Abstract
The cellular cytidine deaminase APOBEC3G (A3G) was first described as an anti-HIV-1 restriction factor by directly deaminating reverse transcripts of the viral genome. HIV-1 Vif neutralizes the activity of A3G, primarily by mediating degradation of A3G to establish effective infection in host target cells. Lymphoma cells, which express high amounts of A3G, can restrict Vif-deficient HIV-1. Interestingly, these cells are more stable in the face of treatments that result in dsDNA damage, such as ionizing irradiation (IR) and chemotherapies. Previously, we showed that the Vif-derived peptide (Vif25-39) efficiently inhibits A3G deamination, and increases sensitivity of lymphoma cells to IR. In the current study, we show that additional peptides derived from Vif, A3G and A3F, which contain the LYYF motif, inhibit deamination activity. Each residue in the Vif25-39 sequence moderately contributes to the inhibitory effect, while, replacing a single amino acid in the LYYF motif completely abrogate inhibition of deamination. Treatment of A3G-expressing lymphoma cells exposed to ionizing radiation with the new inhibitory peptides reduces double-strand break (DSB) repair after radiation. Incubation of cultured irradiated lymphoma cells with peptides that inhibit DSB repair halts their propagation. These results suggest that A3G may be a potential therapeutic target amenable to peptide and peptidomimetic inhibition.
Keywords: APOBEC3G, Peptides, DNA repair, DNA binding, Lymphoma cells
Introduction
A3G is a member of the human APOBEC protein family, comprised of cellular cytidine deaminases, encoded within a cluster of seven APOBEC3 (A3) genes, designated A3A, B, C, D, F, G and H, on chromosome 22 [1, 2]. The human A3 proteins and their homologs in other mammals are members of a multifunctional family of Zn2+-dependent polynucleotide cytosine deaminases acting as potent editors of single-stranded DNA (ssDNA), leading to dC>dU mutations (reviewed by [1-7]). In addition to the A3 deaminases, the APOBEC family includes activation-induced cytidine deaminase (AID), APOBEC1 (A1), APOBEC2 (A2) and APOBEC4 (A4). The protein structure of the catalytic domains of A3G and A3F, as well as the structures of A3A and A3C, were recently elucidated [8-11] and reveal a shared active site architecture and common secondary structural features.
A3 proteins function as innate immune factors, and elicit broad retrotransposon and retrovirus restricting activities [5, 7, 12-14]. However, the physiological functions of the cellular deaminases are not limited to innate immunity, but have roles beyond this setting. APOBEC1 edits apolipoprotein-B mRNA at base C666 to generate an early stop codon, resulting in synthesis of a shorter polypeptide. AID edits immunoglobulin genes to trigger somatic hypermutation (SH) and CSR. Similarly, A3A plays four key roles: clearance of foreign dsDNA from cells [15], insertion of somatic mutations in human mitochondrial and nuclear DNA [16], demethylation of 5-methylcytosine, and induction of DNA breaks in a deaminase-dependent manner.
HIV-1 Vif neutralizes A3G by hijacking a cellular protein complex that includes CBF-β, Cullin5 and ElonginB/ElonginC to degrade A3G via the ubiquitin-proteasome pathway, and controls the deamination threat in virions or in the pre-integration complexes following entry into target cells. Previously, we showed that purified HIV-1 Vif and the Vif-derived peptides Vif25-39 and Vif105-119 inhibit A3G activity in vitro and hamper double-strand break (DSB) repair following exposure to ionizing radiation [17].
Ionizing radiation and the majority of anti-cancer agents inflict deleterious DNA damage on tumor cells, predominantly DNA double-strand breaks (DSBs) and covalent DNA crosslinks. DNA DSBs are highly genotoxic lesions, constituting the most disruptive form of DNA damage. Cells use an intricate set of mechanisms to repair genomic DSBs, based on non-homologous end-joining (NHEJ) or homology-directed repair (HDR) [18, 19]. Conversely, resistance to radiation is a fundamental barrier, limiting the effectiveness of radiation therapy [20]. Recent data strongly imply that pre-exposure to low-dose irradiation (LDIR) enables activation of specific proteins that may increase cellular tolerance to subsequent IR injuries [21].
Several types of cancer cells, such as lymphoma and myeloma cells, e.g., Diffuse large B-cell lymphoma (DLBCL), display efficient repair of genomic DSBs induced by IR or chemotherapies, and enhanced cell survival after these treatments [22-24]. Jais et al. [25] showed that high levels of A3G expression correlate with poor overall survival in patients on R-CHOP therapy (Rituximab; Cyclophosphamide; doxorubicin; vincristine; Prednisolone) [25]. We have extended this finding to demonstrate that A3G is extensively expressed in DLBCL cell lines (Ly-4), as well as in other lymphoma cell lines, e.g., H9 cells, but not leukemic cells [17]. Consistent with a direct role in DSB repair, inhibition of A3G expression or its deaminase activity results in reduced DSB repair, whereas reconstitution of A3G expression in leukemia cells, which are A3G-deficient, enhances DSB repair. We suggest that A3G protein plays a dual role in promoting survival of cancer cells in vivo, first by enhancing DSB repair following genotoxic treatments or spontaneous breaks, thus preventing cell death, and secondly, by promoting a mutator phenotype that drives tumor progression [26-33].
Thus, inhibition of A3G activity could potentially restrain the progressive accumulation of mutations, which is one of the underlying processes that characterizes the cancer phenotype [34, 35], and thereby increase sensitivity of cells to genotoxic agents. Hence, treatment of lymphomas with genotoxic agents such as IR or chemotherapeutic agents may be effectively complemented when combined with anti-A3G inhibitors.
Our current study focuses on inhibiting A3G catalytic activity using variety of peptides to improve the outcome of genotoxic lymphoma therapies. Screening a library of peptides derived from HIV-1 Vif-, A3G- and A3F reveals that the inhibitory sequences are scattered throughout the respective proteins. In the active peptides, several sequence motifs were implicated in efficient inhibition of A3G deaminase activity in vitro. Primarily two motifs in human APOBEC sequences are “LYYF” and “VKHH” were found to have inhibitory activity. The first motif, LYYF, occurs in all primates APOBEC proteins, wherein the sequence conservation implies a functional role for this sequence motif. The second motif, VKHH, is present in both human A3F and A3DE sequences. In the A3F-CTD crystal structure [9], both motifs occur at crystal contacts with the largest intermolecular interface occurring between the two motifs A3F-CTD molecules. This direct interaction may implicate these regions as being critical function. Surprisingly, both sequence motifs are also present in the HIV-1 protein Vif, which is known to directly bind A3G and A3F and targets them for proteasomal degradation using the host-cell machinery [36]. To evaluate the role of each amino acid in VKHH and LYYF motif, mutational studies revealed that all residues in Vif25-39 moderately contributes to the inhibitory effect, while, replacing a single amino acid from LYYF motif completely abrogate inhibition of deamination.
Treatment of IR-resistant lymphoma cells with either of these peptides [12, 17, 37, 38] reduces DSB repair following radiation, leading to a decrease in cell viability. These results indicate that A3G is involved in DSB repair and is a potential therapeutic target that is likely sensitive not only to peptides, but also to therapeutic peptidomimetics.
