Abstract
Inhibitors targeting human glutamate carboxypeptidase II (GCPII) typically consist of a P1’ glutamate-derived binding module, which warrants the high-affinity and specificity, linked to an effector function that is positioned within the entrance funnel of the enzyme. Here we present a comprehensive structural and computational study aimed at dissecting the importance of the effector function for GCPII binding and affinity. To this end we determined crystal structures of human GCPII in complex with a series of phosphoramidate-based inhibitors harboring effector functions of diverse physicochemical characteristics. Our data shows that higher binding affinities of phosphoramidates, when compared to matching phosphonates, are linked to the presence of additional hydrogen bonds between Glu424 and Gly518 of the enzyme and the amide group of the phosphoramidate. While the positioning of the P1’ glutamate-derived module within the S1’ pocket of GCPII is invariant, interaction interfaces between effector functions and residues lining the entrance funnel are highly varied, with the positively charged arginine patch defined by Arg463, Arg534, Arg536, being the only ‘hot-spot’ common to several studied complexes. This variability stems in part from the fact that the effector/GCPII interfaces generally encompass isolated areas of non-polar residues within the entrance funnel and resulting van der Waals contacts lack the directionality typical for hydrogen-bonding interactions. Presented data unravel a complexity of binding modes of inhibitors within non-prime site(s) of GCPII and can be exploited for the design of novel GCPII-specific compounds.
Keywords: NAALADase, prostate-specific membrane antigen, X-ray crystallography, phosphoramidate, molecular modeling
INTRODUCTION
Human glutamate carboxypeptidase II (GCPII; EC 3.4.17.21) is implicated in diverse pathologies and the design and development of novel GCPII-specific ligands attracts research from both academia and industry. Small-molecule ligands targeting GCPII can be used in diagnostic and therapeutic applications in prostate cancer (PCa) and various neurological disorders [1, 2]. As GCPII expression levels in the prostate increase from low levels in the healthy organ to high expression in advanced metastatic disease, it is not surprising that prostate cancer is the main target for imaging/therapy by GCPII-specific ligands [3, 4]. In the nervous system, GCPII modulates neuron-neuron signal transduction and facilitates communication between neurons and support cells (astrocytes, Schwann cells) by hydrolyzing N-acetyl-aspartyl-glutamate (NAAG), the most abundant peptidic transmitter in human brain [5]. Given its intimate involvement in neuronal processes, a pharmacologic modulation of GCPII activity can be beneficial in various neurological disorders including stroke, amyotrophic lateral sclerosis, inflammatory and neuropathic pain, and traumatic brain injury [6-8].
A diversity of settings, in which GCPII inhibitors can be used, mandates designing compounds with varied physicochemical characteristics and ADME (absorption, distribution, metabolism, and excretion) profiles. For example, blood-brain barrier permeable, lipophilic compounds are sought for the neuronal compartment targeting, while more polar inhibitors with rapid renal clearance can be suitable for PCa imaging. In general, GCPII inhibitors can be either simple glutamate-based compounds linked to a zinc-binding group (such as phospho(i)nate, thiol, or hydroxamate), but due to their inherent advantages (e.g. the attachment of effector groups) more "complex" NAAG-derived compounds are gaining prominence in the field [9, 10]. NAAG-based inhibitors typically consist of a binding module that encompasses a zinc-binding group (such as urea, phosphinate, phosphoramidate) connected to a P1’ glutamate moiety. This module warrants high-affinity and specificity for GCPII, and is typically further linked to an effector functionality that spans non-prime positions of a given compound (i.e. putative P1, P2, P3…. parts). In principle, both the binding module and effector parts can be modified to obtain an inhibitor with desired properties, but modifications to the P1’ glutamate inadvertently lead to significant decrease in the inhibitor affinity [11, 12]. This decrease is a consequence of the pronounced selectivity of the S1' (pharmacophore) pocket towards glutamate that is secured by an intricate network of polar interactions between the enzyme and an inhibitor [13, 14].
On the other hand, the effector part of a GCPII-specific ligand is more amenable to even quite complex modifications [15-18] that take advantage of sizeable dimensions and flexibility of the structurally poorly defined non-prime site(s) that are continuous with the entrance funnel. The irregularly shaped funnel is a part of the internal cavity that is approximately 20 Å long and delineated by the active-site zinc ions at the bottom and either capped by the entrance lid (amino acids Trp541-Gly548) in its closed conformation or continuous with the outside space (the entrance lid in the open conformation) [19].
At least three prominent structural features, which can play an important role in interactions with GCPII ligands, have been identified in the entrance funnel. These include the arginine patch, the “S1 hydrophobic accessory pocket“, and the arene-binding site (Fig. 1; reviewed by Pavlicek et al [19]). The arginine patch is an extended, positively charged area at the wall of the entrance funnel defined by the apposition of guanidinium groups of Arg534, Arg536, and Arg463. The electrostatic property of the patch provides a mechanistic explanation for the preference of GCPII for acidic residues at the P1 position of GCPII substrates as well as inhibitors [20]. Consequently, the presence of the P1 carboxylate group is a hallmark of nearly all inhibitors used in the field that take advantage of the above-mentioned fact. Structural studies revealed positional variability for the side chains of Arg536 and Arg463. Upon inhibitor binding, the concerted repositioning of the two arginine side chains can lead to the opening of a “S1 hydrophobic accessory pocket“ that has been shown to accommodate a iodo-benzyl group of several urea-based inhibitors, thus contributing to their high affinity for GCPII [21]. The arene-binding site is a simple structural motif shaped by the side chains of Arg463, Arg511, and Trp541, and is a part of the GCPII entrance lid. We have shown that the engagement of the arene-binding site by a distal inhibitor moiety can result in a substantial increase in the inhibitor affinity for GCPII due to avidity effects [22]. Additionally, studies mapping the folate hydrolyzing activity of GCPII revealed the involvement of the arene-binding site in the binding of the pteridine moiety of dietary folates [23]. The arene-binding site together with the hydrophobic accessory pocket, mentioned earlier, determine the structural plasticity in the S1 site/entrance funnel of GCPII.
Figure 1.
Panel A: Overall architecture of GCPII (cross section of human GCPII, PDB code: 4P45). The protein is shown in gray surface representation in complex with a JRB-4-73. The inhibitor is shown in stick representation with atoms colored green (carbon), red (oxygen), blue (nitrogen), orange (phosphorus), and pale cyan (fluorine). Zinc ions are shown as orange spheres. Approximate positions of the arginine patch, S1’ site, and entrance lid are colored red, cyan, and yellow, respectively Panel B: The superposition of phosphoramidate inhibitors in the internal cavity of GCPII. Complexes of GCPII/phosphoramidate were superimposed on corresponding Cα atoms of the enzyme. Inhibitors are in stick representation, with atoms colored red (oxygen), blue (nitrogen), pale cyan (fluorine), and orange (phosphorus). Carbon atoms are colored magenta (T33), blue (T33D), yellow (MP1C), light pink (MP1D), gray (NC-2-40), cyan (CTT54), green (JRB-4-73), and deep teal (JRB-4-81). The zinc ions are shown as orange spheres. While the conformation of the P1‘ glutamate moiety in the S1’pocket is identical for all inhibitors, there are profound differences in positioning of effector functions in the entrance funnel of GCPII.
In this report, we present a systematic study detailing interactions between effector functionalities of GCPII-specific inhibitors and residues shaping the entrance funnel of the enzyme. Additionally, we compared the applicability of two in silico docking algorithms that can be exploited for the structure-assisted design of novel GCPII-specific compounds.
