Abstract
The hepatic low-density lipoprotein receptor (LDLR) pathway is essential for clearing circulating LDL-cholesterol (LDL-C). While the transcriptional regulation of LDLR is well-characterized, the post-transcriptional mechanisms which govern LDLR expression are just beginning to emerge. Here, we developed a high-throughput genome-wide screening assay to systematically identify microRNAs (miRNAs) that regulate LDLR activity in human hepatic cells. From this screen, we characterize miR-148a as a negative regulator of LDLR expression and activity, and define a novel SREBP1-mediated pathway by which miR-148a regulates LDL-C uptake. Importantly, inhibition of miR-148a increases hepatic LDLR expression and decreases plasma LDL-C in vivo. We also provide evidence that miR-148a regulates hepatic ABCA1 expression and circulating HDL-C levels. Collectively, these studies uncover miR-148a as an important regulator of hepatic LDL-C clearance through direct regulation of LDLR expression, and demonstrate the therapeutic potential of inhibiting miR-148a to ameliorate the elevated LDL-C/HDL-C ratio, a prominent risk factor for cardiovascular disease.
INTRODUCTION
The hepatic low-density lipoprotein (LDL) receptor is crucial for maintaining cholesterol homeostasis. Reduced LDL receptor (LDLR) expression leads to decreased LDL-catabolism and elevated levels of plasma LDL-cholesterol (LDL-C), a strong risk factor for developing cardiovascular disease in humans1,2. As such, the expression of the LDLR is tightly and coordinately regulated. The transcription of LDLR is coupled to intracellular levels of cholesterol by two major regulatory pathways, the sterol regulatory element-binding proteins (SREBPs) and the liver X receptors (LXRs)3,4. SREBPs bind to sterol regulatory elements (SREs) and promote target gene expression3. In mammals there are three isoforms: SREBP1a and SREBP1c, encoded by the SREBF1 gene, and SREBP2, encoded by the SREBF2 gene5–7. While SREBP1c is regulated by insulin, oxysterols, and phosphatidylcholine and preferentially enhances the transcription of genes involved in fatty acid, phospholipid and triacylglycerol synthesis, SREBP2 and SREBP1a are regulated by intracellular cholesterol concentrations3,8,9. SREBP2 is the main regulator of de novo cholesterol biosynthesis and uptake. When the intracellular cholesterol supply is low, the SREBP2 precursor is trafficked from the endoplasmic reticulum (ER) to the Golgi where it is processed to its mature, nuclear form, which then switches on the transcription of genes involved in cholesterol biosynthesis, such as HMGCR, the rate-limiting enzyme of cholesterol biosynthesis, and cholesterol uptake, such as the LDLR3,8. SREBPs also control the expression of proprotein convertase subtilisin/kexin type 9 (PCSK9), a protein involved in the post-transcriptional degradation of LDLR10,11. Conversely, during sterol-replete conditions, the SREBP2 precursor is retained in the ER and can no longer be processed. Under these conditions the nuclear receptor superfamily member liver X receptor (LXR) is activated by oxysterols and induces the expression of genes involved in cholesterol efflux, such as the ATP-binding cassette (ABC) transporters, ABCA1 and ABCG112. Additionally, LXR controls cholesterol homeostasis through upregulating the E3 ubiquitin ligase, IDOL (inducible degrader of LDLR), thereby preventing reuptake of cholesterol and completing the feedback loop4.
MicroRNAs (miRNAs) are short (~22 nt), evolutionarily conserved, single-stranded RNAs that control the expression of complementary target mRNAs, leading to their transcript destabilization, translational inhibition, or both13–15. Recently, multiple laboratories, including ours, have demonstrated that miRNAs are a critical component of the cholesterol regulatory circuitry (e.g. the SREBP/miR-33 loci16,17 and the SREBP-responsive miR-96/182/183 operon18) and have identified a number of miRNAs (miR-122, miR-30c, miR-33a/b, miR-144, miR-223) that control lipid metabolism in vivo. In particular, miR-33, miR-144, and miR-223 demonstrate the critical role of miRNAs in regulating cellular cholesterol efflux and HDL biogenesis19–24, while the liver-restricted miR-122 has been linked to the regulation of cholesterol and fatty acid synthesis through loss-of-function experiments in mice and non-human primates25–27. Additionally, miR-30c was the first miRNA shown to regulate lipoprotein assembly by targeting the microsomal triglyceride transfer protein (MTP), a protein that is crucial for assembly of ApoB-containing lipoproteins28. While these studies highlight the therapeutic potential of manipulating miRNAs to control HDL-cholesterol (HDL-C) levels, cholesterol biosynthesis, and VLDL secretion, the effect of miRNAs on LDLR activity, and thus, LDL-C, remain poorly understood.
RESULTS
Primary miRNA screen design and optimization
To systematically identify miRNAs that regulate LDLR activity, we developed a high-throughput microscope-based screening assay that monitored the effect of miRNA overexpression on DiI-LDL uptake in human hepatic (Huh7) cells (Fig. 1a). In order to avoid confounding effects of lipoproteins in the media, we initially characterized the specific uptake of DiI-LDL in Huh7 cells incubated in 10% lipoprotein deficient serum (LPDS). To this end, we analyzed the LDLR activity in Huh7 cells treated with increasing concentrations of DiI-LDL for 8 h. The cell-associated DiI-fluorescence was determined at the end of the incubation period by flow cytometry. As seen in Supplementary Fig. 1a–b, DiI-LDL uptake kinetics were saturable and showed complete saturation at approximately 20–40 μg/ml DiI-LDL cholesterol, which is in accordance with the well-known kinetic properties of the LDLR29,30. Similar results were observed when we cultured cells in 384-well plates and measured fluorescence intensity with automated fluorescent microscopy (Supplementary Fig. 1c). As expected, LDL uptake was specific, as DiI-LDL accumulation was displaced when cells were incubated in the presence of 30-fold unlabeled LDL (Supplementary Fig. 1d). We further analyzed whether our system was suitable for functional genomic studies by assessing LDLR gene inactivation by RNA interference (RNAi). Importantly, treatment of Huh7 cells with a siRNA directed against the LDLR (siLDLR) significantly reduced LDLR expression at the protein level (Supplementary Fig. 1e). Consistent with this, DiI-LDL uptake was also diminished in siLDLR-treated Huh7 cells (Supplementary Fig. 1f–g). Importantly, the Z′-factor was determined to be greater than 0.5 (Supplementary Fig. 1f), indicative of a robust setup for our screen31.
Figure 1. Genome-wide miRNA screen identifies novel regulators of LDLR activity.
a, Schematic workflow of primary screen and bioinformatic procedures. b, Linear regression analysis between DiI-LDL mean average intensity (MAI) for plate set 1 and 2 (top), plate set 2 and 3 (middle) and plate set 1 and 3 (bottom). The goodness of fit (r2) and regression line (indicative of overall reproducibility of the screen) is indicated in red on each graph. c, DiI-LDL mean average intensity (MAI, open bars) and robust z-score (red dots) comparison for cells transfected with a negative control siRNA (non-silencing, NS) or positive control siRNA (siRNA LDLR, siLDLR). d, Distribution of average robust z-scores for individual miRNAs in the primary screen. Controls are represented by the grey (NS siRNA) and blue (siLDLR) dots. miR-148a, highlighted in green, was chosen for further validation based on predefined criteria (a). All other miRNAs are shown in black. Dashed red line, robust z-score = −1.5.
Identification of miRNAs that regulate LDLR activity
For the genome-wide miRNA screen, Huh7 cells were transfected in triplicate with a library of 1,719 distinct miRNA mimics and incubated with 30 μg DiI-LDL cholesterol/ml. Following 8 h of incubation, cells were washed, fixed and stained with Hoechst Dye (Fig. 1a). In addition to internal controls on each screening replicate (see Methods), previously validated siRNAs against the LDLR and a NS control siRNA were used as positive and negative controls, respectively (Supplementary Fig. 1f–g). Mean average intensity of DiI-LDL uptake was determined on an individual cell basis (Fig. 1a) using automated high-content image analysis software. To standardize measurements from different plates, phenotypic effects of each miRNA (i.e. those that increased or decreased average DiI intensity) were converted to robust z-scores based on the median average intensity of each array plate32. Notably, comparison of plate replicates and internal plate controls suggested high reproducibility of the screen (Fig. 1b, c). Upon normalization, robust z-scores for each individual miRNA were ranked and compared to their respective plate replicates (Supplementary Table 1). While our screen identified miRNAs that both increased and decreased LDL uptake, we chose to focus on those miRNAs whose overexpression decreased receptor activity, as pharmacological inhibitors of this miRNA subset represent potential therapeutic targets to lower LDL-C levels.
In order to narrow down candidates, a multi-step system was designed; specifically, miRNAs were subjected to four screening passes before chosen for further validation (Fig. 1a). In the first pass, miRNAs were considered putative regulators of LDLR activity for which two or more replicate miRNAs yielded activities smaller than 1.5 median absolute deviations (MAD) away from plate medians (deviation ≤ −1.5) (Fig. 1d). Although this criterion is somewhat less stringent than most cut-offs for high-throughput screenings32, this pass was designed to yield a significantly higher hit rate (159 miRNAs, ~9.2% of miRNAs screened) to allow for subsequent passes (Supplementary Table 1).
To minimize the risk of identifying false positives, the selection of miRNAs for follow-up evaluation in the ensuing passes was based on several criteria. Specifically, miRNAs were chosen for further validation if they: 1) were highly expressed in human and/or mouse liver, 2) were previously shown to be active in the liver (i.e. had high miRNA expression versus reduced target gene expression), and 3) were modulated by dietary lipids (Fig. 1a). Out of the 159 miRNAs identified from the initial pass, 5 miRNAs (miR-140, miR-128, miR-148a, miR-148b, and miR-193b; ~0.29% of miRNAs screened) met these cut-offs, with miR-148a emerging as a strong positive hit, showing medium to high expression in human and mouse hepatic tissues33–35, high liver activity36, and differential expression in the livers of mice fed a high fat diet (HFD)35 (Supplementary Table 2). Intriguingly, it was recently shown that single-nucleotide polymorphisms (SNPs) in the promoter region of miR-148a are associated with altered LDL-C and triglyceride levels in humans37–39, suggesting a possible physiological role for this miRNA in regulating lipid metabolism, and therefore, highlighting it for further validation.
miR-148a is regulated by hepatic lipid content
miR-148a is encoded within an intergenic region of human chromosome 7 and is highly conserved among vertebrate species (Supplementary Fig. 2a). In agreement with previous reports35, miR-148a is highly expressed in mouse liver (Supplementary Fig. 2b) and upregulated in the livers of HFD-fed mice (Supplementary Fig. 2c). Additionally, we found that the expression of miR-148a was significantly increased in the livers of HFD-fed rhesus monkeys (Supplementary Fig. 2d). In accordance with this, and consistent with previous observations40, the mature form of miR-148a was also significantly upregulated in the livers of ob/ob mice (Supplementary Fig. 2e).
