated. Other features of the LonA-binding peptides therefore must play a significant role in recognition.
The region of PerR that showed the strongest LonA-interaction signal, residues 49–68 (Fig. 4B), was further analyzed. New peptide blots composed of PerR residues 49–68 were designed to probe the importance of individual residues for Lon binding. Each peptide spot contained this same 20-amino acid peptide carrying a single-residue substitution to either an alanine or aspartate, scanning each position of the peptide. These membranes were probed with 35S-labeled LonA, and the LonA-peptide interactions were quantified (Fig. 4D). The residues that showed larger decreases in LonA binding on both the alanine and aspartate substitution blots were all located on helix 4 of PerR, a region highly conserved among PerR homologs (see Fur, a Paralog of PerR in B. subtilis, Is Not a LonA Substrate) and known to be involved in DNA binding (11, 12).
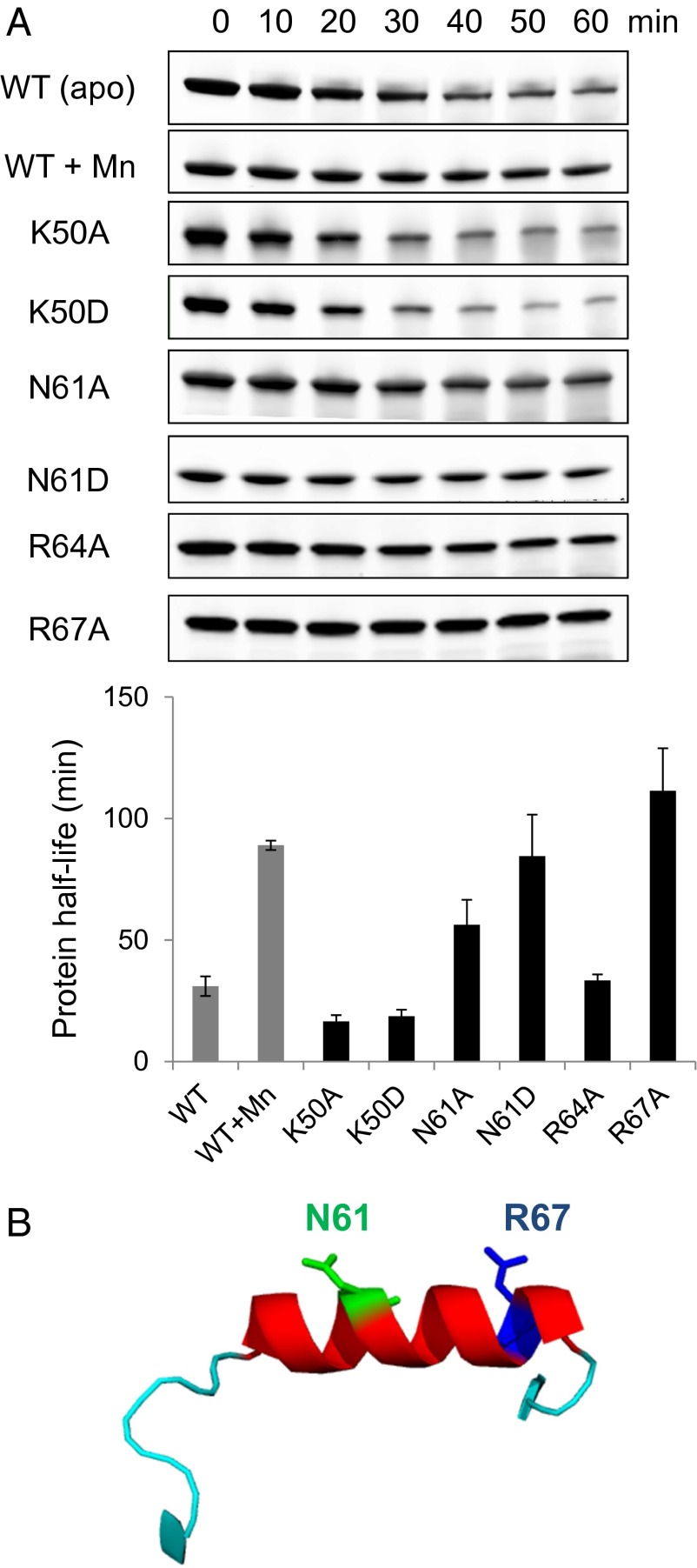
To directly test whether the residues detected as contributing to Lon binding on the blots were important for LonA to recognize and degrade PerR, we introduced Ala or Asp substitutions into full-length PerR at residues K50, N61, R64, and R67. These variants were tested for their ability to be degraded by LonA in vitro in the absence of a sensor-site metal ligand. Among these substitution variants, N61A, N61D, and R67A were degraded at profoundly reduced rates. The half-lives of the N61D and R67A PerR variants were indistinguishable from the stable, Mn-bound wild-type control. In contrast, the K50A and K50D variants did not influence degradation significantly compared with wild-type apo-PerR, and there was only a slightly lengthened half-life for the R64A variant (Fig. 5A). These results reveal that residues N61 and R67 within PerR helix 4 (Fig. 5B) are very important for PerR degradation by LonA and suggest that R64 may also contribute.
