Abstract
Ypt/Rab GTPases are conserved molecular switches that regulate the multiple vesicular transport steps of all intra-cellular trafficking pathways. They are stimulated by guanine-nucleotide exchange factors (GEFs). In yeast, Ypt1 regulates transport from the endoplasmic reticulum (ER) to two alternative pathways: secretion and autophagy. Ypt1 is activated by TRAPP, a modular multi-subunit GEF. Whereas TRAPP I activates Ypt1 to mediate transport through the Golgi, TRAPP III, which contains all the subunits of TRAPP I plus Trs85, activates Ypt1-mediated transport to autophagosomes. The functional pair Ypt31/32 regulates traffic in and out of the trans Golgi and is activated by TRAPP II, which consists of TRAPP I plus two specific subunits, Trs120 and Trs130. To study the interaction of Ypts with specific TRAPP subunits and interactions between the different subunits of TRAPP, including the cellular sites of these interactions, we have employed a number of approaches. One approach that we have recently optimized for the use in yeast is multi-color bimolecular fluorescence complementation (BiFC). BiFC, which employs split fluorescent tags, has emerged as a powerful approach for determining protein interaction in vivo. Because proteins work in complexes, the ability to determine more than one interaction at a time using multi-color BiFC is even more powerful. Defining the sites of protein interaction is possible by co-localization of the BiFC puncta with compartmental markers. Here, we describe a set of plasmids for multi-color BiFC optimized for use in yeast. We combined their use with a set of available yeast strains that express red fluorescence compartmental markers. We have recently used these constructs to determine Ypt1 and TRAPP interactions in two different processes: intracellular trafficking and autophagy.
Keywords: BiFC, Multi-color BiFC, In vivo interaction, Ypt/Rab GTPase, TRAPP, Trs85, Atg11, Trs20, Trs120, Ypt1
1. Introduction
In the last two decades it became apparent that proteins that mediate and regulate intra-cellular trafficking function in large complexes that can be thought of as “molecular machines”. For example, each Ypt/Rab GTPase interacts with upstream regulators, e.g., GEFs, and multiple downstream effectors to regulate and coordinate vesicular trafficking (1, 2). In many cases, both the Ypt/Rab GEFs and their effectors are multiple-protein complexes. These complexes are conserved both among organisms and between the different steps of the intra-cellular pathways.
In yeast, the Ypt1 GTPase regulates two alternative intra-cellular trafficking pathways: in the exocytic pathway it is required for endoplasmic reticulum (ER)-to-Golgi transport, whereas in the autophagic pathway it is required for formation of autophagosomes (APs) (3, 4). The modular complex TRAPP acts as the Ypt1 GEF in both pathways, but in different configurations. The multi-subunit TRAPP I activates Ypt1 in the exocytic pathway and TRAPP III, which in addition to the TRAPP I contains Trs85, activates Ypt1 in autophagy (4–6). In autophagy, activated Ypt1 interacts with its effector Atg11 to form the multi-protein complex pre-autophagosomal structure (PAS) (4). A third TRAPP complex, TRAPP II, activates Ypt31 at the trans Golgi (6). All these players and their functions are conserved from yeast to humans (7–9).
While in-vitro analyses are important for defining protein interactions and complex composition, in vivo studies are crucial for providing the physiological relevance of such analyses and for determining the intra-cellular sites where these interactions occur. One such approach is live-cell microscopy. We have used co-localization analyses of fluorescently tagged Ypts, TRAPP subunits and Ypt effectors to address multiple questions about Ypts and their accessory factors. This approach is even more effective when combined with mutations. For example, using a combination of in vitro and co-localization analyses in wild-type and mutant cells, we have recently shown that the TRAPP III-specific subunit Trs85 is required for recruitment of TRAPP I to PAS via the interaction of Trs85 with the TRAPP subunit Trs20 (10).
Bimolecular fluorescence complementation (BiFC) has emerged as a powerful approach for determining interaction between two proteins in vivo (11). We have used this approach in combination with in vitro studies to determine interactions between two TRAPP subunits, Trs20 and the TRAPP II-specific subunit Trs120 (12). Multi-color BiFC allows to determine interactions of one protein with two other proteins (13). We have used this approach in combination with in vitro studies to determine interactions of Ypt1 with the TRAPP III-specific subunit Trs85 and with its autophagy-specific effector Atg11 (4). In both cases, the effect of mutations on BiFC and co-localization of the BiFC puncta with compartmental markers were used to support the BiFC results. To do this, we constructed a set of plasmids optimized for the use of multi-color BiFC in yeast (4). These plasmids were used in combination with an available set of yeast strains that express RFP-tagged compartmental markers (14). Here, we describe the sets of plasmids and strains optimized for using multi-color BiFC interactions and their intra-cellular localization in yeast, recount examples of their use for determining Ypt1 and TRAPP interactions, and discuss important controls and limitations of this approach.
