Abstract
CYP2A6 genotyping is of clinical importance CYP2A6 gene variants influence nicotine metabolism and are associated with nicotine dependence, cigarettes per day, smoking cessation and the risk for tobacco-associated cancers. CYP2A6 gene variants also influence the metabolism of therapeutic drugs, such as the anti-cancer agents tegafur and letrozole. Over the years, CYP2A6 genotyping methods have evolved to incorporate novel gene variants and to circumvent genotyping errors resulting from the high degree of homology between CYP2A6 and neighboring CYP2A genes. Herein, CYP2A6 genotyping strategies are described for commonly genotyped functionally significant alleles including single nucleotide polymorphisms, small insertions/deletions and more complex structural variants. The methods presented utilize higher throughput SYBR green real time polymerase chain reaction technology in addition to standard thermocycling.
Keywords: CYP2A6, real time PCR, endpoint PCR, SNP, hybrid allele, gene deletion
Background
CYP2A6 genotyping has contributed to the understanding of inter-individual differences in the metabolism of nicotine and other therapeutics, such as the anti-cancer agents tegafur and letrozole, as well as smoking behaviors and the risk for tobacco-associated cancers (reviewed in [1–4]). More recently, CYP2A6 genotype has been established as a significant risk factor for lung cancer among Caucasian, African American and Japanese smokers [5–8], and CYP2A6 enzymatic activity, which is influenced by genetic polymorphisms, has been investigated as a tool for optimizing the selection of smoking cessation pharmacotherapy [9–11].
The CYP2A6 gene is located on chromosome 19q13 within a cluster of CYP2 family genes (CYP2T2P, 2F1P, 2A6, 2A7, 2G1P, 2A18PC, 2B7P, 2B6, 2A18PN, 2G2P, 2A13, 2F1, 2T3P, 2S1), which share a high degree of sequence homology and are thought to have arisen from a single locus through duplication events [12]. To date, 45 CYP2A6 star (*) alleles have been identified and listed in the Human Cytochrome P450 Allele Nomenclature Database [13]; additional polymorphisms continue to be identified and characterized [14]. Polymorphisms include single nucleotide polymorphisms (SNPs), small deletions/insertions, gene deletions and duplications, gene hybrids and gene conversions, which affect CYP2A6 enzymatic activity through diverse mechanisms from altering promoter activity to transcript stability and cofactor binding [13, 15]. Importantly, the prevalence of individual alleles varies greatly across world populations contributing to population differences in CYP2A6 activity [16, 17].
The challenge of CYP2A6 genotyping lies predominantly in the high degree of sequence homology between CYP2A6 and neighboring CYP2A genes (e.g. CYP2A6 and CYP2A7 share 95% sequence homology) and to the presence of many structural variants typically requiring more labor-intensive methods to ensure accurate genotype calls [18–20]. Owing to the complex genetic architecture of variation in CYP2A6 and to the prevalence of large numbers of low frequency alleles, studies using SNP platforms, such as genome wide association studies, have had limited utility identifying significant associations [21, 22]. The increasing number of identified functional variants, often present at low frequencies, also adds to the burden of a genotyping project [14]. To circumvent errors resulting from homology, many assays employ a two-step polymerase chain reaction (PCR) genotyping approach in which a CYP2A6 gene-specific first PCR amplification is performed and then utilized as template for an allele-specific second PCR amplification. Allele calls are typically based on genotyping the ‘defining/functional’ SNP or insertion/deletion, or, in the case of a hybrid allele, the crossover point, as opposed to typing the entire haplotype, which may include numerous additional synonymous or intronic alterations in linkage disequilibrium with the ‘causal’ variant [13], as time, cost and limited DNA are important considerations and high through-put sequencing for this gene is not yet available. In support of this approach, there is agreement between genotype-phenotype associations in vivo and the relationship predicted from investigations of ‘defining’ SNPs and variants in vitro [14, 23–25]. A limitation of this genotyping approach is that individuals may harbour additional variants, which were not screened for, or may not harbour all of the variants as defined in the CYP2A6 nomenclature database.
Herein, PCR-based methods and strategies (i.e. Method 1: Endpoint PCR allele amplification, and Method 2: SYBR green allele amplification) are presented to genotype alleles with altered activity that are found at appreciable frequencies in at least one racial/ethnic populations, CYP2A6*1B, *2, *4, *9, *12 and *1X2A, found predominantly in populations of Asian descent, CYP2A6*7, *8 and *10, and found predominantly in populations of African descent, CYP2A6*17, *20, *23–28, *31, *35, and *1X2B alleles [16, 17, 26]. Assays for the CYP2A6*34 allele are also included – genotypes with this allele are associated with reduced CYP2A6 activity, however the allele has not been extensively genotyped across populations (unpublished findings and [27]). With respect to nicotine metabolism, the CYP2A6*2, *4, *7, *17 and *20 alleles are considered inactive toward nicotine [17, 24, 25, 28–34], the CYP2A6*9 and *12 alleles have decreased nicotine metabolic activity whereby CYP2A6 *1/*9 and *1/*12 individuals have about 75% of the CYP2A6 activity and 80% of the total nicotine clearance of CYP2A6*1/*1 individuals [33, 34], the CYP2A6*23-*28, and *35 alleles also have evidence of reduced CYP2A6 metabolic activity towards nicotine [23, 26, 35, 36], and the CYP2A6*1B and CYP2A6*1X2 duplication alleles have modestly increased activity [20, 37, 38].
Traditional two-step PCR genotyping assays using endpoint PCR and gel electrophoresis to visualize results are described for all of the alleles mentioned above (Method 1). Additionally, for CYP2A6*1B, 9, *12, *17, *20, *23, *24, *31, *34 and *35, two-step PCR genotyping assays using the real time PCR platform and SYBR green to visualize results are provided (Method 2). CYP2A6 genotyping assays using real time PCR and SYBR green expedite the genotyping of large sample sets and minimize the use of genomic DNA, often a precious resource. Aside from the PCR-based methods outlined in this report, a number of commercial TaqMan, SNAPshot and array-based assays are available (e.g. those from Thermo Fisher Scientific, Illumina’s ADME panel, Affymetrix’ DMET panel), with the caveat that these generally work best for SNP genotyping in regions of limited homology. The only assay validated in the authors’ laboratory, is the TaqMan assay (Thermo Fisher Scientific) for the CYP2A6*2 allele (unpublished data). Commercial TaqMan assays, which we have not validated, are also available for the CYP2A6*17 and *20 alleles and to asses copy number variation.
General genotyping strategies for CYP2A6 alleles defined by SNPs or small indels
The general strategy for alleles that are defined by either a SNP or a small insertion or deletion, such as CYP2A6*2, *7, *8, *9, *10, *17, *20, *23-*28, *31, *35 (Figure 1) is a nested PCR approach whereby the first amplification, which is gene-specific, serves as a template for an allele-specific second amplification (Method 1.1 and Method 2.1). The allele-specific amplifications are designed around a ‘defining/functional’ SNP or indel within each allele – for example, the CYP2A6*2 second amplification targets the T>A SNP at genomic position 1799 (rs1801272), which results in the Leu160His amino acid substitution and loss of heme binding [19, 30]; CYP2A6*9 targets the T>G SNP at genomic position −48 (rs28399433), which disrupts the TATA box and results in reduced transcription [39, 40]; while CYP2A6*20 targets the deletion at genomic position 2141–2142, which causes a frameshift that results in an inactive truncated protein [24]. Similarly, allele-specific amplifications for CYP2A6*17 (rs28399454), *23 (rs56256500), *28 (rs8192730) and *31 (rs72549432) target SNPs resulting in amino acid substitutions [25, 35, 36]. For alleles, such as CYP2A6*10, *24, *26 and *27, which are defined as a haplotype of multiple SNPs/indels that may occur either separately or together, haplotypes can be tested directly or inferred based on the co-occurrence of variants in the same individual. For example, the CYP2A6*10 haplotype, which can be inferred based on the presence of the G>T SNP at position 6600 (also known as CYP2A6*8, rs28399468) among those with CYP2A6*7 (rs5031016), can also be determined directly through a haplotyping assay (described in Method 1.1) [28, 41]. Likewise, the CYP2A6*26 and *27 haplotypes can be inferred based on the co-occurrence of the T>G SNP at position 1711 (rs59552350) or the GC>A indel at 2162-3 (rs28399445), respectively, with CYP2A6*25 (rs2839940) [36]. Similarly, the CYP2A6*24 haplotype can be inferred based on the G>C SNP at genomic position 594 (rs72549435) co-occurring with CYP2A6*35 (rs61736436) [36].