To gain insight into the function of miR-148a in regulating cholesterol homeostasis, we analyzed its potential targets using a rigorous bioinformatic algorithm41. For this, predicted targets identified in three target-prediction websites [TargetScan, miRWalk, and miRanda42–44] were assigned to functional annotation clusters using the public gene ontology database, DAVID45. As shown in Supplementary Table 3, miR-148a target genes were enriched (E ≥ 1.0) within 78 clusters and several annotation networks. The functional cluster analysis was combined with data on protein-protein interactions between individual target genes enriched in lipid metabolism using the STRING v946 and PANTHER databases47. The results of this bioinformatic analysis are shown in Supplementary Fig. 3a and indicate that miR-148a targets a vast network of lipid metabolism regulators, including the LDLR.
miR-148a inhibits LDLR expression and regulates LDLR activity
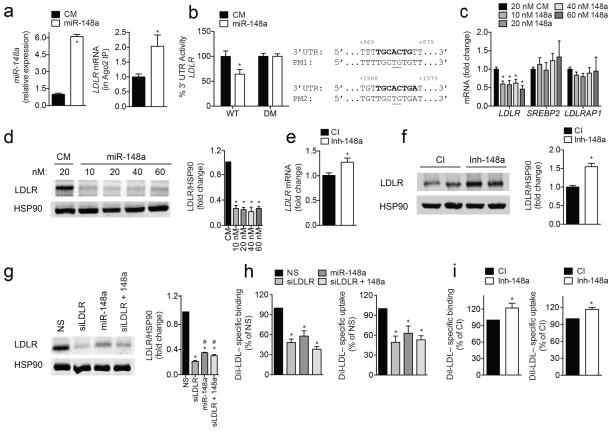
Additional characterization of the aforementioned target genes revealed that miR-148a has two predicted binding sites in the 3′ UTR of the LDLR, one of which is conserved in mammals (Supplementary Fig. 3b). To assess the effect of miR-148a on the LDLR 3′ UTR, we performed ribonucleoprotein immunoprecipitation (RNP-IP) using an antibody against Argonaute-2 (Ago2), a component of the RNA-induced silencing complex (RISC) that mediates miRNA-directed gene silencing. As shown in Fig. 2a, overexpression of miR-148a significantly enriched the association of LDLR mRNA with the Ago2-containing complex, suggesting that the LDLR is directly regulated by miR-148a. Furthermore, we found that overexpression of miR-148a markedly reduced LDLR 3′ UTR activity compared to control-transfected cells (Fig. 2b). Importantly, mutations of both miR-148a target sites relieved miR-148a repression of LDLR 3′ UTR activity (Fig. 2b). We next determined the effect of miR-148a manipulation on LDLR mRNA and protein expression. Transfection of Huh7 cells with miR-148a, but not a control mimic (CM), significantly decreased LDLR mRNA and protein levels (Fig. 2c and d). The effects of miR-148a were seen with concentrations as little as 10 nM (Fig. 2c and d) and using a miR-148a mimic mutated in the seed sequence (CM*) as a negative control (Supplementary Fig. 4a). The inhibition of LDLR expression by miR-148a was highly specific because the expression of other cholesterol-related genes, such as SREBP2 and LDLRAP1, were not influenced by miR-148a overexpression (Fig. 2c). Most importantly, inhibition of endogenous miR-148a significantly increased the expression of LDLR in Huh7 cells (Fig. 2e, f). Similar results were observed in another human hepatic cell line, HepG2, as well as mouse hepatic (Hepa) cells (Supplementary Fig. 4b–e). Finally, we assessed whether miR-148a overexpression caused a further decrease in LDLR levels in Huh7 cells transfected with siLDLR. As seen in Fig. 2g, miR-148a overexpression did not produce an additional effect on LDLR expression.
Figure 2. Post-transcriptional regulation of LDLR expression and activity by miR-148a in human hepatic cells.
a, qRT-PCR analysis of miR-148a and LDLR expression in Huh7 cells transfected with a control mimic (CM) or miR-148a mimic (miR-148a) after Ago2 immunoprecipitation. b, Luciferase reporter activity in COS7 cells transfected with CM or miR-148a and the human 3′ UTR of LDLR. Bold sequences, miR-148a binding sites; underlined nucleotides, point mutations (PM); WT, wild-type; DM, double mutation. c, d, qRT-PCR (c) and representative Western blot analysis (d) of LDLR in Huh7 cells transfected with CM or miR-148a. HSP90 was used as a loading control. e, f, qRT-PCR (e) and representative Western blot analysis (f) of LDLR in Huh7 cells transfected with a control inhibitor (CI) or inhibitor of miR-148a (Inh-148a). HSP90 was used as a loading control. g, h, Representative Western blot (g) and flow cytometry analysis of DiI-LDL binding and uptake (h) in Huh7 cells transfected with a non-silencing (NS) siRNA, miR-148a mimic, siRNA against LDLR (siLDLR) or both. *, P ≤ 0.05 compared to NS-transfected cells by one-way ANOVA. #, P ≤ 0.05 compared to siLDLR-transfected cells by one-way ANOVA with Bonferroni correction for multiple comparisons. i, Flow cytometry analysis of DiI-LDL binding and uptake in Huh7 cells transfected with CI or Inh-148a. In panels (a and c–i), data are the mean ± SEM of ≥ 3 experiments in duplicate. In panel (b), data are the mean ± SEM of ≥ 3 experiments in triplicate. *, P ≤ 0.05 compared to CM- or CI-transfected cells by unpaired t-test (a–f, i).
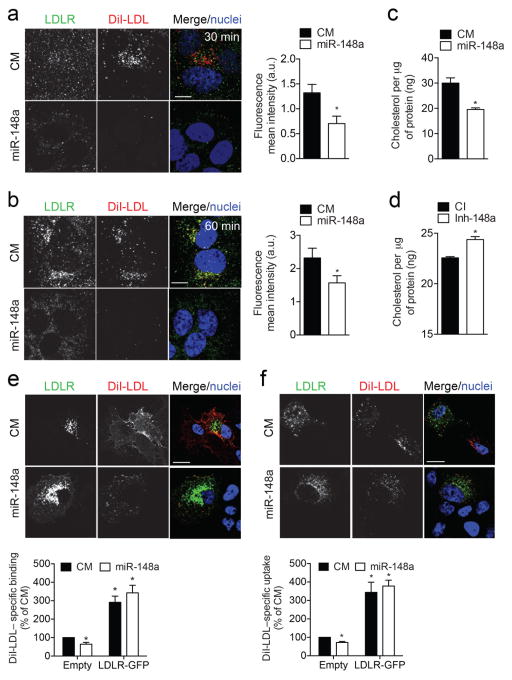
Defective hepatic LDLR activity results in elevated levels of LDL-C in the blood and is associated with an increased risk of coronary artery disease1,2. To assess the role of miR-148a in regulating LDL-C uptake in human and mouse hepatic cells, we overexpressed or inhibited miR-148a and assessed DiI-LDL binding and uptake by flow cytometry. Transfection of Huh7, HepG2 and Hepa cells with miR-148a attenuated specific Dil-LDL binding (Fig. 2h; Supplementary Fig. 4f) and uptake (Fig. 2h; Supplementary Fig. 4g), while antagonists miR-148a (Inh-148a) increased Dil-LDL binding and uptake in these cell types (Fig. 2i; Supplementary Fig. 4h, i). These effects appear to be mediated by the direct targeting of LDLR expression by miR-148a because LDLR silencing abrogates the effect of miR-148a overexpression on the binding and uptake of DiI-LDL (Fig. 2h). Additionally, when we analyzed LDLR-antibody internalization and DiI-LDL uptake by immunofluorescence, we observed reduced LDLR internalization and a concomitant decrease in DiI-LDL uptake in Huh7 cells overexpressing miR-148a compared to cells transfected with a negative control mimic (Fig. 3a, b). Consistent with this, transfection of miR-148a significantly reduced intracellular cholesterol concentration after incubation with unlabeled native LDL (nLDL) compared to CM-treated cells (Fig. 3c). Importantly, intracellular cholesterol levels were also increased in Huh7 cells overexpressing inhibitors of miR-148a (Fig. 3d), thus confirming the endogenous role of miR-148a in modulating cholesterol uptake.
Figure 3. LDLR-GFP overexpression rescues LDLR activity in miR-148a-transfected cells.
a, b, LDLR antibody internalization in Huh7 cells transfected with a control mimic (CM) or miR-148a mimic (miR-148a) and incubated with anti-LDLR and 30 μg/ml DiI-LDL for 40 min at 4 °C. Following internalization at 37 °C for 30 min (a) or 60 min (b) at 37 °C, cells were washed, fixed and stained. Red, DiI-LDL; green, LDLR; blue, TOPRO. Scale bar, 5 μm. Quantification of DiI-LDL mean intensity is shown on the right. c, d, Intracellular cholesterol content in Huh7 cells transfected with CM and miR-148a (c) or control inhibitor (CI) and miR-148a inhibitor (Inh-148a) (d) and incubated with 30 μg/ml native LDL (nLDL) for 2 h. e, f, DiI-LDL binding (e) and uptake (f) in Huh7 cells co-transfected with LDLR-GFP and CM or miR-148a. Red, DiI-LDL; green, LDLR; blue, TOPRO. Scale bar, 10 μm. Quantification by flow cytometry is shown below. *, P ≤ 0.05 compared to cells transfected with CM by one-way ANOVA with Bonferroni correction for multiple comparisons. In panels (a, b, e and f), images are representative of ≥ 3 experiments that gave similar results. In panels (c–f), data are the mean ± SEM and representative of ≥ 3 experiments in duplicate. *, P ≤ 0.05 compared to CM- or CI-transfected cells by unpaired t-test (a–d).
We next determined whether the effect of miR-148a in regulating LDL binding and uptake could be rescued by overexpressing a LDLR-GFP cDNA construct that lacked the 3′ UTR, thereby making it resistant to the inhibitory action of miR-148a. As before, Huh7 cells transfected with miR-148a led to a significant reduction in DiI-LDL binding and uptake when analyzed by immunofluorescence (Fig. 3e, f). Interestingly, this effect was abrogated in cells that expressed the LDLR-GFP construct. While the overexpression of LDLR-GFP construct results in massive overexpression of the LDLR that can influence DiI-LDL uptake independently of the physiological regulation of LDL uptake via LDLR (Fig. 3e, f), these results, together with our previous observation that miR-148a overexpression does not influence DiI-LDL uptake in cells transfected with siLDLR (Fig. 2h), suggest that miR-148a regulates DiI-LDL binding and uptake by direct down-regulation of the LDLR. Alternatively, miR-148a could be acting upstream or downstream of LDLR in the same pathway. In sum, these results demonstrate that manipulation of cellular levels of miR-148a alters LDLR activity.
Transcriptional regulation of miR-148a by SREBP1c
Given that miR-148a is regulated by dietary lipids, we next sought to determine how this miRNA is transcriptionally regulated. Previous computational methods have identified several transcriptional start sites (TSSs) located ~1.1 to ~1.6 Kb upstream of the miR-148a sequence48,49. Importantly, these TSSs correlate with epigenetic signatures and adjacent active promoter and enhancer regions that are involved in the regulation of miR-148a expression50,51 (Supplementary Fig. 5a). Intriguingly, several conserved SREBP1 binding sites (E-box elements, 5′–CANNTG–3′), as well as binding sites for generic transcription factors involved in SREBP activation (Sp1 and Nfy), were previously identified using chromatin immunoprecipitation combined with massive parallel sequencing (ChIP-seq)52 and target-prediction algorithms (Supplementary Fig. 5b). As it has been reported that SREBP1c is increased in the livers of ob/ob mice and under HFD-fed conditions53, we reasoned that SREBP1c, the predominant isoform of SREBP1 in the liver54, is most likely a transcriptional regulator of miR-148a expression.
