Abstract
Mercury is a highly toxic heavy metal and the ability of the neurotoxin methylmercury to biomagnify in the food chain is a serious concern for both public and environmental health globally. Because thousands of tons of mercury are released into the environment each year, remediation strategies are urgently needed and prompted this study. To facilitate remediation of both organic and inorganic forms of mercury, Escherichia coli was engineered to harbor a subset of genes (merRTPAB) from the mercury resistance operon. Protein products of the mer operon enable transport of mercury into the cell, cleavage of organic C-Hg bonds, and subsequent reduction of ionic mercury to the less toxic elemental form, Hg(0). E. coli containing merRTPAB was then encapsulated in silica beads resulting in a biological-based filtration material. Performing encapsulation in aerated mineral oil yielded silica beads that were smooth, spherical, and similar in diameter. Following encapsulation, E. coli containing merRTPAB retained the ability to degrade methylmercury and performed similarly to non-encapsulated cells. Due to the versatility of both the engineered mercury resistant strain and silica bead technology, this study provides a strong foundation for use of the resulting biological-based filtration material for methylmercury remediation.
Introduction
Microbial transformation of metals has a large impact on biogeochemical cycles and can alter metal distribution and partitioning in the environment. Alterations, such as change in redox state and conversion between organic and inorganic states, affect solubility and toxicity of metals and hence have great impact on environmental and public health [1,2]. Toxicity of the metal mercury is a particular concern at present because mono-methylmercury (hereafter, methylmercury), the most common organic form, is a neurotoxin that biomagnifies in the food chain [3]. Five thousand to 8000 tons of mercury are estimated to be emitted into the atmosphere yearly from both human and natural sources, and anthropogenic emissions are expected to increase through 2050 [4]. Current remediation strategies exist for mercury but are prohibitively costly in many environments and other solutions are needed [5].
Bioremediation offers a potentially cost-effective and environmentally conscious approach to the problem of mercury pollution. An attractive biological-based remediation strategy for mercury pollution is utilization of the mercury resistance (mer) operon found in bacteria. The mer operon exists in a variety of structures and organizational forms, and a few key genes have become the central targets for remediation efforts [5]. Essential to remediation of both organic and inorganic forms of mercury are the key enzymes MerB and MerA, respectively. MerB cleaves the C-Hg bond of organomercurials through protonolysis resulting in Hg(II) that is then reduced by MerA, the mercuric reductase, to volatile Hg(0) [2, 6]. Other genes important to the system include merP and merT that encode for an Hg(II) transport system across the periplasm and inner membrane, and merR that encodes for the mercury-specific regulator of the operon [2, 7].
Previous bioremediation approaches for mercury have centered on usage of bacteria with engineered or naturally occurring mer operons and/or a variety of metal binding proteins. Genetic engineering has been used to introduce parts from the mer operon into a variety of hosts proposed for use in mercury removal from contaminated sites [8–10]. Other studies have focused on engineering bio-sorbent strains utilizing Mer proteins and/or metal binding proteins or chelators such as metallothionein and polyphosphate kinase [11–19]. Bio-sorbent strains are limited by their metal retention capacity, and because sorption is a passive process, strains must be regenerated after reaching saturation. Use of bio-sorbent strains also requires methods to separate mercury from biomass for recovery. The only method to date able to recover mercury and work at technical scale is the use of natural mer-containing strains of Pseudomonas adsorbed to silica pumice granules in packed bed bioreactors [20, 21]. Because adsorbed cells can easily be released in effluent water, engineered strains cannot be used with this type of system [20]. Also, the formation of biofilm and exopolysaccharide within pumice material may limit diffusion in flow-through systems. Here we describe the use of a silica gel whole cell encapsulation system to address these challenges.
Silica encapsulation has previously been used in atrazine bioremediation [22, 23], providing protection of the biocatalyst, avoidance of dispersal of organisms, and overall mechanical structure that broadens possible engineering applications. Silica gels are formed by condensation or gelation of a hydrolyzed silicon alkoxide crosslinker into a solid silica matrix. Following cross-linker hydrolysis, cells added during condensation become entrapped within the gel matrix [22]. Recent improvements in encapsulation technology have resulted in methods retaining cell viability, which is imperative for mercury remediation since reduction of Hg(II) by MerA is an NADPH-dependent reaction [2, 23]. Encapsulated cells have been shown to retain high enzymatic activity over a period of months [23]. Optimization and modeling studies are also available to minimize material cost and pressure drop in packed beds while maintaining material strength [24].
