SUMMARY
The molecular changes underlying the higher risk of chronic inflammatory disorders during aging remain incompletely understood. Molecular variations in the innate immune response related to recognition and interaction with microbes at mucosal surfaces could be involved in aging-related inflammation. We developed an ontology analysis of 20 NOD-like receptors (NLRs) and 7 inflammasome-related genes (IRGs) in healthy and inflamed/periodontitis oral mucosal tissues from young, adolescent, adult and aged nonhuman primates (Macaca mulatta) using the GeneChip® Rhesus Macaque Genome array. Validation of some of the significant changes was done by qRT-PCR. The expression of NLRB/NAIP, NLRP12, and AIM2 increased with aging in healthy mucosa whereas NLRC2/NOD2 expression decreased. Although higher expression levels of some NLRs were generally observed with periodontitis in adult mucosal tissues (e.g., NLRB/NAIP, NLRP5, and NLRX1), various receptors (e.g., NLRC2/NOD2, and NLRP2) and the inflammasome adaptor protein ASC, exhibited a significant reduction in expression in aged periodontitis tissues. Accordingly, the expression of NLR-activated innate immune genes, such as HBD3 and IFNB1, was impaired in aged but not adult periodontitis tissues. Both adult and aged tissues showed significant increase in IL-1β expression. These findings suggest that the expression of a subset of NLRs appears to change with aging in healthy oral mucosa, and that aging-related oral mucosal inflammation could involve an impaired regulation of the inflammatory and antimicrobial response associated with down-regulation of specific NLRs and IRGs.
Keywords: Aging, inflammasome, NOD-like receptors, oral mucosa, innate immunity
1. Introduction
Reduced immunity in aging (also named immuno-senescence) is accompanied by chronic inflammation. This is reflected in a higher prevalence of inflammatory disorders or age-related diseases, including periodontal disease, whose prevalence is about 2–3 times higher among those aged >65 years compared to younger individuals (35 to 65 years) (Albandar 2011; Dye et al., 2007). Periodontal disease is a common oral disease that involves inflammation of gingival tissues in response to bacterial species colonizing the oral mucosa, This process, leads to the destruction of the supporting tissues of the teeth (i.e., gingival tissue, periodontal ligament, and alveolar bone) if it is not treated (Van Dyke and Serhan 2003), and is a risk factor for systemic conditions (e.g., cardiovascular disease and diabetes) (Friedewald et al., 2009; Lalla and Papapanou 2011). Humans and microbes have co-evolved during millions of years developing a mutually beneficial relationship with total estimates of about 1014 bacteria including more than 500 different species colonizing a healthy adult human. Similar to other mucosal surfaces, the oral cavity represents an example of this symbiotic relationship between host and bacteria whereby, despite the large and diverse microbial loads constantly challenging the oral mucosa across the life span (Dewhirst et al., 2010), the host generally remains healthy. This is likely due to a balanced host immune response, whereby innate immune components appear to play a critical role (Darveau 2010).
It remains unclear if the higher prevalence of periodontitis seen in the elderly is a natural consequence of aging or the result of unique cellular and molecular changes that occur in the oral mucosa during aging, thus increasing the risk for developing the disease with age. Emerging evidence suggests that perturbations of innate immune mechanisms associated with aging could play a critical role in age-related chronic inflammatory disorders (Qian et al., 2012; Shaw et al., 2011). Accordingly, ex vivo evidence indicates that impaired expression and function of innate immune components (i.e., Toll-like receptors-TLRs, cytokine/chemokine production, neutrophils and monocyte/macrophages) appears to be associated with aging and periodontitis (Hajishengallis 2010). Most recently, it was shown in mice that aging-associated periodontitis is accompanied by lower expression of an endogenous inhibitor of neutrophil adhesion dependent on the integrin LFA-1 (Del-1), which is a negative regulator for the recruitment of inflammatory cells into the tissues (Eskan et al., 2012). Thus, an increased migration of neutrophils into the oral mucosa constantly challenged by bacteria would be associated with periodontitis in aged mice.
The nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs) family of proteins is a growing group of cytosolic pattern recognition receptors (PRRs) involved in the regulation of the innate immune responses against pathogen- and danger-associated molecular patterns (PAMPs and DAMPs) (Geddes et al., 2009). About 22 intracellular NLRs have been reported, which are classified into subfamilies based on their N-terminal domain into: (i) caspase-recruitment domain (CARD)-containing NLRCs or NODs (1 to 5), (ii) pyrin-domain (PYD)-containing NLRPs (1 to 14), and (iii) baculovirus inhibitor of apoptosis repeat (BIR)-containing domain NAIPs (neuronal apoptosis inhibitor proteins) (Franchi et al., 2008). Among these NLRs, NLRC1/NOD1 and NLRC2/NOD2 are perhaps the most studied to date and are activated by the bacterial peptidoglycan motifs diaminopimelic acid (DAP) from Gram-negative bacteria and muramyl dipeptide (MDP) from both Gram-positive and Gram-negative bacteria, which normally leads to NFκB activation and production of cytokines/chemokines (Chamaillard et al., 2003; Girardin et al., 2003). Interestingly, the activation of several members of the NLR gene family (i.e., NLRB/IPAF, NLRC4, NLRPs 1, 3, 5, 6 and 7) by a range of PAMPs such as MDP, poly(I-C), dsRNA from viruses, bacterial RNA and pore-forming toxins, as well as a range of DAMPs such as extracellular ATP, uric acid, asbestos, silica, aluminum hydroxide, and amyloid-β peptide, contributes to the assembly of a macromolecular protein complex termed the inflammasome. This process leads to activation of the cysteine protease, caspase-1, which in turn induces inflammation (i.e., IL-1β, IL-18 and IL-33 secretion), pyroptosis (specialized type of pro-inflammatory cell death to control intracellular infectious niches), and repair and healing (FGF2 secretion and lipid membrane biogenesis) (Mariathasan and Monack 2007). In addition, reactive oxygen species (ROS) and lysosomal damage as well as activation of NLRC4/IPAF4 by flagellin have also shown the ability to enhance inflammasome activation (Martinon 2010; Zhao et al., 2011). Of note, it has been recently suggested that inflammasome activation influences many metabolic disorders, such as atherosclerosis, type 2 diabetes, gout and obesity (Wen et al., 2012).
In addition to some NLRs members, it was recently shown that absent in melanoma 2 (AIM2), a member of the family of hematopoietic IFN-inducible nuclear proteins with a 200-amino acid motif (HIN-200), and the cytoplasmic RIG-I-like helicase (RIG-I) also have the ability to activate the inflammasome in response to cytosolic double stranded DNA from viruses, bacteria or the host itself (Hornung et al., 2009; Poeck et al., 2010). In particular, NLRPs and AIM2 interact with the cytosolic adaptor protein called apoptosis-associated speck-like protein containing a CARD domain (ASC) to further activate caspase-1. In contrast, other NLR members (e.g., NLRP2, NLRP10, NLRP12, and NLRX1) instead of being inflammasome activators, appear to be negative regulators of inflammation, decreasing NFkB activation and reducing type I interferon responses (Allen et al., 2011; Williams et al., 2005), as well as modulators of the adaptive immune response, controlling dendritic cell migration from tissues to lymphoid nodes (Arthur et al., 2010). Thus, this emerging family of cytosolic sensors with the ability to regulate the host responses to bacteria, inflammation, and adaptive immunity appear to play a crucial role at mucosal surfaces highly exposed to PAMPs and DAMPs, to maintain tissue homeostasis. Most recently, it has been suggested that NLRs could be critical innate sensors for discriminating and controlling pathogenic species (i.e., bacteria expressing specialized secretions systems, pore-forming toxins, and increased invasiveness ability) within the context of complex bacterial communities constantly colonizing the mucosal surfaces during health, mainly through a regulated activation of the inflammasome (Blander and Sander 2012). In fact, mutations in some NLRs (e.g., NLRP3, NLRC4, NOD1 and NOD2) have been associated with disorders such as inflammatory bowel disease (e.g., Crohn’s disease), atopic dermatitis and asthma [reviewed in (Geddes et al., 2009)].
There is solid evidence indicating that cytokines belonging to the IL-1 family, such as IL-1β and IL-18, play a critical role in periodontitis (Graves and Cochran 2003; Orozco et al., 2007) and recent in vivo and in vitro studies have shown variation in the expression of some inflammasome genes, in response to oral biofilms and planktonic bacteria, as well as the ability of some oral periodontopathogenic species (e.g., P. gingivalis, A. actinomycetemcomitans) to activate the inflammasome (Belibasakis and Johansson 2012; Bostanci et al., 2009; Yilmaz et al., 2010). Since the aged population is at higher risk for infections and inflammatory disorders, we hypothesized that mucosal changes in the expression of NLRs and inflammasome-related genes occur with aging. In this study, we used the oral cavity as a model of a mucosal surface that naturally becomes constantly exposed to PAMPs and DAMPs across the lifespan to determine the changes in the expression of NLRs and inflammasome-related genes associated with aging during health and inflammation (i.e., periodontitis). Importantly, the nonhuman primates develop periodontal disease naturally with age as it is observed in humans. Thus, in contrast to adult/aged animals, young and adolescent animals develop gingivitis but that does not progress to periodontitis (Schou et al., 1993).