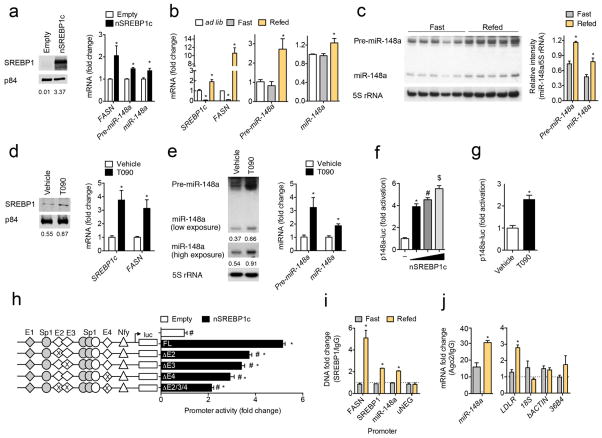
To test whether SREBP1c modulates miR-148a expression, we transfected Huh7 cells with a vector expressing FLAG-tagged nuclear SREBP1c (nSREBP1c) and measured miR-148a expression. As shown in Fig. 4a, overexpression of nSREBP1c significantly increased the expression of miR-148a (mature and precursor forms), as well as the SREBP1c target gene, FASN. To further explore the in vivo relevance of SREBP1c-dependent regulation of miR-148a, we next measured the mature form of miR-148a in the livers of mice that were fasted for 24 h and subsequently refed a high carbohydrate/low-fat diet for 12 h, a dietary condition that increases endogenous SREBP1c expression55. As expected, both precursor and mature miR-148a levels paralleled the refeeding-induced increase in SREBP1c and FASN mRNA levels (Fig. 4b). Similar results were observed when we assessed hepatic pre-miR-148a and miR-148a expression in fasted and refed mice by Northern blotting (Fig. 4c).
Figure 4. SREBP1c modulates miR-148a expression in vitro and in vivo.
a, Representative Western blot and qRT-PCR analysis of SREBP1, FASN, pre-miR-148a, and miR-148a in Huh7 cells transfected with SREBP1c-FLAG (nSREBP1c). p84 was used as a loading control. b, Gene expression analysis in the livers of wild-type (WT) mice fed ad libitum (ad lib, n = 4), fasted for 24 h (Fast, n = 9) or fasted for 24 h and then refed a high carbohydrate/low fat diet for 12 h (Refed, n = 9). *, P ≤ 0.05 compared to ad lib mice by one-way ANOVA with Bonferroni correction for multiple comparisons. c, Northern blot analysis of miR-148a expression in the livers of mice (n = 5 per group) treated as in (b). Ribosomal 5S RNA (5S rRNA) was used as a loading control. d, Western blot and qRT-PCR analysis of SREBP1 and FASN expression in primary mouse hepatocytes treated with 3 μM T090 for 12 h. p84 was used as a loading control. e, Northern blot and qRT-PCR analysis of pre-miR-148a and miR-148a in primary mouse hepatocytes treated as in (d). 5S rRNA was used as a loading control. f, miR-148a promoter activity in HeLa cells transfected with increasing concentrations of nSREBP1c. Statistical comparisons between groups by ANOVA and Student-Newman-Keuls analysis; different symbols denote statistically significant differences (P ≤ 0.05). g, miR-148a promoter activity in Huh7 cells treated with 3 μM T090 for 12 h. h, Luciferase reporter activity in HeLa cells co-transfected with nSREBP1c and the full length miR-148a promoter (p148a-luc FL) or the miR-148a promoter lacking E-box2 (ΔE2), E-box3 (ΔE3), E-box4 (ΔE4), or all three conserved E-box sites (ΔE2/E3/E4). Non-conserved binding sites are shown in grey. Statistical comparisons between groups by one-way ANOVA. *, P ≤ 0.05 compared to empty vector-transfected cells. #, P ≤ 0.05 compared to p148a-luc and nSREBP1-transfected cells. i, Chromatin immunoprecipitation (ChIP) analysis of SREBP1 association with the Fasn, Srebp1 and miR-148a promoters in the livers of mice (n = 2–3 pooled samples per group) treated as in (b). uNEG, negative control. j, qRT-PCR analysis of miR-148a and LDLR expression in the livers of mice treated as in (b) after Ago2 immunoprecipitation (n = 2–3 pooled samples per group). Relative expression levels were normalized to cells immunoprecipitated with a control IgG antibody (dashed line). Data are the mean ± SEM and representative of ≥ 3 experiments in duplicate (a, d–e, and i–j) or triplicate (f–h). Relative intensities of Northern (e) and Western blot bands (a, d) compared to respective loading controls are shown below each blot. *, P ≤ 0.05 compared to empty-vector (a) or vehicle-treated cells (d, e, g) or fasting group (c, i, j) by unpaired t-test.
The LXR activates SREBP1c expression56,57. To ascertain whether miR-148a expression is regulated by LXR, we treated primary mouse hepatocytes and Huh7 cells with T0901317 (T090), a synthetic LXR ligand, and measured miR-148a expression. As shown in Fig. 4d and Supplementary Fig. 6a, the expression of mature SREBP1, as well as the mRNA for FASN and SREBP1c, were both significantly upregulated upon LXR activation. Importantly, the precursor and mature forms of miR-148a were also induced upon T090 treatment (Fig. 4e; Supplementary Fig. 6b).
To determine whether the induction of miR-148a by LXR is dependent on SREBP1c, we silenced SREBP1 using RNA interference. The efficiency of siRNA knockdown was confirmed by qRT-PCR and Western blotting (Supplementary Fig. 6c). As expected, miR-148a levels were significantly increased after treatment with T090 when cells were transfected with a NS siRNA (Supplementary Fig. 6d, e). Conversely, T090 failed to induce the expression of miR-148a in siSREBP1-treated cells, suggesting that SREBP1c is responsible for the induction of miR-148a expression by LXR (Supplementary Fig. 6d, e).
Three E-box motifs facilitate miR-148a promoter activation
We further investigated the role of SREBP1c in directly regulating miR-148a expression by generating luciferase reporter constructs containing a 2.3 Kb region of the human miR-148a promoter (p148a-luc FL). As shown in Fig. 4f, overexpression of nSREBP1c increased miR-148a promoter activity compared to cells transcfected with an empty vector. In agreement with this, LXR-mediated induction of endogenous SREBP1c by T090 also significantly increased miR-148a promoter activity (Fig. 4g), confirming that SREBP1c regulates miR-148a at the transcriptional level. The promoter region of miR-148a contains four E-box elements, three of which are conserved in mice (Supplementary Fig. 5b). To test which E-box elements are responsible for the SREBP1c-mediated induction of miR-148a transcription, we designed miR-148a promoter constructs with deletions in three of the conserved E-box motifs (herein named E-box2, E-box3, and E-box4). As shown in Fig. 4h, SREBP1c-dependent promoter activity was significantly attenuated by deletions in each E-box. Additionally, when all three conserved E-box elements were deleted, miR-148a reporter activity was further diminished upon nSREBP1c overexpression, suggesting that SREBP1c acts through E-box2, E-box3, and E-box4 to fully induce miR-148a transcriptional activity. Indeed, when we transfected cells with a shortened promoter construct (p148a-luc T) lacking the non-conserved E-box1 motif (grey diamond), miR-148a promoter activity was unaffected (Supplementary Fig. 6f). Finally, to assess whether SREBP1 directly binds to the miR-148a promoter, we performed chromatin immunoprecipitation (ChIP) assays in the livers of mice that were fasted for 24 h or fasted for 24 h and subsequently refed a high carbohydrate/low fat diet for 12 h. As shown in Fig. 4i, SREBP1 was significantly enriched in the promoters of Srebp1, Fasn, and miR-148a upon refeeding. Importantly, no SREBP1 association was observed in an upstream region of the miR-148a promoter that lacked SREBP1 binding sites (uNEG), Moreover, we also found an enrichment of miR-148a and LDLR mRNA in the RISC complex of mice that were fasted and subsequently refed (Fig. 4j). Altogether, these results provide compelling evidence that SREBP1c directly regulates the transcriptional expression of the miR-148a gene by binding to the E-box2, E-box3, and E-box4 elements and that SREBP-1c controls miR-148a expression in vivo.
Modulation of miR-148a expression alters plasma lipids in vivo
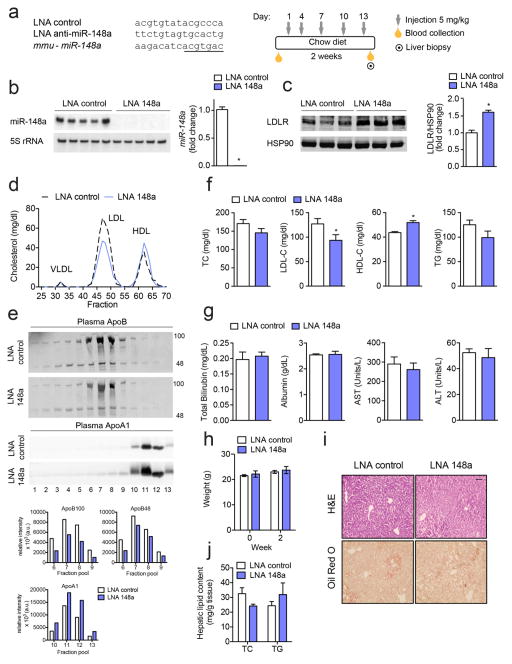
Given the role of miR-148a in negatively regulating LDLR expression and activity in vitro, we next assessed the functional contribution of inhibiting miR-148a in vivo. Because the rate of hepatic LDL-clearance is 40-fold greater in C57BL/6 (wild-type; WT) mice than in humans58, we used ApoBTg;Ldlr−/+ mice, which display a LDL-dominant lipoprotein profile (Supplementary Fig. 7), for our studies. To inhibit miR-148a expression, male ApoBTg;Ldlr−/+ mice were injected every three days for a period of two weeks with 5 mg/kg of DNA/LNA mixmer antisense oligonucleotides against miR-148a (LNA 148a) (Fig. 5a). A scrambled LNA oligonucleotide was used as a control (LNA control). Twenty-four hours following the last injection, mice were sacrificed and serum and livers collected for plasma cholesterol and gene expression analysis, respectively. Treatment with LNA 148a markedly decreased hepatic levels of hepatic miR-148a (Fig. 5b). Importantly, hepatic LDLR expression was significantly increased in LNA 148a-treated mice compared to controls (Fig. 5c). Consistent with this, fractionation of plasma lipoproteins revealed a marked decrease in LDL-C in ApoBTg;Ldlr−/+ mice treated with LNA 148a (Fig. 5d, f). Interestingly, inhibition of miR-148a also significantly increased HDL-C (Fig. 5d, f). As expected by the significant decrease in LDL-C and increase in HDL-C fractions, the expression of ApoB100 and ApoA1 was diminished and enhanced, respectively, in pooled plasma samples isolated from mice treated with LNA 148a compared to controls (Fig. 5e). Similar effects on plasma lipoprotein distribution and apolipoprotein expression were observed in a separate cohort of mice (Supplementary Fig. 8a–c). Total plasma cholesterol levels were slightly, but not significantly, decreased in mice treated with LNA anti-miR-148a (Fig. 5f). This result was predicted as miR-148a antagonism significantly increases circulating HDL-C but decreases LDL-C (Fig. 5d–f; Supplementary Fig. S8). Of note, the effect of LNA miR-148a on LDL-C was mediated by the LDLR because LDL-C levels were similar in Ldlr−/− mice treated with LNA control and LNA 148 (Supplemental Fig. 9). Importantly, the effect of LNA control and LNA 148a injections on lipoprotein metabolism were not influenced by liver toxicity as indicated by the similar blood levels of alanine transaminase (ALT), asparagine transaminase (AST), albumin and total bilirubin observed in both groups of mice (Fig. 5g). Moreover, miR-148a silencing did not influence body weight (Fig. 5h) or hepatic lipid accumulation (Fig. 5i, j).
