Abstract
We investigated the role of genetic, physiological, environmental, and epigenetic factors in regulating CYP2A6 expression and nicotine metabolism. Human livers (n=67) were genotyped for CYP2A6 alleles and assessed for nicotine metabolism and CYP2A6 expression (mRNA and protein). In addition, a subset of livers (n=18), human cryopreserved hepatocytes (n=2), and HepG2 cells were used for DNA methylation analyses. Liver samples with variant CYP2A6 alleles had significantly lower CYP2A6 protein expression, nicotine C-oxidation activity, and affinity for nicotine. Female livers had significantly higher CYP2A6 protein and mRNA expression compared to male livers. Livers exposed to dexamethasone and phenobarbital trended towards having higher CYP2A6 expression and activity. Age and DNA methylation status of the CpG island and a regulatory site were not associated with altered CYP2A6. In conclusion, we identified genotype, gender, and exposure to inducers as sources of variation in CYP2A6 expression and activity, but much variation remains to be accounted for.
Introduction
Cytochrome P450 2A6 (CYP2A6) is primarily expressed in the liver and is involved in the metabolism of coumarin and a number of pharmaceuticals including halothane, valproic acid, tegafur, and SM-12502 (Pelkonen et al., 2000). In addition, this enzyme can activate procarcinogens such as aflatoxin B1 and tobacco specific nitrosamines (e.g. 4-methylnitrosoamino-1-(3-pyridyl)-1-butanone and N-nitrosodiethylamine) (Pelkonen et al., 2000). Clinically, CYP2A6 is of significance due to its major role in the metabolism of nicotine, the main addictive compound in tobacco (Benowitz, 2009). Indeed, multiple genotypic and phenotypic studies have associated variable CYP2A6 with an altered risk for tobacco-related cancers and multiple smoking behaviors such as the risk of being a smoker, the number of cigarettes smoked, and the likelihood of smoking cessation (Ho and Tyndale, 2007).
In vivo, the majority (70–80%) of nicotine is metabolized to cotinine (Benowitz and Jacob, 1994), and CYP2A6 mediates approximately 90% of this reaction (Nakajima et al., 1996; Messina et al., 1997). A common feature among in vivo studies assessing nicotine metabolism and CYP2A6 activity is the large interethnic and interindividual variability observed (Nakajima et al., 2006). Similarly, in human liver microsomes CYP2A6 mRNA, protein, and activity levels have been shown to vary >50-fold (Shimada et al., 1994; Rodriguez-Antona et al., 2001).
To date, 37 variant alleles of the CYP2A6 gene have been reported (http://www.cypalleles.ki.se/cyp2a6.htm), many of which result in altered activity accounting for some of the observed variability in CYP2A6 mediated nicotine metabolism (Mwenifumbo et al., 2008). Although several of these alleles have been studied extensively in vivo and in in vitro cDNA expression systems, their effect on CYP2A6 expression and nicotine pharmacokinetic parameters (Vmax and Km) in human livers has not yet been assessed. One aim of this study was to understand the mechanisms by which some of these variants are associated with altered activity in vivo. For example, do alleles associated with lower in vivo activity encode proteins with lower protein expression, intrinsic activity, and/or apparent affinity for the substrate?
Gender and age have also been shown to influence nicotine pharmacokinetics in vivo. Women have significantly higher rates of nicotine clearance compared to men (Benowitz et al., 2006), while elderly (>65 years) subjects have significantly lower rates of nicotine clearance compared to younger adults (22–44 years) (Molander et al., 2001). The higher rates of nicotine clearance among females may be mediated by higher CYP2A6 activity as measured by the established in vivo phenotypic ratio of trans-3′-hydroxycotinine to cotinine (3HC/COT) (Ho et al., 2009). Another aim of this study was to investigate whether the gender and age related differences in nicotine metabolism were mediated by differences in metabolic factors (i.e. different CYP2A6 expression/activity). Environmental factors could also influence nicotine clearance. For example, grapefruit juice reduces nicotine metabolism by inhibiting CYP2A6 (Hukkanen et al., 2006), while drugs such as phenobarbital and dexamethasone induce CYP2A6 (Maurice et al., 1991).
While CYP2A6 genetic variants, gender, and environment explain some of the variation in CYP2A6 expression and activity, there still remains unaccounted variation (Mwenifumbo et al., 2008). Another source of variation could be epigenetic regulation (Gomez and Ingelman-Sundberg, 2009). Epigenetic processes are heritable, or acquired, modifications of the DNA (i.e. methylation) or its associated proteins such as histones (e.g. histone acetylation) (Schumacher and Petronis, 2006). DNA methylation has been shown to affect the tissue-specific and general expression of several CYPs (e.g. CYP1B1, CYP1A2, CYP2E1, and CYP2W1) (Ingelman-Sundberg et al., 2007). Recently, CYP2A13 was shown to be induced following the co-treatment of NCI-H441 cells with the demethylating agent 5-Aza-2′-deoxycitidine (5-AzaC) and the histone deacetylase inhibitor trichostatin A (Ling et al., 2007). Since CYP2A6 shares a 93.5% amino acid sequence identity with CYP2A13 it is be possible that CYP2A6 is also epigenetically regulated. CYP2A6 has also been shown to contain a putative important CpG island which suggests a possible role for DNA methylation in its regulation (Ingelman-Sundberg et al., 2007). Our final aim was to investigate the potential role of DNA methylation in regulating CYP2A6 expression
In the current study we have used a panel of human liver samples to assess the impact of CYP2A6 genetic variants, gender, age, and exposure to inducers (phenobarbital and dexamethasone) on microsomal CYP2A6 levels and nicotine metabolic parameters (Vmax and Km). In addition, a smaller panel of livers (n=18), human cryopreserved hepatocytes (n=2), and HepG2 cells were utilized to investigate the potential role of DNA methylation in regulating CYP2A6.
Materials and Methods
Human Livers
The tissue samples studied here are a compilation of three liver banks: 1) 27 livers from the L-series (Messina et al., 1997), 2) 17 livers from the K-series (Campbell et al., 1987), and 3) 23 livers from the Biocenter in Basel, Switzerland (Meier et al., 1983). For clarity and consistency the livers from the Biocentre in Basel (Switzerland) will be referred to as the M-series livers. The characteristics and sources of all the livers have been described previously (Meier et al., 1983; Campbell et al., 1987; Messina et al., 1997). The ethnic origin of the liver samples was unknown and the cause of death and drug use was known for some of the samples. Mean age of the organ donors was 30 years (range 2–64) and the gender distribution was as follows: 35 males, 26 females, and 6 unknown.
Cell Culture
Human hepatocarcinoma cell line HepG2 cells were generously provided by Dr. David Riddick’s lab (University of Toronto, ON, Canada). The HepG2 cells were maintained in a humidified incubator (37 °C, 5%CO2) in MEM-α medium (Invitrogen, ON, Canada) supplemented with 10% fetal bovine serum (Invitrogen, ON, Canada). Two lots of human cryopreserved hepatocytes, lot SHM (male, 51 years old, low coumarin hydroxylase activity) and lot VEP (male, 56 years old, high coumarin hydroxylase activity), were purchased from Celsis In Vitro Technologies (MD, USA). The hepatocytes were maintained in InVitroGRO CP medium (Celsis In Vitro Technologies, MD, USA) supplemented with Torpedo Antibiotic Mix (Celsis In Vitro Technologies, MD, USA) on collagen-coated plates (BD Biocoat plates) at 37 °C and 5% CO2.