Results
Vif- and A3F-derived peptides inhibit the catalytic activity of A3G
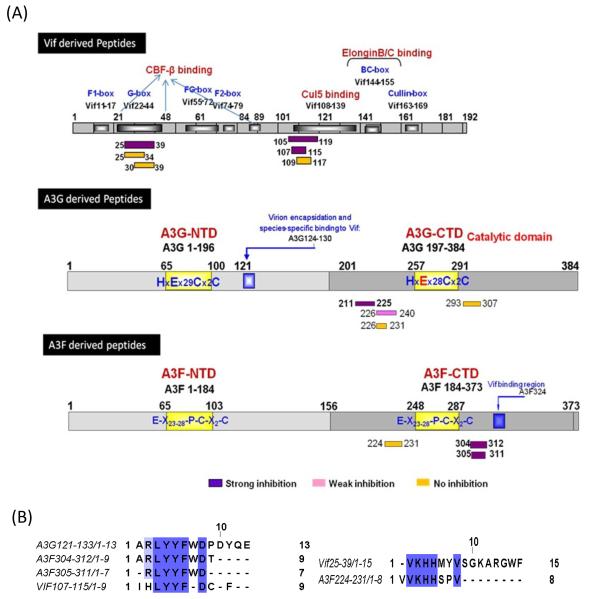
By screening Vif domains responsible for inhibition of A3G deamination, we identified Vif25-39 and Vif105-119 as efficient inhibitors (Table 1) [38]. In this report, 22 new peptides, mostly smaller than previously described, derived from Vif, A3G and A3F (Table 1, Fig. 1) were tested for inhibition. The Vif domain 105-119, which efficiently inhibits A3G, contains the LYYF motif (Fig. 1B). This motif is also present in A3F (position 306-309) and is involved in forming a large crystal contact in the A3F-CTD crystal structure [9].
Table 1.
Inhibition effect of peptides on the deamination activity of A3G
| No. | Peptide | Sequence | IC50 (μM) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Vif25-39 | V | K | H | H | M | Y | V | S | G | K | A | R | G | W | F | 0.6 |
| 2 | Vif 25-39 (27AA28) | V | K | A | A | M | Y | V | S | G | K | A | R | G | W | F | 4.5 |
| 3 | Vif 25-39 (26A) | V | A | H | H | M | Y | V | S | G | K | A | R | G | W | F | 8 |
| 4 | Vif 25-39 (26AAA28) | V | A | A | A | M | Y | V | S | G | K | A | R | G | W | F | 10 |
| 5 | Vif25-36 | V | K | H | H | M | Y | V | S | G | K | A | R | 50 | |||
| 6 | Vif25-34 | V | K | H | H | M | Y | V | S | G | K | NI* | |||||
| 7 | Vif26-39 | K | H | H | M | Y | V | S | G | K | A | R | G | W | F | 80 | |
| 8 | Vif30-39 | Y | V | S | G | K | A | R | G | W | F | NI | |||||
| 9 | A3F224-231 | VV | K | H | H | S | P | V | NI | ||||||||
|
| |||||||||||||||||
| 10 | Vif105-119 | Q | L | I | H | L | Y | Y | F | D | C | F | S | E | S | A | 0.1 |
| 11 | Vif107-115 | I | H | L | Y | Y | F | D | C | F | 0.1 | ||||||
| 12 | Vif107-115(110AA111) | I | H | L | A | A | F | D | C | F | NI | ||||||
| 13 | Vif107-115(112A) | I | H | L | Y | Y | A | D | C | F | NI | ||||||
| 14 | Vif109-117 | L | Y | Y | F | D | C | F | S | E | NI | ||||||
| 15 | Vif109-115 | L | Y | Y | F | D | C | F | NI | ||||||||
| 16 | Vif 107-113 | I | H | L | Y | Y | F | D | 110 | ||||||||
| 17 | A3F304-312 | A | R | L | Y | Y | F | W | D | T | 8 | ||||||
| 18 | A3F305-311 | R | L | Y | Y | F | W | D | 1 | ||||||||
| 19 | Vif107-115 (Reverse) | F | C | D | F | Y | Y | L | H | I | NI | ||||||
| 20 | Vif107-115(scrambled) | I | C | F | L | H | Y | D | F | Y | NI | ||||||
|
| |||||||||||||||||
| 21 | A3G293-307 | Q | E | M | A | K | F | I | S | K | N | K | H | V | S | L | NI |
| 22 | A3G226-240 | R | M | H | N | D | T | W | V | L | L | N | Q | R | R | G | 60 |
| 23 | A3G226-231 | R | M | H | N | D | T | NI | |||||||||
| 24 | A3G211-225 | W | V | R | G | R | H | E | T | Y | L | C | Y | E | V | E | 10 |
denotes No Inhibition.
FIGURE 1.
(A) Schematic representation of Vif-, A3G- and A3F-derived peptides. Purple boxes: strong-, Pink boxes: weak-, Yellow boxes: no-inhibition towards A3G. (B) Sequence alignment of different peptides showing the conserved motives (VKHH and LYYF).
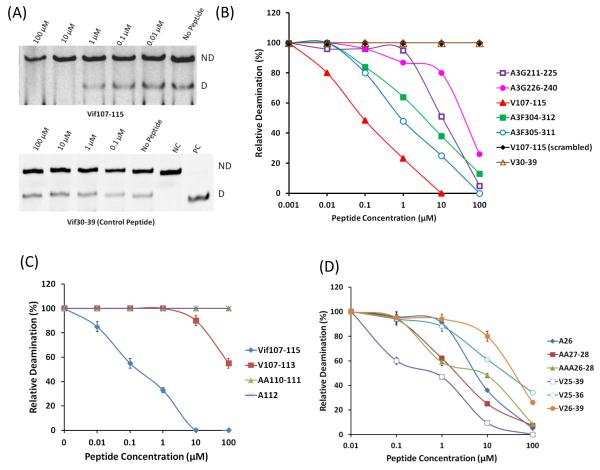
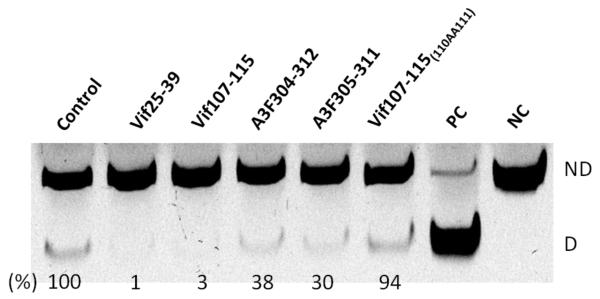
Based on this information novel peptides derived from Vif and A3F including the LYYF or VKHH were synthesized and assessed as A3G inhibitors (Table 1, Fig. 2A and B). Truncation of Vif105-119 to Vif107-115 retains the same efficient inhibitory effect on A3G with the concentrations giving half-maximal inhibition (IC50) of 0.1 μM indicating that the restudies QL and SESA at the N’ and C’ termini respectively, are not essential for A3G inhibition. However, peptides consisting of the Vif107-115 sequence in reverse and scrambled order have no inhibitory effect emphasizing the sequence specificity of this short peptide (Table 1). Similarly, truncation of peptide A3F 304-312 to A3F 305-311, which also contains the LYYF motif, elevates the efficiency of inhibition with IC50 values of 8 μM and 1 μM, respectively (Fig. 2B).
FIGURE 2.
Inhibition of deaminase activity by Vif-, A3G- and A3F-derived peptides. (A) Inhibition of deamination by Vif107-115 peptides in dose-dependent manner. The lower gel represents the negative control using Vif30-39 peptide. NC denotes Negative control; PC denotes Positive control. ND; Not deaminated substrate and D; Deaminated product. (B) The effect of peptides and their concentration on A3G-mediated deamination activity with standard ssDNA substrate (10 fmol). Values were determined as shown in panel A. (C) The effect of Vif107-115 and its mutant-derived peptides. (D) The effect of Vif25-39 and its mutant-derived peptides. Data represent the average of triplicates; SD values were less than ± 2 where not indicated.
To evaluate the contribution of each residue in the Vif 107-115, we synthesized peptides in which two residues from the N’ terminus was removed (Vif 109-115) and peptides in which the 110YY111 or the 112F residues located in LYYF motif were substituted by alanine residue. Truncation of the peptide (Table.1 No.14 and 15), or alterations in the LYYF motif (Table.1 No. 12 and 13) completely abrogate the inhibitory effect of the Vif 107-115 peptides at all the concentrations (Fig. 2C). These results further reinforce the required sequence specificity in the active peptides.