Materials and Methods
Bacterial Strains and Culture Conditions
E. coli strain MG1655 was kindly provided by Dr. Arkady Khodursky (University of Minnesota). E. coli strains UQ950 and WM3064 used for cloning and conjugal transfer have been described previously [25]. For routine propagation of E. coli, single colonies from freshly streaked -80°C stocks were used to inoculate cultures grown for 16 hours in Luria Broth (LB) medium supplemented with 50 μg mL-1 kanamycin when appropriate. Unless specified otherwise, cultures were grown in LB, shaken continuously at 250 rpm, and incubated at 37°C.
Reagents and Materials
Enzymes were purchased from New England Biolabs (Ipswich, MA). Kits for gel purification and plasmid mini preps were purchased from Qiagen (Valencia, CA). All related reactions were carried out according to manufacturer instructions.
Media components, including Noble agar, were purchased from Becton, Dickinson and Company (Sparks, MD). Chemicals for encapsulation including Ludox TM40, alkoxide tetramethyl orthosilicate (TMOS), and Polyethylene Glycol 600 were purchased Sigma-Aldrich (St. Louis, MO). Mercuric chloride and methylmercury chloride were purchased from Fisher Chemical (Pittsburgh, PA). Due to the toxicity of mercury compounds, all safety protocols and operating procedures were reviewed by the Department of Environmental Health and Safety at the University of Minnesota.
Plasmid Construction
Plasmid pDU1358 was kindly provided by Dr. Anne Summers (University of Georgia, Athens) [26]. Plasmid pBBRBB has been described previously [27]. Genes merRTPAB were amplified from pDU1358 in two stages to enable incorporation in the BioBrick compatible vector pBBRBB. First, a portion of merA and merB were amplified using primers merAmut-F (GTCGCGCATGTGAACGGCGAGTTCGTGCTGACCACGGGACA) and merB-R (nnACTAGTTCACGGTGTCCTAGATGACA) to mutate the internal EcoRI restriction site (bp 1024–1029) within merA. The resulting fragment was then gel purified and used to prime the second reaction along with primer merR-F (nnTCTAGACTACACCGCGTCGGCACCAC) to amplify merRTPAB. This fragment was digested with XbaI and SpeI, gel purified, and cloned into the corresponding sites of pBBRBB generating plasmid pBBRBB::mer. Constructs in the pBBRBB backbone were moved into E. coli by conjugal transfer using donor strain WM3064.
Zone of Inhibition Plate Assays
E. coli strains were picked from single colonies into LB medium supplemented with 50 μg mL-1 kanamycin. Overnight cultures were diluted 10-fold, and 3 mL was added to tryptone medium agar plates (containing per liter: 15 g tryptone, 5 g NaCl, 10 g Noble agar, 1 pellet sodium hydroxide). Noble agar was used to limit agar batch variability that can confound heavy metal assays. Excess culture was removed after 5 minutes, a 6 mm paper disc was added to the center of the plate, and 10 μL of a 0.1 M HgCl2 stock solution was added to each disc. HgCl2 stock solutions were made fresh for each experiment. Plates were incubated at 37°C for 16 hours after which zones of inhibition were measured as the diameter of clearing around each disc.
Encapsulation
Methods for encapsulation were adapted from previous sol-gel techniques [23, 28] Hydrolyzed cross-linker was produced by mixing TMOS with ultrapure water and 1 M HCl (1:1:0.001 vol/vol/vol) and incubating for two hours at room temperature. A solution containing colloidal silica nanoparticles (TM40), polyethylene glycol (PEG-600) and phosphate-buffered saline (PBS) was prepared prior to encapsulation (2:2:1 vol/vol/vol). Bacteria re-suspended in PBS between 0.1–0.2 g mL-1 were then introduced to this solution with a 1:1 volumetric ratio to create a homogenous solution with silica interspersed between cells. Hydrolyzed cross-linker was then spiked into this solution and immediately transferred into aerated mineral oil (800 rpm, 15 min) to provide uniform viscosity throughout emulsification. After letting the silica set, beads containing the embedded bacteria were purified by phase-separation. Beads entered the dH2O phase of the oil-water mixture and were isolated using a separatory funnel. Samples were both washed and stored in PBS overnight at 4°C prior to methylmercury testing.
