Significance
Schedules of survival, growth, and reproduction define life-history strategies across species. Understanding how life-history strategies are structured is fundamental to our understanding of the evolution, abundance, and distribution of species. We found that life-history strategies of 418 plant species worldwide are explained by an axis representing the pace of life and another representing the wide range of reproductive strategies. This framework predicts responses to perturbations and long-term population performance, showing great promise as a predictive tool for plant population responses to environmental change.
Keywords: life history strategy, iteroparity, generation time, matrix population model, phylogenetic signal
Abstract
The identification of patterns in life-history strategies across the tree of life is essential to our prediction of population persistence, extinction, and diversification. Plants exhibit a wide range of patterns of longevity, growth, and reproduction, but the general determinants of this enormous variation in life history are poorly understood. We use demographic data from 418 plant species in the wild, from annual herbs to supercentennial trees, to examine how growth form, habitat, and phylogenetic relationships structure plant life histories and to develop a framework to predict population performance. We show that 55% of the variation in plant life-history strategies is adequately characterized using two independent axes: the fast–slow continuum, including fast-growing, short-lived plant species at one end and slow-growing, long-lived species at the other, and a reproductive strategy axis, with highly reproductive, iteroparous species at one extreme and poorly reproductive, semelparous plants with frequent shrinkage at the other. Our findings remain consistent across major habitats and are minimally affected by plant growth form and phylogenetic ancestry, suggesting that the relative independence of the fast–slow and reproduction strategy axes is general in the plant kingdom. Our findings have similarities with how life-history strategies are structured in mammals, birds, and reptiles. The position of plant species populations in the 2D space produced by both axes predicts their rate of recovery from disturbances and population growth rate. This life-history framework may complement trait-based frameworks on leaf and wood economics; together these frameworks may allow prediction of responses of plants to anthropogenic disturbances and changing environments.
Demographic schedules of survival, growth, and reproduction, which comprise life-history strategies, are fundamental to our understanding of a range of ecological and evolutionary processes, such as invasions and local extinctions (1–3), community structure (4, 5), and species diversification (6, 7). Consequently, the development and careful testing of theory on how organisms allocate resources to survival, growth, and reproduction are important goals for evolutionary biology, ecology, and conservation biology (8). Indeed, calls for the development of a “periodic table” to classify species based on their life-history strategies and to predict population dynamics and community composition go back to the early development of evolutionary biology as a discipline (9).
A main axiom of life-history theory is that trade-offs (i.e., budgetary compromises) between different aspects of an organism’s demographic schedules, such as survival, growth, and/or reproduction, constrain and optimize the range of possible life-history strategies that can evolve across the tree of life (10, 11). However, the plant kingdom encompasses a vast amount of life-history variation; plant longevity, for instance, ranges from weeks to millennia (12). Many plant species’ life cycles include cryptic life stages such as seedbanks (13) or dormant adults (similar to animal hibernation) (14). Reproduction also can be highly variable among plants, with seed mass and per-capita seed production ranging across six orders of magnitude (15). Previous classifications of plant life-history strategies have been limited in geographic (16, 17), taxonomic, and phylogenetic scales (17) and in the ability to differentiate life-history trade-offs (17–19).
Here we propose an approach analogous to that developed decades ago for vertebrates (20) to study the drivers behind plant life-history variation. We combine demographic, phylogenetic, and ecological data from natural populations of 418 plant species worldwide (Fig. 1) to address the following questions: (i) What are the main axes of variation of plant life-history strategies? (ii) To what extent do phylogenetic ancestry, habitat, growth form, and size constrain plant life-history variation? We then test whether the position of a species on these axes predicts two important metrics of population performance: population growth rate and speed of recovery from disturbances. If clear patterns emerge, they may form the basis for a satisfactory classification and predictive framework of plant responses to the changing environment and for cross-taxonomic comparisons.
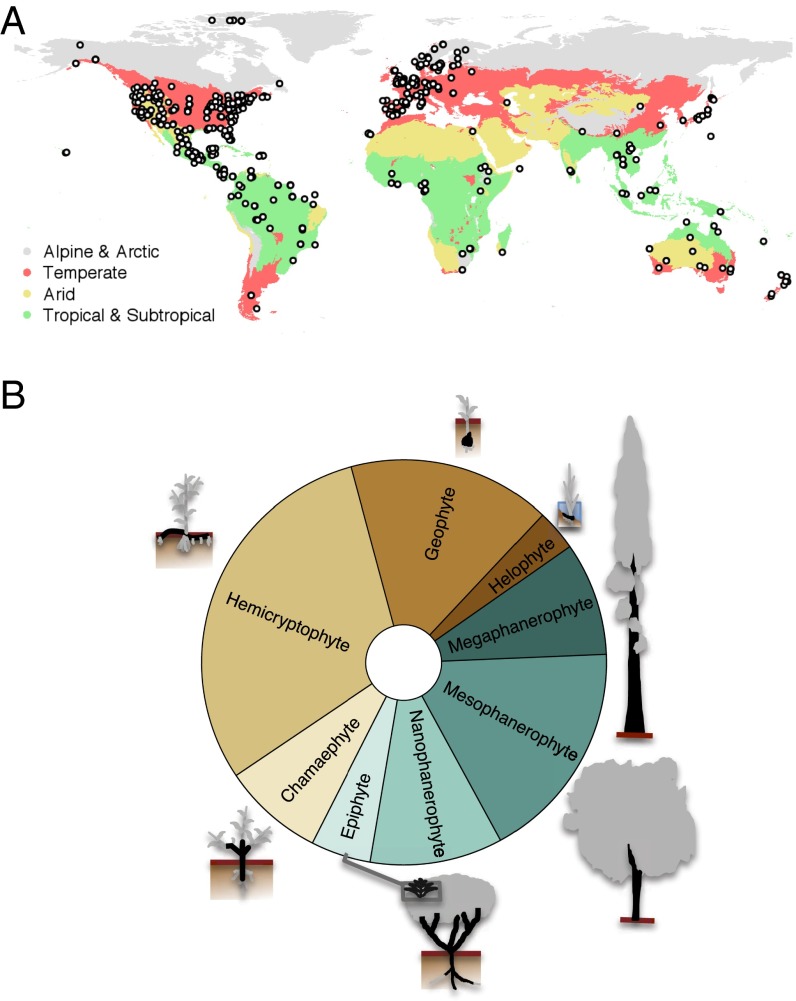
Fig. 1.