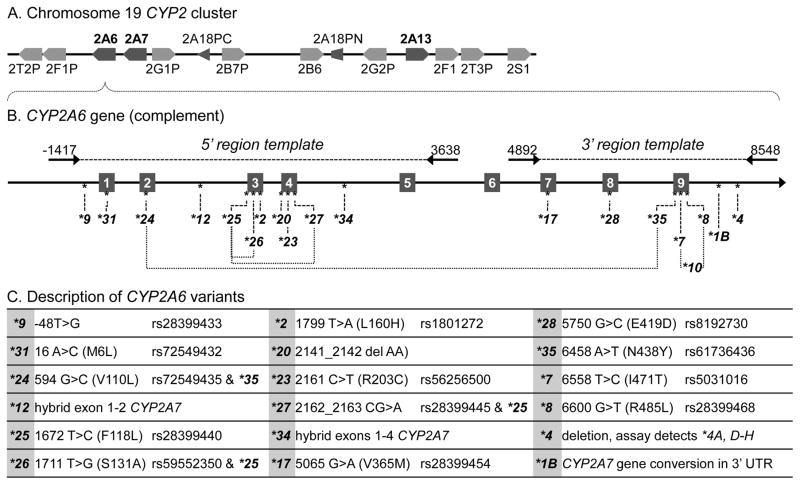
Figure 1.
Location of CYP2A6 gene and commonly genotyped variants. A. CYP2A6 within the CYP2 cluster of genes on chromosome 19 (modified from Hoffman and Hu, 2007). B. CYP2A6 gene indicating exons 1–9 (numbered boxes), commonly genotyped variants (* alleles) and gene-specific 5′ and 3′ region amplicons utilized as template in allele-specific SYBR green assays (Methods 2.1). The CYP2A6*12, *34 genotyping assays use a modified version of the 5′ region amplicon (Method 2.2). CYP2A6-CYP2A7 crossover positions are illustrated for the hybrid/deletion alleles, CYP2A6*12, *34, *4 and *1B. C. Description of commonly genotyped CYP2A6 variants including variation in reference gene template and amino acid sequence (if applicable).
General genotyping strategies for CYP2A6 alleles defined by structural variation
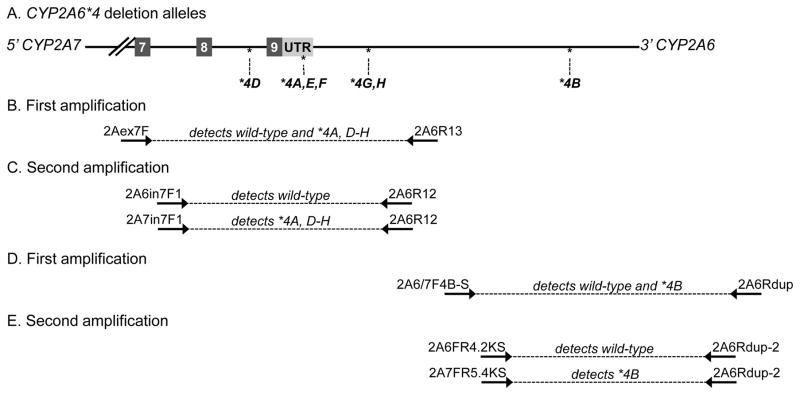
In addition to SNPs and indels, many structural variants have been identified in the CYP2A6 locus: the *1B gene conversion [42], the *4 gene deletions [29], the *1x2 gene duplications [20, 38] and the *12 and *34 hybrid alleles [27, 43] – each of these are hypothesized to result from unequal crossover events between CYP2A6 and the highly homologous CYP2A7 pseudogene during recombination [29, 38, 43]. Nested endpoint PCR assays are employed to genotype these structural variants. The first amplifications employed in these genotyping assays target upstream and/or downstream sequences shared by CYP2A6 and CYP2A7, whereas the second amplifications typically target the CYP2A6-CYP2A7 crossover region. For example, the CYP2A6*4 alleles are a family of CYP2A6 deletion alleles, each with a different amount of the remnant of the 3′ end of CYP2A6 appended to CYP2A7 (Figure 2A). The first amplification detects either wild-type CYP2A6 or the deletion allele using a 5′ primer that anneals to either CYP2A6 or CYP2A7 and a 3′ primer specific for CYP2A6 (Figure 2B & 2D), while the second amplification distinguishes between the wild-type and deletion allele (Figure 2C & 2E).
Figure 2.
CYP2A6*4 as an illustration of a CYP2A6-CYP2A7 hybrid allele. A. Crossover positions of the CYP2A6*4 alleles indicated relative to exons (numbered boxes). B. First amplification of PCR genotyping of CYP2A6*4H detects either wild-type CYP2A6 or all of the deletion alleles (except for *4B which is further downstream) using a 5′ primer that anneals to either CYP2A6 or CYP2A7, 2Aex7F, and a 3′ primer specific for CYP2A6, 2A6R13. C. Second amplification distinguishes between the wild-type allele and the deletion alleles using a 5′ primer that is specific to CYP2A6, 2A6in7F1, or to CYP2A7, 2A7in7F1, and a 3′ primer specific to CYP2A6, 2A6R12. D. First amplification of PCR genotyping of CYP2A6*4B detects either wild-type CYP2A6 or the *4B deletion allele using a 5′ primer that anneals to either CYP2A6 or CYP2A7, 2A6/7F4B-S, and a 3′ primer specific for CYP2A6, 2A6Rdup. E. Second amplification distinguishes between the wild-type allele and the *4B deletion alleles using a 5′ primer that is specific to CYP2A6, 2A6FR4.2KS, or to CYP2A7, 2A7FR5.4KS, and a 3′ primer specific to CYP2A6, 2A6Rdup-2. UTR: untranslated region
Method 1: Endpoint PCR allele amplification
Genotyping assays using endpoint PCR and gel electrophoresis to visualize results are described. All assays employ a two-step PCR genotyping approach in which the first PCR amplification is utilized as template for an allele-specific second PCR amplification that targets either the ‘defining/functional’ SNP or indel (Method 1.1), or, in the case of a hybrid allele, the crossover point (Method 1.2). Over the years as novel genetic variation was discovered which confounded original genotyping strategies (e.g. SNPs underneath first or second amplification primers), many primers were redesigned [29, 36, 37], and references to original or previous versions of primers and genotyping assays are provided (Supplementary Table 1) and a note on the rationale for recent primer modification is provided (Supplementary Table 2).
1.1. Protocols for CYP2A6 SNPs & small indels
First and second amplification primers for the CYP2A6*2, *7, *8, *9, *10, *17, *20, *23-*28, *31, *35 endpoint PCR genotyping assays are presented (Table 1 for primer combinations, Supplementary Table 3 for primer sequences). As indicated (Table 1), typically 2–4 allele-specific second amplifications use a common first amplification as template requiring ~250 ng of genomic DNA per sample to genotype all 15 SNP/indel alleles. The assays presented in Methods 2.1 reduce the genomic DNA requirement by utilizing two common first amplifications for all SNP/indel allele-specific assays. Reaction mixtures, cycling and gel electrophoresis conditions are provided (Table 2A and Table 2B).
Table 1.
