Significance
Circadian rhythms are important in nearly all processes in the brain. Changes in rhythms that come with aging are associated with sleep problems, problems with cognition, and nighttime agitation in elderly people. In this manuscript, we identified transcripts genome-wide that have a circadian rhythm in expression in human prefrontal cortex. Moreover, we describe how these rhythms are changed during normal human aging. Interestingly, we also identified a set of previously unidentified transcripts that become rhythmic only in older individuals. This may represent a compensatory clock that becomes active with the loss of canonical clock function. These studies can help us to develop therapies in the future for older people who suffer from cognitive problems associated with a loss of normal rhythmicity.
Keywords: aging, postmortem, circadian rhythms, gene expression, prefrontal cortex
Abstract
With aging, significant changes in circadian rhythms occur, including a shift in phase toward a “morning” chronotype and a loss of rhythmicity in circulating hormones. However, the effects of aging on molecular rhythms in the human brain have remained elusive. Here, we used a previously described time-of-death analysis to identify transcripts throughout the genome that have a significant circadian rhythm in expression in the human prefrontal cortex [Brodmann’s area 11 (BA11) and BA47]. Expression levels were determined by microarray analysis in 146 individuals. Rhythmicity in expression was found in ∼10% of detected transcripts (P < 0.05). Using a metaanalysis across the two brain areas, we identified a core set of 235 genes (q < 0.05) with significant circadian rhythms of expression. These 235 genes showed 92% concordance in the phase of expression between the two areas. In addition to the canonical core circadian genes, a number of other genes were found to exhibit rhythmic expression in the brain. Notably, we identified more than 1,000 genes (1,186 in BA11; 1,591 in BA47) that exhibited age-dependent rhythmicity or alterations in rhythmicity patterns with aging. Interestingly, a set of transcripts gained rhythmicity in older individuals, which may represent a compensatory mechanism due to a loss of canonical clock function. Thus, we confirm that rhythmic gene expression can be reliably measured in human brain and identified for the first time (to our knowledge) significant changes in molecular rhythms with aging that may contribute to altered cognition, sleep, and mood in later life.
Nearly all processes in the brain and body are controlled by a 24-h circadian rhythm. These rhythms are important in regulating the sleep/wake cycle, metabolism, alertness, cognition, and other processes (1). Environmental or genetic disruptions to circadian rhythms are strongly associated with chronic sleep problems, increased rates of cancer, lowered immune function, metabolic disorders, and psychiatric disorders (2, 3). The molecular clock is controlled by a transcriptional/translational feedback loop with the circadian locomotor output cycles kaput (CLOCK) and brain and muscle Arnt-like protein 1 (BMAL1; also known as ARNTL) proteins acting as the major transcriptional activators, and the Period (PER1, PER2, PER3) and Cryptochrome (CRY1, CRY2) proteins acting as the major repressors (4). This core circadian feedback loop regulates the diurnal expression patterns of many different genes as it is estimated that 10–20% of all transcripts have a circadian rhythm (1, 5). Although the master circadian pacemaker in the suprachiasmatic nucleus (SCN) of the hypothalamus synchronizes rhythms throughout the brain and body, the genes that control circadian rhythms are expressed in nearly every cell (6). In recent years, it has become apparent that these genes serve important functions in specific brain regions, including the control of daily rhythms in neuronal activity and the response to environmental stimuli (7–9).
Evidence from preclinical and clinical studies suggests rhythms in the prefrontal cortex (PFC) are particularly important for cognitive performance and executive function. Several studies in humans have reported diurnal differences in cognitive performance and a significant decrease in performance following circadian rhythm disruption (10–12). Interestingly, these measures vary by age with older adults performing better on cognitive tasks in the morning and getting worse throughout the day (12, 13). In older women, there is a direct correlation between weak circadian activity rhythms and poorer executive function (14). In preclinical studies, mice trained in cortical-driven cognitive tasks show pronounced diurnal differences in performance (15–17). Moreover, mice housed under a shortened day (20-h light–dark cycle of 10-h light, 10-h dark) displayed reduced cognitive flexibility and a loss of dendritic length and complexity in the PFC (18).
Physiological and activity rhythms are generally known to deteriorate with aging and show a phase advance toward early morning wakening (19, 20). Daily rhythms in hormones like melatonin and cortisol are decreased as are sleep and body temperature rhythms in older individuals (21). Interestingly, the recognized stimulation of alertness and cognition by blue light seen in young people is diminished in older people, suggesting decreased input to the clock (22). Moreover, the addition of serum from older people to cultured cells that express a circadian reporter construct (Bmal1-luciferase) leads to a shortening of molecular rhythms and a phase advance that is not seen with serum from younger people (23). This finding suggests the presence of circulating factors that alter molecular rhythms with age.
In the human brain, the investigation of brain region-specific transcriptional rhythms across the life span has been challenging due to the lack of control of environmental factors, such as time of death, sleep–wake cycles, and exposure to other factors known to influence rhythms. A recent study overcame some of these obstacles by ordering postmortem samples around a 24-h clock based on their time of death, essentially reconstructing a pseudo-time series by treating each individual sample as an independently sampled data point over one 24-h cycle (24). In 55 healthy controls and 34 patients with major depressive disorder (MDD), they found robust expression rhythms of hundreds of genes in several brain regions in control subjects, with many of these genes displaying significantly disrupted or altered expression rhythms in subjects with MDD. Here, we used a similar analytical approach to identify rhythmic transcripts in two areas of the PFC [Brodmann’s area 11 (BA11) and BA47] in a large sample of 146 subjects from across the life span with no history of psychiatric illness. This large sample allowed us to investigate the effects of normal aging on molecular rhythms in the human PFC and to test the prediction that older individuals display altered or even a loss of molecular rhythms in this brain region.
Materials and Methods
Human Postmortem Brain Samples.
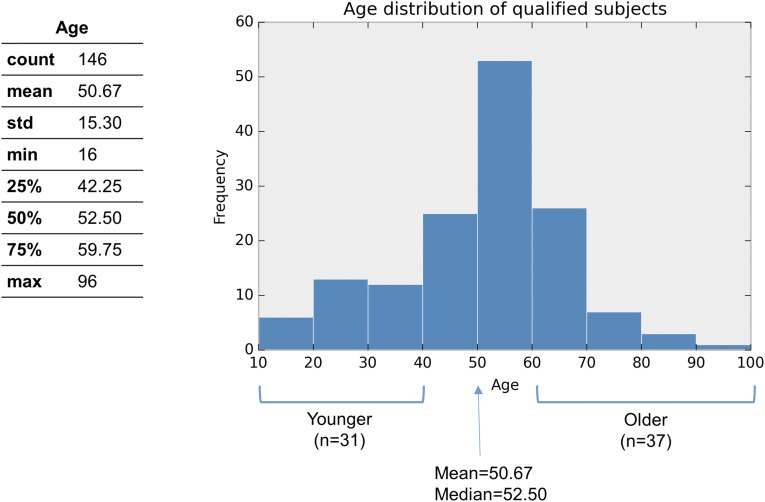
Using the resources of the University of Pittsburgh’s Brain Tissue Donation Program, 210 subjects were identified. Samples were obtained after consent from next of kin during autopsies conducted at the Allegheny County Medical Examiner’s Office (Pittsburgh). The absence of lifetime psychiatric disorders was determined by an independent committee of experienced clinical research scientists using information from clinical records, toxicology results, and a standardized psychological autopsy. Subjects were removed from the study if death was not witnessed because their time of death (TOD) cannot be precisely determined. Subjects that did not meet the criteria of rapid death were also removed from the study. Sixty-four subjects were removed based on these criteria. The final sample consisted of 146 individuals with the following characteristics: mean (range) age of 50.7 (16–96) years, 78% male, 85% Caucasian, mean postmortem interval (PMI) for brain collection of 17.3 h (4.8–28 h), mean pH of 6.7 (5.8–7.6), and RNA integrity number (RIN) of 8.0 (5.9–9.6) (Table S1).
Table S1.