The VKHH sequence is another motif common to Vif and A3F (Fig. 1B). However, while the Vif 25-39 is an efficient A3G inhibitor, the A3F224-231 peptide containing the VKHH motif has no inhibitory effect on the enzyme. Our efforts to increase the inhibitory effect of Vif25-39 by truncating amino acid residues from both termini have so far failed, suggesting that the entire sequence comprising the Vif25-39 peptide is essential for A3G inhibition (Table1). Related mutated peptides, where the 26K, 27HH28 or the 26KHH28 in the Vif25-39 were substituted with alanine residues further corroborated this conclusion. Each of these variants reduced the peptides’s efficiency of the inhibition (Fig. 2D), further supporting that the entire Vif25-39 peptide is needed for effective inhibition of the enzymatic activity of A3G.
In summary, the six amino acid comprise the N’ terminus of the Vif107-115 are essential for A3G inhibition, while each of the 15 residues of the Vif25-39 contribute to the efficiency of inhibition. Table 1 shows that other A3G derived peptides from analogous domains to A3F have marginal or no inhibitory effect on the A3G deamination (Table 1 No. 21-24), indicating the specificity of inhibition of the Vif25-39 and A3F107-115 peptides.
Determining the mode of inhibition of A3G enzyme by Vif- and A3F-derived peptides
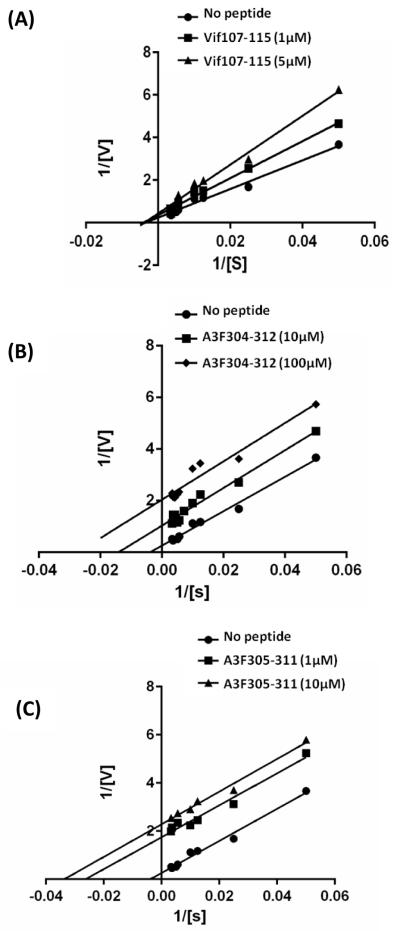
The peptides Vif107-115, A3F304-312 and A3F305-311 all contain the LYYF motif and efficiently inhibit deamination (Table 1). To determine the putative mechanisms, by which these peptides inhibit the deamination activity of A3G in vitro, a double-reciprocal plot (double inverse) for each of the peptides were evaluated. The initial deamination rates for A3G were determined in the presence of 1 and 5 μM of Vif107-115 (Fig. 3A); 10 and 100 μM of A3F304-312 (Fig. 3B); 1 and 10 μM A3F305-311 (Fig. 3C). The double-reciprocal plot of A3G inhibition by Vif107-115 reveals a mixed-mode inhibition, similar to that obtained for Vif105-119 [38], suggesting that these peptides interact both with the free enzyme and the enzyme substrate complex. In contrast, A3F304-312 and A3F305-311 peptides inhibit the A3G enzyme in an uncompetitive mode, like the full-length Vif molecules and the Vif25-39 peptide [38]. The K’i of Vif107-115 is approximately 1.71×10 −6 M while the K’i's of A3F304-312 and A3F305-311 peptides are approximately 7.8×10−6 M and 0.65×10−6 M respectively indicating that subtle alterations around the LYYF motif can affect potency by ten-fold.
FIGURE 3.
Determination of the mode of Inhibition. Deamination of an ss-deoxyoligonucleotide substrate concentration in the presence of Vif-derived peptides Vif107-115 (A), A3F-derived peptides A3F304-312 (B) and A3F305-311(C) was determined and is shown by double-reciprocal plot. The Vif- and A3F-derived peptide concentrations used are indicated. Vif, Vif25-39, A3F304-312 and A3F305-311 reveal an uncompetitive inhibition mode, whereas Vif105-119 and Vif107-115 inhibit A3G in a mixed mode. Values represent the average of triplicates. SD values were less than ± 0.5.
Inhibition of DSB repair by Vif- and A3F-derived peptides
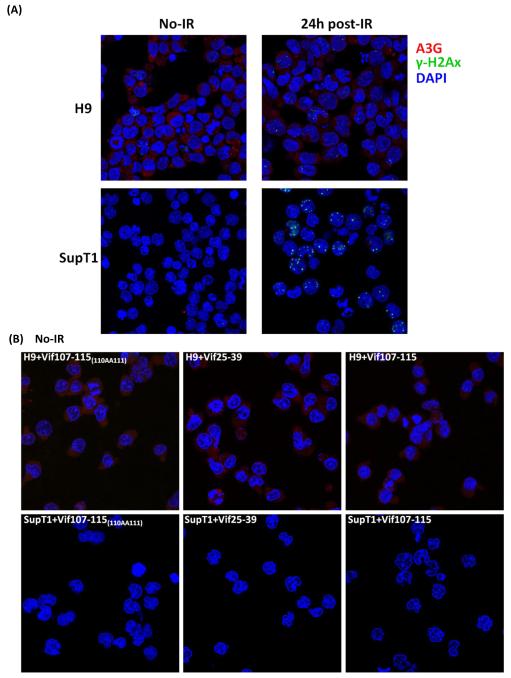
Previously, we have demonstrated the involvement of A3G in DSB repair in lymphoma cells (H9) by knock down of A3G expression with specific shRNA [17]. Here we assess whether the peptides that inhibit A3G cytidine deaminase activity impede DSB repair. The presence of DSB load was assessed in cutaneous T-cell lymphoma (H9) and in diffuse large B cell lymphoma (Ly-4), which both express A3G, and was compared with A3G-deficient T-lymphoblastic leukemia (SupT1) cell line, by the presence of γ-H2AX foci. Fig. 4A shows DSBs induced by IR in H9 cells were repaired in 24hrs, while A3G-devoid SupT1 cells are unable to repair the DNA damage as revealed by the number of γ-H2AX foci.
FIGURE 4.