Primer combinations for CYP2A6 endpoint PCR genotyping (Method 1.1 & 1.2)
| Genotying Assay | Forward primer | Reverse primer | Product size (bp) | Start positionb | End positionb |
|---|---|---|---|---|---|
| First amplifications | |||||
| CYP2A6*9, *31 | 2A65Pr1F | 2A6in1R | 1741 | −1417 | 324 |
| CYP2A6*2, *24, *25, *26 | 2A61F | 2A61R | 2055 | 144 | 2199 |
| CYP2A6*20, *23, *27 | 2A6exin3F | 2A6in5R | 1834 | 1804 | 3638 |
| CYP2A6*12, *34 | 2AinF-L | 2A6in5R | 3322 | 316 | 3638 |
| CYP2A6*1B, *17, *28, *35 | 2A6in6F1 | 2A6R13 | 3656 | 4892 | 8548 |
| CYP2A6*4H | 2Aex7F | 2A6R13 | 3538 | 5010 | 8548 |
| CYP2A6*4B | 2A6/7F4B-S | 2A6Rdup | 3098 | 10679 | 13776 |
| CYP2A6*7, *8, *10 | 2A6in6F1 | 2A6R6 | 3127 | 4892 | 8019 |
| CYP2A6*1X2A | 2Aex7F | 2A7R11 | a | 5010 | 8800 |
| CYP2A6*1X2B | 2A6F3 | 2A6/7R | 6745 | 7076 | 13821 |
| Second amplifications | |||||
| CYP2A6*9 | 2A6-460F | 2A6-17RA | 441 | −466 | −26 |
| 2A6-460F | 2A6-17RC | 441 | −466 | −26 | |
| CYP2A6*31 | 2A6-460F | 2A6ex1Rw | 502 | −466 | 36 |
| 2A6-460F | 2A6ex1Rv | 502 | −466 | 36 | |
| CYP2A6*2 | 2A62wtF | E3R-1 | 96 | 1786 | 1882 |
| 2A62v1F | E3R-1 | 96 | 1786 | 1882 | |
| CYP2A6*24 | 2Aex2Fwt | E3R-1 | 1303 | 579 | 1882 |
| 2Aex2Fv | E3R-1 | 1303 | 579 | 1882 | |
| CYP2A6*25 | 2A6in2ex3Fw | E3R-1 | 229 | 1653 | 1882 |
| 2A6in2ex3Fv | E3R-1 | 229 | 1653 | 1882 | |
| CYP2A6*26 | 2A6ex2Fwt | 2A6ex3R1711w | 1147 | 579 | 1726 |
| 2A6ex2Fwt | 2A6ex3R1711v | 1147 | 579 | 1726 | |
| CYP2A6*20 | 2A6in3F | 2A6ex42144Rw | 189 | 1969 | 2158 |
| 2A6in3F | 2A6ex42144Rv | 189 | 1969 | 2158 | |
| CYP2A6*23 | 2A6ex42161Fw-M | 5M13FOR-H2 | 121 | 2139 | 2260 |
| 2A6ex42161Fv-M | 5M13FOR-H2 | 121 | 2139 | 2260 | |
| CYP2A6*27 | 2A6in3F | 2A6*4171w-M | 213 | 1969 | 2182 |
| 2A6in3F | 2A6*4171v-M | 213 | 1969 | 2182 | |
| CYP2A6*12 | 2A6in/ex2 | 2A6inex5R | 3123 | 436 | 3559 |
| 2A7in/ex2 | 2A6inex5R | a | 954 | 3559 | |
| CYP2A6*34 | 2A6in4F | 2A6inex5R | 986 | 2573 | 3559 |
| 2A7in4F | 2A6inex5R | a | 2977 | 3559 | |
| CYP2A6*1B | 2A6*1Bwt | 2A6R12 | 1559 | 6719 | 8278 |
| 2A6*1Bvar | 2A6R12 | a | 7110 | 8278 | |
| CYP2A6*17 | 2A6*17Fwt-M | 2A6in7AS | 382 | 5044 | 5426 |
| 2A6*17Fv-M | 2A6in7AS | 382 | 5044 | 5426 | |
| CYP2A6*28 | 2A6in7F1 | 2A6ex8R2wt | 569 | 5200 | 5769 |
| 2A6in7F1 | 2A6ex8R2v | 569 | 5200 | 5769 | |
| CYP2A6*35 | 2A6in8ex9F6458w | 2A6R12 | 1835 | 6443 | 8278 |
| 2A6in8ex9F6458v | 2A6R12 | 1835 | 6443 | 8278 | |
| CYP2A6*4H | 2A6in7F1 | 2A6R12 | 3078 | 5200 | 8278 |
| 2A7in7F1 | 2A6R12 | a | 5594 | 8278 | |
| CYP2A6*4B | 2A6FR4.2KS | 2A6Rdup-2 | 2446 | 10770 | 13216 |
| 2A7FR5.4KS | 2A6Rdup-2 | a | 12450 | 13216 | |
| CYP2A6*7 | 2A6*7Fwt-M | 2A6R0 | 1244 | 6539 | 7783 |
| 2A6*7Fv-M | 2A6R0 | 1244 | 6539 | 7783 | |
| CYP2A6*8 | 2A6*8wtF | 2A6R0 | 1201 | 6582 | 7783 |
| 2A6*8vF | 2A6R0 | 1201 | 6582 | 7783 | |
| CYP2A6*10 | 2A6*7Fwt-M | 2A6*8Rwt-L | 81 | 6539 | 6620 |
| 2A6*7Fwt-M | 2A6*8Rv-L | 81 | 6539 | 6620 | |
| 2A6*7Fv-M | 2A6*8Rwt-L | 81 | 6539 | 6620 | |
| 2A6*7Fv-M | 2A6*8Rv-L | 81 | 6539 | 6620 | |
| CYP2A6*1x2A | 2A7in7F1 | 2A7R12 | 3093 | 5594 | 8687 |
| 2A6in7F1 | 2A7R12 | a | 5200 | 8687 | |
| CYP2A6*1x2B | 2A6F0 | 2A6Rdup | 6041 | 7735 | 13776 |
| 2A6F0 | 2A7Rdup | a | 7735 | 15131 |
Multiple slightly different product sizes are possible depending on where the crossover junctions are located. The differences in size between the hybrid alleles were not resolved on our 1.2% agarose gels
CYP2A6 primer position is based on CYP2A6 +1ATG, and CYP2A7 primer position is based on CYP2A7 +1ATG
Table 2A.
Polymerase chain reaction first-step amplification conditions (Method 1.1 & 1.2)
| First-step amplification
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
|
| ||||||||||
| Buffer | 2.5a | 2.5b | 2.5a | 2.5a | 2.5a | 2.5c | 2.5a | 2.5a | 2.5b | 2.5d |
| DNA (ng) | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 75 |
| Primers (each) (nM) | 62.5 | 125 | 62.5 | 125 | 125 | 100 | 125 | 125 | 62.5 | 250 |
| DMSO | – | – | – | – | – | – | – | – | – | 2% |
| dNTPs (uM) | 200 | 200 | 200 | 200 | 200 | 125 | 200 | 200 | 200 | 300 |
| MgCl2 (mM) | 1.3 | 2.0 | 1.5 | 1.5 | 1.5 | – | 2.5 | 1.5 | 1.7 | – |
| Taq (units) | 0.75Ɨ | 0.75Ɨ | 1.25Ɨ | 0.75Ɨ | 1.00Ɨ | 0.5uL£ | 1.25Ɨ | 1.00Ɨ | 1.25Ɨ | 1.25Ŧ |
|
| ||||||||||
| First denaturation (°C:sec) | 95:60 | 95:60 | 95:60 | 95:60 | 95:60 | 95:120 | 95:60 | 95:60 | 95:60 | 94:30 |
| [i]Denaturation (°C:sec) | 95:20 | 95:15 | 95:15 | 95:15 | 95:15 | 95:20 | 95:15 | 95:15 | 95:15 | 94:20 |
| [i]Annealing(°C:sec) | 55:30 | 60:30 | 58:30 | 60:30 | 50:30 | 50:20 | 52:20 | 50:30 | 53:30 | 54:20 |
| [i]Extension(°C:sec) | 72:60 | 72:120 | 72:120 | 72:210 | 72:210 | 72:60 | 72:180 | 72:180 | 72:180 | 68:420 |
| [ii]Denaturation (°C:sec) | – | – | – | – | – | – | – | – | – | 94:20 |
| [ii]Annealing(°C:sec) | – | – | – | – | – | – | – | – | – | 54:20 |
| [ii]Extension(°C:sec) | – | – | – | – | – | – | – | – | – | 68:420e |
| Number of cycles | 35 | 36 | 30 | 35 | 40 | 30 | 35 | 40 | 35 | –g |
| Last extension(°C:min) | 72:7 | 72:7 | 72:7 | 72:7 | 72:7 | 72:3 | 72:7 | 72:7 | 72:7 | 68:10 |
1: First amplification for CYP2A6*9, *31
2: First amplification fo CYP2A6*2, *24, *25, *26
3: First amplification fo CYP2A6*20, *23, *27
4: First amplification fo CYP2A6*12, *34
5: First amplification fo CYP2A6*1B, *17, *28, *35
6: First amplification fo CYP2A6*4H
7: First amplification fo CYP2A6*4B
8: First amplification fo CYP2A6*7,*8,*10
9: First amplification fo CYP2A6*1X2A
10: First amplification fo CYP2A6*1X2B
Taq DNA Polymerase (Thermo Scientific)
Pfu: PfuUltra II Fusion HS DNA Polymerase (Agilent Technologies)
Long PCR Enzyme Mix (Fermentas, Canada)
10X Taq Buffer with MgCl2 (Thermo, Canada)
10X Taq Buffer with (NH4)2SO4 (Thermo, Canada)
10X Pfu Buffer (Agilent Technologies)
10X Long PCR Buffer with MgCl2 (Fermentas, Canada)
+5 seconds per cycle
Ten cycles for [i], 20cycles for [ii]
Table 2B.