Fig. 5.
Single amino acid mutations effect on PerR degradation by LonA. (A) Protein stability of K50A, N61A, R64A, and R67A variants during in vitro degradation assays with LonA. The half-life of each protein variant was calculated from the average of three independent experiments with ±1 SEM. All PerR variants were purified without H2O2-treatment. (B) Important residues for LonA degradation (N61, green; R67, blue) are indicated on helix 4 of PerR, based on the protein structures PDB ID code 2RGV.
PerR Conformation and DNA Binding Directly Affect Accessibility to the Lon Discriminator Residues.
The requirement for LonA to recognize residues from helix 4, especially R67, provides an attractive mechanism to explain the sensitivity of oxidized or apo-PerR and resistance of the Fe or Mn bound PerR to LonA degradation. Inspection of the PerR crystal structures suggests that N61 and R67 might be more accessible and may more easily interact with other proteins in the oxidized, extended conformation than in the metal-bound, compact form (R64 is highly solvent-exposed in both protein conformations). To quantify the change in accessibility of these residues in the extended vs. compact conformation, we calculated the accessible surface area (ASA) (13) in both forms using probe radii of 1.4 Å (the value typically used to represent a water molecule probe) and 7.5 Å (representing a significantly larger molecule but still much smaller than the Lon protease) (Table S1). There was not much difference in the ASA of these residues in the open vs. compact form for any of the three residues when using the smaller probe. However, with the larger probe, the ASA for R67 was reduced substantially in the compact form, dropping from 57.6 to 7.0 Å2 in chain A of the dimer and from 70.6 to 0 Å2 in chain B. Because we demonstrated that R67 is essential for LonA recognition of PerR, the inability of a large molecule such as LonA to contact R67 in the compact form is sufficient to explain why oxidized PerR is degraded, but metal binding in the sensor site renders it resistant. The changes in surface exposure of N61 and R64 were minor compared with those observed for R67. (R64 yields a very high ASA value with the 7.5-Å probe because it is very solvent exposed, and a larger probe on an available residue can generate a large surface-area value.) A recent study, however, revealed that N61 is the critical residue determining DNA-binding specificity of PerR and, thus, very likely makes direct DNA operator contacts (12). Therefore, our results strongly favor the model in which the two residues identified here as most