Abstract
Resistance to therapies targeting the estrogen pathway remains a challenge in the treatment of estrogen-receptor positive breast cancer. To address this challenge, a systems biology approach was used. A library of siRNAs targeting an estrogen receptor- and aromatase-centered network identified 46 genes that are dispensable in estrogen-dependent MCF7 cells, but are selectively required for the survival of estrogen-independent MCF7-derived cells, and multiple additional estrogen-independent breast cancer cell lines. Integration of this information identified a tumor suppressor gene TOB1 as a critical determinant of estrogen-independent estrogen receptor-positive breast cell survival. Depletion of TOB1 selectively promoted G1 phase arrest and sensitivity to AKT and mTOR inhibitors in estrogen-independent cells but not estrogen-dependent cells. Phosphoproteomic profiles from reverse phase protein array analysis supported by mRNA profiling identified a significant signaling network reprogramming by TOB1 that differed in estrogen-sensitive and estrogen-resistant cell lines. These data support a novel function for TOB1 in mediating survival of estrogen-independent breast cancers. These studies also provide evidence for combining TOB1 inhibition and AKT/mTOR inhibition as a therapeutic strategy, with potential translational significance for the management of patients with estrogen receptor-positive breast cancers.
Keywords: Estrogen independence, TOB1, AKT/mTOR, ERBB2/HER2, G1/S transition, Breast cancer, Growth factor receptor signaling
INTRODUCTION
Breast cancer is the most common cancer among women in the United States and is the second leading cause of cancer related deaths in this population [1]. Approximately 70% of newly diagnosed breast cancers express high levels of estrogen receptor alpha (ERα) [2], a nuclear transcription factor that controls cell proliferation, in large part by regulating gene expression. ERα also has been described as having additional functions, including the regulation of growth factor signaling [3]. Anti-estrogens (AEs) and aromatase inhibitors [4] [4] are currently used to treat ER-positive breast cancer in pre-menopausal and post-menopausal women, respectively. AIs such as exemestane or anastrozole block the biosynthesis of the ERα ligand estrogen, while AEs such as tamoxifen or fulvestrant compete with estrogen for binding to ERα, in each case resulting in diminished ERα activity and in some cases diminished nuclear localization, leading to impaired cell proliferation and survival.
De novo and acquired drug resistance to AEs and AIs pose significant challenges to the effective treatment of ERα positive breast cancers. Numerous resistance mechanisms have been identified, including epigenetic changes affecting the ERα promoter [5], mutations activating the ERα protein to ligand independence [6, 7], altered expression or activation of cellular signaling proteins that generally promote survival such as epithelial growth factor receptor (EGFR) [8], insulin-like growth factor receptor (IGFR) [9], PI3K/AKT [10], mTOR signaling [11] and NFκB [12], and altered expression of specific miRNAs [13]. However, in hormone therapy-resistant breast cancer, chemotherapy remains the primary treatment modality [14], and the prognosis of such patients is poor.
To address this problem, we aimed to identify new points of vulnerability in estrogen-independent, AE/AI-resistant breast cancers. A number of studies have demonstrated that changes in the proximal signaling networks to proteins targeted by drugs are particularly common sources of resistance to the targeting agent [15-17]. The goal of this study was to use in silico resources to develop a network centered on ERα and related estrogen receptors and aromatase, and then to create and probe a siRNA library individually targeting genes in this network, to better understand the key mechanisms of estrogen independence and antiestrogen resistance. Interrogation of the functional signaling consequences of this gene targeting was performed using quantitative highly multiplexed protein pathway activation mapping. These studies identified a group of genes with action specifically required for the survival of estrogen-independent cells. Strikingly, this work also demonstrated selective action of the tumor suppressor TOB1 (transducer of c-erbB2) as important for basal growth and drug resistance of estrogen-independent cell lines, based on distinctive regulation of survival and cell cycle signaling in these cell lines. These observations have potential translational significance for the management of estrogen receptor-positive breast cancers.
RESULTS
Estrogen Response- Centered Network
We hypothesized that loss of estrogen dependence would reflect an altered cellular requirement for genes closely linked to core genes regulating estrogen response. A 631-protein estrogen response network (ERN) was developed around 5 seed proteins relevant to estrogen signaling: the estrogen receptor genes ESR1 (ERα) and ESR2 (ERβ), the estrogen-related receptors ESRRA and ESRRG, and CYP19A1 (aromatase) (Figure 1A, Table S1). For network construction, data for each of the 5 seeds was initially collected from public archives reporting protein-protein interactions (PPIs), association in protein complexes, curated pathway information, and estrogen-responsive genes. PPI databases (BIND [18], BioGRID [19], DIP [20], HPRD [21], IntAct [22], and MINT [23]) were mined for first and second neighbors of the 5 seed proteins both directly and via metasearch engines such as MiMI [24] and STRING [25].
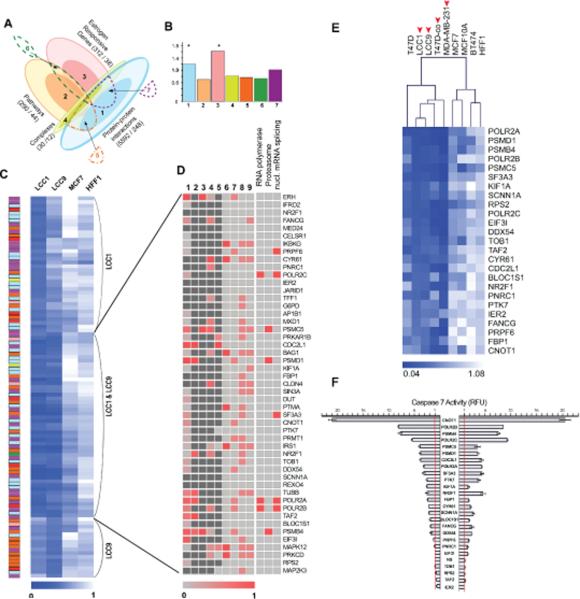
Figure 1. Requirement of a subset of the Estrogen Response Network (ERN) genes for growth of estrogen-independent cell line.
A. Schematic representation of gene inputs (protein-protein interactions (PPIs), pathway maps, estrogen responsive genes, and proteins in complex with network seeds) into ERN library. Light colors represent low confidence dataset, while darker tones represent highest confidence dataset “core”, as defined in Results and Supplemental Table S1; numbers following labels represent total number of genes in category versus in dataset core (e.g. 30/12, 30 genes in category of complexes, 12 genes are dataset cores). Numbers 1-7 indicate sources of validated hits in the ERN discussed in functional studies: 1, core PPIs (5592/248); 2, pathway core (290/44); 3, E2-responsive gene core (312/38); 4, complex core (30/12); 5, both PPI and pathways; 6, E2-responsive core & pathways; 7, PPIs and E2-responsive core. B. Analysis of hit enrichment of the validated hits across sources in the ERN. Number 1-7 refers to categories in (A). Y axis shows fold enrichment over the expected value; asterisks mark significantly enriched categories (p<0.05). C. Cell viability following siRNA depletion. The blue color scale represents viability with the darkest blue indicating the lowest viability, and white representing a viability of 1.0, following siRNA deletion of genes in the indicated cell lines. Colors to left represent relationship of gene to library input as in (B). Annotation to right indicates those considered as specific for LCC1, LCC9, or both cell lines. D. Categorical distribution of validated hits common to LCC1 and LCC9 cells. Categories 1-9 represent publically available data suggesting indicated genes support survival in specific contexts relevant to breast cancer. 1-5, cell line databases; 6-9, gene lists. Sources: 1, http://colt.ccbr.utoronto.ca/, and [46] ; 2, [51]; 3, 4 [52]; 5, [53]; 6, Ingenuity, genes specifically important for proliferation in breast cancer; 7, GO genes [54]; 8, Ingenuity, genes generally important for proliferation; 9, Ingenuity, genes generally important for cell survival. Darker red colors in columns 1-6 indicate higher essentiality for cell survival; dark grey color, data not available. Red color in columns 7-9 indicates that certain gene presents in corresponding list, and light grey colors indicate a certain gene dose not present in the list. E. Heatmap representing viability of indicated breast cancer and immortalized mammary cell lines 144 hr after siRNA transfection. The color scale represents viability with the darkest blue indicating the lowest viability, and white representing a viability of 1.0. Clusters were identified using hierarchical clustering based on average linkage and Euclidean distance. Arrows indicate estrogen-independent breast cancer cells. F. Caspase 7 activity depicted as relative fluorescent units (RFUs) 120 h post-transfection of siRNAs targeting the 25 genes in the EIPS subset, with data normalized to median activity seen with transfection of negative control siRNA. Error bars indicate S.D.