Methods
1.1. Animals and diet
Rhesus monkeys (Macaca mulatta) (n=34; 14 females and 20 males) housed at the Caribbean Primate Research Center (CPRC) at Sabana Seca, Puerto Rico, were used in these studies. Periodontally healthy animals (n=23) were selected by age based on the following criteria: ≤3 years (young; n=5), 3–7 years (adolescent; n=5), 12–16 years (adult; n=7) and 18–23 years (aged; n=6). Only adult (n=5; 3 males and 2 females) and aged (n=6; 5 males and 1 female) animals with periodontitis were used, since periodontitis does not occur naturally in younger animals. The nonhuman primates were typically fed a 20% protein, 5% fat, and 10% fiber commercial monkey diet (diet 8773, Teklad NIB primate diet modified: Harlan Teklad). The diet was supplemented with fruits and vegetables, and water was provided ad libitum in an enclosed corral setting.
1.2. Oral clinical parameters and gingival tissue sample collection
Following a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Puerto Rico, anesthetized animals were examined by a single investigator using a Maryland probe on the facial aspect of the teeth, 2 proximal sites per tooth (mesio- and disto-buccal), excluding the canines and 3rd molars. The clinical examination included probing pocket depth (PD), and bleeding on probing (BOP; 0–3 scale) (Ebersole et al., 2008). Using a standard gingivectomy technique (a crevicular incision followed by an interdental incision at the base of the papillae using a #15 surgical blade), a buccal gingival papillae from either healthy or periodontitis-affected tissue from the premolar/molar maxillary region of each animal was taken and maintained frozen in RNAlater solution until RNA preparation for microarray analysis.
1.3. RNA Extraction, Reverse Transcription, and Gene Chip Hybridizations
Total RNA was isolated from each gingival tissue using Triazol reagent (Invitrogen, CA), and further cleaned up with the Qiagen RNeasy mini kit (Qiagen, Valencia, CA). All microarray RNA expression analyses were done at the University of Kentucky Microarray facility. Tissue RNA samples were submitted to the microarray core and RNA quality was assessed with an Agilent 2100 Bioanalyzer. Reverse transcription of equal amounts of RNA from each sample was performed, followed by hybridization to the GeneChip® Rhesus Macaque Genome Array (Affymetrix) similar to methods we have described previously (Gonzalez et al., 2013; Meka et al., 2010). Briefly, 300ng of total RNA was labeled using Affymetrix GeneChip 3' IVT Express kit, from which 15µg of labeled cDNA was hybridized to the GeneChip Rhesus Macaque Genome Array (Affymetrix) following the Affymetrix protocol. Post-hybridization, washing and staining of arrays were performed in an Affymetrix GeneChip Fluidics FS450 station followed by scanning using an Affymetrix GeneChip 3000 7G Scanner and GeneChip Operating Software MAS 5.0. Individual samples were used for gene expression analyses of 20 NLRs, 7 inflammasome-related genes, and 3 antimicrobial genes (Table 1) in healthy and inflamed gingival tissues.
TABLE 1.
NOD-like receptors (NLR), inflammasome-related genes, and antimicrobial genes evaluated by microarray in gingival tissues and their corresponding Probe Identification numbers.
| NOD-LIKE RECEPTORS | Gene ID | Probe No. |
| NLR family, MHC class II transactivator | NLRA/CIITA | MmugDNA.20.1.S1_at |
| NLR family, apoptosis inhibitory protein | NLRB/NAIP | MmugDNA.16377.1.S1_at |
| NLR family, CARD domain containing 1 | NLRC1/NOD1 | MmugDNA.33152.1.S1_at |
| NLR family, CARD domain containing 2 | NLRC2/NOD2 | MmuSTS.3541.1.S1_at |
| NLR family, CARD domain containing 4 | NLRC4/IPAF | MmugDNA.42771.1.S1_at |
| NLR family, CARD domain containing 5 | NLRC5 | MmugDNA.30929.1.S1_at |
| NLR family, pyrin domain containing 1 | NLRP1 | MmugDNA.34857.1.S1_at |
| similar to NACHT-, LRR- and PYD-containing protein 2 (PYRIN-containing APAF1-like protein 2) | NLRP2 | MmugDNA.16291.1.S1_s_at |
| NLR family, pyrin domain containing 3 | NLRP3 | MmugDNA.4850.1.S1_at |
| NLR family, pyrin domain containing 4 | NLRP4 | MmugDNA.39919.1.S1_at |
| NLR family, pyrin domain containing 5 | NLRP5 | MmuSTS.1762.1.S1_at |
| NLR family, pyrin domain containing 6 | NLRP6 | MmugDNA.28440.1.S1_at |
| NLR family, pyrin domain containing 7 | NLRP7 | MmugDNA.41819.1.S1_at |
| NLR family, pyrin domain containing 8 | NLRP8 | MmugDNA.11176.1.S1_at |
| Similar to NLR family, pyrin domain containing 10 | NLRP10 | MmugDNA.41661.1.S1_at |
| NLR family, pyrin domain containing 11 | NLRP11 | MmugDNA.34108.1.S1_at |
| NLR family, pyrin domain containing 12 | NLRP12 | MmugDNA.25405.1.S1_at |
| NLR family, pyrin domain containing 13 | NLRP13 | MmugDNA.30103.1.S1_at |
| NLR family, pyrin domain containing 14 | NLRP14 | MmuSTS.1761.1.S1_at |
| NLR family member X1 | NLRX1 | MmugDNA.20879.1.S1_at |
| INFLAMMASOME-RELATED GENES | Gene ID | Probe No. |
| absent in melanoma 2 | AIM2 | Mmu.10556.1.S1_at |
| RIG-I-like receptor/ DEAD box polypeptide 58 | RIG-I | MmugDNA.19189.1.S1_at |
| similar to PYD and CARD domain containing isoform b | ASC | MmuSTS.3453.1.S1_at |
| caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | CASPASE-1 | MmugDNA.31375.1.S1_s_at |
| Interleukin 1, beta | IL-1B | MmuSTS.652.1.S1_at |
| Interleukin 18 (interferon-gamma-inducing factor) | IL-18 | MmuSTS.2541.1.S1_at |
| Interleukin 33 | IL-33 | MmugDNA.20693.1.S1_at |
| ANTIMICROBIAL GENES | Gene ID | Probe No. |
| Defensin, Beta 4A/Human beta defensin-2 | DEFB4 | MmugDNA.33367.1.S1_at |
| Defensin, Beta 103B/Human beta defensin-3 | DEF103A/103B | MmugDNA.18488.1.S1_at |
| Interferon, Beta 1 | IFNB1 | MmuSTS.625.1.S1_at |
1.4. Quantitative RT-PCR
For qRT-PCR analysis, three mRNA samples for each group (i.e., healthy or periodontitis) were individually analyzed in two independent experiments (n=6) from adult and aged animals. The cDNA synthesis was carried out starting with 1μg of mRNA and expression of specific transcripts for NLRs and IRGs analyzed using the LightCycler 480 (Roche, IN). Primers for each gene were designed using the software Primer Quest from Integrated DNA Technologies (IDT), and synthesized by the same company (www.idtdna.com, Coralville, IA). The primers sequences (5’-3’) used were: NLRP14: Forward: CAAGATCTCTCCTCTGCTCTTATC and Reverse: CACTTAGGAGACTTCAGGACTTT; NLRP5: Forward: CACTCTCCTTGGCCCTTTC and Reverse: GCTGAACACAGCTTCATCATTC; NOD2: GAGGCAGTTCCATTTCATTTGT and Reverse: TGCTTAGAAGGAAGGGCTTAAT; ASC: Forward: AGGCCTGCACTTTGTAGAC and Reverse: TCCTGGTACTGCTGATCCT; IL-1B: Forward: GACAGGATCTGGAGCAACAA and Reverse: CCCAAGGCCACAGGTATTT; GADPH: Forward: GGTGTGAACCATGAGAAGTATGA and Reverse: GAGTCCTTCCACGATACCAAAG. Concentration ratios for the target genes were calculated by normalizing to the housekeeping gene GADPH.
1.5. Histopathologic Evaluation
Gingival tissues were formalin fixed (4% neutral buffered formalin) for 24 hours at room temperature and further placed in 70% ethanol. Tissues were processed by graded dehydration in alcohols using TissueTek VIP1000 and paraffin embedded. Serial sections of 5μm were obtained using Leica RM2255 Rotary Microtome and tissues were deparaffinized following standard protocol with xylene, and rehydrated with decreasing concentrations of ethanol. Sections of each biopsy were stained with Hematoxylin-Eosin (H&E). The areas of connective tissue under the epithelial basement membrane were analyzed under light microscopy at 200×. Each gingival tissue sample was divided into 10 fields and inflammatory cell infiltrate was determined in each field based on morphological characteristics by a blinded investigator.
1.6. Data analysis
The expression intensities for all genes across the samples were estimated using the Affymetrix PLIER algorithm. The GeneChip® Rhesus Macaque Genome Array contained matched and mismatched pairs allowing the MAS 5 algorithm to be used. For aging-related gene expression changes in healthy gingival tissues, a simple linear regression model was fit to the scatter plot of gene expression by age as a continuous variable. The 95% confidence bands for the fit line were included in the plots. For genes that had significant differences in expression with periodontitis, two sample t-tests were used. Statistical significance was considered by a p value ≤ 0.05. All statistical analysis was performed using the software JMP 10 (SAS, Inc., Cary, NC). Microarray data was uploaded into the ArrayExpress data base (www.ebi.ac.uk) under accession number: E-MTAB-1977.