important for LonA recognition of PerR are expected to be unavailable for LonA interaction in the metal- and operator-bound state. We propose that the accessibility for recognition of these residues is how Lon distinguishes between the oxidized vs. nonoxidized forms of PerR and refer to this critical LonA-binding region as Lon’s discriminator site. This same mechanism would trigger degradation of PerR in the rare cases when cells are depleted for both Mn and Fe, because an empty discriminator site would strongly favor the open conformation of PerR.
Table S1.
Comparison of ASA of N61, R64, and R67 in oxidized vs. metal-bound structures of PerR
| PerR residue | Chain | ASA | |
| Probe radius, Å | |||
| 1.4 | 7.5 | ||
| N61 | |||
| Oxidized (PDB ID code 2RGV) | A | 96 | 133 |
| B | 91 | 126 | |
| Metal-bound (PDB ID code 3F8N) | A | 89 | 125 |
| B | 87 | 118 | |
| R64 | |||
| Oxidized (PDB ID code 2RGV) | A | 137 | 344 |
| B | 138 | 340 | |
| Metal-bound (PDB ID code 3F8N) | A | 118 | 123 |
| B | 136 | 133 | |
| R67 | |||
| Oxidized (PDB ID code 2RGV) | A | 78 | 58 |
| B | 91 | 71 | |
| Metal-bound (PDB ID code 3F8N) | A | 85 | 7 |
| B | 68 | 0 | |
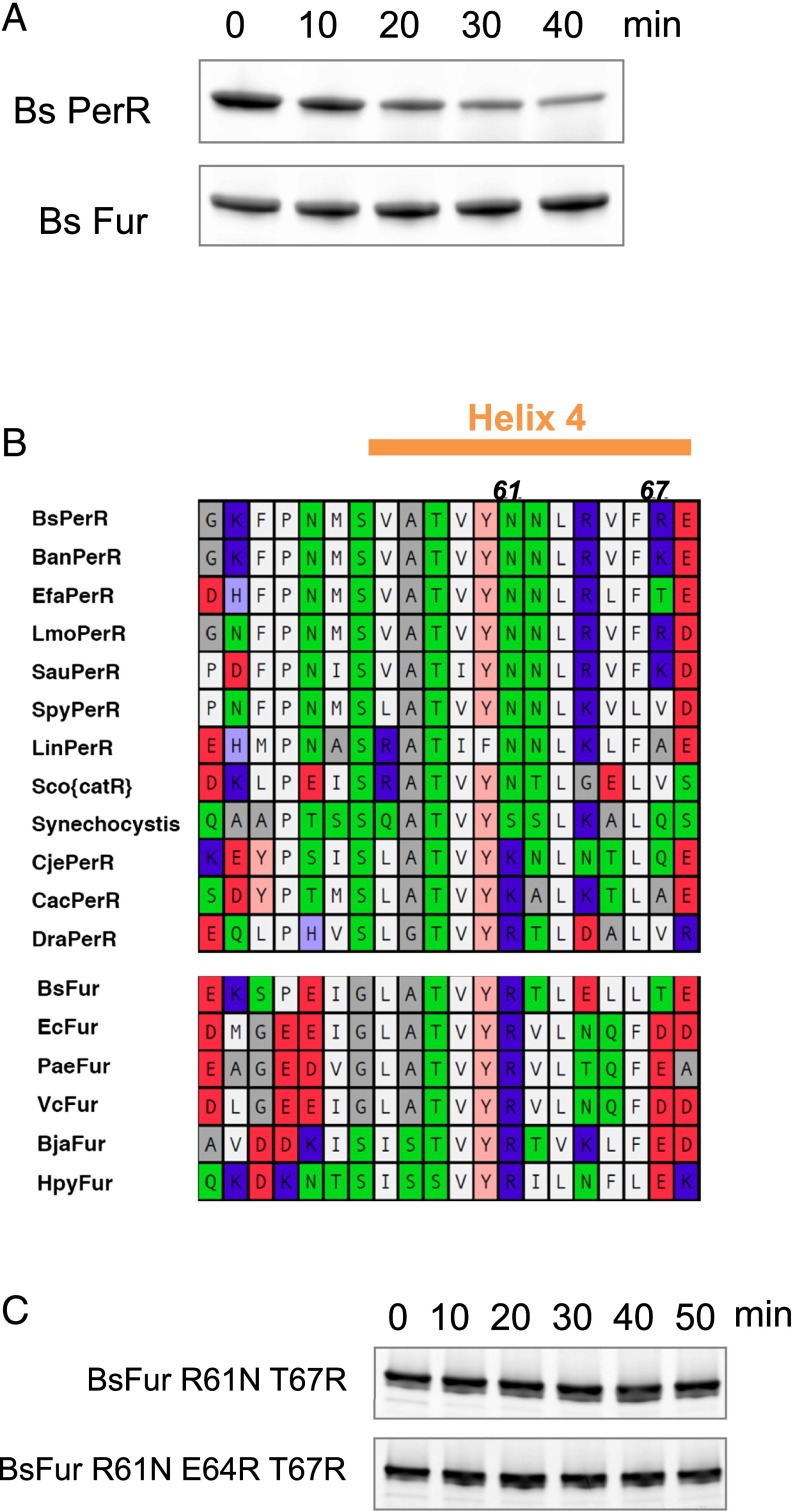
Fur, a Paralog of PerR in B. subtilis, Is Not a LonA Substrate.
PerR is in the Fur (ferric uptake regulator) protein family, a nearly ubiquitous metal-dependent group of bacterial transcriptional regulators. We tested whether B. subtilis Fur, another member of this protein family, was also subject to LonA proteolysis. Under the same conditions that LonA degraded apo-PerR in vitro, no degradation of apo-Fur was evident (Fig. 6A). Although Fur also adopts a more compact conformation upon binding metal, Fur does not undergo irreversible oxidation in response to H2O2 and therefore may be recycled after each use, rather than destroyed (14).
Fig. 6.
B. subtilis Fur is not degraded by LonA in vitro. (A) LonA does not degrade Fur in vitro. Apo-Fur and -PerR were incubated with LonA under the same conditions as for apo-PerR degradation in Fig. 5A. (B) Sequence comparison between PerR and Fur homologs in the helix 4 region (orange bar). Residue colors represent hydrophobicities and charges of each amino acid (hydrophobic, white; amphiphilic, gray; hydrophilic neutral, green; slightly basic, light blue; basic, blue; slightly acidic, pink; acidic, red). Note that the hydrophobic residues at positions 63, 65, and 66 of helix 4 of BsPerR are well conserved between both the PerR and Fur family members. (C) Incubation of B. subtilis apo-Fur R61N and T67R, and R61N, R64E, and T67R variant with LonA was performed as in A.