The 418 plant species examined include all major terrestrial habitats (A) and growth forms (B). Growth form is categorized according to the position of the plant’s shoot apical meristems in relation to ground level. Tissue shown in gray is typically renewed every year; tissue shown in black is perennial.
We use the COMPADRE Plant Matrix Database (21) to address these questions, drawing from the demographic, biogeographic, anatomic, and phylogenetic information of the 418 plant species covering 105 families (Dataset S1). Together, the selected species represent 825 natural populations worldwide across all major terrestrial habitats and vascular plant growth forms (Fig. 1) for which at least 4 years of high-resolution demographic field data exist. For each species, we use their population matrix models (22) to calculate a set of representative life-history traits that inform on schedules of survival, growth, and reproduction (Table 1) (11), and we then evaluate the variation in these traits along major axes using phylogenetically corrected principal component analyses (PCA) (23).
Table 1.
Loading of the life-history traits grouped by their relation to turnover and strategies for longevity, growth, and reproduction onto the first two PCA axes
| Life-history trait | Symbol | Definition | PCA 1 | PCA 2 |
| Turnover | ||||
| Generation time | T | Number of years necessary for the individuals of a population to be fully replaced by new ones | 0.85 | 0.17 |
| Longevity | ||||
| Survivorship curve type | H | Shape of the age-specific survivorship curve lx as quantified by Keyfitz’ entropy (H). H >1, = 1, <1 correspond to survivorship curves types I, II, and III, respectively | 0.55 | 0.23 |
| Age at sexual maturity | Lα | Number of years that it takes an average individual in the population to become sexually reproductive | 0.71 | 0.29 |
| Growth | ||||
| Progressive growth | γ | Mean probability of transitioning to a larger/more developed stage in the life cycle of the species, weighted by the stable stage distribution (SSD) | -0.73 | −0.05 |
| Retrogressive growth | ρ | Mean probability of transitioning to a smaller/less developed stage in the life cycle of the species, SSD-weighted | 0.07 | -0.77 |
| Reproduction | ||||
| Mean sexual reproduction | Φ | Mean per-capita number of sexual recruits across stages in the life cycle of the species, SSD-weighted | -0.83 | 0.30 |
| Degree of iteroparity | S | Spread of reproduction throughout the lifespan of the individual as quantified by Demetrius’ entropy (S). High/low S values correspond to iteroparous/semelparous populations | −0.23 | 0.51 |
| Net reproductive rate | Ro | Mean number of recruits produced during the mean life expectancy of an individual in the population | 0.04 | 0.75 |
| Mature life expectancy | Lω | Number of years from the mean age at sexual maturity (Lα) until the mean life expectancy (ηe) of an individual in the population | 0.15 | 0.27 |
| Explained variation, % | 34.06 | 21.23 | ||
| Cumulative percentage of explained variation | 34.06 | 55.38 | ||
Loadings in bold indicate a high contribution (greater than ±0.50) of the life-history trait to the PCA axis.
Results
Two Life-History Axes: The Fast–Slow Continuum and Reproductive Strategy.
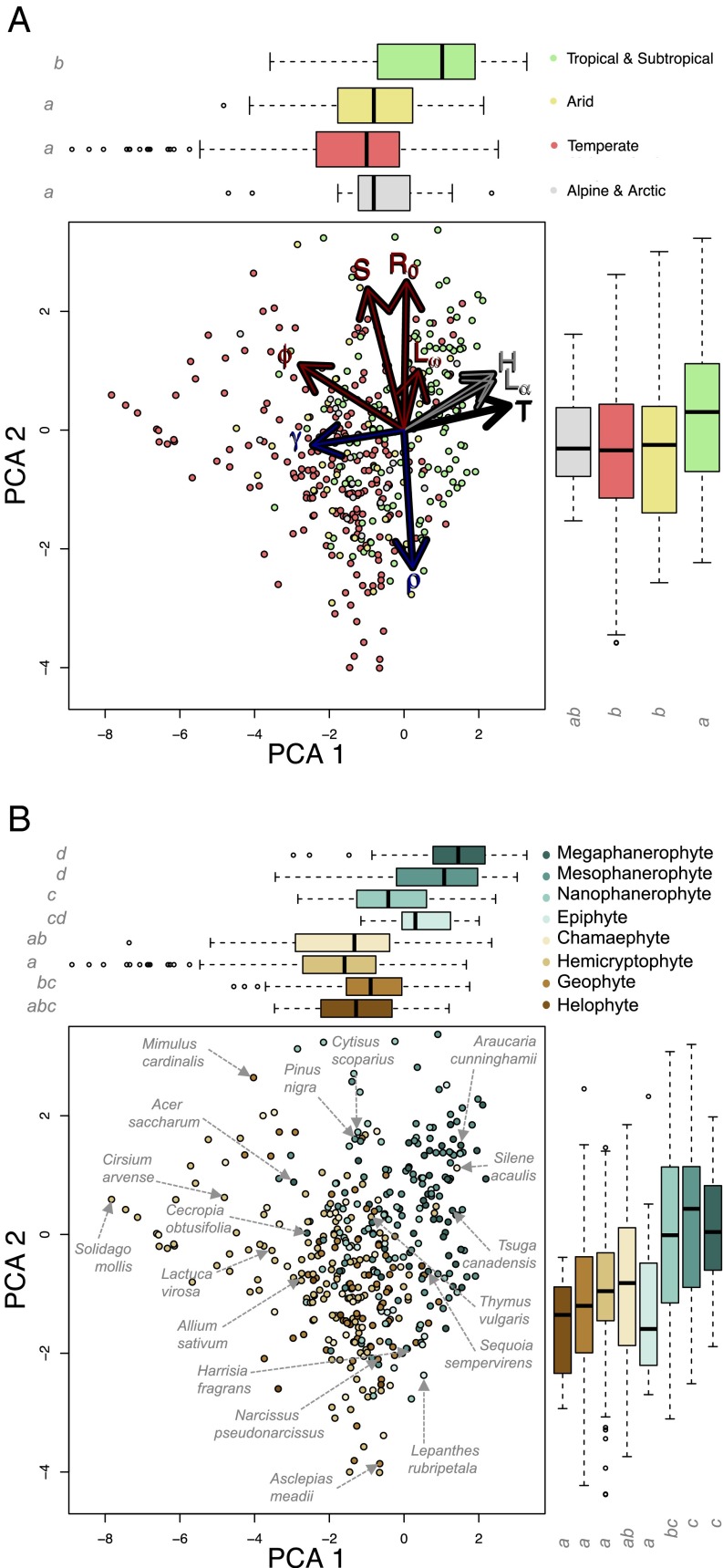
The repertoire of life histories among vascular plants is captured satisfactorily by the first two PCA axes, which together explain 55% of the variation. Following the Kaiser criterion (24), we retain PCA axes 1 and 2 in our global analyses because only for these axes are the associated eigenvalues >1, followed by a sharp drop in amount of variance explained with the third and further axes (SI Appendix, Table S4). PCA 1 and 2 explain 34% and 21% of the variation in plant life-history strategies, respectively. The life-history traits most closely aligned with PCA axis 1 are related to the fast–slow continuum (11): generation time (T) is the life-history trait with greatest loading onto PCA 1 (Table 1), closely followed by the mean sexual reproduction (φ) and the rate of growth of individual plants (progressive growth, γ). The positive loading of T onto PCA 1 had an opposite sign to the negative loadings for both growth and mean sexual reproduction, supporting the well-established trade-off between fast-growing, highly reproductive species and population turnover (11, 25). Two additional life-history traits that inform on longevity and mortality schedules also loaded positively onto PCA 1, i.e., the shape of the survivorship curve (H) and mean age at maturity (Lα) (Fig. 2 and Table 1). The majority of the traits closely aligned with PCA axis 2 represent dimensions of a plant’s reproductive strategy not captured by mean sexual reproduction: The net reproductive rate (Ro) and frequency of reproduction throughout an individual’s life expectancy (i.e., degree of iteroparity; S) are positively loaded onto PCA 2. The rate of shrinking of individual plants (retrogressive growth, ρ) is negatively loaded onto PCA 2. Mature life expectancy (Lω), the period between age of sexual maturity (Lα) and mean life expectancy (21), is a poor contributor to PCA 1 or 2 and is the main driver of PCA 3 (loading = −0.84) (SI Appendix, Table S4).