Polymerase chain reaction second-step amplification conditions (Method 1.1 & 1.2)
| Second-step amplification
| ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CYP2A6*9 | CYP2A6*31 | CYP2A6*2 | CYP2A6*24 | CYP2A6*25 | CYP2A6*26 | CYP2A6*20 | CYP2A6*23 | CYP2A6*27 | CYP2A6*12 | CYP2A6*34 | CYP2A6*1B | CYP2A6*17 | CYP2A6*28 | CYP2A6*35 | CYP2A6*4H | CYP2A6*4B | CYP2A6*7 | CYP2A6*8 | CYP2A6*10 | CYP2A6*1x2A | CYP2A6*1x2B | |
| Buffer | 2.5b | 2.5b | 2.5a | 2.5a | 2.5a | 2.5a | 2.5a | 2.5a | 2.5a | 2.5a | 2.5a | 2.5b | 2.5b | 2.5a | 2.5a | 2.5b | 2.5a | 2.5a | 2.5b | 2.5b | 2.5b | 2.5c |
| First-step product (μL) | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 |
| Primers (each) (nM) | 62.5 | 62.5 | 125 | 75 | 100 | 62.5 | 125 | 150 | 125 | 125 | 62.5 | 125 | 250 | 125 | 125 | 125 | 125 | 250 | 150 | 100 | 125 | 250 |
| DMSO | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 2% |
| dNTPs (μM) | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 300 |
| MgCl2 (mM) | 1.10 | 1.10 | 1.25 | 1.25 | 0.75 | 1.20 | 1.50 | 1.00 | 1.30 | 1.50 | 1.50 | 1.50 | 1.00 | 1.50 | 1.25 | 1.50 | 1.50 | 1.10 | 1.20 | 1.30 | 1.60 | – |
| Taq (units) | 0.4Ɨ | 0.4Ɨ | 0.25Ɨ | 0.3Ɨ | 0.3Ɨ | 0.4Ɨ | 0.3Ɨ | 0.4Ɨ | 0.4Ɨ | 0.5Ɨ | 0.5Ɨ | 0.5Ɨ | 0.5Ɨ | 0.3Ɨ | 0.5Ɨ | 0.5Ɨ | 0.5Ɨ | 0.3Ɨ | 0.4Ɨ | 0.3Ɨ | 0.75Ɨ | 1.25Ŧ |
|
| ||||||||||||||||||||||
| First denaturation (°C:sec) | 95:60 | 95:60 | 95:60 | 95:60 | 95:60 | 95:60 | 95:60 | 95:60 | 95:60 | 95:60 | 95:60 | 95:60 | 95:60 | 95:60 | 95:60 | 95:60 | 95:60 | 95:60 | 95:60 | 95:60 | 95:60 | 94:30:00 |
| [i]Denaturation (°C:sec) | 95:20 | 95:20 | 95:15 | 95:15 | 95:15 | 95:15 | 95:15 | 95:15 | 95:15 | 95:15 | 95:15 | 95:15 | 95:15 | 95:15 | 95:15 | 95:15 | 95:15 | 95:15 | 95:15 | 95:15 | 95:15 | 94:20 |
| [i]Annealing(°C:sec) | 66:40 | 65:30 | 50:20 | 58:10 | 65:10 | 59:20 | 56:20 | 62:10 | 56:20 | 62:20 | 59:20 | 52:30 | 58:30 | 58:20 | 55:40 | 52:30 | 50:20 | 59:20 | 57:20 | 57:30 | 60:30 | 54:20 |
| [i]Extension(°C:sec) | 72:60 | 72:60 | 72:45 | 72:90 | 72:45 | 72:60 | 72:30 | 72:30 | 72:40 | 72:120 | 72:60 | 72:120 | 72:60 | 72:60 | 72:60 | 72:180 | 72:90 | 72:60 | 72:60 | 72:30 | 72:150 | 68:420 |
| [ii]Denaturation (°C:sec) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 94:20 |
| [ii]Annealing(°C:sec) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 54:20 |
| [ii]Extension(°C:sec) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 68:420d |
| Number of cycles | 20 | 20 | 22 | 24 | 16 | 18 | 18 | 23 | 18 | 25 | 22 | 20 | 25 | 20 | 20 | 25 | 20 | 30 | 20 | 30 | 25 | –e |
| Last extension(°C:min) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 68:10 | |
Taq DNA Polymerase (Thermo Scientific)
Long PCR Enzyme Mix (Fermentas, Canada)
10X Taq Buffer with MgCl2 (Thermo, Canada)
10X Taq Buffer with (NH4)2SO4 (Thermo, Canada)
10X Long PCR Buffer with MgCl2 (Fermentas, Canada)
+5 seconds per cycle
Ten cycles for [i], 20cycles for [ii]
1.2. Protocols for CYP2A6 structural variants
First and second amplification primers for the CYP2A6*12 and *34 hybrid alleles, *1B gene conversion, *4 gene deletions, and *1X2A, *1X2B gene duplications endpoint PCR genotyping assays are presented (Table 1 for primer combinations, Supplementary Table 3 for primer sequences). Reaction mixtures, cycling and gel electrophoresis conditions are provided (Table 2). CYP2A6*12 and *34 comprise CYP2A7 sequence from exon 1 to introns 2 and 4, respectively, and CYP2A6 sequence downstream of those crossover points, and a common first amplification is provided as template for both allele specific assays. CYP2A6*1B contains a CYP2A7 gene conversion in the 3′ untranslated region of CYP2A6, which is associated with increased transcript stability but does not alter the enzymatic structure of CYP2A6 (essentially a different version of the *1 “wild-type” allele) [37, 42, 44]. As many reduced/loss of function CYP2A6 SNPs (e.g. *7, *8, *10, *24, *28, *35) occur in haplotype with this gene conversion [13], it is advisable to restrict *1B genotype-phenotype analyses to those subjects without other CYP2A6 variants [37]. Multiple CYP2A6*4 deletion assays exist, as additional crossover points have been discovered between CYP2A6 and CYP2A7 [29]. The CYP2A6*4H assay detects all of the known *4 deletion alleles (i.e. *A, *D-*H) except for the *4B deletion allele, which has a crossover point 3′ of *4H [29, 45]. The CYP2A6*4H first amplification is relatively long at 3,538 base pairs, and call rates can be improved from genomic DNA samples of lower quality using Pfu versus Taq DNA polymerase (Table 2A). An alternative genotyping approach for those exclusively interested in differentiating the CYP2A6*1B and *4C alleles from “wild-type” is a single PCR amplification of the 3′ end of CYP2A6/CYP2A7 followed by digestion with two restriction enzymes [42]. Assays have been developed for two gene duplication alleles, CYP2A6*1X2A and *1X2B, which are believed to be the reciprocal products of the unequal CYP2A6-CYP2A7 crossover events that created the *4D and *4B deletion alleles, respectively [20, 38].
Method 2: SYBR green allele amplification
The real time platform offers immediate time-savings over traditional endpoint PCR by eliminating the need for gel electrophoresis to visualize results. The use of SYBR green to detect amplification has the advantage of ease and low cost of assay modification with the identification of novel genetic variants, which likely will continue to be found within primer sequences. TaqMan technology requires the use of both specialized primers and probes, which may be more costly to modify. A nested PCR strategy is utilized in the real time genotyping protocols to mitigate the high homology between CYP2A6 and other CYP2A genes.
The SYBR green genotyping assays were developed using control samples of known CYP2A6 genotype and validated by genotyping samples drawn from multiple different study populations with both the endpoint PCR assays (Method 1) and the SYBR green assays (Method 2) to assess concordance (Supplementary Table 4). The guiding principle for primer design for SYBR green PCR technology included: melting temperature of 58–60°C, primer length of approximately 20 bases, amplicon size of 50–150 base pairs, GC content of 20–80%, avoidance of runs of identical nucleotides, minimal 3′ Gs and/or Cs, and minimal probability of self- and/or hetero-dimerization (manufacturer’s recommendations). To further minimize off-target amplification (in addition to using a gene-specific first amplification), allele-specific primers were designed to circumvent stretches of CYP2A6 sequence that are 100% homologous to either CYP2A7 or CYP2A13. Non-mismatch and mismatch primers were both tested and the primer pair yielding the greatest allelic discrimination was chosen [46, 47].
2.1. Protocols for CYP2A6 SNPs & small indels
Two separate first amplifications that cover the gene from 5′ of CYP2A6 (-1417) to intron 5 (3638) and from intron 6 (4892) to 3′ of CYP2A6 (8548), referred to as the 5′ and 3′ region templates, respectively (Figure 1B), are utilized as a template for all SNP and indel allele-specific assays to reduce the DNA required for comprehensive CYP2A6 genotyping and to increase throughput (primers Table 3, conditions Table 4A). The 5′ and 3′ region templates were developed using a regular thermocycler. Allele-specific SYBR green assays for the CYP2A6*9, *20, *23, *24 and *31 alleles utilize the amplified 5′ region template while allele-specific SYBR green assays for the CYP2A6*17 and *35 alleles utilize the amplified 3′ region template (primers Table 3, conditions Table 4B).