2. Results
Clinical characterization for healthy and inflamed/periodontitis sites in each group included, young animals (mean PD 1.4±0.4 mm, mean BOP 0.4±0.5); adolescent animals (mean PD 1.9±0.2 mm, mean BOP 0.9±0.7), adult healthy group (mean PD 2.4±0.2 mm, mean BOP 0.9±0.4); aged healthy group (mean PD 2.5±0.4 mm, mean BOP 1.1±0.7); adult periodontitis group (mean PD 3.9±0.2 mm, mean BOP 2.6±0.6); and aged periodontitis group (mean PD 4.6±0.7 mm, mean BOP 2.7±0.4).
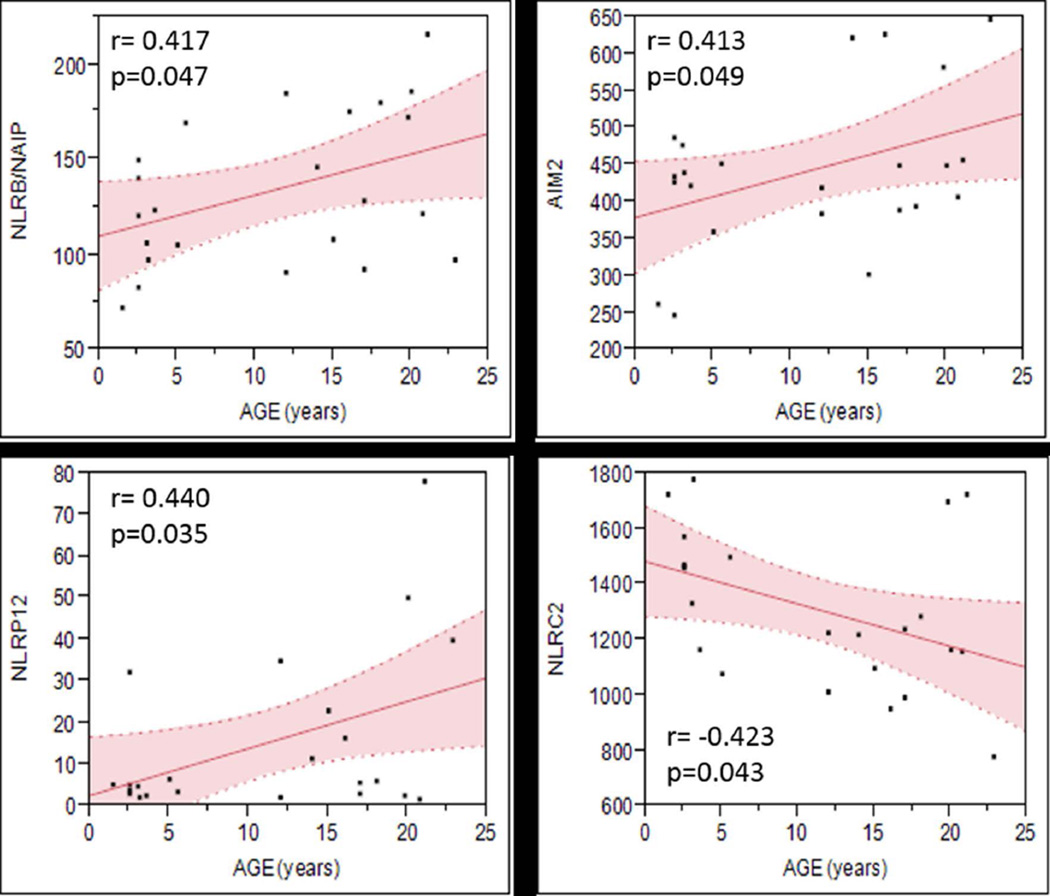
Among the NLRs and inflammasome-related genes evaluated by microarray, only the expression of 4 cytosolic receptors showed significant correlation with age, where the expression of NLRB/NAIP, NLRP12, and AIM2 increased, and NLRC2/NOD2 expression decreased with aging in healthy gingival tissues (Figure 1). Interestingly, the expression of inflammasome related genes including the adaptor protein ASC, as well as the Caspase 1 and its substrates pro-IL1β, and pro-IL18 did not change with age in healthy gingival tissues.
Figure 1. Scatterplot graphs showing the 95% confidence intervals for the regression fitting of NOD-like receptors and inflammasome related genes that significantly correlated with age.
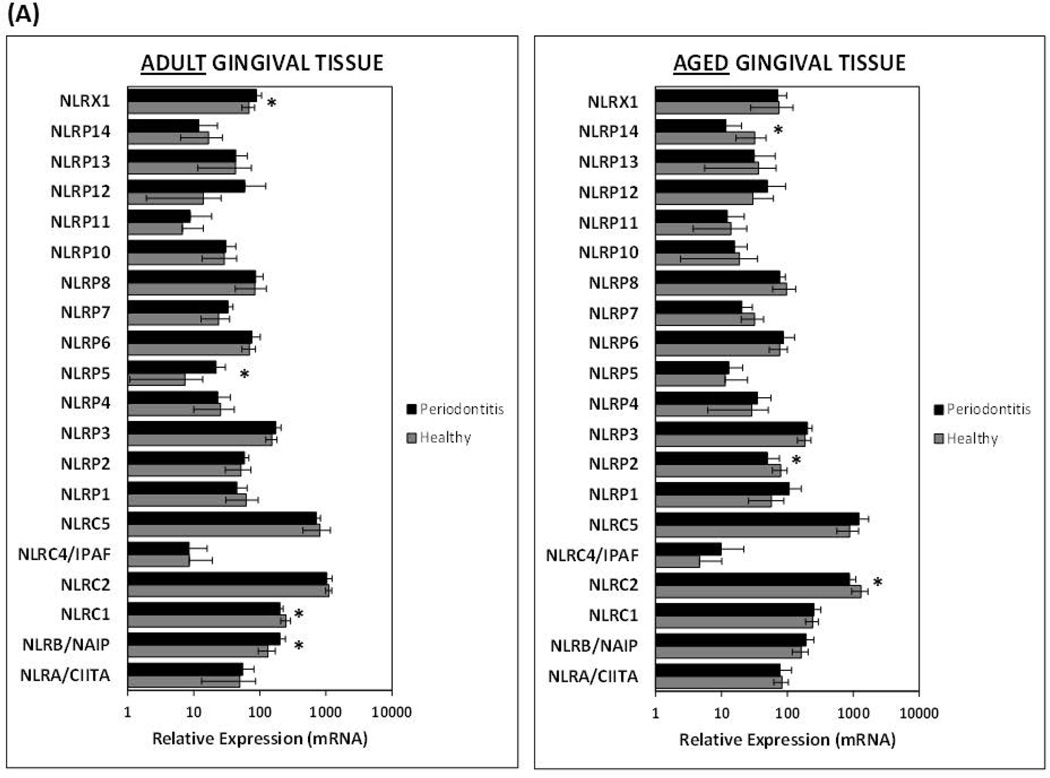
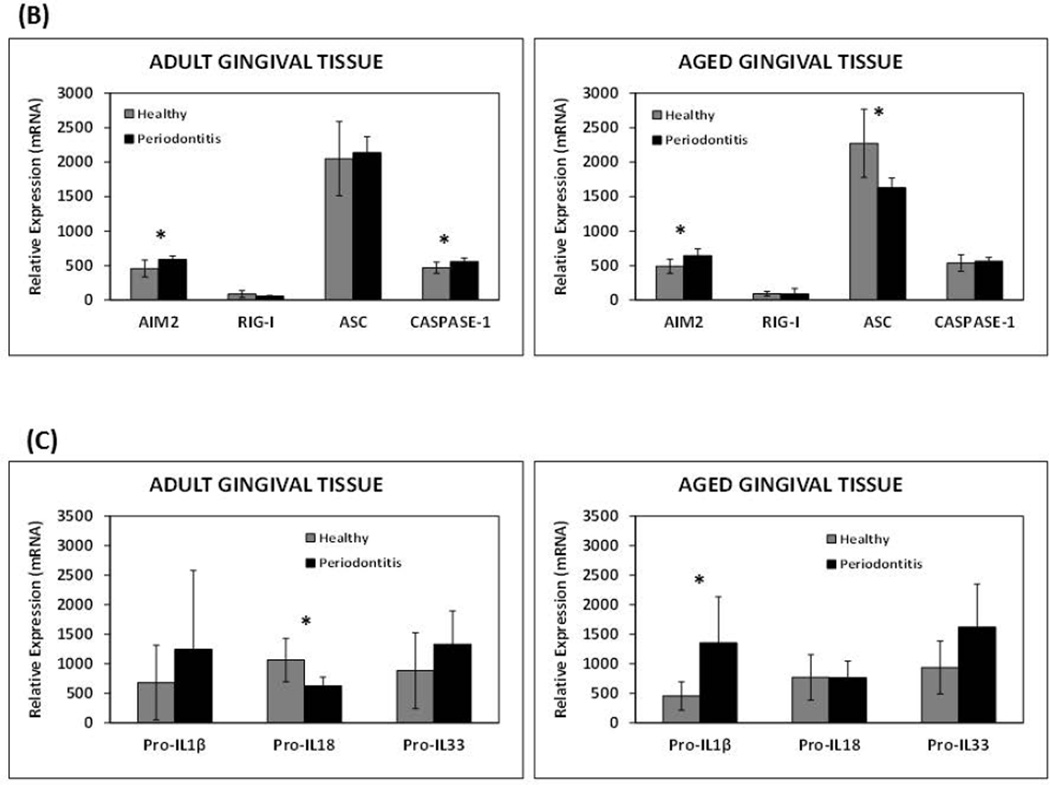
Although there was a similar trend with higher expression of the majority of genes during inflammation/periodontitis in adult and aged gingival tissues compared to healthy tissues, only NLRB, NLRP5, NLRX1, and Caspase-1 reached statistical significance in diseased adult but not aged tissues (Figs. 2A and 2B). Interestingly, inflamed tissues from aged animals, in contrast to the adult counterparts, exhibited a significant reduction in the expression of various NLRs (i.e., NLRC2/NOD2, NLRP2, and NLRP14) compared to healthy tissues of a similar age (Fig. 2A). Only NLRC1/NOD1 decreased expression was observed in periodontitis adult tissues. The presence of periodontitis was also associated with a significant reduction in the expression of the inflammasome adaptor protein ASC in aged, but not adult gingival tissues (Fig. 2B). Finally, the expression of the downstream substrates (i.e., pro-IL-1β, pro-IL-33) of the inflammasome showed similar elevated expression with periodontitis in both adult and aged tissues; however, pro-IL18 mRNA levels were significantly diminished with disease in adults, but not aged gingival tissues (Fig. 2C).