RESULTS
Inhibitors
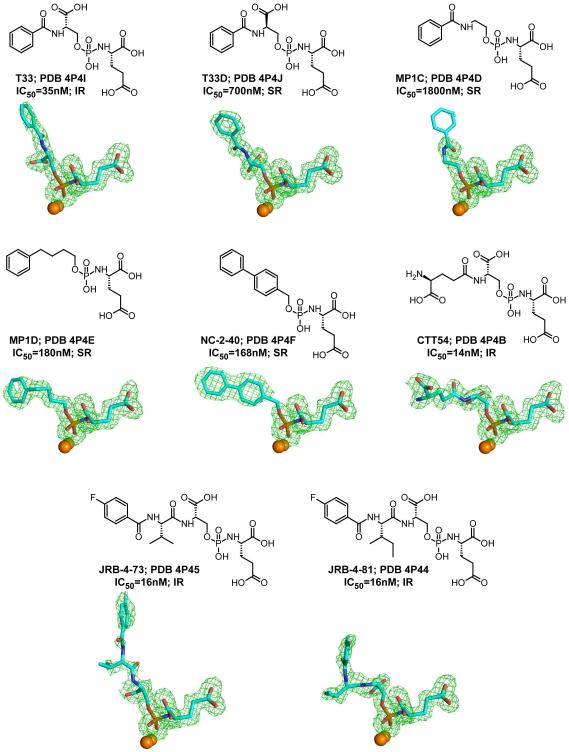
In this study, we structurally and computationally characterized complexes of eight small-molecule inhibitors of GCPII that are non-hydrolyzable transition state analogs of N-acetyl-aspartyl-glutamate, a natural GCPII substrate. All inhibitors feature the conserved C-terminal (P1’) glutamate moiety. The N-acetyl-aspartate part of the natural substrate is substituted by an effector functional group and individual effector functions differ in their structural and physicochemical characteristics. Both non-prime P1 and P1’ parts are linked via the zinc-binding phosphoramidate functionality, which replaces the scissile peptide bond present in the natural substrate. While the P1’ glutamate together with the phosphoramidate function serve as primary high affinity/specificity binding modules, the interactions between an effector function and residues of the entrance funnel of GCPII fine-tune this affinity and help in defining the mode of inhibition (reversible or pseudo-irreversible; Fig 2).
Figure 2.
Chemical formulas, PDB codes, inhibition constants, and the mode of inhibition for inhibitors used in this study. Fo-Fc maps (green) for individual inhibitors are contoured at 3.0 σ and modeled inhibitors are shown in stick representation with atoms colored cyan (carbon), red (oxygen), blue (nitrogen), orange (phosphorus), and pale cyan (fluorine). The active-site zinc ions are shown as orange spheres. IR – pseudo-irreversible mode of binding; SR – slowly reversible mode of binding.
X-ray structures and interactions of the docking module
Crystal structures of GCPII/phosphoramidate complexes were determined with the high resolution limits in the range of 1.65 – 1.93 Å. All complexes belong to the I222 space group with approximate unit cell dimensions a = 101.8 Å, b = 130.4 Å and c = 159.2 Å. For all complexes, the interpretable positive electron density representing the active site-bound ligand was observed, and individual compounds were fitted into the positive peaks of the Fo-Fc density map in the final stages of the refinement (Figure 2). The quality of the final models is documented by > 99.6% residues in allowed regions of the Ramachandran plot (Table 1).
Table 1.
Data Collection and Refinement Statistics.
| Data collection statistics | ||||
| Inhibitor | T33 | T33D | MP1C | MP1D |
| PDB code | 4P4I | 4P4J | 4P4D | 4P4E |
| Wavelength (Å) | 1.000 | 1.000 | 1.000 | 1.000 |
| Resolution limits (Å) | 30.0 – 1.87 (1.94 – 1.87) |
30.0 – 1.66 (1.72 – 1.66) |
30.0 - 1.65 (1.71 - 1.65) |
30.0 - 1.67 (1.73 - 1.67) |
| No of unique refl. | 88725 (8800) | 122688 (11966) | 125052 (11388) | 118838 (9131) |
| Redundancy | 7.1 (6.5) | 6.6 (4.9) | 7.0 (5.6) | 6.5 (3.9) |
| Completeness (%) | 100 (99.9) | 99.8 (98.6) | 98.9 (90.6) | 96.9 (75.4) |
| I/σ(I) | 16.4 (3.9) | 13.8 (3.0) | 24.2 (2.6) | 20.0 (2.5) |
| Rmerge | 0.090 (0.49) | 0.097 (0.48) | 0.065 (0.49) | 0.061 (0.47) |
| Refinement statistics | ||||
| Resolution limits (Å) | 29.69 - 1.86 (1.91 - 1.86) |
29.69 - 1.86 (1.91 - 1.86) |
28.63-1.65 (1.70-1.65) |
28.29-1.67 (1.71-1.67) |
| Total number of reflections |
87244 (5822) |
121038 (7126) |
123560 (8049) |
117420 (6361) |
| Number of reflections in working set |
85939 (5822) |
119811 (7126) |
122305 (8049) |
116226 (6361) |
| Number of reflections in test set |
1305 (92) |
1227 (70) |
1255 (74) |
1194 (66) |
| R/Rfree (%) | 15.2/17.0 (23.3/21.9) |
15.6/17.9 (24.1/28.2) |
15.7/17.4 (29.0/34.2) |
15.4/18.3 (27.8/36.6) |
| Total number of non-H atoms |
6548 | 6624 | 6634 | 6739 |
| Number of non-H protein atoms |
5976 | 5985 | 5991 | 6153 |
| Number of inhibitor molecules |
1 | 1 | 1 | 1 |
| Number of water molecules |
544 | 611 | 618 | 787 |
| Average B-factor (Å2) | 30.5 | 27.7 | 28.8 | 31.8 |
| Protein | 29.8 26.8 | 27.9 | 31.0 | |
| Water molecules | 38.3 | 36.4 | 37.6 | 44.4 |
| Inhibitor | 30.0 | 21.6 | 29.4 | 29.7 |
| &Ramachandran Plot (%) | ||||
| Allowed | 99.7 | 99.9 | 99.7 | 99.6 |
| Disallowed | 0.3 (Val382) |
0.1 (Val382) |
0.3 (Val382) |
0.4 (Val382, Trp541, Asp652) |
| R.m.s. deviations: bon lengths (Å) |
0.019 | 0.018 | 0.017 | 0.019 |
| bond angles (°) | 1.6 | 1.6 | 1.6 | 1.7 |
| Data collection statistics | ||||
| Inhibitor | NC-2-40 | CTT54 | JRB-4-73 | JRB-4-81 |
| PDB code | 4P4F | 4P4B | 4P45 | 4P44 |
| Wavelength (Å) | 1.000 | 1.000 | 0.918 | 0.918 |
| Resolution limits (Å) |
20.0 – 1.85 (1.92 – 1.85) |
20.0 - 1.93 (2.00 - 1.93) |
50.0 - 1.87 (1.98 - 1.87) |
50.0 - 1.75 (1.86 - 1.75) |
| Unique reflections |
88093 (7739) | 79458 (7752) | 86865 (13851) | 105804 (16873) |
| Redundancy | 7.5 (5.2) | 6.2 (5.0) | 5.9 (5.8) | 5.8 (5.8) |
| Completeness (%) |
98.5 (87.7) | 99.8 (98.5) | 99.7 (99.4) | 99.8 (99.4) |
| l/σl | 23.9 (2.1) | 16.1 (2.6) | 18.1 (3.5) | 21.6 (3.4) |
| Rmerge | 0.077 (0.49) | 0.102 (0.50) | 0.088 (0.63) | 0.060 (0.54) |
| Refinement Statistics | ||||
| Resolution limits (Å) |
20.01 - 1.86 (1.91 - 1.86) |
19.82-1.92 (1.97-1.92) |
28.18 - 1.87 (1.92 - 1.87) |
29.51-1.75 (1.80-1.75) |
| Total number of reflections | 85074 (5219) |
76914 (5186) |
82494 (6003) |
100472 (7333) |
| Number of reflections in working set |
83330 (5219) |
74534 (5186) |
78153 (6003) |
95184 (7333) |
| Number of reflections in test set |
1744 (102) |
2380 (154) |
4341 (315) |
5288 (386) |
| R/Rfree (%) | 15.2/17.6 (27.0/26.9) |
15.6/17.8 (24.7/27.9) |
16.4/19.3 (24.1/30.1) |
16.4/18.7 (23.4/26.6) |
| Total number of non-H atoms |
6648 | 6528 | 6634 | 6554 |