Table 3.
Primers combinationsfor CYP2A6 genotyping real-time PCR genotyping (Method 2.1 and 2.2)
| Assay | Primer Name | Locationa | Direction | Sequence |
|---|---|---|---|---|
| First amplifications
| ||||
| 5′ region template: | 2A65Pr1F | 5′ flanking (−1417) | F | ACC TAG ACT TAA TCT TCC CGT ATA C |
| CYP2A6*9, *31, *24, *20, *23 | 2A6in5R | Intron 5 (3638) | R | GGC CTG TGT CAT CTG CCT |
| Modified 5′ region: | 2Ain1F-L | Intron 1 (316) | F | GAT CTT GGG ATG TCC AGC TCC |
| CYP2A6*12, *34 | 2A6in5R | Intron 5 (3638) | R | GGC CTG TGT CAT CTG CCT |
| 3′ region template: | 2A6in6F1 | Intron 6 (4892) | F | ATTTCCTGCTCTGAGACC |
| CYP2A6*17, *35, *1B | 2A6R13 | 3′ flanking (8548) | R | GCC TCC CAT AGT GCT ATA ATT AAC A |
|
| ||||
| Second amplifications
| ||||
| CYP2A6*9 | 2A6*9wtR-Sybr-Mb | 5′ flanking (−29) | R | GCT GGG GTG GTT TGC CTC TA |
| 2A6*9vR-Sybr-Mb | 5′ flanking (−29) | R | GCT GGG GTG GTT TGC CTC TC | |
| 125M13BEV-B | 5′ flanking (−146) | F | CCC AAG CTA GGC AGG ATT CAT G | |
|
| ||||
| CYP2A6*31 | 2A6*31wtR-Sybr | Exon 1 (35) | R | AGC AAG GCC ACC AGA AGC AT |
| 2A6*31vR-Sybr | Exon 1 (35) | R | AGC AAG GCC ACC AGA AGC AG | |
| 2A61F-L | Exon 1 (−155) | F | TGG CTG TGT CCC AAG CTA GGC A | |
|
| ||||
| CYP2A6*24 | 2A6*24wtR-Sybr-Mb | ExIn 2 (619) | R | ACC CCC TCA CCA TAG CCT TTG AAT AC |
| 2A6*24vR-Sybr-Mb | ExIn 2 (619) | R | ACC CCC TCA CCA TAG CCT TTG AAT AG | |
| 2A6ex2-505F | Exon 2 (505) | F | TGC TGT GTG GAC ATG ATG CCG TCA G | |
|
| ||||
| CYP2A6*20 | 2A6*20wtR-sybr-Mb | Exon 4 (2160) | R | CAA CAG TGA CAG GAA CTC TTT GTA CT |
| 2A6*20vtR-sybr-Mb | Exon 4 (2160) | R | CAA CAG TGA CAG GAA CTC TGT ACT | |
| 2A6in3F-L | Intron 3 (1967) | F | CCC TGC CTC CTG GAA TTC TGA C | |
|
| ||||
| CYP2A6*23 | 2A6ex42161Fw-Mb | Exon 4 (2139) | F | CAA AGA GTT CCT GTC ACT GTC GC |
| 2A6ex42161Fv-M-Lb | Exon 4 (2137) | F | GA CAA AGA GTT CCT GTC ACT GTC GT | |
| 5M13FOR-H2 | Intron 4 (2260) | R | GCA GTT GGC AGG TTG TGG TAG G | |
|
| ||||
| CYP2A6*12 | 2A6in1/ex2-L | In1/Ex2 (434) | F | CCACCTCCATCAGATCAGTGAGC |
| 2A7in1/ex2-L | In1/Ex2 (952) | F | TCGCCTCCATCAGTTCAGTGAGT | |
| 2A6R-667 | Intron 2 (667) | R | GAACACTGAGACCTTCGTGTCCA | |
|
| ||||
| CYP2A6*34 | 2A6-2572F | Intron 4 (2572) | F | TCAACCGGCCTCCTGCATA |
| 2A7-2978F | Intron 4 (2978) | F | AAC CCG CCT CCT GCA TG | |
| 2A6-2862R | Intron 4 (2862) | R | GTTTACCTATCCAAGTGGGATGCACT | |
|
| ||||
| CYP2A6*17 | 2A6*17Fwt-Sybr-Mb | Exon 7 (5042) | F | ACG AGA TCC AAA GAT TTG GAG CCG |
| 2A6*17Fv-Sybr-Mb | Exon 7 (5042) | F | ACG AGA TCC AAA GAT TTG GAG CCA | |
| 2A6in7-17R2 | Intron 7 (5198) | R | GGA TGC TGG GGA CAC AGA GAG | |
|
| ||||
| CYP2A6*35 | 2A6*35wtR-Sybr | Exon 9 (6479) | R | CCA GGC CTT CTC CGA AAC AGT T |
| 2A6*35vR-Sybr | Exon 9 (6479) | R | CCA GGC CTT CTC CGA AAC AGT A | |
| 2A6in8-6395F | Intron 8 (6395) | F | CGA GGC TGC ACT GAG AGT GG | |
|
| ||||
| CYP2A6*1B | 2A6*1Bwt | 3′ flanking (6719) | F | ACT GGG GGC AGG ATG GC |
| 2A6*1Bvar-L | 3′ flanking (7101) | F | GGG TAT AAG AAT GGG GGG AAG ATG CG | |
| 2A6R6944 | 3′ flanking (6944) | R | GTG GCA ATT AGG TGA GCG TGC AAT G | |
CYP2A6 primer position is based on CYP2A6 +1ATG, and CYP2A7 primer position is based on CYP2A7 +1ATG
Mismatch primer at third position from 3′end, underlined in sequence
Table 4A.
Polymerase chain reaction first-step amplification conditions (Methods 2.1 & 2.2)
| First-step amplification | |||
|---|---|---|---|
|
| |||
| I | II | *12, *34 | |
|
| |||
| Buffer | 1xPfu buffer | 1xPfu buffer | 1X Taq Buffer with MgCl2 |
| DNA (ng) | 50 | 50 | 50 |
| Primers (each) (nM) | 100 | 100 | 125 |
| dNTPs (uM) | 62.5 | 62.5 | 200 |
| MgCl2 (mM) | na | na | 1.5 |
| TaqƗ | 0.5uL to 25ulƗ | 0.5uL to 25ulƗ | 0.75U to 25ul £ |
|
| |||
| First denaturation (°C:sec) | 95:120 | 95:120 | 95:60 |
| Denaturation (°C:sec) | 95:20 | 95:20 | 95:15 |
| Annealing(°C:sec) | 60:15 | 55:30 | 60:30 |
| Extension(°C:sec) | 72:75 | 72:60 | 72:210 |
| Number of cycles | 40 | 40 | 35 |
| Last extension(°C:sec) | 72:180 | 72:180 | 72:7 |
I: First amplification for 1st region (CYP2A6*9, *20, *23, *24, *31)
II: First amplification for 2nd region (CYP2A6*17, *35, *1B)
Pfu: PfuUltra II Fusion HS DNA Polymerase (Agilent Technologies)
Taq DNA Polymerase (Thermo Scientific)
Table 4B.
SYBR Green Polymerase chain reaction second-step amplification conditions (Methods 2.1 & 2.2)
| Second-step alleles (96 well)Ł | |||
|---|---|---|---|
|
| |||
| *20, *23, *24, *31, *1B | *9, *17, *35 | *12, *34 | |
|
| |||
| First -step prodoct (uL)£ | 1 | 1 | 1 |
| Primers (each) (nM) | 125 | 125 | 125 |
| SYBRGreen supermix (2x)Ŧ | 5uL to 10uL | 5uL to 10uL | 5uL to 10uL |
|
| |||
| Hold stage (°C:min) | 95:10 | 95:10 | 95:10 |
| PCR Stage | |||
| Denaturation (°C:sec) | 95:15 | 95:15 | 95:15 |
| Annealing(°C:sec) | 65:5 | 65:10 | 65:10 |
| Number of cycles | 50 | 50 | 50 |
| Melt Curve stage | |||
| Denaturation (°C:sec) | 95:15 | 95:15 | 95:15 |
| Annealing(°C:sec) | 60:60 | 60:60 | 60:60 |
ViiA 7 Real Time PCR System (Applied Biosystems)
Diluted 10 times
iTaqTM Universal SYBRGreen Supermix (Bio-Rad)
The SYBR green allele-specific second amplifications for the different CYP2A6 alleles utilize similar cycling conditions and reagent concentrations (Table 4B), which enables multiple assays to be run on the same plate. Importantly, the 5′ and 3′ region templates require dilution for use as the template in the allele-specific SYBR green assays. Serial dilutions (in PCR-grade water) from 10X to 1,000X were assayed and the lowest dilution that offered optimal discrimination was chosen (10X). Undiluted first amplifications can be stored at −30°C. Anecdotally, stored first amplification templates have been used for over a year after freezing with no change in assay performance. In addition to 94 well plates, the assays could be re-optimized for 384 well plates, which would require smaller reaction volumes and reagent quantities. As guidance for genotyping calls, typical delta Ct values for each genotype (from subtracting amplification in the ‘variant’ well from the ‘wild-type’ well) are as follows: >-10 for wild-type/wildtype, −2 to +2 for wild-type/variant, and >10 for variant/variant.