Methylmercury Assays
Assays were conducted in acid-cleaned Balch tubes [29] containing 7 mL LB medium and 1 mg L-1 methylmercury chloride. Using previously published viability data for our encapsulation methods, encapsulated cells were inoculated at a cell density within an order of magnitude of that of the non-encapsulated cells [23]. For assays using encapsulated cells, 0.3 g of encapsulation material was added to each tube. For assays with non-encapsulated cells, overnight cultures were washed in minimal medium and inoculated to an initial optical density (OD600) of ~ 0.1. Cultures were incubated at 37°C and shaken at 250 rpm. Samples were removed for analysis at times indicated.
Samples were analyzed for monomethylmercury in the University of Minnesota Mercury Analytical Laboratory using EPA method 1630 modified to eliminate sample distillation. Samples were placed in acid-cleaned 40 mL I-chem glass vials fitted with PTFE/silicone septa and brought to a final volume of 30 mL with distilled deionized water. Samples received 0.225 mL of 2 M pH 4.5 acetate buffer and 0.03 mL of sodium tetraethylborate ethylating solution. Monomethylmercury concentrations were determined from headspace gas analysis by a Tekran model 2700 Automated MethylMercury Analyzer with Hg detection by cold vapor atomic fluorescence spectrometry (CVAFS) following capillary gas chromatography and pyrolization of ethylated Hg species.
Quality Assurance/Quality Control measures included the preparation of a calibration curve from 10 ng L-1 and 500 ng L-1 working standards at the start of each run of samples and the analysis of control check standards (0.1 and 0.5 ng L-1) every 10 samples. Recoveries for control check standards averaged 98%, well within acceptable values.
Microscopy
Samples were fixed in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) overnight at 4°C, rinsed in 0.1 M sodium cacodylate buffer, then placed in 1% osmium tetroxide in 0.1 M sodium cacodylate buffer overnight at 4°C. Specimens were rinsed in ultrapure water (NANOpure Infinity; Barnstead/Thermo Fisher Scientific; Waltham, Maryland), dehydrated in an ethanol series, and processed in a critical point dryer (Autosamdri-814; Tousimis; Rockville, Maryland). Material was mounted on double-sided adhesive carbon tabs on aluminum stubs, sputter-coated with gold-palladium, and observed in a scanning electron microscope (S3500N; Hitachi High Technologies America, Inc.; Schaumberg, Illinois) at an accelerating voltage of 10 kV.
Results and Discussion
To develop a bioremediation strategy for both organic and inorganic forms of mercury, we began by cloning merRTPAB of the mer operon from pDU1358 (originally isolated from Serratia marcescens) into E. coli K12 resulting in strain E. coli pBBRBB::mer [26]. During amplification, the internal EcoRI restriction site in merA was mutated in order to create a mer cassette compatible with the biobrick cloning system and the vector pBBRBB (see Materials and Methods) [27]. Biobrick compatibility creates a plug-and-play vector system and facilitates addition of further remediation cassettes. Use of a modular approach in plasmid design allows the strain to be tailored to the specific needs of future remediation sites. Following construction, zone of inhibition plates were used to assess resistance to ionic forms of mercury by engineered strains. Strains were spread evenly on tryptone medium plates containing discs loaded with an HgCl2 solution. Following overnight incubation, zones of inhibition were measured as the diameter of clearing around discs (Fig 1). E. coli pBBRBB::mer was resistant to ionic mercury as measured zones of clearing for E. coli pBBRBB::mer were comparable to the positive control E. coli pDU1358 and significantly smaller (p-value = 0.0002) than control strains containing empty vector (Fig 1; Table 1).