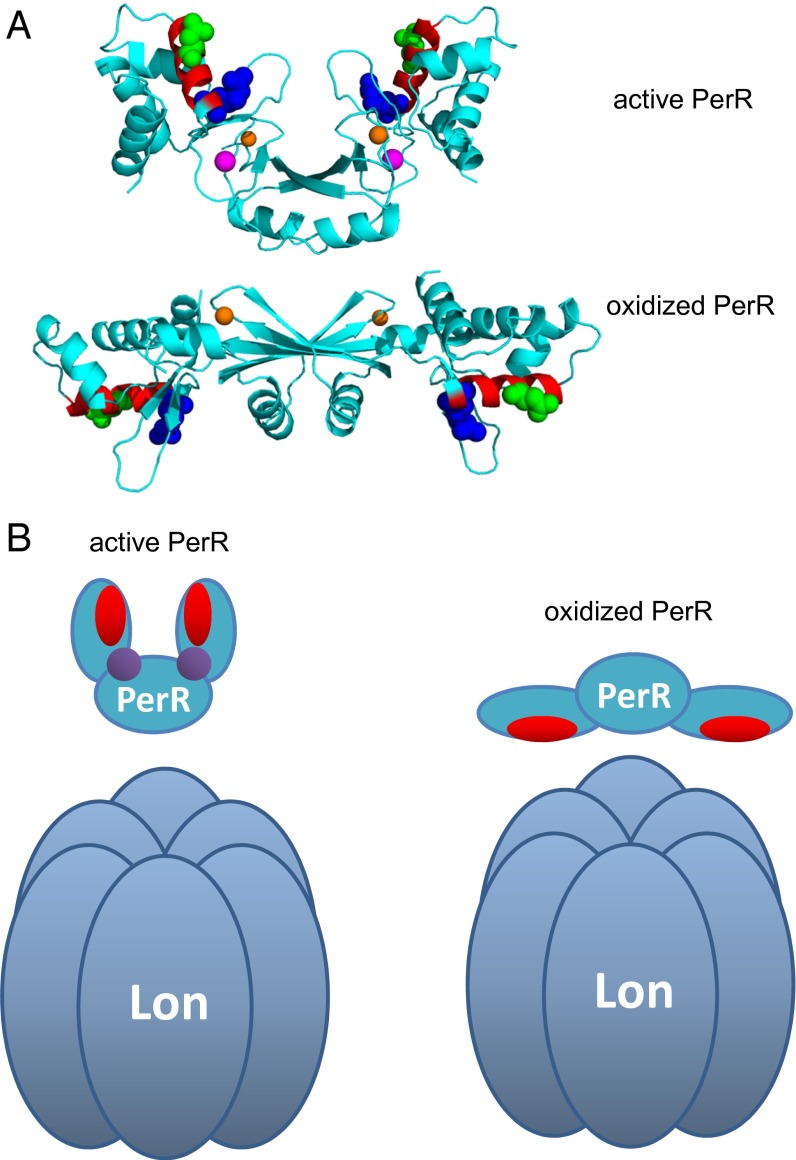
Fig. 7.
Structural position of the Lon-interacting region. (A) The helix 4 LonA-interacting region mapped on the 3D structures solved in the active (Mn-bound form, PDB ID code 3F8N; Upper) (5) and oxidized PerR (PDB ID: 2RGV; Lower) conformations (7). The α-helix (helix 4) that is the main LonA-interacting region is in red with the two critical positions colored green for N61 and blue for R67. The structural zinc is indicated in orange, and regulatory metal (Mn2+ in this case) is indicated in purple. The figure was rendered by using PyMOL (www.pymol.org) based on deposited coordinates. (B) Suggested model of substrate accessibility of PerR to LonA. PerR helix 4 is represented in red, and regulatory metal (iron or manganese) is shown in purple.
Based on the hypothesis that these two Fur homologs share a similar structure, we examined the helix 4 sequence of each protein, because this site contains the critical LonA-discriminator region in PerR. We reasoned that this comparison may reveal features of the region that direct PerR, but not Fur, to LonA for degradation. Hydrophobic amino acids are generally well conserved between the two protein groups. In contrast, the distribution and identity of charged residues at specific positions were clearly distinct (Fig. 6B). This observation also supports the results from the PerR mutational analysis that N61 and R67 are important residues for LonA recognition, because these residues are generally conserved among PerR homologs but uncommon in the Fur group.
We tested whether the difference between PerR and Fur at the two critical discriminator site residues were sufficient to convert Fur into a LonA substrate in its extended apo or oxidized form. Degradation experiments with both our standard LonA concentration and a fivefold higher concentration did not result in detectable degradation of the FurR61N,T67R variant (Fig. 6C). Although R64 was not a critical residue for degradation of intact PerR, it is well-positioned to perhaps assist recognition by LonA. To test whether the positive charge at this position (as is found in PerR) potentially assists substrate recognition by LonA, we also introduced an E64R substitution to the Fur R61N T67R variant. However, this change did not significantly enhance degradation of the Fur–PerR hybrid (Fig. 6C). Thus, these critical residues on helix 4 of PerR are necessary, but not sufficient to impart conformationally controlled degradation on Fur family ROS sensors.
Discussion
Selective elimination of irreversibly oxidized proteins is important for cell fitness, because proteins can be inactivated, even by mild oxidation. Accumulation and aggregation of oxidized proteins can threaten normal cellular function (15). Lon has been long implicated as the major protease with a central role in degrading proteins denatured by oxidative modification (16). The mechanism that Lon uses to recognize damaged, nonnative, or unfolded proteins as substrates is generally thought to be via interactions with exposed residues that are normally buried in the folded proteins (10). One well-studied case of oxidation-stimulated degradation is mitochondrial aconitase, which is preferentially degraded under oxidizing conditions that cause global protein unfolding and therefore exposure of new protease-accessible hydrophobic regions (17).