2.2. Protocols for CYP2A6 structural variants
The CYP2A6*1B SYBR green allele-specific second amplification uses the 3′ region template described in section 2.1 as template (Table 3 for primers and Table 4A for conditions) and targets the CYP2A7 gene conversion the 3′ untranslated region of CYP2A6 (Table 3 for primers, Table 4B for conditions). First and second amplification primers for the CYP2A6*12 and *34 hybrid allele real time genotyping assays are presented (Table 3). Reaction mixtures, cycling and gel electrophoresis conditions are also provided (Table 4A for first amplifications, Table 4B for second amplifications). The common first amplification for the CYP2A6*12 and *34 alleles is a modified version of the 5′ region first amplification (Method 2.1). The CYP2A6*12 and *34 alleles are a hybrid of CYP2A6 and CYP2A7 consisting of CYP2A7 from 5′ through to introns 2 and 4, respectively. As such the 5′ region first amplification forward primer (Method 2.1) was re-designed to detect a region of CYP2A6-CYP2A7 sequence homology.
Detection of the CYP2A6*4A-H deletion alleles by real time with SYBR green technology would require individual assays designed around the breakpoints for each deletion allele, whereas a single endpoint PCR genotyping assay (described above) can detect most of the *4 deletion alleles (Method 1.2). Other real time based PCR approaches have been developed to non-specifically detect CYP2A6 whole deletion alleles [48, 49]. Furthermore, in future, approaches similar to those used by Gaedigk et al. to simultaneously detect CYP2D6 hybrid and duplication alleles could be developed for CYP2A6, such as multiplex long-range PCR [50] and quantitative multiplex PCR amplification [51].
2.3 Using first amplifications as template in Method 1
The 5′ and 3′ region gene specific (first) amplifications presented in Method 2.1 can be utilized as template in the endpoint variant allele specific (second) amplifications in Method 1.1. As a guide, modifications to the endpoint PCR allele-specific amplifications (Method 1.2) include adjustments to the amount of first amplification template (0.6–0.8 uL), an increase in primer concentrations (200–500 nM), a reduction in magnesium chloride (0.75 mM), adjustments to the annealing temperature and time (~20–30 seconds), and the amount of Taq DNA polymerase (~0.3 units per reaction). For the CYP2A6*9 assay, the authors had success modifying the type of Taq DNA polymerase (MgCl2 instead of (NH4)2SO4).
CYP2A6 genotyping project considerations
The genotyping protocols provided may require optimization within each laboratory, as fluctuations in DNA quality and concentration, reagent concentrations, and thermocycling efficiency can have a substantial impact on PCR amplification (refer to reagent manufacturer’s recommendations on optimization). Genotyping assay performance is also a function of upstream sample processing. The source of DNA (e.g. blood, saliva), DNA isolation method, and storage (including freeze/thaw) can all have an impact on the quality of the DNA. The choice of CYP2A6 alleles to genotype depends largely on the allele frequencies within the population under study [4, 16, 17, 26]. For statistical analyses individuals are often separated by genotype into predicted reduced or normal CYP2A6 metabolic activity groups [5, 6] with the caveat that activity predictions may be substrate specific. Gene variants affecting the amount of CYP2A6 protein (e.g. CYP2A6*4 deletion allele) are expected to have a similar impact on all substrates; whereas, gene variants that influence enzymatic affinity and catalytic activity may differentially affect the metabolism of each substrate. For instance, both the CYP2A6*17 and CYP2A6*35 alleles, which are characterized by SNPs resulting in amino acid substitutions, appear to have a limited effect on coumarin metabolism but confer a substantial reduction in nicotine metabolism [35, 52].
Future perspective
The PCR genotyping methods outlined offer the ability to perform comprehensive CYP2A6 genotyping with minimal genomic DNA requirements and can be adjusted at minimal cost as novel variants are discovered. These assays along with commercial TaqMan assays will likely remain the main genotyping methods in the near-term. Higher throughput methods for the simultaneous detection of CYP2A6 copy number and hybrid variants similar to the quantitative multiplex PCR amplification recently developed for CYP2D6 [51] would also be of benefit near-term. Genome wide methods using tag SNPs or sequencing data to investigate the association of genetic variation with drug metabolism and outcomes, while increasingly utilized, appear to have limited utility for genes with complex architecture (i.e. insertion/deletions, copy number variation) and a high degree of homology with neighboring genes, such as CYP2A6, CYP2D6 and other pharmacogenes [21, 22, 53]. In the longer term, with improvements in the accuracy, cost and bioinformatics of sequencing (e.g. computational tools to resolve genotypes [54]), it may be possible to genotype CYP2A6 using sequence data. In the meantime, real time PCR approaches are likely to continue to be utilized for CYP2A6 genotyping, and genotyping is likely to be expanded to include additional regulatory factors, such as miR-126* sites and possibly copy number information for the CYP2A7 pseudogene, which could be influencing CYP2A6 transcript levels [55].
Supplementary Material
Executive Summary.
Background
Genetic variation in CYP2A6 results in altered metabolism of CYP2A6 substrates, such as nicotine and the anti-cancer agents, letrozole and tegafur, and CYP2A6 genotype is associated with cigarette smoking behaviors and the risk of tobacco-related cancers
CYP2A6 polymorphisms include SNPs, small deletions/insertions, gene deletions and duplications, and gene hybrids and conversions
The prevalence of individual alleles varies greatly across world populations contributing to population differences in CYP2A6 activity
Genotyping strategy
The challenge of CYP2A6 genotyping lies predominantly in the high degree of sequence homology between CYP2A6 and neighboring CYP2A genes, and to the presence of many structural variants typically requiring more labor-intensive methods to ensure accurate genotype calls
The general genotyping strategy is a nested PCR approach whereby the first amplification, which is gene-specific, serves as a template for an allele-specific second amplification, typically designed around a ‘defining/functional’ SNP or indel within each allele or the CYP2A6-CYP2A7 crossover region in the case of structural variants
Method 1
Endpoint PCR assays using traditional thermocycling and gel electrophoresis are presented for the SNPs/indels, CYP2A6*2, *7, *8, *9, *10, *17, *20, *23-*28, *31, *35, and for the structural variants, CYP2A6*1B, *1X2A, *1X2B, *4, *12, and *34
Method 2
Real time SYBR green PCR assays are presented for the SNPs/indels, CYP2A6*9, *17, *20, *23, *24, *31 and *35, and for the structural variants, CYP2A6*1B, *12 and *34
The real time platform offers immediate time-savings over traditional endpoint PCR by eliminating the need for gel electrophoresis to visualize results, and the use of SYBR green to detect amplification has the advantage of ease and low cost of assay modification
Genotyping project considerations
The PCR genotyping methods outlined offer the ability to perform comprehensive CYP2A6 genotyping with minimal DNA requirements and can be adjusted at minimal cost as novel variants are discovered
Future Perspective
Until the technical and bioinformatics challenges of genome wide approaches are solved, variation in CYP2A6 is best interrogated with pharmacogenetic versus genomic approaches and will increasingly rely on higher throughput real time based PCR methods
Acknowledgments
The authors thank Ewa B. Hoffmann for her work on refining CYP2A6 genotyping assays over the years. We acknowledge the support of the Endowed Chair in Addictions for the Department of Psychiatry (R.F. Tyndale), CIHR grants MOP86471 (R. F. Tyndale) and TMH-109787 (R. F. Tyndale), the Campbell Family Mental Health Research Institute of CAMH, the CAMH Foundation, the Canada Foundation for Innovation (#20289 and #16014 to R.F. Tyndale) and the Ontario Ministry of Research and Innovation.
Footnotes
Conflict of interest: In the past three years, Dr. Tyndale has consulted for Apotex on issues unrelated to genotyping. The remaining authors declare no conflicts of interest.
Contributor Information
Catherine A. Wassenaar, Email: catherine.wassenaar@utoronto.ca.
Qian Zhou, Email: zhouqian.zhou@utoronto.ca.
Rachel F. Tyndale, Email: r.tyndale@utoronto.ca.