Figure 2. Age-related changes in the expression of NOD-like receptors and inflammasome-related genes in healthy and inflamed/periodontitis gingival tissues.
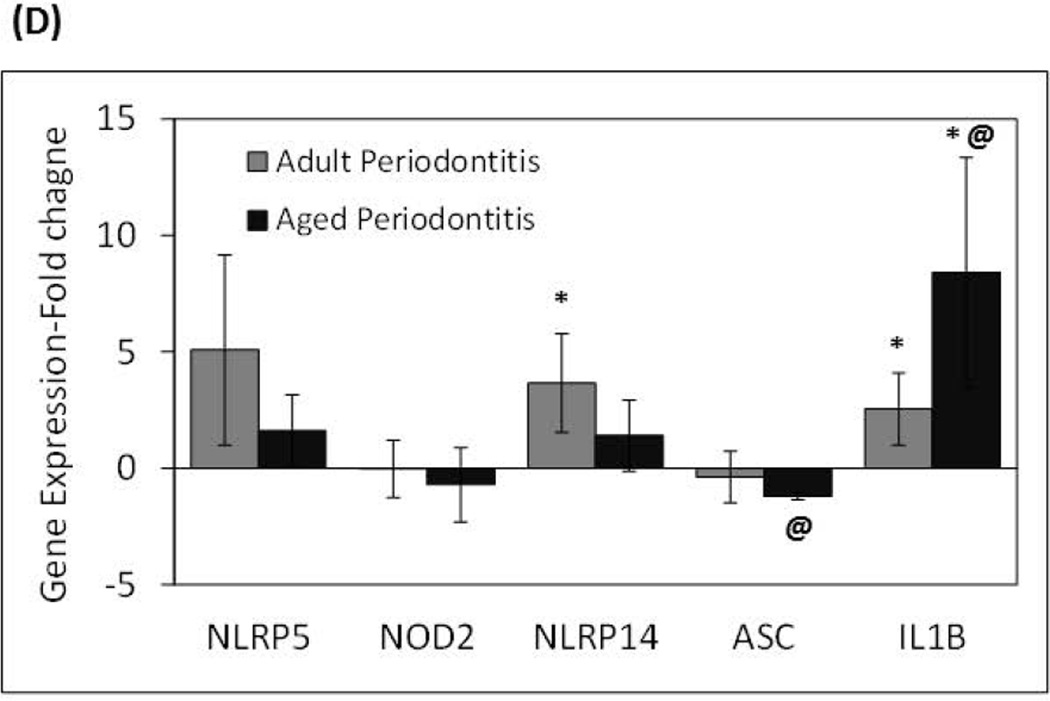
The results shown are the Means ± Standard Deviation of gene expression for (A) NOD-like receptors, and (B and C) inflammasome-related genes of healthy and inflamed/periodontitis gingival tissues obtained from 5–6 animals per each age (i.e., adult or aged non-human primates). Each sample was individually analyzed by microarray (D) Gene expression levels determined by qRT-PCR. Significant changes in gene expression with periodontitis in adult and aged tissues detected by microarray were validated by qRT-PCR in selected genes. Total mRNA from healthy (n=3) and periodontitis (n=3) gingival tissues obtained from adult and aged animals were individually analyzed in two independent experiments. Data are expressed as means ± standard deviations of fold changes in periodontitis vs. healthy tissues. *, p≤0.05 when periodontitis gene expression values where compared to healthy gene expression values, and @, p≤0.01 depicts significance between adult vs. aged periodontitis tissues, as determined by Student’s t test.
Quantitative analyses of selected genes that showed significant differences with periodontitis in adult and aged gingival tissues by microarray were validated using qRT-PCR (Fig. 2D). GADPH expression determined by qRT-PCR was consistent between healthy and periodontitis groups from both adult and aged gingival samples with the following crossing point (Cp) mean values ± standard deviations: adult healthy: 18.69±0.65, adult periodontitis: 18.35±0.35, aged healthy: 19.05±0.72, and aged periodontitis: 18.48±0.63. There were no statistically significant differences (p≥0.05) between healthy and periodontitis tissues. In general, these results are consistent with the overall trend of gene expression detected by microarray analysis, whereby higher levels in the expression of NLRs such as NLRP5 (5-fold) was seen with periodontitis in adult compared with aged tissues. Diminutions, albeit not reaching significance, in ASC and NOD2 were seen during periodontitis in particular related to aged diseased tissues, and increased IL-1β mRNA levels were observed in both adult and aged tissues with periodontitis. There were not significant changes in NLRP14 expression associated with periodontitis in aged tissues using qPCR; however, adult diseased tissues showed a significant increase in the expression of this NLR.
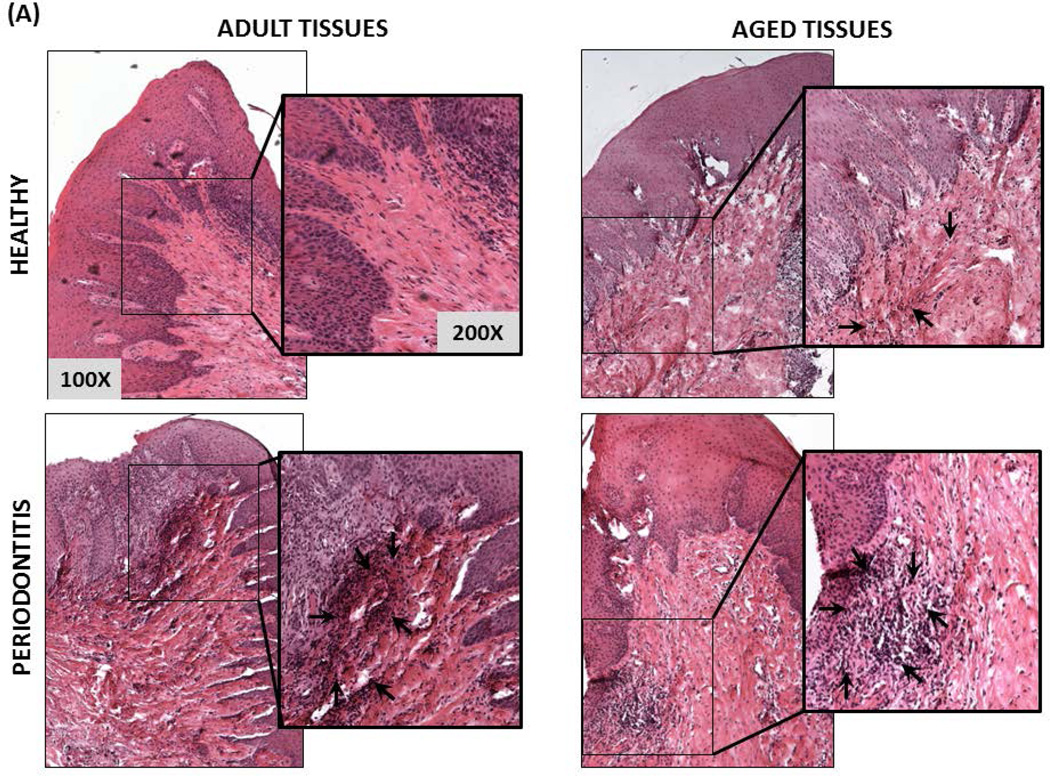
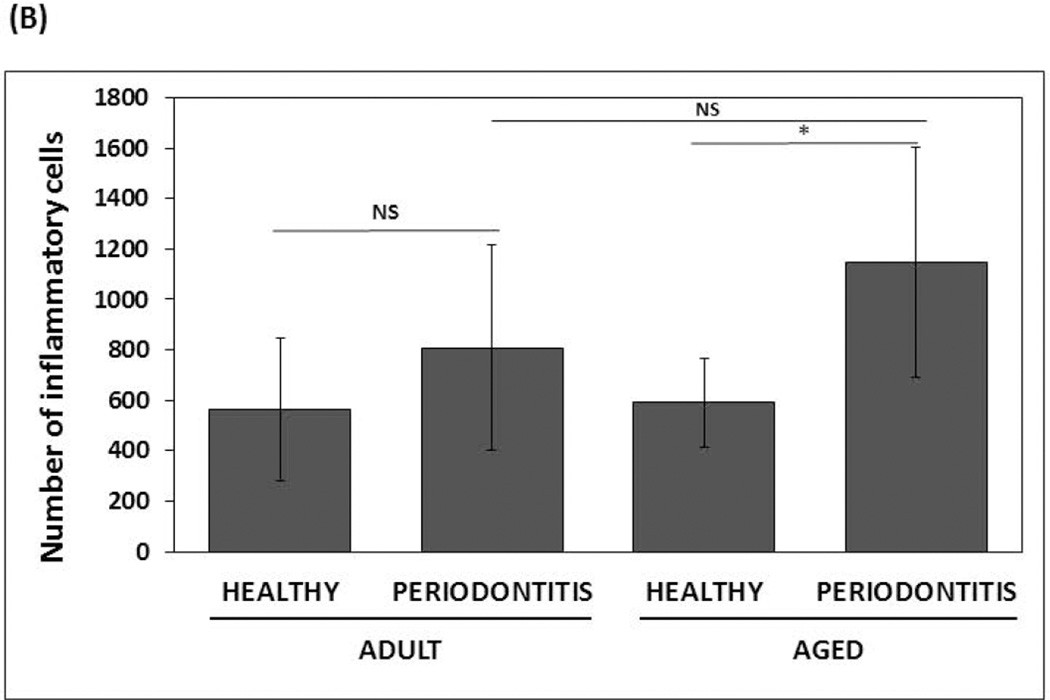
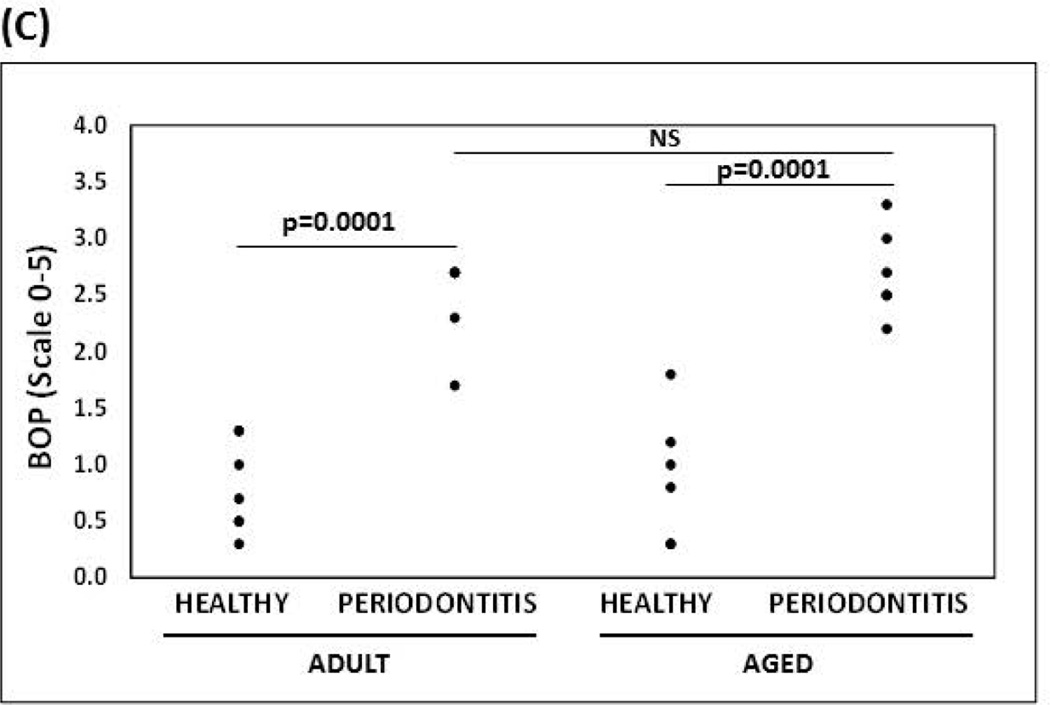
Histological characteristics determined by H&E staining of health and periodontitis gingival tissues from both adult and aged tissues, showed similar cellularity, whereby an increased inflammatory infiltrate was observed in both adult and aged periodontitis tissues compared with the healthy tissues (Figures 3A & 3B). In contrast to adult healthy tissues, inflammatory cells infiltrating clinically healthy aged tissues were more frequently observed, although it was not statistically significant. The histologic results were consistent with the bleeding on probing (BOP) levels as a clinical measure of inflammation with both adult and aged animals exhibiting similar increases in BOP scores with periodontitis (Figure 3C).