Cohort demographic and clinical information
| HU | Age | PMI | pH | RIN | Sex | Race | Cause | Manner | TOD |
| 510 | 63 | 12.4 | 6.5 | 7.5 | M | W | Gastrointestinal bleeding | Natural | −5.83 |
| 516 | 20 | 14 | 7.1 | 8.4 | M | B | Gunshot wound to chest | Homicide | 16.66 |
| 545 | 65 | 13.1 | 6.9 | 6.2 | M | B | Pancreatic carcinoma with pulmonary metastasis | Natural | 14.90 |
| 546 | 37 | 23.5 | 6.7 | 8.6 | F | W | ASCVD | Natural | 6.07 |
| 551 | 61 | 16.4 | 6.6 | 8.3 | M | W | Cardiac tamponade | Natural | 11.35 |
| 567 | 46 | 15 | 6.8 | 8.9 | F | W | Mitral valve prolapse | Natural | 14.11 |
| 568 | 60 | 9.5 | 6.9 | 8.7 | F | W | ASCVD | Natural | −5.47 |
| 575 | 55 | 11.3 | 6.8 | 9.6 | F | B | ASCVD | Natural | 15.78 |
| 585 | 26 | 16 | 6.7 | 8.3 | M | W | Trauma | Accidental | 15.60 |
| 604 | 39 | 19.3 | 7.1 | 8.6 | M | W | Hypoplastic coronary artery | Natural | 9.27 |
| 615 | 62 | 7.2 | 6.4 | 7.8 | M | W | Ruptured abdominal aortic aneurysm | Natural | −3.38 |
| 627 | 43 | 14.1 | 7.1 | 7 | F | B | Chronic obstructive pulmonary disease | Natural | 14.62 |
| 630 | 65 | 21.2 | 7 | 9 | M | W | ASCVD | Natural | 6.39 |
| 634 | 52 | 16.2 | 7 | 8.5 | M | W | ASCVD | Natural | 13.54 |
| 643 | 50 | 24 | 6.2 | 8 | M | W | ASCVD | Natural | 4.34 |
| 645 | 69 | 23 | 6.7 | 8 | M | W | ASCVD | Accidental | 4.91 |
| 681 | 51 | 11.6 | 7.2 | 8.9 | M | W | Hypertrophic cardiomyopathy | Natural | 16.00 |
| 688 | 55 | 21.5 | 7 | 7.2 | M | W | Hypertrophic cardiomyopathy | Natural | 7.61 |
| 694 | 38 | 20.7 | 7 | 7.7 | M | W | Subarachnoid hemorrhage | Natural | 8.95 |
| 700 | 42 | 26.1 | 7 | 8.7 | M | W | ASCVD | Natural | 3.64 |
| 704 | 70 | 20.6 | 7 | 8.7 | M | W | ASCVD | Natural | 10.56 |
| 727 | 19 | 7 | 7.4 | 9.2 | M | B | Multiple trauma | Accidental | −2.83 |
| 731 | 63 | 10.5 | 6.8 | 8.2 | F | W | ASCVD | Natural | −4.11 |
| 737 | 66 | 18.2 | 7.3 | 7.5 | M | W | ASCVD | Natural | 10.81 |
| 739 | 40 | 15.8 | 6.9 | 8.4 | M | W | ASCVD | Natural | 11.25 |
| 746 | 64 | 20.8 | 6.7 | 7.6 | F | B | Peritonitis | Natural | 7.90 |
| 784 | 58 | 5.5 | 7.1 | 9.1 | M | W | ASCVD | Natural | −2.70 |
| 794 | 52 | 20 | 6.8 | 8.4 | M | B | End-stage dilated cardiomyopathy | Natural | 6.68 |
| 795 | 68 | 11.8 | 6.8 | 8.2 | M | W | Ruptured abdominal aortic aneurysm | Natural | 15.13 |
| 806 | 57 | 24 | 6.9 | 7.8 | M | W | Pulmonary embolism | Natural | 4.31 |
| 812 | 55 | 23 | 6.7 | 8.5 | M | W | ASCVD | Natural | 4.37 |
| 818 | 67 | 24 | 7.1 | 8.4 | F | W | Anaphylactic reaction to cef0tan | Accidental | 5.35 |
| 825 | 59 | 16.9 | 7.1 | 7.8 | M | W | Cardiac tamponade | Natural | 12.60 |
| 838 | 58 | 16.5 | 7 | 8.5 | M | W | ASCVD | Natural | 12.43 |
| 839 | 74 | 5.3 | 7.1 | 8.9 | F | W | Massive BFT | Accidental | −1.15 |
| 840 | 41 | 15.4 | 6.8 | 9.1 | F | W | ASCVD | Natural | 13.97 |
| 841 | 70 | 22.3 | 7.2 | 7.2 | M | W | Hypertrophic cardiomyopathy | Natural | 7.69 |
| 855 | 62 | 11.5 | 6.7 | 8.4 | M | B | Hypertrophic cardiomyopathy | Natural | 17.28 |
| 857 | 48 | 16.6 | 6.7 | 8.9 | M | W | ASCVD | Natural | 10.13 |
| 866 | 66 | 12.2 | 7 | 8.5 | M | W | ASCVD | Natural | 15.24 |
| 869 | 51 | 6.5 | 7.2 | 8.9 | M | W | Severe ASCVD | Natural | −3.46 |
| 871 | 28 | 16.5 | 7.1 | 8.5 | M | W | Trauma (chest) | Accidental | 12.04 |
| 895 | 39 | 15.4 | 7.6 | 8.8 | M | W | ASCVD | Natural | 14.25 |
| 902 | 60 | 23.6 | 6.7 | 7.7 | M | W | ASCVD | Natural | 4.45 |
| 920 | 60 | 23 | 7.2 | 8.5 | M | W | ASCVD | Natural | 3.93 |
| 932 | 68 | 25.9 | 6.8 | 8.3 | M | W | Cardiac tamponade | Natural | 2.23 |
| 952 | 62 | 5.5 | 6.6 | 8.7 | M | W | ASCVD | Natural | 4.08 |
| 953 | 54 | 18.6 | 6.2 | 7.1 | M | W | Extensive BFT (head, torso, and limbs) | Accidental | 9.38 |
| 969 | 27 | 10.6 | 7 | 7.7 | M | W | Trauma to trunk and extremities | Accidental | −4.91 |
| 970 | 42 | 25.9 | 6.4 | 7.2 | M | W | ASCVD | Natural | 4.38 |
| 972 | 72 | 16.4 | 6.3 | 7.7 | M | W | Trauma (cervical/upper thoracic vertebra) | Accidental | 11.71 |
| 980 | 52 | 11 | 6.4 | 8.3 | M | W | ASCVD | Natural | −5.96 |
| 998 | 25 | 6.3 | 5.9 | 7.6 | M | W | Pulmonary thromboembolism | Natural | 2.24 |
| 1026 | 59 | 19.8 | 6.3 | 7.4 | M | W | ASCVD | Natural | 9.64 |
| 1030 | 96 | 13.2 | 6.3 | 6.8 | F | W | Fractured cervical spinal column | Accidental | 16.16 |
| 1031 | 53 | 23.2 | 6.8 | 8.9 | M | W | Arteriosclerotic and hypertensive CVD | Natural | 6.71 |
| 1038 | 56 | 14.6 | 6.5 | 7 | M | W | Rupture of dissecting hematoma of aorta | Natural | 14.64 |
| 1047 | 43 | 13.8 | 6.6 | 9 | M | W | ASCVD | Natural | 15.15 |
| 1054 | 44 | 17.1 | 6.6 | 8.8 | F | W | ASCVD | Natural | 10.20 |
| 1065 | 85 | 20.2 | 6.3 | 7.1 | M | B | Pulmonary embolism | Accidental | 7.32 |
| 1067 | 49 | 6 | 6.6 | 8.2 | M | W | Hypertensive heart disease | Natural | −1.95 |
| 1083 | 20 | 19.9 | 6.6 | 8.8 | M | W | Trauma (head and trunk) | Accidental | 9.20 |
| 1091 | 58 | 16.5 | 6.6 | 8.1 | M | W | ASCVD | Natural | 6.67 |
| 1092 | 40 | 16.6 | 6.8 | 8 | F | B | Mitral valve prolapse | Natural | 11.81 |
| 1099 | 24 | 9.1 | 6.5 | 8.6 | F | W | Cardiomyopathy not otherwise specified | Natural | 1.37 |
| 1103 | 89 | 22.9 | 6.8 | 7.8 | M | W | BFT of head and trunk | Accidental | 8.12 |
| 1108 | 60 | 23.3 | 6.3 | 8.1 | M | W | ASCVD | Natural | 6.92 |
| 1114 | 29 | 11.4 | 6.8 | 9.4 | F | W | Trauma (trunk) | Accidental | −5.57 |
| 1119 | 57 | 20.2 | 6.8 | 8 | M | W | ASCVD | Natural | 8.62 |
| 1122 | 55 | 15.4 | 6.7 | 7.9 | M | W | Cardiac tamponade | Natural | 13.18 |
| 1129 | 54 | 21 | 6.8 | 9 | M | W | ASCVD | Natural | 7.91 |
| 1141 | 54 | 19.2 | 6.6 | 8.5 | M | W | Arteriosclerotic and hypertensive CVD | Natural | 9.48 |
| 1142 | 44 | 12.3 | 5.8 | 6.1 | M | B | Hypertensive and arteriosclerotic CVD | Natural | 15.98 |
| 1144 | 75 | 21.4 | 6.7 | 7.3 | M | W | Arteriosclerotic and hypertensive CVD | Natural | 7.75 |
| 1153 | 55 | 28 | 6.6 | 8 | M | W | Atherosclerotic and hypertensive heart disease | Natural | 3.69 |
| 1159 | 51 | 16.7 | 6.5 | 7.6 | M | W | Hypertensive heart disease | Natural | 11.81 |
| 1165 | 67 | 21 | 6.6 | 7.5 | F | W | Cardiac tamponade | Natural | 10.49 |
| 1171 | 56 | 19.2 | 6.5 | 8.5 | M | W | Cardiac tamponade | Natural | 8.55 |
| 1172 | 52 | 16.8 | 6.5 | 7.7 | F | B | Hypertensive heart disease | Natural | 11.79 |
| 1191 | 59 | 19.4 | 6.2 | 8.4 | M | B | Atherosclerotic and hypertensive heart disease | Natural | 9.24 |
| 1193 | 31 | 12.8 | 6.5 | 7.6 | M | W | Myocardial infarction due to ASCVD | Natural | 15.64 |
| 1194 | 27 | 16.4 | 6.4 | 8.8 | M | W | ASCVD | Natural | 12.63 |
| 1196 | 36 | 14.5 | 6.4 | 8.2 | F | W | Positional asphyxia | Accidental | 17.15 |
| 1201 | 52 | 16.4 | 6.2 | 8.1 | F | W | ASCVD | Natural | 12.51 |
| 1212 | 49 | 9.2 | 6.6 | 8.2 | M | B | Massive subarachnoid hemorrhage | Natural | −4.42 |
| 1214 | 57 | 16.4 | 6.4 | 7.5 | M | W | Arteriosclerotic and hypertensive CVD | Natural | 12.56 |
| 1219 | 56 | 14.9 | 6.5 | 8.5 | M | W | ASCVD | Natural | 14.77 |
| 1237 | 64 | 8.7 | 6.4 | 7.7 | M | W | ASCVD | Natural | 0.70 |
| 1239 | 22 | 9.5 | 6.7 | 7.4 | M | W | BFT to the head and neck | Accidental | −5.42 |
| 1245 | 59 | 21.6 | 6.6 | 7.4 | M | W | ASCVD | Natural | 7.37 |
| 1247 | 58 | 22.7 | 6.4 | 8.4 | F | W | ASCVD | Natural | 8.46 |
| 1264 | 38 | 21.1 | 6.7 | 8.6 | M | W | Cardiomyopathy | Natural | 5.60 |
| 1270 | 73 | 19.7 | 6.7 | 7.7 | F | W | BFT of neck and trunk | Accidental | 8.62 |
| 1276 | 57 | 14.9 | 6 | 6.1 | F | W | Arteriosclerotic CVD | Natural | 13.90 |
| 1280 | 50 | 23.5 | 6.7 | 7.7 | F | W | Pulmonary thromboembolism | Natural | 3.95 |