Two hundred and forty-eight “first neighbors”, defined as proteins that directly interacted with a seed protein based on experimental data, constituted a high confidence core. 12 proteins reported in the literature [26, 27] as complexed with ESR1, ESR2 or ESRRA were also included in the ER-centered network as a high confidence core. We used multiple databases reporting ER signaling interactions to identify 44 proteins as a pathway core. The Estrogen Responsive Gene Database (ERGD) [28] listed 38 high confidence genes reported as manifesting altered transcriptional responses to an estrogen stimulus. Beyond these high confidence cores, which in sum contributed 308 genes to the ERN, 323 additional genes were included based on their occurrence in at least two lower confidence sets (Figure 1A).
Screening the ERN library identifies genes specifically required for viability in estrogen-independent cell lines
To compare the cellular requirement for genes in the ERN in the context of decreased estrogen dependence and increased anti-estrogen resistance, we used a siRNA library targeting the 631 genes in the ERN to screen four cell lines: MCF7, LCC1, LCC9, and HFF (Table S2). MCF7 is an estrogen-dependent breast adenocarcinoma cell line and sensitive to treatment with the AEs tamoxifen and fulvestrant. The MCF7-derived cell line LCC1 was selected in vivo for estrogen-independence, which commonly reflects AI resistance, but remains sensitive to tamoxifen and fulvestrant. LCC9, further derived by selection from LCC1 cells, is resistant to both tamoxifen and fulvestrant [29, 30]. Human foreskin fibroblasts (HFF) do not depend on estrogen signaling, and provided a control for non-specific inhibitors of cell viability. Analysis of primary hits indicated that LCC1 and LCC9 were significantly more susceptible to loss of viability following depletion of ERN genes than were MCF7 or HFF cells. Use of a biological cut-off of 50% viability or less identified 190 candidate hits for the LCC1 cells, 117 for LCC9 cells, 9 for MCF7 cells and 1 for HFF1 cells (Figure S1, A-B).
Validation of hits obtained in LCC1 or LCC9 cells was performed using transfection of multiple siRNAs and confirmation of gene depletion by quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) (Figure S1, C-D). Following validation, 85 genes were confirmed as important for viability in LCC1 cells, 65 in LCC9 cells, and 3 in MCF7 cells; of these, 49 were common to LCC1 and LCC9, which included the 3 in MCF7 cells (Figure 1C, Table S1 and S3). Focusing on the genes required in LCC1 and/or LCC9, we assessed if these genes selectively derived from specific categories of input to the ERN. This analysis indicated that this set was enriched for siRNAs targeting genes that either interacted directly with the 5 ERN seeds, or included estrogen-responsive genes (Figure 1B).
A subset of the 49 genes specifically required in LCC1 and LCC9 cells have previously been implicated as influencing viability in breast cancer or untransformed mammary cells (Figure 1D, see references in legend; Table S3). A limited number of these genes have also been identified as contributing to cell proliferation and survival in additional cell types, suggesting they may be similarly important in breast cancers (Figure 1D). Finally, gene ontology assignment [31, 32] suggested that the set of ERN genes specifically required in LCC1 and LCC9 cells was enriched in genes annotated for roles associated with RNA polymerase, the proteasome, and mRNA splicing (Figure 1D).
Estrogen-independent pro-survival (EIPS) genes are selectively required for survival in multiple estrogen-independent cell lines
We selected 25 genes from the LCC1/LCC9 viability group for more in depth analysis (Table S3) and defined them as estrogen-independent pro-survival (EIPS) genes. To determine whether their roles in viability were specific to cells of the MCF7 lineage, or more generally reflected a requirement in estrogen-independent versus estrogen-dependent cell lines, we analyzed the consequences of depletion in five additional breast cancer cell lines: MDA-MB-231 (triple negative, estrogen-independent), T47D (ERα-positive, estrogen-dependent, and tamoxifen/fulvestrant-sensitive), T47D-co (ERα-positive, estrogen-independent, and tamoxifen/fulvestrant-resistant), BT-474 (ERα-positive, estrogen-dependent), and MCF10A (similar to normal mammary epithelial cells) (Figure 1E). Strikingly, this analysis indicated that the group of 5 estrogen-independent breast cancer cell lines MDA-MB231, LCC1, LCC9, T47D-co, and T47D clustered together in requiring EIPS genes for viability, in contrast to the estrogen-dependent cell lines MCF10A, MCF7, and BT474, in which the EIPS genes were generally less necessary for cell survival.
Knockdown of genes in the EIPS set might reduce cell viability by increasing apoptosis, decreasing cell proliferation, or both. Genes required for apoptosis resistance would be of particular interest as potential targets in cancer therapy, in contrast to those that primarily induce cytostasis. Activity levels of caspase 3/7 were assessed as surrogates for apoptotic induction following depletion of each of the EIPS genes. Knockdown of 15 genes in the group increased caspase 3/7 activities at least 2-fold in LCC1 cells, with 16 genes having such activity in LCC9 cells (Figure 1F). In general, the degree of apoptotic induction associated with each gene in the set was comparable in the two lines, with knockdown of the transcription factor CNOT1, the polymerase II subunits POLR2B and POLR2C, and the proteasome component PSMB4 having the most significant effects. Finally, while LCC1 and LCC9 grow in estrogen-independent fashion, they retain ERα and other estrogen receptors, which may retain some biological activities. Potentially, the availability of estrogen might regulate the cell survival effect of genes in the EIPS set. However, no significant difference in viability was observed in knocking down these genes in LCC9 cells grown in the presence or absence of 1 nM estradiol, and only minimal differences were observed for a small number of genes in LCC1 cells (Figure S2).
Among the set of EIPS genes, we noted that TOB1 has been previously described as a transducer of ERBB2 signaling, is upregulated by ERBB2, and is linked to poor prognosis in node-negative breast cancer [33]. These features have considerable relevance to estrogen resistance. Further, by comparing steady state mRNA expression of 45 genes active in LCC1 and/or LCC9 in 9 estrogen-dependent and 8 estrogen-independent breast cancer cell lines (Table S4 for cell line information), TOB1 was one of the top 2 genes in which mRNA expression differed most significantly in estrogen-independent versus estrogen-dependent breast cancer cell lines (Figure 2A), being higher in estrogen-dependent cell lines. Additionally, TOB1 was amplified and/or overexpressed in 14% of 979 invasive breast cancers assembled in The Cancer Genome Atlas (TCGA) (Figure 2B and Figure S3, A). In analysis of TCGA data, TOB1 and ESR1 mRNA expression and promoter methylation correlated positively, suggesting a particular relevance in this setting (Figure S3, B-C). We also found significant correlation in promoter methylation and mRNA expression for TOB1 and ERBB2, further supporting functional link between TOB1 and HER2 signaling.