Figure 3. Histological and clinical measures of periodontal inflammation.
(A) Micrographs of gingival biopsies from adult and aged animals with and without periodontitis stained with Hematoxylin and Eosin (H&E) illustrating cellularity at 100× and 200× magnification. Inflammatory cells are showing by black arrows (B) Scores of inflammatory infiltrate from healthy and periodontitis gingival biopsies (n=4/group) from adult and aged animals. *p≤0.05 healthy compared to periodontitis gingival tissues as determined by Student’s t test. (C) Bleeding on probing scores (scale 0–5) from healthy and periodontitis sites from adult and aged animals (n=5–6) from which gingival biopsies were taken.
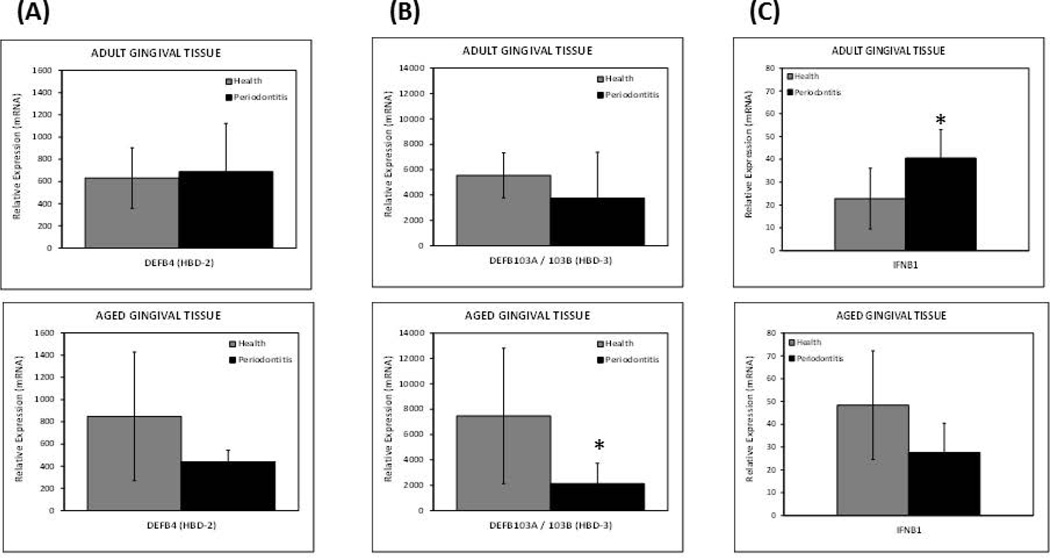
The expression levels of hBD2 were lower in aged but not periodontitis tissues compared to the healthy counterpart, albeit, this difference was not significant (Figure 4A). Although, both adult and aged periodontitis tissues showed lower levels of hBD3 expression with respect to healthy tissues, only aged diseased tissues reached significance with about 4-fold decrease in expression (Figure 4B). A significant increase in the expression of interferon beta (IFNB1) was observed with periodontitis in adult but not in aged periodontitis tissues, which exhibited an approximate 2-fold lower level (Figure 4C).
Figure 4. Expression levels of antimicrobial genes in healthy and inflamed/periodontitis gingival tissues.
The results shown are the Means ± Standard Deviation of gene expression for (A) hBD2, (B) hBD3, and (C) IFNB1, of healthy and inflamed/periodontitis gingival tissues obtained from 5–6 animals per each age (i.e., adult or aged non-human primates). Each sample was individually analyzed by microarray. *p≤0.05 healthy compared to periodontitis gingival tissues as determined by Student’s t test.
3. Discussion
Although there is solid evidence linking environmental factors, such as diet with aging and an increased risk for chronic inflammatory diseases (Gonzalez et al., 2012; Omodei and Fontana 2011), less is known about how molecular variations in the innate immune response related to recognition and interaction with microbes at mucosal surfaces could increase this risk in aged populations. Microbes are a critical component in amplifying and perpetuating inflammation (i.e., enhancing a continuous influx of neutrophils and macrophages), which can be harmful to the tissues as has been shown in several chronic inflammatory disorders, such as atherosclerosis, cardiovascular disease, diabetic chronic wounds, and periodontal disease (Grice and Segre 2012; Rosenfeld and Campbell 2011). Here, we presented a transcriptomic analysis of the age-related changes for an emerging group of innate genes that are crucial regulators of inflammation and infection at epithelial surfaces that are continually exposed to complex microbial communities in healthy and inflamed (i.e., periodontitis) mucosal tissues (i.e., gingiva).