Nicotine C-oxidation kinetic assay
Liver microsomes were prepared by differential centrifugation as previously described (Messina et al., 1997), and protein content was determined by the Bradford protein assay (Bio-Rad Labs, ON, Canada). Nicotine C-oxidation kinetic parameters for the L-livers and two K-livers (K20 and K27) were previously measured by Messina et al. 1997. The remaining K and M-livers were assayed as previously described (Messina et al., 1997), with minor modifications. Nicotine (50, 100, 200, and 500 μM) and 20 minutes incubation was used. One sample (K27) was assessed using both assays yielding similar results (Vmax = 28 vs. 23 nmol/hr/mg of protein, Km = 64 vs. 58 μM). The kinetic parameters (Vmax and Km) for all the livers (n=67) were estimated using the Michaelis-Menten equation by the computer program GraphPad Prism (GraphPad Software Inc., CA, USA, version 5.0 for Windows).
CYP2A6 genotyping
DNA from all liver tissues (n=67) was isolated by phenol/chloroform (Invitrogen, Canada) extraction and ethanol precipitation. The DNA samples were genotyped for the following CYP2A6 alleles: CYP2A6*1B, *2, *4, *5, *6, *9, *12, *14, *17, *20, *21, *23, *24, *25, *26, *27, *28, *35, and the duplication *1x2 (Schoedel et al., 2004; Mwenifumbo et al., 2008; Al Koudsi et al., 2009; Ho et al., 2009). The liver samples were grouped into two genotype groups: 1) Wildtype livers (n=48) and 2) Variant livers (n=17). The wildtype group included samples positively genotyped for CYP2A6*1B, *14, and *21, but not any other allele. Those with CYP2A6*1B were included in the wildtype group for two main reasons. First, CYP2A6*1B is a wildtype allele usually included in the wildtype reference group in the literature (Nakajima et al., 2006; Mwenifumbo et al., 2008), along with other variants of the CYP2A6*1 allele (CYP2A6*1B-*1K, listed at http://www.cypalleles.ki.se/cyp2a6.htm). Second, in this study CYP2A6*1B did not influence CYP2A6 mRNA, protein, or activity levels. CYP2A6*14 was included in the wildtype group since previous studies (Nakajima et al., 2006; Mwenifumbo et al., 2008) and this current study did not find an effect on CYP2A6 expression or activity. To date, only two in vivo studies have assessed the impact of CYP2A6*21, where one study showed no effect (Al Koudsi et al., 2006), the other study associated it with lower activity (Mwenifumbo et al., 2008). Due to the conflicting literature, and that only one liver had a CYP2A6*21 allele, CYP2A6*21 was included in the wildtype group. Of note, the outcomes of all the analyses were not affected by either including or excluding CYP2A6*14 and CYP2A6*21 from the wildtype group.
CYP2A6 protein quantification
The L-livers and two K-livers (K20 and K27) were previously assessed for CYP2A6 protein levels (Messina et al., 1997). CYP2A6 protein levels for the remaining K and M-livers were determined by western blotting as described previously (Messina et al., 1997), with minor modifications. Amount of liver microsomal protein loaded was reduced to 5 μg instead of 30 μg. In addition, the primary (CYP2A6 monoclonal) and secondary (anti-mouse IgG horseradish peroxidase conjugate) antibodies were diluted 1:3000 and 1:17000, respectively. All other procedures were identical to that described by Messina et al., 1997. The CYP2A6 immunoreactivity levels of K20 and K27 obtained by Messina et al. 1997 were used as a normalization factor to provide a relative scale for all three liver banks. Thus, K20 and K27 were used as internal controls when quantifying CYP2A6 protein levels in the K and M-livers.
RNA isolation, cDNA synthesis and relative mRNA quantification
Tissues for only the K and M-livers (n=40) were available for RNA analyses. Following homogenization of ~ 0.1g of liver tissue, total RNA was isolated with TRIzol reagent (Invitrogen, ON, Canada). RNA concentration was determined spectrophotometrically, and the integrity of the 18S and 28S ribosomal bands was confirmed by electrophoresis on a 1.2% agarose gel (OnBio, ON, Canada) stained with ethidium bromide. cDNAs from liver RNAs were synthesized using 1 μg of total RNA, random hexamers (Invitrogen, ON, Canada), RNA guard (Invitrogen, ON, Canada), and M-MLV Reverse Transcriptase (Invitrogen, ON, Canada) according to the manufacturer’s protocols.
Primers for real-time PCR amplification of CYP2A6 and β-actin were as follows. CYP2A6 forward primer (2A6ex2/3F): 5′-TCA AAG GCT ATG GCG TGG TA-3′ and reverse primer (2A6ex3/4R): 5′-CGA TAT TGG CGC CGC CA-3′.β-actin forward primer (ACTBFex3): 5′-CAG AGC AAG AGA GGC ATC CT-3′ and reverse primer (ACTBRex4/3): 5′-GGT CTC AAA CAT GAT CTG GGT C-3′. Primer specificity was assessed using BLAST search.
Amplification and fluorescence detection were performed using the ABI 7500 Real-Time PCR system (Applied Biosystems). Real-time PCR amplification was carried out in a mixture (25μl) containing 1 μl of synthesized cDNA, 12.5 μl of 2 X Power SYBR-Green Madter Mix (Applied Biosystems), and 0.3 μM of each primer. Cycling conditions consisted of two activation steps (50 °C for 2 min, then 95 °C for 10 min) followed by 40 cycles of melting (95 °C for 15 sec) and annealing/extension (60 °C for 1 min). Relative quantifications of CYP2A6 gene expression were obtained by normalizing to β-actin and using the comparative CT method for relative quantification as described by the manufacturer (Real-Time PCR Chemistry Guide, Applied Biosystems, CA, USA). All liver samples were assessed in triplicate.
CYP2A6 DNA Methylation
Analysis of the CYP2A6 gene ± 10kb revealed two CpG sites of potential interest: 1) a CpG dinucleotide within a DR4-like element (−5476 bp) which has been shown to be involved in regulating CYP2A6 expression (Itoh et al., 2006) and 2) a CpG island in intron 2-exon 3 (1656–1889 bp). DNA methylation status of these sites were determined in HepG2 cells, human cryopreserved hepatocytes (n=2), and a subset of liver samples (n=18) that had no CYP2A6 variants (i.e. wildtype) but had extreme CYP2A6 phenotypes (i.e. very high and low CYP2A6 protein and activity levels, >50-fold variability).