Figure 2. Interaction of TOB1 with the signaling landscape of estrogen-dependent and estrogen-independent cells.
A. Comparison of steady state mRNA expression of 45 genes active in LCC1 and/or LCC9 in 9 estrogen-dependent and 8 estrogen-independent breast cancer cell lines (Table S4). Data obtained from Cancer Cell Line Encyclopedia [8] [4]. Unsupervised clustering was performed using MeV. The color scale represents mRNA levels with the darkest blue indicating the lowest level and the darkest yellow indicating the highest level. Red arrows indicate top 2 genes with most significant different expression between estrogen-dependent and –independent cell lines. B. Percentage of gene alteration in Breast Invasive Carcinoma from TCGA (1033 tumors). C-F, TOB1 is not sufficient to induce estrogen independence in MCF7 cells. TOB1-expression plasmid pLOC-TOB1, or vector control, and VSV-Gal vectors were packaged into HEK 293 cells to produce lentivirus containing TOB1. Cells were transduced with virus and selected by Blasticidin S (10 μg/ml for MCF7, 11 μg/mL for LCC1 and LCC9 cells). TOB1 protein level was detected by western blotting. Growth of TOB1-overexpressed cells was determined by CellTiter-Blue. C. Levels of TOB1 protein and p-TOB1 in TOB1-overexpressed MCF7 cells. Representative images of western blots are shown. D. Quantitation of protein levels shown in C. E. Growth of MCF7 cells expressing vector control DNA or TOB1 in the absence of estrogen. Cell growth measurements were determined with CellTiterBlue treatment. F. Proliferation of MCF7 cells expressing vector control or TOB1 in the presence of 1nM estradiol. TOB1 ORF1 and ORF2 were two constructs used to obtain enhanced TOB1 exogenous expression.
Interestingly, overexpression of TOB1 was not sufficient to cause estrogen-independent proliferation of MCF7 cells. When TOB1 was overexpressed by lentiviral infection in MCF7 cells (Figure 2C,D), cellular proliferation was not increased in the absence (Figure 2E) or presence (Figure 2F) of 1 nM estradiol. These results suggest that the interaction of TOB1 with other proteins differing between estrogen-dependent and estrogen-independent cell lines was likely to support its role in estrogen-independent growth. We further investigated the action of TOB1 in this context.
TOB1 control of cell cycle signaling differs in MCF7 versus LCC1 and LCC9 cells
Extending our earlier results, a clonogenic analysis indicated that depletion of TOB1 severely depressed the outgrowth of colonies of LCC1 and LCC9, but not MCF7 cells (Figure 3A,B). Overexpression of TOB1 in LCC1 and LCC9 cells had no effects on cell growth (Figure S4). Moreover, even higher concentrations of estradiol could not rescue the cell proliferation arrest induced by TOB1 knockdown (Figure S5). Interestingly, cell cycle analysis indicated a different basal compartmentalization, with MCF7 having a much higher proportion of cells in G1 phase (57% for MCF7, versus 34% for LCC1 and 37% for LCC9), and LCC1 and LCC9 enriched in S phase (25% for MCF7, versus 46% for LCC1 and 47% for LCC9) (Figure 3C,D). Because TOB1 inhibition has been previously reported to block G0/G1 to S transition [34], we next investigated the effects of TOB1 knockdown on cell cycle distribution. siRNA depletion of TOB1 had a minimal impact on cell cycle distribution in MCF7 cells, but, unexpectedly, caused a significant arrest from G1 to S phase in LCC1 and LCC9 cells (Figure 3C,D). Western analysis indicated that under steady-state growth conditions, the TOB1 protein was overexpressed (2.8 and 2.7 fold) in LCC1 and LCC9 cells as compared with MCF7 cells (Figure 3E). This upregulation was transcriptional and partially post-transcriptional, as TOB1 mRNA levels slightly increased in LCC1 and LCC9 cells (1.5 fold) (Figure S6A). Phosphorylation on TOB1S164 has previously been associated with reduced anti-proliferative activity [34]; however, although there are higher levels of phosphorylated TOB1 in LCC1 and LCC9 cells (Figure 3F), our analysis found no differences in the ratio of this phosphorylation versus total protein in MCF7 and LCC1 cells, but lower in LCC9 cells (Figure 3G). The relative difference in regulatory phosphorylation in the two contexts implied an alternative explanation for differing TOB1 protein functionality associated with acquisition of estrogen independence.
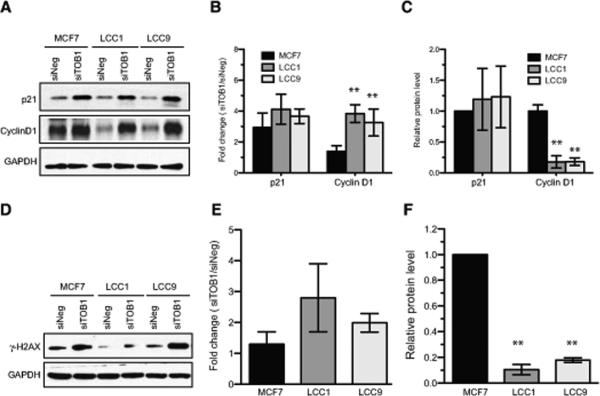
Figure 3. Effects of TOB1 knockdown in MCF7 cells versus LCC1 and LCC9 cells.
A-B. TOB1 knockdown effects on cell proliferation in MCF7, LCC1 and LCC9 cells. 20 nM negative control siRNAs (siNEG) or TOB1 siRNAs (siTOB1) were transfected into cells. After 144 h culture, cells in 96-well plates were stained with crystal violet. A. Representative images of stained cells are shown. B. The cell viability of TOB1 knocked-down cells was quantitated and is presented as average +/− S.D. from 4 independent experiments. Cell viability of siNEG-transfected cells was defined as 100% in each cell line. ** p<0.01 vs. siNEG. C. TOB1 siRNA induces G1/S transition arrest in LCC1 and LCC9, but not in MCF7 cells. 20 nM siNEG or siTOB1 were transfected into cells, and cells were cultured for 48 h and 96 h. BrdU incorporation and flow cytometry were used to measure percentage of replicating cells (numbers shown). 3 independent experiments were repeated. Dot plots shown here were from one typical experiment. D. Cell cycle distribution in siTOB1-transfected cells. Three independent experiments were repeated. Error bar indicates S.D. values. ** p<0.05 vs. siNEG. E. Baseline expression and phosphorylation levels of TOB1 protein in 3 cell lines. Western blots were performed to detect protein levels. Representative images (n=3) of blots are shown. F. Relative protein levels of TOB1 and phospho-TOB1 (Serine 164) were analyzed from 3 independent experiments. Protein level of MCF7 was defined as 1. Error bar indicates S.D. value. * p<0.05 vs. MCF7. G. Relative ratio of phospho-TOB1 (Serine 164) level vs. total TOB1 protein level. The relative ratio of MCF7 was defined as 1. Error bar indicates S.D. value. * p<0.05 vs. MCF7.