The first observation from these analyses is that although the expression of most NLRs in healthy gingival tissues was not affected by aging, a significant positive correlation of the transcripts levels for 3 intracellular receptors (i.e., NLRB/NAIP, NLRP12, and AIM2) with age was observed. Based on the emerging functions of these intracellular microbial receptors, one could hypothesize that the increased ability of gingival tissue to restrict replication of invasive microorganisms through NLRB-, NLRP12 or AIM2-induced inflammasome activation could be crucial for maintaining a healthy state in the periodontium. In particular, a unique feature that distinguishes NLRB/NAIP from other NLRs is the presence of BIR domains, which seems to be related to the ability to suppress apoptosis through activation of MAP kinase (JNK) (Sanna et al., 2002). Thus, over-expression of NLRB/NAIP in healthy gingival tissue could be related to a reduction in apoptotic events normally occurring in gingival tissues from aged animals, as we have previously reported (Gonzalez et al., 2013; Gonzalez et al., 2011). Although the role of NLRP12 in periodontitis remains unknown, the anti-inflammatory effects of NLRP12 suggest that increased levels of this NLR in aged oral mucosa could also play an important role in balancing the host-microbes interactions across the life span with a central role in maintaining health at advanced age. Of note, increased expression of AIM2 also has been reported in senescent cells which could be related with a pro-inflammatory phenotype (i.e., IL-1β secretion) associated with aging-associated inflammatory diseases (Duan et al., 2011). Accordingly, aging-related up-regulation of AIM-2 in healthy gingival tissue could increase the risk for periodontitis in the elderly.
On the other hand, NLRC2/NOD2 expression significantly decreased with aging in healthy oral mucosa. Down-regulation of NLRC2/NOD2 with aging is of special interest, since growing evidence indicates that NOD2 negatively regulates TLR2 signaling in certain settings, it is essential for promoting intestinal epithelial barrier integrity, and NOD2 mutations have been related to reduced IL-10 production by cells from patients with Crohn’s disease when stimulated with MDP and TLR agonists (Lala et al., 2003; Noguchi et al., 2009). Accordingly, aging-related periodontitis could involve an impaired regulation of inflammation and infection associated with decreased expression of some NLRs including NLRC2/NOD2 receptor. This hypothesis is supported by the aging-related decrease in the expression of antimicrobial genes such as hBD2 and hBD3, and the lack of difference in IFNB1 response observed in aged tissues during periodontitis.
It remains unknown, why the oral mucosa at an early age reacts to the accumulation of noxious biofilms with inflammation (i.e., gingivitis), albeit this inflammatory response does not develop into periodontitis as is frequently observed in older individuals. It is tempting to hypothesize that an altered expression of NLRs such as NLRB (lower) and NOD2 (higher) in young/adolescent oral mucosa could be contributing to help maintain homeostatic host-microbes interactions through important regulatory mechanisms that control destructive inflammation and support epithelial barrier integrity, which may be weakened in an age-dependent manner.
In general, a higher expression of several NLRs and inflammasome-related genes was observed in inflamed/periodontitis gingival tissues from adult and aged animals compared to healthy tissues. These observations are consistent with recent evidence in humans showing that some inflammasome-related genes are expressed at higher levels in gingival biopsies from periodontitis sites compared with healthy controls (Bostanci et al., 2009), and the ability of subgingival biofilms to increase the expression of some inflammasome-activating receptors in gingival fibroblasts (Bostanci et al., 2011). Nevertheless, specific aging-related differences were observed with periodontitis, where increased expression of NLRB, NLRP5, and NLRX1 was associated with inflamed tissues from adult but not aged animals when compared with healthy controls, and in contrast, a significant reduction in the expression of NLRC2, NLRP2 and NLRP14 was specifically observed in aged periodontitis tissues. Although the pathogenesis of periodontal disease has been historically considered similar at all ages, these differences in gene expression supports the idea that there could be important molecular differences in the pathogenesis of periodontitis related to age (Gonzalez et al., 2011). Diminution in NLRP14 expression observed by microarray analysis in the aged periodontitis tissues compared to healthy tissues was not observed using the qPCR procedure. Although this discrepancy in gene expression results for this gene could involve fundamental methodological differences in these independent experimental approaches (Etienne et al., 2004), further studies will be necessary to confirm variations in this NLR with disease and aging. Preliminary analysis of gingival transcriptomes of an ongoing longitudinal study conducted by our group using a ligature-induced periodontitis model in the same non-human primate model, indicates that NOD2 expression is significantly reduced, and NLRP14 expression is increased during initiation, and progression of periodontitis (unpublished) irrespective of aging, which re-enforces a potential change in the expression of these intracellular receptors with disease.
Altered expression of some NLRPs has been shown in mice and rhesus macaques (M. mulatta) (i.e., NLRPs 2, 5 and 14) to be mainly in gametes and early embryos, suggesting that these receptors have specific functions in oocyte maturation and early embryonic development (Tian et al., 2009). Our results indicate that some of these receptors are also expressed in healthy gingival tissues and their expression is specifically down-regulated in aged periodontitis tissues. In particular, NLRP2 appears to be a negative regulator of inflammation through the inhibition of TLR-driven NFκB activation in cells of myeloid origin (Fontalba et al., 2007), and play a role as a mediator for the expression of antimicrobial peptides induced by oral bacteria (i.e., F. nucleatum) in epithelial cells (Ji et al., 2009). Therefore, down-regulation of NLRP2 with periodontitis in aged gingival tissue could also be increasing the likelihood for persistent infection and inflammation. Consistently, we found that hBD3 expression was significantly decreased during periodontitis in aged but not adult gingival tissues. The potential immunoregulatory role of NLRPs 5 and 14 at mucosal surfaces still needs to be determined.
As expected and broadly described, pro-IL-1β was up-regulated with disease in both adult and aged gingival tissues; however, pro-IL-18 expression was significantly decreased in adult but not aged periodontitis tissues. This difference in pro-IL-18 expression rather than being a unique characteristic of periodontitis in the adult tissues appears to be likely related to an age-related decrease of IL-18 expression, which although not statistically significant, is reflected in a negative correlation between the expression of this cytokine and age, as well as lower levels of IL-18 in aged healthy tissues compared to the adult tissues. Since both IL-1β and IL-18 play prominent roles in polarizing T-cell helper responses, these age-related transcriptional changes could also be reflecting differences in the T helper responses during periodontal disease.
Finally, ASC expression was reduced with periodontitis in aged gingival tissues. As mentioned above, ASC is a crucial adaptor protein for successful inflammasome activation by NLRPs and AIM2, and most recently it has been shown that ASC-deficient mice exhibited defective antigen presentation by dendritic cells and lymphocyte migration due to impaired actin polymerization mediated by the small GTPase Rac (Ippagunta et al., 2011). Decreased expression of this central adaptor protein for inflammasome assembly and activation observed in periodontitis gingival tissues when compared with healthy controls is consistent with previous evidence demonstrating the ability of the periodontopathogenic P. gingivalis to reduce ASC expression in vitro (Bostanci et al., 2009). Thus, an impaired inflammasome response related to decreased ASC expression driven by oral pathogenic strains could be an interesting pathway to further explore in the pathogenesis of periodontal disease, where selected oral bacterial species could take advantage of this local altered host response leading to overgrowth and contributing to disease. An additional observation was that although both adult and aged periodontitis tissues exhibited a significant increase in mRNA IL-1β levels, aged tissues showed about 3 times higher message levels when compared with the adult periodontitis tissues. It could be hypothesized that a greater accumulation of IL-1β mRNA levels in aged tissues during periodontitis may occur, which would be consistent with a diminution of gingival cell/tissue ability to properly activate the inflammasome for the consequent maturation, translation and release of IL-1β.
It remains to be determined what cell types are involved in the age-related gene expression changes of NLRs and inflammasome-related genes that reflect the tissue milieu in health and periodontitis as reported in this study. Variations in the amount of inflammatory infiltrate with aging and periodontitis could not fully explain these gene expression differences, because despite of the higher presence of inflammatory cells normally observed with periodontitis, we found a significant reduction in the expression of a subset of NLRs and inflammasome-related genes in aging-periodontitis tissues. Moreover, it has been reported that some NLRs are expressed primarily by oral epithelial cells compared with the expression levels of fibroblasts and inflammatory cellular infiltrates in the tissues during health and periodontitis (Sugawara et al., 2006). Thus, variations in gene expression, particularly at the oral epithelium, irrespective of variations in the immunoinflammatory infiltrate may be likely involved.
Age–related changes in the transcriptional profiles of several “sterile” tissues from mice and rhesus monkeys (e.g., skeletal muscle, brain and heart) have shown up-regulation of transcripts involved in inflammation and oxidative stress related to aging (Lee et al., 2002). We have demonstrated changes in gene expression of a group of critical innate regulators of inflammation and infection related with aging in a mucosal tissue constantly exposed to complex microbial communities during health and inflammation (Fig. 5). Whether variation in the expression of these innate genes is a cause or consequence of the disease, and the mechanisms by which these variations could be related to a higher prevalence and/or severity of periodontitis with aging need to be elucidated. The oral microbiome of rhesus monkeys was recently characterized, exhibiting high similarity with the human oral microbiome (Ocon et al., 2013). Therefore, detrimental changes of the immune response associated with aging (i.e., immunosenescence), including those described in this study, could be contributing to impair the symbiotic relationship between oral bacteria and the host leading to dysbiosis, persistent infection and chronic inflammation.