| 1288 | 59 | 19.7 | 6.9 | 7.4 | M | W | Atherosclerotic and hypertensive heart disease | Natural | 9.49 |
| 1293 | 65 | 18.5 | 6.6 | 7 | F | W | BFT of trunk | Accidental | 11.51 |
| 1298 | 48 | 24.5 | 6.8 | 7.9 | M | W | ASCVD | Natural | 4.78 |
| 1300 | 56 | 16.5 | 6 | 6.9 | F | W | Pulmonary thromboembolism | Natural | 13.22 |
| 1306 | 18 | 16 | 6.6 | 7.9 | M | W | Congenital heart anomaly | Natural | 13.54 |
| 1307 | 32 | 4.8 | 6.7 | 7.6 | M | B | Hypertensive cardiomyopathy | Natural | 4.03 |
| 1308 | 58 | 20.2 | 6 | 6.2 | M | W | Hepatocellular carcinoma | Natural | 8.43 |
| 1317 | 56 | 22.9 | 6.5 | 8.8 | M | W | Arteriosclerotic and hypertensive CVD | Natural | 4.80 |
| 1326 | 58 | 16.4 | 6.7 | 8 | M | W | ASCVD | Natural | 13.47 |
| 1333 | 46 | 18.2 | 6.8 | 8.7 | M | W | Arteriosclerotic and hypertensive CVD | Natural | 10.65 |
| 1335 | 18 | 14.6 | 7 | 8.7 | M | W | BFT of head and trunk | Accidental | 16.06 |
| 1350 | 21 | 24.2 | 6.4 | 7.3 | M | W | BFT of trunk | Accidental | 6.62 |
| 1371 | 33 | 13 | 6.2 | 8.5 | M | B | Eosinophilic myocarditis | Natural | −5.61 |
| 1374 | 43 | 21.7 | 6.6 | 7.2 | M | W | Coronary atherosclerotic disease | Natural | 7.91 |
| 1378 | 53 | 20.1 | 6.5 | 7.8 | F | B | Coronary artery disease | Natural | 8.46 |
| 1382 | 73 | 25.3 | 6.6 | 7.9 | F | W | BFT of head with ponto-midbrain transection | Accidental | 2.87 |
| 1391 | 51 | 7.8 | 6.6 | 7.1 | F | W | Atherosclerotic coronary artery disease | Natural | −0.59 |
| 1394 | 45 | 17.3 | 6.6 | 7.3 | M | W | Atherosclerotic coronary artery disease | Natural | 10.12 |
| 1400 | 50 | 16 | 5.8 | 6.8 | M | W | Coronary atherosclerotic disease | Natural | 11.40 |
| 1410 | 54 | 15.3 | 6.9 | 7.8 | M | W | Atherosclerotic coronary artery disease | Natural | 13.11 |
| 1433 | 65 | 24.1 | 6.8 | 7.5 | M | W | Acute subdural hematoma with skull fracture | Accidental | 7.18 |
| 1434 | 25 | 25.5 | 6.7 | 7.5 | M | B | Cardiac arrhythmia while exercising | Natural | 5.35 |
| 1441 | 57 | 22.3 | 6.5 | 8.5 | M | W | Arteriosclerotic CVD | Natural | 7.88 |
| 1444 | 46 | 22 | 6.5 | 8.4 | M | W | Pulmonary thromboembolus | Natural | 5.25 |
| 1447 | 51 | 16.2 | 6.5 | 8.5 | M | W | Calcified atherosclerotic coronary artery disease | Natural | 15.53 |
| 1460 | 53 | 21.3 | 6.2 | 7.3 | M | W | Severe ASCVD | Natural | 6.28 |
| 1462 | 47 | 17.2 | 6.6 | 8.5 | M | W | ASCVD | Natural | 10.58 |
| 1466 | 64 | 20 | 6.7 | 8.8 | F | B | BFT to trunk | Accidental | 9.33 |
| 1482 | 25 | 20.2 | 6.6 | 9.1 | M | W | ASCVD | Natural | 8.26 |
| 1488 | 39 | 21.5 | 6.4 | 8.7 | M | B | Bilateral pulmonary thromboemboli | Natural | 6.38 |
| 1497 | 38 | 8.5 | 6.3 | 9.1 | M | W | ASCVD | Natural | −2.80 |
| 1511 | 48 | 23.5 | 6.8 | 7.2 | M | W | Severe ASCVD | Natural | 4.30 |
| 1514 | 60 | 25 | 6.4 | 6.6 | F | W | Bilateral pulmonary emboli | Natural | 2.91 |
| 1524 | 66 | 9.4 | 6.4 | 8.1 | M | W | Acute small intestinal ischemic infarction | Natural | −4.21 |
| 1541 | 80 | 10.5 | 6.5 | 8 | M | W | Pulmonary fibrosis | Natural | 17.40 |
| 1554 | 50 | 23.2 | 6.5 | 7.6 | M | W | Acute myocardial infarction | Natural | 7.67 |
| 1555 | 17 | 15.1 | 6.9 | 7.9 | M | W | BFT to head, thorax, abdomen, right | Accidental | 16.70 |
| 1558 | 54 | 24.4 | 6.9 | 7.7 | M | W | ASCVD | Natural | 4.60 |
| 1559 | 45 | 8.9 | 6 | 6.9 | M | W | Cardiomegaly | Natural | −1.66 |
| 1564 | 48 | 19.9 | 6.8 | 7.5 | M | W | BFT to trunk | Accidental | 8.99 |
| 1574 | 52 | 26.7 | 6.3 | 7 | M | W | Bilateral pulmonary embolism | Natural | 2.07 |
| 1583 | 58 | 19.1 | 6.8 | 8.2 | M | W | BFT of the trunk | Accidental | 9.10 |
| 1598 | 50 | 23.8 | 6.9 | 7.8 | M | W | Acute myocardial infarction | Natural | 10.12 |
| 1623 | 52 | 25.7 | 6.3 | 6.6 | M | W | BFT of head and neck | Accidental | 4.86 |
| 1637 | 46 | 16.6 | 6.9 | 8.2 | M | W | Coronary artery atherosclerosis | Natural | 15.86 |
| 10003 | 49 | 21.2 | 6.5 | 8.4 | M | W | Multiple blunt force injuries | Accidental | 9.82 |
| 10005 | 42 | 23.5 | 6.7 | 7.4 | M | W | Multiple blunt force injuries | Accidental | 7.43 |
| 10013 | 16 | 9.3 | 6.7 | 9 | F | W | Trauma (multiple blunt force injuries) | Accidental | −3.88 |
| 10016 | 18 | 16.3 | 6.8 | 5.9 | M | B | Gunshot wound of abdomen | Homicide | 15.46 |
| 10021 | 38 | 12.7 | 6.3 | 7.2 | M | W | Multiple blunt force injuries | Accidental | 16.86 |
| 10027 | 53 | 13.1 | 6.2 | 6.6 | F | B | Congestive heart failure | Natural | 17.13 |
ASCVD, atherosclerotic cardiovascular disease; B, African American; BFT, blunt force trauma; CVD, cardiovascular disease; F, female; HU, subject no.; M, male; pH, brain tissue pH; PMI, postmortem interval; RIN, RNA integrity number; TOD, time of death (zeitgeber time); W, Caucasian.
For each subject, the right hemisphere was blocked coronally, frozen, and stored at −80 °C. Blocks containing BA11 or BA47 of the orbital PFC were cut on a cryostat, and cortical gray matter was collected for RNA extraction as previously described (25). All procedures were approved by the University of Pittsburgh’s Institutional Review Board for Biomedical Research and Committee for Research Involving the Dead.
Time of Death Analysis in the Zeitgeber Timescale.
To analyze rhythmic gene expression, the TOD for each subject was normalized to a zeitgeber time (ZT) scale. For each subject, both place and time of death was collected, where the time zone of the place of death was used to adjust the reported TOD from local time to coordinated universal time. Daylight savings time was also incorporated when appropriate. The sunrise time was calculated according to the date and the longitude and latitude of the place of death. Subject’s TOD was set as ZT = t hours after previous sunrise (if t < 18) or before next sunrise (if t ≥ −6).
Microarray Data Preprocessing.
All samples were analyzed using the Affymetrix Human Gene 1.1 ST microarray platform. Gene expression values were corrected, quantile-normalized, and log2-transformed via Affymetrix Expression Console software (build 1.2.1.20) using RMA algorithm. A total of 33,297 probe sets were available on each array; only those probe sets with annotated gene symbols were selected (20,237 gene symbols). If a gene was represented by multiple probe sets, the one with the largest intensity interquartile region was selected to represent that gene. Microarray data for each brain region were analyzed separately. The raw and processed microarray data were deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (GSE71620).
Detection of Circadian Gene Expression Patterns.
Nonlinear regression was used to detect circadian gene expression patterns. All individual samples were ordered by their TODs. Temporal expression was fitted to a sinusoidal curve using the nonlinear least-squares method. The coefficient of determination (R2) was used as a proxy of goodness-of-fit. The empirical P value was estimated from a null distribution of R2 generated from 1,000 TOD-randomized expression datasets for BA11 and BA47 separately. Adaptive weighted Fisher method was adopted to combine multiple P values across brain regions (26). The q value was estimated using R package qvalue (27).
Analysis of Aging Effects on Expression Rhythmicity.
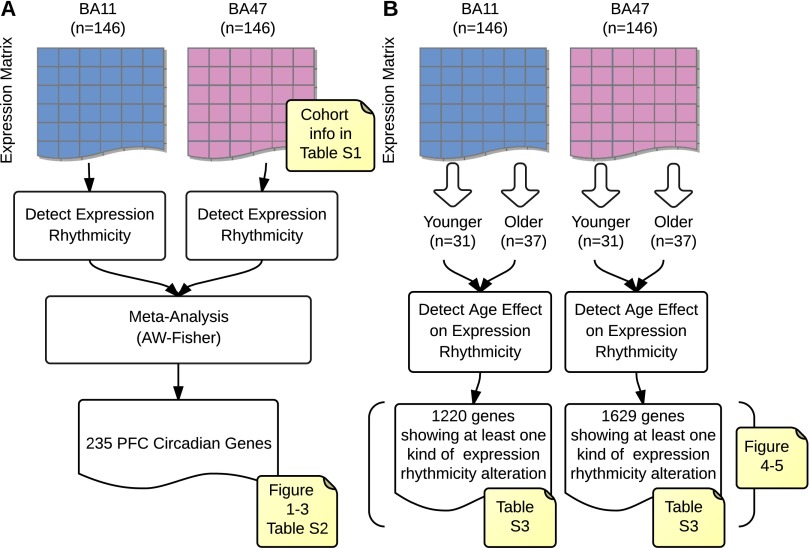
Subjects were classified as younger (<40 y, n = 31) or older (>60 y, n = 37) and analysis of rhythmic gene expression was performed on each group independently as described above. Age effect P values were estimated from 1,000 age-shuffled data matrixes for BA11 and BA47 separately (SI Materials and Methods).