How more mildly oxidized proteins are specifically recognized for destruction is not well understood. In one analyzed example, site-specific oxidation of the bacterial FNR transcriptional regulator damages its [4Fe-4S]2+ clusters, which in turn destabilizes the FNR dimer (18); the resulting monomers are proposed to accessibly display FNR’s ClpXP protease recognition tags, leading to accelerated degradation of the monomers (19). Another example is Irr, a Fe– heme-binding regulator, which is degraded under oxidizing conditions in the presence of heme (20). It is known that the heme-binding site is both sensitive to oxidation and required for Irr degradation; however, the specific protease(s) involved in this degradation is unknown. A eukaryotic iron regulatory protein 2 is also known to be degraded by proteasome via iron-catalyzed oxidation that triggers ubiquitination and subsequent recognition by the 26S proteasome (21). To further elucidate the degradation mechanisms targeting mildly oxidized proteins for destruction, we analyzed the bacterial ROS-sensitive transcriptional regulator, PerR, as a candidate substrate.
Here we show that oxidized PerR is specifically degraded by LonA in vivo and in vitro. We also find that apo-PerR—the form with no metal bound in its sensor site—is also degraded by LonA with similar kinetics to the oxidized protein. Because apo-PerR and oxidized PerR adopt nearly identical conformations (7) and have very similar stabilities (Fig. S2), we propose that the previously described metal-dependent conformational change in PerR structure controls LonA-mediated degradation. This model is strongly supported by the stabilizing effect of Mn and operator DNA, which together essentially render the protein unrecognizable by LonA, in marked contrast to the oxidized form.
In our model, the key transition affecting LonA degradation of PerR is the allosteric change from the compact form to the extended conformation. Oxidation of one or both of the sensor-site histidines, promoted even under mild oxidizing conditions, destroys metal binding and therefore triggers this transition (4). PerR, like FNR, may be a prototypical example of a protein whose oxidation targets it for degradation without oxidation-promoted protein denaturation.
To elucidate how the transition of PerR from the compact to the extended conformation could switch this protein from a stable to an efficiently degradable LonA substrate, we mapped Lon-interacting regions throughout the structure of PerR. A region between residues 49 and 68 bound strongly to LonA, and alterations of specific residues in this segment substantially impaired LonA binding. Furthermore, mutations in N61 and R67 either substantially slowed or abolished PerR degradation in vitro. Thus, these two residues, which lie in helix 4 of PerR, are essential for LonA recognition and degradation of PerR.
Although residues 61 and 67 in PerR are essential Lon “discriminators,” a residue swap experiment between PerR and its cousin Fur reveals that these critical residues are not sufficient to convert B. subtilis Fur into a significant LonA substrate. Perhaps yet-unidentified additional residues from the PerR Lon-binding helix are necessary for a tighter Lon-substrate affinity. Furthermore, the discriminator region does not contain any significant sequence matches to characterized Lon degron tags (10, 22, 23). The degron tag is often the part of the substrate that is engaged by the central pore of the enzyme to begin unfolding and degradation; these degrons are usually considered both necessary and sufficient to target a substrate to its cognate unfoldase/protease. In this manner, the discriminator site may be analogous to the “enhancement tags” found in some other AAA+ unfoldase substrates. These short peptide signals serve to enhance recognition in the overall enzyme–substrate complexes and focus unfoldase action to the proper conformation or stage of the substrate’s lifecycle (24–26). This hypothesis is also consistent with previous observations that Lon contains more than one site for substrate recognition (10, 27). In our peptide blot test, we also observed significant interactions of LonA at regions near the N and C termini of the PerR sequence (Fig. 4A). It is an attractive model that one or both of these regions contain the additional signals that function along with helix 4 and are required for LonA degradation of PerR. These attractive regions include residues ∼13–32 and residues ∼117–136. Unfortunately, our initial attempts to dissect the importance of these regions for degradation were unsuccessful, because the constructed mutant variants overexpressed and purified poorly, presumably due to disruptions in PerR folding. Therefore, further investigations of the Lon sequences that function with the discriminator site to form PerR’s bipartite, ROS sensitive-degron await further systematic study.
In this study, we present the mechanism by which ROS converts PerR from a stable, H2O2-sensitive transcriptional repressor to a protein that is efficiently recognized and degraded by LonA. Key to this mechanism is the large ROS-dependent conformational change from a compact, DNA- and iron-binding molecular arrangement to the ROS oxidized open conformation that first loses specific DNA-binding properties and then is efficiently recognized as a LonA substrate. ROS-dependent loss of iron binding causes exposure of required residues for Lon interaction and degradation. This mechanism provides an elegant example of how oxidative damage, without protein denaturation, can target proteins for destruction. We expect that conformationally hidden or exposed protease-enhancement or discriminator tags will continue to emerge as a mechanism for regulated degradation of critical stress-responsive proteins.