Fig 1. Representative zone of inhibition from filter disc assays for mercury(II) chloride resistance for A) E. coli pDU1358 B) E. coli pBBRBB::mer and C) E. coli pBBRBB.
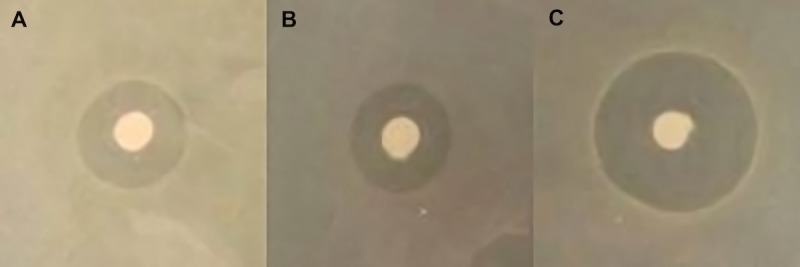
Filter discs in each image are identical, (6 mm in diameter).
Table 1. Filter disc assay for mercury(II) chloride resistance in E. coli.
Results are the average of three independent experiments with error represented as standard error of the mean.
| Strain | Zone of Inhibition Diameter (mm) |
|---|---|
| E. coli pDU1358 | 16.5 ± 0.3 |
| E. coli pBBRBB::mer | 16.7 ± 0.7 |
| E. coli pBBRBB | 26.5 ± 0.3 |
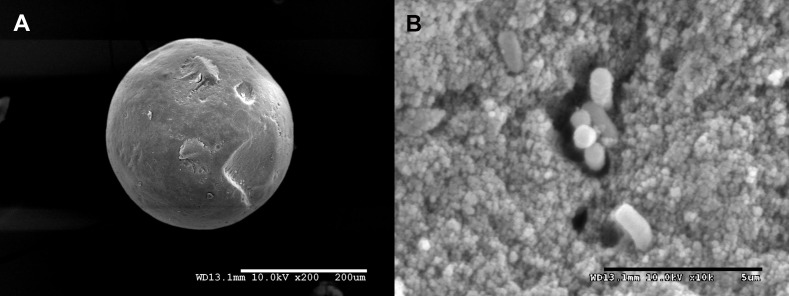
To use engineered E. coli pBBRBB::mer cells in a filtration system, the cells must be fully encapsulated in silica microbeads to prevent release of biological material. Cells were mixed with a colloidal silica nanoparticle/PEG solution and then spiked with a hydrolyzed silicon TMOS solution. Transfer of this solution to aerated mineral oil enabled encapsulation of cells and resulted in the formation of smooth, spherical silica gel microbeads (Fig 2A). Microbead structures were chosen for encapsulation because they increase surface to volume ratio for bioremediation efforts and result in a biological-based filtration material that can be used in packed bed reactors. Measurement of 20 beads using a Hitachi scanning electron microscope indicated an average bead diameter of 210 ± 60 μm. Images also indicated that gel porosity was in the nanometer range, similar to previously characterized hyperporous beads generated using the same sol-gel methods, which limits mobility of encapsulated cells (Fig 2B) [23]. Because of the gel structure and limited space, cellular division is likely also inhibited. Despite limited space, use of PEG in silica bead formation has been shown to retain cell viability for at least three weeks following encapsulation [23].
Fig 2. Scanning Electron Microscopy images of encapsulation silica sol-gel microbeads containing E. coli pBBRBB::mer.
A) Representative image depicting the smooth, spherical shape of silica microbeads following encapsulation in aerated mineral oil. Scale bar represents 200 μm B) Image of engineered E. coli pBBRBB::mer cells within encapsulation beads. Scale bar represents 5 μm.
Importantly, no cells were visualized on the surface of beads, and to image encapsulated cells, beads had to be crushed. Inside the gels, encapsulated cells appeared evenly dispersed within the gel matrix resulting in a highly porous material (Fig 2B). Encapsulated bacteria were found either as small clusters or as single cells throughout the gel. Encapsulated cells also retained normal cellular morphology and dimensions characteristic of E. coli (Fig 2B). Overall, resulting microbeads were within the diameter range and bacterial loading capacity shown in previous models to both maximize diffusion and maintain mechanical strength in flow-through systems [24].