2. Materials
We designed six plasmids that can be used for tagging your proteins of interest for multi-color BiFC analysis in a versatile manner (Table 1). Some of these plasmids were previously used for cloning Ypt1, Atg11 and TRAPP subunits (4, 12). All plasmids are CEN (low copy number), and the tagged proteins are expressed from the constitutive ADH1 promoter and have the CYC1 terminator. The two split fluorophores that we chose are yeast codon-optimized enhanced Venus, yEVenus, because it is the fastest maturing yellow fluorescent protein (YFP) and is yeast-codon optimized (15), and Cerulean, because it is the “bluest” cyan fluorescent protein (CFP) and, importantly, its fluorescence can be separated easily from that of Venus (16). Typically, 6–11 amino acids linkers were designed between the fluorophore fragment and the protein.
Table 1.
A list of plasmids constructed for the use of multi-color BiFC analysis in yeast
| Plasmid | Alias | Used for | Genotype | Source |
|---|---|---|---|---|
| pNS1499 | p416-YFP-N for N | Tag N terminus with YFP-N | CEN, URA3, Ampr | this study |
| pNS1500 | p413-YFP-N for N | Tag N terminus with YFP-N | CEN, HIS3, Ampr | this study |
| pNS1501 | p415-CFP-N for C | Tag C terminus with CFP-N | CEN, LEU2, Ampr | this study |
| pNS1502 | p415-YFP-N for C | Tag C terminus with YFP-N | CEN, LEU2, Ampr | this study |
| pNS1503 | p416-Y/CFP-C for N | Tag N terminus with Y/CFP-C | CEN, URA3, Ampr | this study |
| pNS1504 | p413-Y/CFP-C for N | Tag N terminus with Y/CFP-C | CEN, HIS3, Ampr | this study |
The first two plasmids, pNS1499 and pNS1500, are for tagging proteins at their N terminus with YFP-N: The N terminus of yEVenus, amino acids 1–172, is followed by a multiple cloning site (MCS). These plasmids contain the URA3 and HIS3 selectable markers, respectively. To generate pNS1499, the VF1 fragment in p416-VF1 (17) was replaced by the fragment encoding amino acids 1–172 of yEVenus, which was amplified from pKT103 (15), using the SpeI/XbaI and BspEI restriction sites. To construct pNS1500, the piece containing the ADH1 promoter, amino acids 1–172 of yEVenus, and the CYC1 terminator from pNS1499 was sub-cloned into pRS413 using the PvuII sites.
The next two plasmids, pNS1501 and pNS1502, are for tagging proteins at their C terminus with the N terminus (amino acids 1–172) of Cerulean or yEVenus, respectively. In both plasmids, the MCS is at the N terminus of the fluorophore fragment, and the protein should be cloned without its stop codon. The selectable marker for both plasmids is LEU2. To construct pNS1501 and pNS1502, the VF2 fragment of p415-VF2 (17) was replaced by amino acids 1–172 of Cerulean (16) or yEVenus, respectively, using the BspEI and XhoI sites.
The last two plasmids, pNS1503 and pNS1504, are for tagging proteins at their N terminus with the C terminus of yECFP, amino acids 155–238. Because this fragment does not affect the BiFC color it is termed Y/CFP-C, and it is followed by a MCS. The selectable markers are URA3 and HIS3, respectively. To construct pNS1503, the VF1 fragment in p416-VF1 (17) was replaced by amino acids 155–238 of yECFP, amplified from pKT102 (15), using the SpeI/XbaI and BspEI sites. To generate pNS1504, the piece containing the ADH1 promoter, amino acids 155–238 of yECFP, and the CYC1 terminator from pNS1503 was sub-cloned into pRS413 using the PvuII sites.