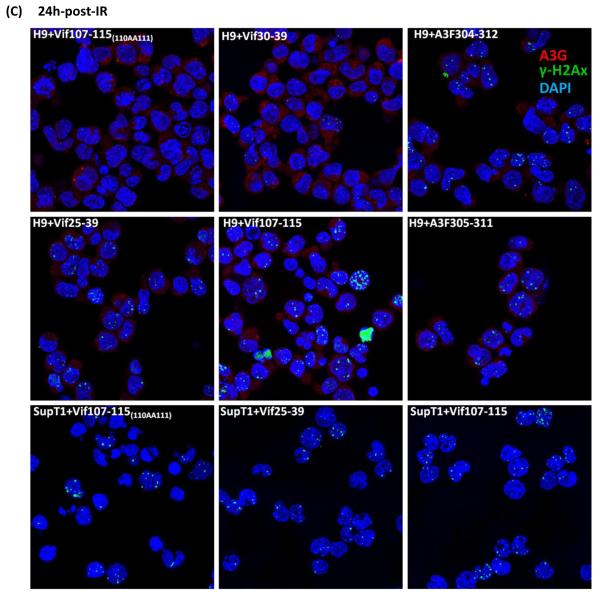
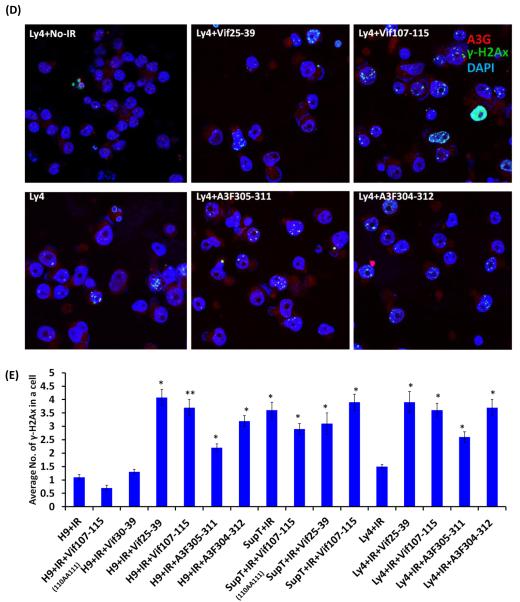
Inhibition of DSB repair by anti-A3G peptides. Cells were irradiated (4 Gy) or mock-irradiated (No-IR) and stained with anti-A3G and anti-γ-H2AX antibody. Nuclei stained by DAPI (Blue); A3G stained by Alexa fluor 647 (red); γH2AX foci by Alexa fluor 488 (Green). (A) H9 and SupT1 cells before and 24h post-irradiation. (B) Non-irradiated H9 and SupT1 cells were cultivates in the presence of indicated peptides. (C) H9 and SupT1 cells were pre-incubated for 2 hours with the indicated peptides, irradiated (4 Gy) and stained after 24 hours. Top panels (from left): H9 cells in the presence of peptides: Mutant of Vif107-115(110AA111); Vif30-39 (Negative control) and A3F304-312. Middle panels (from left): H9 cells irradiated in the presence of peptides, Vif25-39; Vif107-115 and A3F305-311. Bottom panels (from left): SupT1 cells in the presence of peptides: Mutant of Vif107-115(110AA111); Vif25-39 and Vif107-115. (D) Ly-4 cells were pre-incubated for 2 hours with the indicated peptides, irradiated (4 Gy) or mock-irradiated (NoIR), and stained after 24 hours. Top panels (from left): Ly-4 cells non-irradiated; Ly-4 cells irradiated in the presence of peptides, Vif25-39; Vif107-115. Bottom panels (from left): Ly-4 cells irradiated; Ly-4 cells irradiated in the presence of peptides, A3F305-311 and A3F304-312. (E) Quantification of γ-H2AX foci in H9, Ly-4 and SupT1 24 hours post-IR (4 Gy). Values represent mean SD from two independent experiments in each at least five different microscopic fields were analyzed. * P < 0.05, ** P < 0.01, ANOVA, n = 50.
A period of 8 h post irradiation is sufficient to repair DSB in cells expressing A3G [17]. Observation of γ-H2AX foci after radiation indicates significant halt of the repair. H9 cells at 24h post-irradiation are devoid of γ-H2AX foci (Fig. 4A). The peptides have no effect on the non-irradiated H9 and SupT1 cells (Fig. 4B). In contrast, pre-incubation of H9 lymphoma cells in media containing 100 μM of Vif25-39 (positive control), Vif107-115, A3F305-311 or A3F304-312 prior to IR exposure increases the number of γ-H2AX foci (Fig. 4C). The Vif30-39 and Vif107-115(110AA111) peptides (negative controls), which do not inhibit deamination (Table 1 and Fig. 2) by A3G failed to inhibit DSB repair following irradiation in these cells (Fig. 4C).
Non-irradiated and irradiated H9 cells exhibited an average of 0.2 and 1 foci per cell, respectively. H9 cells treated with Vif25-39 contained 4.1 foci per cell, showing that inhibition of deamination by A3G hampers DSB repair. Similar to Vif25-39; Vif107-115, A3F305-311 and A3F304-312 inhibit DSB repair as the average number of γ-H2AX foci per cell were 3.7, 2.2 and 3.2, respectively (Fig. 4E).
The inhibitory effect of DSB repair by Vif- and A3F-derived peptides is not limited to H9 lymphoma cells, which express high amounts of A3G since the same peptides impede DSB repair in Ly-4 lymphoma cells in a similar manner (Fig. 4D). Fig. 4 shows that the four peptides tested in this experiment inhibit DSB repair with similar efficacy.
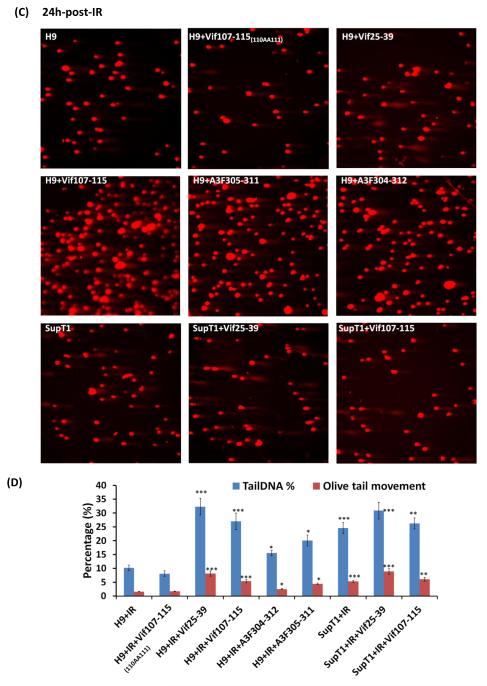
We used the comet assay (Fig. 5) to verify the results obtained by the γ-H2AX foci (Fig. 4). Two effective parameters; Tail DNA % and Olive Tail Movement [39] were taken to evaluate the damaged DNA in the cells. Vif25-39 and Vif107-115 as well as the mutated Vif107-115(110AA111) did not induce any damage to non-irradiated H9 and SupT1 cells (Fig. 5B). Similarly, the peptide A3F305-311 and A3F304-312 did not induce breaks in the non-irradiated H9 and SupT1 cell (results not shown). In contrast, the DNA of irradiated cells treated with the active peptides remained unrepaired in H9 cells at 24 hrs post-IR. The untreated or treated cells with the mutated Vif107-115(110AA111) peptides did not abrogate the DNA repair and most of the cellular DNA is present in the nuclei rather than in the comet (Fig. 5C). Sup T1 cells treated or untreated with the peptides remain sensitive to the genotoxic IR and large portions of their DNA migrate to the comet (Fig. 5C and D).
FIGURE 5.
Comet Assay. Inhibition of DSB repair by anti-A3G peptides in H9 cells. Cells were irradiated (4 Gy) or mock-irradiated (No-IR) and stained with Ethidium Bromide. (A) H9 and SupT1 cells before, 10h and 24h post-irradiation. (B) H9 and SupT1 cells in the presence of indicated peptides without radiation. (C) H9 cells were incubated for 2 hours with or without the indicated peptides, irradiated (4 Gy) and stained after 24 hours. Top panels (from left): H9 cells; H9 cells in the presence of peptides: Mutant of Vif107-115 as negative control; Vif25-39. Middle panels (from left): H9 cells irradiated in the presence of peptides: Vif107-115; A3F305-311and A3F304-312. Bottom panels (from left) SupT1 cells; SupT1 cells irradiated in the presence of peptides: Vif25-39 and Vif107-115. (D) Quantification of Tail DNA % and Olive Tail Movement in H9 and SupT1 cells 24 hours after IR (4 Gy). Values represent mean of two independent experiments; each time 50 cells were scored (50 cells each from two different fields).* P < 0.05, ** P < 0.01, *** P < 0.001, ANOVA, n = 50.