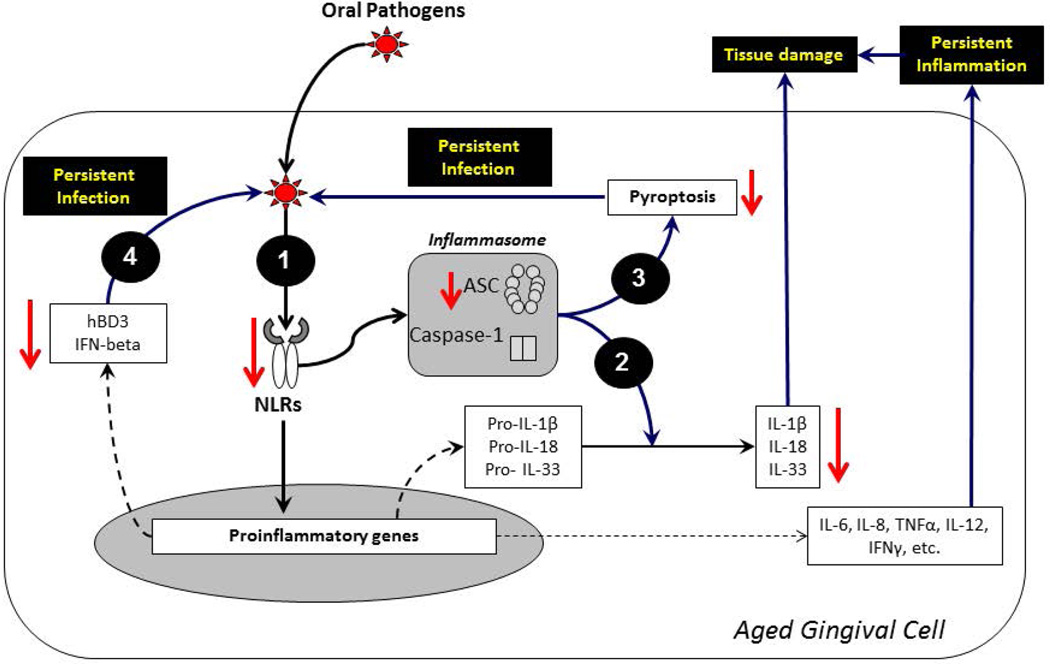
Figure 5. Schematic showing the potential impact of reduced expression of NOD-like receptors in persistent infection and inflammation of the oral mucosa during aging-related periodontitis.
(1) An impaired expression of a subset of NLRs (e.g., NLRPs 2, 5 and NOD2) and ASC during periodontitis in aged gingival tissues could lead to a weakened inflammasome activation which may drive: (2) mucosal tissue damage due to a decrease in the maturation and production of cytokines such as IL1β, IL-18, and IL-33 that are critical to maintain the mucosal integrity, and (3) persistent infection of invasive oral pathogens which could not be removed through pyroptosis (a central type of cell death to control intracellular pathogens). (4) Similarly, the aging-related decrease in NLRs expression during periodontitis could lead to persistent infection through reduced production of central innate antimicrobial factors (e.g., hBDs and Type I IFNs) to protect oral mucosal tissues against invasive pathogens (e.g., P. gingivalis). These molecular changes of the immunoinflammatory response with aging could increase the risk for oral infection/inflammation in aged individuals.
Although up-regulation of some of the genes studied here (e.g., IL-1β) is consistent with previous evidence showing increases at the protein level in the same animal model (Smith et al., 1993), future immunohistochemistry studies to confirm these gingival transcriptional variations associated with aging are clearly necessary, given the fact that protein expression of some of these molecules has been shown to be post-transcriptionally regulated. The approach in this study focused our efforts on determining the inflammasome characteristics within the context of the multiple cellular interactions that would be occurring in the oral mucosa (i.e., gingival tissues). It should be recognized that the literature is replete with studies of oral tissue responses employing reductionist approaches trying to understand disease. As such, these studies isolate single cell types removed from the tissue context and generally challenged with individual bacteria or their components, in attempting to describe the biology of the in situ situation in health and disease. However, as recently described by Dupré (Dupre), biological outcomes are not only dependent upon the activities of their individual constituents, but are critically affected by the complex systems of which they are a part. The evolving field of systems biology emphasizes the importance of moving science towards addressing the micro-environmental milieu that occurs in situ and attempt to reflect the “whole” of the system that defines health or disease.
ACKNOWLEDGMENTS
This work was supported by National Institute of Health (NIH) grants P20GM103538 and UL1TR000117. We express our gratitude to the Caribbean Primate Research Center (CPRC) supported by grant P40RR03640 and the Microarray Core of University Kentucky for their invaluable technical assistance. Dr. Stromberg was also funded by NIH grant 5P20GM103436-13.
REFERENCES
- Albandar JM. Underestimation of periodontitis in NHANES surveys. J Periodontol. 2011;82:337–341. doi: 10.1902/jop.2011.100638. [DOI] [PubMed] [Google Scholar]
- Allen IC, Moore CB, Schneider M, Lei Y, Davis BK, Scull MA, Gris D, Roney KE, Zimmermann AG, Bowzard JB, Ranjan P, Monroe KM, Pickles RJ, Sambhara S, Ting JP. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-kappaB signaling pathways. Immunity. 2011;34:854–865. doi: 10.1016/j.immuni.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JC, Lich JD, Ye Z, Allen IC, Gris D, Wilson JE, Schneider M, Roney KE, O'Connor BP, Moore CB, Morrison A, Sutterwala FS, Bertin J, Koller BH, Liu Z, Ting JP. Cutting edge: NLRP12 controls dendritic and myeloid cell migration to affect contact hypersensitivity. J Immunol. 2010;185:4515–4519. doi: 10.4049/jimmunol.1002227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belibasakis GN, Johansson A. Aggregatibacter actinomycetemcomitans targets NLRP3 and NLRP6 inflammasome expression in human mononuclear leukocytes. Cytokine. 2012;59:124–130. doi: 10.1016/j.cyto.2012.03.016. [DOI] [PubMed] [Google Scholar]
- Blander JM, Sander LE. Beyond pattern recognition: five immune checkpoints for scaling the microbial threat. Nat Rev Immunol. 2012;12:215–225. doi: 10.1038/nri3167. [DOI] [PubMed] [Google Scholar]
- Bostanci N, Emingil G, Saygan B, Turkoglu O, Atilla G, Curtis MA, Belibasakis GN. Expression and regulation of the NALP3 inflammasome complex in periodontal diseases. Clin Exp Immunol. 2009;157:415–422. doi: 10.1111/j.1365-2249.2009.03972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostanci N, Meier A, Guggenheim B, Belibasakis GN. Regulation of NLRP3 and AIM2 inflammasome gene expression levels in gingival fibroblasts by oral biofilms. Cell Immunol. 2011;270:88–93. doi: 10.1016/j.cellimm.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S, Valvano MA, Foster SJ, Mak TW, Nunez G, Inohara N. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Ponomareva L, Veeranki S, Panchanathan R, Dickerson E, Choubey D. Differential roles for the interferon-inducible IFI16 and AIM2 innate immune sensors for cytosolic DNA in cellular senescence of human fibroblasts. Mol Cancer Res. 2011;9:589–602. doi: 10.1158/1541-7786.MCR-10-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre J. Process of Life: Essays in the Philosophy of Biology. Oxford, UK: Oxford Univ. Press; 2012. [Google Scholar]
- Dye BA, Tan S, Smith V, Lewis BG, Barker LK, Thornton-Evans G, Eke PI, Beltran-Aguilar ED, Horowitz AM, Li CH. Trends in oral health status: United States, 1988–1994 and 1999–2004. Vital Health Stat. 2007;11:1–92. [PubMed] [Google Scholar]
- Ebersole JL, Steffen MJ, Gonzalez-Martinez J, Novak MJ. Effects of age and oral disease on systemic inflammatory and immune parameters in nonhuman primates. Clin Vaccine Immunol. 2008;15:1067–1075. doi: 10.1128/CVI.00258-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskan MA, Jotwani R, Abe T, Chmelar J, Lim JH, Liang S, Ciero PA, Krauss JL, Li F, Rauner M, Hofbauer LC, Choi EY, Chung KJ, Hashim A, Curtis MA, Chavakis T, Hajishengallis G. The leukocyte integrin antagonist Del-1 inhibits IL-17-mediated inflammatory bone loss. Nat Immunol. 2012;13:465–473. doi: 10.1038/ni.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne W, Meyer MH, Peppers J, Meyer RA., Jr Comparison of mRNA gene expression by RT-PCR and DNA microarray. Biotechniques. 2004;36:618–620. 622, 624–626. doi: 10.2144/04364ST02. [DOI] [PubMed] [Google Scholar]
- Fontalba A, Gutierrez O, Fernandez-Luna JL. NLRP2, an inhibitor of the NF-kappaB pathway, is transcriptionally activated by NF-kappaB and exhibits a nonfunctional allelic variant. J Immunol. 2007;179:8519–8524. doi: 10.4049/jimmunol.179.12.8519. [DOI] [PubMed] [Google Scholar]
- Franchi L, Park JH, Shaw MH, Marina-Garcia N, Chen G, Kim YG, Nunez G. Intracellular NOD-like receptors in innate immunity, infection and disease. Cell Microbiol. 2008;10:1–8. doi: 10.1111/j.1462-5822.2007.01059.x. [DOI] [PubMed] [Google Scholar]
- Friedewald VE, Kornman KS, Beck JD, Genco R, Goldfine A, Libby P, Offenbacher S, Ridker PM, Van Dyke TE, Roberts WC. The American Journal of Cardiology and Journal of Periodontology editors' consensus: periodontitis and atherosclerotic cardiovascular disease. J Periodontol. 2009;80:1021–1032. doi: 10.1902/jop.2009.097001. [DOI] [PubMed] [Google Scholar]