3. Methods
We have successfully employed BiFC and multi-color BiFC to determine interactions between TRAPP II subunits and of Ypt1 GTPase with its autophagy-specific GEF and effector, including determination of the sites of these interactions. To achieve that, we constructed a set of plasmids (See Materials) and used yeast strains, which were previously employed for GFP-tagged protein localization, for BiFC localization. Both plasmids and strains should be useful for determining other interactions in yeast. The multi-color BiFC principle, rationale of plasmid optimization, BiFC localization, and examples and important controls are described below. Please, see Notes 1–3 for BiFC limitations.
3.1 Multi-color BiFC Principle
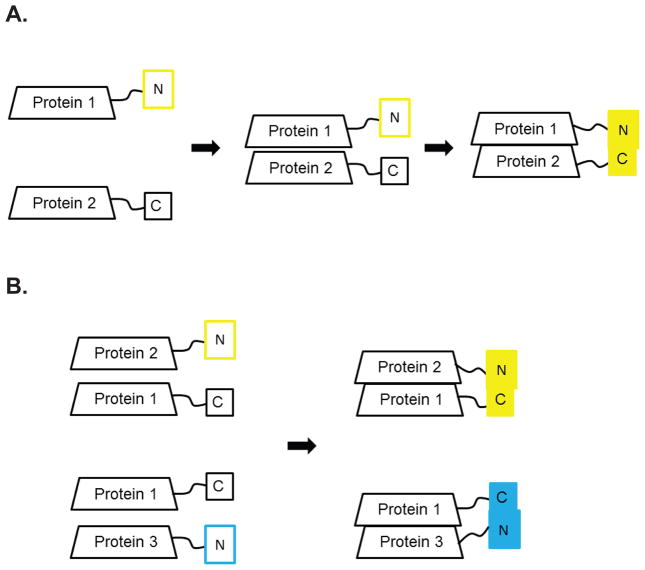
BiFC is a protein fragment complementation assay (PCA). In PCA, two proteins are tagged with two fragments of a reporter, an enzyme or a fluorophore, and the reporter can assemble only if the two proteins interact. The readout of the interaction depends on the nature of the reporter (18). For BiFC analyses, a fluorophore, YFP or CFP, is split into its N and C terminal fragments and each fragment is fused to two different proteins of interest. The two tagged proteins are co-expressed in the same cell. The two fragments can reconstruct the fluorophore only if the two proteins are adjacent and the readout is fluorescence (Figure 1A). Because the C terminal fragment of the YFP and CFP proteins are identical, the N terminal fragment determines the excitation/emission range or color of the fluorophore. The C-terminal Y/CFP fragment will fluoresce when adjacent to either the N-terminal YFP or CFP fragment (Figure 1B). Exploiting this property, interactions of one protein with two other proteins can be visualized using multi-color BiFC.
Figure 1. The principles of BiFC and multi-color BiFC.
A. BiFC analysis between two proteins. Protein 1 and Protein 2 are tagged with the N and C fragments of YFP, respectively. The YFP fluorophore can assemble and fluoresce only if Proteins 1 and 2 come in close proximity. B. Multi-color BiFC analysis of three proteins. Protein 1 is tagged with the C terminal fragment of a fluorophore, which can assemble with the N terminus of either YFP or CFP. Proteins 2 and 3 are tagged with the N terminal fragment of YFP and CFP, respectively. When Protein 1 interacts with Protein 2, the assembled fluorophore fluoresces in the YFP channel. When Protein 1 interacts with Protein 3, the assembled fluorophore fluoresces in the CFP channel. The proteins can be tagged with the fluorophore fragment at either terminus (see text).
3.2 Optimized plasmids for use in yeast
The optimization was done for three purposes. First, the set of plasmids described here was designed for allowing the use of multi-color BiFC in yeast while improving available split YFP BiFC. In addition to construction of plasmids for tagging proteins with split CFP, the following changes were made to the previously reported split yEVenus (17): The length of the split Venus fragments, VF1 and VF2, was changed to allow better assembly of the fluorophore (11). In addition, the C terminal fragment of the fluorophore was cloned from the version of yEVenus that contains the A206K alteration (15). This mutation results in a monomeric fluorescent protein, which should prevent fluorophore aggregation (19).
The second goal was to design plasmids that would allow versatility of tagging proteins at their N or C termini because some proteins can be tagged successfully only at one end; e.g., Ypts can only be tagged at their N terminus because the C terminus is required for their prenylation and membrane attachment. The third goal was to allow transformation of the same yeast cells with three different plasmids. We used plasmids with three different selectable auxotrophic markers, URA3 (pRS416), LEU2 (pRS415) and HIS3 (pRS413) (20, 21); therefore, the yeast strains have to carry the appropriate mutations, ura3, leu2 and his3.