Vif- and A3F-derived peptides inhibit the catalytic activity of endogenous A3G
To define whether the peptides, which inhibit purified A3G in vitro are able to hamper the deamination activity of the natural endogenous deaminase, we treated cultured H9 cells with each peptide (100 μM) for 12 hr prior preparing cell extracts. Peptides Vif25-39, Vif107-115, A3F 304-312 and A3F305-311 inhibit the deamination by the endogenous A3G enzymes (Fig. 6). These experiments also strongly suggest that the four peptides are able to penetrate the cells and inhibit the A3G present in the cell cytoplasm.
FIGURE 6.
Inhibition of the endogenous A3G deaminase activity by Vif-, and A3F-derived peptides. Inhibition of deamination by treating cells with 100 μM of respective peptides 12 hr prior preparation of cytoplasmic extract. NC denotes Negative control; PC denotes Positive control. ND; Not deaminated substrate and D; Deaminated product. The percentage of the deamination product (%) was calculated relative to the control (untreated cells). Data represent the average of duplicates.
The effect of Vif- and A3F-derived peptides on the kinetics of Ly-4 and SupT1 cells propagation following irradiation
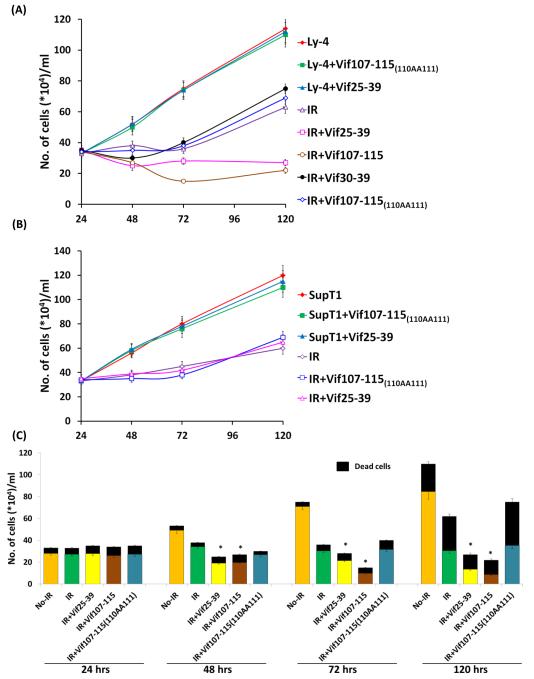
Exposure of cultured Ly-4 cells to IR (4 Gy) decreases their propagation. Pre-treatment of these cells with 100 μM of Vif107-115 or Vif 25-39 inhibits the cell propagation (Fig. 7A), while the control peptides Vif30-39 and Vif107-115(110AA111) do not prevent the recovery of the irradiated cells. SupT1 cells were sensitive to IR, where the active and control peptides could not inhibit their recovery from the genotoxic effect (Fig 7B). The propagation kinetics (Fig. 7A and B) does not reflect the toxicity by radiation with peptides. Double strand break if unrepaired is detrimental for the cells. We expect the peptides that inhibit the repair should increase the number of dead cells. Figure 7C shows the peptides Vif25-39 and Vif 107-115 increase the ratio between dead and live cells comparing the untreated irradiated cells. Note that the peptides have no effect on the propagation of treated or untreated-irradiated SupT1 cells (Fig. 7B). These results strongly suggest that the Vif derived peptides 107-115 and 25-39, which interfere with the DSB repair are also responsible for the reduction in number of cells survived the genotoxic treatment. Overall, these observations indicate that the Vif- and A3F-derived peptides, that inhibit enzymatic activity of A3G, convert IR-resistant H9 and Ly4 lymphoma cells to IR-sensitive.
FIGURE 7.
Kinetics of Ly-4 and SupT1 cells propagation following irradiation. Cultured Ly-4 cells (A) and SupT1 (B) were treated or untreated with peptides (100 μM), irradiated (4Gy) and the number of cells were determined during 5 days post irradiation. (C) Graph describing the ratio between live and dead Ly-4 cells treated with the peptides during 5 days of the experiment. Values represent mean of three independent experiments.* P < 0.01 (Significance was calculated among the irradiated cells versus irradiated cells treated with the peptides).
Conformational plasticity of peptides may influence A3G inhibition
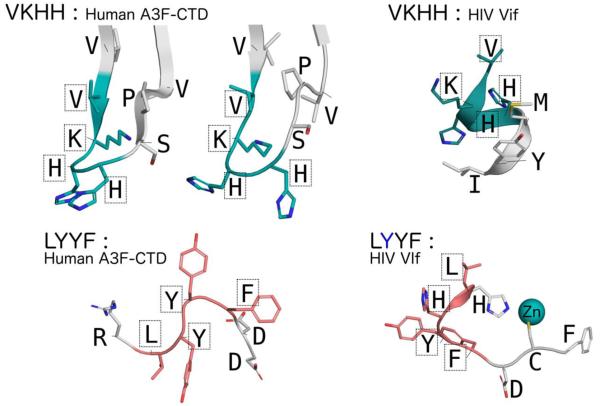
Structural conformations of the various peptides upon A3G binding can be extrapolated from observation of the crystallographic interfaces in the A3F-CTD crystal structure [9] as well as the Vif crystal structure [36]. Comparing the structures of regions containing the two sequence motifs, LYYF and VKHH, reveals wide conformational diversity (Fig. 8). The VKHH motif containing regions adopt two distinct conformations within the A3F-CTD crystal structure and are located at the loop between the β1-β2 strands. The corresponding region in the Vif crystal structure comprises the α1-helix and parts of the subsequent loop region. The LYYF motif region in A3F-CTD comprises the loop between the β4- α4 regions, near the enzyme active-site. The corresponding region in the Vif crystal structure spans the end of the α2-helix and the subsequent loop containing the His108 and Cys114 residues that directly coordinate the Zn atom in the Vif zinc-finger. This conformational diversity implies that these sequences with identical motifs may explore a large conformational space; this structural plasticity likely facilitates their ability to inhibit the A3G activity.
FIGURE 8.
Conformational plasticity of the VKHH and LYYF motifs in A3F-CTD and Vif. (A) With two molecules in the asymmetric unite two distinct conformations of the VKHH motif region are observed in the A3F-CTD crystal structure (PDB: 4IOU), that are strikingly different from the VKHH motif region in the Vif crystal structure (PDB: 4N9F). The lysine side-chain was modeled into the A3F-CTD structure using PyMOL as it was disordered in the crystal structure. (B) Similar conformational diversity of the LYYF motif is also observed between A3F-CTD and Vif.
Discussion
The physiological functions of APOBEC3 (A3) proteins include roles beyond their involvement in innate immunity ([2] and references there in). Lymphoma cells, which express high levels of A3G, are quite resistant to genotoxic treatments [25], because A3G accelerates DSB repair [17, 26]. Inhibition of the cellular endogenous A3G may be key in converting genotoxic-resistant lymphoma cells into being genotoxic-sensitive, pointing to the clinical uses of A3G inhibitors. As the HIV-1 Vif protein antagonizes A3G, allowing HIV-1 multiplication, we scanned the sequence of HIV-1 Vif to isolate peptides that inhibit the enzymatic activity of A3G. Analysis of the Vif domains identified two 15-amino acid peptides, namely Vif25-39 and Vif105-119, which efficiently inhibit the enzymatic activity of A3G (Table 1 and [38]). Truncation of Vif 25-39 from the amino- or carboxy-terminus of the peptide and partial alanine scanning of Vif25-39 reduces its inhibitory efficiency, suggesting that each residue comprised in Vif25-39 peptide contribute for inhibitory-effect (Table 1 and Figure 2C). These results suggest that the entire peptide Vif25-39 is essential for A3G inhibition.