| Number of non-H protein atoms |
6039 | 5987 | 6004 | 5914 |
| Number of inhibitor molecules |
1 | 1 | 1 | 1 |
| Number of water molecules |
582 | 513 | 594 | 603 |
| Average B-factor (Å2) |
30.8 | 29.0 | 27.1 | 26.4 |
| Protein atoms | 30.0 | 28.5 | 26.3 | 25.4 |
| Water molecules | 38.7 | 35.4 | 35.6 | 35.4 |
| Inhibitor | 27.6 | 25.7 | 25.2 | 34.6 |
|
&Ramachandran Plot (%) |
||||
| Allowed | 99.6 | 99.7 | 99.9 | 99.9 |
| Disallowed | 0.4 (Val382, Asn540) |
0.3 (Val382) |
0.1 (Val382) |
0.1 (Val382) |
| R.m.s. deviations: bond lengths (Å) |
0.019 | 0.020 | 0.019 | 0.019 |
| bond angles (°) | 1.6 | 1.7 | 1.6 | 1.7 |
* Values in parentheses are for the highest resolution shells.
Structures were analyzed using the MolProbity package.
Positioning of the P1’ glutamate as well as the phosphoramidate functionality in the S1’ pocket of GCPII is virtually identical for all structures and is consistent with the “canonical” mode of the glutamate-binding as observed in previously reported GCPII complexes (Figure 1) [13, 24]. Here, the glutamate moiety is oriented and bound in the S1’ pocket by the combination of hydrogen bonding interactions with side chains of Arg210, Asn257, Tyr552, Lys699, Tyr700 and of hydrophobic interactions with Phe209 or Leu428. The nitrogen atom of the phosphoramidate function forms hydrogen bonds with the carboxylate of Glu424 (2.7 Å) and the Gly518 carbonyl oxygen (3.0 Å), contributing thus to the increased affinity of phosphoramidates when compared to corresponding phosphinates or phosphonates (Figure 3). Two oxygen atoms of the phosphoramidate function coordinate active-site zinc atoms with distances O1…Zn1 (1.9 Å) and O2…Zn2 (2.1 Å) and are additionally engaged in an intricate network of hydrogen bonds with side chains of His377, Asp387, Glu424, Asp453, Tyr552, and His553.
Figure 3.
Interaction patterns of phosphorus- and urea-based inhibitors within the active-site of human GCPII. Compared to matching phosphorus-based compounds, phosphoramidates are favored by GCPII due to the presence of additional hydrogen bonds between the amide group of the inhibitor and the Glu424 side chain carboxylate and the Gly518 main chain carbonyl (Panel A, distances in Å). Phosphonates (panel B; our unpublished data) and phosphinates (panel C; from pdb code 3BI0) are missing a hydrogen bond donating groups at the position of the phosphoramidate nitrogen. A urea inhibitor (panel D, pdb code 3D7H) is shown for comparison. Individual atoms are colored red (oxygen), blue (nitrogen), orange (phosphorus), and green (carbon). The active-site zinc ions are shown as orange spheres.
Structural details of interactions in the entrance funnel
In contrast to the invariant position of the P1’ glutamate moiety, different non-prime effector functionalities of inhibitors interact with residues lining the entrance funnel with more variability (Figure 1), and these interactions in turn influence the affinities and modes of inhibition of such ligands. These effects can be illustrated by comparing structures of GCPII complexes with T33, T33D and MP1C. While these inhibitors are chemically quite similar, all featuring terminal benzoyl groups attached to the β-serine linker via a peptide bond, they differ by the presence and stereochemistry of the P1 carboxylate group. Inhibitor MP1C lacks the carboxylate altogether and T33 and T33D are diastereomers, (S)P1 and (R)P1, respectively. The absence of the P1 carboxylate in MP1C results in lower affinity (more than 50-fold) when compared to the preferred (S)P1 enantiomer of T33. Additionally, the presence and the “optimal” P1 carboxylate stereochemistry renders the inhibitor T33 “pseudo-irreversible” as compared to the “slowly-reversible” mode of binding observed for the T33D and MP1C [25]. The structural explanation for the importance of the P1 carboxylate is its interaction with the “arginine patch” [20]. In the case of T33, the P1 carboxylate forms ion pairs with Arg534 (2.6 Å) and Arg536 (2.9 and 3.2 Å), and accepts a hydrogen bond from Asn519 (2.9 Å), cumulatively strengthening the binding to GCPII. Similar contacts are observed between GCPII and T33D. These include ion pairs of the P1 carboxylate with Arg534 (2.7 and 3.4 Å) and Arg536 (2.7 and 3.0 Å), and (long) hydrogen bonds to Asn519 (3.2 and 3.4 Å). Additionally, the benzoyl carbonyl group of T33 forms a single H-bond with Arg534 (3.3 Å), while two H-bonds are observed for the T33D benzoyl carbonyl group (Arg536, 2.9 Å; Arg463, 3.4 Å). In addition to these polar interactions, “T-shaped” and “parallel displaced” π-π interactions are observed between the hydroxyphenyl ring of Tyr700 and terminal benzoyl groups of T33D and T33, respectively.
A 5-carbon linker in compound MP1D is identical in the length to linkers in compounds T33, T33D and MP1C, but the presence of the flexible pentenyl chain provides more freedom in the positioning of the terminal phenyl group. Consequently, even with the P1 carboxylate missing in structure of MP1D (compared to T33 and T33D and similar to MP1C), the inhibitor has a relatively high affinity for GCPII (IC50 = 180 nM), which is approximately 10-fold higher compared to the MP1C counterpart (IC50 = 1800 nM) and between P1 carboxylate-containing T33 and T33D, IC50 = 35 nM and IC50 = 700 nM, respectively. Despite the linker flexibility, the P1 part of MP1D is well-defined in the electron density suggesting that only a single conformation of the inhibitor exists within the crystal.