Figure 5. Inhibition of miR-148a alters plasma cholesterol levels in vivo.
a, Experimental outline of eight-week old chow-fed male ApoBTg;Ldlr−/+ mice treated with LNA control or LNA anti-miR-148a (LNA 148a) (n = 10 per group). Underlined nucleotides, miR-148a seed region. b, Northern blot and qRT-PCR analysis of miR-148a in the livers of mice treated as in (a). Ribosomal 5S RNA (5S rRNA) was used as a loading control. c, Representative Western blot analysis of LDLR expression in the livers of mice treated as in (a). HSP90 was used as a loading control. d, Cholesterol content of FPLC-fractionated lipoproteins from pooled plasma (n = 5 per group) of mice treated as in (a). e, Representative Western blot analysis of plasma ApoB and ApoA1 in the FPLC-fractionated lipoproteins in panel (d). Lanes 1–13 correspond to the following pooled fractions: 1 (28–30), 2 (31–33), 3 (34–36), 4 (37–39), 5 (40–42), 6 (43–45), 7 (46–48), 8 (49–51), 9 (52–54), 10 (55–57), 11 (58–60), 12 (61–63) and 13 (64–66). Relative intensities of ApoB100, ApoB48 and ApoA1 are shown below. f, Total cholesterol (TC), LDL-C, HDL-C, and triglycerides (TGs) in the plasma of mice (n = 10 mice per group) treated as in (a). g, h, Total bilirubin, albumin, AST, and ALT (g) and body weight (h) in the plasma of mice treated with LNA control or LNA 148a (n = 5 per group). i, Representative liver sections of mice treated as in (a) and stained with H&E or Oil Red O. Scale bar = 70 μm. j, TC and TGs in the livers of mice treated with LNA control or LNA 148a (n = 5 per group). All data are the mean ± SEM. *, P ≤ 0.05 compared to LNA control-treated mice by unpaired t-test (b, c, f).
Finally, the impact of anti-miR-148a treatment on lipoprotein metabolism was further confirmed in ApoBTg;Ldlr−/+ mice fed a high-fat diet (HFD) for one month (Supplementary Fig. S10a–c). Similar to chow diet-fed mice, HFD-fed mice treated with LNA 148a had increased levels of hepatic LDLR (Supplementary Fig. S10d), which correlated with a respective significant decrease in LDL-C (Supplementary Fig. 10f, g). Interestingly, HDL-C was also significantly increased in the plasma of LNA 148a treated mice (Supplementary Fig. 10f, g). In addition, body weight and hepatic lipid accumulation were similar in both groups of mice (Supplementary Fig. 10h–j).
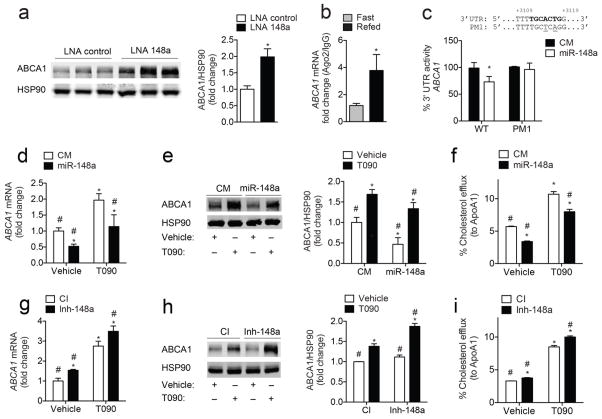
To gain a better understanding of how miR-148a may regulate HDL-C levels in vivo, we further analyzed additional predicted miR-148a target genes. Of note, we found a conserved predicted miR-148a binding site within the 3′ UTR of ABCA1 (Supplementary Fig. 3a, c). Given that ABCA1 plays a major role in regulating HDL biogenesis in the liver59, we hypothesized that the increased HDL-C observed in LNA 148a-treated mice was due to the miR-148a-mediated regulation of ABCA1. Indeed, when we measured ABCA1 by Western blotting, we found a significant increase in hepatic ABCA1 in LNA 148a-treated mice compared to controls (Fig. 6a; Supplementary Fig. S10e).
Figure 6. miR-148a post-transcriptionally regulates ABCA1 expression.
a, Representative Western blot analysis of ABCA1 expression in the livers of mice treated as in (Fig. 5a). HSP90 was used as a loading control. *, P ≤ 0.05 compared to LNA control-treated mice by unpaired t-test. b, qRT-PCR analysis of ABCA1 in the livers of mice that were fasted or refed (n = 2–3 samples per group) after Ago2 immunoprecipitation as in (Fig. 4j). *, P ≤ 0.05 compared to fasted mice by unpaired t-test. c, Luciferase reporter activity in COS7 cells transfected with a control mimic (CM) or miR-148a mimic (miR-148a) and the human 3′ UTR of ABCA1 containing the indicated point mutations (PM1) in the miR-148a target sites. Bold sequences, miR-148a binding sites; underlined nucleotides, PM; wild-type, WT. *, P ≤ 0.05 compared to CM-transfected cells by unpaired t-test. d, e, qRT-PCR (d) and representative Western blot (e) analysis of ABCA1 expression in Huh7 cells transfected with CM or miR-148a in the absence or presence of T090. HSP90 was used as a loading control. f, Cholesterol efflux to ApoA1 in Huh7 cells transfected as in (d). g, h, qRT-PCR (g) and representative Western blot (h) analysis of ABCA1 expression in Huh7 cells transfected with a control inhibitor (CI) or inhibitor of miR-148a (Inh-148a) in the absence or presence of T090. HSP90 was used as a loading control. i, Cholesterol efflux to ApoA1 in Huh7 cells treated as in (g). Data are the mean ± SEM and representative of ≥ 3 experiments in triplicate. Statistical comparisons between groups by one-way ANOVA with Bonferroni correction for multiple comparisons (d–i). *, P ≤ 0.05 compared to cells transfected with CM or CI in vehicle-treated cells. #, P ≤ 0.05 compared to cells transfected with CM or CI in T090-treated cells.
miR-148a directly targets the 3′ UTR of ABCA1
To initially assess whether miR-148a targets ABCA1, we performed RNP-IP using an Ago2 antibody in mice that were fasted or fasted and refed a high carbohydrate diet (Fig. 4j). As expected, miR-148a significantly enriched the association of ABCA1 mRNA within the Ago2-containing complexes after refeeding (Fig. 6b). Consistent with this, luciferase reporter assays revealed a significant down-regulation of ABCA1 3′ UTR activity in cells transfected with a miR-148a mimic, but not a CM (Fig. 6c). Of note, specific point mutations in the predicted binding site (PM1) abolished the inhibitory effect of miR-148a on ABCA1 3′ UTR activity (Fig. 6c). To determine whether miR-148a regulates ABCA1 expression and cholesterol efflux in human hepatic cells, we transfected Huh7 cells with miR-148a mimics or inhibitors and analyzed ABCA1 mRNA/protein levels and cholesterol efflux to ApoA1. As seen in Fig. 6d, e, miR-148a overexpression strongly reduced ABCA1 mRNA and protein levels under basal conditions (vehicle) and when cells were treated with T090 (to directly stimulate LXR-dependent ABCA1 expression). In agreement with this, transfection of Huh7 cells with miR-148a significantly attenuated cholesterol efflux to ApoA1 (Fig. 6f). Importantly, inhibition of endogenous miR-148a in Huh7 cells increased ABCA1 mRNA and protein levels, and resulted in elevated cholesterol efflux to ApoA1 (Fig. 6g–i). Taken together, these experiments identify ABCA1 as a direct and novel target of miR-148a and highlight the physiological role of this miRNA in regulating HDL-C metabolism in vivo.
DISCUSSION
Our data support a role for miRNAs in contributing to the cholesterol regulatory circuitry, particularly with respect to the SREBPs. One of the most extensively studied miRNA families that exemplify this miRNA/SREBP feedback loop includes the miR-33a/b family, which is found within the introns of the SREBF1 and -2 genes, respectively. Both miR-33a/b are co-transcribed with their host genes under conditions that increase SREBP activation and work to control cholesterol and fatty acid homeostasis by targeting genes involved in cellular cholesterol efflux and fatty acid oxidation. miR-185 was also recently shown to regulate cholesterol homeostasis in concert with the SREBF genes. In particular, Yang and colleagues demonstrated that miR-185 is transcriptionally activated by SREBP1c and negatively regulates SREBP2 expression, thereby inhibiting de novo cholesterol biosynthesis and LDL uptake60. Here we show that miR-148a directly controls LDLR activity and is transcriptionally activated by SREBP1c in vitro and in vivo. Similar to miR-185, LXR-mediated induction of SREBP1c results in increased expression of miR-148a. While these results suggest that LDLR expression is regulated by the SREBP1c-dependent induction of miR-148a, one cannot rule out that the decrease in LDLR after T090 treatment is also due to an increase in miR-185 or the LXR-IDOL axis. Given that IDOL is not highly expressed in mouse liver, further studies in non-human primates are needed to assess the physiologic contribution of each pathway to the post-transcriptional regulation of hepatic LDLR expression in humans.
In addition to miR-33 and miR-185, the miR-96/182/183 locus also represents an elegant feedback loop by which miRNAs regulate cholesterol metabolism. The miR-96/182/183 cluster is directly regulated by SREBP2 and works to regulate activation of SREBP2 by controlling its processing (via targeting of INSIG2) and stability (via targeting of FBXW7) in cultured cells18. Interestingly, antagonism of miR-182 in mice had no significant effect on circulating cholesterol and triglyceride levels in mice. Because these studies were conducted in WT mice, which have an HDL-dominant lipoprotein profile, future studies using “humanized” mouse models may provide alternative results. Indeed, we employed the ApoBTg;Ldlr−/+ mouse model, which displays an LDL-dominant lipoprotein profile, for our in vivo analysis and found a significant decrease in LDL-C when hepatic miR-148a levels were antagonized. As a major route of ApoE- and ApoB-containing lipoproteins is by means of LDLR-mediated endocytosis in the liver58, our results suggest that the reduction in LDL-C is mainly due to the miR-148a-mediated repression of hepatic LDLR. While this does not rule out the possibility that miR-148a could be affecting other pathways controlling lipid metabolism, it unequivocally establishes a key role for miR-148a in regulating LDLR activity in vivo. Aside from regulating LDLR expression, we also provide evidence that miR-148a post-transcriptionally controls hepatic ABCA1 expression and cellular cholesterol efflux to ApoA1. Importantly, antagonism of miR-148a significantly increases circulating HDL-C levels in vivo, thus establishing miR-148a as a novel regulator of HDL-C metabolism, as well.