Materials and Methods
Media and Growth.
B. subtilis cells for pulse-chase experiments were grown at 37 °C in Mops-buffered minimal medium containing 40 mM Mops (adjusted to pH 7.4 with NaOH), 2 mM potassium phosphate (pH 7.0), 20 g/L glucose, 2 g/L (NH4)2SO4, 0.2 g/L MgSO4·7H2O, 1 g/L sodium citrate·2H2O, 1 g/L potassium glutamate, 10 mg/L tryptophan, 10 mg/L phenylalanine, 3 nM (NH4)6Mo7O24, 400 nM H3BO3, 30 nM CoCl2, 10 nM CuSO4, 10 nM ZnSO4, and 5 μM FeCl3, as described (8). E. coli cells for protein overexpression were grown in Luria–Bertani (LB) medium (for PerR and Fur, at 37 °C) and 2× tryptone–yeast (TY) medium (for LonA, at 30 °C) with appropriate antibiotics.
Pulse-Chase Labeling and Immunoprecipitation.
For the pulse-chase in vivo degradation experiments, B. subtilis cells were grown in minimal medium described above with appropriate antibiotics added for each of the protease mutant strains. Exponentially growing cells were pulse-labeled at OD600 of 0.5 with 1.11 MBq of l-[35S]methionine per mL for 2 min, followed by chase with 0.5 mM cold methionine; samples of the cultures were removed and immediately flash-frozen in liquid nitrogen at every time point. Frozen samples were resuspended in 5 mg/mL lysozyme-containing buffer for a few seconds and boiled in 1% SDS for 2 min. Polyclonal sera against PerR (provided by the J. D. Helmann laboratory, Cornell University, Ithaca, NY) was used for immunoprecipitation. The amount of 35S in each precipitated protein was then quantified directly from the dried SDS/PAGE gels by using a Typhoon scanner and ImageQuant software (GE Healthcare).
Protein Expression and Purification of PerR, Fur, and LonA for in Vitro Analysis.
To overexpress B. subtilis PerR protein, an E. coli HE9501 strain was used as described (8), with a slight modification. To isolate the H2O2-treated PerR protein preparation, the cell culture carrying the inducible perR gene was incubated with 1 mM isopropyl-beta-d-thiogalactopyranoside for 2 h and then treated with 100 µM H2O2 for 1 min just before harvest. The cell extract used to purify the non-oxidized-dominant protein was treated identically, but H2O2 was omitted. The harvested cells were resuspended in buffer A [20 mM Tris⋅HCl (pH 8.0), 100 mM NaCl, and 5% (vol/vol) glycerol] containing 10 mM EDTA. After lysis by French press, the supernatant was loaded onto a HiTrap Q column (GE Healthcare) and separated by using a linear gradient of 0.1–1 M NaCl. PerR-containing fractions were pooled and loaded onto a HiTrap Heparin column (GE Healthcare) and separated by using a linear gradient of 0.1–1 M NaCl. The PerR-containing factions were further purified on a Superdex 75 10/300 column (GE Healthcare) equilibrated with Chelex-100–treated buffer A without EDTA. The functional activities of purified PerR were tested by DNA binding in the presence or absence of Mn using fluorescence anisotropy as described (4). Only non-H2O2-treated PerR showed DNA binding activity in the presence of 100 μM MnCl2, confirming that the purified proteins have vacant regulatory metal-binding sites and do not lose their structural zinc. All constructs for PerR derivative mutants (K50, N61, R64, and R67) were generated by a site-directed mutagenesis using pJL041 (pET16b::perR) (8) as a template. Mutant proteins were overexpressed and purified by using the same procedure as for the wild-type PerR, and the molecular weight of each variant was verified by electrospray ionization-MS.
To overexpress B. subtilis Fur, an E. coli BL21(DE3) strain containing pHB6505 was used as described (28). The overexpression and purification procedures for Fur and the Fur mutants (R61N and T67R) was the same as used for PerR.
The molecular weights of the purified proteins with altered sequences were analyzed by size-exclusive liquid chromatography (superpose 6; GE Healthcare) to confirm that they formed dimers under native conditions in a manner similar to the wild-type protein. All mutant variants of PerR and Fur had the same retention time as their respective wild-type protein. Far-UV CD spectra from 200 to 260 nm were determined for each of the purified wild-type and mutant proteins, and no change in the secondary structures of the mutant proteins was detected.
His6-tagged B. subtilis LonA protein was overexpressed from the E. coli strain as described (29). The harvested cells were resuspended in 50 mM sodium phosphate buffer (pH 8.0), containing 1 M NaCl, 20 mM imidazole, and 5% (vol/vol) glycerol. The supernatant from the cell lysate after French press was loaded onto Ni–nitrilotriacetic agarose resin (Qiagen), and proteins were eluted with the same buffer containing 500 mM imidazole. The eluted proteins were concentrated up to 100 µM by using centrifugal filters with a 3,000-Da molecular-mass cutoff (Millipore) and loaded onto a HiPrep Sephacryl S300 gel-filtration column (GE Healthcare) equilibrated in 50 mM Hepes (pH 7.5), 1 mM EDTA, and 300 mM NaCl. For purification of radioactive LonA, M9 medium was used instead of 2× TY medium, with 37 mBq of 35S-Met per 100 mL of culture.