When designing multi-color BIFC in yeast, we have two recommendations. It has been our experience that for some protein pairs both the position of the tag and the fluorophore fragment (N or C) can affect the BiFC result: the configuration can affect either the reconstruction of the fluorophore or its localization. Therefore, we suggest trying different configurations. In addition, we find it important for expression of tagged proteins to use yeast codon-optimized tags, especially when tagging proteins at their N terminus. Because the YFP-N and Y/CFP-C fragments in our plasmids are yeast codon optimized, but not the CFP-N, we constructed vectors for tagging proteins at their N or C terminus with YFP, but only at the C terminus with CFP.
3.3 BiFC localization in red strains
One extension of the BiFC analysis is determination of the intra-cellular site where the two proteins interact. At the same time, such co-localization also helps ruling out false positive BiFC interaction due to aggregation in the cytoplasm. Because the BiFC fluorophores fluoresce in the YFP and CFP channels, we used red fluorescence to visualize the compartments. Yeast strains expressing RFP-tagged compartmental markers were previously constructed and are available from the Yeast Resource Center (14). The BiFC plasmids described above have the right auxotrophic markers to be used in this background and can be transformed directly to these strains. When necessary, we tagged a desired cellular site with a red marker. For example, we used mCherry-Atg8 expressed from a plasmid or Atg9-mCherry expressed from the chromosome to visualize autophagosomes (4).
3.4 Examples of using BiFC for studying Ypt1, TRAPP II and beyond
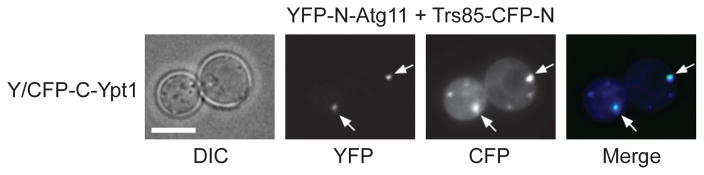
We have reported BiFC interactions in two publications. In the first project, BiFC was used to determine interaction between two TRAPP II subunits, Trs20 and Trs120. This interaction occurs mostly on the trans-Golgi, based on the co-localization of the BiFC puncta with the trans-Golgi marker Chc1-RFP (12). In the second project, multi-color BiFC was used to determine the co-localization of Ypt1 interactions with its autophagy-specific GEF and effector, Trs85 and Atg11, respectively. Whereas only a single Ypt1-Atg11 BiFC punctum per cell was observed, there were multiple Ypt1-Trs85 BiFC puncta. Importantly, in each cell, one of the Ypt1-Trs85 punctum co-localized with the Ypt1-Atg11 punctum (Figure 2). In this case, the BiFC interactions were localized to autophagosomes, based on the co-localization of the Ypt1-Atg11 BiFC punctum and one of the Ypt1-Trs85 BiFC puncta with the autophagosomal marker mCherry-Atg8. The rest of the Ypt1-Trs85 BiFC puncta co-localized with Atg9-mCherry, but not with any other markers of secretory compartments (4).
Figure 2. Co-localization of interactions Ypt1 with its autophagy-specific GEF (Trs85) and effector (Atg11) using multi-color BiFC.
Yeast cells were transformed with three plasmids expressing: i) Y/CFP-C-Ypt1, Ypt1 was tagged at its N terminus with the C terminal fragment of Y/CFP; ii) YFP-N-Atg11, Atg11 tagged at its N terminus with the N terminal fragment of YFP; and iii) Trs85-CFP-N, Trs85 tagged at its C terminus with the N terminal fragment of CFP. Every cell shows multiple puncta in the CFP channel representing the Ypt1-Trs85 interaction. One of these puncta (arrows) co-localizes with the single punctum in the YFP channel representing the Ypt1-Atg11 interaction (merge). Size bar, 5 μm. See details in Lipatova et al, 2012 (4).
Using our constructs, Lee et al., determined a BiFC interaction between the MAPK Hog1 and the glycerol channel Fps1, which was induced by an hyper-osmotic shock (22).