Truncation of Vif105-119 yields the Vif107-115 peptide, which efficiently inhibits A3G enzymatic activity and hinders DSB repair. Alteration of each amino acid in the motif LYYF abrogates the inhibitory effect of the peptide indicating the unique specificity of the Vif07-115 peptide towards A3G inhibition. The Vif107-115 peptide includes the motif LYYF, which is present in A3F306-309. Based on these observations, we synthesized two peptides, A3F304-312 and A3F305-311, and found that both inhibit the deamination activity of A3G and impede DSB repair, suggesting this motif is essential for inhibition of deamination. Although not shown in this study, potentially these sequences may inhibit deamination in part possibly by hampering oligomerization[40].
Overall, our observations indicate that acceleration of DSB repair by A3G is deamination-dependent [17]. A3G interacts with the ssDNA substrate in a processive manner; by sliding and micro-jumping, or non-processively by macro-jumping and inter-segmental transfer [41-43]. Potentially both A3G NTD and CTD, which bind ssDNA, are required for inter-segmental movement, where one A3G molecule holds two substrate molecules or simultaneously binds two distant regions of the same DNA molecule. Oligomerization of A3G may also play a role in this process [44]. Often natural peptides require additional modification in order to increase the efficiency as inhibitors. Peptides are sensitive to proteases inside and outside the target-cells and they often unable to penetrate the cells and may not reach the desire cell compartment. However, the peptides Vif107-115 and Vif25-39 are native, unmodified peptides with IC50 of 0.1 and 0.6 μM respectively. These IC50 value of peptides Vif107-115 is lower than (more efficient) all other A3G inhibitors, while the IC50 of the other active peptides tested are in the range of A3G- and many other native enzyme-inhibitors ([45] and there in). The active peptides described here penetrate the cells, can persist in the cell cytoplasm and inhibit the endogenous A3G deaminase activity (Fig. 6). Thus, these peptides appear to be initial scaffolds for the development of future potent peptide-mimetic inhibitors that could become efficient anti-A3G small molecule-drugs.
Material and Methods
Cell culture
Cutaneous T cell lymphoma (H9 cells), T lymphoblastic leukemia (SupT1 cells) were provided by the National Institutes of Health (NIH) AIDS Reagent Program [Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), NIH, USA], and were grown in RPMI 1640. Ly-4 diffuse large B cell lymphoma cells were provided by Dr. D. Ben-Yehuda (Hadassah Medical School) and cells were maintained in RPMI 1640. Human embryonic kidney 293T adherent cell lines were grown as a sub-confluent monolayer in DMEM. Media were supplemented with 10% fetal calf serum, 100 U/ml penicillin, 100 U/ml streptomycin, and 2 mM L-glutamine (Biological Industries, Beit Haemek, Israel).
Expression and purification of A3G
Wild-type containing a C-terminal His6 tag were expressed in 293T cells and purified as previously described [41]. Briefly, 293T cells were transfected with pcDNA-APO3G expressing wt A3G. Cells (3×108) were harvested 48 h after transfection, washed three times in PBS and suspended in lysis buffer (50 mM Tris, pH 8.0, 1 mM PMSF (Sigma-Aldrich), 10% (v/v) glycerol and 0.8% (v/v) NP-40), to a final concentration of 20,000 cells/μl. Following 10 min incubation in ice, cell debris and nuclei were pelleted by centrifugation at 10,000 g for 20 min. The soluble fraction was adjusted to 0.8 M NaCl and treated with 50 μg/ml RNase A (Sigma-Aldrich) for 30 min at 37°C. The treated lysates were then added to 50 μl of nickel-nitrilotriacetic acid (Ni-NTA) agarose beads (QIAGEN), mixed on shaker for 1 h at 4°C and loaded onto a standard chromatography column (Bio-Rad Laboratories). Following extensive washing with wash buffer (50 mM Tris, pH 8.0, 0.3 M NaCl, 10% (v/v) glycerol) containing 30-50 mM imidazole, bound proteins were eluted seven times in elution buffer containing 120 mM imidazole. Protein samples were resolved by SDS-PAGE and stained with Imperial protein stain (Pierce Biotechnology). A3G concentration was determined by Bradford assay.
Synthetic oligonucleotides and peptides
The sequence of the 80-mer ss-deoxyoligonucleotide substrate used in the deamination assays is shown in table 2. The positive control ss-deoxyoligonucleotide bears the same sequence but has a dU instead of the target dC residue. The following primers were used for PCR amplification of the substrate and positive control oligonucleotides: Forward 5′-GGATTGGTTGGTTATTTGTTTAAGGA-3′; Reverse 5′-CCATCAATCTACCAAACATAACTTCCA-3′. Table 1 shows the amino acid sequences of the peptides assessed in this report.
Table 2.
Table showing the sequence of the substrate DNA oligonucleotide used
| Name | Sequence |
|---|---|
|
Eco147I (HaeIII)
substrate (S80) |
5′ - GGA TTG GTT GGT TAT TTG TTT AAG GAA GGT GGA TTA AAG GCC CAA TAA GGT GAT GGA AGT TAT GTT TGG TAG ATT GAT GG - 3′ |
|
Positive Control -
Eco147I (HaeIII) substrate (S80) |
5′ - GGA TTG GTT GGT TAT TTG TTT AAG GAA GGT GGA TTA AAG GCC UAA TAA GGT GAT GGA AGT TAT GTT TGG TAG ATT GAT GG - 3′ |
Deamination assay using purified A3G preparation
Deamination reactions were performed in a total volume of 10 μl in 25 mM Tris, pH 7.4, 0.1 μg/μl BSA and 1 fmol/μl of 80nt ssDNA substrate (Table 2), containing the underlined CCC deamination motif (Integrated DNA Technologies) at 37°C (standard conditions). Kinetic assays were performed at [E]«[S] ratio so that the overall product formation fell below 15% of the substrate. Note that the A3G enzyme used in this reaction was purified from mammalian cells (see above) and it is a very efficient deaminase. Thus, very low concentration of enzyme and substrate are required. The reactions were terminated by heating to 95°C for 5 min following immediate cooling on ice. One μl of the reaction mixture was used for PCR amplification with Redmix (Larova) in a total volume of 20 μl using the following program: 1 cycle at 95°C for 3 min, followed by 30 cycles of annealing at 61°C for 30 s and denaturing at 94°C for 30 s. PCR products (10 μl) were incubated with the StuI restriction enzyme (NEB, UK) for 1 h at 37°C. Note that the CCC motif is located in the middle of the substrate, yielding similar fragments following cleavage with the restriction enzyme. Completion of the restriction reaction was verified using positive control substrate containing CCU instead of CCC. Restriction reaction products were loaded onto gels and separated by 14% polyacrylamide gel electrophoresis (PAGE). Gels were stained with SYBR gold nucleic acid stain (Molecular Probes; Invitrogen) diluted 1:10,000 in 0.5× Tris-borate-ethylene-diaminetetraacetic acid buffer (pH 7.8), visualized by UV light (460 nm), captured by LAS-3000 (FUJIFILM) and analyzed by densitometry using ImageJ image processing and analysis software.
Deamination assay using cell extracts
Cultured H9 cells were grown in the presence (100 μM) or in the absence of peptide for 12 hr before pelleting and washed with PBS buffer. Pellets were suspended in lysis buffer containing 1% Triton X-100 in phosphate buffer saline. The suspended cells were incubated in ice for 30 minutes and subjected to centrifugation at 10000 rpm for 10 minutes. The supernatant, containing the cytoplasmic fraction was used to check the deamination acivity in the total volume of 10 μl in 25 mM Tris, pH 7.4, 0.1 μg/μl BSA, 0.05 μg/μl RNAse, 1% Triton X-100 and 100 fmol/μl of 80nt ssDNA substrate, containing the underlined CCC deamination motif at 37°C.