The effector functionality of NC-2-40, formed by two phenyl rings, is the least flexible of all inhibitors studied. The proximal phenyl ring packs against a small hydrophobic patch (a shallow pocket) of the entrance funnel, where it is engaged in π-π interactions with the side chains of Tyr552 (parallel displaced; 5.0 Å between ring centers) and Tyr700 (T-shaped; 5.1 Å between ring centers), which form the wall of the patch. It should be noted that the existence of such an “auxiliary hydrophobic register” was proposed by Maung et al and it can be, in principle, exploited to engage non-polar distal functionalities of diverse GCPII inhibitors [26]. The terminal phenyl ring of NC-2-40 is firmly stacked between the Glu457 side chain (4.0 Å) and Gly548 (3.4 Å). The latter forms a hinge of the GCPII entrance lid (amino acids Trp541 - Gly548) and the presence of the terminal phenyl ring seems to stabilize the lid in the closed conformation. Consequently, the GCPII/NC-2-40 complex is one of two complexes reported here with flexible lid in well-defined closed conformation.
CTT54 is the only inhibitor in this series that lacks a non-polar effector functionality. In fact, the free N-terminal group of CTT54 can be derivatized during the construction of probes used for GCPII-specific imaging [17, 27, 28]. Despite its inherent flexibility, the distal γ-glutamate linker of non-functionalized CTT54 is well-defined in the electron density as its position is restricted by a network of interactions with residues lining the internal GCPII pocket (Figure 2). The terminal carboxylate is H-bonded to a single water molecule and the side chains of Tyr234 (2.5 Å) and Ser547 (3.3 Å), and the terminal amino group forms hydrogen bonds with the Ser547 hydroxyl group (2.7 Å) and the main-chain carbonyl of Tyr549 (3.4 Å). Given the flexibility of the γ-glutamate linker, CTT54 can be functionalized by reporter groups with different physicochemical characteristics typically leading to the increase of inhibitor affinity towards GCPII [17, 28]. For example, the attachment of the fluoro-phenyl group is accompanied by decrease in the inhibition constant from 14 nM to 0.7 nM for CTT54 and FB-CTT54 (PDB code 4JZ0, to be published elsewhere), respectively. This decrease is elicited by the additional interactions between the fluoro-phenyl head group with the Tyr700 side chain of the enzyme. The flexibility of the γ-glutamate linker is a key to the productive positioning of the fluoro-phenyl head group (and likely other functions) in the GCPII internal cavity and is illustrated by a diverse set of interactions with GCPII.
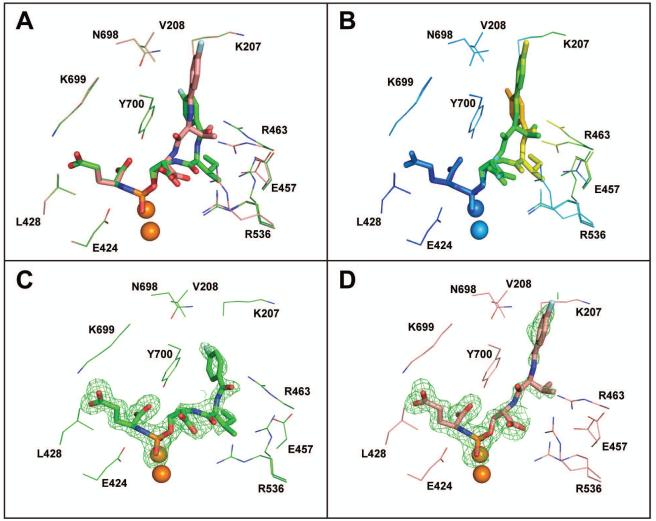
An intricate network of minor contributions by individual functional groups of a given inhibitor, together with inherent conformational constrains, clearly governs the positioning of the distal inhibitor part in the entrance funnel. This conclusion is further illustrated by comparing binding modes of JRB-4-73 and JRB-4-81. The two inhibitors differ only in the Val to Ile substitution at the P2 position and their inhibition constants and pseudo-irreversible binding modes are identical (Fig 2). Yet, the positions of their distal parts are quite different with the relative shift of the fluorine atom of 8.8 Å (Figure 4A). This variability is reflected by the weak Fo-Fc electron density and higher B-factors, suggesting significant positional flexibility of distal fragments of inhibitors (Fig. 4B-D). It is likely that the distal part of an inhibitor in reality adopts several different conformations and only one or two the most populated can be observed and modeled in the crystal structure.
Figure 4.
Comparison of binding poses for compounds JRB-4-73 and JRB-4-81. Panel A; Complexes of GCPII/JRB-4-73 (carbon atoms dark salmon) and GCPII/JRB-4-81 (carbon atoms green) were superimposed on corresponding Cα atoms of the enzyme. Inhibitors are in stick representation, selected GCPII amino acid residues within the 4 Å radius of an inhibitor are shown as lines, and zinc ions are shown as orange spheres. Atoms colored red (oxygen), blue (nitrogen), pale cyan (fluorine), and orange (phosphorus). The different positioning of the two inhibitors in the GCPII internal cavity as well as flexibility of the side chains of Glu457, Arg463, and Arg536 is noticeable. Panel B; Atoms of superposed structures are colored according to their temperature factors. While the P1’ glutamate and surrounding residues have lower B-factors (blue shades) suggesting less positional flexibility, the distal inhibitor parts (together with surrounding residues) are more flexible as reflected in their higher temperature factors. Panels C, D; Fo-Fc maps (green) for individual inhibitors are contoured at 3.0 σ. Weaker, yet clearly interpretable density is observed for the distal part of individual inhibitors (as compared to the P1’ glutamate).
DISCUSSION
Here, we report for the first time crystal structures of human GCPII in complex with phosphoramidate-based inhibitors. Phosphorus-based mimetics of a tetrahedral intermediate or transition state are frequently used to inhibit metallopeptidases (including GCPII) as their interactions with metal ions substantially increase affinity for a target enzyme. It is interesting to note that compared to phosphonates and phosphinates, whose complexes with GCPII were reported previously [13, 14, 20, 24], incorporation of the phosphoramidate functionality can be beneficial in terms of increased affinity and change in the inhibition mode (reversible vs. pseudo-irreversible). These observations were first reported by the Berkman group [25] and are also supported in this work by comparing CTT54 to its phosphonate analog, where the latter is approximately 40-times weaker inhibitor of GCPII (unpublished data). Mechanistic explanation for the increased affinity is the presence of two additional hydrogen bonds between the phosphoramidate nitrogen and carboxylate of Glu424 and the Gly518 carbonyl oxygen, respectively (as compared to phosphinates; Figure 3). In this respect, phosphoramidates might be preferred peptidomimetics functionalities for metallopeptidases that use glutamate as a proton shuttle residue.
The most conspicuous structural feature of the S1 site of GCPII is the arginine patch of Arg534, Arg536 and Arg463 that is responsible for the preference for acidic residues of substrates [20, 24]. Favorable ionic interactions between the arginine patch and negatively charged functionalities of inhibitors (typically carboxylates) are exploited in the design of high-affinity GCPII inhibitors. It has been shown by us and others that the inclusion of the P1 carboxylate into the inhibitor structure increases the inhibitor affinity by several fold [12]. Data reported here corroborate these findings and additionally demonstrate the importance of “proper” stereochemistry at the P1 carboxylate position. In the phosphoramidate series, the (S)P1-diastereomer is clearly favored (compare T33 and T33D). However, it should be stressed that the “(S)-rule” is unlikely to be universal. For compounds with different zinc-binding groups (ZBG) or varying lengths (and chemistries) of the spacer between the ZBG and the P1 carboxylate, the (R)P1-stereoisomer might be preferred. For example, while co-crystallizing GCPII with a diastereomeric mixture of an MTX inhibitor, only the (R) isomer was observed in the crystal suggesting that this is the preferred stereoisomer for this particular compound [20]. Additionally, (R) and (S) enantiomers of several thiol- and hydroxamate-based inhibitors have identical affinity for GCPII, arguing against the simplistic (S)-rule in the inhibitor design [29, 30]. Consequently, if synthetically feasible, both stereoisomers should be prepared and tested for their respective GCPII affinity.