To assess potential for degradation of methylmercury by E. coli pBBRBB::mer, both encapsulated and non-encapsulated cells were incubated in LB medium in the presence of 1 mg L-1 methylmercury chloride, which is a concentration 1000-fold greater than typically seen in contaminated environments and gold mining tailings ponds and thus a stringent test of our approach for methylmercury remediation [30, 31]. Samples were removed at various time points and analyzed for methylmercury concentration using a Tekran model 2700 Automated MethylMercury Analyzer with mercury detection by cold vapor atomic fluorescence spectrometry (CVAFS). Abiotic encapsulation beads and E. coli containing empty vector pBBRBB were included as negative controls.
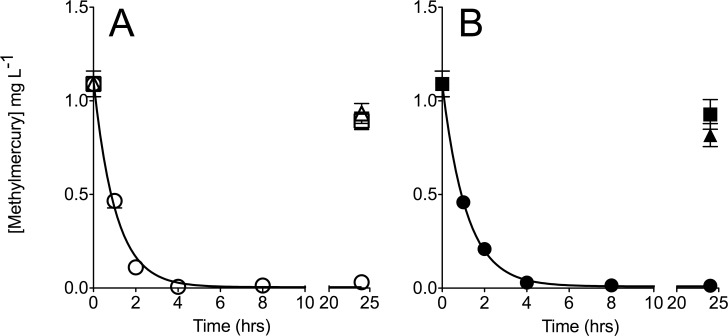
E. coli pBBRBB::mer was efficient at remediation of methylmercury chloride prior to encapsulation. Non-encapsulated E. coli pBBRBB::mer was able to remediate greater than 99% of methylmercury chloride from solution after only 4 hours of incubation (Fig 3A). The rate constant for degradation of methylmercury chloride by E. coli pBBRBB::mer was 0.96 ± 0.07 hr-1 with a measured half-life for methylmercury chloride of 0.72 ± 0.07 hours (Fig 3A). Abiotic samples, as well as tubes containing non-encapsulated E. coli harboring an empty vector, showed slight decreases in the concentration of methylmercury after 24 hours (-0.16 ± 0.05 mg L-1 and -0.20 ± 0.05 mg L-1, respectively), and are likely due to photodecomposition of methylmercury [32] (Fig 3A). Taken together, these results indicate that the dramatically enhanced rate of methylmercury chloride degradation by E. coli pBBRBB::mer is due to Mer-mediated activity in engineered cells.
Fig 3. Degradation of methylmercury chloride by A) Non-encapsulated (open symbols) and B) Encapsulated (closed symbols) E. coli pBBRBB::mer (circles) and E. coli pBBRBB (squares).
Degradation of methylmercury chloride in abiotic medium (open triangle) and sorption by abiotic beads (closed triangle) were included as controls. Data presented is for experiments performed at least in triplicate with error bars represented as SEM.
We next sought to determine if the rate of methylmercury degradation by encapsulated E. coli pBBRBB::mer was inhibited since encapsulation is likely to provide a diffusion barrier to degradation [33]. Methylmercury degradation rates were similar between non-encapsulated and encapsulated cells. The rate constant for degradation of methylmercury chloride by encapsulated E. coli pBBRBB::mer was 0.87 ± 0.04 hr-1 with a measured half-life for methylmercury chloride of 0.80 ± 0.04 hours (Fig 3B). Over 97% remediation of methylmercury chloride was achieved using encapsulated E. coli pBBRBB::mer after 4 hours of incubation (Fig 3B). These results suggest that encapsulation did not diminish the ability of E. coli pBBRBB::mer to degrade methylmercury.
Abiotic encapsulation beads were also incubated in the presence of methylmercury chloride to determine if the beads alone absorbed significant amounts of this compound. Only small concentrations of methylmercury chloride were absorbed by abiotic silica gel beads (-0.27 ± 0.06 mg L-1) as well as beads containing E. coli with the empty vector control (-0.20 ± 0.05 mg L-1) after 24 hours (Fig 3B). Absorption of methylmercury chloride by silica gel beads would aid in remediation efforts but would also hamper efforts to capture and potentially recycle elemental mercury using incorporated activated charcoal filters in scale-up packed bed reactors.