Mass Spectra of PerR Proteins.
Molecular weight of purified PerR was measured by LC-MS with a QSTAR Elite quadrupole time-of–flight mass spectrometer. The sample was loaded onto a reversed-phase C18 capillary HPLC column, and the sample was eluted isocratically. Deconvolution of the electrospray data to generate molecular weight spectra was carried out with the BioAnalyst software that is part of the QSTAR Elite software package.
In Vitro PerR Degradation by LonA.
Degradation of substrates in vitro was performed by using 300 nM Lon6 in 25 mM Tris·HCl (pH 8), 100 mM KCl, and 10 mM MgCl2. Lon and substrate were incubated at 37 °C for 2 min before the addition of the ATP-regeneration mix (4 mM ATP, 50 μg/mL creatine kinase, and 5 mM creatine phosphate). Samples of each reaction were taken at specific time points and were stopped by adding 0.1% SDS. Protein samples from each degradation reaction were loaded on SDS/PAGE and stained with Sypro orange (Life Technology). Signal intensity was measured by scanning with a 473-nm laser (Typhoon FLA 9500; GE Healthcare). Protein half-life and degradation rates of each reaction were calculated by using ImageQuant software (GE Healthcare).
Peptides Blot Screening for LonA-Binding Regions.
The PerR peptide blot was generated by synthesizing 20-mer peptides each from PerR sequence. Adjacent spots differed in from each other by having their N-terminal sequence initiated two residues C-terminal on the PerR sequence, and likewise shifted to carry two additional C-terminal residues of PerR. The blot was incubated with 35S-Met–labeled LonA in PBS, and the radioactive signals were detected by phosphoimager scan. Average surface-burial scores on PerR peptides were calculated with web.expasy.org/protscale/ as described (10, 30) by using the “average area buried” scale with a 19-residue window.
ASA Calculation.
The comparison of solvent accessibilities of specific residues on PerR was calculated by AREAIMOL (www.ccp4.ac.uk/html/areaimol.html) using Protein Data Bank (PDB) data from the oxidized (PDB ID code 2RGV) (7) and active (PDB ID code 3F8N) (5) forms of the B. subtilis PerR structures.
Acknowledgments
We thank John Helmann and his laboratory for PerR overexpressing strains and anti-PerR antibodies; Jin-Won Lee and his laboratory for Fur overexpressing strains; Carol Gross and her laboratory and Alan Grossman and his laboratory for Bacillus mutant strains; and Robert Grant for helping calculate accessible surface area (ASA). This work was supported by Public Health Service Grant GM049224 and the Howard Hughes Medical Institute (HHMI). T.A.B. and B.-E.A. are both employees of HHMI.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522687112/-/DCSupplemental.
References
- 1.Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 2.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279(5357):1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 3.Lee C, et al. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat Struct Mol Biol. 2004;11(12):1179–1185. doi: 10.1038/nsmb856. [DOI] [PubMed] [Google Scholar]
- 4.Lee JW, Helmann JD. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440(7082):363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 5.Jacquamet L, et al. Structural characterization of the active form of PerR: Insights into the metal-induced activation of PerR and Fur proteins for DNA binding. Mol Microbiol. 2009;73(1):20–31. doi: 10.1111/j.1365-2958.2009.06753.x. [DOI] [PubMed] [Google Scholar]
- 6.Traoré DA, et al. Crystal structure of the apo-PerR-Zn protein from Bacillus subtilis. Mol Microbiol. 2006;61(5):1211–1219. doi: 10.1111/j.1365-2958.2006.05313.x. [DOI] [PubMed] [Google Scholar]
- 7.Traoré DA, et al. Structural and functional characterization of 2-oxo-histidine in oxidized PerR protein. Nat Chem Biol. 2009;5(1):53–59. doi: 10.1038/nchembio.133. [DOI] [PubMed] [Google Scholar]
- 8.Lee JW, Helmann JD. Biochemical characterization of the structural Zn2+ site in the Bacillus subtilis peroxide sensor PerR. J Biol Chem. 2006;281(33):23567–23578. doi: 10.1074/jbc.M603968200. [DOI] [PubMed] [Google Scholar]
- 9.Riethdorf S, et al. Cloning, nucleotide sequence, and expression of the Bacillus subtilis lon gene. J Bacteriol. 1994;176(21):6518–6527. doi: 10.1128/jb.176.21.6518-6527.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gur E, Sauer RT. Recognition of misfolded proteins by Lon, a AAA(+) protease. Genes Dev. 2008;22(16):2267–2277. doi: 10.1101/gad.1670908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tiss A, Barre O, Michaud-Soret I, Forest E. Characterization of the DNA-binding site in the ferric uptake regulator protein from Escherichia coli by UV crosslinking and mass spectrometry. FEBS Lett. 2005;579(25):5454–5460. doi: 10.1016/j.febslet.2005.08.067. [DOI] [PubMed] [Google Scholar]