Our prior structural studies revealed the presence of two accessory binding sites that can be used for the inhibitor design. These include a hydrophobic pocket adjacent to the S1 pocket shaped by side chains of Glu457, Asp465, Arg463, and Arg534, Arg536 [21], and the arene-binding site motif located at the entrance lid (amino acids Trp541-Gly548) [20, 22]. Surprisingly, none of the inhibitors from this study exploits any of the two accessory binding sites. Instead, the distal, mostly non-polar effector functionalities take advantage of “isolated” hydrophobic patches forming the wall of the entrance funnel (T33, T33D, MP1C, NC-2-40). Additionally, as in cases of the GCPII/NC-2-40 and GCPII/CTT54 complexes, the distal parts of inhibitors engage the entrance lid, but at the sites that are quite distant from the arene-binding site.
Comparison of X-ray data and in silico docking
In the absence of crystal structures, SAR studies exploit in silico docking to predict binding modes of protein/inhibitor complexes as well as to rationalize inhibitory data. We used a series of structures reported here to assess the usefulness of two widely used docking algorithms implemented in AutoDock and DOCK programs to predict positioning of phosphoramidates in the GCPII internal cavity. Figure 5 illustrates “typical” docking results using the GCPII/MP1C complex as an example. In general, results of calculations with AutoDock, using the default parameters, did not correlate with the crystal structures for any of the compounds described here (Fig. 5A). The glutamate part of the inhibitor did not fit into the S1’ pocket, but replaced the phosphoramidate functionality in the zinc coordination sphere. Apparently, the parameterization of zinc charges in AutoDock overestimates charge-charge energies leading to the incorrect predictions. Slightly better results were obtained when the zinc – phosphorus vicinity was enforced by the covalent map feature of autogrid/AutoDock programs (Fig. 5B). In such a case, however, the information from crystal structures needs to be implicitly incorporated, making a prediction value of the calculations somewhat dubious.
Figure 5.
he summary of docking program performance demonstrated using the GCPII/MP1C complex. Five the most stabilizing/best scored ligand poses are shown for the AutoDock without (A) and with the zinc – phosphorus vicinity enforced (B). Panel (C) summarizes the performance of the DOCK “anchor and grow” grid scoring approach. The crystal position of the ligand is depicted in stick representation while the poses are represented as balls-and-sticks and color coded (red, blue, yellow, green, and violet) with the red being the most stable pose.
To the contrary, the DOCK scoring performed more consistently with predictions obtained for the glutamate and phosphoramidate parts correlating well with crystal structures. At the same time, however, the more pronounced differences were observed for the non-prime distal aromatic/aliphatic moieties of ligands (Fig 5C). The latter finding likely stems from the fact that the effector-enzyme interactions are mostly controlled by weaker and the less direction-specific van der Waals forces as suggested by the B-factors and electron density maps for corresponding inhibitors (Figure 4).
In theory, availability of a crystal structure for a GCPII/inhibitor complex should allow dissection and quantification of the contributions of individual parts of the inhibitor to the overall affinity (interaction energy) by quantum mechanics/molecular mechanics calculations. Recently, we used QM calculations to successfully correlate PM7 interaction energies and experimental inhibition constants in a series of P1’-diversified urea-based GCPII inhibitors [31]. However, replicating this approach for complexes reported here did not yield a significant correlation pattern, suggesting that a more robust approach is needed to obtain a generally applicable protocol. We believe that the main reason for the lack of correlation is pronounced difference in size and positioning of inhibitors in the GCPII structure that in turn mandates the inclusion of variable areas of the enzyme for calculations. In the case of more structurally uniform urea-based compounds reported earlier, the optimal 10-Å selection radius around a given inhibitor was basically identical for all structures compared. To the contrary, a substantial variability and complexity in areas selected here brings about energetic contributions that are difficult to parameterize. Moreover, chemical variability of the studied compounds brings additional energy contributions such as solvation/desolvation and ligand and/or protein deformation energies that might further complicate the quantification of the total interaction energy. As energy calculations assist in SAR campaigns by dissecting and quantifying contributions of individual inhibitor functionalities towards overall potency of a given compound, we are currently trying to develop a generally applicable calculation algorithm that can be used for the design of GCPII-specific ligands.
Conclusions
In conclusion, the data presented here unravel a complexity of interactions between inhibitors and the non-prime site (the entrance funnel) of GCPII. They clearly show that even a minor modification in the inhibitor structure can have a profound effect on inhibitor conformation in the internal GCPII pocket thus presenting major challenges for structure-assisted drug design. Additionally, we show that DOCK scoring is superior to the default AutoDock setup in the prediction of binding modes of small molecules to the internal pocket of GCPII. Together, these findings can be exploited for the structure-assisted design of novel GCPII-specific compounds.
MATERIALS AND METHODS
Unless stated otherwise, all chemicals were purchased from Sigma-Aldrich (Steinheim, Germany).
Protein expression and purification
Cloning, expression and purification of the extracellular part of human GCPII (rhGCPII; amino acids 44 – 750) were carried out as described previously [32]. The protein was overexpressed in S2 cells and purified using concentration/dialysis by tangential flow filtration TFF (Millipore Mosheim France); ion-exchange chromatography (Q and SP Sepharose FF); affinity chromatography on Lentil-Lectin Sepharose and; size-exclusion chromatography on a Superdex 200 column (all resins/columns from GE Healthcare Bio-Sciences, Uppsala, Sweden). Purified rhGCPII (in final buffer 20 mM Tris-HCl, 150 mM NaCl, pH 8.0) was concentrated to 10 mg/mL and kept at −80°C until further use.
Inhibitors
Synthesis and characterization of compounds T33, T33D, MP1C, MP1D, NC-2-40 and CTT54 was reported previously [25, 33].
Synthesis of JRB-4-73 and JRB-4-81.
Reagents and conditions
a) para-fluorobenzoyl chloride, pH 9; b) H.SerOBn, HBTU, TEA, DMF; c) Diphenyl phosphite, pyridine; d) benzyl alcohol; e) H.Glu(OBn)OBn, CCl4, TEA, ACN; f) KHCO3, 10% Pd/C, Dioxane:H2O (1:5 v/v)
General synthesis of FB-X-OH
Unprotected L-amino acid (12.78 mmol) was dissolved in 20 mL of 10% NaOH (wt/wt). Acetone (30 mL) was added to the solution and stirred on ice. A solution of p-Fluorobenzyl chloride (12.780 mmol) in 10 mL of acetone was added dropwise to maintain the reaction at a pH 8-9. The pH was adjusted with 10% NaOH, when needed. Upon complete addition of p-Fluorobenzyl chloride, the mixture was stirred for an hour. The acetone was evaporated and product precipitated out as a white solid once the pH was adjusted to pH=2 (75-83% yield) and was taken on without further purification.
General synthesis of FB-X-SerOBn
FB-X-OH (6.50 mmol) and HBTU (6.50 mmol) was stirred for 30 mins in 20 mL DMF under inert atmosphere. A solution of H-SerOBn (6.50 mmol) and TEA (14.3 mmol) in 10 mLs of DMF was added dropwise to the reaction mixture and stirred until completion, about 1 hr. The reaction mixture was taken up in EtOAc (200 mL) and the organic layer was extracted with 1 N HCl (2X, 50 mL), 10% NaHCO3 (2X, 50 mL), washed with water, followed by brine and dried with MgSO4. The EtOAc layer was filtered and concentrated down to yield the desired compound as a white solid (81% yield).