SI Materials and Methods
Detection of Circadian Gene Expression Patterns.
Nonlinear regression was used to detect circadian gene expression patterns. All individual samples were ordered by their TODs, and for each gene, this temporal expression was fitted to a sinusoidal curve using the nonlinear least-squares method. The sinusoidal curve was defined as follows:
where A represents the amplitude and F represents the frequency, which was fixed at 24-h period. The nonlinear fitting was performed based on the Levenberg–Marquardt algorithm, which was implemented in SciPy’s Optimization package (release 0.13.2). The peak hour of the circadian pattern was calculated directly from the fitted curve.
The Choice of Cohort Age Cutoffs.
We chose age 40 and 60 as the cutoffs in the age stratification analysis for the following reasons:
-
i)
Age 60 or 65 are both widely used cutoffs to define elders, and they generally agree with the typical retirement age in the developed countries (e.g., United Nations has used 60+ to define old age). We also chose >60 because it generates a sample size (n = 37) that is comparable to the younger group (<40) (n = 31).
-
ii)
For this type of analysis, we require strong signals to distinguish different circadian gene expression patterns between the younger and the older groups. Therefore, we have to choose relatively extreme age groups while trying to keep a level of statistical power needed to detect significant differences (i.e., the group sizes cannot be too small and should be approximately equal sizes).
-
iii)
Moreover, current age cutoffs (40, 60) are close to the first quartile (Q1, 25%, 42.45 y) and the third quartile (Q3, 75%, 59.75 y), respectively. In addition, they are of approximately equal distance from the mean (50.67 y) and median (52.50 y).
Analysis of Aging Effects on Expression Rhythmicity.
First, subjects were classified as younger (<40 y, n = 31) or older (>60 y, n = 37) by age as mentioned above. Next, the procedures of detection of circadian gene expression patterns (mentioned in the main text) were applied to each group and each brain region separately. Then, for each gene, the absolute difference of the parameters (i.e., Δbase, Δamplitude, Δphase, and ΔR2) between the younger and the older group can be calculated by comparing their best-fitting circadian curves (P < 0.05) that we obtained at the previous step. After that, we generated 1,000 age-shuffled expression data matrixes (i.e., subjects were randomly assigned to either younger or older group) for BA11 and BA47 separately, by which we repeated all of the procedures to collect a null background of the delta values and estimated the empirical P values of each observation against the null background. The age effect on circadian expression patterns between the younger and the older groups were categorized as follows: change in the level of basal expression (Δbase); change in the amplitude of oscillation (Δamplitude); change in the acrophase of rhythmicity (Δphase); a loss of rhythmicity typically observed as a disruption of circadian rhythmicity of gene expression oscillation (ΔR2); and the gain of rhythmicity typically observed as a gain of circadian rhythmicity of gene expression oscillation (ΔR2). These categories are not mutually exclusive except for the loss and gain of rhythmicity. Note that the comparison of ΔR2 was not limited to rhythmic or arrhythmic genes in either age group. Any gene that showed significant R2 decrease or increase (empirical P < 0.05) in the older group would be annotated as “loss of rhythmicity” or “gain of rhythmicity,” respectively.
Results
Gene Expression Rhythms in Human BA11 and BA47.
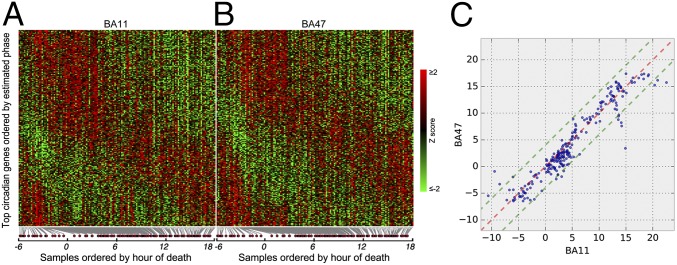
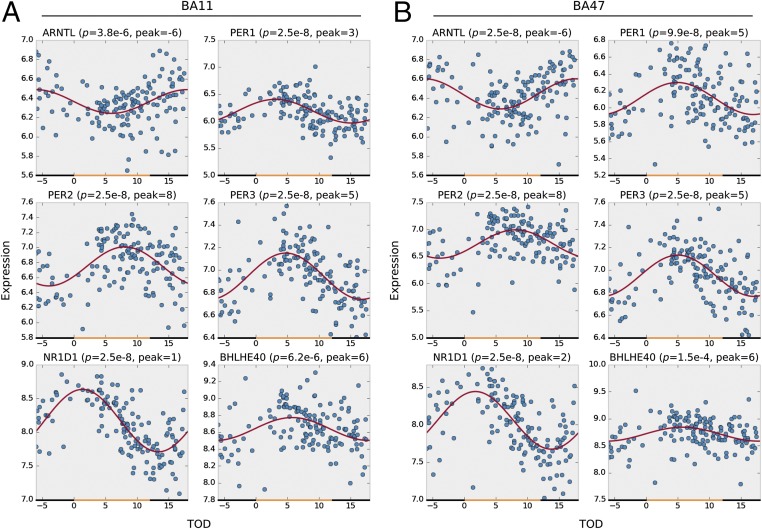
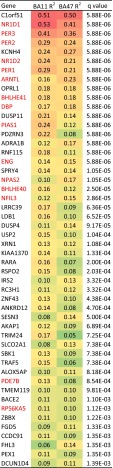
In 146 subjects, we performed a comprehensive gene expression analysis in BA11 and BA47 of the PFC (Fig. S1A). In BA11 2,475 genes (∼12% of the genome) and in BA47 a total of 1,615 (∼8% of the genome) displayed detectable circadian rhythmicity (P < 0.05) (Dataset S1). To identify a set of genes with consistent circadian regulation of gene transcripts across BA11 and BA47, we applied a metaanalysis approach across the brain regions and limited the false-discovery rate to 5% (i.e., q < 0.05). A total of 235 genes with a significant circadian expression pattern were detected at q < 0.05 (Dataset S1 and Table S2). Notably, these genes displayed coordinated temporal expression patterns across BA11 and BA47, as illustrated by the nearly identical patterns observed across the two areas (Fig. 1 A and B). Indeed, the peak or acrophase of expression of those 235 genes was highly concordant (92%) between BA11 and BA47 (Pearson’s r = 0.95, P = 5.8e-120) (Fig. 1C). The core genes that make up the molecular clock showed robust expression rhythms in the PFC (Fig. 2). Their patterns of expression were strikingly similar across the two brain regions and also to those published previously (24), demonstrating the consistency in the use of these analyses to detect significant rhythms in gene expression in human postmortem tissue. The top 50 genes with significant circadian rhythms in expression are listed in Fig. 3. For example, C1ORF51 (more recently known as CIART) has the most robust expression rhythm (Fig. S2). The mouse ortholog of this gene, Gm129, has been recently identified as having a circadian pattern of expression in mice (28, 29). Many of these genes display circadian expression patterns in peripheral tissues of the mouse, such as the liver, heart, and/or skeletal muscle, and in the brain, including the SCN and cerebellum. For example, DUSP4, ANKRD12, TRIM24, and TMEM119 display expression rhythms in the mouse SCN (30), whereas DUSP11, USP2, SESN3, BACE2, and PEX1 are expressed rhythmically in other areas of the brain (30, 31). Notably, some of the genes we identified here as displaying rhythmicity in the human PFC are not known to be involved in circadian rhythms, or even to be expressed in a circadian pattern, suggesting these genes may have previously unidentified roles within or downstream of the molecular clock. For example, KCNH4 (potassium voltage-gated ion channel) is a top-ranked gene with strong circadian expression patterns across BA11 and BA47 (Fig. 3 and Fig. S2) and there are no previous reports of this transcript being regulated in a circadian manner. Other genes with a similarly strong pattern and no prior knowledge of rhythmicity include KIAA1370, RSP02, ZNF43, BACE2, ZBBX, and FGD5.
Fig. S1.
Overview of analysis flows. (A) Detection of clock-regulated genes in human prefrontal cortex (PFC) via metaanalysis. We first estimated the P values of expression rhythmicity for all of the genes in brain regions BA11 and BA47 independently. Then we combined these P values to meta-P values via AW-Fisher and corrected them into q values. A total of 235 cross-region PFC circadian genes were detected at q < 0.05. (B) Detection of age effect on circadian expression rhythmicity. The work flow was independent from A; i.e., the analysis in B did not rely on any a priori results yielded in A. It is possible that a gene shows expression rhythmicity only in younger individuals but not in older individuals (or could be vice versa). Therefore, if younger subjects were mixed with older subjects (e.g., when all 146 subjects were included as how we did in A), we might be unable to detect the expression rhythmicity of certain genes (see Fig. S4 for such examples).
Table S2.
Numbers of significant circadian genes across BA11 and BA47 at different q value cutoffs after metaanalysis
| Cutoff | No. of genes* | Concordant phase (%)† | Average R2 |
| q < 0.01 | 84 | 80 (95%) | 0.13 |
| q < 0.05 | 235 | 217 (92%) | 0.09 |
| q < 0.1 | 465 | 411 (88%) | 0.07 |
The total number of genes is 20,237.
Concordant phase was defined as the difference of peak hour ≤4 h between BA11 and BA47.
Fig. 1.
Heat map of expression levels and comparison of circadian acrophase for the top circadian genes (n = 235, q < 0.05) in BA11 and BA47. (A and B) Expression levels were Z-transformed for each gene. Red indicates higher expression level; green indicates lower expression levels. (C) The circadian phase (peak hours) of 235 circa genes derived from metaanalysis are plotted on TOD axes for BA11 (x axis) and BA47 (y axis). Red dashed line: 1:1 diagonal line. Green dashed lines: ±4-h phase concordance boundaries. Ninety-two percent of circa genes (217 of 235) are located within the concordance interval.
Fig. 2.
Circadian gene expression patterns of six canonical circadian genes in BA11 (A) and BA47 (B). The x axis denotes the time of death (TOD) on ZT scale (−6–18 h). Approximate day–night interval within a pseudoday is represented by yellow (day) and black (night) bands. Data points from 146 subjects are plotted in blue. The best-fitted sinusoidal curves are depicted in red. The empirical P value and the estimated peak hour are also reported above each panel.
Fig. 3.
Top 50 clock-regulated genes across two brain regions. Gene list is sorted by q value after adaptive weighted (AW)-Fisher procedure. Genes in red are known circadian-related genes according to their records in GeneCards database (59, 60).