Conclusion
Because mercury cannot be transformed into a non-toxic state, remediation efforts have focused on conversion of organic and ionic forms to the less toxic elemental form Hg(0). Ultimately, the goal of any mercury remediation strategy is to capture the elemental form, thereby enabling safe disposal and the potential to recycle materials. Encapsulation of bacterial cells containing the mer operon provide a possible answer to the challenges involved in mercury remediation since encapsulation enables use of engineered cells and the filtration material can be incorporated in flow-through systems.
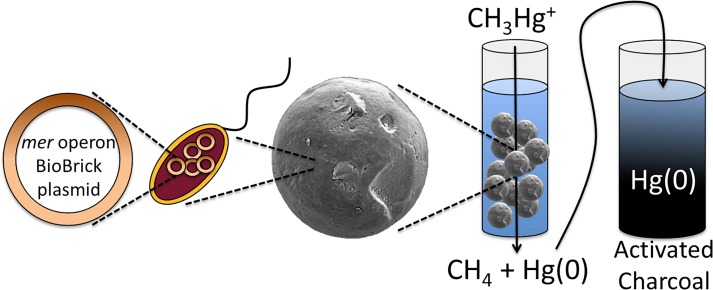
By incorporating a subset of the mer operon in E. coli and encapsulating cells in silica beads, a remediation platform targeting both ionic and organic forms of mercury was developed. Performing encapsulation in aerated mineral oil resulted in the production of smooth, spherical beads (Fig 2) that could be incorporated into packed bed reactor treatment facilities. Encapsulated E. coli pBBRBB::mer performed similarly to non-encapsulated cells, and was able to remediate high methylmercury concentrations to below detection levels after approximately 4 hours (Fig 3). Encapsulation, by providing protection to the biocatalysts and overall mechanical structure, broadens possible engineering applications for the remediation of mercury. A possible scheme for methylmercury remediation based on this engineered, encapsulated system is outlined in Fig 4. Contaminated water is passed through the encapsulation beads where Mer-mediated activity catalyzes cleavage of the C-Hg bond of organic mercury species followed by reduction of Hg(II) to Hg(0) (mediated by MerA, encoded in pBBRBB::mer, but the activity was not tested here). A charcoal filter is incorporated downstream to capture mercury enabling recovery and proper disposal.
Fig 4. Process for utilizing silica-encapsulated E. coli pBBRBB::mer as a bioremediation catalyst in flow-through systems.
E. coli containing pBBRBB::mer are encapsulated in silica beads using sol-gel technology and catalyze the cleavage of organic C-Hg bonds of mercury species and subsequent reduction of Hg(II) to Hg(0). Resulting Hg(0) is then captured downstream by an activated charcoal filter.
Mercury pollution is widespread, and its effects are not limited to areas near the source of pollution. Since mercury can travel thousands of miles through the atmosphere before being deposited back in the environment, it has become an issue of global concern. Remediation methods are needed that can target multiple arenas including industry, mining tailing ponds, and open bodies of water. This study provides a foundation for methods to encapsulate mer-containing bacteria in silica materials to offer a versatile option that can be tailored to various mercury-polluted sites. Further work in this area is being targeted at refactoring the mer operon to increase turnover rates, testing other genera such as encapsulated Pseudomonas species for the remediation of methylmercury, and determination of long-term cell escape rates from beads.
Acknowledgments
This project was conducted by the 2014 iGEM team at the University of Minnesota and was supported by the Office of Naval Research (Award N000141210309 to JAG) and the University of Minnesota MnDRIVE initiative: Advancing industry, conserving our environment. We would like to thank Dr. Anne Summers (University of Georgia) for providing pDU1358, safety protocols, and project guidance. We would also like to thank Professor Alptekin Aksan and Dr. Baris Mutlu (University of Minnesota) for initial discussions concerning encapsulation protocols, and Gail Celio (University of Minnesota Imaging Center) for assistance with microscopy.
Data Availability
All relevant data are within the paper.