Human genetic data suggests that miR-148a predominantly plays a role in governing LDL-C metabolism as a SNP (rs4722551) in the miR-148a promoter region strongly associates with altered LDL-C levels in humans37–39. Here, we provide insight into the mechanism by which this variant alters plasma LDL-C levels, possibly by affecting transcriptional activation of miR-148a and consequently, LDLR expression. Future experiments are warranted to dissect the contribution of this variant to altered lipid levels and cardiovascular disease risk.
In conclusion, our results underscore the importance of miRNAs in post-transcriptionally regulating LDLR activity. Specifically, our data highlight the therapeutic potential of suppressing miR-148a activity to simultaneously reduce circulating levels of LDL-C and increase levels of HDL-C, beneficial outcomes for reducing the global burden of atherosclerosis and related dyslipidemias.
ONLINE METHODS
Materials
The LDLR-GFP plasmid was kindly provided by Dr. Peter Tontonoz (UCLA, Los Angeles, CA). The pcDNA3.1-2xFLAG-SREBP1c vector and empty control vector were from Addgene. Chemicals were obtained from Sigma-Aldrich unless otherwise noted. The synthetic LXR ligand T0901317 (T090) was purchased from Cayman Chemical. Human ApoA1 was obtained from Meridian Life Sciences. Lipoprotein-deficient serum (LPDS) was prepared from FBS delipidated with 4% fumed silica. 1,1′-Dioctadecyl-3,3,3,3′-tetramethylindocarbocyanineperclorate (DiI) was purchased from Molecular Probes (Invitrogen). A mouse monoclonal antibody against ABCA1 (#ab18180) and ApoA1 (#ab20453) were purchased from Abcam. Rabbit polyclonal antibodies against LDLR (#1007665) and SREBP1 (clone C-20, #sc-366) were from Cayman Chemical and Santa Cruz, respectively. Mouse monoclonal antibodies against HSP90 (#610418) and p84 (clone 5E10, #GTX70220) were purchased from BD Bioscience and GeneTex. A mouse monoclonal antibody against LDLR (clone C7, #sc-18823) and SREBP1 (clone 2A4, #NB600-582) were obtained from Santa Cruz and Novus, respectively. ChIP grade rabbit polyclonal antibodies against SREBP1 (clone H-160, #sc-8984X) and normal IgG (#2729) were purchased from Santa Cruz and Cell Signaling, respectively. A mouse monoclonal antibody against ApoB (#K23300R) was purchased from Meridian Life Sciences. A mouse monoclonal antibody against Ago2 (clone 2D4, #014-22023) was purchased from Wako Chemicals. Secondary fluorescently labeled antibodies were from Molecular Probes (Invitrogen). miRNA mimics and inhibitors were obtained from Dharamcon. A scrambled miR-148a mimic (CM*) was designed and purchased from Dharmacon. siRNAs were purchased from Dharmacon and locked nucleic acid (LNA) miRNA detection probes were purchased from Exiqon (Woburn, MA). For in vivo experiments, miRCURY locked nucleic acid (LNA)™ miRNA inhibitors against mmu-miR-148a-3p or scrambled control were purchased from Exiqon.
Cell culture
Human (HepG2) hepatic cells, monkey kidney fibroblast (COS7) cells, and human cervical cancer (HeLa) cells were obtained from American Type Culture Collection (ATCC). The human hepatocellular carcinoma cell line, Huh7, and mouse hepatic cell line (Hepa1–6) were a kind gift from E. Fisher (NYU School of Medicine). Huh7, HepG2, Hepa, HeLa and COS7 cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS) and 2% penicillin-streptomycin in 10 cm2 dishes at 37 °C and 5% CO2. Before performing experiments all cell lines were tested for mycoplasma contamination by PCR. For DiI-LDL uptake and binding experiments, cells were cultured in DMEM containing 10% LPDS and incubated with 30 μg DiI-LDL cholesterol/ml.
miRNA Screen
All steps of the genome-wide miRNA screen, including reverse transfection and image acquisition and analysis, were performed at the NYU RNAi Core Facility (NYU School of Medicine).
Reverse Transfection, Fixation and Staining
Huh7 cells were reverse transfected in triplicate with a library of 1,719 miRNA mimics (Life Technologies mirVana Mimic Library, miRBase release 17.0) in Corning 384-well flat clear-bottom black plates (Fisher Scientific) using a standard reverse transfection protocol. Briefly, Huh7 cells (5,000 cells/well in 30 μl of DMEM media containing 10% LPDS) were seeded into a well containing 30 μl of transfection mix (25 μl of Optimem, 0.07 μl RNAi Max (Invitrogen), and 5 μl of 0.3 μM miRNA or control siRNA). 20 μl of fresh LPDS media was added to all wells 12 h post transfection, giving a final mimic concentration of 18 nM. 48 h later, cells were incubated with 10 μl of fresh LPDS containing 30 μg of DiI-LDL cholesterol/ml for 8 h at 37 °C. Following incubation, cells were washed twice with 1x PBS and fixed with 4% PFA for 15 min. After three subsequent washes with 1x PBS, cells were incubated with PBS containing 1 μg/ml Hoechst (Molecular Probes) for 25 min. Before scanning, a final wash with 1x PBS was performed and plates were spun down to minimize contaminants when imaging with the automated microscope. All liquid handling steps, including seeding, DiI-LDL incubation, fixation, washing, and Hoechst incubation were performed using a Wellmate Microplate Dispenser (Matrix Technologies) and BioTek Plate Washer (PerkinElmer). The triplicate screen consisted of fifteen 384-well plates and was completed over the course of four days.
Image Acquisition and Analysis
Automated high content and throughput images were acquired using an Arrayscan VTI HCS Reader (Thermo Scientific) with a Zeiss 10x objective. 384-well plates were loaded onto the microscope using a Catalyst Express robotic arm and imaged overnight. In each well, cell nuclei and DiI-LDL intensities were imaged in 5 pre-defined fields. Image data was analyzed using BioApplication’s Target Activation V3 image analysis software (Thermo Scientific). Briefly, nuclei were first identified on the Hoechst stain (Channel 1). Following this, cell boundaries were estimated using the geometric segmentation method and used to calculate DiI intensity (Channel 2) within each cell. Valid object count, mean average intensity, and total average intensity of DiI were recorded for each field. For the primary screen, 57,600 images, consisting of on average 533,528 objects/plate, were analyzed.
Hit Classification
miRNAs were scored based on their ability to significantly increase or decrease DiI intensity compared to negative controls. Cytotoxic miRNA overexpression phenotypes were filtered for hit classification by excluding wells in which fewer then 500 cells were identified as valid objects. In addition, 32 validated internal controls, including non-silencing (NS) siRNA and siLDLR (Figure 1), as well as the negative control miRNAs and siRNA KIF11 (Life Technolgogies) were used on each plate to monitor transfection efficiency. After confirming efficient transfection efficiency, mean average intensities of each well were normalized to plate medians and converted to robust z-scores using Matlab, as previously described32. Robust z-scores were compared between each plate replicate and the mean of each score was calculated and used to rank potential candidates. Those miRNAs that had a robust z-score of ≤ −1.5 (159, 9.2% of miRNAs screened) were chosen for further characterization. To narrow down candidate miRNA genes, hits were subjected to several screening passes (Fig. 1a, bottom panel). Briefly, these candidates were filtered based on whether they were highly expressed in mouse or human liver (9 miRNAs, 0.52% of miRNAs screened), were active in the liver (8 miRNAs, 0.46% of miRNAs screened), and previously shown to respond to dietary lipids (5 miRNAs, 0.29% of miRNAs screened).
Bioinformatic analysis of miRNA target genes
Target genes for miR-148a were identified and compared using the online target prediction algorithm, miRWalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/), which provides target interaction information from eight different prediction algorithms. Specifically, the programs miRanda, miRWalk and TargetScan 6.2 were used. Putative targets produced by all three of these algorithms for miR-148a (2,217 targets) were uploaded into DAVID v6.7 for functional annotation clustering (http://david.abcc.ncifcrf.gov). “High” classification stringency settings yielded 398 functional annotation clusters for miR-148a (Supplementary Table 3), of which 78 clusters (miR-148a) were highly enriched (E ≥ 1.0). In another set of analyses, we took the putative targets for miR-148a identified above and uploaded them into the gene classification system, PANTHER v8.0 (http://www.pantherdb.org) to identify gene targets that were mapped to the lipid metabolic process (GO:0006629). The functional interactions of these predicted targets (110 for miR-148a) described in STRING v9.05 (http://string-db.org) were then combined with the functional annotation groups described in DAVID. Matlab and Cytoscape v2.8.3 were used to create the visualization networks, as previously described41. STRING interactions with a confidence score of 0.4 or higher were added and highlighted in grey. Smaller annotation clusters and unconnected genes were left out of the visualization due to space constraints.
siRNA and miRNA mimic/inhibitor transfections
For siRNA transfections, Huh7 cells were transfected with 20 nM of SMARTpool ON-TARGETplus LDLR siRNA or 20 nM of ON-TARGETplus Non-Targeting pool for 48 h in LPDS medium or 60 nM of SMARTpool ON-TARGETplus SREBP1 siRNA or 60 nM of ON-TARGETplus Non-Targeting pool for 60 h as previously described61. Verification of LDLR and SREBP1 knockdown was assessed by Western blotting and qRT-PCR analysis, as described below. For mimic and inhibitor transfections, Huh7, HepG2, and Hepa cells were transfected with 40 nM miRNA mimics (miR-148a) or with 60 nM miRNA inhibitors (Inh-148a) (Dharmacon) utilizing RNAimax (Invitrogen) or Lipofectamine 2000 (Invitrogen) according to previously established protocols16. All experimental control samples were treated with an equal concentration of a non-targeting control mimic sequence (CM) or inhibitor negative control sequence (CI) for use as controls for non-sequence-specific effects in miRNA experiments. Verification of miR-148a overexpression and inhibition was determined using qRT-PCR, as described below. For dose response experiments, Huh7 cells were transfected with 20 nM of a CM or 10, 20, 40 and 60 nM of a miR-148a mimic for 48 h as previously described16. In another set of experiments, Huh7 cells were transfected with 40 nM of a miR-148a mimic or 40 nM of a scrambled miR-148a mimic (CM*, 5′–UCUGAGCUCUACAGAACUUUGU–3′). To assess the combined effect of miR-148a overexpression and knockdown of the LDLR, Huh7 cells were transfected with 60 nM of a NS siRNA, 40 nM of a miR-148a mimic, or both for 48 h in LPDS. Cells were transfected with an equal amount of CM to compensate for total DNA content as previously described23. For overexpression of nSREBP1c, Huh7 cells were transfected with 1 μg of nSREBP1c (pcDNA3.1-2xFLAG-SREBP1c) or 1 μg of empty vector control (pcDNA3.1) for 24 h using Lipofectamine 200062. Overexpression was confirmed by qRT-PCR and Western blotting.
