Abstract
We present data relating to the interactome of MCM9 from the nuclei of human cells. MCM9 belongs to the AAA+ superfamily, and contains an MCM domain and motifs that may confer DNA helicase activity. MCM9 has been shown to bind MCM8, and has been implicated in DNA replication and homologous recombination. However, the mechanistic basis of MCM9’s role in DNA repair is poorly understood, and proteins with which it interacts were hitherto unknown. We performed tandem affinity purification of MCM9 and its interacting proteins from nuclear extracts of human cells, followed by proteomic analysis, thereby generating a set of mass spectrometry data corresponding to the MCM9 interactome [1]. The proteomic data set comprises 29 mass spectrometry RAW files, deposited to the ProteomeXchange Consortium, and freely available from the PRIDE partner repository with the data set identifier PXD000212. A set of 22 interacting proteins identified from the proteomic data was used to create an MCM9-centered interactive network diagram, using the Cytoscape program. These data allow the scientific community to access, mine and explore the human nuclear MCM9 interactome.
Keywords: Proteomics Data, Mass Spectrometry, DNA Replication And Repair, Affinity Purification, Protein-Protein Interactions
Specifications Table
| Subject area | Biology |
| More specific subject area | Proteomics, Molecular Cell Biology |
| Type of data | Mass spectrometry RAW data files from proteomic analysis of protein complexes |
| How data were acquired | MCM9-containing protein complexes were isolated, proteolytically digested, separated by liquid chromatography, and analysed using an LTQ Velos ion-trap mass spectrometer; raw data were searched against the human IPI database using the Sequest algorithm[1]. |
| Data format | Mass spectrometry Thermo RAW data files |
| Experimental factors | Expression of tagged proteins in human cultured cells, preparation of nuclear extracts, tandem protein purification |
| Experimental features | Human MCM9 constructs, tagged at the N- or C-termini with FLAG-HA, were expressed in HeLa S3 cells, and tandem affinity purified. Complexes of MCM9 and associated proteins were analyzed by SDS-PAGE and silver staining, followed by liquid chromatography-mass spectrometry[1]. Peptides and proteins were identified by database searching. |
| Data source location | Institute of Human Genetics (IGH), CNRS, 141 rue de la Cardonille, 34396 Montpellier, France |
| Data accessibility | The data are provided in the public PRIDE repository with the dataset identifier PXD000212. The direct URL to access the data ishttp://www.ebi.ac.uk/pride/archive/projects/PXD000212 |
Value of the data
-
•
The data revealed for the first time the nuclear interactome of human MCM9.
-
•
The data allow the research community to investigate the MCM9 interactome.
-
•
The data act as a benchmark reference of MCM9 interactors for future studies.
-
•
Mining these data could produce additional discoveries about MCM9 interactors.
-
•
Analyzing the data could lead to further insights into MCM9 and its processes.
1. Data
1.1. Mass spectrometry RAW data files
We present 29 mass spectrometry RAW data files that correspond to our proteomic analysis of the human nuclear MCM9 interactome. These data files were deposited to the ProteomeXchange Consortium via the PRIDE partner repository [2] with the data set identifier PXD000212 (direct access: http://www.ebi.ac.uk/pride/archive/projects/PXD000212). Data files with names 24770.RAW to 24784.RAW correspond to N-terminally tagged MCM9; 24786.RAW to 24801.RAW correspond to C-terminally tagged MCM9.
1.2. Protein interaction network
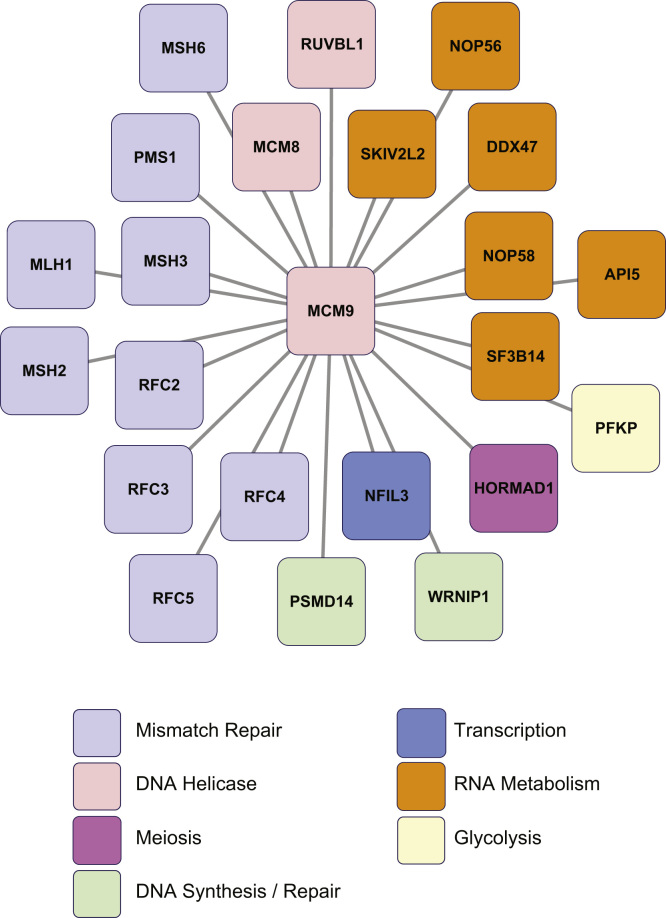
We present a protein interaction network centered on MCM9, generated using the Cytoscape program (http://www.cytoscape.org; version 3.2.1) [3] (Fig. 1), based on our proteomic data. The diagram comprises MCM9 plus 22 interacting proteins (combined from the N-terminal and C-terminal FLAG-HA tagged purifications). Contaminant or background proteins have been removed.