Accordingly, we examined the effects of TOB1 knockdown on G1 regulatory proteins. TOB1 has been reported to inhibit cell cycle progression by negatively regulating cyclin D1 levels [34, 35]. Strikingly, depletion of TOB1 markedly elevated the expression of cyclin D1 in LCC1 and LCC9 cells as compared with MCF7 cells (Figure 4A, B). In contrast, TOB1 knockdown increased p21 expression in all cell lines (Figure 4A-C).
Figure 4. TOB1-dependent molecular signaling changes.
A-C. Induction of p21 and Cyclin D1 following TOB1 knockdown. siNEG or siTOB1 were transfected in MCF7, LCC1 or LCC9 cell lines. A. Western blot analysis was performed 48 hours later to determine protein levels. B. Fold increase of TOB1 knockdown-induced protein level was quantitated and plotted. Data are representative of at least 3 independent experiments and shown as mean values +/− S.D. ** p<0.01 vs. MCF7. C. Basal level of p21 and Cyclin D1 in MCF7, LCC1 and LCC9 cells. ** p<0.01 vs. MCF7. D-F DNA damage (phosphorylation of histone H2AX at Serine 139, γ-H2AX) induced by TOB1 knockdown. siNEG or siTOB1 were transfected into 3 cell lines for 48 h. Cell lysates were analyzed for level of γ-H2AX. D. Representative image of Western blotting. E. Fold changes of siTOB1-induced γ-H2AX were quantitated in 3 independent experiments. Data are shown as mean values ± S.D. F. Basal level of γ-H2AX in MCF7, LCC1 and LCC9 cells. ** p<0.01 vs. MCF7.
A number of studies have indicated that TOB1 expression regulates radio-sensitivity and DNA damage responses in cancer cells (e.g., [36]), based on actions with the DNA repair machinery and in regulation of core survival signaling pathways. We found that basal levels of γ-H2AX (phosphorylated histone H2AX at serine 139)[37], an indicator of DNA damage, were higher in MCF7 than in LCC1 or LCC9 cells, but were equally enhanced by TOB1 knockdown in all three lines (Figure 4D-F).
Defining differences in the TOB1-response network in estrogen-independent versus -dependent cells
To extensively explore the roles of TOB1 in the survival of estrogen-independent cells and the impact on the signaling architecture of cancer-focused pathways, we then used reverse-phase protein array (RPPA) analysis to profile the basal expression and activation/phosphorylation of 76 key signaling proteins in MCF7, LCC1, and LCC9 cells (Figure S7A). Not surprisingly, cultured MCF7 cells exhibited significant differences from their estrogen-independent counterparts, with significantly less activation of important EGFR network proteins such as mTOR, STAT3, S6 kinase Shc, EGFR, ERBB2, PTEN, and ERBB3. MCF7 cells also demonstrated relative activation of other signaling molecules such as Mek1/2, c-Abl, c-Kit, Stat5, IGF-1R and B-Raf. The estrogen-independent cell lines exhibited fewer variations between each other, but there was more activation of Bad, Met, VEGFR2, and Aurora Kinase A/B/C in LCC9 cells than in LCC1 cells. In order to focus the difference of estrogen-independent versus estrogen-dependent cells, molecules that were common in LCC1 and LCC9, but significantly different in MCF7 cells were uploaded for canonical pathway enrichment and network analysis (Figure S7B, C). As shown, the ERBB signaling pathway and its related PI3K/AKT, Erk/MAPK and mTOR signaling pathways were changed in estrogen-independent cells versus MCF7 cells.
We next performed RPPA analysis in MCF7, LCC1, and LCC9 cells 48 hours after siRNA depletion of TOB1 or a negative control siRNA (Figure 5 and Figures S8). Figure 5A describes the expressions of each unmodified or phosphorylated protein in MCF7, LCC1 and LCC9 as a function in the absence (siNeg) and presence (siTOB1) of TOB1 depletion. Figure 5B then describes the fold-changes in protein or phosphorylated expression in each cell line as a specific consequence of TOB1 depletion. Consistent with the data shown in Figure 4, RPPA analysis confirmed that p21 and cyclin D1 were upregulated by siTOB1 knockdown. In order to address the differing responses to TOB1 inhibition in estrogen-independent cells versus estrogen-dependent cells, two groups of molecules were further analyzed. The first group included molecules that were activated by TOB1 knockdown in estrogen-independent cells, but reduced in MCF7 cells, such as Src, ERBB2, PKC, ERBB3, AMPKa, Erk1/2 and IGF-1R/IR (Figure 5B, underlined). The second group contained 4EBP1, MEK1/2, CREB, FADD and Ki67, which were inactivated or downregulated by siTOB1 in estrogen-independent cells, but activated in MCF7 cells (Figure 5B, black frame). The canonical pathway enrichment and network analysis of these genes identified that the TOB1-response network in estrogen-independent cells differed from the parental estrogen-dependent MCF7 cells. The ERBB, growth hormone, IGF-1R and mTOR signaling pathways were selectively activated by TOB1 knockdown in estrogen-independent cells (Figure 5C and E). In contrast, the FGF signaling and death receptor pathways were inactivated by TOB1 inhibition in LCC1 and LCC9 cells (Figure 5D and E).
Figure 5. Definition of signaling networks significantly regulated by TOB1 knockdown in estrogen-independent cells by RPPA analysis.
Forty eight hours after transfection of siTOB1 or siNEG, lysates of MCF7, LCC1 and LCC9 cells were assessed by reverse phase protein array (RPPA) analysis (see Methods). A. Heatmap representing levels of proteins or phosphorylated proteins in negative control or TOB1 siRNA-transfected MCF7, LCC1, and LCC9 cells. Data shown as average from triplicates of each RPPA samples normalized by row-based z-score. B. Heatmap representing log-2 normalized fold change values of proteins or phospho-proteins (siTOB1 vs. siNEG). Blue color indicated decrease and yellow color indicated increase. Purple underlines indicate molecules that were up-regulated by siTOB1 in estrogen-independent cells, but down-regulated in MCF7 cells. The black frame indicates molecules that were down-regulated by siTOB1 in estrogen-independent cells, but up-regulated in MCF7 cells. * FDR-adjusted p-values<0.05, versus MCF7. C-D. The network representing the small group of genes showing differences between MCF7 and estrogen-independent cells as derived by Ingenuity. C. Signaling network derived from proteins SRC, ERBB2, PKCα, ERBB3, Cleaved Caspase 9, AMPKα, ERK1/2, IGF1R (underlined in panel B). D. Signaling network derived from proteins 4EBP1, MEK1/2, CREB, FADD, Ki67 (contained within the frame – panel B). For each colored gene node, the color on the left side indicates regulation in MCF7 cells, and the right, in estrogen-independent cells. Red, up-regulated by siTOB1; green, down-regulated by siTOB1. Non-colored gene nodes were automatically introduced by Ingenuity. E. The canonical pathways enriched in genes differentially regulated by siTOB1 in estrogen-independent cells versus MCF7 cells as identified by Ingenuity. * indicates pathways enriched in molecules down-regulated by siTOB1 in estrogen-independent cells. Others were pathways enriched in molecules up-regulated by siTOB1 in estrogen-independent cells.