DNA methylation status of the DR-4 site (8 CpGs) and the CpG island (25 CpGs) were evaluated using sodium bisulfite genomic sequencing. Genomic DNA was modified using the EpiTect Bisulfite kit (Qiagen, ON, Canada) in accordance with the manufacturer’s protocol. The modified DNA was then subjected to a two-step semi-nested PCR reaction. Primers used to amplify the first PCR product of the DR-4 site and the CpG island were as follows. DR-4 site forward primer (2A6DRF2): 5′-GGT GAT ATA GTT TGG GTT TGT G-3′ and reverse primer (2A6DRR2): 5′-AAC ATA AAA ATA CCT CAA CAT AC-3′. CpG island forward primer (2A6CpGF4): 5′-GGG AGT TTT TTG GAG TTG T-3′ and reverse primer (2A6CpGR): 5′-ACC TAA TCC CCA TCC C-3′. Primers used to amplify the second PCR product of the DR-4 site and the CpG island were as follows. DR-4 site forward primer 2A6DRF2 and reverse primer (2A6CpG5prR): 5′-ATC TCA ACT CAC TAC AAC CTC TA-3′. CpG-island forward primer (2A6PyroCpGF): 5′-TTT TTT AGG YGT GGT ATT TAG TAA-3′ and reverse primer 2A6CpGR.
Cycling conditions for the first amplification consisted of: initial denaturation at 95 °C for 3 min, 40 cycles of denaturation at 95 °C for 30 sec, annealing at 58 °C for 30 sec, and extension at 72 °C for 30 sec, followed by a final extension at 72 °C for 5 min. The reaction mixture (25 μl) contained: 100 ng of modified DNA, 0.2 mM of each dNTPs, 0.5 mM MgCl2, 1 X PCR buffer (Sigma Aldrich, ON, Canada), 1.25 U of Jumpstart Taq DNA polymerase (Sigma Aldrich, ON, Canada), and 0.125 μM of each primer. The cycling conditions for the second PCR reaction were identical to the first, except that the number of cycles was reduced to 25 cycles. The reaction mixture was similar to the first PCR reaction, except that the template was 0.8 μl of 10x diluted first amplification product. In addition, no MgCl2 was added and the final reaction volume was 50 μl.
PCR products were purified using PureLink PCR purification kit (Invitrogen, ON, Canada), cloned into a pGEM-T vector system (Promega, PA, USA), and sequenced using the universal M13-Forward primers. An average of 8–10 clones were sequenced per sample. Sequencing was carried out at Functional Biosciences, Inc (WI, USA). Specific amplification of CYP2A6 was confirmed by aligning the sequences of the PCR products with CYP2A6, CYP2A7, and CYP2A13 using CustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html). The BiQ Analyzer software was used to perform quality control and derive % DNA methylation from the sequencing results (Bock et al., 2005).
Statistical Analyses
Hardy-Weinberg equilibrium was tested using Chi-square, or Fisher’s exact test if five or fewer individuals were in one genotype group. The Kolmogorov-Smirnov and Shapiro-Wilk tests indicated that the phenotypic data (i.e. CYP2A6 mRNA levels, CYP2A6 protein levels, CYP2A6 DNA methylation, Vmax, Km, and Vmax/Km) were not normally distributed. Therefore, nonparametric tests were used in all statistical comparisons unless otherwise stated. Correlation analyses were performed using the Spearman’s rank correlation test (rS, rank correlation coefficient). Comparisons between two independent groups were carried out using the Mann-Whitney test. A linear regression model was formulated to evaluate which factors were associated with either CYP2A6 protein levels or Vmax. In this case the dependent variables (CYP2A6 protein levels or Vmax) were logged in order to normalize their distribution since regression analysis is a parametric test. By use of backward selection, the predictors (CYP2A6 genotype, gender, age, and exposure to inducers) were investigated for model inclusion. A predictor was considered to have a significant influence if p≤0.05. The advantage of a linear regression model is that the effect of each predictor on the outcome is tested while controlling for other variables that might also be affecting the outcome. All statistical analyses were performed using SPSS (SPSS Inc., Il, USA version 15.0 for windows). Graphs were generated using GraphPad Prism (GraphPad Software Inc., CA, USA, version 5.0 for Windows).
Results
Correlations between CYP2A6 mRNA, protein, and nicotine C-oxidation
Extensive interindividual variation in CYP2A6 mRNA levels, protein levels, and nicotine-to-cotinine metabolism (Vmax and Km) was observed. Relative CYP2A6 mRNA and protein levels varied greater than 1000 and 100 fold, respectively. The mean ± S.D. values for Vmax, Km and Vmax/Km were 30±26 nmol/mg protein/hr (range 4.0–120), 62±28 μM (range 16–162), and 0.6±0.5 nl/mg protein/hr (range 0.04–2.6).
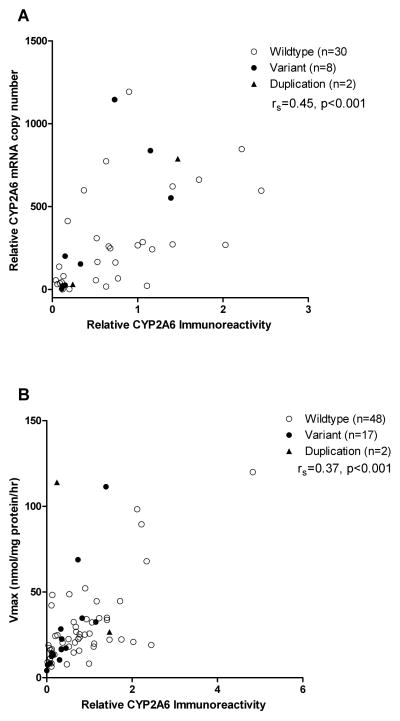
CYP2A6 mRNA correlated significantly with microsomal CYP2A6 protein (Fig. 1A, rS= 0.45, p<0.001). Immunoreactive CYP2A6 protein also correlated significantly with nicotine C-oxidation activity (Vmax) (Fig. 1B, rS= 0.37, p<0.001). The correlation between CYP2A6 mRNA levels and Vmax was less robust (n=40, rS= 0.15, p=0.02).
Figure 1. Correlations between CYP2A6 protein levels, CYP2A6 mRNA levels, and nicotine C-oxidase activity.
Open and closed circles denote livers in the wildtype and variant groups, respectively. Triangles denote livers positively genotyped for the duplication allele (CYP2A6*1X2). The horizontal line denotes the mean value in each group. (A) CYP2A6 protein and mRNA levels correlated significantly (n=40, rS= 0.45, p<0.001). (B) CYP2A6 protein levels and nicotine C-oxidase activity correlated significantly (n=67, rS= 0.37, p<0.001).