- Geddes K, Magalhaes JG, Girardin SE. Unleashing the therapeutic potential of NOD-like receptors. Nat Rev Drug Discov. 2009;8:465–479. doi: 10.1038/nrd2783. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- Gonzalez O, Tobia C, Ebersole J, Novak MJ. Caloric restriction and chronic inflammatory diseases. Oral Dis. 2012;18:16–31. doi: 10.1111/j.1601-0825.2011.01830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez OA, Novak MJ, Kirakodu S, Stromberg AJ, Shen S, Orraca L, Gonzalez-Martinez J, Ebersole JL. Effects of aging on apoptosis gene expression in oral mucosal tissues. Apoptosis. 2013;18:249–259. doi: 10.1007/s10495-013-0806-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez OA, Stromberg AJ, Huggins PM, Gonzalez-Martinez J, Novak MJ, Ebersole JL. Apoptotic genes are differentially expressed in aged gingival tissue. J Dent Res. 2011;90:880–886. doi: 10.1177/0022034511403744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves DT, Cochran D. The contribution of interleukin-1 and tumor necrosis factor to periodontal tissue destruction. J Periodontol. 2003;74:391–401. doi: 10.1902/jop.2003.74.3.391. [DOI] [PubMed] [Google Scholar]
- Grice EA, Segre JA. Interaction of the microbiome with the innate immune response in chronic wounds. Adv Exp Med Biol. 2012;946:55–68. doi: 10.1007/978-1-4614-0106-3_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. Too old to fight? Aging and its toll on innate immunity. Mol Oral Microbiol. 2010;25:25–37. doi: 10.1111/j.2041-1014.2009.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippagunta SK, Malireddi RK, Shaw PJ, Neale GA, Vande Walle L, Green DR, Fukui Y, Lamkanfi M, Kanneganti TD. The inflammasome adaptor ASC regulates the function of adaptive immune cells by controlling Dock2-mediated Rac activation and actin polymerization. Nat Immunol. 2011;12:1010–1016. doi: 10.1038/ni.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S, Shin JE, Kim YS, Oh JE, Min BM, Choi Y. Toll-like receptor 2 and NALP2 mediate induction of human beta-defensins by fusobacterium nucleatum in gingival epithelial cells. Infect Immun. 2009;77:1044–1052. doi: 10.1128/IAI.00449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lala S, Ogura Y, Osborne C, Hor SY, Bromfield A, Davies S, Ogunbiyi O, Nunez G, Keshav S. Crohn's disease and the NOD2 gene: a role for paneth cells. Gastroenterology. 2003;125:47–57. doi: 10.1016/s0016-5085(03)00661-9. [DOI] [PubMed] [Google Scholar]
- Lalla E, Papapanou PN. Diabetes mellitus and periodontitis: a tale of two common interrelated diseases. Nat Rev Endocrinol. 2011;7:738–748. doi: 10.1038/nrendo.2011.106. [DOI] [PubMed] [Google Scholar]
- Lee CK, Allison DB, Brand J, Weindruch R, Prolla TA. Transcriptional profiles associated with aging and middle age-onset caloric restriction in mouse hearts. Proc Natl Acad Sci U S A. 2002;99:14988–14993. doi: 10.1073/pnas.232308999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Monack DM. Inflammasome adaptors and sensors: intracellular regulators of infection and inflammation. Nat Rev Immunol. 2007;7:31–40. doi: 10.1038/nri1997. [DOI] [PubMed] [Google Scholar]
- Martinon F. Signaling by ROS drives inflammasome activation. Eur J Immunol. 2010;40:616–619. doi: 10.1002/eji.200940168. [DOI] [PubMed] [Google Scholar]
- Meka A, Bakthavatchalu V, Sathishkumar S, Lopez MC, Verma RK, Wallet SM, Bhattacharyya I, Boyce BF, Handfield M, Lamont RJ, Baker HV, Ebersole JL, Kesavalu L. Porphyromonas gingivalis infection-induced tissue and bone transcriptional profiles. Mol Oral Microbiol. 2010;25:61–74. doi: 10.1111/j.2041-1014.2009.00555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Homma Y, Kang X, Netea MG, Ma X. A Crohn's disease-associated NOD2 mutation suppresses transcription of human IL10 by inhibiting activity of the nuclear ribonucleoprotein hnRNP-A1. Nat Immunol. 2009;10:471–479. doi: 10.1038/ni.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocon S, Murphy C, Dang AT, Sankaran-Walters S, Li CS, Tarara R, Borujerdpur N, Dandekar S, Paster BJ, George MD. Transcription profiling reveals potential mechanisms of dysbiosis in the oral microbiome of rhesus macaques with chronic untreated SIV infection. PLoS One. 2013;8:e80863. doi: 10.1371/journal.pone.0080863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omodei D, Fontana L. Calorie restriction and prevention of age-associated chronic disease. FEBS Lett. 2011;585:1537–1542. doi: 10.1016/j.febslet.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco A, Gemmell E, Bickel M, Seymour GJ. Interleukin 18 and periodontal disease. J Dent Res. 2007;86:586–593. doi: 10.1177/154405910708600702. [DOI] [PubMed] [Google Scholar]
- Poeck H, Bscheider M, Gross O, Finger K, Roth S, Rebsamen M, Hannesschlager N, Schlee M, Rothenfusser S, Barchet W, Kato H, Akira S, Inoue S, Endres S, Peschel C, Hartmann G, Hornung V, Ruland J. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat Immunol. 2010;11:63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- Qian F, Wang X, Zhang L, Chen S, Piecychna M, Allore H, Bockenstedt L, Malawista S, Bucala R, Shaw AC, Fikrig E, Montgomery RR. Age-associated elevation in TLR5 leads to increased inflammatory responses in the elderly. Aging Cell. 2012;11:104–110. doi: 10.1111/j.1474-9726.2011.00759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld ME, Campbell LA. Pathogens and atherosclerosis: update on the potential contribution of multiple infectious organisms to the pathogenesis of atherosclerosis. Thromb Haemost. 2011;106:858–867. doi: 10.1160/TH11-06-0392. [DOI] [PubMed] [Google Scholar]
- Sanna MG, da Silva Correia J, Ducrey O, Lee J, Nomoto K, Schrantz N, Deveraux QL, Ulevitch RJ. IAP suppression of apoptosis involves distinct mechanisms: the TAK1/JNK1 signaling cascade and caspase inhibition. Mol Cell Biol. 2002;22:1754–1766. doi: 10.1128/MCB.22.6.1754-1766.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schou S, Holmstrup P, Kornman KS. Non-human primates used in studies of periodontal disease pathogenesis: a review of the literature. J Periodontol. 1993;64:497–508. doi: 10.1902/jop.1993.64.6.497. [DOI] [PubMed] [Google Scholar]
- Shaw AC, Panda A, Joshi SR, Qian F, Allore HG, Montgomery RR. Dysregulation of human Toll-like receptor function in aging. Ageing Res Rev. 2011;10:346–353. doi: 10.1016/j.arr.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Braswell LD, Collins JG, Boyd DL, Jeffcoat MK, Reddy M, Li KL, Wilensky S, Vogel R, Alfano M, et al. Changes in inflammatory mediators in experimental periodontitis in the rhesus monkey. Infect Immun. 1993;61:1453–1459. doi: 10.1128/iai.61.4.1453-1459.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara Y, Uehara A, Fujimoto Y, Kusumoto S, Fukase K, Shibata K, Sugawara S, Sasano T, Takada H. Toll-like receptors, NOD1, and NOD2 in oral epithelial cells. J Dent Res. 2006;85:524–529. doi: 10.1177/154405910608500609. [DOI] [PubMed] [Google Scholar]
- Tian X, Pascal G, Monget P. Evolution and functional divergence of NLRP genes in mammalian reproductive systems. BMC Evol Biol. 2009;9:202. doi: 10.1186/1471-2148-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke TE, Serhan CN. Resolution of inflammation: a new paradigm for the pathogenesis of periodontal diseases. J Dent Res. 2003;82:82–90. doi: 10.1177/154405910308200202. [DOI] [PubMed] [Google Scholar]
- Wen H, Ting JP, O'Neill LA. A role for the NLRP3 inflammasome in metabolic diseases--did Warburg miss inflammation? Nat Immunol. 2012;13:352–357. doi: 10.1038/ni.2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KL, Lich JD, Duncan JA, Reed W, Rallabhandi P, Moore C, Kurtz S, Coffield VM, Accavitti-Loper MA, Su L, Vogel SN, Braunstein M, Ting JP. The CATERPILLER protein monarch-1 is an antagonist of toll-like receptor-, tumor necrosis factor alpha-, and Mycobacterium tuberculosis-induced pro-inflammatory signals. J Biol Chem. 2005;280:39914–39924. doi: 10.1074/jbc.M502820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz O, Sater AA, Yao L, Koutouzis T, Pettengill M, Ojcius DM. ATP-dependent activation of an inflammasome in primary gingival epithelial cells infected by Porphyromonas gingivalis. Cell Microbiol. 2010;12:188–198. doi: 10.1111/j.1462-5822.2009.01390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, Liu L, Shao F. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]