Fig. S2.
Top eight rhythmic genes in BA11 (A) and BA47 (B). The x axis denotes the time of death (TOD) on ZT scale (−6–18 h). Approximate day–night interval within a pseudoday is represented by yellow (day) and black (night) bands. Data points from 146 subjects are plotted in blue. The best-fitted sinusoidal curves are depicted in red. The empirical P value and the estimated peak hour are also reported above each panel.
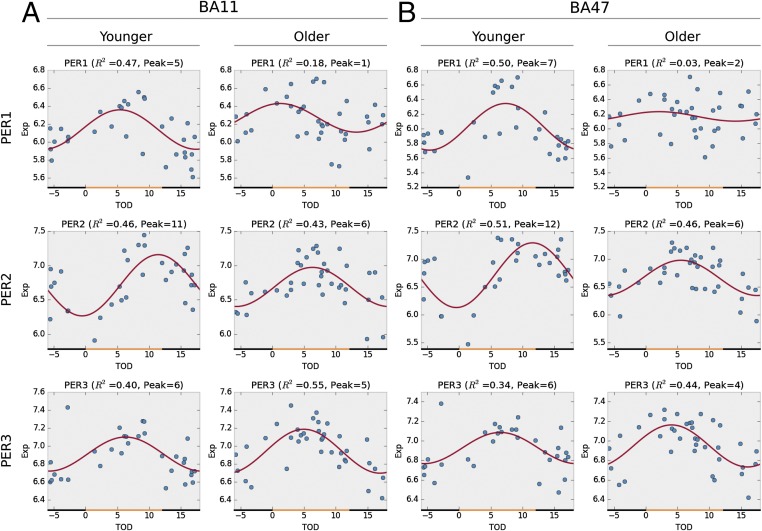
Effects of Aging on the PER Genes Across BA11 and BA47 of Human PFC.
To test the prediction of altered rhythmic patterns of gene expression with aging, we stratified our subjects into younger (<40 y, n = 31) and older (≥60 y, n = 37) groups (Fig. S1B and Fig. S3) and performed an analysis of circadian rhythms of canonical circadian genes of the molecular clock within the two subgroups. Sinusoidal curve analyses were used to investigate differences in total expression levels, amplitude, acrophase, and absolute changes in rhythmicity (loss or gain of rhythm). We found consistent effects of age (younger vs. older) on the circadian pattern of expression for PER1 and PER2, but very minimal changes in PER3, across BA11 and BA47 (Fig. 4). Specifically, the acrophases of PER1 rhythms were shifted from ZT5 (BA11) and ZT7 (BA47) to ZT1–ZT2 with a reduction in amplitude in both regions (Fig. 4 A and B), suggesting significantly disrupted temporal expression of PER1 in older individuals. Similar phase shifts in peak expression were observed for PER2 (Fig. 4 A and B)—acrophase was shifted from sunset (∼ZT12) to the middle of the day (ZT6). However, there were no obvious age effects on expression of PER3 in either BA11 or BA47 (Fig. 4 A and B).
Fig. S3.
Cohort age information in this study.
Fig. 4.
Aging has unique effects on the circadian gene expression patterns of period genes (PER family) in both BA11 (A) and BA47 (B). Older people have disrupted PER1 circadian expression patterns (P = 0.027 in BA11; P = 0.0005 in BA47), together with a phase advance of PER2 expression from sunset to noon (P = 0.005 in BA11; P = 0.004 in BA47), whereas their PER3 expression remained intact.
Effects of Aging on Rhythms of Gene Expression in BA11.
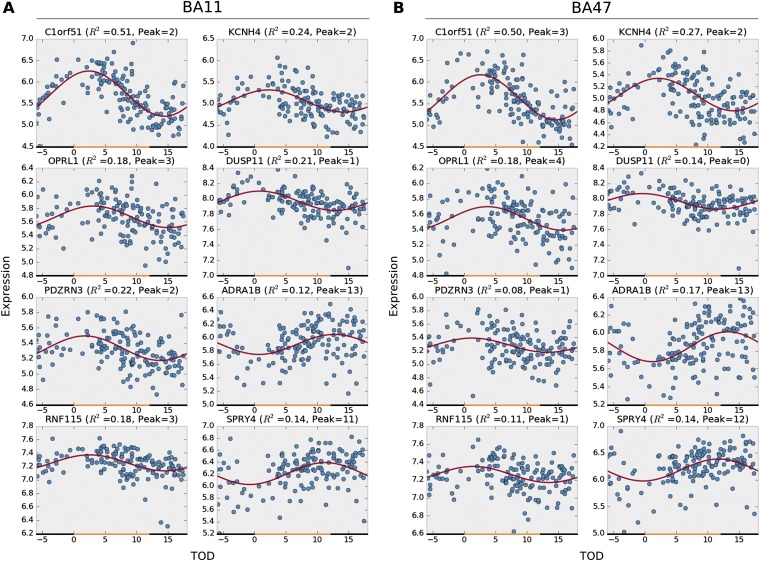
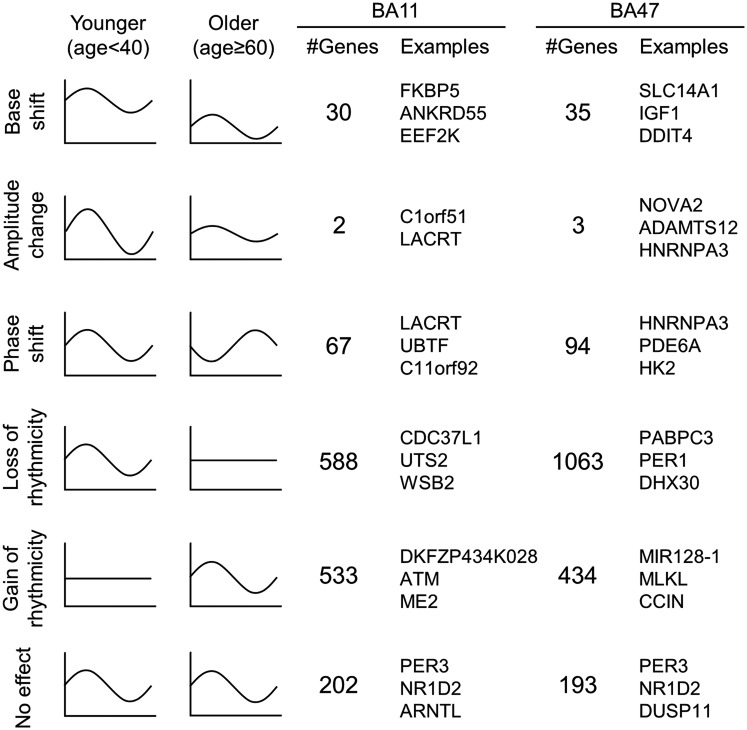
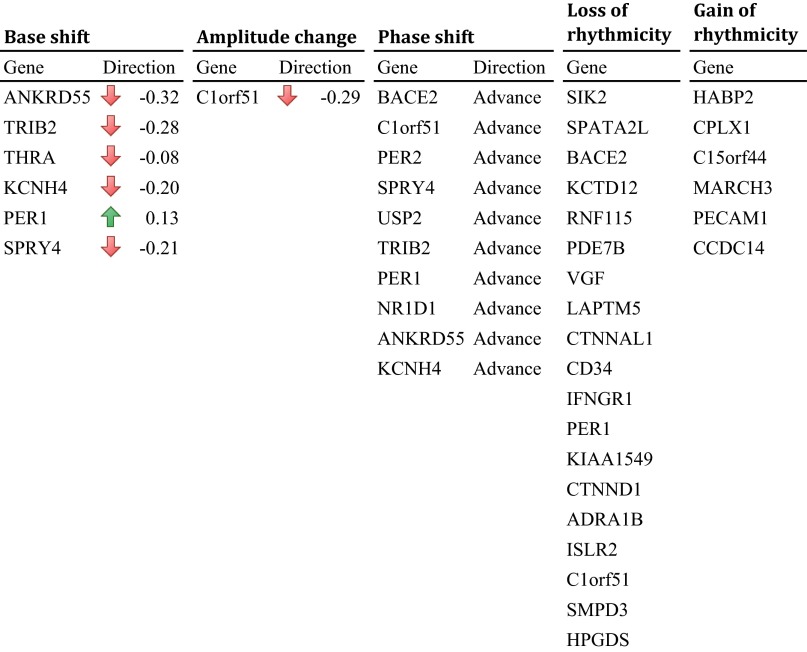
We next conducted separate, whole-genome analyses on BA11 and BA47 to determine whether the age effects on patterns of expression differed between these two regions beyond the canonical circadian genes of the molecular clock. We identified 1,186 genes in BA11 that exhibited age-dependent rhythmicity or alterations in rhythmicity patterns with aging (P < 0.05; Table S3), which can be categorized into five characteristics (Fig. 5): (i) a base shift (meaning a change in levels of expression but not necessarily a change in rhythm); (ii) a decrease (but not complete loss) in amplitude; (iii) a phase shift; (iv) a significant loss of rhythmicity; and (v) a gain of rhythmicity. A total of 201 genes in BA11 do not display any impairment in rhythmicity with aging, including many of the canonical clock genes (Dataset S2). A single gene can display multiple kinds of age-related effects on its rhythmicity pattern. For example, a gene may display both a base shift and a phase shift. Complete lists of genes in each category are in Datasets S2 and S3. When differences between groups were analyzed, we found that, in BA11, there were 30 genes that showed significant changes in level of expression (base shift) while retaining overall rhythmicity from younger to older individuals—for example, several of the most robustly rhythmic genes, PER1, KCNH4, and SPRY4 fell into this category (Fig. S4). There were 67 genes that displayed significant phase shifts in older individuals—for example, among the top circadian genes, PER2, BACE2, and SPRY4 are all significantly phase-advanced in older individuals (Fig. S4). An impressive 588 genes showed a complete loss of rhythmicity due to age, including ADRA1b of the top circadian genes, and a canonical circadian gene, CRY1. Only two genes had dampened amplitudes between age groups. These were C1ORF51 and LACRT. Finally, we identified 533 genes in BA11 that were not rhythmic in younger adults but appeared to gain rhythmicity with age, such as ATM, ME2, and MEPCE (Dataset S2).
Table S3.