Impact of CYP2A6 genetic variants
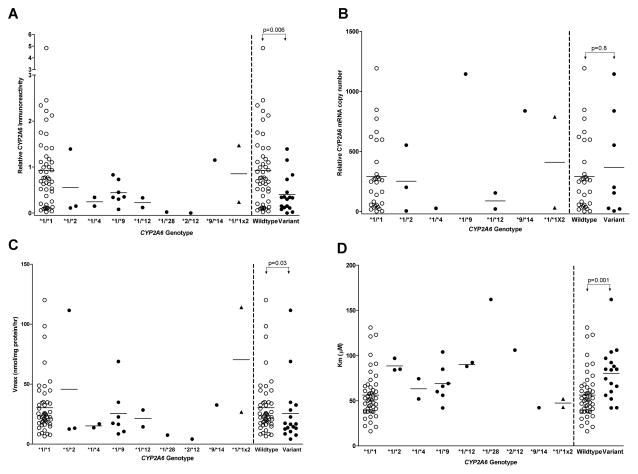
The CYP2A6 variants investigated in this study accounted for some of the variability observed in microsomal CYP2A6 protein expression and nicotine pharmacokinetic parameters (Vmax and Km) (Fig. 2). No livers with CYP2A6*5, *6, *17, *20, *23, *24, *25, *26, *27, *28, or *35 alleles were found. Livers with CYP2A6*2, *4, *9, *12, and *28 were identified and included in the variant group. All genotype frequencies did not deviate significantly from Hardy-Weinberg equilibrium. Liver samples in the wildtype group (n=48) had significantly higher microsomal CYP2A6 protein expression compared to livers in the variant group (n=17) (Fig. 2A; 0.90±0.90 vs. 0.40±0.40, p=0.006). However, relative CYP2A6 mRNA levels were similar between the two groups (Fig. 2B; wildtype (n=30): 291±298 vs. variant (n=8): 368±431, p=0.8). Of note, the clustering of low CYP2A6 mRNA levels in some wildtype samples (Fig. 2B) is not likely to be related to the integrity of the RNA as these samples had relatively high mRNA levels of certain transcription factors (data not shown). The higher CYP2A6 protein levels among the wildtype group (n=48) was associated with higher nicotine C-oxidation activity (Vmax) compared to the variant group (n=17) (Fig. 2C; 30±23 vs. 26±27 nmol/mg protein/hr, p=0.03). In addition, the mean apparent Km value was significantly lower among livers in the wildtype group (n=48) compared to the variant group (n=17) (Fig. 2D; 56±26 vs. 80±29, p=0.001). Finally, the higher Vmax and lower Km values among the wildtype group (n=48) resulted in higher mean Vmax/Km (catalytic efficiency) compared to the variant group (0.6±0.5 vs. 0.4±0.3 nl/mg protein/hr, p=0.002).
Figure 2. Effect of CYP2A6 variants on CYP2A6 protein levels, CYP2A6 mRNA levels, nicotine C-oxidation activity, and the apparent affinity for nicotine.
Open and closed circles denote livers in the wildtype and variant groups, respectively. Triangles denote livers positively genotyped for the duplication allele (CYP2A6*1X2). At least one sample with CYP2A6*1X2 also had other CYP2A6 variants thus livers with CYP2A6*1X2 were not included in the group (wildtype vs. variant) comparisons. The horizontal line denotes the mean value in each group. Liver samples in the variant group had lower CYP2A6 protein expression (A), similar CYP2A6 mRNA levels (B), lower nicotine C-oxidation activity (Vmax) (C), and higher apparent Km (D) compared to the wildtype group.
Impact of gender
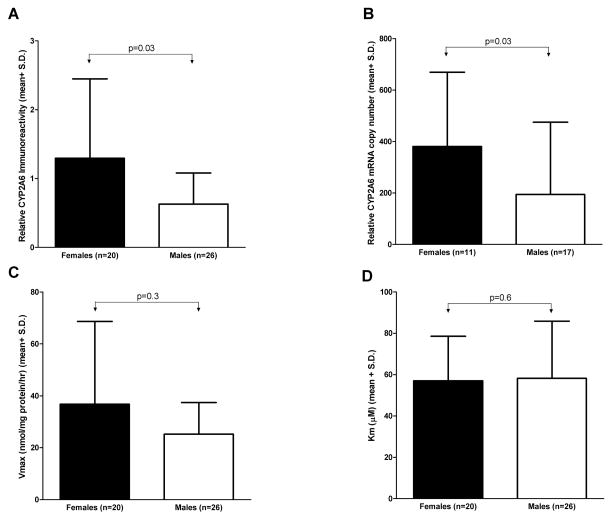
To minimize any potential bias due to known CYP2A6 genetic variants (Fig. 2), gender analyses were restricted to livers from the wildtype group. Females (n=20) had significantly higher microsomal CYP2A6 protein levels compared to males (n=26) (Fig. 3A; 1.3±1.2 vs. 0.6±0.5, p=0.03). In addition, females (n=11) had higher CYP2A6 mRNA levels compared to males (n=17) (Fig. 3B; 381±289 vs. 194±281, p=0.03). Nicotine C-oxidation activity (Vmax) trended towards being higher among females (n=20) compared to males (n=26) (Fig. 3C; 37±32 vs. 25±12 nmol/mg protein/hr, p=0.3). As expected the apparent Km was similar between females (n=20) and males (n=26) (Fig. 3D; 57±22 vs. 58±28 μM, p=0.6). Finally, Vmax/Km was not significantly higher among females (n=20) compared to males (n=26) (0.7±0.6 vs. 0.5±0.3 nl/mg protein/hr, p=0.6). When the variant livers were included in the gender analyses, the results remained the same, however the difference in CYP2A6 protein levels was no longer significant (p=0.07).
Figure 3. Impact of gender on CYP2A6 protein levels, mRNA levels, and nicotine pharmacokinetics.
Females had significantly higher CYP2A6 protein (A) and mRNA (B) levels compared to males. (C) Females trended towards having higher Vmax. (D) The apparent Km was similar between males and females. Two samples with unknown gender were not included in the analyses.
Impact of age
Among wildtype livers, age did not correlate with either CYP2A6 protein (rS= 0.04, p=0.2, n=44) or Vmax (rS= 7.0×10−4, p=0.9, n=44). A similar lack of correlation was observed when all the livers were analyzed.
Impact of CYP2A6 inducers
Dexamethasone and phenobarbital are known inducers of CYP2A6 (Maurice et al., 1991). Five liver samples exposed to these drugs (alone or in combination) trended towards having higher CYP2A6 protein (1.3±1.0 vs. 0.8±0.8, p=0.09), CYP2A6 mRNA (494±356 vs. 287±322, p=0.08) and Vmax (42±28 vs. 29±26 nmol/mg protein/hr, p=0.06) compared to all the other livers.
Modeling of CYP2A6 protein level and nicotine C-oxidation activity
The model developed for CYP2A6 protein levels and nicotine C-oxidation activity included the following predictors, CYP2A6 genetic variants, gender, age, and exposure to inducers. Only CYP2A6 genetic variants (p=0.006) and gender (p=0.008) were significant predictors of CYP2A6 protein levels. The final model accounted for 19% (11% CYP2A6 genotype and 8% gender) of the observed variability in CYP2A6 protein levels. The model developed for Vmax indicated that CYP2A6 genetic variants (p=0.03) and exposure to inducers (p=0.04) were the only significant predictors. The final model accounted for 11% (6% CYP2A6 genotype and 5% inducer exposure) of the observed variability in Vmax.