3.5 Important controls and parameters
When employing BiFC, it is important to use appropriate controls. Negative controls should be used to support the idea that the observed BiFC puncta represent bona fide interactions. One possibility is to use the empty vector control as was done previously by Paquin et al., for the Yck1 and Kdh1 interaction (17). If the interaction between two proteins is dependent on growth conditions, growing the cells under conditions that do not support the interaction can be used as a negative control, as was done for the Hog1-Fps1 BiFC interaction (22). Alternatively, the BiFC interaction could be studied in mutant cells that do not support the specific interaction. Below, we recount several controls and parameters used in our BiFC studies.
a. Interaction mutants
One of the most convincing controls for an interaction is the use of mutations known to abrogate this interaction. It is expected that BiFC interaction would occur with the wild type and not with interaction-defective mutant proteins. For negative BiFC controls, it is important to show that the tagged proteins are expressed, and if possible, that they can interact with other proteins in the BiFC assay.
For the Trs20-Trs120 interaction, we used the trs20-D46Y mutation, which inhibits the Trs20-Trs120 interaction in other assays: yeast two hybrid and co-precipitation of recombinant proteins. We showed that the tagged mutant Trs20-D46Y protein was expressed (using immuno-blot analysis) and interacted in the BiFC assay with another TRAPP subunit, Bet3, but not with Trs120 (12).
For the Ypt1 interaction with Trs85 and Atg11 we used two Ypt1 mutations, one that locks the Ypt in its GTP-bound form, “Ypt1-GTP”, and the other in the effector-binding domain, Ypt1-1. GTPase effectors, but not GEFs, should interact with the GTP-bound form of the GTPase. Conversely, the effector domain mutation should affect the interaction of the Ypt with its effectors, but not with its GEF. As expected for bona fide interactions, Ypt1-Trs85 BiFC was abolished when Ypt1-GTP was used, but remained when Ypt1-1 was used. Conversely, the Ypt1-Atg11 interaction was abolished when Ypt1-1 was used and remained when the Ypt1-GTP was used (4). To show the specificity of the Ypt1-Atg11 interaction, we used Atg1, a known Atg11 interactor (23). While Atg1 could form one BiFC punctum per cell when tested with Atg11 (representing the autophagosome), it did not show interaction with Ypt1. We also used Atg11 interaction mutants deleted for the coil-coiled domains CC2 and CC3, which were independently shown to be important for the Ypt1-Atg11 interaction. The relevance of these Atg11 mutants was supported by the fact that they did show BiFC interaction with another Atg11 interactor, Atg19, but not with Ypt1.
b. Co-localization with compartmental markers
One of the limitations of BiFC is the irreversibility of fluorophore assembly. Once the YFP or CFP fluorophore reconstructs, the proteins that are linked are also held together. This can lead to protein aggregation and signal amplification. To ensure that the signal from the BiFC tagged proteins is pertinent to the protein’s native localization, the BiFC puncta should be localized to compartmental markers. We would like to emphasize that it is important to establish that the marker is relevant for the BiFC interaction by showing the co-localization of each tagged protein with the same compartment.
As explained above, we co-localized each BiFC interaction with a compartmental marker or another relevant protein, and supported the BiFC puncta localization by co-localization of each individual protein. For the Trs20-Trs120 BiFC, which co-localizes mostly with the late Golgi marker Chc1, the co-localization of Trs120-GFP predominantly with this marker was shown independently (12). Trs20-GFP was previously localized to this compartment (14). For the Ypt1-Trs85 BiFC interaction, while the co-localization with the autophagosomal marker, Atg8, was expected, the co-localization with Atg9, but not with any other secretory compartment, was new. Therefore, the co-localization of Trs85-GFP with Atg9 was confirmed independently (4).
c. Quantification
When drawing conclusions about BiFC experiments, quantification should be performed. For example, when comparing the Trs20-Trs120 BiFC with the wild-type Trs20 and the interaction-defective Trs20-D46Y mutant, the portion of cells with BiFC puncta in each case was determined and the number of cells visualized was stated. The same quantification was done also for the Trs20-Bet3 BiFC, which served as a positive control for the Trs20 mutant defective in the interaction with Trs120, but not with Bet3 (12). Another possibility is to determine the increase in the intensity of the signal, as was done for the Fps1-Hog1 BiFC interaction in different growth conditions (22). As a rule, we always perform at least two independent experiments (using two independent transformants) for each experiment. Co-localization of the BiFC puncta with compartmental markers should also be quantified as done for any localization analysis.