Inhibition of A3G deamination assay
The inhibitory peptides derived from Vif (HIV-1 HXBII Vif), APOBEC3G (A3G) and APOBEC3F (A3F) (Table 1) were used to determine their inhibitory effect on deamination activity of purified A3G enzyme. A fluorescein-conjugated peptide was used to assess peptide uptake by H9 cells (not shown). For inhibition of endogenous A3G in H9 cells, cells were incubated with 100 μM of peptides or a control peptide for 2 hours at 37°C before exposure to IR. A3G deaminase activity was assessed as described above.
Cell viability and Kinetics of cell growth
Ly-4 and SupT1 cells were cultivated, irradiated, treated with peptides. The number of cells was determined under light microscope. Ly-4 and SupT1 cells were mixed (1:10) with a solution containing Fluorescein diacetate (20mg/ml) & propidium iodide (2.5mg/ml) to determine the live and dead cells. The cell suspensions were incubated for 5 minutes at room temperature and examined by fluorescent microscope, Nikon Eclipse Ti equipped with an ORCA R2 camera (Hamamatsu Photonics).
Immunofluorescence Microscopy
Cells were irradiated by exposure to a 60Co source producing 1 Gy/second γ-radiation, or mock-irradiated. After incubation at 37°C, cells were washed with PBS, fixed with 4% paraformaldehyde/PBS, attached to glass slides by cytospin for 5 minutes at 200g, permeabilized with 0.2% Triton-X-100/PBS for 10 minutes, and blocked with 10% normal donkey serum 0.1% BSA in PBS for 30 minutes. Cells were then incubated with A3G C-terminal-specific rabbit polyclonal antibody (obtained through the National Institutes of Health AIDSP from J. Lingappa) and anti γ-H2AX- specific mouse monoclonal antibody (abcam, UK), followed by incubation with donkey anti-rabbit Alexa Fluor 647-conjugated antibody, donkey anti-mouse Alexa Fluor 488-conjugated antibody (abcam, UK). Slides were mounted with VECTASHIELD hard set with DAPI (Vector Laboratories) and examined by Zeiss LSM 710 confocal microscope. Data were collected sequentially using an X63 objective with 7-fold averaging at a resolution of 1024 X 1024 pixels. Data were analyzed with the Zen 2009 Light Edition software (Carl Zeiss). For inhibition of DSB repair (endogenous A3G) in irradiated H9 and Ly-4 cells, cells were incubated with 100 μM Vif-, A3G- and A3F-derived peptides or without peptide for 2 h at 37°C before exposure to IR.
Comet assay
The comet assay was carried out under alkaline conditions, as described by Singh et al. [46]. A day following culturing, H9 and SupT1 cells cultures treated, or not, with and without the peptides for 2 h before radiation or mock-irradiated, as described (Fig. 4). Agarose gels were prepared on frosted slides pre-coated with 1% normal melting point agarose (Sigma). Cells were mixed with 0.5% low melting point (LMP) agarose (Sigma), placed on the slides, and covered with additional layer of 0.5% LMP agarose. The slides were immersed for 2 h in freshly prepared ice-cold lysis buffer containing 2.5 M NaCl, 100 mM EDTA, 10 mM Tris (pH 10) with 1% triton-X 100 (sigma) and 10% dimethyl sulfoxide (Sigma). Following lysis, denaturation was carried out at 4°C with freshly prepared electrophoresis buffer (300 mM NaOH, 1 mM EDTA, pH 13). After 20 min of denaturation, the slides were electrophoresed at 45 V for 20 min in a horizontal gel-electrophoresis tank. The slides were then neutralized by washing in buffer containing 400 mM Tris (pH 7.5) and stained with ethidium bromide (5 μg/ml) for 15 min and examined under Nikon Eclipse Ti fluorescence microscope, equipped with an ORCA R2 camera (Hamamatsu Photonics). Random fields were photographed and data were analyzed by using the CASP software (CASP-1.2.2, download in http://casp.sourceforge.net/index.Php), the two parameters including the Tail DNA% (TDNA %) and the Olive tail moment (OTM) were determined [47].
Statistical analysis
Statistical analyses were performed by one-way analysis of variance (ANOVA) followed by Newman-Keuls multiple comparisons test. Each test samples were compared with their respective controls. The analyses were carried out in GraphPad Prism 6 (Graphpad software, Inc., CA, USA). Results were considered as statistically significant at a p-value ≤0.05.
Acknowledgements
We thank Joy Lengyel and Ming Li for A3G plasmid DNA constructs and Dr. Cheryl Balshayi for critical reading of this manuscript. We also thank Dr. Mohan Somasundaran for critical insights. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH: anti-APOBEC3G C terminus from Dr. Jaisri Lingappa and anti-APOBEC3G from Dr. Warner C. Greene. This work was carried out in the PeterA. Krueger Laboratory with the generous support of Nancy and Lawrence Glick and Pat and Marvin Weiss. Dr. Ponnandy Prabhu is a fellow of PBC of council for Higher Education in Israel. This work was supported by an NIH grant (P01 GM091743) and The Ministry of Industry, Trade and Labor (Nofar 49598).
Footnotes
Enzymes: APOBEC3G EC 3.5.4.1
Authors Contribution
P.P. and M.K. designed the research and wrote the paper; P.P., S.S., E.B.R., A.N., performed the experiments; P.P., M.K. and C.A.S. analyzed the data.
References
- 1.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–96. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 2.Refsland EW, Harris RS. The APOBEC3 family of retroelement restriction factors. Current topics in microbiology and immunology. 2013;371:1–27. doi: 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conticello SG, Langlois MA, Yang Z, Neuberger MS. DNA deamination in immunity: AID in the context of its APOBEC relatives. Advances in immunology. 2007;94:37–73. doi: 10.1016/S0065-2776(06)94002-4. [DOI] [PubMed] [Google Scholar]
- 4.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annual review of immunology. 2008;26:317–53. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 5.Goila-Gaur R, Strebel K. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology. 2008;5:51. doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.OhAinle M, Kerns JA, Malik HS, Emerman M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. Journal of virology. 2006;80:3853–62. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imahashi M, Nakashima M, Iwatani Y. Antiviral Mechanism and Biochemical Basis of the Human APOBEC3 Family. Frontiers in microbiology. 2012;3:250. doi: 10.3389/fmicb.2012.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shandilya SM, Nalam MN, Nalivaika EA, Gross PJ, Valesano JC, Shindo K, Li M, Munson M, Royer WE, Harjes E, Kono T, Matsuo H, Harris RS, Somasundaran M, Schiffer CA. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure. 2010;18:28–38. doi: 10.1016/j.str.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bohn MF, Shandilya SM, Albin JS, Kouno T, Anderson BD, McDougle RM, Carpenter MA, Rathore A, Evans L, Davis AN, Zhang J, Lu Y, Somasundaran M, Matsuo H, Harris RS, Schiffer CA. Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. Structure. 2013;21:1042–50. doi: 10.1016/j.str.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byeon IJ, Ahn J, Mitra M, Byeon CH, Hercik K, Hritz J, Charlton LM, Levin JG, Gronenborn AM. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nature communications. 2013;4:1890. doi: 10.1038/ncomms2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitamura S, Ode H, Nakashima M, Imahashi M, Naganawa Y, Kurosawa T, Yokomaku Y, Yamane T, Watanabe N, Suzuki A, Sugiura W, Iwatani Y. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nature structural & molecular biology. 2012;19:1005–10. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- 12.Harris RS, Winkler T, Venegas JG, Medoff B. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2012 [Google Scholar]