Fig. 2.
Life-history variation in vascular plants is characterized by life-history traits associated with the fast–slow continuum and reproductive strategies. Shown are phylogenetically corrected PCA of life-history traits in Table 1 with population turnover (black arrows) and traits related to longevity (gray), growth (dark blue), and reproduction (red). Arrow length indicates the loading of each life-history trait onto PCA axes. Points represent the position of species along the PCA 1 and 2 and are color-coded per major habitat (A) and Raunkiær’s growth form (B). Box-and-whisker plots on the top and right of each panel represent median (thick bar), upper and lower quartiles (edges of rectangle), and maximum and minimum (outer bars) excluding outliers (empty circles; >2/3 of the absolute value of the quartile) of PCAs 1 and 2. The differences between groups marked with different letters display statistically significant differences.
From negative to positive scores along PCA 1 (hereafter, the fast–slow axis), plants increase their allocation to longevity-related life-history traits and decrease in population turnover (i.e., greater generation time) at the expense of growth and production of new recruits (Fig. 2). Transitioning from negative to positive scores on PCA 2 (hereafter, the reproductive strategy axis), plants attain greater lifetime reproductive success and frequency of reproduction and tend to shrink less. The difference in the way size typically is measured in herbs (helophytes, geophytes, and hemicryptophytes) vs. trees (nano-/meso-/megaphanerophytes) (21, 22, 26, 27) does not appear to explain the orientation of retrogressive growth in the PCA space, because this pattern remains consistent in analyses of either group separately (SI Appendix, Table S7). More generally, a robust and consistent association and loading of the life-history traits described above emerges when different subsets of plant growth forms (27), major habitats (28), and taxonomic classes are considered separately (SI Appendix, Tables S6 and S7), suggesting a global pattern throughout the plant kingdom. Interestingly, PCA 3 is retained [its associated eigenvalue >1 (24)] only in certain groups, i.e., herbs, but not in others (shrubs or trees), and for species in the Liliopsida and Magnoliopsida (SI Appendix, Tables S6 and S7). In these groups, mature life expectancy, Lω, is the main driver of PCA 3. Randomization tests suggest that the pattern is robust to spurious correlations that might have been expected from coercing life-history traits onto sequentially orthogonal axes with the PCA (SI Appendix, Figs. S1 and S2 and Tables S9 and S10) (24).
Major habitat is alone a weak predictor of the position of plant species along the reproductive strategy axis (F3, 395 = 2.46; P = 0.06) but is a statistically significant predictor for the fast–slow axis (F3, 395 = 4.83; P = 0.003). Tropical and subtropical species seem to attain greater longevities than species in arid, temperate, and alpine or arctic regions, a result that may be caused by to the dominance of long-lived trees in tropical communities (29) and/or the nonrandom sampling of demographic studies in these habitats (SI Appendix, Table S5) (16, 21). Tall plants such as megaphanerophytes (maximum height >25 m; e.g., Canadian hemlock, Tsuga canadensis) and mesophanerophytes (maximum height 10–25 m; e.g., black pine, Pinus nigra) tend to have greater fast–slow axis scores than smaller species such as hemicryptophytes (whose shoot apical meristems are at ground level; e.g., Mead’s milkweed, Asclepias meadii) and geophytes (whose shoot apical meristems are below ground; e.g., garlic, Allium sativum) (F7, 395 = 34.88; P < 0.001) (Fig. 2B). Growth form also is associated with the reproductive strategy axis (F7, 395 = 17.43; P < 0.001), whereby PCA scores also increase sequentially with growth form size, with helophytes (shoot apical meristems resting below water) and geophytes having the lowest reproductive scores and phanerophytes (shrubs and tall succulent cacti) having the highest reproduction scores (Fig. 2B). Epiphytes (species growing upon other plants; e.g., forest babyboot orchid, Lepanthes rubripetala) do not differ from the other herbs in their reproductive strategy axis scores.