References
- 1**.Mcdonagh EM, Wassenaar C, David SP, et al. PharmGKB summary: very important pharmacogene information for cytochrome P-450, family 2, subfamily A, polypeptide 6. Pharmacogenetics and genomics. 2012;22(9):695–708. doi: 10.1097/FPC.0b013e3283540217. Summary of CYP2A6 genetic variants and clinical associations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raunio H, Rahnasto-Rilla M. CYP2A6: genetics, structure, regulation, and function. Drug metabolism and drug interactions. 2012;27(2):73–88. doi: 10.1515/dmdi-2012-0001. [DOI] [PubMed] [Google Scholar]
- 3.Mwenifumbo JC, Tyndale RF. Genetic variability in CYP2A6 and the pharmacokinetics of nicotine. Pharmacogenomics. 2007;8(10):1385–1402. doi: 10.2217/14622416.8.10.1385. [DOI] [PubMed] [Google Scholar]
- 4*.Tanner JA, Chenoweth MJ, Tyndale RF. Pharmacogenetics of nicotine and associated smoking behaviors. Current topics in behavioral neurosciences. 2015;23:37–86. doi: 10.1007/978-3-319-13665-3_3. Table 1 summarizes CYP2A6 allele frequencies across multiple racial/ethnic populations. [DOI] [PubMed] [Google Scholar]
- 5.Wassenaar CA, Ye Y, Cai Q, et al. CYP2A6 reduced activity gene variants confer reduction in lung cancer risk in African American smokers--findings from two independent populations. Carcinogenesis. 2015;36(1):99–103. doi: 10.1093/carcin/bgu235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wassenaar CA, Dong Q, Wei Q, Amos CI, Spitz MR, Tyndale RF. Relationship between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 variation and smoking behaviors and lung cancer risk. J Natl Cancer Inst. 2011;103(17):1342–1346. doi: 10.1093/jnci/djr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7*.Ariyoshi N, Miyamoto M, Umetsu Y, et al. Genetic polymorphism of CYP2A6 gene and tobacco-induced lung cancer risk in male smokers. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2002;11(9):890–894. Initial report of CYP2A6 genotype and lung cancer risk among cigarette smokers. [PubMed] [Google Scholar]
- 8.Fujieda M, Yamazaki H, Saito T, et al. Evaluation of CYP2A6 genetic polymorphisms as determinants of smoking behavior and tobacco-related lung cancer risk in male Japanese smokers. Carcinogenesis. 2004;25(12):2451–2458. doi: 10.1093/carcin/bgh258. [DOI] [PubMed] [Google Scholar]
- 9*.Lerman C, Schnoll RA, Hawk LW, Jr, et al. Use of the nicotine metabolite ratio as a genetically informed biomarker of response to nicotine patch or varenicline for smoking cessation: a randomised, double-blind placebo-controlled trial. The Lancet. Respiratory medicine. 2015;3(2):131–138. doi: 10.1016/S2213-2600(14)70294-2. Randomised placebo controlled clinical trial demonstrating the utility of a CYP2A6 nicotine metabolism activity marker in optimizing choice of smoking cessation therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lerman C, Jepson C, Wileyto EP, et al. Genetic variation in nicotine metabolism predicts the efficacy of extended-duration transdermal nicotine therapy. Clin Pharmacol Ther. 2010;87(5):553–557. doi: 10.1038/clpt.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lerman C, Tyndale R, Patterson F, et al. Nicotine metabolite ratio predicts efficacy of transdermal nicotine for smoking cessation. Clin Pharmacol Ther. 2006;79(6):600–608. doi: 10.1016/j.clpt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman SM, Nelson DR, Keeney DS. Organization, structure and evolution of the CYP2 gene cluster on human chromosome 19. Pharmacogenetics. 2001;11(8):687–698. doi: 10.1097/00008571-200111000-00007. [DOI] [PubMed] [Google Scholar]
- 13.Cyp Allele Nomenclature Committee: The Human Cytochrome P450 (CYP) Allele Nomenclature Committee: CYP2A6 allele nomenclature.
- 14.Piliguian M, Zhu AZ, Zhou Q, et al. Novel CYP2A6 variants identified in African Americans are associated with slow nicotine metabolism in vitro and in vivo. Pharmacogenetics and genomics. 2014;24(2):118–128. doi: 10.1097/FPC.0000000000000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di YM, Chow VD, Yang LP, Zhou SF. Structure, function, regulation and polymorphism of human cytochrome P450 2A6. Curr Drug Metab. 2009;10(7):754–780. doi: 10.2174/138920009789895507. [DOI] [PubMed] [Google Scholar]
- 16.Binnington MJ, Zhu AZ, Renner CC, et al. CYP2A6 and CYP2B6 genetic variation and its association with nicotine metabolism in South Western Alaska Native people. Pharmacogenetics and genomics. 2012;22(6):429–440. doi: 10.1097/FPC.0b013e3283527c1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakajima M, Fukami T, Yamanaka H, et al. Comprehensive evaluation of variability in nicotine metabolism and CYP2A6 polymorphic alleles in four ethnic populations. Clin Pharmacol Ther. 2006;80(3):282–297. doi: 10.1016/j.clpt.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 18.Oscarson M, Mclellan RA, Gullsten H, et al. Characterisation and PCR-based detection of a CYP2A6 gene deletion found at a high frequency in a Chinese population. FEBS Lett. 1999;448(1):105–110. doi: 10.1016/s0014-5793(99)00359-2. [DOI] [PubMed] [Google Scholar]
- 19.Oscarson M, Gullsten H, Rautio A, et al. Genotyping of human cytochrome P450 2A6 (CYP2A6), a nicotine C-oxidase. FEBS Lett. 1998;438(3):201–205. doi: 10.1016/s0014-5793(98)01297-6. [DOI] [PubMed] [Google Scholar]
- 20.Rao Y, Hoffmann E, Zia M, et al. Duplications and defects in the CYP2A6 gene: identification, genotyping, and in vivo effects on smoking. Mol Pharmacol. 2000;58(4):747–755. doi: 10.1124/mol.58.4.747. [DOI] [PubMed] [Google Scholar]
- 21.Amos CI, Spitz MR, Cinciripini P. Chipping away at the genetics of smoking behavior. Nat Genet. 2010;42(5):366–368. doi: 10.1038/ng0510-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamazon ER, Skol AD, Perera MA. The limits of genome-wide methods for pharmacogenomic testing. Pharmacogenetics and genomics. 2012;22(4):261–272. doi: 10.1097/FPC.0b013e328350ca5f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al Koudsi N, Ahluwalia JS, Lin SK, Sellers EM, Tyndale RF. A novel CYP2A6 allele (CYP2A6*35) resulting in an amino-acid substitution (Asn438Tyr) is associated with lower CYP2A6 activity in vivo. Pharmacogenomics J. 2009;9(4):274–282. doi: 10.1038/tpj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukami T, Nakajima M, Higashi E, Yamanaka H, Mcleod HL, Yokoi T. A novel CYP2A6*20 allele found in African-American population produces a truncated protein lacking enzymatic activity. Biochem Pharmacol. 2005;70(5):801–808. doi: 10.1016/j.bcp.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 25.Fukami T, Nakajima M, Yoshida R, et al. A novel polymorphism of human CYP2A6 gene CYP2A6*17 has an amino acid substitution (V365M) that decreases enzymatic activity in vitro and in vivo. Clin Pharmacol Ther. 2004;76(6):519–527. doi: 10.1016/j.clpt.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 26*.Ho MK, Mwenifumbo JC, Al Koudsi N, et al. Association of nicotine metabolite ratio and CYP2A6 genotype with smoking cessation treatment in African-American light smokers. Clin Pharmacol Ther. 2009;85(6):635–643. doi: 10.1038/clpt.2009.19. Together with REF 29, Mwenifumbo et al 2008, provides genotype-phenotype associations for novel low frequency CYP2A6 alleles predominantly found in populations of African descent. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Iulio J, Fayet A, Arab-Alameddine M, et al. In vivo analysis of efavirenz metabolism in individuals with impaired CYP2A6 function. Pharmacogenetics and genomics. 2009;19(4):300–309. doi: 10.1097/FPC.0b013e328328d577. [DOI] [PubMed] [Google Scholar]
- 28**.Xu C, Rao YS, Xu B, et al. An in vivo pilot study characterizing the new CYP2A6*7, *8, and *10 alleles. Biochem Biophys Res Commun. 2002;290(1):318–324. doi: 10.1006/bbrc.2001.6209. Initial investigation of the nicotine metabolism activity of the CYP2A6*7, *8, and *10 alleles, and the establishment of these alleles as predominantly found in Chinese and Japanese populations. [DOI] [PubMed] [Google Scholar]