In addition, we used RT-PCR to analyze a panel of 84 genes relevant to estrogen receptor signaling for mRNA changes in LCC1 and LCC9 versus MCF7 cells, and specifically associated with TOB1 depletion (Figure S9). We first asked which genes showed statistically distinct changes in mRNA expression in the LCC1 and LCC9 versus the MCF7 cells (Figure S9A, B). We then asked which of these changes were reversed in cells following knockdown of TOB1 (Figure S9C, D); and additionally, which genes showed distinctive patterns of response to TOB1 knockdown, whether they originally differed in expression in the MCF7 versus LCC1 and LCC9 cell lines, or not (Figure S9B, D). As these data indicate, a number of genes relevant to the proliferation of survival of breast tumors have significantly altered expression in the LCC1 and LCC9 versus the MCF7 cell line, including those relevant to TGF-beta/BMP, WNT, mitogen and invasion signaling (TGFB3, IGFBP5, BMP4, WISP2, NRP1, and others); the estrogen receptor itself (ESR1) is induced in the resistant cell lines.
Of this group of genes significantly altered in the ER-resistant derivative lines, knockdown of TOB1 did reverse changes in the expression of some, such as WISP2, FOS, CYP19A and TGFA. However, this effect of TOB1 depletion was only seen in LCC1 cells, not in LCC9 cells, and hence is not directly linked to action of TOB1 in the context of resistance to estrogen. However, in comparing the effect of TOB1 knockdown in the broader list of ER-associated genes, it was notable that this knockdown caused significant reduction of expression of genes such as the ER-interacting protein NROB2, the differentiation-promoting factor WNT5A, and a small set of other genes in MCF7 cells, but not in LCC1 or LCC9 cells. These data suggest that one action of TOB1 may be conditioning the landscape of genes available to support the estrogen signaling apparatus.
TOB1 interaction with AKT/mTOR survival signaling differs in MCF7 versus LCC1 and LCC9 cells, influencing response to pathway-targeted agents
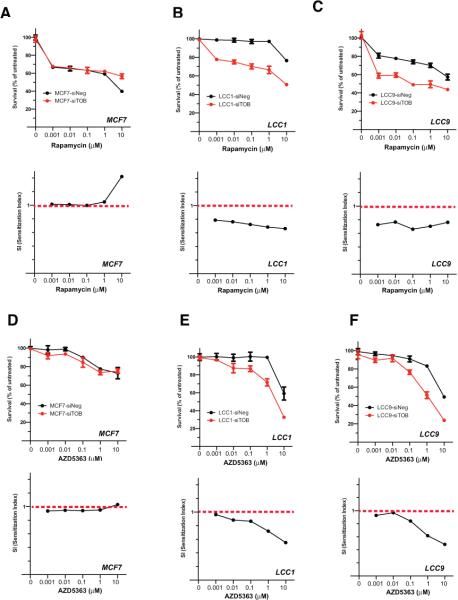
TOB1 overexpression has been reported to reduce MEK/ERK- and AKT-dependent signaling [38], which is potentially relevant to a selective role in estrogen independence [10]. Moreover, RPPA data and canonical pathway analysis identified that the mTOR signaling pathway was activated by TOB1 knockdown in LCC1 and LCC9 cells (Figure 5). We next asked whether TOB1 might influence the response of these cells to inhibitors of mTOR (rapamycin), AKT (AZD5363) or MEK/ERK (U0126). Knockdown of TOB1 significantly sensitized LCC1 and LCC9 cells to rapamycin (Figure 6B, C) and AZD5363 (Figure 6E, F), while having no effect on response to U0126 (Figure S10, and importantly, no effect on response to any drug in MCF7 cells (Figures 6A, D). Therefore, TOB1 knockdown in estrogen-independent breast cancer cells potentiates the anti-tumor effects of clinically relevant signaling inhibitors in this setting. These data also demonstrated that in LCC1 and LCC9 cells, TOB1 plays proliferative functions parallel with AKT and mTOR signaling pathways, but in the same signaling pathway as ERK.
Figure 6. Effects of TOB1 knockdown on the cytotoxicity of mTOR and AKT inhibitors in MCF7, LCC1 or LCC9 cells.
SiNEG or siTOB1-transfected cells were plated into 96-well plates and treated with the mTOR inhibitor, Rapamycin, or the AKT inhibitor, AZD5363, at varying concentrations for 144 h. Cell viability was determined by the CellTiter-Blue method. A-C. Cell viability after rapamycin treatment in negative control or TOB1 siRNAs-transfected MCF7 (A), LCC1 (B) and LCC9 (C) cells. Upper panels, cell viability; bottom panel, sensitization index. D-F. Cell viability after AZD5363 treatment in siNEG or siTOB1 transfected MCF7 (D), LCC1 (E) and LCC9 (F) cells. Upper panels, cell viability; bottom panel, sensitization index.
Finally, TOB1 is known to regulate ERBB2/HER2 signaling, raising the possibility that inhibition of ERBB2 may phenocopy the effect of depleting TOB1. We compared the effect of lapatinib, a small molecule inhibitor of ERBB2 and ERBB1/EGFR, versus other targeted signaling proteins, in MCF7 versus LCC1 and LCC9 cells (Figure S11). Lapatinib had no effect in MCF7 cells, but in both LCC1 and LCC9 cells, 10 μM lapatinib reduced proliferation to 30% of untreated cells, representing an effect similar to that of TOB1 knockdown. However, in the siTOB1-transfected cells, no significant sensitization to lapatinib was observed when compared with siNEG control cells. This pattern was distinctive, and likely due to action of lapatinib against ERBB2/HER2, as we also assessed inhibitors of a group of kinases in the HER2 pathway, including EGFR, RAS, and MEK, and no such distinguishing difference was identified in cells treated with the EGFR inhibitor erlotinib (Figure S11). These data indicated that the proliferative function of TOB1 is involved in ERBBB2 related signaling.
DISCUSSION
The issue of estrogen independence is of great importance in the clinical management of breast cancer. Using a functional genomics screening strategy, we show that numerous genes in an estrogen response-centered network are engaged when estrogen-dependent breast cancer cells develop estrogen-independence. One of those genes, TOB1, was selected for deeper analysis because of its potential interactions with other important signaling proteins relevant to breast cancer, and because of its overexpression in some ER-dependent tumors. A focused protein pathway activation mapping approach underpinned by protein phosphorylation analysis and mRNA profiling then provided quantitative information about the activation levels of canonical cell line-specific signaling events. This approach also assessed the effects of TOB1 knockdown on those targets and pathways, and identified signaling changes caused by TOB1 knockdown in estrogen-independent cells. In initially estrogen-dependent tumors, once anti-estrogen selection is applied, elevated expression of TOB1 based on transcriptional and post-transcriptional mechanisms is associated with the development of resistance.
Intriguingly, besides showing that the ability of TOB1 to influence the activity of canonical effectors such as Cyclin D1 differs between estrogen-dependent and -independent cells, our work identifies an extended chain of signaling proteins culminating with different expression profiles of estrogen-related genes that are differentially influenced by TOB1 in these two environments. Our data also show that depletion of TOB1 selectively sensitizes estrogen-independent cells to clinically important inhibitors of AKT and mTOR. Given growing evidence linking elevated expression of TOB1 to invasive breast cancers (Figure 2B), these studies provide important and unexpected insights into potentially targetable mechanisms that underlie a problem that is at the core of many breast cancer deaths.