Summary of detected age effects on circadian gene expression patterns in BA11 and BA47
| Age effects | No effect | ||||||
| Class | n | Base shift | Amp change | Phase shift | Loss of rhythmicity | Gain of rhythmicity | |
| BA11 | |||||||
| PFC circa genes | 235 | 6 | 1 | 10 | 19 | 6 | 201 |
| Others | 20,002 | 24 | 1 | 57 | 569 | 527 | 18,849 |
| Total | 20,237 | 30 | 2 | 67 | 588 | 533 | 19,051 |
| BA47 | |||||||
| PFC circa genes | 235 | 8 | 0 | 11 | 20 | 7 | 193 |
| Others | 20,002 | 27 | 3 | 83 | 1,043 | 427 | 18,453 |
| Total | 20,237 | 35 | 3 | 94 | 1,063 | 434 | 18,646 |
Note that the age effects are not necessary mutually exclusive. The total numbers of genes with age effects on expression rhythmicity are 20,237 − 19,051 = 1,186 in BA11 and 20,237 − 18,646 = 1,591.
Fig. 5.
Illustration of age effect on circadian gene expression patterns. The age effect on circadian gene expression patterns can be classified into five categories, as illustrated (rows 1–5). The number of instances and the top three most significant examples (sorted by P values) were also reported. See Datasets S2 and S3 for complete lists.
Fig. S4.
List of detected age effects on 235 PFC circadian genes in BA11. Upward arrow represents rise of circadian expression rhythmicity base in the older. Downward arrow represents drop of base or decrease of amplitude in the older. Phase advance means the peak hour of circadian expression rhythmicity comes earlier in the older. See Dataset S2 for full table.
Effect of Aging on Rhythms of Gene Expression in BA47.
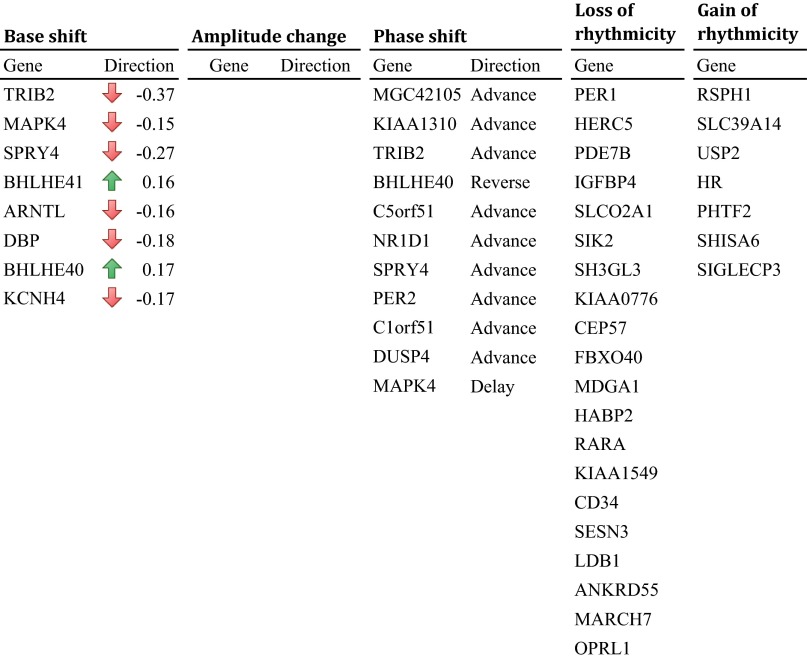
Compared with BA11, there were many more alterations in patterns of gene expression in BA47 between older and younger individuals (1,591 genes), suggesting that this area might be more strongly influenced by age (Table S3 and Dataset S3). We identified 35 genes that had a change in overall level of expression including ARNTL (BMAL1), BHLHE40, DBP, KCNH4, NR1D1, and SPRY4 of the top 50 circadian genes (Fig. S5). Ninety-four genes displayed a significant shift in acrophase due to age, such as BHLHE40, DUSP4, PER2, and SPRY4 of the top circadian genes (Fig. S5). There were three genes that had a reduction in amplitude without a loss of rhythm, NOVA2, ADAMTS12, and HNRNPA3. A large number of genes (1,063) lost rhythmicity due to age in this rain region, with several of these being in the top circadian genes and also having primary roles in molecular clock function, such as PER1, and other genes, including ADRA1B, PDE7B, and RARA. Interestingly, there were 434 genes that gained rhythmicity in older individuals, including two microRNAs, miR128-1 and miR15a. Of note, 30 genes that gained rhythmicity in older individuals in BA47 overlapped with BA11, such as A2M, CDKN1A, and miR30a. Finally, 193 genes in BA47 do not display any impairment in rhythmicity with aging (Dataset S3).
Fig. S5.
List of detected age effects on PFC circadian genes in BA47. Upward arrows and downward arrows represent rise and drop of circadian expression rhythmicity base in the older, respectively. No instance was found with significant amplitude change. Phase advance means the peak hour of circadian expression rhythmicity comes earlier in the older. Phase delay means the peak hour comes later in the older. Phase reverse means the difference of peak hours between the younger and the older groups is nearly 12 h. See Dataset S3 for full table.
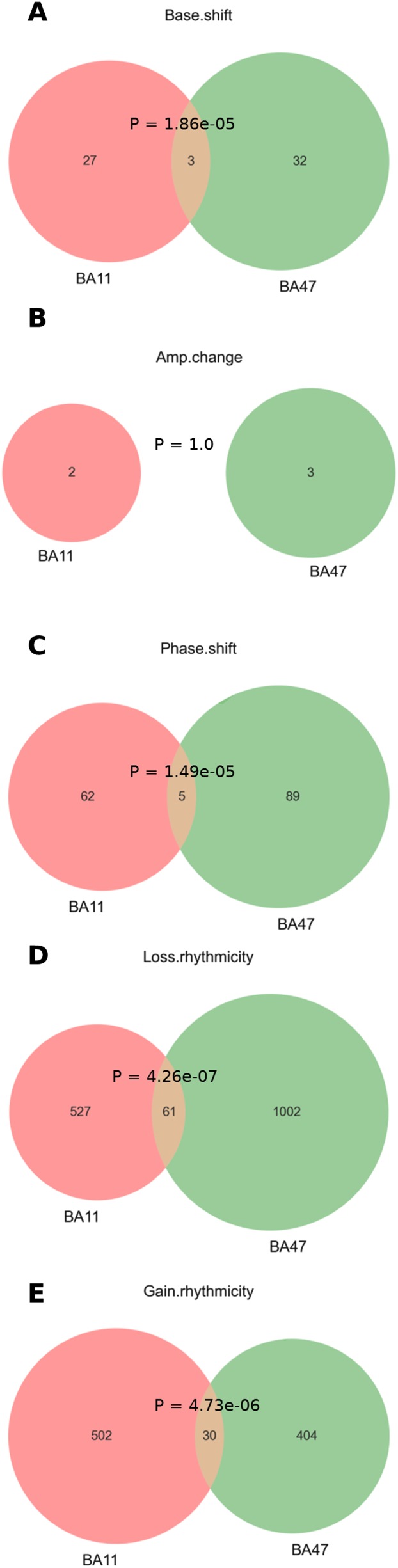
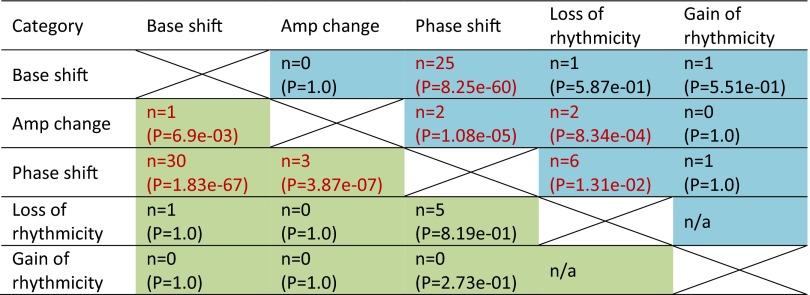
Several genes are overlapped in BA11 and BA47 in each age effect categories (Fig. S6). Most of the overlapped genes have the same direction of changes (Dataset S4). We also found significant co-occurrence between base shift and phase shift (Fig. S7). Notably, the top circadian genes with detectable age effects tend to have a downward base shift (base dropped) and/or an advanced phase (early peak hour) for the older subject group (Figs. S4 and S5).
Fig. S6.
Venn diagram of gene numbers in each age effect category between BA11 and BA47. Only those genes with P < 0.05 in each category were counted here (Datasets S2 and S3). See also Dataset S4 for complete lists. Shown are interactions between BA11 and BA47 in genes that show (A) base shift, (B) amplitude change, (C) phase shift, (D) loss of rhythmicity, or (E) gain of rhythmicity.
Fig. S7.
Co-occurrence of age effects on circadian expression rhythmicity. Shown in each cell is the number of co-occurrence and the associated P value (by Fisher’s exact test) between every two age effect categories. Note that the loss of rhythmicity and the gain of rhythmicity are two mutually exclusive categories by definition. Color code: blue cells, cases in BA11; green cells, cases in BA47.
Discussion
Our data are the first (to our knowledge) to indicate that age significantly alters the circadian rhythms of gene expression in the human PFC. Our data extend and replicate the findings of Li et al. (24) in larger cohort and further demonstrate that rhythms in gene expression in human postmortem tissue are robust and can be accurately measured. Moreover, the phase and amplitude of the clock-regulated genes analyzed here show remarkable consistency with the rhythms measured in these same genes in the Li et al. study. There are limitations to this type of analysis because we do not have information on these subjects regarding their lifestyle, actigraphy, and sleep time before death, which could contribute to changes in molecular rhythms. However, the independent confirmation and consistency between our cohort and the Li et al. study provides a high level of confidence in the overall results.
We decided to focus this study on two regions of the orbitofrontal cortex, BA11 and BA47. These are areas commonly associated with focused attention, executive functioning, and depression (32, 33). Although the Li et al. paper looked at genes that were rhythmic across six brain regions with more diverse cytoarchitectural structure (cortical, subcortical, cerebellum), we find many of the same genes to be rhythmic in BA11 and BA47 including most of the known circadian genes (i.e., ARNTL, PER1, PER2, PER3, NR1D1, NPAS2, etc.). Neither study identified CLOCK as a significantly rhythmic gene, which is consistent with its being constitutively expressed in other brain regions, the SCN in rodents and even in flies (34, 35). Interestingly, some of our most rhythmic genes identified, C1ORF51, KCNH4, and OPRL1, were not found in the Li et al. study, suggesting that perhaps their rhythmicity is specific to the areas of the brain that we investigated. Generally, we find very high concordance between rhythms in BA11 and BA47. This is interesting because studies in rodents have suggested that there can be differences in phase between adjacent brain regions (36, 37).