RNA isolation and quantitative Real-Time PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. For mRNA quantification, cDNA was synthesized using iScript RT Supermix (Bio-Rad), following the manufacturer’s protocol. Quantitative real-time PCR (qRT-PCR) analysis was performed in triplicate using iQ SYBR green Supermix (BioRad) on an iCycler Real-Time Detection System (Biorad). The mRNA level was normalized to GAPDH or 18S as a house keeping gene. For miRNA quantification, total RNA was reverse transcribed using the miScript II RT Kit (Qiagen). Primers specific for human and mouse pre-miR-148a and miR148a (Qiagen) were used and values normalized to SNORD68 (Qiagen) or 18S as a housekeeping gene. For mouse tissues, total liver RNA from WT mice fed a high-fat diet (HFD) were isolated using the Bullet Blender Homogenizer (Next Advance) in TRIzol. 1 μg of total RNA was reverse transcribed and gene/miRNA expression assessed as above. Primer sequences are available upon request.
Western blot analysis
Cells were lysed in ice-cold buffer containing 50 mM Tris-HCl, pH 7.5, 125 mM NaCl, 1% NP-40, 5.3 mM NaF, 1.5 mM NaP, 1 mM orthovanadate and 1 mg/ml of protease inhibitor cocktail (Roche) and 0.25 mg/ml AEBSF (Roche). Cell lysates were rotated at 4 °C for 1 h before the insoluble material was removed by centrifugation at 12,000 x g for 10 min. Nuclear extracts were prepared using the NE-PER Nuclear Protein Extraction Kit (Thermo Scientific) as per the manufacturer’s instructions. After normalizing for equal protein concentration, cell lysates were resuspended in SDS sample buffer before separation by SDS-PAGE. Following overnight transfer of the proteins onto nitrocellulose membranes, the membranes were blocked with 5% BSA (w/v) in wash buffer and probed with the following antibodies overnight at 4 °C: ABCA1 (1:1,000), LDLR (1:1,000), HSP90 (1:1,000), SREBP1 (1:1,000), and p84 (1:1,000). Protein bands were visualized using the Odyssey Infrared Imaging System (LI-COR Biotechnology). Densitometry analysis of the gels was carried out using ImageJ software from the NIH (http://rsbweb.nih.gov/ij/).
For Western blot analysis of ApoB100 and ApoB48 in lipoprotein fractions, an equal volume of three fractions were mixed with reducing SDS sample buffer and separated on a NuPAGE Novex 4–12% Tris-Acetate Mini Gel using 1x NuPAGE Tris-Acetate SDS running buffer (Invitrogen). Following overnight transfer of proteins onto nitrocellulose membranes, the membranes were blocked in 5% (w/v) non-fat milk dissolved in wash buffer. The membranes were probed with an antibody against ApoB (1:2,000) overnight at 4 °C and visualized as above. Western blot analysis of ApoA1 (1:1,000) in pooled lipoprotein fractions was carried out as described in the above paragraph.
Northern blot analysis
miRNA expression was assessed by Northern blot analysis as previously described63. Briefly, total RNA (5 μg) was separated on a 15% acrylamide TBE 8 M urea gel and blotted onto a Hybond N+ nylon filter (Amersham Biosciences). DNA oligonucleotides complementary to mature miR-148a-3p (5′–ACAAAGTTCTGTAGTGCACTGA–3′) were end-labeled with [a-32P] ATP and T4 polynucleotide kinase (New England Biolabs) to generate high-specific activity probes. Hybridization was carried out according to the ExpressHyb (Clontech) protocol. Following overnight membrane hybridization with specific radiolabeled probes, membranes were washed once for 30 min at 42 °C in 4x SSC/0.5% SDS and subjected to autoradiography. Blots were reprobed for 5s rRNA (5′–CAGGCCCGACCCTGCTTAGCTTCCGAGAGATCAGACGAGAT–3′) to control for equal loading.
Ribonucleoprotein immunoprecipitation (RNP-IP)
Ago2 immunoprecipitation (Ago2-IP) experiments after CM or miR-148a transfection were conducted in Huh7 cells as previously described64. Briefly, 1 × 107 cells were transfected with 20 nM miR-148a or CM using RNAimax for 24 h. After 24 h, cells were collected and subjected to Ago2-IP using the RNA isolation kit for human Ago2 (Wako Chemicals) according to the manufacturer’s instructions. The IP pulldown RNA was used to determine the expression levels of miR-148a and LDLR as described above.
In another set of experiments, RISC complexes were immunoprecipitated from the livers of mice that were fasted for 24 h or fasted for 24 h and refed a high-carbohydrate/low fat diet for 12 h using 5 μg of an antibody against mouse Ago2 (2D4) or IgG control as previously described65. Ago2-bound RNA was used to determine the expression levels of miR-148a, ABCA1 and LDLR mRNA as described above. Genes not predicted to be targets of miR-148a (18S, bACTIN and 36B4) were used as negative controls.
Chromatin Immunoprecipitation (ChIP) assays
Equal portions of frozen liver tissue (approximately 100 mg each) from fasted/refed mice were pooled (n = 2–3 mice per group) and crushed into powder in liquid nitrogen. Liver powders were transferred into Eppendorf tubes and homogenized in cold 1x PBS plus protease inhibitors. Following homogenization, samples were filtered, resuspended in 10 ml 1x PBS containing 1% formaldehyde, and rotated on a shaker for 10 min at RT. To quench formaldehyde, glycine was added to a final concentration of 0.125 M and samples were rotated for an additional 5 min at RT. Cells were then collected by centrifugation, washed twice in cold 1x PBS plus protease inhibitors and incubated in 2 ml cold ChIP lysis buffer 1 (50 mM HEPES pH 7.6, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100) for 10 min at 4 °C. The samples were then centrifuged at 3000 x g at 4 °C for 5 min, incubated with 2 ml cold ChIP lysis buffer 2 (10 mM Tris-HCl pH 8, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) for 10 min at 4 °C, and centrifuged at 3000 x g for 5 min at 4 °C. The supernatant was removed and nuclei pellet resuspended in 270 μl ChIP lysis buffer 3 (10 mM Tris-HCl pH 8, 0.5% Sarkosyl, 0.5 mM EGTA, 1 mM EDTA, 100 mM NaCl, 0.1 % Na-Deoxycholate). Nuclear lysates were sonicated 3 × 5 min (30 sec ON/OFF) on high using a Diagenode Biorupter (Diagenode, cat #: UCD-200 TO). After checking chromatin size by agarose gel electrophoresis, extracts were clarified by centrifugation at max speed for 10 min at 4 °C, pre-cleared with 60 μl of Protein G beads (Millipore #16–201) for 1 h at 4 °C, and then incubated overnight at 4 °C with 4 μg of anti-SREBP1 or normal rabbit IgG. Antibody-bound complexes were then captured by incubation with 60 μl of Protein G beads for 1 h at 4 °C. Beads were washed once with low-salt immune complex wash buffer (0.10% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.1, 150 mM NaCl), once with high-salt immune complex wash buffer (0.10% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.1, 500 mM NaCl), twice with LiCl immune complex wash buffer (0.25 M LiCl, 1% NP-40, 1% Deoxycholic acid, 1 mM EDTA, 10 mM Tris-HCl), and once with TE (10 mM Tris-HCl pH 8, 1 mM EDTA). Antibody-bound complexes were then eluted by incubation with 200 μl of Elution buffer (100 mM NaHCO3, 1% SDS) for 15 min with gentle rotation followed by a second 15 min elution with 200 μl of Elution buffer. To reverse crosslinks, the eluates were combined, treated with 5 M NaCl, and incubated overnight at 65 °C. Samples were then incubated with 1 μl RNase A, incubated for 30 min at 37 °C and treated with 4 μl 0.5 M EDTA, 8 μl 1 M Tris-HCl, and 1 μl Proteinase K for 1 h at 45 °C. DNA was purified using the QIAquick PCR Purification Kit (QIAGEN) according to the manufacturer’s instructions and eluted in 50 μl of TE buffer. qPCR was run as described above using primer sets for the following promoters: FASN (5′–GCGCAGCCCCGACGCTCATT–3′ and 5′– CGGCGCTATTTAAACCGCGG–3′) and SREBP1 (5′–GTAGCCAATGGGTGCAAGG–3′ and 5′– CACGTGACCAAAACCAGAGT–3′). Two primer sets were used for the miR-148a promoter, both of which gave similar results (5′–AATAAGAGCGCAGGTCGTC–3′ and 5′– GCTGAGCTAGGCTTCCAGT–3′) and (5′–GGAACCTGCTGACTTGACAC–3′ and 5′– GACGACCTGCGCTCTTATT–3′). The primer set amplifying a region upstream of the predicted SREBP1 binding sites in the miR-148a promoter, uNEG (5′–AAACGCATTGCCATTCTC–3′ and 5′– ATTTCAGTAGCTCAAGCACAG–3′), was used as a negative control. Data was normalized using the % input method and plotted relative to the IgG control.
LDL receptor activity assays
Human LDL was isolated and labeled with the fluorescent probe DiI as previously reported66. Huh7 cells were transfected in 6- or 12-well plates with miRNA mimics and inhibitors in DMEM containing 10% LPDS for 48 h. Then, cells were washed once in 1x PBS and incubated in fresh media containing DiI-LDL (30 μg cholesterol/ml). Non-specific uptake was determined in extra wells containing a 50-fold excess of unlabeled native LDL (nLDL). Cells were incubated for 8 h at 37 °C to allow for DiI-LDL uptake in screening optimization experiments and for 2 h at 37 °C for subsequent validation experiments. In other instances, cells were incubated for 90 min at 4 °C to assess DiI-LDL binding. At the end of the incubation period, cells were washed, resuspended in 1 ml of PBS and analyzed by flow cytometry (FACScalibur, Becton Dickinson), as previously described67. The results are expressed in terms of specific median intensity of fluorescence (M.I.F.) after subtracting autofluorescence of cells incubated in the absence of DiI-LDL.
Fluorescence microscopy
For LDLR-Ab internalization and DiI-LDL uptake assays, Huh7 cells were grown on coverslips and transfected with a miR-148a mimic or negative control mimic (CM) in DMEM containing 10% LPDS. 48 h post transfection, cells were cooled to 4 °C for 20 min to stop membrane internalization. Cells were then incubated with LDLR mAb (clone C7) (Santa Cruz) and 30 μg/ml DiI-LDL for 40 min at 4 °C. Following incubation, cells were gently washed twice with cold medium and shifted to 37 °C to allow for internalization of both LDLR-Ab complexes and DiI-LDL for the indicated times and fixed with 4% PFA. After 5 min of Triton X-100 0.2% permeabilization and 15 min of blocking (PBS BSA 3%), cells were stained with anti-mouse Alexa 488 (Molecular Probes) and TOPRO 3 (Life Technologies) for 1 h at room temperature. After this, cells were washed twice with 1x PBS and mounted on glass slides with Prolong-Gold (Life Technologies). For LDLR-GFP rescue experiments, Huh7 cells were grown on coverslips and co-transfected with 1 μg LDLR-GFP and 40 nM of a CM or miR-148a mimic. 48 h post transfection cells were incubated with 30 μg/ml DiI-LDL for 2 h at 37 °C (uptake) or with 30 μg/ml DiI-LDL for 90 min at 4 °C (binding). Then, cells were washed twice with 1x PBS, fixed with 4% PFA, and blocked (3% BSA in 1x PBS) for 15 min. Following this, cells were washed twice and mounted on glass slides with Prolong-Gold (Life Technologies). All images were analyzed using confocal microscopy (Leica SP5 II) equipped with a 63X Plan Apo Lenses. All gains for the acquisition of comparable images were maintained constant. Analysis of different images was performed using ImageJ (NIH) and Adobe Photoshop CS5.