This is the first study to link TOB1 to the biology of estrogen resistance, since its initial identification was as a growth inhibitory protein that becomes less active in cells overexpressing ERBB2 [39]. Erk1/2 and its downstream substrate p90RSK1 could phosphorylate TOB1, relieving its growth inhibitory activity [34, 40]. In our studies, the activation of ERBB2 and Erk1/2 by TOB1 inhibition indicated a reverse feedback from a downstream target (Figure 7B, right panel). Interestingly, another downstream target of ERBB2 signaling, the PI3K/AKT/mTOR pathway, was activated by TOB1 knockdown in estrogen-independent cells. It is plausible that TOB1 functions in parallel with the PI3K/AKT/mTOR pathway because TOB1 knockdown potentiates the effects of AKT or mTOR inhibitors. Moreover, TOB1 inhibition reduced activation of the cCAMP-response element binding protein (CREB), which is PI3K/Akt dependent [41]. Coupled transcriptional analysis also indicated that TOB1 depletion differentially affected the expression profile of estrogen pathway related genes, some of which were relevant the mitogenic signaling pathways identified in RPPA analysis, further reinforcing the importance of these pathways. In particular, the mRNA expression differences suggest that one action of TOB1 may be conditioning the landscape of genes available to support proliferation and survival signaling.
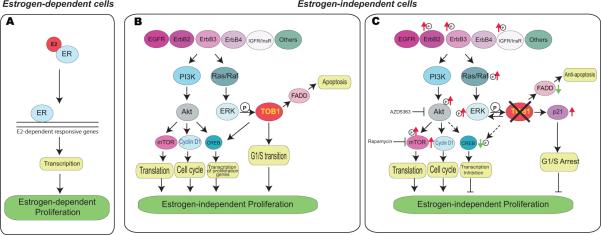
Figure 7. Schematic model of TOB1 function in estrogen-independent cell growth in breast cancer cells.
A. The estrogen receptor-dominated cell growth in estrogen-dependent cells. E2, estradiol. B-C. The TOB1-related signaling pathways in estrogen-independent cells. B: Briefly, Growth factors receptors including EGFR, ERBB family, IGF1R etc. activate two pathways to promote estrogen-independent cell proliferation. One is mediated by PI3K/AKT, which regulates translation and transcription of proliferative genes, and cell cycle progression, then supports estrogen-independent growth. Ras/Raf/MAPK mediates the second signaling pathway downstream of growth factor receptors. TOB1 is phosphorylated by Erk1/2 to promote estrogen-independent proliferation by facilitating G1/S cell cycle phase transition. C: In the presence of TOB1 inhibition, up-regulation of p21 results in G1/S arrest. Additionally, transcription can be inhibited by TOB1 knockdown. Reduction of CreB1 phosphorylation by TOB1 inhibition can reduce its proliferative target gene transcription, and then inhibit cell growth. However, growth factor receptors such as ERBB2 and IGFIR are activated as a reverse feedback mechanism, the PI3/AKT pathway is activated by TOB1 inhibition. Inhibition of AKT or mTOR potentiates the effects of TOB1 inhibition on cell proliferation in estrogen-independent cells.
In addition, the G1/S arrest induced by TOB1 inhibition, which is mediated by up-regulation of p21 indicates TOB1 supports estrogen-independent cell growth by promoting G1/S cell cycle transition and suggests that cells treated with siTOB1s might senesce[42] because TOB1 inhibition could not induce apoptosis (Figure 1F). As summarized in Figure 7, TOB1 plays a proliferative function in a growth factor receptor network in estrogen-independent cells. Briefly, the transition from estrogen-dependence (Figure 7A, left panel) to estrogen-independence (Figure 7B, left panel) utilizes growth factors including EGFR, the ERBB family and IGF1R to activate at least two pathways that promote estrogen-independent cell proliferation. One is mediated by PI3K/AKT, which regulates translation and transcription of proliferative genes (reviewed in [43]), and cell cycle progression to support estrogen-independent growth. The second signaling pathway, which is downstream of these growth factors, is mediated through Ras/Raf/MAPK signaling [44, 45]. TOB1 is phosphorylated by Erk1/2 to promote estrogen-independent proliferation by facilitating G1/S cell cycle transition. TOB1 is also involved in CREB1 activation to regulate its impact on target gene transcription. When TOB1 is knocked down (Figure 7B, right panel) these pro-proliferative activities are reversed, in concert with p21-mediated G1/S arrest.
Various anti-estrogen agents have been employed in sequence to address the estrogen-independence and acquisition of anti-estrogen resistance that lead to relapse and eventual death in many women with ER+ breast cancer. Despite a wealth of information about specific mechanisms that might be attacked [5-13, 46], the context in which those mechanisms are addressed (typically as mono-therapy, and rarely in combinations designed to disable resistance mechanisms) is so complex that additional resistant cell sub-populations almost always emerge to defeat these therapies. Recent studies show that anti-estrogens can be combined with cell cycle-directed treatments with promising anti-tumor activity [47, 48]. Such data are in accord with our studies, and that of many others showing activation of components of the ERBB signaling network in hormone-resistant breast cancer cells. Our data suggest that many proteins are likely to collaborate to generate this estrogen-resistant phenotype, making it imperative to identify critical nodes that can be interdicted alone, or in combination to disable this process. Our work identified that TOB1 supports the estrogen-independent cell survival, and that its function is closely linked to that of ERBB2/HER2, with HER2 inhibition partially phenocopying TOB1 knockdown. However, while TOB1 is not the sole mediator of estrogen-independence, its known anti-proliferative functions within the ERBB signaling network [33, 34, 40] argue for its further study to investigate how it and its collaborating proteins might be disabled to prevent the emergence of estrogen-independence in ER+ breast cancer. In the future, it will be important to extend the analysis of TOB1 and the signaling paradigm identified in this study to the analysis of other independent systems for intrinsic versus acquired estrogen resistance.
These studies demonstrate the diversity and flexibility of survival mechanisms as breast cancer cells develop estrogen independence. Interestingly, many different genes contribute to survival in this setting, challenging previous conventions that estrogen independence and resistance to anti-estrogen drugs involve the gradual accumulation of resistance mechanisms. Because TOB1 is not an identified druggable target, these results cannot be validated in vivo at this time. However, the translational significance of this new molecular mechanism of estrogen independence in human breast cancer will be fully determined when TOB1 inhibitors are developed to enable in vivo validation studies.
MATERIALS AND METHODS
Cell Culture and Chemicals
Cell lines used in these studies are from Lombardi Comprehensive Cancer Center, Georgetown University. Cells are tested for mycoplasma negative. For information on medium and assay reagents used for each cell line see Supplementary Information. HEK293T cells were grown in DMEM containing 10%FBS and 2mM L-glutamine. Cell lines were maintained at 37°C and 5% CO2. Estradiol was obtained from Sigma (St. Louis, MO) and prepared at 1 mM in ethanol. AZD5363 and rapamycin were purchased from Selleckchem (Houston, TX) and prepared at 10 mM in DMSO. Drug stock aliquots were kept at −20 °C. All drugs were diluted to desired concentrations in full medium immediately before each experiment. The final DMSO concentrations did not exceed 0.1%.