Phylogenetic relationships play a rather weak role in explaining the repertoire of life-history strategies. In our analysis, Pagel’s λ, a scaling parameter for the correlation in traits between species ranging from 0 (no correlation) to 1 (the correlation expected under Brownian motion) (30) is 0.20 ± 0.09 (95% confidence interval), suggesting a rather minor role of overall phylogenetic ancestry in our analyses. However, some exceptions exist: Species in the Magnoliopsida have lower fast–slow scores (shorter lives, higher growth) than Cycadophyta and Pinopsida. The phylogenetic signal of species within the same taxonomic class (Liliopsida: 0.18 ± 0.02, Magnoliopsida: 0.20 ± 0.04) is greater than those grouped by growth forms (herbs: 0.03, shrubs: 0.00, or trees: 0.00) (SI Appendix, Table S8), implying some infra-class structuring of life-history strategies.
Life-History Strategies May Overlap Regardless of Plant Growth Form and Size.
Although Raunkiær’s growth forms (27) take somewhat different positions along the fast–slow axis and reproductive strategy axis, the overlap is considerable, so that species with different growth forms may occupy the same life-history space. Similar survival, growth, and reproduction schedules can be realized through different anatomic structures. For example, shorter-lived trees and shrubs (such as Cecropia obtusifolia and Acer saccharum) (Fig. 2B) occupy a life-history space on the fast–slow axis that overlaps with herbaceous perennials (particularly helophytes, geophytes, and epiphytes) (Fig. 2B). The life-history strategies of herbs range from short-lived ephemerals to the tree-like lifestyles of the cushion pink (Silene acaulis) or thyme (Thymus vulgaris).
The amount of variation shown on the reproductive strategy axis for herbs and trees is similar. Both groups display a similar range of life histories in the timing and frequency of reproduction (iteroparity) and lifetime reproductive potential, regardless of their position on the fast–slow axis (Fig. 2). For example, the reproductive strategy axis values of short-lived herbs, such as goldenrod (Solidago mollis) or scarlet monkeyflower (Mimulus cardinalis) are similar to those of woody species such as black pine, scotch broom (Cytisus scoparius), or hoop pine (Araucaria cunninghamii).
In contrast to comparative animal demography (11, 25), a uniform measure of “body size” does not exist for plants across the plant kingdom. Therefore we have presented the results (Fig. 2 and Table 1) without allometric scaling. However, Raunkiær growth forms clearly differ in size, among other crucial functional attributes. By using Raunkiær growth form height thresholds, we have attempted to include size more explicitly in the analyses (SI Appendix, Fig. S4 and Tables S1, S12, and S13). When life-history traits are rescaled by plant height, the results do not change qualitatively. The amount of variation explained by PCA 1 (32.99%) and PCA 2 (19.73%), which also corresponds to the fast–slow continuum and to reproductive strategies, respectively, adds ca. 53%. This result suggests either that plant size does not have a strong effect in the structuring of plant life histories or that using Raunkiær growth form height thresholds as our proxy of plant size is not accurate enough. Compared with the results with no allometric scaling (Fig. 2 and Table 1), the phylogenetic signal is completely lost (Pagel’s λ = 0.00), likely because Raunkiær’s growth forms are phylogenetically conserved in our data (λ = 0.96 ± 0.02, P < 0.001).
Two Orthogonal Axes to Predict Population Performance.
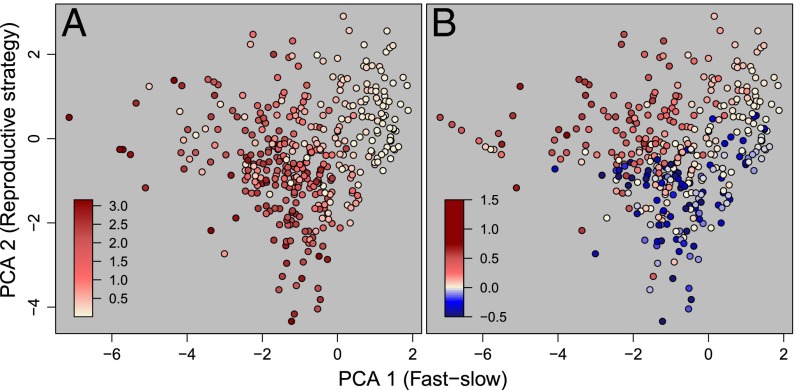
The fast–slow/reproductive strategy framework predicts population performance, including both short-term response to perturbation and long-term asymptotic dynamics (Fig. 3). Although many metrics of short-term (transient) dynamics are available (31, 32), we use the damping ratio here to illustrate the predictive capacity of our life-history framework. The damping ratio can be thought of as the rate at which transient responses to disturbance fade away or as the rate of recovery from asymptotic dynamics (22). A species’ rate of recovery is associated with its scores on the fast–slow (F1, 384 = 96.99, P < 0.001) and the reproductive strategy axis (F1, 384 = 53.3, P < 0.001). Natural populations with faster recovery are found at the upper left and bottom of Fig. 3A, suggesting that rapid recovery can be attained via a strategy of fast growth, high reproduction, and short generation time or, alternatively via a strategy of low reproduction and frequent shrinkage (33). Asymptotic population growth rates [r = log(λ)] are strongly differentiated along both axes (fast–slow axis: F1, 374 = 145.79, P < 0.001; reproductive strategy axis: F1, 374 = 177.80, P < 0.001), with high population growth rates for fast-growing (γ), iteroparous, highly reproductive species (Φ, R0) (Fig. 3B). Lower population growth rates are typical of species that delay maturity (Lα), have low senescence rates as described by their survivorship curve (H), and/or undergo frequent shrinkage (ρ).