General synthesis of FB-X-SerOBn-O-P(O)-OBn-H phosphite
FB-X-SerOBn (0.529 mmol) in 2 mL of freshly distilled pyridine was added dropwise via cannula to a stirring solution of diphenylphosphite (0.634 mmol) in 3 mL of freshly distilled pyridine. The resulting solution was stirred for 2 hrs under a stream of Ar(g), followed by the dropwise addition of benzyl alcohol (1.587 mmol) via syringe. The reaction was stirred for an additional 3.5 hrs. The crude mixture was taken up with 25 mL of EtOAc and extracted with 10% copper sulfate (wt/v) until the pyridine has been removed. The organic was washed with dd H2O, followed by brine and dried with MgSO4, filtered and concentrated down to yield a crude oil. The phosphite was obtained via silica column using EtOAc:Hex as the eluent and taken on to the next step without further purification.
General synthesis of FB-X-SerOBn-O-P(O)-OBn-Glu(OBn)OBn phosphoramidate
The phosphite (0.119 mmol) was dissolved in distilled ACN (2 mL) and CCl4 (1 mL) and stirred for 15 mins at −15°C. H-Glu(OBn)OBn (0.125 mmol) in distilled ACN and TEA (0.369 mmol) was added dropwise. Upon completion, the reaction was concentrated down to yield an oil. The crude was taken up in 50 mL of EtOAc and the organic layer was extracted with 1 N HCl (2X, 25 mL), followed by 10% NaHCO3 (2X, 25 mL) and brine (1X, 25 mL), dried with MgSO4, filtered and concentrated down. The product was obtained as a white foamy solid via prep-Si TLC with 30% ACN:CHCl3 as the elutant (61%).
General deprotection of benzyl esters
The phosphoramidate (0.036 mmol) was dissolved in Dioxane:water (1:5, 2 mL). KHCO3 (0.147 mmol, 1 eq per OBn) was added followed by 10% Pd/C (0.010 g). The reaction flask was purged with N2(g), followed by H2(g), then stirred under H2(g) overnight. The crude mixture was filtered through a 0.2 µm PTFE filter to remove Pd/C and the flow-through was concentrated down to give the final product in quantitative yield.
The identity and purity of final products (compounds JRB-4-73 and JRB-4-81) were verified by mass spectrometry, NMR and analytical HPLC.
IC50 determination
Inhibition studies were performed as described previously [26, 34]. Working solutions of the substrate (N-[4-(phenylazo)-benzoyl]-glutamyl-γ-glutamic acid, PABGγG) and inhibitors were made in the reaction buffer (50 mM Tris-HCl, pH 7.4, containing 1% Triton X-100). Working solutions (50 µg/mL) of purified GCPII [35] were diluted in the reaction buffer to provide from 15% to 20% conversion of substrate to product in the absence of inhibitor. A typical incubation mixture (final volume 250 µL) was prepared by the addition of either 25 µL of an inhibitor solution or 25 µL reaction buffer to 175 µL reaction buffer in a test tube. PABGγG (25 µL, 10 µM) was added to the above solution. The enzymatic reaction was initiated by the addition of 25 µL of the GCPII working solution. In all cases, the final concentration of PABGγG was 1 µM while the enzyme was incubated with five serially diluted inhibitor concentrations providing a range of inhibition from 10% to 90%. The reaction was allowed to proceed for 15 min with constant shaking at 37°C and was terminated by the addition of 25 µL methanolic TFA (2% trifluoroacetic acid by volume in methanol) followed by vortexing. The quenched incubation mixture was quickly buffered by the addition of 25 µL K2HPO4 (0.1 M), vortexed, and centrifuged (10 min at 7000g). An 85 µL aliquot of the resulting supernatant was subsequently quantified by HPLC as previously described [26, 36]. IC50 values were calculated using KaleidaGraph 3.6 (Synergy Software).
Inhibition mode
Mode of inhibition study followed the procedure described in our previous work [25]. Briefly, the concentration of GCPII (2.5 µg/mL) was 100-fold greater than used in the typical enzyme activity assays. The enzyme was pre-incubated for 10 minutes with 40 µL of inhibitor at approximately 10-fold greater than the IC50 value. The solution was diluted with 1 mM of substrate in 50 mM Tris and 1% Triton buffer (100-fold, total volume 3960 µL). The formation of product was monitored every 5 minutes for 1 hour. A control sample was defined as an incubation described here but without inhibitor. Progress curves of product formation were generated to monitor the recovery of enzymatic activity for inhibited PSMA and compared to a control sample in which no inhibitor was added
Crystallization and data collection
Diffracting crystals of GCPII/inhibitor complexes were obtained using procedures described previously [37]. Briefly, GCPII (10 mg/mL) was mixed with a stock solution of a given inhibitor in water (typically 20 – 50 mM inhibitor stock solutions) at the 10:1 (v/v) ratio, the GCPII/inhibitor solution mixed with the same volume of the reservoir solution (33% pentaerythritol propoxylate, (Sigma), 1.5% polyethylene glycol 3350 (Sigma), and 100 mM Tris–HCl, pH 8.0), and then crystallized in the hanging-drop vapor-diffusion setup at 293 K. Monocrystals of GCPII/inhibitor complexes typically appeared within one to two weeks. Crystals were flash frozen in liquid nitrogen directly from the crystallization droplets and diffraction intensities for each complex were collected from a single crystal at 100 K using synchrotron radiation at the SER-CAT beamlines 22-ID and 22-BM at the Advanced Photon Source (Argonne, USA; 1.00 Å) or at the MX 14.2 beamline (BESSYII, Helmholtz-Zentrum Berlin, Germany; 0.9181 Å). The complete dataset for each complex was collected from a single crystal and data was processed using the HKL2000 software package [38] or XDSAPP [39]. The final statistics are shown in Table 1.
Structure determination, refinement, and analysis
Difference Fourier methods were used to determine structures of GCPII/inhibitor complexes with ligand-free GCPII (PDB code 2OOT) used as a starting model [37]. Calculations were performed using Refmac 5.5 [40] and the structure refinement was interspersed with manual corrections to the model employing the program Coot 0.6 [41]. The PRODRG server [42] was used to generate restrains library and coordinate files for individual inhibitors and the inhibitors were fitted into the positive electron density map in the final stages of the refinement. 1% - 5% of the randomly selected reflections were kept aside for cross-validation (Rfree) during the refinement process. The quality of the final models was evaluated using MOLPROBITY [43]. The data collection and refinement statistics are summarized in Table 1.
Docking and computational details
Two docking program suites (AutoDock and DOCK) were tested for accuracy to reproduce the ligand orientation in the crystal of GCPII/ligand complex as well as the binding energy. AutoDock [44] 4.2.3 calculations were prepared and analyzed in AutoDockTools [44] (ADT) version 1.5.6rc3. The protein and ligand charges were assigned by ADT, using the Gasteiger charges. The genetic algorithm (GA) runs of 50 steps were employed for both rigid as well as flexible side-chains surrounding the binding pocket.
The DOCK [45] 6.6 calculations were set up within Chimera [46] graphical interface using the DockPrep. Rigid ligand docking with optimization and flexible ligand “anchor and grow” was tested with grid scoring of 20 conformers in each run. The five best scored inhibitor poses for each complex were further rescored using the amber score.