RNAi Screening
An ER-centered network was developed using the open source software tool, Cytoscape, using methods described in detail in [15]. The ER-centered siRNA library was custom-ordered from Qiagen (Germantown, MD) in a 96-well plate format with a single well representing one of the 631 genes identified as part of the ER-centered network. Each well contained 2-pooled siRNAs, targeting different sequences of the same gene. siRNAs were resuspended in RNase free water at 1 μM. Cell density and transfection reagent for cells were listed in Supplementary Information. Cells were reversed transfected with the ER-centered siRNA library at 20 nM and incubated for 144 hours. Viability for each targeted gene was measured as described below and normalized to median value of 14 non-silencing controls on the plate. In each cell line, RNAi screening were repeated three or four times.
siRNA Validation
Viability data from three or four experimental replicates experiments were statistically analyzed. Those siRNAs with adjusted p values < 0.05 were considered to be significantly different from the siNEG control, and were considered to be hits if they caused at a loss of 50% viability or more in target cell lines, as reflecting a robust biological effect. Using this cut-off, only about 1% of all gene knockdowns met the criteria for hits in the estrogen-dependent MCF7 cell line. For each hit identified, four different siRNAs (Qiagen, MD) targeting the same gene were tested for validation studies in individual wells. Two out of the four siRNAs were the same target sequences as the siRNAs in the screen, when available. The other two siRNAs were new sequences to test. Cells were screened as described above. If at least two out of four of the siRNAs tested reduced viability by at least 50%, the candidate passed validation as a putative hit.
Caspase 7 activity
Cells were reversed transfected with siRNAs at 20nM in triplicate and incubated for 120 hours. Caspase 7 activity was measured using Apo-ONE as described by Promega (#G7790, Madison, WI). Caspase 7 activity for each gene knockdown was calculated by normalizing to median fluorescence values of non-silencing controls on the plate.
Reverse Phase Protein Array
TOB1 or negative scrambled siRNAs with final concentration of 10 nM were reverse-transfected into cells for 48 h. At each sample, triplicates of each transfection were harvested for RPPA analysis [49]. Antibodies used for RPPA were listed in Supplementary Information.
Bromodeoxyuridine Incorporation
After 30 min of pulse labeling with 50 μM bromodeoxyuridine (BrdUrd, BD Biosciences, San Jose, CA), cells were collected, stained with fluorescein isothiocyanate (FITC)-conjugated anti-BrdUrd antibody (BD Biosciences, #347583, San Jose, CA) and analyzed with a FACScan flow cytometer (BD Biosciences)[50].
Western blot
Cells were washed with ice-cold PBS, and lysed. 25~50 μg of protein was used for Western blotting. Primary antibodies against TOB1 (#ab79372) and phosphorylated-TOB1 (#ab78915) were purchased from Abcam Inc. (Boston, MA). Antibodies against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (#5174), p21 (#2947), and Cyclin D1 (#2922) were purchased from Cell Signaling Tech. (Beverly, MA). Anti-phosphorylated-H2AX (γH2AX, clone JBW301) was purchased from Upstate Biotech (Millipore, Billerica, NY). Shown are the representative data from separate experiments. Images were processed by Adobe Photoshop software.
Cell Viability Assay
CellTiter Blue® (Promega, #G8081, Madison, WI) viability assay or CellTiter 96® AQueous One Solution Cell Proliferation assay (Promega, #3582) was used to measure cell viability according to manufacture's manuals. For crystal violet staining method, cells were stained with 0.52% crystal violet for 10 min and rinsed with ddH2O. Crystal violet was dissolved with 100 mM sodium citrate and absorbance at 570 nm was read. Cell viability was calculated as a percentage of the untreated cells.
TOB1 overexpression in MCF7 cells
TOB1 contained within pLOC-TURBO RFP lentiviral constructs were obtained from Thermo Scientific Open Biosystems (#OHS5834-EG10140, Pittsburgh, PA). HEK293T cells were transfected at 60-70% confluence with 6 μg pLOC-TOB1 (or empty vector), 3 μg psPAX2 and 0.3 μg VSV-G plasmids using FuGENE6 transfection reagent (Promega, #E2691). Cells were infected at 30% confluence using 3 mL virus containing media, 1 mL normal growth media and 3.2 μg of polybrene. Media was replaced 24 hrs following transduction with normal growth media and at 48 hrs with media containing blasticidin S (Sigma; #15205, 10 μg/ml for MCF7, 11 μg/mL for LCC1 and LCC9 cells). Selection continued for 7 days and TOB1 overexpression was confirmed by Western blotting.
Network analysis
Canonical pathway enrichment, network analysis and gene interaction networks were generated using IPA (Ingenuity Systems, Redwood City, CA, USA; http://www.Ingenuity.com). Genes of interest were uploaded into the application.
Statistical analysis
All results are presented as mean ± S.D. The results were assessed for appropriate distribution and similar variation prior to significance testing. Differences between samples were assessed using the Student's t-test and p < 0.05 indicates significance.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the Flow Cytometry & Cell Sorting Shared Resource at Lombardi Comprehensive Cancer Center, which is partially supported by NIH/NCI grant P30-CA051008. We also thank Wei Xu, Alan Zwart, David Goldstein, Annie Zuo and Yuri Gusev for their technical assistance. The authors were supported by R01CA050633, CA51880, U54 CA149147 (to LMW), R01CA63366 and R21CA181287 (to EAG), and by the subsidy of the Russian Government to support the program of competitive growth of Kazan Federal University (to IS).
Footnotes
CONFLICT OF INTEREST
All authors declare no conflict of interest with regard to this manuscript.
REFERENCES
- 1.American_Cancer_Society . Cancer Facts & Figures 2013. American Cancer Society; Atlanta: 2013. [Google Scholar]
- 2.Clarke R, et al. Antiestrogen resistance in breast cancer and the role of estrogen receptor signaling. Oncogene. 2003;22(47):7316–39. doi: 10.1038/sj.onc.1206937. [DOI] [PubMed] [Google Scholar]
- 3.Levin ER. Membrane oestrogen receptor alpha signalling to cell functions. The Journal of physiology. 2009;587(Pt 21):5019–23. doi: 10.1113/jphysiol.2009.177097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jansen MP, et al. Hallmarks of aromatase inhibitor drug resistance revealed by epigenetic profiling in breast cancer. Cancer research. 2013;73(22):6632–41. doi: 10.1158/0008-5472.CAN-13-0704. [DOI] [PubMed] [Google Scholar]
- 6.Toy W, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nature genetics. 2013;45(12):1439–45. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson DR, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nature genetics. 2013;45(12):1446–51. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britton DJ, et al. Bidirectional cross talk between ERalpha and EGFR signalling pathways regulates tamoxifen-resistant growth. Breast cancer research and treatment. 2006;96(2):131–46. doi: 10.1007/s10549-005-9070-2. [DOI] [PubMed] [Google Scholar]
- 9.Fox EM, et al. A kinome-wide screen identifies the insulin/IGF-I receptor pathway as a mechanism of escape from hormone dependence in breast cancer. Cancer research. 2011;71(21):6773–84. doi: 10.1158/0008-5472.CAN-11-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun M, et al. Phosphatidylinositol-3-OH Kinase (PI3K)/AKT2, activated in breast cancer, regulates and is induced by estrogen receptor alpha (ERalpha) via interaction between ERalpha and PI3K. Cancer research. 2001;61(16):5985–91. [PubMed] [Google Scholar]