Fig. 1.
Nuclear interactome network for human MCM9. The network diagram, generated with Cytoscape software, shows nuclear proteins (nodes), identified by tandem affinity purification and mass spectrometry analysis, that form a complex with human MCM9. Proteins are represented by their official gene symbols, and attributed a color code according to their associated biological processes or functions, as indicated. Note that connecting lines (edges) between proteins do not imply direct physical association, but merely that proteins are members of a shared interactome centered on MCM9. A “live” interactive version of this Cytoscape figure is available on the Data in Brief web page for this article.
2. Experimental design, materials and methods
Full and detailed methods are described in our recent paper [1]. Here we present a summary for each of the steps.
2.1. Generation of cell lines expressing FLAG-HA-tagged MCM9
Stable cell lines expressing double epitope (FLAG-HA; FH) tagged MCM9 at the N- or C-termini were generated using the pOZ retroviral vectors. Vector design ensures tight coupling between the expression of tagged MCM9 and the selection marker, the interleukin-2 receptor α chain (IL2Rα) expressed at the cell surface [4]. Generation of transduction-competent retroviruses was performed by transfecting the retroviral constructs into HEK-293 cells already expressing the retroviral structural gene products required to release retroviral particles. For transduction, HeLa S3 cells were incubated with viral particles containing pOZ-FH-MCM9, pOZ-MCM9-FH, or pOZ-FH (control). Transduced HeLa S3 cells were selected using magnetic affinity beads coupled to an anti-IL2Rα antibody. Following amplification, cell clones were selected and cultured, and the expression level of double-tagged MCM9 in each clone tested by immunoblotting.
2.2. Tagged protein expression, and tandem affinity purification
For tandem affinity purification, a pool of clones for each construct that showed tagged MCM9 expression levels comparable to those of endogenous MCM9 were used. HeLa S3 cells expressing FLAG-HA-tagged MCM9 at the N- or C-termini, or an empty vector expressing FLAG-HA alone, were grown in exponential culture, and used to prepare soluble extracts of nuclear proteins according to the Dignam method [5]. Isolation of tagged MCM9 and associated proteins was performed using a tandem affinity-purification procedure based on immunoprecipitation of the FLAG and HA tags, according to Nakatani and Ogryzko [4]. In step 1, the nuclear extracts were incubated with anti-FLAG (M2) antibody-conjugated agarose beads. Beads were then washed extensively with FLAG-IP buffer (20 mM Tris–KCl, pH 7.5, 230 mM KCl, 0.03% NP40, 0.07% Tween-20, 1 mM ATP, 5 mM MgCl2), then bound proteins were eluted with two incubations in FLAG peptide. In step 2, the FLAG eluates were incubated with anti-HA antibody-conjugated beads. Beads were washed extensively with HA-IP buffer (20 mM Tris–KCl, pH 7.5, 150 mM KCl, 0.05% NP40, 0.1% Tween-20, 1 mM ATP, 5 mM MgCl2). Bound proteins were eluted with two incubations in HA peptide. MCM9-containing protein complexes were analyzed by SDS-PAGE, and revealed by silver staining.