Funding Statement
This project was supported by the Office of Naval Research (Award N000141210309 to JAG) and the University of Minnesota MnDRIVE initiative: Advancing industry, conserving our environment (https://mndrive.umn.edu/environment). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lin CC, Yee N, Barkay T. Microbial transformation in the mercury cycle In: Liu G, Yong C, O’Driscoll N, editors. Environmental chemistry and toxicology of mercury. Hoboken: John Wiley and Sons; 2012. pp. 155–191. [Google Scholar]
- 2.Barkay T, Miller SM, Summers AO. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol Rev. 2003;27: 355–384. [DOI] [PubMed] [Google Scholar]
- 3.Tchounwou PB, Ayensu WK, Ninashvili N, Sutton D. Environmental exposure to mercury and its toxicopathologic implications for public health. Environ Toxicol. 2003;18: 149–175. [DOI] [PubMed] [Google Scholar]
- 4.United Nations Environment Programme. Global mercury assessment 2013: Sources, emissions, releases and environmental transport Geneva: UNEP Chemicals Branch; 2013. pp. 1–32. [Google Scholar]
- 5.Wagner-Döbler I. Current research for bioremediation of mercury In: Wagner-Dobler I, editor. Bioremediation of mercury: Current research and industrial applications. Norfolk: Caister Academic Press; 2013. pp. 1–13. [Google Scholar]
- 6.Lafrance-Vanasse J, Lefebvre M, Di Lello P, Sygusch J, Omichinski JG. Crystal structure of the organomercurial lyase MerB in its free and mercury-bound forms: insights into the mechanism of methylmercury degradation. J Biol Chem. 2009;284: 938–944. 10.1074/jbc.M807143200 [DOI] [PubMed] [Google Scholar]
- 7.Brown NL, Stoyanov JV, Kidd SP, Hobman JL. The MerR family of transcriptional regulators. FEMS Microbiol Rev. 2003;27: 145–163. [DOI] [PubMed] [Google Scholar]
- 8.Brim H, Venkateswaran A, Kostandarithes HM, Fredrickson JK, Daly MJ. Engineering Deinococcus geothermalis for bioremediation of high-temperature radioactive waste environments. Appl Environ Microbiol. 2003;69: 4575–4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brim H, McFarlan SC, Fredrickson JK, Minton KW, Zhai M, Wackett L. P, et al. Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments. Nat Biotechnol. 2000;18: 85–90. [DOI] [PubMed] [Google Scholar]
- 10.Horn JM, Brunke M, Deckwer WD, Timmis KN. Pseudomonas putida strains which constitutively overexpress mercury resistance for biodetoxification of organomercurial pollutants. Appl Environ Microbiol. 1994;60: 357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruiz ON, Alvarez D, Gonzalez-Ruiz G, Torres C. Characterization of mercury bioremediation by transgenic bacteria expressing metallothionein and polyphosphate kinase. BMC Biotechnol. 2011;11: 82 10.1186/1472-6750-11-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng X, Jia P. Construction and characterization of a photosynthetic bacterium genetically engineered for Hg2+ uptake. Bioresour Technol. 2011;102: 3080–3088. [DOI] [PubMed] [Google Scholar]
- 13.Lin KH, Chien MF, Hsieh JL, Huang CC. Mercury resistance and accumulation in Escherichia coli with cell surface expression of fish metallothionein. Appl Microbiol Biotechnol. 2010;87: 561–569. 10.1007/s00253-010-2466-x [DOI] [PubMed] [Google Scholar]
- 14.Kiyono M, Omura H, Omura T, Murata S, Pan-Hou H. Removal of inorganic and organic mercurials by immobilized bacteria having mer-ppk fusion plasmids. Appl Microbiol Biotechnol. 2003;62: 274–278. [DOI] [PubMed] [Google Scholar]
- 15.Bae W, Wu CH, Kostal J, Mulchandani A, Chen W. Enhanced mercury biosorption by bacterial cells with surface-displayed MerR. Appl Environ Microbiol. 2003;69: 3176–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan-Hou H, Kiyono M, Omura H, Omura T, Endo G. Polyphosphate produced in recombinant Escherichia coli confers mercury resistance. FEMS Microbiol Lett. 2002;207: 159–164. [DOI] [PubMed] [Google Scholar]