Acknowledgment
We thank Petra Baranova (IBT, Czech Republic) for the excellent technical assistance, and Manfred Weiss (BESSYII, Berlin) for the help with data collection and Zsofia Kutil for help with figure preparation. The use of the Advanced Photon Source was supported by the US Department of Energy (Contract No. W-31-109-Eng38). We thank Helmholtz-Zentrum Berlin for the allocation of synchrotron radiation beamtime that received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under BioStruct-X (grant agreement N°283570). C.Barinka acknowledges the support from the Czech Science Foundation (grant No 301/12/1513). Z. Novakova acknowledges support from the program “Biotechnological expert in structural biology and gene expression”, Reg. n.: CZ.1.07/2.3.00/30.0045. C.E. Berkman acknowledges support from the NIH (R01CA140617). This publication is supported by the project BIOCEV (CZ.1.05/1.1.00/02.0109) from the ERDF.
Abbreviations
- GCPII
glutamate carboxypeptidase II
- SAR
structure-activity relationship
- NAAG
N-acetyl-aspartyl-glutamate
Footnotes
PDB ID Codes: Atomic coordinates of the present structures together with the experimental structure factor amplitudes were deposited at the RCSB Protein Data Bank under accession codes 4P44 (complex with JRB-4-81), 4P45 (complex with JRB-4-73), 4P4B (complex with CTT54), 4P4D (complex with MP1C), 4P4E (complex with MP1D), 4P4F (complex with NC-2-40), 4P4I (complex with T33), and 4P4J (complex with T33D).
Author Contributions
CB, JC, CEB, and JL, planned experiments; ZN, JC, and CB performed experiments; ZN, CJC, JN, and JKC contributed reagents or other essential material; ZN, JC, JL, and CB analyzed data; CB, JC, JL, and CEB wrote the paper.
REFERENCES
- 1.Barinka C, Rojas C, Slusher B, Pomper M. Glutamate Carboxypeptidase II in Diagnosis and Treatment of Neurologic Disorders and Prostate Cancer. Curr Med Chem. 2012;19:856–870. doi: 10.2174/092986712799034888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou J, Neale JH, Pomper MG, Kozikowski AP. NAAG peptidase inhibitors and their potential for diagnosis and therapy. Nat Rev Drug Discov. 2005;4:1015–1026. doi: 10.1038/nrd1903. [DOI] [PubMed] [Google Scholar]
- 3.Foss CA, Mease RC, Cho SY, Kim HJ, Pomper MG. GCPII Imaging and Cancer. Curr Med Chem. 2012;19:1346–1359. doi: 10.2174/092986712799462612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bostwick DG, Pacelli A, Blute M, Roche P, Murphy GP. Prostate specific membrane antigen expression in prostatic intraepithelial neoplasia and adenocarcinoma: a study of 184 cases. Cancer. 1998;82:2256–2261. doi: 10.1002/(sici)1097-0142(19980601)82:11<2256::aid-cncr22>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 5.Slusher BS, Robinson MB, Tsai G, Simmons ML, Richards SS, Coyle JT. Rat brain N-acetylated alpha-linked acidic dipeptidase activity. Purification and immunologic characterization. J Biol Chem. 1990;265:21297–21301. [PubMed] [Google Scholar]
- 6.Carozzi VA, Ceresa C. The Role of Glutamate in Diabetic and in Chemotherapy Induced Peripheral Neuropathies and its Regulation by Glutamate Carboxypeptidase II. Curr Med Chem. 2012;19:1261–1268. doi: 10.2174/092986712799462694. [DOI] [PubMed] [Google Scholar]
- 7.Cavaletti G, Slusher B. Regulation of Glutamate Synthesis Via Inhibition of Glutamate Carboxypeptidase II (GCPII): An Effective Method to Treat Central and Peripheral Nervous System Disorders. Curr Med Chem. 2012;19:1259–1260. doi: 10.2174/092986712799462577. [DOI] [PubMed] [Google Scholar]
- 8.Wozniak KM, Rojas C, Wu Y, Slusher BS. The Role of Glutamate Signaling in Pain Processes and its Regulation by GCP II Inhibition. Curr Med Chem. 2012;19:1323–1334. doi: 10.2174/092986712799462630. [DOI] [PubMed] [Google Scholar]
- 9.Ferraris DV, Shukla K, Tsukamoto T. Structure-Activity Relationships of Glutamate Carboxypeptidase II (GCPII) Inhibitors. Curr Med Chem. 2012;19:1282–1294. doi: 10.2174/092986712799462658. [DOI] [PubMed] [Google Scholar]
- 10.Tsukamoto T, Wozniak KM, Slusher BS. Progress in the discovery and development of glutamate carboxypeptidase II inhibitors. Drug Discov Today. 2007;12:767–776. doi: 10.1016/j.drudis.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 11.Plechanovova A, Byun Y, Alquicer G, Skultetyova L, Mlcochova P, Nemcova A, Kim HJ, Navratil M, Mease R, Lubkowski J, Pomper M, Konvalinka J, Rulisek L, Barinka C. Novel substrate-based inhibitors of human glutamate carboxypeptidase II with enhanced lipophilicity. J Med Chem. 2011;54:7535–7546. doi: 10.1021/jm200807m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Byun Y, Barinka C, Pullambhatla M, Bhang HE, Fox JJ, Lubkowski J, Mease RC, Pomper MG. Bioisosterism of urea-based GCPII inhibitors: Synthesis and structure-activity relationship studies. Bioorg Med Chem Lett. 2010;20:392–397. doi: 10.1016/j.bmcl.2009.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barinka C, Rovenska M, Mlcochova P, Hlouchova K, Plechanovova A, Majer P, Tsukamoto T, Slusher BS, Konvalinka J, Lubkowski J. Structural insight into the pharmacophore pocket of human glutamate carboxypeptidase II. J Med Chem. 2007;50:3267–3273. doi: 10.1021/jm070133w. [DOI] [PubMed] [Google Scholar]
- 14.Mesters JR, Henning K, Hilgenfeld R. Human glutamate carboxypeptidase II inhibition: structures of GCPII in complex with two potent inhibitors, quisqualate and 2-PMPA. Acta Crystallogr D Biol Crystallogr. 2007;63:508–513. doi: 10.1107/S090744490700902X. [DOI] [PubMed] [Google Scholar]
- 15.Banerjee SR, Pullambhatla M, Byun Y, Nimmagadda S, Green G, Fox JJ, Horti A, Mease RC, Pomper MG. 68Ga-labeled inhibitors of prostate-specific membrane antigen (PSMA) for imaging prostate cancer. J Med Chem. 2010;53:5333–5341. doi: 10.1021/jm100623e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kularatne SA, Wang K, Santhapuram HK, Low PS. Prostate-specific membrane antigen targeted imaging and therapy of prostate cancer using a PSMA inhibitor as a homing ligand. Mol Pharm. 2009;6:780–789. doi: 10.1021/mp900069d. [DOI] [PubMed] [Google Scholar]
- 17.Liu T, Nedrow-Byers JR, Hopkins MR, Berkman CE. Spacer length effects on in vitro imaging and surface accessibility of fluorescent inhibitors of prostate specific membrane antigen. Bioorg Med Chem Lett. 2011;21:7013–7016. doi: 10.1016/j.bmcl.2011.09.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malik N, Machulla HJ, Solbach C, Winter G, Reske SN, Zlatopolskiy B. Radiosynthesis of a new PSMA targeting ligand ([18F]FPy-DUPA-Pep) Appl Radiat Isot. 2011;69:1014–1018. doi: 10.1016/j.apradiso.2011.03.041. [DOI] [PubMed] [Google Scholar]