Several of the genes that have a significant rhythm in expression have not been described previously as core circadian genes or clock-regulated genes. One example is KCNH4, which is a voltage-gated potassium channel in the ether-a-go-go family (38). Its expression is largely restricted to the brain, although little is known about its function. Another is PDZRN3, a ubiquitin ligase known to be critically involved in cell differentiation via Wnt signaling in a variety of cell types throughout development (39, 40). We also identified certain genes that are known to play a role in rhythm regulation in the SCN or other tissues in rodents. For example, OPRL1 (also known as nociception receptor) is an opiate receptor that is strongly expressed in the mouse SCN where it binds nociception/orphanin FQ, which suppresses 88% of SCN neurons (41). Moreover, OPRL1 activation down-regulates PER2 in the SCN and accelerates reentrainment of rhythms following a shift in the light–dark cycle (42). The importance of the rhythms in this protein in BA11 and BA47 function in human brain is not known. Another example is C1ORF51 (also known as Gm129, CHRONO, and CIART). Annayev et al. (29) recently found that this protein acts as a novel transcriptional repressor with high-amplitude oscillations in the mouse liver that directly interacts with BMAL1 to repress CLOCK/BMAL1 function in this tissue. Here, we find that this transcript has the highest level of rhythmicity of any gene that we identified in human cortex, suggesting it may have a similar important circadian function in human brain. Interestingly, RNF115 [also known as breast cancer-associated gene 2 (BCA2)] was strongly rhythmic in our study. RNF115 is another ubiquitin E3 ligase that is strongly associated with increased tumor growth, particularly in response to estrogen (43). Many studies have found significant correlations between disrupted circadian rhythms and increased risk for certain cancers, including breast cancer (44). Ubiquitination regulates the stability of core clock proteins; thus, it is possible that this protein interacts with core circadian proteins to regulate rhythms in cell growth and differentiation (45). Again, the role of this protein in the human PFC is not clear.
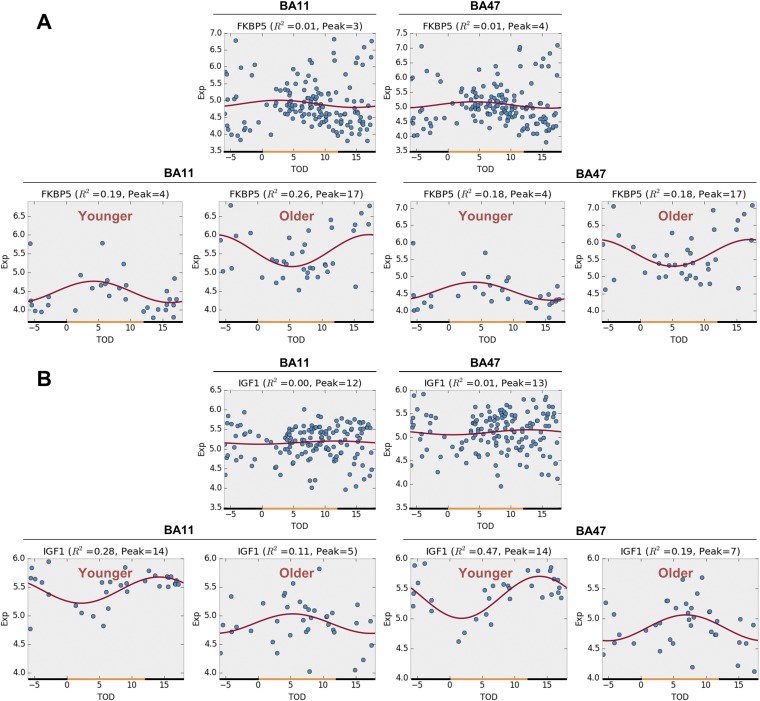
Age-stratified analysis enables us to identify age group-specific circadian genes that are undetectable from a cohort of heterogeneous age composition. For example, FKBP5 shows a reversal of circadian rhythmicity between the younger and the older subjects (Fig. S8A). Its rhythmicity thus cannot be detected from a mixed age cohort composed of both younger and older subjects. Another example is IGF1, which also shows distinct rhythmicity between the younger and the older (Fig. S8B).
Fig. S8.
Rhythmicity expression profiles before and after age stratification. (A) FKBP5. (B) IGF1. (Upper) Before age stratification (n = 146). (Lower) After age stratification (n = 31 in younger and n = 37 in older).
With aging, we found a disruption in several rhythmic transcripts, with the effects somewhat more pronounced in BA47 than in BA11. The reason for this difference is not clear. Samples were processed in parallel from dissection to array hybridization, making technical differences unlikely. Thus, differences may reflect distinct functions and underlying biology of the two regions. The most common deficit that we identified was a complete or partial loss of rhythmicity in transcripts that were rhythmic in younger people. We also identified certain transcripts that had a shift in phase. This is interesting because it is well documented that healthy older individuals tend to experience a shift toward “morningness” where they prefer to wake early in the morning and go to sleep relatively early in the evening (20, 46). There are natural variations in chronotype at all ages with some showing morning preference and some showing evening preference. Extreme chronotypes are associated with multiple psychiatric disorders (47–50), and it would be interesting in future studies to determine how molecular rhythms correlate with chronotype. Interestingly, a condition called “sundowning” or “sundown syndrome” affects some 20–40% of older people with dementia or Alzheimer’s disease where they become confused, delirious, anxious, and agitated in the evening when the sun goes down, causing them to wander, become combative, and have difficulties sleeping (51). Sundowning is thought to be directly related to the breakdown in circadian rhythms in these individuals (52). Although our study examined only healthy individuals, it is possible that some of the genes that experience a loss of rhythmicity with aging might be linked to temporal changes in cognition.
Somewhat surprisingly, we identified a number of transcripts that gain rhythmicity with aging. Very interestingly, two of these transcripts were micro-RNAs miR128 and miR15a. miR128 is involved in the regulation of amyloid-β degradation in Alzheimer’s disease and is involved in tumor suppression (53, 54), and miR15a is also involved in tumor growth and its expression is correlated with plaque score in Alzheimer’s disease (55, 56). A survey of age-related microRNA expression changes in the neuronal stem cell niches of the short-lived annual fish Nothobranchius furzeri found that miR15a was abundant only in older animals (57). It will be interesting in future studies to examine postmortem samples from subjects with conditions such as Alzheimer’s disease to determine whether there are more extreme changes in circadian gene expression. The gain of rhythmicity in these micro-RNAs could impact the cyclic expression of a whole host of proteins, perhaps leading to a novel, compensatory clock that is activated when the canonical clock breaks down. A recent study by Eckel-Mahan et al. (58) had a similar finding in the liver of mice fed a high-fat diet. Mice on regular chow showed normal cycling of canonical circadian genes in the liver and the rhythms in these genes were severely disrupted with a high-fat diet. However, another set of genes became rhythmic with the high-fat diet, suggesting a reprogramming of the liver to allow it to handle this change in diet. It is possible that a similar mechanism occurs in the brain of older individuals, concurrently with a partial breakdown of normal rhythmicity. More studies will need to be done, however, to determine the true significance and potential mechanisms regulating the gain of rhythmicity in these transcripts.
Conclusions
In conclusion, our study demonstrates the power of TOD transcriptomic analysis to identify transcripts with a circadian rhythm in human postmortem brain. We find many genes to have a significant rhythm in BA11 and BA47 with a high concordance in their phase. Furthermore, we have identified age-related changes in rhythmic transcript expression and somewhat surprisingly have uncovered a previously unidentified set of genes that gain rhythmicity in older individuals. These studies will help us better understand how rhythms change during aging and how targeted therapies that enhance molecular rhythmicity might be developed to prevent conditions like sundowning or enhance cognition. Moreover, in the future, we can use this approach to identify changes in molecular rhythms in a variety of brain regions or specific cell types that associate with psychiatric and neurological diseases.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Mental Health Grants R01MH077159 (to E.S. and C.A.M.), R01MH093723 (to E.S.), and MH103204 (to D.A.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE71620).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508249112/-/DCSupplemental.