DNA methylation status of human livers, human cryopreserved hepatocytes, and HepG2 cells
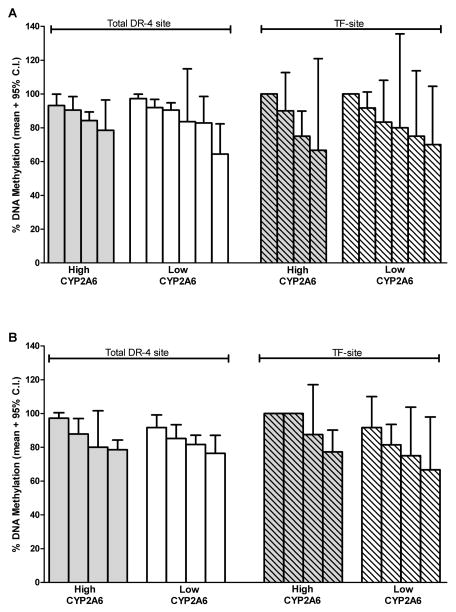
The DR-4 site studied herein contains a total of 8 CpG dinucleotides, one of which is located where the nuclear receptor pregnane X receptor (PXR) and the peroxisome proliferator-activated receptor-γ coactivator (PGC-1α) bind and modulate CYP2A6 expression (Itoh et al., 2006). This specific CpG dinucleotide within the DR-4 site will be referred to as the transcription factor site (TF-site). Among human liver samples, the average DNA methylation levels of the total DR-4 site and the specific TF-site were 85% (range 65–97%) and 84% (range 66–100%), respectively. Liver samples with extreme CYP2A6 phenotype (i.e. very high vs. low CYP2A6 levels/activity) had similar levels of methylation at both the DR-4 site and the TF-site (Fig. 4). Average DNA methylation level (mean ± 95% C.I.) of the DR-4 site (85±1.9 %) was much higher compared to the methylation at the CpG island (5.0±1.1 %) of the same liver samples (n=3). DNA methylation levels (mean ± 95% C.I.) of the CpG island was similar between liver samples with high (n=1) and low (n=2) CYP2A6 levels/activity (5.3±2.2 vs. 4.9±1.3 %, p=0.9).
Figure 4. DNA methylation status of the DR-4 and TF-sites among liver samples with either very high or very low CYP2A6 levels/activity.
Each bar represents a single liver sample. (A) Male liver samples with very high (n=4) and low (n=6) CYP2A6 levels/activity had similar levels of DNA methylation (mean ± 95% C.I.) at the DR-site (86±4 vs. 88±4 %, p=0.2) and the TF-site (81±10 vs. 88±7 %, p=0.2). (B) Female liver samples with very high (n=4) and low (n=4) CYP2A6 levels/activity had similar levels of DNA methylation (mean ± 95% C.I.) at the DR-site (83±4 vs. 83±4 %, p=0.6) and the TF-site (86±8 vs. 80±9 %, p=0.3).
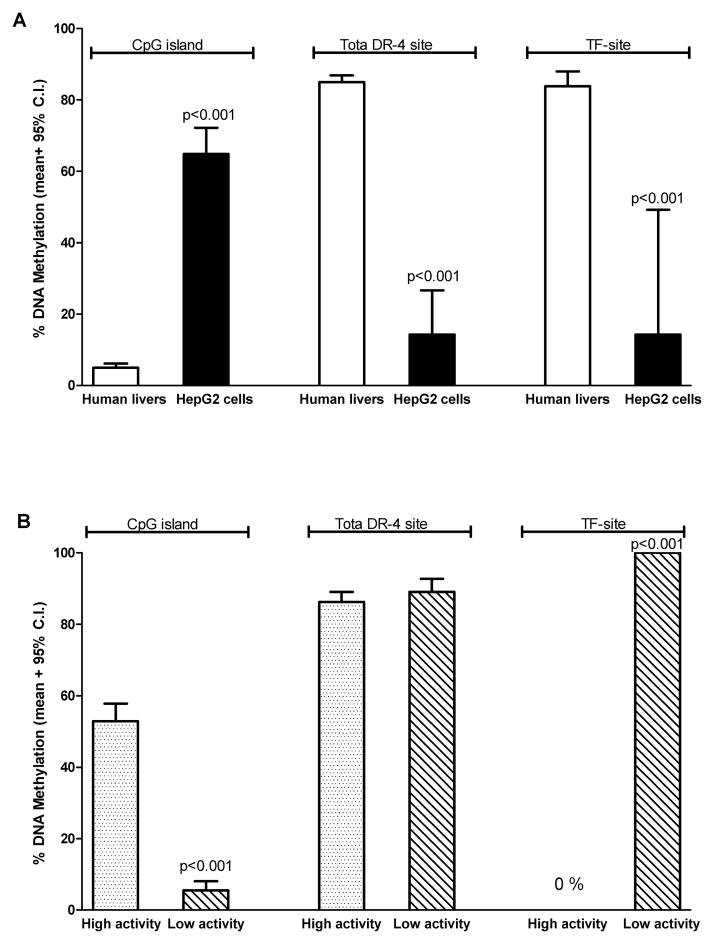
HepG2 cells, which express very low levels of CYP2A6, had an opposite pattern of methylation compared to human livers (i.e. high methylation in the CpG island and low methylation in the DR-4 and TF-sites, Fig. 5A). The cryopreserved hepatocyte lot with high CYP2A6 activity had higher levels of methylation (mean ± 95% C.I.) at the CpG island (53±5.0%) compared to the sample with low CYP2A6 activity (5.5±2.5%), Fig. 5B. The overall methylation (mean ± 95% C.I.) of the DR-4 site was similar between the two cryopreserved hepatocytes (Fig. 6B; 86±2.8 vs. 89±3.7 %, p=0.5). However, the specific CpG dinucleotide within the PXR/PGC-1α binding site (i.e. TF-site) was 0% methylated in the high CYP2A6 activity lot compared to 100% in the low CYP2A6 activity lot (Fig. 5B).
Figure 5. Comparison of DNA methylation levels at the CpG island, DR-4 site, and TF-site between human liver samples and HepG2 cells (A) and two lots of human cryopreserved hepatocytes (B).
Each bar represents the average methylation level. (A) HepG2 cells had significantly higher levels of CpG island DNA methylation (mean ± 95 % C.I.) compared to human livers (n=3) (65±7.4 vs. 5.0±1.1 %, p<0.001). In addition, HepG2 cells had significantly lower levels of DNA methylation compared to human livers (n=18) at both the DR-4 site (14±12 vs. 85±1.9 %, p<0.001) and the TF-site (14±35 vs. 84±4.2 %, p<0.001). (B) The cryopreserved hepatocyte lot with high CYP2A6 activity had significantly higher levels of CpG island methylation (mean ± 95 % C.I.) compared to the low activity lot (53±5.0 vs. 5.5±2.5 %, p<0.001). Although methylation levels (mean ± 95 % C.I.) between the high and low CYP2A6 activity lot were similar at the DR-4 site (86±2.8 vs. 89±3.7 %, p=0.5), the high activity lot was 0% methylated at the TF-site while the low activity lot was 100% methylated.