- 19.Pavlicek J, Ptacek J, Barinka C. Glutamate Carboxypeptidase II: An Overview of Structural Studies and Their Importance for Structure-Based Drug Design and Deciphering the Reaction Mechanism of the Enzyme. Curr Med Chem. 2012;19:1300–1309. doi: 10.2174/092986712799462667. [DOI] [PubMed] [Google Scholar]
- 20.Barinka C, Hlouchova K, Rovenska M, Majer P, Dauter M, Hin N, Ko YS, Tsukamoto T, Slusher BS, Konvalinka J, Lubkowski J. Structural basis of interactions between human glutamate carboxypeptidase II and its substrate analogs. J Mol Biol. 2008;376:1438–1450. doi: 10.1016/j.jmb.2007.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barinka C, Byun Y, Dusich CL, Banerjee SR, Chen Y, Castanares M, Kozikowski AP, Mease RC, Pomper MG, Lubkowski J. Interactions between human glutamate carboxypeptidase II and urea-based inhibitors: structural characterization. J Med Chem. 2008;51:7737–7743. doi: 10.1021/jm800765e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang AX, Murelli RP, Barinka C, Michel J, Cocleaza A, Jorgensen WL, Lubkowski J, Spiegel DA. A remote arene-binding site on prostate specific membrane antigen revealed by antibody-recruiting small molecules. J Am Chem Soc. 2010;132:12711–12716. doi: 10.1021/ja104591m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navratil M, Ptacek J, Sacha P, Starkova J, Lubkowski J, Barinka C, Konvalinka J. Structural and biochemical characterization of the folyl-poly-gamma-l-glutamate hydrolyzing activity of human glutamate carboxypeptidase II. FEBS J. 2014;281:3228–3242. doi: 10.1111/febs.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mesters JR, Barinka C, Li W, Tsukamoto T, Majer P, Slusher BS, Konvalinka J, Hilgenfeld R. Structure of glutamate carboxypeptidase II, a drug target in neuronal damage and prostate cancer. EMBO J. 2006;25:1375–1384. doi: 10.1038/sj.emboj.7600969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu T, Toriyabe Y, Kazak M, Berkman CE. Pseudoirreversible inhibition of prostate-specific membrane antigen by phosphoramidate peptidomimetics. Biochemistry. 2008;47:12658–12660. doi: 10.1021/bi801883v. [DOI] [PubMed] [Google Scholar]
- 26.Maung J, Mallari JP, Girtsman TA, Wu LY, Rowley JA, Santiago NM, Brunelle AN, Berkman CE. Probing for a hydrophobic a binding register in prostate-specific membrane antigen with phenylalkylphosphonamidates. Bioorg Med Chem. 2004;12:4969–4979. doi: 10.1016/j.bmc.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 27.Liu T, Nedrow-Byers JR, Hopkins MR, Wu LY, Lee J, Reilly PT, Berkman CE. Targeting prostate cancer cells with a multivalent PSMA inhibitor-guided streptavidin conjugate. Bioorg Med Chem Lett. 2012;22:3931–3934. doi: 10.1016/j.bmcl.2012.04.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ganguly T, Dannoon S, Hopkins MR, Murphy S, Cahaya H, Blecha JE, Jivan S, Drake CR, Barinka C, Jones EF, VanBrocklin HF, Berkman CE. A high-affinity [(18)F]-labeled phosphoramidate peptidomimetic PSMA-targeted inhibitor for PET imaging of prostate cancer. Nucl Med Biol. 2015;42:780–787. doi: 10.1016/j.nucmedbio.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majer P, Jackson PF, Delahanty G, Grella BS, Ko YS, Li W, Liu Q, Maclin KM, Polakova J, Shaffer KA, Stoermer D, Vitharana D, Wang EY, Zakrzewski A, Rojas C, Slusher BS, Wozniak KM, Burak E, Limsakun T, Tsukamoto T. Synthesis and biological evaluation of thiol-based inhibitors of glutamate carboxypeptidase II: discovery of an orally active GCP II inhibitor. J Med Chem. 2003;46:1989–1996. doi: 10.1021/jm020515w. [DOI] [PubMed] [Google Scholar]
- 30.Stoermer D, Liu Q, Hall MR, Flanary JM, Thomas AG, Rojas C, Slusher BS, Tsukamoto T. Synthesis and biological evaluation of hydroxamate-Based inhibitors of glutamate carboxypeptidase II. Bioorg Med Chem Lett. 2003;13:2097–2100. doi: 10.1016/s0960-894x(03)00407-4. [DOI] [PubMed] [Google Scholar]
- 31.Pavlicek J, Ptacek J, Cerny J, Byun Y, Skultetyova L, Pomper MG, Lubkowski J, Barinka C. Structural characterization of P1'-diversified urea-based inhibitors of glutamate carboxypeptidase II. Bioorg Med Chem Lett. 2014;24:2340–2345. doi: 10.1016/j.bmcl.2014.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barinka C, Mlcochova P, Sacha P, Hilgert I, Majer P, Slusher BS, Horejsi V, Konvalinka J. Amino acids at the N- and C-termini of human glutamate carboxypeptidase II are required for enzymatic activity and proper folding. Eur J Biochem. 2004;271:2782–2790. doi: 10.1111/j.1432-1033.2004.04209.x. [DOI] [PubMed] [Google Scholar]
- 33.Coleman N. The Evaluation of Zinc Binding Domains in Phosphoramidate, Urea and Carbamate Inhibitors of Prostate-specific Membrane Antigen (PSMA) San Francisco State University; 2007. [Google Scholar]
- 34.Wu LY, Anderson MO, Toriyabe Y, Maung J, Campbell TY, Tajon C, Kazak M, Moser J, Berkman CE. The molecular pruning of a phosphoramidate peptidomimetic inhibitor of prostate-specific membrane antigen. Bioorg Med Chem. 2007;15:7434–7443. doi: 10.1016/j.bmc.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu T, Toriyabe Y, Berkman CE. Purification of prostate-specific membrane antigen using conformational epitope-specific antibody-affinity chromatography. Protein Expr Purif. 2006;49:251–255. doi: 10.1016/j.pep.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Anderson MO, Wu LY, Santiago NM, Moser JM, Rowley JA, Bolstad ES, Berkman CE. Substrate specificity of prostate-specific membrane antigen. Bioorg Med Chem. 2007;15:6678–6686. doi: 10.1016/j.bmc.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barinka C, Starkova J, Konvalinka J, Lubkowski J. A high-resolution structure of ligand-free human glutamate carboxypeptidase II. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:150–153. doi: 10.1107/S174430910700379X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minor W, Cymborowski M, Otwinowski Z. Automatic system for crystallographic data collection and analysis. Acta Physica Polonica A. 2002;101:613–619. [Google Scholar]
- 39.Krug M, Weiss MS, Heinemann U, Mueller U. XDSAPP: a graphical user interface for the convenient processing of diffraction data using XDS. Journal of Applied Crystallography. 2012;45:568–572. [Google Scholar]
- 40.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 43.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. Journal of Computational Chemistry. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balius TE, Mukherjee S, Rizzo RC. Implementation and Evaluation of a Docking-Rescoring Method Using Molecular Footprint Comparisons. Journal of Computational Chemistry. 2011;32:2273–2289. doi: 10.1002/jcc.21814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera - A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]