d. Support of interaction by other methods
It is possible to get a positive BiFC interaction between two proteins if they are the same complex, even if they do not interact directly. To support a conclusion about direct interaction between proteins, other methods such as co-precipitation of recombinant proteins or yeast-two-hybrid should be used. For example, to support direct interaction between Trs20 and Trs120, and the effect of the Trs20-D46Y mutation on the interaction with Trs120 and not with Bet3, co-precipitation of bacterially expressed proteins and the yeast two-hybrid assay were used (12). Similarly, identification of the Atg11 domain CC2-CC3 as the Ypt1-interaction domain was shown by co-precipitation of recombinant proteins and yeast two-hybrid assay, in addition to BiFC (4).
Acknowledgments
We thank A.U. Hain for critical reading of the manuscript. This research was supported by grant GM-45444 from NIH to N. Segev.
Footnotes
While extremely informative as in vivo evidence for an interaction and its intracellular localization, it is important to support BiFC results with independent approaches.
In principle, BiFC can be observed between two subunits in a complex even if they do not interact directly. Therefore, to determine a direct interaction, the analysis should be supported by other approaches, e.g., interaction of bacterially expressed proteins, as was done for the interactions described here (4, 12).
Because the fluorophore reconstruction is irreversible (11), BiFC cannot be used to follow dynamics of interactions.
References
- 1.Segev N. Curr Opin Cell Biol. 2001;13:500–11. doi: 10.1016/s0955-0674(00)00242-8. [DOI] [PubMed] [Google Scholar]
- 2.Segev N. Semin Cell Dev Biol. 2011;22:33–8. doi: 10.1016/j.semcdb.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jedd G, Richardson C, Litt R, Segev N. J Cell Biol. 1995;131:583–90. doi: 10.1083/jcb.131.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lipatova Z, Belogortseva N, Zhang XQ, Kim J, Taussig D, Segev N. Proc Natl Acad Sci U S A. 2012;109:6981–6. doi: 10.1073/pnas.1121299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones S, Newman C, Liu F, Segev N. Mol Biol Cell. 2000;11:4403–11. doi: 10.1091/mbc.11.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morozova N, Liang Y, Tokarev AA, Chen SH, Cox R, Andrejic J, Lipatova Z, Sciorra VA, Emr SD, Segev N. Nat Cell Biol. 2006;8:1263–9. doi: 10.1038/ncb1489. [DOI] [PubMed] [Google Scholar]
- 7.Ohsumi Y. Cell Res. 2014;24:9–23. doi: 10.1038/cr.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sacher M, Kim YG, Lavie A, Oh BH, Segev N. Traffic. 2008;9:2032–42. doi: 10.1111/j.1600-0854.2008.00833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segev N. Sci STKE. 2001;2001:re11. doi: 10.1126/stke.2001.100.re11. [DOI] [PubMed] [Google Scholar]
- 10.Taussig D, Lipatova Z, Segev N. Traffic. 2014;15:327–37. doi: 10.1111/tra.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerppola TK. Annu Rev Biophys. 2008;37:465–87. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taussig D, Lipatova Z, Kim JJ, Zhang X, Segev N. Traffic. 2013;14:678–90. doi: 10.1111/tra.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu CD, Kerppola TK. Nat Biotechnol. 2003;21:539–45. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Nature. 2003;425:686–91. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 15.Sheff MA, Thorn KS. Yeast. 2004;21:661–70. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- 16.Rizzo MA, Springer GH, Granada B, Piston DW. Nat Biotechnol. 2004;22:445–9. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 17.Paquin N, Menade M, Poirier G, Donato D, Drouet E, Chartrand P. Mol Cell. 2007;26:795–809. doi: 10.1016/j.molcel.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 18.Michnick SW. Curr Opin Struct Biol. 2001;11:472–7. doi: 10.1016/s0959-440x(00)00235-9. [DOI] [PubMed] [Google Scholar]
- 19.Zacharias DA, Violin JD, Newton AC, Tsien RY. Science. 2002;296:913–6. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 20.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Gene. 1992;110:119–22. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 21.Sikorski RS, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J, Reiter W, Dohnal I, Gregori C, Beese-Sims S, Kuchler K, Ammerer G, Levin DE. Genes Dev. 2013;27:2590–601. doi: 10.1101/gad.229310.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr, Klionsky DJ. J Cell Biol. 2001;153:381–96. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]