Discussion
Consistent with previous studies (reviewed in Pelkonen et al., 2000), we observed substantial interindividual variation in CYP2A6 expression and activity. The significant correlation between CYP2A6 mRNA levels and CYP2A6 protein levels suggests an important role for transcriptional regulation in mediating CYP2A6 protein expression.
One focus of this study was to test the impact of CYP2A6 genetic variants on CYP2A6 expression and nicotine C-oxidation pharmacokinetics in liver tissue. The low gene deletion CYP2A6*4 allele frequency and absence of alleles predominantly present among individuals of African origin (e.g. CYP2A6*17) (Nakajima et al., 2006) suggested that our population is likely of Caucasian origin. Livers in the variant group (CYP2A6*2, *4, *9, *12, and *28) had significantly lower CYP2A6 protein expression compared to the wildtype group. This suggests lower CYP2A6 protein expression as one likely mechanism by which these alleles are associated with lower in vivo CYP2A6 activity. CYP2A6*2 encodes an enzyme that is unable to incorporate heme making it unstable and more likely to be degraded (Yamano et al., 1990). CYP2A6*4 is a gene deletion that results in lower transcript levels and consequently lower protein expression. CYP2A6*9 contains a SNP (−48T>G) in the TATA-box that has been associated with lower CYP2A6 mRNA and activity in human livers (Kiyotani et al., 2003). CYP2A6*12 is a hybrid allele of CYP2A6 and CYP2A7 in which the 5′-prime and exons 1 and 2 are of CYP2A7 origin while exons 3 onwards are of CYP2A6 origin. In vitro, cells expressing CYP2A6.12 had approximately 50% lower immunodetectable CYP2A6, suggesting that it is an unstable enzyme that is more rapidly degraded (Oscarson et al., 2002). CYP2A6*28 is a haplotype allele (N418D and E419D) that has been associated with lower CYP2A6 activity in vivo (Mwenifumbo et al., 2008). It is possible that the two nonsynonymous SNPs in CYP2A6*28 might result in an unstable enzyme that is more readily degraded, however this remains to be tested. Alternatively, the coding SNPs in CYP2A6*28 might be in linkage disequilibrium with polymorphisms in the gene promoter or within introns that affect transcription and mRNA splicing, and as a result might influence protein expression (Mwenifumbo et al., 2008).
Although the variant group had lower CYP2A6 protein levels compared to the wildtype group, CYP2A6 mRNA levels were similar. It is possible that the variants might be affecting CYP2A6 protein levels at the post-transcriptional stage (e.g. enzyme degradation and/or stabilization) as opposed to affecting transcriptional efficiency. This is thought to occur with CYP2A6*2. On the other hand a correlation between the mRNA and protein levels is expected for CYP2A6*4 and CYP2A6*9. CYP2A6*4 (gene deletion) mRNA should not be amplified in our real time PCR reaction, while CYP2A6*9 has been associated with reduced transcriptional efficiency (Pitarque et al., 2001). We observed a trend for lower mRNA transcript levels among livers with CYP2A6*12. Of note, our CYP2A6 primers used in the real time PCR reaction are able to amplify the CYP2A6*12 mRNA. An association of CYP2A6*12 with lower mRNA levels has been previously observed and suggests that this allele is regulated at both the transcriptional and translational level (Haberl et al., 2005).
Consistent with the correlation between CYP2A6 protein levels and Vmax, the variant group had a lower mean Vmax compared to the wildtype group. In terms of the effect of genotype on the apparent Km (i.e. affinity) for nicotine, the variant group had a significantly higher mean Km value (i.e. lower affinity) compared to the wildtype group. More specifically, samples with CYP2A6*2 or CYP2A6*12 had a relatively higher mean Km value compared to the wildtype, while samples with CYP2A6*4 or CYP2A6*9 had roughly a similar mean Km value compared to the wildtype. This was anticipated since Km is an innate characteristic of an enzyme that is altered by structural changes (e.g. CYP2A6*2 and *12) but not by changes in expression levels (e.g. CYP2A6*4 and *9).
Among wildtype livers, female livers had approximately 2.0, 1.9 and 1.5 fold higher levels of CYP2A6 protein, CYP2A6 mRNA, and nicotine C-oxidation activity, respectively, compared to males. This is the first report of significant gender differences in liver CYP2A6 and suggests that the higher in vivo nicotine clearance rates (Benowitz et al., 2006) and CYP2A6 activity (Ho et al., 2009) observed among females is likely due to higher hepatic CYP2A6 expression. Pregnancy and oral contraceptive use have been shown to further increase the rates of nicotine clearance suggesting a hormonal effect on CYP2A6 (Hukkanen et al., 2005). Indeed, CYP2A6 is induced by estrogen in an estrogen receptor-alpha (ER-α) dependent manner (Higashi et al., 2007). The lack of an association in previous studies (Shimada et al., 1994; Messina et al., 1997; Parkinson et al., 2004) is likely due to the fact that these studies did not genotype for CYP2A6 variants. As we have demonstrated, CYP2A6 genetic variants have a significant effect on CYP2A6 expression which might mask gender differences.
There was no significant correlation between age and CYP2A6 expression/activity. This is consistent with previous human liver studies (Shimada et al., 1994; Parkinson et al., 2004). In contrast, an in vivo nicotine pharmacokinetic study found that elderly (>65 years) subjects had significantly lower rates of nicotine clearance compared to younger adults (22–44 years) (Molander et al., 2001). It is possible that we did not find an association between age and CYP2A6 levels as our age range was below that of 65 years; our oldest liver was from a 64 year old organ donor. Alternatively, the lower in vivo nicotine clearance rates among elderly individuals could be due to changes in other factors associated with age such as decrease in liver mass and blood flow (Kinirons and O’Mahony, 2004).
When all the factors (CYP2A6 genetic variants, gender, age, and exposure to inducers) were included in a model to predict variability in CYP2A6 protein levels, only CYP2A6 variants and gender were significant predictors. The final model accounted for only 19% of the observed variability in CYP2A6 levels, suggesting there still remains a large amount of variability that is unaccounted for. We cannot rule out the contribution of other unidentified genetic variants. In addition, variability in the expression of nuclear transcription factors which regulate cytochrome P450 (CYP) expression could also affect CYP2A6 levels. For example, CYP2A6 mRNA levels have been shown to significantly correlate with mRNA levels of the nuclear receptors CAR and HNF4-α (Wortham et al., 2007). We were able to capture a smaller amount of variation (11%) in nicotine C-oxidation activity (Vmax). This is likely due to the fact that other enzymes (CYP2B6) might also play a small role in this metabolic pathway (Yamazaki et al., 1999). In addition, variation in CYP activity could also be mediated by variation in the activity of CYP cofactors (e.g. NADPH-cytochrome P450 oxidoreductase) (Hart et al., 2008).