Fig. 3.
A species’ score on the fast–slow continuum and its reproduction strategy predict population performance including damping ratio (i.e., the rate at which a population returns to equilibrium after disturbance) (A) and the population growth rate (r = log(λ) (i.e., the rate of population size change through time) (B). Redder tones indicate a higher value of these metrics. Bluer colors of r reflect population decline. The damping ratio was quantified for 389 species.
Discussion
Quantifying Life-History Strategies.
The diversity of growth forms, functions, and ecological roles of species have long puzzled biologists. A key question has been whether there are key combinations of survival, growth, and reproduction strategies that can exist only in certain habitats but not in others (9) or whether nature is a random assemblage of traits (34). To address these questions, several frameworks have been developed that aim to classify and predict species’ responses to biotic and abiotic agents (9). Perhaps the most widely acknowledged framework in this respect is the fast–slow continuum (11), which states that, because trade-offs between reproduction and survival are pervasive, the repertoire of life-history strategies are constrained and thus can be described accurately along a single axis with high allocation to reproduction on one end and high allocation to survival on the other. Although the fast–slow continuum has received substantial empirical support, explaining 60–80% of the variation among mammals (25, 35, 36), birds (37), and reptiles (38), analyses going back over 30 years also have pointed out the existence of a secondary axis related to reproductive strategies. For instance, Stearns (20) found that although 68–75% of the covariation in life-history traits of 162 mammals is explained by the first axis, corresponding to the fast–slow continuum, an important second axis describing a continuum from altricial to precocial species explains an additional 12–20%. Gaillard et al. (39) found for 80 mammals and 114 birds that 74–85% of variation is explained by the fast–slow axis but that a second important axis related to iteroparity absorbs 5–15% of the variation.
The results of our global analysis of more than 400 plant species are qualitatively similar to these studies in vertebrates (20, 35–40) albeit suggesting a greater relative importance of the reproductive strategy axis. We find two independent axes of life-history variation in plants, one corresponding to the fast–slow continuum and another to characteristics of reproductive strategy not captured by mean sexual reproduction. When we account for the potential allometric effects of size in these relationships, the percentage of variation explained decreased only minimally (1.1% for PCA 1, and 1.6% for PCA 2), and the phylogenetic signal remained low. In contrast to analogous comparative approaches for animals (20, 39, 40), we find very little phylogenetic signal in our results, nor do we find indications that adult size has a structurally important role.
Typically around 80% of the variation in animal life-history strategies can be captured with two axes, but here we captured just over 50% of the observed variation. The reason for this difference might be that plants typically are characterized by more complex life cycles than vertebrates. For instance, plants often have dormant stages (14) and long-term seedbanks (13); animals usually do not. Furthermore, in contrast to many of the species considered in these animal-based studies, all plants are indeterminate growers (41) in which cellular fate is not determined early in life, so the allocation of meristems to survival (e.g., wood), growth (leaf), or reproduction (flower), as well as overall plant size, can be adjusted continuously. This totipotency has resulted in strategies such as resource-dependent sex-switching (42) and the rejuvenating abilities of some trees (43, 44). Furthermore, all vascular plants are modular constructions based on the repetition of basic units (45–47), enabling some plants to shrink in adverse conditions (33, 47) or to reproduce clonally (48). We find that retrogressive growth (shrinkage) correlates negatively with reproductive traits, in agreement with the frequent increase in reproductive output with plant size (41). However, these complex life-history traits are not exclusive to the plant kingdom; many animals experience dormancy [i.e., hibernation (49), diapause (50), estivation (51), or brumation (52)], clonal reproduction (53), organ/tissue regeneration (54), or modular growth forms [e.g., corals (55)]. Demographic comparative analyses including complex life-history traits across both plant and animal kingdoms will help determine whether, and for which taxa, multiple axes are needed to capture interspecific patterns of life-history variation.
Life-History Analyses and Population Performance.
The life-history traits analyzed here are derived from natural populations examined in the field, and these studies therefore capture population performance as a product of life-history strategy and the particular a/biotic conditions experienced by that population over the course of the study. Clearly, no species can persist indefinitely with populations operating at a population growth rate log(λ) <0. Furthermore, some areas of life-history space remain unfilled; in the 418 plant species of our dataset there are no species with low scores on both the fast–slow axis and the reproductive strategy axis (Fig. 3B, Lower Left), or with high fast–slow scores but low reproductive strategy scores (Fig. 3B, Lower Right), suggesting that such combinations of life-history traits are unviable. Interestingly, we have found species with high scores on both axes (Fig. 3B, Upper Right). Rather than defying basic life-history trade-offs, these species likely represent very successful cases of expanding populations. Several of these species correspond to invasive plants such as black pine in New Zealand (56) and scotch broom in Australia (57). The reproductive strategy axis includes populations of invasive species at the top, where the population growth rate log(λ) >>0 (Fig. 3B), and endangered species such as the fragrant prickly apple (Harrisia fragrans) or Mead’s milkweed (Asclepias meadii) at the low end (Fig. 2B). Given the restricted spatial replication of plant demographic studies (21), we are unable to discern how much the values on the reproductive strategy axis and low population growth rates are driven by habitat quality or other conditions favoring population growth, and this question remains a promising avenue of research. Also, future steps in the applicability of this framework need to focus on the classification of endangered and invasive species along this axis and to take advantage of open access resources (15, 58) to discern the role of propagule quantity vs. quality [e.g., seed mass (59)] in structuring the reproduction strategy axis.