- 12.Caux-Thang C, et al. Single asparagine to arginine mutation allows PerR to switch from PerR box to fur box. ACS Chem Biol. 2015;10(3):682–686. doi: 10.1021/cb500783g. [DOI] [PubMed] [Google Scholar]
- 13.Lee B, Richards FM. The interpretation of protein structures: Estimation of static accessibility. J Mol Biol. 1971;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 14.Parent A, et al. Single glutamate to aspartate mutation makes ferric uptake regulator (Fur) as sensitive to H2O2 as peroxide resistance regulator (PerR) Angew Chem Int Ed Engl. 2013;52(39):10339–10343. doi: 10.1002/anie.201304021. [DOI] [PubMed] [Google Scholar]
- 15.Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11(11):777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- 16.Tsilibaris V, Maenhaut-Michel G, Van Melderen L. Biological roles of the Lon ATP-dependent protease. Res Microbiol. 2006;157(8):701–713. doi: 10.1016/j.resmic.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Bota DA, Davies KJ. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol. 2002;4(9):674–680. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- 18.Khoroshilova N, Popescu C, Münck E, Beinert H, Kiley PJ. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc Natl Acad Sci USA. 1997;94(12):6087–6092. doi: 10.1073/pnas.94.12.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mettert EL, Kiley PJ. ClpXP-dependent proteolysis of FNR upon loss of its O2-sensing [4Fe-4S] cluster. J Mol Biol. 2005;354(2):220–232. doi: 10.1016/j.jmb.2005.09.066. [DOI] [PubMed] [Google Scholar]
- 20.Yang J, Panek HR, O’Brian MR. Oxidative stress promotes degradation of the Irr protein to regulate haem biosynthesis in Bradyrhizobium japonicum. Mol Microbiol. 2006;60(1):209–218. doi: 10.1111/j.1365-2958.2006.05087.x. [DOI] [PubMed] [Google Scholar]
- 21.Iwai K, et al. Iron-dependent oxidation, ubiquitination, and degradation of iron regulatory protein 2: implications for degradation of oxidized proteins. Proc Natl Acad Sci USA. 1998;95(9):4924–4928. doi: 10.1073/pnas.95.9.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shah IM, Wolf RE., Jr Sequence requirements for Lon-dependent degradation of the Escherichia coli transcription activator SoxS: Identification of the SoxS residues critical to proteolysis and specific inhibition of in vitro degradation by a peptide comprised of the N-terminal 21 amino acid residues. J Mol Biol. 2006;357(3):718–731. doi: 10.1016/j.jmb.2005.12.088. [DOI] [PubMed] [Google Scholar]
- 23.Ishii Y, et al. Regulatory role of C-terminal residues of SulA in its degradation by Lon protease in Escherichia coli. J Biochem. 2000;127(5):837–844. doi: 10.1093/oxfordjournals.jbchem.a022677. [DOI] [PubMed] [Google Scholar]
- 24.Abdelhakim AH, Oakes EC, Sauer RT, Baker TA. Unique contacts direct high-priority recognition of the tetrameric Mu transposase-DNA complex by the AAA+ unfoldase ClpX. Mol Cell. 2008;30(1):39–50. doi: 10.1016/j.molcel.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ling L, Montano SP, Sauer RT, Rice PA, Baker TA. Deciphering the roles of multicomponent recognition signals by the AAA+ unfoldase ClpX. J Mol Biol. 2015;427(18):2966–2982. doi: 10.1016/j.jmb.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camberg JL, Viola MG, Rea L, Hoskins JR, Wickner S. Location of dual sites in E. coli FtsZ important for degradation by ClpXP; one at the C-terminus and one in the disordered linker. PLoS One. 2014;9(4):e94964. doi: 10.1371/journal.pone.0094964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebel W, Skinner MM, Dierksen KP, Scott JM, Trempy JE. A conserved domain in Escherichia coli Lon protease is involved in substrate discriminator activity. J Bacteriol. 1999;181(7):2236–2243. doi: 10.1128/jb.181.7.2236-2243.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bsat N, Helmann JD. Interaction of Bacillus subtilis Fur (ferric uptake repressor) with the dhb operator in vitro and in vivo. J Bacteriol. 1999;181(14):4299–4307. doi: 10.1128/jb.181.14.4299-4307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duman RE, Löwe J. Crystal structures of Bacillus subtilis Lon protease. J Mol Biol. 2010;401(4):653–670. doi: 10.1016/j.jmb.2010.06.030. [DOI] [PubMed] [Google Scholar]
- 30.Rose GD, Geselowitz AR, Lesser GJ, Lee RH, Zehfus MH. Hydrophobicity of amino acid residues in globular proteins. Science. 1985;229(4716):834–838. doi: 10.1126/science.4023714. [DOI] [PubMed] [Google Scholar]