3′ UTR luciferase reporter assays
cDNA fragments corresponding to the entire 3′ UTR of human LDLR and ABCA1 were amplified by RT-PCR from total RNA extracted from HepG2 cells with XhoI and NotI linkers. The PCR product was directionally cloned downstream of the Renilla luciferase open reading frame of the psiCHECK2™ vector (Promega) that also contains a constitutively expressed firefly luciferase gene, which is used to normalize transfections. Point mutations in the seed region of the predicted miR-148a binding sites within the 3′ UTR of LDLR were generated using the Multisite-Quickchange Kit (Stratagene), according to the manufacturer’s protocol. All constructs were confirmed by sequencing. COS7 cells were plated into 12-well plates and co-transfected with 1 μg of the indicated 3′ UTR luciferase reporter vectors and miR-148a mimics or control mimics (Life Technologies) utilizing Lipofectamine 2000 (Invitrogen), as previously described16. After 24 h of transfection, luciferase activity was measured using the Dual-Glo Luciferase Assay System (Promega). Renilla luciferase activity was normalized to the corresponding firefly luciferase activity and plotted as a percentage of the control (cells co-transfected with the corresponding concentration of control mimic). Experiments were performed in triplicate wells of a 12-well plate and repeated at least three times.
miR-148a promoter assays
A 2.3 Kb fragment of the human miR-148a promoter was amplified by PCR from BAC clone RP11-184C17 with the following primers: 5′–TGATGGCAGACAATAACTCC–3′ and 5′–AAAGTGCTTTCCCATCTTCC–3′. The PCR product was directionally cloned in a PGL3 promoter vector (Promega) using the KpnI and HindIII linkers. For overexpression assays, HeLa cells were co-transfected with 0.5 μg of the indicated p148a-luc constructs, 0.01 μg of Renilla luciferase reporter plasmid, and 0.5 μg of nuclear SREBP1c or empty vector control using Lipofectamine 2000. For the dose-response experiments, HeLa cells were co-transfected with 0.5 μg of p148a-luc, 0.01 μg of a Renilla luciferase reporter plasmid, and 0.5, 1 or 2 μg of nuclear SREBP1c or empty vector control using Lipofectamine 2000. After 24 h of transfection, luciferase activity was measured using the Dual-Glo Luciferase Assay System (Promega). Renilla luciferase activity was normalized to the corresponding firefly luciferase activity and plotted as a percentage of the control (cells co-transfected with the corresponding concentration of empty vector control) as previously described68. Experiments were performed in triplicate wells of a 12-well plate and repeated at least four times. For assays with T090, Huh7 cells were co-transfected with 0.5 μg of p148a-luc and 0.01 μg of Renilla luciferase reporter plasmid using Lipofectamine 2000. Following 24 h of transfection, cells were washed and treated with vehicle or 3 μM T090 for 12 h. Experiments were performed in triplicate wells of a 12-well plate and repeated at least three times. Luciferase activity was measured as described above and plotted as a percentage of the control (cells treated with vehicle). Experiments were performed in triplicate wells of a 12-well plate and repeated at least three times.
Cholesterol efflux assays
Cholesterol efflux assays were performed as previously described69. Briefly, Huh7 cells were seeded at a density of 2 × 105 cells per well and transfected with either a CM or miR-148a mimic, CI or Inh-148a. Following 48 h of transfection, cells were loaded with 0.5 μCi/ml 3H-cholesterol for 24 h. 12 h after loading, cells were incubated with 3 μM T090 to increase the expression of ABCA1. Then, cells were washed twice with PBS and incubated in DMEM supplemented with 2 mg/ml fatty-acid free BSA (FAFA-media) in the presence of an ACAT inhibitor (2 μmol/L) for 4 h prior to the addition of 50 μg/ml human ApoA1 in FAFA-media with or without the indicated treatments. Supernatants were collected after 6 h and expressed as a percentage of total cell 3H-cholesterol content (total effluxed 3H-cholesterol + cell-associated 3H-cholesterol).
Cellular cholesterol measurements
Huh7 cells were seeded at a density of 5 × 105 cells/well and transfected with either a CM or miR-148a mimic or CI or Inh-148a. Following 48 h transfection, cells were incubated with 30 μg/ml nLDL for 2 h. Intracellular cholesterol content was measured using the Amplex Red Cholesterol Assay Kit (Molecular Probes, Invitrogen), according to the manufacturer’s instructions.
Mouse studies
Male C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, ME) and kept under constant temperature and humidity in a 12 h controlled dark/light cycle. For HFD studies, eight-week old male mice (n = 6 per group) were placed on a chow diet or HFD containing 0.3% cholesterol and 21% (wt/wt) fat (Dyets, Inc) for three weeks. Liver samples were collected as previously described16 and stored at −80 °C until total RNA was harvested for miRNA expression analysis.
For miR-148a inhibition experiments, eight-week old male ApoBTg;Ldlr−/+ mice fed a chow diet were randomized into 2 groups: LNA control (n = 10) and LNA anti-miR-148a (n = 10). Mice received i.p. injections of 5 mg/kg LNA control (5′–ACGTCTATACGCCCA–3′) or LNA anti-miR-148a (5′–TTCTGTAGTGCACTG–3′) oligonucleotides every three days for a total of two weeks. Twenty-four hours after the final injection, mice were sacrificed and hepatic gene expression analyzed (see above). In another set of experiments, eight-week old male ApoBTg;Ldlr′/+ mice (n = 5 per group) were treated and fed a chow diet as above for 2 weeks, at which point mice were switched to a HFD (60% fat, Research Diets D12492) and given weekly i.p. injections of LNA control or LNA anti-miR-148a oligonucleotides for four weeks. One week following the last injection, mice were sacrificed and hepatic gene expression analyzed. All animal experiments were approved by the Institutional Animal Care Use Committee of Yale University School of Medicine. Obese (C57BL/6-ob/ob) mice were maintained as previously described53. For fasting/refeeding experiments, eight-week old male C57BL/6 mice were divided into three treatment groups as previously described: ad libitum (n = 4), fasted for 24 h (n = 9), or fasted for 24 h then refed a high carbohydrate/low fat diet (TD 88122, Harlan Teklad Diets) for 12 h (n = 9) as previously described55. Following sacrifice, hepatic miRNA and gene expression were analyzed as above.
Primary mouse hepatocyte isolation and culture
For analysis of miR-148a expression, non-fasted eight-week old male C57BL/6 mice were sacrificed and primary hepatocytes were isolated by isopycnic centrifugation as previously described70. On day zero, isolated hepatocytes were plated onto 6-well collagen-I-coated dishes (400,00 cells/well) in 2 ml Adherence Medium (Williams’ Medium E containing 5% fetal bovine serum (FBS), 2% penicillin-streptomycin, 10 mM HEPEs buffer, 8 μg/ml gentamicin, 1 μM dexamethasone, and 1 nM insulin). Following 6 h incubation at 37 °C and 5% CO2, the attached cells were washed once with 1 ml 1x PBS and then incubated for 14–16 h in 2 ml Maintenance Medium (Williams’ Medium E containing 5% fetal bovine serum (FBS), 2% penicillin-streptomycin, 8 μg/ml gentamicin, 1 μM dexamethasone, and 1 nM insulin). On day one, cells in each well were washed once with 2 ml 1x PBS, after which cells received 2 ml of fresh Maintenance Medium supplement with 3 μM vehicle or T090. After incubated for 12 h at 37 °C and 5% CO2, RNA and protein were harvested for qRT-PCR and Western blotting analysis as described above.
Non-human primate studies
Male rhesus monkeys (Macaca mulatta), 7–13 years old, were fed a standard (TestDiet #5038; Purina Mills) or high fat/high sucrose diet (42% kcal in fat, Custom formula #07802; Harlan, Teklad, Madison, WI) for two years (n = 4 per group) and maintained as previously described71. Animal procedures were approved by the Animal Care and Use Committee of the NIA Intramural Research Program. Following sacrifice, liver RNA was isolated using the Bullet Blender Homogenizer (Next Advance) in TRIzol. For mRNA quantification, 1 μg of total RNA was reverse transcribed using iScript RT Supermix (BioRad) and iQ SYBR green Supermix (BioRad). Quantification of miR-148a was assessed as described above.
Plasma lipid analysis and lipoprotein profile measurements
Blood samples were collected by retro-orbital venous plexus puncture after a 12 h overnight fast at day 1 and day 14 (for chow diet studies) and at day 1, day 14, and day 43 (for HFD studies) for lipid analysis. Plasma was separated by centrifugation and stored at 4 °C. The lipid distribution in plasma lipoprotein fractions (pooled plasma, n = 5 per group) was assessed by fast-performance liquid chromatography (FPLC) gel filtration with 2 Superose 6 HR 10/30 columns (Pharmacia) as previously described23. Cholesterol in each fraction was enzymatically measured using the Cholesterol Assay Kit (Wako Diagnostics). Total plasma cholesterol, HDL-C, LDL-C and triglycerides were enzymatically measured (Wako Diagnostics) according to manufacturer’s instructions. Additionally, LDL-C levels were confirmed by subtracting HDL-C from total cholesterol (ApoBTg;Ldlr−/+ carry most of the cholesterol in LDL and HDL particles). Plasma alanine aminotransferase (ALT), aspartate aminotransferase (AST), albumin and total bilirubin were analyzed in LNA control and LNA anti-miR-148a treated mice (n = 5 per group) by the Yale University School of Medicine Mouse Metabolic Phenotyping Center (MMPC).
Liver histology and hepatic lipid analysis
For histological analysis, mouse livers were perfused with PBS and fixed overnight with 4% paraformaldehyde (PFA) at 4 °C. After incubation, livers were washed with 1x PBS, incubated in 30% sucrose for 24 h, and embedded in OCT and frozen. Serial sections were cut at 8 μm thickness using a cryostat. Every third slide from the serial sections was stained with hematoxylin and easin (H&E) and each consecutive slide was stained with Oil Red O for visualization of neutral lipids as previously described72. Lipids were extracted from 1 mg of liver tissue from LNA control and LNA anti-miR-148a treated mice (n = 5 per group) as previously described73. Triglyceride and cholesterol content were measured using kits from Wako Diagnostics according to the manufacturers’ protocols.