Population responses to future environmental change and anthropogenic disturbances depend on the species-specific life-history strategy (60, 61). Our analyses reveal that populations from even distantly related plant taxa worldwide can have similar combinations of life-history traits, with a modest influence of habitat and growth form. Therefore, the framework of life histories presented here is a necessary addition to current plant trait-based concepts such as the leaf (62) and wood (63) economics spectra, because traits can be considered truly functional only if they affect the critical fitness components of reproduction and survival (64). This framework, based on the fast–slow continuum and reproductive strategies, presents strong empirical support for the expansion of classical quantification and classifications of life-history strategies of animals well into the plant kingdom. Furthermore, it provides a sound basis for future work untangling the associations in plant and animal functional traits with demographic processes and among physiological and life-history trade-offs.
Materials and Methods
COMPADRE.
We used the COMPADRE Plant Matrix Database (21) to obtain demographic, biogeographic, and growth form data from an initial list of more than 1,000 plant species. The demographic data therein are compiled as state-structured population models that incorporate accurate information on the rates of survival, growth, and reproduction from natural populations in which individuals are typically classified by stage and/or size (22). We only considered whole individual (genetic) demography and omitted studies that treated different parts of the genetic individual as independent units (ramets). Nonnatural vegetation types such as forestry plantations and crop fields were not included. We chose only size-based matrices or ontogeny-based models in which higher stages of development also would correspond to larger sizes. With these and other strict selection criteria used to allow comparative analyses (SI Appendix, section 1.1 and Table S1), we narrowed our initial list to 418 plant species. For each of these species, we calculated the arithmetic element-by-element mean of all available matrices under nonmanipulated conditions, resulting in a single matrix that summarizes the population dynamics of that species under natural conditions.
Phylogeny.
We constructed a species-level phylogenetic tree [www.onezoom.org/FWifhj38wjf/Salguero-Gomez_et_al_2014.htm (65)] with branch length transformations to account for phylogenetic signal, estimated by Pagel’s λ (30). See SI Appendix, section 2 for details.
Analysis.
From each species’ matrix population model, we derived nine life-history traits commonly used in comparative analyses grouped a priori according to their quantification of the timing and magnitude of turnover, longevity, growth, and reproduction (11, 16, 22, 35–40, 67). We calculated these with methods described in detail elsewhere (22) and in SI Appendix, section 2, Table S2. The life-history traits broadly correspond to overall population turnover (T), longevity (H and Lα), growth (γ and ρ), and reproduction (φ, S, Ro, Lω) (Table 1). Life-history traits were log-transformed to fulfill normality assumptions in posterior analyses. Life-history traits were scaled to mean = 0 and SD = 1 for PCA (24). We then carried out a phylogenetically informed PCA (23, 66) on these life-history traits to determine the primary axes of demographic variation while simultaneously assessing nonindependence of lineages. We used the Kaiser criterion (23) after optimization through varimax rotations to determine the number of axes necessary to explain a substantial amount of variation. To explore the role and possible interactions of growth form, matrix dimension (68), and habitat, we used a three-way ANOVA followed by post hoc Tukey's honestly significant difference tests on the phylogenetically informed PCA scores of the species. The major habitat classification (28) informs on the abiotic conditions to which populations are exposed, and the growth form information describes potential anatomical constraints. We used Raunkiær’s growth form classification (27), indicating the distance of the plant’s shoot apical meristems from the ground. Matrix dimension was positively correlated with PCA 1 (t417 = 85.51, P < 0.001, R2 = 16.85%) and PCA 2 (t417 = 17.72, P < 0.001, R2 = 3.85%). However, this effect was driven by the fact that long-lived trees achieve larger sizes (29) and thus require larger matrices to accommodate their dynamics: the ordered ranks of Raunkiær growth forms successfully predicted PCA scores on both PCA 1 (F7, 410 = 34.40, P < 0.001, R2 = 35.93%) and PCA 2 (F7, 410 = 12.13, P < 0.001, R2 = 15.74%) (SI Appendix, Table S5). We checked the consistency of our results by rerunning the analyses on subsets of the data: by plant type (herbaceous perennials, shrubs, and trees), major habitat (temperate, tropical, and subtropical), and taxonomic class (Pinopsida, Liliopsida, Magnoliopsida); other subsets and levels were not tested because of the large data requirements for model convergence. We also tested the robustness of the results to spurious correlations using randomization tests. Finally, to test the usefulness of the suggested framework for plant species classification, we derived the damping ratio [the rate at which populations recover from disturbance (22, 31)] and the rate of change of the population (22) [r = log(λ)] via two-way ANOVAs with PCA 1 and 2 scores as explanatory variables.
Supplementary Material
Acknowledgments
M. Franco provided the phylogenetic tree. We thank H. Possingham, D. Koons, and F. Colchero for feedback and the COMPADRE Plant Matrix Database team for data digitalization and error-checking. This work was supported by the Max Planck Institute for Demographic Research, Australian Research Council Grant DE140100505 (to R.S.-G.), and a Marie-Curie Career Integration Grant (to Y.M.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.C. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the COMPADRE Plant Matrix Database, www.compadre-db.org (accession no. 3.0.0).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506215112/-/DCSupplemental.