- 29**.Mwenifumbo JC, Zhou Q, Benowitz NL, Sellers EM, Tyndale RF. New CYP2A6 gene deletion and conversion variants in a population of Black African descent. Pharmacogenomics. 2010;11(2):189–198. doi: 10.2217/pgs.09.144. Identification of additional CYP2A6 deletion and hybrid alleles in a population of African descent. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamano S, Tatsuno J, Gonzalez FJ. The CYP2A3 gene product catalyzes coumarin 7-hydroxylation in human liver microsomes. Biochemistry. 1990;29(5):1322–1329. doi: 10.1021/bi00457a031. [DOI] [PubMed] [Google Scholar]
- 31.Peamkrasatam S, Sriwatanakul K, Kiyotani K, et al. In vivo evaluation of coumarin and nicotine as probe drugs to predict the metabolic capacity of CYP2A6 due to genetic polymorphism in Thais. Drug Metab Pharmacokinet. 2006;21(6):475–484. doi: 10.2133/dmpk.21.475. [DOI] [PubMed] [Google Scholar]
- 32.Dempsey D, Tutka P, Jacob P, 3rd, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin Pharmacol Ther. 2004;76(1):64–72. doi: 10.1016/j.clpt.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 33.Benowitz NL, Swan GE, Jacob P, 3rd, Lessov-Schlaggar CN, Tyndale RF. CYP2A6 genotype and the metabolism and disposition kinetics of nicotine. Clin Pharmacol Ther. 2006;80(5):457–467. doi: 10.1016/j.clpt.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 34.Malaiyandi V, Lerman C, Benowitz NL, Jepson C, Patterson F, Tyndale RF. Impact of CYP2A6 genotype on pretreatment smoking behaviour and nicotine levels from and usage of nicotine replacement therapy. Mol Psychiatry. 2006;11(4):400–409. doi: 10.1038/sj.mp.4001794. [DOI] [PubMed] [Google Scholar]
- 35.Ho MK, Mwenifumbo JC, Zhao B, Gillam EM, Tyndale RF. A novel CYP2A6 allele, CYP2A6*23, impairs enzyme function in vitro and in vivo and decreases smoking in a population of Black-African descent. Pharmacogenetics and genomics. 2008;18(1):67–75. doi: 10.1097/FPC.0b013e3282f3606e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36*.Mwenifumbo JC, Al Koudsi N, Ho MK, et al. Novel and established CYP2A6 alleles impair in vivo nicotine metabolism in a population of Black African descent. Hum Mutat. 2008;29(5):679–688. doi: 10.1002/humu.20698. Together with REF 23, Ho et al 2009, provides genotype-phenotype associations for novel low frequency CYP2A6 alleles predominantly found in populations of African descent. [DOI] [PubMed] [Google Scholar]
- 37.Mwenifumbo JC, Lessov-Schlaggar CN, Zhou Q, et al. Identification of novel CYP2A6*1B variants: the CYP2A6*1B allele is associated with faster in vivo nicotine metabolism. Clin Pharmacol Ther. 2008;83(1):115–121. doi: 10.1038/sj.clpt.6100246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukami T, Nakajima M, Yamanaka H, Fukushima Y, Mcleod HL, Yokoi T. A novel duplication type of CYP2A6 gene in African-American population. Drug metabolism and disposition: the biological fate of chemicals. 2007;35(4):515–520. doi: 10.1124/dmd.106.013557. [DOI] [PubMed] [Google Scholar]
- 39.Pitarque M, Von Richter O, Oke B, Berkkan H, Oscarson M, Ingelman-Sundberg M. Identification of a single nucleotide polymorphism in the TATA box of the CYP2A6 gene: impairment of its promoter activity. Biochem Biophys Res Commun. 2001;284(2):455–460. doi: 10.1006/bbrc.2001.4990. [DOI] [PubMed] [Google Scholar]
- 40.Kiyotani K, Yamazaki H, Fujieda M, et al. Decreased coumarin 7-hydroxylase activities and CYP2A6 expression levels in humans caused by genetic polymorphism in CYP2A6 promoter region (CYP2A6*9) Pharmacogenetics. 2003;13(11):689–695. doi: 10.1097/00008571-200311000-00005. [DOI] [PubMed] [Google Scholar]
- 41.Mwenifumbo JC, Myers MG, Wall TL, Lin SK, Sellers EM, Tyndale RF. Ethnic variation in CYP2A6*7, CYP2A6*8 and CYP2A6*10 as assessed with a novel haplotyping method. Pharmacogenetics and genomics. 2005;15(3):189–192. doi: 10.1097/01213011-200503000-00008. [DOI] [PubMed] [Google Scholar]
- 42.Ariyoshi N, Takahashi Y, Miyamoto M, et al. Structural characterization of a new variant of the CYP2A6 gene (CYP2A6*1B) apparently diagnosed as heterozygotes of CYP2A6*1A and CYP2A6*4C. Pharmacogenetics. 2000;10(8):687–693. doi: 10.1097/00008571-200011000-00003. [DOI] [PubMed] [Google Scholar]
- 43.Oscarson M, Mclellan RA, Asp V, et al. Characterization of a novel CYP2A7/CYP2A6 hybrid allele (CYP2A6*12) that causes reduced CYP2A6 activity. Hum Mutat. 2002;20(4):275–283. doi: 10.1002/humu.10126. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Pitarque M, Ingelman-Sundberg M. 3′-UTR polymorphism in the human CYP2A6 gene affects mRNA stability and enzyme expression. Biochem Biophys Res Commun. 2006;340(2):491–497. doi: 10.1016/j.bbrc.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 45.Ariyoshi N, Sekine H, Nakayama K, Saito K, Miyamoto A, Kamataki T. Identification of deletion-junction site of CYP2A6*4B allele lacking entire coding region of CYP2A6 in Japanese. Pharmacogenetics. 2004;14(10):701–705. doi: 10.1097/00008571-200410000-00008. [DOI] [PubMed] [Google Scholar]
- 46.Kwok S, Chang SY, Sninsky JJ, Wang A. A guide to the design and use of mismatched and degenerate primers. PCR Methods Appl. 1994;3(4):S39–47. doi: 10.1101/gr.3.4.s39. [DOI] [PubMed] [Google Scholar]
- 47.Liu J, Huang S, Sun M, et al. An improved allele-specific PCR primer design method for SNP marker analysis and its application. Plant Methods. 2012;8(1):34. doi: 10.1186/1746-4811-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimizu M, Sawaya R, Kishimoto I, Yamazaki H. Genotyping of wild-type cytochrome P450 2A6 and whole-gene deletion using human blood samples and a multiplex real-time polymerase chain reaction method with dual-labeled probes. Clinica chimica acta; international journal of clinical chemistry. 2015;441:71–74. doi: 10.1016/j.cca.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 49.Liu JH, Xun XJ, Pang C, et al. Single tube genotyping of CYP2A6 gene deletion based on copy number determination by quantitative real-time PCR. Experimental and molecular pathology. 2014;97(3):529–534. doi: 10.1016/j.yexmp.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 50.Gaedigk A, Fuhr U, Johnson C, Berard LA, Bradford D, Leeder JS. CYP2D7-2D6 hybrid tandems: identification of novel CYP2D6 duplication arrangements and implications for phenotype prediction. Pharmacogenomics. 2010;11(1):43–53. doi: 10.2217/pgs.09.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaedigk A, Twist GP, Leeder JS. CYP2D6, SULT1A1 and UGT2B17 copy number variation: quantitative detection by multiplex PCR. Pharmacogenomics. 2012;13(1):91–111. doi: 10.2217/pgs.11.135. [DOI] [PubMed] [Google Scholar]
- 52.Han S, Choi S, Chun YJ, et al. Functional characterization of allelic variants of polymorphic human cytochrome P450 2A6 (CYP2A6*5, *7, *8, *18, *19, and *35) Biol Pharm Bull. 2012;35(3):394–399. doi: 10.1248/bpb.35.394. [DOI] [PubMed] [Google Scholar]
- 53.Dewey FE, Grove ME, Pan C, et al. Clinical interpretation and implications of whole–genome sequencing. Jama. 2014;311(10):1035–1045. doi: 10.1001/jama.2014.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Numanagic I, Malikic S, Pratt VM, Skaar TC, Flockhart DA, Sahinalp SC. Cypiripi: exact genotyping of CYP2D6 using high-throughput sequencing data. Bioinformatics. 2015;31(12):i27–i34. doi: 10.1093/bioinformatics/btv232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakano M, Fukushima Y, Yokota SI, et al. CYP2A7 Pseudogene Transcript Affects CYP2A6 Expression in Human Liver by Acting as a Decoy for miR-126*. Drug metabolism and disposition: the biological fate of chemicals. 2015 doi: 10.1124/dmd.115.063255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.