- 13.Albin JS, Harris RS. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert reviews in molecular medicine. 2010;12:e4. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shandilya SM, Bohn MF, Schiffer CA. A computational analysis of the structural determinants of APOBEC3's catalytic activity and vulnerability to HIV-1 Vif. Virology. 2014;471-473:105–16. doi: 10.1016/j.virol.2014.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nature structural & molecular biology. 2010;17:222–9. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landry S, Narvaiza I, Linfesty DC, Weitzman MD. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO reports. 2011;12:444–50. doi: 10.1038/embor.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nowarski R, Wilner OI, Cheshin O, Shahar OD, Kenig E, Baraz L, Britan-Rosich E, Nagler A, Harris RS, Goldberg M, Willner I, Kotler M. APOBEC3G enhances lymphoma cell radioresistance by promoting cytidine deaminase-dependent DNA repair. Blood. 2012;120:366–75. doi: 10.1182/blood-2012-01-402123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–9. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 19.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell J, Wheldon TE, Stanton P. A radioresistant variant derived from a human neuroblastoma cell line is less prone to radiation-induced apoptosis. Cancer research. 1995;55:4915–21. [PubMed] [Google Scholar]
- 21.Stecca C, Gerber GB. Adaptive response to DNA-damaging agents: a review of potential mechanisms. Biochemical pharmacology. 1998;55:941–51. doi: 10.1016/s0006-2952(97)00448-6. [DOI] [PubMed] [Google Scholar]
- 22.Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nature cell biology. 2009;11:973–9. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothkamm K, Lobrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5057–62. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aldridge DR, Radford IR. Explaining differences in sensitivity to killing by ionizing radiation between human lymphoid cell lines. Cancer research. 1998;58:2817–24. [PubMed] [Google Scholar]
- 25.Jais JP, Haioun C, Molina TJ, Rickman DS, de Reynies A, Berger F, Gisselbrecht C, Briere J, Reyes F, Gaulard P, Feugier P, Labouyrie E, Tilly H, Bastard C, Coiffier B, Salles G, Leroy K. The expression of 16 genes related to the cell of origin and immune response predicts survival in elderly patients with diffuse large B-cell lymphoma treated with CHOP and rituximab. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2008;22:1917–24. doi: 10.1038/leu.2008.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nowarski R, Kotler M. APOBEC3 Cytidine Deaminases in Double-Strand DNA Break Repair and Cancer Promotion. Cancer research. 2013;73:3494–8. doi: 10.1158/0008-5472.CAN-13-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nik-Zainal S, Wedge DC, Alexandrov LB, Petljak M, Butler AP, Bolli N, Davies HR, Knappskog S, Martin S, Papaemmanuil E, Ramakrishna M, Shlien A, Simonic I, Xue Y, Tyler-Smith C, Campbell PJ, Stratton MR. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nature genetics. 2014;46:487–91. doi: 10.1038/ng.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal SA, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Griffin CA, Burton J, Swerdlow H, Quail MA, Stratton MR, Iacobuzio-Donahue C, Futreal PA. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–13. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 30.Loeb LA, Loeb KR, Anderson JP. Multiple mutations and cancer. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:776–81. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Molecular cell. 2012;46:424–35. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, Menzies A, Martin S, Leung K, Chen L, Leroy C, Ramakrishna M, Rance R, Lau KW, Mudie LJ, Varela I, McBride DJ, Bignell GR, Cooke SL, Shlien A, Gamble J, Whitmore I, Maddison M, Tarpey PS, Davies HR, Papaemmanuil E, Stephens PJ, McLaren S, Butler AP, Teague JW, Jonsson G, Garber JE, Silver D, Miron P, Fatima A, Boyault S, Langerod A, Tutt A, Martens JW, Aparicio SA, Borg A, Salomon AV, Thomas G, Borresen-Dale AL, Richardson AL, Neuberger MS, Futreal PA, Campbell PJ, Stratton MR. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–93. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Miller BF, Furano AV. Repair of naturally occurring mismatches can induce mutations in flanking DNA. eLife. 2014;3:e02001. doi: 10.7554/eLife.02001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding Q, Chang CJ, Xie X, Xia W, Yang JY, Wang SC, Wang Y, Xia J, Chen L, Cai C, Li H, Yen CJ, Kuo HP, Lee DF, Lang J, Huo L, Cheng X, Chen YJ, Li CW, Jeng LB, Hsu JL, Li LY, Tan A, Curley SA, Ellis LM, Dubois RN, Hung MC. APOBEC3G promotes liver metastasis in an orthotopic mouse model of colorectal cancer and predicts human hepatic metastasis. The Journal of clinical investigation. 2011;121:4526–36. doi: 10.1172/JCI45008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loeb LA. Human cancers express mutator phenotypes: origin, consequences and targeting. Nature reviews Cancer. 2011;11:450–7. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo Y, Dong L, Qiu X, Wang Y, Zhang B, Liu H, Yu Y, Zang Y, Yang M, Huang Z. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature. 2014;505:229–33. doi: 10.1038/nature12884. [DOI] [PubMed] [Google Scholar]
- 37.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nature reviews Immunology. 2004;4:868–77. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 38.Britan-Rosich E, Nowarski R, Kotler M. Multifaceted counter-APOBEC3G mechanisms employed by HIV-1 Vif. Journal of molecular biology. 2011;410:1065–76. doi: 10.1016/j.jmb.2011.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benkovic V, Orsolic N, Knezevic AH, Ramic S, Dikic D, Basic I, Kopjar N. Evaluation of the radioprotective effects of propolis and flavonoids in gamma-irradiated mice: the alkaline comet assay study. Biological & pharmaceutical bulletin. 2008;31:167–72. doi: 10.1248/bpb.31.167. [DOI] [PubMed] [Google Scholar]
- 40.Chaurasiya KR, McCauley MJ, Wang W, Qualley DF, Wu T, Kitamura S, Geertsema H, Chan DS, Hertz A, Iwatani Y, Levin JG, Musier-Forsyth K, Rouzina I, Williams MC. Oligomerization transforms human APOBEC3G from an efficient enzyme to a slowly dissociating nucleic acid-binding protein. Nature chemistry. 2014;6:28–33. doi: 10.1038/nchem.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nowarski R, Britan-Rosich E, Shiloach T, Kotler M. Hypermutation by intersegmental transfer of APOBEC3G cytidine deaminase. Nature structural & molecular biology. 2008;15:1059–66. doi: 10.1038/nsmb.1495. [DOI] [PubMed] [Google Scholar]
- 42.Nowarski R, Prabhu P, Kenig E, Smith Y, Britan-Rosich E, Kotler M. APOBEC3G inhibits HIV-1 RNA elongation by inactivating the viral trans-activation response element. Journal of molecular biology. 2014;426:2840–53. doi: 10.1016/j.jmb.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Senavirathne G, Jaszczur M, Auerbach PA, Upton TG, Chelico L, Goodman MF, Rueda D. Single-stranded DNA scanning and deamination by APOBEC3G cytidine deaminase at single molecule resolution. The Journal of biological chemistry. 2012;287:15826–35. doi: 10.1074/jbc.M112.342790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lavens D, Peelman F, Van der Heyden J, Uyttendaele I, Catteeuw D, Verhee A, Van Schoubroeck B, Kurth J, Hallenberger S, Clayton R, Tavernier J. Definition of the interacting interfaces of Apobec3G and HIV-1 Vif using MAPPIT mutagenesis analysis. Nucleic acids research. 2010;38:1902–12. doi: 10.1093/nar/gkp1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M, Shandilya SM, Carpenter MA, Rathore A, Brown WL, Perkins AL, Harki DA, Solberg J, Hook DJ, Pandey KK, Parniak MA, Johnson JR, Krogan NJ, Somasundaran M, Ali A, Schiffer CA, Harris RS. First-in-class small molecule inhibitors of the single-strand DNA cytosine deaminase APOBEC3G. ACS chemical biology. 2012;7:506–17. doi: 10.1021/cb200440y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Experimental cell research. 1988;175:184–91. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 47.Wang JL, Wang PC. The effect of aging on the DNA damage and repair capacity in 2BS cells undergoing oxidative stress. Molecular biology reports. 2012;39:233–41. doi: 10.1007/s11033-011-0731-4. [DOI] [PubMed] [Google Scholar]