References
- 1.Morris WF, Doak DF. Quantitative Conservation Biology: Theory and Practice of Population Viability Analysis. Sinauer Associates; Sunderland, MA: 2002. [Google Scholar]
- 2.Silvertown J, Miguel F, Menges E. Interpretation of elasticity matrices as an aid to the management of plant populations for conservation. Conserv Biol. 1996;10(2):591–597. [Google Scholar]
- 3.Jongejans E, de Vere N, de Kroon H. Demographic vulnerability of the clonal and endangered meadow thistle. Plant Ecol. 2008;198(2):225–240. [Google Scholar]
- 4.Svenning JC, Kinner DA, Stallard RF, Engelbrecht BMJ, Wright SJ. Ecological determinism in plant community structure across a tropical forest landscape. Ecology. 2004;85(9):2526–2538. [Google Scholar]
- 5.West GB, Enquist BJ, Brown JH. A general quantitative theory of forest structure and dynamics. Proc Natl Acad Sci USA. 2009;106(17):7040–7045. doi: 10.1073/pnas.0812294106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chesson P. Mechanisms of maintenance of species diversity. Annu Rev Ecol Syst. 2000;31(1):343–366. [Google Scholar]
- 7.Blair C, Heckman KL, Russell AL, Yoder AD. Multilocus coalescent analyses reveal the demographic history and speciation patterns of mouse lemur sister species. BMC Evol Biol. 2014;14(1):57. doi: 10.1186/1471-2148-14-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Metcalf CJE, Pavard S. Why evolutionary biologists should be demographers. Trends Ecol Evol. 2007;22(4):205–212. doi: 10.1016/j.tree.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Southwood TRE. Tactics, strategies and templets. Oikos. 1988;52(1):3–18. [Google Scholar]
- 10.Lande R. A quantitative genetic theory of life history evolution. Ecology. 1982;63(3):607–615. [Google Scholar]
- 11.Stearns SC. The Evolution of Life Histories. Oxford Univ Press; New York: 1999. p. 249. [Google Scholar]
- 12.Peñuelas J, Munné-Bosch S. Potentially immortal? New Phytol. 2010;187(3):564–567. doi: 10.1111/j.1469-8137.2010.03360.x. [DOI] [PubMed] [Google Scholar]
- 13.Baskin CC, Baskin JM. Seeds. Ecology, Biogeography, and Evolution of Dormancy and Germination. Academic; San Diego, CA: 2001. p. 667. [Google Scholar]
- 14.Shefferson RP. The evolutionary ecology of vegetative dormancy in mature herbaceous perennial plants. J Ecol. 2009;97(5):1000–1009. [Google Scholar]
- 15.Kattge J, et al. TRY - a global database of plant traits. Glob Change Biol. 2011;17(9):2905–2935. [Google Scholar]
- 16.Franco M, Silvertown J. Plant demography: What do we know. Evolutionary Trends in Plants. 1990;4(2):74–76. [Google Scholar]
- 17.Franco M, Silvertown J. Life History Variation in Plants: An Exploration of the Fast-Slow Continuum Hypothesis. Cambridge Univ Press; Cambridge, UK: 1997. pp. 210–227. [Google Scholar]
- 18.Shea K, Rees M, Simon NW. Trade-offs, elasticities and the comparative method. J Ecol. 1994;82(4):951–957. [Google Scholar]
- 19.Franco M, Silvertown J. On trade-offs, elasticities and the comparative method: A reply to Shea, Rees and Wood. J Ecol. 1994;82(4):958. [Google Scholar]
- 20.Stearns SC. The influence of size and phylogeny on paterns of covariation among life-history traits in the mammals. Oikos. 1983;41(2):173–187. [Google Scholar]
- 21.Salguero-Gómez R, et al. The COMPADRE Plant Matrix Database: An open online repository for plant demography. J Ecol. 2015;103(1):202–208. [Google Scholar]
- 22.Caswell H. 2001. Matrix Population Models: Construction, Analysis, and Interpretation (Sinauer Associates, Inc.) 2nd Ed.
- 23.Revell LJ. 2013. R package phytools Available at blog.phytools.org. Accessed March 1, 2015.
- 24.Legendre P, Legendre L. Numerical Ecology. 3rd Ed. Elsevier; London: 2012. p. 1006. [Google Scholar]
- 25.Gaillard JM, et al. Generation time: A reliable metric to measure life-history variation among mammalian populations. Am Nat. 2005;166(1):119–123, discussion 124–128. doi: 10.1086/430330. [DOI] [PubMed] [Google Scholar]
- 26.Crone EE, et al. How do plant ecologists use matrix population models? Ecol Lett. 2011;14(1):1119–1126. doi: 10.1111/j.1461-0248.2010.01540.x. [DOI] [PubMed] [Google Scholar]
- 27.Raunkiær C. The Life Forms of Plants and Statistical Plant Geography. Clarendon; Oxford, UK: 1934. [Google Scholar]
- 28.Olson DM, et al. Terrestrial ecoregions of the worlds: A new map of life on Earth. Bioscience. 2001;51(11):933–938. [Google Scholar]
- 29.Moles AT, et al. Global patterns in plant height. J Ecol. 2009;97(5):923–932. [Google Scholar]
- 30.Freckleton RP, Harvey PH, Pagel M. Phylogenetic analysis and comparative data: A test and review of evidence. Am Nat. 2002;160(6):712–726. doi: 10.1086/343873. [DOI] [PubMed] [Google Scholar]
- 31.Stott I, Franco M, Carslake D, Townley S, Hodgson D. Boom or burst? A comparative analysis of transient population dynamics in plants. J Ecol. 2010;98(2):302–311. [Google Scholar]
- 32.Gamelon M, et al. Influence of life-history tactics on transient dynamics: A comparative analysis across mammalian populations. Am Nat. 2014;184(5):673–683. doi: 10.1086/677929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salguero-Gómez R, Casper BB. Keeping plant shrinkage in the demographic loop. J Ecol. 2010;98(2):312–323. [Google Scholar]
- 34.Hubbell SP. The Unified Neutral Theory of Biodiversity and Biogeography. Princeton Univ Press; Princeton, NJ: 2001. [DOI] [PubMed] [Google Scholar]
- 35.Oli MK. The fast-slow continuum and mammalian life-history patterns: An empirical evaluation. Basic Appl Ecol. 2004;5(5):449–463. [Google Scholar]
- 36.Bielby J, et al. The fast-slow continuum in mammalian life history: An empirical reevaluation. Am Nat. 2007;169(6):748–757. doi: 10.1086/516847. [DOI] [PubMed] [Google Scholar]