In human livers we did not observe a prominent role for DNA methylation in regulating CYP2A6. Interestingly, HepG2 cells, which express low levels of CYP2A6, had an opposite pattern of DNA methylation compared to liver cells, suggesting a possible role for DNA methylation in reducing CYP2A6 expression within this cell line. We treated the HepG2 cells with the demethylating agent 5-AzaC using a previously published paradigm (Dannenberg and Edenberg, 2006) but did not observe any changes in its DNA methylation status. Another interesting observation was that the cryopreserved hepatocyte lot with high CYP2A6 activity was fully demethylated at the TF-site while the low activity lot was fully methylated. The TF-site is a CpG dinucleotide within a DR-4 like element to which PXR/PGC-1α is thought to bind and modulate CYP2A6 expression (Itoh et al., 2006). Thus, the lack of methylation might make the site more accessible to the transcription factors resulting in higher expression/activity. It is important to note that the lack of an association between CYP2A6 expression and DNA methylation among the human livers is not evidence that CYP2A6 is not epigenetically regulated. Our investigation was limited to only two regions; however, there are many CpG-dense sites within the CYP2A6 gene that might be involved in epigenetic regulation. Recently, the allele specific expression of CYP2A7 was reported to be dependent on allele haplotype sequence and its methylation status, suggesting an interesting combinatorial effect between sequence variation and methylation status (Kerkel et al., 2008). In addition, CYP2A13 expression has been shown to be induced by epigenetic modulators (Ling et al., 2007). Given that CYP2A6 is highly homologous to CYP2A7 (96.5% identity) and CYP2A13 (93.5% identity) it is likely that CYP2A6 might be epigenetically regulated, perhaps at other loci.
In conclusion we have shown that CYP2A6 genetic variants markedly influence the expression of CYP2A6 and nicotine pharmacokinetics in human liver microsomes. Moreover, our results indicate that the higher in vivo nicotine clearance rates and CYP2A6 activity observed among females is likely due to higher hepatic CYP2A6 expression. Finally, our study suggests that even with our current state of knowledge, unexplained variation in CYP2A6 expression and activity still remains. Identifying sources of interindividual variation will help refine the relationship between CYP2A6 and phenotypic measures and improve our understanding of the influence of CYP2A6 on clinical outcomes.
Acknowledgments
We thank Bin Zhao, Qian Zhou, Fariba Baghai Wadji, Sharon Miksys, Linda Liu, and Amandeep Mann for their technical assistance and Dr. Arturas Petronis for his valuable input in planning and analyzing the epigenetic experiments.
This work was supported by the Centre for Addiction and Mental Health and Canadian Institutes for Health Research (CIHR) MOP86471. N.K. receives funding from CIHR-Strategic Training Program in Tobacco use in Special Population (TUSP) and Ontario Graduate Scholarship program (OGS). R.F.T. holds a Canada Research Chair in Pharmacogenetics.
Abbreviations
- CYPs
Cytochromes P450
- 5-AzaC
5-Aza-2′-deoxycitidine
- T
Trans-
- 3HC
3′-hydroxycotinine
- COT
Cotinine
References
- Al Koudsi N, Ahluwalia JS, Lin SK, Sellers EM, Tyndale RF. A novel CYP2A6 allele (CYP2A6(*)35) resulting in an amino-acid substitution (Asn438Tyr) is associated with lower CYP2A6 activity in vivo. Pharmacogenomics J. 2009 doi: 10.1038/tpj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Koudsi N, Mwenifumbo JC, Sellers EM, Benowitz NL, Swan GE, Tyndale RF. Characterization of the novel CYP2A6*21 allele using in vivo nicotine kinetics. Eur J Clin Pharmacol. 2006;62:481–484. doi: 10.1007/s00228-006-0113-3. [DOI] [PubMed] [Google Scholar]
- Benowitz NL. Pharmacology of nicotine: addiction, smoking-induced disease, and therapeutics. Annu Rev Pharmacol Toxicol. 2009;49:57–71. doi: 10.1146/annurev.pharmtox.48.113006.094742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benowitz NL, Jacob P., 3rd Metabolism of nicotine to cotinine studied by a dual stable isotope method. Clin Pharmacol Ther. 1994;56:483–493. doi: 10.1038/clpt.1994.169. [DOI] [PubMed] [Google Scholar]
- Benowitz NL, Lessov-Schlaggar CN, Swan GE, Jacob P., 3rd Female sex and oral contraceptive use accelerate nicotine metabolism. Clin Pharmacol Ther. 2006;79:480–488. doi: 10.1016/j.clpt.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–4068. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- Campbell ME, Grant DM, Inaba T, Kalow W. Biotransformation of caffeine, paraxanthine, theophylline, and theobromine by polycyclic aromatic hydrocarbon-inducible cytochrome(s) P-450 in human liver microsomes. Drug Metab Dispos. 1987;15:237–249. [PubMed] [Google Scholar]
- Dannenberg LO, Edenberg HJ. Epigenetics of gene expression in human hepatoma cells: expression profiling the response to inhibition of DNA methylation and histone deacetylation. BMC Genomics. 2006;7:181. doi: 10.1186/1471-2164-7-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez A, Ingelman-Sundberg M. Pharmacoepigenetics: its role in interindividual differences in drug response. Clin Pharmacol Ther. 2009;85:426–430. doi: 10.1038/clpt.2009.2. [DOI] [PubMed] [Google Scholar]
- Haberl M, Anwald B, Klein K, Weil R, Fuss C, Gepdiremen A, Zanger UM, Meyer UA, Wojnowski L. Three haplotypes associated with CYP2A6 phenotypes in Caucasians. Pharmacogenet Genomics. 2005;15:609–624. doi: 10.1097/01.fpc.0000171517.22258.f1. [DOI] [PubMed] [Google Scholar]
- Hart SN, Wang S, Nakamoto K, Wesselman C, Li Y, Zhong XB. Genetic polymorphisms in cytochrome P450 oxidoreductase influence microsomal P450-catalyzed drug metabolism. Pharmacogenet Genomics. 2008;18:11–24. doi: 10.1097/FPC.0b013e3282f2f121. [DOI] [PubMed] [Google Scholar]
- Higashi E, Fukami T, Itoh M, Kyo S, Inoue M, Yokoi T, Nakajima M. Human CYP2A6 is induced by estrogen via estrogen receptor. Drug Metab Dispos. 2007;35:1935–1941. doi: 10.1124/dmd.107.016568. [DOI] [PubMed] [Google Scholar]
- Ho M, Mwenifumbo J, Al Koudsi N, Okuyemi K, Ahluwalia J, Benowitz N, Tyndale R. Association of Nicotine Metabolite Ratio and CYP2A6 Genotype With Smoking Cessation Treatment in African-American Light Smokers. Clin Pharmacol Ther. 2009 doi: 10.1038/clpt.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho MK, Tyndale RF. Overview of the pharmacogenomics of cigarette smoking. Pharmacogenomics J. 2007;7:81–98. doi: 10.1038/sj.tpj.6500436. [DOI] [PubMed] [Google Scholar]