- 11.Bostner J, et al. Activation of Akt, mTOR, and the estrogen receptor as a signature to predict tamoxifen treatment benefit. Breast cancer research and treatment. 2013;137(2):397–406. doi: 10.1007/s10549-012-2376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nehra R, et al. BCL2 and CASP8 regulation by NF-kappaB differentially affect mitochondrial function and cell fate in antiestrogen-sensitive and -resistant breast cancer cells. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24(6):2040–55. doi: 10.1096/fj.09-138305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Becerra R, et al. Mechanisms of Resistance to Endocrine Therapy in Breast Cancer: Focus on Signaling Pathways, miRNAs and Genetically Based Resistance. International journal of molecular sciences. 2012;14(1):108–45. doi: 10.3390/ijms14010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chitre M, Reimers KM. Considerations for payers in managing hormone receptor-positive advanced breast cancer. ClinicoEconomics and outcomes research : CEOR. 2014;6:331–9. doi: 10.2147/CEOR.S57214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Astsaturov I, et al. Synthetic lethal screen of an EGFR-centered network to improve targeted therapies. Science signaling. 2010;3(140):ra67. doi: 10.1126/scisignal.2001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saj A, et al. A combined ex vivo and in vivo RNAi screen for notch regulators in Drosophila reveals an extensive notch interaction network. Developmental cell. 2010;18(5):862–76. doi: 10.1016/j.devcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 17.Kim B, et al. Synthetic lethal screening reveals FGFR as one of the combinatorial targets to overcome resistance to Met-targeted therapy. Oncogene. 2014 doi: 10.1038/onc.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bader GD, Hogue CW. BIND--a data specification for storing and describing biomolecular interactions, molecular complexes and pathways. Bioinformatics. 2000;16(5):465–77. doi: 10.1093/bioinformatics/16.5.465. [DOI] [PubMed] [Google Scholar]
- 19.Stark C, et al. BioGRID: a general repository for interaction datasets. Nucleic acids research. 2006;34(Database issue):D535–9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xenarios I, et al. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic acids research. 2002;30(1):303–5. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mishra GR, et al. Human protein reference database--2006 update. Nucleic acids research. 2006;34(Database issue):D411–4. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orchard S, et al. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic acids research. 2014;42(Database issue):D358–63. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Licata L, et al. MINT, the molecular interaction database: 2012 update. Nucleic acids research. 2012;40(Database issue):D857–61. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tarcea VG, et al. Michigan molecular interactions r2: from interacting proteins to pathways. Nucleic acids research. 2009;37(Database issue):D642–6. doi: 10.1093/nar/gkn722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen LJ, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic acids research. 2009;37(Database issue):D412–6. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tateishi Y, et al. Ligand-dependent switching of ubiquitin-proteasome pathways for estrogen receptor. The EMBO journal. 2004;23(24):4813–23. doi: 10.1038/sj.emboj.7600472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Acevedo ML, Kraus WL. Mediator and p300/CBP-steroid receptor coactivator complexes have distinct roles, but function synergistically, during estrogen receptor alpha-dependent transcription with chromatin templates. Molecular and cellular biology. 2003;23(1):335–48. doi: 10.1128/MCB.23.1.335-348.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang S, Han H, Bajic VB. ERGDB: Estrogen Responsive Genes Database. Nucleic acids research. 2004;32(Database issue):D533–6. doi: 10.1093/nar/gkh083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clarke R, et al. Progression of human breast cancer cells from hormone-dependent to hormone-independent growth both in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(10):3649–53. doi: 10.1073/pnas.86.10.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brunner N, et al. MCF7/LCC9: an antiestrogen-resistant MCF-7 variant in which acquired resistance to the steroidal antiestrogen ICI 182,780 confers an early cross-resistance to the nonsteroidal antiestrogen tamoxifen. Cancer research. 1997;57(16):3486–93. [PubMed] [Google Scholar]
- 31.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Helms MW, et al. TOB1 is regulated by EGF-dependent HER2 and EGFR signaling, is highly phosphorylated, and indicates poor prognosis in node-negative breast cancer. Cancer research. 2009;69(12):5049–56. doi: 10.1158/0008-5472.CAN-08-4154. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki T, et al. Phosphorylation of three regulatory serines of Tob by Erk1 and Erk2 is required for Ras-mediated cell proliferation and transformation. Genes & development. 2002;16(11):1356–70. doi: 10.1101/gad.962802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida Y, et al. Mice lacking a transcriptional corepressor Tob are predisposed to cancer. Genes & development. 2003;17(10):1201–6. doi: 10.1101/gad.1088003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki T, et al. Inhibition of DNA damage-induced apoptosis through Cdc7-mediated stabilization of Tob. The Journal of biological chemistry. 2012;287(48):40256–65. doi: 10.1074/jbc.M112.353805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Redon CE, et al. Histone gammaH2AX and poly(ADP-ribose) as clinical pharmacodynamic biomarkers. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16(18):4532–42. doi: 10.1158/1078-0432.CCR-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Malley S, et al. TOB suppresses breast cancer tumorigenesis. International journal of cancer. Journal international du cancer. 2009;125(8):1805–13. doi: 10.1002/ijc.24490. [DOI] [PubMed] [Google Scholar]
- 39.Matsuda S, et al. Tob, a novel protein that interacts with p185erbB2, is associated with anti-proliferative activity. Oncogene. 1996;12(4):705–13. [PubMed] [Google Scholar]
- 40.Suzuki T, et al. A serine/threonine kinase p90rsk1 phosphorylates the anti-proliferative protein Tob. Genes to cells : devoted to molecular & cellular mechanisms. 2001;6(2):131–8. doi: 10.1046/j.1365-2443.2001.00406.x. [DOI] [PubMed] [Google Scholar]
- 41.Caravatta L, et al. PI3-K/Akt-dependent activation of cAMP-response element-binding (CREB) protein in Jurkat T leukemia cells treated with TRAIL. Journal of cellular physiology. 2008;214(1):192–200. doi: 10.1002/jcp.21186. [DOI] [PubMed] [Google Scholar]
- 42.Childs BG, et al. Senescence and apoptosis: dueling or complementary cell fates? EMBO Rep. 2014;15(11):1139–1153. doi: 10.15252/embr.201439245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hennessy BT, et al. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nature reviews. Drug discovery. 2005;4(12):988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 44.Porter AC, Vaillancourt RR. Tyrosine kinase receptor-activated signal transduction pathways which lead to oncogenesis. Oncogene. 1998;17(11 Reviews):1343–52. doi: 10.1038/sj.onc.1202171. [DOI] [PubMed] [Google Scholar]
- 45.Scaltriti M, Baselga J. The epidermal growth factor receptor pathway: a model for targeted therapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12(18):5268–72. doi: 10.1158/1078-0432.CCR-05-1554. [DOI] [PubMed] [Google Scholar]
- 46.Marcotte R, et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discovery. 2012;2(2):172–89. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gritsch D, et al. Tamoxifen enhances the anti-proliferative effect of roscovitine, a selective cyclin-dependent kinase inhibitor, on human ER-positive human breast cancer cells. Journal of experimental therapeutics & oncology. 2011;9(1):37–45. [PubMed] [Google Scholar]
- 48.Nair BC, et al. Roscovitine confers tumor suppressive effect on therapy-resistant breast tumor cells. Breast cancer research : BCR. 2011;13(3):R80. doi: 10.1186/bcr2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xia W, et al. An heregulin-EGFR-HER3 autocrine signaling axis can mediate acquired lapatinib resistance in HER2+ breast cancer models. Breast cancer research : BCR. 2013;15(5):R85. doi: 10.1186/bcr3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang YW, et al. Implication of checkpoint kinase-dependent up-regulation of ribonucleotide reductase R2 in DNA damage response. The Journal of biological chemistry. 2009;284(27):18085–95. doi: 10.1074/jbc.M109.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105(51):20380–5. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang da W, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server issue):W169–75. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim SY, et al. CK1epsilon is required for breast cancers dependent on beta-catenin activity. PLoS One. 2010;5(2):e8979. doi: 10.1371/journal.pone.0008979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Solimini NL, et al. Recurrent hemizygous deletions in cancers may optimize proliferative potential. Science. 2012;337(6090):104–9. doi: 10.1126/science.1219580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.