References
- 1.Patel VR, Eckel-Mahan K, Sassone-Corsi P, Baldi P. How pervasive are circadian oscillations? Trends Cell Biol. 2014;24(6):329–331. doi: 10.1016/j.tcb.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron KG, Reid KJ. Circadian misalignment and health. Int Rev Psychiatry. 2014;26(2):139–154. doi: 10.3109/09540261.2014.911149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foster RG, Kreitzman L. The rhythms of life: What your body clock means to you! Exp Physiol. 2014;99(4):599–606. doi: 10.1113/expphysiol.2012.071118. [DOI] [PubMed] [Google Scholar]
- 4.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 5.Hogenesch JB, Panda S, Kay S, Takahashi JS. Circadian transcriptional output in the SCN and liver of the mouse. Novartis Found Symp. 2003;253:171–180; discussion 52–55, 102–109, 180–183 passim. [PubMed] [Google Scholar]
- 6.Albrecht U. Timing to perfection: The biology of central and peripheral circadian clocks. Neuron. 2012;74(2):246–260. doi: 10.1016/j.neuron.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Logan RW, Williams WP, 3rd, McClung CA. Circadian rhythms and addiction: Mechanistic insights and future directions. Behav Neurosci. 2014;128(3):387–412. doi: 10.1037/a0036268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukherjee S, et al. Knockdown of Clock in the ventral tegmental area through RNA interference results in a mixed state of mania and depression-like behavior. Biol Psychiatry. 2010;68(6):503–511. doi: 10.1016/j.biopsych.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sidor MM, et al. Daytime spikes in dopaminergic activity drive rapid mood-cycling in mice. Mol Psychiatry. 2015;20(11):1406–1419. doi: 10.1038/mp.2014.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho K, Ennaceur A, Cole JC, Suh CK. Chronic jet lag produces cognitive deficits. J Neurosci. 2000;20(6):RC66. doi: 10.1523/JNEUROSCI.20-06-j0005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marquié JC, Tucker P, Folkard S, Gentil C, Ansiau D. Chronic effects of shift work on cognition: Findings from the VISAT longitudinal study. Occup Environ Med. 2015;72(4):258–264. doi: 10.1136/oemed-2013-101993. [DOI] [PubMed] [Google Scholar]
- 12.Rouch I, Wild P, Ansiau D, Marquié JC. Shiftwork experience, age and cognitive performance. Ergonomics. 2005;48(10):1282–1293. doi: 10.1080/00140130500241670. [DOI] [PubMed] [Google Scholar]
- 13.Anderson JA, Campbell KL, Amer T, Grady CL, Hasher L. Timing is everything: Age differences in the cognitive control network are modulated by time of day. Psychol Aging. 2014;29(3):648–657. doi: 10.1037/a0037243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh CM, et al. Weaker circadian activity rhythms are associated with poorer executive function in older women. Sleep. 2014;37(12):2009–2016. doi: 10.5665/sleep.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mulder CK, Gerkema MP, Van der Zee EA. Circadian clocks and memory: Time-place learning. Front Mol Neurosci. 2013;6:8. doi: 10.3389/fnmol.2013.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulder CK, Papantoniou C, Gerkema MP, Van Der Zee EA. Neither the SCN nor the adrenals are required for circadian time-place learning in mice. Chronobiol Int. 2014;31(9):1075–1092. doi: 10.3109/07420528.2014.944975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roedel A, Storch C, Holsboer F, Ohl F. Effects of light or dark phase testing on behavioural and cognitive performance in DBA mice. Lab Anim. 2006;40(4):371–381. doi: 10.1258/002367706778476343. [DOI] [PubMed] [Google Scholar]
- 18.Karatsoreos IN, Bhagat S, Bloss EB, Morrison JH, McEwen BS. Disruption of circadian clocks has ramifications for metabolism, brain, and behavior. Proc Natl Acad Sci USA. 2011;108(4):1657–1662. doi: 10.1073/pnas.1018375108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Youngstedt SD, Kripke DF, Elliott JA, Klauber MR. Circadian abnormalities in older adults. J Pineal Res. 2001;31(3):264–272. doi: 10.1034/j.1600-079x.2001.310311.x. [DOI] [PubMed] [Google Scholar]
- 20.Yoon IY, et al. Age-related changes of circadian rhythms and sleep-wake cycles. J Am Geriatr Soc. 2003;51(8):1085–1091. doi: 10.1046/j.1532-5415.2003.51356.x. [DOI] [PubMed] [Google Scholar]
- 21.Hofman MA, Swaab DF. Living by the clock: The circadian pacemaker in older people. Ageing Res Rev. 2006;5(1):33–51. doi: 10.1016/j.arr.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Daneault V, et al. Aging reduces the stimulating effect of blue light on cognitive brain functions. Sleep. 2014;37(1):85–96. doi: 10.5665/sleep.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pagani L, et al. Serum factors in older individuals change cellular clock properties. Proc Natl Acad Sci USA. 2011;108(17):7218–7223. doi: 10.1073/pnas.1008882108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li JZ, et al. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc Natl Acad Sci USA. 2013;110(24):9950–9955. doi: 10.1073/pnas.1305814110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seney ML, et al. The role of genetic sex in affect regulation and expression of GABA-related genes across species. Front Psychiatry. 2013;4:104. doi: 10.3389/fpsyt.2013.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang LC, Lin HM, Sibille E, Tseng GC. Meta-analysis methods for combining multiple expression profiles: Comparisons, statistical characterization and an application guideline. BMC Bioinformatics. 2013;14:368. doi: 10.1186/1471-2105-14-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storey JD. The positive false discovery rate: A Bayesian interpretation and the q-value. Ann Stat. 2003;31(6):2013–2035. [Google Scholar]
- 28.Goriki A, et al. A novel protein, CHRONO, functions as a core component of the mammalian circadian clock. PLoS Biol. 2014;12(4):e1001839. doi: 10.1371/journal.pbio.1001839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Annayev Y, et al. Gene model 129 (Gm129) encodes a novel transcriptional repressor that modulates circadian gene expression. J Biol Chem. 2014;289(8):5013–5024. doi: 10.1074/jbc.M113.534651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 31.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pagani M, et al. Imaging the neurobiological substrate of atypical depression by SPECT. Eur J Nucl Med Mol Imaging. 2007;34(1):110–120. doi: 10.1007/s00259-006-0177-4. [DOI] [PubMed] [Google Scholar]
- 33.Nebel K, et al. On the neural basis of focused and divided attention. Brain Res Cogn Brain Res. 2005;25(3):760–776. doi: 10.1016/j.cogbrainres.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 34.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96(2):271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 35.Houl JH, Yu W, Dudek SM, Hardin PE. Drosophila CLOCK is constitutively expressed in circadian oscillator and non-oscillator cells. J Biol Rhythms. 2006;21(2):93–103. doi: 10.1177/0748730405283697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harbour VL, Weigl Y, Robinson B, Amir S. Phase differences in expression of circadian clock genes in the central nucleus of the amygdala, dentate gyrus, and suprachiasmatic nucleus in the rat. PLoS One. 2014;9(7):e103309. doi: 10.1371/journal.pone.0103309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamont EW, Robinson B, Stewart J, Amir S. The central and basolateral nuclei of the amygdala exhibit opposite diurnal rhythms of expression of the clock protein Period2. Proc Natl Acad Sci USA. 2005;102(11):4180–4184. doi: 10.1073/pnas.0500901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyake A, Mochizuki S, Yokoi H, Kohda M, Furuichi K. New ether-à-go-go K+ channel family members localized in human telencephalon. J Biol Chem. 1999;274(35):25018–25025. doi: 10.1074/jbc.274.35.25018. [DOI] [PubMed] [Google Scholar]
- 39.Sewduth RN, et al. The ubiquitin ligase PDZRN3 is required for vascular morphogenesis through Wnt/planar cell polarity signalling. Nat Commun. 2014;5:4832. doi: 10.1038/ncomms5832. [DOI] [PubMed] [Google Scholar]
- 40.Honda T, Yamamoto H, Ishii A, Inui M. PDZRN3 negatively regulates BMP-2-induced osteoblast differentiation through inhibition of Wnt signaling. Mol Biol Cell. 2010;21(18):3269–3277. doi: 10.1091/mbc.E10-02-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allen CN, et al. Orphanin-FQ/nociceptin (OFQ/N) modulates the activity of suprachiasmatic nucleus neurons. J Neurosci. 1999;19(6):2152–2160. doi: 10.1523/JNEUROSCI.19-06-02152.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyakawa K, et al. ORL1 receptor-mediated down-regulation of mPER2 in the suprachiasmatic nucleus accelerates re-entrainment of the circadian clock following a shift in the environmental light/dark cycle. Neuropharmacology. 2007;52(3):1055–1064. doi: 10.1016/j.neuropharm.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 43.Burger AM, et al. Role of the BCA2 ubiquitin E3 ligase in hormone responsive breast cancer. Open Cancer J. 2010;3(1):116–123. doi: 10.2174/1874079001003010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevens RG, Brainard GC, Blask DE, Lockley SW, Motta ME. Breast cancer and circadian disruption from electric lighting in the modern world. CA Cancer J Clin. 2014;64(3):207–218. doi: 10.3322/caac.21218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stojkovic K, Wing SS, Cermakian N. A central role for ubiquitination within a circadian clock protein modification code. Front Mol Neurosci. 2014;7:69. doi: 10.3389/fnmol.2014.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Myers BL, Badia P. Changes in circadian rhythms and sleep quality with aging: Mechanisms and interventions. Neurosci Biobehav Rev. 1995;19(4):553–571. doi: 10.1016/0149-7634(95)00018-6. [DOI] [PubMed] [Google Scholar]
- 47.Abe T, et al. Relation between morningness-eveningness score and depressive symptoms among patients with delayed sleep phase syndrome. Sleep Med. 2011;12(7):680–684. doi: 10.1016/j.sleep.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 48.Adan A. Chronotype and personality factors in the daily consumption of alcohol and psychostimulants. Addiction. 1994;89(4):455–462. doi: 10.1111/j.1360-0443.1994.tb00926.x. [DOI] [PubMed] [Google Scholar]
- 49.Giglio LM, et al. Circadian preference in bipolar disorder. Sleep Breath. 2010;14(2):153–155. doi: 10.1007/s11325-009-0301-3. [DOI] [PubMed] [Google Scholar]
- 50.Merikanto I, et al. Evening types are prone to depression. Chronobiol Int. 2013;30(5):719–725. doi: 10.3109/07420528.2013.784770. [DOI] [PubMed] [Google Scholar]
- 51.Bachman D, Rabins P. “Sundowning” and other temporally associated agitation states in dementia patients. Annu Rev Med. 2006;57:499–511. doi: 10.1146/annurev.med.57.071604.141451. [DOI] [PubMed] [Google Scholar]
- 52.Klaffke S, Staedt J. Sundowning and circadian rhythm disorders in dementia. Acta Neurol Belg. 2006;106(4):168–175. [PubMed] [Google Scholar]
- 53.Tiribuzi R, et al. miR128 up-regulation correlates with impaired amyloid β(1-42) degradation in monocytes from patients with sporadic Alzheimer’s disease. Neurobiol Aging. 2014;35(2):345–356. doi: 10.1016/j.neurobiolaging.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 54.Jin M, et al. miRNA-128 suppresses prostate cancer by inhibiting BMI-1 to inhibit tumor-initiating cells. Cancer Res. 2014;74(15):4183–4195. doi: 10.1158/0008-5472.CAN-14-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo S, et al. miR-15a inhibits cell proliferation and epithelial to mesenchymal transition in pancreatic ductal adenocarcinoma by down-regulating Bmi-1 expression. Cancer Lett. 2014;344(1):40–46. doi: 10.1016/j.canlet.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 56.Bekris LM, et al. MicroRNA in Alzheimer’s disease: An exploratory study in brain, cerebrospinal fluid and plasma. Biomarkers. 2013;18(5):455–466. doi: 10.3109/1354750X.2013.814073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Terzibasi Tozzini E, et al. Regulation of microRNA expression in the neuronal stem cell niches during aging of the short-lived annual fish Nothobranchius furzeri. Front Cell Neurosci. 2014;8:51. doi: 10.3389/fncel.2014.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eckel-Mahan KL, et al. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013;155(7):1464–1478. doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stelzer G, et al. In-silico human genomics with GeneCards. Hum Genomics. 2011;5(6):709–717. doi: 10.1186/1479-7364-5-6-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Safran M, et al. GeneCards Version 3: The human gene integrator. Database (Oxford) 2010;2010:baq020. doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.