- Hukkanen J, Jacob P, 3rd, Benowitz NL. Metabolism and disposition kinetics of nicotine. Pharmacol Rev. 2005;57:79–115. doi: 10.1124/pr.57.1.3. [DOI] [PubMed] [Google Scholar]
- Hukkanen J, Jacob P, 3rd, Benowitz NL. Effect of grapefruit juice on cytochrome P450 2A6 and nicotine renal clearance. Clin Pharmacol Ther. 2006;80:522–530. doi: 10.1016/j.clpt.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther. 2007;116:496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Itoh M, Nakajima M, Higashi E, Yoshida R, Nagata K, Yamazoe Y, Yokoi T. Induction of human CYP2A6 is mediated by the pregnane X receptor with peroxisome proliferator-activated receptor-gamma coactivator 1alpha. J Pharmacol Exp Ther. 2006;319:693–702. doi: 10.1124/jpet.106.107573. [DOI] [PubMed] [Google Scholar]
- Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E, Morris M, Haghighi F, Tycko B. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- Kinirons MT, O’Mahony MS. Drug metabolism and ageing. Br J Clin Pharmacol. 2004;57:540–544. doi: 10.1111/j.1365-2125.2004.02096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyotani K, Yamazaki H, Fujieda M, Iwano S, Matsumura K, Satarug S, Ujjin P, Shimada T, Guengerich FP, Parkinson A, Honda G, Nakagawa K, Ishizaki T, Kamataki T. Decreased coumarin 7-hydroxylase activities and CYP2A6 expression levels in humans caused by genetic polymorphism in CYP2A6 promoter region (CYP2A6*9) Pharmacogenetics. 2003;13:689–695. doi: 10.1097/00008571-200311000-00005. [DOI] [PubMed] [Google Scholar]
- Ling G, Wei Y, Ding X. Transcriptional regulation of human CYP2A13 expression in the respiratory tract by CCAAT/enhancer binding protein and epigenetic modulation. Mol Pharmacol. 2007;71:807–816. doi: 10.1124/mol.106.031104. [DOI] [PubMed] [Google Scholar]
- Maurice M, Emiliani S, Dalet-Beluche I, Derancourt J, Lange R. Isolation and characterization of a cytochrome P450 of the IIA subfamily from human liver microsomes. Eur J Biochem. 1991;200:511–517. doi: 10.1111/j.1432-1033.1991.tb16212.x. [DOI] [PubMed] [Google Scholar]
- Meier PJ, Mueller HK, Dick B, Meyer UA. Hepatic monooxygenase activities in subjects with a genetic defect in drug oxidation. Gastroenterology. 1983;85:682–692. [PubMed] [Google Scholar]
- Messina ES, Tyndale RF, Sellers EM. A major role for CYP2A6 in nicotine C-oxidation by human liver microsomes. J Pharmacol Exp Ther. 1997;282:1608–1614. [PubMed] [Google Scholar]
- Molander L, Hansson A, Lunell E. Pharmacokinetics of nicotine in healthy elderly people. Clin Pharmacol Ther. 2001;69:57–65. doi: 10.1067/mcp.2001.113181. [DOI] [PubMed] [Google Scholar]
- Mwenifumbo JC, Al Koudsi N, Ho MK, Zhou Q, Hoffmann EB, Sellers EM, Tyndale RF. Novel and established CYP2A6 alleles impair in vivo nicotine metabolism in a population of Black African descent. Hum Mutat. 2008;29:679–688. doi: 10.1002/humu.20698. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Fukami T, Yamanaka H, Higashi E, Sakai H, Yoshida R, Kwon JT, McLeod HL, Yokoi T. Comprehensive evaluation of variability in nicotine metabolism and CYP2A6 polymorphic alleles in four ethnic populations. Clin Pharmacol Ther. 2006;80:282–297. doi: 10.1016/j.clpt.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, Inoue K, Funae Y, Shimada N, Kamataki T, Kuroiwa Y. Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metab Dispos. 1996;24:1212–1217. [PubMed] [Google Scholar]
- Oscarson M, McLellan RA, Asp V, Ledesma M, Bernal Ruiz ML, Sinues B, Rautio A, Ingelman-Sundberg M. Characterization of a novel CYP2A7/CYP2A6 hybrid allele (CYP2A6*12) that causes reduced CYP2A6 activity. Hum Mutat. 2002;20:275–283. doi: 10.1002/humu.10126. [DOI] [PubMed] [Google Scholar]
- Parkinson A, Mudra DR, Johnson C, Dwyer A, Carroll KM. The effects of gender, age, ethnicity, and liver cirrhosis on cytochrome P450 enzyme activity in human liver microsomes and inducibility in cultured human hepatocytes. Toxicol Appl Pharmacol. 2004;199:193–209. doi: 10.1016/j.taap.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Pelkonen O, Rautio A, Raunio H, Pasanen M. CYP2A6: a human coumarin 7-hydroxylase. Toxicology. 2000;144:139–147. doi: 10.1016/s0300-483x(99)00200-0. [DOI] [PubMed] [Google Scholar]
- Pitarque M, von Richter O, Oke B, Berkkan H, Oscarson M, Ingelman-Sundberg M. Identification of a single nucleotide polymorphism in the TATA box of the CYP2A6 gene: impairment of its promoter activity. Biochem Biophys Res Commun. 2001;284:455–460. doi: 10.1006/bbrc.2001.4990. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Antona C, Donato MT, Pareja E, Gomez-Lechon MJ, Castell JV. Cytochrome P-450 mRNA expression in human liver and its relationship with enzyme activity. Arch Biochem Biophys. 2001;393:308–315. doi: 10.1006/abbi.2001.2499. [DOI] [PubMed] [Google Scholar]
- Schoedel KA, Hoffmann EB, Rao Y, Sellers EM, Tyndale RF. Ethnic variation in CYP2A6 and association of genetically slow nicotine metabolism and smoking in adult Caucasians. Pharmacogenetics. 2004;14:615–626. doi: 10.1097/00008571-200409000-00006. [DOI] [PubMed] [Google Scholar]
- Schumacher A, Petronis A. Epigenetics of complex diseases: from general theory to laboratory experiments. Curr Top Microbiol Immunol. 2006;310:81–115. doi: 10.1007/3-540-31181-5_6. [DOI] [PubMed] [Google Scholar]
- Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther. 1994;270:414–423. [PubMed] [Google Scholar]
- Wortham M, Czerwinski M, He L, Parkinson A, Wan YJ. Expression of constitutive androstane receptor, hepatic nuclear factor 4 alpha, and P450 oxidoreductase genes determines interindividual variability in basal expression and activity of a broad scope of xenobiotic metabolism genes in the human liver. Drug Metab Dispos. 2007;35:1700–1710. doi: 10.1124/dmd.107.016436. [DOI] [PubMed] [Google Scholar]
- Yamano S, Tatsuno J, Gonzalez FJ. The CYP2A3 gene product catalyzes coumarin 7-hydroxylation in human liver microsomes. Biochemistry. 1990;29:1322–1329. doi: 10.1021/bi00457a031. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Inoue K, Hashimoto M, Shimada T. Roles of CYP2A6 and CYP2B6 in nicotine C-oxidation by human liver microsomes. Arch Toxicol. 1999;73:65–70. doi: 10.1007/s002040050588. [DOI] [PubMed] [Google Scholar]