2.3. Proteomic analysis
The silver-stained gel was sent to the Taplin Biological Mass Spectrometry Facility at Harvard Medical School (http://taplin.med.harvard.edu/) for proteomic analysis. Each SDS-PAGE gel lane was cut into slices, and each processed as a separate sample. Each gel slice was chopped into pieces of approximately 1 mm3, then in-gel trypsin digested using a modification of a standard procedure [6]. Gel pieces were washed and dehydrated with acetonitrile for 10 min, followed by removal of acetonitrile. Pieces were then completely dried in a SpeedVac centrifugal evaporator (Thermo Scientific). Gel pieces were rehydrated using a solution of 50 mM ammonium bicarbonate (pH 8.0-8.2) containing 12.5 ng/μl modified sequencing-grade trypsin (Promega) at 4 °C. After 45 min, excess trypsin was removed and replaced with 50 mM ammonium bicarbonate, just covering the gel pieces, and samples incubated at 37 °C overnight. Extraction of peptides then proceeded by removing the ammonium bicarbonate solution, followed by one wash with a solution containing 50% acetonitrile, 1% formic acid. Extracts were then dried in a SpeedVac for approximately 1 h, then stored at 4 °C until analysis. For analysis, samples were reconstituted in 5–10 μl HPLC solvent A (2.5% acetonitrile, 0.1% formic acid). A nano-scale reverse-phase HPLC capillary column was prepared by packing 5 μm C18 spherical silica beads (Michrom Bioresources) into a fused silica capillary (125 μm inner diameter×~20 cm length) with a flame-drawn tip [7]. After column equilibration, each sample was loaded onto the column using a FAMOS autosampler (LC Packings, Dionex, Thermo Scientific). A gradient was formed and peptides were eluted with increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid). Upon elution, peptides were directly subjected to electrospray ionization and injection into an LTQ Velos ion-trap mass spectrometer (Thermo Scientific). Peptides were detected, isolated and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide.
2.4. Peptide and protein identification
Identification of peptides and proteins was performed by the data analysis pipeline of the Taplin Facility. MS RAW data files were matched to entries in the human International Protein Index (IPI) database [8] using the Sequest algorithm [9]. All accepted peptides have a cross-correlation (Xcorr) score of at least 0.5. Identified peptides, Xcorr scores and corresponding protein identifiers (IDs) for each sample were presented as HTML tables accessible via a web browser.
2.5. Data post-processing and data-mining
Peptide data were copied from HTML tables and pasted into Microsoft Excel spreadsheets for further processing and analysis. Identified proteins were mapped from their IPI to equivalent Swiss-Prot [10] identifiers using the ID mapping functionality of the UniProt website (http://www.uniprot.org/uploadlists/). Percentage sequence coverage, and numbers of non-distinct and distinct peptides were calculated using the Protein Coverage Summarizer program from the Pan-Omics Research division of the Pacific Northwest National Laboratory (http://omics.pnl.gov/software/protein-coverage-summarizer). UniProt IDs were used to generate a hyperlinked data table, as described [11]. A subset of identified proteins was classified as being “background or contaminant” (at the level of gene symbols) based on two reference sources: the set of “contaminant” proteins published by the MitoCheck consortium [12], and the CRAPome database (http://www.crapome.org/) [13]. Protein entries classified as “background or contaminants” were assigned labels in separate columns in the data table, enabling them to be filtered from the dataset, as required. A table of identified proteins, and a spreadsheet containing all identified proteins and peptides for each sample are available as part of our previous publication [1].
Acknowledgments
We thank the Taplin Biological Mass Spectrometry Facility for information and advice. The research leading to the generation of these data received funding from the European Research Council (FP7/2007-2013 Grant agreement no. 233339). This work was also supported by the Fondation ARC pour la Recherche sur le Cancer, and the Fondation pour la Recherche Médicale en France (FRM). P.C. and J.R.A.H. were supported by post-doctoral fellowships from the FRM. P.C. was also supported by a postdoctoral fellowship from the ARC. D.L. was supported by a studentship from the FRM and the Agence Nationale de Recherches sur le Sida et les Hépatites Virales (ANRS).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2015.11.055.
Contributor Information
James R.A. Hutchins, Email: james.hutchins@igh.cnrs.fr.
Marcel Méchali, Email: marcel.mechali@igh.cnrs.fr.
Appendix A. Supplementary material
Supplementary material
References
- 1.Traver S., Coulombe P., Peiffer I., Hutchins J.R.A., Kitzmann M., Latreille D. MCM9 is required for mammalian DNA mismatch repair. Mol. Cell. 2015;59:831–839. doi: 10.1016/j.molcel.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 2.Vizcaino J.A., Cote R.G., Csordas A., Dianes J.A., Fabregat A., Foster J.M. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakatani Y., Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzym. 2003;370:430–444. doi: 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- 5.Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 7.Peng J., Gygi S.P. Proteomics: the move to mixtures. J. Mass Spectrom. 2001;36:1083–1091. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- 8.Kersey P.J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 9.Eng J.K., McCormack A.L., Yates J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 10.UniProt Consortium, UniProt: a hub for protein information, Nucleic Acids Res. 43 (2015) D204–D212. [DOI] [PMC free article] [PubMed]
- 11.Hutchins J.R.A. What׳s that gene (or protein)? Online resources for exploring functions of genes, transcripts, and proteins. Mol. Biol. Cell. 2014;25:1187–1201. doi: 10.1091/mbc.E13-10-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hutchins J.R.A., Toyoda Y., Hegemann B., Poser I., Heriche J.K., Sykora M.M. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mellacheruvu D., Wright Z., Couzens A.L., Lambert J.P., St-Denis N.A., Li T. The CRAPome: a contaminant repository for affinity purification–mass spectrometry data. Nat. Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material