Statistics
Animal sample size for each study was chosen based on literature documentation of similar well-characterized experiments16,53,55,71. The number of animals used in each study is listed in the figure legends and main text. No inclusion/exclusion criteria were used and studies were not blinded to investigators or formally randomized. In vitro experiments were routinely repeated at least three times unless otherwise noted. All data are expressed as mean ± SEM. Statistical differences were measured using an unpaired two-sided Student’s t-test or one-way ANOVA with Bonferroni correction for multiple comparisons or Student-Newman-Keuls analysis when appropriate. Normality was checked using the Kolmogorov-Smirnov test. A non-parametric test (Mann-Whitney) was used when data did not pass the normality test. A value of P 0.05 was considered statistically significant. Data analysis was performed using GraphPad Prism Software Version 5.0a (GraphPad, San Diego, CA).
Supplementary Material
Acknowledgments
We thank C. Yun, J. Recio, S. Katz, and R. DasGupta at the NYU RNAi Core for their advice and assistance with the miRNA screen; K. Harry and members of the Yale University Liver Center for primary mouse hepatocyte isolation; the Iwakiri lab for reagents and advice on primary hepatocyte culture; the Yale University School of Medicine Mouse Metabolic Phenotyping Center (MMPC) for liver toxicity measurements and P. Tontonoz (UCLA) for generously providing the LDLR-GFP plasmid. This work was supported by grants from the National Institutes of Health (R01HL107953, R01HL107953-04S1, and R01HL106063 to CF-H; R01HL105945 to YS; 1F31AG043318 to LG; and P30KD034989 to the Yale University Liver Center), the American Heart Association (15SDG23000025 to CMR), the Howard Hughes Medical Institute International Student Research Fellowship (to EA), the Foundation Leducq Transatlantic Network of Excellence in Cardiovascular Research (to CFH) and the Ministerio de Industria y Comercio, Spain (SAF2011-29951 to MAL). CIBERobn is an initiative of ISCIII, Spain. RdC is supported by the Intramural Research Program of the NIH, National Institute of Aging. AMN and AW are supported by a grant from the National Institutes of Health (R01DK 094184). The NYU RNAi core is supported by the Laura and Isaac Perlmutter Cancer Center (NIH/NCI P30CA16087) and the NYSTEM Contract (C026719). The Yale University School of Medicine MMPC is supported by the NIH (U24 DK059635).
Footnotes
AUTHOR CONTRIBUTIONS
LG and CFH conceived and designed the study. LG optimized and performed the miRNA screen. JFA and ACD performed confocal experiments, assisted with cloning, and analyzed data. LG and ACD performed in vitro experiments and analyzed data. LG, NR, CMR, and ACD performed mouse experiments and analyzed data. NR performed Western blotting for plasma lipoproteins and assessed lipoprotein distribution by FPLC. EA cloned the miR-148a promoter. CSL assisted with Western blotting. NNA analyzed gene expression in the livers of fed/fasted WT mice and ob/ob mice. RdC provided non-human primate samples, while JDH provided mouse samples. MAL provided DiI-LDL and native LDL. AW and AMN assisted with mouse experiments. YS and CFH assisted with experimental design and data interpretation. LG and CFH wrote the manuscript, which was commented on by all authors.
COMPETING FINANCIAL INTERESTS
LG and CFH have a patent (PCT/US2014/042196) on the use of miR-148a inhibitors to treat dyslipidemias and cardiovascular disease.
REFERENCES FOR MAIN TEXT
- 1.Glass CK, Witztum JL. Atherosclerosis. the road ahead. Cell. 2001;104:503–516. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 2.Lusis AJ. Atherosclerosis. Nature. 2000;407:233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 4.Zelcer N, Hong C, Boyadjian R, Tontonoz P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 2009;325:100–104. doi: 10.1126/science.1168974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hua X, et al. SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tontonoz P, Kim JB, Graves RA, Spiegelman BM. ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol Cell Biol. 1993;13:4753–4759. doi: 10.1128/mcb.13.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yokoyama C, et al. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- 8.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 9.Walker AK, et al. A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell. 2011;147:840–852. doi: 10.1016/j.cell.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horton JD, et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maxwell KN, Soccio RE, Duncan EM, Sehayek E, Breslow JL. Novel putative SREBP and LXR target genes identified by microarray analysis in liver of cholesterol-fed mice. Journal of lipid research. 2003;44:2109–2119. doi: 10.1194/jlr.M300203-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Beaven SW, Tontonoz P. Nuclear receptors in lipid metabolism: targeting the heart of dyslipidemia. Annu Rev Med. 2006;57:313–329. doi: 10.1146/annurev.med.57.121304.131428. [DOI] [PubMed] [Google Scholar]
- 13.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature reviews Genetics. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 16.Rayner KJ, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Najafi-Shoushtari SH, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeon TI, et al. An SREBP-responsive microRNA operon contributes to a regulatory loop for intracellular lipid homeostasis. Cell Metab. 2013;18:51–61. doi: 10.1016/j.cmet.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rayner KJ, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121:2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rayner KJ, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rottiers V, et al. Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Sci Transl Med. 2013;5(212):ra162. doi: 10.1126/scitranslmed.3006840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Aguiar Vallim T, et al. MicroRNA-144 Regulates Hepatic ABCA1 and Plasma HDL Following Activation of the Nuclear Receptor FXR. Circ Res. 2013;112:1602–1612. doi: 10.1161/CIRCRESAHA.112.300648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramirez CM, et al. Control of Cholesterol Metabolism and Plasma HDL Levels by miRNA-144. Circ Res. 2013;112:1592–1601. doi: 10.1161/CIRCRESAHA.112.300626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vickers KC, et al. MicroRNA-223 coordinates cholesterol homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:14518–14523. doi: 10.1073/pnas.1215767111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 26.Elmen J, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esau C, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Soh J, Iqbal J, Queiroz J, Fernandez-Hernando C, Hussain MM. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat Med. 2013;19:892–900. doi: 10.1038/nm.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown MS, Dana SE, Goldstein JL. Regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in human fibroblasts by lipoproteins. Proceedings of the National Academy of Sciences of the United States of America. 1973;70:2162–2166. doi: 10.1073/pnas.70.7.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldstein JL, Basu SK, Brunschede GY, Brown MS. Release of low density lipoprotein from its cell surface receptor by sulfated glycosaminoglycans. Cell. 1976;7:85–95. doi: 10.1016/0092-8674(76)90258-0. [DOI] [PubMed] [Google Scholar]
- 31.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 32.Birmingham A, et al. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods. 2009;6:569–575. doi: 10.1038/nmeth.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barad O, et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vickers KC, et al. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2013;57:533–542. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arora A, Simpson DA. Individual mRNA expression profiles reveal the effects of specific microRNAs. Genome Biol. 2008;9:R82. doi: 10.1186/gb-2008-9-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Do R, et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet. 2013;45:1345–1352. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Global Lipids Genetics, C. et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huan T, et al. Genome-wide identification of microRNA expression quantitative trait loci. Nature communications. 2015;6:6601. doi: 10.1038/ncomms7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trajkovski M, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 41.Mercer J, et al. RNAi screening reveals proteasome- and Cullin3-dependent stages in vaccinia virus infection. Cell Rep. 2012;2:1036–1047. doi: 10.1016/j.celrep.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 42.John B, et al. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 44.Dweep H, Gretz N, Sticht C. miRWalk database for miRNA-target interactions. Methods Mol Biol. 2014;1182:289–305. doi: 10.1007/978-1-4939-1062-5_25. [DOI] [PubMed] [Google Scholar]
- 45.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 46.Szklarczyk D, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas PD, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Down TA, Hubbard TJ. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 2002;12:458–461. doi: 10.1101/gr.216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimomura I, Bashmakov Y, Horton JD. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J Biol Chem. 1999;274:30028–30032. doi: 10.1074/jbc.274.42.30028. [DOI] [PubMed] [Google Scholar]
- 54.Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Invest. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horton JD, Bashmakov Y, Shimomura I, Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peet DJ, et al. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- 57.Yoshikawa T, et al. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol Cell Biol. 2001;21:2991–3000. doi: 10.1128/MCB.21.9.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dietschy JM, Turley SD, Spady DK. Role of liver in the maintenance of cholesterol and low density lipoprotein homeostasis in different animal species, including humans. Journal of lipid research. 1993;34:1637–1659. [PubMed] [Google Scholar]
- 59.Oram JF, Vaughan AM. ABCA1-mediated transport of cellular cholesterol and phospholipids to HDL apolipoproteins. Current opinion in lipidology. 2000;11:253–260. doi: 10.1097/00041433-200006000-00005. [DOI] [PubMed] [Google Scholar]
- 60.Yang M, et al. Identification of miR-185 as a regulator of de novo cholesterol biosynthesis and low density lipoprotein uptake. Journal of lipid research. 2014;55:226–238. doi: 10.1194/jlr.M041335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramirez CM, et al. MicroRNA 33 regulates glucose metabolism. Mol Cell Biol. 2013;33:2891–2902. doi: 10.1128/MCB.00016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davalos A, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circulation research. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 64.Goedeke L, et al. A regulatory role for microRNA 33* in controlling lipid metabolism gene expression. Mol Cell Biol. 2013;33:2339–2352. doi: 10.1128/MCB.01714-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Allen RM, Marquart TJ, Jesse JJ, Baldan A. Control of very low-density lipoprotein secretion by N-ethylmaleimide-sensitive factor and miR-33. Circulation research. 2014;115:10–22. doi: 10.1161/CIRCRESAHA.115.303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Calvo D, Gomez-Coronado D, Suarez Y, Lasuncion MA, Vega MA. Human CD36 is a high affinity receptor for the native lipoproteins HDL, LDL, and VLDL. Journal of lipid research. 1998;39:777–788. [PubMed] [Google Scholar]
- 67.Suarez Y, et al. Synergistic upregulation of low-density lipoprotein receptor activity by tamoxifen and lovastatin. Cardiovascular research. 2004;64:346–355. doi: 10.1016/j.cardiores.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 68.Chamorro-Jorganes A, Araldi E, Rotllan N, Cirera-Salinas D, Suarez Y. Autoregulation of glypican-1 by intronic microRNA-149 fine tunes the angiogenic response to FGF2 in human endothelial cells. J Cell Sci. 2014;127:1169–1178. doi: 10.1242/jcs.130518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramirez CM, et al. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arteriosclerosis, thrombosis, and vascular biology. 2011;31:2707–2714. doi: 10.1161/ATVBAHA.111.232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang W, et al. Radixin is required to maintain apical canalicular membrane structure and function in rat hepatocytes. Gastroenterology. 2006;131:878–884. doi: 10.1053/j.gastro.2006.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mattison JA, et al. Resveratrol prevents high fat/sucrose diet-induced central arterial wall inflammation and stiffening in nonhuman primates. Cell Metab. 2014;20:183–190. doi: 10.1016/j.cmet.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goedeke L, et al. Long-term therapeutic silencing of miR-33 increases circulating triglyceride levels and hepatic lipid accumulation in mice. EMBO molecular medicine. 2014;6:1133–1141. doi: 10.15252/emmm.201404046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miller AM, et al. MiR-155 has a protective role in the development of non-alcoholic hepatosteatosis in mice. PloS one. 2013;8:e72324. doi: 10.1371/journal.pone.0072324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.