- 17.Bae W, Mehra RK, Mulchandani A, Chen W. Genetic engineering of Escherichia coli for enhanced uptake and bioaccumulation of mercury. Appl Environ Microbiol. 2001;67: 5335–5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sousa C, Kotrab P, Ruml T, Cebolla A, De Lorenzo V. Metalloadsorption by Escherichia coli cells displaying yeast and mammalian metallothioneins anchored to the outer membrane protein LamB. J Bacteriol. 1998;180: 2280–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S, Wilson DB. Construction and characterization of Escherichia coli genetically engineered for bioremediation of Hg2+-contaminated environments. Appl Environ Microbiol. 1997;63: 2442–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner-Döbler I, von Canstein H, Li Y, Timmis KN, Deckwer W-D. Removal of mercury from chemical wastewater by microorganisms in technical scale. Environ Sci Technol. 2000;34: 4628–4634. [Google Scholar]
- 21.von Canstein H, Li Y, Timmis KN, Deckwer W-D, Wagner-Döbler I. Removal of mercury from chloralkali electrolysis wastewater by a mercury-resistant Pseudomonas putida strain. Appl Environ Microbiol. 1999;65: 5279–5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mutlu BR, Yeom S, Tong H-W, Wackett LP, Aksan A. Silicon alkoxide cross-linked silica nanoparticle gels for encapsulation of bacterial biocatalysts, J Mater Chem A. 2013;1:11051–11060. [Google Scholar]
- 23.Reátegui E, Reynolds E, Kasinkas L, Apparwal A, Sadowsky MJ, Aksan A, Wackett LP. Silica gel-encapsulated AtzA biocatalyst for atrazine biodegradation. Appl Microbiol Biotechnol. 2012;96: 231–240. 10.1007/s00253-011-3821-2 [DOI] [PubMed] [Google Scholar]
- 24.Mutlu BR, Yeom S, Wackett LP, Aksan A. Modeling and optimization of a bioremediation system utilizing silica gel encapsulated whole-cell biocatalyst. Chem Eng J. 2015;259: 574–580. [Google Scholar]
- 25.Saltikov CW, Newman DK. Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci U S A. 2003;100: 10983–10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffin HG, Foster TJ, Silver S, Misra TK. Cloning and DNA sequence of the mercuric- and organomercurial-resistance determinants of plasmid pDU1358. Proc Natl Acad Sci U S A. 1987;84: 3112–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vick JE, Johnson ET, Choudhary S, Bloch SE, Lopez-Gallego F, Srivastava P, Tikh IB, Wawrzyn GT, Schmidt-Dannert C. Optimized compatible set of BioBrickTM vectors for metabolic pathway engineering. Appl Microbiol Biotechnol. 2011;92: 1275–1286. 10.1007/s00253-011-3633-4 [DOI] [PubMed] [Google Scholar]
- 28.Aukema KG, Kasinkas L, Aksan A, Wackett LP. Use of silica-encapsulated Pseudomonas sp. strain NCIB 9816–4 in biodegradation of novel hydrocarbon ring structures found in hydrolic fracturing waters. Appl Environ Microbiol. 2014;80: 4968–4976. 10.1128/AEM.01100-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balch WE, Wolfe RS. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl Environ Microbiol. 1976;32: 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guedron S, Cossa D, Grimaldi M, Charlet C. Methylmercury in tailings ponds of Amazonian gold mines (French Guiana): Field observations and an experimental flocculation method for in situ remediation. Appl Geochem. 2011;26: 222–229. [Google Scholar]
- 31.Leopold K, Foulkes M, Worsfold P. Methods for the determination and speciation of mercury in natural waters–A review. Anal Chem Acta. 2010;663: 127–138 [DOI] [PubMed] [Google Scholar]
- 32.Zhang T, Hsu-Kim H. Photolytic degradation of methylmercury enhanced by binding to natural organic ligands. Nature. 2010;3: 473–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeom S, Mutlu BR, Aksan A, Wackett LP. Bacterial cyanuric acid hydrolases for water treatment. Appl Environ Microbiol. Epub 2015. July 17 10.1128/AEM.02175-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.