- 37.Sæther BE. The influence of body weight on the covariation between reproductive traits in European birds. Oikos. 1987;48(1):79–88. [Google Scholar]
- 38.Bauwens D, Diaz-Uriarte R. Covariation of life-history traits in lacertid lizards: A comparative study. Am Nat. 1997;149(1):91–111. [Google Scholar]
- 39.Gaillard JM, et al. An analysis of demographic tactics in birds and mammals. Oikos. 1989;56(1):59–76. [Google Scholar]
- 40.Western D. Size, life history and ecology in mammals. Afr J Ecol. 1979;17(4):185–204. [Google Scholar]
- 41.Harper JL, White J. The demography of plants. Annu Rev Ecol Syst. 1974;5:419–463. [Google Scholar]
- 42.Bierzychudek P. The demography of Jack-in-the-pulpit, a forest perennial that changes sex. Ecol Monogr. 1982;52(4):335–351. [Google Scholar]
- 43.Del Tredici P. Aging and rejuvenation in trees. Arnoldia. 1999;(Winter):11–16. [Google Scholar]
- 44.Chen YT, et al. Small RNAs of Sequoia sempervirens during rejuvenation and phase change. Plant Biol (Stuttg) 2013;15(1):27–36. doi: 10.1111/j.1438-8677.2012.00622.x. [DOI] [PubMed] [Google Scholar]
- 45.Watkinson AR, White J. Some life-history consequences of modular construction in plants. Philos Trans R Soc Lond B Biol Sci. 1986;313(1159):31–51. [Google Scholar]
- 46.Espino S, Schenk HJ. Hydraulically integrated or modular? Comparing whole-plant-level hydraulic systems between two desert shrub species with different growth forms. New Phytol. 2009;183(1):142–152. doi: 10.1111/j.1469-8137.2009.02828.x. [DOI] [PubMed] [Google Scholar]
- 47.Salguero-Gómez R, Casper BB. A hydraulic explanation for size-specific plant shrinkage: Developmental hydraulic sectoriality. New Phytol. 2011;189(1):229–240. doi: 10.1111/j.1469-8137.2010.03447.x. [DOI] [PubMed] [Google Scholar]
- 48.de Kroon H, van Groenendael J. The Ecology and Evolution of Clonal Growth in Plants. Backhuys Publishers; Leiden, The Netherlands: 1997. [Google Scholar]
- 49.Tøien Ø, et al. Hibernation in black bears: Independence of metabolic suppression from body temperature. Science. 2011;331(6019):906–909. doi: 10.1126/science.1199435. [DOI] [PubMed] [Google Scholar]
- 50.Sandrelli F, et al. A molecular basis for natural selection at the timeless locus in Drosophila melanogaster. Science. 2007;316(5833):1898–1900. doi: 10.1126/science.1138426. [DOI] [PubMed] [Google Scholar]
- 51.Besansky NJ. Malaria: How vector mosquitoes beat the heat. Nature. 2014;516(7531):334–336. doi: 10.1038/nature14073. [DOI] [PubMed] [Google Scholar]
- 52.Brasfield SM, Talent LG, Janz DM. Reproductive and thyroid hormone profiles in captive Western fence lizards (Sceloporus occidentalis) after a period of brumation. Zoo Biol. 2008;27(1):36–48. doi: 10.1002/zoo.20159. [DOI] [PubMed] [Google Scholar]
- 53.Matsuura K, et al. Queen succession through asexual reproduction in termites. Science. 2009;323(5922):1687. doi: 10.1126/science.1169702. [DOI] [PubMed] [Google Scholar]
- 54.Sánchez Alvarado A. Developmental biology: A cellular view of regeneration. Nature. 2009;460(7251):39–40. doi: 10.1038/460039a. [DOI] [PubMed] [Google Scholar]
- 55.Hall VR, Hughes TP. Reproductive strategies of modular organisms: Comparative studies of reef-building corals. Ecology. 1996;77(3):950–963. [Google Scholar]
- 56.Buckley YM, et al. Slowing down a pine invasion despite uncertainty in demography and dispersal. J Appl Ecol. 2005;42(6):1020–1030. [Google Scholar]
- 57.Neubert MG, Parker IM. Projecting rates of spread for invasive species. Risk Anal. 2004;24(4):817–831. doi: 10.1111/j.0272-4332.2004.00481.x. [DOI] [PubMed] [Google Scholar]
- 58.Moles AT, Westoby M. Seed survival and seed size: A synthesis of the literature. J Ecol. 2004;92:372–383. [Google Scholar]
- 59.Rees M. Trade-offs among dispersal strategies in British plants. Nature. 1993;366(6451):150–152. [Google Scholar]
- 60.Morris WF, et al. Longevity can buffer plant and animal populations against changing climatic variability. Ecology. 2008;89(1):19–25. doi: 10.1890/07-0774.1. [DOI] [PubMed] [Google Scholar]
- 61.Doak DF, Morris WF. Demographic compensation and tipping points in climate-induced range shifts. Nature. 2010;467(7318):959–962. doi: 10.1038/nature09439. [DOI] [PubMed] [Google Scholar]
- 62.Wright IJ, et al. The worldwide leaf economics spectrum. Nature. 2004;428(6985):821–827. doi: 10.1038/nature02403. [DOI] [PubMed] [Google Scholar]
- 63.Chave J, et al. Towards a worldwide wood economics spectrum. Ecol Lett. 2009;12(4):351–366. doi: 10.1111/j.1461-0248.2009.01285.x. [DOI] [PubMed] [Google Scholar]
- 64.Adler PB, et al. Functional traits explain variation in plant life history strategies. Proc Natl Acad Sci USA. 2014;111(2):740–745. doi: 10.1073/pnas.1315179111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosindell J, Harmon LJ. OneZoom: A fractal explorer for the tree of life. PLoS Biol. 2012;10(10):e1001406. doi: 10.1371/journal.pbio.1001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Revell LJ. Size-correction and principal components for interspecific comparative studies. Evolution. 2009;63(12):3258–3268. doi: 10.1111/j.1558-5646.2009.00804.x. [DOI] [PubMed] [Google Scholar]
- 67.Dunham AE, Miles DB. Patterns of covariation in life history traits of squamate reptiles: The effects of size and phylogeny reconsidered. Am Nat. 1985;126:231–257. [Google Scholar]
- 68.Salguero-Gómez R, Plotkin JB. Matrix dimensions bias demographic inferences: Implications for comparative plant demography. Am Nat. 2010;176(6):710–722. doi: 10.1086/657044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.